Abstract
Stem cells preserve tissue homeostasis by replacing the cells lost through damage or natural turnover. Thus, stem cells and their daughters can adopt two identities, characterized by different programs of gene expression and metabolic activity. The composition and regulation of these programs have been extensively studied, particularly by identifying transcription factor networks that define cellular identity and the epigenetic changes that underlie the progressive restriction in gene expression potential. However, there is increasing evidence that post-transcriptional mechanisms influence gene expression in stem cells and their progeny, in particular through the control of mRNA translation. Here, we review the described roles of translational regulation in controlling all aspects of stem cell biology, from the decision to enter or exit quiescence to maintaining self-renewal and promoting differentiation. We focus on mechanisms controlling global translation rates in cells, mTOR signaling, eIF2ɑ phosphorylation, and ribosome biogenesis and how they allow stem cells to rapidly change their gene expression in response to tissue needs or environmental changes. These studies emphasize that translation acts as an additional layer of control in regulating gene expression in stem cells and that understanding this regulation is critical to gaining a full understanding of the mechanisms that underlie fate decisions in stem cells.
Keywords: stem cell, self-renewal, differentiation, translation, protein synthesis, mTOR, eIF2 kinase, ribosome biogenesis
Introduction
Stem cells share the unique property of being able to both self-renew and differentiate, generating progeny with specialized functions. Nonetheless, stem cells encompass a wide variety of cells with a broad range of behaviors, from multipotent embryonic stem cells, which give rise to all cell types in an embryo, to lineage-restricted adult stem cells. For instance, in some mammalian tissues such as the blood, muscle, or brain, stem cells are mostly quiescent and proliferate only when activated by environmental signals, while in other tissues such as the intestine or epidermis, stem cells are highly proliferative to maintain tissue integrity despite continued turnover of differentiated cells. Even in those tissues, differentiation or proliferation can be modulated in response to stimuli from dying cells, or systemic signals including nutrition. Thus, adult stem cells are capable of rapidly altering their behavior and fate in response to the needs of the tissue or the organism, emphasizing the flexibility in their gene expression programs.
How gene expression programs controlling quiescence, proliferation, self-renewal, and differentiation can be both stable and plastic is the subject of much study, often focusing on understanding the transcriptional networks that maintain cell identity and the inputs that destabilize these networks and allow cells to change fate. Our understanding of these networks has grown and is continually being refined, showing that various stable network states exist and explaining transitions between these states (Kim et al., 2008; Moignard et al., 2013; Theunissen and Jaenisch, 2017; Kim et al., 2020; Sagner et al., 2021). Furthermore, we have gained considerable understanding of the epigenetic changes that reinforce these transcriptional changes and ensure that stem cells maintain plasticity in gene expression while differentiating cells gradually become restricted in potential (Lunyak and Rosenfeld, 2008; Ohbo and Tomizawa, 2015; Theunissen and Jaenisch, 2017; Ding et al., 2021).
However, technological advances enabling the comparison of the proteins in cells with their transcriptome led to the discovery that the two are often poorly correlated and that changes in the proteome can occur without accompanying transcriptional changes (Unwin et al., 2006; de Sousa Abreu et al., 2009; Lu et al., 2009; Maier et al., 2009; Schwanhausser et al., 2011), indicating that there are additional layers of regulation of gene expression beyond transcription. This mismatch has been described in many cell types, including stem cells (Lu et al., 2009; Ingolia et al., 2011; Baser et al., 2019; Habowski et al., 2020; Spevak et al., 2020), suggesting that post-transcriptional control of gene expression is common. The protein content of a cell depends on both synthesis and degradation: work describing extensive links between protein degradation and stem cell fate has been reviewed elsewhere (Strikoudis et al., 2014; Suresh et al., 2016; Yan et al., 2020); here, we will focus on the mechanisms affecting stem cell fate through the regulation of protein synthesis.
Bulk Translation Rates Change During Stem Cell Activation and Differentiation
Until recently, precise measurement of translation rates was mostly restricted to cell culture models where newly synthesized proteins could be labeled by providing a pulse of radioactive amino acids. For instance, in ex vivo cultures, differentiating murine embryonic stem cells (mESCs) into structures known as embryoid bodies resulted in a ∼2-fold increase in their translation rate, as indicated by [35S] methionine incorporation (Sampath et al., 2008). Consistently, embryoid bodies display an increased content of the Golgi apparatus and rough endoplasmic reticulum (ER) and an increased proportion of polysomes (multiple ribosomes bound to the same mRNA), indicating higher rates of protein synthesis. Similarly, cultured human embryonic stem cells (hESCs) show immature Golgi and rough ER and much lower translation rates than differentiated derivatives (Easley et al., 2010).
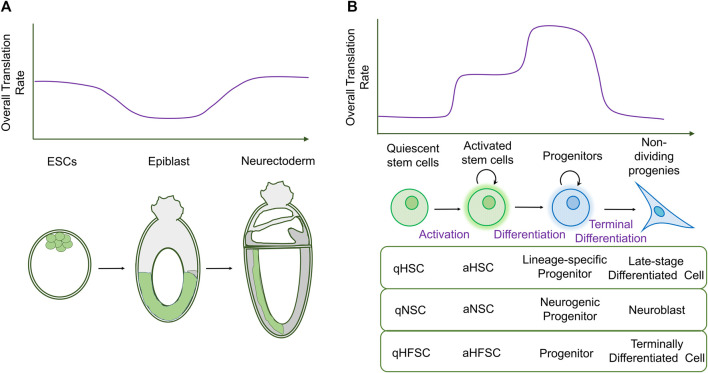
However, new techniques, known as bio-orthogonal non-canonical amino acid tagging (BONCAT) (Dieterich et al., 2007; Dieterich et al., 2006) and fluorescent non-canonical amino acid tagging (FUNCAT) (Dieterich et al., 2010), have enabled direct visualization of translation in tissue samples and comparison between cell types in situ, leading to a different conclusion. In this case, a transient decrease in the overall translation rate was observed as mESCs differentiated into epiblasts, increasing again during neuroectodermal differentiation (Figure 1A) (Corsini et al., 2018). One likely explanation for the discrepancy in translation rates between cultured and in vivo mESCs is that protein synthesis is artificially repressed by the factors added to maintain pluripotency in ex vivo cultures, leukemia inhibitory factor (LIF) and bone morphogenetic protein 4 (BMP4) (Friend et al., 2015).
FIGURE 1.
Changes in global translation during stem cell differentiation. (A) Diagram schematizing bulk translation rates during early mammalian embryogenesis (top), with stages shown later. Bulk translation rates are initially high in ESCs and decrease during differentiation into the epiblast. Global translation levels rise again during neurectodermal development. (B) In adult stem cells, quiescent stem cells have low rates of bulk translation. Translation increases in activated stem cells and is highest in proliferating progenitors that initiate differentiation. Terminally differentiated cells display low rates of translation. The table shows examples of stem cells in which translation rates follow this pattern: hematopoietic stem cells (HSCs), neural stem cells (NSCs), and hair follicle stem cells (HFSCs). q: quiescent; a: activated.
In adult stem cells, however, a clearer picture emerges of how the translation rate changes during differentiation. In most stem cell models, translation is low in stem cells, increasing their differentiating progeny, but this increase is reversed when cells terminally differentiate and become postmitotic (Figure 1B).
This was first shown in one of the best characterized adult stem cell populations, hematopoietic stem cells (HSCs), which show low translation rates. As the progeny of HSCs progress along the differentiation pathway through highly proliferative transit-amplifying stages, translation rates increase, remaining high as these progenitors become more lineage-restricted, but eventually dropping in further differentiated cell types (Signer et al., 2014). This latter observation was consistent with previous work showing that the polysome fraction decreased during myeloid differentiation from a promyelocytic cell line in culture, indicating an overall decrease in protein synthesis during terminal differentiation (Krichevsky et al., 1999). Similarly, adult neural stem cells (NSCs) in the sub-ventricular zone have lower translation rates than the neuronal progenitors they give rise to, while differentiation of the latter into postmitotic neuroblasts correlates with decreased levels of protein synthesis (Llorens-Bobadilla et al., 2015; Baser et al., 2019). Hair follicle stem cells (HFSCs) also show a similar pattern of increasing translation rates during differentiation into progenitors, followed by a decrease in terminally differentiated cells (Blanco et al., 2016).
Intriguingly, HSCs, NSCs, and HFSCs can all exist in a quiescent state, in which they do not proliferate; in all cases, quiescent stem cells have significantly lower translation rates than activated stem cells (Signer et al., 2014; Llorens-Bobadilla et al., 2015; Blanco et al., 2016). Although this observation, together with the fact that postmitotic cells tend to have lower translation rates than proliferative progenitors, suggests a link between the translation rate and cell proliferation, proliferation only accounts for part of the difference in protein synthesis rates observed, at least in both blood and hair follicle lineages (Signer et al., 2016; Blanco et al., 2016).
One interesting exception to the general trend that stem cells have lower translation rates than their differentiating offspring is seen in intestinal stem cells (ISCs) in Drosophila, which give rise to daughters that are postmitotic, without a transit-amplifying stage (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Obata et al. (2018) showed that ISCs have the highest bulk translation rate of all cell types in the Drosophila intestine, suggesting that differentiating stem cell daughters that immediately become postmitotic does not increase their translation rate, consistent with observations of lower protein synthesis in non-dividing cells in other tissues.
Altogether, these studies indicate that global translation is dynamically regulated during stem cell activation and differentiation and suggest a general pattern (Figure 1B): each step from the activation of quiescent stem cells to differentiation into proliferative progenitors results in an increase in the translation rate. This high rate of protein synthesis is sustained in proliferating progenitors as they continue to mature, until differentiation into postmitotic cells is associated with a decrease in translation. Given this tight control of overall translation rates during development, we focus on the regulation of global protein synthesis in stem cells and their differentiated offspring and how this contributes to gene expression and the maintenance of cell identity.
Several mechanisms have been described to regulate the translation of individual mRNAs, and these include regulating mRNA splicing, stability, and methylation, as well as microRNAs (Jackson et al., 2010; Zhang M. et al., 2020). The Drosophila germline provides an excellent example to understand how RNA-binding proteins can affect the translation of key factors regulating self-renewal and differentiation (Slaidina and Lehmann, 2014; Blatt et al., 2020); however, in this review, we will focus on mechanisms that affect global translation rates, in particular translation initiation and ribosome biogenesis, and how they influence stem cell identity.
