Abstract
In certain conditions, dye-conjugated icosahedral virus shells exhibit suppression of concentration quenching. The recently observed radiation brightening at high fluorophore densities has been attributed to coherent emission, i.e., to a cooperative process occurring within a subset of the virus-supported fluorophores. Until now, the distribution of fluorophores among potential conjugation sites and the nature of the active subset remained unknown. With the help of mass spectrometry and molecular dynamics simulations, we found which conjugation sites in the brome mosaic virus capsid are accessible to fluorophores. Reactive external surface lysines but also those at the lumenal interface where the coat protein N-termini are located showed virtually unrestricted access to dyes. The third type of labeled lysines was situated at the intercapsomeric interfaces. Through limited proteolysis of flexible N-termini, it was determined that dyes bound to them are unlikely to be involved in the radiation brightening effect. At the same time, specific labeling of genetically inserted cysteines on the exterior capsid surface alone did not lead to radiation brightening. The results suggest that lysines situated within the more rigid structural part of the coat protein provide the chemical environments conducive to radiation brightening, and we discuss some of the characteristics of these environments.
Graphical Abstract
INTRODUCTION
Luminescent probes are an enabling factor of progress in cutting-edge areas of biotechnology including nonintrusive biomedical imaging and sensing of biomechanical stresses at the submicroscopic scale.1-3 High signal-to-background ratio (contrast), small footprint, photostability, and biocompatibility are key attributes when designing luminescent imaging agents.4 Further progress in luminescence imaging technologies is expected from a combination of brighter probes along with lower noise and higher sensitivity detection strategies.5-7 For brighter probes and higher contrast, a common approach has been to load polymeric nanoparticles with a large number of fluorophores.4,8 However, this approach is limited by a phenomenon known as concentration fluorescence quenching: at high fluorophore densities, the emission quantum yield decreases significantly due to homo-Förster resonance energy transfer (homo-FRET).9 This issue, common in multiemitter nanoparticles, can be partially circumvented if the fluorophores are grafted at well-defined locations on the surface of symmetric virus shells.10 In this case, the fixed, well-defined separation distance between fluorophores afforded by the precise locations on the virus surface can limit homo-FRET quenching, which has a rate that rapidly increases with the inverse of the interfluorophore distance.9 Moreover, under strong pumping conditions (fs and ps pulsed excitation), due to coupling, molecular fluorophores can emit collectively at an accelerated rate with respect to a system of incoherent fluorophores.11 At high numbers of fluorophores, the rate of this collective effect exceeds that of homo-FRET, thus leading to suppression of quenching. Such radiation brightening via collective relaxation can be achieved in solution and at room temperature.11
In previous work,11 the brome mosaic virus (BMV), a nonenveloped icosahedral virus with a diameter of ~28 nm, provided the structurally precise molecular scaffold required for the conjugation of hundreds of fluorophores at well-defined locations. After conjugation, BMV became more stable against changes in pH and storage temperature, but no differences could be observed between the native and the fluorophore-conjugated virus structures by cryoelectron microscopy and single-particle reconstruction at a resolution of ≈7 Å.11 Because of the stability enhancement, we concluded that at least some of the fluorophores interact strongly with the virus shell. The brightening, and the extra stability, vanished when a longer linker connecting the fluorophore and the capsid protein was used.11 Radiation brightening was not observed when the virus template was replaced by a glass nanoparticle.11 The latter suggests a relationship between the virus scaffold structure and the brightening effect. The current experimental study is an exploration of this relationship, with the goal of better understanding the role of the virus as a template.
We note that although the development of functional multifluorophore architectures with high absorbance has begun more than a decade ago and currently expanded to DNA assemblies, viral capsids, protein templates, polymers, dendrimers, quantum dots, and antibodies,12-17 only very recently, other reports have emerged on the occurrence and/or mechanisms of radiation brightening via interfluorophore coupling.18,19 Moreover, while super-radiant molecular aggregates have been extensively studied in relation to coherent energy transfer,20-22 they are qualitatively different from the virus-templated systems discussed here. The nearest-neighbor distance in virus-templated fluorophores is an order of magnitude greater than in molecular aggregates, which precludes the same coupling mechanism.
The relatively rigid virus scaffold affords the placement of fluorophores in a consistent manner. Rigidity alleviates dephasing of coherently excited emitters, promoting collective behavior.23 A recent study by Rolczynski and co-workers indicated that a structured nuclear environment within the photosynthetic complex promotes coherent energy transfer.24 Another factor that promotes collective behavior is the size of the coupled set of emitters. This is because the radiative decay rate of a coherent ensemble of N emitters scales as N2, while for an uncorrelated ensemble, it scales with N.25 Moreover, icosahedral symmetry has been predicted to be a determinant of unique collective photophysical properties in a system of coupled quantum dots.26 With viruses, fluorophore attachment can be achieved in a versatile manner, either through covalent modification (as in this work) or by genetic insertion of fluorescent amino acids, aptamers, or proteins.14,19,27,28
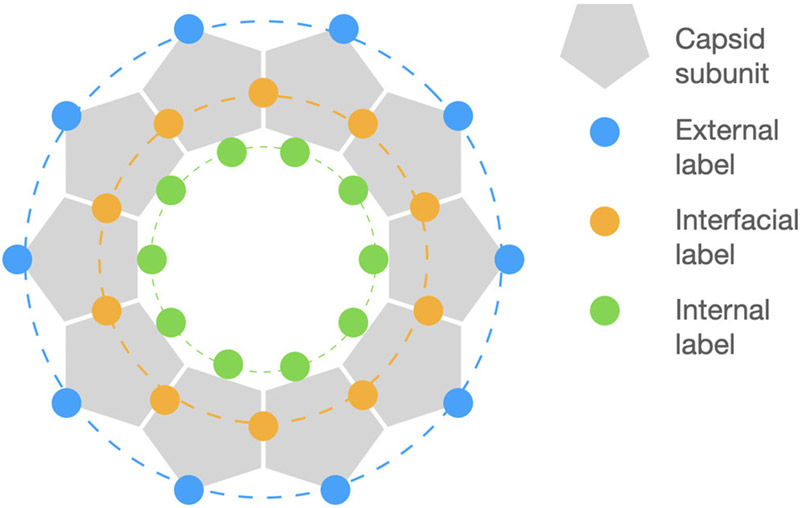
The reactive lysine residues in BMV can be categorized according to three distinct label environments differentiated with respect to radial location, backbone dynamics, and exposure to solvent. From the center to the virus surface, we distinguish among “internal” labels, mainly on the N-terminal coat protein domain, “interfacial” labels, at the intercapsomer interface, and “external” labels, on the outer virus surface (Figure 1).11,29 The reactivity of these residues is influenced by accessibility, local pKa,30-32 and the nature of the label.33
Figure 1.
BMV-OG schematic. There are three distinct types of fluorophore locations separated into three radial layers.
Amide hydrogen exchange studies suggest that the BMV capsid is characterized by rich dynamics.34,35 The relative solvent accessibility of BMV lysines was explored by means of chemical labeling and tandem mass spectrometry.36 Such bottom-up proteomics approach involves enzymatic digestion combined with peptide mapping37 and has proven successful for rapid virus identification38 and elucidation of fluorophore modification sites in proteins.39,40 Here, we probe the fluorophore conjugation sites on the BMV capsid by employing a similar proteomics method to elucidate the active distribution of the fluorescein derivative commercially known as Oregon Green (OG). By selectively removing label layers and examining the changes in photophysical properties, we evaluate the role of each layer in the radiation brightening phenomenon. We further employ all-atom molecular dynamics (MD) simulations to investigate relevant physical properties of the BMV capsid structure. Altogether, the results may prove useful in designing future enhanced performance luminescent probes.
METHODS
BMV Purification.
wtBMV was expressed in Nicotiana benthamiana via Agrobacterium-mediated gene delivery according to the previously described protocol.41 Seven days after infection, N. benthamiana leaves were homogenized in virus buffer [250 mM NaOAc, 10 mM MgCl2 (pH 4.5)], followed by sucrose cushion and CsCl centrifugation. The resulting virus band was collected and dialyzed against SAMA buffer [50 mM NaOAc, 8 mM Mg(OAc)2(pH 4.6)], followed by purification using liquid chromatography, and determination of virus concentration using NanoDrop spectrophotometer.
Fluorescent Modification of the Virus.
The lysines of wtBMV were modified by bioconjugation with Oregon Green 488 carboxylic acid, succinimidyl ester. wtBMV in SAMA buffer was mixed with sodium bicarbonate buffer (100 mM, pH 8.2, keep it cold) in a 1:1 ratio to obtain a concentration of 1013 particles/ml (final pH ~ 6.5). Dye stock was freshly prepared in dimethyl sulfoxide (DMSO) and was added to virus solution at varied ratios ranging from 500 to 8500 dyes/capsid. The reaction buffer should have ~6.5% DMSO in total. The reaction was left for 1–2 h at room temperature, followed by the removal of free dyes via filter wash and dialysis with SAMA buffer [50 mM NaOAc, 8 mM Mg(OAc)2(pH 4.6)]. The OG-labeled BMV is referred to as BMV-OG, and UV–vis spectroscopy was performed to confirm the absence of free dyes in the solution.
