Abstract
Objective
This study was to explore the most appropriate radiomics modeling method to predict the progression-free survival of EGFR-TKI treatment in advanced non-small cell lung cancer with EGFR mutations. Different machine learning methods may vary considerably and the selection of a proper model is essential for accurate treatment outcome prediction. Our study were established 176 discrimination models constructed with 22 feature selection methods and 8 classifiers. The predictive performance of each model were evaluated using the AUC, ACC, sensitivity and specificity, where the optimal model was identified.
Results
There were totally 107 radiomics features and 7 clinical features obtained from each patient. After feature selection, the top-ten most relevant features were fed to train 176 models. Significant performance variations were observed in the established models, with the best performance achieved by the logistic regression model using gini-index feature selection (AUC = 0.797, ACC = 0.722, sensitivity = 0.758, specificity = 0.693). The median R-score was 0.518 (IQR, 0.023–0.987), and the patients were divided into high-risk and low-risk groups based on this cut-off value. The KM survival curves of the two groups demonstrated evident stratification results (p = 0.000).
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-022-06019-x.
Keywords: Non-small cell lung cancer, EGFR-TKI, Radiomics, Machine learning
Introduction
The EGFR-TKIs (epidermal growth factor receptor-Tyrosine kinase inhibitors) such as gefitinib and erlotinib are used as the first-line treatment for NSCLC (non-small cell lung cancer) patients with EGFR mutation. However, acquired resistance is usually observed after 9–13 months of evident tumor response [1]. Moreover, primary resistance would tend to affect the therapeutic efficacy of TKIs by leaving with only a few months of reaction time. In this sense, exploring an effective prognostic marker to predict the development of TKIs resistance of EGFR positive patients will be clinically meaningful to allow physicians to timely adjust the treatment strategies.
Radiomics is able to extract quantitative imaging biomarker regarding the biological, prognostic and predictive hidden information from medical images. Successful application of CT-based (computed tomography) radiomics has witnessed in NSCLC prediction and prognosis [2], in particular for prediction of EGFR mutations [3, 4], and EGFR-TKI prognosis [5, 6]. These studies empirically adopted different ML (machine learning) methods to build the predictive model, e.g., RF (random forest) [3, 4] or LASSO regression [5–7], it is still unclear how to choose an appropriate ML algorithm for modeling and which model is more superior over the other.
The goal of current study is to explore the most appropriate radiomics modeling method to predict the progression-free survival of EGFR-TKI treatment in advanced non-small cell lung cancer with EGFR mutations. In order to screen patients suitable for EGFR- TKI therapy.
Main text
Patients
A total of 100 patients with stage IIIB-IV EGFR positive NSCLC patients treated with EGFR-TKIs in the Affiliated Cancer Hospital of Guangzhou Medical University between January 2016 and December 2019 met the inclusion criteria and were finally recruited. The inclusion criteria were as follows: (1) stage IIIB-IV lung adenocarcinoma confirmed by histopathology; (2) patients of EGFR mutations with confirmed 19del or 21L858R; (3) no treatments received prior to EGFR TKIs therapy; (4) the EGFR-TKIs therapy was used as the first-line treatment; (5) with complete CT images within 2 weeks before EGFR-TKIs treatment. The exclusion criteria included: (1) age less than 18 years old; (2) underwent any other antitumor therapies; and (3) with incomplete clinical records or CT images.
Patients with 19del or 21L858R mutations have received gefitinib, erlotinib, or other EGFR-TKIs as first-line treatment. Drugs were orally administrated daily until disease progressed. Treatment efficacy evaluations included routine laboratory tests and chest CT scans at least every 4–12 weeks. PFS was estimated from the beginning time of EGFR-TKIs therapy to the date of disease progression or death. The outcome is assessed within 3 months of the targeted therapy via the RECIST1.1 criterion. Patients’ clinical characteristics including sex, age, stage, smoking status, mutations, TKIs and outcome are summarized in Additional file 1: Table S1.
Methods
Image acquisition and feature extraction
Pretreatment CT scans were acquired after intravenous injection of 100 ml ioversol (Heng Rui Pharmaceuticals, Jiangsu China), with scanning parameters of 120 kV, 160 mAs, 0.6 s rotation time, and image matrix size of 512 × 512. All patient images were stored in DICOM format. The VOI (volume of interest) on CT images was delineated independently by two radiologists using 3D-slicer software (slicer4.10.2, https://www.slicer.org). The VOI delineation was performed slice-by-slice on the CT images with standard mediastinal (window width, 400 HU; window level, 40 HU) and lung (window width, 1500 HU; window level, − 700 HU) window settings. The conformity of delineated VOIs were measured by the Dice similarity coefficient. The two delineated VOIs with Dice index greater than 0.9 were averaged to yield the final VOI. Discrepancies on the lesion boundary (Dice < 0.9) were resolved by further discussions until mutual consensus were reached.
