Abstract
A new natural TEM-2 derivative, named TEM-72, was identified in a Proteus mirabilis strain and in a Morganella morganii strain isolated in Italy in 1999. Compared to TEM-1, TEM-72 contains the following amino acid substitutions: Q39K, M182T, G238S, and E240K. Kinetic analysis showed that TEM-72 exhibits an extended-spectrum activity, including activity against oxyimino-cephalosporins and aztreonam. Expression of blaTEM-72 in Escherichia coli was capable of decreasing the host susceptibility to the above drugs.
Extended-spectrum β-lactamases (ESBLs) are a group of enzymes that confer resistance to oxyimino cephalosporins and monobactams (7, 8, 11, 18). Most ESBLs found in clinical isolates of the Enterobacteriaceae are plasmid-borne variants of the original TEM-1 and SHV-1 enzymes in which one or more amino acid substitutions expand the substrate specificity (2–4, 21). Currently, almost 100 of these variants have been described (K. Bush and G. Jacoby, http://www.lahey.org/studies/webt.htm).
In this work we report on the characterization of a new natural TEM derivative with ESBL activity, named TEM-72, identified during a survey that was recently undertaken to evaluate the prevalence of ESBL-producing isolates of Enterobacteriaceae in Italian hospitals.
Proteus mirabilis (isolate FI-14) and Morganella morganii (isolate FI-13) were simultaneously isolated from a decubitus ulcer of an inpatient at the Careggi Hospital of Florence, Italy, in 1999. Escherichia coli DH5α (Life Technologies, Milan, Italy) was used as a recipient for the ESBL-carrying plasmids. Media were from Difco Laboratories (Detroit, Mich.). Piperacillin and tazobactam were from Wyeth-Lederle (Catania, Italy), aztreonam and cefepime were from Bristol-Myers Squibb (Wallingford, Conn.), cefotaxime and cefpirome were from Hoechst Marion Roussel (Romainville, France), ceftazidime was from Glaxo-Wellcome (Verona, Italy), nitrocefin was from Unipath (Milan, Italy), clavulanic acid was from SmithKline Beecham (Brentford, United Kingdom), and other antibiotics were from Sigma Chemical Co. (St. Louis, Mo.). MICs were determined by a broth macrodilution procedure as recommended by the National Committee for Clinical Laboratory Standards (13). Colony and Southern blot hybridizations were performed as described (16) using random-primed 32P-labeled DNA probes comprised of PCR-generated amplicons containing the blaTEM-1 (19) or the blaSHV-1 (1) genes. Plasmids were purified by the alkaline lysis method (16). Electroporation was performed using a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) according to the manufacturer's instructions. PCR amplification of blaTEM alleles was carried out as described previously (15) using the AmpliTaq Gold DNA polymerase (Perkin-Elmer, Milan, Italy). Sequencing was performed on both strands using a dRhodamine Terminator Cycle Sequencing Ready reaction kit, an ABI PRISM 377 DNA Sequencer (Perkin-Elmer), and custom sequencing primers. Sequencing was performed in triplicate on PCR products derived from three independent reactions. TEM-72 was purified from E. coli DH5α(pPM-14) grown overnight at 37°C, aerobically, in 6 liters of brain heart infusion broth containing 30 μg of ceftazidime per ml. Cells were harvested, and a sonic extract (sonicated five times for 30 s each time, at 60 W) was prepared in 80 ml of 50 mM Tris-HCl buffer (pH 8.0) (TB) and cleared by high-speed centrifugation (105,000 × g for 30 min at 4°C). The cleared extract was loaded onto a Q-Sepharose FF column (2 by 20 cm; Amersham Pharmacia Biotech, Milan, Italy) equilibrated with TB, and the β-lactamase was eluted with a linear gradient of NaCl (0.2 to 0.8 M) in TB. The fractions containing cefotaxime-hydrolyzing activity were pooled, dialyzed against TB, and loaded onto a Superdex G75 FF column (1 by 60 cm; Amersham Pharmacia Biotech) equilibrated with TB. The active fractions were dialyzed against 25 mM bis(2-hydroxethyl)iminotris(hydroxymethyl)methane (Bis–Tris buffer (pH 7.1), and loaded onto a Mono P HR 5/20 column (Amersham Pharmacia Biotech) equilibrated with the same buffer. The proteins were eluted with 25 ml of 10-fold-diluted Polybuffer 74 in the pH range of 7.1 to 4. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by the method of Laemmli (12). Analytical isoelectric focusing and zymogram detection of β-lactamase bands using the chromogenic substrate nitrocefin were performed as described previously (14). Kinetic parameters were determined by monitoring the hydrolysis of each substrate at 30°C in 50 mM sodium phosphate buffer (pH 6.7) with a Lambda 2 spectrophotometer (Perkin-Elmer). The total reaction volume was 1 ml, and the enzyme concentration in the reaction was in the range 190 to 900 nM. Kinetic parameters were determined by analyzing either the complete hydrolysis time courses (5) or under initial-rate conditions (17). Km values lower than 25 μM were calculated as Ki as described previously (5), using 80 μM nitrocefin as the reporter substrate. Inhibition by clavulanic acid and tazobactam was monitored using 80 μM cefazolin as the reporter substrate.
