Abstract
Rationale
The biochemical mechanisms underlying lung function are incompletely understood.
Objectives
To identify and validate the plasma metabolome of lung function using two independent adult cohorts: discovery—the European Prospective Investigation into Cancer-Norfolk (EPIC-Norfolk, n=10 460) and validation—the VA Normative Aging Study (NAS) metabolomic cohort (n=437).
Methods
We ran linear regression models for 693 metabolites to identify associations with forced expiratory volume in one second (FEV1) and the ratio of FEV1 to forced vital capacity (FEV1/FVC), in EPIC-Norfolk then validated significant findings in NAS. Significance in EPIC-Norfolk was denoted using an effective number of tests threshold of 95%; a metabolite was considered validated in NAS if the direction of effect was consistent and p<0.05.
Measurements and main results
Of 156 metabolites that associated with FEV1 in EPIC-Norfolk after adjustment for age, sex, body mass index, height, smoking and asthma status, 34 (21.8%) validated in NAS, including several metabolites involved in oxidative stress. When restricting the discovery sample to men only, a similar percentage, 18 of 79 significant metabolites (22.8%) were validated. A smaller number of metabolites were validated for FEV1/FVC, 6 of 65 (9.2%) when including all EPIC-Norfolk as the discovery population, and 2 of 34 (5.9%) when restricting to men. These metabolites were characterised by involvement in respiratory track secretants. Interestingly, no metabolites were validated for both FEV1 and FEV1/FVC.
Conclusions
The validation of metabolites associated with respiratory function can help to better understand mechanisms of lung health and may assist the development of biomarkers.
INTRODUCTION
Metabolomics, the systematic profiling of the small molecules in a biological system,1 represents a powerful tool to increase the understanding of the mechanisms of respiratory health, providing a downstream ‘snapshot’ of the status of a biological system reflecting phenotype as well as upstream genetic and environmental influences. As such, it is ideally suited to examine alterations in biological pathways that accompany phenotypical changes.2 Several studies have successfully used metabolomics to explore diseases including asthma and COPD,3 4 suggesting there are measurable and biologically informative alterations in the metabolome that reflect perturbations in the respiratory system. However, only a small number of studies have investigated the metabolome of forced expiratory volume in one second (FEV1) or the ratio of FEV1 to forced vital capacity (FEV1/FVC).5-10 These metrics, which can be considered as measures of airflow obstruction, represent two of the most important indicators of pulmonary function in adults.11 More broadly, given the known relationship between pulmonary function with systemic inflammation and a range of clinical pathologies from diabetes to cardiovascular disease, FEV1 and FEV1/FVC can also inform on overall health status.5 Improving our understanding of the mechanisms of FEV1 and FEV1/FVC is therefore paramount, and consequently, metabolomic profiling of spirometric lung function warrants further investigation.
In this study, we aimed to identify and validate metabolites associated with FEV1 and FEV1/FVC, in two independent cohorts: the European Prospective Investigation into Cancer–Norfolk (EPIC-Norfolk)12 and the VA Normative Aging Study (NAS).13
METHODS
Study populations
Discovery population
EPIC-Norfolk is a prospective population-based cohort12 which recruited 25 639 men and women aged 40–79 years residing in Norfolk, England, between 1993 and 1997. Information was collected on lifestyle variables and dietary habits; anthropometrics and blood samples were taken; and spirometry was performed. The majority (~93%) of blood samples were non-fasted14 15 (online supplemental methods).
Validation population
NAS is a longitudinal study of ageing based in Boston, USA, that recruited 2280 men aged 21–80 between 1961 and 1970.13 Participants have routine physical examinations and laboratory tests every 3–5 years, including fasting blood collection and spirometry. A subset of men with plasma samples collected during follow-up that were suitable for metabolomic profiling were selected to create the MAS Metabolomic Cohort.16 All samples in this cohort were collected between 2000 and 2008. Samples collected prior to 2000 were not suitable for metabolomic profiling due to their storge conditions.
Metabolomic profiling
Metabolomic profiling was conducted by independently in each population by Metabolon (Durham, North Carolina, USA) using four non-targeted liquid chromatography–mass spectroscopy (LC-MS), platforms.17 Metabolites were identified by mass-to-charge ratio, retention time and through a comparison to a library of purified known standards. Metabolites were matched to standards using LC-MS peaks and quantified using area under the curve. Data were processed according to the established quality control pipeline for each cohort15 16 18-23 (online supplemental methods). As the metabolomic profiling was conducted independently in both cohorts, a slightly different subset of metabolites was measured in each. For validation purposes, these analyses were restricted to only those metabolites that were present and passed quality control (QC) in both cohorts. These common metabolites were matched between cohorts according to the unique Metabolon COMP.ID identifier.
Statistical analysis
Analyses were conducted in EPIC-Norfolk, and then validation of significant findings was performed in NAS. Linear regression models for two outcome variables, FEV1 (as measured in litres) and FEV1/FVC (expressed as a proportion), were run for each metabolite with adjustment for sex, age, body mass index (BMI), height, smoking status (never and current former) and asthma status. As NAS was all men, sex-stratified models were also run in EPIC-Norfolk.
The ‘number of effective tests approach’ was employed to account for multiple testing.24 25 This method performs principal components analysis (PCA) on the full dataset in order to reduce the dimensionality of the dataset by creating new uncorrelated variables (principal components) that successively maximize variance. It then determines the number of principal components (PCs) required to explain a given percentage of variance in the data (ie, the number of effective tests). The adjusted p value threshold is calculated as m, which denotes the nominal p value threshold of 0.05, and m denotes the number of effective tests. For these analyses, we set a threshold of 95% variance explained (ENT95%). In EPIC-Norfolk, 415 PCs were required to explain 95% of the variance in the data, corresponding to a p value threshold of 1.205×10−4. In EPIC-Norfolk men, it was 410 PCs and p<1.220×10−4, and in women 407 PCs and p<1.229×10−4. In the validation analyses, we considered a metabolite validated if the p value is <0.05 and the direction of effect was consistent.
We conducted several sensitivity analyses to further explore the robustness of our findings. These included stratification of the EPIC-Norfolk subjects by BMI category and smoking status, additional adjustment for fasting status and prevalent diabetes and the inclusion of participants with self-reported emphysema or bronchitis at blood draw.
All analyses were conducted using R V.3.6.0 and Stata V.14.2. Significant metabolites were further explored using the ‘Pathway Analysis’ functionality in MetaboAnalyst V.4.0.26 Pathway analysis tests for overenrichment of a given set of metabolites within a curated list of KEGG (Kyoto Encyclopedia of Genes and Genomes) defined metabolomic pathways while also taking pathway topology into account, that is, the positional importance of a metabolite within a pathway, to provide an overall enrichment p value and pathway impact score. All 693 metabolites that could be measured in both EPIC-Norfolk and in NAS were input as the reference metabolome. The hypergeometric test was specified for the over-representation analysis and relative ‘betweenness’ centrality for the pathway topology analysis.
RESULTS
Study population
The mean age of the 10 460 EPIC-Norfolk participants eligible for these analyses was 59.73 years (SD=8.95 years), and >99% were of white British/European descent (online supplemental table E1). Eleven per cent reported current smoking at the time of blood sample collection. The majority of the population had normal lung function as defined by their spirometry measures. Mean FEV1 and FEV1/FVC were 2.52 L (SD=0.72 L), and 0.82 (SD=0.11), respectively. The correlation between FEV1 and FEV1/FVC was r=0.183 (p<2.2×10−16). Within EPIC-Norfolk, men were slightly older than women and were more likely to have a higher FEV1 and a lower FEV1/FVC.
