Abstract
The coronavirus disease 2019 (COVID-19), caused by the SARS-CoV-2 virus, is a current pandemic that has resulted in nearly 250 million cases and over 5 million deaths. While vaccines have been developed to prevent infection, and most COVID-19 cases end up being fairly light, there are severe cases of COVID-19 that may end up in death, even with adequate healthcare treatment. New options to combat this disease’s effects, therefore, could prove to be invaluable in saving lives. Adamalysins are proteins that have several roles in regulating different functions in the human body but are also known to have functions in inflammation. They are also known to have roles in several different diseases, including COVID-19, where ADAM17, in particular, is now well-known to have a prominent role, but also several diseases which include comorbidities that may worsen cases of COVID-19. Therefore, investigating the functions of adamalysins in disease may give us clues to the molecular workings of COVID-19 as well as potentially new therapeutic targets. Understanding these molecular mechanisms may also allow for an understanding of the mechanisms behind the rare severe side effects that occur in response to current COVID-19 vaccines, which may lead to better monitoring measures for people who may be more at risk of developing these side effects. This review investigates the known roles and functions of adamalysins in disease, including what is currently known of their involvement in COVID-19, and how these functions might be involved.
Keywords: SARS-CoV-2, COVID-19, Inflammation, ADAM, ADAMTS, ADAMTSL
Graphical Abstract
1. Introduction
The current pandemic that the world is facing known as coronavirus disease 2019 (COVID-19), has had 247,472,724 confirmed cases throughout the world, with 5012,337 reported deaths, as of the 4th of November of 2021, according to the World Health Organization (WHO) [1].
In this review, we investigate the known roles of the adamalysin family of proteins, particularly those in COVID-19, and whether other adamalysins could have roles in COVID-19 progression and be new therapeutic targets.
1.1. SARS-CoV-2, COVID-19 and Symptoms
COVID-19 is caused by the severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2), an enveloped RNA virus whose genome sequence is 79.5% identical to SARS-CoV, the virus responsible for SARS [2], [3], [4]. This virus is first known to have appeared in December 2019, when pneumonia cases of unknown etiology appeared in Wuhan, China. Spreading from there, this disease was officially declared a pandemic by the WHO in March 2020 [4], [5].
After entry into the cell, SARS-CoV-2 utilizes the host cell’s ribosomes to replicate and induces cytotoxicity, which may lead to organ failure [3]. Typically, the virus first infects the upper airways, remaining restricted there in most infected individuals, causing either asymptomatic or mild respiratory disease. Severe disease occurs when SARS-CoV-2 spreads and infects the distal airways, where inflammation and injury to the gas exchange areas of the lung leads to complications [6]. Upon viral replication, the production and exocytosis of new viral particles can induce the host cell to release damage-associated molecular pattern (DAMP) signals which, in the lung, can be recognized by adjacent epithelial and endothelial cells and induce activation of nuclear factor-κB (NF-κB) and interferon regulatory factor 3 (IRF3), resulting in production and secretion of pro-inflammatory cytokines. The acute inflammatory response that has been observed in severe COVID-19 patients involves the production and secretion of many pro-inflammatory mediators such as interleukin-1β (IL-1β), IL-2, IL-6, IL-7, IL-8, interferon (IFN)-γ-induced protein 10 (IP-10), granulocyte colony-stimulating factor (G-CSF), monocyte chemoattractant protein 1 (MCP1), macrophage inflammatory protein (MIP)− 1α, platelet-derived growth factor (PDGF), tumor necrosis factor (TNF)-α and vascular endothelial growth factor (VEGF), among others. This cytokine storm appears to correlate with severity of the disease [7], [8].
After infection, it normally takes an average of 5 days for symptoms to appear, if any, although patients will become infectious an average of 3–4 days after initial infection, creating a period when a patient is infectious for COVID-19 but displays no symptoms. Mild cases may take approximately 2 weeks to recover, but serious or critical cases average 3–6 weeks until recovery. A study of three independent approaches calculated that COVID-19 has an infection fatality rate estimated at a crude range of ≈ 0.3–1.3% [9].
Common symptoms of COVID-19 include fever, cough, fatigue, dyspnoea, chest tightness and sputum production. Less common symptoms are anorexia, diarrhoea and myalgia.[4], [5] In severe cases, the disease may progress and develop microvascular thrombosis, acute respiratory distress syndrome (ARDS) and hypoxic respiratory failure, the latter being a main cause of death in severe COVID-19 patients,[5], [10] along with acute inflammation and disseminated intravascular coagulation [11]. COVID-19-induced effects on the lungs include increased neutrophil and mononuclear cell count, diffuse alveolar injury, epithelial type II cell hyperplasia, thickened alveolar aorta, hyaline formation, thrombosis, fibroblast proliferation and fibrosis. In some cases, multinucleated giant cells were also found. Thrombosis cases include deep venous thrombosis and pulmonary embolism [8].
While pulmonary manifestations of symptoms are the most common in COVID-19, this disease can induce disorders in other organs, such as gastrointestinal complications (such as abdominal pain, nausea, diarrhea and vomiting), cardiovascular injury, coagulopathy (such as thrombosis), liver damage, renal dysfunction (such as acute kidney injury) and neurological disorders [5], [8], [12], [13].
In the cardiovascular system, ACE2 has been demonstrated to be highly expressed in smooth muscle and arterial and venous endothelium in almost all organs, indicating these tissues are potential SARS-CoV-2 targets, inducing endothelial dysfunction, to which hypoxia resulting from COVID-19-induced pulmonary dysfunction contributes. ACE2 high expression in cardiomyocytes makes the heart susceptible to direct SARS-CoV-2-induced cytotoxicity, as well as further myocardial injury due to COVID-19-induced cytokine storm and the hypoxic state resulting from pulmonary dysfunction. Heart disease such as palpitations and chest tightness have been observed with high incidence in severe COVID-19. Other COVID-19-induced cardiovascular dysfunctions include cardiomegaly and mild fibrosis. Early acute myocardial injury is associated to higher mortality risk in COVID-19 [8].
SARS-CoV-2 infection is known to cause coagulation dysfunction in a high proportion of COVID-19 patients, such as thrombotic manifestations like macro- and micro-thrombosis,[8] and thrombotic thrombocytopenia [14], [15], [16], [17], [18] Also contributing to these coagulopathies is the excessive production of pro-inflammatory mediators which leads to imbalance between pro- and anti-coagulant factors and to induction of platelet aggregation, along with increased levels of thrombin, tissue factor V and VIII, fibrinogen and neutrophil extracellular trap (NET) formation [8].
Neurological dysfunction in COVID-19 manifests in symptoms such as headache, confusion, loss of smell and taste, and visual impairment. Autopsies of COVID-19 patients have found cerebral edema, partial neuronal degeneration, lymphocytic encephalitis, meningitis and massive intracranial hemorrhage [8].
Inflammatory conditions may also occur due to SARS-CoV-2 infection, potentially due to the pro-inflammatory overreaction, such as the cytokine storm, that may occur in COVID-19 and can potentially trigger autoimmune inflammatory disorders [19], [20], [21] Cases of adult-onset Still’s disease (AOSD), a rare multisystem inflammatory disorder of as-yet-unknown causes, have been reported after recovery from COVID-19. It is believed to involve aberrant activation of the innate immune system [19], [20], [22] A few cases of anti-citrullinated peptide antibody (ACPA)-positive rheumatoid arthritis were also detected post-COVID-19 and associated with having been triggered by SARS-CoV-2 infection due to the lack of any prior history of inflammatory joint disease and negative testing during the COVID-19 illness period [21], [23]. Furthermore, there have been reports of autoantibodies being detected post-COVID-19.[21] While aberrant pathogenic B- and T-cell-activating CD4 + T-cells that co-express IFN-γ and G-CFS have been found in some severe COVID-19 cases, other severe COVID-19 patients display instead lymphopenia, characterized by a significant reduction of peripheral CD4 + T-cells and CD8 + T-cells.[8].
Asides from virus-induced cytotoxicity, dysregulation of immune responses and thrombo-inflammation, COVID-19 progression also involves endothelial cell injury, dysregulation of the renin-angiotensin-aldosterone system (RAAS) and tissue fibrosis [8].
2. Adamylisins
Adamalysins are a zinc protease superfamily characterized by a multidomain structure that confers their multiple functions. Proteins of the adamalysin family are subdivided into three categories: A disintegrin and metalloproteinase (ADAM), ADAM with thrombospondin motifs (ADAMTS) and ADAMTS-like (ADAMTSL). Of the known adamalysins in humans, there are 20 ADAMs, 19 ADAMTSs and 7 ADAMTSLs. These proteins’ multidomain structure is organized into signal peptide, pro-peptide and metalloprotease, disintegrin and cysteine-rich domains. ADAMs are further distinguished by the presence of additional epidermal growth factor (EGF)-like, transmembrane and cytoplasmic domains, while ADAMTS proteins are characterized by an ancillary domain containing a thrombospondin type 1 repeat (TSR) domain, a spacer domain and additional motifs (excepting ADAMTS4) which determines their function as secreted proteins. ADAMTSLs, on the other hand, lack catalytic and disintegrin domains and are likely to be involved in ECM assembly and may modulate ADAMTS activity [14], [24] Fig. 1 illustrates generalized diagrams of the structure of the adamalysins.
Fig. 1.
General structure and domains of ADAM, ADAMTS and ADAMTSL proteins.
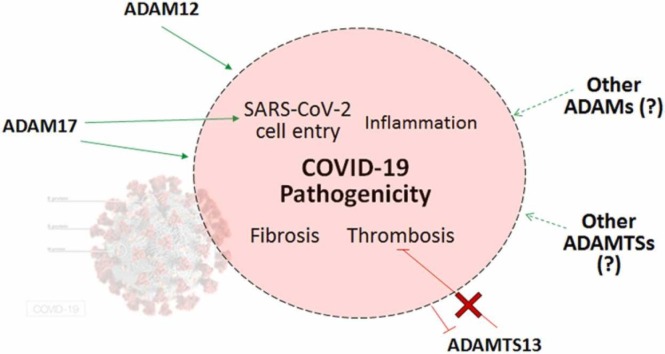
A few adamalysin proteins have been demonstrated to have roles in COVID-19,[5], [12], [13], [19], [20], [21], [25], [26], [27] especially ADAM17 which has been demonstrated to have a key role in this disease’s pathogenesis [25], [26]. Our search of the literature also identified many roles that adamalysins have in pathogenic processes that are also found in COVID-19,[8], [14], [15], [16], [17], [18], [19], [20], [21] and thus could potentially have roles in this disease and prove to be viable therapeutic targets.
2.1. ADAM proteins
ADAM metalloproteinases, which are typically membrane-anchored enzymes [28] and closely related to snake venom metalloproteases, [29] are also known as sheddases due to most of their substrates being membrane-bound precursors, such as growth factors, chemokines, adhesion molecules and their receptors and only a very few ECM components[24] which, upon shedding, their now soluble ectodomains are available to perform their functions. The stub remaining on the cell surface may also be processed further, through regulated intramembrane proteolysis (RIP) by γ-secretases which releases the intracellular domain of the cleaved protein to function as a signaling molecule or transcription factor [28], [30]. Due to there being no known ADAM-specific motif that could be used to predict which molecules might be more likely to be cleaved by ADAM proteins, there is a long and complex list of potential substrates [28].
