Abstract
Background
SARS-CoV-2 Omicron variant of concern (VOC) has evolved multiple mutations within the spike protein, raising concerns of increased antibody evasion. In this study, we assessed the neutralization potential of COVID-19 convalescent sera and sera from vaccinated individuals against ancestral SARS-CoV-2 and VOCs.
Methods
The neutralizing activity of sera from 65 coronavirus disease (COVID-19) vaccine recipients and convalescent individuals against clinical isolates of ancestral SARS-CoV-2 and Beta, Delta, and Omicron VOCs was assessed using a micro-neutralization assay.
Findings
Convalescent sera from unvaccinated individuals infected by the ancestral virus demonstrated reduced neutralization against Beta and Omicron VOCs. Sera from individuals that received three doses of the Pfizer or Moderna vaccines demonstrated reduced neutralization of the Omicron variant relative to ancestral SARS-CoV-2. Sera from individuals that were naturally infected with ancestral SARS-CoV-2 and subsequently received two doses of the Pfizer vaccine induced significantly higher neutralizing antibody levels against ancestral virus and all VOCs. Infection alone, either with ancestral SARS-CoV-2 or the Delta variant, was not sufficient to induce high neutralizing antibody titers against Omicron.
Conclusions
In summary, we demonstrate that convalescent and vaccinated sera display varying levels of SARS-CoV-2 VOC neutralization. Data from this study will inform booster vaccination strategies against SARS-CoV-2 VOCs.
Funding
This research was funded by the Canadian Institutes of Health Research (CIHR). VIDO receives operational funding from the Government of Saskatchewan through Innovation Saskatchewan and the Ministry of Agriculture and from the Canada Foundation for Innovation through the Major Science Initiatives for its CL3 facility.
Keywords: SARS-CoV-2, neutralization, variants of concern, Omicron, vaccine, neutralizing antibodies, convalescent, microneutralization, Delta, long-term
Graphical abstract
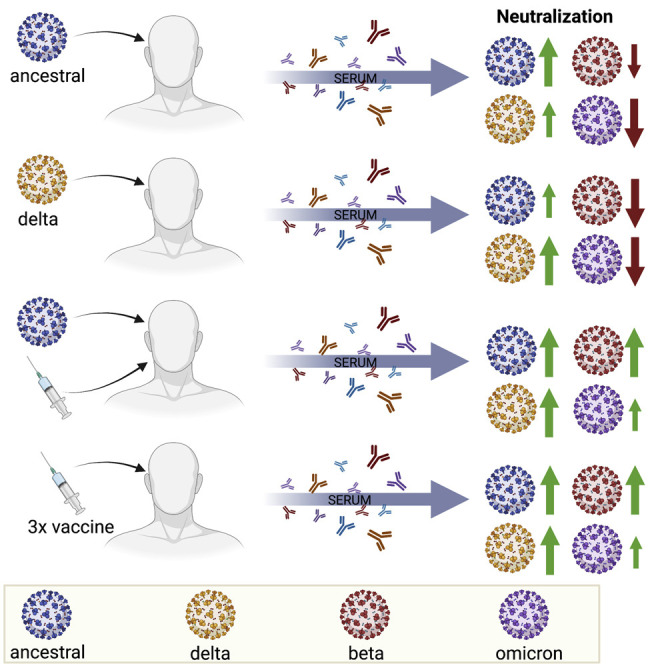
Context and significance
SARS-CoV-2 Omicron variant of concern (VOC) has evolved multiple mutations within the spike protein, raising concerns of increased antibody evasion. In this study, we quantified neutralizing antibody levels in convalescent and vaccinated sera against ancestral SARS-CoV-2 and VOCs. Convalescent sera had lower neutralizing antibody levels against the Omicron VOC. Two doses of an mRNA vaccine following infection induced high levels of neutralizing antibodies against all VOCs, including Omicron. Three doses of authorized mRNA vaccines induced detectable but lower levels of neutralizing antibodies against VOCs in long-term care residents. Data from our study, along with other published studies, support the utility of third vaccine doses and will help inform future booster vaccination strategies to tackle the ongoing COVID-19 pandemic.
Banerjee et al. demonstrate that sera from convalescent individuals following ancestral SARS-CoV-2 or Delta infection have low neutralizing activity against Omicron. Three doses of mRNA vaccines induce detectable levels of neutralizing antibodies against all VOCs in long-term care residents. Omicron is more resistant to antibody-mediated neutralization compared to other VOCs.
Introduction
SARS-CoV-2 has continued to evolve since its emergence in December 2019.1 , 2 Variants of SARS-CoV-2 that demonstrate potential for interference with diagnostics, therapies, and vaccine efficacy, along with evidence for increased transmissibility or disease severity are termed variants of concern (VOCs). The most recent VOC, Omicron, was first reported in November 2021 in Botswana and South Africa.3 , 4 The Omicron variant has evolved multiple mutations within the spike protein and the receptor binding domain (RBD) that raise concerns regarding a possible increased ability to evade pre-existing antibodies, both from prior infection and from vaccination.5 The Omicron variant has demonstrated increased transmission and a higher level of resistance to antibody-mediated neutralization.4 , 5 However, little is known about its pathogenicity and whether disease severity is altered in convalescent, vaccinated, or unvaccinated individuals. In Canada, long-term care (LTC) residents were prioritized for third vaccine doses against SARS-CoV-2 based on the observation that antibody titers in older adults waned within 6 months of their second vaccine dose.6 , 7 The neutralizing potential of antibodies generated in LTC residents against VOCs such as Delta and Omicron after three doses of mRNA vaccines remain unknown. Thus, to better assess the efficacy of antibody-mediated neutralization against ancestral SARS-CoV-2 and VOCs (Beta, Delta, and Omicron) in naturally infected and vaccinated individuals, we collected sera from multiple cohorts and tested their neutralization ability against clinical isolates of ancestral SARS-CoV-2 and VOCs.