NAS (online supplemental table E2) was older than EPIC-Norfolk, with a mean age of 75.12 years (SD=6.66) among the 437 men eligible for the NAS metabolomic cohort, but was similarly predominantly white with few current smokers (4.3%). The mean FEV1 was 2.50 L (SD=0.60 L), and FEV1/FVC was 0.74 (SD=0.08), both of which were lower than in EPIC-Norfolk, indicating slightly worse lung function in this older population. The correlation between FEV1 and FEV1/FVC was r=0.518, p<2.2×10−16.
After data QC and processing, 1002 and 858 metabolites were available in EPIC-Norfolk and NAS, respectively. Of these, 693 were common to both datasets and included in these analyses.
Forced expiratory volume in one second
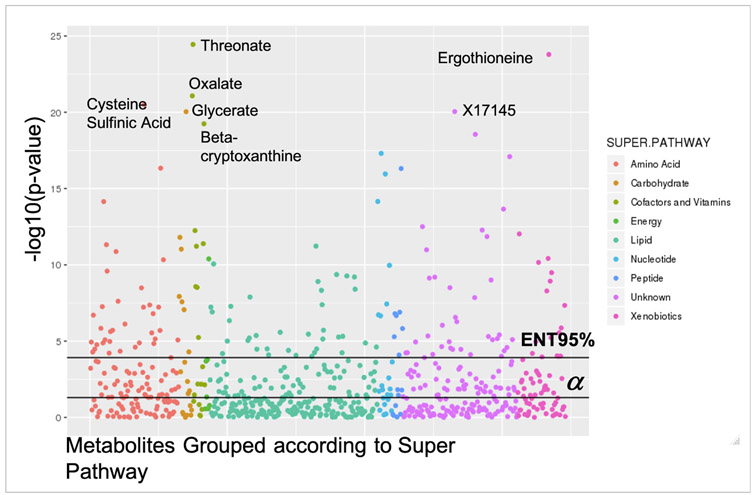
A total of 156 (22.5%) of 693 metabolites were associated (p<ENT95%) with FEV1 in EPIC-Norfolk (figure 1). The largest percentages were lipids (n=36, 23.1%) and amino acids (n=36, 23.1%). Half (n=78, 50%) were positively associated with FEV1. The top hits were threonate, where a 1-unit increase in threonate levels results in an increase in of 0.049 L in FEV1 (β=0.049, 95% CI 0.040 to 0.058, p=3.57×10−25), oxalate (β=0.046, 95% CI 0.036 to 0.055, p=8.46×10−22) and ergothioneine (β=0.047, 95% CI 0.038 to 0.056, p=1.62×10−24). The most significant finding for inverse associations was cysteine sulfinic acid where a 1-unit increase in cysteine sulfinic acid levels was associated with a drop of 0.046 L in FEV1 volume (β=−0.046, 95% CI −0.056 to −0.037, p=3.16×10−21). Pathway analyses determined the 156 metabolites were enriched for metabolic pathways including glycine, serine and threonine metabolism (p for enrichment=3.33×10−5), aminoacyl-tRNA biosynthesis (p for enrichment=5.52×10−4), caffeine metabolism (p for enrichment=0.007), and cysteine and methionine metabolism (p for enrichment=0.010) (online supplemental table E3).
Figure 1.
Manhattan plot demonstrating the strength of association between FEV1 and 693 metabolites in European Prospective Investigation into Cancer–Norfolk, coloured according to Metabolon Superpathway. Top metabolite hits are identified according to their common name. Metabolites with the format Xnnnnn are of unknown identity but can be tracked and quantified, and therefore Metabolon is confident they represent biologically relevant molecules and not analytical artefacts. The black horizontal lines indicate significance thresholds. FEV1, forced expiratory volume in one second.
In sex-stratified analyses, 79 (11.4%) and 74 (10.7%) metabolites were associated with FEV1 in men and women, respectively. Forty were significant in both sexes (online supplemental table E4 and figure E1). Four metabolites were significant in men but not women, three of which were inversely associated with FEV1: 1-stearoyl-2-arachidonoyl-Glycerophosphoethanolamine (18:0/20:4) and 1-palmitoyl-2-arachidonoyl-GPE (16:0/20:4) (both phosphatidylethanolamines), and gamma-tocopherol/beta-tocopherol. One, 5alpha-pregnan-3beta,20alpha-diol monosulfate, was positively associated with FEV1 in litres. Four metabolites, cysteine, 3-methoxycatechol sulfate and two metabolites of unknown identify were significant only in women, all were inversely associated with FEV1. However, it should be noted, for both the male-only and female-only metabolites, the direction of effect was consistent in the other sex.
Validation in the NAS
Thirty-four (21.8%) of the 156 metabolites significant in the total EPIC-Norfolk sample were validated in the NAS (table 1), including several top hits, such as ergothioneine and beta-cryptoxanthine (online supplemental figure E2). It should be noted that due to differences in the metabolite normalisation and scaling procedures implemented prior to analysis, the scales of the beta-coefficients differed between the two studies.
Table 1.
FEV1-associated metabolites validated in the VA NAS
| EPIC-Norfolk total |
EPIC-Norfolk men |
VA NAS |
|||||
|---|---|---|---|---|---|---|---|
| Metabolite | Super pathway | Change in FEV1 litres with a 1-unit increase in metabolite level β (95% CI) |
P value | Change in FEV1 litres with a 1-unit increase in metabolite level β (95% CI) |
P value | Change in FEV1 litres with a 1-unit increase in metabolite level β (95% CI) |
P value |
| 2-aminobutyrate * | Amino acid | 0.04 (0.03 to 0.04) | 7.12E-15 | 0.03 (0.02 to 0.05) | 1.59E-05 | 0.17 (0.03 to 0.31) | 1.81E-02 |
| Imidazole propionate * | Amino acid | −0.03 (−0.04 to −0.02) | 1.35E-11 | −0.05 (−0.06 to −0.03) | 1.62E-08 | −0.08 (−0.15 to −0.01) | 3.04E-02 |
| Vanillylmandelate * | Amino acid | −0.03 (−0.04 to −0.02) | 4.63E-11 | −0.03 (−0.05 to −0.02) | 5.64E-05 | −0.21 (−0.32 to −0.1) | 1.97E-04 |
| N-formylmethionine* | Amino acid | −0.03 (−0.04 to −0.02) | 5.97E-08 | −0.03 (−0.04 to −0.01) | 1.64E-03 | −0.23 (−0.38 to −0.08) | 3.73E-03 |
| N-acetylputrescine | Amino acid | −0.02 (−0.03 to −0.02) | 1.58E-07 | −0.03 (−0.05 to −0.02) | 5.97E-05 | −0.24 (−0.37 to −0.1) | 5.34E-04 |
| 5-oxoproline | Amino acid | 0.02 (0.01 to 0.03) | 1.45E-06 | 0.02 (0.01 to 0.03) | 5.77E-03 | 0.14(0.06 to 0.23) | 1.10E-03 |
| 5-methylthioadenosine | Amino acid | −0.02 (−0.03 to −0.01) | 2.88E-06 | −0.03 (−0.04 to −0.01) | 7.92E-04 | −0.12 (−0.21 to −0.04) | 5.66E-03 |
| Kynurenine | Amino acid | −0.02 (−0.03 to −0.01) | 7.20E-06 | −0.03 (−0.05 to −0.01) | 4.93E-04 | −0.19 (−0.34 to −0.04) | 1.21E-02 |
| Threonine | Amino acid | 0.02 (0.01 to 0.03) | 8.20E-06 | 0.03 (0.02 to 0.05) | 8.02E-05 | 0.21 (0.05 to 0.37) | 1.10E-02 |