To check the electrophoretic mobility, purified wtBMV and BMV-OG were loaded in a 1% agarose gel. Electrophoresis was performed in SAMA buffer (pH 4.6) at 80 V for 1.5 h at 4 °C. The bands were visualized by an Amersham Typhoon 9210 Variable Mode Imager after Coomassie Blue staining.
Fluorescent Modification of BMV Mutant.
BMV mutant V168C has 180 cysteine residues on the capsid structure.42 First, the disulfide bonds on the mutant (dispersed in pH 6 SAMA buffer) were reduced with a 3-fold molar excess of Tris (2-carboxyethyl)phosphine (TCEP) for an hour under oxygen-reduced conditions (by buffer degassing and bubbling nitrogen for a few minutes before the reaction). Then, 200–5000 times excess of Oregon Green 488 Maleimide could react with the virus overnight at 4 °C, followed by free dye removal using membrane dialysis (10–12 kDa).
Modification of BMV with SMTA.
S-Methylthioacetimi-date (SMTA) was prepared by Thumm’s method;43,44 1 mg/mL of 100 μL virus was dialyzed (with 3 kDa Amicon spin filters) to pH 6 and 8 using 100 mM of MES and ammonium bicarbonate NH4HCO3 with 5 mM MgCl2, respectively, in each of these buffers. The protein concentration was measured by Bradford assay. Then, 43.4 μg/μL of SMTA (~500 mM) was added to the solution at room temperature and the reaction was monitored for 30 min. Excess SMTA was then removed by 3 KDa MWCO Amicon filters with 25 mM NH4HCO3 (pH 8).
Fluorescent Modification of Model Peptide.
Peptide RVVPV (2 mg/mL, molecular weight = 568.72 g/mol) was prepared in 100 mM sodium bicarbonate buffer (pH 7.5). Then, the N-terminus of the peptide was subjected to modification with 2X Oregon Green 488 carboxylic acid, succinimidyl ester for 2 h. For comprehending the ionization efficiency, an equal amount of unlabeled and dye-labeled peptides was mixed and injected into an Orbitrap Fusion Lumos.
Limited Proteolysis of BMV and BMV-OG.
wtBMV and BMV-OGs were dialyzed against swelling buffer45 for 4 h (100 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA) using a Slide-A-Lyzer MINI dialysis device, 10 K MWCO. wtBMV was treated with 1:20 of trypsin for 5 and 10 min at 37 °C. Similarly, BMV-OG was incubated with different ratios of trypsin ranging from 1:10 to 1:1 from 30 min to 2 h depending upon the number of dyes/virus in a sample. These samples were quenched with 10 mM acetic acid, followed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) to monitor the released peptides. They were also visualized using 4–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to confirm the proteolysis. The samples were further dialyzed back to SAMA pH 4.6 using 100 kDa Amicon filters and were imaged under transmission electron microscopy (TEM).
Enzymatic Digestion and Liquid Chromatography–Mass Spectrometry (LC–MS)/MS.
wtBMV and BMV-OG samples were buffer-exchanged to 25 mM NH4HCO3(pH 8) with 3 kDa Amicon spin filters. To remove the RNA, 0.75 μg/μL of RNase A (from bovine pancreas) was added to each aliquot and the mixtures were incubated for 30 min at 30 °C. Then, the samples were kept in water bath for a few minutes at 70 °C, followed by digestion with different enzyme: substrate amounts: trypsin = 1:20 (w/w), chymotrypsin = 1:20 (w/w) for 18 h, and proteinase K = 1:100 (w/w) for 30 min. The initial step of thermal denaturation increases the flexibility of the structure and enhances the propensity for proteolytic cleavage.46 The resultant peptide mixtures were loaded onto the LC instrument.
HPLC/Nano-ESI MS/MS Analysis of wtBMV and BMV-OG Proteolytic Digests.
Approximately 1 μg of the proteolytic digests was loaded onto the trap column, which was separated by an Acclaim PepMap RSLC C18 column (75 μm × 25 cm, 2 μm, 100 Å) with an EASY-nLC 1200 liquid chromatography to supply the mobile phases. The gradient used is detailed in Table S1 of the SI document. Eluting peptides were detected with an Orbitrap Fusion Lumos Tribrid mass spectrometer. Full-scan spectra were recorded in an Orbitrap at a resolution of 120 K with a maximum ion injection time of 50 ms and an automatic gain control (AGC) of 4E5. The data-dependent mode was used in the MS/MS experiments to fragment the most abundant peaks with a cycle time of 1 s. Precursor ions in charge states of +1 to +7 were subjected to fragmentation by higher-energy collisional dissociation. An isolation window was set at 2.5 m/z with an offset of 0.5 m/z. All MS/MS spectra were recorded in an ion trap operating in rapid mode. Dynamic exclusion was prohibited to obtain better correlation between peptide abundance and spectrum counts.
MS Data Analysis.
Raw data files were converted to *.mgf files by Proteome Discoverer 2.1 software. Spectra were interpreted with the Batch-Tag Web of Protein Prospector against the database of the BMV capsid protein. Spectra resulted from precursors in charge states of +1 to +7 were searched with a precursor mass tolerance of 10 ppm and a fragment mass tolerance of 0.8 Da. Loss of N-terminal methionine with acetylation of the successive residue and oxidation of methionine were allowed as variable modifications. Up to three missed cleavages were considered with tryptic and chymotryptic digests. Proteinase K digests were searched as no enzyme specificity at two termini with a maximum of 20 missed cleavages. For the OG488-labeled samples, a user-defined variable modification was set on lysine residue with mass additions of 375–396 Da, whereas for the SMTA-labeled samples, this mass range was set as 40–43 Da. A maximum of three modifications per peptide was considered. All peptide spectrum matches were required to have scores higher than 15, with the E-values lower than 0.001.
Transmission Electron Microscopy.
Electron-transparent samples were prepared by placing 10 μL of dilute sample onto a carbon-coated copper grid. After 10 min, the excess solution on the grid was removed with filter paper. It was stained with 10 μL of 2% uranyl acetate for 10 min, and the excess solution was removed by blotting with filter paper. The sample was then left to dry for several minutes. Images were acquired at an accelerating voltage of 80 kV on a JEOL JEM 1010 transmission electron microscope and analyzed with the ImageJ Processing Toolkit.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Spectrometry.
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry was done on a Bruker Autoflex III in the reflectron mode. A 355 nm frequency-tripled beam Nd-YAG laser was incident on the 384-well steel plate of a 1 μL spot size. Trypsin-treated samples were desalted and purified with Pierce C18 zip tips and further mixed with α-cyano-4-hydroxycinnamic acid (CCA) matrix in a 1:5 ratio (varies depending upon the concentration of the sample). A total of 4700 mix, which is a mixture of peptides with masses from 379 to 3659 m/z, was used as the calibrant. The peptide fragments were identified with the MS-Digest function in Protein Prospector.
Steady-State Absorption and Fluorescence Emission Spectroscopy.
The concentration of wtBMV and the labeling efficiency of dyes were determined from the UV–visible absorbance spectra using the Varian Cary 100 Bio instrument. Fluorescence measurements were conducted on a Quanta-Master fluorescence spectrometer (Horiba) with the following parameters: excitation wavelength: 488 nm; emission wave-length: 515 nm. The excitation and steady-state emission spectra of 100 nM solutions were an average of at least three independent measurements. wtBMV and OG488 were used as reference standards whose optical characteristics were compared against BMV-OGs.
Total Internal Reflectance Fluorescence Microscopy (TIRF).
A Revolution XD optical microscopy system (Andor Technology, South Windsor, CT) based on an inverted Nikon Ti microscope and fitted with a Yokogawa spinning disk unit and TIRF optics and detection was used for single-particle fluorescence imaging. Samples were excited through a high-numerical-aperture 100× oil-immersed objective (CFI APO TIRF, NA: 1.49, Nikon) with a 488 nm laser at an angle of 1400 to achieve total reflection. Fluorescence was collected back through the objective, passed through an emission filter 525/30, and recorded on a CCD camera iXon DU-897-BV (Andor Technology), imaging frame rate being 10 Hz. Images were processed and analyzed using Andor iQ, ImageJ, and IgorPro.
Fluorescence Lifetime Imaging Microscopy (FLIM).
The lifetime measurements were carried out with a Leica TCS SP8 SMD in FLIM configuration. The sample was excited at a wavelength of 488 nm from a filtered (10 nm bandwidth) supercontinuum laser emitting pulses of 120 ps at 40 MHz. The time-average maximum power was 500 μW at the sample. The collection spectral window was 500–550 nm. Data analysis was done with SymPhoTime 64 (PicoQuant) software, ImageJ Processing Toolkit, and IgorPro.
Pulsed Excitation Fluorescence Spectroscopy.
Light pulses with a 170 fs pulse width from an optical parametric amplifier (Coherent Inc.) pumped by a regenerative amplifier operating at a 250 kHz repetition rate were used as an excitation source. The output pulse was tuned to λ = 490 nm, with an average power of <P> = 500 μW at the sample, focused using a fused silica lens of a 5 cm focal distance. The emission spectra were analyzed using an Acton SpectraPro-300i monochromator fit with a TE-cooled CCD camera (iDus, Andor). Data collection was done using Andor Solis spectroscopy software, followed by data processing in IgorPro (Wavemetrics).