Translation Initiation and Its Regulation
Initiation is thought to be the rate-limiting step of protein synthesis, determining which transcripts are translated and how much protein is produced (Duncan et al., 1987; Shah et al., 2013), and it is subjected to regulation by multiple upstream inputs, including nutrient-responsive signals, growth factor signaling, and the amount of ribosomes available in the cell (Sonenberg and Hinnebusch, 2009; Jackson et al., 2010; Roux and Topisirovic, 2018). These regulatory interactions determine both the total rate of translation and the specificity of translated mRNAs. Many signals governing translation rates converge either on controlling the rate of assembly of initiation factors at the m7G 5’ cap of the mRNA or the availability of the initiator tRNA carrying methionine (Met-tRNAi Met).
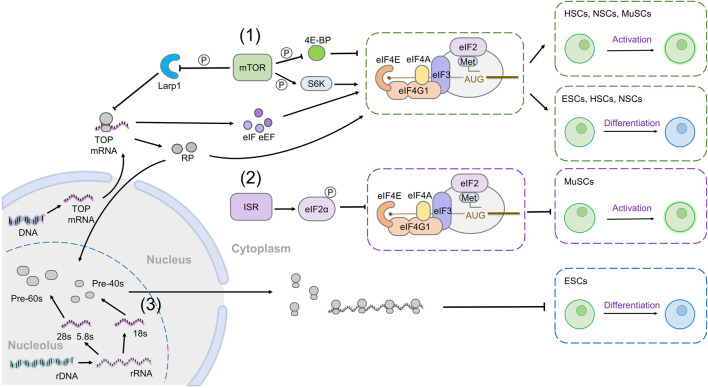
Canonical translation begins with the eukaryotic initiation factor (eIF) 4E binding to the 5′cap of mRNAs, and assembling a complex known as eIF4F, composed of eIF4E, the helicase eIF4A which unwinds secondary structure and eIF4G. eIF4G acts as a scaffold to bring other initiation complexes, together with the 40S small ribosome subunit to the 5′ end of the mRNA, from where the ribosome will begin scanning for a start codon (Figure 2). Initiation of translation requires Met-tRNAi Met, which is brought to the ribosome as part of the so-called ternary complex formed of eIF2 bound to GTP and Met-tRNAi Met.
FIGURE 2.
Global mechanisms of translation regulation and their roles in stem cell maintenance. (1) mTOR activity increases global translation through its effectors 4E-BP and S6K and promotes cap-dependent translation. In addition, mTOR increases the translation of TOP mRNAs, which include most components of the translation machinery including translation initiation factors (eIFs), translation elongation factors (eEFs), and ribosomal proteins (RPs). Increased mTOR signaling leads to the activation of HSCs, NSCs, and MuSCs from quiescence and induces differentiation of ESCs, HSCs, and NSCs (green boxes). (2) The integrated stress response (ISR) pathway promotes eIF2ɑ phosphorylation, which reduces eIF2ɑ association with Met-tRNAi Met and impairs global translation. P-eIF2ɑ prevents MuSC activation from quiescence (purple box). (3) Ribosomes are assembled in the nucleolus. Decreasing the rate of ribosome biogenesis results in ESC differentiation (blue box).
A major regulator of global cellular translation rates is the mechanistic target of rapamycin (mTOR) pathway (Albert and Hall, 2015; Liu and Sabatini, 2020; Ma and Blenis, 2009), which regulates cell growth and metabolism in response to extracellular growth factors and amino acid levels (Showkat et al., 2014). Two of the best characterized effectors of mTOR are ribosomal protein S6 kinase 1 (S6K1) and eIF4E-binding protein (4E-BP), both of which are phosphorylated upon mTOR activation. 4E-BP binds and sequesters eIF4E, preventing it from interacting with the 5′ mRNA cap, but phosphorylation of 4E-BP by mTOR inactivates it, releasing eIF4E and promoting cap-dependent translation (Figure 2 (1)) (Brunn et al., 1997; Hara et al., 1997; Gingras et al., 1999). Phosphorylated S6K1 increases the activity of several proteins involved in mRNA translation, including eIF4B and ribosomal protein S6 (RpS6), and inactivates translational repressors such as eukaryotic elongation factor-2 kinase (eEF2K), programmed cell death 4 (PDCD4), and La ribonucleoprotein 1 (Larp1) (Ferrari et al., 1991; Wang et al., 2001; Yang et al., 2003; Raught et al., 2004; Fonseca et al., 2015; Hong et al., 2017). Through various targets, activation of mTOR specifically increases the translation of mRNAs containing 5′ terminal oligopyrimidine (TOP) or TOP-like motifs, which consist of a 5’ cytidine at the cap immediately followed by a stretch of 4–15 pyrimidines (Hsieh et al., 2012; Thoreen et al., 2012; Meyuhas and Kahan, 2015; Hong et al., 2017; Iezaki et al., 2018; Philippe et al., 2018; Jia et al., 2021). Interestingly, many mRNAs encoding components of the translation machinery, including but not limited to translation elongation factors (eEFs), some translation initiation factors (eIFs) and most ribosomal proteins, have a TOP motif (Iadevaia et al., 2008; Meyuhas and Kahan, 2015; Hong et al., 2017). Thus, activation of mTOR and its effectors dramatically increases the synthesis of the translation machinery itself, as well as increasing the efficiency of existing translation factors.
Another critical regulator of cellular translation rates is a signaling pathway known as the integrated stress response (ISR). The ISR dramatically decreases mRNA translation following cellular stresses by phosphorylating eIF2ɑ, which prevents its assembly into the ternary complex with GTP and Met-tRNAi Met (Figure 2 (2)) (Wek et al., 2006; Pakos-Zebrucka et al., 2016; Costa-Mattioli and Walter, 2020). There are four known kinases that phosphorylate eIF2ɑ in response to various physiological or environmental stresses: PKR-like ER kinase (PERK) is activated downstream of ER stress; general control non-derepressible 2 (GCN2) is responsive to amino acid deprivation; protein kinase RNA-activated (PKR) senses infection-derived dsRNA; and heme-regulated inhibitor (HRI) binds hemin and is disinhibited upon cellular heme deficiency. ISR activation is thought to restore homeostasis and save energy under adverse conditions, particularly by restraining translation; however, a subset of transcripts is specifically translated when eIF2ɑ is phosphorylated, providing a mechanism for the upregulation of stress response genes when most translation is inhibited.
Finally, in addition to these pathways which are dedicated to growth control, other signaling pathways that control patterning during development and influence self-renewal decisions in stem cells can also affect translation. In particular, the Ras/mitogen-activated protein kinase (MAPK) pathway also promotes cellular growth and translation through promoting the activity of eIF4F, via its effector MAPK-interacting kinase 1 (Mnk1) (Waskiewicz et al., 1999). In sum, translation initiation is under the control of multiple signaling pathways, enabling the coordination of protein synthesis rates with other inputs into cell identity.
Ribosomal Biogenesis
Another critical parameter affecting the amount of protein produced in a cell is the number of ribosomes available for translation. Ribosome biogenesis is a complex process bringing together the ribosomal RNAs (rRNAs) and ribosomal proteins into the small and large ribosomal subunits, with the cooperation of non-ribosomal factors, such as small nucleolar ribonucleoproteins (SnoRNPs) (Figure 2 (3)). The amount of rRNA and protein available and the rate of assembly depend on several factors, including cellular stress, nutrient availability, and signaling (de la Cruz et al., 2018; Pelletier et al., 2018; Klinge and Woolford, 2019). The rRNAs are transcribed from nuclear DNA by two specific RNA polymerases (RNA Pol), RNA Pol I, which transcribes most rRNAs and RNA Pol III, which transcribes the 5s rRNA and tRNAs. However, rRNA synthesis and ribosome biogenesis also requires the action of RNA Pol II (Abraham et al., 2020). mTOR has emerged as a critical regulator of ribosome assembly, as its activity coordinately increases the transcription of rRNA and ribosomal proteins. Indeed, mTOR directly regulates the activity of RNA Pol I and RNA Pol III (Powers and Walter, 1999). Similarly, Ras/MAPK signaling increases rRNA synthesis to mediate its effects on growth (Stefanovsky et al., 2001).
Finally, although ribosomes were assumed to be equal and identical, recent work has identified that the composition of ribosomal proteins can change from cell-to-cell, and that, in turn, this composition can affect the mRNAs which are translated (Genuth and Barna, 2018). Thus, both abundance and specificity of ribosomes can be regulated to control overall translation rates and specificity in cells.
Changes in Bulk Translation Influence Stem Cell Maintenance and Differentiation
mTOR Promotes Stem Cell Activation and Differentiation Through Increased Translation
mTOR activity changes during stem cell differentiation or activation, and in many cases increased mTOR signaling is sufficient to induce differentiation. The differentiation of ex vivo-cultured ESCs, derived from both human and mouse, is coupled with the activation of mTOR activation indicated by phosphorylation of 4E-BP1, RPS6, and eIF4B (Sampath et al., 2008; Easley et al., 2010; Zhou et al., 2020). Similarly, mTOR activity is reduced during the early stages of reprogramming somatic cells into induced pluripotent stem cells (iPSCs) (Wang et al., 2013; Wu et al., 2015). Importantly, while mTOR activity is not required to maintain ESC self-renewal, activating mTOR or its effector S6K primes ESCs to differentiate and mTOR hyperactivity prevents the reprogramming of somatic cells into iPSCs (Murakami et al., 2004; Easley et al., 2010; He et al., 2012; Wang et al., 2013; Wu et al., 2015). Reprogramming also requires the presence of 4E-BPs (Tahmasebi et al., 2014), further implicating translation as one of the key cellular processes by which mTOR activity leads to the loss of pluripotency (Figure 2 (1)).
