Abstract
Studies are needed to better understand the genomic evolution of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This study aimed to describe viral quasispecies population of upper and lower respiratory tract by next-generation sequencing in patients admitted to intensive care unit. A deep sequencing of the S gene of SARS-CoV-2 from 109 clinical specimens, sampled from the upper respiratory tract (URT) and lower respiratory tract (LRT) of 77 patients was performed. A higher incidence of non-synonymous mutations and indels was observed in the LRT among minority variants. This might be explained by the ability of the virus to invade cells without interacting with ACE2 (e.g. exploiting macrophage phagocytosis). Minority variants are highly concentrated around the gene portion encoding for the Spike cleavage site, with a higher incidence in the URT; four mutations are highly recurring among samples and were found associated with the URT. Interestingly, 55.8% of minority variants detected in this locus were T>G and G>T transversions. Results from this study evidenced the presence of selective pressure and suggest that an evolutionary process is still ongoing in one of the crucial sites of spike protein associated with the spillover to humans.
Keywords: SARS-CoV-2, Spike protein, Deep sequencing, Minority variants, Deleterious mutations, Spillover
1. Introduction
Since coronavirus disease 2019 (COVID-19) was initially reported in China on 30th December 2019 (Dong et al., 2020; Wu et al., 2020), SARS-CoV-2 has been spreading worldwide. As of 8th March 2022, there have been 447 million confirmed infections and more than 6 million deaths have been reported worldwide (Dong et al., 2020, https://ourworldindata.org/coronavirus). The origin of SARS-CoV-2 is still debated but a hypothesis suggests the Malayan pangolins (Manis javanica) to be the possible intermediate host for the virus and recombination signals between pangolin, bat and human coronavirus sequences have been identified (Lam et al., 2020; Wong et al., 2020; Wu et al., 2020; Xiao et al., 2020). In fact, the SARS-CoV-2 genome sequence showed a high percentage of genomic identity (around 96%) with BatCoV-RaTG13 virus as well as (around 88%) with two other SARS-like bat viruses (Bat-SL-CoV-ZC45 and Bat-SL-CoV-ZXC21) (Zhou et al., 2020). Instead, the comparison with the SARS-CoV genome sequences showed an overall lower identity, approximately 79.6% (Gralinski and Menachery 2020; Zhou et al., 2020). Similar identity scores were observed when comparison analyzes were focused on the Spike (S) protein (around 75%) (Gralinski and Menachery 2020; Zhou et al., 2020). The S protein is the main determinant of viral tropism and is responsible for receptor binding and membrane fusion (Belouzard et al., 2012). For this reason, amino acid changes on this protein might have effects on infectivity, viral pathogenesis as well as transmissibility. It was initially reported that mutation D614G in the S protein, which has emerged and has become dominant, might have induced an enhancement of viral replication and viral fitness (Shi et al., 2020). Monitoring of emerging mutations, especially in the S protein, has been performed extensively with the establishment of Virus Evolution Expert Working Group (VEWG) by the WHO (World Health Organization WHO, 2021). The great effort on this concern is also highlighted by the huge number of SARS-CoV-2 sequences submitted on public repositories (e.g. GISAID). Initial report suggested a little viral diversity for SARS-CoV-2 (Karamitros et al., 2020; Simmonds, 2020), however, since December 2020 a process of positive selection with presumed advantages such as increased transmission rates has been documented for a series of variants of concern (VOCs) such as alpha, delta and omicron (Harvey et al., 2021; Tao et al., 2021; Volz et al., 2021). These VOCs have demonstrated a significant public health impact, with changes in the virus transmissibility and reduce the efficacy of vaccines (Harvey et al., 2021; Tao et al., 2021). Quasispecies is believed to be a strategy of virus evolution (Domingo and Perales, 2019) and although the viral kinetics of SARS-CoV-2 infection from the upper respiratory tract (URT) to the lower respiratory tract (LRT) have been gradually clarified more work is still needed to explore the inter-host and intra-host variations of SARS-CoV-2. Overall, studies aimed at investigating intra-host evolution or the dynamic of SARS-CoV-2 quasispecies have been mainly focused on samples collected from upper respiratory tract (URT) (Al Khatib et al., 2020; Jary et al., 2020; Pérez-Lago et al., 2021; Shen et al., 2020; Siqueira et al., 2020; Sun et al., 2021; To et al., 2020). In fact, the dynamic of SARS-CoV-2 population in the lower respiratory tract (LRT) of patients showing severe acute respiratory infections (SARIs) is poorly investigated. It would be important to elucidate the role of specific mutations in the progression of SARS-CoV-2 from upper to lower respiratory tract or identify specific mutational patterns associated with severe infection. In the present study, high-depth next-generation sequencing (NGS) of the S gene was performed in a set of 109 respiratory samples from URT (n = 58) and LRT (n = 51) in order to: (i) evaluate the genetic diversity in two different body compartments; (ii) identify minority variants potentially associated with progression from upper to lower respiratory tract on paired samples from patients admitted to intensive care unit (ICU) with severe infection.
2. Material and methods
2.1. Patients and samples
A total of 109 clinical specimens from URT (nasopharyngeal swabs; NPS) and LRT (bronchoalveolar lavage; BAL or broncho aspirate; Brasp) were collected and analyzed from 77 COVID-19-positive patients (Appendix Table S1). Respiratory samples were prospectively collected from patients hospitalized at ICUwith severe to critical COVID-19 disease and from patients with mild symptoms not requiring hospitalization according to the WHO clinical management of COVID-19 guide (World Health Organization WHOa, 2020). Among patients admitted to ICU with severe to critical COVID-19 infection, whenever possible, paired URT and LRT samples were collected. All specimens were collected from late February 2020 to January 2021 at the Microbiology and Virology Department of Fondazione IRCCS Policlinico San Matteo in Pavia as Regional Reference laboratory for COVID-19 diagnosis. The presence of SARS-CoV-2 RNA in respiratory specimens was assessed by using specific real-time reverse transcriptase–polymerase chain reaction (RT-PCR) targeting the RNA-dependent RNA polymerase and E genes, following the WHO guidelines and the Corman et colleagues’ protocols (Corman et al., 2020; World Health Organization WHOb, 2020). Quantification cycle (Cq) values according to MIQE guidelines (Bustin et al., 2009), were used as a semiquantitative measure of SARS-CoV-2 viral load. The sequence investigation of patient samples was approved by the Ethics Committee of our institution (P_20200085574).
