Abstract
Colorectal cancer (CRC), a common malignancy, is one of the leading cause of cancer death in adults. AT-rich interaction domain 1A (ARID1A), a critical portion of the SWItch/sucrose non-fermentation (SWI/SNF) chromatin remodeling complexes, shows one of the most frequent mutant genes across different human cancer types. Deleterious variations of ARID1A has been recognized to be correlated the tumorigenesis and the poor prognosis of CRC. Here, we summarize recent advances in the clinical implications and molecular pathogenesis of ARID1A variations in CRC. According to independent data of 23 included studies, ARID1A is mutated in 3.6–66.7%. Consistently, all of the 23 relevant studies report that ARID1A functions as a specific tumor suppressor in CRC. Clinically, ARID1A variation status serves as a biomarker for survival prognosis and various therapies for CRC. Mechanistically, the pathophysiologic impacts of ARID1A variations on CRC may be associated with the co-occurrence variations of other genes (i.e., TP53, KRAS, APC, FBXW7, and PIK3CA) and the regulation of several signaling pathways being affected (i.e., WNT signaling, Akt signaling, and MEK/ERK pathway), leading to cell cycle arrest, chromatin remodeling, chromosome organization, and DNA hypermethylation of the cancer cells. The present review highlights ARID1A serving as a potent tumor suppressor and an important prognostic factor in CRC. ARID1A variations hint towards a promising tool for diagnostic tumor profiling and individualized therapeutic targets for CRC in the future.
Keywords: ARID1A variations, Colorectal cancer (CRC), Biomarker, Prognosis, Pathogenesis
Introduction
Colorectal cancer (CRC) is one of the top-common malignancies worldwide, nearly 1,850,000 incidences, and is the second leading cause of cancer deaths, with an approximate 881,000 fatalities (9.2% of all fatal cancer cases) in the world yearly (Bray et al. 2018). CRC with locoregional lymph node diffuse has a 5-year overall survival (OS) of 70% while disuse to distant organs carries a substantially worse prognosis with a 5-year OS of 12% (Siegel et al. 2018). Metastasis to the liver is the most common site of distant spread (Fong et al. 1997) while the peritoneal surface is the second most common site of metastasis, involving roughly 10% of patients with CRC at the very beginning of the presentation and the sole site of recurrence in as much as 25% of patients with CRC (Dawson et al. 1983; Russell et al. 1983). Peritoneal metastasis (PM) is associated with a poor prognosis, and the survival period for systemic chemotherapy alone is 5–7 months (Chu et al. 1989; Koppe et al. 2006). Compared to other site transfers, PM is associated with a greatly shorter progression-free survival (PFS) and OS (Franko et al. 2012). The molecular underlying mechanisms of CRC is driven by the continuous acquisition of epigenetic and genetic abnormalities, which is related to the repression of the tumor suppressor and the activation of pro-oncogenic factors (Lao and Grady 2011). The low effectiveness of conventional therapeutic interventions to prolong life span in CRC patients needs new and effective targeted therapies.
The heterogeneity of CRC tumor aggressiveness and prognosis might be prompted by differences in genetic variation. According to some reports, the gene encoding the SWItch/sucrose non-fermentation (SWI/SNF) chromatin remodeling complex is one of the most common mutant genes in a variety of malignant tumors. SWI/SNF chromatin remodeling complex play role in the transcription and DNA reproduction and repair (Wilson and Roberts 2011). Among the family of the SWI/SNF genes, AT-rich interaction domain 1A (ARID1A) is a common-mutated gene in human cancers, which contributes to the binding of protein and DNA (Kadoch and Crabtree 2013; Wang et al. 2004). ARID1A, a gene located on chromosome 1p36.11, is a core component of the mammalian SWI/SNF complex (Megaridis et al. 2018). ARID1A encodes a protein with nuclear/cytosolic localization. Nuclear ARID1A is speedily degraded by the nuclear ubiquitin–proteasome system unstable due to the nuclear ARID1A is unstable (Mao and Shih 2013). In-frame deletions disrupting the nuclear export signal cause a declination of ARID1A expression, due to the nuclear retention of the protein and its subsequent degradation (Mao and Shih 2013; Guan et al. 2012). ARID1A exhibits its biological function by interacting with DNA and recruiting associated transcriptional co-activators, while ARID1A variation commonly cause the dysregulation of BAF complex-mediated chromatin remodeling (Chandler et al. 2013). ARID1A contains an ARID domain, which interacts with DNA in a sequence-nonspecific manner modulating cellular processes (e.g., proliferation and differentiation) (De and Dey 2019). Thus, ARID1A has been found to be contributed to the tumorigenesis of multiple cancers.
ARID1A has lately been recognized as a crucial tumor suppressor gene in diverse cancer types. Ovarian cancer, stomach cancer, and pancreatic cancer have the highest mutation (or variation) frequency (29–57%), while CRC (13%), liver cancer (10–17%), bladder cancer (13%), esophageal cancer (9%), breast cancer (3%) and childhood retinoblastoma (6%) have somewhat lower variation frequencies (Cornen et al. 2012; Dulak et al. 2013; Fujimoto et al. 2012; Gui et al. 2011; Guichard et al. 2012; Jones et al. 2012, 2010; Sausen et al. 2013; Shain et al. 2012; Wiegand et al. 2010). Also, Ogiwara et al. (2019) summarized that ARID1A is mutated in about 46% of ovarian clear cell carcinomas, 43% of uterine corpus endometrial carcinomas, 33% of gastric carcinomas, 30% of ovarian endometrioid carcinomas, 28% of bladder carcinomas, 27% of cholangiocarcinomas, 15% of pancreatic carcinomas, 12% of lung adenocarcinomas, and 10% of CRC. The frequency of ARID1A variations in ovarian clear cell carcinomas is up to 60% in the US, Canada, and Japan, indicating that ARID1A deficiency may be a potential biomarker for precision medicine of ovarian cancer (Takahashi et al. 2021). It was reported that ARID1A variations was observed in up to 40% of low-grade endometrioid carcinomas (Toumpeki et al. 2019). The reported ARID1A mutant prevalence in gastric cancer among different studies was 8–27% (Wang et al. 2021). Dugas et al. (2019) demonstrated that ARID1A variation was observed in 3.6% of the non-muscle-invasive bladder cancer and 10% of the muscle-invasive bladder cancer. Zhao et al. showed that the variation rate of ARID1A in cholangiocarcinomas ranged from 5% to 68.2% (Zhao et al. 2021). Though ARID1A may be not the most highly mutated gene in the aforementioned malignancies, it can synergize with other mutant genes to promote the pathogenesis and the development of cancers.
Most of the ARID1A variations are inactive condition that result in the loss of the protein expression of ARID1A (Kishida et al. 2019). In current years, mounting evidence revealed that ARID1A variation is related to the clinicopathologic characteristics of CRC (Wei et al. 2014; Ye et al. 2014). At present, in different clinical studies, the specific role of ARID1A on the prognosis and clinicopathological features of CRC is widely debated. According to published data, most studies indicate that ARID1A serves as an important tumor suppressor gene. For example, Lee et al. (2016) demonstrated that no connection was evident between ARID1A expression and 5-year OS. However, a recent study conducted by Jiang et al. (2020) showed that disease-free or PFS of patients with ARID1A variations [DFS/PFS, HR = 0.74 (0.64–0.91), P = 0.0026]. The OS of patients with ARID1A variations was significantly prolonged by 28 months, compared with 18 months in those with wild-type ARID1A [HR = 0.73 (0.61–0.93), P = 0.0092]. The role of ARID1A in CRC is currently uncertain. In this narrative review, we aim to overview all the current evidence that ARID1A variation or expression is associated with the development of CRC, and reveal the potential molecular mechanisms.
