Abstract
Background
Burkitt’s lymphoma is an aggressive B-cell lymphoma characterized by typical morphological, immunophenotypic and molecular features. Gene expression profiling provided a molecular signature of Burkitt’s lymphoma, but also demonstrated that a subset of aggressive B-cell lymphomas not fulfilling the current World Health Organization criteria for the diagnosis of Burkitt’s lymphoma nonetheless show a molecular signature of Burkitt’s lymphoma (‘discrepant Burkitt’s lymphoma’). Given the different treatment of Burkitt’s lymphoma and diffuse large B-cell lymphomas we investigated molecular differences within gene expression-defined Burkitt’s lymphoma.
Design and Methods
We studied tumors from 51 Burkitt’s lymphoma patients, comprising 26 with classic Burkitt’s lymphoma, 17 with atypical Burkitt’s lymphoma and 8 with ‘discrepant Burkitt’s lymphoma’, by comparative genomic hybridization and gene expression profiling.
Results
Classic and atypical Burkitt’s lymphoma (excluding ‘discrepant Burkitt’s lymphoma’), in adult and pediatric cases do not differ in underlying genomic imbalances or gene expression suggesting that these subgroups are molecularly homogeneous. ‘Discrepant Burkitt’s lymphoma’, however, differ dramatically in the absolute number of alterations from classic/atypical Burkitt’s lymphoma and from diffuse large B-cell lymphoma. Moreover, this category includes lymphomas that carry both the t(14;18) and t(8;14) translocations and are clinically characterized by presentation in adult patients and an aggressive course.
Conclusions
Pediatric and adult Burkitt’s lymphoma are molecularly homogeneous, whereas ‘discrepant Burkitt’s lymphoma’ differ in underlying genetic and clinical features from typical/atypical Burkitt’s lymphoma. ‘Discrepant Burkitt’s lymphoma’ may therefore form a distinct genetic subgroup of aggressive B-cell lymphomas, which show poor response to multi-agent chemotherapy.
Keywords: Burkitt’s lymphoma, comparative genomic hybridization, DLBCL, gene expression
Introduction
Burkitt’s lymphoma (BL) is an aggressive B-cell lymphoma characterized by specific morphological and immunophenotypic features, a high proliferation rate (Ki-67 index exceeding 90%) and, on the molecular level, by a chromosomal translocation involving the MYC oncogene.1 However, the characteristic immunophenotype of BL and MYC translocations can also be detected in a subset of diffuse large B-cell lymphomas (DLBCL)2 making the distinction between these two entities sometimes problematic. This is of importance clinically, since the therapeutic approaches for BL and DLBCL differ substantially.3 A further complication is introduced by the diagnostic category of atypical BL, which is used for aggressive B-cell lymphomas carrying the immunophenotype of classic BL and a MYC translocation, but in which the morphological and/or immunophenotypic features are slightly atypical.1 Two major gene expression profiling studies recently sharpened the molecular distinction between BL and DLBCL.4,5 Whereas virtually all lymphomas diagnosed as classic or atypical BL according to current World Health Organization (WHO) criteria1 showed gene expression profiles characteristic of BL, a subset of DLBCL had the typical gene expression signature of BL suggesting that these cases correspond to BL at the molecular level.4 Moreover, some of the cases with a clear-cut gene expression profile of BL showed expression of BCL-2 and a Ki-67 index below 90% by immunohistochemistry thus extending the molecular category of BL beyond currently accepted criteria.4
In recent years, global genetic analyses including conventional karyotyping, comparative genomic hybridization (CGH) and array-based CGH have described secondary genomic alterations in BL.6–13 One of the larger studies using CGH described gains of 12q, 22q, Xq and losses of 13q as the most frequent alterations in BL.11 Moreover, abnormalities in 1q and 7q were associated with inferior outcome.11 A recent combined cytogenetic study including CGH and spectral karyotyping (SKY) identified frequent gains of chromosome 7 and losses of 17p in a panel of ten BL cell lines.12 Using classical cytogenetic analysis in 39 Burkitt-like lymphomas, however, gains of chromosome 12 and losses of chromosomes 13 and 15 appeared to be the most frequent secondary genetic alterations in addition to the MYC translocation.13
This study was designed to investigate secondary genomic alterations in gene expression-defined BL4 using conventional CGH. In particular, we addressed the questions of whether: (i) morphological subgroups of molecularly defined BL (classic BL, atypical BL, discrepant BL) differ in underlying genetic alterations, (ii) pediatric and adult BL show differences in their genetic and gene expression profiles, and (iii) genetic alterations in BL lead to consequences in locus-specific or global gene expression profiles.
Design and Methods
Characteristics of the patients
We studied 51 patients with a gene expression signature of BL.4 Information on these patients’ Epstein-Barr virus status was not available. According to currently established WHO criteria, these 51 patients comprised 26 with classic BL, 17 with atypical BL, 7 with diffuse large B-cell lymphoma (DLBCL) and one patient with a high grade B-cell lymphoma, not otherwise specified. Importantly, all cases were reviewed by a panel of eight expert hematopathologists, with consensus reached in all cases. The pathology review was blinded to age information. The eight cases with a gene expression signature of BL, but a histomorphological diagnosis of DLBCL or high-grade B-cell lymphoma, not otherwise specified, are referred to as discrepant BL. Clinical data were obtained according to a protocol approved by the National Cancer Institute Institutional Review Board (Table 1). CGH results in BL were compared with published data from 161 cases of untreated DLBCL (84 with germinal center B-cell-like (GCB) DLBCL and 77 with activated B-cell-like (ABC) DLBCL).14
Table 1.