2.2. S gene amplification and sequencing
Total RNA was extracted using the QIAamp Viral RNA Mini Kit according to the manufacturer's instructions using a starting volume of 400 µL elute in a final volume of 60 µL.
The extracted RNA was subjected to a one-step RT-PCR using the SuperScript IV One-Step RT-PCR System (Thermo Fisher Scientific, USA). Two strategies were adopted: a “long PCR” for the amplification of the entire S gene (near 4000 bp) using the primer pairs SARS-2-S-F3 (tatcttggcaaaccacgcgaacaa) and SARS-2-S-R3 (acccttggagagtgctagttgccatctc) or using a semi-nested approach with the following two primers pairs: the first step with SARS-2-S-F3 and SARS-2-R6 (ttctgcaccaagtgacatagtgtaggca), followed by a second step with SARS-2-F6 (tcaggatgttaactgcacagaagtcc) and SARS-2-S-R3 (complete list of primers and their position are reported on Appendix Table S2). The thermal profile for the retro transcription was 55 °C for 10 min, followed by the “long PCR” with an initial denaturation/RT inactivation step at 98 °C for 2 min, the amplification for 42 cycles including the initial denaturation (98 °C for 10 s), the annealing step at 60 °C for 10 s, and the extension at 72 °C for 3 min; a final extension of 5 min at 72 °C. The semi-nested PCR was performed using the Platinum SuperFi DNA Polymerase with 5 µl of the first-step DNA and the following thermal program: 98 °C for 30 s, the amplification for 40 cycles including the initial denaturation (98 °C for 10 s), the annealing step at 60 °C for 10 s, and the extension at 72 °C for 2 min and 30 s. The proper PCR products were purified with AMPure Beads, with elution in TE buffer. Enriched DNA samples were used to prepare sequencing libraries with the Nextera XT kit. Sequencing was performed on an Illumina MiSeq machine, aiming for ∼1,000,000 250 bp paired-end reads per sample.
2.3. In silico analysis of sequences
Quality control of sequencing reads was performed using the program FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads in each sample were filtered and trimmed for quality using fastp (Chen et al., 2018). In addition, 28 bases were cut from each end of all reads, to remove sequences generated from the semi-nested PCR primers. Filtered reads were aligned to the S gene of the Wuhan-hu-1 reference genome (NC_045512.2) (Wu et al., 2020) using bowtie2 (Langmead and Salzberg, 2012). Haplotype sequences for each sample were obtained from the alignment SAM files, using the software CliqueSNV (Knyazev et al., 2018). The hedgehog algorithm was used on the most abundant haplotype of each sample to classify the SARS-CoV-2 strains using only the S protein sequences (O'Toole et al., 2022). In parallel, alignment data was processed with samtools (Li et al., 2009) and bam-readcount (https://github.com/genome/bam-readcount) in order to calculate the number of occurrences of each nucleotide and indel in all positions of the reference. Only nucleotides and indels with at least 1% prevalence were considered in the following analyzes.
Python and R scripting (scripts are available from github link) was used to extract and classify all mutations using the following algorithm:
-
(a)
For each position in all samples, the nucleotide or indel with the highest prevalence was called the “majority variant”.
-
(b)
All other bases with at least 1% prevalence were called “minority variants” (MVs).
-
(c)
The correlation between the presence of MVs and the respiratory tract district of sampling was tested for all positions of the gene using the Fisher exact test (p < 0.05).
-
(d)
MVs in each sample were counted and classified by mutation type (synonymous, non-synonymous or indel), gene sub-domain (Huang et al., 2020), and mutation pattern (from which majority base to which low prevalence base). The differential distribution of the number of MVs between URT and LRT samples was tested using the Wilcoxon rank sum test (p < 0.05). The test was repeated for all classes determined, weighting each count on the total number of MVs of the sample.
This allowed us to test their association with LRT or URT (Fisher exact test) and to measure the gene variability sampled in this work. Sequencing reads are available on the SRA database under BioProject ID PRJNA686083. The scripts generated to perform this project are available on GitHub at https://github.com/SteMIDIfactory/DeepSpike.
2.4. Statistical analyzes
Comparisons of Cq, number of minority variants and haplotypes were performed with Mann-Witney test for continuous unpaired variables and in paired respiratory samples with the Wilcoxon rank sum test for continuous paired variables. Correlations between two quantitative variables were measured by the Spearman correlation test. Fisher's exact test for categorical variables was used for analysing mutation frequencies between groups of patients. Descriptive statistics and linear regression lines were performed using Graph Pad Prism software (version 8.3.0).
3. Results
3.1. Patients population
A total of 77 patients were included in the study. Fifty-five (71.4%) of them had severe presentations and were admitted to the ICU, while 22 (28.6%) had mild infections not requiring hospitalization. Among 55 ICU patients, for 28 (50.9%) of them paired URT and LRT samples were available, for 19 (34.5%) patients only LRT samples were analyzed (two of them with two and one with three serially collected BAL, respectively) and for 8 (14.6%) patients only URT samples were available. Among 28 patients with paired URT and LRT samples, the LRT sample was collected at the same time for a great majority of paired samples (range -4 to 9 days of difference). From the 22 patients with mild disease only URT samples were collected and analyzed.
3.2. Dataset description
A total of 109 samples were included in the NGS analyzes, of which 58 (53.2%) were collected from the URT and 51 (46.8%) from the LRT. The DNA of the S gene was enriched with PCR methods and deep short-read sequencing was performed. A total of 69279628 reads were obtained from sequencing, with an average of 635593 reads for sample (range 56214-3346036). Reads were mapped to the SARS-CoV-2 reference strain and the average depth obtained was 6923x (range 1301x-7954x). Reads mapping allowed to extract, classify, and count all genomic variants present in the samples with a prevalence of at least 1% of the sequencing depth (corresponding to ∼10x depth in the samples with the worst read yield).
