Abstract
Alzheimer's disease (AD) is the result of abnormal processing of the amyloid precursor protein (APP) by β-secretase and γ-secretase, which leads to the formation of toxic β-amyloid peptides. The toxic β-amyloid peptides induce neuron death, memory problems, and AD development. Several APP mutations increase the risk of developing early-onset AD. However, the A673T mutation identified in the Icelandic population prevents AD development by reducing the cleavage of APP by β-secretase. In this study, we inserted the A673T mutation in human cells using the CRISPR prime editing (PE) technique. Repeated PE treatments resulted in the insertion of the A673T mutation in up to 49.2% of APP genes when a second nick was induced in the other DNA strand. When the protospacer adjacent motif used for PE was also mutated, up to 68.9% of the APP genes contained the protective A673T mutation. PE is a promising approach to introduce the A673T mutation precisely without mutating nearby nucleotides.
Introduction
Dr. Alois Alzheimer first described Alzheimer's disease (AD) in 1906. Since then, this disease has become the most prevalent neurodegenerative disease in the world.1 Despite several decades worth of intensive research to develop treatments for AD, current treatments only manage to reduce symptoms but fail to stop or reverse the progression of the disease. While some drugs such as aducanumab, a monoclonal antibody targeting β-amyloid, could potentially curb the disease, continuous treatments are required to maintain the beneficial effects, and more studies are needed to conclude its efficacy.2
The etiology of AD is extremely complex and involves multiple intrinsic and extrinsic factors.3 To develop a more permanent treatment for this disease, it will be important to target the fundamental problem leading to AD: the formation of toxic β-amyloid (Aβ) peptides.
Aβ peptides are generated by the metabolism of the amyloid precursor protein (APP). APP is an integral membrane glycoprotein that is expressed ubiquitously.3 It can undergo two competitive cleavage pathways: the nonamyloidogenic and the amyloidogenic. In the nonamyloidogenic pathway, the processing of APP by α-secretase followed by the γ-secretase cleavage generates the soluble APPα, the P3 peptide, and the APP intracellular domain (AICD).4
In the amyloidogenic pathway, APP is successively cleaved by β-secretase (also called the β-site APP cleaving enzyme 1) and γ-secretase. β-Secretase cleaves APP between Met671 and Asp672. This forms a soluble APP-β ectodomain and a carboxy-terminal counterpart of 99 aa termed C99, which remains in the membrane. The subsequent cleavage of C99 by γ-secretase generates the Aβ peptides (containing 39–43 aa) and the AICD. The Aβ1-40 and Aβ1-42 peptides are the most abundant peptides. Aβ1-42 is the most toxic peptide generated by amyloidogenic processing.
The amyloid hypothesis, according to which these Aβ peptides play a causative role in AD pathology, is supported by early genetic studies. These studies demonstrated that several autosomal-dominant mutations or duplications of the APP gene, as well as mutations in PSEN1 or PSEN2 (the genes that encode γ-secretase subunits), lead to early-onset familial AD.5
These Aβ peptides form oligomers, which co-localize with axonal voltage gated calcium channels, facilitate calcium influx and impair fast axonal transport of cargos such as brain-derived neurotrophic factor.6 Aβ peptides aggregate and accumulate at synapses.7,8 Aβ oligomers also form larger deposits called senile plaques. Furthermore, toxic Aβ peptides induce hyperphosphorylation of the tau protein, leading to intracellular accumulation of neurofibrillary tangles. This leads to the loss of neuronal synapses at an early stage of the AD pathology, which is correlated with and is probably responsible for the cognitive decline.1–9
Thirty coding mutations have been identified in the APP gene. Several of them (e.g., the London mutation V717I, the Swedish mutation KM670/671NL, and the A673V mutation) promote Aβ accumulation or increase the Aβ42/Aβ40 ratio.10,11 The majority of these coding mutations are autosomal dominant, except for A673V.12,13 As a result, these mutations are responsible for hereditary forms of AD, also called familial AD (FAD). However, in 2012, Jonsson et al. identified a rare APP coding mutation A673T in the Icelandic population, which protects against sporadic late-onset AD.14
The A673T mutation has also been found with a relatively low prevalence in the Danish, Finnish, Norwegian, and Swedish populations.15–17 The carriers of the A673T polymorphism have a higher cognitive level than noncarriers based on the Cognitive Performance Scale, suggesting that the effect of this mutation may reach beyond the boundaries of protection against AD.17 A very low accumulation of amyloid plaques was found in a 104.8-year-old person with this mutation.15 The A673T mutation is located very close to the β-secretase cut site, and it reduces the cleavage of APP by this enzyme.
Moreover, the A673T mutation is in position 2 of the Aβ(1-XX) peptide cleavage product (A2T), and it was also found that it reduces the aggregation of these peptides.18 Another study demonstrated that the plasma levels of Aβ40 and Aβ42 of the healthy heterozygous carriers of the A673T mutation were reduced by 28% compared to noncarriers.19 Kimura et al. demonstrated that A673T reduced C99 generation and decreased the C99/C89 ratio by changing the cleavage site selection of BACE1 from the β-site (between Met671 and Asp672) to the β′-site (between Thy681 and Gln682), resulting in decreased production of Aβ1-40 and an increase in Aβ11-40, which is more metabolically labile.20
Given the available evidence that the A673T mutation prevents AD development in those who carry this mutation, our research group hypothesized that the insertion of this mutation in patients' neurons could be an effective and sustainable method for slowing down or even stopping the progression of some hereditary and sporadic forms of AD.
Our research group has previously shown that the presence of the protective co-dominant mutation A673T in the APP gene of SH-SY5Y neuroblastomas decreases Aβ peptide production in 14/29 forms of FAD.21,22 The reduction in the production of Aβ peptides for the London mutation (V717I) with the co-mutation A673T even reached the same level as the control containing only the A673T mutation. These preliminary results suggested that the insertion of A673T in APP genes containing one of these 14 FAD mutations could confer a clinical benefit in preventing or delaying the onset of the disease.
We also modified the APP gene in HEK293T cells and in SH-SY5Y neuroblastomas using the CRISPR-Cas9 base editing technique.23 We used a Cas9 nickase (Cas9n) fused with a cytidine deaminase enzyme to convert the alanine anti-sense codon (5′TGC3′) to a threonine anti-sense codon (5′TGT3′).24 We tested several Cas9n-deaminase variants to compare their efficiency of conversion. The results were characterized and quantified by deep sequencing. We successfully introduced the A673T mutation in 53% of the APP gene of HEK293T cells.
However, another nearby cytosine was also modified into a thymine, resulting in the frequent insertion of an additional mutation (E674K), which seemed to reduce Aβ peptide accumulation further. This approach provided a new strategy for the treatment of AD and, by doing so, demonstrated the capacity of base editing techniques for treating genetic diseases.
Following our experiences with the cytidine base editing technique, a new and more versatile base editing technique called prime editing was developed.25 This technique uses a Cas9n fused to an engineered reverse transcriptase (RT) and a modified single-guide RNA (sgRNA) called a prime editing guide RNA (pegRNA). This pegRNA contains a protospacer sequence of 20 nt that specifies the target site. It also contains a primer binding site (PBS) and a RT template (RTT) that encodes the desired nucleotide edition.
When the protospacer sequence binds to DNA, the noncomplementary strand of the DNA is simply cut by the Cas9n three bases upstream from the protospacer adjacent motif (PAM). The PBS binds with the 3′ OH group exposed from the DNA flap, serving as a primer for RT, which extends the 3′ flap by copying the editing RTT sequence of the pegRNA. The unedited 5′ flap is subsequently removed by a 5′ endonuclease (e.g., FEN1) or by a 5′ exonuclease (e.g., EXO1). Mismatch repair corrects the DNA, resulting in either the original variant sequence or precisely modified DNA.