Results
Isolation of viruses
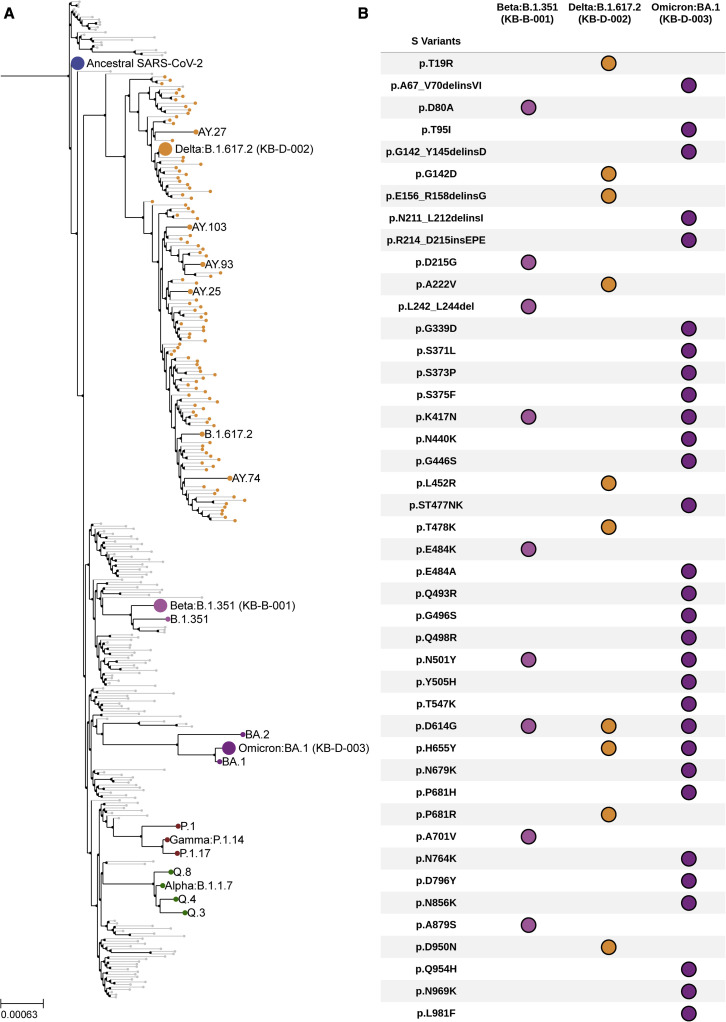
Isolates of VOCs used in this study were derived from clinical specimens. Nasopharyngeal swabs were collected from PCR-positive patients, and virus isolation was performed on African Green monkey kidney cells (Vero’76) as previously described.8 We confirmed the whole genome sequence of the isolates and determined their phylogenetic relationship with other SARS-CoV-2 isolates (Figure 1A and see Table S1). Beta, Delta, and Omicron isolates used in this study aligned with their expected lineages (Figure 1A). SARS-CoV-2 can rapidly adapt in cell culture and evolve adaptive mutations. We confirmed mutations across the full-length viral genome, including the spike protein for all variants prior to using the viruses in a micro-neutralization assay (Figure 1B and see Table S2).
Figure 1.
Spike mutations and phylogenetic analyses of clinical isolates of VOCs
(A) Clinical isolates of VOCs used in this study (Delta, B.1.617.2 kB-D-002; Beta, B.1.351 kB-B-001; and Omicron, BA.1 kB-D-003) were sequenced and assigned lineages by phylogeny analysis.
(B) Mutations within the spike (S) protein of each variant are shown here. Delins, deletion + insertion; ins, insertion; p, amino acid position. See also Tables S1 and S2.
Neutralization of Omicron by convalescent sera from individuals infected with ancestral SARS-CoV-2 or the Delta variant
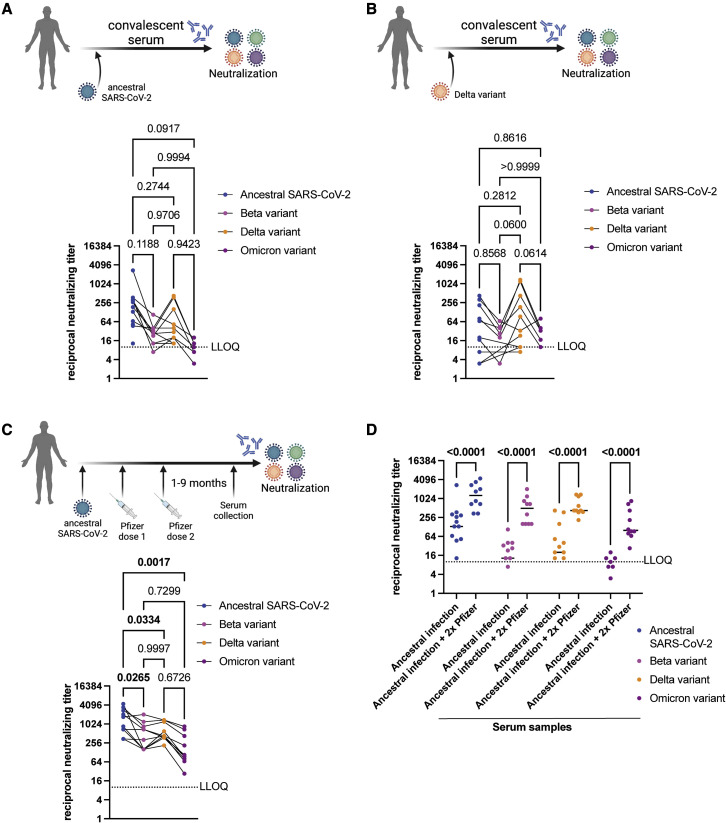
To determine the neutralizing titer of sera from individuals that were naturally infected with SARS-CoV-2, we tested convalescent sera from individuals that were infected with ancestral SARS-CoV-2 during the first wave of coronavirus disease (COVID-19) in Canada (July 2020; see Table S3). Serum samples were collected 1–5 months after the onset of COVID-19 (see Table S3). Convalescent sera (n = 15) from individuals infected with ancestral SARS-CoV-2 during the first wave of COVID-19 in Canada contained lower neutralizing antibodies against both Beta and Omicron VOCs, relative to the ancestral virus, albeit the difference was not statistically significant (Figure 2A). Neutralizing antibody titers in these serum samples were not significantly different between ancestral SARS-CoV-2 and the Delta variant (Figure 2A). Next, the neutralizing antibody titers in convalescent sera (n = 10) from individuals who were infected with the Delta variant in Canada between May and June 2021 were determined (see Table S3). Serum samples were collected 1–2 months after the date of onset of COVID-19 (see Table S3). Convalescent Delta sera contained lower levels of neutralizing antibodies against both Beta and Omicron variants, relative to the Delta variant (Figure 2B), while titers against ancestral SARS-CoV-2 and Delta were comparable (Figure 2B). These data suggest that infection with either ancestral SARS-CoV-2 or the Delta variant induces cross-neutralizing antibodies with comparable titers against both viruses. However, natural infection with either ancestral SARS-CoV-2 or the Delta variant induces lower levels of neutralizing antibodies against both Beta and Omicron variants.
Figure 2.
Detection of neutralizing antibodies in convalescent sera and sera from naturally infected individuals who subsequently received two doses of an mRNA vaccine