The hedgehog algorithm was used in order to classify the SARS-CoV-2 strains using only the S protein sequences. The great majority of SARS-CoV-2 included in this study belonged to A_1 lineage harbouring mainly the D614G change (84/109; 77.1%). Thirteen (11.9%) strains belonged to B.1.177_1 lineage (A222V, D614G), 7 (6.4%) to alpha VOC (lineage A_9; del69-70, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H), 4 (3.7%) strains to lineage B.1_14 (D614G, D839Y) and 1 (0.9%) strain to lineage B.1.177.52_1 (A222V, D614G, P1162R) (Supplementary Material Table S2)
3.3. Viral load and correlation between Cq and intra-host variability
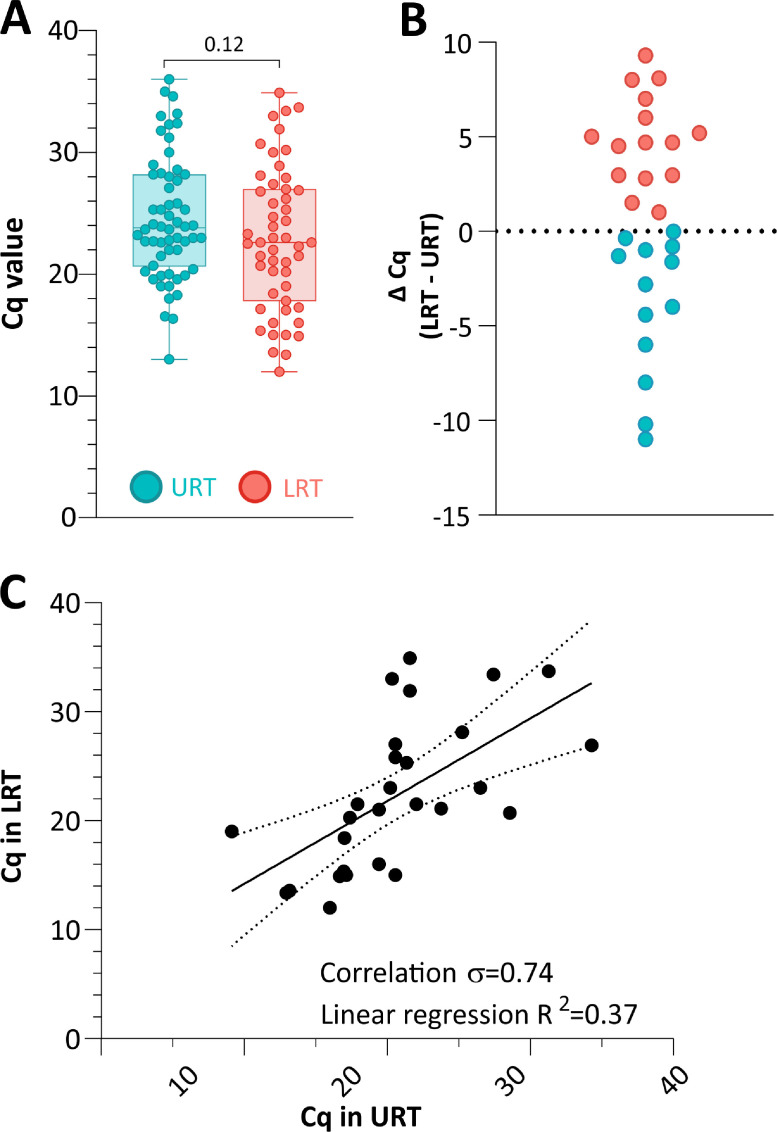
The median viral load measured as Cq value observed in URT samples (23.8; range 13.0–36.0) was similar to those observed in the LRT samples (22.6; range 12.0-34.9; p = 0.12) (Fig. 1 A). Similarly, among 28 paired samples no difference in the median Cq value was observed (p = 0.83). In order to describe the vial load variability on paired samples, we performed a correlation analysis and a plot on difference in Cq value between paired URT and LRT samples. In 13/28 (46.4%) paired samples the Ct value was lower in URT (higher viral load) as compared to LRT samples with a median ΔCq of 2.8 (range 0.04–11.0 ΔCq) (pink circles on Fig.1B), while in 15/28 (53.6%) paired samples, Ct value was lower (higher viral load) in LRT as compared to URT samples with a median ΔCq of 4.7 (range 0.1 to 9.3 ΔCq) (light blue circles on Fig. 1B). In addition, an overall correlation was observed among paired samples (rho=0.74, Fig. 1C).
Fig. 1.
(A) Distribution of Cq in the URT and LRT samples. (B, C) Differences and correlation plots for Cq values in 28 paired URT and LRT samples. The statistic Spearman's correlation coefficient and linear regression R2 value are also reported.
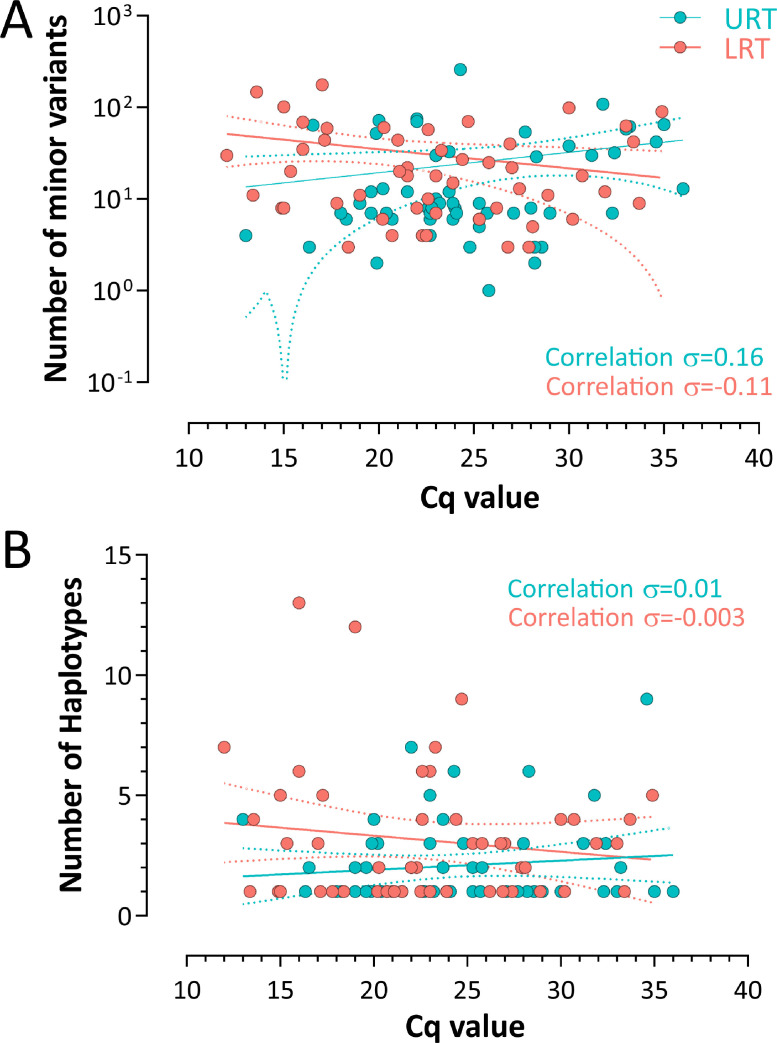
In general, viral replication has been associated with the intra-host diversification of viral population. For this reason, we compared the Cq values, as expression of the viral load, and the number of MVs and haplotypes observed. No evidence of correlation between Cq and number of MVs in both URT (σ=0.16) and LRT samples (σ = 0.11) was observed (Fig. 2 A). Similar findings were observed in the comparison of Cq and the number of haplotypes in URT samples (σ = 0.01) as well as in the LRT (σ = -0.003) (Fig. 2B).