Similarly, increasing mTOR activity is sufficient to promote differentiation and loss of self-renewal ability in a variety of adult stem cell types across organisms, from HSCs and NSCs in mouse to Drosophila intestinal stem cells and both somatic and germline stem cells in the gonads (Zhang et al., 2006; Chen et al., 2009; Sun et al., 2010; Magri et al., 2011; Kapuria et al., 2012; Quan et al., 2013; Yuen et al., 2021). In HSCs, the deletion of Pten, a repressor of mTOR, or constitutive activation of mTOR, lead to ectopic proliferation of transit-amplifying progenitors, resulting in leukemia. However, despite this over-proliferation, increases in mTOR activity result in a depletion of HSCs, as determined by a deficiency in reconstituting the blood lineage upon transplantation into an immunodeficient host (Yilmaz et al., 2006; Zhang et al., 2006; Guo et al., 2008; Chen et al., 2009; Magee et al., 2012). In elegant genetic experiments, Signer et al. (2014) showed that Pten mutant HSCs had higher translation rates than control, and, importantly, that introducing a mutant copy of the belly spot and tail (Ferretti et al., 2017), encoding the ribosomal protein Rpl24, could decrease overall translation and restore the self-renewal and reconstitutive capacity of Pten mutant HSCs. Further work identified 4E-BP1 and 4E-BP2 as mediators of translational repression in HSCs, and their loss results in a similar decrease in long-term self-renewal ability to that of Pten mutant HSCs (Signer et al., 2016). Thus, an increased translation downstream of mTOR activation results in the loss of quiescence, increased proliferation, and eventual loss of the stem cell pool (Figure 2 (1)).
mTOR plays similar roles in regulating NSC quiescence and differentiation, in two different NSC populations, in the sub-ventricular zone (SVZ) of the lateral ventricle and the dentate gyrus. In both postnatal and adult SVZ NSCs, activating mTOR through loss of function of the negative regulators Pten or TSC1, or gain of function of the activator Rheb, led to increased production of neurons, at the expense of stem cell maintenance (Groszer et al., 2006; Gregorian et al., 2009; Magri et al., 2011; Hartman et al., 2013; Mahoney et al., 2016). Loss of NSCs was attributed to an increased frequency of symmetric divisions generating two proliferative progenitors, rather than self-renewing asymmetric divisions. Similarly, in the dentate gyrus, Pten loss mobilizes quiescent NSCs and induces them to proliferate through symmetric self-renewing divisions, but eventually results in increased terminal differentiation and stem cell loss (Bonaguidi et al., 2011). As in HSCs, 4E-BP2 is a critical downstream effector of mTORC1, controlling cap-dependent translation during neuronal differentiation in the SVZ (Hartman et al., 2013; Mahoney et al., 2016) (Figure 2 (1)).
The function of mTOR in controlling exit from quiescence is remarkably conserved across tissues and even species. Indeed, TOR activation in quiescent Drosophila NSCs arrested in either G0 or G2 leads to cell cycle entry and differentiation (Chell and Brand, 2010; Sousa-Nunes et al., 2011; Otsuki and Brand, 2018). In muscle stem cells (MuSCs, also known as satellite cells), mTOR activity promotes an “alert” state of quiescence in which cells are primed for reactivation, leading them to re-enter the cell cycle upon injury or stress (Rodgers et al., 2014). Moreover, in the intestine of both flies and mice, mTOR controls the ability of the population of quiescent stem cells to contribute to the regenerative response following fasting and refeeding (Richmond et al., 2015); however, repeated regenerative episodes and bouts of mTOR activity lead to eventual loss of stem cell maintenance (Haller et al., 2017). In sum, the mTOR pathway is widely associated with stem cell activation and differentiation, and persistent activation leads to the loss of self-renewing potential. In several instances, the effects of mTOR are mediated through its effects on translation through its effectors, 4E-BP, and S6K (Figure 2 (1)).
Of note, although the global translation rate correlates with mTOR activity throughout differentiation in the neural lineage, this is not true in all tissues (Paliouras et al., 2012; Cloetta et al., 2013; Baser et al., 2019). In myeloid progenitors derived from HSCs, mTOR is degraded by the proteasome, yet translation is still regulated by the mTOR target 4E-BP1 (Spevak et al., 2020). In this case, the cell cycle-dependent kinase CDK1 phosphorylates 4E-BP1 to promote eIF4E-dependent translation and maintain high translation rates. Thus, mTOR activity does not always linearly correlate with the overall translation rate and many other regulators may independently regulate translation initiation factors to achieve a precise rate of global translation.
eIF2α Phosphorylation in Stem Cell Maintenance
eIF2ɑ phosphorylation has also emerged as an important regulator of stem cell maintenance through effects on global translation. High levels of p-eIF2ɑ are observed in both ex vivo-cultured mESCs and murine MuSCs (Friend et al., 2015; Zismanov et al., 2016). Although in mESCs, there are conflicting data as to whether p-eIF2ɑ levels decrease with differentiation, the signaling factors BMP4 and LIF, which maintain pluripotency in ESCs, both increase eIF2ɑ phosphorylation (Friend et al., 2015). Indeed, preventing dephosphorylation of eIF2ɑ is sufficient to prevent differentiation even in the absence of LIF. Similarly, in intestinal stem cells in Drosophila, eIF2ɑ is phosphorylated by PERK in response to ER stress, and promotes stem cell proliferation. Continued eIF2ɑ phosphorylation results in tissue dysplasia with an accumulation of undifferentiated cells, consistent with a role for p-eIF2 in maintaining stem cell identity (Wang et al., 2015).
In MuSCs, eIF2ɑ is highly phosphorylated in quiescent cells. Indeed, replacing endogenous eIF2ɑ with a non-phosphorylatable mutant, results in the short-term activation of quiescent stem cells, increased translation and proliferation, and myogenic differentiation. In the long term, however, MuSCs unable to phosphorylate eIF2ɑ are lost from the stem cell population (Zismanov et al., 2016) (Figure 2 (2)). Thus, in MuSCs at least, eIF2 is a critical regulator of both overall translation rates and quiescence, precise regulation of which is essential to maintain long-term self-renewal potential.
Ribosome Biogenesis Is Highly Regulated and Required for Stem Cell Maintenance
Given the importance of translation in regulating stem cell biology, ribosome biogenesis has also emerged as a critical factor controlling self-renewal and differentiation. In hematopoietic and muscle lineages, rRNA transcription follows a similar pattern to the bulk translation rate, increasing during the differentiation of stem cells into proliferative progenitor cells and decreasing in terminally differentiated cells (Larson et al., 1993; Hayashi et al., 2014; Stedman et al., 2015; Gayraud-Morel et al., 2018). In addition to rRNA, the expression of regulators controlling rRNA transcription or maturation also correlates with the bulk translation rate during stem cell differentiation. In zebrafish, the expression of ddx27, encoding a regulator of rRNA maturation, is detected in activated MuSCs and proliferating myoblasts, and decreases when cells terminally differentiate (Bennett et al., 2018). In mESCs and hESCs, the expression of rRNA and the regulators of ribosome biogenesis correlate with the overall translation rate both in vivo and in vitro (Watanabe-Susaki et al., 2014; Zaidi et al., 2016; Corsini et al., 2018).
Despite this broad correlation, it is notable that ribosome biogenesis is proportionally higher in stem cells than that in differentiating cells, relative to the rate of translation (Stedman et al., 2015; Zaidi et al., 2016; Gayraud-Morel et al., 2018). Strikingly, in the Drosophila germline, rRNA transcription is the highest in germline stem cells (GSCs), while translation is lower in GSCs than in differentiated offspring (Zhang et al., 2014; Sanchez et al., 2016). These observations suggest a specific requirement for increased ribosome biogenesis in stem cells.
Indeed, disrupting ribosomal biogenesis in many stem cell models leads to defects in both survival and self-renewal (Stedman et al., 2015; Sanchez et al., 2016; Bennett et al., 2018; Baral et al., 2020; Farooq et al., 2020; Saez et al., 2020), while fully differentiated somatic cells demonstrate less dependency on ribosome biogenesis (Bennett et al., 2018; Gayraud-Morel et al., 2018; Saez et al., 2020). Impairing rRNA transcription induces differentiation in Drosophila GSCs and mouse hematopoietic progenitor cells (Hayashi et al., 2014; Zhang et al., 2014). Importantly, this effect on hematopoiesis is not mediated by a global repression of translation or a cell cycle arrest as inhibiting overall protein synthesis by cycloheximide and puromycin, or inhibiting cell cycle by roscovitine, a CDK inhibitor, does not have the same effect (Pilz et al., 1987; Hayashi et al., 2014). Similarly, disrupting ribosomal biogenesis in mESCs or hESCs by either repressing rRNA maturation or transcription triggers the expression of differentiation-related genes, and this is coupled with a reduced expression of pluripotent mRNAs such as OCT4 or SOX2 (You et al., 2015; Woolnough et al., 2016; Zhang H. et al., 2020). Importantly, the overexpression of fibrillarin, an important regulator of ribosomal RNA processing, can sustain pluripotency in the absence of LIF(Watanabe-Susaki et al., 2014) (Figure 2 (3)).
Altogether, ribosomal biogenesis has begun to be recognized as a major factor maintaining pluripotency and self-renewal potential. Little work to date has sought to identify the upstream regulators ensuring the coordinated production of high levels of rRNA, ribosomal proteins, and assembly factors in stem cells. Nonetheless, it is clear that elevated ribosome levels are required to maintain stem cell potential. Together with strong evidence showing lower translation in stem cells, this suggests that ribosome levels and translation rates are not correlated in stem cells; one suggestion is that a large pool of ribosomes is required to prepare cells to rapidly increase their translation rates and change their proteome during differentiation (Saba et al., 2021). However, this is hard to reconcile with the fact that decreasing ribosome biogenesis promotes differentiation, indicating that either ribosome biogenesis itself or the availability of large numbers of ribosomes relative to the amount of transcripts is in itself important for stem cell biology.
From Global Translational Control to Specific Protein Expression: Mechanisms Ensuring Selectivity in Translation in Stem Cells
How do changes in global translation rates or ribosome biogenesis affect stem cell maintenance? At least in part, the answer to this question lies in the selective translation of specific transcripts in response to changes that globally alter translation rates. Indeed, accumulating evidence show that specific mRNAs are translated in stem or differentiated cells, without always being accompanied by changes in mRNA abundance (Unwin et al., 2006; Lu et al., 2009; Habowski et al., 2020). In other words, mRNA translation is a regulatory mechanism allowing gene expression changes independently of transcription.
