Abstract
Norovirus infection causing acute gastroenteritis could lead to adverse effects on the gut microbiome. We assessed the association of microbiome diversity with norovirus infection and secretor status in patients from Veterans Affairs medical centers. Alpha diversity metrics were lower among patients with acute gastroenteritis but were similar for other comparisons.
Keywords: norovirus, microbiome, secretor status, acute gastroenteritis
The human digestive tract is populated by a diverse array of bacterial species, though these are highly variable, differing with diet, geography, and genetics [1–4]. Dysbioses in the gut microbiome have been associated with a wide range of chronic and infectious diseases [3, 5, 6].
Norovirus causes 19–21 million cases of acute gastroenteritis (AGE) in the United States annually [7], with persons >65 years of age at greatest risk for norovirus-associated deaths [8]. Norovirus infection can cause diarrhea and vomiting, disrupting the intestinal environment and reducing gut microbiome diversity [9, 10]. Additionally, the microbiome may offer protection against symptoms of norovirus infection [11]. Further examination of the association between norovirus and the microbiome could explain differences between symptomatic and asymptomatically infected individuals.
Host genetics may also affect microbiome diversity [4]. Individuals with a functional fucosyltransferase-2 (FUT2) gene have histo-blood group antigens (HBGAs) present in the mucosa of the gut and are known as “secretors” [12]. Gut microbiome diversity and composition may differ between secretors and nonsecretors [13–15] or may not [16, 17]. Susceptibility to norovirus infection is strongly associated with secretor status, with nonsecretors being resistant to some norovirus genotypes [12, 18, 19]. Noroviruses use HBGAs as attachment factors for cellular entry, though the exact mechanism is unknown [20, 21].
To date, few studies have examined the complex relationships between secretor status, norovirus infection, and the microbiome. Due to the increased risk of norovirus-associated morbidity and mortality in the older adult population and adverse effects of potential microbiome disruptions, further exploration of these relationships in this population is needed.
The ongoing SUrveillance Platform for Enteric and Respiratory iNfectious Organisms in the VA (SUPERNOVA) is a collaboration between Emory University, the Centers for Disease Control and Prevention, and Department of Veterans Affairs Medical Centers (VAMCs) in Atlanta, Georgia, Houston, Texas, Bronx, New York, Los Angeles, California, and Palo Alto, California. We analyzed the microbiomes of 100 SUPERNOVA patients to assess differences in microbiome diversity by recent norovirus infection and secretor status. Additional analyses explored diversity by recent AGE symptoms and microbiome composition between norovirus and secretor status groups.
METHODS
Recruitment into the SUPERNOVA study has been described elsewhere [22]. In brief, individuals were enrolled as cases in the SUPERNOVA study if they met the definition for AGE (Supplementary Methods) when presenting as outpatients or when hospitalized at participating VAMCs (Atlanta, Houston, Bronx, and Los Angeles). Controls without AGE presented to the same VAMC with an admission or outpatient visit date within a week of a matched case.
Patients provided stool and saliva samples within 10 days of symptom onset (cases) or enrollment (controls). All stool specimens were tested using the BioFire FilmArray Gastrointestinal Panel for 22 pathogens [22]. Saliva samples were tested for secretor status using established methods [23].
For our analysis, we sampled 51 norovirus-positive patients and 49 norovirus-negative patients enrolled between December 1, 2016, and December 31, 2017. The 49 norovirus-negative patients included both AGE cases and non-AGE controls, allowing microbiome comparisons that assessed differences between norovirus AGE and non-norovirus AGE as well as the combined effect of norovirus and AGE together. Stool specimens were sequenced and analyzed using established methods (Supplementary Methods) [24–26].
Microbiome diversity was assessed through sample richness (total number of amplicon sequence variants [ASVs]) and using the Shannon diversity index [27]. Statistical analyses were performed using R, version 4.0.4 [28], including the packages “phyloseq” [29], “vegan” [30], and “ldm” [31]. ASV counts within each sample were normalized to the median number of total reads per sample (97 183). Due to the limited sample size, we chose not to statistically compare diversity metrics using P values.
To examine differences in overall microbiome composition (ie, which specific genera are in a sample) between groups, we used ordination by nonmetric multidimensional scaling (NMDS) based on Bray-Curtis distances using only genera that accounted for at least 10% of reads in a sample. One patient was excluded from all ordination analyses due to having <1000 reads and <1% of median reads.
To evaluate the impact of the separate and combined effects of secretor status, norovirus infection, and AGE on microbiome composition, we constructed linear decomposition models (LDMs) [31]. Separate models for each variable of interest were run as well as combined models including age (in years) and anti-infective drug use as confounders, giving P values of associations for each variable with microbiome composition. Recent anti-infective medication use was a binary variable defined as any inpatient or outpatient prescription dispensed in VA records for specific drug classes in the 3 months before stool collection (Supplementary Table 5).