Classification of Burkitt’s lymphoma and clinical data.
| Classic BL | Atypical BL | Discrepant BL | |
|---|---|---|---|
|
| |||
| Patients | |||
| Number | 29 | 21 | 8 |
| Median age, years (range) | 18 (3–67) | 31 (3–73) | 61 (21–91)a |
| Male/female | 19/4 | 11/2 | 4/4 |
| Ann Arbor stage III/IVb | 10/14 | 7/8 | 4/4 |
| Extranodal sites ≥ 2b | 4/15 | 5/7 | 4/4 |
| Intensive chemotherapy regimensb | 12/15 | 5/7 | 2/5 |
| Lactate dehydrogenase level > normalb | 9/14 | 4/7 | 3/4 |
| Median overall survival | Not reached | Not reached | 0.58 years |
p<0.01.
Number of patients/number of patients for whom clinical information was available.
Comparative genomic hybridization
CGH was performed using a commercially available CGH kit provided by Vysis (Downers Grove, IL, USA). Hybridizations and digital image acquisition, processing, and evaluation were performed as previously described.15 Signals were visualized with a Zeiss Axiophot fluorescence microscope and analyzed with the ISIS digital image analysis system (MetaSystems, Altlussheim, Germany). Signal ratios greater than 1.25 or less than 0.75 were considered as chromosomal gains or losses, respectively. Ratios exceeding 1.5 and/or strong focal signals with the ratio profile showing over-representation were considered as high-level DNA amplifications. Complete CGH data are available at http://www.ncbi.nlm.nih.gov/sky.
Statistical analysis
CGH alterations in individual cytobands were treated as categorical variables and their associations with various BL and DLBCL subgroups or gene-expression signatures were analyzed as detailed below. Preliminary analyses did not reveal significant differences in the effect of gains and amplifications; thus, these were treated as equivalent chromosomal abnormalities. Since a large number of individual chromosomal abnormalities (290 bands) were analyzed, there was a possibility that some of the observed differences would turn out to be significant purely by chance. To avoid such false positives, we used a stepwise permutation test,16 which corrected for multiple hypothesis testing, while accounting for the very strong correlation between the consecutive bands. All p-values presented have been so adjusted. Differences in the frequency of observed genomic imbalances between the subgroups were detected using a χ2 test. Comparisons of the overall numbers of alterations, gains, losses and amplifications between the different subgroups of BL, and differences between adult and pediatric BL cases were analyzed using a t test.
To determine the relationship between chromosomal imbalances and the expression levels (as a continuous variable) of the genes located in the corresponding altered chromosomal regions (gain/amplification versus normal copy number and loss versus normal copy number) the non-parametric Mann-Whitney test was performed. To further reduce the effects of multiple comparisons, we analyzed only those chromosomal abnormalities that were detected with a frequency of greater than 20% (39 bands) in one or more of the BL subgroups.
The correlation between chromosomal alterations and the previously described gene expression signatures in BL4 was established as follows. The signatures “MYC and target genes”, “NF-κB target genes”, “MHC class I genes” and the germinal center B-cell gene signatures “BL-high”, “BL-low” and “BL=GCB4” were each averaged. For each band which included four or more losses, a t-test was computed between those BL samples with losses, and those BL samples with a wild type copy number. These results were then adjusted for multiple comparisons using the stepwise permutation test. An identical analysis was performed for BL cases showing gains/amplifications versus those harboring a wild type copy number.
Results
Chromosomal imbalances in Burkitt’s lymphoma
The clinical characteristics of the patients with gene expression-defined BL are summarized in Table 1. The median age of the patients was 29 years (range, 3–91 years). Interestingly, patients with morphologically discrepant BL were significantly older (median age 61; range, 27–91) than patients with classic BL (median age 18; range, 3–67) or atypical BL (median age 31; range, 3–73) (p<0.01). The male/female ratio was 37:6 in patients with classic and atypical BL, but 4:4 in discrepant BL. Twenty-seven patients received multi-agent chemotherapy, including 20 patients who received intensive chemotherapy regimens, and no information on the treatment was available for 24 patients. Among the 51 BL cases studied, 36 (71%) showed genetic alterations by CGH. The total number of alterations was 122 (68 gains, 13 amplifications and 41 losses) (Figure 1A). Overall, the most frequent gains were 1q, 7/7q, 8q24-qter, 13q31-q32, and the most frequent loss occurred in 17p. Among the 26 cases of classic BL, 17 (65%) showed aberrant profiles by CGH analysis, whereas no chromosomal imbalances were detected in 9 cases. Among aberrant cases, 25 gains, 4 amplifications and 10 losses of chromosomal material were observed, the most frequent chromosomal alterations being gains of different regions of 1q and chromosome 7 (Figure 1B, Table 2). Twelve of 17 atypical BL (59%) showed aberrant CGH profiles demonstrating 13 gains, 3 amplifications and 10 losses. Most frequently, we observed losses of 17p12-pter and amplifications of 13q31-q32 (Figure 1C). Finally, all of the discrepant BL showed CGH alterations with a total of 30 gains, 6 amplifications and 21 losses. This subset showed complex karyotypes with a median of 6.9 alterations per case (Figure 1D) and the presence of the t(14;18) translocation was detected in three out of five cases investigated (besides an underlying translocation involving MYC that was present in all cases). Correlations between CGH alterations and clinical parameters (overall survival) were not performed due to the small size of the respective subgroups and the fact that not all patients received intensified treatment regimens.