Most ADAM proteins have a multidomain structure in common, specifically an amino-terminal extracellular domain that includes a chaperone-like prodomain which self-inhibits the ADAM protein, a zinc-dependent metalloproteinase catalytic domain, a disintegrin domain, a cysteine-rich domain with a hypervariable region, an epidermal growth factor (EGF)-like repeat domain, a transmembrane domain and a carboxy-terminal SH3-binding cytoplasmic domain [28]. ADAM10 and ADAM17, however, lack the EGF-like repeat domain, instead having what is referred to as a stalk domain [28]. There are 24 ADAM genes in humans but 4 of these are pseudogenes [29]. Furthermore, of the 20 ADAM proteins in humans, only 12 have a functional catalytic domain. These include ADAM8, 9, 10, 12, 15, 17, 19, 20, 21, 28, 30 and 33 [24]. ADAM2, 7, 11, 18, 22, 23, 29 and 32 lack a functional active site and may instead play roles in protein folding and protein-protein interactions [29]. ADAM1, ADAM3, ADAM5 and ADAM6 are pseudogenes in humans [30], [31], [32], [33]. ADAM27 and 31 are better known by their official designation of ADAM18 and 21, respectively [34], [35]. The prodomain is not only important for correct protein folding of the ADAM protein and intracellular transport through the secretory pathway,[30] but also responsible for self-inhibiting the ADAM proteins, and it is progressively cleaved off as the proteins move from the ER and trans-Golgi network to the cell surface. The ADAM proteins are expressed at the cell surface as dimers, which causes blockage of the catalytic sites due to the close packing putting them near the disintegrin and cysteine-rich domains, causing an auto-inhibitory effect [28].
Expression of ADAMs varies from tissue to tissue and depends on the ADAM in question. For example, ADAM9, 10, 12, 15, 17 and 19 are expressed broadly in somatic tissues [29], [36], [37], [38], [39], [40], [41] while ADAM28 and 33 have a more restricted range [29], [42] and ADAM8 has been found primarily in hematopoietic cells [29] but has also been found to be expressed in cartilage, bones, gonads, thymus and the central nervous system [37]. ADAM8, 9, 10, 12, 15, 17, 19, 28 and 33 have been found to be expressed in immune cells, with ADAM10 and 17 being widely expressed [28], [42].
ADAM proteins have a key role in maintaining tissue homeostasis due to their multiple functions in regulating cell-cell and cell-matrix communication [24] and regulating cell phenotype via their effects on cell adhesion, migration, proteolysis and signaling.[29] They are also well documented to have an important role in regulating the inflammatory processes due to their shedding activity which targets key molecular factors of the immune system [24]. Dysregulation of these functions is often associated with diseases such as cardiovascular disease and asthma, among others [24], [29].
We discuss the ADAM proteins that have been verified to influence COVID-19 severity before discussing some that we propose should be studied in the COVID-19 context to determine if their roles in other diseases could be involved in COVID-19. These ADAM proteins have all been summarized in Table 1.
Table 1.
ADAM proteins that are known to have roles in COVID-19 and those that could potentially be involved in COVID-19 due to their roles in pathogenic processes.
| Name | Expression | (Potential) Functions | Known Roles in COVID-19 |
|---|---|---|---|
| ADAM8 | Hematopoietic cells such as immune cells, cartilage, bones, gonads, thymus, central nervous system | Anti-inflammation | ? |
| ADAM9 | Broadly in somatic tissues (including immune cells such as monocytes, macrophages and neutrophils, fibroblasts, keratinocytes, lungs, colon, kidneys, vascular smooth muscle, secretory organs such as pancreas, nervous system such as hippocampus and the cerebellum, reproductive system) | Inflammation, monocyte/macrophage conversion into multinucleated giant cells, neutrophil activation, preventing recognition by NK cells, shedding ADAM10 |
? |
| ADAM10 | Broadly in somatic tissues (including immune cells, mesenchymal stem cells, urinary bladder) | Most relevant Notch-cleaving protease, ACE2 cleavage, inflammation, cytokine and chemokine processing, antigen-induced T cell proliferation, suppression of T cell activation in exhausted T cells, B cell activation, evasion of NK cell cytotoxicity, inhibition of apoptosis, myeloid cell migration, neutrophil recruitment into the alveolar space | ? |
| ADAM12 | Broadly in somatic tissues (including immune cells, cartilage, bone, liver, muscle, uterus, placenta, brain) and mesenchymal stem cells | Inflammation, neutrophil recruitment, endothelial cell permeability, fibrogenesis-related ECM remodeling, TGF-β signaling activation | Lung injury, pro-inflammatory ephrin-A1 upregulation |
| ADAM15 | Broadly in somatic tissues (including immune cells, hippocampus, cerebellum, chondrocytes, endothelial cells such as in vasculature, mesenchymal stem cells, urogenital system) | Inflammation, monocyte migration, cytokine and chemokine secretion | ? |
| ADAM17 | Broadly in somatic tissues (including immune cells) | Pro- and anti-inflammation, main sheddase of IL-6R, IL-6 trans-signaling pathway, thymocyte development, activation and migration of myeoloid cells such as T cell regulation, B cell activation and leukocyte transendothelial migration, evasion of NK cell cytotoxicity | SARS-CoV-2 cell entry and inflammation |
| ADAM19 | Broadly in somatic tissues (including immune cells, lymphatic system, heart, lungs, bones, brain, spleen, liver, kidneys, testes, placenta, skeletal muscle, mesenchymal stem cells) | Inflammation, fibrosis | ? |
| ADAM28 | Restricted in somatic tissues (including immune cells, epithelial cells, lymphoid tissues, pancreas, gastrointestinal system, respiratory system, urinary bladder) | Inflammation, cell adhesion such as B cell adhesion, T cell immune response, vWF cleavage | ? |
| ADAM33 | Restricted in somatic tissues (including immune cells, smooth muscle cells, fibroblasts, uterus, urogenital system, respiratory system, gastrointestinal system, tongue, endocrine system) | Inflammation, airway remodeling | ? |
2.2. ADAM Involvement in COVID-19
2.2.1. ADAM17
ADAM17 is a phylogenic neighbor of ADAM10, lacking the EGF-like domain and shares many substrates with it [24], [28], [29]. Substrates of ADAM17 include, for example, cytokines such as TNF-α and IL-6, growth factors like EGF and other EGF receptor (EGFR) ligands, adhesion proteins such as L-selectin and e-cadherin, and receptors like IL-6 receptor (IL-6R), TNF receptor 1 (TNFR1), TNFR2, IL-15Rα and EGFR [43]. Further substrates include, but are not limited to, Notch, its ligand Delta-like 1 (DLL1), the pro-apoptotic TNF receptor CD30 and receptor activator of nuclear factor κ-B ligand (RANKL) [24], [29], [30].
Phosphatidylserine (PtdSer) is one of the main acidic and highly negatively charged phospholipid in mammalian cell membranes and its exposure, that is, its trafficking from the inner side of the cell membrane to the outer side also plays a role as a docking site for enzymes, such as protein kinase C (PKC), an enzyme that can induce ADAM17 activity. PtdSer exposure is an important requirement for ADAM17 to exert its sheddase activity. ADAM17’s enzymatic activity can be regulated at the cell membrane through interaction with PKC-induced regulated exposed PtdSer, which induces a conformational change in ADAM17 that pulls its catalytic domain closer to the cell surface in order to cleave substrates. SARS-CoV-2 has been demonstrated to mediate PtdSer exposure, which could be a mechanism for the cleavage of ACE2, TNF-α, IL-6R and other pro-inflammatory molecules that are critical components of COVID-19’s inflammatory processes [11], [28].
2.2.1.1. RAAS, ADAM17 and COVID-19
RAAS has a pivotal role in COVID-19. It normally has a role in maintaining blood pressure homeostasis as well as regulating other important functions dependent on the cellular context, including inflammation and proliferation [2], [26]. In the classic pathway, renin converts angiotensinogen to angiotensin I (Ang I), which is, in turn, cleaved into Ang II by angiotensin-converting enzyme (ACE). Other enzymes are also capable of forming Ang II in tissue-specific RAAS. Ang II then binds to angiotensin type 1 receptors (AT1Rs) in order to induce vasoconstriction and reduce renal sodium excretion via aldosterone stimulation. Ang II is also capable of promoting microvascular thrombosis, coagulopathy, hypofibrinolysis and pro-inflammatory mechanisms. Another enzyme, ACE2, counteracts Ang II’s effects by converting Ang I into Ang 1–9 and Ang II into Ang 1–7, which then binds to the Mas receptor (MasR) to counteract Ang II’s effects. ACE2 ectodomain is recognized as a cleavage site by the sheddases ADAM17 and transmembrane serine protease 2 (TMPRSS2). ADAM17 cleavage results in a biologically active soluble ACE2 (sACE2), while TMPRSS2 cleavage results in an inactive form of sACE2. [5], [26]. TMPRSS2 is also the main pathway by which SARS-CoV-2 enters the cell, in conjunction with the TMPRSS2-cleaved membrane-bound ACE2 [2], [26], [44]. Thus, the balance between the ACE2 forms likely plays a role in COVID-19 progression [26]. In fact, the infection progression of SARS-CoV-2 appears to parallel the ACE2 expression levels in the lungs, with the upper airways’ cells, which are the first infected in COVID-19, expressing larger baseline levels of ACE2 than the cells in the distal airways, such as ATII cells, which have a lower baseline ACE2 expression [6].
An increase in Ang II has been observed to occur as one of the early responses during inflammatory processes, inducing chemotaxis and leading to the massive neutrophilic infiltration seen in the lungs of critically ill patients suffering from infections while also cooperating in T-cell activation. AT1R activation leads to IFN-γ and TNF-α production, enhancing Ang II signaling and antiviral mechanisms such as promoting apoptosis of infected cells. Inflammation is negatively regulated by sACE2 levels, with Ang II-induced AT1R activation upregulating ADAM17 and the functional ADAM17-cleaved sACE2, causing sACE2 to cleave the pro-inflammatory Ang II and modulating inflammation to prevent further tissue damage. As an infectious disease, COVID-19’s inflammatory process in the lungs likely acts in a similar way [26]. Indeed, the spike (S) protein of SARS-CoV-2 induces AT1R-mediated activation of MAPK, inducing transcriptional molecules NF-κB and AP-1/c-Fos, resulting in increased IL-6 secretion. Furthermore, S protein-mediated AT1R signaling also induces ADAM17, which is also the main sheddase for IL-6R, increasing soluble IL-6R (sIL-6R) that, upon binding to IL-6, which can also be shed by ADAM17, and gp130 activates the alternative IL-6 trans-signaling pathway [25], [28], [43] This IL-6 trans-signaling pathway induces the acute phase of the hyper-inflammatory response observed in COVID-19 patients [25]. sACE2 also binds to SARS-CoV-2 and may either block or promote viral entry [26].
The S protein of SARS-CoV-2, an envelope glycoprotein that creates a distinctive corona-like shape,[2] has been demonstrated to decrease full-length ACE2 protein levels and activity, and increase expression of a shorter-length ACE2 in cells and of sACE2, as well as ADAM17 activity [45] and cytokine (such as TNF-α, IL-1β and IL-6) [45], [46] and chemokine (such as fibrosis-associated CCL2) levels in alveolar type-II pneumocytes [45]. While apparently not necessary for cell entry, cleavage of the S protein by TMPRSS2, or, to a lesser extent, by furin proteases or cathepsins, primes the S protein and may facilitate binding to ACE2 by exposing the ACE2-binding domain in S protein [2], [3].
