ABSTRACT
For decades, biologist have exploited the near boundless advantages that molecular and genetic tools and analysis provide for our ability to understand biological systems. One of these genetic tools, suppressor analysis, has proven invaluable in furthering our understanding of biological processes and pathways and in discovering unknown interactions between genes and gene products. The power of suppressor analysis lies in its ability to discover genetic interactions in an unbiased manner, often leading to surprising discoveries. With advancements in technology, high-throughput approaches have aided in large-scale identification of suppressors and have helped provide insight into the core functional mechanisms through which suppressors act. In this review, we examine some of the fundamental discoveries that have been made possible through analysis of suppressor mutations. In addition, we cover the different types of suppressor mutants that can be isolated and the biological insights afforded by each type. Moreover, we provide considerations for the design of experiments to isolate suppressor mutants and for strategies to identify intergenic suppressor mutations. Finally, we provide guidance and example protocols for the isolation and mapping of suppressor mutants.
KEYWORDS: bacterial genetics, genetic mapping, screening, selection, suppressor mutant
Genetic alteration and manipulation are helpful tools for deepening our understanding of biological concepts. Within this genetic approach lies a class of mutations known as suppressor mutations. These mutations have been observed in both eukaryotic and prokaryotic systems. Suppressor mutations are secondary mutations that alter the phenotypes caused by an existing mutation. This is not to be confused with reversion mutations in which the original mutation has been lost (1). Instead, suppressor mutations return the phenotype caused by the original mutation to a more wild-type phenotype despite the presence of the original mutation (2). The suppressor mutation’s effect on the phenotype can vary from partial restoration to full restoration of the wild-type phenotype.
The two largest categories of suppressors, which specify where the secondary mutation occurs, are “intragenic” or “intergenic” suppressors (3). Intragenic suppression occurs when the original mutation and the suppressor mutation occur within the same gene (1, 3). Suppressor analysis using intragenic suppressors can identify regions, specific amino acids or interactions necessary for functional activity, stability, and more (3). If the goal, however, is to investigate the relationship between genes, then intergenic suppressors are ideal. Intergenic or extragenic suppression occurs when the original mutation and the suppressor mutation are located in different genes. These suppressors can help identify interacting genes and the specific interactions between the genes. In this way, the phenotypes of the original mutation allow for the identification of new interacting genes through suppressor selection, often demonstrating a functional relationship between genes that might not have been discovered by other means (1, 2, 4–10).
Investigating suppressors can shed light on scientific problems researchers once believed to be nearly impossible to research. With the advancement of genetic tools such as whole genome sequencing (WGS) and the availability of mapping and classical genetic tools, the usefulness of these mutants only grows. Much of our understanding of transcription, transcriptional regulation, and translation can be ascribed to analysis of suppressor mutations. Suppressors have also helped us learn about processes beyond gene expression. For example, they were critical for studying protein secretion. Thus, suppressor mutations play a critical role in genetics and biological research. In this review, we cover some of the advances made possible by suppressor analysis, types of suppressors, screens and selections for suppressor isolation, approaches and example protocols for mapping and whole genome sequencing, and examples of suppressor use in modern science.
HISTORICAL EXAMPLES OF DISCOVERIES MADE WITH SUPPRESSORS
Suppressor mutations have been established in the scientific literature for close to a century. The earliest reports of genetic suppression trace back to what, at the time, was interpreted as gene duplications in Drosophila (6). Although they were misinterpreted at first, the reoccurrence of suppressors in the field solidified their importance in genetic research. As research continued, some suppressor mutations showed interactions between nonallelic genes (4) and, by the 1950s, microorganism genetics and biochemical analysis allowed the mechanisms underlying nonallelic suppressor activity to be unraveled (8). Although early suppressors were found in a eukaryotic system, we will focus mainly on the impact suppressors have had on our understanding of prokaryotic systems.
Nonsense suppressors.
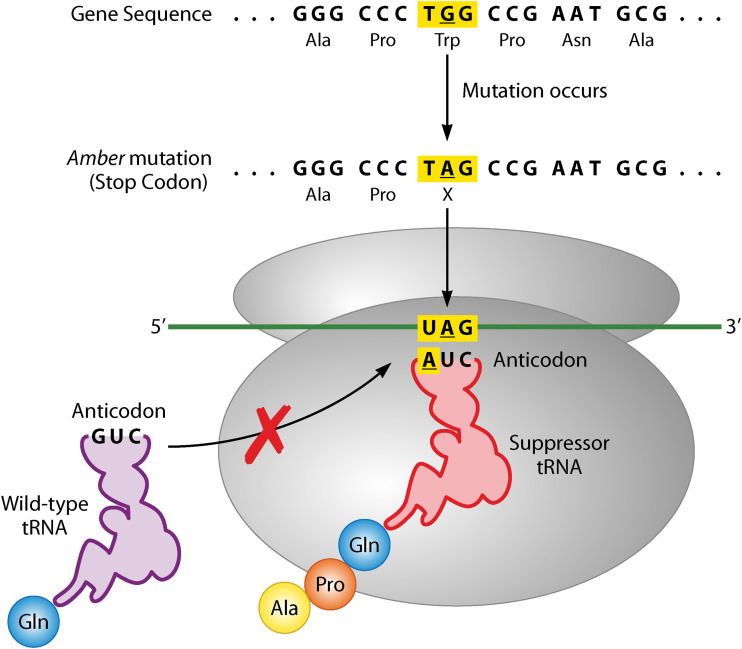
Nonsense suppressors are a type of intergenic suppressor that has played an important role in our understanding of translation and translational termination. Two major classes of nonsense suppressors are amber and ochre suppressors. Amber and Ochre suppressors are differentiated by which termination codons they suppress. The discoveries began with the observation that T4 bacteriophages with amber mutations in essential genes can infect particular strains of Escherichia coli: these strains carry amber suppressors (5). For instance, a mutation to a tRNA that allows it to read the amber stop codon and insert an amino acid will bypass the stop codon and allow translation to read through the codon producing full-length protein (Fig. 1). Thus, these suppressor strains carry their own mutation, which allows for restoration of the original function of the phage genes (7). These amber mutants were instrumental in the discovery of stop codons (11, 12). Amber suppressors were also used to show that genes are colinear with their polypeptide chain (i.e., the sequence of a gene determines the sequence of the protein it produces) (13). Moreover, using amber suppressors provided the phage field with genetic tools and insights that led to phage biology discoveries, such as the mapping of the T4 genome (9, 10).
FIG 1.
Mechanism of amber suppression. A primary mutation occurs changing the original codon sequence from UGG (Trp) to UAG (Stop). This is an amber mutation in the middle of the protein coding sequence. A secondary mutation occurs in the gene for a glutamine tRNA leading to the anticodon switch from a GUC to AUC. This mutant tRNA can recognize the UAG amber stop codon and insert a glutamine in place of the stop, allowing translation to continue past the stop and suppressing the amber mutation.
At around the same time, researchers performed experiments with the lac operon in E. coli (see below) to find suppressors of LacOo mutants (14). These mutants had mutations that removed the activity of lacZ and lacY and were thought to be in the operator site where LacI binds to repress transcription. Selection of lacOo suppressors for growth on lactose generated mutants that each expressed various levels of β-galactosidase (15, 16). The secondary mutations were originally thought to be in the operator or promoter leading to changes in the level of expression of the operon. However, it was quickly discovered that intergenic suppressor mutations were responsible for the restoration of lac operon function (17, 18). These suppressor mutations were found at different loci around the Escherichia coli chromosome. At the time, the concept of trans-effects restoring gene transcription was new to the field and, thus, the function of the suppressors continued to be investigated.
The researchers hypothesized that the lacOo mutants were a novel class of protein chain-terminating mutations (15). This led to the discovery that lacOo suppressors restored translation and were what are now known as ochre suppressors, which cause read through of the stop codon UAA (18). The discovery of lacOo mutations as chain terminating mutations called into question their classification as operator mutations. In fact, these ochre mutations were located early in lacZ and caused strong polar effects on the downstream lacY gene (19). In addition to the lac operon, ochre mutants were also studied in phage. Phage containing ochre mutants and their suppressors have very similar properties to amber suppressors: phages with ochre mutations can recover viability in strains of bacteria that have a ochre suppressor (20). The ochre suppressors discovered in this manner have been used to learn about the properties of tRNA and played a pivotal role in the characterization of lysV, which codes for a lysine tRNA (21).
Nonsense suppressors have also been demonstrated to function in eukaryotes. For instance, the anti-codon in a human serine tRNA has been mutated using site-directed mutagenesis to recognize either amber or ochre stop codons (22). These mutated tRNA genes were used with a Simian Virus 40 recombinant vector to show that tRNA genes produce tRNAs that suppress amber and ochre codons, respectively. Although not the first of its kind, this was one of the first successful studies to pave the way for the use of nonsense suppressor mutations in a eukaryotic context. The availability of nonsense suppressors has led to the development of several important technologies for molecular biology, such as methods to incorporate unnatural amino acids into proteins for fluorescence and cross-linking purposes (23–29).
Suppressors and transcription.
Suppressor mutations have been imperative to understanding fundamental concepts of gene expression—specifically through investigation of the lac operon. The lac system is commonly used in gene expression research and much of our insight of the lac operon has come with analysis of suppressor mutants, more specifically, lacO suppressors. The work of Dr. Jon Beckwith on the lac operon has been vital to our current knowledge of gene expression regulation and of translation termination. Determining a gene’s level of expression, specifically expression of constitutively expressed genes, was central to understanding transcriptional regulation and the role of promoters and operators.
The idea that gene expression could be regulated, specifically by repressors and inducers, stems from the work of Pardee, Jacob, and Monod studying the synthesis of β-galactosidase in E. coli (14). This work and further work related to the fundamentals of molecular biology led to the Nobel Prize in Physiology or Medicine being awarded to François Jacob, Jacques Monod, and André Lwoff in 1965. Pardee, Jacob, and Monod postulated the idea of an operator, which would be located at the start of an operon, and would serve as the site where the repressor blocks initiation of transcription (30). Investigation of two classes of mutations lead to the idea of an operator. The first class was operator-constitutive mutations (LacOc), which are resistant to repression. The second class was LacOo mutants that mapped early in the operon and removed the activity of lacZ and lacY (30). These studies originated the concept of a promoter where RNA polymerase would bind and start transcription, which was essential to the further study of gene expression.
As mentioned above, characterization of LacOo suppressor mutants determined the mutant’s phenotypes were caused by nonsense mutations early in lacZ (19). However, interest in using suppressor mutations of lacOo mutations to characterize the gene expression of lac operon continued. Restoration lacY expression could be selected for using growth on melibiose as a carbon source, since this α-galactoside requires the LacY permease but not β-galactosidase for its metabolism (17). Thus, suppressors could be found that would abolish the polarity of lacOo mutations but not restore β-galactosidase. Within these suppressors, a set of mutations that mapped in another locus distant from the lac region, was found (17). In fact, the suppressor mutations were found in rho, which encodes the factor responsible for Rho-dependent transcriptional termination (31).
Rho-dependent polarity is caused by premature translational termination leading to early transcriptional termination (32). It has been traditionally thought that Rho binding to RNA is blocked by ribosomes. When ribosomes confront a stop codon, they fall off the RNA, allowing Rho to bind rut (Rho utilization) sites in uncovered RNA. Rho will then move down the RNA faster than RNA polymerase, eventually allowing interaction with RNA polymerase. This interaction causes transcriptional termination and prevents downstream genes from being expressed. However, it has now been demonstrated using cryo-electron microscopy that the process of Rho-dependent termination begins with direct binding of Rho to NusA, NusG, and RNA polymerase, before contacting the nascent RNA and well before termination (33, 34). Termination begins when Rho contacts an emerging rut site in the RNA. Ribosomes may affect the conformations and interactions of the components of this transcriptional complex, allowing termination to only occur in untranslated regions of the RNA. Rho was shown to be the gene product of suA, the gene mutated in the lacOo suppressors, and rho-dependent termination was lowered in suppressor strains. In this way, suppressor mutants were important to the understanding of Rho-dependent polarity (31).
Eventually, the work done with lacOo mutations led to the acquisition of transcription-down mutations of the lac operon (35), and these mutations were later found to be positioned in the binding site for the transcriptional activator of cAMP binding protein, CRP (36, 37). This allowed the authors to propose the current model for CRP-cAMP function of binding to promoters and stimulating transcriptional initiation. Through this work and many more studies, investigation of suppressors of lac operon mutants led to the discovery of many fundamental aspects of transcriptional and translational.
Suppressors in protein secretion.