If the PAM sequence has not been changed, the Cas9n/pegRNA complex can bind to the sequence and attempt the prime editing again.26 The prime editing technique has the great advantage that it can modify any standard nucleotide into any other standard nucleotide (i.e., adenine, guanine, thymine, and cytosine). Thus, it is an interesting tool to insert the A673T mutation and also to correct the V717I London mutation or other point mutations responsible for FADs directly.
Methods
PE2 plasmid
The prime editor 2 (PE2) plasmid coding for the SpCas9 nickase gene fused with an engineered murine leukmia virus RT was obtained from Addgene (Watertown, MA; pCMV-PE2 #132775). The plasmid used to express a pegRNA was also obtained from Addgene (pU6-pegRNA-GG-acceptor #132777). The cloning of the pegRNA into the BsaI digested pU6-pegRNA-GG-acceptor was performed as described by Anzalone et al.25
PE3 plasmid
The plasmid pBSU6 used to express a sgRNA was obtained from Dirk Grimm labs (Heidelberg University, Heidelberg, Germany; pBSU6_FE_Scaffold_RSV_GFP). The cloning of the sgRNA into the BbsI digested pBSU6 was performed using the protocol for the NEB T4 DNA Ligase enzyme where 50 ng of the vector was used at a ratio of 1:3 for the insert (using NEBioCalculator®). Oligonucleotides used to construct all sgRNAs were purchased from Integrated DNA Technologies (Coralville, IA).
Cell culture and transfection
HEK293T cells were cultured in high glucose Dulbecco's modified Eagle's medium (Wisent, Quebec, Canada) supplemented with 10% fetal bovine serum (Wisent) and 1% penicillin-streptomycin (Wisent). The day before transfection, the cells were detached from the flask with a Trypsin-EDTA solution (Sigma–Aldrich, St. Louis, MO) and counted. Cells were plated on a 24-well plate at a density of 60,000 cells per well with 1 mL of culture medium.
On the day of the transfection, solutions A and B were prepared. Solution A contained 48 μL of Opti-MEM-1™ and 2 μL of the transfection reagent (Lipofectamine™ 2000) to give a final volume of 50 μL. Solution B was prepared with 1 μg of DNA solution (500 ng of PE2 plasmid, 250 ng of pegRNA plasmid, and 250 ng of sgRNA plasmid) and completed with Opti-MEM-1™ to give a final volume of 50 μL. Solutions A and B were then mixed and incubated at room temperature for 20 min before being added to each well. The medium was changed for 1 mL of fresh medium 12 h later, and cells were incubated for 72 h before genomic DNA extraction.
For multiple treatment experiments, cells were plated on day 0 and transfected as described above. On day 3, 50% of cells were recovered for genomic DNA extraction, and the remaining 50% were plated in one well of a six-well plate containing 2 mL of fresh culture medium. The plate was put back into the incubator, and the cells were allowed to grow until day 6. Cells were detached from the well with a trypsin-EDTA solution and counted. The DNA was then extracted from cell samples. The remaining cells were plated at 60,000 cells per well on a 24-well plate. The following day, cells were transfected as described above. We used the same procedure for each treatment.
Genomic DNA preparation and polymerase chain reaction amplification
Cells were detached from wells directly with up and down pipetting in the culture medium and transferred to 1.5 mL Eppendorf tubes. Cells were centrifuged for 5 min at 7800 g at room temperature. Cell pellets were washed once with 1 mL of phosphate-buffered saline 1 × and centrifuged again for 5 min at 7800 g. Genomic DNA was prepared using the DirectPCR Lysis Reagent (Viagen Biotech, Los Angeles, CA). Briefly, 50 μL of DirectPCR Lysis Reagent containing 0.5 μL of proteinase K solution (20 mg/mL) was added to each cell pellet and incubated overnight at 56°C followed by another incubation at 85°C for 45 min.
The sample was then centrifuged at 17,000 g for 5 min. One microliter of each genomic DNA preparation (supernatant) was used for the polymerase chain reaction (PCR). For each primer set (Table 1), PCR temperature cycling was performed as follows: pre-denaturation at 98°C for 30 s, following by 35 cycles of 98°C for 10 s (denaturation), 60°C for 20 s (annealing), and 72°C for 45 s (extension). A final extension at 72°C for 5 min was also performed.
Table 1.
Primer Sequences for PCR, Sanger, and Deep Sequencing
| Sanger sequencing of APP exon 16 | TCAAGCCTGTAATCCCAGCA |
| Sanger sequencing of EMX1 exon 3 | TGAGTTTCTCATCTGTGCCC |
| PCR of APP exon 16 Fwd | AAATCCTTGCCAACCTCTCAACCA |
| PCR of APP exon 16 Rev | GTTCTTTCAGACTACAAGGCAGCA |
| PCR of EMX1 exon 3 Fwd | CAGCTCAGCCTGAGTGTTGA |
| PCR of EMX1 exon 3 Rev | CTCGTGGGTTTGTGGTTGC |
| Deepseq of APP exon 16 Fwd | ACACTGACGACATGGTTCTACA GGTAGGCTTTGTCTTACAGTGTTA |
| Deepseq of APP exon 16 Rev | TACGGTAGCAGAGACTTGGTCT TGGTAATCCTATAGGCAAGCATTG |
| Deepseq of APP exon 17 Fwd | ACACTGACGACATGGTTCTACA ATCCAAATGTCCCCTGCATTTAAG |
| Deepseq of APP exon 17 Rev | TACGGTAGCAGAGACTTGGTCT TTTACCTACCTCCACCACACCATG |
| Deepseq of EMX1 exon 3 Fwd | ACACTGACGACATGGTTCTACA CAGCTCAGCCTGAGTGTTGA |
| Deepseq of EMX1 exon 3 Rev | TACGGTAGCAGAGACTTGGTCT CTCGTGGGTTTGTGGTTGC |
PCR, polymerase chain reaction.
We used Phusion™ High-Fidelity DNA Polymerase from Thermo Fisher Scientific (Waltham, MA) for all PCR reactions. Five microliters of the sample was electrophorized in 1 × TBE buffer on 1% agarose gel to control the PCR quality and to make sure that only one specific band was present. The remaining 45 μL of the PCR was sequenced with an internal primer (Table 1).
Sanger and deep-sequencing analysis
Deep-sequencing samples were prepared by a PCR (as described above), with special primers for deep sequencing containing a bar code sequence to permit the subsequent deep sequencing (Table 1). PCR samples were sent to the Sanger sequencing platform at the research center of CHU de Québec-Université Laval and the Genome Quebec Innovation Centre at McGill University to sequence amplicons with an Illumina sequencing platform. Roughly 6,000–10,000 reads were obtained per sample.
Sanger sequencing results were analyzed with the EditR online program.27 Deep-sequencing results were analyzed online with the CRISPResso2 online program.28 For all prime editing yield quantification, prime editing efficiency was calculated as the percentage resulting from the number of reads with the desired edition that did not contain insertions or deletions (indels) divided by the total number of reads of amplicons. For all experiments, indel yields were calculated as the numbers of indel-containing reads in the amplicons divided by the total reads.
Statistical analyses
The data were analyzed using GraphPad Prism v5.0 (GraphPad Software, La Jolla, CA). The means of different pegRNAs and negative controls were compared with a one-way analysis of variance (ANOVA) test and Dunnett's multiple comparisons test. A p-value of <0.05 was considered statistically significant for a 95% confidence interval.