Xavier et al. (2021) suggested that the RAAS pathway could have a significant role in modulating the physiological response to SARS-CoV-2 infection and proposed two hypotheses for the molecular mechanisms and expression changes of the RAAS pathway, based on the available information: one for the best prognosis observed in COVID-19, related to an increase in Ang II/AT1R-mediated ADAM17 upregulation due to increased transmembrane ACE2 shedding by both TMPRSS2 and ADAM17, resulting in limited viral entry and maintained active sACE2-mediated Ang 1–7 levels; another for the worst prognosis, related to a lower Ang II presence and/or AT1R activity due to the presence of a higher level of transmembrane ACE2, resulting in lower ADAM17 activation and higher viral entry. Lower Ang II levels may also result due to use of drug treatments, such as ACE inhibitors, [26] or due to genetic differences, such as people with the blood group A who exhibit lower plasmatic ACE levels and consequently lower Ang II levels, and are at higher risk of developing severe COVID-19 [47].
Despite obesity, diabetes, hypertension, chronic lung disease and cardiovascular diseases being risk groups for developing the worst outcomes of COVID-19, the increased sACE2 levels found in patients with these disorders would seem to contradict the above suggested patterns [2], [4], [26], [48] Xavier et al. put forward two hypothesis to resolve this issue: first, that patients with diabetes and cardiovascular diseases have been found to have high plasmatic levels of Ang II, indicating that the RAAS pathway is highly activated and, consequently, it would be unlikely to produce a new peak of Ang II that would lead to the best case scenario for COVID-19; second, that the increased Ang II levels may lead to AT1R downregulation as a compensatory mechanism, thereby preventing the ADAM17 upregulation needed that would lead to the best case scenario [26]. However, this appears to contrast with studies that reported a protective role of therapeutic sACE2 in COVID-19 [2].
2.2.1.2. RAS/RAAS, ADAM17 and SARS-CoV-2 cell entry
ADAM17 plays a major role in the entry mechanisms of several viruses, including SARS-CoV viruses. ADAM17 activation has not only a major role in ACE2 shedding [45] but also enhances viral entry [49], [50].
SARS-CoV-2 is able to enter cells via 2 main uptake pathways: through membrane fusion and through endocytosis, which could be receptor-dependent (i.e. ACE2-dependent) or receptor-independent endocytosis. In both uptake pathways, the SARS-CoV-2 S protein plays an important role, through either its internal fusion peptide being revealed upon S protein cleavage (membrane fusion), or through the ACE2-binding domain (receptor-dependent endocytic pathway) [2]. It should be noted that another receptor-dependent endocytosis pathway exists via the S protein binding to CD147 which allows entry of SARS-CoV-2 into the cell via ACE2-independent endocytosis. Thus, CD147 potentially allows viral entry into ACE2-deficient cell types [51].
Rahman et al. suggested a few possible events regarding sACE2-mediated COVID-19 severity. sACE2 binds to viral S proteins in the extracellular space, forming a virus-sACE2 complex. This binding potentially induces conformational shifting of S proteins, allowing floating TMPRSS2 or furin catalytic domains to prime the S proteins for membrane fusion cell entry. A virus-sACE2 complex that is only partially covered in sACE2 also means that this conformational shifting may also facilitate the opening of neighboring unbound S proteins to enhance ACE2 binding when the virus-sACE2 complex randomly rolls over the membrane and any available transmembrane ACE2. This binding stops the complex and may either result in membrane fusion or may result in endocytosis. This primed virus-sACE2 may also be able to evade host immune cells, thanks to the sACE2 coating, and travel via the circulatory system to distant organs, whereupon the viral complex can enter the organ’s cells, spreading COVID-19 throughout the body. The binding of sACE2 and membrane ACE2 to the virus and their subsequent entry into the cell also translates to a rapid decrease in available ACE2 in the RAAS pathway, potentially leading to an imbalance in RAAS homeostasis.[2].
2.2.1.3. ADAM17 in Immunity and Inflammation
ADAM17 also has many roles in immunity. For example, asides from the previously mentioned release of TNF-α receptors, TNFR1 and TNFR2, whether or not ADAM17 sheds TNF-α regulates the immune system signaling pathways activated, with TNFR1, which activates inflammatory- and apoptosis-related pathways, being stimulated mainly by soluble TNF-α, while TNFR2, which activates anti-inflammatory and protective pathways, is mainly stimulated by the membrane-bound form [43]. It is involved in the shedding of Notch [30] and can also cleave lymphocyte activating gene 3 (LAG3), preventing LAG3-inhibition of activated T cells, and Fas ligand (FasL), preventing T cell apoptosis. ADAM17 can cleave CD40L on the surface of activated T cells, dendritic cells, mast cells, platelets and stromal cells, leading to prolonged B cell activation, production of pro-inflammatory factors, increase in the number of Treg cells and inducing the immunosuppressive indoleamine 2–3-dioxygenase (IDO) pathway. In acute inflammation, ADAM17 sheds enzymes that control transendothelial migration, cytokine secretion and signaling cascades that regulate the activation and migration of innate myeoloid cells, such as macrophages, dendritic cells and neutrophils [28]. Leukocyte transendothelial migration, a process necessary in leukocyte recruitment to sites of inflammatory stimuli, occurs, in part, via ADAM17-mediated shedding of L-selectin and via shedding of chemokines[29], [52]. ADAM17 causes shedding of the tyrosine kinase receptor Met, releasing the soluble form (sMet) in HCC, resulting in liver damage and inflammation [24].
Given its prominent role in shedding of pro-inflammatory factors, ADAM17 is involved in several human inflammatory diseases, such as rheumatoid arthritis (a systemic inflammatory autoimmune disease), multiple sclerosis (an inflammatory autoimmune demyelinating disease of the central nervous system), alcoholic hepatitis and systemic lupus erythematosus (an autoimmune disease affecting almost all organs) [24], [30]. In lung diseases like asthma, COPD and cystic fibrosis, ADAM17-mediated EGFR signaling is aberrantly activated, which leads to pathological remodeling of the airways. In COPD and cystic fibrosis, this aberrant EGFR activation results in airway epithelial cell wound healing, abnormal airway proliferation and progressive lung tissue scarring.[52] Further mechanisms of ADAM17 action in COPD include ADAM17-mediated activation of TGF-β and thrombin signaling which leads to upregulation and enhanced activity of ADAM17 in a positive feedback loop. Enhanced ADAM17 activity results in cleavage of EGFR ligands, such as HB-EGF, which leads to α-SMA-mediated ECM production and fibrosis. ADAM17-mediated cleavage of IL-6Rα in the lungs may also lead to pulmonary fibrosis[53]. These roles in fibrotic developments could possibly be some of the mechanisms by which fibrosis develops in COVID-19.
ADAM17 may also have a role in shedding of major histocompatibility complex class I-related chain A (MICA) in order to prevent tumor cell recognition by natural killer (NK) cells, thus evading anti-tumor immunity [24]. ADAM17 further reduces the antitumor activity of immune cells by shedding T-cell immunoglobulin and mucin domain containing protein 3 (TIM3), a cell surface receptor of CD4 + CD8 + T-cells, shedding CD16, which results in a decrease of expression of IFN-γ and other cytokines necessary for immune cell activation, and shedding PD-L1 from tumor cells, leading to T-cell apoptosis and compromising the killing ability of CD8 + T-cells [43]. While these immune evasion mechanisms by ADAM17 have been demonstrated in cancers, it is not out of the question that they may play a role in SARS-CoV-2’s ability to evade antiviral mechanisms [54].
2.2.2. ADAM12
This adamalysin’s wide expression in several tissues tends to mostly involve roles in cell adhesion and fusion, ECM restructuring and cell signaling [41]. ADAM12 interacts and stabilizes TGFBRII receptor, promoting TGF-β signaling pathways through increased receptor trafficking,[24] which could potentially be involved in COVID-19 fibrosis.
miR-29 suppresses transcription of ADAM12 but NF-κB activation represses transcription of miR-29, which in turn upregulates ADAM12 transcription, suggesting that NF-κB-mediated inflammation, such as occurs in COVID-19, may upregulate expression of this protein. In fact, ADAM12 has been found to be upregulated in response to TNF-α [41]. ADAM12, as well as one of its substrates, ephrin-A1, are known to be involved in inflammation, particularly in the lungs for ephrin-A1, and regulate endothelial cell permeability [27]. In COVID-19, some patients have been identified to have significant elevated blood serum levels of ADAM12 and ephrin-A1, which likely play a role in COVID-19-induced lung injury given their role in inflammation. The clinical outcome of COVID-19 has been found to correlate with the serum levels of these two proteins, with the critical patients having the highest serum levels and the milder patients having the lowest. Ephrin-A1-mediated inflammation may be an important factor in COVID-19 morbidity and mortality [27].
Given ADAM12’s role in tissue and ECM remodeling, which are effects observed in chronic asthma, it is believed that ADAM12 contributes to bronchial airway remodeling and recruitment and accumulation of neutrophils in the airway mucosa [41]. Since ADAM12 activity is already know to play a role in COVID-19, these effects could also potentially play a part in COVID-19 progression.
2.3. ADAMs with potential roles in COVID-19
Other ADAM proteins have been found to have roles in pathogenic processes in other diseases that also occur in COVID-19. Therefore, they could potentially have roles in COVID-19 progression and severity and may prove to be viable therapeutic targets.
Several ADAMs have been shown to promote inflammation or other immune responses. Among these is ADAM9, which can mediate conversion of monocytes-macrophages into multinucleated giant cells as a response to foreign bodies or bacteria, resulting in granulomatous lesions that help to isolate pathogens and enhance phagocytosis. It is expressed in neutrophils, where, upon neutrophil activation and trafficking of ADAM9 to the cell surface, it can promote ECM degradation and interact with integrins to activate neutrophils, which can lead to formation of NETs. It also sheds TNF-α and IL-11 receptor, which play a role in inflammation [36]. Its role in inflammation and the involvement observed in lung injury and in cardiovascular and lung diseases [36] indicate a possibility of a role in COVID-19-related similar processes. ADAM10 is a phylogenic neighbor of ADAM17 [24], [28] and thus shares many substrates in common with it, including Notch, [24], [29], [30] and has been observed to have a prominent role in the immune system. In vivo, ADAM10 is the sheddase most relevant in Notch cleavage, [28], [30] where, among the many functions ADAM10-mediated Notch shedding has in the immune system is, for example, antigen-induced T cell proliferation.[28] ADAM10 displays roles in TCR ligation, like ADAM17, suppresses T cell activation in exhausted CD8 + T cells, regulates T cell survival and effector function, prolongs B-cell activation via shedding CD40L (like ADAM17), as well as produces pro-inflammatory factors. Another role for ADAM10 in immunity that it shares with ADAM17, specifically in acute inflammation, is in the recruitment and tissue infiltration of innate myeloid cells, where it is one of the essential shedding enzymes (along with ADAM17) that controls transendothelial migration, cytokine production and intracellular signaling cascades in these cells. ADAM10 is also necessary for recruitment of neutrophils to the alveolar space [28]. ADAM15 has been observed to mediate release of pro-inflammatory cytokines, such as TNF-α, IL-6 and IL-15, and chemokines, such as CXCL12, in rheumatic fibroblast-like synoviocytes, displaying a role in rheumatoid arthritis [55]. ADAM15 has also been demonstrated to have a role in impaired endothelial barrier dysfunction, transmigration of monocytes and inflammation in atherosclerosis as well as in sepsis, [39] and has also been linked to COPD pathogenesis.[56] ADAM19 is capable of cleaving KL1 and TNF-α, albeit at a reduced rate compared to ADAM17, thus suggesting a role in inflammation [37], [40]. ADAM28 sheds TNF-α and growth factors such as IGF binding protein-3 and connective tissue growth factor (CTGF). It has also been shown to have a function as an adhesion factor, such as binding to lymphocyte integrins, [57] and aiding B cell adhesion, [58] has been reported to promote T cell immune response.[24] Lastly, ADAM33 has also displayed potential in inflammation by having been demonstrated to cleave some canonical ADAM substrates such as KL-1 and TNF-α, albeit this has so far only been observed in vitro [37]. Accumulating evidence suggests a key role for ADAM33 in the pathogenesis of diseases that involve airway remodeling, [59] thus it could play a role in promoting COVID-19 progression.