Identification of a new natural TEM-derived ESBL in M. morganii FI-13 and in P. mirabilis FI-14.
M. morganii FI-13 and P. mirabilis FI-14 were resistant to amoxicillin, piperacillin, cefotaxime, ceftazidime, and aztreonam but susceptible to piperacillin-tazobactam and carbapenems. FI-14 was also susceptible to amoxicillin-clavulanate and cefoxitin, while FI-13 was resistant to these compounds (Table 1). In both isolates ESBL production was suspected on the basis of the susceptibility pattern and a positive double-disk synergy test (9) with clavulanate and ceftazidime (data not shown).
TABLE 1.
MICs of various β-lactams for strains used in this study
| Antibiotic(s) | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| M. morganii FI-13 | P. mirabilis FI-14 | E. coli DH5α(pMN-13) | E. coli DH5α(pPM-14) | E. coli DH5α | |
| Amoxicillin | >64 | >64 | >64 | >64 | 4 |
| Amoxicillin-clavulanatea | >64 | 8 | 8 | 8 | 4 |
| Piperacillin | >256 | >256 | 256 | 256 | 1 |
| Piperacillin-tazobactamb | 1 | 0.25 | 0.5 | 1 | 1 |
| Cefoxitin | >64 | 8 | 8 | 16 | 8 |
| Cefotaxime | >128 | 64 | 16 | 16 | ≤0.12 |
| Ceftazidime | >64 | 64 | >64 | >64 | ≤0.12 |
| Cefepime | 8 | 64 | 2 | 4 | ≤0.12 |
| Cefpirome | >128 | >128 | 16 | 8 | ≤0.12 |
| Aztreonam | >64 | 32 | 64 | 32 | 0.12 |
| Imipenem | 2 | 4 | 0.12 | 0.12 | 0.12 |
| Meropenem | 0.03 | 0.125 | ≤0.03 | ≤0.03 | ≤0.03 |
Amoxicillin-clavulanate at a 2:1 ratio.
Tazobactam at a fixed concentration of 4 μg/ml.
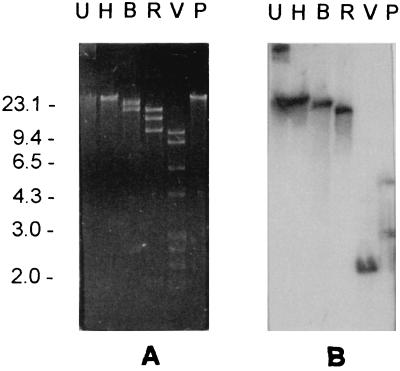
A colony-blot hybridization revealed the presence of blaTEM- but not of blaSHV-related sequences in either strain (data not shown). A plasmid of approximately 40 kb was detected in either strain. These plasmids, named pMN-13 and pPM-14, respectively, exhibited apparently identical restriction profiles with HindIII, BglII, EcoRI, EcoRV, and PstI (Fig. 1 and data not shown). A Southern blot hybridization revealed that the blaTEM-related sequences were plasmid borne. With both plasmids the probe hybridized to single restriction fragments after digestion with HindIII, BglII, EcoRI, and EcoRV and with two restriction fragments after digestion with PstI (Fig. 1 and data not shown).