Searching strategy
Four common data bases were searched to find the eligible studies prior to January 1, 2022. The searching strategy these databases was: ((((((((((((((ARID1A) OR (B120)) OR (BAF250)) OR (BAF250a)) OR (BM029)) OR (C1orf4)) OR (CSS2)) OR (ELD)) OR (MRD14)) OR (OSA1)) OR (P270)) OR (SMARCF1)) OR (hELD)) OR (hOSA1)) AND ((((((((((((((((((((“Colorectal Neoplasms”[Mesh]) OR (Colorectal Neoplasm)) OR (Neoplasm, Colorectal)) OR (Neoplasms, Colorectal)) OR (Colorectal Tumors)) OR (Colorectal Tumor)) OR (Tumor, Colorectal)) OR (Tumors, Colorectal)) OR (Colorectal Cancer)) OR (Cancer, Colorectal)) OR (Cancers, Colorectal)) OR (Colorectal Cancers)) OR (Colorectal Carcinoma)) OR (Carcinoma, Colorectal)) OR (Carcinomas, Colorectal)) OR (Colorectal Carcinomas)) OR (Colonic Neoplasm)) OR (Colon Cancer)) OR (Rectal Neoplasms)) OR (Rectum Cancer)). For identifying more eligible studies, we manually inspected the reference lists in the related articles. According to the data collection form, the following information in each study was extracted, including the first authors’ names, the publication year, study area, type of CRC, ARID1A variations in CRC, and some details of clinical and molecular aspects.
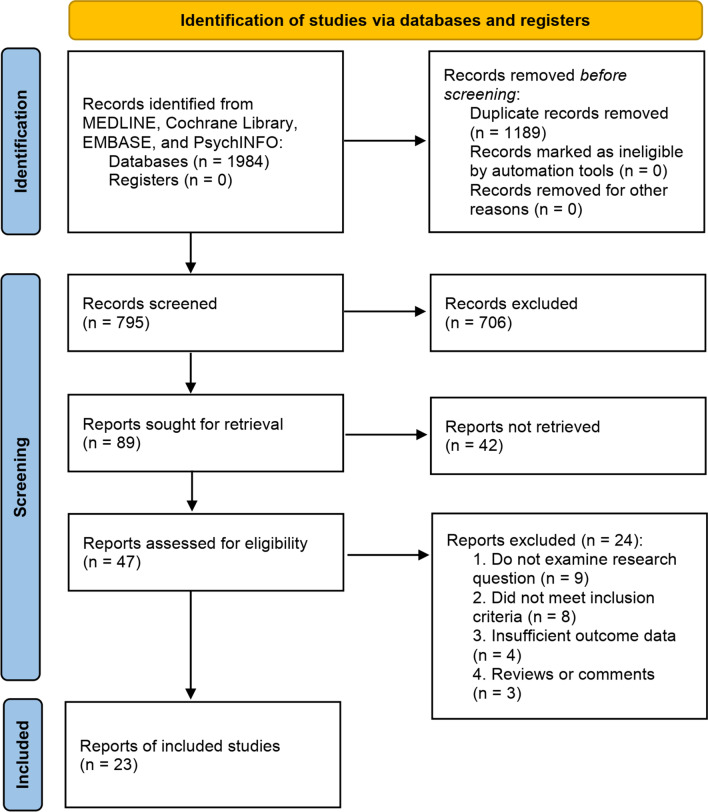
Figure 1 showed the search flowchart. Finally, 23 eligible studies (16, 23–44) with a total of 15,580 subjects were included. The characteristics of the 23 eligible studies were listed in Table 1. According to the available information from the 23 included studies, ARID1A variation was defined as the loss or low expression levels of ARID1A. Therefore, though “ARID1A mutation” could be found several previous related studies, ARID1A is in fact variant rather than mutated. Besides, according to the ACMG guidelines for nomenclature of the genomic variations, it is recommended to use the term “variation” instead of “mutation” (Richards et al. 2015). As shown in Fig. 2, the frequency of ARID1A variations among the 23 studies ranged from 3.6% to 66.7%. The mutant type is defined as the loss or low of ARID1A protein expression. Jones et al. (2012), suggested that ARID1A has a tumor suppressor function in the pathogenesis of CRC, and reported that ARID1A expression reduction and/or somatic variations are associated with the progression of CRC. In Cajuso et al.’s study (Cajuso et al. 2014), they used exome sequencing data to investigate the variation frequency of all genes containing the drought domain in 25 cases of microsatellite unstable (MSI) CRC. The authors identified 47 different somatic variations, including 18 frameshift (38%, c.5548delG), 3 nonsense (6%), 18 missense (38%), 1 splice site (2%), and 7 silent mutation (15%).
Fig. 1.
Flow chart of study selection
Table 1.
The characteristics of the 23 eligible studies
| Author and country | Publication year | Type of CRC | ARID1A variation (%) | ARID1A expression | Roles of ARID1A | Clinicopathologic features or biological effects of ARID1A | Antibodies of ARID1A | References |
|---|---|---|---|---|---|---|---|---|
| Jones, USA | 2012 | CRC | 12/119, 10% | Downregulated | Suppressor | ARID1A inactivation promoted CRC development | NA | Jones et al. (2012) |
| Chou, Australia | 2014 | CRC | 110/1876, 5.9%; | Loss of expression | Suppressor | No significant relationship between loss expression of ARID1A and the OS of CRC; Loss expression of ARID1A was associated with multiple clinical features, BRAFV600E variation and loss of mismatch repair protein expression (all P < 0.01) | Sigma 1: 100 | Chou et al. (2014) |
| Cajuso, Finland | 2014 | CRC | 18/46, 39% | Downregulated | Suppressor | Exome sequencing showed that ARID1A play an important role in microsatellite-unstable CRC via DNA binding activity and transcription coactivator activity | Santa Clara | Cajuso et al. (2014) |
| Xie, China | 2014 | CRC | 26/86, 30.2% | Loss of expression | Suppressor | Loss of ARID1A significantly associated with poor differentiation of CRC (P = 0.0009) | Rabbit antibodies Sigma 1:500 | Xie et al. (2014) |
| Ye, USA | 2014 | CRC | 22/257, 9% | Loss of expression | Suppressor | ARID1A loss was significantly associated with various clinicopathological features of CRC (all P < 0.05), and with a trend toward a worse OS (P > 0.05) | polyclonal antibody Sigma- 1:100 | Ye et al. (2014) |
| Wei, China | 2014 | CRC | 54/209, 25.8% | Loss of expression | Suppressor | ARID1A loss was correlated to late TNM stage, distant metastasis, and poor pathological classification (all P < 0.05) | Santa Cruz Biotechnology | Wei et al. (2014) |
| Lee, Korea | 2015 | CRC | 12/196, 6.1% | Loss of expression | Suppressor | Loss of ARID1A expression was significantly correlated with negative lymphatic invasion (P = 0.003) in CRC, and with expanding tumor border (CRC, P = 0.010) | Rabbit polyclonal, Sigma 1:100 | Lee et al. (2015) |
| Lee, USA | 2016 | CRC | 49/552, 8.9% | Loss of expression | Suppressor | ARID1A loss was associated with mismatch-repair protein deficiency. poor differentiation, lymphovascular invasion, and higher pT stage (all P < 0.05) | Rabbit polyclonal, Sigma, 1:300 | Lee et al. (2016) |
| Agaimy, Germany | 2016 | Colon, small bowel, and stomach cancer | 2/13, 15.4% | Loss of expression | Suppressor | NA | Rabbit polyclonal Abcam, 1:100 | Agaimy et al. (2016) |
| Fountzilas, USA | 2018 | CRC | 16/36, 44% | Loss of expression | Suppressor | ARID1A variations independently predicted for unfavorable DFS (HR = 1.99, 95%CI 1.11–3.54, P = 0.020) | NA | Fountzilas et al. (2018) |
| Wan, China | 2018 | CRC | 3/16, 18.8% | Loss of expression | Suppressor | NA | MygeneSeq technology | Wan et al. (2018) |
| Sen, USA | 2019 | CRC | 24/164, 14.6% | Loss of expression | Suppressor | The expression of ARID1A plays a key role in KRAS-mutated CRC cells | Cell Signaling, 1:500 | Sen et al. (2019) |
| Kishida, Japan | 2019 | CRC | 10/218, 4.6% | Loss of expression | Suppressor | Loss expression of ARID1A was significantly correlated to younger age, lymphatic invasion, and lymph node metastasis | Rabbit monoclonal, 1:500 | Kishida et al. (2019) |
| Xu, China | 2020 | sCRC | 1/28, 3.6% | Frameshift variation | Suppressor | ARID1A variations and the deficiency of its protein expression were significantly involved in advanced tumor depth, poor differentiation, lymphatic metastasis, BRAF V600E variation, MMR deficiency and MSI phenotype in tumors of CRC patients | NA | Xu et al. (2020) |
| Tokunaga-1,USA | 2020 | CRC | 468/5726, 8% | Downregulated | Suppressor | ARID1A variations could regulate DNA repair pathways | NA | Tokunaga et al. (2020) |