ADAM8 also displays a role in inflammation, although instead of promoting inflammation, this ADAM protein dampens inflammation by shedding TNFR1 and thus inhibiting the pro-inflammatory TNF-α-induced signaling. Given that ADAM8 is also upregulated by TNF-α, [37] this adamalysin may already be found to be upregulated in COVID-19 but is insufficient to stop inflammation. Targeting other mechanisms that mediate ADAM8 expression to promote upregulation may be a potential therapeutic approach, not just in COVID-19, but also in other diseases where TNF-α plays a major role in their pathogenesis. Also, much like ADAM17, ADAM9 and ADAM10 contribute to ADAM-dependent shedding of MICA, preventing tumor cell recognition by NK cells and thus evading anti-tumor immunity, [24] which could play a role in SARS-CoV-2’s immune evasion. ADAM10 is also able to induce the immunosuppressive IDO pathway [28].
ADAM8 is also able to cleave ECM components, such as fibronectin and the soluble form of its catalytic domain may aid in ECM degradation, [37] indicating a potential role in ECM remodeling. ADAM19 has also been associated with fibrosis of the lung and kidney, [40] thus potentially playing a role in the fibrosis observed in COVID-19 patients.
Given ADAM10’s substrate list significantly overlapping that of ADAM17 which has a significant role in COVID-19, this indicates that ADAM10 and its regulation could have significant influence in COVID-19 progression. ADAM9 and ADAM15 have been observed to be able to shed ADAM10, [36], [37] thus displaying some regulation over it.
Interestingly, ADAM28 has been demonstrated to be able to cleave von-Willebrand factor (vWF), a protein involved in coagulation that is produced as large multimeric proteins [42]. Its ability to cleave vWF suggests a potential involvement for this ADAM protein in thrombotic events in COVID-19.
ADAM9 may potentially be upregulated in COVID-19 patients via COVID-19-induced IL-6 secretion, [25] as IL-6 enhances ADAM9 expression through activating the JNK pathway [36]. Just as ADAM17, ADAM10 sheddase activity has been observed to require exposure of PtdSer, thus SARS-CoV-2-mediated PtdSer exposure likely plays a role in ADAM10’s activity during COVID-19 [11]. ADAM10 has also been correlated to ACE2 cleavage regulation in human airway epithelial cells and has also been found to be upregulated in several comorbidities linked to severe COVID-19 [60]. ADAM15 has also been demonstrated to be upregulated in response to Ang II and protecting against abdominal aortic aneurysm [39]. Given the increase in Ang II observed in COVID-19 [25], [26], this suggests ADAM15 may be upregulated by this disease.
2.4. ADAMTS and ADAMTSL proteins
The ADAMTS proteins are a superfamily of proteases that are secreted, multi-domain matrix-associated zinc metallopeptidases[14], [15] which typically classified according to their substrates. These classifications include the aggrecanases and proteoglycanases (ADAMTS1, 4, 5, 8, 9, 15 and 20), the pro-collagen N-propeptidases (ADAMTS2, 3 and 14), the cartilage oligomeric matrix protein-cleaving (COMP) proteinases (ADAMTS7 and 12) and the vWF proteinase (ADAMTS13). There is also a group of orphan enzymes (ADAMTS6, 10, 16, 17, 18 and 19) [14], [24]. However, new substrates have been identified for many of these proteins, expanding the pool of substrates cleaved by ADAMTS proteins [24]. The designation ADAMTS11 is no longer used due to having been assigned to a gene that was identified to be ADAMTS5. ADAMTS expression can be found in a wide variety of tissues and cells in the body [14], [24], [61]. The 7 ADAMTSL proteins in humans include ADAMTSL1 to 6 and papilin [15], [24], [62], [63] Protein expression of ADAMTSLs appears to differ from protein to protein but mRNA expression of them has been found ubiquitously throughout the body [15], [64], [65], [66], [67].
Along with ADAMTSL proteins, ADAMTS proteins have a critical role in microfibril formation, stabilization and functions, and therefore ECM assembly and regulation of growth factors bioavailability.[14], [15], [24], [62], [63] Much like ADAM proteins, ADAMTS proteins also have important functions in cell-cell and cell-matrix regulation, maintaining tissue homeostasis, tissue remodeling, vascular biology, inflammation and cell migration [14], [24]. ADAMTSL proteins may also have a function in modulating activity of ADAMTS proteins, which might be related to their ECM assembly role [14], [15].
The structure of ADAMTS is composed of three major domains: a proteinase domain, which is responsible for the cleavage action of the protein, a central thrombospondin type 1 sequence repeat (TSR) motif and the ADAMTS hallmark non-catalytic ancillary domain, which is responsible for substrate recognition, ADAMTS localization, interaction partners and shares homology with ADAMTSL proteins [14], [15], [24] In more detail, the proteinase domain is composed of a signal peptide, a pro-domain of variable length, a metalloproteinase domain and a disintegrin-like domain, while the ancillary domain is composed of a cysteine-rich domain and following that a spacer region, which might then be followed by further modules, including additional TSRs, depending on the ADAMTS.[14], [24] Secretion, localization or activation of ADAMTS proteins may be further regulated by post-translational modifications such as, among others, pro-domain excision, glycosylation or proteolytically processing their ancillary domains and releasing C-terminal fragments [14], [15] Like ADAM proteins, ADAMTS proteins display limited susceptibility to inhibition by the four TIMPs, with TIMP3 being the most effective inhibitor [14]. ADAMTSLs resemble ADAMTS ancillary domains and lack proteolytic activity, and similar to ADAMTS proteins, post-translational modifications, such as fucosylation, regulates ADAMTSL localization, secretion, activity and functions.[15].
ADAMTSL proteins also have the ability to modulate activity of ADAMTS proteins, such as papilin being able to inhibit ADAMTS2 [68]. However, it is ADAMTS13’s ability to cleave vWF that has been found to have a role in COVID-19 [17], [18], [69].
A summary of the ADAMTS and ADAMTSL proteins with potential roles in COVID-19 is available in Table 2, while a summary of all of the existing ADAMTS proteins and of the ADAMTSL proteins, their known roles and the diseases they influence can be found in Supplementary Table 2 and Supplementary Table 3, respectively.
Table 2.
ADAMTS and ADAMTSL proteins’ expression in the human body and their functions, known and potential, in COVID-19.
| Name | Expression | (Potential) Functions | Known Roles in COVID-19 |
|---|---|---|---|
| ADAMTS1 | Ovaries, placenta, uterus, fetal lungs, bronchial epithelial cells, smooth muscle, adrenal cortex, adipocytes, ciliary ganglions, prostate, olfactory bulb, breast stromal fibroblasts, myoepithelial cells | TGF-β activation, fibrosis | ? |
| ADAMTS2 | Adipocytes, skeletal muscle, smooth muscle, uterus, placenta, heart, liver, lungs, tongue, breast stromal fibroblasts | TGF-β pathway regulation | ? |
| ADAMTS3 | Pineal gland, cartilage, bone, skeletal muscle, tendons, endothelial cells, CD34 + cells, breast myoepithelial cells | TGF-β pathway regulation | ? |
| ADAMTS7 | Trigeminal ganglion, adrenal cortex, intervertebral discs, liver, heart, skeletal muscle, breast stromal fibroblasts | Positive feedback loop with TNF-α, delay of artery repair | ? |
| ADAMTS8 | Superior cervical ganglion, adrenal cortex, skeletal muscle, heart, liver, luminal epithelial cells, breast stromal fibroblasts | Pro-inflammation | ? |
| ADAMTS10 | Brain, uterus, CD8 + T cells, breast stromal fibroblasts | TGF-β signaling regulation | ? |
| ADAMTS13 | Lungs, thyroid, CD71 + early erythroid cells, breast myoepithelial cells, predominantly expressed in liver | vWF protease, coagulation | Thrombotic events such as microvascular thrombosis and thrombotic thrombocytopenic purpura |
| ADAMTS14 | Thalamus, cerebellum, bone, bone marrow, fetal thyroid, skin, adipocytes, fibroblasts, breast myoepithelial cells, luminal epithelial cells | TGF-β pathway regulation | ? |
| ADAMTS19 | Dorsal root ganglion, breast myoepithelial cells | NF-κB inhibition | ? |
| ADAMTSL2 | Blood plasma, heart (such as cardiac myofibroblasts), adipocytes, eyes, pancreas, testes, urine | TGF-β pathway negative regulation | ? |
2.4.1. ADAMTS13
ADAMTS13’x main role is cleaving vWF, processing large multimeric vWF precursor proteins to generate vWF proteins of optimal size to be used for blood coagulation[14], [15].
Autoantibodies against ADAMTS13 lead to decreased activity result in acquired thrombotic thrombocytopenic purpura, a rare and rapidly fatal disorder if left undiagnosed and untreated, which is caused by the inability to cleave pro-thrombogenic large vWF multimers in circulation, leading to vWF-platelet aggregation, vessel occlusion and microvascular thrombosis due to endothelial injury [14], [15], [16] This disorder is also characterized by the levels of ADAMTS13 being less than 10% of normal [69].
The pro-inflammatory molecules IL-1β and C reactive protein, which are upregulated in COVID-19 patients, mediate the accumulation of large vWF multimers in plasma, likely by these pro-inflammatory molecules inhibiting activity of ADAMTS13,[11] seeing as in a study no anti-ADAMTS13 antibodies have been found in COVID-19 patients, although excess of vWF has been observed to induce consumption of ADAMTS13 [13]. This potentially plays a role in the higher thrombotic risk observed in COVID-19 patients, which is characterized in many patients by a rise in the vWF:ADAMTS13 ratio [17], [18], [69] This ratio was also found to correlate with disease severity, being the highest in patients with the worst severity of COVID-19 or those who died from it [17], [69] One study revealed that, at least in a subset of COVID-19 patients admitted to hospital, mortality inversely correlated with ADAMTS13 activity, with the lower the level of ADAMTS13 activity, the higher the risk of death.[13] Another study, however, detected no significant change in ADAMTS13 activity in COVID-19 patients although vWF levels were substantially increased, leading to the suggestion that the massive amounts of multimeric vWF lead to a relative deficiency of ADAMTS13 and to coagulopathy disorders typically observed in cases of absolute deficiency of ADAMTS13. The massive levels of vWF observed in COVID-19 is likely as a result of endothelial injury commonly observed in this disease, as endothelial activation is known to be associated with increased production of vWF [70].
2.4.2. Potential roles of other ADAMTS proteins in COVID-19
ADAMTS7 appears to work in a positive feedback loop with TNF-α, thus may contribute to inflammation in COVID-19 in this manner, although the mechanism behind this feedback loop is not yet understood [71]. ADAMTS8 has also been proven to have a pro-inflammatory role in nasopharyngeal carcinoma,[61] thus it is not out of the question that it could also play a role in driving the inflammatory processes of COVID-19. ADAMTS19, on the other hand, has demonstrated to be able to bind to cytoplasmic p65 and decrease nucleus phosphorylation of NF-κB, one of the key transcription factors in mediating expression of inflammatory factors,[72] thus it could it have some influence in COVID-19 if its expression is affected by the disease.
