Abstract
Ubiquitin-specific peptidase 10 (USP10) can sustain cellular functions and regulate cellular processes. It plays an essential role in cancer inhibition or facilitation by reversing ubiquitin-proteasome degradation. Studies have identified USP10 to be involved in tumor progression in various cancers. However, the pan-cancer expression pattern of USP10, its prognostic value, and the association between tumor immune cell infiltration and USP10 expression remain to be discussed and thus comprised the aims of the present study. Based on clinical samples and bioinformatic analyses, high USP10 expression was observed in most cancer tissues except for ovarian cancer. High USP10 expression correlated with pathological stage and node metastasis and predicted poor patient prognosis. In addition, further analyses at the TIMER and GEPIA databases showed that USP10 is involved in the infiltration of multiple immune cells and regulated the infiltration levels of specific immune cell subpopulations, particularly in pancreatic adenocarcinoma (PAAD) and liver hepatocellular carcinoma (LIHC). Importantly, USP10 might influence survival by modulating immune infiltration in patients with PAAD and LIHC. These results identified USP10 as a potential biomarker for pan-cancer prognosis, and in certain cancers, USP10 could identify clinical prognosis linked to tumor immune infiltration.
Keywords: USP10, prognosis, immune infiltration, pan-cancer, biomarker
Introduction
Currently, in many human populations and regions, the leading cause of death is cancer, representing a severe threat to human health (1). The malignant phenotype of neoplasms usually correlates with dysregulation of protein synthesis (2). Proteostasis disorders and aberrant gene expression in cancer affect the patients’ clinical outcomes. In addition, there has been increasing interest in the role of the tumor microenvironment (TME) in cancer metastasis, in which the infiltration levels of dendritic cells, neutrophils, macrophages, T cells, and B cells, vary significantly. Targeting of immune cells that infiltrate the TME using immunotherapy has become a landmark in the history of tumor therapy and has dramatically advanced the development of oncological immunology (3).
There are various types of post-translational modifications of proteins. Among them, ubiquitination and deubiquitination, which add or remove ubiquitin from target proteins to promote protein degradation or stabilization, are essential to regulate cell cycle processes, cell signaling, the DNA damage response, and the nuclear factor kappa B (NF-κB) pathway (4, 5). Protein ubiquitination is the result of the covalent modification of substrate proteins by ubiquitin-activating enzyme E1, ubiquitin-binding enzyme E2, and ubiquitin-ligase E3.
Deubiquitinating enzymes (DUBs) can mediate and regulate the reversible deubiquitination of substrate proteins and are important factors in regulating the ubiquitin system. On the one hand, they are involved in the recycling of ubiquitin molecules, processing ubiquitin precursors, and editing of ubiquitin chains to regulate the function of conjugated proteins. On the other hand, they can influence proteostasis by removing ubiquitin from substrates and exert pro- or anti-cancer effects (4, 6). In addition, the function of DUBs in cells can be regulated by modifications such as phosphorylation, ubiquitination, and sumoylation, which in turn affect their catalytic activity, cellular localization, or protein abundance.
Ubiquitin-specific peptidase 10 (USP10) is a crucial DUB that is primarily localized in the cytoplasm. The USP10 gene is situated on chromosome 16q24.1 and encodes a protein product comprising 798 amino acids (relative molecular mass = ~ 93 KDa) (7, 8). Its molecular structure is mainly that of a cysteine-type endopeptidase and a ubiquitin sulfhydryl esterase. USP10 acts as a regulator of the cell cycle and autophagy by deubiquitinating various proteins that are post-translationally transferred to the cytoplasm. USP10 specifically deubiquitinates and stabilizes P53. Under DNA damage stress conditions, USP10 is stabilized by Ataxia Telangiectasia mutated (ATM) kinase phosphorylation modification at Thr42 and Ser337, which drives its entry into the nucleus where it deubiquitinates P53, thereby regulating the P53 downstream network functions (9). In addition, by acting on wild-type P53, USP10 can exercise cancer suppressive functions; however, for some mutant P53s, USP10 might exert cancer-promoting functions.
Studies have shown that the abnormal expression of USP10 in different types of cancer correlates strongly with patient prognosis. High USP10 expression in prostate cancer, breast cancer, non-small cell lung cancer, colon cancer, and melanoma is associated with poor patient prognosis (10–14). However, low expression of USP10 predicts a poor prognosis in patients with ovary cancer (15). Furthermore, USP10 could deubiquitinate sirtuin 6 (SIRT6) to antagonize transcriptional activation of c-Myc oncogenes to inhibit tumor formation (16). In non-small cell lung cancer with mutant P53, targeting USP10 could boost drug sensitivity in patients with lung cancer (17). By contrast, USP10 is closely associated with tumor immunity. USP10 has been identified to be involved in metastasis and can drive tumor-associated macrophage polarization in colorectal cancer (18). Depletion of USP10 markedly reduced apoptosis and immune cell infiltration (19). It also stimulates the production of reactive oxygen species (ROS) in T cells, thereby promoting malignant mutations (20). These studies suggest that USP10 has a critical function in the initiation and progression cancer, and in tumor immunity. Therefore, the design of highly selective inhibitors might bring new hope for anti-cancer immunotherapy.
However, to date, USP10 has not been studied in pan-cancer. Its potential prognostic value and the relevance of immune infiltration are unclear, and there is a lack of macroscopic presentation and discussion. Therefore, this study aimed to determine whether USP10 influences the prognosis of patients with cancer and if such an influence is associated with immune cell infiltration.
Materials and Methods
Oncomine Database
The Oncomine cancer database was used to analyze USP10 mRNA expression levels in different cancer types (https://www.oncomine.org/resource/login.html) (21). The threshold was set as a P-value of 0.001, a fold-change of 1.5, and the gene rank of ‘all’.
TIMER Database
TIMER (https://cistrome.shinyapps.io/timer/) (22) enables the systematic analysis of immune cell infiltration in various types of cancer. Using TIMER, the association between USP10 expression in different tumors and six types of immune infiltrates (including B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells) was explored, as well as the correlation between USP10 expression and immune cell gene markers after determining the tumor purity. Finally, we determined the associations between the expression of USP10 and the genetic markers of specific subpopulations of immune infiltrating cells.
HPA Database
The Human Protein Atlas (HPA) (http://www.proteinatlas.org/) was used to assess differences in USP10 expression at the protein level. This database contains immunohistochemical (IHC) data for USP10 protein levels in eight cancer tissues and their normal counterparts (breast, liver, lung, skin, colon, kidney, ovarian, and prostate cancers).
GEPIA Database
Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) (23) is an online interactive web server for the analysis of tumor samples from The Cancer Genome Atlas (TCGA) and RNA sequencing expression data from the Genotype-Tissue Expression (GTEx) project. We used GEPIA to evaluate the association of USP10 expression with prognosis in various tumor types, including overall survival (OS) and relapse-free survival (RFS). We further estimated the interaction between USP10 expression and specific markers related to tumor immune cell infiltration.
PrognoScan Database
The PrognoScan database (http://www.abren.net/PrognoScan/) (24) was used to assess the correlation between USP10 expression and survival rate in different types of cancer, and to explore the prognostic value of USP10.
Kaplan-Meier Plotter Database
The Kaplan-Meier plotter database (http://kmplot.com/analysis/) (25) uses meta-analysis data to discover and validate prognosis-related biomarkers. The association between USP10 expression and patient survival in pan-cancer was investigated.
cBioPortal Database
The open web resource cBioPortal database (http://www.cbioportal.org/) (26) is used to explore multidimensional cancer genome datasets. We used cBioPortal to analyze the impact of USP10 mutations and copy number variation on various cancers.
UALCAN Database
The UALCAN (http://ualcan.path.uab.edu/) (27) database contains valuable cancer OMICS data and information to help understand tumor staging and node metastasis related to the USP10 protein in a diverse range of cancers, which could inform mechanistic studies.
Tissue Specimens
From May 2008 to Oct 2015, we collected tumors and adjacent non-tumor tissues from 12 patients suffering from different types of cancer who underwent surgical treatment in the Shanghai East Hospital Affiliated to Tongji University. The Ethics Committee of Shanghai East Hospital Affiliated to Tongji University approved this study (No. 2019tjdx110). Exemption from informed consent was granted because of the retrospective nature of the study.
Hematoxylin and Eosin (H&E) Staining
Cancerous and control tissues from the breast, liver, lung, colorectum, kidney, ovary, prostate, stomach, skin, cerebrum, esophagus, and uterus were fixed in 4% paraformaldehyde. After deparaffinization and rehydration, sections were cuts at 4 μm thick and stained with H&E according to the manufacturer’s protocol (C0105S, Beyotime, Jiangsu, China). The sections were finally dehydrated and sealed for viewing under a light microscope (×200, Leica DM3000; Wetzlar, Germany).
Immunofluorescence Staining
All tumor sections (4 μm) were deparaffinized and hydrated, quenched in 3% H2O2, immersed in citrate buffer, heated to retrieve the antigen, and then stained using immunofluorescence. USP10 primary antibodies (Invitrogen, Waltham, MA, USA; Cat. #PA5-52334) and fluorescently-conjugated secondary antibodies were applied, followed by 4′,6-diamidino-2-phenylindole (DAPI) staining to visualize the nuclei. The sections were observed under a microscope (×200, Leica DM3000).
Statistical Analysis
Data in Oncomine are presented based on gene ranking, fold-change, and P-values. Survival curves were plotted using PrognoScan, Kaplan-Meier plotter, and GEPIA, and the results are displayed as the hazard ratio (HR) and P-value, or the P-value alone, from a log-rank test. Gene expression correlation was estimated using Spearman’s correlation, and r values were used to determine the magnitude of the correlation. P-values less than 0.05 were considered statistically significant.