Specific Targets of mTOR Activity
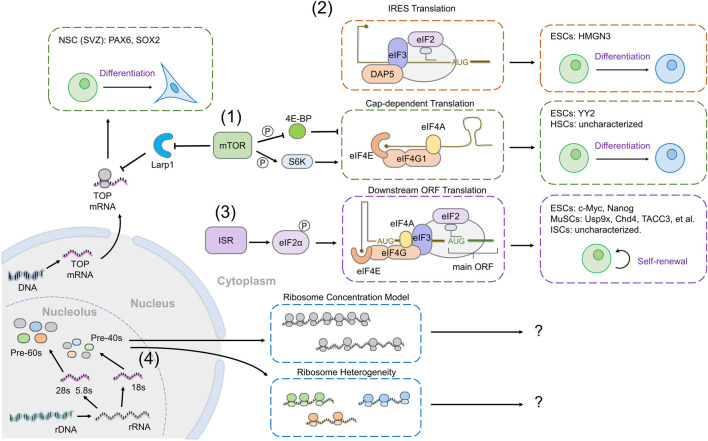
Although mTOR activity increases bulk translation by increasing the activity of initiation complexes, it disproportionally targets mRNAs containing TOP motifs for increased translation (Hsieh et al., 2012; Thoreen et al., 2012; Meyuhas and Kahan, 2015; Hong et al., 2017; Iezaki et al., 2018; Philippe et al., 2018; Jia et al., 2021). For instance, during the differentiation of SVZ neurogenic progenitors into neurons, both mTOR activity and bulk translation levels decrease (Baser et al., 2019). Notably, transcripts containing a pyrimidine-rich motif, similar to the TOP motif, are specifically repressed during differentiation; these encode both ribosomal proteins and transcription factors regulating stem cell identity such as Pax6 and Sox2, providing a mechanism by which mTOR activity correlates both with translation rates and with fate acquisition (Baser et al., 2019) (Figure 3 (1)).
FIGURE 3.
Global translation mechanisms result in selective translation of specific transcripts to regulate stem cell fate. (1) Increased cap-dependent translation downstream of mTOR activity can result in selective translation of transcripts that have low translation efficiency. In ESCs, the mRNA encoding YY2 is one such target, in other tissues (HSCs), whether mTOR activity results in specific target expression is unknown (green box). mTOR also promotes the translation of mRNAs with TOP-like motifs, such as PAX6 and SOX2, which are translationally repressed during neuronal differentiation, in response to reduced mTOR activity. (2) Internal ribosome entry site (IRES)-mediated translation occurs when cap-binding is inhibited and can direct transcript-specific translation. In ESCs, DAP5 replaces eIF4G to promote IRES-dependent translation and ESC differentiation, through translation of HMGN3 (orange boxes). (3) p-eIF2ɑ selectively promotes the translation of mRNAs with an upstream open reading frame (uORF). In ESCs, p-eIF2ɑ promotes the translation of the mRNAs encoding c-Myc and Nanog. In MuSCs, p-eIF2ɑ promotes the translation of mRNAs coding for Usp9x, Chd4, TACC3, etc. (purple box). (4) Ribosomes can selectively regulate mRNA translation. Two models have been proposed, the ribosome concentration model in which the amount of ribosomes available affects the translation of transcripts with high or low translation efficiency differently, or ribosome heterogeneity in which different ribosomal subunit composition directs specific translation of particular transcripts (blue boxes). Although ribosomes impact stem cell fate, no evidence directly supports either model to date.
eIF4F-Mediated Cap-Dependent Translation and Non-Canonical Translation
Regulation of the activity and ability of the eIF4F complex to bind the mRNA cap also provides a means to achieve specificity in translation (Hernández et al., 2020). 4E-BP is a major regulator of eIF4F activity, and its regulation by mTOR and other signals makes it an ideal modulator to act as a switch for gene expression. Surprisingly, in mESCs, loss of function of 4E-BP1/2 does not influence the global translation rate, but results in the loss of pluripotency marker expression (Tahmasebi et al., 2016). This effect is mediated by the selective translation of Yin Yang 2 (YY2) upon ablation of 4E-BP. The YY2 mRNA retains an intron in its 5’ UTR, making its translation acutely sensitive to eIF4E activity due to a complex secondary structure (Figure 3 (1)).
Other regulators of the assembly of the eIF4F complex also contribute to specific gene expression. In SVZ neural precursors, 4E-T competes with eIF4G for binding to eIF4E1, forming a complex which represses the translation of neurogenic mRNAs (Yang et al., 2014). Knock down of eIF4E1 or 4E-T promotes precursor differentiation while knocking down eIF4G1, on the contrary, impairs differentiation, indicating that in the SVZ, the main function of eIF4E1 in neural precursors is to repress the translation of neurogenic mRNAs.
Another regulator of eIF4F function is eIF4G2 (also named death-associated protein 5 (DAP5) or the novel APOBEC1 target 1 (NAT1)). eIF4G2 contains a similar C-terminal region to eIF4G1, enabling it to interact with eIF3 and eIF4A, but lacks an N-terminal eIF4E-binding domain, meaning that eIF4G2 promotes translation independently of eIF4F, and instead stimulates the translation of mRNAs containing an element known as an internal ribosome entry site (IRES) (Henis-Korenblit et al., 2002). DAP5 is required for neural and mesodermal differentiation of hESCs (Yoffe et al., 2016). The block in differentiation observed upon DAP5 depletion is not the consequence of a global translational repression, but instead it is due to selective IRES-driven translation by DAP5, in particular of the chromatin modifier HMGN3. Similarly, NAT1, the mouse homolog of DAP5, is required for the differentiation of mESCs (Sugiyama et al., 2017; Yamanaka et al., 2000). This was ascribed to NAT1 promoting the translation of two components of the ERK signaling pathway, which is required for ESC differentiation (Figure 3 (2)). However, the role of DAP5 in ESCs is still not fully understood, and may differ between mouse and humans, as loss of DAP5 in primed mESCs results in reduced self-renewal and defects in neural differentiation, in contrast to loss of DAP5 in naïve mESCs, which prevents differentiation into all cell types (Takahashi et al., 2020).
Thus, the eIF4F complex is a central node through which multiple regulators can control bulk protein synthesis and the translation of specific subsets of mRNAs. Indeed, due to the presence of the eIF4A helicase in the eIF4F complex, mRNAs with long and/or complex secondary structures are particularly sensitive to eIF4F activity. Thus, changes in eIF4F activity (in the absence of some of the more specific regulations described earlier) can result in a binary regulation of individual mRNA translation (Leppek et al., 2018), enabling the fine control of gene expression. It is highly likely that in other situations where bulk translation is increased during stem cell differentiation, such as in HSCs, the effects of translation increase on cell identity are mediated by such mechanisms.
eIF2α-p Selectively Regulates mRNAs With uORFs
Although eIF2ɑ phosphorylation dramatically reduces bulk translation, a subset of mRNAs is translated under these conditions (Baird et al., 2014). The best characterized example is the translation of the mRNA encoding ATF4 (Vattem and Wek, 2004; Asano, 2021), which contains two upstream open reading frames (uORFs), preventing the translation of the main open reading frame. Phosphorylation of eIF2 delays re-initiation of translation at the second uORF, resulting in initiation and translation at the main ATF-coding open reading frame.
Ribosome profiling in mESCs has revealed higher translation of uORFs in ESCs than EBs (Ingolia et al., 2011). Intriguingly, transcripts encoding the pluripotency factors, c-Myc and Nanog, have multiple uORFs (Figure 3 (3)). Whether this change in uORF translation during ESC differentiation is related to eIF2 activity, and whether it plays a role in fate determination is yet to be established.
A more direct example of p-eIF2ɑ-dependent expression of specific transcripts is seen in MuSCs, in which quiescence and self-renewal depend on eIF2ɑ phosphorylation (Zismanov et al., 2016). A study of proteins upregulated by eIF2 phosphorylation without accompanying changes in mRNA levels identified several genes encoding mitotic spindle assembly factors, in particular TACC3. The TACC3 transcript contains multiple uORFs and the protein is present in stem cells but downregulated in differentiating myoblasts. Importantly, TACC3 is required for MuSC expansion and self-renewal, demonstrating the functional importance of selective translation of uORF-containing transcripts in stem cell maintenance (Vattem and Wek, 2004; Fujita et al., 2021) (Figure 3 (3)).
Translational Specificity From Ribosomes: Effects of Ribosome Concentration and Subunit Composition
Stem cells require high levels of ribosome biogenesis for maintenance, despite lower translation rates, raising the possibility that ribosome numbers may play a role in specifically regulating stem cell gene expression. One model put forward to explain this is that different transcripts are differentially sensitive to ribosome concentration; mRNAs that are less efficiently translated would require a higher concentration of ribosomes to be expressed (Gabut et al., 2020; Lodish, 1974; Mills and Green, 2017) (Figure 3 (4)). Evidence in support of this model has been found in the case of a mutation in a ribosomal protein chaperone that causes Diamond-Blackfan anemia, which leads to reduced ribosome numbers but specifically alters the translation of a susbset of transcripts (Khajuria et al., 2018). This study linked a lineage commitment decision in progenitors with ribosome levels for the first time, but as yet, the same findings have not been reproduced in a stem cell model. Future work will determine whether this model does indeed apply to stem cells, and importantly, what determines the sensitivity of particular mRNAs to ribosome concentration.
Another means by which specificity in transcript translation can be achieved by ribosomes is through the specific subunit composition of each ribosome (Figure 3 (4)). Although ribosomes were initially assumed to be equivalent and to translate all mRNAs equally, work in the past decade has established that different ribosomes incorporate different ribosomal proteins. Different ribosomal proteins can confer mRNA sequence recognition (Genuth and Barna, 2018) and direct specific translation through IRES-dependent mechanisms. Intriguingly, mESCs display different ribosome subunit stoichiometries in monosomes and polysomes, and these associate with different mRNAs (Shi et al., 2017). Recent work in the Drosophila germline has shown that a paralogue of RpS5 is required for normal progression of differentiation and preferentially promotes translation of a subset of transcripts (Kong et al., 2019; Jang et al., 2021). These tantalizing observations raise the possibility that different incorporation of ribosomal subunits into ribosomes may regulate stem cell behavior; however, this has not yet been demonstrated.
Conclusions and Perspectives
From an initial view of mRNA translation as a “housekeeping” function that is performed equally in all cells and for all transcripts, our understanding has evolved to grasp the complexity and precision of translational regulation and its ability to tune cell fate. This is especially evident in stem cells where the decision to self-renew and differentiate is exquisitely sensitive to changes in protein synthesis. This raises the important question as to why translational regulation is such a pervasive mechanism to control identity across stem cells. One possible explanation is that stem cell differentiation requires a large remodeling of the cell’s proteome. Indeed, another important cellular function in stem cell biology is protein degradation, emphasizing the importance of accurate regulation of the cellular protein content in stem cell fate decisions (Llamas et al., 2020). Additionally, transcription is an inherently noisy process (Elowitz et al., 2002; Raj et al., 2010; Raser and O'Shea, 2005); this noise may play important roles in enabling cell decisions (Eldar and Elowitz, 2010). However, overlaying selective translation onto noisy gene expression could be a way to ensure that cells with the potential to adopt two different fates can only commit to one of these.