Fig. 2.
Comparison of number of haplotypes and minority variants in upper vs lower respiratory tract samples. (A) Correlation between the number of minority variants and viral load expressed in cycle of quantification (Cq). (B) Correlation between the number of haplotypes and viral load expressed in Cq.
3.4. Haplotype and minority variant counts
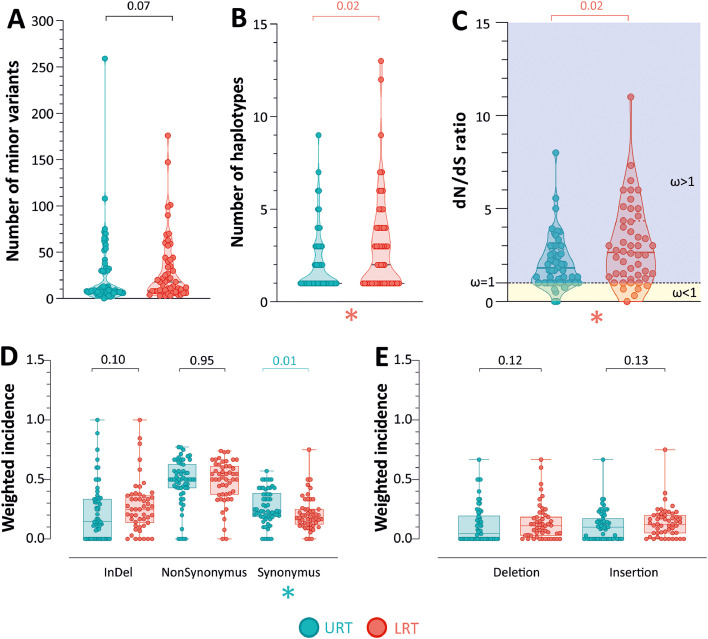
The number of MVs is slightly higher in LRT (median 13.5; range 3–99) than in URT samples (median 8; range 0–263), but with no statistical significance (p = 0.07, Fig. 3 A). Conversely, the number of haplotypes identified was significatively greater in LRT samples (median 2; range 1–13), compared to URT ones (median 1; range 1-9; p = 0.02, Fig. 3B). The ratio of non-synonymous (dN) to synonymous (dS) substitutions (dN/dS) was calculated. The median value observed in LRT samples (median 2.65, range 0–11) was greater than those observed in UTR samples (median 1.81, range 0–8; p = 0.02), suggesting a higher positive selective pressure in the lung environment (Fig. 3C).
Fig. 3.
Distribution of the number of (A) minority variants, (B) haplotypes and (C) dN/dS ratio in the URT and LRT samples. Values are represented as a boxplot with all points inscribed. (D) Distribution of the weighted incidence of synonymous, non-synonymous, and insertions and deletions (Indel) in the URT and the LRT samples. (E) Distribution of the weighted incidence of frameshifting insertions and deletions in the URT and the LRT. Values are weighted by dividing them by the total number of minority variants in the sample.
With a more in-depth analysis, the weighted incidence of synonymous mutations was higher in the URT samples compared to LRT samples (p = 0.01, Fig. 3D), while, although not significant, a greater number of indels was observed in the LRT samples as compared to URT samples, (p = 0.10, Fig. 3D). Lastly, no difference was observed in the number of non-synonymous mutations. The analysis was repeated considering only deletions and frameshifting insertions; both of them had greater incidence in the LRT, without significant difference (p = 0.12 and p = 0.13, Fig. 3E).
The weighted incidence of MVs was calculated in all regions of the gene corresponding to the functional and structural domains of the protein. We observed that MVs in the N-terminal Domain (NTD) are more likely to occur in the LRT (p < 0.001). On the other hand, mutations in the Fusion Peptide (FP, p = 0.006) and in Subdomain 2 (SD2, p < 0.001) are more common in the URT (Fig. 4 A). Furthermore, MVs in the region coding for the protein subunit S1 are more common in the LRT, while in subunit S2 we measured a significantly higher abundance in the URT (Fig. 4B). This result is expected as it is the reflection of the values observed in the functional domains.
Fig. 4.
(A) Distribution of the weighted incidence of minority variants in the S gene subdomains (NTD: N-Terminal Domain; RBD: Receptor-Binding Domain; SD1: Structural Domain 1; SD2: Structural Domain 2; FP: Fusion Peptide; HR1: Heptad Repeat 1; HR2: Heptad Repeat 2; TM: TransMembrane domain) in the URT and LRT samples. (B) Distribution of the weighted incidence of minority variants in the two S gene subunits in the URT and the LRT (S1: Subunit 1; S2: Subunit 2). (C) Distribution in the URT and in the LRT of the weighted incidence of minority variants, classified by mutation patterns. Values are weighted by dividing them by the total number of minority variants in the sample.
