Abstract
Rapid spread of SARS-CoV-2 variants C.1.2 and B.1.621 (Mu variant) in Africa and the Americas, respectively, as well as a high number of mutations in the viral spike proteins raised concerns that these variants might pose an elevated threat to human health. Here, we show that C.1.2 and B.1.621 spike proteins mediate increased entry into certain cell lines but do not exhibit increased ACE2 binding. Further, we demonstrate that C.1.2 and B.1.621 are resistant to neutralization by bamlanivimab but remain sensitive to inhibition by antibody cocktails used for COVID-19 therapy. Finally, we show that C.1.2 and B.1.621 partially escape neutralization by antibodies induced upon infection and vaccination, with escape of vaccine-induced antibodies being as potent as that measured for B.1.351 (Beta variant), which is known to be highly neutralization resistant. Collectively, C.1.2 and B.1.621 partially evade control by vaccine-induced antibodies, suggesting that close monitoring of these variants is warranted.
Keywords: SARS-CoV-2, COVID-19, spike, variants, C.1.2, B.1.621, Mu, neutralization, antibody
Graphical abstract
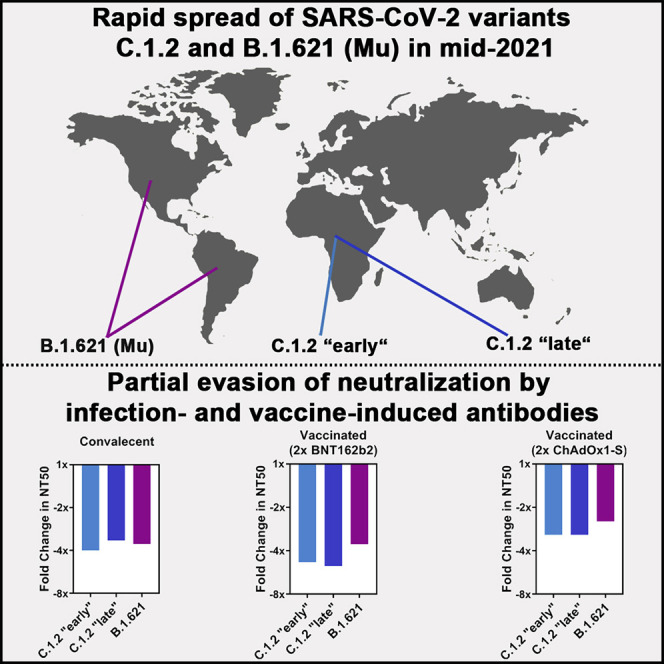
SARS-CoV-2 variants C.1.2 and B.1.621 (Mu) rapidly spread in Africa and the Americas, respectively. Arora et el. show that C.1.2 and B.1.621 spike proteins enable increased entry into certain cell lines, are resistant to neutralization by the therapeutic antibody bamlanivimab, and partially escape neutralization by infection- or vaccination-induced antibodies.
Introduction
The repeated emergence of SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) variants with increased transmissibility and/or reduced neutralization sensitivity threatens our efforts to combat the COVID-19 (coronavirus disease 2019) pandemic though vaccination. To date, five SARS-CoV-2 variants are classified as variants of concern (VOCs)—Alpha (B.1.1.7 and sublineages), Beta (B.1.351 and sublineages), Gamma (P.1 and sublineages), Delta (B.1.617.2 and sublineages), and Omicron (B.1.1.529 and sublineages)—since they are more transmissible, virulent, and/or resistant to antibodies elicited upon infection or vaccination compared with the original virus (Arora et al., 2021; Cele et al., 2021; Hoffmann et al., 2021a, 2022; Lucas et al., 2021; Mlcochova et al., 2021). Further, additional SARS-CoV-2 variants have been listed as variants of interest (VOIs) because they exhibit certain characteristics (e.g., mutations) of VOCs and thus might become a VOC in the future (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
At present, vaccination represents our main weapon to combat the COVID-19 pandemic, with several vaccines being available (Baden et al., 2021; Jara et al., 2021; Polack et al., 2020; Sadoff et al., 2021; Voysey et al., 2021). Although some SARS-CoV-2 variants partially evade neutralization by antibodies and may thus cause vaccine breakthrough infections (Arora et al., 2021; Chin et al., 2021; Hoffmann et al., 2021a; Lucas et al., 2021; Mlcochova et al., 2021), current vaccines are highly effective in protecting from severe disease and death (Reis et al., 2021; Sheikh et al., 2021a, 2021b).
It is crucial to identify and contain emerging SARS-CoV-2 variants with VOC-like traits before they become globally disseminated. Thus, we investigated host cell entry and neutralization of two SARS-CoV-2 variants, C.1.2 and B.1.621 (Mu variant), that emerged and spread (mainly) in Africa and the Americas, respectively, making use of pseudoviruses bearing the C.1.2 or B.1.621 spike (S) protein. The SARS-CoV-2 S protein mediates viral entry into target cells. For this, it engages the cellular receptor, angiotensin converting enzyme 2 (ACE2), via its receptor-binding domain (RBD) and, following activation (priming) by host cell proteases, fuses viral and cellular membranes, allowing delivery of the viral genome into the cytoplasm (Jackson et al., 2021).
Here we show that amino acid substitutions in the C.1.2 and B.1.621 S proteins augment entry into certain cell lines in an ACE2-independent manner. Further, they confer full resistance to bamlanivimab and partial resistance to casirivimab and etesevimab, antibodies used for COVID-19 therapy. Finally, we show that C.1.2 and B.1.621 evade neutralization by antibodies elicited upon infection or vaccination with similar potency as the highly neutralization-resistant B.1.351 (Beta) variant.