(A) Neutralizing antibody titers in convalescent sera collected from individuals that were infected with ancestral SARS-CoV-2 during the first wave in Canada (July 2020; n = 15) tested against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants. Three samples had undetectable neutralizing antibody titers against ancestral SARS-CoV-2. Six samples had undetectable neutralizing antibody titers against Beta VOC. Five samples had undetectable neutralizing antibody titers against Delta VOC. Eight samples had undetectable neutralizing antibody titers against Omicron VOC.
(B) Neutralizing antibody titers in convalescent sera collected from individuals that were infected with the Delta variant of SARS-CoV-2 in Canada (May–June 2021; n = 10) tested against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants. Four samples had undetectable neutralizing antibody titers against Beta VOC. Four samples had undetectable neutralizing antibody titers against Omicron VOC.
(C) Neutralizing antibody titers in sera collected from individuals that were infected with ancestral SARS-CoV-2, followed by two doses of the Pfizer BNT162b2 vaccine tested against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants (n = 10).
(D) Neutralizing antibody titers in convalescent sera from individuals infected with ancestral SARS-CoV-2 compared to neutralizing antibody levels in sera from individuals who were infected with the ancestral virus and subsequently received two doses of the Pfizer mRNA vaccine. Data compiled and replotted from (A) and (C) for comparison. Median values are indicated by horizontal black bars. p values are indicated in the figure (Mann-Whitney test). In Figures 1A–1C, individual data points are shown and titers for matching serum samples are shown across different virus isolates. n = 15 or 10, p values are indicated in the figures (Tukey’s multiple comparisons test with alpha = 0.05 or Sidak’s multiple comparisons test with alpha = 0.05). Significant p values are highlighted in bold. Samples with neutralizing titer of 0 are not shown. LLOQ, lower limit of quantitation. Schematic made with BioRender.com. See also Table S3.
Neutralization of Omicron by sera from individuals who received two doses of the Pfizer BNT162b2 mRNA vaccine post COVID-19 infection
We determined levels of neutralizing antibodies in sera (n = 10) from individuals who had received two doses of the Pfizer BNT162b2 vaccine after being naturally infected with ancestral SARS-CoV-2 (Table S3). Two doses of the Pfizer BNT162b2 vaccine after natural infection led to higher levels of neutralizing antibodies against ancestral SARS-CoV-2 that were significantly higher than levels against the Beta (p = 0.0265), Delta (p = 0.0334), and Omicron (p = 0.0017) variants (Figure 2C). Indeed, infection and subsequent two-dose vaccination with Pfizer BNT162b2 induced higher levels of neutralizing antibodies against ancestral virus and all VOCs (Figure 2D).
Neutralization of Omicron by sera from individuals that received one dose of the Pfizer BNT162b2 mRNA vaccine
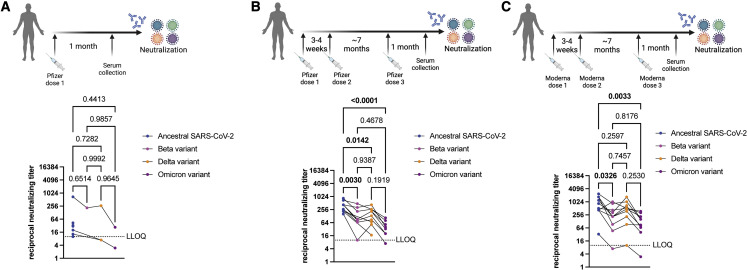
To determine neutralizing antibody titers in sera from individuals that received one dose of the Pfizer BNT162b2 mRNA vaccine, we collected sera (n = 10) 1 month after the first dose of the vaccine and tested neutralizing antibody titers against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants (Table S3). Low neutralizing antibody titers against the ancestral virus were detected in some samples; however, no neutralization of the three VOCs was observed, except for one serum sample that had detectable levels of neutralizing antibodies against all VOCs (Figure 3A).
