Abstract
In this paper, the first robust experimental evidence of in vitro and in vivo concentration-dependent selection of low-level antibiotic-resistant genetic variants is described. The work is based on the study of an asymmetric competition assay with pairs of isogenic Escherichia coli strains, differing only (apart from a neutral chromosomal marker) in a single amino acid replacement in a plasmid-mediated TEM-1 beta-lactamase enzyme, which results in the new TEM-12 beta-lactamase. The mixture was challenged by different antibiotic concentrations, both in vitro and in the animal model, and the selective process of the variant population was carefully monitored. A mathematical model was constructed to test the hypothesis that measured growth and killing rates of the individual TEM variants at different antibiotic concentrations could be used to predict quantitatively the strength of selection for TEM-12 observed in competition experiments at these different concentrations.
The bacterial development of antibiotic resistance is one of the best-documented examples of contemporary biological evolution. After half a century of massive, largely uncontrolled release of industrial antibiotics around the world, microbial populations have developed a wide variety of mechanisms of resistance. TEM-1 (and TEM-2) beta-lactamases were detected among resistant gram-negative organisms in the 1960s, shortly after the introduction of ampicillin in the clinical armamentarium (9). During the last two decades, nearly 60 new molecular variants of these early enzymes, showing altered substrate specificity, have been described (http://www.lahey.org/studies/temtable.htm). This variety represents a unique example of protein evolution in “real time” (26). Such diversification was probably a consequence of an equivalent diversification of selective challenges resulting from the introduction of multiple beta-lactam antibiotic molecules designed to resist hydrolysis by the TEM-1 enzyme, in particular broad-spectrum cephalosporins (such as cefotaxime) and beta-lactamase inhibitors.
Among the more efficient new TEM variants that have evolved to hydrolyze cefotaxime are those which differ from the earlier molecules by several amino acids. Assuming that mutation rates in Escherichia coli are on the order of 10−10 per base pair per generation, it is unlikely that two or more point mutations would appear simultaneously in a beta-lactamase gene. Therefore, if the TEM-1 beta-lactamase is the ancestor of these multiply mutated variants, it is most likely that the variants arose by a process of sequential point mutation and selection of singly mutated intermediates. For such a scenario to be plausible, each mutation would need to confer a selective advantage over the ancestral strain. In many cases, strains with monomutated TEM-1 enzymes (such as TEM-12, resulting from a single substitution of arginine for serine at position 164) exhibit only a very small increase in resistance to cefotaxime. Typically, TEM-1-producing E. coli is inhibited by 0.008 μg/ml, and TEM-12-producing E. coli is inhibited by 0.015 μg/ml. Despite such a small phenotypic difference, TEM-12-containing strains may have been selected by cefotaxime use, thereby providing the genetic background for double-mutated, more efficient enzymes, such as TEM-10 (in which glutamic acid replaces lysine at position 240), which confers resistance at cefotaxime concentrations up to 0.25 μg/ml (6).
The hypothesis that the selection of resistant genetic variants is driven by precise selective antibiotic concentrations occurring at particular spatial locations (selective compartments) has been previously proposed by our group (1, 2, 3, 24). In the work described in this report, we hypothesized that selection of strains with increased levels of resistance to a drug will occur at a particular drug concentration in proportion to the difference in the growth (or killing) rates between the two strains at that concentration and in proportion to the duration of exposure. Furthermore, we hypothesized that independent measurements of the growth and killing rates of two bacterial strains could be used to predict quantitatively the selective increase in the more resistant strain when the strains were competing at a particular drug concentration. One prediction of these hypotheses is that there would be a range of concentrations, corresponding to the maximum differences in bacterial growth and killing rates, at which selection would be the most intense; we call this range of concentrations a “selective window.” This paper describes experimental and mathematical modeling work designed to test these hypotheses. The first robust experimental evidence of in vitro and in vivo concentration-dependent selection of low-level antibiotic-resistant genetic variants was obtained.
MATERIALS AND METHODS
Bacterial strains and plasmids.
For in vitro studies, the baseline strain was E. coli B REL606, which has been used in several other evolutionary studies (17, 18, 19, 30). This strain is prototrophic but unable to grow on l-arabinose (Ara−). A spontaneous Ara+ mutant, REL607, was previously obtained by Lenski (18). The Ara− and Ara+ clones form red and light-pink colonies, respectively, on tetrazolium-arabinose (TA) indicator plates (19), making it easy to score the relative frequencies of these markers when a mixed population is plated. The neutrality of the arabinose marker in our experimental conditions was confirmed; as in the original work of Lenski (18), the ratio of fitnesses between the two clones was 1.00 ± 0.01 (95% confidence interval). The baseline plasmid was the nonconjugative plasmid pBGS19− (30); the derivatives pBGTEM-1 and pBGTEM-12 were obtained, respectively, carrying the blaTEM-1 and blaTEM-12 beta-lactamase genes. pBGTEM-1 was constructed by cloning a BamHI-BamHI fragment from the plasmid pKT254Ω-Ap (11) containing the blaT1 (TEM-1 beta-lactamase) gene from Tn3 into the BamHI site of the phage M13mp19. An EcoRI-SalI fragment from the hybrid phage M13mpΩ-Ap was cloned in plasmid pBGS19−, digested with the same restriction enzymes. This new hybrid plasmid was named pBGTEM-1. The Arg164→Ser (pBGTEM-12) mutant was constructed by site-directed mutagenesis (4). The plasmids pBGTEM-1 and pBGTEM-12 were transformed into both the REL606 and the REL607 strains. The cefotaxime MICs for these REL606 and REL607 derivatives were repeatedly studied (in eight independent experiments) by conventional susceptibility testing procedures (23); for strains containing TEM-1, MICs repeatedly were 0.008 μg/ml, and for TEM-12 strains, MICs were 0.015 to 0.03 μg/ml. A different pair of E. coli strains was used for animal experiments. The thigh model (see below) of infection in mice required a challenge with E. coli derivatives lacking the capsular K1 antigen in order to reduce massive bacterial invasion and mortality. The K1− strain E. coli CAB281 (a K1-K12 chimera) (28), which is nalidixic acid-susceptible (Nals), and its nalidixic acid-resistant derivative CAB281 (Nalr) were transformed with the plasmids pBGTEM-1 and pBGTEM-12, respectively. The cefotaxime MICs for the resulting strains were 0.12 and 0.25 μg/ml, respectively.