Results
Transcriptional and Translational Levels of USP10 in Pan-Cancer
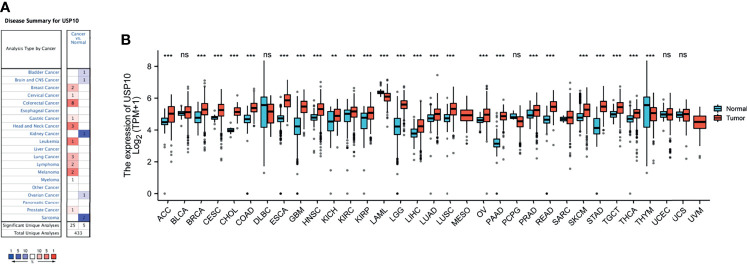
In the Oncomine database, the levels of USP10 mRNA in various cancers and normal tissues were analyzed. The results showed that USP10 expression in breast cancer, cervical cancer, colorectal cancer, gastric cancer, head and neck cancer, leukemia, lung cancer, lymphoma, melanoma, myeloma, and prostate cancer was higher than that in normal tissues (Figure 1A). By contrast, in some datasets, lower USP10 expression was observed for brain and CNS cancer, bladder cancer, kidney cancer, ovarian cancer, and sarcoma. Supplementary Table 1 shows the detailed results for the expression of USP10 in various cancer types.
Figure 1.
The mRNA expression level of USP10 in pan-cancer. (A) Oncomine data showing enhanced or reduced USP10 expression in various tumor tissue types and in normal tissues. (B) TGCA data showing the expression levels of human USP10 in different types of cancer, with GTEx data as a control. **P < 0.01; ***P < 0.001. NS, no significance.
Next, TCGA RNA-seq data from multiple malignancies was analyzed for USP10 expression (Figure 1B). Significantly higher USP10 expression was detected in adrenocortical carcinoma (ACC), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), (esophageal carcinoma (ESCA), glioblastoma multiforme (GMB), (head and neck cancer (HNSC), (kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), and thyroid carcinoma (THCA) tissues compared with that in adjacent normal tissues. However, significantly lower USP10 expression was observed in acute myeloid leukemia (LAML) and thymoma (THYM) compared with that in adjacent normal tissues. The analyses of the two databases were relatively consistent, except for KIRC and OV. These analytical differences were mainly due to the different databases and different sample sizes.
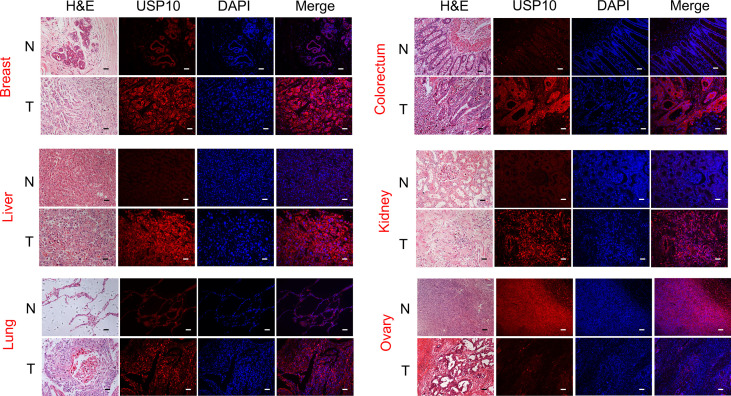
We used immunofluorescence (IF) to examine the expression of USP10 protein in various cancer tissues and their normal counterparts. USP10 protein expression was higher in breast cancer, liver cancer, lung cancer, colorectal cancer, kidney cancer, prostate cancer, stomach cancer, skin cancer, cerebrum cancer, esophagus cancer, and uterus cancer tissues than in normal tissues (Figure 2 and Supplementary Figure 1). However, USP10 protein levels were lower in ovarian cancer tumor tissues compared with those in normal tissues (Figure 2), which was in line with the results for the mRNA levels from the Oncomine database (Figure 1A). Furthermore, the immunohistochemistry (IHC) results from the HPA database shown in Supplementary Figure 2 showed similar results. Normal lung, prostate, and liver tissues showed moderate USP10 IHC staining, whereas tumor tissues showed intense staining. Normal breast, colon, kidney, and skin tissue samples showed weak USP10 staining, while tumor tissues showed intense staining. Interestingly, the normal ovary tissue sample had moderate USP10 staining, while tumor tissue had low staining (Supplementary Figure 2). These results suggested that USP10 protein levels are generally upregulated in the above tumor tissues but downregulated in ovarian cancer. The transcriptional and translational levels of USP10 in these cancer types were broadly consistent.
Figure 2.
The protein level of USP10 in different types of cancer. Representative images of H&E stained normal and tumor slides (left 1st). Immunofluorescence staining analysis (left 2nd to 4th) showing that an evident fluorescent signal in the tumor tissue sections compared with that in the control group. However, in ovarian cancer, normal tissue has a stronger fluorescent signal than tumor tissue. All scale bars = 50 μm. H&E, hematoxylin-eosin staining. N, normal tissues; T, tumor tissues.
Diagnostic Value of USP10 in Representative Tumors
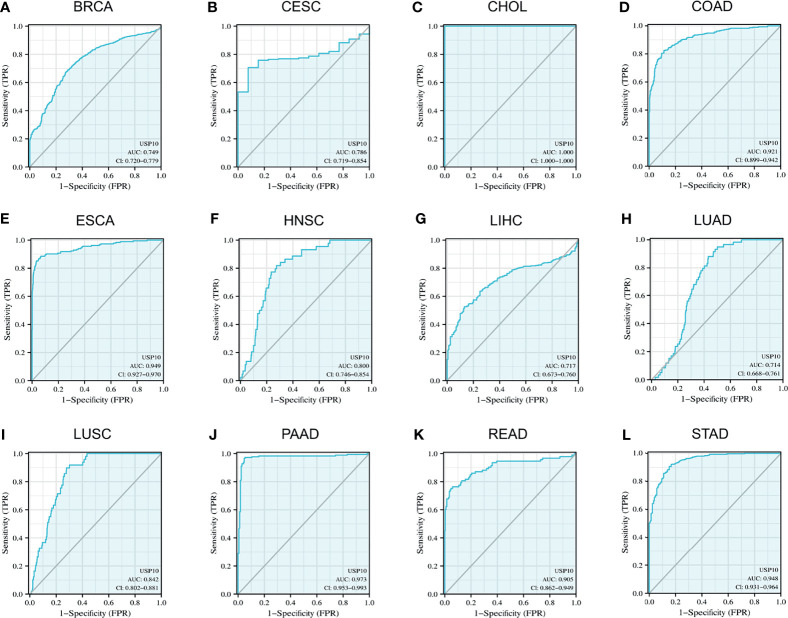
Given that USP10 expression was upregulated in a variety of cancers, we used ROC curves to assess the diagnostic value of USP10 for pan-cancer. The results revealed that USP10 had a certain accuracy (area under the ROC curve (AUC) > 0.7) to predict 12 representative tumors, including BRCA (AUC = 0.749) (Figure 3A), CESC (AUC = 0.786) (Figure 3B), CHOL (AUC = 1.000) (Figure 3C), COAD (AUC = 0.921) (Figure 3D), ESCA (AUC = 0.949) (Figure 3E), HNSC (AUC = 0.800) (Figure 3F), LIHC (AUC = 0.717) (Figure 3G), LUAD (AUC = 0.714) (Figure 3H), LUSC (AUC = 0.842) (Figure 3I), PAAD (AUC = 0.973) (Figure 3J), READ (AUC = 0.905) (Figure 3K), and STAD (AUC = 0.948) (Figure 3L). Among them, USP10 had high accuracy (AUC > 0.9) for PAAD, CHOL, ESCA, STAD, and READ. These results suggested that USP10 has a different diagnostic value depending on the type of cancer.
Figure 3.
Receiver operating characteristic (ROC) curves for USP10 in representative tumors. (A) BRCA, (B) CESC, (C) CHOL, (D) COAD, (E) ESCA, (F) HNSC, (G) LIHC, (H) LUAD, (I) LUSC, (J) PAAD, (K) READ, (L) STAD. Values under the ROC curve ranges from 0.5 to 1. The closer the area under the curve (AUC) is to 1, the better the diagnosis. The horizontal coordinate is the False Positive Rate (FPR), and the vertical coordinate is the True Positive Rate (TPR).
Prognostic Value of USP10 in Different Cancers
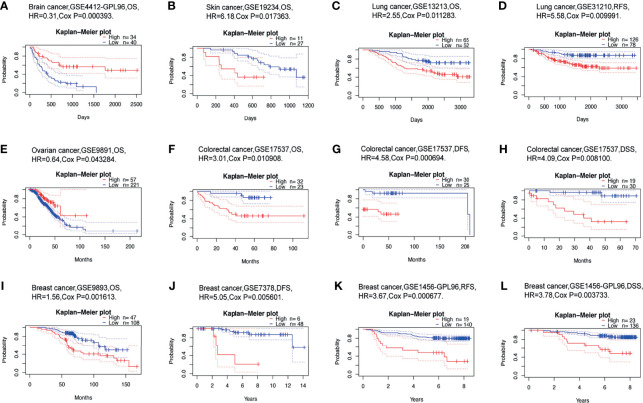
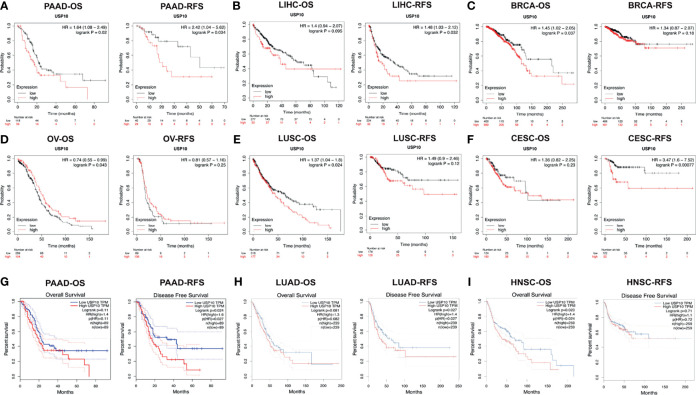
We than investigated whether USP10 expression was linked to the prognosis of patients with cancer. The impact of USP10 expression on survival rates was evaluated using PrognoScan, primarily using Gene Expression Omnibus (GEO) data. Notably, the expression of USP10 had a marked effect on the prognosis of six types of cancers, including brain cancer, skin cancer, lung cancer, ovarian cancer, colorectal cancer, and breast cancer (Figures 4A–L). In patients with these cancers, high USP10 expression could be an independent risk factor. Details of the association between USP10 expression and the prognosis of different cancers are shown in Supplementary Table 2.