The role that ADAMTS and ADAMTSL proteins play in ECM assembly could potentially have important relevance in COVID-19,[14], [24], [68], [73], [74] particularly in fibrosis observed in patients. In fact, all the ADAMTSL proteins have been demonstrated to be upregulated in fibrotic hearts,[65] suggesting a potential role in fibrosis-related processes, although only ADAMTSL2 has been demonstrated to negatively regulate the TGF-β pathway, and thus possess anti-fibrotic properties [15], [65] ADAMTS1 has also been demonstrated to have a role in TGF-β activation and fibrosis in the liver while ADAMTS2, ADAMTS3 and ADAMTS14 are other proteins found to mediate the TGF-β pathway [24] and ADAMTS10 is a component of a TGF-β-regulating extracellular network centered in fibrillin microfibrils.[15] It is therefore possible that these ADAMTS proteins may play a role in fibrosis in COVID-19.
Expression levels of ADAMTS7 have been linked to risk of cardiovascular events, such as coronary artery disease [66]. This ADAMTS could have a potential role in cardiovascular disorder events that may occur in more severe COVID-19.
Pro-inflammatory cytokines such as TNF-α, IL-1 and IL-6 have been observed to upregulate expression of several ADAMTS proteins, such as ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS6, ADAMTS7 and ADAMTS9, [14], [71] and thus it is a possibility that COVID-19-induced inflammatory cytokines may also cause upregulation of these ADAMTSs and their function may contribute to the progress of the disease.
2.5. Adamalysin expression in COVID-19 patients
We have performed a search of the GEO DataSets database from the National Center for Biotechnology Information website for any gene expression studies performed on human samples derived from COVID-19 patients that tested for adamalysin expression. For this purpose, we utilized the keyword ‘COVID-19’ and the filter ‘Expression profiling by array’, resulting in 11 results, of which only 2 fit our criteria (dataset series GSE177477 [75] and GSE164805 [76]).
The dataset series GSE177477, part of a study of blood leukocyte transcriptomes of COVID-19 patients and healthy controls done by Masood et al., involves a comparison between COVID-19 patients exhibiting symptoms, asymptomatic (or very mild) COVID-19 patients and healthy controls. This dataset reveals that, in a comparison between asymptomatic COVID-19 cases against healthy individuals, ADAM28 was found to be significantly downregulated, while in a comparison between symptomatic versus asymptomatic cases, ADAM8, ADAM9, ADAM17 and ADAM32 were found to be significantly upregulated [75].
The GEO dataset series GSE164805 is a microarray analysis of the whole genome transcriptome of peripheral blood mononuclear cells of patients suffering from severe COVID-19, patients suffering from mild COVID-19 and healthy controls. It is part of a study investigating transcriptional signatures of severe COVID-19 by Zhang et al. ADAMTS6 and ADAMTS16 expressions are elevated in both mild and severe COVID-19 patients compared to healthy controls while ADAM28 and ADAM30 expressions are downregulated. ADAMTS3 and ADAMTS9 are upregulated and ADAMTS13 is downregulated in the mild cases of this study but the same is not observed in severe cases. ADAM17, ADAMTS7, ADAMTS15 and papilin are upregulated and ADAM11, ADAM15 and ADAMTSL3 are downregulated in the severe cases compared to healthy controls, but this is not observed to a significant effect in mild cases. In comparing adamalysin expression in severe cases versus the mild cases, ADAM8 was also found to be upregulated in this comparison, despite not being found to be significantly upregulated (at least 2-fold according to the authors of the study's choice) in comparison against healthy controls.[76].
Both dataset series reveal a downregulation of ADAM28 and an upregulation of ADAM8 and ADAM17 in COVID-19 patients, although it should be noted that the sample pool was relatively small [75], [76] (11 symptomatic patients, 18 asymptomatic patients and 18 healthy controls for the Masood et al. study [75], and 5 severe COVID-19 patients, 5 mild patients and 5 healthy controls for the Zhang et al. study [76]) and involved only leukocytes in the blood.[75], [76] Given ADAM28’s ability to cleave vWF, the downregulation of this adamalysin observed in both studies could be an indication of higher risk of thrombotic events in these patients. The observed ADAM8 upregulation may be indication of TNF-α-induced upregulation as an attempt to inhibit inflammation, and, given ADAM17’s known important role in COVID-19 entry into the cell and in promoting inflammation, it is unsurprising to find its upregulation detected in both studies.[25], [28], [37], [42], [43], [75], [76] These two studies also identified modulation of expression of different adamalysins from each other. In the Masood et al. study, ADAM9 upregulation was detected in severe patients, which is in agreement with previous suggestion that this upregulation may occur in this disease due to IL-6 secretion seen in COVID-19. This ADAM may also play a role in promoting NET formation and immunity [24], [25], [75] In the Zhang et al. study, the downregulation of ADAMTS13 in mild cases compared to healthy controls suggests a potential for higher thrombosis events in these patients, while in severe cases, ADAMTS7, which has been linked to promoting inflammation and risk of cardiovascular events, has been found to be upregulated. This could be potential indicators of higher risk for cardiovascular disorders in these patients. [14], [15], [24], [57], [66], [71], [76].
3. COVID-19, comorbidities and the role of adamalysins
COVID-19 severity increases in patients also suffering from other comorbidities, such as hypertension, cardiovascular disease, obesity, diabetes, respiratory disorders and kidney disease [3], [4], [8], [12], [48], [60]. These comorbidities have a common denominator in the fact that they induce endothelial dysfunction, which has also been observed to be caused by COVID-19, a fact that leads to multi-organ immunothrombosis, contributing to severe COVID-19 [5]. Adamalysins also play several roles in many such comorbidities and in COVID-19 itself [11], [13], [17], [18], [24], [27], [29], [69].
An infection by other viruses, in addition to SARS-CoV-2, is likely to exacerbate the severity of COVID-19. Indeed, a study on infection of influenza A virus demonstrated an upregulation of mRNA of ACE2, and expression of TMPRSS2 and ADAM17, in other words, the molecules required for SARS-CoV-2 uptake. Expression of ACE2 was found to be downregulated in the cells but sACE2 expression was upregulated, which diminishes the available membrane ACE2 to convert Ang II.[6].
Sufferers of cardiovascular disease may also suffer severely from COVID-19. Estrogen affects RAAS, a critical pathway in cardiovascular disease. Estrogen increases ACE2 gene expression in the heart in an estrogen receptor alpha (ERα)-dependent manner, and in the hearts of patients suffering from some cardiovascular diseases, such as left heart valvular disease or ischemic heart disease, ACE2 has been found to correlate positively with ERα. Through its conversion of Ang II to Ang 1–7, ACE2 protects the body from the pro-inflammatory Ang II. Therefore, an increase in expression of ACE2 should ameliorate cardiovascular disease, such as through RAS inhibitors. However, infection with COVID-19 means that SARS-CoV-2 has a greater amount of ACE2 to facilitate viral entrance into the cells. In addition to ACE2, ADAM17 and TMPRSS2 gene expression levels also correlated positively with ERα gene expression, with TMPRSS2 levels showing the highest level of association with ERα levels [77]. In congestive heart failure, a study identified overexpression of ACE2 and furin mRNA expression in pulmonary, cardiac and renal tissues, while ADAM17 and TMPRSS2 mRNA expression was downregulated. The upregulation of ACE2 mRNA along with the downregulation of ADAM17 mRNA suggests greater levels of membrane-bound ACE2 on the cell surface, which is potentially a significant factor in the increased susceptibility of people with this disease to SARS-CoV-2. The observed downregulation of TMPRSS2 mRNA is potentially compensated by the increase of furin mRNA expression [78].
Coagulation abnormalities can also enhance severity of COVID-19. SARS-CoV-2 induces exposure of PtdSer on the cell membrane. This exposure of PtdSer in infected cells may be a possible mechanism by which a coagulation cascade is activated, potentially resulting in disseminated intravascular coagulation (DIC) and thrombi formation [11]. As previously mentioned, SARS-CoV-2 can induce immune factors that inhibit ADAMTS13, [16] and inhibition of ADAMTS13 activity, which prevents cleavage of vWF, results in thromboembolic complications, such as microvascular thrombosis and thrombotic thrombocytopenic purpura, a rapidly fatal disorder if left untreated [14], [15], [16], [17], [18].
COVID-19 patients who also suffer from cancer have been found to have a higher mortality rate [79]. Cancer patients that contract COVID-19 must also contend with the effects of their current cancer therapeutic treatments on the disease. This may be a case of the cancer treatment having beneficial effects against SARS-CoV-2, such as anti-androgens used in prostate cancer treatment which inhibit androgens and prevent their consequent upregulation of expression of TMPRSS2, the latter of which has a key role in prostate cancer. Another situation would be having prejudicial effects in COVID-19 treatment, such as the use of anti-EGFR tyrosine kinase inhibitors, like vandetanib, which prevents EGFR signaling upregulation of ADAM17. Immunosuppressive chemotherapies are also problematic when a patient contracts COVID-19 (or indeed, any other infectious disease) [80]. The tumor microenvironment has a major role to play in tumor progression, including growth, invasion/metastasis and resistance to chemotherapy, involving a complex number of actors. Proteins of the adamalysin family play important roles in this space by modulating cell-to-cell and cell-to-extracellular matrix (ECM) communications [24].
Chronic kidney disease is one of the comorbidities that in COVID-19 may lead to acute kidney injury, which is predictive of mortality and organ dysfunction in patients. Other comorbidities that may lead to this kidney dysfunction include diabetes, obesity and hypertension. ACE2 is found highly expressed in kidney cells, with proximal renal tubular cells and kidney podocytes not only expressing high levels of ACE2 but also co-expressing TMPRSS2, which may facilitate SARS-CoV-2 entry into the cells, driving the RAS system towards its pro-inflammatory branch, resulting in deleterious effects, such as inflammatory cell infiltration, Ang II-induced renal lesions, and ADAM17-mediated proteinuria, tubular hyperplasia and fibrosis. SARS-CoV-2 may also enter the cells via CD147, resulting in activation of partner proteins of CD147, such as cyclophilins and integrins, which results in arousing inflammation and inducing renal tubular damage [12].
Obesity and diabetes are more comorbidities that have been shown to cause susceptibility to develop severe COVID-19. Obesity has an impact on pulmonary function, leading to decreased functional capacity and respiratory system compliance, and may enhance pro-inflammatory conditions, contributing to the increased morbidity associated with obesity in COVID-19 cases. Adipocytes and adipocyte-like cells, such as pulmonary lipofibroblasts, may be the main cells involved in exacerbating COVID-19 severity in obese and diabetic patients, seeing as in these patients the expression of ACE2 is upregulated. Thus, adipose tissue may turn into a potential viral reservoir. In a study of human subcutaneous adipocytes, genes that are known to favor viral infection have been found to be expressed, such as ADAM10, which can play a role in driving inflammation in COVID-19, FURIN, which is one of the main proteases responsible for cleaving the pro-domain that is inhibitory in most ADAMs, including ADAM10 and ADAM17, as well as being capable of cleaving SARS-CoV-2 spike protein, TLR3, which is important for the innate response to viral infection and regulates ACE2 cleavage, KDM5B, which upregulates ACE2, and SIRT1, which has been found upregulated in the lungs of severe COVID-19 patients with comorbidities [60].
Both current and former smokers, as well as individuals suffering from chronic obstructive pulmonary disease (COPD), have been found to overexpress ACE2 in airway cells, explaining, at least in part, the increased risk of severe COVID-19 in these individuals [8].
