Abstract
Background:
The ascending aorta is a common location for aneurysm and dissection. This aortic region is populated by a mosaic of medial and adventitial cells that are embryonically derived from either the second heart field (SHF) or the cardiac neural crest. SHF-derived cells populate areas that coincide with the spatial specificity of thoracic aortopathies. The purpose of this study was to determine whether and how SHF-derived cells contribute to ascending aortopathies.
Methods:
Ascending aortic pathologies were examined in patients with sporadic thoracic aortopathies and angiotensin II (AngII)-infused mice. Ascending aortas without overt pathology from AngII-infused mice were subjected to mass spectrometry assisted proteomics, and molecular features of SHF-derived cells were determined by single cell transcriptomic analyses. Genetic deletion of either low-density lipoprotein receptor-related protein 1 (Lrp1) or transforming growth factor-β receptor 2 (Tgfbr2) in SHF-derived cells was conducted to examine the impact of SHF-derived cells on vascular integrity.
Results:
Pathologies in human ascending aortic aneurysmal tissues were predominant in outer medial layers and adventitia. This gradient was mimicked in mouse aortas following AngII infusion that was coincident with the distribution of SHF-derived cells. Proteomics indicated that brief AngII infusion, prior to overt pathology, evoked downregulation of SMC proteins and differential expression of extracellular matrix proteins, including several LRP1 ligands. LRP1 deletion in SHF-derived cells augmented AngII-induced ascending aortic aneurysm and rupture. Single cell transcriptomic analysis revealed that brief AngII infusion decreased Lrp1 and Tgfbr2 mRNA abundance in SHF-derived cells and induced a unique fibroblast population with low abundance of Tgfbr2 mRNA. SHF-specific Tgfbr2 deletion led to embryonic lethality at E12.5 with dilatation of the outflow tract and retroperitoneal hemorrhage. Integration of proteomic and single cell transcriptomics results identified plasminogen activator inhibitor 1 (PAI1) as the most increased protein in SHF-derived SMCs and fibroblasts during AngII infusion. Immunostaining revealed a transmural gradient of PAI1 in both ascending aortas of AngII-infused mice and human ascending aneurysmal aortas that mimicked the gradient of medial and adventitial pathologies.
Conclusion:
SHF-derived cells exert a critical role in maintaining vascular integrity through LRP1 and TGF-β signaling associated with increases of aortic PAI1.
Keywords: angiotensin, embryonic origin, smooth muscle cell, thoracic aortic aneurysm, mouse
Introduction
Thoracic aortopathies are a spectrum of lethal diseases associated with aneurysm, dissection, and rupture.1, 2 Despite the devastating consequences of these diseases, current medical therapy has limited efficacy. Therefore, there is an urgent need to investigate mechanisms of thoracic aortopathies to facilitate development of effective therapeutics.
Aortopathies occur throughout the thoracic aorta with the ascending aorta being a frequently affected region.3, 4 Aortic remodeling, including medial and adventitial thickening, is a histological feature of thoracic aortopathies that shows a transmedial gradient towards the outer medial layers and the adjacent adventitia.5–10 Therefore, thoracic aortopathies have regional and transmural specificities with outer medial layers in the ascending aorta being a disease-prone location. However, the mechanism by which outer medial layers of the ascending aorta are prone to aortopathy remains unclear.
The major cellular components of the aortic wall are smooth muscle cells (SMCs) and fibroblasts (FBs). These two cell types are thought to exert a pivotal role in the pathogenesis of thoracic aortopathies.11 SMCs in the ascending aorta are derived from two embryonic origins: cardiac neural crest (CNC) and second heart field (SHF).12 Select FBs are also derived from the CNC and SHF.13 Lineage tracking studies have demonstrated that CNC-and SHF-derived SMCs occupy inner and outer medial layers of the ascending aorta, respectively.13 The region-specific distribution of thoracic aortopathies coincides with the distribution of SMCs from the SHF origin. Therefore, we hypothesized that SHF-derived cells play an important role in ascending aortopathies. To test this hypothesis, we investigated whether SHF-derived cells colocalized with the evolution of aortic pathology produced by angiotensin II (AngII) infusion in mice, and investigated the molecular basis of AngII-mediated ascending aortopathies using proteomic and single cell transcriptomic approaches. Our findings using omics analyses were validated by two SHF-specific genetic deletion studies in mice.
Methods
Additional detailed methods are presented in Supplemental Methods. Numerical data are available in Supplemental Excel File I–III. All raw data and analytical methods are available from the corresponding author upon appropriate request.
Human aortic samples
Ascending aortas were acquired from patients undergoing aortic surgery for sporadic thoracic aortic aneurysms at Baylor College of Medicine (n=10, mean age=66±7 years, 5 males and 5 females, mean aortic diameters=5.3±0.4 cm). Aortic tissues were fixed with formalin (10% wt/vol) and then incubated with ethanol (70% vol/vol), as described previously.14 Subsequently, tissues were embedded in paraffin blocks, and sliced into 5 μm sections. The protocol for collecting human tissue samples was approved by the Institutional Review Board at Baylor College of Medicine.
Mice
The following mice were purchased from The Jackson Laboratory (Supplemental Table I): ROSA26RLacZ (#003474), ROSA26RmT/mG (#007676), Lrp1 floxed (#012604), Tgfbr2 floxed (#012603), and Wnt1-Cre (#022501, also known as B6-Wnt1-Cre2). Mef2c-Cre mice (#030262) were purchased from the Mutant Mouse Resource and Research Center. For cell tracking studies of CNC and SHF origins, either Wnt1- or Mef2c-Cre male mice were bred to ROSA26RLacZ female mice, respectively. For the single cell RNA sequencing (scRNAseq), Mef2c-Cre male mice were bred to ROSA26RmT/mG female mice. Mef2c-Cre male mice were crossbred to Lrp1 floxed female mice to delete Lrp1 in SHF-derived cells. Mef2c-Cre male mice were also used for lineage-specific deletion of Tgfbr2 in SHF-derived cells. Because of a modest induction of AngII-induced thoracic aortic aneurysms (TAAs) in female mice, only male mice were studied.15 All experiments were approved by the IACUC at either the University of Kentucky or Baylor College of Medicine in accordance with the guidelines of the National Institutes of Health.
Subcutaneous infusion using an osmotic pump
After random assignment, either saline or AngII (1,000 ng/kg/min, H-1705, Bachem) was infused via a subcutaneously implanted osmotic pump (Alzet model 2001 for 3 days and model 2004 for 28 days of infusion, respectively, Durect) into male mice at 10 to 14 weeks of age.16 Surgical staples, used to close incision sites, were removed 7 days after surgery. Post-operative pain was alleviated by application of topical lidocaine cream (4% wt/wt, LMX4, #0496–0882-15, Eloquest Healthcare, Inc).
Statistical analyses
Non-omics data are presented as the mean ± standard error of the mean (a diamond-shaped symbol with error bars) or the median and 25th/75th percentiles (a box plot). Normality and homogeneity of variance were assessed by Shapiro-Wilk and Brown-Forsythe tests, respectively. Because the original data of ascending aortic diameter, Western blotting for Serpine1, and collagen deposition in human TAAs did not pass Shapiro-Wilk or Brown-Forsythe tests, Log transformation was applied for these data. After confirming homogeneous variances and normality, two-group or multi-group comparisons for means were performed by two-sided Student’s t-test or two-way analysis of variance (ANOVA) with Holm-Sidak multiple comparison test, respectively. The log-rank test was used to compare the incidence of ascending aortic rupture between groups. Other causes of death, such as abdominal aortic rupture, were treated as censored events. P<0.05 was considered statistically significant. Statistical analyses mentioned above were performed using SigmaPlot version 14.0 (SYSTAT Software Inc.).
Statistical analyses for proteomic data were performed with R (version 4.1.0).17 Proteins in the principal component analysis (PCA) plot were clustered by k-means algorithm (k=2). Differentiated proteins, transformed by “log1p” function in R, between saline and AngII were identified with a Student’s t-test based on the false discovery rate (FDR) adjusted P-value (Q-value) with less than 0.05 as a threshold. Enrichment analyses for gene ontology (biological process) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed using “clusterProfiler (4.0.2)” R package.18 Heatmaps for comparisons in the standardized protein abundances (i.e. Z scores) between saline vs AngII were generated for differentiated proteins related to SMC contraction, matrix metalloproteinase (MMP), TGFβ signaling, extracellular matrix (ECM)-related molecules. Protein-protein interaction networks were examined using the STRING database (version 11.0) in Cytoscape (version 3.8.0).19 Interactions were acquired using the following thresholds: confidence interaction scores≥0.4, active interaction sources=text mining, experiments, databases, co-expression, neighborhood, gene fusion, co-occurrence.