| N-acetylalanine | Amino acid | −0.02 (−0.03 to −0.01) | 1.15E-05 | −0.02 (−0.04 to −0.004) | 1.27E-02 | −0.21 (−0.39 to −0.04) | 1.57E-02 |
| N-acetylserine* | Amino acid | −0.02 (−0.03 to −0.01) | 6.11E-05 | −0.02 (−0.03 to −0.001) | 5.77E-02 | −0.22 (−0.37 to −0.08) | 2.53E-03 |
| Mannose | Carbohydrate | −0.02 (−0.03 to −0.02) | 8.73E-08 | −0.03 (−0.04 to −0.01) | 1.36E-04 | −0.1 (−0.16 to −0.03) | 3.53E-03 |
| Beta-cryptoxanthin * | Cofactors and vitamins | 0.04 (0.03 to 0.05) | 5.68E-20 | 0.05 (0.03 to 0.06) | 4.27E-09 | 0.14 (0.06 to 0.21) | 2.36E-04 |
| 1-(1-enyl-palmitoyl)-2-linoleoyl-GPC(P-16:0/18:2) * † | Lipid | 0.03 (0.02 to 0.04) | 4.31E-10 | 0.05 (0.03 to 0.07) | 2.94E-10 | 0.2 (0.07 to 0.33) | 2.93E-03 |
| Glycochenodeoxycholate sulfate | Lipid | −0.03 (−0.04 to −0.02) | 6.24E-10 | −0.04 (−0.06 to −0.03) | 1.30E-07 | −0.09 (−0.16 to −0.02) | 1.56E-02 |
| Linoleoyl ethanolamide | Lipid | 0.02 (0.01 to 0.03) | 4.57E-07 | 0.03 (0.02 to 0.05) | 4.30E-05 | 0.12 (0.04 to 0.2) | 4.69E-03 |
| 1-palmitoyl-GPC (16:0) | Lipid | 0.02 (0.01 to 0.03) | 2.63E-06 | 0.02 (0.01 to 0.04) | 4.84E-03 | 0.18 (0.02 to 0.35) | 2.88E-02 |
| Linoleoylcarnitine (C18:2) † | Lipid | 0.02 (0.01 to 0.03) | 9.07E-06 | 0.03 (0.02 to 0.05) | 1.26E-05 | 0.11 (0.02 to 0.2) | 2.35E-02 |
| 1-(1-enyl-palmitoyl)-GPC (P-16:0)*† | Lipid | 0.02 (0.01 to 0.03) | 8.63E-05 | 0.02 (0.01 to 0.04) | 1.92E-03 | 0.17 (0.04 to 0.3) | 1.05E-02 |
| N2,N2-dimethylguanosine * | Nucleotide | −0.04 (−0.05 to −0.03) | 4.92E-18 | −0.04 (−0.06 to −0.03) | 2.72E-07 | −0.29 (−0.43 to −0.15) | 6.58E-05 |
| N1-methyladenosine * | Nucleotide | −0.04 (−0.05 to −0.03) | 7.05E-15 | −0.04 (−0.06 to −0.02) | 2.54E-06 | −0.18 (−0.34 to −0.02) | 2.47E-02 |
| Pseudouridine * | Nucleotide | −0.03 (−0.04 to −0.02) | 1.08E-10 | −0.04 (−0.05 to −0.02) | 3.86E-05 | −0.22 (−0.37 to −0.07) | 3.55E-03 |
| N6-carbamoylthreonyl adenosine* | Nucleotide | −0.03 (−0.04 to −0.02) | 1.89E-07 | −0.02 (−0.04 to −0.004) | 1.58E-02 | −0.16 (−0.26 to −0.06) | 2.21E-03 |
| 7-methylguanine* | Nucleotide | −0.02 (−0.03 to −0.02) | 2.20E-07 | −0.03 (−0.04 to −0.01) | 1.14E-03 | −0.19 (−0.36 to −0.02) | 2.63E-02 |
| X-12117 * | Unknown | −0.05 (−0.06 to −0.04) | 2.81E-19 | −0.06 (−0.08 to −0.05) | 8.22E-13 | −0.13 (−0.2 to −0.05) | 7.32E-04 |
| X-11315 * | Unknown | 0.03 (0.03 to 0.04) | 2.21E-14 | 0.03 (0.02 to 0.05) | 2.89E-05 | 0.14 (0.04 to 0.23) | 5.87E-03 |
| X-18901 | Unknown | 0.02 (0.01 to 0.03) | 2.76E-07 | 0.04 (0.02 to 0.05) | 8.54E-07 | 0.09 (0.01 to 0.17) | 3.53E-02 |
| X-23590* | Unknown | −0.02 (−0.03 to −0.01) | 3.87E-06 | −0.02 (−0.03 to −0.003) | 2.26E-02 | −0.11 (−0.2 to −0.02) | 2.02E-02 |
| X-16087* | Unknown | −0.02 (−0.03 to −0.01) | 4.95E-06 | −0.02 (−0.04 to −0.01) | 7.39E-03 | −0.13 (−0.21 to −0.06) | 7.52E-04 |
| X-15 728 | Unknown | 0.02 (0.01 to 0.03) | 2.85E-05 | 0.03 (0.01 to 0.05) | 6.33E-04 | 0.09 (0.04 to 0.14) | 7.37E-04 |
| X-15 486 | Unknown | −0.02 (−0.03 to −0.01) | 5.71E-05 | −0.02 (−0.04 to −0.01) | 4.93E-03 | −0.14 (−0.23 to −0.05) | 2.07E-03 |
| X-21 258 | Unknown | 0.02 (0.01 to 0.03) | 5.90E-05 | 0.03 (0.01 to 0.04) | 4.09E-04 | 0.07 (0.001 to 0.13) | 4.88E-02 |
| 4-allylphenol sulfate | Xenobiotics | 0.03 (0.02 to 0.04) | 3.81E-11 | 0.04 (0.02 to 0.06) | 2.76E-07 | 0.09 (0.01 to 0.16) | 2.72E-02 |
| Ergothioneine * | Xenobiotics | 0.05 (0.04 to 0.06) | 1.62E-24 | 0.06 (0.04 to 0.07) | 9.80E-14 | 0.1 (0.01 to 0.19) | 2.43E-02 |
18 metabolites in bold are those that are also significant at an ENT95% threshold in EPIC-Norfolk men and validated in NAS.
Metabolites with the format Xnnnnn are of unknown identity but can be tracked and quantified, and therefore, Metabolon is confident they represent biologically relevant molecules and not analytical artefacts.
ENT95% significant in EPIC-Norfolk women.
Indicates a metabolite has not been confirmed using an analytical standard, but Metabolon is confident in its identity based on its analytical parameters.
EPIC-Norfolk, European Prospective Investigation into Cancer–Norfolk; FEV1, forced expiratory volume in one second; NAS, Normative Aging Study.
Given NAS is all-male, we then explored the 79 metabolites significant in the EPIC-Norfolk men. Of these, 18 validated (22.8%). This was only a slightly greater level of validation than observed for the total population and a majority of these 18 (n=10, 55.6%) were also significant in EPIC-Norfolk women, suggesting little difference in the metabolome of FEV1 by sex.
Ratio of FEV1 to forced vital capacity
A total of 65 (9.4%) metabolites were significantly (p<ENT95%) associated with FEV1/FVC in EPIC-Norfolk, of which the majority (n=52, 80.0%) were inversely associated with ratio (online supplemental table E5 and figure E3). The top hits included lactate, which was associated with a decrease in FEV1/FVC proportion of 0.014 for every unit increase in lactate level (β=−0.014, 95% CI −0.016 to −0.012, p=2.19×10−4), 5-oxoproline (β=−0.014, 95% CI −0.016 to −0.011, p=3.33×10−38), sphingosine 1-phosphate (β=− 0.013, 95% CI −0.015 to −0.011, p=1.81×10−37) and inosine (β=0.011, 95% CI 0.009 to 0.013, p=7.09×10−24). The 65 metabolites were enriched for metabolites of 12 pathways, including alanine, aspartate and glutamate metabolism (p for enrichment=7.62×10−5), arginine biosynthesis (p for enrichment=4.21×10−4), the citrate cycle (p for enrichment=0.001), butanoate metabolism (p for enrichment=0.007) and aminoacyl tRNA biosynthesis (p for enrichment=0.009) (online supplemental table E3).