Incidence of mutation patterns were tested as well, both counting the total events, and weighting them on the total number of MVs of each sample. Fig. 4C shows the weighted incidence of each mutation pattern in both respiratory tract compartments, while Appendix Fig. S3 shows the absolute one. Both analyzes show a higher presence of A>C and T>G mutations in the MVs of URT samples, while A>T, T>A, and T>C have a higher incidence in the LRT.
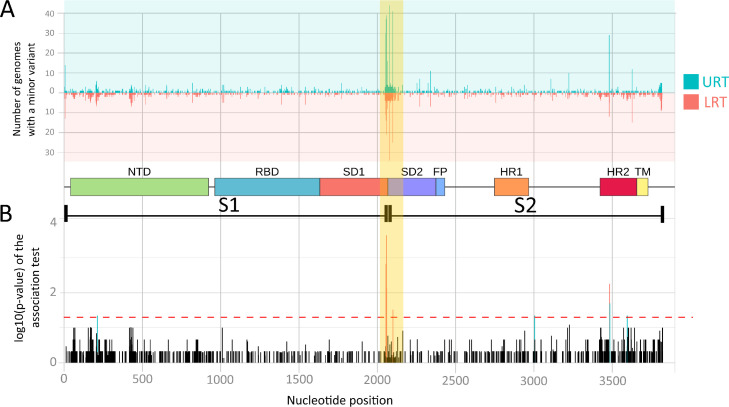
Lastly, we tested the correlation in all nucleotide positions between the presence of MVs (binary value) and the two respiratory tract districts (shown in Fig. 5 A). Fig. 5B, instead, shows the incidence of MVs along the gene. Mutations are equally distributed on the sequence, with the exception of the area around the cleavage site between the two subunits of the gene. Here mutation sites are highly concentrated both in the upper and in the lower respiratory tract samples, especially in the former, as already seen in Fig. 5A. In addition, the presence of mutations is associated with the respiratory district in nine codons, five correlated with URT, four with LRT (see Table 1 ). Four of the MVs associated with URT are located around the cleavage site of the two subunits. In this area, we detected a high concentration of MV transversions between T andG: from position 2000 to position 2150, 248 out of 643 (38.6%) mutations are T>G and 11 (17.3%) are G>T (total = 359; 55.8%)..
Fig. 5.
Graphical distribution of changes along S protein gene. (A) Number of samples containing minority variants in each position. Two separate histograms are used for URT and LRT samples, which are indicated upside down for image clarity. (B) Correlation of the presence of minority variants with URT and LRT in each position of the gene. Bar height represents the log10 (p-value) of the Fisher exact test. In the middle, a scheme of the gene subdomains and subunits is used as separator. NTD: N-Terminal Domain; RBD: Receptor-Binding Domain; SD1: Structural Domain 1; SD2: Structural Domain 2; FP: Fusion Peptide; HR1: Heptad Repeat 1; HR2: Heptad Repeat 2; TM: TransMembrane domain; S1: Subunit 1; S2: Subunit 2.
Table 1.
Description of the 9 Minority Variants (MVs) positions associated with either URT or LRT compared to the reference sequence (NC_045512.2). Global frequency is referred to the frequency of the mutation in the same amino acid in GISAID global database. Data are accessible at www.cov.lanl.gov .
| Codon position | Amino acid | Reference | Mutation | Mutation type | Associated with |
|---|---|---|---|---|---|
| 212 | 71 | C(S) | A(Y)/T(F) | NotSyn | LRT |
| 2055 | 685 | T(R) | G(R) | Syn | URT |
| 2058 | 686 | T(S) | G(R) | NotSyn | URT |
| 2060 | 687 | T(V) | G(G) | NotSyn | URT |
| 2100 | 700 | T(G) | G(G) | Syn | URT |
| 3005 | 1002 | A(Q) | T(L) / - | NotSyn/Del | LRT |
| 3483 | 1161 | A(S) | C(S) | Syn | URT |
| 3485 | 1162 | C(P) | T(L)/G(R) | NotSyn | LRT |
| 3596 | 1199 | A (D) | G(G)/T(G) | NotSyn | LRT |
4. Discussion
The evolution of SARS-CoV-2 was initially quite slow, when compared to other RNA viruses (van Dorp et al., 2020). Yet, its rapid global spread has allowed to record thousands of mutations in public databases; some of those were favourable and have emerged worldwide (Long et al., 2020). The emergence of VOCs was favored by more than 400 million of infections worldwide (Parra-Lucares et al., 2022) with a more significant number of mutations observed in S sequences as compared to other genomic regions (Yusof et al., 2021). Generally, mutations in viral structural such as S glycoprotein can play a crucial role in their virulence by possibly determining changes in their cellular tropism and the generation of antibody escape variants as reported for Delta and Omicron variants (Andrews et al., 2022; Dejnirattisai et al. 2022; Mlcochova et al., 2021). The emergence of these variants has been promoted by the viral quasispecies evolution and the severity of SARS-CoV-2 infection is driven by progression from URT to LRT (Ke et al., 2020). In this perspective, our study has investigated the genetic diversity in SARS-CoV-2 quasispecies focusing on structural S protein sequences in two body compartments in order to: (i) evaluate the genetic diversity in two different body compartments; (ii) identify minority variants potentially associated with progression from upper to lower respiratory tract on paired samples from patients admitted to intensive care unit with severe infection.