As our ability to probe translation increases, it is becoming more apparent that regulatory mechanisms-controlling global translation do not affect all transcripts equally; translation efficiency varies for individual mRNAs in different conditions. Thus, whether bulk translation changes are relevant to stem cell differentiation, or whether all the effects of changes in translation are mediated by the altered translation of a few key transcripts is still an open question.
In addition to contributing to our understanding of the mechanisms underlying self-renewal and differentiation and to our ability to manipulate those processes, and studying translation in stem cells will yield important advances in the study of aging. Reducing mTOR activity has long been known to extend the lifespan and promote continued health of organisms (Liu and Sabatini, 2020). Although other targets of mTOR have been implicated, S6K or eIF4E reduction, or 4E-BP overexpression, can contribute to lifespan extension, suggesting that decreased translation rates are at least partly responsible (Hansen et al., 2007; Syntichaki et al., 2007; Selman et al., 2009; Zid et al., 2009). Moreover, recent work has shown that both RNA Pol I and RNA Pol III, which synthesize rRNAs, mediate lifespan control downstream of mTOR, and that, in Drosophila, they exert their effects on lifespan specifically in intestinal stem cells (Filer et al., 2017; Martinez Corrales et al., 2020). As we deepen our understanding of how translational regulation influences stem cell behavior, new avenues for interventions that mitigate the effects of aging will be opened up.
Acknowledgments
The authors thank members of the Amoyel lab for discussions.
Author Contributions
RW and MA contributed to conceptualization of the manuscript, wrote the draft, and edited it.
Funding
This work was funded by an MRC Career Development Award MR/P009646/2.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abraham K. J., Khosraviani N., Chan J. N. Y., Gorthi A., Samman A., Zhao D. Y., et al. (2020). Nucleolar RNA Polymerase II Drives Ribosome Biogenesis. Nature 585, 298–302. 10.1038/s41586-020-2497-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert V., Hall M. N. (2015). mTOR Signaling in Cellular and Organismal Energetics. Curr. Opin. Cel Biol. 33, 55–66. 10.1016/j.ceb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Asano K. (2021). Origin of Translational Control by eIF2α Phosphorylation: Insights from Genome-wide Translational Profiling Studies in Fission Yeast. Curr. Genet. 67, 359–368. 10.1007/s00294-020-01149-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird T. D., Palam L. R., Fusakio M. E., Willy J. A., Davis C. M., McClintick J. N., et al. (2014). Selective mRNA Translation during eIF2 Phosphorylation Induces Expression of IBTKα. MBoC 25, 1686–1697. 10.1091/mbc.e14-02-0704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral S. S., Lieux M. E., DiMario P. J. (2020). Nucleolar Stress in Drosophila Neuroblasts, a Model for Human Ribosomopathies. Biol. Open 9. 10.1242/bio.046565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baser A., Skabkin M., Kleber S., Dang Y., Gülcüler Balta G. S., Kalamakis G., et al. (2019). Onset of Differentiation Is post-transcriptionally Controlled in Adult Neural Stem Cells. Nature 566, 100–104. 10.1038/s41586-019-0888-x [DOI] [PubMed] [Google Scholar]
- Bennett A. H., O’Donohue M.-F., Gundry S. R., Chan A. T., Widrick J., Draper I., et al. (2018). RNA Helicase, DDX27 Regulates Skeletal Muscle Growth and Regeneration by Modulation of Translational Processes. Plos Genet. 14, e1007226. 10.1371/journal.pgen.1007226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., et al. (2016). Stem Cell Function and Stress Response Are Controlled by Protein Synthesis. Nature 534, 335–340. 10.1038/nature18282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt P., Martin E. T., Breznak S. M., Rangan P. (2020). Post-transcriptional Gene Regulation Regulates Germline Stem Cell to Oocyte Transition during Drosophila Oogenesis. Curr. Top. Dev. Biol. 140, 3–34. 10.1016/bs.ctdb.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Bonaguidi M. A., Wheeler M. A., Shapiro J. S., Stadel R. P., Sun G. J., Ming G.-l., et al. (2011). In Vivo clonal Analysis Reveals Self-Renewing and Multipotent Adult Neural Stem Cell Characteristics. Cell 145, 1142–1155. 10.1016/j.cell.2011.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunn G. J., Hudson C. C., Sekulić A., Williams J. M., Hosoi H., Houghton P. J., et al. (1997). Phosphorylation of the Translational Repressor PHAS-I by the Mammalian Target of Rapamycin. Science 277, 99–101. 10.1126/science.277.5322.99 [DOI] [PubMed] [Google Scholar]
- Chell J. M., Brand A. H. (2010). Nutrition-responsive Glia Control Exit of Neural Stem Cells from Quiescence. Cell 143, 1161–1173. 10.1016/j.cell.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu Y., Zheng P. (2009). mTOR Regulation and Therapeutic Rejuvenation of Aging Hematopoietic Stem Cells. Sci. Signal. 2, ra75. 10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloetta D., Thomanetz V., Baranek C., Lustenberger R. M., Lin S., Oliveri F., et al. (2013). Inactivation of mTORC1 in the Developing Brain Causes Microcephaly and Affects Gliogenesis. J. Neurosci. 33, 7799–7810. 10.1523/jneurosci.3294-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsini N. S., Peer A. M., Moeseneder P., Roiuk M., Burkard T. R., Theussl H.-C., et al. (2018). Coordinated Control of mRNA and rRNA Processing Controls Embryonic Stem Cell Pluripotency and Differentiation. Cell Stem Cell 22, 543–558 e512. 10.1016/j.stem.2018.03.002 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M., Walter P. (2020). The Integrated Stress Response: From Mechanism to Disease. Science 368. 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Gómez-Herreros F., Rodríguez-Galán O., Begley V., de la Cruz Muñoz-Centeno M., Chávez S. (2018). Feedback Regulation of Ribosome Assembly. Curr. Genet. 64, 393–404. 10.1007/s00294-017-0764-x [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R., Penalva L. O., Marcotte E. M., Vogel C. (2009). Global Signatures of Protein and mRNA Expression Levels. Mol. Biosyst. 5, 1512–1526. 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich D. C., Hodas J. J. L., Gouzer G., Shadrin I. Y., Ngo J. T., Triller A., et al. (2010). In Situ visualization and Dynamics of Newly Synthesized Proteins in Rat Hippocampal Neurons. Nat. Neurosci. 13, 897–905. 10.1038/nn.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich D. C., Lee J. J., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. (2007). Labeling, Detection and Identification of Newly Synthesized Proteomes with Bioorthogonal Non-canonical Amino-Acid Tagging. Nat. Protoc. 2, 532–540. 10.1038/nprot.2007.52 [DOI] [PubMed] [Google Scholar]
- Dieterich D. C., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. (2006). Selective Identification of Newly Synthesized Proteins in Mammalian Cells Using Bioorthogonal Noncanonical Amino Acid Tagging (BONCAT). Proc. Natl. Acad. Sci. 103, 9482–9487. 10.1073/pnas.0601637103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Liu Z., Liu F. (2021). Transcriptional and Epigenetic Control of Hematopoietic Stem Cell Fate Decisions in Vertebrates. Dev. Biol. 475, 156–164. 10.1016/j.ydbio.2021.03.003 [DOI] [PubMed] [Google Scholar]
- Duncan R., Milburn S. C., Hershey J. W. (1987). Regulated Phosphorylation and Low Abundance of HeLa Cell Initiation Factor eIF-4F Suggest a Role in Translational Control. Heat Shock Effects on eIF-4F. J. Biol. Chem. 262, 380–388. 10.1016/s0021-9258(19)75938-9 [DOI] [PubMed] [Google Scholar]
- Easley C. A., Ben-Yehudah A., Redinger C. J., Oliver S. L., Varum S. T., Eisinger V. M., et al. (2010). mTOR-Mediated Activation of P70 S6K Induces Differentiation of Pluripotent Human Embryonic Stem Cells. Cell Reprogramming 12, 263–273. 10.1089/cell.2010.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A., Elowitz M. B. (2010). Functional Roles for Noise in Genetic Circuits. Nature 467, 167–173. 10.1038/nature09326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. (2002). Stochastic Gene Expression in a Single Cell. Science 297, 1183–1186. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Farooq M., Lindbæk L., Krogh N., Doganli C., Keller C., Mönnich M., et al. (2020). RRP7A Links Primary Microcephaly to Dysfunction of Ribosome Biogenesis, Resorption of Primary Cilia, and Neurogenesis. Nat. Commun. 11, 5816. 10.1038/s41467-020-19658-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Bandi H. R., Hofsteenge J., Bussian B. M., Thomas G. (1991). Mitogen-activated 70K S6 Kinase. Identification of In Vitro 40 S Ribosomal S6 Phosphorylation Sites. J. Biol. Chem. 266, 22770–22775. 10.1016/s0021-9258(18)54634-2 [DOI] [PubMed] [Google Scholar]
- Ferretti M. B., Ghalei H., Ward E. A., Potts E. L., Karbstein K. (2017). Rps26 Directs mRNA-specific Translation by Recognition of Kozak Sequence Elements. Nat. Struct. Mol. Biol. 24, 700–707. 10.1038/nsmb.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer D., Thompson M. A., Takhaveev V., Dobson A. J., Kotronaki I., Green J. W. M., et al. (2017). RNA Polymerase III Limits Longevity Downstream of TORC1. Nature 552, 263–267. 10.1038/nature25007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca B. D., Zakaria C., Jia J.-J., Graber T. E., Svitkin Y., Tahmasebi S., et al. (2015). La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1). J. Biol. Chem. 290, 15996–16020. 10.1074/jbc.m114.621730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K., Brooks H. A., Propson N. E., Thomson J. A., Kimble J. (2015). Embryonic Stem Cell Growth Factors Regulate eIF2α Phosphorylation. PLoS One 10, e0139076. 10.1371/journal.pone.0139076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R., Jamet S., Lean G., Cheng H. C. M., Hebert S., Kleinman C. L., et al. (2021). Satellite Cell Expansion Is Mediated by P-eIF2alpha-dependent Tacc3 Translation. Development, 148. 10.1242/dev.194480 [DOI] [PubMed] [Google Scholar]