In sex-stratified analyses, 34 (4.9%) and 36 (5.2%) metabolites were significant in men and women, respectively. As with FEV1, there was strong concordance between the sexes for FEV1/FVC (online supplemental figure E4). Twenty-six metabolites were common to men and women: lactate, 5-oxoproline, sphingosine 1-phosphate and inosine were the most strongly associated in both sexes.
Validation in the NAS
Of the 65 metabolites significant in the total EPIC-Norfolk sample, 6 (9.2%) were validated in NAS (online supplemental figure E5): stearoylcarnitine (C18), oleoylcarnitine (C18:1), palmitoylcarnitine (C16), 5-methyluridine, cysteine s-sulfate and N-acetylglucosamine/N-acetylgalactosamine. All but 5-methyluridine were inversely associated with ratio (table 2).
Table 2.
FEV1:FVC-associated metabolites validated in the NAS
| EPIC-Norfolk total |
EPIC-Norfolk men |
VA NAS |
|||||
|---|---|---|---|---|---|---|---|
| Change in FEV1/FVC with a 1- unit increase in metabolite level |
Change in FEV1/FVC with a 1-unit increase in metabolite level |
Change in FEV1/FVC with a 1-unit increase in metabolite level |
|||||
| Metabolite | Super pathway | β (95% CI) | P value | β (95% CI) | P value | β (95% CI) | P value |
| Cysteine s-sulfate | Amino acid | −0.005 (−0.007 to −0.003) | 1.70E-05 | −0.004 (−0.008 to 0.001) | 1.88E-02 | −0.011 (−0.02 to −0.001) | 3.03E-02 |
| N-acetylglucosamine/N-acetylgalactosamine | Carbohydrate | −0.006 (−0.008 to −0.003) | 3.02E-07 | −0.007 (−0.01 to −0.003) | 8.92E-05 | −0.022 (−0.042 to −0.001) | 3.98E-02 |
| Oleoylcarnitine (C18:1) * | Lipid | −0.009 (−0.011 to −0.007) | 1.83E-16 | −0.01 (−0.013 to 0.007) | 2.86E-09 | −0.016 (−0.031 to 0.001) | 3.15E-02 |
| Stearoylcarnitine (C18) | Lipid | −0.005 (−0.007 to −0.003) | 1.88E-06 | −0.006 (−0.01 to −0.003) | 1.98E-04 | −0.017 (−0.034 to −0.001) | 4.27E-02 |
| Palmitoylcarnitine (C16) | Lipid | −0.005 (−0.007 to −0.002) | 2.55E-05 | −0.005 (−0.008 to 0.002) | 2.08E-03 | −0.019 (−0.037 to −0.002) | 2.96E-02 |
| 5-methyluridine (ribothymidine) | Nucleotide | 0.004 (0.002 to 0.006) | 4.97E-05 | 0.005 (0.002 to 0.009) | 2.63E-03 | 0.023 (0.001 to 0.046) | 4.15E-02 |
2 Metabolites in bold are those that are also significant at an ENT95% threshold in EPIC-Norfolk men and validated in NAS.
ENT95% significant in EPIC-Norfolk women.
EPIC-Norfolk, European Prospective Investigation into Cancer–Norfolk; FEV1/FVC, ratio of FEV1 to forced vital capacity; NAS, Normative Aging Study.
Of the 34 significant in EPIC-Norfolk men, 2 (5.9%) were validated in NAS: oleoylcarnitine (C18:1) and N-acetylglucosamine/N-acetylgalactosamine.
Consistency of metabolite associations between FEV1 and FEV1/FVC
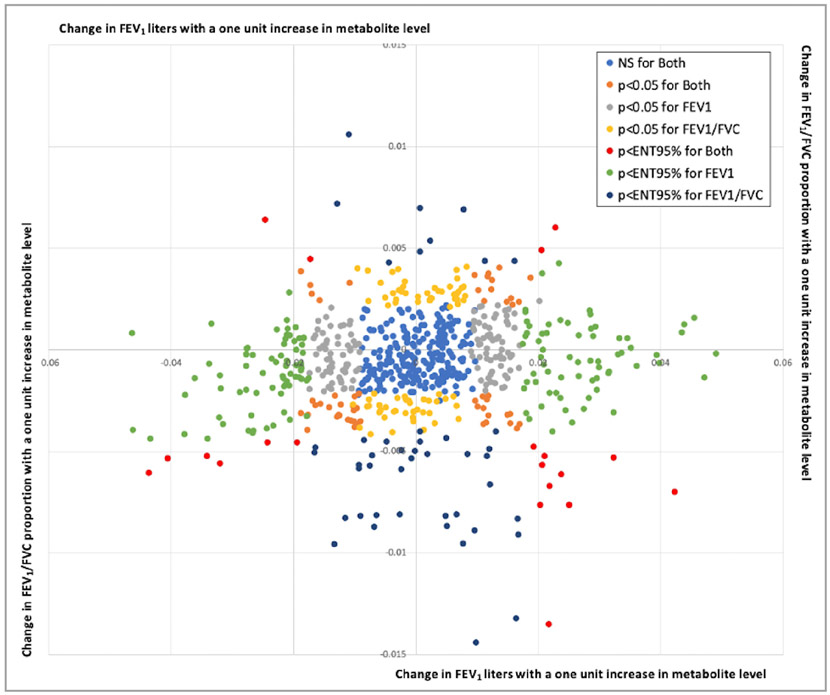
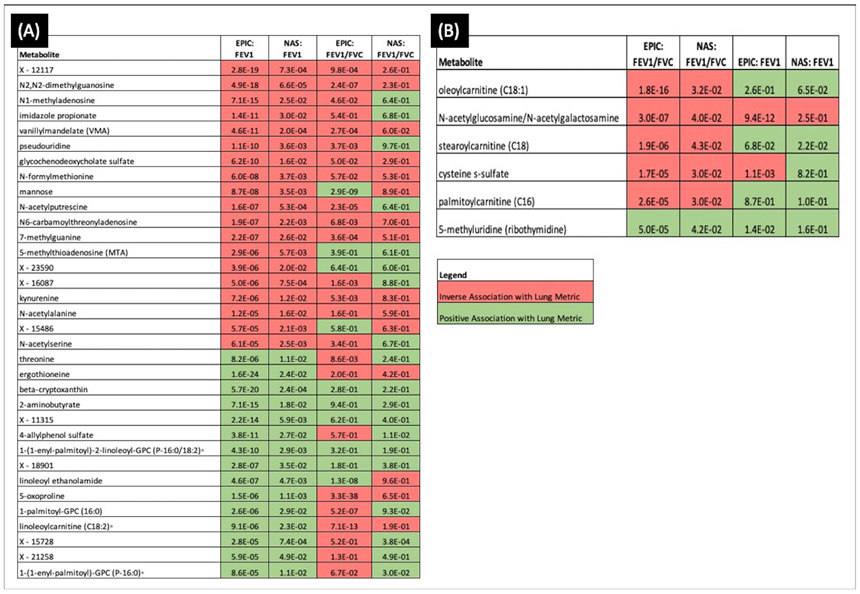
Figure 2 shows the unit changes for FEV1 litres and FEV1/FVC proportion with a 1-unit increase in metabolite level for every measured metabolite. When restricting to those that were validated in NAS, 34 metabolites were validated for FEV1 and six for FEV1/FVC. The two sets of metabolites were distinct, with no metabolite validated for both phenotypes. Figure 3 shows the direction of effect and p value from both EPIC-Norfolk and NAS for the 34 metabolites that were validated for FEV1 and the six validated for ratio. While for many metabolites the directions of effect were consistent for the two phenotypes and approached ENT95% significance, for others such as 1-palmitoyl-GPC (16:0) and 5-oxoproline, the directions of effect were discordant between phenotypes.