scRNAseq data analyses were also performed with R (version 4.1.0).17 Seurat package was used to identify cell types.20 Two Seurat objects for each of the mapped unique molecular identifier (UMI) count datasets from mGFP positive cells (Ctrl-SHF and AngII-SHF) were built using the “CreateSeuratObject” function with the following criteria: ≥3 cells and ≥200 detected genes. Cells expressing less than 200 or more than 5,000 genes were filtered out for exclusion of non-cell or cell aggregates, respectively. Cells with more than 10% mitochondrial genes were also excluded. Each of the UMI counts was then normalized as the following: counts for each cell were divided by total counts, multiplied by 10,000, and transformed to natural-log. “FindIntegrationAnchors” and “IntegrateData” functions were used to remove batch effects and integrate the two normalized UMI count datasets. Uniform manifold approximation and projection (UMAP) dimensionality reduction to 20 dimensions for the first 30 principal components (PCs) with 0.1 or 0.5 resolution was applied to identify cell clusters using the normalized- and scaled-UMI count data. “FindAllMarkers” and “FindConcervedMarkers” functions were used to identify conserved marker genes to determine cell types of each of the clusters. The difference of cellular proportions between infusions were examined by Chi-square test. Using “zinbwave (1.14.1)” and “edgeR (3.34.0)” R packages,21, 22 differentially expressed genes (DEGs) were identified with a zero-inflated negative binomial regression model based on Q-value<0.05 as a threshold. Gene ontology enrichment analyses in scRNAseq was performed using “clusterProfiler (4.0.2)” in genes with Q-value<0.05 and |Log2fold change|>0.5. Monocle package was used for trajectory pseudotime analysis.23
Cell-cell interaction was evaluated using a list containing 2,009 ligand-receptor pairs, developed by Skelly et al.24 The presence of ligand-receptor pairs within and between each cell type was defined if more than 20% of cells in that cell type had non-zero read counts for the gene encoding ligand or receptor. Then, two cell types were linked and network plots were generated using “igraph” package on R.25
For human scRNAseq analyses, read count data were downloaded from Gene Expression Omnibus (GSE155468).26 TAA samples were harvested from the ascending region of patients who underwent a surgery for aortic replacement (n=8, mean age=56–78 years, 4 males, 4 females, mean aortic diameters= 4.9–5.8 cm) and control samples were obtained from recipients of heart transplants or lung donors (n=3, mean age=61–63 years, 1 male, 2 females). mRNA abundance was evaluated in SMC and FB clusters. DEG analyses in SMCs and FBs were performed on R using the Hurdle model adjusted for age implemented in the MAST R package.27
Results
Medial gradients of aortic pathologies in patients with TAAs and AngII-infused mice
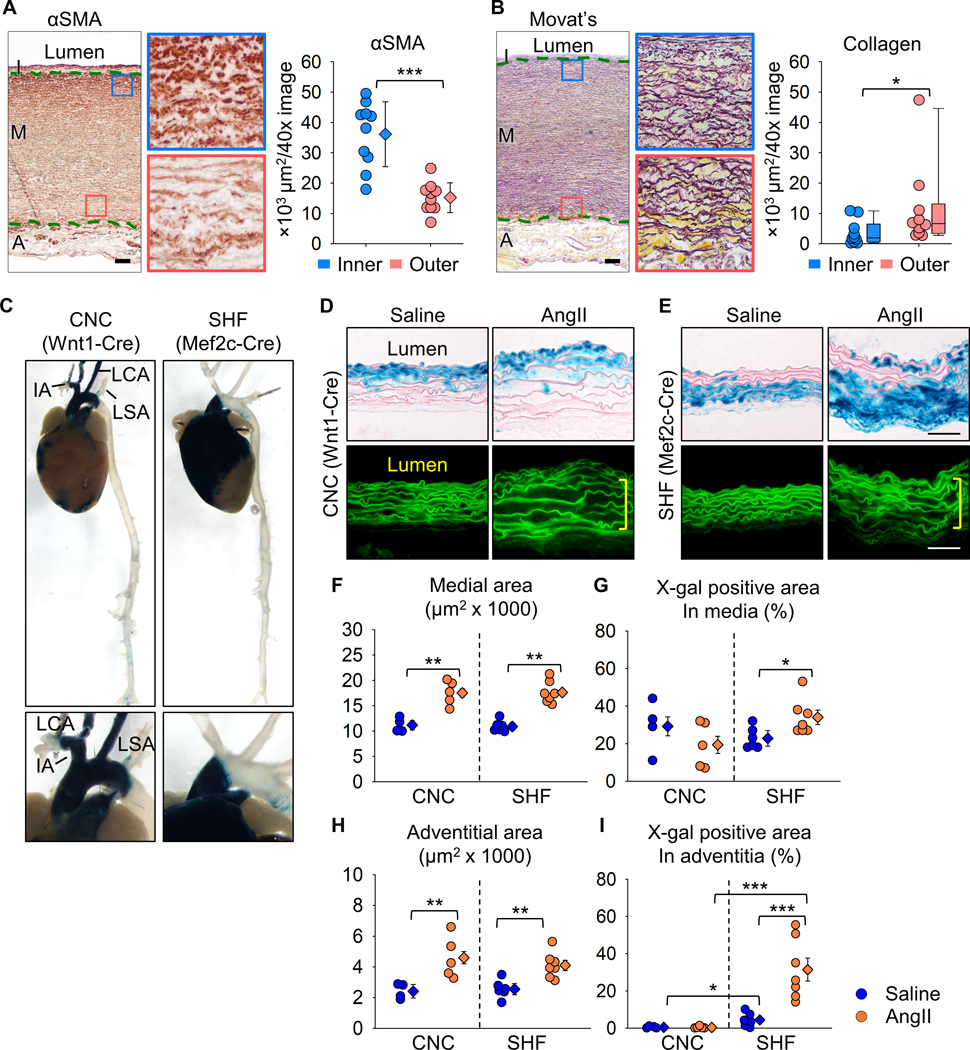
We first performed histological analyses to determine the pathological patterns in human sporadic TAA tissues. Immunostaining for α-smooth muscle actin and Movat’s pentachrome staining were performed to detect SMCs and ECM, respectively. Immunostaining for α-smooth muscle actin revealed that the outer media had significantly lower positive areas for α-smooth muscle actin than the inner media in human TAA tissues (Figure 1A). Movat’s pentachrome staining identified higher collagen deposition in the outer media (Figure 1B).
Figure 1. Transmural gradients of aortic pathologies in patients with sporadic TAAs and AngII-infused mice.
Representative images of (A) immunostaining for α-smooth muscle actin (red) and (B) Movat’s pentachrome staining (elastin: dark purple, collagen: yellow). High magnification images are captured from the inner (blue box) and outer (red box) media (n=10). I indicates intima; M, media; A, adventitia. Green dotted lines indicate internal and external elastic layers. αSMA positive and collagen deposited areas were quantified in inner and outer media. Log transformation was applied for the data of collagen deposition. (C) Representative images of X-gal stained (blue) aortas from Wnt1- and Mef2c-Cre ROSA26RLacZ mice. IA indicates innominate artery; LCA, left common carotid artery; LSA, left subclavian artery. High magnification images of the proximal thoracic aortas are captured. Representative X-gal staining and FITC images of aortas from either saline- or AngII-infused (D) Wnt1- and (E) Mef2c-Cre ROSA26RLacZ mice (n=5 to 7 per group). Yellow square brackets indicate medial thickening. Scale bar, 50 μm. Dot plots for (F) medial and (H) adventitial areas and X-gal positive area in the (G) media and (I) adventitia. Diamonds and error bars indicate the mean and SEM, respectively. *P<0.05, **P<0.01, ***P<0.001 by Student’s t-test or two-way ANOVA followed by Holm-Sidak test.
A cell lineage study was subsequently performed to investigate the interaction of SMC origins to medial pathologies. CNC- and SHF-derived cells were tracked using Wnt1- or Mef2c-Cre ROSA26RLacZ mice infused with AngII, respectively. Consistent with our previous study,13 the ascending aorta was composed of both CNC- and SHF-derived SMCs (Figure 1C). SHF-derived cells were restricted to the ascending aorta, while CNC-derived cells extended into the aortic arch. Although not altering distribution of cells of SMC origins (Figure 1D, E), AngII increased medial area (Figure 1F). Of interest, X-gal positive areas were increased significantly in medial SHF-, but not CNC-, derived cells (Figure 1G). Because selected adventitial FBs are derived from the SHF,13 we also evaluated the effects of AngII infusion on cells in the adventitia. Similar to the media, the adventitial area was also increased by AngII infusion (Figure 1H) and increased X-gal positive areas were observed in SHF-derived, but not CNC-derived, cells (Figure 1I). Thus, TAAs have a transmural gradient of aortic pathologies in humans and mice. The linear tracing data support the notion that SHF-derived cells are susceptible to AngII-induced outer medial and adventitial thickenings.