- Gabut M., Bourdelais F., Durand S. (2020). Ribosome and Translational Control in Stem Cells. Cells 9. 10.3390/cells9020497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B., Le Bouteiller M., Commere P. H., Cohen-Tannoudji M., Tajbakhsh S. (2018). Notchless Defines a Stage-specific Requirement for Ribosome Biogenesis during Lineage Progression in Adult Skeletal Myogenesis. Development, 145. 10.1242/dev.162636 [DOI] [PubMed] [Google Scholar]
- Genuth N. R., Barna M. (2018). The Discovery of Ribosome Heterogeneity and its Implications for Gene Regulation and Organismal Life. Mol. Cel 71, 364–374. 10.1016/j.molcel.2018.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Gygi S. P., Raught B., Polakiewicz R. D., Abraham R. T., Hoekstra M. F., et al. (1999). Regulation of 4E-BP1 Phosphorylation: a Novel Two-step Mechanism. Genes Dev. 13, 1422–1437. 10.1101/gad.13.11.1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorian C., Nakashima J., Le Belle J., Ohab J., Kim R., Liu A., et al. (2009). Pten Deletion in Adult Neural Stem/progenitor Cells Enhances Constitutive Neurogenesis. J. Neurosci. 29, 1874–1886. 10.1523/jneurosci.3095-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M., Erickson R., Scripture-Adams D. D., Dougherty J. D., Le Belle J., Zack J. A., et al. (2006). PTEN Negatively Regulates Neural Stem Cell Self-Renewal by Modulating G0-G1 Cell Cycle Entry. Proc. Natl. Acad. Sci. 103, 111–116. 10.1073/pnas.0509939103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Lasky J. L., Chang C.-J., Mosessian S., Lewis X., Xiao Y., et al. (2008). Multi-genetic Events Collaboratively Contribute to Pten-Null Leukaemia Stem-Cell Formation. Nature 453, 529–533. 10.1038/nature06933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habowski A. N., Flesher J. L., Bates J. M., Tsai C.-F., Martin K., Zhao R., et al. (2020). Transcriptomic and Proteomic Signatures of Stemness and Differentiation in the colon Crypt. Commun. Biol. 3, 453. 10.1038/s42003-020-01181-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller S., Kapuria S., Riley R. R., O’Leary M. N., Schreiber K. H., Andersen J. K., et al. (2017). mTORC1 Activation during Repeated Regeneration Impairs Somatic Stem Cell Maintenance. Cell Stem Cell 21, 806–818 e805. 10.1016/j.stem.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.-J., Kenyon C. (2007). Lifespan Extension by Conditions that Inhibit Translation in Caenorhabditis elegans . Aging Cell 6, 95–110. 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Hara K., Yonezawa K., Kozlowski M. T., Sugimoto T., Andrabi K., Weng Q.-P., et al. (1997). Regulation of eIF-4E BP1 Phosphorylation by mTOR. J. Biol. Chem. 272, 26457–26463. 10.1074/jbc.272.42.26457 [DOI] [PubMed] [Google Scholar]
- Hartman N. W., Lin T. V., Zhang L., Paquelet G. E., Feliciano D. M., Bordey A. (2013). mTORC1 Targets the Translational Repressor 4E-BP2, but Not S6 Kinase 1/2, to Regulate Neural Stem Cell Self-Renewal In Vivo . Cel Rep. 5, 433–444. 10.1016/j.celrep.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Kuroda T., Kishimoto H., Wang C., Iwama A., Kimura K. (2014). Downregulation of rRNA Transcription Triggers Cell Differentiation. PLoS One 9, e98586. 10.1371/journal.pone.0098586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Kang L., Wu T., Zhang J., Wang H., Gao H., et al. (2012). An Elaborate Regulation of Mammalian Target of Rapamycin Activity Is Required for Somatic Cell Reprogramming Induced by Defined Transcription Factors. Stem Cell Dev. 21, 2630–2641. 10.1089/scd.2012.0015 [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S., Shani G., Sines T., Marash L., Shohat G., Kimchi A. (2002). The Caspase-Cleaved DAP5 Protein Supports Internal Ribosome Entry Site-Mediated Translation of Death Proteins. Proc. Natl. Acad. Sci. 99, 5400–5405. 10.1073/pnas.082102499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G., García A., Sonenberg N., Lasko P. (2020). Unorthodox Mechanisms to Initiate Translation Open Novel Paths for Gene Expression. J. Mol. Biol. 432, 166702. 10.1016/j.jmb.2020.10.035 [DOI] [PubMed] [Google Scholar]
- Hong S., Freeberg M. A., Han T., Kamath A., Yao Y., Fukuda T., et al. (2017). LARP1 Functions as a Molecular Switch for mTORC1-Mediated Translation of an Essential Class of mRNAs. Elife 6. 10.7554/eLife.25237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh A. C., Liu Y., Edlind M. P., Ingolia N. T., Janes M. R., Sher A., et al. (2012). The Translational Landscape of mTOR Signalling Steers Cancer Initiation and Metastasis. Nature 485, 55–61. 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V., Caldarola S., Tino E., Amaldi F., Loreni F. (2008). All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA 14, 1730–1736. 10.1261/rna.1037108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezaki T., Horie T., Fukasawa K., Kitabatake M., Nakamura Y., Park G., et al. (2018). Translational Control of Sox9 RNA by mTORC1 Contributes to Skeletogenesis. Stem Cel Rep. 11, 228–241. 10.1016/j.stemcr.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N. T., Lareau L. F., Weissman J. S. (2011). Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 147, 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U. T., Pestova T. V. (2010). The Mechanism of Eukaryotic Translation Initiation and Principles of its Regulation. Nat. Rev. Mol. Cel Biol 11, 113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Lee J., Mathews J., Ruess H., Williford A. O., Rangan P., et al. (2021). The Drosophila Ribosome Protein S5 Paralog RpS5b Promotes Germ Cell and Follicle Cell Differentiation during Oogenesis. Development, 148. 10.1242/dev.199511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J.-J., Lahr R. M., Solgaard M. T., Moraes B. J., Pointet R., Yang A.-D., et al. (2021). mTORC1 Promotes TOP mRNA Translation through Site-specific Phosphorylation of LARP1. Nucleic Acids Res. 49, 3461–3489. 10.1093/nar/gkaa1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapuria S., Karpac J., Biteau B., Hwangbo D., Jasper H. (2012). Notch-mediated Suppression of TSC2 Expression Regulates Cell Differentiation in the Drosophila Intestinal Stem Cell Lineage. Plos Genet. 8, e1003045. 10.1371/journal.pgen.1003045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria R. K., Munschauer M., Ulirsch J. C., Fiorini C., Ludwig L. S., McFarland S. K., et al. (2018). Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173, 90–103. 10.1016/j.cell.2018.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Osteil P., Humphrey S. J., Cinghu S., Oldfield A. J., Patrick E., et al. (2020). Transcriptional Network Dynamics during the Progression of Pluripotency Revealed by Integrative Statistical Learning. Nucleic Acids Res. 48, 1828–1842. 10.1093/nar/gkz1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008). An Extended Transcriptional Network for Pluripotency of Embryonic Stem Cells. Cell 132, 1049–1061. 10.1016/j.cell.2008.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S., Woolford J. L., Jr. (2019). Ribosome Assembly Coming into Focus. Nat. Rev. Mol. Cel Biol 20, 116–131. 10.1038/s41580-018-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong J., Han H., Bergalet J., Bouvrette L. P. B., Hernández G., Moon N.-S., et al. (2019). A Ribosomal Protein S5 Isoform Is Essential for Oogenesis and Interacts with Distinct RNAs in Drosophila melanogaster . Sci. Rep. 9, 13779. 10.1038/s41598-019-50357-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A. M., Metzer E., Rosen H. (1999). Translational Control of Specific Genes during Differentiation of HL-60 Cells. J. Biol. Chem. 274, 14295–14305. 10.1074/jbc.274.20.14295 [DOI] [PubMed] [Google Scholar]
- Larson D. E., Xie W., Glibetic M., O'Mahony D., Sells B. H., Rothblum L. I. (1993). Coordinated Decreases in rRNA Gene Transcription Factors and rRNA Synthesis during Muscle Cell Differentiation. Proc. Natl. Acad. Sci. 90, 7933–7936. 10.1073/pnas.90.17.7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K., Das R., Barna M. (2018). Functional 5′ UTR mRNA Structures in Eukaryotic Translation Regulation and How to Find Them. Nat. Rev. Mol. Cel Biol 19, 158–174. 10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. Y., Sabatini D. M. (2020). mTOR at the Nexus of Nutrition, Growth, Ageing and Disease. Nat. Rev. Mol. Cel Biol 21, 183–203. 10.1038/s41580-019-0199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas E., Alirzayeva H., Loureiro R., Vilchez D. (2020). The Intrinsic Proteostasis Network of Stem Cells. Curr. Opin. Cel Biol. 67, 46–55. 10.1016/j.ceb.2020.08.005 [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. (2015). Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell 17, 329–340. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Lodish H. F. (1974). Model for the Regulation of mRNA Translation Applied to Haemoglobin Synthesis. Nature 251, 385–388. 10.1038/251385a0 [DOI] [PubMed] [Google Scholar]
- Lu R., Markowetz F., Unwin R. D., Leek J. T., Airoldi E. M., MacArthur B. D., et al. (2009). Systems-level Dynamic Analyses of Fate Change in Murine Embryonic Stem Cells. Nature 462, 358–362. 10.1038/nature08575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak V. V., Rosenfeld M. G. (2008). Epigenetic Regulation of Stem Cell Fate. Hum. Mol. Genet. 17, R28–R36. 10.1093/hmg/ddn149 [DOI] [PubMed] [Google Scholar]
