Abstract
Efavirenz is a potent and selective nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT). Nucleotide sequence analyses of the protease and RT genes (coding region for amino acids 1 to 229) of multiple cloned HIV-1 genomes from virus found in the plasma of patients in phase II clinical studies of efavirenz combination therapy were undertaken in order to identify the spectrum of mutations in plasma-borne HIV-1 associated with virological treatment failure. A K103N substitution was the HIV-1 RT gene mutation most frequently observed among plasma samples from patients for whom combination therapy including efavirenz failed, occurring in at least 90% of cases of efavirenz-indinavir or efavirenz-zidovudine (ZDV)-lamivudine (3TC) treatment failure. V108I and P225H mutations were observed frequently, predominantly in viral genomes that also contained other nonnucleoside RT inhibitor (NNRTI) resistance mutations. L100I, K101E, K101Q, Y188H, Y188L, G190S, G190A, and G190E mutations were also observed. V106A, Y181C, and Y188C mutations, which have been associated with high levels of resistance to other NNRTIs, were rare in the patient samples in this study, both before and after exposure to efavirenz. The spectrum of mutations observed in cases of virological treatment failure was similar for patients initially dosed with efavirenz at 200, 400, or 600 mg once a day and for patients treated with efavirenz in combination with indinavir, stavudine, or ZDV-3TC. The proportion of patients carrying NNRTI resistance mutations, usually K103N, increased dramatically at the time of initial viral load rebound in cases of treatment failure after exposure to efavirenz. Viruses with multiple, linked NNRTI mutations, especially K103N-V108I and K103N-P225H double mutants, accumulated more slowly following the emergence of K103N mutant viruses.
The reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) is critical to the life cycle of HIV and is without a homologue in eukaryotic organisms. As such, it is an attractive target for selective antiviral therapy. Among inhibitors of RT, a large class of chemically diverse, generally HIV-1-specific, nonnucleoside reverse transcriptase inhibitors (NNRTIs) has been identified. These inhibitors generally act by binding to a site on the RT that is distinct from the polymerase catalytic site. While NNRTIs can be potent inhibitors of HIV-1 replication, with favorable safety and pharmacokinetic parameters, rapid emergence of resistant viruses both in vitro (20, 24) and in vivo (11, 14, 15, 25, 30), often as the result of single nucleotide changes, has limited the therapeutic utility of these compounds as monotherapy. However, recent clinical trials of the use of NNRTIs in combination with other antiretroviral agents have demonstrated an added benefit from inclusion of an NNRTI in such combination regimens (5, 6, 12, 21, 29, 30; S. Green, M. F. Para, P. W. Daly, W. W. Freimuth, L. D. Getchel, C. A. Greenwald, and L. K. Wathen, 12th World AIDS Conference, Geneva, Switzerland, abstr. 12219, 1998). A variety of mutations in the HIV-1 RT gene associated with resistance to NNRTIs have been identified (2, 23, 27). In models of the three-dimensional structure of HIV-1 RT (28), these mutations cluster around the NNRTI binding site.
Efavirenz is a potent inhibitor of HIV-1 replication. In clinical studies, efavirenz used once daily in combination with nucleoside analogs or protease inhibitors has shown potent antiviral activity and significant clinical efficacy (29, 30). In cell culture, efavirenz retains significant activity against a variety of mutant strains of HIV-1 with single amino acid substitutions in the RT gene which have been associated with resistance to other NNRTIs (4, 33; L. Bacheler, unpublished data). Cell culture selection experiments (32) demonstrated that passage of the RF strain of HIV-1 in MT-2 cell or peripheral blood mononuclear cell culture led to the selection of mutations at amino acid positions 100, 108, 179, and 181 of the HIV-1 RT gene. Young et al. (33) reported the selection in cell culture of an L100I-K103N double mutant that was highly resistant to efavirenz. In both cases, high-level resistance to efavirenz (≥100-fold increase in 90% inhibitory concentration) appeared to be associated with multiple mutations in the RT gene of HIV-1.
In the studies reported here, we determined the nucleotide sequence of the protease and RT genes of plasma-borne HIV-1 from patients in three phase II dose-ranging studies of efavirenz combination therapy. These studies allowed us to assess the potential for selection of drug-resistant virus variants in vivo during efavirenz combination therapy and to identify the nature and time course of the emergence of such mutations relative to rebounds in plasma virus load, which might be indicative of loss of therapeutic efficacy.
MATERIALS AND METHODS
Selection of plasma samples and clonal sequencing approach.
Plasma samples collected at baseline and at intervals before and after a significant rebound in plasma virus load from efavirenz-exposed patients in clinical studies DMP 266-003, -004, and -005 (Table 1) were selected for sequencing. As per clinical study protocol, cases where a patient's plasma HIV RNA level, as determined by the Roche Amplicor HIV-1 Monitor assay, rose above 1,000 copies/ml of plasma on two successive occasions after reaching a nadir of 400 or fewer copies/ml or, if the plasma HIV RNA level never dropped below 400 copies/ml, where the level rose ≥0.75 log units relative to the lowest RNA copy number previously measured on two successive visits were defined as virological treatment failures. In addition, baseline plasma samples from some patients who experienced a sustained suppression in viral load were selected for sequencing.
TABLE 1.
Phase II clinical studies of efavirenz combination therapy in which virological treatment failures were characterizeda
| Study | Entry criteria | Study medications | Comments |
|---|---|---|---|
| DMP 266-003 | HIV RNA level, >20,000 copies/ml; 100 to 500 CD4 cells/mm3; NNRTI and PI naïve | 200/600 mg of EFV or placebo q.d. plus 800/1,000 mg of IDV q8h | Two cohorts began with a 2-week monotherapy lead-in period prior to the initiation of combination therapy. |
| Most patients were initially dosed at 200 mg of efavirenz q.d. (later raised to 600 mg q.d.) | |||
| DMP 266-004 | HIV RNA level, >2,500 copies/ml; >50 CD4 cells/mm3; ≥8 weeks of prior ZDV-3TC treatment; NNRTI and PI naïve | 400 or 600 mg of EFV or placebo q.d.; continued on ZDV-3TC | Presently recognized as suboptimal therapy since a single agent was added to a failing drug regimen. |
| DMP 266-005 | HIV RNA level, >10,000 copies/ml; >50 CD4 cells/mm3; antiretroviral therapy naïve | 200, 400, or 600 mg of EFV or placebo q.d. in combination with double NRTIs |
EFV, efavirenz; IDV, indinavir; q8h, every 8 h.
In this study a clonal sequencing approach was adopted. RNA extracted from a patient plasma sample was used as the template in a single RT reaction. The cDNA products of this reaction were divided into 12 separate nested PCRs. PCR products from eight or more reactions were independently cloned, and a single clone from each PCR was sequenced; thus, each sequence reflects the genotype of an independent viral genome. The nucleotide sequence of a 1.1-kb segment of the HIV-1 genome including the entire protease coding region and the region of RT encoding amino acid residues 1 to 229 was determined. The translated amino acid sequences of cloned viral genomes were compared to the clade B consensus sequence, and variations at amino acid positions previously described as being associated with resistance to inhibitors of the NNRTI class were tabulated. In addition, a visual examination of multiple aligned amino acid sequences of the RT region of virus from each patient was also undertaken in order to identify the emergence of novel mutations.