| Tokunaga-2,USA | 2020 | CRC | 50/619, 8% | Downregulated | Suppressor | ARID1A variation was significantly associated with a favourable immune profile indicative of a higher likelihood of response to immune checkpoint inhibitors | NA | Tokunaga et al. (2020) |
| Tokunaga-3,USA | 2020 | CRC | 104/1099, 10% | Downregulated | Suppressor | ARID1A variation was associated with right-sided primary tumor location and earlier tumor stage | NA | Tokunaga et al. (2020) |
| Tokunaga-4,USA | 2020 | CRC | 58/534, 11% | Downregulated | Suppressor | ARID1A variations lead to strong immune activation in CRC | NA | (Tokunaga et al. 2020) |
| Erfani, Iran | 2020 | CRC | 12/18, 66.7% | Loss or low expression | Suppressor | No significant relationship was found between the loss of ARID1A and the OS or the clinicopathological features in CRC | Rabbit antibody Sigma 1:200 | Erfani et al. (2020) |
| Villatoro, USA | 2020 | Colorectal adenocarcinoma | 16/338, 4.7%; | Deficiency | Suppressor | No difference in disease-specific or disease-free survival was found for ARID1A deficiency (all P > 0.05) | Abcam | Villatoro et al. (2020) |
| Stein, USA | 2020 | pCRC PM | pCRC: 179/617, 29%, PM: 42/348, 12% | Variation | Suppressor | NA | Primary antibody clones | Stein et al. (2020) |
| Wang-1, China | 2020 | CRC | 76/156, 48.7% | Downregulated | Suppressor | NA | NA | Wang et al. (2020) |
| Wang-2, China | 2020 | CRC | 17/225, 7.6% | Downregulated | Suppressor | NA | NA | Wang et al. (2020) |
| Jiang, China | 2020 | CRC | 89/1234, 7.2% | Variation | Suppressor | CRC patients with ARID1A variation showed a significantly longer DFS/PFS (HR = 0.74, P = 0.0026) | NA | Jiang et al. (2020) |
| Huang, China | 2021 | CRC | 65/630, 10.3% | Variation | Suppressor | NA | NA | (Huang et al. 2021) |
| Perna, Spain | 2021 | HG-CRCs | 12/29, 41.4% | Loss of expression | Suppressor | The differences in survival were not statistically significant (HR = 0.58, 95% CI = 0.23–1.49, P = 0.257) | Polyclonal Sigma, 1:500 | Perna et al. (2021) |
| Kamori, Japan | 2021 | CRC | 20/201, 10% | Variation | Suppressor | Tumor histological grade was significantly correlated with ARID1A variation status in those patients with right-sided CRC | Rabbit polyclonal, | Kamori et al. (2021) |
ARID1A AT-rich interactive domain 1A, CRC colorectal cancer, HR Hazard ratio, OR odds ratio, OS overall survival, DFS disease-free survival, HG-CRC high grade colorectal carcinomas, RCC right-sided colorectal cancer, LCC left-sided colorectal cancer, pCRC primary colorectal cancer, NA not available, PFS progression-free survival, RFS recurrence-free survival
Fig. 2.
Variation rate of ARID1A in CRC among different studies
Since the variation rate, clinical significance, and biological function of ARID1A are different in 23 qualified studies, we have therefore conducted an in-depth review of these eligible studies as follows.
ARID1A expression and variations in CRC
Variation rate of ARID1A in CRC among different studies
Comprehensive genome analysis is one useful tool to identify variations of various oncogenes and tumor-suppressor genes, particularly in those genes that code for chromatin remodeling factors (Centore et al. 2020; Goswami et al. 2020; Mao et al. 2013; Mathur 2018; Wei et al. 2014; Ye et al. 2014) One of such genes is ARID1A. However, the variation rate of ARID1A in CRC is low, Jones et al. (Jones et al. 2012) reported 10%, and Kim et al. (49) did not find variations. Based on this evidence, the effect of ARID1A loss in CRC is still underestimated. Wei et al. (2014) found that the ARID1A protein loss caused by immunohistochemistry occurred in 25.8% of primary CRC tumors, and the proportion was higher in stage IV CRC, which was 35.2%, suggesting that ARID1A protein loss is not very common in CRC.
In this narrative review, the evidence related to the ARID1A variants in CRC was comprehensively summarized. According to the data of 23 eligible studies, great different has been identified on the ARID1A variation rate varies greatly. As shown in Fig. 2, ARID1A variation in pCRC, PM-CRC, and HG-CRC was recorded at 29%, 12%, and 41.4%, respectively. Based on the previous publications, the highest variation rate of ARID1A in ovarian clear cell carcinoma, which is as high as 46–57% (Yoshino et al. 2020). But CRC did not make any comments or conclusions.
CRCs have two genetic and clinically distinct subtypes: chromosomal instability tumors (CIN) and microsatellite instability tumors (MSI). Most tumors show CIN, and about 15% are MSI. In MSI tumors, the mismatch repair (MMR) system is defective, which can usually correct a large number of errors that occur during DNA replication. This leads to a large number of small insertions and deletions in the repetitive regions surrounding the genome, especially in the short tandem repeat regions called microsatellites. The overall variation rate of MSI-CRC is estimated to be about 10 times that of microsatellite stable (MSS)-CRC (2012; Vogelstein et al. 2013). Tokunaga et al. (2020) compared the relationship between ARID1A variations and the molecular characteristics in CRC by using the next-generation sequencing, RNA sequencing, and the immunohistochemistry methods. They found that ARID1A variations were more common in primary and early age tumors on the right site of CRC. ARID1A mutant tumors mainly have gene variations related to chromatin modification, DNA repair, WNT signaling pathway, and EGFR inhibitor resistance pathway at the same time, and ARID1A variations have a strong regulatory effect on DNA repair pathways. CMS1, one of the consensus molecular subtypes of the CRC classification system, plays an essential role in immune response (Guinney et al. 2015). It was reported that ARID1A mutant samples proved a higher prevalence of CMS1 than ARID1A wild-type samples, which indicates ARID1A variation could result in strong immune activation (Tokunaga et al. 2020).
The data from the 23 eligible studies showed that the ARID1A variation rates and expression levels were different among different studies. These discrepancies might be related to various factors, e.g., different demographic features (i.e., sample size and race), CRC stage (early or advanced), different antibodies of anti-ARID1A, the assessments of the ARID1A protein expression (i.e., IHC, western blot, targeted sequencing analysis, qRT-PCR, tissue microarrays, and chromatin immunoprecipitation), and multifarious co-present or targeted genes being affected.