Results
Design of various pegRNAs and optimization of the PE2 technique
The efficacy of inducing nucleotide mutations with the prime editing technique depends on the components of pegRNA and on the target DNA. The frequency of installation for the desired mutation rapidly decreases with increasing distance from the single-stranded cut induced by the Cas9 nickase.25
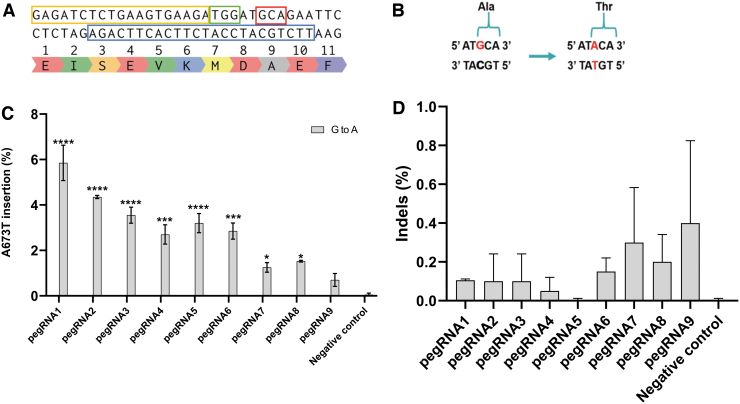
The SpCas9 enzyme requires an NGG PAM. Fortunately, there is such a PAM at position Met671-Asp672 of the APP gene only 3 nt away from the target nucleotide guanine (G; Fig. 1A). This was used to exchange the alanine codon (5′GCA3′) for a threonine codon (5′ACA3′; Fig. 1B). The desired G → A modification is +9 nt from the site that the SpCas9n will nick. Since the length of the PBS and of the RTT could affect the prime editing efficiency, we designed nine different pegRNAs (Table 2) and tested their efficacy with the PE2 method.
FIG. 1.
Test of various prime editing guide RNAs (pegRNAs) targeting the APP gene in HEK293T cells using the prime editor 2 (PE2) method. (A) Section of APP exon 16 with codon of interest (Ala673) in the red box. In the blue box, the unmodified sequence used to design the primer binding site (PBS) and the reverse transcriptase (RT) template (RTT). The protospacer adjacent motif (PAM) is in the green box, and the pegRNA spacer sequence is in the yellow box. (B) The target nucleotide G (guanine) in red is modified into an A (adenine) in red to change the alanine codon (5′GCA3′) into a threonine codon (5′ACA3′). (C) Nine different pegRNAs with various PBS and RTT were transfected in HEK293T cells with Lipofectamine™ 2000. DNA was extracted 72 h later, and exon 16 of the APP gene was polymerase chain reaction (PCR) amplified. The sequences were then analyzed using the Illumina deep-sequencing method. The negative control did not receive any treatment. The results were analyzed with a one-way analysis of variance (ANOVA) test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. *p < 0.0387; ***p < 0.0002; ****p < 0.0001. (D) Insertion or deletion (indel) levels resulting from prime editing of the APP gene exon 16 using the PE2 method. There were no indels in the control sample. The level of indels varied from 0% to 0.8% for the PE2 method. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. The p-value was not significant for all samples.
Table 2.
Sequences of pegRNAs Used in Experiments
| pegRNA | Spacer sequence | 3′ extension (5′ to 3′) | PBS length (nt) | RTT length (nt) |
|---|---|---|---|---|
| pegRNA1 | GGAGATCTCTGAAGTGAAGA | TTCTGTATCCATCTTCACTTCAGAG | 11 | 14 |
| pegRNA2 | GGAGATCTCTGAAGTGAAGA | GAATTCTGTATCCATCTTCACTTCAGAG | 11 | 17 |
| pegRNA3 | GGAGATCTCTGAAGTGAAGA | TCGGAATTCTGTATCCATCTTCACTTCAGAG | 11 | 20 |
| pegRNA4 | GGAGATCTCTGAAGTGAAGA | TTCTGTATCCATCTTCACTTCAGAGATC | 14 | 14 |
| pegRNA5 | GGAGATCTCTGAAGTGAAGA | GAATTCTGTATCCATCTTCACTTCAGAGATC | 14 | 17 |
| pegRNA6 | GGAGATCTCTGAAGTGAAGA | TCGGAATTCTGTATCCATCTTCACTTCAGAGATC | 14 | 20 |
| pegRNA7 | GGAGATCTCTGAAGTGAAGA | TTCTGTATCCATCTTCACTTCAGAGATCTC | 16 | 14 |
| pegRNA8 | GGAGATCTCTGAAGTGAAGA | GAATTCTGTATCCATCTTCACTTCAGAGATCTC | 16 | 17 |
| pegRNA9 | GGAGATCTCTGAAGTGAAGA | TCGGAATTCTGTATCCATCTTCACTTCAGAGATCTC | 16 | 20 |
| pegRNA_PM1 | GGAGATCTCTGAAGTGAAGA | TTCTGTATCGATCTTCACTTCAGAG | 11 | 14 |
| pegRNA_PM2 | GGAGATCTCTGAAGTGAAGA | TTCTGTATGCATCTTCACTTCAGAG | 11 | 14 |
| pegRNA1_London | GTTCTTCTTCAGCATCACCA | TCATCATCACCTTGGTGATGCTGAAGAA | 13 | 15 |
| pegRNA2_London | GTTCTTCTTCAGCATCACCA | GATCATCATCACCTTGGTGATGCTGAAGAAGAA | 16 | 17 |
| pegRNA1_PM_London | GTTCTTCTTCAGCATCACCA | TCATCATCACATTGGTGATGCTGAAGAA | 13 | 15 |
| pegRNA2_PM_London | GTTCTTCTTCAGCATCACCA | GATCATCATCACATTGGTGATGCTGAAGAAGAA | 16 | 17 |
| pegRNA_EMX1 | GAGTCCGAGCAGAAGAAGAA | GTGATGGGAGCCCTTGTTCTTCTGCTCGG | 13 | 16 |
| pegRNA1_PAM2 | CTTCATATCCTGAGTCATGT | ACAGAATTCCGACATGACTCAGGAT | 11 | 14 |
| pegRNA2_PAM2 | CTTCATATCCTGAGTCATGT | TACAGAATTCCGACATGACTCAGGAT | 11 | 15 |
| pegRNA3_PAM2 | CTTCATATCCTGAGTCATGT | ATACAGAATTCCGACATGACTCAGGAT | 11 | 16 |
| pegRNA1_PM_PAM2 | CTTCATATCCTGAGTCATGT | ACAGAATTCAGACATGACTCAGGAT | 11 | 14 |
| pegRNA2_PM_PAM2 | CTTCATATCCTGAGTCATGT | TACAGAATTCAGACATGACTCAGGAT | 11 | 15 |
| pegRNA3_PM_PAM2 | CTTCATATCCTGAGTCATGT | ATACAGAATTCAGACATGACTCAGGAT | 11 | 16 |
pegRNA, prime editing guide RNA; PBS, primer binding site; RRT, reverse transcriptase template.
We also identified a second NGG PAM in the antisense strand at position Phe675-Arg676. However, the nick-to-edit distance is 14 nt, which decreases the probability that the intended mutation will be integrated into the genome following the repair processes. The results of the insertion of the A673T mutation were therefore very low, and this PAM was not used for our experiments (Supplementary Fig. S1).
The plasmid (pCMV-PE2) coding for the SpCas9 nickase fused with a RT (SpCas9n-RT) was co-transfected with the plasmid (pU6-pegRNA-GG-acceptor) coding for one of the pegRNA in HEK293T cells. Exon 16 of the APP gene was amplified by PCR, and amplicons were sequenced by Illumina deep sequencing. The guanine-to-adenine modification frequency with the PE2 editing technique varied from only 0.5% to 6.4% for the different pegRNAs (Fig. 1C). The best results were obtained with pegRNA1, having a PBS of 11 nt and a RTT of 14 nt.