Extraction of RNA from plasma samples.
RNA was extracted from 200-μl plasma samples using the RNAgents Total RNA Isolation System kit (Promega Corp.). RNA precipitated by isopropanol was washed twice with 80% ethanol and dried for 5 min under vacuum at room temperature.
RT-PCR amplification.
The RNA pellet was resuspended in 120 μl of RT solution (0.5 U of Superscript II RT per ml, 1× Superscript II buffer [Gibco/BRL, Gaithersburg, Md.], 10 mM dithiothreitol [Gibco/BRL], 0.75 mM [each] dATP, dCTP, dGTP, and dTTP [Perkin-Elmer], 1 U of human placental RNase inhibitor [Boehringer Mannheim] per μl, and 1 μM primer P8 [5′ TAAATCTGACTTGCCCAATTCAATTTT 3′, corresponding to nucleotides 3335 to 3361 of HIV strain HXB2; Midland Certified Reagent Co.]). The mixture was allowed to dissolve for 5 min at room temperature and then incubated at 50°C (or at 42°C in some experiments) for 1 h.
The cDNA products of the reverse transcription reaction were used as the template in 12 nested PCRs to amplify a 1.1-kb region of HIV-1 containing both the protease gene and the 5′ portion of the RT gene. In the first PCR, 10 μl of the RT reaction mixture was added to 40 μl of PCR I mix consisting of 1 μl of TaqStart antibody (Clonetech), 1 μl of TaqPlus Long DNA polymerase (Stratagene) (these two reagents were mixed for 5 min before being added to the remaining reagents), 4 μl of 10× TaqPlus Long low-salt buffer (Stratagene), 1 μl of 12.4 mM primer P7 (5′ AGACCAGAGCCAACAGCCCCAC 3′, corresponding to nucleotides 2143 to 2164 of HIV strain HXB2; Midland Certified Reagent Co.), and 33 μl of water. The cycling conditions were an initial incubation at 95°C for 5 min, followed by 35 cycles of 94°C for 15 s, 56°C for 1 min, and 72°C for 2 min, with a final 6 min at 72°C. Second-round PCRs were performed using primer P9-amp1 (5′ CUACUACUACUAAGCCGATAGACAAGGAACTGTA 3′, corresponding to nucleotides 2219 to 2240 of HIV strain HXB2; Midland Certified Reagent Co.) and P10-amp1 (5′ CAUCAUCAUCAUCAGGATGGAGTTCATAACCCAT 3′, corresponding to nucleotides 3237 to 3258 of HIV strain HXB2; Midland Certified Reagent Co.). Fifty microliters of PCR II mix (1 μl of TaqStart antibody plus 1 μl of TaqPlus Long DNA polymerase (mixed as described above), 5 μl of 10× TaqPlus Long low-salt buffer, 1 μl each of 10 mM dATP, 10 mM dCTP, 10 mM dGTP, and 10 mM dTTP, 1 μl of 100 mM P9-amp1 and 1 μl of 100 mM P10-amp1) was added to the initial 50-μl PCR mixture and amplified for 35 cycles of 94°C for 15 s, 52°C for 1 min, and 72°C for 2 min, with a final 6 min at 72°C. Five microliters of each PCR mixture were run on a 1% agarose gel to confirm the presence of a 1.1-kb product and the absence of products of similar sizes in extracted and amplified samples of seronegative human plasma. Multiple (usually 12) nested PCRs were performed with the cDNA from each plasma sample, with the goal of obtaining one HIV-1 sequence from each of eight independently amplified viral genomes.
Purification of PCR products.
PCR amplification products were purified using solid-phase, reversible immobilization on carboxy-coated magnetic particles (PerSeptiveBiosystems, Inc., Framingham, Mass.) as described by DeAngelis et al. (10). Briefly, 45 μl of each PCR mixture was mixed with 50 μl of binding buffer (2.5 M NaCl–20% polyethylene glycol) and 10 μl of SPRI beads at a concentration of 10 mg/ml (previously washed 3 times with 0.5 M EDTA [pH 8.0]). Samples were incubated at room temperature for 10 min, washed twice with 150 μl of 70% ethanol, and air dried for 2 min. DNA was eluted from the beads with 20 μl of elution buffer (10 mM Tris-acetate [pH 7.8]). The particles were magnetically separated, and the supernatants were removed and retained. For selected samples, PCR products were purified following electrophoresis on 1% agarose gels. The desired band (approximately 1,200 bp) was excised from the gel with the tip of a sterile Pasteur pipette. The resulting 5- to 10-μl agarose gel plug containing the PCR product was expelled into 7.5 μl of 10 mM Tris-acetate (pH 7.8).
Cloning of PCR products.
Purified PCR products were inserted into the cloning vector pAMP-1 (CloneAmp System; Gibco/BRL), which utilizes uracil DNA glycosylase to facilitate directional cloning of PCR products. Annealed DNA samples were subsequently transformed into Escherichia coli strain DH5α (DH5 maximum efficiency; Gibco/BRL) using the procedure of Hanahan (13).
Preparation of plasmid DNA.
Purification of plasmid DNA from individual E. coli clones was performed on a BioRobot 9600 (Qiagen Inc., Chatsworth, Calif.) using Qiawell Ultra plasmid kits based on the optimized alkaline lysis method of Birnboim (3). Following cell growth in Luria broth containing 150 μg of ampicillin per ml, cells were pelleted and loaded onto the BioRobot 9600 for processing using Qiasoft version 2.0 software.
Nucleic acid sequencing.
The cloned DNAs were sequenced from double-stranded plasmids using six primers that spanned the cloned protease and RT genes (all primers were from Midland Certified Reagent Co.). Three primers (pSP8 [5′ GGTGACACTATAGAAGAGCT 3′, complementary to the cloning vector], p517F [5′ GGATGGCCCAAAAGTTA 3′, corresponding to nucleotides 2597 to 2613 of the HXB2 strain of HIV-1], and p743F [5′ CAATTAGGAATACCACATCC 3′, corresponding to nucleotides 2820 to 2839 of the HXB2 strain of HIV-1]) were used to sequence in the forward direction, and three primers (pT7 [5′ TAATACGACTCACTATAGGG 3′, complementary to the cloning vector], p461R [5′ TGGTACAGTTTCAATAGGAC 3′, corresponding to nucleotides 2557 to 2576 of the HXB2 strain of HIV-1], and p2A [5′ CCACATCCAGTACTGTTACTG 3′, corresponding to nucleotides 2863 to 2883 of the HXB2 strain of HIV-1]) were used to sequence in the reverse direction.
Thermal cycle sequencing reactions were carried out using 0.5-μg samples of the DNA templates and 20 ng of each primer. Polymerase TaqFS and either Big Dye or d-rhodamine dye terminators were used for sequencing; purified sequencing reaction were run on a Perkin-Elmer/Applied Biosystems 377 DNA sequencer. Consensus sequences for each clone were assembled using Sequencher software (Gene Codes Corporation).
Database.