All-Atom Modeling.
An all-atom model of the intact BMV T = 3 capsid was constructed based on the crystal structure PDB 1JS9 (3.4 Å).47 Residues 1–40 of the A chain were not resolved in the crystal. These missing residues were reconstructed using RosettaCM48 to perform fragment-based loop modeling with the cyclic coordinate descent algorithm.49 The five N-termini of an A-chain pentamer were folded simultaneously with symmetry constraints to produce 10 000 candidate conformations. The seven coil residues preceding the modeled region were allowed flexibility, while all other crystallographic residues were held fixed. The ensemble of N-termini conformations was clustered based on the root-mean-square deviation (RMSD) of the protein’s Cα trace, employing a partitioning around the medoids algorithm;50 the optimal number of clusters for the ensemble was determined using the gap statistic.51 The medoid for the most populated cluster was taken as the final model for the A chain. Residues 1–24 of the B chain were not resolved in the crystal. Given the sixfold symmetry relationship of these subunits, the missing N-terminus of the B chain was modeled based on homology to the C chain, for which a complete backbone trace was resolved in the crystal structure. The side chains for residues 1–24 for both B and C chains were packed using SCWRL4.52 Minimal resulting structural clashes were resolved with molecular mechanics minimization.
MD Simulations.
The simulation system was designed to mimic the BMV capsid in SAMA buffer. Protonation states for titratable residues were assigned for pH 4.5 using APBS-PDB2PQR.53 Local Na+ and Cl− ions were placed around the capsid with cionize in VMD 1.9.3;54 solvate and autoionize were used to construct a 320 Å2 box of explicit solvent containing 50 mM NaCl and 8 mM MgCl2. The system was described with the AMBER ff14SB force field55 and TIP3P water model56 with compatible ion parameters.57,58 Simulation files were prepared with psfgen in VMD, and the final system size was 3.2 million atoms.
MD simulations were performed with NAMD 2.14.59 The system energy was brought to a local minimum using the steepest descent protocol over 20 000 cycles. The system temperature was increased from 50 to 300 K over 5 ns. Protein backbone restraints were removed over an additional 5 ns. The system was allowed to equilibrate in the isothermal–isobaric ensemble for a final 5 ns, and production sampling was carried out for 25 ns, saving trajectory frames every 10 ps. The r-RESPA integrator was employed with a time step of 2 fs. Bonds to hydrogen were constrained using SHAKE. The nonbonded interaction cutoff was 12 Å. Electrostatic forces were split between short and long ranges using a quintic polynomial splitting function and computed with particle-mesh Ewald beyond the cutoff; full electrostatic evaluation was performed every other time step. Temperature control used the Langevin thermostat with a damping coefficient of 1.0 ps−1. Pressure control at 1.0 bar used the Nosé–Hoover Langevin piston with an oscillation period of 2000 fs and a damping time scale of 1000 fs.
MD Trajectory Analysis.
MD simulation trajectory analysis was carried out with VMD. The average per-residue root-mean-square fluctuation (RMSF) was calculated based on the protein’s Cα trace, aligning on residues 41–189. The solvent accessible surface area and dye accessible surface area were calculated for the amino group of each lysine using a probe radius of 1.4 and 5.0 Å, respectively. Salt bridges were detected as electrostatic interactions within a distance cutoff of 3.2 Å. Hydrogen bonds were detected based on a distance cutoff of 3.0 Å and an angle cutoff of 30.0 Å degrees.
RESULTS AND DISCUSSION
X-ray crystallography and cryoelectron microscopy studies have provided detailed structural information for the BMV particle.47,60 BMV encapsulates a multipartite single-stranded RNA genome inside a cavity of 18 nm diameter.47 The capsid wall is approximately 5 nm thick, and the capsid consists of 180 identical self-assembled proteins forming a T = 3 icosahedral structure.47 There are 12 lysines per BMV coat protein. Capsid symmetry, identity of protein subunits, and chemical addressability are responsible for the BMV appeal for bioconjugation and bioengineering applications.61
In this work, a common NHS ester coupling reaction was used to covalently attach Oregon Green 488 carboxylic acid succinimidyl ester to accessible lysines.11 The average number of dyes per virus particle was estimated from the UV–vis absorption spectra of BMV-OG conjugates (see Supporting Information, Figure S1). Varying the average number of dyes was achieved by changing the reagent concentration at a constant reaction time. BMV-OG particles loaded with 30 to ~300 dyes/virus on average could be obtained this way.
Transmission electron microscopy (TEM) indicated that BMV-OG particles are structurally similar to wtBMV.11 However, a change in the particle surface charge with OG conjugation was observed by electrophoretic mobility in agarose gel image (Figure S2A). As a possible consequence of this apparent change in the surface charge, at high dye loads (N > 200), some of the samples showed a propensity to aggregate. The samples that exhibited such aggregation were discarded from the study, and the results presented here only concern stable suspensions.
As one may expect from this particular conjugation approach, actual loading varies from particle to particle in magnitude and in distribution over the reactive lysine set. Single-particle intensities of labeled particles were obtained via total internal reflection fluorescence microscopy (TIRF). The box-and-whisker plot in Figure S2B illustrates the labeling heterogeneity. As expected, the dye load variance decreases toward saturation, i.e., at higher dye numbers. In addition, matrix-assisted laser desorption/ionization assays (MALDI) suggest that certain lysine reactive sites are more accessible than others.11
The bottom-up proteomics approach was applied with three variants of proteolytic digestion, employing three different enzymes: trypsin, chymotrypsin, and proteinase K (PK). The cleavage sites for trypsin and chymotrypsin on the BMV capsid protein are indicated in Figures S3 and S4 and PK digests adjacent to the carboxyl group of aliphatic and aromatic amino acids. The multienzyme approach reduces the uncertainties created by the possibility of dyes blocking some of the tryptic cleavage sites in a protein, thus leading to longer peptides, which are difficult to identify via MS. Since the cleavage sites are different for the three enzymes, by comparing the MS data in the three cases, one can eliminate dye blocking artifacts.
It is known that the nature of the peptide, i.e., charge, mass, and fragment length, may influence the ionization efficiency of the fragment in the MS assay.29,62 As a control experiment, to determine the dependence of ionization efficiency on the presence of the dye label, we have investigated a short synthetic peptide, RVVPV, by comparing results from equal concentrations of unlabeled and labeled peptides. The extracted ion chromatogram (Figure S5) shows a slight change in the peak areas, which suggests that the ionization efficiency was affected by labeling. This is consistent with reports on other dyes, which have been shown to intensify the signal with respect to the unlabeled peptide.63 Thus, instead of a quantitative assessment, here we will resort to a qualitative data analysis.
For analysis of the MS/MS data, we initially employed the spectral counting method, where the ratio of the number of dye-labeled peptide counts with a particular lysine to the total number of peptide counts was considered. We also performed manual integration of peptide ion chromatogram peak areas for few data sets. Here, the relative ratio of the area of chromatogram for the labeled and unlabeled peptides was calculated. The results from the two methods were in good agreement.
Peptides produced as a result of the enzymatic digestion are reported in Tables S2-S6 along with their corresponding masses and retention times. The experimental molecular mass of the identified peptides matched the expected mass within 10 ppm. Table 1 summarizes 13 different peptides (compiled from all three enzymatic digests) that were identified as carrying OG dye, and the corresponding lysines are colored in red. Dye labeling increases the mass of a peptide by 394.02 Da.
Table 1.
Cumulative List of Peptides Conjugated to the OG Dyea
| Lysine | Classification | Peptide location | Sequence | Peptide mass, Da |
|---|---|---|---|---|
| 8 | Internal | 1-11 | M (Met loss+acetyl) STSGTGKMTR | 1462.50 |
| 41 | Internal | 27-44 | VQPVIVEPLAAGQGKAIK | 2213.12 |
| 44 | Internal | 42-53 | AIKAIAGYSISK | 1616.75 |
| 64 | External | 54-81 | WEASSDAITAKATNAMSITLPHELSSEK | 3384.51 |
| 54-81 | WEASSDAITAKATNAM (ox)SITLPHELSSEK | 3400.50 | ||
| 81 | Interface | 65-83 | ATNAMSITLPHELSSEKNK | 2467.09 |
| 86 | Internal | 82-89 | NKELKVGR | 1339.61 |
| 84-89 | ELKVGR | 1096.46 | ||
| 105 | External | 104-111 | IKACVAEK | 1256.52 |
| 165 | External | 157-171 | YASEAVPAKAVVVHL | 1948.91 |
| 158-180 | ASEAVPAKKAVVVHLEVEHVRPTF | 2881.40 | ||
| 158-183 | ASEAVPAKKAVVVHLEVEHVRPTFDDF | 3259.53 | ||
| 158-171 | ASEAVPAKAVVVHL | 1785.84 |
Peptides were detected after digestion with trypsin, chymotrypsin, and PK. Labeled lysines are colored in red.