Results
C.1.2 and B.1.621 harbor multiple mutations in the S protein and showed exponential spread in mid-2021
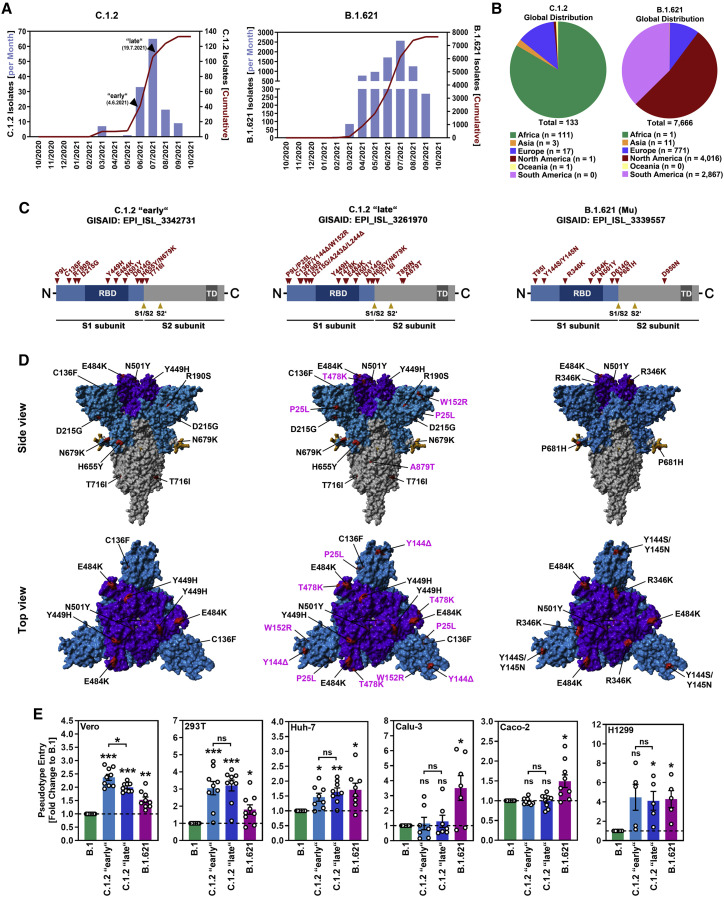
Between March and September 2021, the number of sequences belonging to SARS-CoV-2 variants C.1.2 and B.1.621 deposited in the GISAID (Global Initiative on Sharing All Influenza Data) database increased exponentially (Figure 1A). While the majority of C.1.2 sequences was derived from the African continent, B.1.621 sequences mainly originated from North and South America (Figure 1B). However, for both variants infections have also been detected in other countries. Both variants harbor mutation D614G in their S proteins, which emerged in the early phase of the pandemic (Korber et al., 2020) and augments SARS-CoV-2 host cell entry and transmission by enabling the RBD to more frequently adopt the so-called open/up conformation required for ACE2 binding (Yurkovetskiy et al., 2020). Further, when compared with the S protein of the virus present at the start of the pandemic (Wuhan-Hu-1 isolate), the S proteins of SARS-CoV-2 variants B.1.621 (which has been declared a VOI by WHO and is also known as Mu variant) and C.1.2 harbor several mutations, some of which have been previously detected in other SARS-CoV-2 variants and are associated with increased affinity for ACE2 or reduced sensitivity to antibody-mediated neutralization. This includes RBD mutation N501Y, which is also found in VOCs Alpha, Beta, and Gamma, and which is associated with stronger binding to ACE2 (Tian et al., 2021). In addition, RBD mutation T478K, found in VOC Delta, and RBD mutation E484K, found in VOCs Beta and Gamma, have been reported to contribute to evasion from antibody-mediated neutralization (Arora et al., 2021; Hoffmann et al., 2021a). Moreover, both variants harbor mutations in their N-terminal domain (NTD), which contains an antigenic supersite that is targeted by most neutralizing antibodies not directed against the RBD (McCallum et al., 2021).
Figure 1.
Spike proteins of SARS-CoV-2 variants C.1.2 and B.1.621 harbor several mutations and enable enhanced entry into certain cell lines
(A) Epidemiology of SARS-CoV-2 variants C.1.2 and B.1.621 (as of September 10, 2021; based on data deposited in the GISAID database). Arrowheads indicate the time points when isolates C.1.2 “early” and C.1.2 “late” were detected.
(B) Global distribution of SARS-CoV-2 variants C.1.2 and B.1.621 (as of September 10, 2021; based on data deposited in the GISAID database).
(C) Schematic overview and S protein domain organization. Mutations compared with SARS-CoV-2 Wuhan-Hu-1 are highlighted in red. Abbreviations: RBD, receptor-binding domain; TD, transmembrane domain.
(D) Location of C.1.2 “early,” C.1.2 “late,” and B.1.621-specific mutations in the context of the trimeric spike protein. Color code: light blue, S1 subunit with RBD in dark blue; gray, S2 subunit; orange, S1/S2 and S2′ cleavage sites; red, mutated amino acid residues (compared with SARS-CoV-2 Wuhan-Hu-1; pink labels highlight differences between the two C.1.2 S proteins).
(E) Pseudotyped particles harboring the indicated S proteins were inoculated onto the indicated cell lines. Transduction efficiency was quantified at 16–18 h post inoculation and normalized against B.1 S (set as 1). Shown are the average (mean) data from five to nine biological replicates (each performed with technical quadruplicates). Error bars indicate the standard error of the mean (SEM). Statistical significance was analyzed by two-tailed Student’s t test with Welch correction (p > 0.05, not significant [ns]; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗).
See also Figure S1.
We focused on a single B.1.621 sequence and on two C.1.2 S protein sequences derived from the early (C.1.2 “early”) and late (C.1.2 “late”) phase of exponential spread (between June and July 2021; Figure 1A). This selection was based on the observation that C.1.2 appeared to have accumulated several additional mutations during this period, whereas B.1.621 remained genetically stable with respect to its S protein (apart from minor, low-frequency mutations found in individual isolates) (Figure 1C). The S protein of C.1.2 “early” contains eleven amino acid substitutions (Figures 1C and 1D), ten (P9L, C136F, R190S, D215G, Y449H, E484K, N501Y, D614G, H655Y, N679K) of which are present in the S1 subunit, while one (T716I) is found in the S2 subunit. Within the S1 subunit, four substitutions (P9L, C136F, R190S, D215G) are located in the NTD, three (Y449H, E484K, N501Y) in the RBD, and three (D614G, H655Y, N679K) between the RBD and S1/S2 cleavage site (Figures 1C and 1D). The S protein of C.1.2 “late” contains all amino acid substitutions present in C.1.2 “early” plus eight additional changes (P25L, Y144Δ, W152R, A243Δ, L244Δ, T478K, T859N, A879T). These additional changes include two substitutions (P25L, W152R) and three deletions (Y144Δ, A243Δ, L244Δ) in the NTD, one substitution (T478K) in the RBD, and two substitutions (T859N, A879T) in the S2 subunit (Figures 1C and 1D). The B.1.621 S protein contains nine amino acid substitutions, eight of which can be found in the S1 subunit (T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H), while one (D950N) is present in the S2 subunit. Of the eight amino acid substitutions in the S1 subunit, three (T95I, Y144S, Y145N) are present in the NTD, another three (R346K, E484K, N501Y) reside in the RBD, and the remaining two (D614G, P681H) are located between the RBD and S1/S2 cleavage site (Figures 1C and 1D).