Effect of cefotaxime on bacterial growth rate in vitro.
To measure the activity of different cefotaxime concentrations on the growth rates of REL606(pBGTEM-1) and REL607(pBGTEM-12), overnight cultures of both strains in Mueller-Hinton broth (Oxoid Ltd., Basingstoke, England) were prepared. Two 250-ml flasks containing 40 ml of Mueller-Hinton broth and cefotaxime concentrations increasing in twofold increments from 0.004 to 0.5 μg/ml (and the appropriate control without antibiotic) were inoculated with 106 CFU/ml and incubated with shaking (170 rpm) at 37°C for 6 h. Every hour, aliquots were obtained, appropriately diluted, and plated onto TA indicator plates. After 24 h of incubation at 37°C, the colonies were counted. Resulting colony counts for the 2-, 3-, 4-, and 5-h time points, corresponding to the exponential phase of the organisms grown with and without antibiotic, were converted into growth rates, calculated as the linear regression of the natural logarithm of bacterial density versus time. Exactly the same procedure was applied to the OmpF− derivatives of REL606(pBGTEM-1) and REL607(pBGTEM-12) that were obtained during the experiment.
In vitro selection models.
In the basic selection assays, the two competing REL606 and REL607 E. coli strains, containing either pBGTEM-1 or pBGTEM-12, were grown separately overnight in Mueller-Hinton broth, reaching ca. 5 × 108 CFU/ml. After this time, the mixtures were prepared. The proportions of the mixtures were intended to reflect the predominance of the wild TEM-1-harboring population, with a lesser representation of the single-mutated TEM-12 population (volumetric ratio, 99:1). Tubes containing 5 ml of Mueller-Hinton broth with different cefotaxime concentrations (range: 0 [control] and 0.004 to 0.5 μg/ml) were inoculated with 0.1 ml of a dilution of the bacterial mixture to obtain an inoculum of approximately 106 CFU/ml. In a series of experiments, the two competitors were allowed to grow together for 4 h at 37°C to mimic the expected mean contact period of bacterial populations with such a range of concentrations in the human body. Note that very low concentrations are expected to be maintained for a much longer time than high concentrations after a single antibiotic dosage. A 200-μl aliquot was taken, and selection as measured by changes in the frequencies of TEM-1- and TEM-12-bearing strains was then scored as described below. The original mixture was maintained in antibiotic-containing culture tubes (and controls) for a total of 24 h. Cultures were propagated during 5 days by transferring daily 0.1 ml from each tube to fresh Mueller-Hinton tubes containing the same cefotaxime concentrations. At the fifth day, aliquots were obtained, and selection was scored. Selection was measured as follows. At each time point (4 h and 5 days), a 200-μl aliquot from each tube was taken and treated with 100 mU of beta-lactamase type IV from Enterobacter cloacae (Sigma Chemical) for 20 min to prevent antibiotic carryover effects. The surviving cells of the original populations were then allowed to grow overnight in a fresh, antibiotic-free Mueller-Hinton medium to obtain countable numbers of cells. This amplification period might imitate the expected recolonization process after chemotherapy. After 18 h of incubation (in the 4-h case), and subsequently after every 24 h of incubation (in the 5-day case), the relative densities of each competitor were estimated by seeding dilutions of the mixed culture on TA indicator plates. The differences in survival rates among competitors were estimated in percentages of each Ara+ and Ara− strain and expressed as total selection rates (r) for TEM-12-harboring strains, calculated according to the formula:
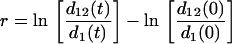 |
1 |
where d1 and d12 represent the densities of TEM-1- and TEM-12-bearing bacteria, respectively. Because of the practical limitations in colony counting, the maximal observable r value (100 colonies of one of the competitors and less than 1 colony from the other) was considered to be 9.2. The data were obtained from six replicate experiments at each concentration.
Every experiment was done in a direct and an inverse way, using the mixtures of E. coli strains REL606(pBGTEM-1) + REL607(pBGTEM-12) and REL606(pBGTEM-12) + REL607(pBGTEM-1), to ascertain the neutrality of bacterial hosts and plasmids. Moreover, these strains were cocultivated (at a 1:1 volumetric ratio) and transferred daily for 5 days in antibiotic-free Mueller-Hinton broth; no significant change from the original densities was detectable. In each experiment, a control tube was incubated without antibiotic, and the original 99:1 proportion was also maintained after the different periods of observation (4 h and 5 days). In a complementary and independent four-series experiment, directed to evaluate the effect of the initial inoculum density, the initial cell density was reduced from 104 to 103 CFU/ml in the strains harboring pBGTEM-12 and from 106 to 105 CFU/ml in the strains with pBGTEM-1. Finally, in a two-series experiment, the same bacterial mixtures were allowed to grow on cefotaxime-containing tubes for 24 h, and then 0.1 ml from each tube was transferred directly to fresh tubes containing the same antibiotic concentrations; this procedure was repeated for three days. At 4, 24, 48, and 72 h, 0.2 ml from each tube was treated with beta-lactamase (to prevent carryover), appropriately diluted, and then plated on TA plates. The same basic protocol was applied to control the strains to be used in the animal model, now using experiments that were replicated three times. The E. coli CAB281 strain and its nalidixic acid-resistant derivative CAB281 Nalr harbored the plasmids pBGTEM-1 and pBGTEM-12, respectively. Also, in this case, the nalidixic acid resistance marker proved to be essentially neutral for in vivo competition experiments.