Consensus DNA sequences were exported from Sequencher as text files. These files were translated into protein sequences for the HIV protease and RT genes. The region sequenced covers the coding region for amino acids 1 to 99 of protease and amino acids 1 to 229 of RT. The protein and nucleotide sequence information was imported into an Oracle database that linked the sequence information from each clone to the corresponding patient, clinical trial, and time point at which the sample was isolated. Sequences were analyzed following alignment of the protease and RT sequences from individual clones; comparisons were made between these sequences and the clade B consensus sequence (22). Sequences were analyzed by database query for the presence of mutations previously reported to be associated with the development of resistance to any NNRTI (2, 23, 27) and were visually examined for the emergence of novel mutations after exposure to efavirenz.
BLAST searches.
Each nucleotide sequence was compared (by a BLASTN [version 1.4.7] [1] search) to the entire set of HIV sequences derived from patient samples, as well as to sequences of laboratory strains of HIV-1, including those handled in the laboratory in which plasma samples were extracted and amplified. Individual sequences were automatically flagged for further scrutiny if (i) among 10 or more sequences from the same patient in the database, 5 or fewer were among the top 10 matches to the test sequence; (ii) a sequence from a wild-type laboratory strain was among the top 10 matches to the test sequence, and the wild-type strain matched the test sequence more closely than it did other sequences from the same patient; or (iii) the test sequence was divided into two high-scoring segment pairs (an n = 2 in the BLASTN search results), indicative of a frameshift or deletion in one of the two sequences. Flagged sequences, as well as a random sampling of nonflagged sequences, were reviewed. Sequences with insertions, deletions, or frameshifts were allowed if the amino acid translation of the remainder of the sequence showed high homology to other sequences from the same patient. Sequences which showed closer homology to sequences from a different patient or to sequences of laboratory strains of HIV-1, suggestive of cross-contamination, were tagged in the Oracle database and excluded from the set of sequences queried to calculate the data reported here.
Nucleotide sequence accession numbers.
Nucleotide sequences determined in this study have been deposited in GenBank under accession numbers AY000001 to AY003708.
RESULTS
The emergence of genotypic resistance to efavirenz was studied in samples collected during three clinical studies of efavirenz combination therapy summarized in Table 1. These phase II studies tested efavirenz in combination with the protease inhibitor indinavir or the nucleoside analog RT inhibitors (NRTIs) zidovudine (ZDV, also known as AZT) and lamivudine (3TC). Both NRTI-naïve and -experienced patients were enrolled in these studies, but all patients were required to be NNRTI and protease inhibitor (PI) therapy naïve. Two cohorts (21 patients) of study DMP 266-003 began with a 2-week efavirenz monotherapy lead-in period before combination therapy was started.
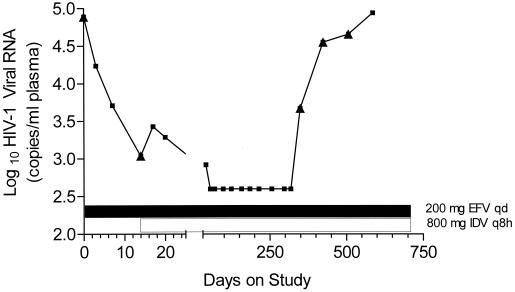
As an example of the clonal sequencing approach, results from a single patient are shown. The viral load response of patient 22, who received efavirenz monotherapy at 200 mg once a day (q.d.) for 14 days before the initiation of efavirenz-indinavir combination therapy, is shown in Fig. 1. Table 2 shows variations from the clade B amino acid consensus sequence observed at selected positions in the RT gene of virus isolated from the plasma of patient 22 and cloned. At baseline, this patient's mixed plasma virus population included variations at amino acids 98 and 211 of RT and at amino acid 77 of protease (protease gene data not shown). After only 14 days of efavirenz monotherapy, K103N mutations in RT were observed in 2 out of 10 cloned viral genomes. Despite the emergence of a K103N mutant virus, initiation of efavirenz-indinavir combination therapy led to a further reduction in viral load (to fewer than 400 copies/ml of plasma), which reduced level was maintained until day 349. At this time, HIV-1 RNA in plasma began to increase, eventually returning to close to baseline levels. At day 349, sequencing revealed a virus population carrying K103N mutations in RT and V82A mutations in protease in all 4 clones sequenced. At day 424, as the viral load continued to rebound, K101E and P225H mutations in RT, in combination with K103N or K103E mutations, were observed, while L63P and M46L mutations were observed in combination with V82A mutations in protease. By day 507, approximately 158 days after the beginning of viral load rebound, K103N-P225H double RT mutants were more prevalent (observed in 4 of 8 clones). In protease, additional mutations at positions 54 and 71 of the protease gene were detected.
FIG. 1.
Viral load (plasma-borne HIV-1) in patient 22 in study DMP 266-003. The log10 of the plasma HIV-1 viral load, as measured by the Roche Amplicor HIV-1 Monitor assay, is plotted against the number of days of therapy (two different scales). ▴, time point at which plasma virus genotype was determined. The timing of efavirenz monotherapy and efavirenz plus indinavir combination therapy is indicated by the bars. EFV, efavirenz; IDV, indinavir; q8h, every 8 h.
TABLE 2.
Variations from HIV-1 clade B amino acid consensus sequence at selected positions in RT of cloned viral genomes isolated from plasma of patient 22 in study DMP 266-003a
| Study day | Log RNA | Amino acid that in the consensus sequence was:
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M41 | A62 | K65 | D67 | T69 | K70 | L74 | F77 | A98 | L100 | K101 | K103 | V106 | V108 | E138 | Y181 | M184 | Y188 | V189 | G190 | L210 | R211 | F214 | T215 | K219 | P225 | F227 | ||
| 0 | 4.89 | — | — | — | — | — | — | — | — | S | — | — | — | — | — | — | — | — | — | — | — | — | K | — | — | — | — | — |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | K | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| 14 | 3.04 | — | — | — | — | — | — | — | — | S | — | — | — | — | — | G | — | — | — | — | — | — | — | — | — | — | — | — |
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | K | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | S | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | S | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | K | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| 349 | 3.67 | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| 424 | 4.55 | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | K | — | — | — | H | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | + | E | E | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | V | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| 507 | 4.66 | — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | H | — |
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | K | — | — | — | H | — | ||
| — | V | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | K | — | — | — | H | — | ||
| — | V | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | H | — | ||
| — | — | — | — | — | — | — | — | — | — | — | N | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | ||
The residues listed are those at which specific substitutions have been associated with the development of resistance. The study day at which each plasma sample was collected and the number of HIV-1 RNA copies/ml of plasma (log10) are indicated for each set of sequences. +, frameshift mutation; —, sequence same as consensus.
In Table 3, a summary of the frequencies of mutations previously reported to be associated with resistance to one or more NNRTIs among plasma samples collected at baseline (prior to exposure to any study medications) and during the study for patients experiencing virological treatment failure in study DMP 266-003 is provided. In this study, patients were treated with the PI indinavir plus efavirenz or efavirenz placebo. Some patients, including patient 22 (described above), received efavirenz monotherapy for 2 weeks before efavirenz-indinavir combination therapy was initiated. Other patients initially randomized to indinavir monotherapy were later allowed to switch to efavirenz-D4T combination therapy. Some of these patients failed this second drug regimen, and a genotypic characterization of virus from such “switched” patients is included in Table 3.
TABLE 3.