RESULTS
Participant Characteristics
Specimens from 100 enrolled patients were selected for the microbiome analysis from the SUPERNOVA surveillance system. Fifty-one norovirus-positive patients and 49 norovirus-negative patients were included, with norovirus-negative patients selected based on secretor status and AGE symptoms. A majority of norovirus-positive patients were secretors (45/51; 88%). Norovirus-positive patients had a lower average age compared with norovirus-negative patients in the study (55.7 years vs 60.6). Nearly all norovirus-positive patients had AGE symptoms (48/51; 94%), and 12 (24%) had other AGE pathogens detected (Supplementary Table 1). The most common capsid genotype among norovirus-positive patients was GII.4 (20/51; 39%), though not all norovirus-positive samples were genotyped.
Among norovirus-negative patients with AGE, 16/23 (70 %) did not have pathogens detected on BioFire. Other detected pathogens included C. difficile (2 patients), Rotavirus, Shigella, multiple pathogens (2 patients), and 1 inconclusive result. Three norovirus-negative patients without AGE had pathogens detected, including C. difficile, enteropathogenic E. coli, and multiple pathogens (1 patient each).
Recent AGE Illness Associated With Reduced Microbiome Diversity
Across all samples, secretors had a lower mean richness compared with nonsecretors, with a mean number of detected ASVs of 224.6 among secretors and 247.7 among nonsecretors (Figure 1; Supplementary Table 2). Mean richness was similar between norovirus-positive secretors and norovirus-positive nonsecretors, with 228.1 and 223.3 ASVs, respectively. Shannon indices were similar in all comparisons (Supplementary Table 3).
Figure 1.
Strip plots comparing microbiome richness (A) and Shannon diversity index (B) between patient show greatest differences between patients with AGE and those without. Strip plots for microbiome richness (total number of species detected in a sample) and Shannon diversity index (defined as:, where R is the total number of species and pi is the proportion of individuals belonging to the ith species in a sample) for each comparison discussed in Supplementary Tables 2 and 3. Positive patients for each comparison are in red, while negative patients are in black (1 inconclusive secretor status is excluded in those comparisons). Comparisons are as follows: (1) Secretor positive to secretor negative. (2) Secretor positive to secretor negative among norovirus-positive patients. (3) Secretor positive to secretor negative among norovirus-negative patients with AGE. (4) Secretor positive to secretor negative among norovirus-negative patients without AGE. (5) Norovirus-positive to all norovirus-negative patients. (6) Norovirus-positive to norovirus-negative patients with AGE. (7) Norovirus-positive to norovirus-negative patients without AGE. (8) AGE positive to AGE negative. (9) AGE positive to AGE negative among norovirus-negative patients. Most comparisons are similar. The greatest differences are between patients with AGE and those without AGE, including when restricting to norovirus-negative patients.
Overall, richness and Shannon indices were similar between patients with and without norovirus (Figure 1; Supplementary Tables 2 and 3). However, norovirus-positive patients had a lower richness when compared with norovirus-negative patients without AGE (227.5 and 268.5, respectively), but higher when compared with norovirus-negative patients with non-norovirus AGE (227.5 and 200.3). Shannon diversity indices had a similar pattern.
Patients with AGE had substantially lower mean richness (216.2) and a lower Shannon index (3.773) than patients without AGE (270.2 and 4.096, respectively) (Figure 1; Supplementary Tables 2 and 3). This held true when restricting to norovirus-negative individuals.