Figure 4.
Correlation between USP10 expression and the prognosis of various types of cancer using data from the PrognoScan database. (A) Overall survival (OS) of a cohort of 74 patients with brain cancer (GSE4412-GPL96). (B) OS of a cohort of 38 patients with skin cancer (GSE19234). (C, D) OS and relapse-free survival (RFS) survival curves for patients in two lung cancer cohorts [GSE13213 (n=117) and GSE31210 (n=204)]. (E) OS of a cohort of 278 patients with ovarian cancer (GSE9891). (F–H) OS (n = 55), disease-free survival (DFS) (n = 55), disease free survival (DSS) (n = 49) survival curves for a cohort of patients with colorectal cancer (GSE17537). (I, J) OS and DFS survival curves for two cohorts of patients with breast cancer [GSE9893 (n =155) and GSE7378 (n =54)]. (K, L) RFS (n =159) and DSS (n =159) survival curves for a cohort of patients with breast cancer (GSE1456-GPL96). Patients with high USP10 expression are represented by the red curve. Significance is indicated by a P-value < 0.05.
To further investigate USP10’s prognostic potential various cancers, Kaplan-Meier Plotter was employed, which mainly uses TCGA data from Affymetrix microarrays. Figures 5A–F show the details of the expression of USP10 in the different types of cancer. High USP10 expression correlated with unfavorable prognosis of in terms of OS and RFS in PAAD; OS in BRCA and LUSC; and RFS in LIHC and CESC. However, a favorable prognosis in terms of OS was related to high USP10 expression in OV. The analysis revealed that USP10 mRNA and protein levels were downregulated significantly in ovarian tumor tissues compared with those in normal tissues, but high USP10 expression improved the OS of patients with ovarian cancer significantly (Figures 4E, 5D). In addition, we examined USP10-related survival (OS and RFS) using the GEPIA database (Figures 5G–I). Poor prognosis in PAAD (RFS, HR = 1.6, P = 0.027), LUAD (RFS, HR = 1.4, P = 0.027), and HNSC (OS, HR = 1.4, P = 0.024) correlated with higher USP10 expression. These results suggest that USP10 expression has different prognostic values depending on the cancer type.
Figure 5.
Correlation between USP10 expression and the prognosis of various types of cancer in the Kaplan–Meier Plotter (A–F) and GEPIA (G–I) databases. Overall survival (OS) and relapse-free survival (RFS) survival curves of patients with (A) pancreatic cancer (PAAD), (B) liver cancer (LIHC), (C) breast cancer (BRCA), (D) ovarian cancer (OV), (E) lung squamous cell carcinoma (LUSC), (F) cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), (G) pancreatic cancer (PAAD), (H) lung adenocarcinoma (LUAD), (I) head and neck squamous cell carcinoma (HNSC). Patients with high USP10 expression are represented by the red curve. Significance is indicated by a P-value < 0.05.
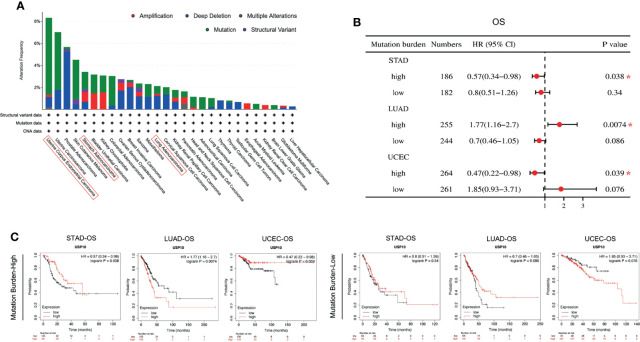
Genetic Alterations of USP10 in Pan-Cancer and TMB-Based Survival Analysis of USP10 Expression in Patients With Tumors
Tumorigenesis is usually accompanied by genetic alterations. Therefore, the genetic alterations of USP10 were examined in various tumor samples in the TCGA database (Figure 6A). USP10 showed the highest alteration frequency (6.99%) in patients with endometrial tumors with “mutation” as the primary type. Copy number alterations (CAN) of the “deep deletion” type were the predominant type of mutation in prostate adenocarcinoma cases, with a frequency of 5.26%. Noticeably, almost all the tumor cases with genetic alterations had deletions or mutations of USP10 (Figure 6A). USP10 gene mutations were observed in numerous cancers; therefore, we next explored the potential relationship between the tumor mutational burden (TMB) and USP10 expression in the clinical survival prognosis of different types of cancer (Figure 6B). USP10 expression correlated significantly with the OS of patients with STAD (HR = 0.57, P = 0.038) and UCEC (HR = 0.47, P = 0.039) with a high TMB, showing a better prognosis compared with subjects with a low TMB (Figure 6C). By contrast, the expression of USP10 correlated significantly with OS in patients with LUAD (HR = 1.77, P = 0.0074) with a high TMB; however, their prognosis was worse compared with patients with a low TMB (Figure 6C). These results suggested that patient prognosis is affected by the association between USP10 expression and the TMB in certain cancers. Moreover, genetic alterations in USP10 might play an important role in the genomes of endometrial and prostate cancers. These findings warrant further in-depth investigation.
Figure 6.
cBioPortal access of TGCA data showing USP10 mutation features in pan-cancer correlation of the TMB and USP10 in Kaplan–Meier Plotter. (A) Mutation type alteration frequencies. (B) The potential associations between the TMB and USP10 expression in the OS of patients with cancer, shown as a forest plot. (C) Survival curves between the TMB and USP10 expression in the OS of patients with cancer. TMB, tumor mutation burden. STAD, stomach adenocarcinoma; LUAD, lung adenocarcinoma; UCEC, uterine corpus endometrial carcinoma. OS, overall survival. *P < 0.05.
USP10 Expression and Clinical Parameters of Patients With Different Cancers
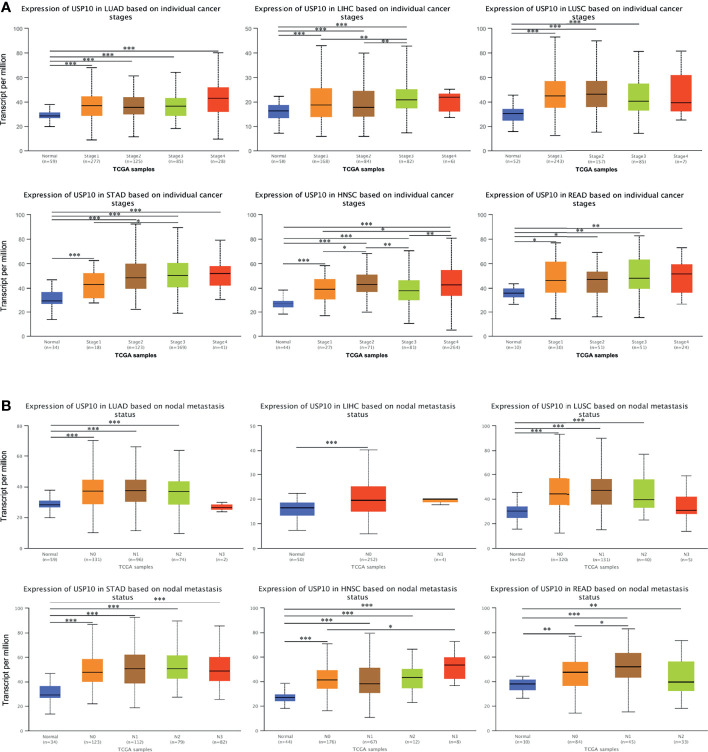
To obtain a more detailed understanding of the role of USP10 in the progression of cancers, USP10 expression was analyzed in patients with multiple cancers based on different clinical parameters using UALCAN. In terms of tumor staging, the expression of USP10 was increased significantly in patients with LUSC at stages 1, 2, and 3; and in patients with READ and LUAD at stages 1, 2, 3, and 4 (Figure 7A). Moreover, patients with STAD had markedly elevated USP10 expression in stages 1, 2, 3, and 4; and the changes at stage 1 and 3 were statistically significance (P < 0.05) (Figure 7A). The expression of USP10 was upregulated in patients with LIHC. Interestingly, with the deterioration of LIHC, USP10 expression increased gradually (Figure 7A). Notably, patients with HNSC showed significant overexpression of USP10 in stages 1, 2, 3, and 4. In addition, the comparisons of stage 1 vs. 2, stage 1 vs. 4, stage 2 vs. 3, and stage 3 vs. 4 were also statistically significant. However, USP10 was downregulated significantly in stage 3, possibly influenced by different molecular signaling pathways or different molecular subtypes (Figure 7A). For lymph node metastasis, patients with LUSC, READ, and LUAD with N0, N1, or N2 metastasis, USP10 expression was higher than that in patients with other stages of metastasis (Figure 7B). The same USP10 expression pattern was observed for patients with STAD and HNSC N0, N1, N2, or N3 metastasis and in patients with LIHC with N0 metastasis (Figure 7B). Thus, the expression of USP10 correlated closely with the proliferation and lymph node metastasis of tumors.
Figure 7.
UALCAN database analysis of correlation between clinicopathological parameters and USP10 expression in multiple cancer types. (A) Association between USP10 expression and tumor stages in patients with different types of cancer. (B) The association between USP10 expression and nodal metastasis status in patients with different types of cancer. LUAD, lung adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma; HNSC, head and neck squamous cell carcinoma; READ, rectal adenocarcinoma. *P < 0.05; **P < 0.01; ***P < 0.001.