Prevalence of NNRTI resistance mutations in virus from plasma of patients in study DMP 266-003: comparison of baseline, placebo, and efavirenz-exposed patientsa
| RT gene mutation | % of patients (no. of patients, no. of clones)
|
|||
|---|---|---|---|---|
| Patients at baseline | Patients with significant rebounds in viral load who:
|
|||
| Underwent indinavir monotherapy | Underwent efavirenz plus indinavir combination therapy | Switched to efavirenz combination therapy | ||
| L74V | — | — | 9.4 (3, 5) | — |
| V75I | 2.1 (2, 7) | — | 3.1 (1, 1) | 7.1 (1, 1) |
| A98G | 1.1 (1, 8) | — | 6.3 (2, 2) | 7.1 (1, 1) |
| L100I | — | — | 9.4 (3, 6) | 21.4 (3, 4) |
| K101E | — | — | 12.5 (4, 38) | 7.1 (1, 3) |
| K101Q | 1.1 (1, 1) | — | 18.8 (6, 44) | 28.6 (4, 9) |
| K103N | 1.1 (1, 1) | 8.3 (1, 9) | 93.8 (30, 712) | 100.0 (14, 241) |
| V106A | — | — | 3.1 (1, 1) | 7.1 (1, 1) |
| V106I | 2.1 (2, 5) | — | 3.1 (1, 17) | 7.1 (1, 1) |
| V108I | — | — | 37.5 (12, 65) | 21.4 (3, 18) |
| E138K | — | — | 9.4 (3, 4) | — |
| T139I | 1.1 (1, 1) | — | 3.1 (1, 1) | — |
| G141E | — | — | 3.1 (1, 1) | — |
| V179E | — | — | 3.1 (1, 1) | 7.1 (1, 1) |
| Y181C | 2.1 (2, 2) | — | 9.4 (3, 3) | — |
| Y188L | — | — | 6.3 (2, 67) | 14.3 (2, 32) |
| Y188H | 2.1 (2, 2) | — | 18.8 (6, 7) | 7.1 (1, 7) |
| Y188C | — | 8.3 (1, 1) | — | — |
| V189I | 2.1 (2, 2) | — | 3.1 (1, 3) | 7.1 (1, 1) |
| G190A | — | — | 6.3 (2, 16) | 7.1 (1, 3) |
| G190E | 1.1 (1, 1) | — | 3.1 (1, 7) | — |
| G190S | — | — | 18.8 (6, 57) | 14.3 (2, 23) |
| P225H | — | — | 40.6 (13, 117) | 50.0 (7, 65) |
| F227L | — | 8.3 (1, 1) | 9.4 (3, 3) | — |
Baseline samples were collected on study day 0, before any exposure to study medications, and include samples derived from the plasma virus of patients (94 patients, 677 clones) who achieved a sustained suppression of viral load as well as from patients for whom subsequent efavirenz combination therapy failed. Indinavir monotherapy samples are those collected after study day 0 from patients (12 patients, 170 clones) initially randomized to efavirenz placebo and before any switch to active efavirenz-containing drug combinations. Efavirenz samples are those collected after day 0 from patients (32 patients, 717 clones) initially randomized to active efavirenz-containing drug combinations. Switched to efavirenz samples were those collected after patients (14 patients, 336 clones) initially randomized to efavirenz placebo (indinavir monotherapy) were switched to active efavirenz plus 3TC and/or D4T. Some NNRTI resistance mutations that were not detected in any sample are excluded from the table. —, no clones detected.
As shown in Table 3, mutations previously described as associated with resistance to one or more NNRTIs were infrequently detected in plasma virus from the NNRTI-naïve patients enrolling in this study. The mutations that were detected were frequently seen in a single clone per patient sample. Similarly, mutations associated with resistance to one or more NNRTIs were infrequent among samples collected during the study from patients receiving efavirenz placebo (i.e., indinavir monotherapy) who experienced a rebound in viral load. In contrast, a variety of mutations previously described as associated with NNRTI resistance was detected in plasma samples collected during the study from patients who experienced virological treatment failure of an efavirenz-containing drug regimen. K103N was the most frequently observed NNRTI resistance mutation in samples from efavirenz-exposed patients who experienced a significant rebound in viral load, detected in more than 90% of cases of efavirenz treatment failure. This amino acid substitution can be achieved by a single base pair change or point mutation (AAG or AAA to AAC or AAT). V108I and P225H mutations were observed in 37.5 and 40.6%, respectively, of efavirenz-exposed patients who experienced treatment failure and at similar frequencies (21.4 and 50%, respectively) among patients initially randomized to efavirenz placebo who later switched to an efavirenz-containing regimen and for whom the second regimen failed. Additional mutations observed in multiple clones from a significant number of patients for whom combination therapy with efavirenz failed included L100I, K101E, K101Q, Y188H, Y188L, and G190S. Mutations at Y181C, Y188C, or V106A, which are associated with high levels of resistance to nevirapine and/or delavirdine, were observed only rarely, often in a single cloned viral genome from a given patient.
Mutations associated with resistance to one or more PIs were more frequently detected at baseline than those associated with resistance to NNRTIs, although most of the mutations detected are considered secondary resistance mutations (16) and are recognized as naturally occurring polymorphisms in the highly variable HIV-1 protease gene (17). Primary PI resistance mutations (D30N, M46I, M46L, M46V, G48V, I50V, V82A, V82F, and V82T, as defined by Hirsch et al. [16]) were only infrequently detected, usually in a single clone per patient sample. Viral sequences collected from patients who experienced treatment failure with either indinavir monotherapy or efavirenz-indinavir combination therapy had an increased prevalence of a variety of mutations associated with resistance to PIs, including mutations at amino acid positions 46 and 82, which have been associated with phenotypic resistance to indinavir (8). Secondary PI mutations detected at increased frequency in viral genomes from patients who had failed indinavir therapy included L10F, L10I, L10R, L10V, L24I, A71V, G73S, and L90M. The spectrum of PI resistance mutations observed did not appear to vary between patients for whom indinavir monotherapy failed and those for whom efavirenz-indinavir combination therapy failed.
Since nucleotide sequence information was determined for cloned HIV-1 genomes, the linkage of multiple mutations in the same DNA clone could be identified. These linked mutations represent sequence variations present in the same viral RNA genome prior to RT-PCR amplification. Table 4 compares the prevalence of specific, single NNRTI mutations and linked, multiple NNRTI mutations in 161 baseline samples collected from patients entering studies DMP 266-003, -004, and -005 to the prevalence of the same mutations in samples collected from 104 of these patients after a significant rebound in viral load. As in study DMP 266-003, the prevalence of NNRTI mutations was low at study entry. Among cases of efavirenz treatment failure, the K103N mutation was identified in cloned HIV-1 genomes both as a single NNRTI mutation and in combination with other NNRTI mutations. Many patients had viruses with K103N both as a single mutation (88.5% of patients) and in combination with other NNRTI resistance mutations (64.4% of patients), either in sequential samples or within a single plasma sample. In contrast, L100I, K101Q, V108I, Y188H, and P225H mutations were seen largely or exclusively in combination with other NNRTI resistance mutations, not as single NNRTI resistance mutations. While a variety of viral genotypes combining one or more mutations previously associated with NNRTI resistance were identified, combinations containing K103N were by far the most prevalent. The most frequently observed double NNRTI mutants include K103N-V108I and K103N-P225H, each observed in approximately one-third of efavirenz treatment failures. Other double mutants observed in more than 10% of efavirenz treatment failures include L100I-K103N and K101Q-K103N. A triple mutant virus carrying K103N, V108I and P225H mutations was observed in 9.6% of patients for whom efavirenz combination therapy failed. K101E mutations were observed in 13.8% of efavirenz treatment failure patients overall, either as a single NNRTI resistance mutation (4.8% of patients) or linked to a variety of other NNRTI resistance mutations (not just K103N). Viruses with Y188L mutations, often linked to V106I mutations, were observed in a small percentage of patients. Y188L mutated viruses appear to represent an alternate, infrequently utilized path to efavirenz resistance; K103N mutant viruses were infrequently observed in patients who developed Y188L mutant viruses.