Among SARS-CoV-2 evolution, G614 variant in S protein has become worldwide predominant since April 2020 and was associated with an increased fitness advantage (Korber at al., 2020). This finding was also confirmed by other studies that compared D614 and G614 variants and found that G614 was associated with increased replication in human lung epithelial cells (Shi et al., 2020). On the contrary, G614 variant was not associated with an increased disease severity and its role in pathogenesis has yet to be elucidated (Long et al., 2020; Shi et al., 2020). More than 60% of patients included in this study had severe infections developing severe pneumonia and requiring oxygen therapy. Mutations associated with these symptoms were explored by obtaining sequences from the LRT samples. All SARS-CoV-2 sequences generated in this study harboured the G614 variant, while the original D614 variant was found neither among majority nor among minority variants. Thus, we do not report any evidence of the G614 variant favouring the severe presentation. Moreover, no evidence of mutations on S gene associated with progression from upper to lower respiratory tract emerged from our analysis. This result was also observed in a series of paired samples and is consistent with the finding, previously reported by Rueca et al. in a lower number of patients (Rueca et al., 2020). Our data are in keeping with Wylezich et colleagues reporting no evidence of compartment-specific pattern of mutations between different respiratory compartments (Wylezich et al., 2021). This finding suggests us that disease severity could be mainly determined by host factor such as comorbidities, age and absence of pre-existing immunity (Al Khatib et al., 2020). In addition, deletions have been observed as MVs in a few samples but with a lower frequency than that observed in a recent publication (Andrés et al. 2020).
Overall, the S gene sequences originated in our study showed a greater variability (number of haplotypes) in LRT as compared to URT samples. This difference was unrelated to the viral load (measured as Ct) since comparable Ct values were observed in URT and LRT samples. Indeed, no correlation between viral load and viral diversity was observed and this is consistent with the finding previously reported by Siqueira et colleagues, who investigated quasispecies variation in cancer patients (Siqueira et al., 2020). The difference in viral population between URT and LRT could be explained by the hypothesis of an independent replication in the two respiratory districts also suggested by Wölfel et al. (2020). In addition, a higher positive selective pressure in the lung environment has been observed as compared to upper respiratory tract (dN/dS>1). Similar observation was reported by Sun et al suggesting that diversifying of the quasispecies mutants indicated potential independent virus replication in different tissues or organs (Sun et al. 2021). The great variability in the LRT samples resulted also in an increased number of frameshifting deletion and insertions. The presence of deleterious mutations could indicate a loss of function of the S protein in a fraction of the viral population. This subpopulation might be maintained thanks to replication and cell invasions events that do not imply the ACE2 receptor (e.g. within syncytia or in macrophages after phagocytosis)(Abassi et al., 2020). However, this and other theories on how such mutations can influence viral replication in lung tissues might be elucidated and extensively investigated with additional studies.
Nucleotides changes arise during the virus replication and persistence, in particular A>G. These were shown to be related to the host editing mechanisms such as the APOBEC and ADARs (Carpenter et al., 2009; Di Giorgio et al., 2020). The A>G transition is caused by the deamination from Adenosine to Inosine (A>I) generated by the ADARs. Thus, the significant rate of T>C observed in the LRT both by Di Giorgio et al. (2020) and in this study agrees with the hypothesis that T>C in SARS-CoV-2 could also be related to the ADARs mechanism32. Although not associated with host editing mechanism, the T>G pattern in MVs is also of particular interest in this study, as they are the most prevalent pattern observed in the entire dataset and they were found associated with the URT.
Finally, the cleavage site between S1 and S2 of the S protein corresponds to one of the two genetic sites in which Andersen and colleagues found crucial mutations associated with the spillover to humans (Andersen et al., 2020). For this reason, we can hypothesize the presence of an evolutionary selective pressure in this site and that the driving force of this evolution might reside in the URT. Modifications (mutations or deletion) in the S1/S2 junction site has been associated with virus attenuation in hamsters (Wang et al., 2021). Alternatively, these observations might be explained by an absence of negative selection on random occurring mutations: this is a cleavage site; thus, SNPs affect the protein structure much less than in other sites. This last hypothesis explains the high density of mutations in the cleavage site (S1/S2 junction) both in URT and LRT samples. However, it does not explain the higher incidence in the URT and the presence of associated mutations. Moreover, other studies found specific low frequency mutations around the cleavage site, i.e. deletions that were associated with milder symptoms (Andrés et al., 2020; Lau et al., 2020). Whereas, in our study mutations are mainly T>G and G>T transversions in this site (359/643 MVs). Since such mutations are usually rare changes in nucleic acids, this observation underlines further the presence of a selective pressure. Interestingly, the T>G pattern was also identified as an inexplicable intra-host mutational signature in HPV (Zhu et al., 2020).
In conclusion, the results of the present study indicate that severe SARS-CoV-2 infections are not associated with a specific mutational pattern. However, a great variability was observed on viral population in LRT also associated with a positive selective pressure. How the difference may impact immune response escape, tissue tropism and pathogenicity is still to be elucidated. Moreover, we observed the evidence of possible ongoing evolution in one of the gene loci that were crucial for the spillover to humans. This highlights the importance of genomic surveillance to predict and avoid vaccine escape mutants.
CRediT authorship contribution statement
Stefano Gaiarsa: Writing – original draft, Software, Formal analysis, Data curation. Federica Giardina: Writing – original draft, Formal analysis, Investigation. Gherard Batisti Biffignandi: Methodology, Software, Formal analysis, Data curation. Guglielmo Ferrari: Investigation. Aurora Piazza: Validation, Investigation, Data curation. Monica Tallarita: Investigation. Federica Novazzi: Investigation. Claudio Bandi: Writing – review & editing. Stefania Paolucci: Resources, Visualization. Francesca Rovida: Investigation, Resources. Giulia Campanini: Writing – review & editing. Antonio Piralla: Conceptualization, Methodology, Writing – original draft, Project administration, Funding acquisition. Fausto Baldanti: Supervision, Project administration, Writing – review & editing.
Declaration of Competing of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgments
We thank Daniela Sartori for manuscript editing. This study was supported by Ricerca Finalizzata from Ministry of Health, Italy (grant no. GR-2013-02358399 and COVID-2020-12371817).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198786.