- Ma X. M., Blenis J. (2009). Molecular Mechanisms of mTOR-Mediated Translational Control. Nat. Rev. Mol. Cel Biol 10, 307–318. 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- Magee J. A., Ikenoue T., Nakada D., Lee J. Y., Guan K.-L., Morrison S. J. (2012). Temporal Changes in PTEN and mTORC2 Regulation of Hematopoietic Stem Cell Self-Renewal and Leukemia Suppression. Cell Stem Cell 11, 415–428. 10.1016/j.stem.2012.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L., Cambiaghi M., Cominelli M., Alfaro-Cervello C., Cursi M., Pala M., et al. (2011). Sustained Activation of mTOR Pathway in Embryonic Neural Stem Cells Leads to Development of Tuberous Sclerosis Complex-Associated Lesions. Cell Stem Cell 9, 447–462. 10.1016/j.stem.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Mahoney C., Feliciano D. M., Bordey A., Hartman N. W. (2016). Switching on mTORC1 Induces Neurogenesis but Not Proliferation in Neural Stem Cells of Young Mice. Neurosci. Lett. 614, 112–118. 10.1016/j.neulet.2015.12.042 [DOI] [PubMed] [Google Scholar]
- Maier T., Güell M., Serrano L. (2009). Correlation of mRNA and Protein in Complex Biological Samples. FEBS Lett. 583, 3966–3973. 10.1016/j.febslet.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Martínez Corrales G., Filer D., Wenz K. C., Rogan A., Phillips G., Li M., et al. (2020). Partial Inhibition of RNA Polymerase I Promotes Animal Health and Longevity. Cel Rep. 30, 1661–1669 e1664. 10.1016/j.celrep.2020.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O., Kahan T. (2015). The Race to Decipher the Top Secrets of TOP mRNAs. Biochim. Biophys. Acta (Bba) - Gene Regul. Mech. 1849, 801–811. 10.1016/j.bbagrm.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Micchelli C. A., Perrimon N. (2006). Evidence that Stem Cells Reside in the Adult Drosophila Midgut Epithelium. Nature 439, 475–479. 10.1038/nature04371 [DOI] [PubMed] [Google Scholar]
- Mills E. W., Green R. (2017). Ribosomopathies: There's Strength in Numbers. Science 358. 10.1126/science.aan2755 [DOI] [PubMed] [Google Scholar]
- Moignard V., Macaulay I. C., Swiers G., Buettner F., Schütte J., Calero-Nieto F. J., et al. (2013). Characterization of Transcriptional Networks in Blood Stem and Progenitor Cells Using High-Throughput Single-Cell Gene Expression Analysis. Nat. Cel Biol 15, 363–372. 10.1038/ncb2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Ichisaka T., Maeda M., Oshiro N., Hara K., Edenhofer F., et al. (2004). mTOR Is Essential for Growth and Proliferation in Early Mouse Embryos and Embryonic Stem Cells. Mol. Cel Biol 24, 6710–6718. 10.1128/mcb.24.15.6710-6718.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata F., Tsuda-Sakurai K., Yamazaki T., Nishio R., Nishimura K., Kimura M., et al. (2018). Nutritional Control of Stem Cell Division through S-Adenosylmethionine in Drosophila Intestine. Dev. Cel 44, 741–751. 10.1016/j.devcel.2018.02.017 [DOI] [PubMed] [Google Scholar]
- Ohbo K., Tomizawa S.-i. (2015). Epigenetic Regulation in Stem Cell Development, Cell Fate Conversion, and Reprogramming. Biomol. Concepts 6, 1–9. 10.1515/bmc-2014-0036 [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2006). The Adult Drosophila Posterior Midgut Is Maintained by Pluripotent Stem Cells. Nature 439, 470–474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Otsuki L., Brand A. H. (2018). Cell Cycle Heterogeneity Directs the Timing of Neural Stem Cell Activation from Quiescence. Science 360, 99–102. 10.1126/science.aan8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A. M. (2016). The Integrated Stress Response. EMBO Rep. 17, 1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paliouras G. N., Hamilton L. K., Aumont A., Joppe S. E., Barnabe-Heider F., Fernandes K. J. L. (2012). Mammalian Target of Rapamycin Signaling Is a Key Regulator of the Transit-Amplifying Progenitor Pool in the Adult and Aging Forebrain. J. Neurosci. 32, 15012–15026. 10.1523/jneurosci.2248-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Thomas G., Volarević S. (2018). Ribosome Biogenesis in Cancer: New Players and Therapeutic Avenues. Nat. Rev. Cancer 18, 51–63. 10.1038/nrc.2017.104 [DOI] [PubMed] [Google Scholar]
- Philippe L., Vasseur J.-J., Debart F., Thoreen C. C. (2018). La-related Protein 1 (LARP1) Repression of TOP mRNA Translation Is Mediated through its Cap-Binding Domain and Controlled by an Adjacent Regulatory Region. Nucleic Acids Res. 46, 1457–1469. 10.1093/nar/gkx1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz R. B., Van den Berghe G., Boss G. R. (1987). Induction of HL-60 Differentiation by Starvation for a Single Essential Amino Acid but Not by Protein Synthesis Inhibitors. J. Clin. Invest. 79, 1006–1009. 10.1172/jci112867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Walter P. (1999). Regulation of Ribosome Biogenesis by the Rapamycin-Sensitive TOR-Signaling Pathway inSaccharomyces Cerevisiae. MBoC 10, 987–1000. 10.1091/mbc.10.4.987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan Z., Sun P., Lin G., Xi R. (2013). TSC1/2 Regulates Intestinal Stem Cell Maintenance and Lineage Differentiation through Rheb-TORC1-S6k but Independently of Nutritional Status or Notch Regulation. J. Cel Sci 126, 3884–3892. 10.1242/jcs.125294 [DOI] [PubMed] [Google Scholar]
- Raj A., Rifkin S. A., Andersen E., van Oudenaarden A. (2010). Variability in Gene Expression Underlies Incomplete Penetrance. Nature 463, 913–918. 10.1038/nature08781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser J. M., O'Shea E. K. (2005). Noise in Gene Expression: Origins, Consequences, and Control. Science 309, 2010–2013. 10.1126/science.1105891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B., Peiretti F., Gingras A.-C., Livingstone M., Shahbazian D., Mayeur G. L., et al. (2004). Phosphorylation of Eucaryotic Translation Initiation Factor 4B Ser422 Is Modulated by S6 Kinases. EMBO J. 23, 1761–1769. 10.1038/sj.emboj.7600193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C. A., Shah M. S., Deary L. T., Trotier D. C., Thomas H., Ambruzs D. M., et al. (2015). Dormant Intestinal Stem Cells Are Regulated by PTEN and Nutritional Status. Cel Rep. 13, 2403–2411. 10.1016/j.celrep.2015.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers J. T., King K. Y., Brett J. O., Cromie M. J., Charville G. W., Maguire K. K., et al. (2014). mTORC1 Controls the Adaptive Transition of Quiescent Stem Cells from G0 to GAlert. Nature 510, 393–396. 10.1038/nature13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux P. P., Topisirovic I. (2018). Signaling Pathways Involved in the Regulation of mRNA Translation. Mol. Cel Biol 38. 10.1128/MCB.00070-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba J. A., Liakath-Ali K., Green R., Watt F. M. (2021). Translational Control of Stem Cell Function. Nat. Rev. Mol. Cel Biol 22, 671–690. 10.1038/s41580-021-00386-2 [DOI] [PubMed] [Google Scholar]
- Saez I., Gerbracht J. V., Koyuncu S., Lee H. J., Horn M., Kroef V., et al. (2020). The E3 Ubiquitin Ligase UBR 5 Interacts with the H/ACA Ribonucleoprotein Complex and Regulates Ribosomal RNA Biogenesis in Embryonic Stem Cells. FEBS Lett. 594, 175–188. 10.1002/1873-3468.13559 [DOI] [PubMed] [Google Scholar]
- Sagner A., Zhang I., Watson T., Lazaro J., Melchionda M., Briscoe J. (2021). A Shared Transcriptional Code Orchestrates Temporal Patterning of the central Nervous System. Plos Biol. 19, e3001450. 10.1371/journal.pbio.3001450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P., Pritchard D. K., Pabon L., Reinecke H., Schwartz S. M., Morris D. R., et al. (2008). A Hierarchical Network Controls Protein Translation during Murine Embryonic Stem Cell Self-Renewal and Differentiation. Cell Stem Cell 2, 448–460. 10.1016/j.stem.2008.03.013 [DOI] [PubMed] [Google Scholar]
- Sanchez C. G., Teixeira F. K., Czech B., Preall J. B., Zamparini A. L., Seifert J. R. K., et al. (2016). Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation. Cell Stem Cell 18, 276–290. 10.1016/j.stem.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., et al. (2011). Global Quantification of Mammalian Gene Expression Control. Nature 473, 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Selman C., Tullet J. M. A., Wieser D., Irvine E., Lingard S. J., Choudhury A. I., et al. (2009). Ribosomal Protein S6 Kinase 1 Signaling Regulates Mammalian Life Span. Science 326, 140–144. 10.1126/science.1177221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J. B. (2013). Rate-limiting Steps in Yeast Protein Translation. Cell 153, 1589–1601. 10.1016/j.cell.2013.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Fujii K., Kovary K. M., Genuth N. R., Röst H. L., Teruel M. N., et al. (2017). Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol. Cel 67, 71–83. 10.1016/j.molcel.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showkat M., Beigh M. A., Andrabi K. I. (2014). mTOR Signaling in Protein Translation Regulation: Implications in Cancer Genesis and Therapeutic Interventions. Mol. Biol. Int. 2014, 686984. 10.1155/2014/686984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer R. A. J., Magee J. A., Salic A., Morrison S. J. (2014). Haematopoietic Stem Cells Require a Highly Regulated Protein Synthesis Rate. Nature 509, 49–54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer R. A. J., Qi L., Zhao Z., Thompson D., Sigova A. A., Fan Z. P., et al. (2016). The Rate of Protein Synthesis in Hematopoietic Stem Cells Is Limited Partly by 4E-BPs. Genes Dev. 30, 1698–1703. 10.1101/gad.282756.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaidina M., Lehmann R. (2014). Translational Control in Germline Stem Cell Development. J. Cel Biol 207, 13–21. 10.1083/jcb.201407102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A. G. (2009). Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell 136, 731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R., Yee L. L., Gould A. P. (2011). Fat Cells Reactivate Quiescent Neuroblasts via TOR and Glial Insulin Relays in Drosophila. Nature 471, 508–512. 10.1038/nature09867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak C. C., Elias H. K., Kannan L., Ali M. A. E., Martin G. H., Selvaraj S., et al. (2020). Hematopoietic Stem and Progenitor Cells Exhibit Stage-specific Translational Programs via mTOR- and CDK1-dependent Mechanisms. Cell Stem Cell 26, 755–765. 10.1016/j.stem.2019.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman A., Beck-Cormier S., Le Bouteiller M., Raveux A., Vandormael-Pournin S., Coqueran S., et al. (2015). Ribosome Biogenesis Dysfunction Leads to P53-Mediated Apoptosis and Goblet Cell Differentiation of Mouse Intestinal Stem/progenitor Cells. Cell Death Differ 22, 1865–1876. 10.1038/cdd.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovsky V. Y., Pelletier G., Hannan R., Gagnon-Kugler T., Rothblum L. I., Moss T. (2001). An Immediate Response of Ribosomal Transcription to Growth Factor Stimulation in Mammals Is Mediated by ERK Phosphorylation of UBF. Mol. Cel 8, 1063–1073. 10.1016/s1097-2765(01)00384-7 [DOI] [PubMed] [Google Scholar]