There are several reports of skin lesions developed in COVID-19 patients. Symptoms from these include lesions found in other viral and non-viral infectious diseases which include, in most reports, maculopapular exanthema, vesicular exanthema and urticarial eruptions. There can also appear skin lesions possibly related to virus-induced or indirectly induced vascular dysfunctions, such as livedo reticularis, petechiae and cutaneous acro-ischemia. Many viral skin lesion symptoms from COVID-19 likely appear due to mechanisms of signaling cascades and immune system activation induced by SARS-CoV-2, however some skin lesions that appear could potentially occur due to more direct interaction between the virus and epithelial cells, causing tissue damage. There is likely overlap between virus-induced symptoms and drug allergy-induced symptoms from the drugs used in COVID-19 treatment. In children, it has been postulated that there is an association between COVID-19 and a Kawasaki-like disease, which is a systemic vasculitis of medium sized vessels. SARS-CoV-2 might also reactivate or worsen pre-existing skin diseases including urticaria, psoriasis and autoimmune diseases [81].
4. COVID-19 therapy
As previously mentioned, reports have claimed that addition of sACE2 decreases Ang II and blocks viral entry both in vitro and in treated patients. However, the opposite effect has been observed in comorbid COVID-19 patients, who have an increased level of sACE2 but suffer the most severe outcomes. A possible reason for this could be the reports themselves, specifically that the experiment parameters applied do not quite mimic host cell infection. This includes, in cell-based experiments, pre-incubating SARS-CoV-2 with sACE2 prior to incubation with selected cell lines, thereby exposing a virus saturated by sACE2 binding to the cells and preventing its binding to membrane ACE2, which does not mimic the competition between membrane ACE2, native sACE2 and therapeutic sACE2 for the virus that would be present in a real case scenario [2]. Other studies have admitted their own limitations [82], [83] Therefore, the inconsistency observed between reports on therapeutic sACE2 administration and the clinical data on COVID-19 patients with comorbidities may be due to, at least in part, the limitations of the reports’ experimental parameters.
Given the role played by RAAS in COVID-19 progression, drugs targeting this system used as treatment options in other diseases (e.g. hypertension, cardiovascular diseases, diabetes), such as AT1R blockers (e.g. candesartan) and ACE inhibitors (e.g. captopril), have the potential to be problematic in regards to the severity of COVID-19 in the lungs [2], [45], [48] This is especially true when considering the double-edged sword that is the key protease ACE2, whose activity is important to switch the RAS pathway from the pro-inflammatory arm to the anti-inflammatory arm but that is also the entry receptor for SARS-CoV-2. It is therefore a major question on whether ACE2 levels in tissues should be promoted, in order to reduce inflammation, or decreased, in order to reduce viral entry. There is also an issue on whether this may increase or decrease the severity of COVID-19. There is, unfortunately, a lack of experimental data regarding this question and what is available is considered controversial [45].
Pedrosa et al. tested the lungs of healthy control rats treated with either captopril or candesartan and found they enhance the anti-inflammatory arm of RAAS. Candesartan and captopril were also shown to inhibit ADAM17 activation, which may be responsible for lowering ADAM17-induced shedding of ACE2 and ADAM17-induced production of TNF-α, IL-6 and CCL2 [45]. However, the effects observed in rats may not be the same observed in humans. Indeed, patients with existing comorbidities are likely to be those who also utilize AT1R blockers and ACE inhibitors and yet still suffer from a higher severity of COVID-19.[2], [84] Despite this, use of these types of drugs has been shown to be associated with improved clinical outcomes and decreased mortality in COVID-19 patients compared to those patients not using these drugs [5], [85], [86], [87].
Use of other established therapies in order to treat other aspects of severe COVID-19 may also present problems. For example, when it comes to managing the inflammatory aspect of COVID-19, utilizing anti-TNF-α monoclonal antibodies, a standard treatment for autoimmune diseases, might be an apparently obvious treatment option. However, a study on this possibility indicated that, at least in some patients that may already be suffering from inflammatory conditions prior to SARS-CoV-2 infection, this type of treatment could instead increase susceptibility to infection due to anti-TNF-α antibody treatment decreasing ACE2 and ADAM17 expression and shedding through Notch-1/IL-6 signaling [88].
Recently, there have been clinical trials of new oral antivirals that have demonstrated to be effective in treating COVID-19 but only if administered in the early days post SARS-CoV-2 infection, prior to hospitalization, [89], [90], [91] with two of them recently receiving emergency approval from the FDA in the USA.[91]. Paxlovid, from Pfizer, inhibits the activity of the SARS-CoV-2–3-chymotrypsin-like SARS-CoV-2–3CL protease which is required for viral replication [89], [92] Molnupiravir, from Merck, is metabolized in the body into a ribonucleoside analogue (β-D-N4-hydroxycytidine triphosphate) which is incorporated into SARS-CoV-2 RNA and induces lethal mutagenesis during viral replication. It is significantly more effective than other similar antivirals already in use for COVID-19 treatment, such as remdesivir which is administered intravenously instead [90], [91].
Despite these new effective therapies, there is still a need to understand the molecular machinery affected by SARS-CoV-2 infection and for new targets for COVID-19 therapy as the new antivirals are effective only if administered early on in SARS-CoV-2 infection. New therapies that can treat those who develop severe forms of COVID-19 are still needed. Given their role in many processes affected by COVID-19, such as cell signaling, inflammation or fibrosis, [24], [29] the adamalysin proteins may prove potential new targets for COVID-19 therapy.
Furthermore, as most current vaccines for COVID-19 introduce either a part of the SARS-CoV-2 virus (usually S protein) [2], [93], [94], [95], [96], [97], [98] or the whole (inactivated) virus,[95], [99], [100] adamalysins may also prove to influence the serious side effects observed in some rare people [16], [101] Many of these vaccines have been linked to at least some cases of thrombotic events post-vaccination, [16], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112] and cases of cardiovascular dysfunction such as myocarditis and pericarditis [113], [114], [115], [116]. Also, cases of renal dysfunction, particularly in the form of glomerulonephritis, [117], [118], [119], [120], [121], [122], [123], [124], [125], [126], [127] and autoimmune disorders such as rheumatoid arthritis and AOSD have been linked to COVID-19 vaccines [19], [128], [129], [130]. These are similar to cases that may occur in COVID-19, [8], [19], [20], [21] potentially indicating a common link between the vaccines and COVID-19 and these dysfunctions. In a post-vaccine development of the multisystem inflammatory disorder AOSD, the encoded viral S protein originating from protein translation of the mRNA in the vaccine may be inducing inflammatory signaling cascades in a similar way to the SARS-CoV-2 virus itself [19]. One of the adamalysins has been linked with some of these potentially vaccine-related side effects. The BNT162b2 mRNA vaccine has been suggested to have a potential link to increased risk of suffering thrombotic thrombocytopenia purpura due to extremely low levels of ADAMTS13 and testing for low levels of ADAMTS13 prior to vaccine administration has been suggested in patients who previously suffered from this disorder or other thrombosis [16]. Adenovirus vector-based vaccines may also induce ADAMTS13-related thrombotic events as a study on replication-deficient adenovirus-induced thrombocytopenia identified that ultra-large vWF multimers played a major role in the development of thrombosis [131]. If the same is occurring in the COVID-19 adenovirus vector-based vaccines, enhancing activity of ADAMTS13 may be a viable therapeutic avenue for vaccine-induced thrombosis in individuals who received these vaccines. Vaccine-/S protein-induced inflammation may also serve as the cause of these thrombotic events by inducing production of vWF fibers, [132] thus adamalysins involved in inflammation, such as ADAM17, could potentially be involved in promoting these severe side effects, potentially in a similar manner as to what is found to occur in COVID-19.
5. Perspective and conclusion
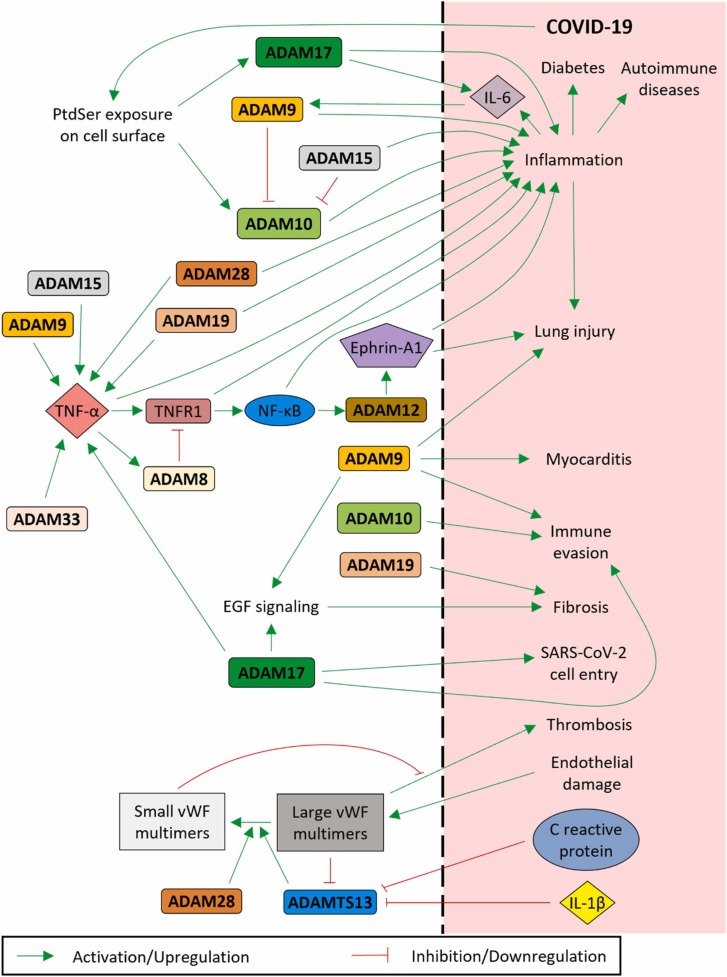
Adamalysins play roles in several different functions, such as different types of cell signaling and inflammatory pathways, and in many conditions and disorders, including COVID-19 and some of the comorbidities that have been observed to worsen the severity and lethality of COVID-19. As such, identifying the roles of the adamalysins may provide new therapeutic targets to treat COVID-19. A summary of the adamalysins known to have roles in COVID-19 and those we have suggested as having potential roles has been illustrated in Fig. 2.
Fig. 2.
Summary of adamalysins and their pathways that could have potential effects in COVID-19 pathology.
We suggest in this review some of the adamalysins that could be involved in COVID-19 and that might serve as potential viable therapeutic targets for this widespread, debilitating disease. As has also been mentioned, COVID-19 vaccines have also demonstrated some rare cases of severe side effects, some of which have been witnessed in severe COVID-19 cases, which, together with the fact that these vaccines involve either coding the SARS-CoV-2 S protein or inactivated SARS-CoV-2, suggests the strong possibility that the same mechanisms that involve these events in COVID-19 are also those involved in vaccine-related severe side effects. These adamalysins may, in some cases, serve to indicate those who might be at risk of suffering a vaccine-related side effect or even to help treat these situations. Further study into these adamalysins and their potential roles in COVID-19 could yield valuable data and new therapeutic options for treatment of this disease.
CRediT authorship contribution statement
IRSRD and HFK conceived and designed the review; IRSRD performed the data analysis; IRSRD, ZC and HFK discussed the data and wrote the manuscript; ZC and HFK revised the manuscript and supervised the entire project.
Funding
This study was supported by the Science and Technology Development Fund of Macau SAR (FDCT) [File no. 0055/2019/A1] and the joint research fund of National Science Fund of China (NSFC)–FDCT [File no. 0011/2021/AFJ for FDCT and 32161160303 for NSFC).
Authors' contributions
IRSRD and HFK conceived and designed the review; IRSRD performed the data analysis; IRSRD, ZC and HFK discussed the data and wrote the manuscript; ZC and HFK revised the manuscript and supervised the entire project.