Figure 2.
Effect estimates of metabolite association with FEV1 and FEV1/FVC in European Prospective Investigation into Cancer–Norfolk, coloured according to significance of effect. FEV1, forced expiratory volume in one second; FVC, forced vital capacity; NS, not significant.
Figure 3.
Direction of effect and p value in the validated lung function-associated metabolites for (A) EPIC-Norfolk men and NAS for 34 FEV1-validated metabolites, (B) EPIC-Norfolk men and NAS for 6 FEV1-validated metabolites. To allow comparison between FEV1 and FEV1/FVC, the effect estimates for both are given for each set of metabolites in each cohort. ° indicates a metabolite has not been confirmed using an analytical standard, but Metabolon is confident in its identity based on its analytical parameters. Metabolites with the format Xnnnnn are of unknown identity but can be tracked and quantified, and therefore, Metabolon is confident they represent biologically relevant molecules and not analytical artefacts. EPIC, European Prospective Investigation into Cancer; EPIC-Norfolk, European Prospective Investigation into Cancer–Norfolk; FEV1, forced expiratory volume in one second; FVC, forced vital capacity; NAS, Normative Aging Study.
When considering the EPIC-Norfolk men as the discovery sample, 18 and 2 metabolites were validated for FEV1 and FEV1/FVC, respectively. Again, there were no common validated metabolites between the two phenotypes (online supplemental figure E6).
Sensitivity analyses
Individuals with self-reported emphysema or bronchitis at baseline were initially excluded from analyses. When running sensitivity analyses including the 1050 individuals in EPIC-Norfolk with these diagnoses, there was little change in the results. For both FEV1 and FEV1/FVC, the vast majority of significant metabolites were robust to the inclusion of the new subjects, including 100% of the validated metabolites. There were also a small number of novel metabolites that were validated when including the emphysema and bronchitis cases (online supplemental table E6).
Additional adjustment for fasting status and diabetes status in EPIC-Norfolk also had little influence on our findings. In the fasting adjusted models, 154 of the 156 (98.7%) metabolites were associated with FEV1 in the primary analysis retained significance with adjustment for fasting status, including all 34 of the metabolites that could be validated in NAS. For FEV1/FVC, 64 of 65 retained significance with adjustment for fasting status, including all 6 of the validated metabolites (results not shown). There were 371 cases of prevalent diabetes in the EPIC-Norfolk dataset. When diabetes status was adjusted for, 154 of the 156 (98.7%) metabolites associated with FEV1 in the primary analysis retained significance with adjustment for fasting status. All but 1 of the 34 validated metabolites, (1-(1-enyl-palmitoyl)-GPC (P-16:0)), retained significance, and it should be noted this metabolite almost reached ENT95% significance. For FEV1/FVC, again 64 of 65 retained significance with adjustment for fasting status, including all 6 of the validated metabolites (results not shown).
We further ran our EPIC-Norfolk models stratified by WHO-defined BMI category, for healthy (n=4189), overweight (n=4750) and obese (n=1477) participants. We note that the number of individuals in the underweight category (n=44) was too small to run analyses. Seventy (10.1%), fifty-four (7.8%) and four (0.6%) metabolites were associated with FEV1 at ENT95% in healthy, overweight and obese individuals, respectively (online supplemental table E7). Of these, 28 were common between the healthy and overweight individuals. Six of the 28 common metabolites were also among those validated in NAS; these included a number of the top hits from the primary analysis: ergothioneine, beta-cryptoxanthin, X-12117, N2,N2-dimethylguanosine, 2-aminobutyrate and N1-methyladenosine.
Twenty-nine (4.2%), 28 (4.0%) and 10 (1.4%) metabolites were associated with FEV1/FVC at ENT95% in healthy, overweight and obese participants, respectively (online supplemental table E8). Of these, seven were significant in all three groups. The top validated hit for FEV1/FVC in the primary analysis, oleoylcarnitine (C18:1), was significant in the both healthy and overweight individuals. Even where ENT95% significance was not reach for both FEV1 and FEV1/FVC, the direction of effect was consistent and at least p<0.05 was reached for most metabolites across all the groups.
Finally, we ran our EPIC-Norfolk models stratifying by smoking status at blood draw: current (n=4867), former (n=4750) and never (n=1145). Fifty-eight (8.4%), 65 (9.4%) and 20 (2.9%) metabolites were associated with FEV1 at ENT95% in never, former and current smokers, respectively. Never and former smokers were more alike in their associations than current smokers. However, three metabolites were ENT95% significant in all groups and were among the validated hits: ergothioneine, beta-cryptoxanthin and N1-methyladenosine (online supplemental table E9). For FEV1/FVC, 32 (4.6%), 30 (4.3%) and 2 (0.3%) metabolites were significant at ENT95% in never, former and current smokers, respectively. The top hit, oleoylcarnitine (C18:1), was ENT95% significant in never and former smokers (online supplemental table E10). Again, even where ENT95% significance was not reached for both FEV1 and FEV1/FVC, the direction of effect was consistent and at least p<0.05 was reached for most metabolites across all the groups.
DISCUSSION
Pulmonary function, as measured by FEV1 and FEV1/FVC, reflects the physiological state of the lung.5 Common chronic lung diseases such as asthma, COPD, emphysema, chronic bronchitis and pulmonary fibrosis are characterised by impairments in pulmonary function. Measures of lung function as assessed by spirometry can be used for diagnosis, assessment of severity and monitoring.11 Furthermore, there is evidence that lung function and its associated diseases have far-reaching systemic effects beyond the lung, including on the cardiovascular, nervous and immune systems.27 Poor lung function has been linked to chronic inflammation, as well as the development of other chronic conditions including diabetes, kidney and cardiovascular disease.5 28 29 What’s more, reduced FEV1 has been shown to be correlated with higher mortality, irrespective of disease and smoking status.30 As such, elucidating the underlying biological mechanisms of lung function is crucial to improve our understanding not only of the lung, but of the pathogenesis of multiple diseases and morbidity.
Individuals’ pulmonary function is a product of the interactions between their underlying genetics and their environmental exposures.31-33 Consequently, it is ideally suited to metabolomics profiling, which reflects the downstream products of these interactions across the life course.2
In this study, we identified and validated metabolites measured in the circulating blood that were associated with lung function. One hundred and fifty-six metabolites were significantly associated with FEV1 with stringent correction for multiple testing in over 10 000 subjects from EPIC-Norfolk, a large population-based cohort of men and women residing in the UK. Of these, 34 (21.8%) could be validated in 437 men from the US based NAS. A slightly higher proportion, 22.8%, validated when restricting the EPIC-Norfolk discovery sample to men only. For FEV1/FVC, a much smaller number of metabolites were found to be significant in the total EPIC-Norfolk population, 65 (9.4%) of which 6 were validated. Only 2 of 34 were validated when restricting to EPIC-Norfolk men. Interestingly, there were no metabolites that were validated for both FEV1 and FEV1/FVC. This is in agreement with existing metabolomics studies of lung function5 and corresponds to other omic studies which report a smaller number of genetic associations for FEV1/FVC compared with FEV1.33 Nevertheless, validated metabolites were found for both metrics that inform on underlying biology. Given the unique position of the metabolome closest to phenotype and the relative ease and cost-effectiveness of measuring the metabolome in blood, metabolites that reflect a physiological state may have translational utility in biomarker development.
Few studies have explored the global metabolome of FEV1 or FEV1/FVC. In a multiethnic study of lung function, Yu et al identified 95 serum metabolites associated with FEV1 that were characterised by branched chain amino acids and glutamine.5 As in this current analysis, they identified fewer FEV1/FVC-associated metabolites, and these were primarily lipids. Two studies by Menni et al within the TWINS UK study also reported several associations between FEV1 and serum levels of amino acids, particularly of the glycine, serine and threonine metabolism pathway,6 10 while other studies in blood report strong correlations between FEV1 with kynurenine, several sphingomyelins and sphingolipids.9 34 In a cohort of patients with asthma–COPD overlap, urinary histidine was negatively correlated with both FEV1 and FEV1/FVC.7 Finally, Halper-Stromberg et al explored plasma and bronchoalveolar lavage (BAL) fluid, observing there was little correlation between the results for the two biosamples. The greatest number of associations were found for BAL, but that both BAL and plasma provided distinct biologically important information.