C.1.2 and B.1.621 show increased cell entry but not augmented ACE2 binding
We first investigated entry of C.1.2 “early,” C.1.2 “late,” and B.1.621 into five human (293T, Huh-7, Calu-3, Caco-2, and H1299) cell lines and one nonhuman primate (Vero) cell line using vesicular stomatitis virus-based pseudoviruses bearing the respective S proteins. For comparison, we used pseudoviruses harboring the SARS-CoV-2 B.1 S protein (identical to the Wuhan-Hu-1 S protein except for mutation D614G). Cell lines were chosen based on their different organ origin (colon: Caco-2; kidney: Vero, 293T; liver: Huh-7; lung: Calu-3, H1299) and availability of known SARS-CoV-2 entry factors, i.e., cellular proteases required for spike protein activation, TMPRSS2 (Caco-2, Calu-3, H1299) or cathepsin B/L (Vero, 293T, Huh-7). All S proteins mediated robust entry into the cell lines tested (Figure 1E and S1A). While S proteins from both C.1.2 isolates as well as B.1.621 S allowed for augmented cell entry compared with B.1 S, the enhanced entry phenotype was not uniform. For instance, while B.1.621 S showed ∼3.5× and ∼1.5× increased ability to drive entry into Calu-3 and Caco-2 cells, respectively, both C.1.2 S proteins were more efficient in mediating entry into Vero (∼2.5×/∼2.0× C.1.2 “early”/C.1.2 “late”) and 293T (∼3.0×/∼3.1× C.1.2 “early”/C.1.2 “late”) cells (Figures 1E and S1A). For the remaining two cell lines (Huh-7 and H1299), C.1.2 and B.1.621 S proteins displayed ∼1.5× (Huh-7) and ∼4.0× (H1299) higher entry efficiency compared with B.1 S (Figures 1E and S1A).
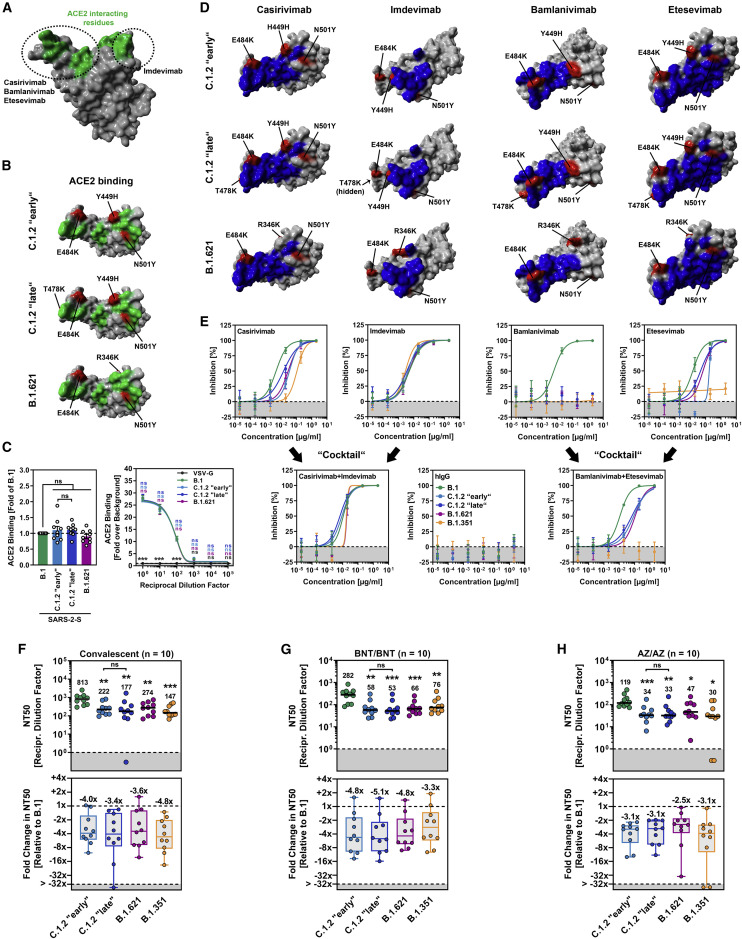
Since C.1.2 “early,” C.1.2 “late,” and B.1.621 harbor several mutations at RBD residues that directly interact with ACE2 (Figures 2 A and 2B), we further investigated whether augmented cell entry stems from increased S protein binding to ACE2. For this, we employed a flow cytometric assay and a cell-based ACE2 binding assay (CABA) with soluble ACE2 and S proteins transiently expressed in 293T-cells. Using these techniques, we did not observe significant differences in ACE2 binding between the S proteins studied (Figures 2C and S1B) despite comparable cell surface expression levels (Figure S1C).
Figure 2.
Spike proteins of SARS-CoV-2 variants C.1.2 and B.1.621 robustly bind ACE2 and efficiently evade antibody-mediated neutralization
(A) Protein model of the SARS-CoV-2 RBD in which the ACE2-interacting interface (green) and target regions of therapeutic monoclonal antibodies (circles) are highlighted.
(B) Location of C.1.2 “early,” C.1.2 “late,” and B.1.621-specific mutations (red) in the context of the RBD (gray). RBD residues that interact with ACE2 are colored in green.
(C) 293T cells expressing the indicated S proteins (or VSV-G) were incubated with soluble ACE2-Fc and secondary antibody and analyzed by flow cytometry (left) and CABA (right). Shown are the average (mean) data from nine (flow cytometry; single samples) or three (CABA, four technical replicates) biological replicates for which ACE2 binding was normalized against B.1 (flow cytometry; set as 1) or the background (CABA; no soluble ACE2-Fc, set as 1). Error bars indicate the SEM. Statistical significance was analyzed by two-tailed Student’s t test with Welch’s correction (flow cytometry) or two-way analysis of variance with Dunnett’s post-hoc test (CABA) (p > 0.05, ns; p < 0.05, ∗). See also Figure S1.
(D) Location of C.1.2 “early,” C.1.2 “late,” and B.1.621-specific mutations (red) in the context of the RBD (gray) epitopes targeted by casirivimab, imdevimab, bamlanivimab, and etesevimab (blue).
(E) Pseudotyped particles bearing the indicated S protein were pre-incubated in with the indicated antibodies or antibody cocktails and subsequently inoculated onto Vero cells. At 16–18 h post inoculation, pseudotype entry was quantified and normalized against samples that did not contain antibody (= 0% inhibition). Shown are the average (mean) data from a single biological replicate (performed with technical quadruplicates), and data were confirmed in a separate independent experiment. Error bars indicate the standard deviation. See also Figure S1.
(F–H) Pseudotyped particles bearing the indicated S proteins were pre-incubated in the presence of convalescent plasma (F) or serum from individuals either vaccinated with BioNTech/Pfizer’s BNT162b2 vaccine (BNT/BNT (G) or AstraZeneca’s ChAdOx1 nCoV-19 vaccine (AZ/AZ) (H), and subsequently inoculated onto Vero cells. At 16–18 h post inoculation, pseudotype entry was quantified and used for the calculation of the neutralizing titer 50 (NT50). Presented are the data from ten plasma/serum samples per group (black lines and numerical values indicate the median NT50). Further, the median fold reduction in NT50 between SARS-CoV-2 B.1 (set as 1) and the indicated variants was calculated (boxplots indicate the median, quartiles, and range; circles indicate individual samples). Statistical significance was analyzed by two-tailed Mann-Whitney test with 95% confidence level (p > 0.05, ns; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗).
See also Figure S2.