Radiomics features extraction were conducted within the VOIs utilized an open-source python package Pyradiomics [8]. Extracted features (n = 107, Additional file 1: Table S2) included: (1) first order features (n = 18); (2) shape features (n = 14); (3) GLCM (gray level co-occurrence matrix) features (n = 24); (4) GLSZM (gray level size zone matrix) features (n = 16); (5) GLRLM (gray level run length matrix) features (n = 16); (6) NGTDM (neighboring gray tone difference matrix) features (n = 5); (7) GLDM (gray level dependence matrix) features (n = 14).
Prediction modeling
Noted that our goal was to identify the most significant variables that could discriminate patients with fast and slow progression. Thus, the dimension reduction was necessary to improve the accuracy in the later step of building the machine learning model for classification. Therefore, the feature selection was conducted before constructing the models, and the most related features to this study were selected.
Prediction modeling was performed on 107 radiomics features combined with 7 clinical features (including sex, age, stage, smoking status, mutations, TKI, and outcome). A specific model was constructed by a feature selection procedure followed by a particular classifier. In this study, 22 feature selection methods and 8 classifiers (Additional file 1: Table S3) were studied, and therefore resulting in 176 prediction models (different combinations of ‘feature selection’ + ‘classifier’) to be evaluated. In each prediction model, we empirically set the number of selected features to 10 to balance the patient sample size vs. feature numbers.
The prediction model was evaluated via a repeated (5 times) five-fold CV (cross-validation), where 80% and 20% of the dataset were respectively reserved for model training and validation. To reduce the effect of the imbalance, the SMOTE (synthetic minority oversampling) [9] technique was applied on the training set in each fold of the CV. The prediction performances were quantified by the area under the AUC (receiver operating characteristic (ROC) curve), ACC (accuracy), SEN (sensitivity), and SPE (specificity).
Statistical analysis
All statistical analyses were conducted using the SPSS (version 22.0). Comparison between groups of classified variables used chi-square test. The Kaplan–Meier algorithm was used to estimate the survival curves that were compared by the log-rank test. Normality of data distribution was assessed by the Kolmogorov–Smirnov test. The student’s t-test was used for normally distributed continuous variables and the Mann–Whitney U test was used for non-normally distributed continuous variables. A p-value of < 0.05 was regarded as statistically significant.
Results
Patient characteristics
A median PFS (10 months) of the whole patient cohort was used to divide patients into a rapidly progressing group (n = 49) and a slowly progressing group (n = 51). The median age of all patients was 59 years. More patients with 19del mutation were found in the slow-progression group (19del vs 21L858R, 59.6% vs 37.5%); In contrast, more 21L858R mutation patients were found in the fast-progression group (19del vs 21L858R, 40.4% vs 62.5%). Significant difference in mutation site of the two groups was observed (p = 0.03). In the slow progression group, 44 (63.8%) patients exhibited ‘CR (complete remission)’ or ‘PR (partial remissions)’ after EGFR-TKIs therapy, while only 5 (16.1%) patients were categorized as ‘SD (stable diseases)’ or ‘PD (progressive diseases)’ (by RECIST1.1 criteria). In contrast, in the rapid-progression group, only 25 patients (36.2%) achieved CR or PR after EGFR-TKIs therapy, and 26 patients (83.9%) were classified as SD or PD. The clinical factor of ‘CR + PR’ outcome was found to be associated with better prognosis (p = 0.03) by the univariate analysis.
Model performance comparisons
The 107 radiomics features and 7 clinical features were obtained from each patient. In each of the 176 model, the top-ten most relevant features were selected by feature selection and fed to train a classifier in each model. The performances of the 176 models are depicted as a AUC heatmap as shown in Additional file 1: Fig. S1. The other quantitative performance metrics as such ACC, SEN, SPE of each model were detailed in Additional file 1: Fig. S2. Evident prediction performance variations were observed in the evaluated models. The average AUC of all 176 models is 0.591 with the maximum AUC of 0.797 achieved by the model built with LR (Logistic Regression) and gini-index feature selection (ACC = 0.722, SEN = 0.758, SPE = 0.693). While the worst model (SVM (support vector machine) and JMI (joint mutual information) feature selection) only yield AUC = 0.371, ACC = 0.559, SEN = 0.611 and SPE = 0.507. The performance of the best 10 models was listed in Table 1. The mean AUC tends to significantly (p = 0.000) decline to 0.524 if only radiomics features were used for modeling, as compared with the AUC heatmap shown in Additional file 1: Fig. S3. Each of the 176 models were respectively trained by all the features (n = 114, including 107 radiomics and 7 clinical features) and cross-validated by the five-fold cross-validation. The most relevant features had been selected in each model, and we can count and rank their frequencies of being selected as the top feature, as shown in Table 2. The top-ten features most selected were marked in blue in Additional file 1: Fig. S4. Among these top-ten features, four features including two texture features (glcm-difference variance: 14.59 ± 6.63 vs. 18.16 ± 8.71, p = 0.024; glszm-small area emphasis: 0.68 ± 0.04 vs. 0.70 ± 0.03, p = 0.003) and two clinical features (mutation, p = 0.03; outcome, p = 0.000) showed a statistically significant difference between the slow progression group and the fast progression group.