Negative control cells were transfected with a plasmid containing a reporter eGFP gene. No mutations were detected in the negative control. The results were statistically significant, except for pegRNA9, when comparing the different pegRNAs to the negative control using a one-way ANOVA test. Moreover, detailed analysis of the nucleotides around the A673T mutation and cleavage sites did not reveal any major unintended modifications of other nucleotides. The frequency of indels remained low, varying between 0% and 0.8%. The indel frequencies from one pegRNA to another compared to the negative control were not statistically significant (Fig. 1D).
Insertion of the A673T mutation using the PE3 technique
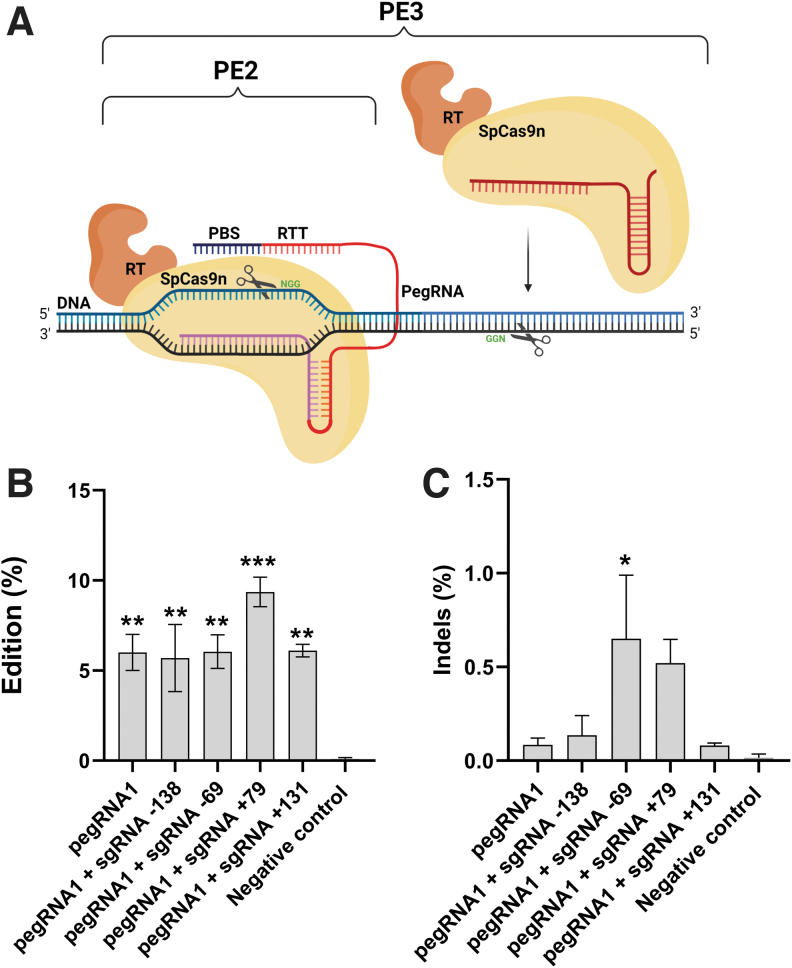
Since the editing rate was rather low with the PE2 technique, we decided to test the PE3 technique described by Anzalone et al. (Fig. 2A).25 For this technique, HEK293T cells were co-transfected with three plasmids coding for the SpCas9n-RT, pegRNA1, and one of four potential sgRNAs. This combination of plasmids induced an additional single-strand nick in the DNA strand that was not cut by the complex pegRNA1/SpCas9n-RT.
FIG. 2.
PE3 Prime editing method. (A) Scheme of the PE2 and the PE3 prime editing methods. The PE2 method uses (1) an SpCas9 H840A nickase (SpCas9n) to induce a nick in the DNA strand containing the NGG PAM fused with an engineered RT and (2) a pegRNA (i.e., a single-guide RNA [sgRNA] extended by a RTT and a PBS). In addition to the PE2 components, the PE3 method uses an sgRNA to induce a nick in the nonedited strand containing another PAM, which will induce repair processes and increase editing. (B) Tests of the pegRNA1 targeting the APP gene in combination with various sgRNAs. HEK293T cells were transfected with plasmids coding for the SpCas9n-RT, the pegRNA1, and four different sgRNAs, which induced a nick in the other DNA strand that was not previously nicked by pegRNA1. These additional nicks were induced at various base pair distances from the pegRNA1 nick. DNA was extracted 72 h later. APP exon 16 was PCR amplified, and amplicons were sequenced by Illumina deep sequencing. The negative control did not receive any treatment. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 3) compared to the negative control. **p < 0.0050; ***p = 0.0004. (C) Indel levels resulting from prime editing of the APP gene exon 16 using the PE3 method. There were 0.03% indels in the control sample. The level of indels varied from 0.06% to 0.89% for the PE3 method. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 3) compared to the negative control. The p-value was significant only for pegRNA1 + sgRNA-69. *p = 0.0225.
Whereas PE2 used an engineered RT to increase editing efficiency, PE3 nicks the nonedited strand to force its replacement and to boost editing efficiency. The positions of these additional nicks are limited by the presence of adequate SpCas9 PAM sequences. We tested four potential sgRNAs (sgRNA-138, sgRNA-69, sgRNA+79, and sgRNA+131 sequences in Table 3) that were able to induce a nick at −138, −69, +79, or +131 nt from the nick induced by the pegRNA1.
Table 3.
Sequences of sgRNAs Used in Experiments
| sgRNA | Spacer sequence |
|---|---|
| sgRNA −138 | CCTACCCAAAACTTCTTTCT |
| sgRNA −69 | ACATCATAATTAAAGTATGC |
| sgRNA +79 | AGACAAACAGTAGTGGAAAG |
| sgRNA +131 | CTTAATTTGATTTCTAGCAC |
| sgRNA_London +48 | AGGTGCAATCATTGGACTCA |
| sgRNA_PAM2 − 92 | GCCAAATGACCTATTAACTC |
| sgRNA_PAM2 + 72 | CTTTAATTATGATGTAATAC |
| sgRNA_PAM2 PE3b | ACAGAATTCCGACATGACTC |
| sgRNA_PAM2 PE3b_pm | ACAGAATTCAGACATGACTC |
sgRNA, single-guide RNA.
Another sgRNA hybridized with the APP exon 16 only following a modification of the target gene by the pegRNA (a method called PE3b) could not be tested due to the absence of NGG PAM near the site of the mutation in the nonedited strand.25 As in the previous experiments, exon 16 of the APP gene was amplified by PCR, and amplicons were sequenced by Illumina deep sequencing.
Among the four sgRNAs tested, only sgRNA+79 minimally increased the frequency of the desired A673T mutation to 9.9%. The results were statistically significant compared to the negative control (Fig. 2B). The frequency of indels remained low, varying between 0.06% and 0.89%. The indel frequencies from one pegRNA to another compared to the negative control were not statistically significant, except for pegRNA1 + sgRNA-69 (Fig. 2C).