C.1.2 and B.1.621 are resistant against neutralization by bamlanivimab
Single monoclonal antibodies (mAbs) and cocktails thereof showed therapeutic benefit in clinical settings (Dougan et al., 2021; Weinreich et al., 2021) and have received emergency use authorization for COVID-19 treatment. In order to assess whether C.1.2 “early,” C.1.2 “late,” and B.1.621 can be potently neutralized by therapeutic mAbs, we conducted neutralization experiments with four therapeutic mAbs (casirivimab, imdevimab, bamlanivimab, etesevimab) or antibody cocktails consisting of two therapeutic mAbs (casirivimab and imdevimab; bamlanivimab and etesevimab) as they are given to COVID-19 patients. For comparison, we included the S protein of SARS-CoV-2 B.1.351 (Beta), which is known to be resistant against bamlanivimab and etesevimab, and less potently neutralized by casirivimab (Hoffmann et al., 2021a, 2021b). Since the RBD epitopes to which the mAbs bind to are known, mapping the mutated amino acid residues of C.1.2 “early,” C.1.2 “late,” and B.1.621 suggested that neutralization by casirivimab, bamlanivimab, and etesevimab might be compromised (Figure 2D).
Consistent with the findings from the in silico analysis, we observed partial loss of neutralization by casirivimab (∼3.0–7.4× increase in IC50, likely due to mutation E484K) and etesevimab (∼3.2–11.4× increase in IC50, likely due to mutations Y449H [only C.1.2 S proteins] and N501Y), and complete resistance against bamlanivimab (likely due to mutation E484K). In contrast, neutralization by imdevimab was unaffected (Figures 2D, 2E, and S1D). However, although individual mAbs (partially) lost their neutralizing potential, mAb cocktails remained highly potent (Figures 2E and S1D). In keeping with published data, particles bearing B.1.351 S were completely resistant against neutralization by bamlanivimab and etesevimab (Hoffmann et al., 2021a, 2021b), while a control antibody not directed against the SARS-CoV-2 S protein (hIgG) did not block entry driven by any S protein tested (Figures 2D, 2E, and S1D). In sum, therapeutic mAb cocktails but not single mAbs should represent useful therapeutic options for treatment of patients infected with SARS-CoV-2 variants C.1.2 and B.1.621.
C.1.2 and B.1.621 partially evade neutralization by antibodies induced upon infection or vaccination
Finally, we investigated whether the amino acid substitutions present in C.1.2 and B.1.621 S proteins affect neutralization by antibodies present in the serum/plasma of convalescent COVID-19 patients or individuals who received two shots of either the BNT162b2 (BNT/BNT; BioNTech/Pfizer) or ChAdOx1 nCoV-19 vaccine (AZ/AZ; AstraZeneca) (Table S1). Again, we used particles bearing the S protein of either SARS-CoV-2 B.1 or B.1.351 (Beta) for comparison, since the B.1 S protein is more or less identical to the S protein that is the basis of the vaccines, while the B.1.351 S protein shows a high level of antibody evasion (Hoffmann et al., 2021a).
Particles bearing C.1.2 and B.1.621 S proteins efficiently escaped from neutralization by antibodies present in convalescent plasma as evidenced by lower NT50 (neutralizing titer 50) values compared with B.1 S (B.1, NT50 = 813; C.1.2 “early,” NT50 = 222, ∼4.0× reduced neutralization compared with B.1; C.1.2 “late,” NT50 = 177, ∼3.4× reduced; B.1.621, NT50 = 274, ∼3.6× reduced), but neutralization resistance was less compared with B.1.351 S (NT50 = 147, ∼4.8× reduced neutralization compared with B.1) (Figures 2F and S2). Similar observations were made for sera from BNT/BNT-vaccinated (B.1, NT50 = 282; C.1.2 “early,” NT50 = 58, ∼4.8× reduced neutralization compared with B.1; C.1.2 “late,” NT50 = 53, ∼5.1× reduced; B.1.621, NT50 = 66, ∼4.8× reduced) (Figures 2G and S2) or AZ/AZ-vaccinated individuals (B.1, NT50 = 119; C.1.2 “early,” NT50 = 34, ∼3.1× reduced neutralization compared with B.1; C.1.2 “late,” NT50 = 33, ∼3.1× reduced; B.1.621, NT50 = 47, ∼2.5× reduced) (Figures 2H and S2). However, different to the trend that was observed for convalescent plasma, neutralization resistance was either comparable (AZ/AZ) or slightly enhanced compared with B.1.351 S (BNT/BNT: NT50 = 76, ∼3.3× reduced neutralization compared with B.1; AZ/AZ: NT50 = 30, ∼3.1× reduced) when vaccinee sera were investigated. Of note, no significant differences in neutralization sensitivity were observed for particles bearing the different C.1.2 S proteins (Figures 2F–2H and S2). Thus, amino acid substitutions in C.1.2 and B.1.621 S proteins substantially reduce sensitivity to neutralizing antibodies present in the plasma/serum of convalescent COVID-19 patients and BNT/BNT- or AZ/AZ-vaccinated individuals.
Discussion
To identify emerging SARS-CoV-2 variants that have the potential to become VOCs in the future, it is crucial to characterize these variants at an early stage and install proper countermeasures to contain their spread.
Compared with host cell entry mediated by the S protein of SARS-CoV-2 variant B.1, cellular entry mediated by C.1.2 and B.1.621 S proteins was increased in a target cell-specific fashion. The observation that neither C.1.2 nor B.1.621 S proteins displayed increased ACE2 binding suggests that augmented cell entry may have resulted from improved interactions with attachment-promoting factors like heparan sulfate proteoglycans (Clausen et al., 2020) or S protein-activating proteases. The SARS-CoV-2 variants C.1.2 and B.1.621 were less potently neutralized by some antibodies used for the treatment of COVID-19 patients. Thus, both C.1.2 and B.1.621 S proteins were completely resistant to bamlanivimab, likely due to the presence of RBD mutation E484K (Starr et al., 2021), while neutralization by casirivimab was moderately reduced. Neutralization by etesevimab was also reduced, likely due to RBD mutations E484K and N501Y (and possibly also RBD mutation Y449H in case of C.1.2) (Starr et al., 2021). However, antibody cocktails consisting of casirivimab/imdevimab or bamlanivimab/etesevimab still robustly neutralized both variants and thus constitute therapeutic options.
Both C.1.2 and B.1.621 S proteins evaded neutralization by antibodies elicited upon infection or vaccination with high efficiency, reaching (or exceeding) levels measured for variant B.1.351 (Beta), which was previously reported to show a high level of neutralization resistance (Garcia-Beltran et al., 2021; Hoffmann et al., 2021a; Lucas et al., 2021). These results are in line with two recent studies, which also showed that B.1.621 escaped neutralization from infection- and vaccination-elicited antibodies with comparable or higher potency as B.1.351 (Beta) (Tada et al., 2022; Uriu et al., 2021). Finally, it is noteworthy that the SARS-CoV-2 Delta variant (B.1.617.2) was shown to enter Calu-3 lung cells ∼2× better than B.1 (which is comparable to B.1.621) but displayed only moderate potency to evade neutralization by infection- or vaccination-induced antibodies (Arora et al., 2021), which may partially explain why B.1.621 maintained its prevalence even when B.1.617.2 (Delta) was spreading globally.