Characterization of strains after cefotaxime challenge.
Samples of at least 10 colonies isolated from cultures exposed to each of the different cefotaxime concentrations (five Ara+ and five Ara−) were screened for the OmpF− phenotype. A reduction of at least 30% of the diameter of the inhibition zone around a cefoxitin 30-μg disk (Oxoid) in comparison with the original strain was considered a presumptive OmpF− phenotype. In these strains, diminished susceptibility to microcin B17 (this antibiotic peptide requires OmpF for internalization) and phage TuIa (OmpF is the phage receptor in the bacterial outer membrane) were also screened as secondary markers (8, 21). Inhibition-resistant cultures are expected to be OmpF−. In all cases, known OmpF− and OmpF+ control strains were used. For a comprehensive sample of 20 presumptive mutant strains, direct detection of absence of the OmpF band in polyacrylamide gel electrophoresis of outer membrane preparations (25, 27) was performed. With these strains, plasmid DNA was obtained and transformed in the wild-type REL606 or REL607 strain to exclude the presence of changes in cefotaxime and/or ceftazidime susceptibility that may result from new mutations in the TEM enzyme. With an identical purpose, with at least two strains that were recovered from the three higher cefotaxime concentrations of each experiment, isoelectric focusing of extracted beta-lactamases was performed. A pattern of decreased susceptibility to cefoxitin, cefotaxime, and ceftazidime, in the absence of an OmpF− phenotype (see above), was considered as indicative of AmpC hyperproduction. Both the REL606 and the REL607 strains are OmpC− (31), so changes in phenotype cannot be attributed to this porin. With a pair of well-characterized OmpF− mutants of the REL606 strain containing either pBGTEM-1 or pBGTEM-12, growth rates in the presence of different cefotaxime concentrations were determined, in a way identical to that reported above.
In vivo selection experiments.
To test the effect of concentration-dependent selection in vivo, competition experiments were performed in ICR outbred mice. A mixture of approximately 108 CFU of CAB281(pBGTEM-1) and approximately 105 CFU/ml of CAB281Nalr (pBGTEM-12) (ratio, 1,000:1) was inoculated into the right thighs of four mice per group, and the exact inoculum composition was verified by selective plating. Six doses of cefotaxime sodium were administered intraperitoneally at 2-h intervals following the inoculation. Doses ranged from 1.56 μg to 6.4 mg q2h, in fourfold intervals, corresponding to doses of 0.06 to 256 mg/kg of body weight. Six hours after the last dose, each mouse was sacrificed, and its entire right gastrocnemius muscle was removed aseptically and homogenized in 2 ml of 0.85% saline. Bacterial densities in this suspension were estimated by serial dilution. Antibiotic efficacy against each strain (containing either TEM-1 or TEM-12) at each concentration was calculated as the difference in the natural logarithm of bacterial density in animals given each antibiotic concentration compared to controls. Selection was calculated as the change in the natural logarithm of the ratio of TEM-12- to TEM-1-bearing bacteria in the ex vivo samples compared to the inoculum.
The mathematical model.
A mathematical model was used to determine the extent to which differential growth and killing rates for the TEM-1 and TEM-12 variants, measured separately, could account for the outcomes observed in the selection (competition) experiments. The growth rates were estimated as ordinary regression coefficients of ln (cell density) versus time measured for TEM-1 and TEM-12 growth in Mueller-Hinton broth with antibiotics at various concentrations. Time points from 2, 3, 4, and 5 h were used to obtain exponential growth rates, free from the effects of lag or resource depletion. The resulting growth rates are shown in Fig. 4. Analogous experiments and calculations were performed for TEM-1 OmpF− and TEM-12 Omp-F− variants.
FIG. 4.
Growth rates of the strains REL606(pBGTEM-1) (black rhombus), REL607(pBGTEM-12) (black squares), REL606(pBGTEM-1) OmpF (dashed line and black triangles), and REL607(pBGTEM-12) OmpF (dashed line and black squares) at different cefotaxime concentrations. The points correspond to the means of two separate experiments.
Competition experiments were modeled by assuming initial densities of TEM-1 and TEM-12 bacteria of 106 and 104, respectively. At the start of the first day of the serial transfer “experiment,” bacteria were assumed to begin growing (or dying) immediately at rates specified by the parameter estimates described above (lag time was ignored) and to continue until the end of the transfer (24 h) or until they reached a total density of 109 bacteria. Serial transfer was simulated by a reduction of 50-fold in both bacterial populations, which were then restarted in the same volume of medium. Stochastic loss of rare subpopulations was simulated by setting any population whose size was less than 1 at the time of transfer equal to 0 (extinction).
Total selection coefficients were calculated in the mathematical model for both 4 h and 5 days, using equation 1.
The lack of fit of this basic model with the data from 5 days, in conjunction with the observation of OmpF− bacterial mutants in many of the experiments at higher concentrations, led us to expand the model to include the possibility of emergence of such mutants under cefotaxime selection. In the expanded model, the TEM-1 and TEM-12 populations each consisted of a majority subpopulation of OmpF+ cells and a minority population, present at a frequency of 10−4, of OmpF− cells. Concentration-dependent growth (or killing) rates for the TEM-1 OmpF− and TEM-12 OmpF− cells were estimated from growth experiments as before, and their growth rates were incorporated into the competition model, so that four separate populations were competing at each concentration. As before, selection was calculated at 4 h and 5 days, this time using the total TEM-1-bearing cells (OmpF− and OmpF+) and the total TEM-12-bearing cells (OmpF− and OmpF+).
RESULTS
Selective window: a short range of cefotaxime concentrations selects TEM-12- over TEM-1-harboring strains after 4 h of challenge.