Figure 1.
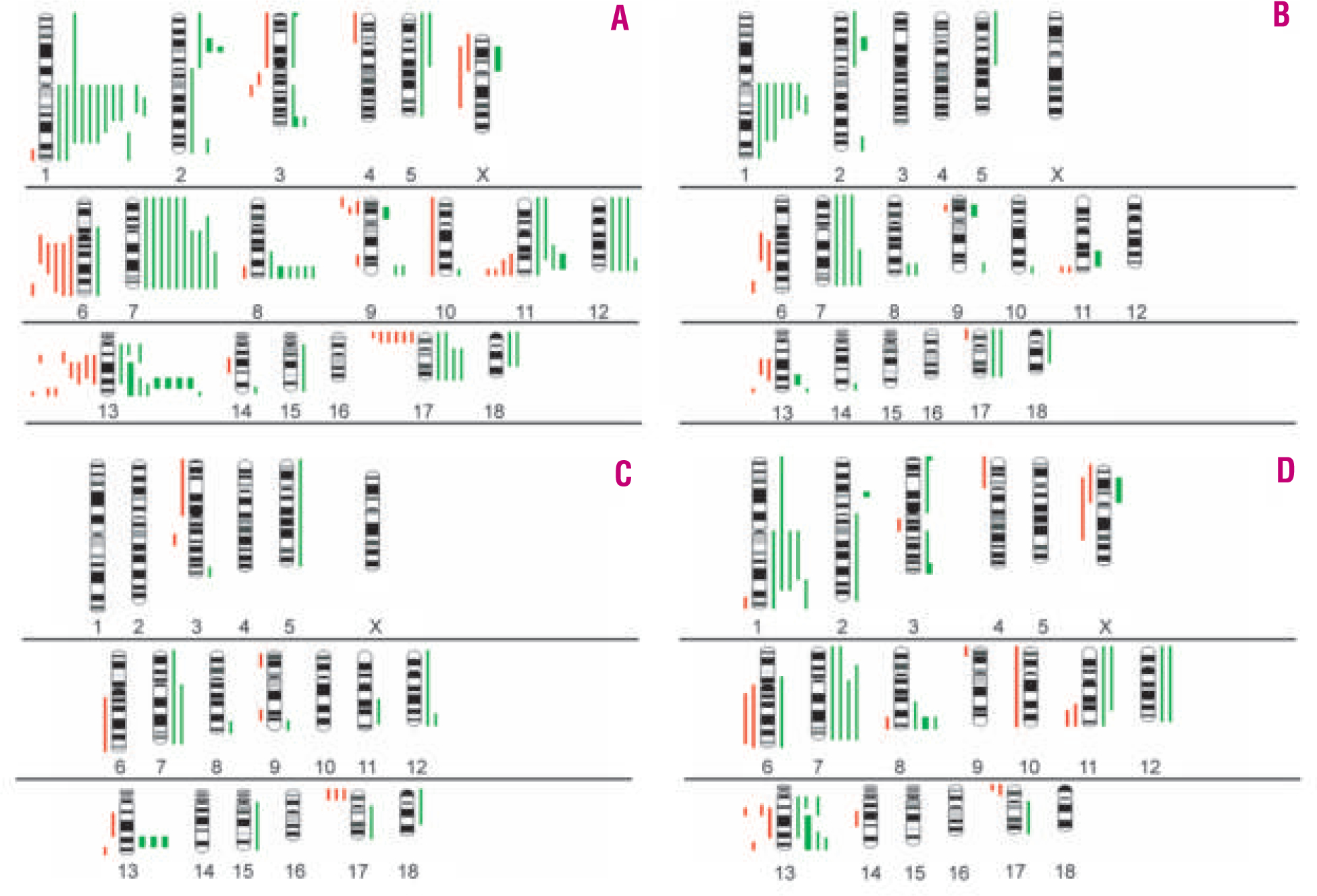
Chromosomal imbalances detected by comparative genomic hybridization in molecularly defined Burkitt’s lymphoma (BL). Ideograms of gains and losses of genetic material in (A) the total series of BL, (B) classic BL, (C) atypical BL and (D) ‘discrepant BL’. Red bars on the left side of each chromosome correspond to genetic losses. Green bars on the right side indicate genetic gains. Bold green bars highlight areas of amplifications.
Table 2.
Results of comparative genomic hybridization of cases of Burkitt’s lymphoma.
| Genetic alterations | Chromosomal region | Classic BL n =26 (%) | Atypical BL n=17 (%) | Discrepant BL n =8 (%) |
|---|---|---|---|---|
|
| ||||
| Gains | 1cen-q25 | 7(27) | – | 4(50) |
| 1q31-q32* | 4(15) | – | 4(50) | |
| 7/7q | 4(15) | 2(12) | 4(50) | |
| 8q24-qter | 2(8) | 1(6) | 3(27) | |
| 13q11-q13* | – | – | 3(27) | |
| 13q31-q32 | 1(4) | 3(18) | 3(27) | |
| 13q33* | 1(4) | – | 3(27) | |
| Losses | 13q14* | – | 1(6) | 3(27) |
| 17p12-pter | 1(4) | 3(18) | 2(25) | |
Alterations associated with ‘discrepant BL’ (p ≤0.05).