Table 1.
Feature selection methods and classifiers for the top ten models
| Classifier | Feature selection method | AUC | ACC | Sensitivity | Specificity |
|---|---|---|---|---|---|
| Logistic | gini-index | 0.797 | 0.722 | 0.758 | 0.693 |
| Logistic | ll-l21 | 0.763 | 0.681 | 0.716 | 0.653 |
| Bagging | CIFE | 0.764 | 0.671 | 0.636 | 0.707 |
| Logistic | reliefF | 0.759 | 0.662 | 0.676 | 0.655 |
| Bagging | MRMR | 0.743 | 0.662 | 0.616 | 0.729 |
| Bagging | MIFS | 0.742 | 0.670 | 0.656 | 0.673 |
| Adaboosting | f-score | 0.740 | 0.671 | 0.756 | 0.593 |
| SVM | MRMR | 0.739 | 0.712 | 0.733 | 0.695 |
| SVM | CIFE | 0.734 | 0.692 | 0.713 | 0.676 |
| SVM | MIFS | 0.734 | 0.712 | 0.733 | 0.695 |
Table 2.
The top ten features and the corresponding mean (± SD) value (or median (IQR)) and the p-value between the slow and fast progression groups
| Feature category | Feature | Slow-progress | Fast-progress | p-value | |
|---|---|---|---|---|---|
| Shape-based (n = 4) | Elongation | 0.76 ± 0.12 | 0.71 ± 0.13 | 0.067a | |
| Least Axis Length | 30.06 ± 11.91 | 20.53 ± 8.85 | 0.234a | ||
| Flatness | 0.59 ± 0.14 | 0.55 ± 0.13 | 0.229a | ||
| Major Axis Length | 52.25 (19.52,110.61) | 46.56 (23.28,145.60) | 0.858b | ||
| First-order based (n = 1) | Inter quartile range | 140 (36,560) | 154 (42,385) | 0.962b | |
| Texture | GLSZM (n = 1) | Small Area Emphasis | 0.68 ± 0.04 | 0.70 ± 0.03 | 0.003a |
| GLCM (n = 1) | Difference variance | 14.59 ± 6.63 | 18.16 ± 8.71 | 0.024a | |
| Clinical based (n = 3) | Smoke | – | – | 0.238c | |
| Mutation | – | – | 0.030c | ||
| Outcome | – | – | 0.000c | ||
at-test
bMann–Whitney U test
cChi-square test
Prognostic performance
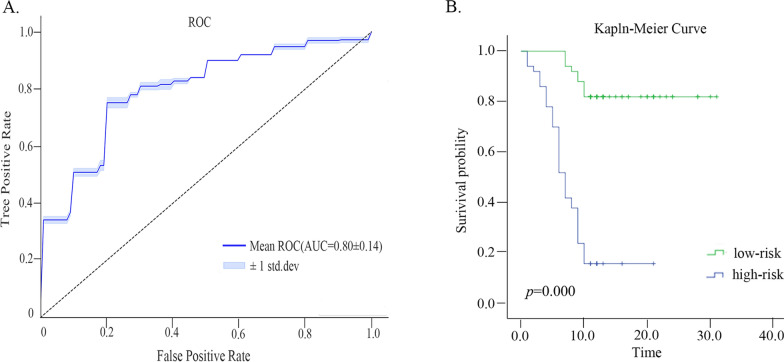
We stratified patients by the R-score (risk score) given by the best model, i.e., the Gini-index-LR model evaluated from the 176 models. The estimated median R-score 0.518 (IQR, 0.023–0.987) was used as the cut-off value to stratify the high-risk group from the low-risk group. The higher the R value, the higher the likelihood of rapid progression. The KM (Kaplan–meier) method and the Log-Rank test were used to evaluate and compare the survival curves of the high-risk group and the low-risk group. The ROC and KM survival curves (p = 0.000) of Gini-index-LR method were shown in Fig. 1.