We neither observed significant differences in cell entry between the two C.1.2 S proteins nor major changes in neutralization sensitivity. Therefore, it remains speculative whether the additional mutations found in C.1.2 “late” may endow the virus with an advantage that could not be identified in this study, potentially usage of additional attachment-promoting factors not expressed in the tested cell lines. Alternatively, C.1.2 “late” may have spread as the result of a founder effect, e.g., because C.1.2 “late” may have been introduced into regions that are more populated or populations with more social contacts.
In sum, this study provides evidence for increased host cell entry, resistance to therapeutic options (full/partial resistance against therapeutic antibodies), and partial evasion of preexisting immunity (antibodies elicited upon infection or vaccination) by SARS-CoV-2 variants C.1.2 and B.1.621. However, while a less robust preexisting immunity may still protect against infection and/or developing severe disease upon infection with C.1.2 and B.1.621, the chance of becoming infected with these variants is higher compared with variants that cannot escape antibody neutralization with high potency. As a consequence, efforts to contain the spread of these variants are warranted, and surveillance programs should screen for both variants to detect small C.1.2- and B.1.621-related infection clusters before they grow too big to be contained.
Limitations of the study
Our study has the following limitations: we employed VSV (vesicular stomatitis virus) pseudotyped with SARS-CoV-2 S proteins to study cell entry and its neutralization by antibodies. Although these vectors faithfully recapitulate SARS-CoV-2 cell entry and thus represent a widely accepted surrogate model, our results await formal confirmation with authentic virus. Furthermore, host cell entry was studied using immortalized cell lines, and these analyses should be extended to primary cell cultures and in vivo models.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Casirivimab | Laboratory of Hans-Martin Jäck | N/A |
| Imdevimab | Laboratory of Hans-Martin Jäck | N/A |
| Bamlanivimab | Laboratory of Hans-Martin Jäck | N/A |
| Etesevimab | Laboratory of Hans-Martin Jäck | N/A |
| hIgG | Laboratory of Hans-Martin Jäck | N/A |
| Goat anti-Human IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11013; RRID: AB_2534080 |
| Goat anti-Human IgG (Fc)-HRPO | Dianova | Cat# 109-035-098; RRID: AB_2337586 |
| Anti-VSV-G antibody (I1, produced from CRL-2700 mouse hybridoma cells) | ATCC | Cat# CRL-2700; RRID: CVCL_G654 |
| Bacterial and virus strains | ||
| VSV∗ΔG-FLuc | Laboratory of Gert Zimmer | N/A |
| One Shot™ OmniMAX™ 2 T1R Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C854003 |
| Biological samples | ||
| Convalescent plasma (ID15) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID18) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID20) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID22) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID23) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID24) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID27) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID33) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID51) | Laboratory of Martin S. Winkler | N/A |
| Convalescent plasma (ID56) | Laboratory of Martin S. Winkler | N/A |
| Vaccinee serum (L3) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L4) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L9) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L11) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L12) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L13) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L16) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L22) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L25) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (L26) | Laboratory of Martin Lier | N/A |
| Vaccinee serum (ID6205) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6272) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6236) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6239) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6243) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6262) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6297) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6321) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6358) | Laboratory of Georg M. N. Behrens | N/A |
| Vaccinee serum (ID6365) | Laboratory of Georg M. N. Behrens | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Soluble human ACE2 (sol-hACE2-Fc) | Laboratory of Stefan Pöhlmann | N/A |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668019 |
| Critical commercial assays | ||
| Beetle-Juice Kit | PJK | Cat# 102511 |
| Deposited data | ||
| N/A | N/A | N/A |
| Experimental models: cell lines | ||
| 293T | DSMZ | Cat# ACC-635; RRID: CVCL_0063 |
| BHK-21 | Laboratory of Georg Herrler | ATCC Cat# CCL-10; RRID:CVCL_1915 |
| Caco-2 | Laboratory of Stefan Pöhlmann | ATCC Cat# HTB-37; RRID: CVCL_0025 |
| Calu-3 | Laboratory of Stephan Ludwig | ATCC Cat# HTB-55; RRID: CVCL_0609 |
| Huh-7 | Laboratory of Thomas Pietschmann | JCRB Cat# JCRB0403; RRID: CVCL_0336 |
| NCI-H1299 | Laboratory of Stefan Pöhlmann | ATCC Cat# CRL-5803; RRID:CVCL_0060 |
| Vero76 | Laboratory of Andrea Maisner | ATCC Cat# CRL-1586; RRID: CVCL_0574 |
| Experimental models: organisms/strains | ||
| N/A | N/A | N/A |
| Oligonucleotides | ||
| Please see Table S3 | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| Plasmid: pCG1 | Laboratory of Roberto Cattaneo | N/A |
| Plasmid: pCAGGS-VSV-G | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCAGGS-DsRed | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (B.1), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (B.1.351), codon-optimized | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (B.1.621), codon-optimized | This study | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (C.1.2 “early”), codon-optimized | This study | N/A |
| Plasmid: pCG1-SARS-2-SΔ18 (C.1.2 “late”), codon-optimized | This study | N/A |
| Plasmid: pCG1-solACE2-Fc | Laboratory of Stefan Pöhlmann | N/A |
| Software and algorithms | ||
| Hidex Sense Microplate Reader Software | Hidex Deutschland Vertrieb GmbH | https://www.hidex.de |
| YASARA (version 19.1.27) | YASARA Biosciences GmbH | http://www.yasara.org |
| SWISS-MODEL online tool | Protein Structure Bioinformatics Group, Swiss Institute of Bioinformatics Biozentrum, University of Basel |
https://swissmodel.expasy.org |
| Adobe Photoshop CS5 Extended (version 12.0 x 32) | Adobe | https://www.adobe.com/ |
| GraphPad Prism (version 8.3.0(538)) | GraphPad Software | https://www.graphpad.com/ |
| FloJo (version 10.8) | Becton Dickinson | https://www.flowjo.com/ |
| Flowing Software (version 2.5.1) | Turku Bioscience | https://bioscience.fi/services/cell-imaging/flowing-software/ |
| Adobe Photoshop CS5 Extended (version 12.0 x 32) | Adobe | https://www.adobe.com/ |
| Microsoft Office Standard 2010 (version 14.0.7232.5000) | Microsoft Corporation | https://products.office.com/ |
| Other | ||
| Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies (PDB: 6XDG) | (Hansen et al., 2020) | https://www.rcsb.org/structure/6XDG |
| SARS-CoV 2 Spike Protein bound to LY-CoV555 (PDB: 7L3N) | (Jones et al., 2021) | https://www.rcsb.org/structure/7L3N |
| Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD (PDB: 7C01) | (Shi et al., 2020) | https://www.rcsb.org/structure/7C01 |
| Distinct conformational states of SARS-CoV-2 spike protein (PDB: 6XR8) | (Cai et al., 2020) | https://www.rcsb.org/structure/6XR8 |
Resource availability
Lead contact
Requests for material can be directed to Stefan Pöhlmann (spoehlmann@dpz.eu) and the lead contact, Markus Hoffmann (mhoffmann@dpz.eu).