Comparison of genomic imbalances between Burkitt’s lymphoma subgroups
No differences in genomic imbalances were observed between classic and atypical BL. Although chromosomal gains in 1q appeared to be more common in classic BL, this was not statistically significant when accounting for multiple testing. Discrepant BL did however, have more gains (3.6±2.6 vs. 0.9±1.4; p=0.02), losses (2.5±2.3 vs. 0.5±0.8, p=0.039), and a higher total number of alterations (6.9±4.4 vs. 1.5±1.8; p=0.01) compared to both classic and atypical BL. Additionally, discrepant BL samples showed more frequent gains in 1q31-q32 (p=0.05), 13q11-q13 (p=0.02) and 13q33 (p=0.02) as well as losses of 13q14 (p=0.03). Cases classified as discrepant BL in general showed an aggressive clinical course (median survival of 0.58 years) (Table 1).
Comparison of genomic imbalances between Burkitt’s lymphoma and DLBCL
A comparison of genomic imbalances between BL and 161 previously studied DLBCL14 showed that losses of 11q24-q25 were present in BL but uncommon in DLBCL (p=0.03) whereas there was a trend towards more gains/amplifications of 18q21-q23 in DLBCL compared to in BL (p=0.1) Comparing BL with the molecular subgroups of DLBCL, the frequently observed gains of chromosome 3 in ABC DLBCL were infrequent in BL (p=0.035), as were gains in 18q21-q23 (p=0.01) and losses of 6q16-q27 (p=0.02). No statistically significant differences were found regarding the genetic constitution of BL and the GCB DLBCL subgroup.
Comparison of genomic imbalances between adult and pediatric Burkitt’s lymphoma cases
To elucidate whether certain chromosomal alterations were associated with pediatric (age ≤ 18 years) or adult BL, we compared CGH alterations between these different age groups (27 adult and 24 pediatric cases). Nineteen of the 27 adult patients (70%) and 18 of the 24 pediatric cases (75%) displayed detectable alterations. Overall, adult BL patients had a significantly higher complexity of abnormalities than pediatric cases (3.1 abnormalities per tumor in adults versus 1.5 in the pediatric group). Further analysis showed that this was entirely accounted for by the group of eight discrepant BL. Excluding these cases from the adult group, the complexity (both 1.5 abnormalities per tumor) and also the pattern of alterations was similar in adult and pediatric BL (Figure 2A and 2B). Based on these results, we tested whether differences in overall gene expression exist between pediatric and adult BL. Searching the gene expression data generated previously on an Affymetrix custom oligonucleotide microarray with 2524 unique genes,4 no statistically significant differences could be detected and the attempt to establish a gene expression-based predictor of pediatric and adult BL failed (data not shown).
Figure 2.
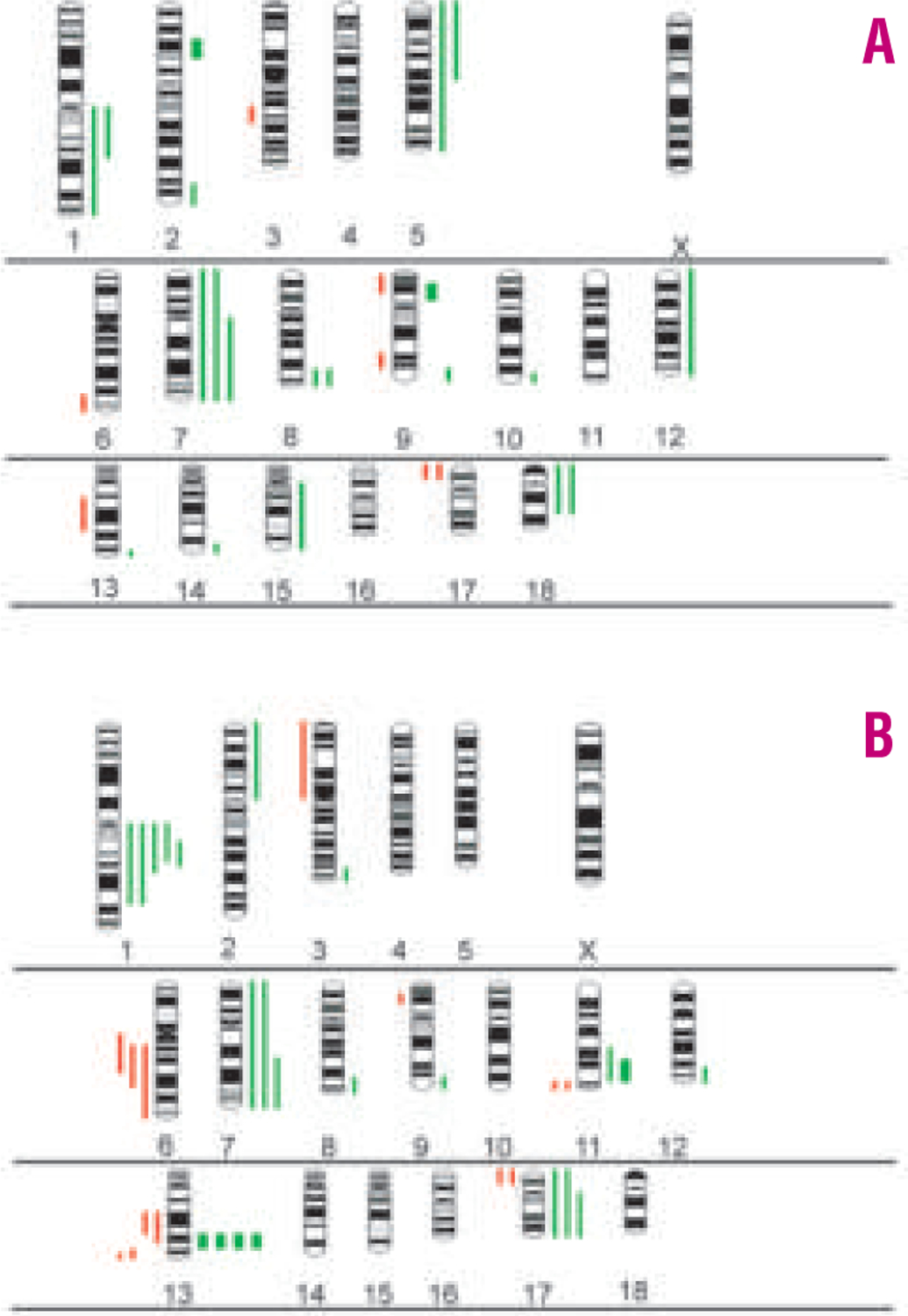
Ideograms of the distribution of gains and losses of genetic material in adult (A) and pediatric (B) Burkitt’s lymphoma.