Fig. 1.
A Time-dependent ROC curves of the “gini-index-Logistic regression” model of using the top-10 features at 10 months. B Kaplan Meier survival curves of EGFR positive NSCLC patients. The p-values were calculated using the log-rank tests
Discussion
A particular prediction model can be built with a feature selection followed by a classifier. Given a large pool of available feature selection methods and different classifier algorithm, their combinations may result in a large number of prediction models to choose from. In this study, we constructed and comprehensively evaluated 176 models using 22 feature selection methods and 8 classifiers. The best model method was gini-index-logistic (AUC = 0.797). The R-score derived from the gini-index-logistic model was validated on the KM curve, and had also achieved satisfactory stratification result. We have observed that the other models, e.g., the CIFE-Bagging (AUC = 0.764), the ll-l21-LR (AUC = 0.763), and the ReliefF-LR (AUC = 0.759) also demonstrated high predictive performances. In general, the predictive models built with the LR classifier seemed to perform better than those models built with the RF classifier, which has been used in previous investigations for EGFR mutations prediction modeling [4].
In this study, we have identified two CT image textural features, i.e., the GLCM-derived feature "DV (Difference Variance)" and the GLSZM-based feature "SAE (Small Area Emphasis)" to be significantly associated with progression (p = 0.003 and p = 0.024). The DV is a measure of heterogeneity that places higher weights on differing intensity level pairs that deviate more from the mean. The lesions on the fast-progression group (DV: 18.16 ± 8.71) seemed to be more heterogeneous than the slow-progression group (DV: 14.59 ± 6.63). While the SAE is a measure of the distribution of small size zones [8]. The fast-progression group had greater SAE values than the slow-progression group (0.70 ± 0.03 vs. 0.68 ± 0.04), which was indicative of more smaller size zones and more fine textures from the lesion on CT image that might be correlated with progression.
Similar to Hong et al. [10] study, we also found that the EGFR mutation type was indicative of patient prognosis. In addition, we identified complete or partial remission (CR or PR) within 3 months after EGFR treatment as a prognostic factor. This finding was consistent with clinical observations and previous studies, e.g., Mizuki et al. [7] claimed that the proportional volume change at 8 weeks was related with overall survival in EGFR-mutant advanced NSCLC patients treated with first-line EGFR-TKIs.
In summary, our study screened out an optimal model to predict progression-free survival time of NSCLC patients treated with first-line EGFR-TKI within a machine learning based framework.
Limitations
This study has several limitations need to be addressed. First, this was a single institutional study where the patient sample size was relatively small. An independent external validation cohort was lacked to confirm the generalization capability of the model and the associated findings presented here. Second, we didn't delve into the effect of the number of features on the model. Third, the number of the clinical factors studied in the model were relatively small. We have observed increased AUC values when clinical features were incorporated into the model rather than using image texture feature alone. Improved model performance might be expected if more clinical or pathological factors were embedded.
Supplementary Information
Additional file 1: Table S1 Demographics and characteristics of the 100 patients. Table S2 Radiomics features. Table S3 Feature selection methods and classifiers used for discrimination models. Figure S1 Heatmap representing AUC of 176 models was constructed from the clinical features and radiomics features. Figure S2 Heatmap representing the ACC(A),SEN(B) and SPE(C) of 176 models constructed from clinical features and radiomic features. Figure S3 Heatmap representing AUC of 176 models constructed from radiomic features. Figure S4 Histogram of the number of times that each feature is selected in the five-fold cross validation.
Acknowledgements
Not applicable.
Abbreviations
- EGFR-TKIs
Epidermal growth factor receptor-Tyrosine kinase inhibitors
- NSCLC
Non-small cell lung cancer
- PFS
Progression-free survival
- CT
Computed tomography
- ML
Machine learning
- RF
Random forest
- VOI
Volume of interest
- GLCM
Gray level co-occurrence matrix
- GLSZM
Gray level size zone matrix
- GLRLM
Gray level run length matrix
- NGTDM
Neighboring gray tone difference matrix
- GLDM
Gray level dependence matrix
- CV
Cross-validation
- SMOTE
Synthetic minority oversampling
- AUC
Receiver operating characteristic (ROC) curve
- ACC
Accuracy
- SEN
Sensitivity
- SPE
Specificity
- CR
Complete remission
- PR
Partial remissions
- SD
Stable diseases
- PD
Progressive diseases
- LR
Logistic Regression
- SVM
Support vector machine
- JMI
Joint mutual information
- R-score
Risk score
- KM
Kaplan–Meier
- DV
Difference Variance
- SAE
Small Area Emphasis
Author contributions
JMZ and LS are responsible for writing the article. ZWL and XZ are responsible for the modification and quality control of the articles. LJW, TCZ and LS are responsible for the image acquisition and analysis. YWY, ZWL and JMZ are responsible for compiling the medical records. All authors read and approved the final manuscript.