Suppressor mutations have also enhanced our knowledge of the function and characteristics of protein secretion. Fusing the N-terminus of a secreted protein to the enzyme β-galactosidase allowed the mechanism of protein secretion across the cytoplasmic membrane to be explored. The goal was to determine the portion of the exported protein needed to promote secretion of β-galactosidase in order to characterize the signal necessary for secretion (now known as the signal sequence or signal peptide). The proteins LamB and maltose-binding protein (MBP, MalE) were subjected to this strategy (38, 39). These fusion proteins resulted in a Lac- phenotype in the absence of the inducer (maltose), which activates transcription of the gene fusion. This is due to the secretion of the fusion construct resulting in LacZ’s improper folding and localization. The cells were sensitive to the inducer (i.e., maltose treatment was lethal) as higher expression allowed LacZ to fold in the cytoplasm clogging the secretion machinery (38–41). In order to find mutants that expressed the fusion protein but did not secrete it efficiently, maltose resistant mutants were isolated on maltose minimal medium which contained X-gal (allowing observation of Lac+ phenotypes) (42–45). Thus, the maltose resistance suggested the protein was not secreted, while the Lac+ phenotype ensured the protein was still expressed. This approach led to the characterization of the signal sequence which is necessary for Sec-dependent protein secretion.
The lacZ-fusion mutants were then used to discover the machinery necessary for protein secretion using two approaches. First, a screen for Lac+ suppressors of the LacZ-fusion proteins generated extragenic suppressor mutations (sec [secretion] mutants) with a partial loss of function in components of the secretion machinery. Second, strains with lamB or malE signal sequence mutations were selected for suppressors that restored the protein’s secretion (prl [protein localization] mutants). The restoration of secretion was selected for through fermentation of maltodextrin for lamB fusions and through growth on maltose for malE fusions. Together, these strategies led to the discovery of the secA, secB, secD, secE, secF, secG, and secY genes, which constitute the Sec translocon, the major pathway for bacterial protein secretion (46–53). Suppressor analysis also allowed for structure-function investigation of the Sec translocon. The SecYEG complex translocates unfolded proteins through the inner membrane to the periplasm or inserts them into the inner membrane. The SecB protein recognizes post-translationally secreted polypeptides and interactions with SecA to bring them to the SecYEG complex (54). SecD and SecF are accessory proteins. Thus, suppressors played a key role in the discovery of the mechanisms of protein secretion (see reference [54] for a thorough review).
TYPES OF SUPPRESSOR MUTATIONS
As described above, suppressor mutations have been used to make many fundamental discoveries. Their analysis has led to identification of new genes, proteins, interactions, and pathways. With knowledge of the possible types and mechanisms of suppression, interpretation of the relationship between two gene products is made possible, allowing the researcher to establish unknown genetic relationships and provide insight into a wide variety of interaction mechanisms. Here, we will detail the underlying differences in the mechanisms through which intragenic and intergenic suppressors function, the classes of suppressors contained within these larger groups, and the specific insights that can be gained from each type of suppressor.
Reversion.
A molecular definition of suppressor mutations classifies revertants as secondary mutations that occur in the codon of the original mutation and intragenic suppressor as mutations that occur elsewhere in the originally mutated gene (55). Thus, a revertant, or reverse mutation, is a mutation that restores the phenotype of the original mutant to wild-type or wild-type like expression by altering the amino acid produced at the site of the original mutation (1, 3, 55–58). There are three types of revertants: true, partial, and pseudo-revertants. A true revertant is a same site mutant that restores the original DNA or protein sequence (55). Both partial and pseudo revertants occur when a mutation changes the amino acid coded for at the site of the original mutation to a different amino acid than the wild-type or parent mutant. Partial and pseudo revertants restore the wild-type phenotype partially and fully, respectively (56, 57).
A common way that revertants can be isolated is by using a missense mutation which causes loss of function by altering protein folding, degradation, or activity and selecting for restoration of wild-type function (57). A classic example of a missense mutation used for genetic screening was performed by Yanofsky et al. who elucidated codons coding for various amino acids (59). Using single nucleotide substitutions to change from wild type to mutant and mutant to revertant phenotypes, the authors discovered the RNA codons associated with several amino acids at specific loci in tryptophan synthetase protein A (TrpA) in E. coli (59). They characterized 12 missense mutations that contained different amino acids at residue 210 of TrpA (59, 60). After analyzing the various true and partial revertants, the authors used these mutants to determine the nucleotide sequences that specify a given amino acid (59).
When analyzing suspected revertants, it is necessary to be wary of classifying pseudo-revertants as true revertants. In a study by Sherman et al. the authors isolated the first reported example of pseudo-revertants. Phenotypically wild type, these revertants contained a same site mutation that led to an amino acid compatible with wild-type function, but different from the original amino acid (61). This study and others (62–65) contributed to our modern day understanding of the genetic code and inspired caution in classification of genetic changes (61, 65).
Experimental usefulness varies between the types of revertants. There are very specific circumstances when true revertants are useful such as the study of mutation rates. However, pseudo- or partial revertants can provide information about what amino acids at the site of the original mutation can restore function to the protein. When the goal is to isolate pseudo-revertants, partial revertants, or intragenic suppressors without the isolation of true revertants, it is helpful to design the original missense mutant so that multiple nucleotides changes are needed to restore the amino acid sequence to wild type. This reduces the chances of isolating true revertants as a spontaneous reversion of two or three nucleotides is less likely than one.
Intragenic suppression.
An intragenic suppressor mutation occurs when an original mutation is either fully or partially compensated for by a second mutation at a different location in the same gene (1, 3, 66). The mechanism through which this second mutation acts can vary depending on the original mutation type and its phenotype. Some possible mechanisms include restoration of reading frame, correcting folding to reduce degradation rates, changing protein conformation to allow catalytic activity, and restoration of interactions with other proteins (56).
The elucidation of the triplet reading frame for translation involved intragenic suppressors that restored function to frameshift mutations by either inserting or deleting nucleotide(s) to restore the original reading frame (60, 67–72). Crick et al. utilized an original insertion mutation that caused a +1 frameshift at the 5′ end of the T2 phage rIIB gene (67). They then isolated intragenic suppressors that restored partial or full function through a nearby –1 frameshift mutation that restored the proper reading frame (67). Through further experiments using combinations of their mutants and suppressors, this classic experimental design allowed the authors to determine that DNA is read in a triplet reading frame (or at least in a reading frame that was a multiple of three) (67). Intragenic suppression of frameshift mutations was also employed by Douglas et al. (73). The authors characterize a frameshift mutation of +1 nucleotide located in rpoB (the beta-subunit of RNA polymerase) of E. coli. They report that E.coli with this mutation are viable and rifampicin resistant due to a spontaneous intragenic suppressor downstream that deletes one nucleotide in a string of repeated nucleotides. The change in amino acid sequence between these two mutations is sufficient to cause rifampicin resistance while still maintaining viability (73).
On the other hand, if a parental mutation causes incorrect folding of the original protein or instability, a suppressor mutation could restore proper folding by making a compensatory change in another part of the protein that restores interaction between two regions or domains. This type of suppressor is useful for determining which regions of a protein interact and the determinants of their interaction. Thus, this type of suppressor study, facilitated by site directed mutagenesis, can be used to confirm interaction predictions made by structural studies (74). An early example of analysis of secondary site suppressors was performed by Helinsky and Yanofsky (75, 76). The authors selected a glycine to glutamic acid mutant of trpA in E. coli for growth without tryptophan and found a suppressor that changed a tyrosine to a cysteine in a separate region of trpA. This second-site intragenic suppressor thus was able to partially restore function of TrpA likely by partially restoring TrpA’s conformation and enzymatic activity (75, 76).
If, rather than causing misfolding, a mutation causes a protein to be locked into a given conformation (active or inactive) and prevents full functional cycling from occurring, intragenic suppressors can be isolated that restore cycling between the conformations or lock the protein into another conformation. For instance, Park et al. performed a study of mutants in the Min system: a biological oscillator consisting of the MinC, MinD, and MinE proteins which controls the site of cell division. In this study, the authors analyzed the intragenic suppressor of a mutant minE in order to study its role within the Min system. Analysis of their intragenic suppressor in minE showed the mutant had difficulty releasing the interaction between two protein domains in one conformation to adopt a different membrane interacting conformation. The results uncovered a dynamic interaction between the domains that allows MinE to balance sensing of MinD localization with cytoplasmic diffusion to facilitate oscillation (77). Thus, this type of suppressor can provide insight into the conformational states of a protein. Similar to interaction between domains, if a mutation disrupts interactions between two proteins, an intragenic suppressor can restore the binding surface of the mutated protein, restoring the interaction between the proteins.
The last form intragenic suppressor is mutations to promoters that can be thought of as similar to informational suppressors (see below). This form of suppressor mutation arises when the promoter of a mutated gene is altered in a way that increases expression of the gene and restores wild-type phenotype due to increased accumulation of the partially active protein (78).
Intragenic suppressors can be easily identified and distinguished from intergenic suppressors through sequence analysis of the originally mutated gene, while intergenic suppressors often require genetic mapping or whole genome sequence to locate. Intragenic suppressors can lead to insights into the structure-function relationship of a given protein. However, they do not allow for analysis of relationships between proteins. For this, intergenic suppressors are necessary.
Intergenic suppression.
(i) Informational suppressors.
Informational suppressors are suppressors that alter the expression of genes, most commonly at the translational level (56). These suppressors are generally ribosomal or tRNA mutations that suppress either point mutations in the 5′ UTR, nonsense mutations, or frameshift mutations (79). The original mutated gene and mRNA remain; however, the mutation is sometimes “misread” in the suppressed strain causing a functional protein to be formed (3, 55, 80). Informational suppressors will suppress any mutant that has the same effect on translation as the original mutant: They are allele specific, gene nonspecific suppressors (3, 56, 79). The amber and ochre suppressors discussed above are classic examples of informational suppressors (see “Nonsense Suppressors”).
In addition to classical informational suppressors affecting translation, non-classical informational suppressors have been described that increase a transcript's abundance by altering transcription machinery or affecting RNA degradation (81). An example of a non-classical informational suppressor was elucidated by Modolell et al. (82). This study used a suppressor mutant of the hairy-wing allele (su(Hw)) of Drosophila melanogaster that could suppress an unknown class of spontaneous mutants found at 11 loci throughout the genome. The authors discovered that mutations suppressible by su(Hw) are due to insertion of a mobile element, named gypsy (82). It was later discovered that the gypsy mobile element contained a transcriptional insulator that required su(Hw) in order to function and so loss of function mutants of su(Hw) restored transcription of the effected gene (83, 84).
In summary, informational suppressors afford the best insights when the aim is to study the processes of gene expression rather than the function of a particular gene. Therefore, when the goal is to elucidate the function of a gene, suppression of a frameshift or nonsense mutation should be avoided, as large numbers of information suppressors will be isolated. However, if the goal is to enrich for informational suppressors, selecting for suppressors of nonsense or frameshift mutations is an effective strategy (45, 85).
(ii) Functional suppression.
The role of functional suppressors is to either replace or restore the function of a mutated gene (56). Functional suppressors result in genetic changes that alter the post-translational modification, activation state, localization, and/or degradation of the protein, or that change its interaction with activators, inhibitors, regulatory factors, or other pathway members. In addition, a mutation to a second gene can allow it to replace the function of the original gene product through changes in either the specificity or abundance of the second product. If the mutant gene is part of a regulatory pathway, suppression may occur by activation of a downstream gene in the pathway. Below, we expand on the mechanisms and classifications of functional suppressors.
(iii) Bypass suppression.
A bypass suppressor is a type of functional suppressor that works by “bypassing” the original mutated gene, using an alternative pathway or means to provide the function of that gene (56, 81, 86–89). The major characteristic that distinguishes bypass suppressors from other functional suppressors is that they are not dependent on the original allele and suppress both missense mutations and null mutations. Bypass suppressors typically arise from one of two distinct mechanisms.
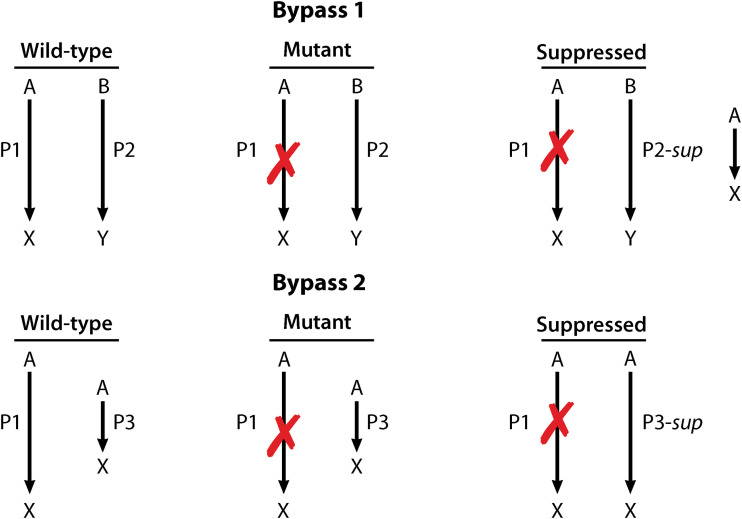
First, a protein in an alternative biochemical pathway can be altered to broaden its range of function compensating for the original mutation (Fig. 2, Bypass 1). For instance, if two proteins (P1 and P2) carry out related but functionally separate processes, a mutation that occurs in the gene for P1 would stop all further reactions in the first pathway. However, a mutation in gene two producing P2 could allow P2 to gain a new function that substitutes for P1, bypassing the original mutation in P1.