TABLE 4.
Prevalence and linkage of RT gene mutations associated with NNRTI resistance in the plasma virus of study subjectsa
| Mutation(s) | % of patients (no. of patients, no. of clones)
|
|
|---|---|---|
| Baseline | Efavirenz-exposed treatment failures | |
| V75I | 1.2 (2, 5) | — |
| A98G | 0.6 (1, 8) | 2.9 (3, 3) |
| A98G K103N | — | 1.9 (2, 2) |
| A98G G190S | — | 1.9 (2, 4) |
| A98G K103N V108I | — | 3.8 (4, 7) |
| K101E | — | 4.8 (5, 7) |
| K101E K103N | — | 2.9 (3, 8) |
| K101E Y188L | — | 1.9 (2, 3) |
| K101E P225H | — | 1.0 (1, 2) |
| K101E K103R P225H | — | 1.0 (1, 5) |
| K101Q | 2.5 (4, 8) | — |
| K101Q K103N | — | 16.3 (17, 59) |
| K101Q K103N P225H | — | 3.8 (4, 15) |
| K103N | — | 88.5 (92, 1,110) |
| K103N L100I | — | 10.6 (11, 68) |
| K103N V108I | — | 28.8 (30, 138) |
| K103N Y181C | — | 2.9 (3, 3) |
| K103N Y188H | — | 4.8 (5, 11) |
| K103N Y188L | — | 3.8 (4, 20) |
| K103N V189I | — | 3.8 (4, 5) |
| K103N P225H | — | 32.7 (34, 225) |
| K103N L100I P225H | — | 1.0 (1, 2) |
| K103N L100I Y188L | — | 1.9 (2, 2) |
| K103N L100I L74V | — | 1.9 (2, 9) |
| K103N P225H Y188H | — | 1.9 (2, 2) |
| K103N P225H V106A | — | 1.9 (2, 2) |
| K103N P225H V108I | — | 9.6 (10, 13) |
| K103R | 2.5 (4, 20) | — |
| K103R V75I | 0.6 (1, 2) | — |
| V106I | 1.2 (2, 7) | 1.9 (2, 2) |
| V106I Y188L | — | 4.8 (5, 39) |
| V108I | — | 5.8 (6, 7) |
| E138K | — | 2.9 (3, 3) |
| E138K K103N | — | 1.9 (2, 2) |
| Y181C | — | 1.9 (2, 2) |
| Y188L | — | 6.7 (7, 141) |
| G190A | — | 3.8 (4, 10) |
| G190A K101E | — | 1.0 (1, 15) |
| G190A K103N | — | 3.8 (4, 6) |
| G190A K101E K103R | — | 1.0 (1, 5) |
| G190A K101Q K103N | — | 1.0 (1, 2) |
| G190E | — | 1.0 (1, 5) |
| G190E L74V | — | 1.0 (1, 2) |
| G190S | — | 10.6 (11, 64) |
| G190S K101E | — | 2.9 (3, 22) |
| G190S K101Q | — | 1.0 (1, 2) |
| G190S K103N | — | 3.8 (4, 4) |
| G190S K101E V106I | — | 1.9 (2, 2) |
| G190S K101Q K103N | — | 1.9 (2, 2) |
| G190S K103N V108I | — | 1.9 (2, 2) |
| P225H | — | 2.9 (3, 3) |
| F227L K103N | — | 4.8 (5, 5) |
| F227L K103N V108I | — | 1.0 (1, 2) |
Combinations of mutations associated with resistance to one or more NNRTIs that were observed in two or more cloned viral genomes. HIV-1 plasma virus sequences from study subjects in studies DMP 266-003, -004, and -005 are included. Baseline samples were collected on study day 0, before any exposure to study medications, and include samples derived from the plasma virus of patients (161 patients, 1,100 clones) who achieved a sustained suppression of viral load as well as from patients (104 patients, 2,191 clones) for whom subsequent efavirenz combination therapy failed. —, one or fewer genomes with the listed combination of NNRTI resistance mutations was detected among clones derived from patients in the category listed. Linked mutations associated with resistance to NRTIs of HIV-1 RT or PIs were observed in some samples but are not considered in this table.
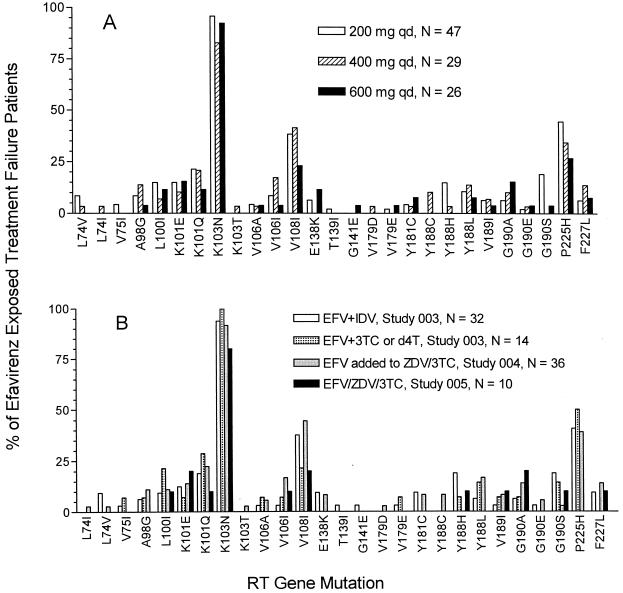
In the three phase II clinical trials studied, patients were dosed with three different doses of efavirenz (200, 400, or 600 mg q.d.). Some patients initially dosed at 200 mg in study DMP 266-003 had the dose of efavirenz increased to 600 mg q.d. during the study. The pattern of NNRTI mutations observed in cases of efavirenz treatment failure was examined for the three doses of efavirenz tested (Fig. 2A). K103N was the most frequent NNRTI resistance mutation observed in the plasma virus of patients who experienced virological treatment failure and who were initially dosed at 200, 400, or 600 mg q.d. of efavirenz. V108I and P225H mutations, most often in combination with K103N, were frequent at each dose of efavirenz studied. K101E and K101Q mutations were observed for more than 10% of cases of efavirenz treatment failure at each efavirenz dose. The types and relative frequency of NNRTI mutations selected in patients for whom efavirenz combination therapy failed was also examined as a function of the coadministered study medications. Data were available for patients treated with efavirenz in combination with indinavir, with D4T or 3TC, or with ZDV-3TC in both NRTI-experienced and -naïve patients. Figure 2B illustrates that a similar pattern of selected NNRTI resistance mutations was observed for each drug combination.