All studies indicate that ARID1A is low or absent in CRC, and ARID1A acts as a tumor suppressor, which is consistent with the function of ARID1A in other types of cancer (Wu and Roberts 2013). According to the current evidence, ARID1A variants are only expected to play a tumor suppressor effect CRC development.
Clinical significance of ARID1A in CRC
Although the high frequency of ARID1A variants has been observed in CRC, the prognostic value of ARID1A in CRC is still controversial. Jiang et al. (2020) found that DFS or PFS of patients with variations in ARID1A was significantly prolonged (HR = 0.74, 95%CI: 0.64–0.91, P = 0.0026). The OS of patients with ARID1A variation was significantly prolonged than those with wild-type (28 months vs. 18 months, P = 0.0092). In other words, ARID1A deletion predicts superior OS in stage IV CRC. Wei et al. (2014) analyzed 209 primary CRC tumor samples by IHC and discovered that ARID1A loss was detected in fifty-four (25.8%) primary CRC tumors. Moreover, the authors also observed that the distant metastasis rate was higher in patients with ARID1A loss than those without ARID1A loss (46.3% vs. 29.7%). In addition, Wei et al. further observed that ARID1A loss was related to the late TNM stage (P = 0.020) and poor pathological classification (P = 0.035). However, this study highlighted that positive ARID1A was associated with worse OS as compared to those with negative ARID1A in stage IV CRC (HR = 2.49, 95% CI: 1.13–5.51), indicating that ARID1A loss predicted superior OS in stage IV CRC. Largely consistent with Wei et al.’s findings, (Ye et al. 2014) found that ARID1A was related to tumor staging, lymphatic invasion, and tumor recurrence of CRC. ARID1A-deficient CRC has a higher proportion of lymph node and distant metastasis, and the overall 5-year survival rate shows a downward trend. Kishida et al. (2019) proved that lymphatic invasion is independently related to ARID1A expression. The above studies confirmed that the prognostic value of ARID1A variants in CRC is related to ARID1A defect or low expression. Tokunaga et al. (2020) believed that the ARID1A variation was related to the location of the primary tumor on the right side and the stage of the early tumor. The above data suggest that both low and high expression of ARID1A variants are related to the prognostic significance of CRC.
However, other studies did not support a positive association between the ARID1A variant or expression level and the prognosis of the disease in the CRC. Chou et al. (2014) found that there is a strong correlation between ARID1A expression loss and older age, right-sided tumors, larger tumor size, medullary morphology, high histological grade, BRAFV600E variation, and loss of mismatch repair protein expression (all P < 0.01), however, no significant association was found between loss of ARID1A expression and overall survival. Similarly, Lee et al. (2015) have also found that the loss of ARID1A expression was significantly related to the negative lymphatic invasion of CRC (P = 0.003), and tumor boundary expansion (CRC, P = 0.010). But there is no obvious correlation between ARID1A expression and 5-year OS. Lee et al. (2016) suggested that at a median follow-up of 49 months, ARID1A deletion was not associated with the OS, disease-specific survival, or recurrence-free survival in CRC patients.
Based on these studies, there is no significant correlation between ARID1A variants and the survival of CRC. One potential explanation for this observation may be due to the low number of CRC cases in some studies. For instance, the study by Erfani et al. (2020) reported that the ARID1A variation rate in CRC was as high as 66.7%. The authors found that among the 18 CRC tumors studied, 7 cases (38.8%) and 5 cases (27.7%) had no or low ARID1A expression, respectively. The limited number of patients may limit the study’s results. Conversely, studies involving a relative large number of patients are more likely to determine the poor prognostic significance of ARID1A variants in CRC (Fountzilas et al. 2018; Jiang et al. 2020; Xie et al. 2014). Certainly, various anti-ARID1A antibodies being used in every study conducted by IHC (e.g., antibody’s clone, manufacturer, dilution rate, IHC score, and cut-off value). These factors may be the underlying reasons behind the different results obtained in each study.
In summary, ARID1A variants may be predictive of metastasis, recurrence, and death of CRC patients, which indicates that ARID1A may play a crucial role in the development of CRC. It is worth noting that because some studies do not support the prognostic value of ARID1A, further studies are needed to verify the prognostic significance of ARID1A variants in CRC.
Molecular mechanisms of ARID1A variations on CRC
Since the causal association between ARID1A variation and CRC has been observed in multiple clinical studies, an exhaustive comprehension of the molecular functions of ARID1A is of great significance to researchers. ARID1A is a driver gene that encodes the DNA binding subunit of the SWI/SNF chromatin-remodeling complex. ARID1A provides specificity for the SWI/SNF complex and promotes protein–protein or protein-DNA molecular interactions. ARID1A inactivation may activate the cell cycle process, resulting in uncontrolled cell proliferation of cancer cells, indicating that ARID1A is a potential tumor suppressor function and the correlation between ARID1A deletion and tumorigenesis (Nagl et al. 2005). ARID1A might exert its biological functions and pathological impact on CRC by interacting with multiple mutated genes, affected signaling pathways, and some other factors.
ARID1A variation was associated with the co-occurrence variation of TP53 and some other genes
Some authors believe that there is a link between ARID1A and TP53 variations. TP53 (also named P53) is one of the most common genetic variants in human cancers and plays an important role in the regulation of the apoptosis, cell cycle, and DNA repair (Pinto et al. 2020). The variation of TP53 has become a critical biomarker of cancer prognosis due to its cancerous biological function. Guan et al. (2011) proposed the theory that ARID1A and p53 inhibit tumor growth synergistically at the molecular level. Other researchers suggested that ARID1A and TP53 variations are reciprocally exclusive and in charge of alternative pathways of tumorigenesis (Jones et al. 2012; Wang et al. 2011). In gastric cancer and gynecological cancer, ARID1A variation or loss of ARID1A protein expression is closely related to microsatellite instability, and negatively related to the variation of TP53 (Bosse et al. 2013). Tokunaga et al. (2020) reported that among the 20 genes assessed in the CRC cohort, only TP53 variations and ARID1A variations were reciprocally exclusive. ARID1A variation cause defects in cell cycle control point activation and TP53 variation in answer to DNA damage (Watanabe et al. 2014). ARID1A and TP53 jointly prevent tumorigenesis by inhibiting the transcriptional activation of genes downstream of tumors. As a result, the prognostic significance and biological effects of ARID1A in CRC may partly depend on the variation of TP53.
TP53 and ARID1A are considered to be the most common mutant genes in CRC (Stein et al. 2020). In addition to TP53, ARID1A variations can also occur simultaneously and may interact with some other genes (such as APC, FBXW7, PIK3CA, PD-L1, and KRAS), which may be involved in the development of CRC. Numerous studies have shown that ARID1A variations are often accompanied by Adenomatous polyposis coli (APC) variations in CRC. It was reported that the APC tumor suppressor is mutated in 27–71.7% of the CRC cases (Ashktorab et al. 2019; Huang et al. 2021). ARID1A and APC variations could increase the proliferation and survival of the CRC cells (Sen et al. 2019). It was reported that FBXW7 was one of the most frequently mutated genes of Chinese CRC patients (Liu et al. 2018). ARID1A variations are frequently accompanied by FBXW7 variations. Huang et al. (2021) found that both FBXW7 (17.5%) and ARID1A (10.3%) were the most common mutated genes in CRC patients via a genomic alteration analysis. Wang et al. (2020) showed that ARID1A (7.6%) and FBXW7 (6.2%) frequently mutated in the deficient mismatch repair CRC. In a study of the African Americans population, Ashktorab et al. (2019) demonstrated that ARID1A (7%) and FBXW7 (4%) were the common variants in CRC patients. PIK3CA is an oncogene in CRC. A comparative genomic analysis demonstrated that variations in ARID1A and PIK3CA (6.7%) genes between primary CRC and metastatic liver tumors of CRC (Lee et al. 2014). A genes exome sequencing study (Ashktorab et al. 2019) reported that the variation rate of ARID1A is 7% (8/121), while in PIK3CA is 6% (7/121) in CRC, and both two genes contributed to the carcinogenic process of CRC.