FIG 2.
Example mechanisms of bypass suppression. In wild-type cells, expression leads to the normal production of P1 and P2. The primary mutation leads to loss of P1 expression or function. In the first situation (illustrated as Bypass 1), suppression occurs from a secondary gain-of-function mutation in the gene for P2 that allows P2 to substitute for P1. In the second situation (illustrated as Bypass 2), suppression occurs from increasing expression of another protein, P3, in order to allow it to take over the functions of P1. (Republished from reference 168 with permission of the publisher.)
The second mechanism of bypass suppression is when expression of a gene is increased, allowing the increased abundance of the protein to facilitate a new function (Fig. 2, Bypass 2) (56). For example, in the case above, the expression of another gene is increased to a point that its product, P3, substitutes for the function of P1. Here, P3 carries out functions similar to wild-type P1 but at rates inadequate to support wild-type physiology. This is usually due to a preference for the protein’s canonical substrate (56). However, increased abundance of P3 allows it to compensate for P1 function by creating a pool of free enzyme (also see “Overproduction/Multicopy Suppressors”).
One pioneering example of the usefulness of bypass suppressors is the work performed by Spencer Benson and colleagues investigating bypass suppressors of lamB null mutants in E. coli. To begin these studies, Benson and Decloux took advantage of the necessity of LamB for uptake of maltodextrins to design a genetic selection for suppressors of lamB null mutations (90). The authors found mutations in two major porin genes, ompF and ompC, that created larger pore openings. These mutants greatly decreased the cell’s antibiotic resistance, demonstrating the fundamental role the outer membrane plays in antibiotic resistance (90). Continuing this work led to the identification of more mutations in ompF and ompC that suppressed lamB null alleles allowing structural characterization of OmpF and OmpC (91, 92).
Beyond mutants in ompC and ompF, Sampson, Misra, and Benson isolated two suppressor mutants which they termed imp-4213 and imp-208 that did not map to ompC or ompF and resulted in increased membrane permeability (93). Eventually, this work led to the identification of imp as lptD, the gene for the outer membrane protein component of Lpt system that transports lipopolysaccharide to the cell surface (94), and to the characterization of the remainder of the Lpt system (95–97). Furthermore, characterization of suppressors of imp-4213 membrane permeability led to the identification of an outer membrane lipoprotein (YfgL/BamB), which was found to be part of the multi-protein BAM complex responsible for inserting proteins into the OM (98, 99). Not only was imp-4213 fundamental to the discovery of the Bam complex, it has also been an invaluable tool for the investigation of the mechanism of action of BamA, the essential outer membrane protein component of the Bam complex (100, 101).
(iv) Allele specific suppression.
Allele specific suppressors are intergenic suppressors that suppress only a particular allele or select group of non-null alleles of a gene. Thus, this form of suppressor is characterized by the ability to suppress a subset of missense mutations of a particular gene and an inability to suppress gene deletions or null alleles. Allele-specific suppression generally indicates direct interaction or physical contact between the two gene products (56). This interaction could indicate an enzyme-substrate interaction, interaction between catalytic and regulatory proteins, or interaction between proteins in a heteromultimer (56).
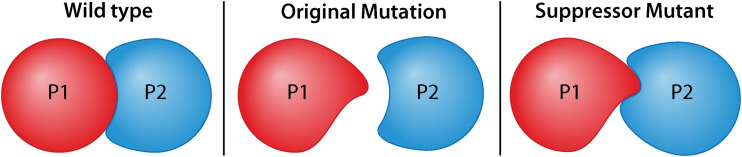
As illustrated in Fig. 3, the primary mutations tend to alter binding strength and interaction between the two proteins by either weakening or strengthening it. In the indicated case, a primary mutation in the gene for P1 decreases the binding affinity of P1 for P2. A suppressor mutation in the gene for P2 then changes the P2 binding surface and restores binding between P1 and P2. Restoration of function will only occur if the suppressor mutation causes alteration of the surface region of P2 that makes direct contact with P1. It can also be expected that the suppressor mutation can only suppress mutations in gene 1 that causes similar alterations to the region involved in P1–P2 interaction.
FIG 3.
Example mechanism of allele specific suppression. P1 and P2 represent products produced from two separate genes. In the wild-type condition, P1 and P2 physically interact. A mutation in the gene for P1 causes a change to the binding interface preventing interaction between P1 and P2. A suppressor mutation in the gene for P2 alters the binding interface of P2 and restores interaction between the mutant proteins.
As an example, investigation of allele-specific suppressors was performed by Ricci et al. (102). The authors were investigating the BamABCDE complex responsible for inserting β-barrel proteins into the outer membrane of E. coli and isolated a temperature-sensitive mutation of bamA that disrupted BamAB and BamCDE subcomplex interaction (102). They isolated an allele-specific suppressor mutant in bamD that restored Bam function but did not restore Bam complex stability. Instead, the suppressor bypassed the requirement for stable complex formation (102). Later work by McCabe et al. characterized several bamA mutants’ ability to suppress the mutation in bamD and found some mutations that suppressed the lethal phenotypes of the bamD mutant and some that restored complex formation but little correlation between the two (103). Together, these works demonstrated that both BamA and BamD need to be activated in order for the Bam complex to function and that this activation is more important than complex stability.
Allele-specific suppression has been used to investigate DNA- or RNA-protein interactions as well as protein-protein interactions (88, 104). As noted in the example above, it is possible that suppression can result from changing a gene product’s conformational state rather than a direct restoration of binding. Thus, the characteristics of the suppressors need to be further investigated to verify their specific function.
(v) Epistatic suppression.
Epistatic suppression occurs when genes are part of a regulatory pathway or a metabolic pathway with toxic intermediates. Often, a regulatory pathway is controlled by a signal that activates the pathway producing a downstream response. This allows mutations that decrease or increase signaling at one step within the pathway to be epistatically suppressed by secondary mutations acting on a subsequent step. Generally, these secondary mutations result in constitutive activation of the pathway. However, this depends on whether the regulatory system is positively or negatively regulated (81, 87, 105, 106).
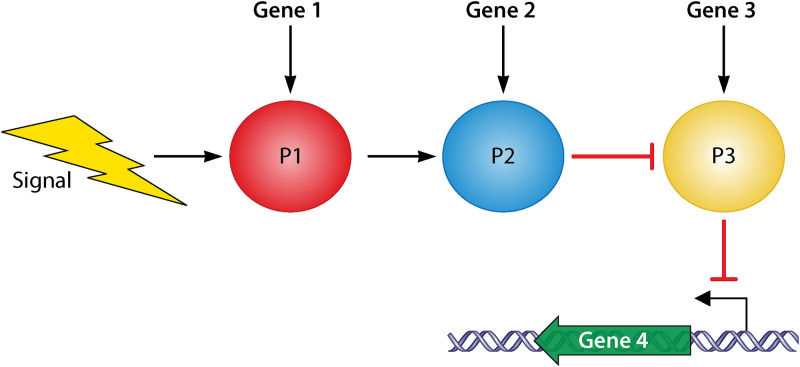
In the regulatory model shown in Fig. 4, a signal can activate the expression of P1, which in turns activates P2. P2 acts as an inhibitor of P3, while P3 is an inhibitor of gene 4 expression. A loss-of-function mutation in gene 1 or gene 2 would lead to repression of gene 4. A loss-of-function mutation in gene 3 would lead to constitutive expression of gene 4. Thus, a loss-of-function mutation in gene 3 will suppress a mutation in gene 1 or gene 2 and restore expression of gene 4. However, in the above example, suppression does not result in the normal regulation of gene 4 expression, but rather in unregulated expression. This form of suppression can occur with both null and non-null alleles of either the original or the suppressor gene: any loss-of- function mutant of gene 3 will suppress either a deletion or a missense mutation in gene 1 or 2. Suppression can also happen if the repressor-binding site found upstream of gene 4 is mutated.
FIG 4.
Example pathway in which epistatic suppression could occur. As a result of an outside stimulus or signal, gene products, P1, P2, and P3, are produced from Gene 1, Gene 2, and Gene 3, respectively. P1 leads to the expression of gene 2 to make its product P2. P2 inhibits the expression of gene 3 and preventing the expression of P3. P3 in turn prevents the expression of gene 4, the output of the pathway. Mutations that occur earlier in this pathway can be suppressed by mutations occurring latter in the pathway that mask the effect of the original mutation. (Republished from reference 168 with permission of the publisher.)
Epistatic suppressors in a regulatory pathway were isolated by Seo et al. (89). The authors were investigating the molecular mechanisms of fluG-dependent asexual development program in Aspergillus nidulans. The authors start by performing an unbiased selection for spontaneous suppressors that overcome ΔfluG sporulation defects. They isolated 14 suppressors in four genes they named sfgA, sfgB, sfgC, and sfgD (89). These suppressors resulted in hyperactive asexual spore formation and acted as epistatic suppressors. For instance, SfgA is a negative regulator of asexual sporulation that is inhibited by FluG (89). Loss of sfgA allows the asexual developmental program to proceed in the absence of fluG (89, 107). Thus, the suppressors altered the activation of the pathway overcoming the defects caused by the original mutation.
Epistatic suppressors also occur in non-regulatory pathways when a mutation in the pathway causes the accumulation of a toxic intermediate. A mutation earlier in the pathway can suppress this accumulation by preventing the intermediate from being made. A classic example is the isolation of epistatic suppressor mutants that prevented arabinose sensitivity caused by disruption of the araD gene, which codes for L-ribulose 5-phosphate 4-epimerase (108). The suppressors relieved the arabinose sensitivity by preventing the accumulation of L-ribulose-5-phosphate through mutation of araB, which is responsible for the previous step in arabinose catabolism. These results demonstrated that accumulation of L-ribulose-5-phosphate is responsible for the arabinose sensitivity in araD mutants (108).
(vi) Overproduction/multicopy suppression.
With many plasmids available for common bacterial model organisms, the expression of genes can be increased both by expression in trans and by promoter alteration in cis (56). Overproduction or multicopy suppression is characterized by the use of multicopy plasmids to overexpress a gene. Genes can be expressed under different inducible or repressible promoters on either high, medium, or low copy number plasmids allowing for a wide range of expression. A large advantage of plasmid-based multicopy suppressor approaches is that the suppressing insert can be easily sequenced without the need for mapping or whole genome sequencing (56).
One approach to find multicopy suppressors is to partially digest genomic DNA with a restriction enzyme and clone the fragments in a certain size range to make a library. For, instance, Ueguchi et al. used this approach to identify genes that bypassed defective secretion mutants within the Sec translocon (109). The authors isolated several multicopy suppressors, namely, chaperones and proteases, demonstrating that maintaining secreted proteins in an unfolded state was sufficient to overcome the defects (109). Another approach for multicopy suppression is to take advantage of preexisting collections of overexpression plasmids. For instance, the ASKA collection comprises a near complete sets of E. coli K-12 open reading frames (ORFs) in the pCA24N plasmid (110). In this high copy number plasmid, the cloned ORFs are expressed under the IPTG-inducible T5-lac promoter (110).
By utilizing multicopy suppression, it is possible to isolate all of the aforementioned functional suppressor types. Multicopy expression of a gene can cause bypass suppression through mechanisms similar to other bypass through overexpression mutants except that the overexpression is from a plasmid (i.e., the excess protein takes on a new role due to lack of its canonical substrates, binding partners, etc.). LpxL (HtrB) is a responsible for adding laurate to lipid A, the lipid component of lipopolysaccharide, as its second to last acyl chain and mutants of lpxL are temperature sensitive (111, 112). Clementz et al. investigated multicopy suppressors of temperature sensitivity in lpxL deletion strains (113). They found that msbB (lpxM) could act as a multicopy suppressor of ΔlpxL. Through analyzing the role of LpxM in vitro and in vivo, the authors discovered that LpxM is responsible for transferring myristate to lipid A as its final acyl chain (the step after LpxL). LpxM could also act on the substrate for LpxL’s reaction but with efficiency 100-times less than its native substrate (113). Thus, overexpressing lpxM in the absence of lpxL could partially compensate for the loss of lpxL. A more recent example of the versatility of multicopy suppressors is the characterization of multicopy suppressors for peptidyl-prolyl isomerase (PPIase) mutants (114). Through this approach, the authors identified chaperones, transcriptional factors, and replication proteins as multicopy suppressors of PPIase mutants. After further analysis, the authors suggested PPIase functions in three E. coli proteins, DksA, MetL, and Cmk, not previously known to be PPIases (114).