Materials availability
All materials and reagents will be made available upon installment of a material transfer agreement (MTA).
Experimental model and subject details
Cell culture
All cell lines were incubated at 37°C in a humidified atmosphere containing 5% CO2. 293T (human, female, kidney; ACC-635, DSMZ; RRID: CVCL_0063), BHK-21 (Syrian hamster, kidney, male; ATCC Cat# CCL-10; RRID: CVCL_1915, kindly provided by Georg Herrler), Vero (African green monkey kidney, female, kidney; CRL-1586, ATCC; RRID: CVCL_0574, kindly provided by Andrea Maisner) and Huh-7 cells (human, male, liver; JCRB Cat# JCRB0403; RRID: CVCL_0336, kindly provided by Thomas Pietschmann) were maintained in Dulbecco’s modified Eagle medium (DMEM, PAN-Biotech). Calu-3 (human, male, lung; HTB-55, ATCC; RRID: CVCL_0609, kindly provided by Stephan Ludwig) and Caco-2 cells (human, male, colon; HTB-37, ATCC, RRID: CVCL_0025) were maintained in minimum essential medium (GIBCO). NCI-H1299 (human, male, lung; CRL-5803, ATCC, RRID: CVCL_0060) were maintained in DMEM/F-12 medium (GIBCO). All media were supplemented with 10% fetal bovine serum (Biochrom) and 100 U/mL penicillin and 0.1 mg/mL streptomycin (PAA). Furthermore, Calu-3 and Caco-2 cells received 1× non-essential amino acid solution (from 100× stock, PAA) and 1 mM sodium pyruvate (GIBCO). Cell lines were validated by STR-typing, amplification, and sequencing of a fragment of the cytochrome c oxidase gene, microscopic examination and/or according to their growth characteristics. In addition, all cell lines were regularly tested for mycoplasma contamination. Transfection of cells was carried out by calcium-phosphate precipitation (293T) or using Lipofectamine 2000 (Thermo Fisher Scientific; BHK-21).
Human subjects
Convalescent plasma was obtained during the first and second waves of the COVID-19 pandemic from COVID-19 patients (n = 10, seven male and three female patients, median age group: 65–74 years) treated at the intensive care unit of the University Medicine Göttingen (UMG) under approval given by the ethic committee of the UMG (SeptImmun Study 25/4/19 Ü). Patient details can be found in Table S1 and elsewhere (Hoffmann et al., 2021a). Unfortunately, no information on the infecting SARS-CoV-2 lineage is available for these samples. Sera from individuals vaccinated with BNT162b2/BNT162b2 (n = 10, five male and five female patients, median age group: 18–24 years) or ChAdOx1 nCoV-19/BNT162b2 (n = 10, zero male and ten female patients, median age group: 55–64 years) were collected 72–204 days after receiving the second dose under the approval given by the ethic committee of the UMG (reference number: 8/9/20, patient details can be found in Table S2). Sera from individuals vaccinated with ChAdOx1 nCoV-19/ChAdOx1 nCoV-19 were collected 27–42 days after receiving the second dose under the approval given by the ethic committee of the Hannover Medical School (COVID-19 Contact Study, reference number: 8973_BO-K_2020, patient details can be found in Table S2). Before analysis, all serum and plasma samples were heat-inactivated at 56°C for 30 min. Further, all plasma/serum samples were pre-screened for their ability to neutralize transduction of Vero cells by pseudotype particles bearing SARS-CoV-2 S B.1.
Method details
Plasmids
Plasmids encoding dsRed, VSV-G (vesicular stomatitis virus glycoprotein), SARS-CoV-2 S B.1 (codon optimized, contains C-terminal truncation of the last 18 amino acid) and SARS-CoV-2 S B.1.351 have been described earlier (Brinkmann et al., 2017; Hoffmann et al., 2021a, 2021c). To introduce spike (S) mutations in the expression vector of SARS-CoV-2 variants C.1.2 “early” (GISAID Accession ID: EPI_ISL_3342731), C.1.2 “late” (GISAID Accession ID: EPI_ISL_3261970) and B.1.621 (GISAID Accession ID: EPI_ISL_3339557) the respective mutations were inserted into the expression plasmid for the S protein of SARS-CoV-2 B.1 by hybrid PCR. For this, overlapping primers harboring the respective mutations were used to amplify several regions of the B.1 S protein vector. PCR products were subsequently purified from an agarose gel using a commercial kit (NucleoSpin Gel and PCR Clean-up, Macherey-Nagel), mixed at equimolar concentrations before finally the full-length S protein open reading frames were amplified by PCR with primers corresponding to the 3′ and 5′ ends full-length S protein sequence (primer sequences can be found in Table S3). The resulting open reading frames were further inserted into the pCG1 vector (kindly provided by Roberto Cattaneo, Mayo Clinic College of Medicine, Rochester, MN, USA), using restriction sites BamHI and XbaI. The integrity of all sequences was analyzed by using a commercial service provider (Microsynth SeqLab). Specific details on the cloning strategy and procedure can be attained upon request.
Sequence analysis and protein models
S protein sequences and information on collection date and global distribution of SARS-CoV-2 isolates belonging to variants C.1.2 and B.1.621 were retrieved from the GISAID (Global Initiative on Sharing All Influenza Data) database (https://www.gisaid.org/). S Protein models were generated employing the YASARA software (http://www.yasara.org/index.html) or UCSF Chimera version 1.14 (https://www.cgl.ucsf.edu/chimera/) and are based on a template that was constructed by modeling the SARS-2 S sequence on PDB: 6XR8 (Cai et al., 2020) using the SWISS-MODEL online tool (https://swissmodel.expasy.org/), or on the following published crystal structures; PDB: 6XDG (Hansen et al., 2020), PDB: 7L3N (Jones et al., 2021) or PDB: 7C01 (Shi et al., 2020).
Production of pseudotype particles
Production of rhabdoviral pseudotypes bearing SARS-CoV-2 spike protein was achieved using a published protocol (Kleine-Weber et al., 2019). In brief, 293T cells were transfected with expression plasmid for SARS-CoV-2 S protein, VSV-G or empty plasmid (control). At 24 h posttransfection, cells were inoculated with, VSV∗ΔG-FLuc (Berger Rentsch and Zimmer, 2011), a replication-deficient vesicular stomatitis virus that lacks the genetic information for VSV-G and instead codes for two reporter proteins, enhanced green fluorescent protein and firefly luciferase (FLuc) (kindly provided by Gert Zimmer) at a multiplicity of infection of 3. Following 1 h of incubation, the inoculum was removed and cells were washed with phosphate-buffered saline (PBS). Subsequently, cells received culture medium containing anti-VSV-G antibody (culture supernatant from I1-hybridoma cells; ATCC no. CRL-2700; except for cells expressing VSV-G, which received only medium) in order to neutralize residual input virus. After 16–18 h, the culture supernatant was harvested, separated from cellular debris by centrifugation for 10 min at 4,000 × g at room temperature (RT), and the clarified supernatants were aliquoted and stored at −80°C.