AngII altered aortic protein profiles related to extracellular matrix organization
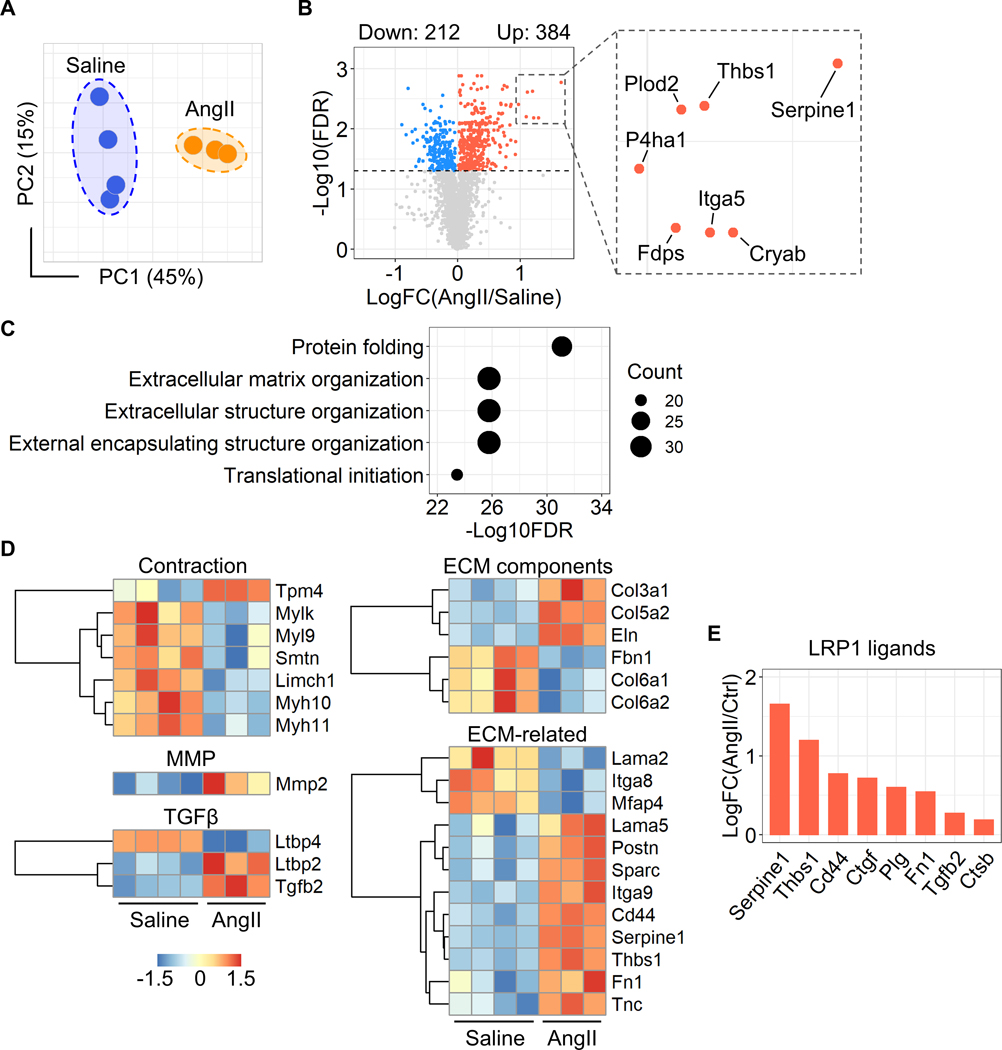
To uncover the molecular mechanisms of AngII-induced ascending aortic pathologies, we performed mass spectrometry-assisted proteomics of the ascending aorta of AngII-infused mice. Ascending aortic tissues were harvested at day 3 of either saline or AngII infusion. Aortic samples showing intramural hemorrhage or dissection were excluded. Proteomic analysis identified 2,644 proteins among all aortic samples. In PCA using the unfiltered proteome, AngII infusion altered protein profiles significantly (45% variation along PC1, Figure 2A). Compared to saline-infused mice, AngII infusion changed 596 protein abundances (Figure 2B, up- and downregulated: 384 and 212 proteins, respectively). Gene ontology and KEGG pathway enrichment analyses were also performed using these differentially expressed proteins. In addition to terms related to translation and ribosome, ECM related terms, such as “Extracellular matrix organization” and “Proteoglycan in cancer”, were identified in both gene ontology and KEGG analyses (Figure 2C, Supplemental Figure IA). A protein-protein interaction analysis also demonstrated the presence of ECM-related molecules in the differentially expressed proteins (Supplemental Figure IB). Therefore, we assessed ECM-related molecules comprehensively (Figure 2D). Several contractile proteins, including Myh11, were downregulated by AngII, while Mmp2 (matrix metalloproteinase 2) was upregulated in AngII-infused mouse aortas. In TGFβ signaling molecules, AngII upregulated Ltbp2 (latent TGFβ binding protein 2) and Tgfb2 (TGFβ−2). AngII also increased several ECM components, Col3a1 (collagen type III alpha 1 chain), Col5a2 (collagen type V alpha 2 chain), and Eln (elastin). Other ECM-related molecules, Thbs1 (thrombospondin 1), Fn1 (fibronectin 1) and Tnc (tenascin C), were also upregulated by AngII infusion. Thus, during the short interval of 3 days of infusion, AngII changed ECM-related proteome significantly in the ascending aorta.
Figure 2. AngII altered proteins related to ECM in the ascending aorta of mice.
(A) Principal component analysis of the unfiltered proteome. PC1/PC2 indicate first/second principal components (n=3 or 4 per group). (B) Volcano plots for differentiated proteins between in ascending aortas of saline and AngII-infused mice. (C) Top 5 annotations of enrichment analyses in gene ontology - biological process. (D) Heatmap with Z score coloring for proteins related to SMC contraction, MMP, TGFβ signaling, ECM-related molecules compared between saline and AngII infusion. (E) Log fold change of LRP1 ligands.
Low-density lipoprotein receptor-related protein 1 (LRP1) is a multifunctional protein that plays a role in ECM maturation.28 LRP1 deletion in pan SMCs causes aortic pathologies including aneurysm and tortuosity with ECM degradation.29–32 Thus, we examined protein abundances of LRP1 and its ligands. Aortic LRP1 protein abundance was not changed by AngII infusion (data not shown, P=0.89); however, multiple LRP1 ligands, including Serpine1 (plasminogen activator inhibitor 1, PAI1), were increased by AngII (Figure 2E). Of note, a volcano plot demonstrates that Serpine1 was the most significantly increased molecule by AngII infusion (Figure 2B). Since these ligands are internalized into the cytosol through LRP1 for their degradation,28 these data suggest the impairment of aortic LRP1 function. Taken together, the aortic proteome related to ECM was altered as early as day 3 by AngII infusion with the increase of multiple LRP1 ligands.
LRP1 deletion in SHF-derived cells augmented AngII-induced thoracic aortopathies
As described above, our proteomic analysis, in combination with our previous findings that deletion of LRP1 in pan SMCs augmented TAA formation with ECM degradation in AngII-infused mice,30 prompted us to determine whether LRP1 deletion in SHF lineage affects AngII-induced TAA formation. LRP1 was deleted in SHF-derived cells using a Mef2c promoter driven Cre. In Mef2c-Cre-expressing mice, Cre activity was observed mainly in the outer media (Supplemental Figure IIA), and the Lrp1 delta flox PCR fragment was detected in addition to a band of native Lrp1 (Supplemental Figure IIB–D), confirming DNA recombination of LRP1 in SHF-derived cells. LRP1 protein abundance was reduced by 37% in the ascending aortic media of SHF-specific LRP1 deleted mice compared to Cre negative littermates with LRP1 deletion restricted to the outer media of the ascending aorta (Supplemental Figure IIE–F). Mass spectrometry-assisted proteomics also verified reduction of LRP1 peptide intensity in the ascending aorta of Mef2c-Cre expressing mice (Supplemental Figure IIG).
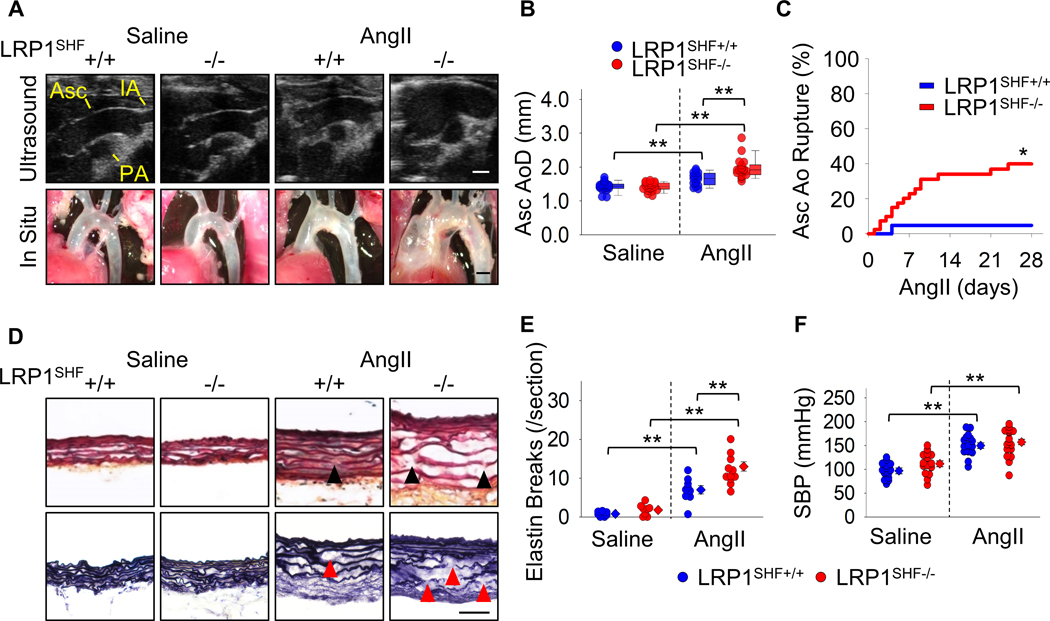
SHF-specific LRP1 deletion did not affect growth or development of mice. To promote TAAs, AngII was infused subcutaneously into mice for 4 weeks. LRP1 deletion in SHF-derived cells augmented AngII-induced aortic dilation in the ascending aorta as demonstrated by both ultrasound and in situ imaging (Figure 3A, B). SHF-specific LRP1 deletion also exacerbated AngII-induced ascending aortic rupture and elastin fragmentation (Figure 3C–E). Systolic blood pressure was not different between genotypes in response to AngII infusion (Figure 3F), demonstrating that augmented aortopathy was independent of systolic blood pressure. These data support that SHF-derived cells exert a critical role in the structural integrity of the ascending aorta.
Figure 3. LRP1 deletion in SHF-derived cells augmented AngII-induced TAA formation and aortic rupture in mice.