Allele-specific multicopy suppression can occur through increased concentration stabilizing binding between gene products (56). For example, in Fig. 3, overexpression of P2 has the potential to kinetically drive formation of a P1–P2 complex despite the altered binding affinity of P1. In a regulatory pathway, activator or repressor overproduction could alter the activation of the pathway, regardless of the upstream signaling state. For instance, if an activator within the pathway is normally expressed at a low level, its overproduction could potentially increase that activation level of the pathway (115). Given the wide variety of useful information that can be obtained from different types of suppressor mutations, it is useful to consider how best to design experiments to enrich the desired type of suppressor.
CONSIDERATIONS FOR DESIGN OF SUPPRESSORS SCREENS AND SELECTIONS
Suppressor screens and selections are used to identify mutations capable of reversing the phenotypes associated with a primary mutation. Success in discovering useful information is highly dependent on the type of suppressors isolated and therefore the experimental design. Below, we discuss experimental designs that maximize the recovery of a given type of suppressor mutation, confirmation methods for suppressor mutants, and some pitfalls to avoid while performing the aforementioned experiments.
Effect of mutation on type of suppressors isolated.
The better characterized the selectable or screenable phenotypes of the mutation are, the better the chance of designing an efficient experiment to isolate desirable suppressors. This does not, however, mean that knowledge of the molecular nature of a mutation or even the gene in which the mutation occurs is necessary for successful isolation of suppressors. Nevertheless, if a specific type of suppressor is desired from a screen or selection, choosing the most appropriate starting mutation is helpful, as the choice of mutation (e.g., a deletion, nonsense, or missense allele, a loss-of-function or gain-of-function mutation, or a mutation outside the ORF) will ultimately affect the type of suppressor mutations isolated.
For example, to determine the suppressors that could be isolated using a missense mutation that causes a full or partial loss of protein function, it is important to know whether a stable protein is produced. If a stable protein that cannot function were produced, then the mutation would be ideal to isolate allele-specific or intragenic suppressors and could also allow the isolation of bypass and epistatic suppressors. A partial loss-of-function mutant could lead to the isolation of similar mutants, but would be ideal for the isolation of mutants that alter the gene expression of the mutated gene leading to increased expression.
If the gene with the loss-of-function mutation does not produce a stable protein, bypass, epistatic, or intragenic suppressors could be recovered while recovery of allele-specific suppressor would be unlikely. However, this is not to say that allele specific mutations in this case cannot be isolated. If a protein relies on an interaction with a binding partner for stability, a secondary mutation in the binding partner that restores interaction could restore stability of the primary protein. To force the recovery of bypass or epistatic suppressors, a null mutation, such as a gene deletion, is ideal. In contrast, informational suppressors can be isolated by selecting for suppressors of nonsense or frameshift mutations. Therefore, when looking for other mechanistic classes, these types of mutations should be avoided.
Finally, when a gain-of-function mutant is the basis for a suppressor screen, it is possible to isolate and identify many intragenic loss-of-function mutations, including null mutations. Often, these suppressors are less instructive about the biological process being investigated, although the exact location of the intragenic mutations can show which amino acids are required for protein function. To avoid intragenic suppressors, the gain-of-function mutation can be expressed in diploid.
Strategies for isolating suppressor mutations.
To efficiently isolate suppressor mutants, a selection or screen needs to be designed to identify mutations that have lost the phenotypes of the original mutation. Phenotypes that can be used in these experiments include sensitivity or resistance to a phage, antibiotic, or growth condition (e.g., temperature, pH, ion concentration) and various auxotrophies (i.e., reliance on specific growth factors or nutrients for growth) (115). Given multiple phenotypes, it is always preferable to design a selection for a positive phenotype (i.e., growth of the suppressors in a condition where the original mutant cannot grow) as selections allow for millions of cells to be assayed simultaneously. If the original mutant was isolated through a selection, then investigation of other phenotypes might be necessary in order to design a selection for suppressors. However, it is often the case that a mutant that is resistant in one growth condition might be sensitive in another, allowing for selection of both the original mutant and the suppressor (116).
If a screen is necessary because the desired suppressor has only negative phenotypes (i.e., failure to grow in a condition where the original mutant can grow), the likelihood of isolating suppressors can be increased, and the labor necessary decreased, through mutagenesis and enrichment of mutants of interest. Depending on the goal of the suppressor screen, either specific genes or the whole genome can be mutagenized to increase the frequency of suppressor mutations in the population (74, 106, 117, 118). Even with mutagenesis, the frequency of mutants answering a screen can still be quite low. Therefore, it is helpful to employ an enrichment, such as ampicillin or penicillin treatment in conditions where suppressors will not grow to increase the frequency of these mutants in the population (119). This method has been successfully employed with both bacteria in culture and for bacteria growing in mammalian cells (120–122).
Besides these traditional techniques, high-throughput sequencing technologies have made it possible to find mutants with negative phenotypes while avoiding labor-intensive screening. In this case, the pool of mutants before and after selection is sequenced, allowing the identification of mutants that have been enriched or depleted in the library (123, 124). This technique can be applied to pools of transposon mutants (transposon-insertion sequencing [TIS]) or to mutants generated by chemical mutagenesis (mutational enrichment analysis after phenotypic selection [MEAPS]) (124–128). These techniques have been thoroughly reviewed elsewhere (123, 124, 129).
An additional strategy for the generation of suppressors when the original mutation does not have tractable phenotypes is the use of fusions to generate the phenotypes. For instance, the interaction of two binding partners could be assayed through a bacterial two-hybrid assay, an ampicillin reconstitution assay, split GFP assay, or POLAR (PopZ-linked apical recruitment) assay depending on the cellular compartment of the target proteins (130–134). If the goal is to generate suppressors that increase the stability of a protein, then an easily assayable fusion with LacZ, a fluorescent protein, or an antibiotic resistance protein could aid in this goal (115, 135–137). Finally, if a change in gene expression is expected because of a suppressor mutation, a transcriptional fusion could allow easy monitoring of the expression the gene of interest (138–140). The use of lac fusions in genetic screens and selections has been thoroughly reviewed (115).
Avoiding pitfalls.
One of the major pitfalls researchers face when working with suppressor mutants is the inadvertent isolation of many true revertants or pseudo-revertants when suppressor mutants are desired. Analysis can be further complicated when revertants are misclassified as suppressors. The gene with the original mutation should be sequenced to avoid this misclassification. To avoid isolation of high numbers of true revertants, the desired amino acid change can be made in such a way that more than one nucleotide needs to be altered to return the protein to the original sequence, reducing the chance of a single base pair change resulting in a true revertant. Pseudo-revertants are harder to avoid but can be more interesting to investigate.
Another pitfall is isolation of spontaneous resistance mutant instead of suppressors specific to the original mutation. For example, in situations where a mutant is sensitive to an antibiotic and resistant suppressors are selected, it is possible to isolate both suppressors that affect the activity of the mutant and mutants with resistance that affects the mechanism of action of the antibiotic. To avoid the latter situation, it is best to select or screen for multiple phenotypes of the original mutant as suppressor should affect most or all of them.
It is also important that the method of mutagenesis employed fits the goal of the experiment. For example, spontaneous mutations are rare, and so are best for selections. While chemical mutagenesis increases the probability of finding a mutant of interest, it also increases the likelihood of multiple mutations that could need to be investigated to identify the suppressor mutation. Finally, transposon insertions are easy to locate but only cause gain-of-function phenotypes in rare circumstances, such as by insertion into a promoter increasing expression or through loss of specific 3′ gene regions, and are unlikely to produce an allele-specific suppressor.
Confirmation of suppressor mutants.
After isolation of putative suppressor mutants, it is necessary to confirm the secondary mutation is in fact responsible for suppressing the original mutant phenotype. There are several initial confirmation steps that should be taken before critical analysis and interpretation of results. First, it is important to investigate the phenotypes of the suppressors to differentiate between suppressors and resistance mutants and to classify suppressors into categories with similar phenotypes. Multiple suppressors with similar phenotypes may be mutations in the same gene. Therefore, if one suppressor mutation is located, the candidate gene should be sequenced in other suppressors with similar phenotypes to determine whether they also carry mutations in the gene. Additionally, depending on the method of suppressor isolation and whether whole genome sequencing has been performed, the suppressor mutation should be moved to a clean background to determine whether it is sufficient to suppress the original mutant (141). The mutation identified in the suppressor strain can also be removed through linkage or complemented to determine whether the mutation is necessary for suppression (142). It is also beneficial to check whether the suppressor mutation has phenotypes independent of the original mutation.
SUPPRESSOR MAPPING AND WHOLE GENOME SEQUENCING
Mechanisms for identifying suppressor mutations have evolved from early studies that relied on mapping to current day research where sequencing can be used for identification suppressors or aid in the mapping process. Though sequencing and mapping each have drawbacks, both tools can aid in analyzing intergenic suppressors. Here, we discuss the use of mapping and/or whole genome sequencing (WGS) to locate suppressor mutants.
Genetic mapping describes the location of mutants in relation to known markers, determining where these mutations lie on the chromosome (143). On the other hand, WGS determines an organism’s entire genome sequence at a one time (144), providing every mutation in the genome regardless of whether it is related to suppression. This can become challenging if mutagenesis has been used to increase the frequency of suppressors. As both mapping and sequencing have advantages and disadvantages, using these tools to complement one another can lead to more efficient suppressor identification. Here, we will cover the use and drawbacks of WGS in suppressor identification and detail the role mapping still plays in identification of suppressor mutants.
Mapping of suppressor mutations.
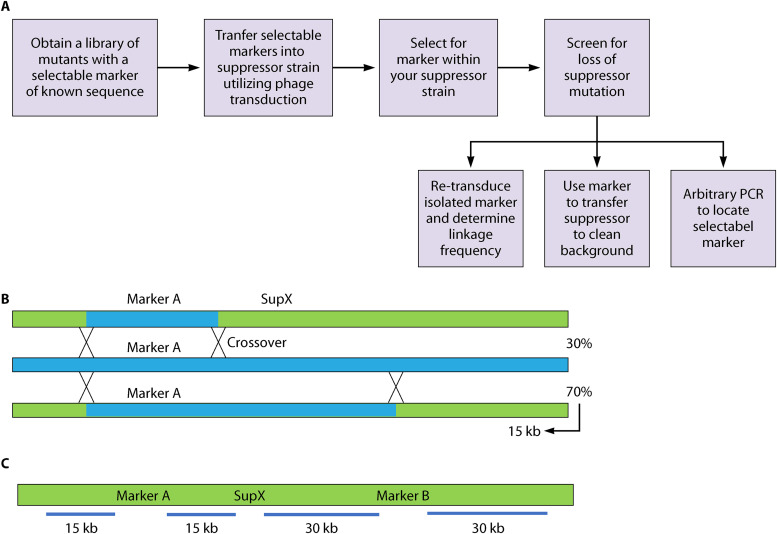
General approaches for mapping include crossing strains, transfer of chromosomal segments, integration, genetic recombination, and phenotypic expression for differentiation between strains (145). Current approaches to map suppressors often use phage to transfer selectable markers to the suppressed strain. In general, the first step is to obtain a library of selectable mutants containing antibiotic resistance cassettes with known sequence, along with a method of transferring the mutants into the suppressor strains with relatively large pieces of genomic DNA. This transfer method is often transduction, such as via P1 phage or P22 phage, for E. coli and Salmonella enterica serovar Typhimurium (S. Typhimurium), respectively. After identification of selectable markers linked to the suppressor mutation, the linkage frequency between the maker and the suppressor mutation is used to determine the physical distance between the suppressor and the marker (Fig. 5) (146, 147). The linked resistance cassette or transposon isolated for mapping can also be used to transfer the suppressor to a clean background for further analysis. Arbitrary PCR is used to provide the location of the selectable marker, and so determine the whereabouts of the suppressor. The suppressor mutation is then identified through Sanger sequencing or WGS.
FIG 5.
Locating suppressor mutations through mapping. (A) Flow chart showing the general steps necessary for genetic mapping of suppressor mutations. Please see text for detailed approaches. (B) The distance between a marker and a suppressor mutation can be determined by co-transduction frequency. Crossover events to transfer a marker, A, are shown. The region recombined can include the suppressor mutant or not. The frequency with which the suppressor is lost due to recombination can be used to calculate the distance between the marker and the suppressor. In this case, the co-transduction frequency shows the suppressor is 15 Kbp, either upstream or downstream (i.e., to the left or right) of A. (C) To determine the precise location of the suppressor, two selectable genetic markers, A and B, genetically linked to the suppressor mutation with known locations are needed. Their distances from the suppressor are calculated as in (B). With two markers, comparing the locations pinpointed by each maker allows the location of the suppressor mutation to be determined. In this case, the distances align between the two genetic markers, A and B, at one possible location upstream of A and downstream of B.