Dual installation of the A673T mutation and a mutation in the PAM sequence
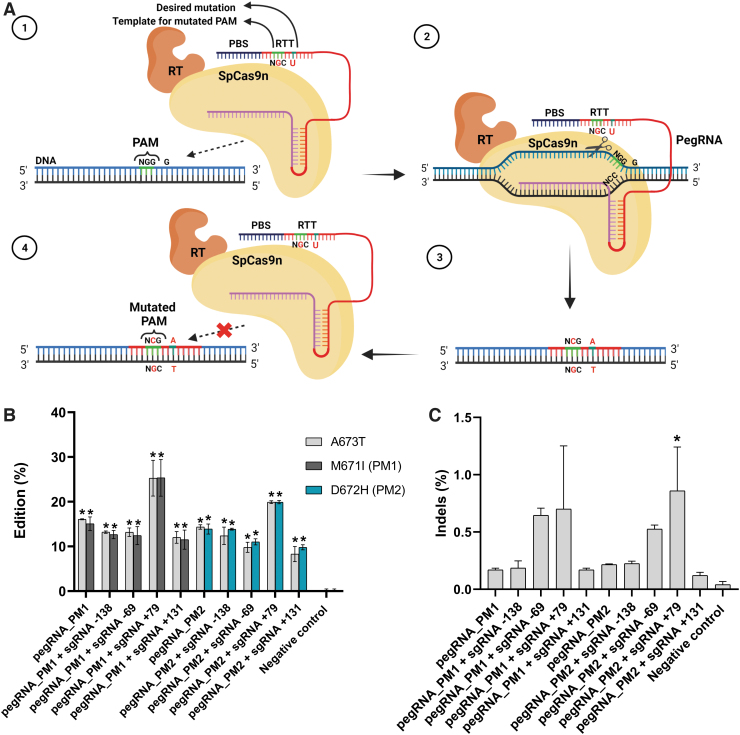
For this experiment, we designed two new pegRNAs called pegRNAPM1 and pegRNAPM2 (Table 2). Each was able to mutate one of the guanine nucleotides in the PAM while also introducing the desired A673T mutation (Fig. 3A). By inducing a mutation in the PAM of the SpCas9n, these guides could prevent the prime editor from targeting APP genes that already contained the A673T mutation.
FIG. 3.
Simultaneous mutation of the target codon and of the PAM by a pegRNA (A) Scheme illustrating the method for mutating the target codon and the PAM by a pegRNA. (1) The pegRNA contains the desired mutation in the RTT sequence to induce the A673T mutation in the upper strand of APP gene (G→A). The RTT sequence also includes an NGC sequence to mutate one of the guanines (G) of the PAM. (2) The pegRNA hybridizes with the lower DNA strand not containing the PAM, and a nick is induced in the upper DNA strand 3 nt upstream of the PAM. (3) Following DNA repair the upper DNA strand contain an adenine (A) for the A673T mutation and a mutated PAM (NCG). (4) The mutation of the PAM prevents a subsequent binging of the SpCas9n-RT to the DNA. Therefore, this method increases the editing rate by preventing a new nick that could remove the previously inserted mutation. (B) Simultaneous insertion of the A673T and the PAM mutation (PM) in the APP gene by PE2 and PE3. HEK293T cells were transfected with plasmids coding for the SpCas9n-RT and for a pegRNA (either pegRNAPM1 or pegRNAPM2), which were able to induce both the A673T mutation as well as mutate one of the guanines (G) of the PAM. In addition, some cells were also transfected with a sgRNA (sgRNA-138, sgRNA-69, sgRNA+79, or sgRNA+131). DNA was extracted 72 h later. APP exon 16 was PCR amplified, and amplicons were sequenced by Illumina deep sequencing. The figure illustrates the insertion percentage of the A673T mutation and the percentage of the PAM mutation (either PM1, i.e., mutation of the first G in the PAM inducing the M671I mutation, or PM2, i.e., mutation of the second G in the PAM inducing the D672H mutation). The negative control did not receive any treatment. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 3) compared to the negative control. *p < 0.0001. (C) Indel levels resulting from prime editing of the APP gene exon 16 using the PAM mutation. There were 0.02% indels in the control sample. The level of indels varied from 0.10% to 1.13% for the PAM mutation method. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 3) compared to the negative control. The p-value was significant only for pegRNAPM2 + sgRNA+79. *p = 0.0141.
Thus, in addition to the A673T mutation, pegRNAPM1 induced the M671I mutation, and pegRNAPM2 induced the D672H mutation. PegRNAPM1 and pegRNAPM2 were used with the basic PE2 technique and in the PE3 technique alongside sgRNA-138, sgRNA-69, sgRNA+79, and sgRNA+131. PegRNAPM1 induced the PAM mutation (PM) in 10.0–28.2% of the APP gene. However, 28.2% of the PAM mutation was observed when pegRNAPM1 was used in combination with sgRNA+79. PegRNAPM2 mutated the PAM slightly less frequently (in 9.4–20.2%), but the percentage of PAM mutation reached 20.2% when pegRNAPM2 was used in combination with sgRNA+79.
The A673T mutation was therefore also favored by reaching 28.1% and 20.1% using pegRNAPM1 and pegRNAPM2 respectively, and the results were statistically significant compared to the negative control (Fig. 3B). The frequency of indels remained low, varying between 0.10% and 1.13%. The indel frequencies from one pegRNA to another compared to the negative control were not statistically significant, except for pegRNAPM2 + sgRNA+79 (Fig. 3C).
Insertion of the A673T mutation by repeated prime editing treatments
Since the pegRNAs (pegRNAPM1 and pegRNAPM2), which induce the A673T mutation, also install a mutation in the PAM, repeated treatments of the cells should not lead to a loss of the already introduced A673T mutation. Again, this is because the SpCas9n-RT protein cannot bind to previously mutated APP genes containing a modified PAM sequence. As such, this favors the installation of the A673T mutation in cells where the PAM and the target APP 673 codon are not yet mutated.
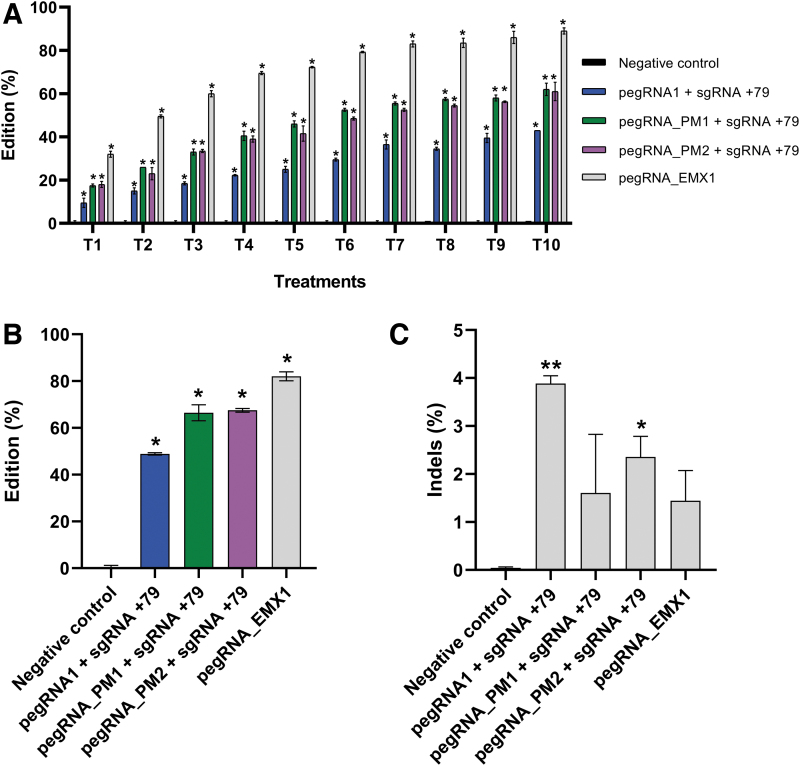
We thus attempted to treat the HEK293T cells repeatedly with the PE3 method over the course of 10 weeks using the sgRNA+79 with pegRNA1, pegRNAPM1, or pegRNAPM2. As an additional control, we used a pegRNAEMX1 (Table 2) targeting the EMX1 gene. The EMX1 G to C mutation at position Lys263 is ideal as a positive control gene because it can be reliably installed at a fairly high rate (∼25%), and this allowed us to confirm that the transfections were effective.