Figure 3.
Detection of neutralizing antibodies in sera from vaccinated individuals
(A) Neutralizing antibody titers in sera collected from individuals that received one dose of the Pfizer BNT162b2 vaccine (n = 10) tested against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants. Three samples had undetectable neutralizing antibody titers against ancestral SARS-CoV-2. Nine samples had undetectable neutralizing antibody titers against Beta VOC. Seven samples had undetectable neutralizing antibody titers against Delta VOC. Eight samples had undetectable neutralizing antibody titers against Omicron VOC.
(B) Neutralizing antibody titers in sera collected from individuals that received three doses of the Pfizer BNT162b2 vaccine (n = 10) tested against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants. Two samples had undetectable neutralizing antibody titers against Beta VOC. Two samples had undetectable neutralizing antibody titers against Omicron VOC.
(C) Neutralizing antibody titers in sera collected from individuals that received three doses of the Moderna mRNA-1273 vaccine (n = 10) tested against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants. Individual data points are shown, and titers for matching serum samples are shown across different virus isolates. n = 10, p values are indicated in the figures (Tukey’s multiple comparisons test with alpha = 0.05). Significant p values are highlighted in bold. Samples with neutralizing titer of 0 are not shown. LLOQ, lower limit of quantitation. Schematic made with BioRender.com. See also Table S3.
Neutralization of Omicron by sera from triple-vaccinated individuals
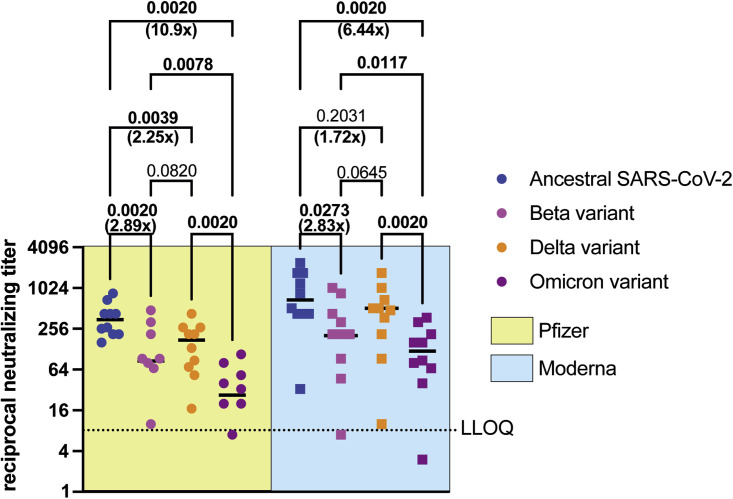
Additional booster vaccinations have been deemed critical in protecting us from VOCs in part by inducing higher levels of neutralizing antibodies. Thus, we tested the levels of neutralizing antibodies in sera collected from LTC residents that received three doses of the Pfizer BNT162b2 (n = 10)9 or the Moderna mRNA-1273 (n = 10)10 vaccines (see Table S3). For both vaccine recipients, doses one and two were received 3–4 weeks apart. The third vaccine dose was received ∼7 months after dose 2, and serum samples were collected 1 month after the third dose. Sera from individuals that received three doses of the Pfizer BNT162b2 vaccine induced high neutralizing titers against ancestral SARS-CoV-2, but levels of neutralizing antibodies were significantly lower against Beta (p = 0.0030), Delta (p = 0.0142), and Omicron (p < 0.0001) variants, compared to ancestral SARS-CoV-2 (Figure 3B). Sera from individuals that received three doses of the Moderna mRNA-1273 vaccine induced high neutralizing titers against ancestral SARS-CoV-2, but levels of neutralizing antibodies were significantly lower against the Beta (p = 0.0326) and Omicron (p = 0.0033) variants, relative to ancestral SARS-CoV-2 (Figure 3C). Neutralizing antibody titers against ancestral SARS-CoV-2 were not significantly different from the Delta variant. Serum samples from individuals that received three doses of the Pfizer BNT162b2 vaccine contained 2.89×, 2.25×, and 10.9× lower median neutralizing antibody titers against Beta, Delta, and Omicron VOCs, respectively, relative to ancestral SARS-CoV-2 (Figure 4 ). Serum samples from individuals that received three doses of the Moderna mRNA-1273 vaccine contained 2.83x, 1.72x, and 6.44x lower median neutralizing antibody titers against Beta, Delta, and Omicron VOCs, respectively, relative to ancestral SARS-CoV-2 (Figure 4).
Figure 4.
Neutralizing antibody levels in individuals vaccinated with three doses of the Pfizer BNT162b2 or Moderna mRNA-1273 vaccines against ancestral SARS-CoV-2 and VOCs
To compare neutralizing antibody titers in sera collected from long-term care residents that received three doses of either the Pfizer BNT162b2 or Moderna mRNA-1273 vaccines, we reanalyzed the data in Figure 3. Neutralizing antibody levels against ancestral SARS-CoV-2, Beta, Delta, and Omicron variants are shown here. Median values are indicated by horizontal black bars. Data are represented as median, n = 15 or 10, p values are indicated in the figure (two-tailed Wilcoxon matched-pairs signed rank test). Significant p values are highlighted in bold. Fold reduction in median neutralizing antibody titers against VOCs relative to ancestral SARS-CoV-2 is indicated. Samples with neutralizing titer of 0 are not shown. LLOQ, lower limit of quantitation. See also Table S3.