On the BMV coat protein structure,47 5 out of the 12 lysines can be classified as external and solvent accessible,36 and these were expected to be most susceptible to labeling. Interestingly, the results compiled in Table 1 suggest that even the lumenal (internal) surface lysines are readily accessible to the dye. Specifically, the internal lysines K8, K41, K44, and K86 and the external lysines K64, K105, and K165 have all shown similarly high probability of labeling (see further discussion and Table 3). In addition, even some of the partially buried lysines, i.e., K81 at the intercapsomer interface, could be labeled. As a side note, sufficient data on the K165-containing fragment was challenging to obtain using trypsin because, when K165 is labeled, the cleavage site becomes unavailable for trypsin action. The mass of the larger fragment is difficult to detect. As anticipated, in this case, chymotrypsin and PK were found to be useful in complementing trypsin proteolysis.
Table 3.
Reactivity of OG488 Dye vs SMTAa
| lysine | location | OG | SMTA (pH 6) | SMTA (pH 8) |
|---|---|---|---|---|
| 8 | internal | very high | very high | very high |
| 41 | internal | high | high | high |
| 44 | internal | high | high | low |
| 53 | interface | zero | zero | low |
| 64 | external | high | high | high |
| 81 | interface | low | low | low |
| 83 | external | zero | low | zero |
| 86 | internal | high | high | very high |
| 105 | external | high | high | very high |
| 111 | external | zero | zero | zero |
| 130 | interface | zero | low | high |
| 165 | external | medium | medium | very high |
Classification of lysines as “external”, “internal”, or “interface” was done by inspecting the intact capsid structure. Reactivity was evaluated from the percentage of spectral counts (labeled peptides vs total observations): >80 %: very high, 50–80 %: high, 30–50 %: medium, 10–30 %: low, <10 %: zero.
The above results corroborate previous reports concerning the accessibility of labeling agents to the internal sites for a few different other icosahedral viruses, which suggest that their capsids have very dynamic structures, in general, with transient pores opening and closing constantly.64 Specifically, modifications of the cowpea mosaic virus (CPMV) have been explored fairly well.65,66 Similar to this work, Wang et al. found that interior cysteine residues in CPMV are accessible to maleimidofluorescein, stilbene derivatives, and ethyl mercury phosphate (EMP).32,67 Partial proteolysis experiments confirmed enzymatic access to the N-terminal regions of cowpea chlorotic mottle virus (CCMV), which bears an ~70% protein sequence homology to BMV.45
Another possible reason for the efficient labeling of internal residues in BMV is the virus conjugation reaction being performed at pH > 4.6. BMV is known to swell at pH ≥ 6 and salt concentrations >200 mM.68 The cryo-EM structure of the CCMV at neutral pH69 shows that the hexamers and pentamers are intact at this pH, but their mutual orientation is different from that of the structure at acidic pH. As a consequence, the CCMV capsid undergoes radial expansion by ~10% and 2 nm pores appear at the quasi 3-fold axes.34,69,70 These pores can act as conduits for the dye transport toward the interior. Moreover, the N-terminal tail is thought to gain the ability to undergo a flip-flop motion.71,72 Transport in and out swollen BMV capsids was previously studied by H/D exchange mass spectrometry, and significant deuterium exchange rates were measured for the internal lysines.34
To estimate the importance of steric hindrance for the labeling reaction, we compared the OG tag with labeling by a thioimidate reagent i.e., S-methylthioacetimidate (SMTA),43 which is less bulky and less hydrophobic. Reilly and co-workers have previously utilized SMTA to evaluate the extent of reactivity of BMV lysines across a pH range.36 Note that in the previous work, the amidination reaction was performed at near-saturation conditions achieved through 10 cycles of reagent addition.36 In our case, the reaction was performed at pH 6 and 8 with just one cycle of reagent addition. Therefore, in the present experiments, only the most accessible lysines were modified and the samples were further evaluated with LC–MS/MS.
Table 2 summarizes 16 different peptides (compiled from all enzymatic digests) with SMTA-modified lysines. The mass of the fragment before and after the conjugation of SMTA (+41.02 Da) is provided alongside. None of the peptides were fully labeled—both labeled and unlabeled peptides were observed in the same sample. Thus, 11 lysines are accessible to SMTA but with varying probabilities. The peptide fragments produced by enzymatic digestion are tabulated in Tables S7-S10.
Table 2.
List of Peptides Modified with SMTAa
| Lysine | Classification | Peptide location | Sequence | Peptide mass, Da |
|---|---|---|---|---|
| 8 | Internal | 1-11 | M (Met loss+acet) STSGTGKMTR | 1109.50 |
| 41 | Internal | 27-44 | VQPVIVEPLAAGQGKAIK | 1861.10 |
| 44 | Internal | 42-53 | AIKAIAGYSISK | 1263.80 |
| 42-64 | AIKAIAGYSISKWEASSDAITAK | 2425.30 | ||
| 53 | Interface | 45-64 | AIAGYSISKWEASSDAITAK | 2112.10 |
| 64 | External | 54-81 | WEASSDAITAKATNAM (oxid)SITLPHELSSEK | 3048.50 |
| 81 | Interface | 65-83 | ATNAMSITLPHELSSEKNK | 2113.10 |
| 83 | External | 82-89 | NKELKVGR | 985.60 |
| 86 | Internal | 84-89 | ELKVGR | 742.46 |
| 86-91 | KVGRVL | 713.49 | ||
| 105 | External | 104-111 | IKACVAEK | 903.52 |
| 130 | Interface | 124-137 | AVADSSKEVVAAMY | 1482.73 |
| 165 | External | 124-137 | AVADSSKEVVAAM(Oxid)Y | 1498.73 |
| 158-183 | ASEAVPAKAVVVHLEVEHVRPTFDDF | 2906.53 | ||
| 158-180 | ASEAVPAKAVVVHLEVEHVRPTF | 2529.40 | ||
| 158-184 | ASEAVPAKAVVVHLEVEHVRPTFDDFF | 3052.59 |
Labeled lysines are colored in red.
A qualitative comparison of the reactivity of SMTA and OG toward various BMV coat protein lysines is provided in Table 3. While differences in the reactivity of SMTA at pH 6 and 8 are expected due to the structural changes that occur as a result of pH alteration, labeling of residues K8, K41, K64, and K81 appears to be unaffected by pH changes. Further, the residues that are protected from amidination can either be buried at the protein–protein or protein–nucleic acid interface or engaged in salt bridges or hydrogen-bonding interactions.36,73,74
Both SMTA and OG show high labeling coverage toward the external lysines K64, 105, and 165. From the structural model, the distance between these external lysines ranges from ~11 to 35 Å, while the size of OG is ~10 Å. Thus, due to steric constraints, not all of the external residues can be labeled at saturation and the surface dye distribution is necessarily inhomogeneous.
Among the external group, K83 and K111 show zero to low labeling efficiency. Both are located at the trimeric interface, which undergoes a visible conformational transformation during pH change.69 As a consequence, at high pH, labeling of these residues is sterically hindered. Another possibility for the low labeling coverage of these residues is that they may be less reactive with respect to free amines due to involvement in salt bridges;65 e.g., K83 can exhibit ionic interaction with neighboring glutamic acid residues (as evident from the MD simulation results, which will be discussed in the subsequent section). Unlabeled peptides containing K111 were frequently detected. Note that if K111 was actually labeled with the dye, then trypsin would digest at the next available residue, i.e., at K130. This would make the mass of the peptide 4928.53 Da, which is harder to detect. However, other enzymes created shorter K111-containing peptides that were also not labeled. Thus, the OG label does not react efficiently with the K111 residue.
The high reactivity of K8, K41, and K44 internal residues (Figure 2B) confirms that, as already mentioned, capsids are sufficiently dynamic to allow label access.76,77 This is true especially for the N-terminal region (which contains the above residues)36 and is a general feature for similar RNA viruses. For instance, Barnhill et al. showed that K32, at the N-terminus of the turnip yellow mosaic virus (TYMV), is highly reactive toward fluorescein and rhodamine derivatives.78 K86, another labeled internal residue, is located near trimeric openings in the capsid below K83 and K111 residues and likely becomes easily accessible to the dye during the swelling transition.
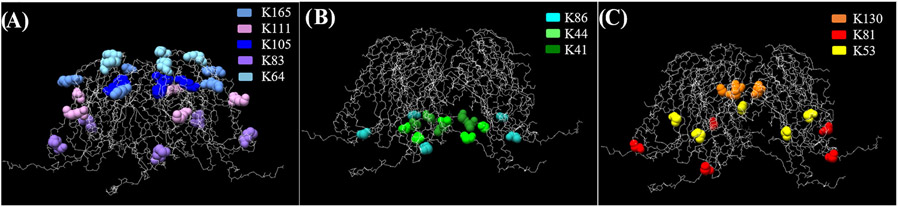
Figure 2.
Depiction of lysines on the pentameric face of capsid proteins. Image made with Chimera.75 Side view of (A) external residues K165, K111, K105, K83, and K64, (B) internal residues K41, K44, and K86 (K8 is not visible in the structure), and (C) interfacial residues K130, K81, and K53.
Among the interfacial residues, K130 is highly reactive toward SMTA at pH 8 and less at pH 6. However, K130 is inaccessible to the dye, presumably due to steric hindrance. The reactivity of K53 is very low for both molecules. On the structure model, the site is masked by tight protein–protein interactions at the quasi 2-fold axis. K81, another residue at the interface, does show reactivity, albeit low, presumably because the site is located at the trimeric interface. Moreover, since K86 shows high reactivity toward OG and the distance of K86 to K81/K83 is ~11.3–11.9 Å, charge–charge repulsion and sterics may prevent the dye from further binding at the trimeric interface.