The treatments were repeated every 7 days and were performed up to 10 times (Fig. 4A). The editing of the EMX1 and of APP genes progressively increased with each treatment repetition. After the 10th treatment (T10), the successful EMX1 mutation rate reached 83.4%. The A673T mutation rate increased to 49.2% with the PE3 treatment alone and up to 68.9% when using pegRNAPM1 and 68.1% when using pegRNAPM2. The results were statistically significant compared to the negative control (Fig. 4B). The frequency of indels remained low, varying between 0.74% and 4%. The indel frequencies from one pegRNA to another compared to the negative control were not statistically significant, except for pegRNA1 + sgRNA+79 and pegRNAPM2 + sgRNA+79 (Fig. 4C).
FIG. 4.
Repeated PE2 treatments of the EMX1 gene and PE3 treatments of the APP gene. (A) HEK293T cells were transfected with plasmids coding for the SpCas9n-RT and a pegRNA (pegRNAEMX1, pegRNA1, pegRNAPM1 or pegRNAPM2). Cells were also transfected with a plasmid coding for the sgRNA+79. These treatments were repeated 10 times at weekly intervals. The DNA was extracted from cell samples 72 and 144 h after each treatment. The remaining cells were replated for expansion and for a subsequent treatment. For each DNA sample, the APP exon 16 and the EMX1 exon 3 were PCR amplified and analyzed by Sanger sequencing. The figure illustrates in gray the edition percentages of the desired mutation in the EMX1 gene (as a positive control) and, in blue, green, and purple, the edition percentages for the A673T mutations. The negative control in black (barely visible because close to zero) did not receive any treatment. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. *p < 0.0001. (B) For T10, the APP exon 16 and the EMX1 exon 3 were PCR amplified, and amplicons were also sequenced by Illumina deep sequencing. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. *p < 0.0001. (C) Indel levels resulting from the repeated PE2 treatments of EMX1 gene and repeated PE3 treatments of the APP gene exon 16. There were 0.03% indels in the control sample. The level of indels varied from 0.74% to 4%. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 2) compared to the negative control. The p-value was significant for pegRNA1 + sgRNA+79 and pegRNAPM2 + sgRNA+79. *p = 0.0459; **p = 0.0064.
Insertion of the London V717I mutation by PE2, PE3, and mutated PAM
Prime editing now makes it possible to correct almost any point mutation if a PAM is located near it. Hence, FAD mutations could also theoretically be targeted by prime editing. We did not have cells containing one FAD mutation. We therefore decided to insert the London mutation into a normal APP gene to demonstrate that a precise point mutation can be inserted and could eventually be reversed by changing one nucleotide in the pegRNA.
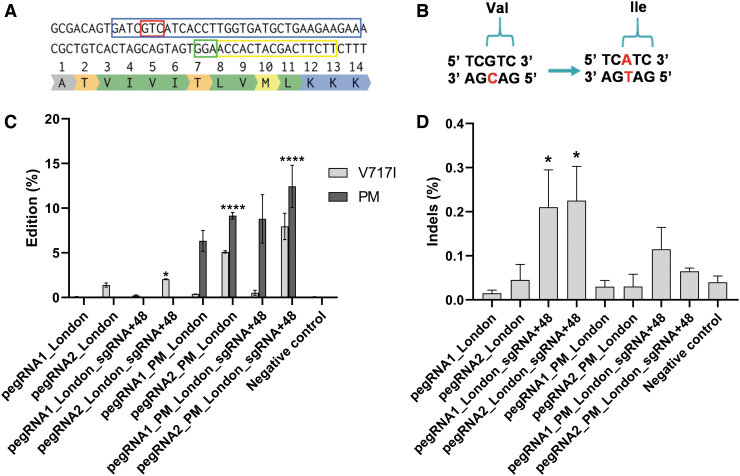
We selected a PAM at position Thr719 of the APP gene exon 17 at 6 nt from the target nucleotide cytosine (C) in the antisense strand to modify the valine codon (5′GTC3′) into an isoleucine (5′ATC3′) in the sense strand (Fig. 5A and B). We designed two different pegRNAs (pegRNA1-London, pegRNA2-London) with PBS and RTT sections of different lengths (Table 2), and we tested their efficacy with the PE2 method.
FIG. 5.
Simultaneous insertion of the V717I and the PAM mutations in the APP gene by PE2, PE3, and PAM mutation methods. (A) Section of APP exon 17 with codon of interest (Val717) in the red box. In the blue box, the unmodified sequence used to design the PBS and the RTT. The PAM is in a green box, and the pegRNA spacer sequence is in a yellow box. (B) The change in the antisense strand of the C (cytosine in red) into a T (thymine in red) modifies the valine (5′GTC3′) codon in the sense strand into an isoleucine (5′ATC3′). (C) Four different pegRNAs with various PBS, RTT inducing or not a PAM mutation (PM) were transfected in HEK293T cells with Lipofectamine™ 2000. In addition, some cells were also transfected with a sgRNA (sgRNA-London +48). DNA was extracted 72 h later, exon 17 of the APP gene was PCR amplified, and amplicons were sequenced by Illumina deep sequencing. The negative control did not receive any treatment. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 3) compared to the negative control. For the V717I mutation, *p = 0.0201 and ****p < 0.0001. For the PM, p < 0.0001 for all samples. (D) Indel levels resulting from prime editing of the APP gene exon 17 using PE2, PE3, and PAM mutation methods. There were 0.03% indels in the control sample. The level of indels varied from 0.01% to 0.28%. The results were analyzed with a one-way ANOVA test and Dunnett's multiple comparisons test (n = 3) compared to the negative control. The p-value was significant only for pegRNA1-London + sgRNA-London+48 and pegRNA2-London + sgRNA-London+48. *p < 0.0234.
We also tested the PE3 method using the sgRNA (sgRNA-London+48; Table 3) with the different pegRNAs. In addition to the V717I mutation, the pegRNA1PM-London and the pegRNA2PM-London induced a silent mutation in the PAM (ACC to ACG) where the amino acid stayed the same (Thr719). The frequency of the desired guanine to adenine modification of the APP gene was 0.1% and 1.6% for pegRNA1-London and pegRNA2-London, respectively, with PE2.
The combination of PAM mutation and PE3 method increased the mutation rate up to 9.0% for V717I and up to 14.1% for the PAM mutation induced by pegRNA2PM-London, having a PBS of 16 nt and a RTT of 17 nt. For the V717I mutation, the results were statistically significant for pegRNA2-London + sgRNA-London+48, pegRNA2PM-London, and pegRNA2PM-London + sgRNA-London+48 compared to the negative control (Fig. 5C). The frequency of indels remained low, varying between 0.01% and 0.28%. The indel frequencies from one pegRNA to another compared to the negative control were not statistically significant, except for pegRNA1-London + sgRNA-London+48 and pegRNA2-London + sgRNA-London+48 (Fig. 5D).
Discussion
This study focused on the use of the prime editing technique to introduce the protective Icelandic mutation (A673T) in cultured cells. Our results clearly indicate that it is possible to introduce this mutation in HEK293T cells in culture with interesting editing rates. In a previous article, we used the cytidine base editing technique to introduce the A673T mutation, and Target-AID_SpCas9VQR was the most efficient base editor.24
However, while it did preferentially deaminate the targeted cytosine, it also weakly deaminated the other four nearby cytosines. Thus, the base editing technique also induced the mutation of the glutamine codon in position 674 into a lysine (E674K). Previously, with the base editing, the A673T mutation was created when the cytosine (C2) at position 673 in the antisense strand had been deaminated. The highest C2 deamination obtained through Target-AID_SpCas9VQR demonstrated a C2 deamination of 53.3% when including changes in surrounding cytosines and 42.6% when excluding all but the E674K mutation. Only 3.8% of the DNA contained the desired C2 mutation without altering other surrounding cytosines.