Discussion
The emergence of yet another SARS-CoV-2 VOC, Omicron, has led to increasing speculation about the ability of this variant to escape vaccine and natural infection-mediated immunity. The current generation of COVID-19 mRNA vaccines are designed using the spike gene sequence of ancestral SARS-CoV-2.9 , 10 The Omicron variant has accumulated 29 amino acid substitutions, three amino acid deletions, and a three-residue insertion within the spike protein compared to the ancestral SARS-CoV-2 Wuhan isolate.5 Accumulating data suggest that the Omicron variant is at least partially resistant to neutralization by antibodies in vaccinated individuals, along with partial or complete resistance to neutralization by therapeutic monoclonal antibodies.5 Emerging data demonstrate that T cell-mediated immunity generated upon infection or vaccination likely remain effective against the Omicron variant,11 and an additional booster vaccine dose results in higher levels of antibodies against the Omicron variant when tested using pseudotyped viruses.12 Despite these recent advances, considerable gaps currently exist in our knowledge regarding the ability of Omicron to cause severe COVID-19 and whether partial or complete escape of vaccine or natural infection-mediated immunity occurs and if escape is age dependent. In addition, it is not known if Omicron has altered host range, or if transmissibility is increased and whether there are changes in cellular tropism. Furthermore, data on neutralizing antibody titers against clinical isolates of Omicron are limited. Thus, as part of this study, we determined the levels of neutralizing antibodies in individuals that were naturally infected, infected and subsequently vaccinated, or vaccinated with three doses of mRNA vaccines using clinical isolates of ancestral SARS-CoV-2, Beta, Delta, and Omicron variants.
When Omicron was first detected, multiple laboratories reported difficulties in isolating and generating laboratory stocks of this variant. In this study, we used Vero’76 cells to isolate the Omicron variant from a clinical specimen (nasopharyngeal swab) that was collected from a Canadian patient. We also confirmed the whole genome sequences of the Omicron variant, along with Beta and Delta variants (Figure 1 and Table S1). Thus, we report that Vero’76 cells are sufficient to facilitate the isolation and propagation of the Omicron variant.
Next, we tested the levels of neutralizing antibody titers in convalescent sera against the ancestral virus, Beta, Delta, and Omicron variants (Figure 2). Infection with both the ancestral virus and the Delta variant induced high levels of neutralizing antibodies against each other. However, the levels of neutralizing antibodies in convalescent sera against the Omicron variant were lower compared to both the ancestral virus and the Delta variant (Figures 2A and 2B). Thus, our data suggest that infection alone, either with the ancestral virus or the Delta variant, may not be sufficient to induce high levels of neutralizing antibodies against the Omicron variant. Indeed, our data demonstrate that two doses of the Pfizer BNT162b2 mRNA vaccine following infection with ancestral SARS-CoV-2 induced significantly higher levels of neutralizing antibodies against ancestral SARS-CoV-2 and Beta, Delta, and Omicron variants (Figure 2D).
Our data highlight that one dose of the Pfizer vaccine is not sufficient to induce high levels of neutralizing antibodies against ancestral virus or variants (Figure 3A), so the second vaccine dose appears to be critically required to induce neutralizing antibodies. Three doses of either the Pfizer BNT162b2 or Moderna mRNA-1273 vaccine induced comparable neutralizing antibodies against the Beta and Omicron variants (Figures 3B and 3C). However, levels of neutralizing antibodies against Omicron were significantly lower compared to ancestral SARS-CoV-2 in serum samples from individuals vaccinated with three doses of either mRNA vaccine (Figure 3, Figure 4B, 3C, and 4).
In summary, our data demonstrate that infection alone, either with ancestral SARS-CoV-2 or the Delta variant, is not sufficient to induce high levels of neutralizing antibodies against Omicron, a finding which was also recently reported by Rossler et al. 13 However, two doses of the Pfizer vaccine in previously infected individuals induces higher levels of neutralizing antibodies. While we did not test the effect of two doses of the Moderna mRNA-1273 vaccine in previously infected and recovered individuals, we speculate that the results will be comparable to the Pfizer BNT162b2 vaccine. Our data also show that while three doses of either mRNA vaccines induce neutralizing titers against Omicron variant in LTC residents, the levels of neutralizing antibodies remain low compared to ancestral SARS-CoV-2. While antibody-mediated neutralization, and as a corollary, spike binding antibodies are associated with protection from COVID-19,14 there are other immune mechanisms that contribute. Fc-mediated functionality of antibodies15, 16, 17, 18 and cellular immunity also contribute to protection, and they are potentially less affected by the evolution of VOCs.19 Thus, our data support the ongoing third vaccine dose booster strategy for LTC residents in Canada. Indeed, there is a need for studies to establish a correlation between levels of neutralizing antibodies (or other protective immune functions) that result in protection against infection, disease, hospitalization, and/or severe COVID-19, which will allow better policy decisions to be made regarding vaccine boosters and enable equitable distribution of vaccines.