We have seen so far that the internal lysines show significant reactivity toward the dye. However, the first 25 amino acid region of the N-terminus domain is flexible, lacking a well-defined structure.47 Are the dyes bound to this domain involved in the radiation brightening phenomenon? To answer this question, we selectively truncated N-terminal residues and measured the changes in the photophysical properties of the VLPs at radiation brightening conditions. Removal of N-termini was achieved by capsid swelling at pH > 6 in the absence of divalent cations. This was followed by limited proteolysis.45,71 MALDI-TOF peptide mapping helped pinpointing the cleavage site. A similar characterization method was previously employed in studying the coat protein N-terminus and C terminus dynamics of other icosahedral viruses.35,45,79
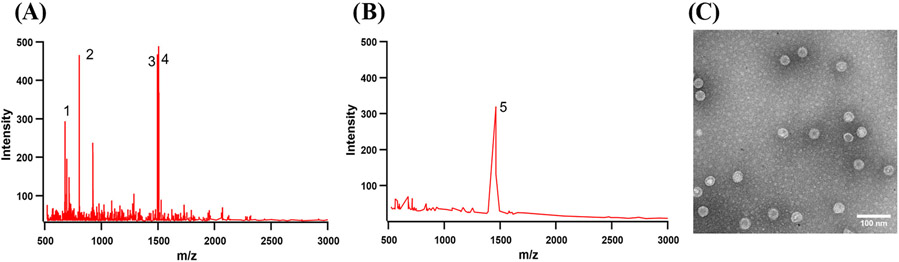
Most of the proteolysis fragments analyzed by MALDI-TOF were from the N-terminal domain, i.e., 2–8, 21–26, and 27–41. In a lesser measure, the 90–103 peptide fragment was also evident (Figure S6). In Figure 3A, the four main peaks can be matched to the peptide sequences of BMV in Table 4. Some of the unassigned peaks in the MALDI spectra are due to trypsin autolysis. To determine the adequate time interval and the trypsin concentration, we have examined the result of trypsin exposure with electron microscopy (Figure S7) and an SDS-PAGE gel, followed by Coomassie staining (Figure S8A).
Figure 3.
MALDI-TOF analysis of peptides released from (A) wtBMV and (B) BMV-OG with 317 dyes/virus after trypsin treatment. The masses and the identity of labeled ions can be seen in Table 4. (C) TEM image of BMV-OG 317 particles after digestion.
Table 4.
Peptide Sequence and Their Corresponding Masses Obtained after Trypsin Digestion Identified by MALDI-TOF Mass Analysis
| ion no. | amino acid residues | molecular weight of peptide, Da |
|---|---|---|
| 1 | 2(acetyl)–8 | 679.32 |
| 2 | 21–26 | 803.42 |
| 3 | 90–103 | 1493.92 |
| 4 | 27–41 | 1505.86 |
| 5 | 2(acetyl)–11+dye | 1461.51 |
It was shown previously that virus gains thermodynamic stability after dye labeling.11 Specifically, even after extended exposure to BMV disassembly conditions, the labeled-virus morphology remained unaltered.11 In addition, dye-labeled virions are much more resistant to digestion by trypsin. As labeling proceeds, the virus shell tightens. This is why the concentration of trypsin was increased to a 1:1 (w/w) ratio and the digestion times ranged from 30 min to 3 h from samples scaling from 56 to 317 dyes/virus, respectively. Tryptic peptides (Figure S6B-D) mostly showing ions 1, 2, and 5 correspond to the fragments 2–8, 21–26, and 2–11, where K8 was tagged with OG. However, samples loaded with 317 dyes/particle (see Figure 3B) only produced ion 5, which supports the idea of trypsin digestion being blocked. Indeed, coomassie-stained gel electrophoresis confirmed that at high dye numbers, most capsids remained intact after trypsination (Figure S8B). The above observations suggest that dye conjugation diminishes the BMV-OG protein cleavage by trypsin mostly in the structural part, while allowing the selective removal of the N-terminus, with capsid morphology remaining largely intact.
To estimate the degree of labeled N-termini removal, UV–visible absorption spectroscopy was performed post proteolysis (see Figure S9 for before and after digestion). The overall spectral shape remained the same after proteolysis, although the number of dyes remaining on the virus had changed. Ratiometric measurements allowed us to estimate that 15–30% dyes have been removed after proteolysis. This is consistent with the number of labeling sites on the N-termini.
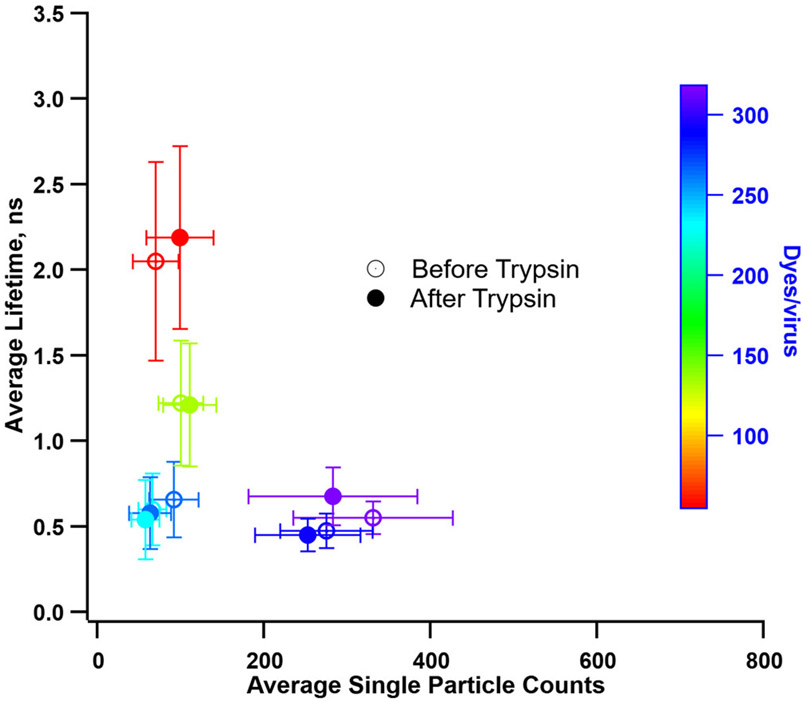
Further, single particles were characterized by fluorescence lifetime imaging microscopy (FLIM).11 The average number of detected photons from single particles was plotted against the average excited-state lifetime for several average dye numbers per particle (Figure 4). In previous experiments with BMV-OG, bright particles with shorter lifetime tend to dominate above a certain average number of dyes per particle.11 Similarly, the brightness of BMV-OG particles with truncated N-termini is retained, even after the removal of most of the K8-labeled residues. Thus, the results suggest that the floppy N-terminus, although abundantly labeled, does not contribute significantly toward radiation brightening. The remaining lysine residues must play the dominant role in the accelerated, collective emission.
Figure 4.
Average plot of the estimated lifetime vs fluorescence photon count collected from hundreds of OG-labeled BMV virions by single-particle FLIM. The color gradient legend maps the average number of dyes/virus. Open and filled circles represent samples before and after trypsin digestion, respectively.
To investigate the role of the dyes in the structural part of the coat protein, we designed a system where dyes can access only certain reactive sites on the virus coat surface. In this case, we employed a V168C mutant of the BMV coat protein.42,61 From previous work, the cysteine BMV mutant (cBMV) has a stable capsid, which allows for thiol–maleimide chemistry and hydrazone ligation for attaching fluorescent molecules, PEG moieties, and cell-penetrating peptides.61 cBMV has 180 cysteine residues on the external capsid surface.42 cBMV was covalently modified with Oregon Green 488 Maleimide via a thiol–maleimide coupling reaction (Methods section). The construct will be henceforth named cBMV-OG.
The number of dyes on cBMV-OG was estimated from UV–visible absorption spectra of conjugates (see Figure S10A). The average number of dyes per particle could be varied from 14 to 174 dyes/virus by adjusting the dye concentration in the reaction mixture. We found that the number of dyes per particle could be significantly increased with respect to previous works61 by tuning the labeling conditions (see the Methods section) and that even at the highest dye loading there is still measurable fluorescence. The structural integrity of particles was checked with TEM (Figure S10B), and the distance between adjacent cysteine residues was measured to be 2.6 ± 0.2 nm in the X-ray crystallographic structure (PDB 1JS9). Interestingly, unlike the lysine-labeled particles, cBMV and cBMV-OG particles readily disassemble when dialyzed to BMV disassembly conditions.