The Association of USP10 Expression With Immune Cell Infiltration in Various Cancers
Numerous inflammatory and immune cells infiltrate cancer tissue, and recent studies have revealed the importance of tumor-infiltrating lymphocytes (TILs) in predicting the prognosis of patient survival (28–31). Therefore, the relationship between USP10 expression and immune cell infiltration in 39 types of cancer was assessed using the TIMER database. (Supplementary Figure 3). Notably, in 14 cancers, the expression of USP10 correlated significantly with B cell infiltration levels. Moreover, the expression of USP10 correlated strongly with the infiltration level of dendritic cells in 10 cancers, with neutrophils in 24 cancers, with macrophages in 18 cancers, with CD4+ T cells in 13 cancers, and with CD8+ T cells in 15 cancers. Notably, USP10 expression levels in ACC, CHOL, HNSC-HPV+, UCEC, and UCS were not significantly associated with the infiltration of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells (Supplementary Figure 3). Thus, USP10 might exert a fundamental function in the TME of cancers.
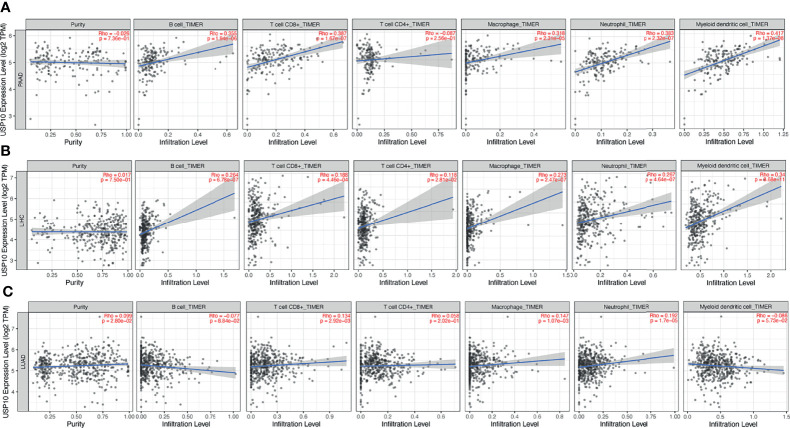
The correlation between the expression of USP10 and immune cell infiltration in various cancers prompted us to identify those cancers in which prognosis and immune infiltration were associated with USP10 expression. The effect of immunotherapy and immune cell infiltration can be better evaluated in cancers for which the tumor purity has been determined (32, 33). Therefore, we selected PAAD, LIHC, and LUAD cancers for validation. After determining the tumor purity, we found that for PAAD, high expression of USP10 correlated significantly and positively with the infiltration levels of CD8+ T cells (r = 0.387, P = 1.67e-07), B cells (r = 0.355, P = 1.94e-06), macrophages (r = 0.318, P = 2.31e-05), neutrophils (r = 0.383, P = 2.32e-07), and dendritic cells (r = 0.417, P = 1.37e-08) (Figure 8A). Similarly, there were significant positive correlations with the infiltration levels of dendritic cells (r = 0.34, P = 8.68e-11), neutrophils (r = 0.267, P = 4.64e-07), macrophages (r = 0.273, P = 2.47e-07), CD4+ T cells (r = 0.118, P = 2.81e-02), CD8+ T cells (r = 0.188, P = 4.46e-04), and B cells (r = 0.264, P = 6.78e-07), in LIHC (Figure 8B). However, USP10 expression correlated weakly with immune cell infiltration in LUAD, including only CD8+ T cells (r = 0.134, P = 2.92e-03), macrophages (r = 0.147, P = 1.07e-03), and neutrophils (r = 0.192, P = 1.7e-05) (Figure 8C). These findings suggested that USP10 might play an important part in immune cell infiltration in these cancers.
Figure 8.
TIMER database analysis of the correlation between USP10 expression and immune infiltration level in LUAD, LIHC, and PAAD. (A) In PAAD, USP10 expression was not related to tumor purity, but correlated significantly and positively with infiltration levels of dendritic cells, neutrophils, macrophages, CD8+ T cells, and B cells. (B) In LIHC, USP10 expression was not related to tumor purity, but correlated significantly and positively with dendritic cells, neutrophils, macrophages, CD4+ T cells, CD8+ T cells, and B cells. (C) In LUAD, USP10 expression correlated weakly with tumor purity and correlated significantly and positively with CD8+ T cells, macrophages, and neutrophils, but had no significant relationship with the immune infiltration of B cells, CD4+ T cells, and dendritic cells. PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma. Significance is indicated by a P-value < 0.05.
Assessment of Correlations Between Immune Cell Markers and USP10 Expression
Next, we used the TIMER database to investigate potential correlations between USP10 and immune cell marker genes in PAAD, LIHC, and LUAD, such as CD8+ T cells, B cells, tumor-associated macrophages (TAMs), monocytes, M1/M2 macrophages, natural killer (NK) cells, DCs, neutrophils, general T cells, and T cell with different functions, e.g., T follicular helper (Tfh) cells, T helper type 1 (Th1) cells, T helper type 2 (Th2) cells, T helper type 17 (Th17) cells, regulatory T cells (Tregs), and exhausted T cells (Table 1). The results, which were adjusted for tumor purity, showed that in PAAD and LIHC, the expression level of USP10 correlated significantly with most immune markers for the various immune cells. However, in LUAD, the expression level of USP10 was only associated with 23 marker genes (Table 1).
Table 1.
Analysis of correlations between USP10 expression and immune cell markers by TIMER.
| Description | Gene markers | PAAD | LIHC | LUAD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | None | Purity | |||||||||
| Cor | P | Cor | P | Cor | P | Cor | P | Cor | P | Cor | P | |||
| CD8+ T cells | CD8A | 0.214 | * | 0.189 | 0.013 | 0.173 | ** | 0.191 | ** | 0.071 | 0.106 | 0.13 | * | |
| CD8B | 0.251 | ** | 0.22 | * | 0.038 | 0.388 | 0.107 | 0.047 | 0.038 | 0.388 | 0.073 | 0.107 | ||
| T cells (general) | CD3D | 0.154 | 0.039 | 0.12 | 0.118 | 0.067 | 0.196 | 0.082 | 0.127 | -0.082 | 0.062 | -0.031 | 0.494 | |
| CD3E | 0.194 | * | 0.162 | 0.035 | 0.129 | 0.013 | 0.156 | * | -0.005 | 0.910 | 0.063 | 0.161 | ||
| CD2 | 0.202 | * | 0.174 | 0.023 | 0.1 | 0.055 | 0.121 | 0.024 | -0.028 | 0.520 | 0.038 | 0.398 | ||
| B cells | CD19 | 0.146 | 0.051 | 0.115 | 0.134 | 0.143 | * | 0.128 | 0.018 | -0.036 | 0.412 | 0.002 | 0.968 | |
| CD79A | 0.157 | 0.035 | 0.118 | 0.123 | 0.077 | 0.136 | 0.085 | 0.114 | -0.052 | 0.236 | -0.019 | 0.681 | ||
| Monocytes | CD86 | 0.289 | *** | 0.267 | ** | 0.279 | *** | 0.329 | *** | -0.055 | 0.213 | 0 | 0.991 | |
| CD115 (CSF1R) | 0.257 | ** | 0.255 | ** | 0.266 | *** | 0.315 | *** | -0.003 | 0.938 | 0.051 | 0.255 | ||
| TAMs | CCL2 | 0.016 | 0.832 | -0.012 | 0.873 | 0.184 | ** | 0.223 | *** | -0.02 | 0.656 | 0.019 | 0.675 | |
| CD68 | 0.325 | *** | 0.312 | *** | 0.225 | *** | 0.245 | *** | -0.015 | 0.741 | 0.037 | 0.416 | ||
| IL10 | 0.201 | * | 0.173 | 0.024 | 0.245 | *** | 0.27 | *** | -0.037 | 0.408 | 0.014 | 0.765 | ||
| M1 Macrophages | INOS (NOS2) | 0.224 | * | 0.184 | 0.016 | 0.149 | * | 0.158 | * | 0.085 | 0.055 | 0.098 | 0.030 | |
| IRF5 | 0.261 | ** | 0.237 | * | 0.33 | *** | 0.323 | *** | -0.042 | 0.346 | -0.007 | 0.872 | ||
| COX2 (PTGS2) | 0.337 | *** | 0.33 | *** | 0.285 | *** | 0.353 | *** | 0.089 | 0.044 | 0.081 | 0.072 | ||
| M2 Macrophages | CD163 | 0.342 | *** | 0.337 | *** | 0.277 | *** | 0.317 | *** | 0.092 | 0.037 | 0.158 | ** | |
| VSIG4 | 0.26 | ** | 0.261 | ** | 0.218 | *** | 0.259 | *** | -0.097 | 0.028 | -0.053 | 0.239 | ||