The programmed death-1 (PD-1)/programmed death-ligand 1 (PD-L1) axis is one of the effective therapeutic targets for immune checkpoint blockade therapy. Kamori et al. (2021) reported that CRC with ARID1A variations was likely to have a higher tumor mutational burden, while ARID1A-deficient CRC was frequently accompanied by enhanced PD-L1 expression by stromal cells. Kirsten rat sarcoma viral oncogene homolog (KRAS) and ARID1A variants have also been found by many researchers to coexist in CRC development. Several activation-type KRAS variations are observed in the group positive for the protein expression of ARID1A. The existence of ARID1A variations (44%) and KRAS variations (48%) has been demonstrated in stage I–III CRC (Fountzilas et al. 2018). Sen et al. (2019) suggested that ARID1A might facilitate KRAS signaling-regulated enhancer activity in CRC. They found that KRAS variations were particularly dependent on the presence of ARID1A. According to several reports, along with ARID1A variations, the KRAS variations rate were recorded ranging from 4.3% to 50% (Ashktorab et al. 2019; Cajuso et al. 2014; Huang et al. 2021). In KRAS mutant cells, after ARID1A is deleted, the enhancer co-occupied by ARID1A and AP1 transcription factors become inactive, resulting in a decrease in target gene expression (Sen et al. 2019). Therefore, in CRC with KRAS variation, mSWI/SNF complex may provide a unique and context-dependent treatment option.
Roles of the ARID domain-containing gene family
The ARID domain-containing gene family might also contribute to the tumorigenesis mechanisms of CRC, and act collectively with ARID1A variations. It was described ARID1A as belonging to the ARID domain-containing gene family (Cajuso et al. 2014). ARID1B (13%, 6/46), ARID2 (13%, 6/46), ARID4A (20%, 9/46) and ARID1A (39%, 18/46) was reported to frequently have variations in tumors. The results show that besides ARID1A, other members of the ARID gene family might also play a part in MSI CRC. Jones et al. (2012) evaluated 759 malignant tumors, including pancreas, breast, colon, stomach, lung, prostate, brain, and blood (leukemia). And truncated variations were found in 6% of the tumors studied; non-truncated cell variations were found in another 0.4% of tumors. Variations are most common in gastrointestinal samples, and 12 of 119 (10%) colon samples have ARID1A variations. The majority of the mutant colorectal tumors show microsatellite instability (MSI). The variations in these tumors are insertions or deletions of single nucleotide repeats outside the frame.
Roles of the affected signaling pathways
The pro-oncogenic roles of ARID1A variation on CRC development may also associate with its regulation on the activity of several affected signaling pathways. Some investigators even believe that variations in some tumor pathways are involved in the first step of progress from normal to CRC (Suleiman et al. 2015). Crosstalk between ARID1A and PI3K/Akt pathway has been detected in multiple cancers (Sun et al. 2021). Xie et al. (2014) believed that ARID1A depletion could promote CRC cell proliferation, enhance chemoresistance, and inhibit cell apoptosis by regulating the activity of the Akt signaling pathway. MTT experiments showed that overexpression of ARID1A in SW620 cells led to decreased cell proliferation, and depletion of ARID1A could increase cell growth rate. Sen et al. (2019) found that ARID1A has a previously unknown background-dependent tumor support function in CRC downstream of the KRAS signal and MEK/ERK pathway, showing that the absence of ARID1A enhances the proliferation of CRC cells. In addition, at the transcriptional level, the authors also detected a strong colocalization of ARID1A and TCF7L2, a downstream effector of the Wnt pathway. Aurora kinase A (AURKA) commonly functions in mitosis and non-mitotic biological processes. Wu et al. (Wu et al. 2018) demonstrated that ARID1A loss contributed to the growth and survival of the CRC cells via negatively regulating AURKA-mediated signaling and the downstream genes, such as PLK1 and CDC25C. A gene set enrichment analysis conducted by Tokunaga et al. showed that ARID1A mutant status was closely correlated to the DNA repair pathway, mediating chemotherapy/radiotherapy sensitivity of CRC (Tokunaga et al. 2020). Since intestinal deletion of ARID1A was tightly associated with CRC development, Hiramatsu et al. believed the underlying molecular mechanisms might be related to the disruption of the intestinal homeostasis, and pointed out that the Wnt signaling pathway crucially involved this action.
ARID1A variation associated with MMR deficiency and hypermethylation
MMR deficiency is one of the important prognostic factors in CRC. The significant association between ARID1A deletion and MMR defect in CRC has been fully demonstrated in the literature, showing that loss ARID1A expression in 15–25% of MMR-deficient versus 4–6% of MMR-intact CRC cases, respectively (Agaimy et al. 2016). Lee et al. (Lee et al. 2016) reported that ARID1A loss was significantly more prevalent in the MMR-deficient CRC cases than in the MMR-proficient CRC cases (18.7% vs 6.3%, P < 0.001). A previous study (Ye et al. 2014) indicated that ARID1A variations were associated with a worse outcome among the MMR-abnormal CRC cases. This study also demonstrated that the main mechanism of MMR deficiency in ARID1A-deficient tumors was hypermethylation of the mutL homolog 1 (MLH1) gene promoter (Ye et al. 2014). BRAF V600E variations are frequently shown in these MMR-deficient tumors with ARID1A deletion. By comparison, MMR defects due to germline variations (i.e., Lynch syndrome) appear to occur mainly in ARID1A-preserving cases (Chou et al. 2014; Ye et al. 2014). The association between ARID1A deletion and MMR defect co-exists in the early CRC. In addition, most of these MMR-deficient ARID1A deletion tumors do show simultaneous deletion of MLH1 and PMS2, and this pattern is expected in tumors where the MLH1 promoter is methylated (Lee et al. 2016). Chou et al. (Chou et al. 2014) believe that, considering these associations, ARID1A may be used as a marker of somatic hypermethylation for the classification genetic testing of Lynch syndrome. It is worth noting that the MMR defect pattern that suggests Lynch syndrome can also occur in tumors with ARID1A deletion. Promoter hypermethylation is one of the main reasons for ARID1A variations. ARID1A loss leads to epigenetic alterations by a deficient SWI/SNF complex with subsequent MLH1 promoter methylation. Chou et al. reported that a low level of ARID1A was closely associated with larger tumor size, right-sided tumors, and high histological grade of CRC, which were features of somatic hypermethylation (Chou et al. 2014). Erfani et al. (2020) found that promoter DNA hypermethylation significantly promoted the silencing or down-regulation of ARID1A in CRC cell lines. The authors also suggested that ARID1A might be an effective tumor suppressor gene in certain subtypes of CRCs because it affects many genes through its role in chromatin remodeling expression (Erfani et al. 2020). Based on the above evidence, promoter hypermethylation may serve as a down-regulation mechanism of ARID1A in CRC.