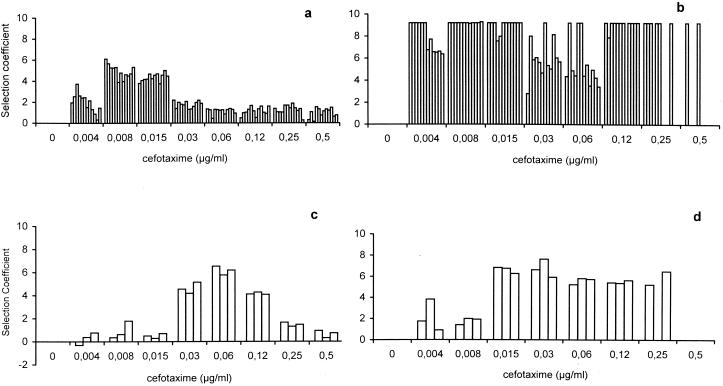
The strains REL606(pBGTEM-1) and REL607(pBGTEM-12) were mixed at a 99:1 proportion, and the mixture was challenged with different cefotaxime concentrations. The same experiment was done in reverse, mixing REL607(pBGTEM-1) with REL606(pBGTEM-12) at a 99:1 proportion. Increases in the proportions of the E. coli organisms carrying TEM-12 were observed at all concentrations after a challenge of 4 h with the antibiotic, followed by 18 h of culturing in drug-free medium. In contrast, control cultures without antibiotic challenge maintained the original proportions of the inoculum. Selection of the TEM-12-bearing strain was particularly strong at very low antibiotic concentrations (0.008 to 0.015 μg/ml) (Fig. 1a). At these concentrations, the original percentage of strains carrying pBGTEM-12 in the final population rose from 1 to about 50 (r = 4 to 6). At a slightly higher cefotaxime concentration, 0.03 μg/ml, the selection of pBGTEM-12-harboring strains appeared to be much less effective, and the pBGTEM-1-harboring population was maintained at a very high proportion in the mixture. The TEM-1-harboring strain remained predominant at higher cefotaxime concentrations, up to 0.5 μg/ml. Below the selective concentrations, at 0.004 μg/ml, pBGTEM-12 strains also were selected with low efficiency. Overall reproducibility of the results was good, not only in direct and inverse experiments but also among the six replicate experiments carried out with each pair of strains. A qualitatively identical effect was obtained with the pair of E. coli CAB281 strains harboring pBGTEM-1 or pBGTEM-12 (Fig. 1c). In this case, the TEM-12-containing population was selected within a short range of cefotaxime concentrations of 0.03 to 0.12 μg/ml. These experiments confirm the existence of selective windows, which may differ for different strains and genotypes, for resistant populations that were originally in the minority. Within these windows, small phenotypic differences between competing populations can be selected with extremely high efficiency. The appearance of a single sharp selective peak appears to be distinctive of the antibiotic selective window for a particular mutant strain.
FIG. 1.
The top graphs show selection rates for an E. coli strain (REL606 or REL607) harboring the plasmid pBGTEM-12 (size of inoculum, 104 CFU/ml) in competition with the isogenic strain harboring pBGTEM-1 (REL607 or REL606; size of inoculum, 106 CFU/ml) in the presence of cefotaxime. For each antibiotic concentration, 12 replicated experiments are represented (6 for the direct and 6 for the inverse competitions). (a) Selection after 4 h of cefotaxime exposure. (b) Selection after 5 days of continuous challenge with each cefotaxime concentration. The bottom graphs show selection rates for the E. coli CAB281Nalr strain harboring pBGTEM-12, in competition with CAB281Nals harboring pBGTEM-1. (c) Selection after 4 h of cefotaxime exposure. (d) Selection after 5 days of continuous challenge with each cefotaxime concentration.
A second peak of selection of the TEM-12 strain may emerge after prolonged challenge with higher cefotaxime concentrations.
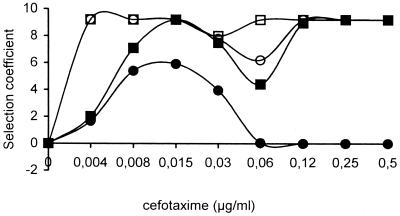
As one would expect, 5-day passages of the mixed REL606 and REL607 populations harboring TEM-1 or TEM-12 in tubes containing cefotaxime resulted in heavier selection of the strain with TEM-12 at the selective window concentrations (0.008 to 0.015 μg/ml). At these concentrations, there was an almost complete replacement of TEM-1 strains by TEM-12 strains. Some selection of the TEM-12 variant also occurred at the concentrations just below and just above this window, 0.004 and 0.03 μg/ml, albeit at lower rates (r = 6 to 7). (Fig. 1b). Unexpectedly, a second peak of high-efficiency selection of the TEM-12 strain appeared at higher cefotaxime concentrations (0.12 to 0.25 μg/ml). The first version of the mathematical model (see below) totally failed to predict the appearance of this second peak, using just the growth parameters of the original TEM-1- and TEM-12-bearing strains. According to the selective window hypothesis, such a second peak should correspond to the selection of a different bacterial subpopulation with a higher level of antibiotic resistance than the original TEM-1- or TEM-12-harboring strains. A similar result was obtained with the CAB281 strains (Fig. 1d). Experiments were carried out to ascertain the dynamics of emergence of this second TEM-12-harboring population. The degree of selection for TEM-12 over time in the presence of cefotaxime is shown in Fig. 2. The secondary peak is already evident after 24 h of exposure with relatively high cefotaxime concentrations (0.12 to 0.5 μg/ml) and tends to move to the left with further incubation in the presence of the antibiotic.
FIG. 2.