| MS4A4A | 0.255 | ** | 0.239 | * | 0.24 | *** | 0.284 | *** | -0.08 | 0.068 | -0.026 | 0.564 | ||
| Neutrophils | CD66b (CEACAM8) | 0.149 | 0.046 | 0.124 | 0.106 | 0.034 | 0.514 | 0.046 | 0.398 | -0.063 | 0.153 | -0.052 | 0.251 | |
| CD11b (ITGAM) | 0.264 | ** | 0.244 | * | 0.361 | *** | 0.397 | *** | -0.053 | 0.230 | -0.001 | 0.984 | ||
| CCR7 | 0.157 | 0.036 | 0.128 | 0.096 | 0.143 | * | 0.161 | * | -0.012 | 0.785 | 0.047 | 0.295 | ||
| Natural killer cells | KIR2DL1 | 0.021 | 0.779 | 0.023 | 0.763 | 0.091 | 0.080 | 0.073 | 0.177 | 0.031 | 0.486 | 0.047 | 0.298 | |
| KIR2DL3 | 0.064 | 0.398 | 0.03 | 0.693 | 0.176 | ** | 0.189 | ** | 0.122 | * | 0.162 | ** | ||
| KIR2DL4 | 0.215 | * | 0.115 | 0.019 | 0.189 | ** | 0.198 | ** | 0.111 | 0.012 | 0.141 | * | ||
| KIR3DL1 | 0.029 | 0.699 | 0.006 | 0.938 | 0.174 | ** | 0.197 | ** | 0.069 | 0.120 | 0.093 | 0.040 | ||
| KIR3DL2 | 0.226 | * | 0.198 | * | 0.063 | 0.230 | 0.077 | 0.154 | 0.121 | * | 0.166 | ** | ||
| KIR3DL3 | 0.188 | 0.012 | 0.166 | 0.030 | 0.04 | 0.437 | -0.016 | 0.774 | 0.081 | 0.067 | 0.095 | 0.035 | ||
| KIR2DS4 | 0.105 | 0.163 | 0.078 | 0.313 | 0.101 | 0.052 | 0.114 | 0.034 | 0.056 | 0.208 | 0.087 | 0.053 | ||
| Dendritic cells | HLA-DPB1 | 0.159 | 0.034 | 0.126 | 0.101 | 0.211 | *** | 0.238 | *** | -0.198 | *** | -0.17 | ** | |
| HLA-DQB1 | 0.212 | * | 0.192 | 0.012 | 0.126 | 0.015 | 0.147 | * | -0.197 | *** | -0.167 | ** | ||
| HLA-DRA | 0.265 | ** | 0.241 | * | 0.287 | *** | 0.328 | *** | -0.205 | *** | -0.173 | ** | ||
| HLA-DPA1 | 0.259 | ** | 0.234 | * | 0.274 | *** | 0.312 | *** | -0.139 | * | -0.109 | 0.015 | ||
| BDCA-1 (CD1C) | 0.137 | 0.068 | 0.116 | 0.132 | 0.124 | 0.017 | 0.142 | * | -0.234 | *** | -0.208 | *** | ||
| BDCA-4 (NRP1) | 0.362 | *** | 0.388 | *** | 0.541 | *** | 0.552 | *** | 0.142 | * | 0.164 | ** | ||
| CD11c (ITGAX) | 0.15 | 0.045 | 0.104 | 0.177 | 0.312 | *** | 0.355 | *** | -0.083 | 0.065 | 0.034 | 0.454 | ||
| Th1 cells | T-bet (TBX21) | 0.108 | 0.149 | 0.091 | 0.237 | 0.139 | * | 0.161 | * | 0.077 | 0.082 | 0.152 | ** | |
| STAT4 | 0.097 | 0.196 | 0.117 | 0.128 | 0.06 | 0.245 | 0.07 | 0.195 | -0.073 | 0.096 | -0.026 | 0.571 | ||
| STAT1 | 0.412 | *** | 0.396 | *** | 0.385 | *** | 0.392 | *** | 0.263 | *** | 0.322 | *** | ||
| IFN-γ (IFNG) | 0.091 | 0.227 | 0.078 | 0.308 | 0.139 | * | 0.148 | * | 0.076 | 0.084 | 0.133 | * | ||
| TNF-α (TNF) | 0.083 | 0.269 | 0.069 | 0.372 | 0.26 | *** | 0.289 | *** | -0.001 | 0.987 | 0.058 | 0.200 | ||
| Th2 cells | GATA3 | 0.213 | * | 0.2 | * | 0.221 | *** | 0.267 | *** | 0.088 | 0.046 | 0.15 | ** | |
| STAT6 | 0.436 | *** | 0.417 | *** | 0.429 | *** | 0.404 | *** | 0.137 | * | 0.147 | * | ||
| STAT5A | 0.372 | *** | 0.344 | *** | 0.316 | *** | 0.315 | *** | 0.081 | 0.066 | 0.146 | * | ||
| IL13 | -0.019 | 0.799 | -0.025 | 0.749 | 0.039 | 0.451 | 0.016 | 0.771 | -0.073 | 0.097 | -0.035 | 0.440 | ||
| Tfh cells | BCL6 | 0.458 | *** | 0.444 | *** | 0.432 | *** | 0.427 | *** | 0.155 | ** | 0.156 | ** | |
| IL21 | 0.183 | 0.015 | 0.161 | 0.036 | 0.087 | 0.096 | 0.103 | 0.057 | 0.138 | * | 0.168 | ** | ||
| Th17 cells | STAT3 | 0.528 | *** | 0.524 | *** | 0.476 | *** | 0.483 | *** | 0.33 | *** | 0.324 | *** | |
| IL17A | 0.1 | 0.182 | 0.1 | 0.194 | 0.086 | 0.097 | 0.096 | 0.075 | 0.001 | 0.981 | 0.018 | 0.686 | ||
| Tregs | FOXP3 | 0.235 | * | 0.213 | * | 0.257 | *** | 0.278 | *** | 0.019 | 0.671 | 0.08 | 0.075 | |
| CCR8 | 0.348 | *** | 0.324 | *** | 0.409 | *** | 0.438 | *** | 0.09 | 0.041 | 0.153 | ** | ||
| STAT5B | 0.39 | *** | 0.427 | *** | 0.523 | *** | 0.512 | *** | 0.321 | *** | 0.331 | *** | ||
| TGFβ (TGFB1) | 0.129 | 0.086 | 0.095 | 0.216 | 0.294 | *** | 0.334 | *** | 0.01 | 0.818 | 0.048 | 0.287 | ||
| Exhausted T cells | PD-1 (PDCD1) | 0.18 | 0.016 | 0.144 | 0.060 | 0.129 | 0.013 | 0.13 | 0.016 | 0.047 | 0.286 | 0.103 | 0.022 | |
| CTLA4 | 0.178 | 0.017 | 0.145 | 0.058 | 0.136 | * | 0.154 | * | -0.009 | 0.845 | 0.063 | 0.163 | ||
| LAG3 | 0.122 | 0.102 | 0.114 | 0.138 | 0.079 | 0.128 | 0.086 | 0.109 | 0.108 | 0.015 | 0.156 | ** | ||
| TIM-3 (HAVCR2) | 0.247 | ** | 0.226 | * | 0.287 | *** | 0.342 | *** | -0.093 | 0.034 | -0.04 | 0.379 | ||
| GZMB | 0.196 | * | 0.156 | 0.042 | 0.084 | 0.105 | 0.078 | 0.146 | 0.089 | 0.042 | 0.147 | * | ||
PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma. TAM, tumor-associated macrophage; Treg, regulatory T cell; Tfh, follicular helper T cell; Th, T helper cell. Purity, adjusted correlation according to tumor purity; None, non-adjusted correlation. Cor, R value of Spearman’s correlation. *P < 0.01; **P < 0.001; ***P < 0.0001.
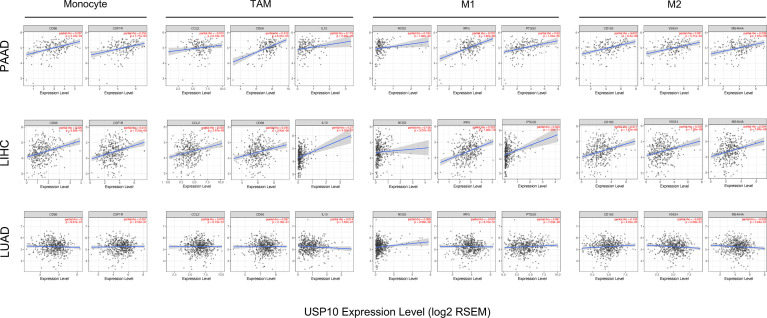
Interestingly, USP10 expression correlated significantly with markers of Tregs (CCR8 (encoding C-C motif chemokine receptor 8) and STAT5B (encoding signal transducer and activator of transcription 5B)) in LUAD, LIHC, and PAAD ( Table 1). The expression of USP10 was statistically significant for TAMs, monocytes, and M1/M2 macrophages in PAAD and LIHC, but not in LUAD (Table 1). Specifically, it was markedly correlated with marker genes of monocytes (CD86, CSF1R (encoding colony stimulating factor 1 receptor)), marker genes of TAMs (CD68, IL10 (encoding interleukin-10)), marker genes of M1 macrophages (NOS2 (encoding nitrous oxide synthase 2), IRF5 (encoding interferon regulatory factor 5), and PTGS2 (encoding prostaglandin-endoperoxide synthase 2)), marker genes of M2 macrophages (CD163, VSIG4 (encoding V-set and immunoglobulin domain containing 4), and MS4A4A (encoding membrane spanning 4-domains A4A)) in PAAD and LIHC (Figure 9). Given the homologous data in GEPIA and TIMER from the TCGA, we used the GEPIA database to further assess the associations between the expression of USP10 and markers genes of monocytes and TAMs in tumor tissues of PAAD, LIHC, and LUAD. Similar results to those obtained using TIMER were observed (Table 2). These results indicated that USP10 might participate in immune cell infiltration and regulate the polarization of macrophages in both PAAD and LIHC TMEs. The precise mechanism requires confirmation in further studies.
Figure 9.
The correlation between the expression of USP10 and marker genes for M2 macrophages, M1 macrophages, TAMs, and monocytes in LUAD, LIHC, and PAAD, as shown by scatter plots. The monocyte markers were CSF1R and CD86; the markers for TAMs were CCL2, CD68, and IL10; the markers for M1 macrophages were PTGS2, IRF5, and NOS2; and the markers for M2 macrophages were MS4A4A, VSIG4, and CD163. PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma. TAMs, tumor-associated macrophages. Significance is indicated by a P-value < 0.05.
Table 2.