(A) Representative ultrasound images and in situ gross appearances of proximal thoracic aortas (n=21 to 40 per group). Asc indicates ascending aorta; IA, innominate artery; PA, pulmonary artery. Scale bar, 1 mm. (B) Ascending aortic diameter (Asc AoD) measured by ultrasonography. (C) Cumulative incidence of ascending aortic (Asc Ao) rupture. (D) Representative Movat’s and Verhoeff iron hematoxylin staining. Scale bar, 50 μm. Arrow heads indicate elastin fragmentation. (E) Elastin break counts in Movat’s stained sections. (F) Systolic blood pressure. Diamonds and error bars indicate the mean and SEM, respectively. *P<0.01 by log-rank test, **P<0.001 by two-way ANOVA followed by Holm-Sidak test. Log transformation was applied for the data of ascending aortic diameters.
AngII infusion compromised TGFβ signaling in SHF-derived SMCs and FBs
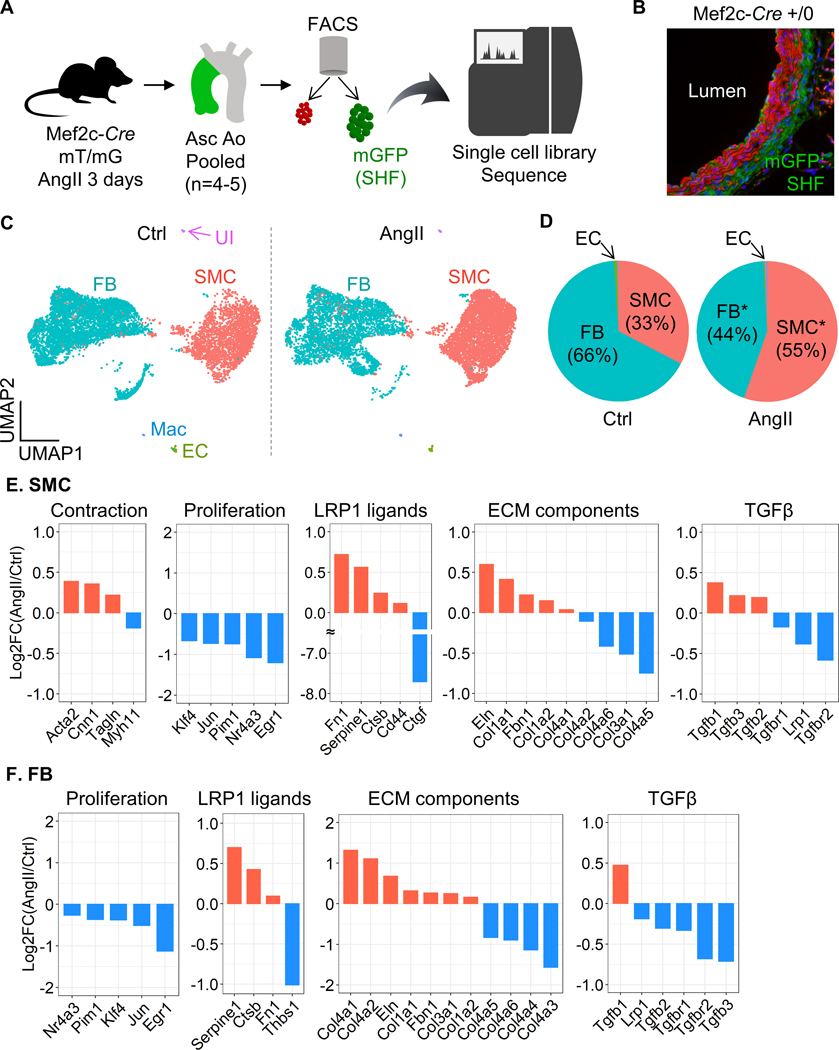
Since using tissues of fate mapping mice enables unbiased transcriptomics profiling in a lineage-specific manner, we investigated molecular roles of SHF-derived cells in AngII-induced TAA formation by scRNAseq in aortas from Mef2c-Cre ROSA26RmT/mG mice. Aortic tissues were harvested from Mef2c-Cre ROSA26RmT/mG male mice at baseline and after 3 days of AngII infusion. Four to five aortic tissues were pooled to obtain sufficient number of aortic cells (Figure 4A). Since mGFP protein was present on Mef2c-Cre expressing cells of this mouse strain, mGFP positive cells were derived from the SHF (Figure 4B). After sorting cells based on mGFP signal, scRNAseq was performed using mGFP positive cells.
Figure 4. Significant reduction of Tgfbr2 mRNA in SHF-derived SMCs and FBs of AngII-infused mice.
(A) Experimental design of scRNAseq using Mef2c-Cre +/0 ROSA26RmT/mG mice. (B) Representative fluorescent image of the ascending aorta from Mef2c-Cre +/0 ROSA26RmT/mG mice. (C) UMAP plots of aortic cells at baseline (Ctrl) and after 3 days of AngII infusion. SMC indicates smooth muscle cells; FB, fibroblasts; EC, endothelial cells; Mac, macrophages; UI, unidentified cells. (D) Pie charts for the composition of each cell type. *P<0.0001 by Chi-square test. Log2 fold change of mRNA abundance related to SMC contraction, cell proliferation, LRP1 ligands, ECM components, and TGFβ ligands and receptors between Ctrl vs AngII in (E) SMCs and (F) FB clusters.
The two normalized datasets were properly integrated (Figure 4C). scRNAseq analysis revealed multiple cell types, including SMC, FB, and endothelial cells, in the mGFP cell population (Figure 4C, D, Supplemental Figure IIIA, B). At baseline, SHF-derived SMCs were composed of 33% SHF-derived cells and it was increased to 55% by AngII infusion (Figure 4D). Meanwhile, FB population was decreased from 66% to 44%. Since SMCs and FBs were comprised the majority, DEG analysis was performed in these clusters. AngII altered 3,848 (up: 1,862; down: 1,986) and 3,398 (up: 1,931; down: 1,467) genes in SMCs and FBs, respectively (Supplemental Figure IVA, VA). Although the top 10 up- and downregulated genes were variable between SMCs and FBs, except for Ptrf (Supplemental Figure IVB, VB), enrichment analysis identified “extracellular matrix organization” as a common term in SMC and FB clusters (Supplemental Figure IVC, VC). Therefore, similar to the proteomic analyses, we next examined ECM-related molecules in the scRNAseq data (Figure 4E, F). In SMCs, Acta2, Cnn1, and Tagln, contractile genes, were increased after 3 days of AngII infusion. Of note, proliferation genes, such as Klf4 and Egr1, were downregulated in both SMCs and FBs by AngII infusion. As LRP1 ligands, Serpine1, Ctsb (cathepsin B), and Fn1 (fibronectin 1) were increased by AngII in SHF-derived SMCs and FBs. Multiple ECM components, Eln, Col1a1, Col1a2, and Col4a1, were increased in both SMC and FB clusters in response to AngII infusion. However, TGFβ ligands demonstrated variable responses to AngII between cell types. AngII increased Tgfb1–3 in SMCs, but Tgfb2–3 were decreased in FBs. It is noteworthy that TGFβ receptors, including Lrp1 and Tgfbr2, were significantly downregulated by AngII in both SMCs and FBs.
mRNA abundance of TGFβ ligands and receptors were also examined in human scRNAseq data (GSE155468).26 SMCs and FBs were extracted and mRNA abundance of TGFB1–3, TGFBR1–2, and LRP1 were compared between control and TAA samples. In SMCs, TGFβ ligands and receptors, including LRP1, were decreased modestly in TAAs compared to control aortas (Supplemental Figure VIA). In contrast, TGFβ ligands and receptors were increased in FBs of TAAs, except for TGFB1 (Supplemental Figure VIB).