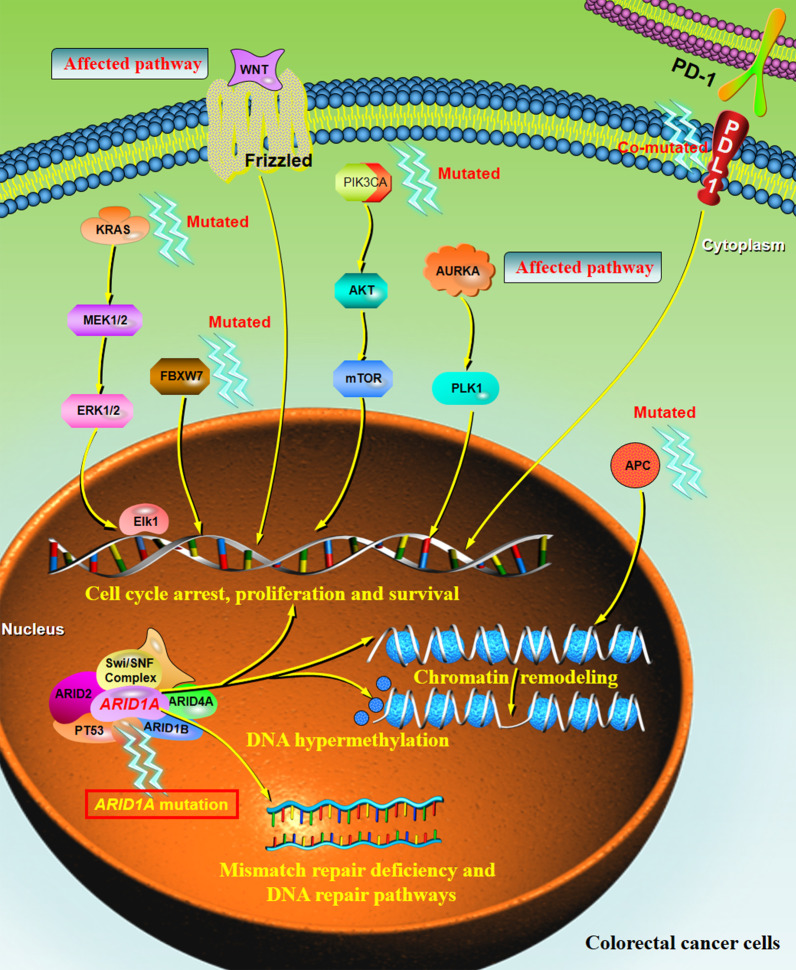
In summary, ARID1A variants seem to play an important role in the occurrence and progression of CRC tumors. As illustrated in Fig. 3, this schematic diagram summarizes the multi-factor mechanisms that may be involved in the development of ARID1A-driven CRC, including cell cycle arrest, chromatin remodeling and chromosome organization, and DNA hypermethylation. The interactions of multiple genes (i.e., TP53, APC, FBXW7, PIK3CA, PD-L1, and KRAS) and the affected signaling pathways (i.e., PI3K/Akt, MEK/ERK pathway, Wnt pathway, AURKA-mediated signaling, and DNA repair pathway) enhance the process of cell proliferation and anti-apoptosis. Nevertheless, further relevant studies are still needed to better clarify the potential mechanism of ARID1A variations that trigger the development of CRC.
Fig. 3.
The mechanism by which the ARID1A variation contributes to the pathogenesis of CRCs. ARID1A, a subunit of the chromatin remodeling protein SWI/SNF, is considered to be associated with the tumorigenesis and the progression of CRCs. The process is initiated by the mutation of multiple genes (i.e., TP53, ARID domain-containing gene family, APC, FBXW7, PIK3CA, PD-L1, and KRAS), the dysregulation of several signaling pathways (i.e., PI3K/Akt signaling, MEK/ERK pathway, WNT pathway, AURKA-mediated signaling, and DNA repair pathways), chromatin remodeling, mismatch repair deficiency, and DNA hypermethylation, leading to the cell cycle arrest, proliferation, and survival of the CRC cells. ARID1A AT-rich interaction domain 1A, APC adenomatous polyposis coli, FBXW7 F-Box and WD repeat domain containing 7, PIK3CA phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α, SWI/SNF SWItch/Sucrose non-fermenting, PD-L1 programmed death ligand 1, KRAS Kirsten rat sarcoma viral oncogene homolog, AURKA aurora kinase A
Limitations and perspectives
This is the first study to comprehensively review ARID1A variations associated specifically with CRC from the clinical through the molecular level. However, several drawbacks in the present study should be acknowledged. First, though ARID1A variation is closely associated with the clinicopathologic features of CRC (i.e., TNM stage, tumor location, and histological grade), its role on the prognostic significance of CRC remains controversial among the 23 eligible studies, especially on the survival. Second, large differences in the variation rate of ARID1A in CRC were observed among different included studies, ranging between 3.6 and 66.7%. This heterogeneity might be partly due to various geographic populations, study design, sample size, different tumor staging, gender, age, and the assessments for the expression level of ARID1A. Third, the biomarker role, the potential antitumor effect, and the underlying biological mechanisms for the participation of ARID1A variants in the tumorigenesis of CRC, development, prediction, and therapy need to be further studied.
Conclusions
In the present review, all of the 23 included studies consistently suggest that ARID1A is a tumor suppressor in CRC. The loss of ARID1A expression may represent the ARID1A-driven carcinogenesis in CRC. However, the rate of ARID1A variation in CRC cases is diverse across different studies, ranging from 3.6 to 66.7%. Though ARID1A variation status has several clinical impacts on CRC, such as serving as a biomarker for survival prognosis and various therapies, no significant differences were observed between the variation and wild type of ARID1A in a few studies. The biological functions and pathological impacts of ARID1A variations on CRC might be correlated to the co-occurrence variation of other genes (i.e., TP53 and KRAS) and the regulation of signaling pathways (i.e., Akt signaling and WNT signaling). Upon further validation with the clinical and biological features of ARID1A variations in CRC by future studies, ARID1A has the potential to serve as an important prognostic factor and individualized therapeutic target for CRC.
Acknowledgements
Not applicable.
Abbreviations
- CRC
Colorectal cancer
- ARID1A
AT-rich interaction domain 1A
- SWI/SNF
SWItch/sucrose non-fermenting
- APC
Adenomatous polyposis coli
- FBXW7
F-box and WD repeat domain containing 7
- PIK3CA
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
- PD-L1
Programmed death ligand 1
- KRAS
Kirsten rat sarcoma viral oncogene homolog
- AURKA
Aurora kinase A
Author contributions
RL and WW wrote the manuscript. XW summarized the table. ZJ and TF designed the figures. ZS, DL, and XW participated in the revision of manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the grants from the Zhejiang Medical and Health Science and Technology Program (No. 2022RC297, 2021PY085, and 2022KY1402); the Natural Science Foundation of Zhejiang Province (No. LQ22H040009); the Clinical Research Funding of Zhejiang Medical Association (NO. 2019ZYC-A182); the Science and Technology Planning Project of Taizhou City, Zhejiang Province (No. 20ywb40 and 1902ky37); the Project of Taizhou Central Hospital (Taizhou University Hospital) (NO. 2019KT015); the Scientific Research Project of Taizhou University (NO. 2017PY047); and the High-level Hospital Construction Research Project of Maoming People’s Hospital.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors approved for the publication.
Competing interests
Shankun Zhao, Weizhou Wu, Zufu Jiang, Fuqin Tang, Lingzhi Ding, Weifang Xu, and Libin Ruan declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shankun Zhao and Weizhou Wu have contributed equally to this work.
Contributor Information
Weifang Xu, Email: weifangxu2019@163.com.
Libin Ruan, Email: libinruan@126.com.