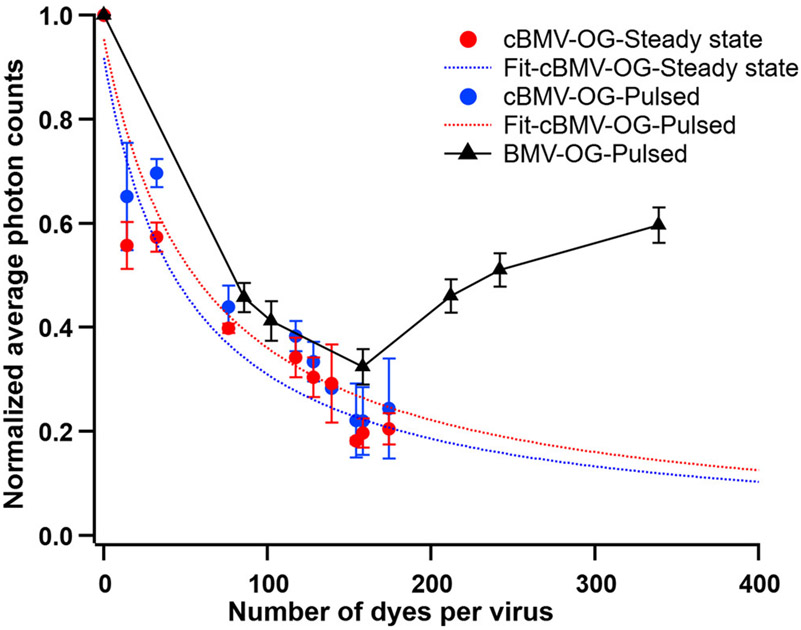
We next determined whether cBMV-OG particles can display radiation brightening under pulsed and focused excitation conditions. Figure 5 depicts the ensemble-averaged fluorescence intensity vs the number of dyes per particle (N). Quantitative analysis shows an exponential decrease in the photon counts with increasing N, consistent with fluorescence quenching behavior, both at steady-state and pulsed laser settings. For comparison, data from BMV-OGs is also shown, for which the photon counts tend to take off at N ~ 170.11 There are several possible reasons for the absence of radiation brightening in this case: smaller total number of dyes than the threshold observed on BMV-OG, a different interaction of the dye with the virus template reflected in the absence of an effect on capsid stability, and a larger interchromophore distance could all affect coupling and thus the collective radiative rate. If the latter decreases, dephasing processes have a chance to destroy coherence.
Figure 5.
Integrated photon counts normalized to free dye vs <N> for cBMV-OGs at steady-state and pulsed excitation conditions. The dotted curves show the fitting corresponding to the Stern–Volmer quenching equation,9,11 which describes concentration quenching with increase in <N>. For comparison, the experimental data obtained for BMV-OGs by Tsvetkova et al. at pulsed excitation has also been shown in black.
To gain further insight into capsid flexibility and lysine accessibility, we performed all-atom MD simulations of an intact BMV capsid. The capsid model was based on PDB 1JS9,47 which was selected for its resolution and relative completeness compared to other available experimental structures. When aligned on residues 49–176 to exclude the disordered and incomplete N- and C-termini, 1JS9 and alternative cryo-EM structures60,80 produced a total root-mean-square deviation (RMSD) of less than 1.0 Å The simulation system, with unresolved protein regions reconstructed, was designed to mimic the capsid in SAMA buffer at room temperature.
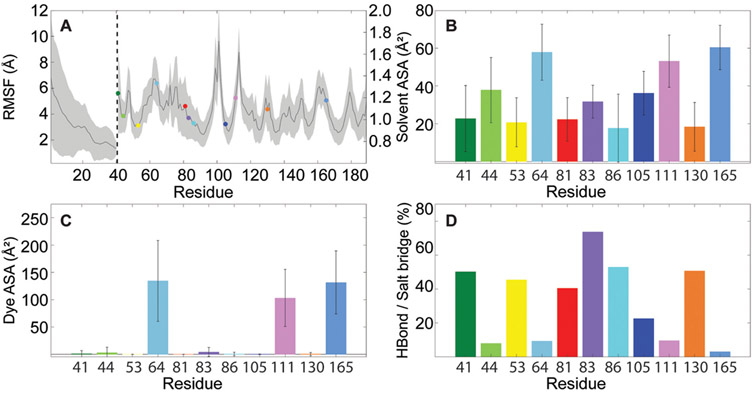
Root-mean-square fluctuation (RMSF) of the capsid’s Cα trace indicates the extent of motion observed along the protein scaffold (Figure 6a). Beyond the expected high flexibility of the N-termini, the core of the capsid is relatively stable, with the range of values observed for lysines falling well within 1.0 Å. Although flexibility measurements do not strongly correlate with internal, interfacial, and external designations, molecular accessibility is clearly dependent on the lysine position and, in some cases, quasi-equivalence (Figures S11-S13). Solvent accessible surface area (Figure 6b) is generally low for interfacial residues and higher for those exposed on the external face of the capsid. Dye accessible surface area, calculated for an OG size of ~10 Å (Figure 6c), reveals that most lysines are not sufficiently exposed to accommodate covalent attachment of the fluorophore. Notably, only K64, K111, and K165 were observed to be consistently accessible over the time scale of the simulation; these residues also exhibit relatively high Cα flexibility. For most lysines, inaccessibility to OG arises from frequent involvement in hydrogen bonds or salt bridges with neighboring residues (Figure 6d). A list of lysine salt bridge interactions is provided in Table S11.
Figure 6.
MD simulation results. (a) Average Cα RMSF over 60 copies of all chains in the capsid. The axis is shown on the left for highly flexible residues 1–39 and on the right for the remaining residues. Lysines are highlighted according to the color scheme used in Figure 2. (b) Average solvent accessible surface area for each lysine calculated with a probe radius of 1.4 Å. (c) Average dye accessible surface area for each lysine calculated with a probe radius of 5.0 Å. (d) Percentage of trajectory frames in which each lysine was involved in hydrogen bonds and/or salt bridges. Averages were calculated over 4.5 μs of cumulative simulation sampling. Error bars represent standard deviation. Data is presented by quasi-equivalent chain in Figure S11.
Experimental observations and computational measurements agree that K64 and K165 are highly probable sites for fluorescent modification with OG. However, experiments indicate K105 as a third site for labeling, while simulations indicate K111. Likely, the discrepancy between experimental and computational results can be attributed to pH. In the case of K111, as with K64 and K165, simulations at pH 4.5 show the lysine side chain protruding away from the protein surface when not forming intermolecular interactions. K105, on the other hand, sits within a depression of the protein surface, underneath K64, in the opening of the fivefold pore. While K105 can occasionally form a salt bridge with D127, its inaccessibility arises primarily from its near-interfacial location within the protein. Structural shifts (e.g., swelling and changes in protonation states that alter intermolecular interactions) expected to occur at pH 6.0 could cause K105 to become more exposed; however, it is yet unclear from simulations how such shifts would lower the accessibility of K111. Although the labeling reaction was carried out at pH 6.0, BMV-OG was returned to pH 4.5 for experimental characterization. Interestingly, simulations show that at pH 4.5, K105 exhibits minimal protein flexibility. As such, fluorescent modification at this position may be an important contributor to radiation brightening since rigidity of the scaffold alleviates dephasing of coherently excited emitters.
CONCLUSIONS
With the aid of a bottom-up proteomics approach and all-atom MD simulations, we have determined which lysines in BMV are accessible to labeling by OG488. Intriguingly, upon OG488 labeling, the virus becomes more stable relative to changes in thermodynamic conditions such as pH, salinity, and temperature. We also determined the effects of selective alteration of the labeled sites on the collective photophysical properties of BMV-OG complexes. Our results suggest that (1) not all of the fluorophores participate in accelerated emission after pulsed excitation. In particular, the fluorophores located on the flexible N-termini can be removed without an effect on the radiative decay rate. (2) The set of fluorophores involved in the previously observed radiation brightening phenomenon is located in the structural part of the protein. (3) Placing the fluorophores on the external surface, even at well-defined locations, does not guarantee radiation brightening, although concentration quenching is partially suppressed.
To continue unveiling design principles for bright coherent particles, the chemical and physical factors involved in achieving robust coherent effects at room temperature will be further explored. Additional studies through cryo-EM and more extensive MD simulations are currently in progress, which will assist in elucidating the orientation and dynamics of fluorophores bound to the structural part of the coat protein. Once this central issue is solved, it may open venues for the development of more efficient subwavelength photonic sources for biological applications by genetic modification.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Army Research Office, under awards W911NF2010072 and W911NF2010071, and by the National Science Foundation, under award CBET-1803440. J.P.R. acknowledges support from the National Science Foundation grant CHE-1904749 and the National Institutes of Health grant ROI GM135264-01. J.A.H.-P. acknowledges support from the University of Delaware Research Foundation. The authors are grateful to the Center for Bioanalytical Metrology (CBM), an NSF Industry-University Cooperative Research Center, for providing funding for this project under grant NSF IIP 1916645 and to members of the industry advisory board of the CBM for valuable discussions and feedback. The authors gratefully acknowledge Indiana University at Bloomington for access to the Electron Microscopy Center, Mass Spectrometry Facilities in the Chemistry and Biology departments, the Light Microscopy Imaging Center, and the Nanoscience Characterization Facility.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jpcb.1c06395.
Supplemental figures supporting the virus modification, characterization, and peptide labeling; tables depict the peptides produced as a result of enzymatic digestion, as well as HPLC gradient for the LC–MS/MS; and detailed MD simulations of A, B, and C chains of BMV protein are also presented alongside (PDF)
The authors declare no competing financial interest.
Contributor Information
Arathi Anil Sushma, Department of Chemistry, Indiana University, Bloomington, Indiana 47405, United States.
Bingqing Zhao, Department of Chemistry, Indiana University, Bloomington, Indiana 47405, United States.