Impact of chromosomal imbalances on locus-specific gene expression in Burkitt’s lymphoma
The six chromosomal regions that were most frequently altered were studied to determine the impact of specific chromosomal gains or losses on locus-specific gene expression levels. These were the regions 1cen-q22, 1q31-q32, 7q22-qter, 8q24-qter, 13q31-q32, and 17p13-pter. As demonstrated in previous analyses,14,17 chromosomal imbalances were associated with elevated or reduced expression of a subset of genes within the involved chromosomal regions. In the chromosomal region 1cen-q22, for example, 10 out of 23 genes (43%) represented on the custom microarray in the respective region, showed increased expression levels in cases with gains as compared to cases with two copies. In particular, in this region the levels of expression of NOTCH, BCL9, CKIP-1, MCL1, PSMB4, ILF2, JTB, HAX1, ATP8B2, and CKS1B were found to be elevated (Figure 3A). Significantly fewer genes (20%) were upregulated in the chromosomal region 1q31-q32 (Figure 3B). The results for the remaining regions are presented in Figures 3C–F. In BL cases with a chromosomal loss in 17p13-pter, approximately half of the genes present on the microarray had reduced expression, demonstrating a major impact of this genetic event on locus-specific gene expression (Figure 3F). No significantly overexpressed genes were observed in areas of a genomic loss and, vice versa, no genes were significantly downregulated in any of the regions with chromosomal gains.
Figure 3.
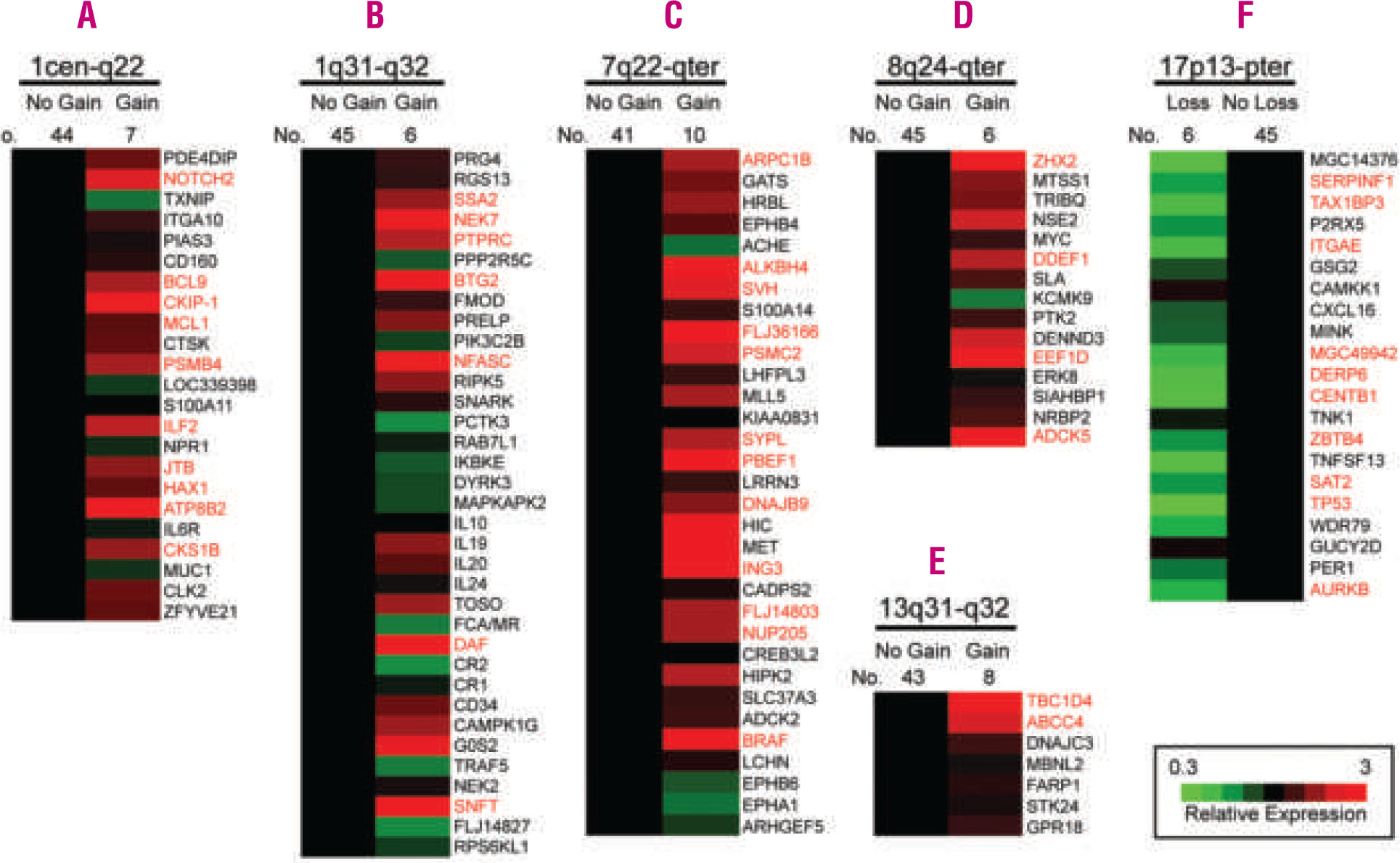
Influence of gains and losses on locus-specific gene expression levels in the chromosomal regions 1cen-q22 (A), 1q31-q32 (B), 7q22-qter (C), 8q24-qter (D), 13q31-q32 (E) and 17p13-pter (F) Changes in gene expression levels are depicted for each gene (averaged in each cohort) with regard to the locus-specific genetic status. Genes are ordered according to their chromosomal position shown on the right. The comparison between chromosomal imbalances and the expression levels of the genes located in the corresponding chromosomal region (gain vs. normal copy number or loss vs. normal copy number) was performed using the non-parametric Mann-Whitney test. Genes with significant differences (p<0.05) are highlighted in red.