Evolution in time of the selection rates for E. coli REL606 strains harboring pBGTEM-12 in competition with REL607 harboring pBGTEM-1 in the presence of cefotaxime. Proportion in the original inoculum was 104/106 CFU/ml. Black circles, 4 h of exposure; black squares, 24 h; white circles, 48 h; white squares, 72 h.
The second peak of selection of the TEM-12 strain corresponds to a new subpopulation formed by OmpF− secondary mutants.
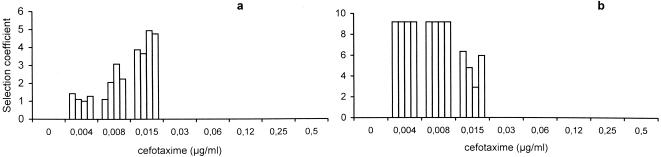
If the second peak of selection originated from the progressive enrichment of a minority subpopulation of secondary mutants among the original TEM-12-harboring population, a reduction in the original density of this population should diminish the probability of emergence of the secondary peak. Competition experiments were repeated with smaller starting inocula of the mixture of E. coli REL607(pBGTEM-12) plus REL606(pBGTEM-1) that was reduced 10 times, respectively, from 104 + 106 to 103 + 105. Under these circumstances, the first REL607(pBGTEM-12) selection peak at 0.004 to 0.015 μg/ml was again reproduced, after a challenge of 4 h (Fig. 3). After 5 days of continuous challenge at the same cefotaxime concentrations, the first peak of TEM-12 selection was maintained, but the second peak of selection at higher antibiotic concentrations (0.12 to 0.25 μg/ml) completely disappeared in the low-population-density bacterial mixture, consistent with the hypothesis of the secondary mutant subpopulation.
FIG. 3.
Effect of a low inoculum on selection rates of E. coli strain REL606 harboring the plasmid pBGTEM-12 (size of inoculum, 103 CFU/ml), in competition with the isogenic REL607 strain harboring pBGTEM-1 (size of inoculum, 105 CFU/ml) in the presence of different cefotaxime concentrations. Selection rates after 4 h of challenge (a) and after 5 days of continuous challenge (b) with cefotaxime are shown. For each antibiotic concentration, results from four replicated experiments are represented.
To further test this hypothesis, a comprehensive sample (see Materials and Methods) of REL606 and REL607 strains harboring pBGTEM-12 and corresponding to the first and second peaks of selection was examined for the presence of the OmpF− phenotype using a cefoxitin susceptibility screening test. All positive cases were confirmed by testing for reduced susceptibility to microcin B17 and phage TuIa. The absence of the OmpF band in electrophoretic analysis of outer membrane proteins was documented in all 20 strains that were analyzed among those exhibiting the OmpF− phenotype. All strains with the OmpF− phenotype had reduced cefoxitin susceptibility. Conversely, no reduction in cefoxitin (or cefotaxime and ceftazidime) susceptibility was found among strains lacking the OmpF− phenotype, indicating that AmpC hyperproduction was not responsible for the observed decrease in susceptibility. All transformants in REL606 or REL607 wild strains harboring pBGTEM-1 or pBGTEM-12 plasmids obtained from OmpF− strains retained the original susceptibility. No changes in the original pI of the beta-lactamase in experimental strains recovered from the three higher cefotaxime concentrations were detected, and therefore the presence of mutations in the original TEM enzyme was ruled out. In conclusion, every isolate obtained from any TEM-12-harboring population at the second peak of selection (but not at the first peak) was an OmpF− variant. Thus, the second peak corresponds to the selection of a new bacterial genotype.
Growth rates of TEM-1- and TEM-12-harboring strains challenged by low cefotaxime concentrations.
Hourly cell counts of the strains REL606(pBGTEM-1) and REL607(pBGTEM-12) challenged with different cefotaxime concentrations were obtained during the period corresponding to exponential phase (2 to 5 h), and the results were expressed as growth rates (k) (Fig. 4). This experiment clearly shows a differential activity of cefotaxime against REL strains harboring TEM-1 and those harboring TEM-12 that occurs exclusively along a narrow range of low-level antibiotic concentrations (0.008 to 0.06 μg/ml). Two secondary confirmed OmpF− REL mutants presented a different growth curve profile. The REL606(pBGTEM-1)-OmpF− strain was able to survive at one dilution higher than REL607(pBGTEM-12). The OmpF− derivative of REL606(pBGTEM-12) was much more fit and maintained a constant growth rate along the studied cefotaxime concentrations. The MIC of cefotaxime for this variant strain (equivalent to a 0 growth rate) was 4 μg/ml.
Mathematical model: predictions and realities.
A mathematical model was initially constructed simply to predict changes in TEM-1- and TEM-12-bearing populations in competition experiments based on their growth rates in separate cultures (as shown in Fig. 5). The model qualitatively predicted the window selection of the TEM-12-harboring strain after 4 h of antibiotic exposure but totally failed to predict the emergence of a second peak of selection after 5 days of cefotaxime challenge (model results not shown). This result indicated that the initial model was too simple, and it was part of the impetus to look further for heterogeneity in the bacterial populations. When the OmpF− strains were identified and their growth rates were measured, these were incorporated into the model, under the assumption that a single OmpF− mutant was present in the TEM-12 population at the start of the experiments (a mutation rate corresponding to 100 such mutants in the TEM-1 population). The predicted selection rates at 4 h and 5 days from this expanded model are shown in Fig. 5a and 5b, respectively, along with the experimental values.
FIG. 5.
Comparison of selection coefficients of the strain containing TEM-12 as predicted for the mathematical model (squares) and those obtained from data of experimental competition experiments. Competition between homologous E. coli strains containing either pBGTEM-12 or pBGTEM-1 was done in the presence of cefotaxime for 4 h (a) and 5 days (b). Mean (± 1 SD) experimental selection rates for six experiments with TEM-1 in REL606 (triangles) or REL607 (circles) are shown. White squares, model predictions.