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Acknowledgements
We thank Dr. Hiu-Fung Yuen and members of Kwok lab for discussion and valuable comments on manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2022.112970.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
No data was used for the research described in the article.
References
- 1.WHO. WHO Coronavirus (COVID-19) Dashboard. 2021 [cited 2021 04/11/2021]; Available from: 〈https://covid19.who.int/〉.
- 2.Rahman M.M., Hasan M., Ahmed A. Potential detrimental role of soluble ACE2 in severe COVID-19 comorbid patients. Rev. Med. Virol. 2021 doi: 10.1002/rmv.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma R.K., et al. Angiotensin-converting enzyme 2 and COVID-19 in cardiorenal diseases. Clin. Sci. 2021;135(1):1–17. doi: 10.1042/CS20200482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen T., et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard I., et al. Endothelium infection and dysregulation by SARS-CoV-2: evidence and caveats in COVID-19. Viruses. 2020;13:1. doi: 10.3390/v13010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schweitzer K.S., et al. Influenza virus infection increases ACE2 expression and shedding in human small airway epithelial cells. Eur. Respir. J. 2021;58:1. doi: 10.1183/13993003.03988-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C., et al. Cytokine release syndrome in severe COVID-19: interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopes-Pacheco M., et al. Pathogenesis of multiple organ injury in COVID-19 and potential therapeutic strategies. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.593223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar-On Y.M., et al. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020:9. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serebrovska Z.O., et al. Hypoxia, HIF-1alpha, and COVID-19: from pathogenic factors to potential therapeutic targets. Acta Pharm. Sin. 2020;41(12):1539–1546. doi: 10.1038/s41401-020-00554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arganaraz G.A., Palmeira J.D.F., Arganaraz E.R. Phosphatidylserine inside out: a possible underlying mechanism in the inflammation and coagulation abnormalities of COVID-19. Cell Commun. Signal. 2020;18(1):190. doi: 10.1186/s12964-020-00687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chueh T.I., et al. Novel evidence of acute kidney injury in COVID-19. J. Clin. Med. 2020;9:11. doi: 10.3390/jcm9113547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sweeney J.M., et al. Low ADAMTS13 activity correlates with increased mortality in COVID-19 patients. TH Open. 2021;5(1):e89–e103. doi: 10.1055/s-0041-1723784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelwick R., et al. The ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16:113. doi: 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apte S.S. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J. Biol. Chem. 2009;284(46):31493–31497. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maayan H., et al. Acquired thrombotic thrombocytopenic purpura: a rare disease associated with BNT162b2 vaccine. J. Thromb. Haemost. 2021;19(9):2314–2317. doi: 10.1111/jth.15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisman A., et al. Involvement of ADAMTS13 and von Willebrand factor in thromboembolic events in patients infected with SARS-CoV-2. Int J. Lab Hematol. 2020;42(5):e211–e212. doi: 10.1111/ijlh.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morici N., et al. Role of von willebrand factor and ADAMTS-13 in the pathogenesis of thrombi in SARS-CoV-2 infection: time to rethink. Thromb. Haemost. 2020;120(9):1339–1342. doi: 10.1055/s-0040-1713400. [DOI] [PubMed] [Google Scholar]
- 19.Magliulo D., et al. Adult-onset Still’s disease after mRNA COVID-19 vaccine. Lancet Rheuma. 2021;3(10):e680–e682. doi: 10.1016/S2665-9913(21)00219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bamidis A.D., et al. First manifestation of adult-onset Still’s disease after COVID-19. Lancet Rheuma. 2021;3(5):e319–e321. doi: 10.1016/S2665-9913(21)00072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baimukhamedov C., Barskova T., Matucci-Cerinic M. Arthritis after SARS-CoV-2 infection. Lancet Rheuma. 2021;3(5):e324–e325. doi: 10.1016/S2665-9913(21)00067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alshablan A., et al. Diagnosis of adult onset still’s disease in a patient who has recovered from coronavirus-19. Clin. Med Insights Case Rep. 2021;14 doi: 10.1177/1179547621996306. 1179547621996306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrot L., et al. First flare of ACPA-positive rheumatoid arthritis after SARS-CoV-2 infection. Lancet Rheuma. 2021;3(1):e6–e8. doi: 10.1016/S2665-9913(20)30396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theret N., et al. ADAM and ADAMTS proteins, new players in the regulation of hepatocellular carcinoma microenvironment. Cancers (Basel) 2021;13:7. doi: 10.3390/cancers13071563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patra T., et al. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xavier L.L., et al. Does angiotensin II peak in response to SARS-CoV-2? Front Immunol. 2021;11 doi: 10.3389/fimmu.2020.577875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendoza R., et al. Ephrin-A1 and the sheddase ADAM12 are upregulated in COVID-19. Heliyon. 2021;7(6) doi: 10.1016/j.heliyon.2021.e07200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambrecht B.N., Vanderkerken M., Hammad H. The emerging role of ADAM metalloproteinases in immunity. Nat. Rev. Immunol. 2018;18(12):745–758. doi: 10.1038/s41577-018-0068-5. [DOI] [PubMed] [Google Scholar]
- 29.Edwards D.R., Handsley M.M., Pennington C.J. The ADAM metalloproteinases. Mol. Asp. Med. 2008;29(5):258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giebeler N., Zigrino P. A disintegrin and metalloprotease (ADAM): historical overview of their functions. Toxins (Basel) 2016;8(4):122. doi: 10.3390/toxins8040122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gene. ADAM3A ADAM metallopeptidase domain 3A (pseudogene) [Homo sapiens (human)] - Gene - NCBI. 2021 [cited 2021 27/10/2021]; Available from: 〈https://www.ncbi.nlm.nih.gov/gene/1587〉.
- 32.Gene. ADAM5 ADAM metallopeptidase domain 5 (pseudogene) [Homo sapiens (human)] - Gene - NCBI. 2021 [cited 2021 27/10/2021]; Available from: 〈https://www.ncbi.nlm.nih.gov/gene/255926〉.
- 33.Gene. ADAM6 ADAM metallopeptidase domain 6 (pseudogene) [Homo sapiens (human)] - Gene - NCBI. 2021 [cited 2021 27/10/2021]; Available from: 〈https://www.ncbi.nlm.nih.gov/gene/8755〉.
- 34.Gene. ADAM21 ADAM metallopeptidase domain 21 [Homo sapiens (human)] - Gene - NCBI. 2021 [cited 2021 27/10/2021]; Available from: 〈https://www.ncbi.nlm.nih.gov/gene/8747〉.
- 35.Gene. ADAM18 ADAM metallopeptidase domain 18 [Homo sapiens (human)] - Gene - NCBI. 2021 [cited 2021 27/10/2021]; Available from: 〈https://www.ncbi.nlm.nih.gov/gene/8749〉.
- 36.Chou C.W., et al. An overview of ADAM9: structure, activation, and regulation in human diseases. Int J. Mol. Sci. 2020;21:20. doi: 10.3390/ijms21207790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsia H.E., et al. Functions of ‘A disintegrin and metalloproteases (ADAMs)’ in the mammalian nervous system. Cell Mol. Life Sci. 2019;76(16):3055–3081. doi: 10.1007/s00018-019-03173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang C.Y., et al. Interleukin 13 (IL-13)-regulated expression of the chondroprotective metalloproteinase ADAM15 is reduced in aging cartilage. Osteoarthr. Cartil. Open. 2020;2(4) doi: 10.1016/j.ocarto.2020.100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jana S., et al. ADAM (a disintegrin and metalloproteinase) 15 deficiency exacerbates Ang II (Angiotensin II)-induced aortic remodeling leading to abdominal aortic aneurysm. Arterioscler. Thromb. Vasc. Biol. 2020;40(8):1918–1934. doi: 10.1161/ATVBAHA.120.314600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi B., Newcomer R.G., Sang Q.X. ADAM19/adamalysin 19 structure, function, and role as a putative target in tumors and inflammatory diseases. Curr. Pharm. Des. 2009;15(20):2336–2348. doi: 10.2174/138161209788682352. [DOI] [PubMed] [Google Scholar]
- 41.Nyren-Erickson E.K., et al. A disintegrin and metalloproteinase-12 (ADAM12): function, roles in disease progression, and clinical implications. Biochim Biophys. Acta. 2013;1830(10):4445–4455. doi: 10.1016/j.bbagen.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin Q., et al. ADAM28 from both endothelium and gastric cancer cleaves von Willebrand factor to eliminate von willebrand factor-induced apoptosis of gastric cancer cells. Eur. J. Pharm. 2021;898 doi: 10.1016/j.ejphar.2021.173994. [DOI] [PubMed] [Google Scholar]
- 43.Yang G., et al. Molecular switch in human diseases-disintegrin and metalloproteinases, ADAM17. Aging (Albany NY) 2021;13(12):16859–16872. doi: 10.18632/aging.203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedrosa M.A., et al. Experimental data using candesartan and captopril indicate no double-edged sword effect in COVID-19. Clin. Sci. (Lond. ) 2021;135(3):465–481. doi: 10.1042/CS20201511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Severe Covid G.G., et al. Genomewide association study of severe Covid-19 with respiratory failure. New Engl. J. Med. 2020;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richardson S., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323(20):2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haga S., et al. TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antivir. Res. 2010;85(3):551–555. doi: 10.1016/j.antiviral.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haga S., et al. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105(22):7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target Ther. 2020;5(1):283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calligaris M., et al. Strategies to target ADAM17 in disease: from its discovery to the iRhom revolution. Molecules. 2021;26:4. doi: 10.3390/molecules26040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J.Y., et al. Abnormal ADAM17 expression causes airway fibrosis in chronic obstructive asthma. Biomed. Pharm. 2021;140 doi: 10.1016/j.biopha.2021.111701. [DOI] [PubMed] [Google Scholar]
- 54.Taefehshokr N., et al. Covid-19: perspectives on innate immune evasion. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao J., et al. A disintegrin and metallproteinase 15 knockout decreases migration of fibroblast-like synoviocytes and inflammation in rheumatoid arthritis. Mol. Med Rep. 2015;11(6):4389–4396. doi: 10.3892/mmr.2015.3302. [DOI] [PubMed] [Google Scholar]
- 56.Wang X., et al. ADAM15 expression is increased in lung CD8(+) T cells, macrophages, and bronchial epithelial cells in patients with COPD and is inversely related to airflow obstruction. Respir. Res. 2020;21(1):188. doi: 10.1186/s12931-020-01446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bridges L.C., Sheppard D., Bowditch R.D. ADAM disintegrin-like domain recognition by the lymphocyte integrins alpha4beta1 and alpha4beta7. Biochem J. 2005;387(Pt 1):101–108. doi: 10.1042/BJ20041444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bridges L.C., et al. All-trans-retinoic acid induces integrin-independent B-cell adhesion to ADAM disintegrin domains. Biochemistry. 2008;47(15):4544–4551. doi: 10.1021/bi702447u. [DOI] [PubMed] [Google Scholar]