Analysis of spike protein-mediated cell entry
For experiments assessing S protein-driven cell entry, target cells were seeded in 96-well plates. At 24 h post seeding, the culture medium was aspirated and cells were inoculated with equal volumes of pseudotype particles. At 16–18 h post inoculation, pseudotype entry efficiency was quantified by measuring the activity of virus-encoded luciferase. For this, cells were lysed using PBS containing 0.5% Triton X-100 (Carl Roth) for 30 min at RT. Afterward, cell lysates were transferred into white 96-well plates and mixed with luciferase substrate (Beetle- Juice, PJK) before luminescence was documented using a Hidex Sense Plate luminometer (Hidex).
Production of soluble ACE2
The production of soluble ACE2 has been described elsewhere in detail (Hoffmann et al., 2021c). Briefly, 293T cells were seeded and transfected with expression plasmid for soluble ACE2. After overnight incubation, the medium was replaced and the cells further incubated for 38 h before the supernatant was collected and centrifuged to remove any cell debris. Further, the culture supernatant was concentrated 100x using a Vivaspin protein concentrator column (molecular weight 30 kDa; Sartorius). The concentrated soluble ACE2 was aliquoted and stored at −80°C for further use.
Analysis of ACE2 binding by flow cytometry
In order to test ACE2 binding of soluble ACE2 to S protein, 293T cells were seeded in 6-well plates and transfected with expression plasmid for the respective SARS-CoV-2 S protein. Cells transfected with empty plasmid served as a negative control. At 24 h posttransfection, the medium was replaced. At 48 h posttransfection, the culture medium was removed and cells were resuspended in PBS and transferred into 1.5 mL reaction tubes before being pelleted by centrifugation All centrifugation steps were carried out at room temperature at 600 × g for 5 min. Subsequently, the supernatant was aspirated and the cells were washed with PBS containing 1% bovine serum albumin (BSA, PBS-B) and pelleted by centrifugation. Next, the supernatant was removed and cell pellets were resuspended in 250 μL PBS-B containing soluble ACE2-Fc (1:500) and rotated for 60 min at 4°C using a Rotospin test tube rotator disk (IKA). Following incubation, cells were pelleted, resuspended in 250 μL PBS-B containing anti-human AlexaFluor-488-conjugated antibody (1:200; Thermo Fisher Scientific) and rotated again for 60 min at 4°C. Finally, the cells washed with PBS-B, fixed by incubation in 4% paraformaldehyde solution for 30 min at RT, washed again and resuspended in 150 μL PBS-B before being subjected to flow cytometric analysis using an ID7000 Spectral Analyser (Sony) or an LSR II flow cytometer (BD Biosciences). Data were further analyzed using the FlowJo and Flowing software (https://bioscience.fi/services/cell-imaging/flowing-software/) in order to determine the geometric mean channel fluorescence.
Analysis of ACE2 binding by cell-based ACE2 binding assay (CABA)
BHK-21 cells were seeded in 96-well plates and transfected with expression plasmid for the respective SARS-CoV-2 S protein or empty plasmid using the Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. At 24 h posttransfection, the medium was removed, cells were washed with PBS and fixed by incubation in 4% paraformaldehyde solution for 30 min at RT. Next, cells were washed once with PBS and incubated at 4°C with PBS containing 3% BSA for additional 30 min for blocking. After an additional washing step with PBS, cells were incubated for 1h at 4°C with 10-fold serial dilutions of soluble ACE2-Fc (diluted in PBS-B) or PBS-B alone (control). Subsequently, cells were washed three times with PBS and incubated for 30 min at 4°C with horseradish peroxidase-conjugated anti-human antibody (1:10,000; Dianova). Finally, cells were washed three times with PBS and ACE2 binding was measured. For this, an in house-made luminescent peroxidase substrate (0.1 M Tris-HCl [pH 8.6], 250 μg/mL luminol, 0.1 mg/mL para-hydroxycoumaric acid, 0.3% hydrogen peroxide) was added and luminescence was recorded using a Hidex Sense Plate luminometer (Hidex).
Analysis of S protein expression on the cell surface
293T cells expressing the respective SARS-CoV-2 S protein (or no S protein, control) upon transfection were analyzed for cell surface S protein expression by flow cytometry at 48h posttransfection. For this, cells were washed with PBS, resuspended in PBS-B and pelleted (600 × g for 5 min). Next, the supernatant was removed and cell pellets were resuspended in 250 μL PBS-B containing the monoclonal anti-SARS-CoV-2 S antibody imdevimab (or an isotype human control antibody, hIgG) at a concentration of 1 μg/mL. Following an incubation for 60 min at 4°C in a Rotospin test tube rotator disk (IKA) cells were pelleted, washed with PBS-B and in 250 μL PBS-B containing anti-human AlexaFluor-488-conjugated antibody (1:200; Thermo Fisher Scientific). Following an additional incubation for 60 min at 4°C, cells were pelleted, washed with PBS-B, fixed by incubation in 4% paraformaldehyde solution for 30 min at RT, washed again and resuspended in 150 μL PBS-B before being subjected to flow cytometric analysis using an LSR II flow cytometer (BD Biosciences). Data were further analyzed using the Flowing software version 2.5.1 (https://bioscience.fi/services/cell-imaging/flowing-software/) in order to determine the geometric mean channel fluorescence.
Neutralization assay
For neutralization experiments, S protein bearing pseudotype particles were pre-incubated for 30 min at 37°C with different concentrations of casirivimab, imdevimab, bamlanivimab, etesevimab, casirivimab + imdevimab, bamlanivimab + etesevimab or unrelated control human IgG (2, 0.2, 0.02, 0.002, 0.0002, 0.00002 μg/mL). Alternatively, pseudotype particles were pre-incubated with different dilutions of convalescent plasma (dilution range: 1:50 to 1:12,800) or serum from vaccinated individuals (dilution range: 1:25 to 1:6,400). Following incubation, mixtures were inoculated onto Vero cells with particles incubated only with medium serving as controls. Transduction efficiency was determined at 16–18 h postinoculation as described above.