With the addition of the OmpF mutants, the model qualitatively predicts the single large peak at low concentrations after 4 h, and it also correctly predicts the second peak after 5 days, which represents the ascent of the TEM-12 OmpF variants. The region between the peaks at 5 days represents a narrow range of concentrations at which OmpF variants are selected in both the TEM-1 and TEM-12 populations. In this range, although the TEM-12 OmpF variants have a higher degree of fitness than the TEM-1 OmpF variants, their fitness advantage is not sufficient after 5 days to compensate fully for their initial numerical disadvantage.
While the fit of the model at 5 days is satisfactory (albeit largely in a range near the limits of detection), the fit at 4 h is qualitative only, rather than quantitative. In particular, the model predicts more selection than actually occurs, and it begins to show a second peak of selection at high concentrations even by 4 h. This overestimation of the amount of selection is not surprising, since the model does not take into account the lag time which typically occurs before bacterial growth and before beta-lactams begin killing. While this lag may be negligible over a period of days, it may be a substantial portion of 4 h.
TEM-12 is selected over TEM-1 in an animal model, with evidence of a selective window.
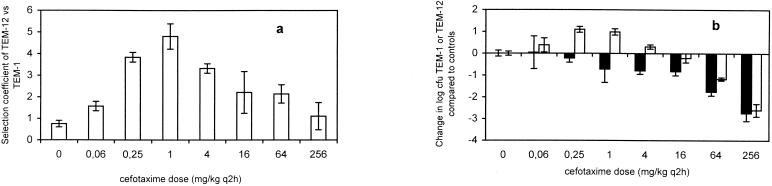
Mice inoculated in the right thigh with an approximately 1,000:1 mixture of TEM-1- and TEM-12-carrying CAB281 strains were treated every 2 h from 2 to 14 h postinoculation with defined doses of cefotaxime. The extent of selection of TEM-12 was calculated as in equation 1 as the change in the natural logarithm of the ratio of TEM-12 to TEM-1 variants among bacteria recovered from the thigh at 20 h postinoculation, compared to the inoculum. Figure 6a shows the average in vivo selection coefficients (± the standard deviation [SD]) for controls and each cefotaxime dose. TEM-12 variants increased in frequency at all doses in the 4,000-fold range tested, and selection was greatest at an intermediate dose, 1 mg/kg. These results are consistent with the existence of an in vivo selective window, albeit one encompassing a wider range of concentrations than the selective window observed in vitro (12).
FIG. 6.
(a) Selection coefficients r (calculated according to equation 1) ± the SD of the E. coli CAB281NalR-TEM-12 strain under in vivo competition experiments with CAB281-TEM-1. Cefotaxime doses are given as mg/kg/dose; six doses were administered q12h. (b) Changes (log10 CFU) in bacteria containing TEM1 (black bars) and TEM12 (white bars) ± the SD at various cefotaxime doses, relative to controls. Most bars are negative, indicating that fewer bacteria were recovered in treated animals. Positive bars indicate that more TEM-12 bacteria were recovered from treated animals than from controls at specified concentrations.
Figure 6b shows the change in the final count of log10 CFU/ml of TEM-1- and TEM-12-carrying CAB281 organisms obtained at different concentrations, relative to results for the controls. Interestingly, at doses of 0.25 and 1 mg/kg, there was approximately a 10-fold increase in the absolute number of TEM-12 variants over that of the controls. This would be unexpected from the action of the antibiotic alone; one would expect that low concentrations of antibiotics would reduce growth rates of both populations but would preferentially reduce the growth rate of TEM-1 organisms. These results suggest that total in vivo bacterial growth was limited by factors other than the antibiotic at low concentrations and in controls, so that antibiotic-mediated killing of TEM-1 bacteria permitted absolute increases in the number of TEM-12 bacteria.
DISCUSSION
The usual pattern for natural selection is to act on very small differences in fitness (29). The evolution of the mechanisms involved in bacterial resistance to antibiotics has probably occurred in many cases by sequential selection of small differences. Unfortunately, clinical microbiologists have frequently disregarded small differences in susceptibility, believing that if the phenotypic effect of a mutation is low, its contribution to the selective advantage should be similarly low, or at best believing that “we do not know how small an effect constitutes a selective advantage” (13). Results presented in this paper indicate that weak selective processes (low antibiotic concentrations) are indeed strong in changing gene frequencies when acting on small differences (small increases in the MIC). It is clear that the environmental pressure of new beta-lactam agents has probably been the cause for the current multiple-enzyme polymorphism in TEM beta-lactamases (5, 21). Nevertheless, the kind of selection pressures involved in the fixation of the different alleles remains obscure. We suggest that they may have occurred within particular selective compartments in the environment, where particular antibiotic concentrations have selected the most fit variant genotypes (1, 2, 3). Such concentrations may occur in the human body during antibiotic therapy.
The experimental results obtained in this work indicate that an E. coli strain harboring a monomutated variant of TEM-1 beta-lactamase, giving rise to the enzyme TEM-12, may be heavily selected over a majority ancestor TEM-1 population along a very short range of very low cefotaxime concentrations after a 4-h exposure. At these cefotaxime concentrations (ranging from 0.008 to 0.015 μg/ml), the strain with TEM-12 maintains a constant growth rate (Fig. 4). At slightly higher concentrations (0.06 to 0.12 μg/ml), the growth rate of the TEM-12 strain also decreases, and the selective activity of cefotaxime has ceased. If at the starting time the TEM-1 strain was in excess (99:1 in the experiments), this susceptible population could still predominate after 4 h of exposure at cefotaxime concentrations that are decreasing in an equivalent way the growth rates of the TEM-1 and TEM-12 strains. When the surviving cells are allowed to recover in drug-free medium after a challenge, the susceptible population will be dominant over the resistant one, in a paradoxical way, at the higher concentrations tested. In short, antibiotic selection is only exerted at precise concentrations (selective concentrations, forming a selective window). During antibiotic therapy, the period during which the antibiotic concentration falls below the MIC provides selective windows in which resistant variants can outcompete their sensitive wild-type counterparts. Indeed, very low antibiotic concentrations may exert a selective effect in wide compartments of the human body after most administrations of an antibiotic. These compartments, where the concentrations correspond to the selective windows, can be considered selective compartments.