Funding
This study were supported by the Guangzhou Key Medical Discipline Construction Project Fund, the Key Clinical Technology of Guangzhou (2019ZD17), Guangzhou Key Medical Discipline Construction Project Fund (02-412-B205002-1004042), Natural Science Foundation of Guangdong Province (2022A1515012104) and Guangdong Provincial Medical Scientific Research Fund Project (A2022213).
Availability of data and materials
The data that support the findings of our study are available from the Affiliated Cancer Hospital of Guangzhou Medical University but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the Affiliated Cancer Hospital of Guangzhou Medical University.
Declarations
Ethics approval and consent to participate
This study was approved by the ethics review board of the Affiliated Cancer Hospital of Guangzhou Medical University, considered as a retrospective study and therefore waived the need for patient informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jian-man Zhu and Lei Sun contributed equally to this work
Contributor Information
Xin Zhen, Email: xinzhen@smu.edu.cn.
Zhi-Wei Liao, Email: vigo310@126.com.
References
- 1.Chee-Khoon Lee Wu, Yi-Long D-N, et al. Impact of specific epidermal growth factor receptor (EGFR) mutations and clinical characteristics on outcomes after treatment with EGFR tyrosine kinase inhibitors versus chemotherapy in EGFR-mutant lung cancer: a meta-analysis. J Clin Oncol. 2015;33(17):1958–1965. doi: 10.1200/JCO.2014.58.1736. [DOI] [PubMed] [Google Scholar]
- 2.Fornacon-Wood I, Faivre-Finn C, O’Connor JP, et al. Radiomics as a personalized medicine tool in lung cancer: separating the hope from the hype. Lung Cancer. 2020;146:197–208. doi: 10.1016/j.lungcan.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Dong X, Wang J, et al. Computed tomography-based radiomics signature: a potential indicator of epidermal growth factor receptor mutation in pulmonary adenocarcinoma appearing as a subsolid nodule. Oncologist. 2019;24(11):e1156–e1164. doi: 10.1634/theoncologist.2018-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia T-Y, Jun-Feng X, Xiao-Yang Li, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol. 2019;29(9):4742–4750. doi: 10.1007/s00330-019-06024-y. [DOI] [PubMed] [Google Scholar]
- 5.Song J, Jingyun S, Di D, et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin Cancer Res. 2018;24(15):3583–3592. doi: 10.1158/1078-0432.CCR-17-2507. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Rui Z, Siwen W, et al. CT-based radiomic signature as a prognostic factor in stage IV ALK-positive non-small-cell lung cancer treated with TKI Crizotinib: a proof-of-concept study. Front Oncol. 2020;10:57. doi: 10.3389/fonc.2020.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishino M, Suzanne-E D, Stephanie C, et al. Tumor volume decrease at 8 weeks is associated with longer survival in EGFR-mutant advanced non–small-cell lung cancer patients treated with EGFR TKI. J Thorac Oncol. 2013;8(8):1059–1068. doi: 10.1097/JTO.0b013e318294c909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Griethuysen J-J-M, Andriy F, Chintan P, et al. Computational radiomics system to decode the radiographic phenotype. Can Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chawla N-V, Kevin-W B, Lawrence-O H. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 10.Weiwei Hong Wu, Qiuji ZJ, et al. Prognostic value of EGFR 19-del and 21–L858R mutations in patients with non-small cell lung cancer. Oncol Lett. 2019;18(4):3887–3895. doi: 10.3892/ol.2019.10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1 Demographics and characteristics of the 100 patients. Table S2 Radiomics features. Table S3 Feature selection methods and classifiers used for discrimination models. Figure S1 Heatmap representing AUC of 176 models was constructed from the clinical features and radiomics features. Figure S2 Heatmap representing the ACC(A),SEN(B) and SPE(C) of 176 models constructed from clinical features and radiomic features. Figure S3 Heatmap representing AUC of 176 models constructed from radiomic features. Figure S4 Histogram of the number of times that each feature is selected in the five-fold cross validation.
Data Availability Statement
The data that support the findings of our study are available from the Affiliated Cancer Hospital of Guangzhou Medical University but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of the Affiliated Cancer Hospital of Guangzhou Medical University.