Quantification and statistical analysis
The results on S protein-driven cell entry represent average (mean) data acquired from five to nine biological replicates, each conducted with four technical replicates. The transduction was normalized against SARS-CoV-2 S B.1 (= 1). Alternatively, transduction was normalized against the background signal (luminescence measured for cells inoculated with particles bearing no viral glycoprotein; set as 1). For ACE2 binding analyzed by flow cytometry, presented are the average (mean) normalized (B.1 set as 1) data from nine biological replicates, each conducted with single samples, while for ACE2 binding analyzed by CABA, the average (mean) data of three biological replicates (each performed with four technical replicates), for which binding was normalized against the background (no soluble ACE2-Fc = 1), is shown. The results on neutralization of spike protein-driven cell entry by monoclonal antibodies and IgG represent average (mean) data from a single biological replicate (conducted with technical quadruplicates) for which transduction was normalized against samples that did not contain any antibody (= 0% inhibition). The data were confirmed in a separate independent experiment. The results on neutralization of spike protein-driven cell entry by convalescent plasma or serum from vaccinated individuals are based on a single experiment, which was conducted with technical quadruplicates. For data normalization, the plasma/serum dilution factor that leads to 50% reduction in S protein-driven cell entry (neutralizing titer 50, NT50) was calculated. In addition, for each plasma/serum the fold change in NT50 between SARS-CoV 2 B.1 (set as 1) and the indicated variants was calculated.
Error bars are defined as either standard deviation (SD) or standard error of the mean (SEM). Data were analyzed using Microsoft Excel (as part of the Microsoft Office software package, version 2019, Microsoft Corporation) and GraphPad Prism 8 version 8.4.3 (GraphPad Software). Statistical significance was analyzed by two-tailed Student’s t test with Welch correction (pseudotype entry, ACE2 binding), two-tailed Mann-Whitney test with 95% confidence level (neutralization), or two-way analysis of variance (ANOVA) with Dunnett’s post-hoc test (ACE2 binding). Only p values of 0.05 or lower were considered statistically significant (p > 0.05, not significant [ns]; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗). Details on the statistical test and the error bars can be found in the figure legends.
Acknowledgments
The authors thank Roberto Cattaneo, Georg Herrler, Stephan Ludwig, Andrea Maisner, Thomas Pietschmann, and Gert Zimmer for providing reagents. S.P. acknowledges funding by BMBF (01KI2006D, 01KI20328A, 01KI20396, and 01KX2021), the Ministry for Science and Culture of Lower Saxony (14-76103-184, MWK HZI COVID-19), and the German Research Foundation (DFG; PO 716/11-1, PO 716/14-1). M.S.W. received unrestricted funding from Sartorius AG, Lung research. H.-M.J. received funding from BMBF (01KI2043, NaFoUniMedCovid19-COVIM: 01KX2021), Bavarian State Ministry for Science and the Arts, and Deutsche Forschungsgemeinschaft through the research training groups RTG1660 and TRR130. G.B. acknowledges funding by German Center for Infection Research (grant no. 80018019238).
Author contributions
S.P. and M.H. conceived the project. P.A. and M.H. designed the experiments. S.P. and M.H. wrote the manuscript, all authors revised the manuscript. P.A., A.K., I.N., and L.G. performed experiments. P.A., M.H., and S.P. analyzed the data. M.S.W, M.L., S.S., H.-M.J., A.C., M.V.S., and G.M.N.B. provided essential reagents.
Declaration of interests
M.S.W. received unrestricted funding for independent research projects from Sartorius.
Published: May 3, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110754.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the Lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
References
- Arora P., Sidarovich A., Kruger N., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., Schulz S., Jack H.M., et al. B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep. 2021;37:109825. doi: 10.1016/j.celrep.2021.109825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 2011;6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann C., Hoffmann M., Lubke A., Nehlmeier I., Kramer-Kuhl A., Winkler M., Pohlmann S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS One. 2017;12:e0189073. doi: 10.1371/journal.pone.0189073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin E.T., Leidner D., Zhang Y., Long E., Prince L., Li Y., Andrews J.R., Studdert D.M., Goldhaber-Fiebert J.D., Salomon J.A. Effectiveness of the mRNA-1273 vaccine during a SARS-CoV-2 delta outbreak in a prison. N. Engl. J. Med. 2021;385:2300–2301. doi: 10.1056/NEJMc2114089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan M., Nirula A., Azizad M., Mocherla B., Gottlieb R.L., Chen P., Hebert C., Perry R., Boscia J., Heller B., et al. Bamlanivimab plus etesevimab in mild or moderate Covid-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e2379. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Gross R., Seidel A., Hornich B.F., Hahn A.S., Kruger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Hofmann-Winkler H., Kruger N., Kempf A., Nehlmeier I., Graichen L., Arora P., Sidarovich A., Moldenhauer A.S., Winkler M.S., et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36:109415. doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kruger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Zhang L., Kruger N., Graichen L., Kleine-Weber H., Hofmann-Winkler H., Kempf A., Nessler S., Riggert J., Winkler M.S., et al. SARS-CoV-2 mutations acquired in mink reduce antibody-mediated neutralization. Cell Rep. 2021;35:109017. doi: 10.1016/j.celrep.2021.109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2021;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara A., Undurraga E.A., Gonzalez C., Paredes F., Fontecilla T., Jara G., Pizarro A., Acevedo J., Leo K., Leon F., et al. Effectiveness of an inactivated SARS-CoV-2 vaccine in Chile. N. Engl. J. Med. 2021;385:875–884. doi: 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Wang L., Graham B.S., Muller M.A., Drosten C., Pohlmann S., Hoffmann M. Mutations in the spike protein of middle east respiratory syndrome coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J. Virol. 2019;93 doi: 10.1128/JVI.01381-18. e01381–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e9. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., Vogels C.B.F., Yildirim I., Rothman J.E., Lu P., Monteiro V., Gelhausen J.R., Campbell M., Silva J., Tabachikova A., et al. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature. 2021;600:523–529. doi: 10.1038/s41586-021-04085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I., Datir R., Collier D.A., Albecka A., Singh S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Perez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis B.Y., Barda N., Leshchinsky M., Kepten E., Hernan M.A., Lipsitch M., Dagan N., Balicer R.D. Effectiveness of BNT162b2 vaccine against delta variant in adolescents. N. Engl. J. Med. 2021;385:2101–2103. doi: 10.1056/NEJMc2114290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cardenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh A., McMenamin J., Taylor B., Robertson C., Public Health S., the E.I.I.C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh A., Robertson C., Taylor B. BNT162b2 and ChAdOx1 nCoV-19 vaccine effectiveness against death from the delta variant. N. Engl. J. Med. 2021;385:2195–2197. doi: 10.1056/NEJMc2113864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Cornelius A., Herati R.S., Mulligan M.J., Landau N.R. High-titer neutralization of Mu and C.1.2 SARS-CoV-2 variants by vaccine-elicited antibodies of previously infected individuals. Cell Rep. 2022;38:110237. doi: 10.1016/j.celrep.2021.110237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. Elife. 2021;10:e69091. doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.A., Kaneda A., Nakagawa S., Sato K., Genotype to phenotype Japan, Consortium Neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine serum. N. Engl. J. Med. 2021;385:2397–2399. doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the Lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.