Irina B. Tsvetkova, Department of Chemistry, Indiana University, Bloomington, Indiana 47405, United States
Carolina Pérez-Segura, Department of Chemistry and Biochemistry, University of Delaware, Newark, Delaware 19716, United States.
Jodi A. Hadden-Perilla, Department of Chemistry and Biochemistry, University of Delaware, Newark, Delaware 19716, United States.
James P. Reilly, Department of Chemistry, Indiana University, Bloomington, Indiana 47405, United States.
Bogdan Dragnea, Department of Chemistry, Indiana University, Bloomington, Indiana 47405, United States.
REFERENCES
- (1).Tipirneni KE; Warram JM; Moore LS; Prince AC; de Boer E; Jani AH; Wapnir IL; Liao JC; Bouvet M; Behnke NK; et al. Oncologic procedures amenable to fluorescence-guided surgery. Ann. Surg 2017, 266, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Stepanenko OV; Stepanenko OV; Shcherbakova DM; Kuznetsova IM; Turoverov KK; Verkhusha VV Modern fluorescent proteins: from chromophore formation to novel intracellular applications. Biotechniques 2011, 51, 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ueno T; Nagano T Fluorescent probes for sensing and imaging. Nat. Methods 2011, 8, 642–645. [DOI] [PubMed] [Google Scholar]
- (4).Reineck P; Gibson BC Near-Infrared Fluorescent Nanomaterials for Bioimaging and Sensing. Adv. Opt. Mater 2017, 5, 1600446. [Google Scholar]
- (5).Letokhov V Laser biology and medicine. Nature 1985, 316, 325–330. [DOI] [PubMed] [Google Scholar]
- (6).Kuroiwa T; Kajimoto Y; Ohta T Development of a fluorescein operative microscope for use during malignant glioma surgery: a technical note and preliminary report. Surg. Neurol 1998, 50, 41–49. [DOI] [PubMed] [Google Scholar]
- (7).Reineck P; Francis A; Orth A; Lau DWM; Nixon-Luke RDV; Rastogi ID; Razali WAW; Cordina NM; Parker LM; Sreenivasan VKA; et al. Brightness and Photostability of Emerging Red and Near-IR Fluorescent Nanomaterials for Bioimaging. Adv. Opt. Mater 2016, 4, 1549–1557. [Google Scholar]
- (8).Oesterling I; Müllen K Multichromophoric polyphenylene dendrimers: toward brilliant light emitters with an increased number of fluorophores. J. Am. Chem. Soc 2007, 129, 4595–4605. [DOI] [PubMed] [Google Scholar]
- (9).Valeur B; Berberan-Santos MN Molecular Fluorescence: Principles and Applications, 2nd ed.; John Wiley & Sons: New York, 2013. [Google Scholar]
- (10).Soto CM; Blum AS; Vora GJ; Lebedev N; Meador CD; Won AP; Chatterji A; Johnson JE; Ratna BR Fluorescent signal amplification of carbocyanine dyes using engineered viral nanoparticles. J. Am. Chem. Soc 2006, 128, 5184–5189. [DOI] [PubMed] [Google Scholar]
- (11).Tsvetkova IB; Anil Sushma A; Wang JC-Y; Schaich WL; Dragnea B Radiation Brightening from Virus-like Particles. ACS Nano 2019, 13, 11401–11408. [DOI] [PubMed] [Google Scholar]
- (12).Balzani V; Ceroni P; Maestri M; Vicinelli V Light-harvesting dendrimers. Curr. Opin. Chem. Biol 2003, 7, 657–665. [DOI] [PubMed] [Google Scholar]
- (13).Zhou X; Mandal S; Jiang S; Lin S; Yang J; Liu Y; Whitten DG; Woodbury NW; Yan H Efficient Long-Range, Directional Energy Transfer through DNA-Templated Dye Aggregates. J. Am. Chem. Soc 2019, 141, 8473–8481. [DOI] [PubMed] [Google Scholar]
- (14).Nam YS; Shin T; Park H; Magyar AP; Choi K; Fantner G; Nelson KA; Belcher AM Virus-templated assembly of porphyrins into light-harvesting nanoantennae. J. Am. Chem. Soc 2010,132, 1462–1463. [DOI] [PubMed] [Google Scholar]
- (15).Chen L; Honsho Y; Seki S; Jiang D Light-harvesting conjugated microporous polymers: rapid and highly efficient flow of light energy with a porous polyphenylene framework as antenna. J. Am. Chem. Soc 2010, 132, 6742–6748. [DOI] [PubMed] [Google Scholar]
- (16).Krieg F; Sercel PC; Burian M; Andrusiv H; Bodnarchuk MI; Stöferle T; Mahrt RF; Naumenko D; Amenitsch H; Rainò G; Kovalenko MV Monodisperse long-chain sulfobetaine-capped CsPbBr3 nanocrystals and their superfluorescent assemblies. ACS Cent. Sci 2021, 7, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Spillmann CM; Medintz IL Use of biomolecular scaffolds for assembling multistep light harvesting and energy transfer devices. J. Photochem. Photobiol., C 2015, 23, 1–24. [Google Scholar]
- (18).Schröder T; Scheible MB; Steiner F; Vogelsang J; Tinnefeld P Interchromophoric Interactions Determine the Maximum Brightness Density in DNA Origami Structures. Nano Lett. 2019, 19, 1275–1281. [DOI] [PubMed] [Google Scholar]
- (19).Helmerich DA; Beliu G; Sauer M Multiple-labeled antibodies behave like single emitters in photoswitching buffer. ACS Nano 2020, 14, 12629–12641. [DOI] [PubMed] [Google Scholar]
- (20).Sorokin AV; Ropakova IY; Grankina II; Borovoy IA; Yefimova SL; Malyukin YV Unusual enhancement of dye luminescence by exciton resonance of J-Aggregates. Opt. Mater 2019, 96, 109263. [Google Scholar]
- (21).Monshouwer R; Abrahamsson M; Van Mourik F; Van Grondelle R Superradiance and exciton delocalization in bacterial photosynthetic light-harvesting systems. J. Phys. Chem. B 1997, 101, 7241–7248. [Google Scholar]
- (22).Noriega R; Finley DT; Haberstroh J; Geissler PL; Francis MB; Ginsberg NS Manipulating excited-state dynamics of individual light-harvesting chromophores through restricted motions in a hydrated nanoscale protein cavity. J. Phys. Chem. B 2015, 119, 6963–6973. [DOI] [PubMed] [Google Scholar]
- (23).Gross M; Haroche S Superradiance: An essay on the theory of collective spontaneous emission. Phys. Rep 1982, 93, 301–396. [Google Scholar]
- (24).Rolczynski BS; Zheng H; Singh VP; Navotnaya P; Ginzburg AR; Caram JR; Ashraf K; Gardiner AT; Yeh S-H; Kais S; Cogdell RJ; Engel GS Correlated protein environments drive quantum coherence lifetimes in photosynthetic pigment-protein complexes. Chem 2018, 4, 138–149. [Google Scholar]
- (25).Cong K; Zhang Q; Wang Y; Noe GT; Belyanin A; Kono J Dicke superradiance in solids. J. Opt. Soc. Am. B 2016, 33, C80–C101. [Google Scholar]
- (26).Mallawaarachchi S; Premaratne M; Maini PK Superradiant cancer hyperthermia using a buckyball assembly of quantum dot emitters. IEEE J. Sel. Top. Quantum Electron 2019, 25, 1–8. [Google Scholar]
- (27).Miller RA; Presley AD; Francis MB Self-assembling light-harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J. Am. Chem. Soc 2007, 129, 3104–3109. [DOI] [PubMed] [Google Scholar]
- (28).Dutta PK; Varghese R; Nangreave J; Lin S; Yan H; Liu Y DNA-directed artificial light-harvesting antenna. J. Am. Chem. Soc 2011, 133, 11985–11993. [DOI] [PubMed] [Google Scholar]
- (29).Stoyanov AV; Ahmadzadeh H; Krylov SN Heterogeneity of protein labeling with a fluorogenic reagent, 3-(2-furoyl) quinoline-2-carboxaldehyde. J. Chromatogr. B 2002, 780, 283–287. [DOI] [PubMed] [Google Scholar]
- (30).Šantrůček J; Strohalm M; Kadlcík V; Hynek R; Kodícek M Tyrosine residues modification studied by MALDI-TOF mass spectrometry. Biochem. Biophys. Res. Commun 2004, 323, 1151–1156. [DOI] [PubMed] [Google Scholar]
- (31).Suckau D; Mak M; Przybylski M Protein surface topologyprobing by selective chemical modification and mass spectrometric peptide mapping. Proc. Natl. Acad. Sci. U.S.A 1992, 89, 5630–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wang Q; Raja KS; Janda KD; Lin T; Finn M Blue fluorescent antibodies as reporters of steric accessibility in virus conjugates. Bioconjugate Chem. 2003, 14, 38–43. [DOI] [PubMed] [Google Scholar]
- (33).Ruan Q; Zhao C; Ramsay CS; Tetin SY Characterization of Fluorescently Labeled Protein with Electrospray Ionization-MS and Fluorescence Spectroscopy: How Random is Random Labeling? Anal. Chem 2018, 90, 9695–9699. [DOI] [PubMed] [Google Scholar]
- (34).Wang L; Lane LC; Smith DL Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci. 2001, 10, 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chakravarty A; Reddy VS; Rao A Unravelling the stability and capsid dynamics of the three virions of brome mosaic virus assembled autonomously in vivo. J. Virol 2020, 94, No. e01794–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Running W; Ni P; Kao C; Reilly J Chemical reactivity of brome mosaic virus capsid protein. J. Mol. Biol 2012, 423, 79–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Hunt DF; Yates JR; Shabanowitz J; Winston S; Hauer CR Protein sequencing by tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A 1986, 83, 6233–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yao Z-P; Demirev PA; Fenselau C Mass spectrometry-based proteolytic mapping for rapid virus identification. Anal. Chem 2002, 74, 2529–2534. [DOI] [PubMed] [Google Scholar]