Limitations of the study
In this study, we have assessed the levels of neutralizing antibodies in convalescent and vaccinated sera against ancestral SARS-CoV-2 and Beta, Delta, and Omicron VOCs. The correlates of protection against COVID-19 remain poorly understood. The protective role of cell-mediated immunity during COVID-19 has been demonstrated. Thus, for a holistic understanding of immune-mediated protection in naturally infected and vaccinated individuals, it is necessary to assess both antibody- and cell-mediated immunity. We did not categorize data by sex, race, age, or ethnicity. In the future, it will be important to identify whether sex, race, age, and ethnicity play a significant role in natural and vaccine-mediated immunity against SARS-CoV-2 and VOCs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Ancestral SARS-CoV-2 VIDO-01 isolate | Laboratory of Dr. Darryl Falzarano | (20) |
| SARS-CoV-2 delta VOC (B.1.617.2) | This study | N/A |
| SARS-CoV-2 beta VOC (B.1.351) | This study | N/A |
| SARS-CoV-2 omicron VOC (BA.1) | This study | N/A |
| Critical commercial assays | ||
| LunaScript RT Super-Mix | New England Biolabs | E3010L |
| ARTIC V4 primer pools | Artic-network and IDT | https://github.com/artic-network/artic-ncov2019 |
| Q5 High-Fidelity 2x Master Mix | New England Biolabs | M0492L |
| Deposited data | ||
| Raw data and scripts | This study | https://github.com/fmaguire/voc_neutralisation_sc2_phylogenomics |
| Code | This study | Zenodo Data: https://doi.org/10.5281/zenodo.5817727 |
| Full genomic sequences of viruses | This study | NCBI BioProject PRJNA794206 |
| Supplementary data | This study | Mendeley Data: https://doi.org/10.17632/mfvm8fwf3s.1 |
| Experimental models: Cell lines | ||
| Vero’76 | ATCC | CRL-1587 |
| Software and algorithms | ||
| Prism 9 | GraphPad | www.graphpad.com |
| BioRender | BioRender.com | www.biorender.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by lead contact, Dr. Darryl Falzarano (darryl.falzarano@usask.ca).
Materials availability
This study generated multiple virus isolates. The reagents will be made available on request through institutional Material Transfer Agreements for organizations that have a compliant BSL3 laboratory.
Experimental model and subject details
Viruses
Briefly, the fluid from PCR-positive nasopharyngeal swabs received from Sunnybrook Research Institute (R.K, S.M. – ancestral SARS-CoV-2), omicron was identified by SPAR-Seq (PMID: 33658502) at the joint MSH/UHN Microbiology clinical diagnostic laboratory (J.L.W., T.M., S.M. – omicron) and beta at the Roy Romanow Provincial Laboratory (A.L., R.M.) were centrifuged at 8000xg for 15 min and 50μL removed and mixed with vDMEM containing 1 μg/ml of TPCK trypsin. The mixture was added to a 24 well plate of Vero’76 cells (CRL-1587, ATCC) and centrifuged for 1 h at 37°C at 800xg and then placed at 37°C for 30 min. The inoculum was removed and replaced with fresh vDMEM containing 1 μg/ml of TPCK trypsin. Cells were monitored daily for cytopathic effect and on day 3 or 4, supernatant was passaged to fresh Vero’76 cells in a 6 well plate. Supernatant was subsequently collected on day 3 or 4 and passaged to T175 flasks to generate a p.1 virus stock. Virus stocks were subsequently titered on Vero’76 cells by TCID50 assay. Delta was obtained as a virus stock from the National Microbiology Laboratory (D. Safronetz) and used to generate a stock as described.
For ancestral SARS-CoV-2, we used SARS-CoV-2/VIDO-1, the sequence for which has been previously reported (>hCoV-19/Canada/ON_ON-VIDO-01-2/2020|EPI_ISL_425177|2020-01-23).20 All work with infectious SARS-CoV-2 isolates were performed in a containment level 3 laboratory at the Vaccine and Infectious Disease Organization using approved protocols. Use of clinical specimen for virus isolation and use of human serum samples for micro-neutralization assays were approved by the University of Saskatchewan’s Biomedical Research Ethics Board (REB# 2591).
Cells
Vero’76 cells (CRL-1587, ATCC) were used to isolate and/or propagate all virus isolates using a previously published protocol.8
Human subjects
Serum samples were acquired from a series of different cohorts (see Table S3). Sex and age were reported by the participants. Information on gender, race, ethnicity, and socioeconomic status was not collected. Cohort participants provided informed consent for sharing of serum, and studies were approved by the Sunnybrook Research Institute (REB# 149–1994) and/or the Mount Sinai Hospital (REB# 02-0118-U, 20-0339-E, and 21-0069-E) Research Ethics Board.21 For samples from each cohort, samples were selected to have a representative range of anti-spike trimer and anti-RBD antibodies as measured by enzyme-linked immunosorbent assay.22
Method details
Sequencing and bioinformatic analyses
cDNA was synthesized from extracted RNA. In brief, 4 μL LunaScript RT Super-Mix 5X (New England Biolabs, NEB, USA) and 8 μL nuclease free water, were added to 8 μL extracted RNA. cDNA synthesis was performed using the following conditions: 25°C for 2 min, 55°C for 20 min, 95°C for 1 min, and holding at 4°C.
Amplicons were generated from cDNA using ARTIC V4 primer pools (https://github.com/artic-network/artic-ncov2019). Two multiplex PCR tiling reactions were prepared by combining 2.5 μL cDNA with 12.5 μL Q5 High-Fidelity 2X Master Mix (NEB, USA), 6μL nuclease free water, and 4 μL of respective 10 μM ARTIC v4 primer pool (Integrated DNA Technologies). PCR cycling was then performed in the following manner: 98°C for 30 s followed by 35 cycles of 98°C for 15 s and 63°C for 5 min.
Both PCR reactions were combined and cleaned with 1X ratio Sample Purification Beads (Illumina) at a 1:1 bead to sample ratio. The quantity of amplicons was measured with the Qubit 4.0 fluorometer using the 1X dsDNA HS Assay Kit (Thermo Fisher Scientific, USA) and the sequencing libraries were prepared using the Nextera DNA Flex Prep kit (Illumina, USA) as per manufacturer’s instructions. Paired-end (2 × 150 bp) sequencing was performed on a MiniSeq with a 300–cycle reagent kit (Illumina, USA) with a negative control library with no input SARS-CoV-2 RNA extract.