FIG. 1.
(A) Agarose gel electrophoresis of the plasmid pPM14, purified from P. mirabilis FI-14. Lanes: U, uncut; H, B, R, V, P, after digestion with HindIII, BglII, EcoRI, EcoRV, and PstI, respectively. (B) Results of Southern blot analysis of the gel shown in panel A using the blaTEM probe. DNA size standards (in kilobase pairs) are shown on the left. Identical results were obtained with plasmid pMN-13 purified from M. morganii FI-13 and with plasmids purified from the respective E. coli transformants.
Ceftazidime resistance could be transferred to E. coli DH5α by electroporation with the plasmid preparations and selection on medium containing 50 μg of the antibiotic per ml. The E. coli transformants exhibited a positive double-disk synergy test with clavulanate and ceftazidime and showed a plasmid profile identical to that of FI-13 and FI-14. Analytical isoelectric focusing of the crude extracts of DH5α(pMN-13) and of DH5α(pPM-14) revealed, in either case, the presence of a single band of β-lactamase activity of pI 5.9 (data not shown).
The sequences of the blaTEM alleles carried by pMN-13 and pPM-14 were determined and turned out to be identical. Compared to TEM-1 (19), the encoded enzyme showed a unique array of amino acid changes (Q39K, M182T, G238S, and E240K). The enzyme was named TEM-72.
Purification and characterization of TEM-72.
TEM-72 was purified from E. coli DH5α(pPM-14) by three chromatographic steps (Table 2), yielding a preparation that was >95% pure as evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The isoelectric pH of the purified enzyme was 5.9 (data not shown).
TABLE 2.
Summary of the purification steps of TEM-72 β-lactamase from E. coli DH5α(pPM-14)
| Purification step | Total protein (mg) | Sp act (U/mg protein)a | Total activity (U) | Purification factor | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 980 | 1.4 | 1,372 | 1 | 100 |
| Q-Sepharose FF | 120 | 8 | 960 | 5.7 | 70 |
| Superdex G75 FF | 36 | 21 | 756 | 15 | 55 |
| Fast chromatofocusing | 3.2 | 136 | 435 | 97 | 32 |
Measured against cefotaxime 100 μM; 1 U is defined as the activity that hydrolyzed 1 μmol of the substrate/min.
Measurement of the kinetic parameters revealed that, in addition to penicillins and older cephalosporins, TEM-72 also hydrolyzes oxyimino-cephalosporins and aztreonam, but not cefoxitin (Table 3). The highest catalytic efficiencies (kcat/Km, where kcat is the catalytic rate constant) were observed with penicillins, narrow-spectrum cephalosporins, and cefotaxime. Lower values were observed with other oxyimino-cephalosporins. Aztreonam was slowly hydrolyzed, but the very high affinity of TEM-72 for this compound resulted in a kcat/Km ratio that was in the range of those observed with oxyimino-cephalosporins. Tazobactam behaved as a competitive inhibitor (Ki, 3.1 nM). With clavulanate a progressive inactivation was observed: at a molar ratio of 100:1 (clavulanate/enzyme) no reactivation of the enzyme was detected after prolonged (18-h) incubation.
TABLE 3.