Genetic mapping of suppressors was imperative to early discoveries made using suppressors, such as use of ochre mutants (18) to determine that genes are colinear with their polypeptide chain (13) and the description of conditional lethal mutants of bacteriophage T4D (148), among many others. In more recent work, mapping of suppressors has helped to investigate mechanisms of cell death caused by loss σE (149), led to discovery of a gain-of-function mutant in a regulator of outer membrane asymmetry (150), and allowed discovery of the role of cyclic enterobacterial common antigen (ECA) in maintenance of the OM permeability barrier (151).
Mapping strategies are often quick, inexpensive, and successful without the need for WGS to identify mutations. However, if only the general location of a suppressor is determined through mapping and no obviously related genes are in the region, WGS can be useful to identify the mutation. These strategies can also be useful when WGS is followed by mapping. For instance, in a genome with many mutations identified by WGS, mapping can provide insight into the location of suppressor mutation without testing each mutation in the genome. Furthermore, identification of suppressors by mapping can be difficult if the suppressor lacks a selectable phenotype or in organisms without genetic tools. In these cases, WGS is useful.
Whole genome sequencing.
WGS is an important tool to identify gene function when complemented with computational tools to predict homology and proteins structure. Invented in the 1970s, DNA sequencing was a breakthrough for biological research and led to an increase in knowledge of genes’ identity, structures, function, and regulation (152). WGS analysis is performed with next generation sequencing technologies and these technologies require computational and bioinformatic means to analyze abundant sequence data (153). WGS has become useful in the identification of suppressor mutations. For instance, WGS was used to locate suppressors of a loss of motility in a Salmonella strain with impaired Type III secretion system (154). A deletion in fliO, one of the transmembrane proteins in the secretion system, caused poor motility. Using WGS, the authors then located suppressor mutations that partially rescued motility. Through analysis of these suppressors, the authors concluded that FliO plays a role in regulation of FliP, another member of the secretion machinery, during flagellum assembly (154).
WGS was also used to locate high frequency suppressors of dGTP starvation in a mutant of E. coli that quickly degrades dGTP (155). Many of these suppressors upregulate de novo purine synthesis through altering a transcriptional regulator or duplicating a large region of DNA including purine biosynthesis genes. Thus, WGS led to the identification of suppressors that can evade starvation by simply upregulating purine synthesis de novo, leading to restoration of dGTP to adequate levels (155). High throughput sequencing technology has also been used in Vibrio cholerae to pinpoint spontaneously occurring suppressor mutations which facilitated disruption of rpoE (the gene for σE) (156). σE is activated by envelope stress and aids in preserving cell envelope integrity. Over 75% of the V. cholerae rpoE suppressor mutations reduced OmpU production, V. cholerae’s principal outer membrane porin. Thus, OmpU seems to be an imperative to determining the essentiality of σE in V. cholerae (156).
While WGS can identify suppressor mutations, it also has drawbacks. WGS can identify multiple mutations necessary for suppression. However, if many unrelated mutations are present, such as when chemical mutagenesis has been used, WGS will not identify which mutation is the suppressor. In addition, short-read sequencing can have difficulty identifying duplications, gene movement, and movement of insertion elements and transposons. When a genome contains multiple copies of the same insertion element, assembly of short read sequencing data becomes complicated and insertion sites become difficult to identify. Although, this problem can be solved using long read sequencing technologies, these approaches generally have lower accuracy (157). With these types of suppressor mutants, mapping may be ideal. Nevertheless, in many cases, WGS is the only way to identify a suppressor in organisms where linkage-mapping tools are not available.
PROTOCOLS
Design of suppressor selections and screens.
Suppressor screens and selections are potent tools to uncover additional information about a known gene or protein and for the identification of other unknown interacting components. The success of a suppressor screen and selection will depend in large part on its design. The key to designing an effective suppressor screen and selection lies in having a mutation with well-characterized, strong phenotypes to suppress that is of a type that will yield the suppressor types of interest (see above). Depending on the phenotypes either a selection or a screen can be setup to isolate suppressor mutants.
Selections are easily setup when a parent mutant causes a growth defect such as an auxotrophy or sensitivity to a growth condition, antibiotic, or phage: suppressor mutants can be selected through growth in these conditions. Thus, a selection could be readily setup for suppressors of conditional lethal mutants, catabolic mutants, metabolic mutants, or temperature sensitive mutants. For example, if the goal is to isolate suppressors of a mutation that causes antibiotic sensitivity, a selection can be used to isolate mutants that are now resistant to the antibiotic.
The key to a successful selection strategy is optimizing the conditions so that the parent strain will have no growth, but the suppressed strain will thrive. The first step is determining the correct growth conditions. These conditions could include temperature, nutrient concentrations, media type, selective agent concentration, if applicable, and time of growth. Comparing the phenotypes of the parent mutant and the wild-type strain can provide a useful starting point for designing the selection. The second factor for optimization is the concentration of cells that will be tested in the selection. The proper number of cells depends on the effect of cell number on selection (for instance, some antibiotics are less effective at high cell numbers), possible cross feeding between auxotrophic mutants, and the expected frequency of suppressors. Depending on the number of genes and types of mutations that could answer the selection, the frequency of suppressors expected can vary by several orders of magnitude (155, 158). Optimizing these conditions will ensure a successful selection with a low rate of both false positives and false negatives.
Some mutants do not have a phenotype that will allow for selection of suppressor mutants, necessitating a screen. By performing a screen, suppressor mutants that cannot grow in a condition where the parent mutant can grow can be isolated. In addition, screens allow for differentiation of the original mutant and the suppressive mutant based on other characteristics such as colony morphology or color of an indicator. The feasibility of suppressor screens is limited by the number of colonies that need to be screened to find a rare suppressor; however, enrichment of mutants can help decrease the labor involved in such a screen (see below). A common experimental strategy when performing suppressor screens for negative phenotypes is replica plating. Cells with a screenable phenotype are grown in non-selective conditions. The resulting colonies are then transferred to selective plates producing replicates of the first. This allows the identification of colonies that are suppressors of the original mutation by their lack of growth on selective plates.
Another common experimental screen strategy is to use a lacZ fusion for screening. This approach can be utilized when the mutant to be suppressed does not have any phenotypes that allow for a selection or screen to be performed. Several β-galactosidase (LacZ) indicators are available which differ in their sensitivity and choosing the correct indicator can greatly aid in the success of a suppressor screen utilizing lacZ fusions (thoroughly reviewed in [116]). For instance, when LacZ breaks down X-gal, a highly sensitive LacZ substrate, an insoluble blue pigment is released. The amount of pigment produced depends on the amount of LacZ present. Depending on the desired phenotype of the suppressor, a transcriptional or translational lacZ fusion can be used. For example, if a suppressor should turn off the expression of a given gene, a lacZ transcriptional reporter can be constructed and used for screening. Pools of mutants would be screened with X-gal for white colonies where lacZ is not expressed.
The frequency of suppressors in a population, which depends on the types of mutation that can lead to suppression, can be very low making direct screening for spontaneous suppressors impractical. For instance, while some mutants may generate suppressors at a frequency as high as 1 in 10,000 other mutants that require a specific change to a small number of possible nucleotides can be as infrequent less than 1 in 1,000,000,000 (155, 158). This makes screening for spontaneous suppressors unrealistically labor-intensive. Two approaches, which are not mutually exclusive, can be taken to decrease the number of colonies that need to be screened in order to identify suppressors: mutagenesis and enrichment.
There are many different mechanisms to induce mutations when isolating suppressors. Two widely used and studied methods are physical (e.g., UV light) and chemical (e.g., alkylating agents and azides) mutagenesis. The effectiveness of chemical mutagenesis is dependent on dose of the mutagenic agent. The increased rates of mutations caused by increasing dose must be balanced with the decreased survival of the treated strain. The most commonly used form of physical mutagenesis is UV radiation, which causes the formation pyrimidine dimers. However, physical radiation can also induce indirect mutations due to the production of reactive oxygen species that cause genetic damage (141, 155, 158). When targeting specific gene(s) for the suppressor screen, PCR mutagenesis can be used, which is unique in the largely unbiased nature of commercial error-prone polymerases, and cause transitions and transversions at nearly equal levels.
Enrichment increases the frequency of mutants in a population. A classic example of an enrichment was detailed by Bernard Davis who used a penicillin enrichment strategy to isolate auxotrophic mutants of E. coli, using the ability of penicillin to kill only growing cells (159). This enrichment works by first growing a population of mutants in nonselective conditions. After growth, the population is then transferred to conditions where the desired mutants cannot grow and treated with ampicillin or penicillin. The surviving population of bacteria, which has a higher proportion of cells that cannot grow in the selective condition, is then plated in nonselective conditions before screening, often through replica plating. Once suppressors are isolated, additional studies can be conducted to verify the nature of the suppressor. If intergenic suppressors have been isolated, this analysis can involve genetic mapping to locate the suppressor.
Mapping of suppressor mutations.
(i) Overview.
When mapping suppressors, there are two main goals: finding a selectable marker linked to the suppressor mutant and using distance from that marker to determine the location of the suppressor (Fig. 5A). The first step is to obtain a library of selectable mutants, along with a method of transferring the mutants into the suppressor strains. Single gene deletion libraries are available for both S. Typhimurium 14028s and E. coli K-12, which can be used for mapping suppressor mutants (160, 161). However, if these resources are not available in the lab, obtaining them for mapping is unnecessary, as transposon libraries are easily constructed. Although there are many options for constructing a transposon library, the EZ-Tn5 transposon has been successfully used to create high density libraries (128).
The first step in locating the suppressor mutation is transferring the selectable markers to the suppressor strain and finding mutants that have lost the suppressor. Using generalized transducing phages for this transfer is ideal, as the segments of DNA transferred are large enough to allow a reasonable number of mutants to be screened while small enough to limit analysis to confined areas of the genome. For instance, using P1vir and a pooled library of mutants, a clone that has lost the suppressor mutation should be found within the first 400 to 500 mutants screened (121). If a transducing phage is not available, conjugation can also be used to map suppressor mutants, although precise mapping is made difficult by the large segments of DNA transferred (146, 162, 163). With the isolation of selectable markers linked to the suppressor mutation, the linkage frequency between the two makers can be calculated. The frequency of recombination allows for the mapping of the suppressor to a specific location on the chromosome by providing evidence of the suppressor's distance from known genetic markers. Here, we describe the process for using a library and a generalized transducing phage to map suppressor mutants.
(ii) Isolating selectable markers linked to a suppressor mutation.
As described above, mapping requires a pooled library of selectable markers and a way to transfer them to the suppressor strain. To create the pool of selectable markers, the mutations in the Keio collection for E. coli or the single gene deletion library in Salmonella can be pooled or a pool of transposon mutants can be used (160, 161, 164). For the transduction, the phage used will depend on the organism being used. P1vir phage will transduce E.coli K-12 and P22 phage can be used if working with S. Typhimurium. Lysates should be made from the pool of selectable mutants and used to transduce the strain containing the suppressor mutation. After transduction of the markers into the suppressor strain, the colonies should be replica plated to plates with the same antibiotic from the transduction and on plates selective for the suppressor mutation. From the replicate plating, the clones of interest will have lost the phenotype of the suppressor mutant and returned to the phenotype of the parent mutant.
These colonies should be restreaked from the control plate that selects for the marker but not the phenotype of interest to confirm the phenotypes. A new lysate for transduction is then made for this marker to confirm linkage to the suppressor mutation and to determine linkage frequency (see below). Using this lysate to transduce the original suppressor mutant will generate two types of transductants: those that have lost the suppressor and those that now have the marker and the suppressor mutation linked. The loss of suppression in the transductants without mutation demonstrates that the mutation is necessary for suppression. A lysate made from a transductant that has the marker linked to the suppressor mutation can be used to move the suppressor mutation to a clean strain with the mutation that is being suppressed and determine whether the suppressor mutation is sufficient for suppression. In addition, moving the suppressor to other strain backgrounds allows the phenotypes of the mutant to be more broadly examined.
Arbitrary PCR and Sanger sequencing, as described by Malinverni et al. (165), allows the selectable marker to be located. Arbitrary PCR works by performing PCR from primers near the end of the selectable marker and an arbitrary location outside the marker, which will then isolate the junction between the marker and the chromosome. In the example of Malinverni et al. (165), primers for miniTn10 transposon were used but primers can be designed to work with any desired marker. With at least one marker linked to the suppressor, a region of genome in which the suppressor mutation is located can be determined (within about 100 Kb from the marker for P1vir and 44 Kb for P22) (166, 167). Isolation of two linked markers decreases the size of the possible genome location of the suppressor and facilitates determination of the suppressor mutation’s the exact location by linkage frequency.