A unique sub-cluster of SHF-derived fibroblasts in ascending aortas of AngII-infused mice
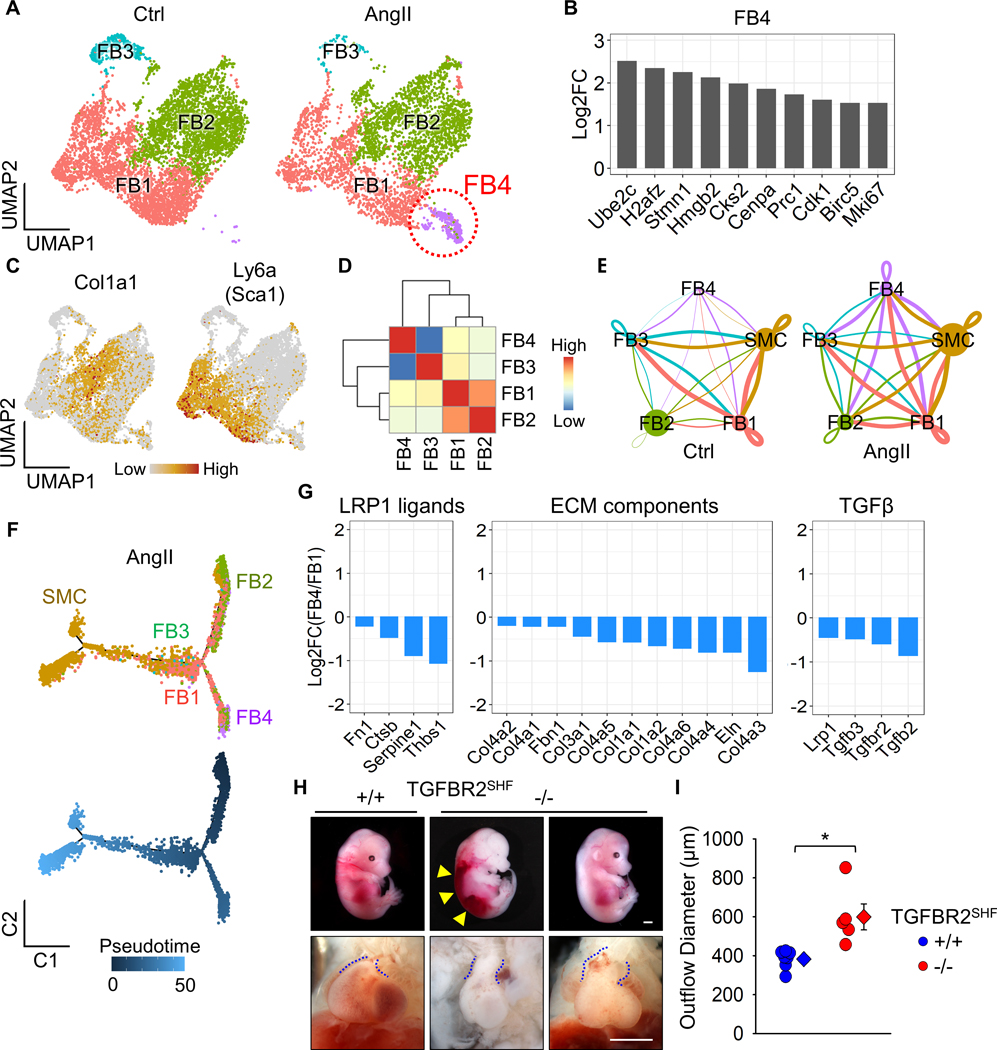
Since the aortic wall comprises mostly SMCs and FBs (Figure 4C, D), we performed further sub-clustering analyses in SHF-derived SMCs and FBs. SHF-derived SMCs had 3 distinct clusters, but there was no remarkable alteration during AngII infusion in transcriptomic distributions of these cells (Supplemental Figure VII). Meanwhile, a unique sub-cluster was observed in SHF-derived FBs of AngII-infused mice (FB4, Figure 5A). The FB4 sub-cluster was composed of 10 and 544 cells at baseline and after 3 days of AngII infusion, respectively (Supplemental Figure VIIIA). AngII infusion altered 586 genes in the FB4 sub-cluster and these genes were significantly associated with cytokinesis, as identified by gene ontology analysis (Supplemental Figure VIIIB, C). To further characterize the FB4 sub-cluster, featured genes were determined by comparing the transcriptomes among all FB sub-clusters. Genes related to cell division, such as H2afz, Cks2, Cenpa, Cdk1, and Mki67, were identified uniquely in the FB4 sub-cluster (Figure 5B, Supplemental Figure IX). These results indicate that FB4 cells had enhanced proliferative features. However, FB4 cells were not highly abundant for Ly6a, as a marker for Sca1+ FBs (Figure 5C). While the transcriptome of FB4 differed from other FB sub-clusters (Figure 5D), the ligand-receptor interaction analyses in SMC and FB clusters demonstrated accelerated cell-cell interactions among cell types by AngII infusion (Figure 5E). In particular, FB4 sub-cluster had more apparent interactions with SMC and FB1 clusters under AngII infusion. Trajectory analysis demonstrated that FB4 cells were derived from FB1 cells in a pseudotime (Figure 5F). Because of these interactions, we subsequently compared DEGs related to ECM maturation between FB1 and FB4 sub-clusters (Figure 5G). LRP1 ligands, including Serpine1, were all downregulated in FB4. ECM component genes, such as Eln and Col1a1, were also lower in FB4. TGFβ receptors were further downregulated in FB4 by AngII infusion. Alongside the scRNAseq analyses in SMCs and FBs, these data suggest that AngII compromises the TGFβ signaling pathway in SHF-derived cells, which may contribute to the susceptibility of SHF-derived cells to vascular pathologies.
Figure 5. A distinct sub-cluster with decreased Tgfbr2 mRNA in SHF-derived fibroblasts of AngII-infused mice.
(A) UMAP plots at baseline (Ctrl) and after 3 days of AngII infusion in SHF-derived FBs. (B) Log2 fold change of mRNA abundance for top 10 unique genes in FB4 sub-cluster. (C) Featured plots for Col1a1 and Ly6a in FB sub-clusters. (D) Heatmap for cluster similarity among FB sub-clusters. (E) Ligand-receptor interactions among aortic cells. Lines connect to cell populations that express cognate ligands or receptors. The line thickness and node size correspond to the number of cognate ligand-receptor expression and cell number, respectively. (F) Trajectory analysis in SMC and FB sub-clusters from AngII-infused mice. (G) Log2 fold change of mRNA abundance related to LRP1 ligands, ECM components, and TGFβ molecules between FB1 and FB4 sub-clusters after AngII infusion. (H) Representative images of gross appearance and the outflow tract of wild type littermates (left), dead (middle), and survived (right) fetuses with SHF-specific TGFBR2 deletion at E12.5. Blue dotted lines indicate the edge of the outflow tract. Yellow triangles indicate retroperitoneal hemorrhage. (I) Outflow diameter was measured at 300 to 400 μm distal to the root of viable fetuses at termination (n=5 to 7 per group). Scale bar, 1 mm. Diamonds and error bars indicate the mean and SEM, respectively. *P<0.001 by two-tailed Student’s t-test.
To investigate the impact of TGFβ signaling pathway in SHF-derived cells on vascular integrity, we deleted TGFBR2 in SHF-derived cells in mice. Of note, SHF-specific TGFBR2 deletion led to embryonic lethality with retroperitoneal hemorrhage beginning at E12.5 (Figure 5H). In addition, the outflow tract was dilated significantly in fetuses with SHF-specific TGFBR2 deletion compared to wild type littermates (Figure 5H, I). Thus, TGFBR2 deficiency in SHF-derived cells led to prenatal vasculopathies, suggesting the importance of SHF-derived cells through TGFBR2 for vascular development and maintaining of its integrity.
Transmedial gradient of PAI1 in ascending aortas of AngII-infused mice and sporadic ascending TAAs in humans
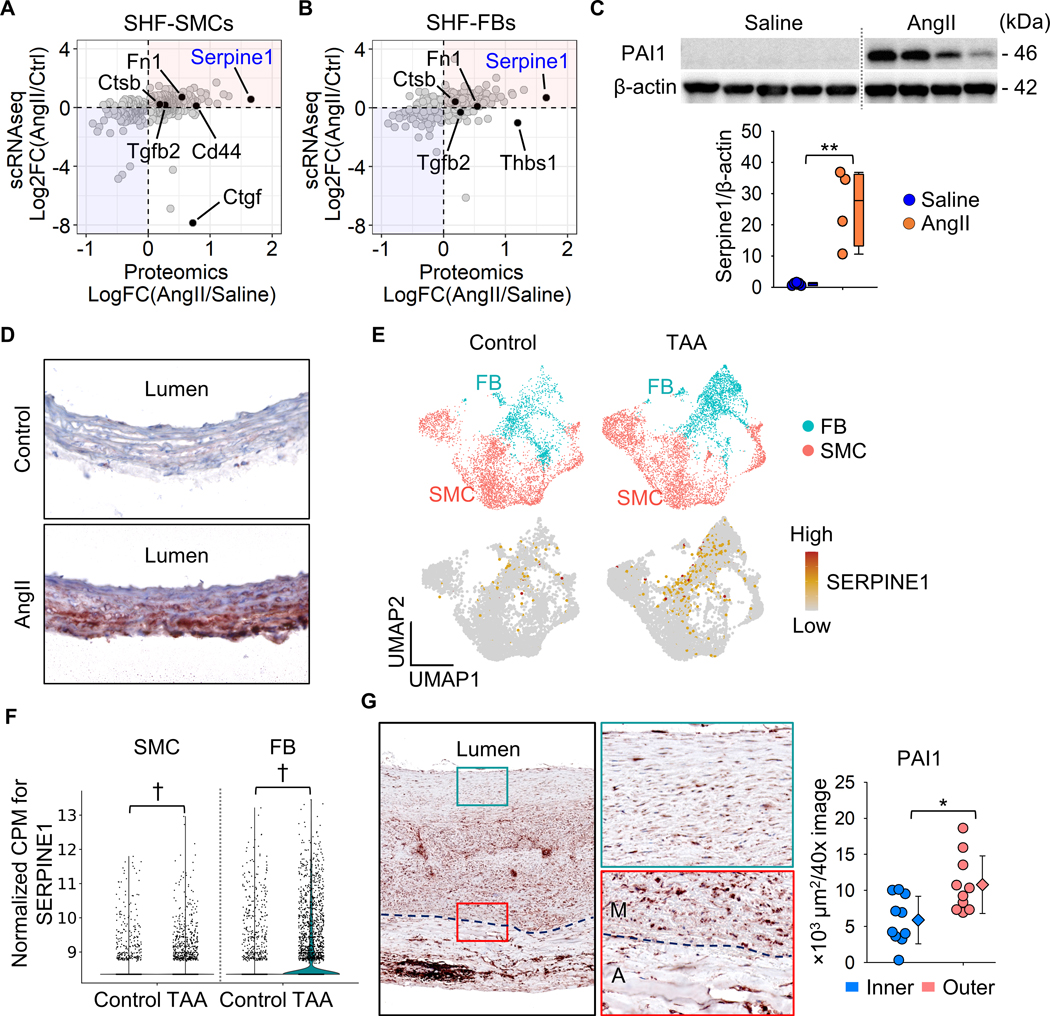
We integrated DEG results of proteomics and scRNAseq data from mice. There were 412 overlapped genes between proteomics and scRNAseq using the SHF-SMC cluster, and 198 molecules were upregulated in both datasets (highlighted in red, Figure 6A). In SHF-FBs, 375 genes were overlapped, and 205 genes were increased (Figure 6B). As LRP1 ligands, Serpine1, Fn1, and Ctsb were consistently greater abundance in AngII-infused mice, as shown in proteomics and scRNAseq analyses of SMCs and FBs (Figure 6A, B). In protein analyses, PAI1 (Serpine1) was the most significantly increased molecule by AngII (Figure 6A, B). Western blot analysis showed a striking increase (~26 fold) of PAI1 in ascending aortas from mice infused with AngII (Figure 6C). Immunostaining of PAI1 was hardly detected in aortas from control mice, whereas it was intense in ascending aortas of AngII-infused mice (Figure 6D). In ascending aortas of AngII infused mice, PAI1 was present through the aortic wall; aortic PAI1 was dominant in the outer media and the adventitia, coincident with the gradient of medial pathologies (Figure 6D).