Analysis of the correlations between USP10 expression and genetic markers of monocytes and macrophages using GEPIA data.
| Description | Gene markers | PAAD | LIHC | LUAD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor | Normal | Tumor | Normal | Tumor | Normal | |||||||||
| R | P | R | P | R | P | R | P | R | P | R | P | |||
| Monocytes | CD86 | 0.340 | *** | -0.700 | 0.300 | 0.320 | *** | 0.380 | * | -0.033 | 0.470 | -0.130 | 0.35 | |
| CD115 (CSF1R) | 0.320 | *** | 0.230 | 0.770 | 0.280 | *** | 0.410 | * | 0.034 | 0.450 | 0.220 | 0.091 | ||
| TAMs | CCL2 | 0.130 | 0.079 | -0.180 | 0.820 | 0.210 | *** | 0.170 | 0.220 | 0.005 | 0.910 | 0.097 | 0.46 | |
| CD68 | 0.400 | *** | -0.940 | 0.058 | 0.230 | *** | 0.430 | * | 0.064 | 0.160 | 0.017 | 0.9 | ||
| IL10 | 0.180 | 0.015 | -0.910 | 0.093 | 0.260 | *** | 0.130 | 0.350 | -0.042 | 0.360 | -0.074 | 0.58 | ||
| M1 Macrophages | INOS (NOS2) | 0.130 | 0.080 | 0.360 | * | 0.015 | 0.770 | 0.110 | 0.460 | 0.068 | 0.140 | 0.290 | 0.026 | |
| IRF5 | 0.320 | *** | -0.470 | ** | 0.320 | *** | 0.240 | 0.088 | 0.009 | 0.840 | 0.058 | 0.66 | ||
| COX2 (PTGS2) | 0.110 | 0.140 | -0.850 | 0.150 | 0.170 | * | 0.120 | 0.400 | 0.097 | 0.034 | 0.350 | * | ||
| M2 Macrophages | CD163 | 0.310 | *** | 0.037 | 0.960 | 0.190 | ** | 0.330 | 0.018 | 0.027 | 0.550 | -0.029 | 0.83 | |
| VSIG4 | 0.290 | *** | -0.980 | 0.017 | 0.210 | *** | 0.350 | 0.013 | -0.066 | 0.150 | -0.200 | 0.13 | ||
| MS4A4A | 0.280 | ** | -0.950 | 0.055 | 0.220 | *** | 0.370 | * | -0.054 | 0.240 | -0.200 | 0.14 | ||
PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma. TAM, tumor-associated macrophage. Tumor, association analysis in tumor tissues from TCGA; Normal, association analysis in normal tissues from TCGA. *P < 0.01; **P < 0.001; ***P < 0.0001.
Analysis of Survival Related to USP10 Expression Based on Immune Cell Infiltration
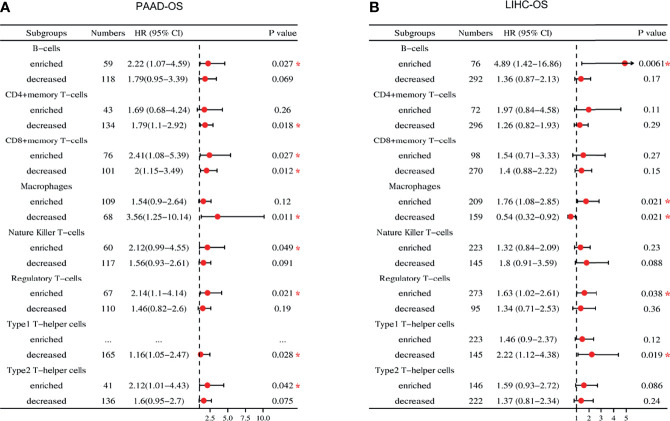
USP10 expression correlated significantly with poor prognosis and immune infiltration of patients with PAAD and LIHC; therefore, we investigated whether USP10 expression could affect the prognosis of patients with PAAD and LIHC via immune infiltration (Figure 10). Based on USP10 expression in relevant immune cell subsets, we found that in PAAD, high USP10 expression was linked to increased infiltration of B cells (OS, HR = 2.22, P = 0.027), NK cells (OS, HR = 2.12, P = 0.049), Tregs (OS, HR = 2.14, P = 0.021), and Th2 cells (OS, HR = 2.12, P = 0.042); and with decreased infiltration of CD4+ T cells, macrophages, and Th1 cells (all P < 0.05), and predicted inferior prognostic survival in patients with PAAD (Figure 10A and Supplementary Figure 4A). In LIHC, overexpression of USP10 and abundant infiltration of B cells (OS, HR = 4.89, P = 0.0061) and Treg cells (OS, HR = 1.63, P = 0.038) or reduced infiltration of Th1 cells (OS, HR = 2.22, P = 0.019) predicted a worse prognosis (Figure 10B and Supplementary Figure 4B).
Figure 10.
Forest plot of the prognostic value of the expression of USP10 in immune cell subpopulations of patients with PAAD and LIHC. (A) In patients with PAAD, the prognostic benefit of USP10 expression depends on the patients’ different immune cell subsets, according to the forest plot. (B) In patients with LIHC, there was an association between the expression of USP10 and OS for the infiltration of different immune cell subgroups, according to a forest plot. PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma. OS, overall survival. *P < 0.05.
Interestingly, USP10 expression with increased (OS, HR = 2.41, P = 0.027) or decreased (OS, HR = 2, P = 0.012) CD8+ T cell infiltration had a marked impact on the survival of patients with PAAD and predicted a worse survival outcome (Figure 10A and Supplementary Figure 4A). In addition, USP10 expression in LIHC correlated statistically with enriched infiltration of macrophages (OS, HR = 1.76, P = 0.021), indicating a worse prognosis. In contrast, USP10 expression was associated with reduced infiltration of macrophages (OS, HR = 0.54, P = 0.021), predicting a better survival outcome (Figure 10B and Supplementary Figure 4B). These results further demonstrated that USP10 might regulate macrophage polarization in the TME of LIHC and have an important impact on prognosis. Critically, the potential explanation for how USP10 influences the prognosis of patients with PAAD and LIHC might stem in part from immune infiltration.
Discussion
USP10 specifically cleaves ubiquitin from ubiquitin-conjugated protein substrates and thus affects cellular processes. Although the aberrant expression of USP10 has been reported in many cancers (10–12, 14, 34, 35), and the role of USP10 in tumorigenesis and prognosis has been partially confirmed in several cancers (10–14), a systematic bioinformatic analysis is still lacking. Based on bioinformatics, this study investigated USP10 expression in pan-cancer and its correlation with prognosis and analyzed the importance of USP10 in the development of different cancers. Moreover, the association between USP10 gene expression in the TME and immune cell infiltration was determined. Our findings provide useful insights to further explore the role of USP10 in tumorigenesis and progression via mechanistic studies.
In this study, we analyzed the differential expression of USP10 and its prognostic value in different types of cancer. The results showed that USP10 mRNA was highly expressed in most tumor types, except for bladder cancer (BLCA), brain and central nervous system cancer, OV, sarcoma (SARC), acute myeloid leukemia (AML), and THYM. The results of the current study suggest that USP10 acts as a therapeutic target for PRAD and AML (36, 37). In addition, IF analysis of 12 clinical samples and the IHC images available through an online database confirmed this trend at the protein level. The results for liver, breast, lung, and colon cancers were similar to those in previous research (11–13, 34). Takayama et al. (10) showed that high expression of USP10 is related significantly to poor prognosis in patients with prostate cancer, which is consistent with our experimental validation. However, Wang et al. (38) showed reduced expression of USP10 in human LUAD tissues, which contradicts the results of the present study, possibly because most of the samples analyzed in Wang’s study were derived from metastatic tumor tissues rather than in situ tumors. Furthermore, we found that USP10 was overexpressed in gastric cancer, which is inconsistent with Zang et al.’s (7) findings and might be caused by the different subtypes of cancer and sample differences. Therefore, the sample size needs to be further expanded. Another exciting finding of this study was that the USP10 protein level was significantly lower in ovarian tumor tissues than in adjacent tissues. Furthermore, survival analysis found that low USP10 expression predicted poor prognosis in patients with ovarian cancer. A previous study by Han et al. (15) agreed with our findings and showed that differential USP10 expression correlated with promoter hypermethylation. Whether USP10 has utility as an independent biomarker of prognosis in OV requires further biological experiments.
Kaplan–Meier survival analysis using the TCGA database demonstrated that in most cancer types (PAAD, LIHC, LUAD, and BRCA), high USP10 expression was associated with poor prognosis. Similarly, as previously reported, USP10 overexpression was shown to be associated with a shorter patient survival time (11, 12).. Moreover, USP10 mutations are closely associated with the development of cancers. We postulated that USP10 expression might be linked to the TMB in various cancers to influence patient survival and is a useful immunotherapy biomarker for checkpoint blockade selection in many types of cancer (39, 40). Analysis revealed that USP10 expression was obviously associated with a high TMB in STAD, LUAD, and UCEC, which influenced patient OS. Previous studies have shown that the TMB predicted prognosis in patients with non-small-cell lung and colorectal cancers (41, 42). Further studies showed that high USP10 expression correlated closely with the stage of cancer and the presence of lymph node metastasis in patients suffering from various types of cancer. These findings suggested that USP10 might serve as a predictable biomarker to determine the prognosis of different cancers. However, more in-depth molecular experimental evidence is needed to verify this.
Importantly, we found that in different types of cancer, USP10 expression was associated with immune cell infiltration levels and has a critical function in cancer immunity, particularly in PAAD and LIHC. Thus, the results of the present study revealed the possible use of USP10 as a cancer biomarker and its function in tumor immunology. We revealed that the infiltration of dendritic cells, neutrophils, macrophages, CD4+ T cells, CD8+ T cells, and B cells in PAAD and LIHC are associated significantly with USP10 expression; whereas, there was only a weak correlation between immune cell infiltration and USP10 expression in LUAD. Interestingly, the association between the expression of USP10 and the expression levels of marker genes of immune cells (e.g., CD19, CD79A, CCL2, CD66b, HLA-DQB1, CD1C, and ITGAX) was not always consistent with the overall trend, suggesting that specific interactions exist between USP10 and certain subtypes of immune cells. Furthermore, the close association between USP10 expression and immune cell marker gene expression suggested that USP10 might function in PAAD and LIHC tumor immune regulation. DCs and macrophages, which are important antigen-presenting cells (APCs), were most related to the expression of USP10 in LIHC. Tumor metastasis is promoted by DCs via their effects on Treg levels and the reduction in the CD8+ T cell response (43). However, in PAAD, DCs correlated weakly with USP10. These differences suggested heterogeneity between cancers that recruit APCs to the TME. A recent study showed that USP10 promotes tumor progression and TAM polarization in colorectal cancer (18). Therefore, our results revealed that USP10 might regulate TAM polarization.