However, prime editing allowed us to insert the A673T mutation precisely with a maximum rate of 49.2% using the PE3 method. Unintended mutations of nearby nucleotides were not observed in the present study with the prime editing technique. Prime editing also has the advantage of having less stringent PAM positioning requirements due to the variable length of the RTT compared to base editing.29 We have to keep in mind that the off-target effects of Cas9 and base editors may differ; cytosine base editors generate more indels, off-target editing, and unwanted changes than adenine base editors.30
Expanding PAM flexibility through the development of genetically engineered Cas9 proteins and the discovery of novel Cas9 nucleases would widen the spectrum of genome targeting while preserving editing efficiency and specificity for prime editing and base editing. More studies are needed to understand better and improve these editing techniques in a variety of cell types and organisms, as well as in vivo characterization for therapeutic applications.29 Overall, because multiple cytosines are present near the A673T mutation site and bystander edits are undesirable, the prime editing technique offers more advantages for the potential treatment of AD requiring precise edits than the cytidine base editing technique.
Our results also clearly demonstrated that repeated treatment increased the frequency of the intended mutation. The first explanation for this increased mutation frequency is that the repeated treatments permitted the simultaneous transfection of the three plasmids in HEK293T cells that were not previously simultaneously transfected. We have therefore shown that repeated prime editing can increase the desired mutation when more than one delivery vector is used.
This method of repeated treatment could also be used to create a pool of highly modified cells and thus more easily screened cells to find a clone with the mutation installed. It is also possible to consider repeating the treatment with prime editing to increase the editing rate of a specific mutation for future gene therapy with a vector that does not induce an immunogenic response such as extracellular vesicles.31
The optimization of the PBS and RTT sequences, as well as the addition of the PE3 method with the mutation of the PAM, maximized the editing percentages. However, the mutation of the PAM cannot be used in all cases because it can cause a change of an amino acid. It is therefore preferable to use the PAM mutation technique only when it does not induce an amino acid change from the mutation of the nucleotide, as was the case in the experiments that we did to insert the London mutation.
At the present time, there are no data suggesting that the additional PAM mutation (M671I or D672H) can alter the protective effects of the A673T mutation. These mutations are not linked to a familial form of AD. Furthermore, the latest experiments with base editing have shown that the addition of a mutation (E674K) near A673T seemed to have a positive synergistic effect with A673T.24 In the future, if we want to use the PAM mutation for the development of gene therapy, it will be necessary to verify whether these additional mutations affect peptide aggregation or have other effects in vivo.
The conformation of the amyloid peptide has changed following the insertion of the A673T mutation. Therefore, it remains necessary to ensure that the resulting product will not be recognized as nonself and induce an immune response with future studies. An immune response against the mutated protein could ultimately negate the therapeutic effect.32
Moreover, the London mutation (V717I) is one of the most common forms of FAD worldwide. We have already had interesting results showing that the insertion of the A673T mutation by base editing with the London mutation reduces the amount of toxic peptides Aβ1-40 and Aβ1-42.22–24 Because the cells used in the current experiments are wild type, we have shown that it is possible to induce the mutation responsible for the V717I form of AD. In future experiments, we will use cell lines including the London FAD, and the opposite mutation, I717V correction, will be made with prime editing.
Although the indels of APP gene amplicons were analyzed in this study, it remains necessary to assess off-target prime editing in a genome-wide manner before proceeding to possible clinical trials. These measurements will thus confirm that our approach with prime editing does not induce undesirable effects on other genes. Interestingly, a recent study looking at genome-wide specificity with prime editing came to the conclusion that prime editing provides a highly specific method of precise genome editing with a very low number of off-target events.33
The main problem for developing a treatment for AD using this prime editing approach will be to deliver the components of the prime editing technology to neurons efficiently in the brains of patients. The SpCas9n-RT gene is ∼6.3 kb, and this is too large to be delivered by a single adeno-associated virus (AAV), which contains at most ∼5 kb. One possibility is to use a dual AAV delivery system, each encoding one of the fragments of the protein to be delivered flanked by short split-inteins, resulting in protein trans-splicing and full-length protein reconstitutions.34
The AAV-PHP.eB has the great advantage of crossing the blood–brain barrier (BBB) and thus of delivering genes to neurons in some mouse species.35 Other AAV serotypes, such as the AAV-F, have been proven effective for delivery to the brain in nonhuman primates, and may one day be used in humans.36 Another potential delivery vector is extracellular vesicles.31 They have been shown to cross the BBB and can be loaded with genes or proteins.37–39
The AAV is the vector most frequently used in the field of gene transfer therapy in vivo. Indeed, the gene delivered by this vector is not integrated into the patient genome and has low immunogenicity.40 There are now a few treatments approved for commercialization and currently available with AAVs. A Phase I study of bilateral intracerebral delivery of AAV2-NGF to the basal forebrain of patients with mild to moderate AD-associated dementia has already been performed. This study proved that surgical delivery was safe and well tolerated.41
Human data on the protective effect of the A673T mutation are derived mostly from heterozygous carriers.14,15 In our study, the insertion of the A673T mutation was analyzed by amplicon sequencing. So, it is impossible to determine the homozygous or heterozygous distribution of the A673T mutation within the modified cells.
Consequently, the percentage of insertions of the A673T mutation in the samples of cells could vary, considering those having undergone a homozygous insertion. Additional experiments on single cells would clarify this issue. Nevertheless, it would be unlikely for the homozygous character to influence the protective effect of the A673T mutation negatively. Jonsson et al. have already identified three homozygous carriers of A673T in Icelandic samples, and none of these homozygous carriers had a history of dementia.14
The next part of our project is to develop a gene editing treatment for AD that will aim to deliver the prime editing agents using a dual AAV to neurons of AD patients (obtained by direct conversion of fibroblasts) in vitro.42 Subsequently, we will attempt to insert the A673T mutation in mouse models of AD.43
Although our results are still only preliminary, we hope to develop a treatment that will slow down the formation of amyloid plaques in AD patients. Prime editing may also be used to treat many other hereditary diseases due to point mutations. However, some specific optimizations will be required to adjust for each different target in the genome.
Conclusion
We have used the prime editing technique to insert the APP A673T mutation directly by modifying the guanine in the alanine codon (5′GCA3′) into an adenine, resulting in the formation of a threonine codon (5′ACA3′) in HEK293T cells. Following repeated treatments, we obtained a A673T mutation rate of up to 49.2% with the PE3 technique and up to 68.9% with a simultaneous PAM mutation. These results clearly show that it is possible to introduce the Icelandic protective mutation with the prime editing technique in cells in vitro, and they raise the possibility that with an effective delivery system, this mutation may be inserted directly in patient neurons in vivo to prevent hereditary AD and eventually sporadic AD.
Supplementary Material
Acknowledgments
Figures 2A and 3A were created with BioRender.com. We thank Gabriel Lamothe for reviewing this article.
Author Disclosure Statement
J.R. and J.P.T. have a U.S. patent and a pending U.S. patent application on the potential treatment of Alzheimer's disease by the insertion of the Icelandic mutation.
Funding Information
G.T. has a studentship from the Canadian Institutes of Health Research (CIHR) and from Laval University. This work was supported by a grant from the Weston Brain Institute (grant TR150002).