Prolonged exposure at the same cefotaxime concentrations increased the proportion of the TEM-12 strain at the selective concentrations and also extended the window in which selection is exerted. After a contact period of 24 h, a second peak of cefotaxime selection emerged at higher concentrations (0.12 to 0.5 μg/ml). This peak corresponded to the TEM-12 strain in which a new mutation arose, now impeding the penetration of the antibiotic throughout the cell outer membrane (OmpF−). This mutation significantly increases the effectiveness of TEM enzymes hydrolyzing beta-lactam agents (4, 27). The emergence of this new mutant population is probably the result of the selection of a preexisting OmpF− variant among the original TEM-12 population. This is supported by the fact that a 10-fold decrease in the starting TEM-12 strain inoculum completely prevents the emergence of the second peak of OmpF− mutants. The rate of mutation for the OmpF phenotype (which may result from mutations in various independent genes) from wild strains is variable, but certainly high, within the range of 10−5 and 10−7. We obtained OmpF mutants by cefoxitin selection (plates containing 8 μg/ml) at this range, confirming classic results (14). OmpF− mutants are also obtained at the same rate, using microcin B17 as selector (16). In the basic experiments described in this paper, total cell numbers of the TEM-12-harboring population in the tubes may be approaching the required number. Moreover, we recently observed that increased rates of mutation may occur in E. coli under stress by sublethal concentrations of broad-spectrum cephalosporins (preliminary results), and such a factor cannot be ruled out from our experiments, in particular at the concentrations where the TEM-12 strain was driven (or nearly driven) to total extinction, precisely where the OmpF mutants produce the second peak. The valley between TEM-12 peaks is probably formed by the concentrations at which OmpF− variants from the TEM-1-harboring strain are selected.
The selection of the different TEM genotypes was considered here as a concentration-dependent phenomenon. Indeed, pharmacodynamics of beta-lactam drugs shows that the antibacterial activity of these compounds is proportional to the time at concentrations above the MIC or to the area under the concentration-time curve above the MIC (7). As shown in Fig. 1, extinctions did not occur in 4 h of challenge with 0.5 μg of cefotaxime per ml but happened after prolonged exposure. As shown in Fig. 2, the shift to the left of the peak composed by the OmpF− TEM-12-harboring population probably occurred because of the differential inhibition of the OmpF− TEM-1 population by more than 24 h of exposure to 0.06 or 0.12 μg/ml. Similarly, the shift to the left of the first OmpF+ TEM-12 population peak shows that low cefotaxime concentrations (such as 0.004 μg/ml) exert a killer effect on the OmpF+ TEM-1 population when maintained for more than 48 h. Nevertheless, the basic concept proposed here is that for every fixed period of time, selection of resistant variants is not a linear function of the drug concentration but occurs at certain selective concentrations, which are specific for each particular genotype.
During antibiotic therapy, gradients of antibiotic concentrations are formed in the human body. These gradients are due to pharmacokinetic factors, such as the different rates of diffusion into various cells or tissues, metabolization, local binding or inactivation, or variation in the rate of elimination from different body compartments. Antibiotic-detoxifying microbial mechanisms (such as beta-lactamases) also contribute to the gradient formation. Due to the extreme compartmentalization of the human body, a diversity of environmental antibiotic pressures is expected to occur in different places, creating different selective compartments, which may lead to the emergence of spatial genetic polymorphisms (2). We have recently suggested (5) that drug concentration heterogeneity may facilitate the evolution of drug resistance, because relatively narrow windows of drug concentrations may allow the local evolution of resistant variants. The concept of the role of selective antibiotic concentrations in the evolution of bacterial drug resistance was previously presented by our group (1, 2, 3, 24) and by others (10), but this work shows the first experimental evidence of strong concentration-dependent selection of a monomutated variant with small phenotypic differences from the wild-type strain.
Some practical consequences may be inferred from our results. The evolution of TEM beta-lactamases provides a relevant example of stepwise increases in resistance, in which consecutive amino acid substitutions code for progressive increases in minimal inhibitory concentrations of broad-spectrum cephalosporins (21). Multistep resistance depends on the selection of intermediate variants with very small phenotypic differences from each previous genetic ancestor. Antibiotic dosages that produce low antibiotic concentrations in particular (colonized) body compartments will be sufficient to select these intermediates. Even though our data were obtained with models and not under standard clinical conditions, the prediction of our work is that antibiotic dosages should be optimized (i.e., should be sufficiently high and continuous) during therapy to avoid selection of low-level resistant variants. Indeed, the evolution of antibiotic resistance during treatment depends on a temporal variation of drug concentration (19). Low concentrations of antibiotics may serve as stepping-stones for the ascent of clinical resistance. In more general terms, our results suggest that the study of selection of very small differences may provide some clues about the evolution of complex biological systems when a fine-grained variation between individuals exists.
ACKNOWLEDGMENTS
We thank Maria-Rosario Baquero and Jose C. Pérez-Díaz, from the Department of Microbiology of the Ramón y Cajal Hospital, for help with the characterization of OmpF mutants and Richard Lenski for valuable discussions.