- (39).Schnaible V; Przybylski M Identification of Fluorescein-5 ‘-Isothiocyanate-Modification Sites in Proteins by Electrospray-Ionization Mass Spectrometry. Bioconjugate Chem. 1999, 10, 861–866. [DOI] [PubMed] [Google Scholar]
- (40).Manikwar P; Zimmerman T; Blanco FJ; Williams TD; Siahaan TJ Rapid identification of fluorochrome modification sites in proteins by LC ESI-Q-TOF mass spectrometry. Bioconjugate Chem. 2011, 22, 1330–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gopinath K; Kao CC Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 2007, 19, 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Hema M; Murali A; Ni P; Vaughan RC; Fujisaki K; Tsvetkova I; Dragnea B; Kao CC Effects of amino-acid substitutions in the Brome mosaic virus capsid protein on RNA encapsidation. Mol. Plant-Microbe Interact 2010, 23, 1433–1447. [DOI] [PubMed] [Google Scholar]
- (43).Thumm M; Hoenes J; Pfleiderer G S-Methylthioacetimidate is a new reagent for the amidination of proteins at low pH. Biochim. Biophys. Acta, Gen. Subj 1987, 923, 263–267. [Google Scholar]
- (44).Beardsley RL; Reilly JP Quantitation using enhanced signal tags: a technique for comparative proteomics. J. Proteome Res 2003, 2, 15–21. [DOI] [PubMed] [Google Scholar]
- (45).Speir JA; Bothner B; Qu C; Willits DA; Young MJ; Johnson JE Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J. Virol 2006, 80, 3582–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Bark SJ; Muster N; Yates JR; Siuzdak G High-temperature protein mass mapping using a thermophilic protease. J. Am. Chem. Soc 2001, 123, 1774–1775. [DOI] [PubMed] [Google Scholar]
- (47).Lucas RW; Larson SB; McPherson A The crystallographic structure of brome mosaic virus. J. Mol. Biol 2002, 317, 95–108. [DOI] [PubMed] [Google Scholar]
- (48).Song Y; DiMaio F; Wang RY-R; Kim D; Miles C; Brunette T; Thompson J; Baker D High-resolution comparative modeling with RosettaCM. Structure 2013, 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Canutescu AA; Dunbrack RL Jr, Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Sci. 2003, 12, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kaufman L; Rousseeuw PJ Finding Groups in Data: an Introduction to Cluster Analysis; John Wiley & Sons: New York, 2009; Vol. 344. [Google Scholar]
- (51).Tibshirani R; Walther G; Hastie T Estimating the number of clusters in a data set via the gap statistic. J.R. Stat. Soc. B 2001, 63, 411–423. [Google Scholar]
- (52).Krivov GG; Shapovalov MV; Dunbrack RL Jr, Improved prediction of protein side-chain conformations with SCWRL4. Proteins 2009, 77, 778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Jurrus E; Engel D; Star K; Monson K; Brandi J; Felberg LE; Brookes DH; Wilson L; Chen J; Liles K; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Humphrey W; Dalke A; Schulten K VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- (55).Maier JA; Martinez C; Kasavajhala K; Wickstrom L; Hauser KE; Simmerling C ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Jorgensen W; Chandrasekhar J; Madura J; Impey R; Klein M Comparison of simple model potentials for simulating liquid water. J. Chem. Phys 1983, 79, 926. [Google Scholar]
- (57).Li P; Song LF; Merz KM Jr. Systematic parameterization of monovalent ions employing the nonbonded model. J. Chem. Theory Comput 2015, 11, 1645–1657. [DOI] [PubMed] [Google Scholar]
- (58).Li P; Roberts BP; Chakravorty DK; Merz KM Jr, Rational design of particle mesh Ewald compatible Lennard-Jones parameters for +2 metal cations in explicit solvent. J. Chem. Theory Comput 2013, 9, 2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Phillips JC; Hardy DJ; Maia JDC; Stone JE; Ribeiro JV; Bernardi RC; Buch R; Fiorin G; Hénin J; Jiang W; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys 2020, 153, 044130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Wang Z; Hryc CF; Bammes B; Afonine PV; Jakana J; Chen D-H; Liu X; Baker ML; Kao C; Ludtke SJ; Schmid MF; Adams PD; Chiu W An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun 2014, 5, No. 4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Yildiz I; Tsvetkova I; Wen AM; Shukla S; Masarapu M; Dragnea B; Steinmetz NF Engineering of Brome mosaic virus for biomedical applications. RSC Adv. 2012, 2, 3670–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Kiontke A; Oliveira-Birkmeier A; Opitz A; Birkemeyer C Electrospray ionization efficiency is dependent on different molecular descriptors with respect to solvent pH and instrumental configuration. PLoS One 2016, 11, e0167502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Faller T; Hutton K; Okafo G; Gribble A; Camilleri P; Games DE A novel acridone derivative for the fluorescence tagging and masss pectrometric sequencing of peptides. Chem. Commun 1997, 1529–1530. [Google Scholar]
- (64).Taylor DJ; Wang Q; Bothner B; Natarajan P; Finn M; Johnson JE Correlation of chemical reactivity of Nudaurelia capensis ω virus with a pH-induced conformational change. Chem. Commun 2003, 2770–2771. [DOI] [PubMed] [Google Scholar]
- (65).Chatterji A; Ochoa WF; Paine M; Ratna B; Johnson JE; Lin T New addresses on an addressable virus nanoblock: uniquely reactive Lys residues on cowpea mosaic virus. Chem. Biol 2004, 11 , 855–863. [DOI] [PubMed] [Google Scholar]
- (66).Wang Q; Kaltgrad E; Lin T; Johnson JE; Finn M Natural supramolecular building blocks: wild-type cowpea mosaic virus. Chem. Biol 2002, 9, 805–811. [DOI] [PubMed] [Google Scholar]
- (67).Wang Q; Lin T; Tang L; Johnson JE; Finn M Icosahedral virus particles as addressable nanoscale building blocks. Angew. Chem. Int. Ed 2002, 41, 459–462. [DOI] [PubMed] [Google Scholar]
- (68).Incardona N; Kaesberg P A pH-induced structural change in bromegrass mosaic virus. Biophys. J 1964, 4, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Speir JA; Munshi S; Wang G; Baker TS; Johnson JE Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 1995, 3, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Konecny R; Trylska J; Tama F; Zhang D; Baker NA; Brooks CL III; McCammon JA Electrostatic properties of cowpea chlorotic mottle virus and cucumber mosaic virus capsids. Biopolymers 2006, 82, 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Bothner B; Dong XF; Bibbs L; Johnson JE; Siuzdak G Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J. Biol. Chem 1998, 273, 673–676. [DOI] [PubMed] [Google Scholar]
- (72).Miao Y; Johnson JE; Ortoleva PJ All-atom multiscale simulation of cowpea chlorotic mottle virus capsid swelling. J. Phys. Chem. B 2010, 114, 11181–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Running WE; Reilly JP Variation of the chemical reactivity of Thermus thermophilus HB8 ribosomal proteins as a function of pH. Proteomics 2010, 10, 3669–3687. [DOI] [PubMed] [Google Scholar]
- (74).Liu X; Reilly JP Correlating the chemical modification of Escherichia coli ribosomal proteins with crystal structure data. J. Proteome Res 2009, 8, 4466–4478. [DOI] [PubMed] [Google Scholar]
- (75).Pettersen EF; Goddard TD; Huang CC; Couch GS; Greenblatt DM; Meng EC; Ferrin TE UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- (76).Bothner B; Hilmer JK Structural Virology; Agbandje-McKenna M; McKenna R, Eds.; Royal Society of Chemistry: Cambridge, UK, 2010; pp 41–61. [Google Scholar]
- (77).Morton VL; Stockley PG; Stonehouse NJ; Ashcroft AE Insights into virus capsid assembly from non-covalent mass spectrometry. Mass Spectrom. Rev 2008, 27, 575–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Barnhill HN; Reuther R; Ferguson PL; Dreher T; Wang Q Turnip yellow mosaic virus as a chemoaddressable bionanoparticle. Bioconjugate Chem. 2007, 18, 852–859. [DOI] [PubMed] [Google Scholar]
- (79).Hilmer JK; Zlotnick A; Bothner B Conformational equilibria and rates of localized motion within hepatitis B virus capsids. J. Mol. Biol 2008, 375, 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Beren C; Cui Y; Chakravarty A; Yang X; Rao A; Knobler CM; Zhou ZH; Gelbart WM Genome organization and interaction with capsid protein in a multipartite RNA virus. Proc. Natl. Acad. Sci. U.S.A 2020, 117, 10673–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.