(iii) Mapping of suppressor location by linkage frequency.
The principle behind linkage frequency is that the distance between genetic loci determines their co-transduction frequency with closer loci co-transducing more frequently. The phenotype of one genetic marker will be selected and the resulting colonies are then screened for the unselected marker. In order to map the suppressor location by linkage frequency, at least two selectable markers close to the suppressor mutation must be located. Using lysates for the markers, the suppressor mutant is transduced and the proportion of the transductants that lose the suppressor mutation is calculated (Fig. 5B). This proportion can be easily calculated by replica plating 100 colonies onto conditions that differentiate between the phenotype of the suppressor mutant and the parent strain. Counting the number of colonies that lose the suppressor mutation will provide the percent linkage.
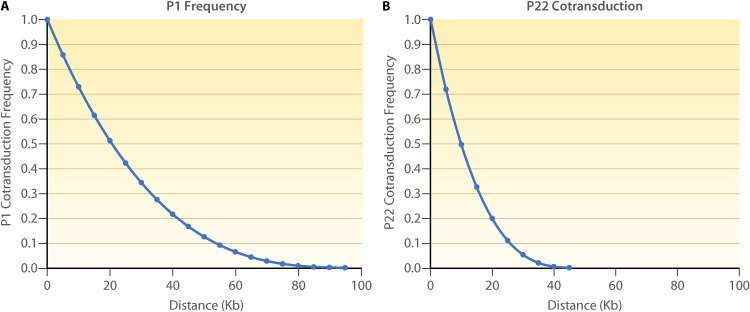
The Wu formula, which takes into account the size of the transducing fragment, is used to find the genetic distance between the markers and the suppressor (Fig. 6) (147). As insertions and deletions in the donor DNA will alter the co-transduction frequency, there is a modified version of Wu’s formula that accounts for this variation (166). The relationship between co-transduction frequency and physical distance will depend on the phage used for the transduction (147, 166). With the location of the suppressor mutation now determined (Fig. 5C), Sanger sequencing of nearby genes can identify the suppressor mutation. Alternatively, whole genome sequencing can be performed to locate the mutation. In this case, the mapping can determine which mutations in the genome is the suppressor. This is especially helpful in cases where a mutagen has been used leading to a high rate of mutations. With the suppressor mutation identified, the mechanism of suppression can be investigated leading to significant insights into the genetic interaction relating the original mutant gene and the suppressor.
FIG 6.
Correlation between distance and linkage frequency for P1 and P22 transduction. The Wu formula [C = (1 – D/L)3] can be used to be used to find the genetic distance between two markers based on their co-transduction frequency (147). C, co-transduction frequency; d, distance between markers; L, size of the transducing fragment. (A) Frequency of P1 cotransduction by distance based on the 100 Kbp P1 transducing fragment (167). (B) Frequency of P22 cotransduction by distance based on the 44 Kbp P22 transducing fragment (166).
CONCLUDING REMARKS
Suppressor mutants are incredibly powerful tools for discovering and investigating the genetic interactions between genes, proteins, and pathways. Their true strength is their ability to provide new and surprising insights due to the potentially unbiased nature of suppressor screens and selections. The characterization of suppressor mutants has played a key role in many of the fundamental discoveries in molecular biology and microbiology and they continue to be an important tool in mechanistic investigations.
The types of insights that can be gained from suppressor screens depends greatly on the types of mutations used for suppressor generation and so the types of suppressors generated. These categories of suppressor mutants act through very different mechanisms from mechanisms directly related to the function of the gene or pathway under investigation to mechanisms that affect alternative pathways to mechanisms that alter the fundamental processes of gene expression. Thus, suppressors, although powerful, are also highly susceptible to misinterpretation. This makes knowledge of the types of suppressor mutations and the proper design of experiments imperative. Current molecular and biochemical tools allow for the identification of a suppressor to be the fundamental observation that leads to many new discoveries. Investigation of the suppressor can extend beyond selectable phenotypes to in depth analysis of the changes that occur in the cell with suppression, greatly advancing our understanding of often complex genetic interactions. Thus, with full consideration of pitfalls and interpretation, suppressor mutants will continue to be an essential tool for genetic investigations for years to come.
ACKNOWLEDGMENTS
We thank Ashutosh K. Rai for critical reading and insightful comments on our manuscript.
This work was supported by the National Institute of Allergy and Infectious Diseases under Award Number R01-AI155915 (to A.M.M.) and by startup funds from Texas A&M University.
Contributor Information
Angela M. Mitchell, Email: amitchell@bio.tamu.edu.
James M. Slauch, University of Illinois at Urbana Champaign
REFERENCES
- 1.Rosenberg SM. 2013. Reverse mutation, p 220–221. In Maloy S, Hughes K (ed), Brenner's encyclopedia of genetics (2nd edition). Academic Press, San Diego. doi: 10.1016/B978-0-12-374984-0.01325-5. [DOI] [Google Scholar]
- 2.Dronamraju K. 2017. Decoding the language of genetics by David Botstein. The Quarterly Rev of Biology 92:107–107. doi: 10.1086/690885. [DOI] [Google Scholar]
- 3.Hartman PE, Roth JR. 1973. Mechanisms of suppression, p 1–105. In Caspari EW (ed), Advances in genetics, vol 17. Academic Press. [DOI] [PubMed] [Google Scholar]
- 4.Bonnier G. 2010. Note on the so-called Vermilion-duplication. Hereditas 7:229–232. doi: 10.1111/j.1601-5223.1926.tb03155.x. [DOI] [Google Scholar]
- 5.Karam JD, O’Donnell PV. 1973. Suppression of amber mutations of bacteriophage T4 gene 43 (DNA polymerase) by translational ambiguity. J Virol 11:933–945. doi: 10.1128/JVI.11.6.933-945.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan LV. 1925. Polyploidy in Drosophila melanogaster with two attached X chromosomes. Genetics 10:148–178. doi: 10.1093/genetics/10.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl FW. 1995. The amber mutants of phage T4. Genetics 141:439–442. doi: 10.1093/genetics/141.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner RP, Mitchell HK. 1955. Genetics and metabolism. Wiley. 2nd edition. [Google Scholar]
- 9.Stahl FW, Edgar RS, Steinberg J. 1964. The linkage map of bacteriophage T4. Genetics 50:539–552. doi: 10.1093/genetics/50.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streisinger G, Edgar RS, Denhardt GH. 1964. Chromosome structure in phage T4, I. Circularity of the linkage map. Proc Natl Acad Sci U S A 51:775–779. doi: 10.1073/pnas.51.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benzer S, Champe SP. 1962. A change from nonsense to sense in the genetic code. Proc Natl Acad Sci U S A 48:1114–1121. doi: 10.1073/pnas.48.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanofsky C, Lawrence PS. 1960. Gene action. Annu Rev Microbiol 14:311–340. doi: 10.1146/annurev.mi.14.100160.001523. [DOI] [PubMed] [Google Scholar]
- 13.Sarabhai AS, Stretton AOW, Brenner S, Bolle A. 1964. Co-linearity of the gene with the polypeptide chain. Nature 201:13–17. doi: 10.1038/201013a0. [DOI] [PubMed] [Google Scholar]
- 14.Pardee AB, Jacob F, Monod J. 1959. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase by E. coli. J Molecular Biology 1:165–178. doi: 10.1016/S0022-2836(59)80045-0. [DOI] [Google Scholar]
- 15.Beckwith J. 2009. Genetic suppressors and recovery of repressed biochemical memory. J Biol Chem 284:12585–12592. doi: 10.1074/jbc.X800017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pardee AB, Beckwith JR. 1962. Genetic determination of constitutive enzyme levels. Biochim Biophys Acta 60:452–454. doi: 10.1016/0006-3002(62)90435-3. [DOI] [PubMed] [Google Scholar]
- 17.Beckwith J. 1963. Restoration of operon activity by suppressors. Biochim Biophys Acta 76:162–164. doi: 10.1016/0926-6550(63)90026-4. [DOI] [PubMed] [Google Scholar]
- 18.Brenner S, Beckwith JR. 1965. Ochre mutants, a new class of suppressible nonsense mutants. J Molecular Biology 13:629–637. doi: 10.1016/S0022-2836(65)80131-0. [DOI] [Google Scholar]
- 19.Beckwith JR. 1964. A deletion analysis of the lac operator region in Escherichia coli. J Mol Biol 8:427–430. doi: 10.1016/s0022-2836(64)80206-0. [DOI] [PubMed] [Google Scholar]
- 20.Brenner S, Stretton AO, Kaplan S. 1965. Genetic code: the “nonsense” triplets for chain termination and their suppression. Nature 206:994–998. doi: 10.1038/206994a0. [DOI] [PubMed] [Google Scholar]
- 21.Uemura H, Thorbjarnardóttir S, Gamulin V, Yano J, Andrésson OS, Söll D, Eggertsson G. 1985. supN ochre suppressor gene in Escherichia coli codes for tRNALys. J Bacteriol 163:1288–1289. doi: 10.1128/jb.163.3.1288-1289.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capone JP, Sharp PA, RajBhandary UL. 1985. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J 4:213–221. doi: 10.1002/j.1460-2075.1985.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. 1989. A general method for site-specific incorporation of unnatural amino acids into proteins. Science 244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez EA, Lester HA, Dougherty DA. 2007. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: minimizing misacylation. RNA 13:1703–1714. doi: 10.1261/rna.666807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin JW, Martin AB, King DS, Wang L, Schultz PG. 2002. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci U S A 99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen BE, McAnaney TB, Park ES, Jan YN, Boxer SG, Jan LY. 2002. Probing protein electrostatics with a synthetic fluorescent amino acid. Science 296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 27.Hohsaka T, Muranaka N, Komiyama C, Matsui K, Takaura S, Abe R, Murakami H, Sisido M. 2004. Position-specific incorporation of dansylated non-natural amino acids into streptavidin by using a four-base codon. FEBS Lett 560:173–177. doi: 10.1016/S0014-5793(04)00099-7. [DOI] [PubMed] [Google Scholar]
- 28.Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. 2006. A genetically encoded fluorescent amino acid. Proc Natl Acad Sci U S A 103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taki M, Hohsaka T, Murakami H, Kuno A, Hasegawa T, Taira K, Sisido M. 2002. A novel fluorescent nonnatural amino acid that can be incorporated into a specific position of streptavidin. Nucleic Acids Res Suppl 2:203–204. doi: 10.1093/nass/2.1.203. [DOI] [PubMed] [Google Scholar]
- 30.Jacob F, Perrin D, Sanchez C, Monod J. 1960. Operon: a group of genes with the expression coordinated by an operator. C R Hebd Seances Acad Sci 250:1727–1729. [PubMed] [Google Scholar]
- 31.Richardson JP, Grimley C, Lowery C. 1975. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci U S A 72:1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossi L, Schwartz A, Guillemardet B, Boudvillain M, Figueroa-Bossi N. 2012. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev 26:1864–1873. doi: 10.1101/gad.195412.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao Z, Epshtein V, Kim KH, Proshkin S, Svetlov V, Kamarthapu V, Bharati B, Mironov A, Walz T, Nudler E. 2021. Pre-termination transcription complex: structure and function. Mol Cell 81:281–292.e8. doi: 10.1016/j.molcel.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Said N, Hilal T, Sunday ND, Khatri A, Bürger J, Mielke T, Belogurov GA, Loll B, Sen R, Artsimovitch I, Wahl MC. 2021. Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Science 371. doi: 10.1126/science.abd1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scaife J, Beckwith JR. 1966. Mutational alteration of the maximal level of Lac operon expression. Cold Spring Harbor Symp Quant Biol 31:403–408. doi: 10.1101/sqb.1966.031.01.052. [DOI] [PubMed] [Google Scholar]
- 36.Beckwith J, Grodzicker T, Arditti R. 1972. Evidence for two sites in the lac promoter region. J Mol Biol 69:155–160. doi: 10.1016/0022-2836(72)90031-9. [DOI] [PubMed] [Google Scholar]
- 37.Eichenberger P, Déthiollaz S, Fujita N, Ishihama A, Geiselmann J. 1996. Influence of the location of the cAMP receptor protein binding site on the geometry of a transcriptional activation complex in Escherichia coli. Biochemistry 35:15302–15312. doi: 10.1021/bi961377d. [DOI] [PubMed] [Google Scholar]
- 38.Bassford PJ, Jr, Silhavy TJ, Beckwith JR. 1979. Use of gene fusion to study secretion of maltose-binding protein into Escherichia coli periplasm. J Bacteriol 139:19–31. doi: 10.1128/jb.139.1.19-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silhavy TJ, Shuman HA, Beckwith J, Schwartz M. 1977. Use of gene fusions to study outer membrane protein localization in Escherichia coli. Proc Natl Acad Sci U S A 74:5411–5415. doi: 10.1073/pnas.74.12.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Stelten J, Silva F, Belin D, Silhavy TJ. 2009. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325:753–756. doi: 10.1126/science.1172221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cosma CL, Danese PN, Carlson JH, Silhavy TJ, Snyder WB. 1995. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol Microbiol 18:491–505. doi: 10.1111/j.1365-2958.1995.mmi_18030491.x. [DOI] [PubMed] [Google Scholar]