Despite differences between these studies and ours, there was high consistency with our findings. The strongest association with FEV1 in the EPIC-Norfolk total population was with thre-onante, a major metabolite of the antioxidant ascorbic acid,10 higher levels of which were associated with increased FEV1, indicating a better oxidative state. Although threonante was not validated in the NAS, it has previously been associated with FEV1 in other studies.5 10 Our results were also consistent with existing literature for many of our validated hits. Metabolites of glycine, serine and threonine metabolism, demonstrated some of the strongest associations with FEV1 in our analyses and have been implicated in lung function,5 7 10 as well as in COPD35 and asthma,4 due to their role in oxidative stress. Oxidative stress is a product of increased exposure to free radicals and/or decreased antioxidant defence mechanisms, and given its proximity to ambient air, the respiratory system is particularly susceptible. Exposure to reactive oxidant species causes mitochondrial DNA damage and disruption of the electron transport chain, leading to overall reduced pulmonary function.36
In common with previous literature,5 we also observed an enrichment of metabolites of the aminoacyl-tRNA biosynthesis pathway, a pathway that is similarly involved in oxidative stress and which plays a central role in protein biosynthesis by catalysing the attachment of an amino acid to the 3′ end of its cognate tRNA.37 The susceptibility of the lungs to oxidative stress38 may also explain our validated association with beta-cryptoxanthin, an oxygenated carotenoid that acts as an antioxidant.39 Dietary supplementation of beta-cryptoxanthin has been shown to improve pulmonary function,40 and our findings may provide mechanistic support role the role of beta-crytoxanthin as a supplement. It is also worth noting that the association between this metabolite and FEV1 was observed across all smoking categories and in both healthy-weight and overweight individuals, strengthening its potential as therapeutic strategy. Our validated hits for FEV1 also included other metabolites previously reported in the literature,5 for example, two nucelosides, N2, N2-dimethylguanosine and pseudouridine, and the carbohydrate mannose.
We further validated several novel associations with FEV1, the majority of which involve metabolites previously associated with COPD, asthma or oxidative stress. Our top validated hit was the xenobiotic ergothionine, a sulfur-containing derivative of histidine, suggested to have antioxidant properties,41 which was also robust to stratification by smoking status and BMI category. We validated 4-allylphenol sulfate, which like ergothionine is a xenobiotic originating from food and is positively associated with FEV1. Interestingly, levels of these two metabolites were increased with a decrease in sodium intake in the Dietary Approaches to Stop Hypertension trial,42 and lower sodium intake has been associated with improved lung function.43 This again suggests that supplementation with these metabolites may be beneficial for the improvement of FEV1. Clinical trials are necessary to explore this question further.
Among the six metabolites that validated for FEV1/FVC were three acylcarnitines, which were associated with a lower ratio and, therefore, greater airway obstruction. Accumulation of acyl carnitines in the pulmonary surfactant of the lung has previously been shown to sensitise lungs to injury by inhibiting the surface adsorption of pulmonary surfactant and its ability to reduce surface tension.44 Levels of acylcarnitines have also been shown to be higher in the plasma of individuals with pulmonary hypertension, who tend to have a lower FEV1/FVC.45 Further, our top hit, oleoylcarnitine (C18:1), was also identified by Yu et al as associated with both FEV1 and FEV1:FVC5 and was robust to stratification by smoking status and BMI in these analyses. Interestingly, we also validated N-acetylglucosamine/N-acetylgalactosamine, a glycoprotein which forms an important component of airway mucins, another respiratory tract secretant.46 Finally, we validated a positive association between 5-methyluridine (ribothy-midine) and FEV1/FVC, in agreement with our previous work demonstrating that higher levels of ribothymidine can attenuate the decline in bronchodilator response that occurs with age.47
There were a number of limitations to these analyses. There was a large discrepancy in power between our discovery and validation populations, and it is possible that those metabolites which we were able to validate in the smaller NAS cohort represent those with the largest effect estimates. It is likely other associations in EPIC-Norfolk represent true positives that the NAS cohort was too underpowered to detect. Validation was likely also hindered by the differences in the underlying populations of the two cohorts. Our discovery population was younger, and in addition to the known influence of age on the metabolome,48 lung function also changes as an individual ages due to physiological changes in lung elasticity and muscle tone and decreased surface area for alveolar gas exchange, in addition to immunological changes.49 As such, the metabolomic changes associated with FEV1 and FEV1/FVC may also differ by age, again decreasing the potential for replication between the cohorts.
EPIC-Norfolk included both men and women, while our validation population was all-male, and sex is also known to influence the metabolome and lung function.50 51 Nevertheless, there was high concordance in the significantly associated metabolites by sex for both FEV1 and FEV1/FVC in EPIC-Norfolk. Even when significance in one sex was not met in the other, the directions of effect were consistent. Of the 39 metabolites that were significant in men but not women for FEV1, 31 were nominally significant (p<0.05). Similarly, for FEV1/FVC, seven of the eight ‘male-only’ metabolites were nominally significant in women. When attempting to validate our findings in the NAS, there was a similar level of validation when using the metabolites identified in the EPIC-Norfolk total and all-male populations. Taken together, these results suggest little difference in metabolomics of lung function by sex. Although our cohort was almost entirely Caucasian, and for this reason, to maintain stability of our models, we did not adjust for race; results from the previous multiethnic studies5 and the consistency of our findings with these studies suggest the metabolome of lung function also does not differ greatly by race.
Our results were also largely consistent, particularly with regard to the validated metabolites across healthy and overweight individuals and former and current smokers. There were some differences in the metabolite –lung function associations in the individuals who were obese and those who were current smokers. There is a biological rationale as to why these relationships may differ; both BMI and smoking have metabolomic consequences52 53; and smoking has a well-characterised determinantal effect on spirometrics.54 The association with obesity was less well characterised; although it reduces lung function overall due to both mechanical effects on the chest wall and immunological changes, FEV1 and FEV1/FVC have been reported to be only mildly affected except in cases of extreme obesity (62 kg/m2).55 Obese subjects and current smokers were also our smallest groups and had reduced power; therefore, further work is needed to disentangle these findings.
In both populations, the majority of participants had good to moderate lung function, potentially limiting the generalisability of our findings to populations with very poor lung function. However, sensitivity analyses including individuals with emphysema and bronchitis did not manifestly affect our results and had little influence on our overall conclusions. We did validate a small number of additional metabolites with the additional participants, providing further support for the roles of glutamate metabolism and kynurenine in lung function.9 56
These analyses were based on two existing datasets that were generated independently and have been published on previously.15 16 18-21 The spirometry measures in EPIC-Norfolk were not specifically defined according to American Thoracic Society or European Respiratory Society standards. However, we note that the spirometer used in EPIC Norfolk has demonstrated good sensitivity and specificity for identifying COPD compared with the gold standard, and the EPIC-Norfolk spirometry measures have previously been published in peer-reviewed publications.57 58 As we used pre-existing blood samples for these analyses collected prior to widespread use of untargeted metabolomic profiling, neither of the cohorts’ blood collection procedures were optimised to metabolomics. Time of day of collection varied across the cohorts and diurnal variation in the metabolome may also have influenced our findings. We note that, to date, there are no gold standards in the QC or data processing of metabolomic data.59 As such, there were some differences in the QC pipelines of the two datasets, and this may have hampered validation of a larger number of metabolites. Samples in Epic-Norfolk were between 17 and 24 years old at profiling, and the NAS samples were between 10 and 18 years old. It has been shown that long-term storage can influence the levels of certain metabolites,60 which may have affected our results. Our discovery and replication approach, including samples with a range of storage times, limits the potential for false-positive findings; however, we cannot rule out the possibility we may have missed metabolites of interest due to the age of our samples.