References
- (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407): 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed]
- Agaimy A, Daum O, Markl B, Lichtmannegger I, Michal M, Hartmann A. Swi/snf complex-deficient undifferentiated/rhabdoid carcinomas of the gastrointestinal tract: a series of 13 cases highlighting mutually exclusive loss of smarca4 and smarca2 and frequent co-inactivation of smarcb1 and smarca2. Am J Surg Pathol. 2016;40(4):544–553. doi: 10.1097/PAS.0000000000000554. [DOI] [PubMed] [Google Scholar]
- Ashktorab H, Azimi H, Varma S, Lee EL, Laiyemo AO, Nickerson ML, et al. Driver genes exome sequencing reveals distinct variants in African Americans with colorectal neoplasia. Oncotarget. 2019;10(27):2607–2624. doi: 10.18632/oncotarget.26721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse T, ter Haar NT, Seeber LM, Diest Pv, Hes FJ, Vasen HF, et al. Loss of arid1a expression and its relationship with pi3k-akt pathway alterations, tp53 and microsatellite instability in endometrial cancer. Mod Pathol. 2013;26(11):1525–1535. doi: 10.1038/modpathol.2013.96. [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Cajuso T, Hanninen UA, Kondelin J, Gylfe AE, Tanskanen T, Katainen R, et al. Exome sequencing reveals frequent inactivating mutations in arid1a, arid1b, arid2 and arid4a in microsatellite unstable colorectal cancer. Int J Cancer. 2014;135(3):611–623. doi: 10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- Centore RC, Sandoval GJ, Soares L, Kadoch C, Chan HM. Mammalian swi/snf chromatin remodeling complexes: emerging mechanisms and therapeutic strategies. Trends Genet. 2020;36(12):936–950. doi: 10.1016/j.tig.2020.07.011. [DOI] [PubMed] [Google Scholar]
- Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. Arid1a-dna interactions are required for promoter occupancy by swi/snf. Mol Cell Biol. 2013;33(2):265–280. doi: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou A, Toon CW, Clarkson A, Sioson L, Houang M, Watson N, et al. Loss of arid1a expression in colorectal carcinoma is strongly associated with mismatch repair deficiency. Hum Pathol. 2014;45(8):1697–1703. doi: 10.1016/j.humpath.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Chu DZ, Lang NP, Thompson C, Osteen PK, Westbrook KC. Peritoneal carcinomatosis in nongynecologic malignancy. A prospective study of prognostic factors. Cancer-Am Cancer Soc. 1989;63(2):364–367. doi: 10.1002/1097-0142(19890115)63:2<364::aid-cncr2820630228>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Cornen S, Adelaide J, Bertucci F, Finetti P, Guille A, Birnbaum DJ, et al. Mutations and deletions of arid1a in breast tumors. Oncogene. 2012;31(38):4255–4256. doi: 10.1038/onc.2011.598. [DOI] [PubMed] [Google Scholar]
- Dawson LE, Russell AH, Tong D, Wisbeck WM. Adenocarcinoma of the sigmoid colon: sites of initial dissemination and clinical patterns of recurrence following surgery alone. J Surg Oncol. 1983;22(2):95–99. doi: 10.1002/jso.2930220208. [DOI] [PubMed] [Google Scholar]
- De P, Dey N. Mutation-driven signals of arid1a and pi3k pathways in ovarian carcinomas: alteration is an opportunity. Int J Mol Sci. 2019 doi: 10.3390/ijms20225732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas SG, Muller DC, Le Magnen C, Federer-Gsponer J, Seifert HH, Ruiz C, et al. Immunocytochemistry for arid1a as a potential biomarker in urine cytology of bladder cancer. Cancer Cytopathol. 2019;127(9):578–585. doi: 10.1002/cncy.22167. [DOI] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45(5):478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfani M, Hosseini SV, Mokhtari M, Zamani M, Tahmasebi K, Alizadeh NM, et al. Altered arid1a expression in colorectal cancer. BMC Cancer. 2020;20(1):350. doi: 10.1186/s12885-020-6706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y, Cohen AM, Fortner JG, Enker WE, Turnbull AD, Coit DG, et al. Liver resection for colorectal metastases. J Clin Oncol. 1997;15(3):938–946. doi: 10.1200/JCO.1997.15.3.938. [DOI] [PubMed] [Google Scholar]
- Fountzilas E, Kotoula V, Tikas I, Manousou K, Papadopoulou K, Poulios C, et al. Prognostic significance of tumor genotypes and cd8+ infiltrates in stage i–iii colorectal cancer. Oncotarget. 2018;9(86):35623–35638. doi: 10.18632/oncotarget.26256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko J, Shi Q, Goldman CD, Pockaj BA, Nelson GD, Goldberg RM, et al. Treatment of colorectal peritoneal carcinomatosis with systemic chemotherapy: a pooled analysis of north central cancer treatment group phase iii trials n9741 and n9841. J Clin Oncol. 2012;30(3):263–267. doi: 10.1200/JCO.2011.37.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- Goswami S, Chen Y, Anandhan S, Szabo PM, Basu S, Blando JM, et al. Arid1a mutation plus cxcl13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci Transl Med. 2020 doi: 10.1126/scitranslmed.abc4220. [DOI] [PubMed] [Google Scholar]
- Guan B, Wang TL, Shih I. Arid1a, a factor that promotes formation of swi/snf-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71(21):6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Gao M, Wu CH, Wang TL, Shih I. Functional analysis of in-frame indel arid1a mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia. 2012;14(10):986–993. doi: 10.1593/neo.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Li H, Shi X, Lin M, Liao C, Zhang S, et al. Characterization of genomic alterations in Chinese colorectal cancer patients. Jpn J Clin Oncol. 2021;51(1):120–129. doi: 10.1093/jjco/hyaa182. [DOI] [PubMed] [Google Scholar]
- Jiang T, Chen X, Su C, Ren S, Zhou C. Pan-cancer analysis of arid1a alterations as biomarkers for immunotherapy outcomes. J Cancer. 2020;11(4):776–780. doi: 10.7150/jca.41296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Wang TL, Shih I, Mao TL, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene arid1a in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, et al. Somatic mutations in the chromatin remodeling gene arid1a occur in several tumor types. Hum Mutat. 2012;33(1):100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Crabtree GR. Reversible disruption of mswi/snf (baf) complexes by the ss18-ssx oncogenic fusion in synovial sarcoma. Cell. 2013;153(1):71–85. doi: 10.1016/j.cell.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamori T, Oki E, Shimada Y, Hu Q, Hisamatsu Y, Ando K, et al. The effects of arid1a mutations on colorectal cancer and associations with pd-l1 expression by stromal cells. Cancer Rep (hoboken) 2021 doi: 10.1002/cnr2.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida Y, Oishi T, Sugino T, Shiomi A, Urakami K, Kusuhara M, et al. Associations between loss of arid1a expression and clinicopathologic and genetic variables in t1 early colorectal cancer. Am J Clin Pathol. 2019;152(4):463–470. doi: 10.1093/ajcp/aqz062. [DOI] [PubMed] [Google Scholar]
- Koppe MJ, Boerman OC, Oyen WJ, Bleichrodt RP. Peritoneal carcinomatosis of colorectal origin: incidence and current treatment strategies. Ann Surg. 2006;243(2):212–222. doi: 10.1097/01.sla.0000197702.46394.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8(12):686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Haq F, Kim D, Jun C, Jo HJ, Ahn SM, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS ONE. 2014;9(3):e90459. doi: 10.1371/journal.pone.0090459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kim DW, Lee HS, Ihn MH, Oh HK, Park DJ, et al. Loss of at-rich interactive domain 1a expression in gastrointestinal malignancies. Oncology. 2015;88(4):234–240. doi: 10.1159/000369140. [DOI] [PubMed] [Google Scholar]