Figure 6. Integration of proteomic and scRNAseq results identified PAI1 (Serpine1) as a key contributor to the medial gradient of aortic pathologies.
Scatter plots for Log or Log2 fold change of protein and mRNA abundances. Data were compared between proteomics and scRNAseq in SHF-derived (A) SMC and (B) FB clusters. Selected LRP1 ligands were highlighted. (C) Western blot analysis for PAI1 (Serpine1) and β-actin in the whole ascending aorta from saline or AngII-infused mice (n=4–5 per group). **P<0.001 by Student’s t-test after Log transformation. (D) Representative images of immunostaining for PAI1 (red) in ascending aortas from control or AngII-infused mice (n=7 per group). (E) UMAP and (F) violin plots for SERPINE1 in SMC and FB clusters of control subjects and patients with ascending TAAs (n=3 and 8 per group, scRNAseq data were downloaded from GSE155468). †P<0.001 analyzed using the Hurdle model adjusted for age implemented. (G) Representative images of immunostaining for PAI1 in patients with sporadic ascending TAA (n=10) and its quantification. Dotted lines indicate external elastic laminae; M, media; A, adventitia. Diamonds and error bars indicate the mean and SEM, respectively. *P<0.01 by Student’s t-test.
For clinical relevance, we evaluated SERPINE1 mRNA abundance and protein distribution in human aortic samples. In the human scRNAseq data (GSE155468),26 SERPINE1 abundance was assessed in SMC and FB clusters. UMAP plots showed the presence of SERPINE1 in both SMC and FB clusters of control and TAA samples (Figure 6E); SERPINE1 was significantly higher in SMCs and FBs of human TAA samples compared to controls (Figure 6F). Immunostaining demonstrated that PAI1 was expressed in the aortic media and adventitia of human TAA samples (Figure 6G). Consistent with AngII-infused mouse aortas (Figure 6D), highly abundant areas of PAI1 were observed in the outer media adjacent to the external elastic lamina in most TAA tissues (Figure 6G).
Discussion
Our study demonstrated that (1) aortic pathologies were mainly observed in the outer media of the ascending aorta in patients with TAAs and AngII-infused mice, (2) SHF-derived cells were responsible for AngII-induced medial and adventitial thickening in the ascending aorta, (3) AngII altered protein profiles related to ECM and LRP1 ligands, (4) AngII decreased Tgfbr2 mRNA in SHF-derived SMCs and FBs, (5) AngII led to a distinct sub-cluster of SHF-derived FBs that had lower Tgfbr2 mRNA abundance, (6) Either LRP1 or TGFBR2 deletion in SHF-derived cells led to vasculopathies in mice, and (7) proteomic and scRNAseq analyses identified PAI1 as a key molecule that showed a transmedial gradient in the ascending aorta of AngII-infused mice or human TAA tissues. Collectively, this study provides multiple evidence to support the importance of SHF-derived cells in maintaining the integrity of aortic wall.
A number of studies have shown an important role for TGFβ signaling in maintaining aortic wall integrity.9, 33–35 Both LRP1 and TGFBR2 are related to the TGFβ signaling pathways,28, 33 and genetic deletions of these molecules in pan-SMCs promotes TAA formation in mice.8, 9, 29–31, 36 Therefore, in the present study, the biological effects of SHF-derived cells in aortic integrity was investigated by SHF-specific deletion of either LRP1 or TGFBR2. Of note, SHF-specific TGFBR2 deletion led to vascular malformation and SHF-specific LRP1 deletion recapitulated aortic phenotypes in mice with its deletion in pan-SMCs. In the ascending aorta, SHF-derived cells comprise approximately half of SMCs and selected FBs.13 Thus, we concluded that SHF-derived cells were functionally important to maintain the structural integrity of vascular wall.
The present study revealed that deletion of either TGFBR2 or LRP1 in SHF-derived cells promotes vasculopathies. These data are consistent with the outer layers of SMCs derived cells being crucial for development of vascular pathologies related to TGFβ signaling. A previous study using mice expressing a fibrillin1 mutation (Fbn1C1041G/+) reported that heterozygous deletion of Notch1 in SHF-derived cells had a trend for augmenting luminal dilatation of the aortic sinus.37 In contrast, another study demonstrated that Smad2 deletion in CNC-derived cells reduced dilation of the aortic sinus in a mouse model of Loeys-Dietz syndrome.38 Since there are aortic region-specific responses to TGFβ,39 these two developmental origins may have divergent pathophysiological functions in different aortic regions.
Elastin fragmentation is a pathological feature of TAAs and is associated with decrease of aortic resilience.40, 41 Elastic fibers are one of the major matrix components and vital for maintaining aortic structure.42 Mutations of the elastin gene cause aortic stenosis due to overgrowth of SMCs.40, 41 Also, SMC-specific elastin deletion induces aortic extension and coarctation in mice.43 Our proteomic and scRNAseq analyses demonstrated increased aortic elastin in response to AngII infusion. These data suggest that de novo elastin synthesis is a compensatory reaction for maintaining aortic integrity during AngII stimulation. It is of note that major TGFβ receptors, Tgfbr2 and Lrp1, were downregulated during AngII infusion for 3 days in SHF-derived SMCs and FBs, although the TGFβ signaling pathway plays an important role in ECM maturation.44 Hence, increase of aortic elastin in SHF-derived cells may not be regulated through the TGFβ signaling pathway directly during this interval. In addition, multiple collagen genes showed variable responses to AngII infusion. Therefore, it remains to be clarified which ECM component molecules are regulated during AngII infusion.
Our proteomic and scRNAseq analyses identified PAI1 as a key molecule in AngII-induced TAA formation. PAI1 has been implicated in thoracic aortic diseases by omics approaches in several previous studies.45, 46 A microarray of aortas from patients with aortic dissections revealed an increase of PAI1 mRNA abundance in dissected aortas.45 scRNAseq using aortas from patients with Marfan syndrome and a mouse model of this disease demonstrated the presence of PAI1 in aortic SMCs.46 Consistent with these reports, the present study also identified PAI1 in the ascending aorta shortly after initiation of AngII infusion. Of interest, aortic PAI1 was increased dramatically by AngII. Furthermore, our histological analyses found a transmedial gradient of PAI1 in AngII-infused mice and human TAA samples. The distinct distribution of PAI1 was matched with the medial gradient of aortic pathologies, consistent with PAI1 contributing to the complex mechanism of TAA formation.
Several articles have investigated a role of PAI1 in abdominal aortic aneurysms.47–49 Focal overexpression of PAI1 inhibit aneurysm formation in the abdominal aorta of a rat model using xenograft.47 Adventitial overexpression of PAI1 also protect abdominal aneurysm formation in AngII-infused mice.48 In addition, systemic deletion of PAI1 augmented elastase-induced abdominal aortic aneurysms in mice.49 Therefore, PAI1 plays a protective role in development of abdominal aortic aneurysms. However, molecular mechanisms underlying TAAs and abdominal aortic aneurysms are distinct. There is a lack of evidence showing biological function of PAI in thoracic aortopathies. PAI1 is a ligand for LRP1 and involved in the TGFβ signaling pathway.50–52 In subsequent studies, we will elucidate the role of PAI1 in SHF-derived cells on TAA formation as well as TGFβ signaling pathway through LRP1.
SHF-specific TGFBR2 deletion led to dilatation of the outflow tract with post peritoneal hemorrhage. Thus, TGFBR2 in SHF-derived cells is essential for vascular development and integrity. However, due to the embryonic lethality by constitutive deletion of TGFBR2 in SMCs, the role of TGFBR2 in SHF-derived cells on thoracic aortopathies remains unclear. The Mef2c gene is not sufficiently expressed in the cardiovascular system at the postnatal phase, especially in normal conditions during the adult phase.53 Therefore, it is not feasible to activate Mef2c-Cre using the ERT2 system in adult mice. Development of research methods are needed to overcome the barrier to postnatal gene manipulation in the SHF origin.
Previous studies have reported that prenatal deletion of TGFBR2 in CNC-derived cells results in cleft palate and a congenital heart defect, persistent truncus arteriosus.54, 55 Therefore, there is compelling evidence that CNC-derived cells exert a pivotal role in the integrity of the cardiovascular system.54, 55 However, the present study focused on the role of SHF-derived cells in thoracic aortopathies, since scRNAseq data revealed the important role of these cells on the regulation of elastin and TGFβ signaling genes. Furthermore, our mouse studies demonstrated vascular phenotypes by gene manipulation in SHF-derived cells. These results support the notion that SHF-derived cells are crucial in maintaining aortic wall integrity. In fact, a recent study reported that a gene TGFBR1 mutation suppressed muscle contractility in SMCs from the SHF, but not CNC, origin.56 However, it is still desirable to directly compare the role of two different SMC origins on formation of thoracic aortopathies.