- Strikoudis A., Guillamot M., Aifantis I. (2014). Regulation of Stem Cell Function by Protein Ubiquitylation. EMBO Rep. 15, 365–382. 10.1002/embr.201338373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H., Takahashi K., Yamamoto T., Iwasaki M., Narita M., Nakamura M., et al. (2017). Nat1 Promotes Translation of Specific Proteins that Induce Differentiation of Mouse Embryonic Stem Cells. Proc. Natl. Acad. Sci. USA 114, 340–345. 10.1073/pnas.1617234114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Quan Z., Zhang B., Wu T., Xi R. (2010). TSC1/2 Tumour Suppressor Complex maintainsDrosophilagermline Stem Cells by Preventing Differentiation. Development 137, 2461–2469. 10.1242/dev.051466 [DOI] [PubMed] [Google Scholar]
- Suresh B., Lee J., Kim H., Ramakrishna S. (2016). Regulation of Pluripotency and Differentiation by Deubiquitinating Enzymes. Cel Death Differ 23, 1257–1264. 10.1038/cdd.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K., Tavernarakis N. (2007). eIF4E Function in Somatic Cells Modulates Ageing in Caenorhabditis elegans . Nature 445, 922–926. 10.1038/nature05603 [DOI] [PubMed] [Google Scholar]
- Tahmasebi S., Alain T., Rajasekhar V. K., Zhang J.-P., Prager-Khoutorsky M., Khoutorsky A., et al. (2014). Multifaceted Regulation of Somatic Cell Reprogramming by mRNA Translational Control. Cell Stem Cell 14, 606–616. 10.1016/j.stem.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Tahmasebi S., Jafarnejad S. M., Tam I. S., Gonatopoulos-Pournatzis T., Matta-Camacho E., Tsukumo Y., et al. (2016). Control of Embryonic Stem Cell Self-Renewal and Differentiation via Coordinated Alternative Splicing and Translation of YY2. Proc. Natl. Acad. Sci. USA 113, 12360–12367. 10.1073/pnas.1615540113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Jeong D., Wang S., Narita M., Jin X., Iwasaki M., et al. (2020). Critical Roles of Translation Initiation and RNA Uridylation in Endogenous Retroviral Expression and Neural Differentiation in Pluripotent Stem Cells. Cel Rep. 31, 107715. 10.1016/j.celrep.2020.107715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen T. W., Jaenisch R. (2017). Mechanisms of Gene Regulation in Human Embryos and Pluripotent Stem Cells. Development 144, 4496–4509. 10.1242/dev.157404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., Sabatini D. M. (2012). A Unifying Model for mTORC1-Mediated Regulation of mRNA Translation. Nature 485, 109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin R. D., Smith D. L., Blinco D., Wilson C. L., Miller C. J., Evans C. A., et al. (2006). Quantitative Proteomics Reveals Posttranslational Control as a Regulatory Factor in Primary Hematopoietic Stem Cells. Blood 107, 4687–4694. 10.1182/blood-2005-12-4995 [DOI] [PubMed] [Google Scholar]
- Vattem K. M., Wek R. C. (2004). Reinitiation Involving Upstream ORFs Regulates ATF4 mRNA Translation in Mammalian Cells. Proc. Natl. Acad. Sci. 101, 11269–11274. 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Ryoo H. D., Qi Y., Jasper H. (2015). PERK Limits Drosophila Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress. Plos Genet. 11, e1005220. 10.1371/journal.pgen.1005220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Xia P., Ye B., Huang G., Liu J., Fan Z. (2013). Transient Activation of Autophagy via Sox2-Mediated Suppression of mTOR Is an Important Early Step in Reprogramming to Pluripotency. Cell Stem Cell 13, 617–625. 10.1016/j.stem.2013.10.005 [DOI] [PubMed] [Google Scholar]
- Wang X., Li W., Williams M., Terada N., Alessi D. R., Proud C. G. (2001). Regulation of Elongation Factor 2 Kinase by p90RSK1 and P70 S6 Kinase. EMBO J. 20, 4370–4379. 10.1093/emboj/20.16.4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waskiewicz A. J., Johnson J. C., Penn B., Mahalingam M., Kimball S. R., Cooper J. A. (1999). Phosphorylation of the Cap-Binding Protein Eukaryotic Translation Initiation Factor 4E by Protein Kinase Mnk1 In Vivo . Mol. Cel Biol 19, 1871–1880. 10.1128/mcb.19.3.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Susaki K., Takada H., Enomoto K., Miwata K., Ishimine H., Intoh A., et al. (2014). Biosynthesis of Ribosomal RNA in Nucleoli Regulates Pluripotency and Differentiation Ability of Pluripotent Stem Cells. Stem Cells 32, 3099–3111. 10.1002/stem.1825 [DOI] [PubMed] [Google Scholar]
- Wek R. C., Jiang H.-Y., Anthony T. G. (2006). Coping with Stress: eIF2 Kinases and Translational Control. Biochem. Soc. Trans. 34, 7–11. 10.1042/bst0340007 [DOI] [PubMed] [Google Scholar]
- Woolnough J. L., Atwood B. L., Liu Z., Zhao R., Giles K. E. (2016). The Regulation of rRNA Gene Transcription during Directed Differentiation of Human Embryonic Stem Cells. PLoS One 11, e0157276. 10.1371/journal.pone.0157276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li Y., Zhang H., Huang Y., Zhao P., Tang Y., et al. (2015). Autophagy and mTORC1 Regulate the Stochastic Phase of Somatic Cell Reprogramming. Nat. Cel Biol 17, 715–725. 10.1038/ncb3172 [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Zhang X. Y., Maeda M., Miura K., Wang S., Farese R. V., Jr., et al. (2000). Essential Role of NAT1/p97/DAP5 in Embryonic Differentiation and the Retinoic Acid Pathway. EMBO J. 19, 5533–5541. 10.1093/emboj/19.20.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P., Ren J., Zhang W., Qu J., Liu G.-H. (2020). Protein Quality Control of Cell Stemness. Cell Regen 9, 22. 10.1186/s13619-020-00064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Smibert C. A., Kaplan D. R., Miller F. D. (2014). An eIF4E1/4E-T Complex Determines the Genesis of Neurons from Precursors by Translationally Repressing a Proneurogenic Transcription Program. Neuron 84, 723–739. 10.1016/j.neuron.2014.10.022 [DOI] [PubMed] [Google Scholar]
- Yang H.-S., Jansen A. P., Komar A. A., Zheng X., Merrick W. C., Costes S., et al. (2003). The Transformation Suppressor Pdcd4 Is a Novel Eukaryotic Translation Initiation Factor 4A Binding Protein that Inhibits Translation. Mol. Cel Biol 23, 26–37. 10.1128/mcb.23.1.26-37.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz Ö. H., Valdez R., Theisen B. K., Guo W., Ferguson D. O., Wu H., et al. (2006). Pten Dependence Distinguishes Haematopoietic Stem Cells from Leukaemia-Initiating Cells. Nature 441, 475–482. 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- Yoffe Y., David M., Kalaora R., Povodovski L., Friedlander G., Feldmesser E., et al. (2016). Cap-independent Translation by DAP5 Controls Cell Fate Decisions in Human Embryonic Stem Cells. Genes Dev. 30, 1991–2004. 10.1101/gad.285239.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You K. T., Park J., Kim V. N. (2015). Role of the Small Subunit Processome in the Maintenance of Pluripotent Stem Cells. Genes Dev. 29, 2004–2009. 10.1101/gad.267112.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen A. C., Hillion K.-H., Wang R., Amoyel M. (2021). Germ Cells Commit Somatic Stem Cells to Differentiation Following Priming by PI3K/Tor Activity in the Drosophila Testis. Plos Genet. 17, e1009609. 10.1371/journal.pgen.1009609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S. K., Boyd J. R., Grandy R. A., Medina R., Lian J. B., Stein G. S., et al. (2016). Expression of Ribosomal RNA and Protein Genes in Human Embryonic Stem Cells Is Associated with the Activating H3K4me3 Histone Mark. J. Cel. Physiol. 231, 2007–2013. 10.1002/jcp.25309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu Z., Lu J. Y., Huang B., Zhou H., Xie W., et al. (2020a). DEAD-box Helicase 18 Counteracts PRC2 to Safeguard Ribosomal DNA in Pluripotency Regulation. Cel Rep. 30, 81–97. 10.1016/j.celrep.2019.12.021 [DOI] [PubMed] [Google Scholar]
- Zhang J., Grindley J. C., Yin T., Jayasinghe S., He X. C., Ross J. T., et al. (2006). PTEN Maintains Haematopoietic Stem Cells and Acts in Lineage Choice and Leukaemia Prevention. Nature 441, 518–522. 10.1038/nature04747 [DOI] [PubMed] [Google Scholar]
- Zhang M., Zhai Y., Zhang S., Dai X., Li Z. (2020b). Roles of N6-Methyladenosine (m6A) in Stem Cell Fate Decisions and Early Embryonic Development in Mammals. Front. Cel Dev. Biol. 8, 782. 10.3389/fcell.2020.00782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Shalaby N. A., Buszczak M. (2014). Changes in rRNA Transcription Influence Proliferation and Cell Fate within a Stem Cell Lineage. Science 343, 298–301. 10.1126/science.1246384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Lv X., Zhang L., Yan J., Hu R., Sun Y., et al. (2020). Ketamine Promotes the Neural Differentiation of Mouse Embryonic Stem Cells by Activating mTOR. Mol. Med. Rep. 21, 2443–2451. 10.3892/mmr.2020.11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid B. M., Rogers A. N., Katewa S. D., Vargas M. A., Kolipinski M. C., Lu T. A., et al. (2009). 4E-BP Extends Lifespan upon Dietary Restriction by Enhancing Mitochondrial Activity in Drosophila. Cell 139, 149–160. 10.1016/j.cell.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V., Chichkov V., Colangelo V., Jamet S., Wang S., Syme A., et al. (2016). Phosphorylation of eIF2α Is a Translational Control Mechanism Regulating Muscle Stem Cell Quiescence and Self-Renewal. Cell Stem Cell 18, 79–90. 10.1016/j.stem.2015.09.020 [DOI] [PubMed] [Google Scholar]