We did not see any overlap in terms of specific metabolites associated with FEV1/FVC between our plasma study and Halper-Stromberg’s BAL analysis.8 However, there are a number of disadvantages to using BAL including its cost, discomfort to the patient, difficulty of obtaining the sample and the related risk of contamination.61 While blood may not be the biofluid closest to the lung, given lung function has shown to be reflected systemically,5 27-29 blood can still inform on the processes occurring there. As such, recognising both BAL samples and blood provide vital insights into the biochemical mechanisms of lung function, we found that blood, which is simple and cost-effective to collect, may be superior for the development of future clinically translatable biomarkers of lung function.
This study also had several strengths: it represents the largest study to date to focus on the metabolome of FEV1 and FEV1/FVC, and the first to consider differences by sex. Crucially, we were able to validate our findings in an independent sample, profiled using the same platform as the discovery population.17 The differences between our two cohorts would only have biased our results to the null, and we note that additional adjustment for fasting status in EPIC-Norfolk did not alter our results or conclusions. Additionally, our results were robust to additional adjustment for prevalent diabetes status, a common comorbidity in populations in the age range of those included in these analyses. We validated multiple biologically relevant metabolites, confirming findings from prior literature and identifying novel ones. We used a stringent correction for multiple testing to obtain the most robust results possible. At a less stringent discovery p value in EPIC-Norfolk, there would have been even greater validation in the NAS. Therefore, additional validation of these metabolites in other populations may be warranted. Given the cross-sectional nature of this study, we cannot determine whether the observed metabolomic perturbations are the cause of reduced lung function or an effect. Therefore, follow-up studies should consider repeated longitudinal sampling, as well as absolute quantification to determine their biomarker potential. We included unnamed metabolites in our analysis to reflect the fact that Metabolon is continually updating its library and may in the future be able to assign names to these metabolites.
In conclusion, this study is among the first to validate metabolites significantly associated with lung function parameters. These findings indicate that oxidative stress is correlated with an individual’s FEV1, while the composition of respiratory secretants seems to be of importance for FEV1/FVC. It is of note that two dietary compounds were strongly associated with FEV1, indicating the potential of simple dietary interventions to improve lung health. We can therefore use these results to both better understand the mechanisms of lung function and support the development of future biologically informative biomarkers or therapeutic strategies for the overall improvement of pulmonary care.
Supplementary Material
What is the key question?
To determine the, as yet not fully understood, systemic effects of lung function.
What is the bottom line?
This is the largest study to identify validated metabolomic associations with forced expiratory volume in one second (FEV1) and ratio of FEV1 to forced vital capacity, providing novel insights into the mechanisms underlying lung function.
Why read on?
These findings pave the way for the development of novel biomarkers or therapeutic strategies for improved lung function.
Acknowledgements
We thank all participants of EPIC-Norfolk and the VA Normative Aging study, and the many members of the study teams at the University of Cambridge, the VA and the various study sites who have enabled this research.
Funding
This study was supported by PR161204 W81XWH-17-1-0533 from the Congressionally Directed Medical Research Programmes, USAMRDC. The VA Normative Aging Study is a research component of the Massachusetts Veterans Epidemiology Research and Information Centre at VA Boston Healthcare System and is supported by the Cooperative Studies ProgramProgramme/Epidemiology Research and Information Centres, Office of Research and Development, US Department of Veterans Affairs (grant number not applicable). AS was supported by a Senior Research Career Scientist award from the Clinical Science Research and Development Service, Office of Research and Development, US Department of Veterans Affairs (grant number not applicable). RSK was supported by K01HL146980 from the National Heart, Lung, and Blood Institute (NHLBI). PK was supported by P01HL132825 from the National Institutes of Health/NHLBI. JL-S was supported by R01HL123915, R01HL141826 and P01HL132825. HMK was supported by the Jane and Aatos Erkko Foundation, the Paulo Foundation and the Paediatric Research Foundation (grant number not applicable). The EPIC-Norfolk study (https://doi.org/10.22025/2019.10.105.00004) has received funding from the Medical Research Council (MR/N003284/1 and MC-UU_12015/1) and Cancer Research UK (C864/A14136). Metabolite measurements in the EPIC-Norfolk study were supported by the MRC Cambridge Initiative in Metabolic Science (MR/L00002/1) and the Innovative Medicines Initiative Joint Undertaking under EMIF grant agreement number 115 372.
Footnotes
Competing interests None declared.
Ethics approval The study was approved by the Norwich Local Ethics Committee (REC Ref. 98CN01), and all participants provided signed informed consent. The Normative Aging Study received written consent from all study participants at each visit, and approval from the Rreview Bboards of all institutions was involved.
Additional supplemental material is published online only. To view, please visit the journal online (http://dx.doi.org/10.1136/thoraxjnl-2020-216639).
Data availability statement
Data are available upon reasonable request. Data are available on request with agreement from all coauthors and funding bodies.
REFERENCES
- 1.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol 2012;13:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fiehn O Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol 2002;48:155–71. [PubMed] [Google Scholar]
- 3.Stringer KA, McKay RT, Karnovsky A, et al. Metabolomics and its application to acute lung diseases. Front Immunol 2016;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly RS, Dahlin A, McGeachie MJ, et al. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest 2017;151:262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, Flexeder C, McGarrah RW, et al. Metabolomics identifies novel blood biomarkers of pulmonary function and COPD in the general population. Metabolites 2019;9. doi: 10.3390/metabo9040061. [Epub ahead of print: 01 Apr 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menni C, Kastenmüller G, Petersen AK, et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int J Epidemiol 2013;42:1111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh JY, Lee YS, Min KH, et al. Increased urinary L-histidine in patients with asthma-COPD overlap: a pilot study. Int J Chron Obstruct Pulmon Dis 2018;13:1809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halper-Stromberg E, Gillenwater L, Cruickshank-Quinn C, et al. Bronchoalveolar lavage fluid from COPD patients reveals more compounds associated with disease than matched plasma. Metabolites 2019;9. doi: 10.3390/metabo9080157. [Epub ahead of print: 25 Jul 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilk K, Aug A, Ottas A, et al. Phenotyping of chronic obstructive pulmonary disease based on the integration of metabolomes and clinical characteristics. Int J Mol Sci 2018;19. doi: 10.3390/ijms19030666. [Epub ahead of print: 27 Feb 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menni C, Metrustry SJ, Mohney RP, et al. Circulating levels of antioxidant vitamins correlate with better lung function and reduced exposure to ambient pollution. Am J Respir Crit Care Med 2015;191:1203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranu H, Wilde M, Madden B. Pulmonary function tests. Ulster Med J 2011;80:84–90. [PMC free article] [PubMed] [Google Scholar]
- 12.Day N, Oakes S, Luben R, et al. Epic-Norfolk: study design and characteristics of the cohort. European prospective investigation of cancer. Br J Cancer 1999;80:95–103. [PubMed] [Google Scholar]
- 13.Spiro AIB R The Normative Aging Study. In: The encyclopedia of aging. 3 edn. New York: Springer Press, 2001: 744–6. [Google Scholar]
- 14.Jakes RW, Day NE, Patel B, et al. Physical inactivity is associated with lower forced expiratory volume in 1 second : European Prospective Investigation into Cancer-Norfolk Prospective Population Study. Am J Epidemiol 2002;156:139–47. [DOI] [PubMed] [Google Scholar]