In addition, Tregs are the most important cell type in the TME. Tregs are believed to suppress the excessive immune response by expressing cytotoxic T-lymphocyte associated protein 4 (CTLA4) and secreting IL-10 and transforming growth factor beta (TGFβ), thereby promoting the immune escape of tumor cells (44, 45). TGFβ signaling can be activated by USP10 depletion (46). Recently, researchers have found that depletion of Tregs does not prevent their suppressive activity. Moreover, the therapeutic effect of programmed cell death-1 (PD-1) and PD-1 ligand-1 (PD-L1) signaling blockade therapy on patients with tumors is still not as beneficial as expected. However, Maj et al. (47) showed that in the TME, Tregs are highly apoptotic and can greatly reduce the efficacy of PD-L1 anti-tumor immunotherapy. We further found a significant positive correlation between USP10 and genetic markers of Tregs (CCR8 (encoding C-C motif chemokine receptor 8) and STAT5B (encoding signal transducer and activator of transcription 5B)) ( Table 1), suggesting that USP10 could be involved in activating the immunosuppressive activity of Tregs in PAAD and LIHC. Moreover, in PAAD and LIHC, USP10 levels correlated significantly with several T helper cell markers (STAT1, STAT6, STAT5A, and STAT3) (Table 1). Studies have shown that STAT signaling is involved in numerous aspects of immune regulation, including immune escape and shaping the epigenetic structure of immune cells (48, 49). These findings suggest that USP10 might be closely related to STAT signaling to regulate tumor immune responses. Importantly, the prognosis of PAAD and LIHC was influenced by USP10 through immune cell infiltration. Taken together, USP10 is closely associated with immune cell activity in the TME and might affect patient prognosis through immune infiltration. These findings suggested that USP10 is an immune-related therapeutic target. Nevertheless, USP10’s exact role in tumor immunity requires further exploration.
In conclusion, we determined the universal applicability of USP10 in pan-cancer and found that high expression of USP10 is usually associated with poor clinical prognosis. Furthermore, USP10 is intimately linked to immune cell infiltration in certain cancers and might affect the overall survival of patients with PAAD and LIHC via immune infiltration. These results will enhance our understanding USP10’s vital function in tumorigenesis and serve as a useful basis for future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Shanghai East Hospital Affiliated to Tongji University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
DG designed this study. DG, ZZ, RX, ZH, FL, YH, HC, JL, and XC collected the data and performed the bioinformatic analyses and visualization. DG, YH, and HC performed the hematoxylin and eosin staining and immunofluorescence staining experiments. YL and ZX provided study supervision and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the National Natural Science Foundation of China [grant number 81972183].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.829705/full#supplementary-material
The protein level of USP10 in different types of cancer. Representative images of H&E stained normal and tumor slides (left 1st). Immunofluorescence staining analysis (left 2nd to 4th) showing an evident fluorescence signal in the tumor tissue sections compared with that in the control group. All scale bars = 50 μm. H&E, hematoxylin-eosin staining. N, normal tissues; T, tumor tissues.
Protein expression of USP10 in cancers versus normal tissues as assessed in the HPA database. Compared to normal tissues, USP10 was significantly overexpressed in breast cancer, liver cancer, lung cancer, skin cancer, colon cancer, kidney cancer, and prostate cancer tissues. However, in ovarian cancer, there was a significant low expression of USP10 in tumor tissues compared with that in normal tissues. N, normal tissues; T, tumor tissues.
TIMER database analysis of the correlation between USP10 expression and immune cell infiltration levels in different cancer types. USP10 expression levels in PAAD, LIHC, PCPG, PRAD, THCA, KIRC, and BLCA generally correlated positively with immune cell infiltration. However, in ACC, CHOL, HNSC-HPV+, UCEC, and UCS, USP10 expression levels were not significantly correlated with infiltration of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells. Red box, positive Spearman’s correlation; blue box, negative Spearman’s correlation; gray box, no significant correlation. Significance is indicated by a P-value < 0.05.
Analysis of the prognostic value of USP10 expression in immune cell subsets in patients with PAAD and LIHC using Kaplan–Meier Plotter. (A) In PAAD, high USP10 expression was linked to increased infiltration of B cells, NK cells, Tregs, and Th2 cells (all P < 0.05); and with decreased infiltration of CD4+ T cells, macrophages, and Th1 cells (all P < 0.05), and predicted inferior prognostic survival in patients with PAAD. However, USP10 expression with increased (OS, HR = 2.41, P = 0.027) or decreased (OS, HR = 2, P = 0.012) CD8+ T cell infiltration had a marked impact on the survival of patients with PAAD and predicted a worse survival outcome. (B) In LIHC, overexpression of USP10 and abundant infiltration of B cells and Treg cells (all P < 0.05) or reduced infiltration of Th1 cells (P < 0.05) predicted a worse prognosis. Interestingly, USP10 expression in LIHC correlated statistically with enriched infiltration of macrophages (OS, HR = 1.76, P = 0.021), indicating a worse prognosis. In contrast, USP10 expression was associated with reduced infiltration of macrophages (OS, HR = 0.54, P = 0.021), predicting a better survival outcome. PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma. OS, overall survival. HR, hazard ratio. Significance is indicated by a P-value < 0.05.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Hagner PR, Schneider A, Gartenhaus RB. Targeting the Translational Machinery as a Novel Treatment Strategy for Hematologic Malignancies. Blood (2010) 115(11):2127–35. doi: 10.1182/blood-2009-09-220020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y, Zhang Z. The History and Advances in Cancer Immunotherapy: Understanding the Characteristics of Tumor-Infiltrating Immune Cells and Their Therapeutic Implications. Cell Mol Immunol (2020) 17(8):807–21. doi: 10.1038/s41423-020-0488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amerik AY, Hochstrasser M. Mechanism and Function of Deubiquitinating Enzymes. Biochim Biophys Acta (2004) 1695(1-3):189–207. doi: 10.1016/j.bbamcr.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 5. Grabbe C, Husnjak K, Dikic I. The Spatial and Temporal Organization of Ubiquitin Networks. Nat Rev Mol Cell Biol (2011) 12(5):295–307. doi: 10.1038/nrm3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Popovic D, Vucic D, Dikic I. Ubiquitination in Disease Pathogenesis and Treatment. Nat Med (2014) 20(11):1242–53. doi: 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- 7. Zeng Z, Wu HX, Zhan N, Huang YB, Wang ZS, Yang GF, et al. Prognostic Significance of USP10 as a Tumor-Associated Marker in Gastric Carcinoma. Tumour Biol (2014) 35(4):3845–53. doi: 10.1007/s13277-013-1509-1 [DOI] [PubMed] [Google Scholar]
- 8. Soncini C, Berdo I, Draetta G. Ras-GAP SH3 Domain Binding Protein (G3BP) Is a Modulator of USP10, a Novel Human Ubiquitin Specific Protease. Oncogene (2001) 20(29):3869–79. doi: 10.1038/sj.onc.1204553 [DOI] [PubMed] [Google Scholar]
- 9. Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 Regulates P53 Localization and Stability by Deubiquitinating P53. Cell (2010) 140(3):384–96. doi: 10.1016/j.cell.2009.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takayama KI, Suzuki T, Fujimura T, Takahashi S, Inoue S. Association of USP10 With G3BP2 Inhibits P53 Signaling and Contributes to Poor Outcome in Prostate Cancer. Mol Cancer Res (2018) 16(5):846–56. doi: 10.1158/1541-7786.Mcr-17-0471 [DOI] [PubMed] [Google Scholar]
- 11. Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, et al. Over-Expression of Genes and Proteins of Ubiquitin Specific Peptidases (USPs) and Proteasome Subunits (PSs) in Breast Cancer Tissue Observed by the Methods of RFDD-PCR and Proteomics. Breast Cancer Res Treat (2007) 104(1):21–30. doi: 10.1007/s10549-006-9393-7 [DOI] [PubMed] [Google Scholar]
- 12. Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, et al. Gene Expression-Based Classification of Non-Small Cell Lung Carcinomas and Survival Prediction. PloS One (2010) 5(4):e10312. doi: 10.1371/journal.pone.0010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally Derived Metastasis Gene Expression Profile Predicts Recurrence and Death in Patients With Colon Cancer. Gastroenterology (2010) 138(3):958–68. doi: 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo J, Zhang J, Liang L, Liu N, Qi M, Zhao S, et al. Potent USP10/13 Antagonist Spautin-1 Suppresses Melanoma Growth via ROS-Mediated DNA Damage and Exhibits Synergy With Cisplatin. J Cell Mol Med (2020) 24(7):4324–40. doi: 10.1111/jcmm.15093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han GH, Chay DB, Yi JM, Cho H, Chung JY, Kim JH. Loss of Both USP10 and P14arf Protein Expression Is an Independent Prognostic Biomarker for Poor Prognosis in Patients With Epithelial Ovarian Cancer. Cancer Genomics Proteomics (2019) 16(6):553–62. doi: 10.21873/cgp.20157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, et al. USP10 Antagonizes C-Myc Transcriptional Activation Through SIRT6 Stabilization to Suppress Tumor Formation. Cell Rep (2013) 5(6):1639–49. doi: 10.1016/j.celrep.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu C, Zhang M, Moses N, Hu C-l, Polin L, Chen W, et al. The USP10-HDAC6 Axis Confers Cisplatin Resistance in Non-Small Cell Lung Cancer Lacking Wild-Type P53. Cell Death Dis (2020) 11(5):328. doi: 10.1038/s41419-020-2519-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li B, Qi ZP, He DL, Chen ZH, Liu JY, Wong MW, et al. NLRP7 Deubiquitination by USP10 Promotes Tumor Progression and Tumor-Associated Macrophage Polarization in Colorectal Cancer. J Exp Clin Cancer Res (2021) 40(1):126. doi: 10.1186/s13046-021-01920-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boumil EF, Castro N, Phillips AT, Chatterton JE, McCauley SM, Wolfson AD, et al. USP10 Targeted Self-Deliverable siRNA to Prevent Scarring in the Cornea. Mol Ther Nucleic Acids (2020) 21:1029–43. doi: 10.1016/j.omtn.2020.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahashi M, Higuchi M, Makokha GN, Matsuki H, Yoshita M, Tanaka Y, et al. HTLV-1 Tax Oncoprotein Stimulates ROS Production and Apoptosis in T Cells by Interacting With USP10. Blood (2013) 122(5):715–25. doi: 10.1182/blood-2013-03-493718 [DOI] [PubMed] [Google Scholar]