No Differences in Microbiome Composition Seen by Any Variables of Interest
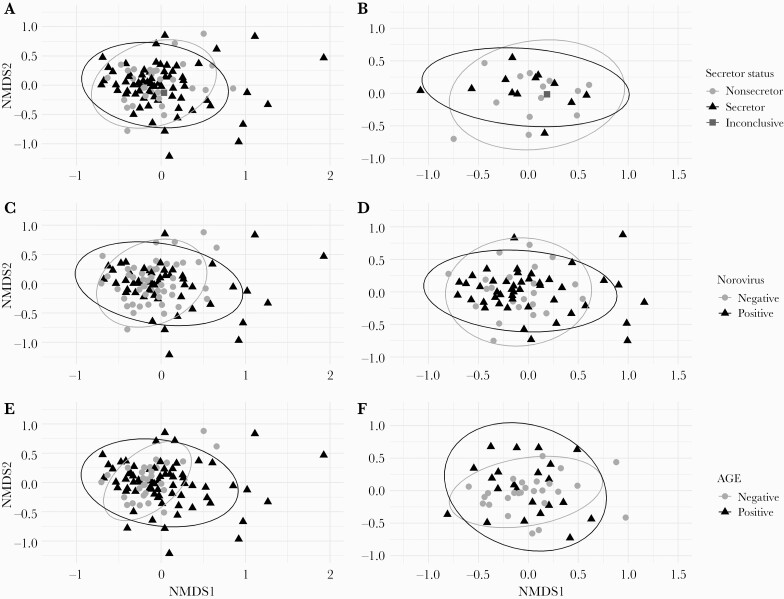
There were no differences found in microbiome composition between secretors and nonsecretors via ordination using the most abundant genera, including when restricting to individuals without AGE (Figure 2A and B). Points representing secretors were slightly more graphically dispersed than nonsecretors, but there was substantial overlap in the 2 groups. Norovirus-positive patients had similar microbiome compositions to norovirus-negative patients on ordination plots (Figure 2C). Further, overall compositions were similar when comparing norovirus AGE with non-norovirus AGE (Figure 2D). Patients with AGE appeared similarly in microbiome composition to patients without AGE, including when restricting to only norovirus-negative patients (Figure 2E and F).
Figure 2.
Non-metric multidimensional scaling ordination shows no difference in microbiome composition by secretor status and norovirus infection, with possible greater differences in composition among patients with AGE. Ordination using non-metric multidimensional scaling based on Bray-Curtis distances was used to compare overall microbiome composition between secretors and non-secretors (A and B), norovirus-positive patients and norovirus-negative patients (C and D), as well as patients with AGE and those without AGE (E and F). Included populations by panel: A, all individuals, with secretor points appearing slightly more dispersed. B Only patients without AGE, where microbiome compositions appear similar across both secretor status groups. C, all individuals, with points for norovirus-positive patients appearing slightly more dispersed. D, only patients with AGE, where points for norovirus-positive patients also appeared more dispersed than norovirus-negative patients. E, all individuals, with points for AGE-free individuals more clustered together compared to points for patients with AGE. F, only patients without norovirus and there is little apparent difference between the two groups. Ellipses in all panels are based on multivariate t-distribution. One individual (square point, norovirus-negative and AGE-free) with an inconclusive secretor status is shown on secretor status plots.
Individual LDMs for secretor status, norovirus infection, and recent AGE did not show significant associations with microbiome composition (Supplementary Table 4). In the full model with all variables and controlling for age and prescription drug use, no significant associations were detected (Supplementary Table 4).
DISCUSSION
Secretor status and norovirus infection were not independently associated with differences in either microbiome richness or Shannon diversity index. Additionally, no differences by secretor status or norovirus infection in microbiome composition were seen on NMDS ordination or LDM regression. No differences were detected despite a larger sample size compared with some similar studies [13–15, 17].
Due to the disruption of the gut environment during AGE [32], we expected to see lowered microbiome diversity for patients with AGE; this was true for both richness and Shannon index. However, no difference was seen in microbiome composition between AGE groups upon ordination. As composition analyses only included the most abundant genera, the reduction in diversity metrics may be borne by only less abundant genera.
Our secretor status results support previous findings of no differences in microbiome composition between secretor status groups [16, 17] Though previous studies found a relationship between secretor status, norovirus IgA titer (past norovirus infection), and microbiome composition, we did not see relationships between analogous variables in our data with a larger sample size examining recent infections [17]. Additional work incorporating pre-and postinfection stool samples, such as challenge studies [11], could better explore microbiome changes following infection.
Our study had at least 4 limitations, including lack of serial pre-infection stools, a limited sample size, potential nonrepresentativeness of the VA population compared with the general US population, and additional factors (eg, unreported medication, dietary differences, norovirus genotype, or co-infecting pathogens) that were not explored here.
In summary, we did not detect an association between secretor status or norovirus infection and microbiome diversity or composition. Though findings suggest that there might not be a relationship between norovirus infection and the microbiome, longitudinal studies would be better suited to capture any causal effect of secretor status and norovirus infection on the microbiome.
Supplementary Material
Acknowledgments
We sincerely thank the collaborators and study teams at our SUPERNOVA sites for their continued time and efforts that made this study possible.
Financial support. This work was supported by the Foundation for Atlanta Veterans Education and Outreach (FAVER; formerly the Atlanta Research and Education Foundation or AREF) and the Centers for Disease Control and Prevention, Atlanta, Georgia.
Potential conflicts of interest. The authors have no conflicts of interest to disclose. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Patient consent. This study does not include factors necessitating patient consent.
Disclaimer. The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the US Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors’ affiliated institutions. Use of trade names is for identification only and does not imply endorsement by the Public Health Service or the US Department of Health and Human Services.