- 15.Hertel J, Harms AC, Heinken A, et al. Integrated analyses of microbiome and longitudinal metabolome data reveal Microbial-Host interactions on sulfur metabolism in Parkinson’s disease. Cell Rep 2019;29:1767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly RS, Bayne H, Spiro A, et al. Metabolomic signatures of lead exposure in the Va normative aging study. Environ Res 2020;190:110022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans AB, Bridgewater B, Liu Q, et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 2014;4. [Google Scholar]
- 18.Nassan FL, Kelly RS, Kosheleva A, et al. Metabolomic signatures of the long-term exposure to air pollution and temperature. Environ Health 2021;20:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nassan FL, Wang C, Kelly RS, et al. Ambient PM2.5 species and ultrafine particle exposure and their differential metabolomic signatures. Environ Int 2021;151:106447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Do KT, Wahl S, Raffler J, et al. Characterization of missing values in untargeted MS-based metabolomics data and evaluation of missing data handling strategies. Metabolomics 2018;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann JP, Pietzner M, Wittemans LB, et al. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum Mol Genet 2020;29:3451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lotta LA, Pietzner M, Stewart ID, et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet 2021;53:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietzner M, Stewart ID, Raffler J, et al. Plasma metabolites to profile pathways in noncommunicable disease multimorbidity. Nat Med 2021;27:471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M-X, Yeung JMY, Cherny SS, et al. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet 2012;131:747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004;74:765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong J, Soufan O, Li C, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 2018;46:W486–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agustí AGN, Noguera A, Sauleda J, et al. Systemic effects of chronic obstructive pulmonary disease. Eur Respir J 2003;21:347–60. [DOI] [PubMed] [Google Scholar]
- 28.Shaaban R, Kony S, Driss F, et al. Change in C-reactive protein levels and FEV1 decline: a longitudinal population-based study. Respir Med 2006;100:2112–20. [DOI] [PubMed] [Google Scholar]
- 29.Seeger W, Adir Y, Barbera JA, et al. Pulmonary hypertension in chronic lung diseases. J Am Coll Cardiol 2013;62:D109–16. [DOI] [PubMed] [Google Scholar]
- 30.Hole DJ, Watt GC, Davey-Smith G, et al. Impaired lung function and mortality risk in men and women: findings from the Renfrew and Paisley prospective population study. BMJ 1996;313:711–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hancock DB, Eijgelsheim M, Wilk JB, et al. Meta-Analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 2010;42:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hallberg J, Iliadou A, Anderson M, et al. Genetic and environmental influence on lung function impairment in Swedish twins. Respir Res 2010;11:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melén E, Guerra S. Recent advances in understanding lung function development. F1000Res 2017;6:726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowler RP, Jacobson S, Cruickshank C, et al. Plasma sphingolipids associated with chronic obstructive pulmonary disease phenotypes. Am J Respir Crit Care Med 2015;191:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubhi BK, Riley JH, Shaw PA, et al. Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur Respir J 2012;40:345–55. [DOI] [PubMed] [Google Scholar]
- 36.Okeleji LO, Ajayi AF, Adebayo-Gege G, et al. Epidemiologic evidence linking oxidative stress and pulmonary function in healthy populations. Chronic Dis Transl Med 2021;7:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Topp H, Fusch G, Schöch G, et al. Noninvasive markers of oxidative DNA stress, RNA degradation and protein degradation are differentially correlated with resting metabolic rate and energy intake in children and adolescents. Pediatr Res 2008;64:246–50. [DOI] [PubMed] [Google Scholar]
- 38.Ochs-Balcom HM, Grant BJB, Muti P, et al. Oxidative stress and pulmonary function in the general population. Am J Epidemiol 2005;162:1137–45. [DOI] [PubMed] [Google Scholar]
- 39.Burri BJ, La Frano MR, Zhu C. Absorption, metabolism, and functions of β-cryptoxanthin. Nutr Rev 2016;74:69–82. doi: 10.1093/nutrit/nuv064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schünemann HJ, McCann S, Grant BJB, et al. Lung function in relation to intake of carotenoids and other antioxidant vitamins in a population-based study. Am J Epidemiol 2002;155:463–71. [DOI] [PubMed] [Google Scholar]
- 41.Paul BD, Snyder SH. The unusual amino acid L-ergothioneine is a physiologic cytoprotectant. Cell Death Differ 2010;17:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derkach A, Sampson J, Joseph J, et al. Effects of dietary sodium on metabolites: the Dietary Approaches to Stop Hypertension (DASH)-Sodium Feeding Study. Am J Clin Nutr 2017;106:1131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scoditti E, Massaro M, Garbarino S, et al. Role of diet in chronic obstructive pulmonary disease prevention and treatment. Nutrients 2019;11. doi: 10.3390/nu11061357. [Epub ahead of print: 16 Jun 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otsubo C, Bharathi S, Uppala R, et al. Long-Chain acylcarnitines reduce lung function by inhibiting pulmonary surfactant. J Biol Chem 2015;290:23897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo N, Craig D, Ilkayeva O, et al. Plasma acylcarnitines are associated with pulmonary hypertension. Pulm Circ 2017;7:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lillehoj EP, Kato K, Lu W, et al. Cellular and molecular biology of airway mucins. Int Rev Cell Mol Biol 2013;303:139–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly RS, Sordillo JE, Lutz SM, et al. Pharmacometabolomics of bronchodilator response in asthma and the role of Age-Metabolite interactions. Metabolites 2019;9. doi: 10.3390/metabo9090179. [Epub ahead of print: 07 Sep 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivastava S. Emerging insights into the metabolic alterations in aging using metabolomics. Metabolites 2019;9. doi: 10.3390/metabo9120301. [Epub ahead of print: 13 Dec 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas ET, Guppy M, Straus SE, et al. Rate of normal lung function decline in ageing adults: a systematic review of prospective cohort studies. BMJ Open 2019;9:e028150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slupsky CM, Rankin KN, Wagner J, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem 2007;79:6995–7004. [DOI] [PubMed] [Google Scholar]
- 51.Carey MA, Card JW, Voltz JW, et al. It’s all about sex: gender, lung development and lung disease. Trends Endocrinol Metab 2007;18:308–13. doi: 10.1016/j.tem.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cirulli ET, Guo L, Leon Swisher C, et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab 2019;29:488–500.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsu P-C, Lan RS, Brasky TM, et al. Metabolomic profiles of current cigarette smokers. Mol Carcinog 2017;56:594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee PN, Fry JS. Systematic review of the evidence relating FEV1 decline to giving up smoking. BMC Med 2010;8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dixon AE, Peters U. The effect of obesity on lung function. Expert Rev Respir Med 2018;12:755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowerman KL, Rehman SF, Vaughan A, et al. Disease-Associated gut microbiome and metabolome changes in patients with chronic obstructive pulmonary disease. Nat Commun 2020;11:5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ching S-M, Chia Y-C, Lentjes MAH, et al. FEV1 and total cardiovascular mortality and morbidity over an 18 years follow-up population-based prospective EPIC-Norfolk study. BMC Public Health 2019;19:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Canoy D, Luben R, Welch A, et al. Abdominal obesity and respiratory function in men and women in the EPIC-Norfolk study, United Kingdom. Am J Epidemiol 2004;159:1140–9. [DOI] [PubMed] [Google Scholar]
- 59.Lasky-Su J, Kelly RS, Wheelock CE, et al. A strategy for advancing for population-based scientific discovery using the metabolome: the establishment of the metabolomics Society metabolomic epidemiology task group. Metabolomics 2021;17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner-Golbs A, Neuber S, Kamlage B, et al. Effects of long-term storage at −80 °C on the human plasma metabolome. Metabolites 2019;9. doi: 10.3390/metabo9050099. [Epub ahead of print: 17 May 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bowler RP, Wendt CH, Fessler MB, et al. New strategies and challenges in lung proteomics and metabolomics. An official American thoracic Society workshop report. Ann Am Thorac Soc 2017;14:1721–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request. Data are available on request with agreement from all coauthors and funding bodies.