- 21. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: Genes, Pathways, and Networks in a Collection of 18,000 Cancer Gene Expression Profiles. Neoplasia (2007) 9(2):166–80. doi: 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res (2020) 48(W1):W509–w514. doi: 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–w102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A New Database for Meta-Analysis of the Prognostic Value of Genes. BMC Med Genomics (2009) 2:18. doi: 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nagy Á, Munkácsy G, Győrffy B. Pancancer Survival Analysis of Cancer Hallmark Genes. Sci Rep (2021) 11(1):6047. doi: 10.1038/s41598-021-84787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The Cbio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov (2012) 2(5):401–4. doi: 10.1158/2159-8290.Cd-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohtani H. Focus on TILs: Prognostic Significance of Tumor Infiltrating Lymphocytes in Human Colorectal Cancer. Cancer Immun (2007) 7:4. [PMC free article] [PubMed] [Google Scholar]
- 29. Ravelli A, Roviello G, Cretella D, Cavazzoni A, Biondi A, Cappelletti MR, et al. Tumor-Infiltrating Lymphocytes and Breast Cancer: Beyond the Prognostic and Predictive Utility. Tumour Biol (2017) 39(4):1010428317695023. doi: 10.1177/1010428317695023 [DOI] [PubMed] [Google Scholar]
- 30. Yusuf M, Gaskins J, Mandish S, May ME, Wall W, Fisher W, et al. Tumor Infiltrating Lymphocyte Grade in Merkel Cell Carcinoma: Relationships With Clinical Factors and Independent Prognostic Value. Acta Oncol (2020) 59(12):1409–15. doi: 10.1080/0284186x.2020.1794033 [DOI] [PubMed] [Google Scholar]
- 31. Azimi F, Scolyer RA, Rumcheva P, Moncrieff M, Murali R, McCarthy SW, et al. Tumor-Infiltrating Lymphocyte Grade is an Independent Predictor of Sentinel Lymph Node Status and Survival in Patients With Cutaneous Melanoma. J Clin Oncol (2012) 30(21):2678–83. doi: 10.1200/jco.2011.37.8539 [DOI] [PubMed] [Google Scholar]
- 32. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring Tumour Purity and Stromal and Immune Cell Admixture From Expression Data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aran D, Sirota M, Butte AJ. Systematic Pan-Cancer Analysis of Tumour Purity. Nat Commun (2015) 6:8971. doi: 10.1038/ncomms9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu H, Yan F, Yuan T, Qian M, Zhou T, Dai X, et al. USP10 Promotes Proliferation of Hepatocellular Carcinoma by Deubiquitinating and Stabilizing YAP/TAZ. Cancer Res (2020) 80(11):2204–16. doi: 10.1158/0008-5472.Can-19-2388 [DOI] [PubMed] [Google Scholar]
- 35. Kim K, Huh T, Park Y, Koo DH, Kim H, Hwang I, et al. Prognostic Significance of USP10 and P14arf Expression in Patients With Colorectal Cancer. Pathol Res Pract (2020) 216(6):152988. doi: 10.1016/j.prp.2020.152988 [DOI] [PubMed] [Google Scholar]
- 36. Liao Y, Guo Z, Xia X, Liu Y, Huang C, Jiang L, et al. Inhibition of EGFR Signaling With Spautin-1 Represents a Novel Therapeutics for Prostate Cancer. J Exp Clin Cancer Res (2019) 38(1):157. doi: 10.1186/s13046-019-1165-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weisberg EL, Schauer NJ, Yang J, Lamberto I, Doherty L, Bhatt S, et al. Inhibition of USP10 Induces Degradation of Oncogenic FLT3. Nat Chem Biol (2017) 13(12):1207–15. doi: 10.1038/nchembio.2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X, Xia S, Li H, Wang X, Li C, Chao Y, et al. The Deubiquitinase USP10 Regulates KLF4 Stability and Suppresses Lung Tumorigenesis. Cell Death Differ (2020) 27(6):1747–64. doi: 10.1038/s41418-019-0458-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of Tumor Mutation Burden as an Immunotherapy Biomarker: Utility for the Oncology Clinic. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fumet JD, Truntzer C, Yarchoan M, Ghiringhelli F. Tumour Mutational Burden as a Biomarker for Immunotherapy: Current Data and Emerging Concepts. Eur J Cancer (2020) 131:40–50. doi: 10.1016/j.ejca.2020.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee DW, Han SW, Bae JM, Jang H, Han H, Kim H, et al. Tumor Mutation Burden and Prognosis in Patients With Colorectal Cancer Treated With Adjuvant Fluoropyrimidine and Oxaliplatin. Clin Cancer Res (2019) 25(20):6141–7. doi: 10.1158/1078-0432.Ccr-19-1105 [DOI] [PubMed] [Google Scholar]
- 42. Devarakonda S, Rotolo F, Tsao MS, Lanc I, Brambilla E, Masood A, et al. Tumor Mutation Burden as a Biomarker in Resected Non-Small-Cell Lung Cancer. J Clin Oncol (2018) 36(30):2995–3006. doi: 10.1200/jco.2018.78.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sawant A, Hensel JA, Chanda D, Harris BA, Siegal GP, Maheshwari A, et al. Depletion of Plasmacytoid Dendritic Cells Inhibits Tumor Growth and Prevents Bone Metastasis of Breast Cancer Cells. J Immunol (2012) 189(9):4258–65. doi: 10.4049/jimmunol.1101855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Izcue A, Coombes JL, Powrie F. Regulatory T Cells Suppress Systemic and Mucosal Immune Activation to Control Intestinal Inflammation. Immunol Rev (2006) 212:256–71. doi: 10.1111/j.0105-2896.2006.00423.x [DOI] [PubMed] [Google Scholar]
- 45. Boonpiyathad T, Satitsuksanoa P, Akdis M, Akdis CA. Il-10 Producing T and B Cells in Allergy. Semin Immunol (2019) 44:101326. doi: 10.1016/j.smim.2019.101326 [DOI] [PubMed] [Google Scholar]
- 46. Gillespie SR, Tedesco LJ, Wang L, Bernstein AM. The Deubiquitylase USP10 Regulates Integrin β1 and β5 and Fibrotic Wound Healing. J Cell Sci (2017) 130(20):3481–95. doi: 10.1242/jcs.204628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maj T, Wang W, Crespo J, Zhang H, Wang W, Wei S, et al. Oxidative Stress Controls Regulatory T Cell Apoptosis and Suppressor Activity and PD-L1-Blockade Resistance in Tumor. Nat Immunol (2017) 18(12):1332–41. doi: 10.1038/ni.3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Owen KL, Brockwell NK, Parker BS. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers (Basel) (2019) 11(12):2002. doi: 10.3390/cancers11122002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Verhoeven Y, Tilborghs S, Jacobs J, De Waele J, Quatannens D, Deben C, et al. The Potential and Controversy of Targeting STAT Family Members in Cancer. Semin Cancer Biol (2020) 60:41–56. doi: 10.1016/j.semcancer.2019.10.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The protein level of USP10 in different types of cancer. Representative images of H&E stained normal and tumor slides (left 1st). Immunofluorescence staining analysis (left 2nd to 4th) showing an evident fluorescence signal in the tumor tissue sections compared with that in the control group. All scale bars = 50 μm. H&E, hematoxylin-eosin staining. N, normal tissues; T, tumor tissues.
Protein expression of USP10 in cancers versus normal tissues as assessed in the HPA database. Compared to normal tissues, USP10 was significantly overexpressed in breast cancer, liver cancer, lung cancer, skin cancer, colon cancer, kidney cancer, and prostate cancer tissues. However, in ovarian cancer, there was a significant low expression of USP10 in tumor tissues compared with that in normal tissues. N, normal tissues; T, tumor tissues.
TIMER database analysis of the correlation between USP10 expression and immune cell infiltration levels in different cancer types. USP10 expression levels in PAAD, LIHC, PCPG, PRAD, THCA, KIRC, and BLCA generally correlated positively with immune cell infiltration. However, in ACC, CHOL, HNSC-HPV+, UCEC, and UCS, USP10 expression levels were not significantly correlated with infiltration of B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells. Red box, positive Spearman’s correlation; blue box, negative Spearman’s correlation; gray box, no significant correlation. Significance is indicated by a P-value < 0.05.
Analysis of the prognostic value of USP10 expression in immune cell subsets in patients with PAAD and LIHC using Kaplan–Meier Plotter. (A) In PAAD, high USP10 expression was linked to increased infiltration of B cells, NK cells, Tregs, and Th2 cells (all P < 0.05); and with decreased infiltration of CD4+ T cells, macrophages, and Th1 cells (all P < 0.05), and predicted inferior prognostic survival in patients with PAAD. However, USP10 expression with increased (OS, HR = 2.41, P = 0.027) or decreased (OS, HR = 2, P = 0.012) CD8+ T cell infiltration had a marked impact on the survival of patients with PAAD and predicted a worse survival outcome. (B) In LIHC, overexpression of USP10 and abundant infiltration of B cells and Treg cells (all P < 0.05) or reduced infiltration of Th1 cells (P < 0.05) predicted a worse prognosis. Interestingly, USP10 expression in LIHC correlated statistically with enriched infiltration of macrophages (OS, HR = 1.76, P = 0.021), indicating a worse prognosis. In contrast, USP10 expression was associated with reduced infiltration of macrophages (OS, HR = 0.54, P = 0.021), predicting a better survival outcome. PAAD, pancreatic adenocarcinoma; LIHC, liver hepatocellular carcinoma. OS, overall survival. HR, hazard ratio. Significance is indicated by a P-value < 0.05.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.