FIG. 2.
Prevalence of NNRTI resistance mutations in the plasma-borne virus of patients for whom efavirenz treatment failed. (A) Prevalence as a function of initial efavirenz dose. Shown are the percentages of patients for whom efavirenz treatment failed in whose plasma virus the listed NNRTI mutation was detected by clonal sequencing as a function of initial efavirenz dose. Linkage of mutations is not considered. Genotypic data from patients in studies DMP 266-003, -004, and -005 are included. The efavirenz dose was changed for some patients during the study. (B) Prevalence as a function of drug regimen. Shown are the percentages of patients for whom efavirenz treatment failed in whose plasma virus the listed NNRTI mutation was detected by clonal sequencing in different efavirenz-containing drug regimens. Linkage of mutations is not considered. Genotypic data from patients in studies DMP 266-003, -004, and -005 are included.
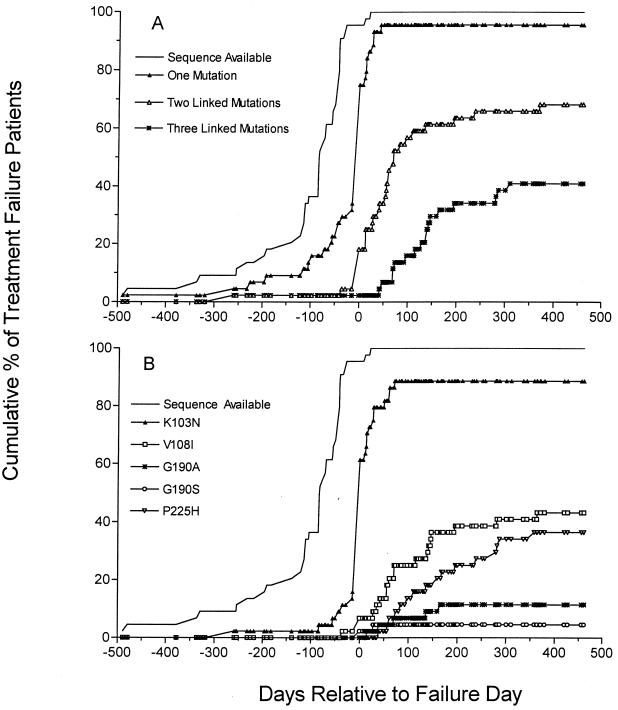
Since multiple plasma samples from patients for whom efavirenz combination therapy failed collected before and at various times during the rebound in viral load were analyzed, it was possible to examine the time of appearance of viruses with single versus multiple mutations. Figure 3A shows the time of appearance of virus with one, two, or three linked NNRTI mutations relative to the time of initial rebound in viral load used to define virological treatment failure for patients in study DMP 266-004. The proportion of treatment failure patients whose plasma virus carried at least one NNRTI resistance mutation increased dramatically around the time of initial rebound in viral load, from 20% of patient samples tested 57 or more days prior to the beginning of viral load rebound to approximately 75% of patient samples tested at the time of initial viral load rebound (failure day). By 100 days after the beginning of viral load rebound, more than 50% of patients had viruses with two NNRTI mutations in the same viral genome, and 15% of patients had viruses with three linked NNRTI mutations. A similar pattern of early emergence of single and later emergence of double or triple NNRTI resistance mutations was also observed in cases of efavirenz treatment failure in studies DMP 266-003 and -005, although the number of patients who experienced treatment failure in study DMP 266-005 whose plasma virus was characterized was small (10 patients).
FIG. 3.
Time of appearance of NNRTI resistance mutations relative to rebound in plasma virus load in study DMP 266-004 patients for whom efavirenz combination therapy failed. (A) Time of appearance of single NNRTI resistance mutations and multiple, linked NNRTI resistance mutations. The cumulative percentage of patients with sequence data available and with linked NNRTI mutations in one or more cloned viral genomes in study DMP 266-004 is plotted as a function of the interval before or after the initial rebound in viral load which defined the treatment failure day. (B) Time of appearance of specific efavirenz-selected NNRTI resistance mutations. The cumulative percentage of patients with sequence data available and with specific NNRTI mutations in one or more cloned viral genomes in study DMP 266-004 is plotted as a function of the interval before or after the initial rebound in viral load which defined the treatment failure day. Linkage of mutations is not considered.
In study DMP 266-003, the proportion of patients that had viruses with NNRTI resistance mutations before the viral load rebounded and who experienced treatment failure was larger than that in study DMP 266-004 (data not shown). This early appearance of NNRTI resistance mutations was observed mainly among patients in the two efavirenz monotherapy cohorts. Sequence data from plasma samples collected after 2 weeks of efavirenz monotherapy at 200 mg q.d. were available for 16 patients. At the end of this short monotherapy period, viruses with one or more NNRTI mutations were detected, although often as a minority of the clones sequenced, in 11 of 16 patients. For many of these monotherapy patients, addition of indinavir at the end of 2 weeks led to further suppression of plasma virus load. Overall, however, the proportion of patients who began with short-term efavirenz monotherapy and eventually experienced virological treatment failure was higher than that of patients who always received efavirenz in combination with other antiretroviral drugs.
Figure 3B shows the accumulation of specific NNRTI resistance mutations as a function of time before or after the beginning of viral load rebound in cases of efavirenz treatment failure in study DMP 266-004. The K103N mutation was detected in the majority of patients at or shortly after the initial rebound in viral load. V108I and P225H mutations, arising largely in combination with K103N mutations, accumulated more gradually with increasing time after the beginning of viral load rebound. As demonstrated in vitro (32, 33; L. T. Bacheler, unpublished data), these combinations of multiple NNRTI mutations are likely to confer higher levels of resistance to efavirenz beyond that associated with the initial K103N mutation.
DISCUSSION
In the studies reported here, we adopted a clonal sequencing approach for the detection of mutations in plasma-borne HIV-1 that might be associated with resistance to efavirenz. Multiple independently amplified and cloned viral genomes were sequenced from each plasma sample. This methodology offers some unique advantages for the characterization of resistance to a new antiretroviral agent. Sequencing multiple clones facilitates the detection of minority viral species, including those associated with emerging viral resistance (7, 19). In many cases, we first identified a K103N mutation in one or two clones out of eight derived from a single plasma sample and then identified a K103N mutation in all clones derived from a second, later plasma sample. Sequencing a single clone from each independent PCR amplification avoids founder effects that could distort the representation of specific genotypes in the collection of sequences derived from a plasma sample. The linkage of specific mutations in a single viral genome is readily detected by this approach. Certain mutations associated with resistance to efavirenz were largely or exclusively detected linked to other mutations. The L100I mutation, while conferring resistance to efavirenz in constructed viruses as a single mutation (34; L. T. Bacheler, unpublished data), was never observed as the only NNRTI resistance mutation in plasma virus from patients for whom efavirenz therapy failed; rather, this mutation was found linked to a K103N mutation. The frequently observed V108I and P225H mutations, which do not confer detectable resistance to efavirenz in vitro as single mutations (L. T. Bacheler, unpublished data) were also found predominantly in viral genomes also carrying K103N mutations. In some plasma samples, a diverse collection of NNRTI resistance mutations was detected. Only through a clonal sequencing approach could associations between specific mutations be discerned. There are, however, several disadvantages to the clonal sequencing method. First, it is very labor intensive. Second, since single cloned viral genomes are sequenced, this method may identify mutations introduced during PCR amplification, which do not reflect the initial plasma virus population. The viral origin of rare mutations, such as Y181C in patients for whom efavirenz therapy failed or a K103N mutation in a plasma sample collected before exposure to efavirenz, must remain in some doubt because of this technical drawback. For this reason we have chosen to focus on mutations detected either in virus from plasma of at least two patients or in multiple clones in virus from plasma of a single patient. Even rare mutations were, however, embedded in viral sequence with the highest homology by BLASTN search to other sequences derived from the same patient and are therefore unlikely to represent PCR contamination from other patient samples or laboratory strains.