Our scRNAseq identified a distinct FB sub-cluster, and AngII infusion further decreased Tgfbr2 mRNA abundance in the distinct FB sub-cluster compared to other FBs. Since TGFBR2 deletion in SHF-derived cell led to vascular malformation, significant reduction of Tgfbr2 mRNA abundance by AngII in this cluster may exert a deleterious role in the pathophysiology of TAAs. scRNAseq enabled us to evaluate cellular heterogeneity and identify a unique cellular cluster, which is a great advantage of scRNAseq. However, it is still difficult to investigate cluster-specific molecular mechanisms. A technical breakthrough is needed to manipulate specific sub-clusters for understanding its role in the pathophysiology of TAAs.
scRNAseq demonstrated that Tgfbr2 and Lrp1 were downregulated by AngII infusion in both SMCs and FBs derived from the SHF of mouse aortas. While in human aortas, TGFBR2 and LRP1 were decreased in SMCs of TAAs, but increased in FBs of TAAs. TGFBR2 and LRP1 in FBs in human scRNAseq conflict with the mouse scRNAseq results. In the mouse scRNAseq, aortic samples were harvested at day 3 of AngII infusion prior to TAA formation, but human scRNAseq used aortas at an advanced stage of diseases that were acquired during surgical repair. As the role of TGFβ signaling changes depending on disease stage,57 this may contribute to the difference of scRNAseq results seen between humans and mice in the present study. In addition, since it was not feasible to discern embryonic origins in the human scRNAseq dataset, the data were analyzed using all SMCs and FBs rather than isolated SHF-derived cells. This may also contribute to the difference seen in these scRNAseq results.
In conclusion, our study highlights the functional importance of SHF-derived cells in maintaining vascular integrity using multiple TAA mouse models. This study provides strong evidence that SHF-derived cells exert a role during development of thoracic aortopathies. Heterogeneity of SHF-derived cells contributes to complex mechanisms of aortopathy formation, which should be taken into consideration when investigating the pathogenesis of thoracic aortopathies.
Supplementary Material
Clinical Perspective.
What is new?
SHF-derived SMCs and fibroblasts associate with AngII-induced aortic pathologies.
AngII induces a distinct fibroblast sub-cluster that is less abundant for mRNAs related to major extracellular components and TGFβ ligands and receptors, but more abundant for proliferative genes.
TGFBR2 deletion in SHF-derived cells are embryonic lethal with significant dilatation of the outflow tract in mice.
SHF-specific deletion of LRP1 leads to aortic pathologies in mice, supporting the importance of SHF-derived cells in maintaining ascending aortic wall integrity.
What are the clinical implications?
Heterogeneity of the embryonic origins of SMCs and fibroblasts contributes to complex mechanisms of vasculopathy formation, which should be considered when investigating the pathogenesis of thoracic aortopathies.
Acknowledgments
Study design: HS, HH, CZ, YL, SM, SAS, JZC, DLR, YHS, MA, MWM, HSL, AD
Implementation of animal experiments: HS, CZ, JJM, DAH, DLR
Histological analyses: HS, MKF, DAH, DLR
Data analyses: HS, YK, DLR, HSL, AD
Proteomics and associated informatics: HS, YK, HH, SM, LHL, SAS
scRNAseq and associated informatics: HS, YK, CZ, YL, YHS
Supervising and data verification: SAS, YHS, SAL, MA, MWM, HSL, AD
Writing the draft manuscript: HS
Editing the manuscript: all authors
Sources of Funding
The authors’ research work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health [R01HL133723, R35HL155649 (AD), R01HL126901 (MA), R01HL149302 (MA), R01HL121877 (MWM)] and the American Heart Association SFRN in Vascular Disease (18SFRN33960163 and 33960114). HS was supported by an AHA postdoctoral fellowship (18POST33990468). JZC was supported by NCATS UL1TR001998 and NHLBI F30HL143943. SAL is supported in part by the Jimmy and Roberta Howell Professorship in Cardiovascular Surgery at Baylor College of Medicine. The content in this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosures
The following authors have declared that no conflict of interest exists: HS, YK, CZ, YL, SM, LHL, SAS, JZC, MKF, JJM, DAH, DLR, YHS, SAL, MA, MWM, HSL, AD. HH is an employee of Kowa Company, Ltd. (Nagoya, Japan) and was a visiting scientist at Brigham and Women’s Hospital when the study was conducted.
Non-standard Abbreviations and Acronyms
- AngII
angiotensin II
- CNC
cardiac neural crest
- ECM
Extracellular matrix
- FB
fibroblast
- LRP1
low density lipoprotein-related protein 1
- PAI1
plasminogen activator inhibitor 1
- SHF
second heart field
- SMC
smooth muscle cell
- TAA
thoracic aortic aneurysm
- TGF-β
transforming growth factor beta
- Tgfbr2
transforming growth factor beta receptor type 2
References
- 1.Goldfinger JZ, Halperin JL, Marin ML, Stewart AS, Eagle KA and Fuster V. Thoracic aortic aneurysm and dissection. J Am Coll Cardiol. 2014;64:1725–1739. [DOI] [PubMed] [Google Scholar]
- 2.Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE Jr., Eagle KA, Hermann LK, Isselbacher EM, Kazerooni EA, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM Guidelines for the diagnosis and management of patients with thoracic aortic disease. A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology,American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons,and Society for Vascular Medicine. J Am Coll Cardiol. 2010;55:e27–e129. [DOI] [PubMed] [Google Scholar]
- 3.Isselbacher EM. Thoracic and abdominal aortic aneurysms. Circulation. 2005;111:816–828. [DOI] [PubMed] [Google Scholar]
- 4.Albornoz G, Coady MA, Roberts M, Davies RR, Tranquilli M, Rizzo JA and Elefteriades JA. Familial thoracic aortic aneurysms and dissections--incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg. 2006;82:1400–1405. [DOI] [PubMed] [Google Scholar]
- 5.Trachet B, Piersigilli A, Fraga-Silva RA, Aslanidou L, Sordet-Dessimoz J, Astolfo A, Stampanoni MF, Segers P and Stergiopulos N. Ascending aortic aneurysm in angiotensin II-infused mice: Formation, progression, and the role of focal dissections. Arterioscler Thromb Vasc Biol. 2016;36:673–681. [DOI] [PubMed] [Google Scholar]
- 6.Daugherty A, Rateri DL, Charo IF, Owens AP, Howatt DA and Cassis LA. Angiotensin II infusion promotes ascending aortic aneurysms: attenuation by CCR2 deficiency in apoE−/− mice. Clin Sci (Lond). 2010;118:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wanga S, Hibender S, Ridwan Y, van Roomen C, Vos M, van der Made I, van Vliet N, Franken R, van Riel LA, Groenink M, et al. Aortic microcalcification is associated with elastin fragmentation in Marfan syndrome. J Pathol. 2017;243:294–306. [DOI] [PubMed] [Google Scholar]
- 8.Yang P, Schmit BM, Fu C, DeSart K, Oh SP, Berceli SA and Jiang Z. Smooth muscle cell-specific Tgfbr1 deficiency promotes aortic aneurysm formation by stimulating multiple signaling events. Sci Rep. 2016;6:35444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Li Q, Jiao Y, Qin L, Ali R, Zhou J, Ferruzzi J, Kim RW, Geirsson A, Dietz HC, et al. Tgfbr2 disruption in postnatal smooth muscle impairs aortic wall homeostasis. J Clin Invest. 2014;124:755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawada H, Chen JZ, Wright BC, Sheppard MB, Lu HS and Daugherty A. Heterogeneity of aortic smooth muscle cells: A determinant for regional characteristics of thoracic aortic aneurysms? J Transl Int Med. 2018;6:93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quintana RA and Taylor WR. Cellular mechanisms of aortic aneurysm formation. Circ Res. 2019;124:607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. [DOI] [PubMed] [Google Scholar]
- 13.Sawada H, Rateri DL, Moorleghen JJ, Majesky MW and Daugherty A. Smooth muscle cells derived from second heart field and cardiac neural crest reside in spatially distinct domains in the media of the ascending aorta-Brief report. Arterioscler Thromb Vasc Biol. 2017;37:1722–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo W, Wang Y, Zhang L, Ren P, Zhang C, Li Y, Azares AR, Zhang M, Guo J, Ghaghada KB, et al. Critical role of cytosolic DNA and its sensing adaptor STING in aortic degeneration, dissection, and rupture. Circulation. 2020;141:42–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinet P, Milewicz DM, Cassis LA, Leeper NJ, Lu HS and Smith JD. Consideration of sex differences in design and reporting of experimental arterial pathology studies-statement from ATVB council. Arterioscler Thromb Vasc Biol. 2018;38:292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu H, Howatt DA, Balakrishnan A, Moorleghen JJ, Rateri DL, Cassis LA and Daugherty A. Subcutaneous angiotensin II infusion using osmotic pumps induces aortic aneurysms in mice. J Vis Exp. 2015:doi: 10.3791/53191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2021: https://www.R-project.org/. [Google Scholar]
- 18.Kanehisa M and Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler A, Hoffman P, Smibert P, Papalexi E and Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risso D, Perraudeau F, Gribkova S, Dudoit S and Vert JP. A general and flexible method for signal extraction from single-cell RNA-seq data. Nat Commun. 2018;9:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MD, McCarthy DJ and Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS and Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skelly DA, Squiers GT, McLellan MA, Bolisetty MT, Robson P, Rosenthal NA and Pinto AR. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018;22:600–610. [DOI] [PubMed] [Google Scholar]