Impact of chromosomal imbalances on biologically relevant gene expression signatures in Burkitt’s lymphoma
In a previous study,4 we had defined gene expression signatures that distinguish BL from DLBCL. In particular, the signatures “NF-κB target genes” and “MHC class I genes” were expressed at very low levels in BL, whereas, by the very nature of the pathogenesis of BL, the “MYC and target genes” signature is increased. Germinal center B cell-associated genes showed a heterogeneous picture including genes expressed more highly in BL (“BL-high” signature), genes with reduced expression levels (“BL-low” signature) and genes with equal expression between BL and DLBCL (BL=GCB signature).4 Here, we sought to determine whether certain genetic alterations in BL were associated with increased or decreased expression of any of these signatures, since the altered chromosomal regions may harbor master regulators of these pathways. Indeed, gains of the 17q11-q25 region were associated with increased expression of the NF-κB signature (p=0.046), whereas gains of different regions of chromosome 1 (1cen-q12 and 1q21-q32) were associated with the specific “BL-high” signature (p=0.035) and the more general germinal center associated “BL=GCB” signature (p=0.04).
Discussion
Although a number of previous studies described genetic alterations in BL,6–13 two recent major gene expression profiling studies4,5 provided a molecular signature of BL that - at the same time - sharpened, but also broadened the diagnostic category of BL. Specifically, gene expression-defined BL appear to also include some aggressive B-cell lymphomas that do not fulfill the current strict WHO criteria1 (e.g. a subgroup of DLBCL and BL cases with BCL2 expression or a Ki-67 index below 90%). In view of these findings, the establishment of the global profile of genomic imbalances in molecularly defined BL deserves renewed attention.