Many of the Phase II patients in whom efavirenz resistance has been characterized were treated with regimens which would be considered suboptimal by present standards. Two weeks of efavirenz monotherapy prior to the initiation of combination therapy, suboptimal doses of efavirenz (e.g., 200 or 400 mg q.d.), and addition of efavirenz as a single new antiretroviral agent to a submaximally suppressive regimen (e.g., ZDV-3TC-experienced patients with active viral replication in study DMP 266-004) each led to virological treatment failure and the emergence of efavirenz-resistant virus in some patients. Nevertheless, the pattern of NNRTI resistance mutations selected in these patients appears to be similar to that observed in patients treated with the recommended clinical dose of 600 mg of efavirenz q.d. In addition, the pattern of resistance mutations associated with efavirenz treatment failure was similar in regimens combining efavirenz with a PI (indinavir) or dual nucleoside analogs (ZDV-3TC or D4T-3TC). In contrast, different patterns of resistance mutations have been reported for patients for whom nevirapine combination therapy failed, depending upon the coadministered nucleoside analogs. While nevirapine monotherapy rapidly selects for Y181C mutations, mutations at RT positions 103, 106, 188, and 190 were observed in patients failing nevirapine plus ZDV combination therapy, while fewer mutations were found at position 181 (6, 15). This result may be explained in part by the observation that the Y181C mutation significantly enhances susceptibility to AZT in molecular constructs carrying this mutation in an AZT resistant background (4, 18). Efavirenz is fully active in vitro against virus constructs with a Y181C mutation (33; L. T. Bacheler, unpublished data). Thus, Y181C mutations are unlikely to be selected by efavirenz combination therapy, with or without coadministered ZDV.
Like other NNRTIs studied in vivo, efavirenz appears to rapidly select for resistant viruses when used in suboptimally suppressive regimens. More than half of the patients treated with efavirenz monotherapy (200 mg q.d.) for 14 days had developed resistance mutations at the end of this short monotherapy period; the mutations were detected by our clonal sequencing approach, albeit as minority components of the virus population. Some patients in study DMP 266-004, who added efavirenz at 200 or 400 mg q.d. to a preexisting dual nucleoside regimen which was not maximally suppressing HIV replication also experienced early viral load rebound associated with the emergence of NNRTI resistance mutations. Both of these observations highlight the importance of avoiding the use of actual or functional efavirenz monotherapy, such as when efavirenz is added as a single agent to a failing drug regimen, in order to avoid the selection of resistant viruses.
While the point mutation K103N was most often the first NNRTI resistance mutation observed in patients for whom efavirenz combination therapy failed, the majority of patients studied went on to develop viruses carrying multiple mutations. Indeed, a number of the most prevalent mutations observed (V108I, P225H, K101E or K101Q, and L100I) were almost exclusively observed as double mutants in combination with K103N. In cases where the effect on drug susceptibility of such double mutants has been tested, the double mutants are associated with higher levels of resistance to efavirenz. (L. T. Bacheler, unpublished data). These data suggest that efavirenz continues to exert selective pressure on K103N mutant strains, resulting in the eventual selection of double- and even triple-mutant viruses which have higher levels of drug resistance than that of a K103N single-mutant virus.
The spectrum of resistance mutations observed in vivo in patients for whom efavirenz combination therapy failed, as well as their relative frequency, was substantially different from that observed in cell culture selection experiments. The L100I-K103N double mutant selected in cell culture (33) was relatively infrequently observed in vivo, occurring in 10.6% of virological failures. In the selection experiments described by Winslow et al. (32) viruses with mutations at L100, V108, V179, and Y181 were described. In vivo, the L100I mutation was seen only in combination with K103N, and V108I mutations were observed largely in combination with K103N. V179D and Y181C mutations were extremely rare in plasma virus from patients exposed to efavirenz. The results of our characterization of the genotypic correlates of resistance to efavirenz in vivo emphasize that cell culture selection experiments are often not predictive of the types or frequencies of resistance mutations selected in patients receiving antiretroviral therapy. Similar discordancies between resistance mutations selected in vivo and in vitro have been described for delavirdine (11) and the experimental NNRTI HBY 097 (26).
ACKNOWLEDGMENTS
We thank Jon Condra and Emilio Emini of Merck Research Laboratories for discussions and initial sequencing.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Balzarini J, Pelemans H, Esnouf R, De Clercq E. A novel mutation (F227L) arises in the reverse transcriptase of human immunodeficiency virus type 1 on dose-escalating treatment of HIV type 1-infected cell cultures with the nonnucleoside reverse transcriptase inhibitor thiocarboxanilide UC-781. AIDS Res Hum Retrovir. 1998;14:255–260. doi: 10.1089/aid.1998.14.255. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 1983;100:243–255. doi: 10.1016/0076-6879(83)00059-2. [DOI] [PubMed] [Google Scholar]
- 4.Byrnes V W, Sardana V V, Schleif W A, Condra J H, Waterbury J A, Wolfgang J A, Long W J, Schneider C L, Schlabach A J, Wolanski B S. Comprehensive mutant enzyme and viral variant assessment of human immunodeficiency virus type 1 reverse transcriptase resistance to nonnucleoside inhibitors. Antimicrob Agents Chemother. 1993;37:1576–1579. doi: 10.1128/aac.37.8.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr A, Vella S, de Jong M D, Sorice F, Imrie A, Boucher C A, Cooper D A. Controlled trial of nevirapine plus zidovudine versus zidovudine alone in p24 antigenaemic HIV-infected patients. The Dutch-Italian-Australian Nevirapine Study Group. AIDS. 1996;10:635–641. doi: 10.1097/00002030-199606000-00009. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman S H, Havlir D, McLaughlin M M, Greenough T C, Sullivan J L, Hall D, Hattox S E, Spector S A, Stein D S, Myers M, Richman D D. Phase I/II evaluation of nevirapine alone and in combination with zidovudine for infection with human immunodeficiency virus. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:141–151. [PubMed] [Google Scholar]