Appendix. Supplementary materials
References
- Abassi Z., Knaney Y., Karram T., Heyman S.N. The lung macrophage in SARS-CoV-2 infection: a friend or a foe? Front. Immunol. 2020;11:1312. doi: 10.3389/fimmu.2020.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Khatib H.A., Benslimane F.M., Elbashir I.E., Coyle P.V., Al Maslamani M.A., Al-Khal A., Al Thani A.A., Yassine H.M. Within-host diversity of SARS-CoV-2 in COVID-19 patients with variable disease severities. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.575613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés C., Garcia-Cehic D., Gregori J., Piñana M., Rodriguez-Frias F., Guerrero-Murillo M., Esperalba J., Rando A., Goterris L., Codina M.G., Quer S., Martín M.C., Campins M., Ferrer R., Almirante B., Esteban J.I., Pumarola T., Antón A., Quer J. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg. Microbes Infect. 2020;9(1):1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., Gower C., Kall M., Groves N., O'Connell A.M., Simons D., Blomquist P.B., Zaidi A., Nash S., Iwani Binti Abdul Aziz N., Thelwall S., Dabrera G., Myers R., Amirthalingam G., Gharbia S., Barrett J.C., Elson R., Ladhani S.N., Ferguson N., Zambon M., Campbell C.N.J., Brown K., Hopkins S., Chand M., Ramsay M., Lopez Bernal J. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N. Engl. J. Med. 2022 doi: 10.1056/NEJMoa2119451. 10.1056/NEJMoa2119451. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carpenter J.A., Keegan L.P., Wilfert L., O'Connell M.A., Jiggins F.M. Evidence for ADAR-induced hypermutation of the drosophila sigma virus (Rhabdoviridae) BMC Genet. 2009;10:75. doi: 10.1186/1471-2156-10-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. (Oxford, England) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. Bull. Eur. Mal. Transm. Eur. Commun. Dis. Bull. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., Nutalai R., Wang B., Dijokaite A., Khan S., Avinoam O., Bahar M., Skelly D., Adele S., Johnson S.A., Amini A., Ritter T.G., Mason C., Dold C., Pan D., Assadi S., Bellass A., Omo-Dare N., Koeckerling D., Flaxman A., Jenkin D., Aley P.K., Voysey M., Costa Clemens S.A., Naveca F.G., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Baillie V., Serafin N., Kwatra G., Da Silva K., Madhi S.A., Nunes M.C., Malik T., Openshaw P.J.M., Baillie J.K., Semple M.G., Townsend A.R., Huang K.A., Tan T.K., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Constantinides B., Webster H., Crook D., Pollard A.J., Lambe T., OPTIC Consortium. ISARIC4C Consortium. Paterson N.G., Williams M.A., Hall D.R., Fry E.E., Mongkolsapaya J., Ren J., Schreiber G., Stuart D.I., Screaton G.R. SARS-CoV-2 omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–484. doi: 10.1016/j.cell.2021.12.046. e15Epub 2022 Jan 4. PMID: 35081335; PMCID: PMC8723827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio S., Martignano F., Torcia M.G., Mattiuz G., Conticello S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Science Advances. 2020;6(25):eabb5813. doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E., Perales C. Viral quasispecies. PLoS Genet. 2019;15(10) doi: 10.1371/journal.pgen.1008271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet. Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., COVID-19 Genomics UK (COG-UK) Consortium. Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nature reviews. Microbiology. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jary A., Leducq V., Malet I., Marot S., Klement-Frutos E., Teyssou E., Soulié C., Abdi B., Wirden M., Pourcher V., Caumes E., Calvez V., Burrel S., Marcelin A.G., Boutolleau D. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020;26(11):1560.e1–1560.e4. doi: 10.1016/j.cmi.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamitros T., Papadopoulou G., Bousali M., Mexias A., Tsiodras S., Mentis A. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J. Clin. Virol. 2020;131 doi: 10.1016/j.jcv.2020.104585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke R., Zitzmann C., Ribeiro R.M., et al. Kinetics of SARS-CoV-2 infection in the human upper and lower respiratory tracts and their relationship with infectiousness. medRxiv. 2020 doi: 10.1101/2020.09.25.20201772. 2020.09.25.20201772. [DOI] [Google Scholar]
- Knyazev S., Tsyvina V., Shankar A., Melnyk A., Artyomenko A., Malygina T., Porozov Y.B., Campbell E.M., Switzer W.M., Skums P., Zelikovsky A., Knyazev S. CliqueSNV: an efficient noise reduction technique for accurate assembly of viral variants from NGS data. biorXiv. 2018 doi: 10.1101/264242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group. McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., Li W.J., Jiang B.G., Wei W., Yuan T.T., Zheng K., Cui X.M., Li J., Pei G.Q., Qiang X., Cheung W.Y., Li L., Sun F., Qin S., Huang J., Leung G.M., Holmes E.C., Hu Y., Guan Y., Cao W.C. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583(7815):282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.Y., Wang P., Mok B.W., Zhang A.J., Chu H., Lee A.C., Deng S., Chen P., Chan K.H., Song W., Chen Z., To K.K., Chan J.F., Yuen K.Y., Chen H. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020;9(1):837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. & 1000 Genome project data processing subgroup. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. The Sequence Alignment/Map format and SAMtools. (Oxford, England)10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.W., Olsen R.J., Christensen P.A., Bernard D.W., Davis J.J., Shukla M., Nguyen M., Saavedra M.O., Yerramilli P., Pruitt L., Subedi S., Kuo H.C., Hendrickson H., Eskandari G., Nguyen H., Long J.H., Kumaraswami M., Goike J., Boutz D., Gollihar J., McLellan J.S., Chou C.W., Javanmardi K., Finkelstein I.J., Musser J.M. Molecular architecture of early dissemination and massive second wave of the SARS-CoV-2 virus in a major Metropolitan area. mBio. 2020;11(6):e02707–e02720. doi: 10.1128/mBio.02707-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I., Datir R., Collier D.A., Albecka A., Singh S., Pandey R., Brown J., Zhou J., Goonawardane N., Mishra S., Whittaker C., Mellan T., Marwal R., Datta M., Sengupta S., Ponnusamy K., Radhakrishnan V.S., Abdullahi A., Charles O., Chattopadhyay P., Devi P., Caputo D., Peacock T., Wattal C., Goel N., Satwik A., Vaishya R., Agarwal M., Indian SARS-CoV-2 Genomics Consortium (INSACOG) Genotype to Phenotype Japan (G2P-Japan) Consortium. CITIID-NIHR BioResource COVID-19 Collaboration. Mavousian A., Lee J.H., Bassi J., Silacci-Fegni C., Saliba C., Pinto D., Irie T., Yoshida I., Hamilton W.L., Sato K., Bhatt S., Flaxman S., James L.C., Corti D., Piccoli L., Barclay W.S., Rakshit P., Agrawal A., Gupta R.K. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature, 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. Epub 2021 Sep 6. PMID: 34488225; PMCID: PMC8566220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole Á., Pybus O.G., Abram M.E., Kelly E.J., Rambaut A. Pango lineage designation and assignment using SARS-CoV-2 spike gene nucleotide sequences. BMC Genom. 2022;23(1):121. doi: 10.1186/s12864-022-08358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Lucares A., Segura P., Rojas V., Pumarino C., Saint-Pierre G., Toro L. Emergence of SARS-CoV-2 variants in the world: how could this happen? Life. 2022;12(2):194. doi: 10.3390/life12020194. (Basel, Switzerland) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Lago L., Aldámiz-Echevarría T., García-Martínez R., Pérez-Latorre L., Herranz M., Sola-Campoy P.J., Suárez-González J., Martínez-Laperche C., Comas I., González-Candelas F., Catalán P., Muñoz P., García de Viedma D., On Behalf Of Gregorio Marañón Microbiology-Id Covid Study Group . Different within-host viral evolution dynamics in severely immunosuppressed cases with persistent SARS-CoV-2. Biomedicines. 2021;9(7):808. doi: 10.3390/biomedicines9070808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueca M., Bartolini B., Gruber C., Piralla A., Baldanti F., Giombini E., Messina F., Marchioni L., Ippolito G., Di Caro A., Capobianchi M.R. Compartmentalized replication of SARS-Cov-2 in upper vs. lower respiratory tract assessed by whole genome quasispecies analysis. Microorganisms. 2020;8(9):1302. doi: 10.3390/microorganisms8091302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., Guo L., Zhang G., Li H., Xu Y., Chen M., Gao Z., Wang J., Ren L., Li M. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020;71(15):713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P.Y., Plante J., Liu Y., Liu J., Xia H., Johnson B., Lokugamage K., Zhang X., Muruato A., Zou J., Fontes-Garfias C., Mirchandani D., Scharton D., Kalveram B., Bilello J., Ku Z., An Z., Freiberg A., Menachery V., Xie X., Weaver S. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. Res. Sq. 2020:70482. doi: 10.21203/rs.3.rs-70482/v1. rs.3.rs. [DOI] [Google Scholar]
- Simmonds P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5(3):e00408–e00420. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira J.D., Goes L.R., Alves B.M., de Carvalho P.S., Cicala C., Arthos J., Viola J., de Melo A.C., Soares M.A. SARS-CoV-2 genomic and quasispecies analyses in cancer patients reveal relaxed intrahost virus evolution. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.08.26.267831. 2020.08.26.267831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Wang X., Tan S., Dan Y., Lu Y., Zhang J., Xu J., Tan Z., Xiang X., Zhou Y., He W., Wan X., Zhang W., Chen Y., Tan W., Deng G. SARS-CoV-2 quasispecies provides an advantage mutation pool for the epidemic variants. Microbiol. Spectr. 2021;9(1) doi: 10.1128/Spectrum.00261-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Richard D., Tan C., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11(1):5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O'Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., COVID-19 Genomics UK (COG-UK) consortium. Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593(7858):266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- Wang P., Lau S.Y., Deng S., Chen P., Mok B.W., Zhang A.J., Lee A.C., Chan K.H., Tam R.C., Xu H., Zhou R., Song W., Liu L., To K.K., Chan J.F., Chen Z., Yuen K.Y., Chen H. Characterization of an attenuated SARS-CoV-2 variant with a deletion at the S1/S2 junction of the spike protein. Nat. Commun. 2021;12(1):2790. doi: 10.1038/s41467-021-23166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHOa). 2020. Clinical management of severe acute respiratory infection when novel coronavirus (2019-nCoV) infection is suspected: interim guidance. World Health Organization. Available at: https://apps.who.int/iris/handle/10665/330893 [Accessed on 19 October 2020].
- World Health Organization (WHOb). 2020. Diagnostic detection of 2019-nCoV by real-time RT-PCR. Available at: https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf. [Accessed 30 January 2020].
- World Health Organization (WHO). 2021. Terms of Reference for the Technical Advisory Group on SARS-CoV-2 Virus Evolution (TAG-VE). COVID-19: Surveillance, case investigation and epidemiological protocols. Available at: https://www.who.int/publications/m/item/terms-of-reference-for-the-technical-advisory-group-on-sars-cov-2-virus-evolution-(tag-ve) [Accessed 1 December 2021].
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wong M.C., Javornik Cregeen S.J., Ajami N.J., Petrosino J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.02.07.939207. 2020.02.07.939207. [DOI] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylezich C., Schaller T., Claus R., Hirschbühl K., Märkl B., Kling E., Spring O., Höper D., Schlegel J., Beer M., Dintner S. Whole-genome analysis of SARS-CoV-2 samples indicate no tissue specific genetic adaptation of the virus in COVID-19 patients' upper and lower respiratory tract. Diagn. Microbiol. Infect. Dis. 2021;101(4) doi: 10.1016/j.diagmicrobio.2021.115520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., Zhang Z., Shu F., Huang W., Li Y., Zhang Z., Chen R.A., Wu Y.J., Peng S.M., Huang M., Xie W.J., Cai Q.H., Hou F.H., Chen W., Xiao L., Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583(7815):286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Yusof W., Irekeola A.A., Wada Y., Engku Abd Rahman E., Ahmed N., Musa N., Khalid M.F., Rahman Z.A., Hassan R., Yusof N.Y., Yean Yean C. A global mutational profile of SARS-CoV-2: a systematic review and meta-analysis of 368,316 COVID-19 patients. Life. 2021;11(11):1224. doi: 10.3390/life11111224. (Basel, Switzerland) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wangm Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Xiao Y., Yeager M., Clifford G., Wentzensen N., Cullen M., Boland J.F., Bass S., Steinberg M.K., Raine-Bennett T., Lee D., Burk R.D., Pinheiro M., Song L., Dean M., Nelson C.W., Burdett L., Yu K., Roberson D., Lorey T., Franceschi S., Castle P.E., Walker J., Zuna R., Schiffman M., Mirabello L. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 2020;11(1):886. doi: 10.1038/s41467-020-14730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.