REFERENCES
- 1.Baquero F, Blázquez J. Evolution of antibiotic resistance. Trends Ecol Evol. 1997;12:482–487. doi: 10.1016/s0169-5347(97)01223-8. [DOI] [PubMed] [Google Scholar]
- 2.Baquero F, Negri M C. Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays. 1997;19:731–736. doi: 10.1002/bies.950190814. [DOI] [PubMed] [Google Scholar]
- 3.Baquero F, Negri M C, Morosini M I, Blázquez J. Antibiotic selective environments. Clin Infect Dis. 1998;27(Suppl. 1):5–11. doi: 10.1086/514916. [DOI] [PubMed] [Google Scholar]
- 4.Blázquez J, Morosini M I, Negri M C, González-Leiza M, Baquero F. Single amino acid replacements at positions altered in naturally occurring extended-spectrum TEM β-lactamases. Antimicrob Agents Chemother. 1995;39:145–149. doi: 10.1128/aac.39.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blázquez J, Morosini M I, Negri M C, Baquero F. Selection of naturally occurring extended-spectrum TEM β-lactamase variants by fluctuating β-lactam pressure. Antimicrob Agents Chemother. 2000;44:2182–2184. doi: 10.1128/aac.44.8.2182-2184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Jacoby G, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig W A. Pharmacokinetic/pharmacodynamic parameters: rationale for antibacterial dosing of mice and men. Clin Infect Dis. 1998;26:1–12. doi: 10.1086/516284. [DOI] [PubMed] [Google Scholar]
- 8.Datta D B, Arden B, Henning U. Major proteins of Escherichia coli outer cell envelope membrane as bacteriophage receptors. J Bacteriol. 1977;131:821–829. doi: 10.1128/jb.131.3.821-829.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta N, Kontomichalou P. Penicillinase synthesis controlled by infectious R-factors in Enterobacteriaceae. Nature. 1965;208:239–241. doi: 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 10.Dong Y, Zhao X, Domalaga J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellay R, Frey J, Kirsch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 12.Gavaldá J, Torres C, Tenorio C, López P, Zaragoza M, Capdevila J A, Almirante B, Ruiz F, Borrell N, Gomis X, Pigrau C, Baquero F, Pahissa A. Efficacy of ampicillin plus ceftriaxone in treatment of experimental endocarditis due to Enterococcus faecalis strains highly resistant to aminoglycosides. Antimicrob Agents Chemother. 1999;43:639–646. doi: 10.1128/aac.43.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Le K K, LaRocco M, Palzkill T. Effect of threonine-to-methionine substitution at position 265 on structure and function of TEM-1 β-lactamase. Antimicrob Agents Chemother. 1994;38:2266–2269. doi: 10.1128/aac.38.10.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaffe A, Chabbert Y A, Semonin O. Role of porin proteins OmpF and OmpC in the permeation of β-lactams. Antimicrob Agents Chemother. 1982;22:942–948. doi: 10.1128/aac.22.6.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kepler T B, Perelson A S. Drug concentration heterogeneity facilitates the evolution of drug resistance. Proc Natl Acad Sci USA. 1998;95:11514–11519. doi: 10.1073/pnas.95.20.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laviña M, Pugsley A P, Moreno F. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J Gen Microbiol. 1986;132:1685–1693. doi: 10.1099/00221287-132-6-1685. [DOI] [PubMed] [Google Scholar]
- 17.Lenski R E, Levin B R. Constraints on the co-evolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am Nat. 1985;125:585–602. [Google Scholar]
- 18.Lenski R E. Experimental studies of pleiotrophy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution. 1988;42:425–433. doi: 10.1111/j.1558-5646.1988.tb04149.x. [DOI] [PubMed] [Google Scholar]
- 19.Levin B R, Stewart F M, Chao L. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am Nat. 1977;111:3–24. [Google Scholar]
- 20.Levin B R, Lipsitch M, Perrot V, Schrag S, Antia R, Simonsen L, Moore Walker N, Stewart F M. The population genetics of antibiotic resistance. Clin Infect Dis. 1997;24(Suppl. 1):9–16. doi: 10.1093/clinids/24.supplement_1.s9. [DOI] [PubMed] [Google Scholar]
- 21.Medeiros A A. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin Infect Dis. 1997;24(Suppl. 1):19–45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 22.Moreno F, San Millan J L, Hernandez-Chico C, Kolter R. Microcins. Biotechnology. 1995;28:307–321. doi: 10.1016/b978-0-7506-9095-9.50019-8. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 3rd approved standard, NCCLS document M7A3. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1993. [Google Scholar]
- 24.Negri M C, Morosini M I, Loza E, Baquero F. In vitro selective concentrations of β-lactams for penicillin-resistant Streptococcus pneumoniae populations. Antimicrob Agents Chemother. 1994;38:122–125. doi: 10.1128/aac.38.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozawa Y, Mizushima S. Regulation of outer membrane porin protein synthesis in Escherichia coli K12: ompF regulates the expression of ompC. J Bacteriol. 1983;154:669–675. doi: 10.1128/jb.154.2.669-675.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrosino J, Cantu III C, Palzkill T. Beta-lactamases: protein evolution in real time. Trends Microbiol. 1998;6:323–327. doi: 10.1016/s0966-842x(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 27.Reguera J A, Baquero F, Pérez-Díaz J C, Martínez J L. Factors determining resistance to beta-lactams combined with beta-lactamase inhibitors in Escherichia coli. J Antimicrob Chemother. 1991;27:569–575. doi: 10.1093/jac/27.5.569. [DOI] [PubMed] [Google Scholar]
- 28.Schrag S J, Perrot V. Reducing antibiotic resistance. Nature. 1997;381:120–121. doi: 10.1038/381120b0. [DOI] [PubMed] [Google Scholar]
- 29.Sober E. The nature of selection: evolutionary theory in philosophical focus. Chicago, Ill: The University of Chicago Press; 1993. p. 136. [Google Scholar]
- 30.Spratt B G, Hedge P I, Heesen S, Edelman A, Broome-Smith J K. Kanamycin resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 31.Travisano M, Lenski R E. Long term experimental evolution in Escherichia coli. IV. Targets of selection and specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]