- 7.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yand T, Teppler H, Squires K E, Deutsch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 8.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Yang T, Chodakewitz J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E A. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Aquila R T, Hughes M D, Johnson V A, Fischl M A, Sommadossi J P, Liou S H, Timpone J, Myers M, Basgoz N, Niu M, Hirsch M S. Nevirapine, zidovudine, and didanosine compared with zidovudine and didanosine in patients with HIV-1 infection. A randomized, double-blind, placebo-controlled trial. National Institute of Allergy and Infectious Diseases AIDS Clinical Trials Group Protocol 241 Investigators. Ann Intern Med. 1996;124:1019–1030. doi: 10.7326/0003-4819-124-12-199606150-00001. [DOI] [PubMed] [Google Scholar]
- 10.DeAngelis M M, Wang D G, Hawkins T L. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demeter L M, Shafer R W, Meehan P M, Holden-Wiltse J, Fischl M A, Freimuth W W, Para M F, Reichman R C. Delavirdine susceptibilities and associated reverse transcriptase mutations in human immunodeficiency virus type 1 isolates from patients in a phase I/II trial of delavirdinemonotherapy (ACTG 260) Antimicrob Agents Chemother. 2000;44:794–797. doi: 10.1128/aac.44.3.794-797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedland G H, Pollard R, Griffith B, Hughes M, Morse G, Bassett R, Freimuth W, Demeter L, Connick E, Nevin T, Hirsch M, Fischl M. Efficacy and safety of delavirdine mesylate with zidovudine and didanosine compared with two-drug combinations of these agents in persons with HIV disease with CD4 counts of 100 to 500 cells/mm3 (ACTG 261). ACTG 261 Team. J Acquir Immune Defic Syndr. 1999;21:281–292. doi: 10.1097/00126334-199908010-00005. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Havlir D, Cheeseman S H, McLaughlin M, Murphy R, Erice A, Spector S A, Greenough T C, Sullivan J L, Hall D, Myers M, Lamson M, Richman D D. High-dose nevirapine: safety, pharmacokinetics, and antiviral effect in patients with human immunodeficiency virus infection. J Infect Dis. 1995;171:537–545. doi: 10.1093/infdis/171.3.537. [DOI] [PubMed] [Google Scholar]
- 15.Havlir D, McLaughlin M M, Richman D D. A pilot study to evaluate the development of resistance to nevirapine in asymptomatic human immunodeficiency virus-infected patients with CD4 cell counts of >500/mm3: AIDS Clinical Trials Group Protocol 208. J Infect Dis. 1995;172:1379–1383. doi: 10.1093/infdis/172.5.1379. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch M S, Conway B, D'Aquila R, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobson D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 17.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 18.Larder B A. 3′-Azido-3′-deoxythymidine resistance suppressed by a mutation conferring human immunodeficiency virus type 1 resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1992;36:2664–2669. doi: 10.1128/aac.36.12.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Picado J, Sutton L, De Pasquale M P, Savara A V, D'Aquila R. Human immunodeficiency virus type 1 cloning vectors for antiretroviral resistance testing J. Clin Microbiol. 1999;37:2943–2951. doi: 10.1128/jcm.37.9.2943-2951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellors J W, Dutschman G E, Im G J, Tramontano E, Winkler S R, Cheng Y C. In vitro selection and molecular characterization of human immunodeficiency virus-1 resistant to non-nucleoside inhibitors of reverse transcriptase. Mol Pharmacol. 1992;41:446–451. [PubMed] [Google Scholar]
- 21.Montaner J S, Reiss P, Cooper D, Vella S, Harris M, Conway B, Wainberg M A, Smith D, Robinson P, Hall D, Myers M, Lange J M. A randomized, double-blind trial comparing combinations of nevirapine, didanosine, and zidovudine for HIV-infected patients: the INCAS trial. Italy, The Netherlands, Canada and Australia Study. JAMA. 1998;279:930–937. doi: 10.1001/jama.279.12.930. [DOI] [PubMed] [Google Scholar]
- 22.Myers G, Korber B, Wain-Hobson S, Smith R F, editors. Human retroviruses and AIDS 1993: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.M: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1993. [Google Scholar]
- 23.Pelemans H, Esnouf R, Dunkler A, Parniak M A, Vandamme A M, Karlsson A, De Clercq E, Kleim J P, Balzarini J. Characteristics of the Pro225His mutation in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase that appears under selective pressure of dose-escalating quinoxaline treatment of HIV-1. J Virol. 1997;71:8195–8203. doi: 10.1128/jvi.71.11.8195-8203.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richman D, Shih C K, Lowy I, Rose J, Prodanovich P, Goff S, Griffin J. Human immunodeficiency virus type 1 mutants resistant to nonnucleoside inhibitors of reverse transcriptase arise in tissue culture. Proc Natl Acad Sci USA. 1991;88:11241–11245. doi: 10.1073/pnas.88.24.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubsamen-Waigmann H, Huguenel E, Shah A, Paessens A, Ruoff H J, von Briesen H, Immelmann A, Dietrich U, Wainberg M A. Resistance mutations selected in vivo under therapy with anti-HIV drug HBY 097 differ from resistance pattern selected in vitro. Antivir Res. 1999;42:15–24. doi: 10.1016/s0166-3542(99)00010-8. [DOI] [PubMed] [Google Scholar]
- 27.Schinazi R F, Larder B A, Mellors J W. Mutations in retroviral genes associated with drug resistance. Int Antivir News. 1996;4:95–107. [Google Scholar]
- 28.Smerdon S J, Jager J, Wang J, Kohlstaedt L A, Chirino A J, Friedman J M, Rice P A, Steitz T A. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1994;91:3911–3915. doi: 10.1073/pnas.91.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starr S E, Fletcher C V, Spector S A, Yong F H, Fenton T, Brundage R C, Manion D, Ruiz N, Gersten M, Becker M, McNamara J, Mofenson L M. Combination therapy with efavirenz, nelfinavir, and nucleoside reverse-transcriptase inhibitors in children infected with human immunodeficiency virus type 1. N Engl J Med. 1999;341:1874. doi: 10.1056/NEJM199912163412502. [DOI] [PubMed] [Google Scholar]
- 30.Staszewski S, Morales-Ramirez J, Tashima K T, Rachlis A, Skiest D, Stanford J, Stryker R, Johnson P, Labriola D F, Farina D, Manion D J, Ruiz N M. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir, and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. N Engl J Med. 1999;341:1865. doi: 10.1056/NEJM199912163412501. [DOI] [PubMed] [Google Scholar]
- 31.Vandamme A M, Debyser Z, Pauwels R, De Vreese K, Goubau P, Youle M, Gazzard B, Stoffels P A, Cauwenbergh G F, Anne J, et al. Characterization of HIV-1 strains isolated from patients treated with TIBO R82913. AIDS Res Hum Retrovir. 1994;10:39–46. doi: 10.1089/aid.1994.10.39. [DOI] [PubMed] [Google Scholar]
- 32.Winslow D L, Garber S, Reid C, Scarnati H, Baker D, Rayner M M, Anton E D. Selection conditions affect the evolution of specific mutations in the reverse transcriptase gene associated with resistance to DMP 266. AIDS. 1996;10:1205–1209. doi: 10.1097/00002030-199609000-00005. [DOI] [PubMed] [Google Scholar]
- 33.Young S D, Britcher S F, Tran L O, Payne L S, Lumma W C, Lyle T A, Huff J R, Anderson P S, Olsen D B, Carroll S S. L-743, 726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39:2602–2605. doi: 10.1128/aac.39.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]