Supplementary Material
References
- 1. Querfurth HW, LaFerla FM. Alzheimer's disease. N Engl J Med 2010;362:329–344. DOI: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 2. Yang P, Sun F. Aducanumab: the first targeted Alzheimer's therapy. Drug Discov Ther 2021;15:166–168. DOI: 10.5582/ddt.2021.01061. [DOI] [PubMed] [Google Scholar]
- 3. Sosa LJ, Cáceres A, Dupraz S, et al. The physiological role of the amyloid precursor protein as an adhesion molecule in the developing nervous system. J Neurochem 2017;143:11–29. DOI: 10.1111/jnc.14122. [DOI] [PubMed] [Google Scholar]
- 4. Sheng M, Sabatini BL, Südhof TC. Synapses and Alzheimer's disease. Cold Spring Harb Perspect Biol 2012;4:a005777. DOI: 10.1101/cshperspect.a005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002;297:353–356. DOI: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 6. Gan KJ, Silverman MA. Dendritic and axonal mechanisms of Ca2+ elevation impair BDNF transport in Aβ oligomer-treated hippocampal neurons. Mol Biol Cell 2015;26:1058–1071. DOI: 10.1091/mbc.E14-12-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bilousova T, Miller CA, Poon WW, et al. Synaptic amyloid-β oligomers precede p-tau and differentiate high pathology control cases. Am J Pathol 2016;186:185–198. DOI: 10.1016/j.ajpath.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klementieva O, Willén K, Martinsson I, et al. Pre-plaque conformational changes in Alzheimer's disease-linked Aβ and APP. Nat Commun 2017;8:14726. DOI: 10.1038/ncomms14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forner S, Baglietto-Vargas D, Martini AC, et al. Synaptic impairment in Alzheimer's disease: a dysregulated symphony. Trends Neurosci 2017;40:347–357. DOI: 10.1016/j.tins.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 10. Bertram L, Tanzi RE. The genetics of Alzheimer's disease. Prog Mol Biol Transl Sci 2012;107:79–100. DOI: 10.1016/B978-0-12-385883-2.00008-4. [DOI] [PubMed] [Google Scholar]
- 11. Hatami A, Monjazeb S, Milton S, et al. Familial Alzheimer's disease mutations within the amyloid precursor protein alter the aggregation and conformation of the amyloid-β peptide. J Biol Chem 2017;292:3172–3185. DOI: 10.1074/jbc.M116.755264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Di Fede G, Catania M, Morbin M, et al. A recessive mutation in the APP gene with dominant-negative effect on amyloidogenesis. Science 2009;323:1473–1477. DOI: 10.1126/science.1168979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lanoiselée HM, Nicolas G, Wallon D, et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: a genetic screening study of familial and sporadic cases. PLoS Med 2017;14:e1002270. DOI: 10.1371/journal.pmed.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jonsson T, Atwal JK, Steinberg S, et al. A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature 2012;488:96–99. DOI: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 15. Kero M, Paetau A, Polvikoski T, et al. Amyloid precursor protein (APP) A673T mutation in the elderly Finnish population. Neurobiol Aging 2013;34:1518.e1–3. DOI: 10.1016/j.neurobiolaging.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 16. Mengel-From J, Jeune B, Pentti T, et al. The APP A673T frequency differs between Nordic countries. Neurobiol Aging 2015;36:2909.e1–4. DOI: 10.1016/j.neurobiolaging.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xia Q, Yang X, Shi J, et al. The protective A673T mutation of amyloid precursor protein (APP) in Alzheimer's disease. Mol Neurobiol 2021;58:4038–4050. DOI: 10.1007/s12035-021-02385-y. [DOI] [PubMed] [Google Scholar]
- 18. Benilova I, Gallardo R, Ungureanu AA, et al. The Alzheimer disease protective mutation A2T modulates kinetic and thermodynamic properties of amyloid-β (Aβ) aggregation. J Biol Chem 2014;289:30977–30989. DOI: 10.1074/jbc.M114.599027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martiskainen H, Herukka SK, Stančáková A, et al. Decreased plasma β-amyloid in the Alzheimer's disease APP A673T variant carriers. Ann Neurol 2017;82:128–132. DOI: 10.1002/ana.24969. [DOI] [PubMed] [Google Scholar]
- 20. Kimura A, Hata S, Suzuki T. Alternative selection of β-site APP-cleaving enzyme 1 (BACE1) cleavage sites in amyloid β-protein precursor (APP) harboring protective and pathogenic mutations within the Aβ sequence. J Biol Chem 2016;291:24041–24053. DOI: 10.1074/jbc.M116.744722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guyon A, Rousseau J, Bertin T, et al. Protective A673T mutation decreases amyloid-beta peptide production for a majority of APP genes containing a mutation responsible for a Familial Alzheimer Disease. bioRxiv 2020. DOI: 10.1101/2020.07.22.215616. [DOI] [Google Scholar]
- 22. Guyon A, Rousseau J, Lamothe G, et al. The protective mutation A673T in amyloid precursor protein gene decreases Aβ peptides production for 14 forms of familial Alzheimer's disease in SH-SY5Y cells. PLoS One 2020;15:e0237122. DOI: 10.1371/journal.pone.0237122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komor A, Kim Y, Packer M, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420–424. DOI: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guyon A, Rousseau J, Bégin FG, et al. Base editing strategy for insertion of the A673T mutation in the APP gene to prevent the development of AD in vitro. Mol Ther Nucleic Acids 2021;24:253–263. DOI: 10.1016/j.omtn.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019;576:149–157. DOI: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scholefield J, Harrison PT. Prime editing—an update on the field. Gene Ther 2021;28:396–401. DOI: 10.1038/s41434-021-00263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kluesner MG, Nedveck DA, Lahr WS, et al. EditR: a method to quantify base editing from Sanger sequencing. CRISPR J 2018;1:239–250. DOI: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Clement K, Rees H, Canver MC, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 2019;37:224–226. DOI: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA base-editing and prime-editing. Int J Mol Sci 2020;21:6240. DOI: 10.3390/ijms21176240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zuo E, Sun Y, Wei W, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019;364:289–292. DOI: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duan L, Xu L, Xu X, et al. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale 2021;13:1387–1397. DOI: 10.1039/D0NR07622H. [DOI] [PubMed] [Google Scholar]
- 32. Pardridge WM. Blood–brain barrier and delivery of protein and gene therapeutics to brain. Front Aging Neurosci 2020;11:373. DOI: 10.3389/fnagi.2019.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim DY, Moon SB, Ko JH, et al. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res 2020;48:10576–10589. DOI: 10.1093/nar/gkaa764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tornabene P, Trapani I, Minopoli R, et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci Transl Med 2019;11:eaav4523. DOI: 10.1126/scitranslmed.aav4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chan KY, Jang MJ, Yoo BB, et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 2017;20:1172–1179. DOI: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beharry A, Gong Y, Kim JC, et al. The AAV9 variant capsid AAV-F mediates widespread transgene expression in nonhuman primate spinal cord after intrathecal administration. Hum Gene Ther 2021. Aug 26 [Epub ahead of print]; DOI: 10.1089/hum.2021.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin T, Gu J, Li Z, et al. Recent advances on extracellular vesicles in central nervous system diseases. Clin Interv Aging 2021;16:257–274. DOI: 10.2147/CIA.S288415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA loading into extracellular vesicles via electroporation is size-dependent and enables limited gene delivery. Mol Pharm 2015;12:3650–3657. DOI: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Busatto S, Iannotta D, Walker SA, et al. A simple and quick method for loading proteins in extracellular vesicles. Pharmaceuticals (Basel) 2021;14:356. DOI: 10.3390/ph14040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, et al. Current clinical applications of in vivo gene therapy with AAVs. Mol Ther 2021;29:464–488. DOI: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rafii MS, Baumann TL, Bakay RA, et al. A phase1 study of stereotactic gene delivery of AAV2-NGF for Alzheimer's disease. Alzheimers Dement 2014;10:571–581. DOI: 10.1016/j.jalz.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 42. Drouin-Ouellet J, Pircs K, Barker RA, et al. Direct neuronal reprogramming for disease modeling studies using patient-derived neurons: what have we learned? Front Neurosci 2017;11:530. DOI: 10.3389/fnins.2017.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saito T, Matsuba Y, Mihira N, et al. Single APP knock-in mouse models of Alzheimer's disease. Nat Neurosci 2014;17:661–663. DOI: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.