- 25.Csardi G and Nepusz T. The igraph software package for complex network research. InterJournal. 2006;Complex Systems:1695. [Google Scholar]
- 26.Li Y, Ren P, Dawson A, Vasquez HG, Ageedi W, Zhang C, Luo W, Chen R, Li Y, Kim S, et al. Single-Cell transcriptome analysis reveals dynamic cell populations and differential gene expression patterns in control and aneurysmal human aortic tissue. Circulation. 2020;142:1374–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strickland DK, Au DT, Cunfer P and Muratoglu SC. Low-density lipoprotein receptor-related protein-1: role in the regulation of vascular integrity. Arterioscler Thromb Vasc Biol. 2014;34:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boucher P, Gotthardt M, Li WP, Anderson RG and Herz J. LRP: role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–332. [DOI] [PubMed] [Google Scholar]
- 30.Davis FM, Rateri DL, Balakrishnan A, Howatt DA, Strickland DK, Muratoglu SC, Haggerty CM, Fornwalt BK, Cassis LA and Daugherty A. Smooth muscle cell deletion of low-density lipoprotein receptor-related protein 1 augments angiotensin II-induced superior mesenteric arterial and ascending aortic aneurysms. Arterioscler Thromb Vasc Biol. 2015;35:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basford JE, Koch S, Anjak A, Singh VP, Krause EG, Robbins N, Weintraub NL, Hui DY and Rubinstein J. Smooth muscle LDL receptor-related protein-1 deletion induces aortic insufficiency and promotes vascular cardiomyopathy in mice. PLoS One. 2013;8:e82026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muratoglu SC, Belgrave S, Hampton B, Migliorini M, Coksaygan T, Chen L, Mikhailenko I and Strickland DK. LRP1 protects the vasculature by regulating levels of connective tissue growth factor and HtrA1. Arterioscler Thromb Vasc Biol. 2013;33:2137–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindsay ME and Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmit BM, Yang P, Fu C, DeSart K, Berceli SA and Jiang Z. Hypertension overrides the protective effect of female hormones on the development of aortic aneurysm secondary to Alk5 deficiency via ERK activation. Am J Physiol Heart Circ Physiol. 2015;308:H115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallo EM, Loch DC, Habashi JP, Calderon JF, Chen Y, Bedja D, van Erp C, Gerber EE, Parker SJ, Sauls K, et al. Angiotensin II-dependent TGF-beta signaling contributes to Loeys-Dietz syndrome vascular pathogenesis. J Clin Invest. 2014;124:448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu JH, Wei H, Jaffe M, Airhart N, Du L, Angelov SN, Yan J, Allen JK, Kang I, Wight TN, et al. Postnatal deletion of the type II transforming growth factor-beta receptor in smooth muscle cells causes severe aortopathy in mice. Arterioscler Thromb Vasc Biol. 2015;35:2647–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koenig SN, LaHaye S, Feller JD, Rowland P, Hor KN, Trask AJ, Janssen PM, Radtke F, Lilly B and Garg V. Notch1 haploinsufficiency causes ascending aortic aneurysms in mice. JCI Insight. 2017;2: 10.1172/jci.insight.91353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacFarlane EG, Parker SJ, Shin JY, Kang BE, Ziegler SG, Creamer TJ, Bagirzadeh R, Bedja D, Chen Y, Calderon JF, et al. Lineage-specific events underlie aortic root aneurysm pathogenesis in Loeys-Dietz syndrome. J Clin Invest. 2019;129:659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Topouzis S and Majesky MW. Smooth muscle lineage diversity in the chick embryo. Two types of aortic smooth muscle cell differ in growth and receptor-mediated transcriptional responses to transforming growth factor-beta. Dev Biol. 1996;178:430–445. [DOI] [PubMed] [Google Scholar]
- 40.Li DY, Brooke B, Davis EC, Mecham RP, Sorensen LK, Boak BB, Eichwald E and Keating MT. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998;393:276–280. [DOI] [PubMed] [Google Scholar]
- 41.Curran ME, Atkinson DL, Ewart AK, Morris CA, Leppert MF and Keating MT. The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis. Cell. 1993;73:159–168. [DOI] [PubMed] [Google Scholar]
- 42.Shen YH and LeMaire SA. Molecular pathogenesis of genetic and sporadic aortic aneurysms and dissections. Curr Probl Surg. 2017;54:95–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CJ, Staiculescu MC, Hawes JZ, Cocciolone AJ, Hunkins BM, Roth RA, Lin CY, Mecham RP and Wagenseil JE. Heterogeneous cellular contributions to elastic laminae formation in arterial wall development. Circ Res. 2019;125:1006–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verrecchia F and Mauviel A. Transforming growth factor-beta signaling through the Smad pathway: role in extracellular matrix gene expression and regulation. J Invest Dermatol. 2002;118:211–215. [DOI] [PubMed] [Google Scholar]
- 45.Kimura N, Futamura K, Arakawa M, Okada N, Emrich F, Okamura H, Sato T, Shudo Y, Koyano TK, Yamaguchi A, et al. Gene expression profiling of acute type A aortic dissection combined with in vitro assessment. Eur J Cardiothorac Surg. 2017;52:810–817. [DOI] [PubMed] [Google Scholar]
- 46.Pedroza AJ, Tashima Y, Shad R, Cheng P, Wirka R, Churovich S, Nakamura K, Yokoyama N, Cui JZ, Iosef C, et al. Single-cell transcriptomic profiling of vascular smooth muscle cell phenotype modulation in Marfan syndrome aortic aneurysm. Arterioscler Thromb Vasc Biol. 2020;40:2195–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allaire E, Hasenstab D, Kenagy RD, Starcher B, Clowes MM and Clowes AW. Prevention of aneurysm development and rupture by local overexpression of plasminogen activator inhibitor-1. Circulation. 1998;98:249–255. [DOI] [PubMed] [Google Scholar]
- 48.Qian HS, Gu JM, Liu P, Kauser K, Halks-Miller M, Vergona R, Sullivan ME, Dole WP and Deng GG. Overexpression of PAI-1 prevents the development of abdominal aortic aneurysm in mice. Gene Ther. 2008;15:224–232. [DOI] [PubMed] [Google Scholar]
- 49.DiMusto PD, Lu G, Ghosh A, Roelofs KJ, Su G, Zhao Y, Lau CL, Sadiq O, McEvoy B, Laser A, et al. Increased PAI-1 in females compared with males is protective for abdominal aortic aneurysm formation in a rodent model. Am J Physiol Heart Circ Physiol. 2012;302:H1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otsuka G, Agah R, Frutkin AD, Wight TN and Dichek DA. Transforming growth factor beta 1 induces neointima formation through plasminogen activator inhibitor-1-dependent pathways. Arterioscler Thromb Vasc Biol. 2006;26:737–743. [DOI] [PubMed] [Google Scholar]
- 51.Samarakoon R, Higgins SP, Higgins CE and Higgins PJ. TGF-beta1-induced plasminogen activator inhibitor-1 expression in vascular smooth muscle cells requires pp60(c-src)/EGFR(Y845) and Rho/ROCK signaling. J Mol Cell Cardiol. 2008;44:527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaughan DE. PAI-1 and TGF-beta: unmasking the real driver of TGF-beta-induced vascular pathology. Arterioscler Thromb Vasc Biol. 2006;26:679–680. [DOI] [PubMed] [Google Scholar]
- 53.Edmondson DG, Lyons GE, Martin JF and Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. [DOI] [PubMed] [Google Scholar]
- 54.Ito Y, Yeo JY, Chytil A, Han J, Bringas P Jr., Nakajima A, Shuler CF, Moses HL and Chai Y. Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. [DOI] [PubMed] [Google Scholar]
- 55.Choudhary B, Ito Y, Makita T, Sasaki T, Chai Y and Sucov HM. Cardiovascular malformations with normal smooth muscle differentiation in neural crest-specific type II TGFbeta receptor (Tgfbr2) mutant mice. Dev Biol. 2006;289:420–429. [DOI] [PubMed] [Google Scholar]
- 56.Zhou D, Feng H, Yang Y, Huang T, Qiu P, Zhang C, Olsen T, Zhang J, Chen YE, Mizrak D, et al. hiPSC modeling of lineage-specific smooth muscle cell defects caused by TGFBR1(A230T) variant, and its therapeutic implications for Loeys-Dietz syndrome. Circulation. 2021;144:1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cook JR, Clayton NP, Carta L, Galatioto J, Chiu E, Smaldone S, Nelson CA, Cheng SH, Wentworth BM and Ramirez F. Dimorphic effects of transforming growth factor-beta signaling during aortic aneurysm progression in mice suggest a combinatorial therapy for Marfan syndrome. Arterioscler Thromb Vasc Biol. 2015;35:911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rateri DL, Davis FM, Balakrishnan A, Howatt DA, Moorleghen JJ, O’Connor WN, Charnigo R, Cassis LA and Daugherty A. Angiotensin II induces region-specific medial disruption during evolution of ascending aortic aneurysms. Am J Pathol. 2014;184:2586–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sawada H, Chen JZ, Wright BC, Moorleghen JJ, Lu HS and Daugherty A. Ultrasound imaging of the thoracic and abdominal aorta in mice to determine aneurysm dimensions. J Vis Exp. 2019:doi: 10.3791/59013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen JZ, Sawada H, Moorleghen JJ, Weiland M, Daugherty A and Sheppard MB. Aortic strain correlates with elastin fragmentation in fibrillin-1 hypomorphic mice. Circ Rep. 2019;1:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daugherty A, Rateri D, Hong L and Balakrishnan A. Measuring blood pressure in mice using volume pressure recording, a tail-cuff method. J Vis Exp. 2009:doi: 10.3791/1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.