- 42.Emr SD, Schwartz M, Silhavy TJ. 1978. Mutations altering the cellular localization of the phage lambda receptor, an Escherichia coli outer membrane protein. Proc Natl Acad Sci U S A 75:5802–5806. doi: 10.1073/pnas.75.12.5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidson AL, Shuman HA, Nikaido H. 1992. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci U S A 89:2360–2364. doi: 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedouelle H, Bassford PJ, Jr, Fowler AV, Zabin I, Beckwith J, Hofnung M. 1980. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature 285:78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- 45.Emr SD, Hedgpeth J, Clément J-M, Silhavy TJ, Hofnung M. 1980. Sequence analysis of mutations that prevent export of λ receptor, an Escherichia coli outer membrane protein. Nature 285:82–85. doi: 10.1038/285082a0. [DOI] [PubMed] [Google Scholar]
- 46.Oliver DB, Beckwith J. 1982. Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli. J Bacteriol 150:686–691. doi: 10.1128/jb.150.2.686-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito K, Wittekind M, Nomura M, Shiba K, Yura T, Miura A, Nashimoto H. 1983. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell 32:789–797. doi: 10.1016/0092-8674(83)90065-x. [DOI] [PubMed] [Google Scholar]
- 48.Fikes JD, Bassford PJ. 1989. Novel secA alleles improve export of maltose-binding protein synthesized with a defective signal peptide. J Bacteriol 171:402–409. doi: 10.1128/jb.171.1.402-409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliver DB, Beckwith J. 1981. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell 25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 50.Stader J, Gansheroff LJ, Silhavy TJ. 1989. New suppressors of signal-sequence mutations, prlG, are linked tightly to the secE gene of Escherichia coli. Genes Dev 3:1045–1052. doi: 10.1101/gad.3.7.1045. [DOI] [PubMed] [Google Scholar]
- 51.Schatz PJ, Riggs PD, Jacq A, Fath MJ, Beckwith J. 1989. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev 3:1035–1044. doi: 10.1101/gad.3.7.1035. [DOI] [PubMed] [Google Scholar]
- 52.Bost S, Belin D. 1997. prl mutations in the Escherichia coli secG gene. J Biol Chem 272:4087–4093. doi: 10.1074/jbc.272.7.4087. [DOI] [PubMed] [Google Scholar]
- 53.Kumamoto CA, Beckwith J. 1983. Mutations in a new gene, secB, cause defective protein localization in Escherichia coli. J Bacteriol 154:253–260. doi: 10.1128/jb.154.1.253-260.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silhavy TJ, Mitchell AM. 2019. Genetic analysis of protein translocation. Protein J 38:217–228. doi: 10.1007/s10930-019-09813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorini L, Beckwith JR. 1966. Suppression. Annu Rev Microbiol 20:401–422. doi: 10.1146/annurev.mi.20.100166.002153. [DOI] [PubMed] [Google Scholar]
- 56.Michels C. 2002. Mutant hunts — to select or to screen (perhaps even by Brute Force), p 65–72. In Michels CA (ed), Genetic techniques for biological research. John Wiley & Sons, Ltd, West Sussex, England. doi: 10.1002/0470846623.ch4. [DOI] [Google Scholar]
- 57.di Rago JP, Netter P, Slonimski PP. 1990. Pseudo-wild type revertants from inactive apocytochrome b mutants as a tool for the analysis of the structure/function relationships of the mitochondrial ubiquinol-cytochrome c reductase of Saccharomyces cerevisiae. J Biol Chem 265:3332–3339. doi: 10.1016/S0021-9258(19)39771-6. [DOI] [PubMed] [Google Scholar]
- 58.Prelich G. 1999. Suppression mechanisms: themes from variations. Trends Genet 15:261–266. doi: 10.1016/s0168-9525(99)01749-7. [DOI] [PubMed] [Google Scholar]
- 59.Yanofsky C. 1965. Possible RNA codewords for the eight amino acids that can occupy one position in the tryptophan synthetase a protein. Biochem Biophys Res Commun 18:898–909. doi: 10.1016/0006-291X(65)90867-3. [DOI] [Google Scholar]
- 60.Yanofsky C, Crawford IP. 1959. The effects of deletions, point mutations, reversions and suppressor mutations on the two components of the tryptophan synthetase of Escherichia coli. Proc Natl Acad Sci U S A 45:1016–1026. doi: 10.1073/pnas.45.7.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hampsey DM, Das G, Sherman F. 1988. Yeast iso-l-cytochromec: Genetic analysis of structural requirements. FEBS Lett 231:275–283. doi: 10.1016/0014-5793(88)80834-2. [DOI] [PubMed] [Google Scholar]
- 62.Weigert MG, Garen A. 1965. Amino acid substitutions resulting from suppression of nonsense mutations. I. Serine insertion by the su-1 suppressor gene. J Mol Biol 12:448–455. doi: 10.1016/s0022-2836(65)80267-4. [DOI] [PubMed] [Google Scholar]
- 63.Weigert MG, Lanka E, Garen A. 1967. Amino acid substitutions resulting from suppression of nonsense mutations. 3. Tyrosine insertion by the Su-4 gene. J Mol Biol 23:401–404. doi: 10.1016/S0022-2836(67)80114-1. [DOI] [PubMed] [Google Scholar]
- 64.Sarabhai A, Brenner S. 1967. Further evidence that UGA does not code for tryptophan. J Mol Biol 26:141–142. doi: 10.1016/0022-2836(67)90269-0. [DOI] [PubMed] [Google Scholar]
- 65.Brenner S, Barnett L, Katz ER, Crick FHC. 1967. UGA: A third nonsense triplet in the genetic code. Nature 213:449–450. doi: 10.1038/213449a0. [DOI] [PubMed] [Google Scholar]
- 66.Wu Z, Gumport RI, Gardner JF. 1997. Genetic analysis of second-site revertants of bacteriophage lambda integrase mutants. J Bacteriol 179:4030–4038. doi: 10.1128/jb.179.12.4030-4038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crick FHC, Barnett L, Brenner S, Watts-Tobin RJ. 1961. General nature of the genetic code for proteins. Nature 192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 68.Hatfield D, Lee BJ, Smith DWE, Oroszlan S. 1990. Role of nonsense, frameshift, and missense suppressor tRNAs in mammalian cells, p 115–146. In Jeanteur P, Kuchino Y, Müller WEG, Paine PL (ed), Progress in molecular and subcellular biology. Springer Berlin Heidelberg, Berlin, Heidelberg. doi: 10.1007/978-3-642-75178-3_5. [DOI] [Google Scholar]
- 69.Mack KD, Goetz MV, Lin M, Venegas M, Barnhart J, Lu Y, Lamar B, Stull R, Silvin C, Owings P, Bih F-Y, Abo A. 2005. Functional identification of kinases essential for T-cell activation through a genetic suppression screen. Immunol Lett 96:129–145. doi: 10.1016/j.imlet.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 70.Nonet ML, Young RA. 1989. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riddle DL, Carbon J. 1973. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol 242:230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- 72.Riddle DL, Roth JR. 1972. Frameshift suppressors: III. Effects of suppressor mutations on transfer RNA. J Molecular Biology 66:495–506. doi: 10.1016/0022-2836(72)90429-9. [DOI] [PubMed] [Google Scholar]
- 73.Huseby DL, Brandis G, Praski Alzrigat L, Hughes D. 2020. Antibiotic resistance by high-level intrinsic suppression of a frameshift mutation in an essential gene. Proc Natl Acad Sci U S A 117:3185–3191. doi: 10.1073/pnas.1919390117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davis JE, Voisine C, Craig EA. 1999. Intragenic suppressors of Hsp70 mutants: Interplay between the ATPase- and peptide-binding domains. Proc Natl Acad Sci U S A 96:9269–9276. doi: 10.1073/pnas.96.16.9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helinski DR, Yanofsky C. 1962. Correspondence between genetic data and the position of amino acid alteration in a protein. Proc Natl Acad Sci U S A 48:173–183. doi: 10.1073/pnas.48.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Helinski DR, Yanofsky C. 1963. A genetic and biochemical analysis of second site reversion. J Biol Chem 238:1043–1048. doi: 10.1016/S0021-9258(18)81256-X. [DOI] [PubMed] [Google Scholar]
- 77.Park K-T, Villar MT, Artigues A, Lutkenhaus J. 2017. MinE conformational dynamics regulate membrane binding, MinD interaction, and Min oscillation. Proc Natl Acad Sci U S A 114:7497–7504. doi: 10.1073/pnas.1707385114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJM, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hill CW, Combriato G, Dolph W. 1974. Three different missense suppressor mutations affecting the tRNA GGG Gly species of Escherichia coli. J Bacteriol 117:351–359. doi: 10.1128/jb.117.2.351-359.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Stasio EA. 2012. Suppressors, screens, and genes: an educational primer for use with “A network of genes antagonistic to the LIN-35 retinoblastoma protein of Caenorhabditis elegans.” Genetics 191:1031–1035. doi: 10.1534/genetics.112.142943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van Leeuwen J, Pons C, Boone C, Andrews BJ. 2017. Mechanisms of suppression: The wiring of genetic resilience. BioEssays: news and Rev in Molecular, Cellular and Developmental Biology 39:1700042. doi: 10.1002/bies.201700042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Modolell J, Bender W, Meselson M. 1983. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A 80:1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roseman RR, Pirrotta V, Geyer PK. 1993. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO j 12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Georgiev PG, Corces VG. 1995. The su(Hw) protein bound to gypsy sequences in one chromosome can repress enhancer-promoter interactions in the paired gene located in the other homolog. Proc Natl Acad Sci U S A 92:5184–5188. doi: 10.1073/pnas.92.11.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gocke CD, Benko FA, Kopreski MS, Evans DB. 2000. Enrichment methods for mutation detection. Ann N Y Acad Sci 906:31–38. doi: 10.1111/j.1749-6632.2000.tb06587.x. [DOI] [PubMed] [Google Scholar]
- 86.Bailis JM, Smith AV, Roeder GS. 2000. Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins. Mol Cell Biol 20:4838–4848. doi: 10.1128/MCB.20.13.4838-4848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X, Zhang Y. 2016. Suppressor screens in Arabidopsis. Methods Mol Biol 1363:1–8. doi: 10.1007/978-1-4939-3115-6_1. [DOI] [PubMed] [Google Scholar]
- 88.Manson MD. 2000. Allele-specific suppression as a tool to study protein–protein interactions in bacteria. Methods 20:18–34. doi: 10.1006/meth.1999.0902. [DOI] [PubMed] [Google Scholar]
- 89.Seo JA, Guan Y, Yu JH. 2003. Suppressor mutations bypass the requirement of fluG for asexual sporulation and sterigmatocystin production in Aspergillus nidulans. Genetics 165:1083–1093. doi: 10.1093/genetics/165.3.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benson SA, Decloux A. 1985. Isolation and characterization of outer membrane permeability mutants in Escherichia coli K-12. J Bacteriol 161:361–367. doi: 10.1128/jb.161.1.361-367.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benson SA, Occi JL, Sampson BA. 1988. Mutations that alter the pore function of the OmpF porin of Escherichia coli K12. J Mol Biol 203:961–970. doi: 10.1016/0022-2836(88)90121-0. [DOI] [PubMed] [Google Scholar]
- 92.Misra R, Benson SA. 1988. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J Bacteriol 170:3611–3617. doi: 10.1128/jb.170.8.3611-3617.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sampson BA, Misra R, Benson SA. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491–501. doi: 10.1093/genetics/122.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu T, McCandlish AC, Gronenberg LS, Chng S-S, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 103:11754–11759. doi: 10.1073/pnas.0604744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sperandeo P, Cescutti R, Villa R, Di Benedetto C, Candia D, Dehò G, Polissi A. 2007. Characterization of lptA and lptB, two essential genes implicated in lipopolysaccharide transport to the outer membrane of Escherichia coli. J Bacteriol 189:244–253. doi: 10.1128/JB.01126-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Dehò G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruiz N, Falcone B, Kahne D, Silhavy TJ. 2005. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 99.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 100.Lee J, Xue M, Wzorek JS, Wu T, Grabowicz M, Gronenberg LS, Sutterlin HA, Davis RM, Ruiz N, Silhavy TJ, Kahne DE. 2016. Characterization of a stalled complex on the β-barrel assembly machine. Proc Natl Acad Sci U S A 113:8717–8722. doi: 10.1073/pnas.1604100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee J, Tomasek D, Santos TMA, May MD, Meuskens I, Kahne D. 2019. Formation of a β-barrel membrane protein is catalyzed by the interior surface of the assembly machine protein BamA. Elife 8:e49787. doi: 10.7554/eLife.49787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. 2012. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci U S A 109:3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McCabe AL, Ricci D, Adetunji M, Silhavy TJ. 2017. Conformational changes that coordinate the activity of BamA and BamD Allowing beta-barrel assembly. J Bacteriol 199:eoo373. doi: 10.1128/JB.