In a global view, the CGH-defined genetic alterations in all BL cases in our series are well in accordance with the published literature. With regards to the morphological subgroups of gene expression-defined BL, however, the finding that classic and atypical BL did not differ in underlying genomic imbalances in concert with the presence of an identical gene expression profile4,5 provides further evidence that classic and atypical BL are, except for minor differences in morphological features, highly similar on the molecular level. These results may, therefore, support the view that classic and atypical BL comprise one molecular entity and provide a molecular rationale for the current practice of including patients with both classic and atypical BL in the same prospective trials. Despite showing a gene expression signature of BL, some of the discrepant BL (i.e. B-cell lymphomas that currently do not fulfill the WHO criteria of BL) may be genetically distinct from classic and atypical BL. First, this subgroup contains aggressive B-cell non-Hodgkin’s lymphomas carrying both the translocations t(14;18) and t(8;14)(dual translocation or double-hit lymphomas) as did three of the discrepant BL in our study. These lymphomas had been described in earlier reports and are characterized by an extremely aggressive clinical course that is fatal in almost all cases.13,18 It is unlikely that in these dual translocation cases the potentially secondary rearrangement of MYC alone accounts for the BL gene expression signature, since the gene expression predictor is clearly able to discriminate BL with MYC rearrangement from DLBCL with MYC rearrangement. From a different perspective, a number of DLBCL carrying a MYC rearrangement are not identified as having a BL gene expression signature in the published studies.4,5
In addition, our results demonstrate complex genetic alterations in discrepant BL with significantly higher numbers of chromosomal gains, and losses and total number of alterations than classic and atypical BL. Interestingly, discrepant BL - although corresponding morphologically to DLBCL in most cases – do not appear to carry some of the alterations typically observed in DLBCL.14 Instead, discrepant BL frequently carry aberrations not seen in either BL or DLBCL (gains of 1q31-q32, 13q11-q13, 13q33 and losses of 13q14) suggesting that some discrepant BL may represent a new genetic entity distinct from classic and atypical BL as well as from DLBCL. This view is further supported by the findings that patients with discrepant BL were significantly older and that this disease was equally distributed between males and females, in contrast to classic and atypical BL that predominantly affect males. Finally, the clinical course of discrepant BL appears to be dramatically different and usually rapidly fatal, although this finding must be viewed with caution, since some patients in our series did not receive intensified treatment regimens. At first glance, the dismal clinical course may appear contradictory to the findings in the study by Hummel et al.5 in which cases with a molecular BL signature had a favorable prognosis, regardless of their morphological appearance of classic/atypical BL or DLBCL. However, a straightforward comparison between the discrepant BL in our series and the potentially corresponding 11 DLBCL cases with a molecular BL profile described by Hummel et al. is not easy at present, since detailed information on the genetic alterations and clinical course for each case awaits publication. Nevertheless, there appears to be a general agreement between the two studies. First, the core and molecular BL categories in the study by Hummel et al. also contained lymphomas with increased genetic complexity and the frequency of these cases does not differ between the two studies. Second, the molecular BL cases5 also included a dual translocation case. A comparison of clinical findings is, however, compromised by the fact, that molecular BL cases in the study by Hummel et al. were enriched for younger patients, which may explain the more favorable outcome.
Another major aim of this study was to compare gene expression and genetic features between pediatric and adult cases of BL. Currently, pediatric BL patients are treated in prospective clinical trials separately from adult BL patients. However, the adaptation of therapeutic protocols initially designed for pediatric patients to adults has led to improved survival times for adult BL patients in recent years.19,20 Gene expression data4 and, in addition, genetic data obtained in the current report failed to provide evidence that molecular differences exist between pediatric and adult BL. Our CGH analysis revealed genetic imbalances in 75% of pediatric and 70% of adult BL patients and the spectrum of genetic alterations did not differ between the two groups. Moreover, no gene expression-based predictor could be constructed that was able to distinguish between patients with molecular BL in different age groups. It should be noted that the resolution of conventional CGH for detecting genomic alterations is limited and that high resolution techniques, such as array CGH, will reveal smaller alterations and more complex findings, e.g. a small biallelic loss in a chromosomal region that shows a gain on a larger scale. However, our finding that pediatric and adult BL patients show no significant differences in underlying genetic alterations is supported by unpublished data from the German Network Project Molecular Mechanisms in Malignant Lymphomas (W. Klapper, personal communication). Taken together, these data provide the rationale that future targeted therapeutic approaches in BL may be aimed at both pediatric and adult age groups and may be modified according to clinical factors that can limit therapy (e.g. dose intensity).
Finally, we demonstrated that chromosomal imbalances in frequently affected regions lead to locus-specific as well as global gene expression changes. For example, in the chromosomal region 1q, upregulation of NOTCH2, MCL1 and BCL9 may be pathogenetically relevant by altering apoptotic properties of the malignant B cells. On a global level, gains in 1q correlated with an increased expression of germinal center-associated genes that are characteristically expressed at high levels in BL (“BL-high” signature) suggesting that this BL-specific phenotype may be orchestrated by important regulators in this genetic region. Likewise, genetic losses in the chromosomal region 17p13-pter lead to major gene expression changes affecting almost half of the genes located in this region. Besides alterations of the tumor suppressor TP53, other genes, such as Aurora kinase B (AURKB) may influence the biological behavior as a consequence of deregulated expression.
In summary, we show that classic and atypical BL carrying a molecular profile of BL, as defined by gene expression profiling, do not differ in gene expression or underlying chromosomal alterations, whereas a subset of discrepant BL, despite its BL-characteristic gene expression profile, may form a genetic entity distinct from classic and atypical BL as well as DLBCL. Furthermore, we failed to identify gene expression or genetic differences between molecularly defined cases of pediatric and adult BL supporting the notion that BL in both age groups should be viewed as one molecular entity.
Acknowledgments:
we are indebted to the following investigators, who contributed specimens from patients or clinical data to this study: Dr. J.H.J.M. van Krieken, Department of Pathology, and Dr. P. Hoogerbrugge, Department of Pediatric Oncology, Radboud University Medical Center, Nijmegen; Dr. W. Kamps, Department of Pediatric Oncology, University Medical Center, Groningen; and Dr. K. Lam, Department of Pathology, and Dr. I.M. Appel, Department of Pediatrics, Sophia Children’s Hospital, Erasmus University Medical Center, Rotterdam, the Netherlands.
Funding:
supported by grants from the Spanish Comisión Interministerial de Ciencia y Tecnología (CICYT) SAF05/5855, Instituto de Salud Carlos III, Red Temática de Investigacion Cooperativa de Cáncer, by the Interdisciplinary Center for Clinical Research (IZKF) of the University of Würzburg, Germany (AR, EH), and by an NIH grant (UO1-CA84967) from the National Cancer Institute, Bethesda, MD, USA, (WCC, AR, EC, GO, HKMH).