Data availability. Sequence data are available at the NBI SRA database under BioProject accession PRJNA741048.
References
- 1. Khanna S, Tosh PK.. A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin Proc 2014; 89:107–14. [DOI] [PubMed] [Google Scholar]
- 2. Lloyd-Price J, Abu-Ali G, Huttenhower C.. The healthy human microbiome. Genome Med 2016; 8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynch SV, Pedersen O.. The human intestinal microbiome in health and disease. N Engl J Med 2016; 375:2369–79. [DOI] [PubMed] [Google Scholar]
- 4. Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell 2014; 159:789–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clemente JC, Ursell LK, Parfrey LW, Knight R.. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148:1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thaiss CA, Zmora N, Levy M, Elinav E.. The microbiome and innate immunity. Nature 2016; 535:65–74. [DOI] [PubMed] [Google Scholar]
- 7. Kirk MD, Pires SM, Black RE, et al. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 2015; 12:e1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hall AJ, Lopman BA, Payne DC, et al. Norovirus disease in the United States. Emerg Infect Dis 2013; 19:1198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nelson AM, Walk ST, Taube S, et al. Disruption of the human gut microbiota following norovirus infection. PLoS One 2012; 7:e48224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen SY, Tsai CN, Lee YS, et al. Intestinal microbiome in children with severe and complicated acute viral gastroenteritis. Sci Rep 2017; 7:46130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patin NV, Pena-Gonzalez A, Hatt JK, Moe C, Kirby A, Konstantinidis KT.. The role of the gut microbiome in resisting norovirus infection as revealed by a human challenge study. mBio 2020; 11:e02634-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindesmith L, Moe C, Marionneau S, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548–53. [DOI] [PubMed] [Google Scholar]
- 13. Gampa A, Engen PA, Shobar R, Mutlu EA.. Relationships between gastrointestinal microbiota and blood group antigens. Physiol Genomics 2017; 49:473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wacklin P, Makivuokko H, Alakulppi N, et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 2011; 6:e20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wacklin P, Tuimala J, Nikkila J, et al. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One 2014; 9:e94863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davenport ER, Goodrich JK, Bell JT, et al. ABO antigen and secretor statuses are not associated with gut microbiota composition in 1,500 twins. BMC Genomics 2016; 17:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rodriguez-Diaz J, Garcia-Mantrana I, Vila-Vicent S, et al. Relevance of secretor status genotype and microbiota composition in susceptibility to Rotavirus and norovirus infections in humans. Sci Rep 2017; 7:45559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nordgren J, Svensson L.. Genetic susceptibility to human norovirus infection: an update. Viruses 2019; 11:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nordgren J, Kindberg E, Lindgren PE, Matussek A, Svensson L.. Norovirus gastroenteritis outbreak with a secretor-independent susceptibility pattern, Sweden. Emerg Infect Dis 2010; 16:81–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marionneau S, Ruvoen N, Le Moullac-Vaidye B, et al. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 2002; 122:1967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopman BA, Trivedi T, Vicuna Y, et al. Norovirus infection and disease in an Ecuadorian birth cohort: association of certain norovirus genotypes with host FUT2 secretor status. J Infect Dis 2015; 211:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cardemil CV, Balachandran N, Kambhampati A, et al. Incidence, etiology, and severity of acute gastroenteritis among prospectively enrolled patients in 4 Veterans Affairs hospitals and outpatient centers, 2016-18. Clin Infect Dis 2021; 73:e2729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reeck A, Kavanagh O, Estes MK, et al. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis 2010; 202:1212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bushnell B. BBMap. 2014. Available at: https://sourceforge.net/projects/bbmap/. Accessed 4 September 2019. [Google Scholar]
- 25. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP.. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013; 41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spellerberg IF, Fedor PJ.. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’ Index. Glob Ecol Biogeogr 2003; 12:177–9. [Google Scholar]
- 28. R: A Language and Environment for Statistical Computing. R Foundation; for Statistical Computing; 2021. Available at: https://www.R-project.org/. Accessed February 2021. [Google Scholar]
- 29. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oksanen J, Blanchet FG, Kindt R, et al. Package “Vegan.” Community Ecology Package, Version. 2013. Version 2.5-7. Available at: http://CRAN.R-project.org/package=vegan. Accessed 10 December 2019.
- 31. Hu YJ, Satten GA.. Testing hypotheses about the microbiome using the linear decomposition model (LDM). Bioinformatics 2020; 36:4106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y, Xia S, Jiang X, et al. Gut microbiota and diarrhea: an updated review. Front Cell Infect Microbiol 2021; 11:625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.