Raw reads underwent adapter/quality trimming (trim-galore v0.6.5),23 host filtering and read mapping to reference [bwa v0.7.17,24 samtools v.1.7]25 , 26 trimming of primers (iVar v1.327) and variant/consensus calling (freebayes v1.3.2)28 using the SIGNAL workflow21 (https://github.com/jaleezyy/covid-19-signal) v1.4.4dev (#60dd466) with the ARTICv4 amplicon scheme (from https://github.com/artic-network/artic-ncov2019) and the MN908947.3 SARS-CoV-2 reference genome and annotations. Additional quality control and variant effect annotation (SnpEff v5.0-0)29 was performed using the ncov-tools v1.8.0 (https://github.com/jts/ncov-tools/). Finally, PANGO lineages were assigned to consensus sequences using pangolin v3.1.17 (with the PangoLEARN v2021-12-06 models),30 scorpio v0.3.16 (with constellations v0.1.1) [citation: https://github.com/cov-lineages/scorpio], and PANGO-designations v1.2.117.31 Variants were summarised using PyVCF v0.6.8 [citation: https://github.com/jamescasbon/PyVCF] and pandas v1.2.4.32 Phylogenetic analysis was performed using augur v13.1.033 with IQTree (v2.2.0_beta)34 and the resulting phylogenetic figure generated using ETE v3.1.2.35 Contexual sequences were incorporated into the phylogenetic analysis by using Nexstrain’s ingested GISAID metadata and pandas to randomly sample a representative subset of sequences (jointly deposited in NCBI and GISAID) that belonged to lineages observed in Canada (see also Tables S1 and S2 for metadata).
Micro-neutralization assay
Serum samples were heat-inactivated at 56°C for 30 min and then serially diluted 1:2 in DMEM supplemented with 2% FBS and 1% P/S (vDMEM) in 96-well blocks. The final volume of diluted serum per well was 180 μL. Each virus was diluted to 500 TCID50/mL (25 TCID50 per well) in vDMEM, from which 180 μL was added to each well containing serum. Positive controls containing only media and negative controls containing only virus were also included. The serum-virus mixture was incubated at 37 °C for 1 h and then added to cultured Vero’76 cells in 96 well plates in triplicate, with each dilution replicate being placed on a separate plate. On days one, three and five, the cells were evaluated by phase contrast microscopy (100X) for cytopathic effect (CPE), and the endpoint neutralization titer (the highest dilution of sera without CPE) was recorded. Any observation of CPE scores the well as positive; however, by day 5, wells either have widespread CPE or are negative. The final neutralization titer is reported as the average of the three replicates per sample. As a control, each virus isolate that was diluted for our micro-neutralization assay was also back-titered using 2-fold serial dilutions. This micro-neutralization assay was qualified for intra-assay reproducibility, intermediate precision, accuracy, and robustness, including performance by multiple operators. The assay reports lower overall titers than other 50% micro-neutralization, plaque-reduction neutralization 50 and reporter pseudovirus assays, however, the assay may not be sensitive for samples with very low neutralizing activity.
Quantification and statistical analysis
Statistical analysis
Statistical analyses were performed using Tukey’s multiple comparisons test with alpha = 0.05, Sidak’s multiple comparisons test with alpha = 0.05, Mann-Whitney test or Wilcoxon matched-pairs signed rank test. Statistical analyses were performed using GraphPad Prism (version 9.3.1). To compare median values in Figures 2D and 4, non-parametric tests were used as data were not normally distributed as tested using a Shapiro-Wilk normality test. Statistical details can be found in the figure legends. ‘n’ represents the number of serum samples collected from individual humans.
Acknowledgments
SARS-CoV-2 research is supported in the laboratory of D.F. by the Canadian Institutes of Health Research (CIHR; OV5-170349, VRI-173022 and VS1-175531). Research in the laboratory of A.B. is funded by CIHR, Saskatchewan Health Research Foundation (SHRF) and Lung Saskatchewan. Studies from which clinical samples were collected were funded by CIHR grants to A.J.M. and S.M. (#439999, #465038); from the Canadian COVID-19 Immunity Task Force to S.E.S. and A.J.M.; and from the Toronto COVID Action Initiative Fund from the University of Toronto to J.L.W. and T.M. A.B., A-.C.G., J.L.W., S.M., and D.F. are members of the CIHR-funded Coronavirus Variants Rapid Response Network (CoVaRR-Net). R.K. is supported by an Ontario Together grant. We acknowledge contributions by Dr. Andrew G. McArthur who connected our teams to facilitate virus sequencing. We acknowledge the help of Dr. Akarin Asavajaru who handled shipping and receiving of samples.
Author contributions
Conceptualization, A.B., F.M., S.M., and D.F.; sample collection and selection: S.E.S., L.G., A.X.L., M.M., S.W., and A.C.G; methodology, A.B., J.L., A.K., K.B., P.A., F.M., and D.F.; formal analysis, A.B., J.L., A.K., F.M., and D.F.; reagents, J.L., K.N., R.K., R.M., A.L., S.E.S., L.G., A.X.L., M.M., S.W., A.-C.G., J.L.W., T.M., K.C., A.J.M., and S.M.; unrestricted access to all data, A.B., J.L., A.K., and D.F.; funding acquisition, A.B. and D.F.; writing – reviewing and editing, A.B., J.L., A.K., F.M., V.G., S.M., and D.F.; all authors read and approved the final manuscript and take responsibility for its content; supervision, A.B. and D.F.
Declaration of interests
S.W. has served on advisory boards, speaking engagements, attended meetings and symposiums, and conducted clinical studies for ViiV Health Care, GSK, Merck, Janssen, and Gilead Sciences outside of the submitted work. A.J.M. reports income from advisory board membership from Astra-Zeneca, GlaxoSmithKline, Janssen, Medicago, Merck, Moderna, Pfizer, and Sanofi-Pasteur, and research grant funds paid to her institution from Pfizer and Sanofi-Pasteur.