Kinetic parameters determined with the TEM-72 β-lactamase purified from E. coli DH5α(pPM-14)
| Substrate | Km (μM)c | kcat (s−1)c | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Benzylpenicillin | 1.6 ± 0.1 | 13 ± 0.8 | 8.1 × 106 |
| Piperacillin | 14 ± 1.2 | 67 ± 2.0 | 4.8 × 106 |
| Nitrocefin | 4.0 ± 0.2 | 14 ± 0.6 | 3.5 × 106 |
| Cephalothin | 5.0 ± 0.1 | 5.0 ± 0.2 | 1.0 × 106 |
| Cephaloridine | 3.7 ± 0.2 | 13 ± 0.6 | 3.5 × 106 |
| Cefazolin | 24 ± 0.9 | 45 ± 1.8 | 1.9 × 106 |
| Cefotaxime | 22 ± 1.2 | 37 ± 1.5 | 1.7 × 106 |
| Ceftazidime | 75 ± 2.0 | 6.0 ± 0.3 | 8.0 × 104 |
| Cefoxitin | NDa | NHb | ND |
| Cefepime | 51 ± 1.8 | 6.4 ± 0.5 | 1.2 × 105 |
| Cefpirome | 246 ± 12 | 25 ± 1.0 | 1.0 × 105 |
| Aztreonam | 2.1 ± 0.1 | 0.5 ± 0.03 | 2.3 × 105 |
ND, not determined.
NH, no hydrolysis detected.
Km and kcat values are the means of three different measurements ± standard deviations.
E. coli DH5α(pMN-13) and E. coli DH5α(pPM-14) showed comparable patterns of decreased susceptibility to penicillins, narrow-spectrum cephalosporins, oxyimino-cephalosporins and aztreonam. The MICs of cefotaxime and cefepime, although increased, remained below the breakpoint for resistance. In these cases, however, a resistant phenotype was observed using a larger bacterial inoculum (5 × 107 CFU) (data not shown). Clavulanic acid and tazobactam were able to restore the susceptibility to amoxicillin and piperacillin, respectively (Table 1).
These results were overall consistent with kinetic data and indicate that a spread of TEM-72 could be very dangerous in the clinical setting.
Concluding remarks.
TEM-72 is a new natural TEM-type ESBL apparently derived from TEM-2, on the basis of the presence of the Q39K mutation. Of the three additional mutations present in TEM-72, M182T is known to act as a global suppressor of missense mutations (6), while G238S and E240K are known to be involved in the extension of substrate specificity to oxyimino-cephalosporins and monobactams (10, 20). It should be noted, however, that G238S and E240K have rarely been found together in natural TEM variants and that, in those instances when they have been (TEM-42, TEM-47, TEM-48, and TEM-49), the two mutations were always associated with additional ones (http://www.lahey.org/studies/webt.htm) of uncertain significance (11). This suggests that the presence of additional mutations could be relevant for the stability and/or function of a mutant enzyme in which G238S and E240K are simultaneously present and that, in TEM-72, a similar role could be played by the M182T global suppressor. Interestingly, kinetic parameters of TEM-72 with cephaloridine, cefotaxime, and ceftazidime were apparently different (both Km and kcat values were lower) from those previously reported for a simple G238S-E240K mutant of TEM-1 (21), suggesting that in a similar background one or both of the additional mutations found in TEM-72 (Q39K and/or M182T) could be relevant to the conformation of the active site. It will be interesting to further investigate this point by comparative kinetic analysis of the appropriate TEM variants constructed by site-directed mutagenesis.
Nucleotide sequence accession numbers.
(The nucleotide sequences reported in this paper have been submitted to the EMBL/GenBank database and assigned the accession numbers AF157413 and AF157553.)
Acknowledgments
We acknowledge Wyeth-Lederle Italia S.p.A. for a generous educational grant. Part of this work was also supported by grants from the Italian National Research Council (C.N.R.) and the Italian Ministry of Education (M.U.R.S.T.). We also acknowledge the contribution of the Italian ESBL Study Group: A. Toniolo and F. Luzzaro (Varese), P. Nicoletti (Florence), G. Gesu and R. Vaiani (Milan), G. Fortina (Novara), G. Nicoletti and G. Bonfiglio (Catania), G. Fadda and T. Spanu (Rome), G. Miragliotta (Bari), F. Menichetti (Pisa), and E. Magliano and G. Ortisi (Milan).