The authors wish to thank Theodora Nedeva for superb technical assistance.
Footnotes
The authors reported no potential conflicts of interest.
References
- 1.Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press, 2001. [Google Scholar]
- 2.Lossos IS. Molecular pathogenesis of diffuse large B-cell lymphoma. J Clin Oncol 2005;23:6351–7. [DOI] [PubMed] [Google Scholar]
- 3.Bishop PC, Rao VK, Wilson WH. Burkitt’s lymphoma: molecular pathogenesis and treatment. Cancer Invest 2000;18:574–83. [DOI] [PubMed] [Google Scholar]
- 4.Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, et al. Molecular diagnosis of Burkitt’s lymphoma. N Engl J Med 2006;354:2431–42. [DOI] [PubMed] [Google Scholar]
- 5.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med 2006;354:2419–30. [DOI] [PubMed] [Google Scholar]
- 6.Lones MA, Sanger WG, Le Beau MM, Heerema NA, Sposto R, Perkins SL, et al. Chromosome abnormalities may correlate with prognosis in Burkitt/Burkitt-like lymphomas of children and adolescents: a report from Children’s Cancer Group Study CCG-E08. J Pediatr Hematol Oncol 2004; 26: 169–78. [DOI] [PubMed] [Google Scholar]
- 7.Onciu M, Schlette E, Zhou Y, Raimondi SC, Giles FJ, Kantarjian HM, et al. Secondary chromosomal abnormalities predict outcome in pediatric and adult high-stage Burkitt lymphoma. Cancer 2006; 107:1084–92. [DOI] [PubMed] [Google Scholar]
- 8.Pienkowska-Grela B, Witkowska A, Grygalewicz B, Rymkiewicz G, Rygier J, Woroniecka R, et al. Frequent aberrations of chromosome 8 in aggressive B-cell non-Hodgkin lymphoma. Cancer Genet Cytogenet 2005;156:114–21. [DOI] [PubMed] [Google Scholar]
- 9.Barth TF, Muller S, Pawlita M, Siebert R, Rother JU, Mechter-sheimer G, et al. Homogeneous immunophenotype and paucity of secondary genomic aberrations are distinctive features of endemic but not of sporadic Burkitt’s lymphoma and diffuse large B-cell lymphoma with MYC rearrangement. J Pathol 2004; 203:940–5. [DOI] [PubMed] [Google Scholar]
- 10.Wessendorf S, Schwaenen C, Kohlhammer H, Kienle D, Wrobel G, Barth TF, et al. Hidden gene amplifications in aggressive B-cell non-Hodgkin lymphomas detected by microarray-based comparative genomic hybridization. Oncogene 2003;22:1425–9. [DOI] [PubMed] [Google Scholar]
- 11.Garcia JL, Hernandez JM, Gutierrez NC, Flores T, Gonzalez D, Calasanz MJ, et al. Abnormalities on 1q and 7q are associated with poor outcome in sporadic Burkitt’s lymphoma. A cytogenetic and comparative genomic hybridization study. Leukemia 2003;17:2016–24. [DOI] [PubMed] [Google Scholar]
- 12.Karpova MB, Schoumans J, Blennow E, Ernberg I, Henter JI, Smirnov AF, et al. Combined spectral karyotyping, comparative genomic hybridization, and in vitro apoptyping of a panel of Burkitt’s lymphoma-derived B cell lines reveals an unexpected complexity of chromosomal aberrations and a recurrence of specific abnormalities in chemoresistant cell lines. Int J Oncol 2006;28:605–17. [PubMed] [Google Scholar]
- 13.Macpherson N, Lesack D, Klasa R, Horsman D, Connors JM, Barnett M, et al. Small noncleaved, non-Burkitt’s (Burkitt-like) lymphoma: cytogenetics predict outcome and reflect clinical presentation. J Clin Oncol 1999;17:1558–67. [DOI] [PubMed] [Google Scholar]
- 14.Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood 2005; 106:3183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zettl A, Ott G, Makulik A, Katzenberger T, Starostik P, Eichler T, et al. Chromosomal gains at 9q characterize enteropathy-type T-cell lymphoma. Am J Pathol 2002;161:1635–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Design and Analysis of DNA Microarray Investigations. New York: Springer-Verlag, 2003. [Google Scholar]
- 17.Salaverria I, Zettl A, Bea S, Moreno V, Valls J, Hartmann E, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J Clin Oncol 2007;25:1216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanungo A, Medeiros LJ, Abruzzo LV, Lin P. Lymphoid neoplasms associated with concurrent t(14;18) and 8q24/c-MYC translocation generally have a poor prognosis. Mod Pathol 2006;19:25–33. [DOI] [PubMed] [Google Scholar]
- 19.Smeland S, Blystad AK, Kvaloy SO, Ikonomou IM, Delabie J, Kvalheim G, et al. Treatment of Burkitt’s/Burkitt-like lymphoma in adolescents and adults: a 20-year experience from the Norwegian Radium Hospital with the use of three successive regimens. Ann Oncol 2004;15:1072–8. [DOI] [PubMed] [Google Scholar]
- 20.Divine M, Casassus P, Koscielny S, Bosq J, Sebban C, Le Maignan C, et al. Burkitt lymphoma in adults: a prospective study of 72 patients treated with an adapted pediatric LMB protocol. Ann Oncol 2005;16: 1928–35. [DOI] [PubMed] [Google Scholar]