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sandrock TM, O'Dell JL, Adams AE. 1997. Allele-specific suppression by formation of new protein-protein interactions in yeast. Genetics 147:1635–1642. doi: 10.1093/genetics/147.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lagator M, Igler C, Moreno AB, Guet CC, Bollback JP. 2016. Epistatic interactions in the Arabinose Cis-regulatory element. Mol Biol Evol 33:761–769. doi: 10.1093/molbev/msv269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Slauch JM, Silhavy TJ. 1989. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol 210:281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- 107.Lee M-K, Kwon N-J, Lee I-S, Jung S, Kim S-C, Yu J-H. 2016. Negative regulation and developmental competence in Aspergillus. Sci Rep 6:28874. doi: 10.1038/srep28874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Englesberg E, Anderson RL, Weinberg R, Lee N, Hoffee P, Huttenhauer G, Boyer H. 1962. L-Arabinose-sensitive, L-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J Bacteriol 84:137–146. doi: 10.1128/jb.84.1.137-146.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ueguchi C, Ito K. 1992. Multicopy suppression: an approach to understanding intracellular functioning of the protein export system. J Bacteriol 174:1454–1461. doi: 10.1128/jb.174.5.1454-1461.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 111.Karow M, Fayet O, Cegielska A, Ziegelhoffer T, Georgopoulos C. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33 degrees C in rich media. J Bacteriol 173:741–750. doi: 10.1128/jb.173.2.741-750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clementz T, Bednarski JJ, Raetz CRH. 1996. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J Biol Chem 271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 113.Clementz T, Zhou Z, Raetz CH. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A: acylation by MsbB follows laurate incorporation by HtrB. J Biol Chem 272:10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- 114.Wojtkiewicz P, Biernacka D, Gorzelak P, Stupak A, Klein G, Raina S. 2020. Multicopy suppressor analysis of strains lacking cytoplasmic peptidyl-prolyl cis/trans isomerases identifies three new PPIase activities in Escherichia coli that includes the DksA transcription factor. Int J Mol Sci 21:5843. doi: 10.3390/ijms21165843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shuman HA, Silhavy TJ. 2003. The art and design of genetic screens: Escherichia coli. Nat Rev Genet 4:419–431. doi: 10.1038/nrg1087. [DOI] [PubMed] [Google Scholar]
- 116.Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, Shales M, Lovett S, Winkler ME, Krogan NJ, Typas A, Gross CA. 2011. Phenotypic landscape of a bacterial cell. Cell 144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ramesh V, Gite S, Li Y, RajBhandary UL. 1997. Suppressor mutations in Escherichia coli methionyl-tRNA formyltransferase: Role of a 16-amino acid insertion module in initiator tRNA recognition. Proc Natl Acad Sci U S A 94:13524–13529. doi: 10.1073/pnas.94.25.13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanna MG, Simon MI. 1996. Isolation and in vitro characterization of CheZ suppressors for the Escherichia coli chemotactic response regulator mutant CheYN23D. J Biol Chem 271:7357–7361. doi: 10.1074/jbc.271.13.7357. [DOI] [PubMed] [Google Scholar]
- 119.Bigger J. 1944. Treatment of Staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 120.Fitzgerald G, Williams LS. 1975. Modified penicillin enrichment procedure for the selection of bacterial mutants. J Bacteriol 122:345–346. doi: 10.1128/jb.122.1.345-346.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leung KY, Finlay BB. 1991. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci U S A 88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mortin MA. 1990. Use of second-site suppressor mutations in Drosophila to identify components of the transcriptional machinery. Proc Natl Acad Sci U S A 87:4864–4868. doi: 10.1073/pnas.87.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cain AK, Barquist L, Goodman AL, Paulsen IT, Parkhill J, van Opijnen T. 2020. A decade of advances in transposon-insertion sequencing. Nat Rev Genet 21:526–540. doi: 10.1038/s41576-020-0244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Davey L, Valdivia RH. 2020. Bacterial genetics and molecular pathogenesis in the age of high throughput DNA sequencing. Curr Opin Microbiol 54:59–66. doi: 10.1016/j.mib.2020.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bae S, Mueller O, Wong S, Rawls JF, Valdivia RH. 2016. Genomic sequencing-based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proc Natl Acad Sci U S A 113:14127–14132. doi: 10.1073/pnas.1612753113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Galarneau A, Primeau M, Trudeau L-E, Michnick SW. 2002. β Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein–protein interactions. Nat Biotechnol 20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 131.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lim HC, Bernhardt TG. 2019. A PopZ-linked apical recruitment assay for studying protein-protein interactions in the bacterial cell envelope. Mol Microbiol 112:1757–1768. doi: 10.1111/mmi.14391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Roggo C, Carraro N, van der Meer JR. 2019. Probing chemotaxis activity in Escherichia coli using fluorescent protein fusions. Sci Rep 9:3845. doi: 10.1038/s41598-019-40655-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wehrman T, Kleaveland B, Her J-H, Balint RF, Blau HM. 2002. Protein–protein interactions monitored in mammalian cells via complementation of β-lactamase enzyme fragments. Proc Natl Acad Sci U S A 99:3469–3474. doi: 10.1073/pnas.062043699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malik A, Mueller-Schickert A, Bardwell JCA. 2014. Cytosolic selection systems to study protein stability. J Bacteriol 196:4333–4343. doi: 10.1128/JB.02215-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu F, Van Rijn E, Van Schie BGC, Keymer JE, Dekker C. 2015. Multi-color imaging of the bacterial nucleoid and division proteins with blue, orange, and near-infrared fluorescent proteins. Front Microbiol 6. doi: 10.3389/fmicb.2015.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhou Y, Gottesman S. 2006. Modes of regulation of RpoS by H-NS. J Bacteriol 188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guo Y, Hui C-Y, Liu L, Zheng H-Q, Wu H-M. 2019. Improved monitoring of low-level transcription in Escherichia coli by a β-galactosidase α-complementation system. Front Microbiol 10:1454. doi: 10.3389/fmicb.2019.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hui C-Y, Guo Y, Zhang W, Huang X-Q. 2018. Rapid monitoring of the target protein expression with a fluorescent signal based on a dicistronic construct in Escherichia coli. AMB Express 8:81. doi: 10.1186/s13568-018-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Iqbal M, Doherty N, Page AML, Qazi SNA, Ajmera I, Lund PA, Kypraios T, Scott DJ, Hill PJ, Stekel DJ. 2017. Reconstructing promoter activity from Lux bioluminescent reporters. PLoS Comput Biol 13:e1005731. doi: 10.1371/journal.pcbi.1005731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kodym A, Afza R. 2003. Physical and chemical mutagenesis. Methods Mol Biol 236:189–204. doi: 10.1385/1-59259-413-1:189. [DOI] [PubMed] [Google Scholar]
- 142.Anonymous. 2008. Physical mutagens, p 1493–1493. In Rédei GP (ed), Encyclopedia of genetics, genomics, proteomics and informatics. Springer, Netherlands, Dordrecht. doi: 10.1007/978-1-4020-6754-9_12828. [DOI] [Google Scholar]
- 143.Tiwari VK, Faris JD, Friebe B, Gill BS. 2016. Genome mapping, reference module in food science. Elsevier. doi: 10.1016/B978-0-08-100596-5.00220-1. [DOI] [Google Scholar]
- 144.Nakagawa H, Fujita M. 2018. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci 109:513–522. doi: 10.1111/cas.13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jacob F, Wollman EL. 1958. Genetic and physical determinations of chromosomal segments in Escherichia coli. Symp Soc Exp Biol 12:75–92. [PubMed] [Google Scholar]
- 146.Sanderson KE. 1972. Linkage map of Salmonella typhimurium, edition IV. Bacteriol Rev 36:558–586. doi: 10.1128/br.36.4.558-586.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu TT. 1966. A model for three-point analysis of random general transduction. Genetics 54:405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Epstein RH, Bolle A, Steinberg CM, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar RS, Susman M, Denhardt GH, Lielausis A. 1963. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp Quant Biol 28:375–394. doi: 10.1101/SQB.1963.028.01.053. [DOI] [Google Scholar]
- 149.Button JE, Silhavy TJ, Ruiz N. 2007. A suppressor of cell death caused by the loss of sigmaE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J Bacteriol 189:1523–1530. doi: 10.1128/JB.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. 2016. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113:E1565–E1574. doi: 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitchell AM, Srikumar T, Silhavy TJ. 2018. Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. mBio 9. doi: 10.1128/mBio.01321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Watson JD. 2007. Recombinant DNA: Genes and genome - a short course. W.H. Freeman, New York, NY. [Google Scholar]
- 153.Yin R, Kwoh CK, Zheng J. 2019. Whole genome sequencing analysis, p 176–183. In Ranganathan S, Gribskov M, Nakai K, Schönbach C (ed), Encyclopedia of bioinformatics and computational biology. Academic Press, Oxford, United Kingdom. doi: 10.1016/B978-0-12-809633-8.20095-2. [DOI] [Google Scholar]
- 154.Barker CS, Meshcheryakova IV, Inoue T, Samatey FA. 2014. Assembling flagella in Salmonella mutant strains producing a type III export apparatus without FliO. J Bacteriol 196:4001–4011. doi: 10.1128/JB.02184-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Itsko M, Schaaper RM. 2017. Suppressors of dGTP Starvation in Escherichia coli. J Bacteriol 199:e00142-17. doi: 10.1128/JB.00142-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Davis BM, Waldor MK. 2009. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: one fewer porin is enough. Nucleic Acids Res 37:5757–5767. doi: 10.1093/nar/gkp568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hawkey J, Hamidian M, Wick RR, Edwards DJ, Billman-Jacobe H, Hall RM, Holt KE. 2015. IS Mapper: identifying transposase insertion sites in bacterial genomes from short read sequence data. BMC Genomics 16:667. doi: 10.1186/s12864-015-1860-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Spagnolo F, Rinaldi C, Sajorda DR, Dykhuizen DE. 2015. Evolution of resistance to continuously increasing streptomycin concentrations in populations of Escherichia coli. Antimicrob Agents Chemother 60:1336–1342. doi: 10.1128/AAC.01359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Davis BD. 1948. Isolation of biochemically deficient mutants of bacteria by penicillin. J Am Chem Soc 70:4267–4267. doi: 10.1021/ja01192a520. [DOI] [PubMed] [Google Scholar]
- 160.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang H-J, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M. 2014. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv typhimurium. PLoS One 9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology 2. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Arnardóttir A, Thorbjarnardóttir S, Eggertsson G. 1980. Mapping of the supP (Su6+) amber suppressor gene in Escherichia coli. J Bacteriol 141:977–978. doi: 10.1128/jb.141.2.977-978.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Michalka J, Margolin P. 1977. Ochre suppression in Salmonella typhimurium. Genetics 86:237–260. doi: 10.1093/genetics/86.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Winterberg KM, Luecke J, Bruegl AS, Reznikoff WS. 2005. Phenotypic screening of Escherichia coli K-12 Tn5 insertion libraries, using whole-genome oligonucleotide microarrays. Appl Environ Microbiol 71:451–459. doi: 10.1128/AEM.71.1.451-459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Malinverni JC, Silhavy TJ. 2009. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci U S A 106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sanderson KE, Roth JR. 1988. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev 52:485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sternberg N. 1990. Bacteriophage P1 cloning system for the isolation, amplification, and recovery of DNA fragments as large as 100 kilobase pairs. Proc Natl Acad Sci U S A 87:103–107. doi: 10.1073/pnas.87.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Michels CA. 2002. Suppression analysis, p 91–98. In Michels CA (ed), Genetic Techniques for Biological Research. John Wiley & Sons, Ltd, West Sussex, England. doi: 10.1002/0470846623.ch8.. [DOI] [Google Scholar]