REFERENCES
- 1.Barthélémy M, Peduzzi J, Labia R. Complete amino acid sequence of p453-plasmid-mediated PIT-2 β-lactamase (SHV-1) Biochem J. 1988;251:73–79. doi: 10.1042/bj2510073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnet R, De Champs C, Sirot D, Chanal C, Labia R, Sirot J. Diversity of TEM mutants in Proteus mirabilis. Antimicrob Agents Chemother. 1999;43:2671–2677. doi: 10.1128/aac.43.11.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford P A, Jacobus N V, Bhachech N, Bush K. TEM-28 from an Escherichia coli clinical isolate is a member of the His-164 family of TEM-1 extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1996;40:260–262. doi: 10.1128/aac.40.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford P A, Cherubin C E, Idemyor V, Rasmussen B A, Bush K. Multiply resistant Klebsiella pneumoniae strains from two Chicago hospitals: identification of the extended-spectrum TEM-12 and TEM-10 ceftazidime-hydrolyzing β-lactamases in a single isolate. Antimicrob Agents Chemother. 1994;38:761–766. doi: 10.1128/aac.38.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Meester F, Joris B, Reckinger G, Bellefroid-Bourguignon C, Frère J M, Waley S G. Automated analysis of enzyme inactivation phenomena. Biochem J. 1987;36:2393–2403. doi: 10.1016/0006-2952(87)90609-5. [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Palzkill T. A natural polymorphism in β-lactamase is a global suppressor. Proc Natl Acad Sci USA. 1997;94:8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby G A, Medeiros A A. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacoby G A. Extended-spectrum β-lactamases and other enzymes providing resistance to oxyimino-β-lactams. Infect Dis Clin N Am. 1997;11:875–887. doi: 10.1016/s0891-5520(05)70395-0. [DOI] [PubMed] [Google Scholar]
- 9.Jarlier V, Nicholas M H, Fournier G, Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 10.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labia R, Morand A, Tiwari A, Sirot D, Petit A. Interactions of new plasmid-mediated β-lactamases with third generation cephalosporins. Rev Infect Dis. 1988;10:885–891. doi: 10.1093/clinids/10.4.885. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard. NCCLS document M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 14.Oliva B, Marinucci M C, Amicosante G, Oratore A, Bennet P M. Citrobacter diversus ULA27 produces two forms of a chromosomal β-lactamase. J Antimicrob Chemother. 1987;20:23–35. doi: 10.1093/jac/20.1.23. [DOI] [PubMed] [Google Scholar]
- 15.Perilli M, Felici A, Franceschini N, De Santis A, Pagani L, Luzzaro F, Oratore A, Rossolini G M, Knox J R, Amicosante G. Characterization of a new TEM-derived β-lactamase produced in a Serratia marcescens strain. Antimicrob Agents Chemother. 1997;41:2374–2382. doi: 10.1128/aac.41.11.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Segel I H. Biochemical calculations. 2nd ed. New York, N.Y: John Wiley & Sons; 1976. pp. 236–241. [Google Scholar]
- 18.Sirot J, Chanal C, Petit A, Sirot D, Labia R, Gerbaud G. Klebsiella pneumoniae and other Enterobacteriaceae producing novel plasmid-mediated β-lactamases markedly active against third-generation cephalosporins: epidemiologic studies. Rev Infect Dis. 1988;10:850–859. doi: 10.1093/clinids/10.4.850. [DOI] [PubMed] [Google Scholar]
- 19.Sutcliffe J G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci USA. 1978;75:3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Venkatachalam K V, Huang W, La Rocco M, Palzkill T. Characterization of TEM-1 β-lactamase mutants from positions 238 to 241 with increased catalytic efficiency for ceftazidime. J Biol Chem. 1994;269:23444–23450. [PubMed] [Google Scholar]
- 21.Yang Y, Bhachech N, Bradford P A, Jett B D, Sahm D F, Bush K. Ceftazidime-resistant Klebsiella pneumoniae and Escherichia coli isolates producing TEM-10 and TEM-43 β-lactamases from St. Louis, Missouri. Antimicrob Agents Chemother. 1998;42:1671–1676. doi: 10.1128/aac.42.7.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]