Published: May 4, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.04.002.
Supplemental information
Data and code availability
Full sequences of the viral isolates have been deposited to NCBI BioProject PRJNA794206. All data and scripts associated with analyses of the virus sequences have been deposited at github and is publicly available as of the date of publication at Github Data: https://github.com/fmaguire/voc_neutralisation_sc2_phylogenomics and Zenodo Data: https://doi.org/10.5281/zenodo.5817727. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/mfvm8fwf3s.1. Any additional information required to reanalyze the data reported in this work paper is available from the Lead contact upon request.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh J., Pandit P., McArthur A.G., Banerjee A., Mossman K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol. J. 2021;18:166. doi: 10.1186/s12985-021-01633-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 doi: 10.1038/d41586-021-03832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021 doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 6.Abe K.T., Hu Q., Mozafarihashjin M., Samson R., Manguiat K., Robinson A., Rathod B., Hardy W.R., Wang J.H., Iskilova M., et al. Neutralizing antibody responses to SARS-CoV-2 variants in vaccinated Ontario long-term care home residents and workers. medRxiv. 2021 doi: 10.1101/2021.08.06.21261721. Preprint at. [DOI] [Google Scholar]
- 7.Eliakim-Raz N., Leibovici-Weisman Y., Stemmer A., Ness A., Awwad M., Ghantous N., Stemmer S.M. Antibody titers before and after a third dose of the SARS-CoV-2 BNT162b2 vaccine in adults aged >/=60 Years. Jama. 2021;326:2203–2204. doi: 10.1001/jama.2021.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee A., Nasir J.A., Budylowski P., Yip L., Aftanas P., Christie N., Ghalami A., Baid K., Raphenya A.R., Hirota J.A., et al. Isolation, sequence, infectivity, and replication kinetics of severe acute respiratory syndrome coronavirus 2. Emerging Infect. Dis. 2020;26:2054–2063. doi: 10.3201/eid2609.201495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. SARS-CoV-2 spike T cell responses induced upon vaccination or infection remain robust against Omicron. medRxiv. 2021 doi: 10.1101/2021.12.26.21268380. Preprint at. [DOI] [Google Scholar]
- 12.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. medRxiv. 2021 doi: 10.1101/2021.12.14.21267755. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossler A., Riepler L., Bante D., von Laer D., Kimpel J. SARS-CoV-2 omicron variant neutralization in serum from vaccinated and convalescent Persons. N. Engl. J. Med. 2022;386:698–700. doi: 10.1056/NEJMc2119236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 15.Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e16. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson S.I., Manamela N.P., Motsoeneng B.M., Kaldine H., Ayres F., Makhado Z., Mennen M., Skelem S., Williams N., Sullivan N.J., et al. SARS-CoV-2 Beta and Delta variants trigger Fc effector function with increased cross-reactivity. Cell Rep. Med. 2022;3:100510. doi: 10.1016/j.xcrm.2022.100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussieres G., Brassard N., Laumaea A., Vezina D., Prevost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplonek P., Fischinger S., Cizmeci D., Bartsch Y.C., Kang J., Burke J.S., Shin S.A., Dayal D., Martin P., Mann C., et al. mRNA-1273 vaccine-induced antibodies maintain Fc effector functions across SARS-CoV-2 variants of concern. Immunity. 2022;55:355–365.e4. doi: 10.1016/j.immuni.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geers D., Shamier M.C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Schafer A., Kulkarni S.S., Liu X., Martinez D.R., Chen C., Sun Z., Leist S.R., Drelich A., Zhang L., et al. High potency of a Bivalent human VH domain in SARS-CoV-2 animal models. Cell. 2020;183:429–441.e16. doi: 10.1016/j.cell.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasir J.A., Kozak R.A., Aftanas P., Raphenya A.R., Smith K.M., Maguire F., Maan H., Alruwaili M., Banerjee A., Mbareche H., et al. A comparison of whole genome sequencing of SARS-CoV-2 using amplicon-based sequencing, random Hexamers, and Bait capture. Viruses. 2020;12 doi: 10.3390/v12080895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isho B., Abe K.T., Zuo M., Jamal A.J., Rathod B., Wang J.H., Li Z., Chao G., Rojas O.L., Bang Y.M., et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krueger F. Trim Galore. 2019. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
- 24.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- 25.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20 doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 2012 doi: 10.48550/arXiv.1207.3907. Preprint at. [DOI] [Google Scholar]
- 29.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms. Snpeff. Fly. 2014;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021 doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinney W. 2010. Data Structures for Statistical Computing in Python. [Google Scholar]
- 33.Huddleston J., Hadfield J., Sibley T., Lee J., Fay K., Ilcisin M., Harkins E., Bedford T., Neher R., Hodcroft E. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. J. Open Source Softw. 2021;6 doi: 10.21105/joss.02906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanfear R., von Haeseler A., Woodhams M.D., Schrempf D., Chernomor O., Schmidt H.A., Minh B.Q., Teeling E. IQ-TREE 2: new models and Efficient methods for phylogenetic inference in the genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huerta-Cepas J., Serra F., Bork P. Ete 3: reconstruction, analysis, and Visualization of Phylogenomic data. Mol. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Full sequences of the viral isolates have been deposited to NCBI BioProject PRJNA794206. All data and scripts associated with analyses of the virus sequences have been deposited at github and is publicly available as of the date of publication at Github Data: https://github.com/fmaguire/voc_neutralisation_sc2_phylogenomics and Zenodo Data: https://doi.org/10.5281/zenodo.5817727. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/mfvm8fwf3s.1. Any additional information required to reanalyze the data reported in this work paper is available from the Lead contact upon request.