- Lee LH, Sadot E, Ivelja S, Vakiani E, Hechtman JF, Sevinsky CJ, et al. Arid1a expression in early stage colorectal adenocarcinoma: an exploration of its prognostic significance. Hum Pathol. 2016;53:97–104. doi: 10.1016/j.humpath.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang C, Li X, Luo W, Roy B, Xiong T, et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget. 2018;9(44):27412–27422. doi: 10.18632/oncotarget.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao TL, Shih I. The roles of arid1a in gynecologic cancer. J Gynecol Oncol. 2013;24(4):376–381. doi: 10.3802/jgo.2013.24.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R. Arid1a loss in cancer: towards a mechanistic understanding. Pharmacol Ther. 2018;190:15–23. doi: 10.1016/j.pharmthera.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Megaridis MR, Lu Y, Tevonian EN, Junger KM, Moy JM, Bohn-Wippert K, et al. Fine-tuning of noise in gene expression with nucleosome remodeling. APL Bioeng. 2018;2(2):26106. doi: 10.1063/1.5021183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl NJ, Patsialou A, Haines DS, Dallas PB, Beck GJ, Moran E. The p270 (arid1a/smarcf1) subunit of mammalian swi/snf-related complexes is essential for normal cell cycle arrest. Cancer Res. 2005;65(20):9236–9244. doi: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- Ogiwara H, Takahashi K, Sasaki M, Kuroda T, Yoshida H, Watanabe R, et al. Targeting the vulnerability of glutathione metabolism in arid1a-deficient cancers. Cancer Cell. 2019;35(2):177–190. doi: 10.1016/j.ccell.2018.12.009. [DOI] [PubMed] [Google Scholar]
- Perna C, Navarro A, Ruz-Caracuel I, Caniego-Casas T, Cristobal E, Leskela S, et al. Molecular heterogeneity of high grade colorectal adenocarcinoma. Cancers (basel) 2021 doi: 10.3390/cancers13020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto EM, Zambetti GP. What 20 years of research has taught us about the tp53 p.r337h mutation. Cancer-Am Cancer Soc. 2020;126(21):4678–4686. doi: 10.1002/cncr.33143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AH, Tong D, Dawson LE, Wisbeck WM, Griffin TW, Laramore GE, et al. Adenocarcinoma of the retroperitoneal ascending and descending colon: sites of initial dissemination and clinical patterns of recurrence following surgery alone. Int J Radiat Oncol Biol Phys. 1983;9(3):361–365. doi: 10.1016/0360-3016(83)90297-3. [DOI] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, et al. Integrated genomic analyses identify arid1a and arid1b alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45(1):12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen M, Wang X, Hamdan FH, Rapp J, Eggert J, Kosinsky RL, et al. Arid1a facilitates kras signaling-regulated enhancer activity in an ap1-dependent manner in colorectal cancer cells. Clin Epigenetics. 2019;11(1):92. doi: 10.1186/s13148-019-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, et al. Convergent structural alterations define switch/sucrose nonfermentable (swi/snf) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109(5):E252–E259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- Stein MK, Williard FW, Xiu J, Tsao MW, Martin MG, Deschner BW, et al. Comprehensive tumor profiling reveals unique molecular differences between peritoneal metastases and primary colorectal adenocarcinoma. J Surg Oncol. 2020;121(8):1320–1328. doi: 10.1002/jso.25899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleiman SH, Koko ME, Nasir WH, Elfateh O, Elgizouli UK, Abdallah MO, et al. Exome sequencing of a colorectal cancer family reveals shared mutation pattern and predisposition circuitry along tumor pathways. Front Genet. 2015;6:288. doi: 10.3389/fgene.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Teng F, Xing P, Li J. Arid1a serves as a receivable biomarker for the resistance to egfr-tkis in non-small cell lung cancer. Mol Med. 2021;27(1):138. doi: 10.1186/s10020-021-00400-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Takenaka M, Okamoto A, Bowtell D, Kohno T. Treatment strategies for arid1a-deficient ovarian clear cell carcinoma. Cancers (basel) 2021 doi: 10.3390/cancers13081769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga R, Xiu J, Goldberg RM, Philip PA, Seeber A, Battaglin F, et al. The impact of arid1a mutation on molecular characteristics in colorectal cancer. Eur J Cancer. 2020;140:119–129. doi: 10.1016/j.ejca.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toumpeki C, Liberis A, Tsirkas I, Tsirka T, Kalagasidou S, Inagamova L, et al. The role of arid1a in endometrial cancer and the molecular pathways associated with pathogenesis and cancer progression. In Vivo. 2019;33(3):659–667. doi: 10.21873/invivo.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villatoro TM, Ma C, Pai RK. Switch/sucrose nonfermenting nucleosome complex-deficient colorectal carcinomas have distinct clinicopathologic features. Hum Pathol. 2020;99:53–61. doi: 10.1016/j.humpath.2020.03.009. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LJ, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan JF, Li XQ, Zhang J, Yang LF, Zhu J, Li GC, et al. Aneuploidy of chromosome 8 and mutation of circulating tumor cells predict pathologic complete response in the treatment of locally advanced rectal cancer. Oncol Lett. 2018;16(2):1863–1868. doi: 10.3892/ol.2018.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nagl NG, Wilsker D, Van Scoy M, Pacchione S, Yaciuk P, et al. Two related arid family proteins are alternative subunits of human swi/snf complexes. Biochem J. 2004;383(Pt 2):319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of arid1a in molecular subtypes of gastric cancer. Nat Genet. 2011;43(12):1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- Wang J, Li R, He Y, Yi Y, Wu H, Liang Z. Next-generation sequencing reveals heterogeneous genetic alterations in key signaling pathways of mismatch repair deficient colorectal carcinomas. Mod Pathol. 2020;33(12):2591–2601. doi: 10.1038/s41379-020-0612-2. [DOI] [PubMed] [Google Scholar]
- Wang R, Chen M, Ye X, Poon K. Role and potential clinical utility of arid1a in gastrointestinal malignancy. Mutat Res Rev Mutat Res. 2021;787:108360. doi: 10.1016/j.mrrev.2020.108360. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T, Kohno T, et al. Swi/snf factors required for cellular resistance to dna damage include arid1a and arid1b and show interdependent protein stability. Cancer Res. 2014;74(9):2465–2475. doi: 10.1158/0008-5472.CAN-13-3608. [DOI] [PubMed] [Google Scholar]
- Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL, Zeng ZL, et al. Clinicopathologic and prognostic relevance of arid1a protein loss in colorectal cancer. World J Gastroenterol. 2014;20(48):18404–18412. doi: 10.3748/wjg.v20.i48.18404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, et al. Arid1a mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BG, Roberts CW. Swi/snf nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- Wu JN, Roberts CW. Arid1a mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3(1):35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Lyu J, Yang EJ, Liu Y, Zhang B, Shim JS. Targeting aurka-cdc25c axis to induce synthetic lethality in arid1a-deficient colorectal cancer cells. Nat Commun. 2018;9(1):3212. doi: 10.1038/s41467-018-05694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Fu L, Han Y, Li Q, Wang E. Decreased arid1a expression facilitates cell proliferation and inhibits 5-fluorouracil-induced apoptosis in colorectal carcinoma. Tumour Biol. 2014;35(8):7921–7927. doi: 10.1007/s13277-014-2074-y. [DOI] [PubMed] [Google Scholar]
- Xu HX, Zhu P, Zheng YY, Zhang X, Chen YQ, Qiao LC, et al. Molecular screening and clinicopathologic characteristics of lynch-like syndrome in a Chinese colorectal cancer cohort. Am J Cancer Res. 2020;10(11):3920–3934. [PMC free article] [PubMed] [Google Scholar]
- Ye J, Zhou Y, Weiser MR, Gonen M, Zhang L, Samdani T, et al. Immunohistochemical detection of arid1a in colorectal carcinoma: loss of staining is associated with sporadic microsatellite unstable tumors with medullary histology and high tnm stage. Hum Pathol. 2014;45(12):2430–2436. doi: 10.1016/j.humpath.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Yoshino J, Akiyama Y, Shimada S, Ogura T, Ogawa K, Ono H, et al. Loss of arid1a induces a stemness gene aldh1a1 expression with histone acetylation in the malignant subtype of cholangiocarcinoma. Carcinogenesis. 2020;41(6):734–742. doi: 10.1093/carcin/bgz179. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu Y, Wu W, Wang P, Wang Y, Jiang H, et al. Arid1a variations in cholangiocarcinoma: clinical significances and molecular mechanisms. Front Oncol. 2021;11:693295. doi: 10.3389/fonc.2021.693295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.