- 59.Yan F., et al. Silencing a disintegrin and metalloproteinase33 attenuates the proliferation of vascular smooth muscle cells via PI3K/AKT pathway: Implications in the pathogenesis of airway vascular remodeling. Mol. Med. Rep. 2021;24:1. doi: 10.3892/mmr.2021.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Oliveira M., et al. Irisin modulates genes associated with severe coronavirus disease (COVID-19) outcome in human subcutaneous adipocytes cell culture. Mol. Cell Endocrinol. 2020;515 doi: 10.1016/j.mce.2020.110917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santamaria S., de Groot R. ADAMTS proteases in cardiovascular physiology and disease. Open Biol. 2020;10(12) doi: 10.1098/rsob.200333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bader H.L., et al. A disintegrin-like and metalloprotease domain containing thrombospondin type 1 motif-like 5 (ADAMTSL5) is a novel fibrillin-1-, fibrillin-2-, and heparin-binding member of the ADAMTS superfamily containing a netrin-like module. Matrix Biol. 2012;31(7–8):398–411. doi: 10.1016/j.matbio.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elbitar S., et al. Pathogenic variants in THSD4, encoding the ADAMTS-like 6 protein, predispose to inherited thoracic aortic aneurysm. Genet Med. 2021;23(1):111–122. doi: 10.1038/s41436-020-00947-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stelzer G., et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 2016;54:1 30 1–1 30 33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 65.Rypdal K.B., et al. The extracellular matrix glycoprotein ADAMTSL2 is increased in heart failure and inhibits TGFbeta signalling in cardiac fibroblasts. Sci. Rep. 2021;11(1):19757. doi: 10.1038/s41598-021-99032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mead T.J., Apte S.S. ADAMTS proteins in human disorders. Matrix Biol. 2018;71–72:225–239. doi: 10.1016/j.matbio.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arakawa A., et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J. Exp. Med. 2015;212(13):2203–2212. doi: 10.1084/jem.20151093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tola E.N., et al. The role of ADAMTS-2, collagen type-1, TIMP-3 and papilin levels of uterosacral and cardinal ligaments in the etiopathogenesis of pelvic organ prolapse among women without stress urinary incontinence. Eur. J. Obstet. Gynecol. Reprod. Biol. 2018;231:158–163. doi: 10.1016/j.ejogrb.2018.10.043. [DOI] [PubMed] [Google Scholar]
- 69.Favaloro E.J., Henry B.M., Lippi G. Increased VWF and decreased ADAMTS-13 in COVID-19: creating a milieu for (micro)thrombosis. Semin Thromb. Hemost. 2021;47(4):400–418. doi: 10.1055/s-0041-1727282. [DOI] [PubMed] [Google Scholar]
- 70.Doevelaar A.A.N., et al. von willebrand factor multimer formation contributes to immunothrombosis in coronavirus disease 2019. Crit. Care Med. 2021;49(5):e512–e520. doi: 10.1097/CCM.0000000000004918. [DOI] [PubMed] [Google Scholar]
- 71.Yang C.Y., Chanalaris A., Troeberg L. ADAMTS and ADAM metalloproteinases in osteoarthritis - looking beyond the ‘usual suspects’. Osteoarthr. Cartil. 2017;25(7):1000–1009. doi: 10.1016/j.joca.2017.02.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang Y., et al. ADAMTS19 suppresses cell migration and invasion by targeting S100A16 via the NF-kappaB pathway in human gastric cancer. Biomolecules. 2021;11:4. doi: 10.3390/biom11040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Balic Z., et al. Alternative splicing of the metalloprotease ADAMTS17 spacer regulates secretion and modulates autoproteolytic activity. FASEB J. 2021;35(2) doi: 10.1096/fj.202001120RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karoulias S.Z., et al. The ADAMTS/fibrillin connection: insights into the biological functions of ADAMTS10 and ADAMTS17 and their respective sister proteases. Biomolecules. 2020;10:4. doi: 10.3390/biom10040596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masood K.I., et al. Upregulated type I interferon responses in asymptomatic COVID-19 infection are associated with improved clinical outcome. Sci. Rep. 2021;11(1):22958. doi: 10.1038/s41598-021-02489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Q., et al. Inflammation and antiviral immune response associated with severe progression of COVID-19. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.631226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang H., et al. Estrogen receptors are linked to angiotensin-converting enzyme 2 (ACE2), ADAM metallopeptidase domain 17 (ADAM-17), and transmembrane protease serine 2 (TMPRSS2) expression in the human atrium: insights into COVID-19. Hypertens. Res. 2021;44(7):882–884. doi: 10.1038/s41440-021-00626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khoury E.E., et al. Pulmonary, cardiac and renal distribution of ACE2, furin, TMPRSS2 and ADAM17 in rats with heart failure: Potential implication for COVID-19 disease. J. Cell Mol. Med. 2021;25(8):3840–3855. doi: 10.1111/jcmm.16310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saini K.S., et al. Mortality in patients with cancer and coronavirus disease 2019: a systematic review and pooled analysis of 52 studies. Eur. J. Cancer. 2020;139:43–50. doi: 10.1016/j.ejca.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brest P., et al. More light on cancer and COVID-19 reciprocal interaction. Br. J. Cancer. 2021;124(8):1344–1345. doi: 10.1038/s41416-020-01246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Novak N., et al. SARS-CoV-2, COVID-19, skin and immunology - what do we know so far? Allergy. 2021;76(3):698–713. doi: 10.1111/all.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Monteil V., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181(4):905–913. doi: 10.1016/j.cell.2020.04.004. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zoufaly A., et al. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir. Med. 2020;8(11):1154–1158. doi: 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dalan R., et al. The ACE-2 in COVID-19: foe or friend? Horm. Metab. Res. 2020;52(5):257–263. doi: 10.1055/a-1155-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Meng J., et al. Renin-angiotensin system inhibitors improve the clinical outcomes of COVID-19 patients with hypertension. Emerg. Microbes Infect. 2020;9(1):757–760. doi: 10.1080/22221751.2020.1746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y., et al. The use of renin-angiotensin-aldosterone system (RAAS) inhibitors is associated with a lower risk of mortality in hypertensive COVID-19 patients: A systematic review and meta-analysis. J. Med Virol. 2021;93(3):1370–1377. doi: 10.1002/jmv.26625. [DOI] [PubMed] [Google Scholar]
- 87.Zhang P., et al. Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin ii receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ. Res. 2020;126(12):1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Keewan E., Beg S., Naser S.A. Anti-TNF-alpha agents modulate SARS-CoV-2 receptors and increase the risk of infection through notch-1 signaling. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.641295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pfizer. Pfizer's novel COVID-19 oral antiviral treatment candidate reduced risk of hospitalization or death by 89% in interim analysis of phase 2/3 EPIC-HR study. 2021 5/11/2021 [cited 2021 31/12/2021]; Available from: 〈https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate〉.
- 90.Kabinger F., et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28(9):740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gumbrecht, J., A. Sealy, J. Howard. Covid pill: FDA authorizes two antivirals to treat coronavirus infection - CNN. 2021 24/12/2021 [cited 2021 31/12/2021]; Available from: 〈https://edition.cnn.com/2021/12/22/health/pfizer-antiviral-pill-authorized/index.html〉.
- 92.Vandyck K., Deval J. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr. Opin. Virol. 2021;49:36–40. doi: 10.1016/j.coviro.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baden L.R., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. New Engl. J. Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Polack F.P., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. New Engl. J. Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mascellino M.T., et al. Overview of the main Anti-SARS-CoV-2 vaccines: mechanism of action, efficacy and safety. Infect. Drug Resist. 2021;14:3459–3476. doi: 10.2147/IDR.S315727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Heath P.T., et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. New Engl. J. Med. 2021;385(13):1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Capone S., et al. Immunogenicity of a new gorilla adenovirus vaccine candidate for COVID-19. Mol. Ther. 2021;29(8):2412–2423. doi: 10.1016/j.ymthe.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Basta, N.E. E.E.M. Moodie. COVID19 Vaccine Tracker. 2021 [cited 2021 26/10/2021]; Available from: covid19.trackvaccines.org.
- 99.Jara A., et al. Effectiveness of an Inactivated SARS-CoV-2 Vaccine in Chile. New Engl. J. Med. 2021;385(10):875–884. doi: 10.1056/NEJMoa2107715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21(2):181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shimabukuro T., Nair N. Allergic reactions including anaphylaxis after receipt of the first dose of Pfizer-BioNTech COVID-19 vaccine. JAMA. 2021;325(8):780–781. doi: 10.1001/jama.2021.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee E.J., et al. Thrombocytopenia following Pfizer and Moderna SARS-CoV-2 vaccination. Am. J. Hematol. 2021;96(5):534–537. doi: 10.1002/ajh.26132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Welsh K.J., et al. Thrombocytopenia including immune thrombocytopenia after receipt of mRNA COVID-19 vaccines reported to the vaccine adverse event reporting system (VAERS) Vaccine. 2021;39(25):3329–3332. doi: 10.1016/j.vaccine.2021.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kantarcioglu B., et al. An update on the pathogenesis of COVID-19 and the reportedly rare thrombotic events following vaccination. Clin. Appl. Thromb. Hemost. 2021;27 doi: 10.1177/10760296211021498. 10760296211021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Greinacher A., et al. Thrombotic thrombocytopenia after ChAdOx1 nCov-19 vaccination. N. Engl. J. Med. 2021;384(22):2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schultz N.H., et al. Thrombosis and thrombocytopenia after ChAdOx1 nCoV-19 vaccination. N. Engl. J. Med. 2021;384(22):2124–2130. doi: 10.1056/NEJMoa2104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.MacNeil J.R., et al. Updated recommendations from the advisory committee on immunization practices for use of the Janssen (Johnson & Johnson) COVID-19 vaccine after reports of thrombosis with thrombocytopenia syndrome among vaccine recipients - United States, April 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70(17):651–656. doi: 10.15585/mmwr.mm7017e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shay D.K., et al. Safety monitoring of the Janssen (Johnson & Johnson) COVID-19 Vaccine - United States, March-April 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70(18):680–684. doi: 10.15585/mmwr.mm7018e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kragholm K., et al. Thrombocytopenia after COVID-19 vaccination. J. Autoimmun. 2021;123 doi: 10.1016/j.jaut.2021.102712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abbattista M., Martinelli I., Peyvandi F. Comparison of adverse drug reactions among four COVID-19 vaccines in Europe using the EudraVigilance database: Thrombosis at unusual sites. J. Thromb. Haemost. 2021;19(10):2554–2558. doi: 10.1111/jth.15493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ozdemir I.H., et al. Type 1 kounis syndrome induced by inactivated SARS-COV-2 Vaccine. J. Emerg. Med. 2021 doi: 10.1016/j.jemermed.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Erdem N.S., et al. Acute transverse myelitis after inactivated COVID-19 vaccine. Ideggyogy. Sz. 2021;74(7–8):273–276. doi: 10.18071/isz.74.0273. [DOI] [PubMed] [Google Scholar]
- 113.Gargano J.W., et al. Use of mRNA COVID-19 vaccine after reports of myocarditis among vaccine recipients: update from the advisory committee on immunization practices - United States, June 2021. MMWR Morb. Mortal. Wkly Rep. 2021;70(27):977–982. doi: 10.15585/mmwr.mm7027e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muthukumar A., et al. In-Depth evaluation of a case of presumed myocarditis after the second dose of COVID-19 mRNA Vaccine. Circulation. 2021;144(6):487–498. doi: 10.1161/CIRCULATIONAHA.121.056038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Diaz G.A., et al. Myocarditis and pericarditis after vaccination for COVID-19. JAMA. 2021;326(12):1210–1212. doi: 10.1001/jama.2021.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sulemankhil I., Abdelrahman M., Negi S.I. Temporal association between the COVID-19 Ad26.COV2.S vaccine and acute myocarditis: a case report and literature review. Cardiovasc Revasc Med. 2021 doi: 10.1016/j.carrev.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klomjit N., et al. COVID-19 vaccination and glomerulonephritis. Kidney Int Rep. 2021 doi: 10.1016/j.ekir.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.D'Agati V.D., et al. Minimal change disease and acute kidney injury following the Pfizer-BioNTech COVID-19 vaccine. Kidney Int. 2021;100(2):461–463. doi: 10.1016/j.kint.2021.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Komaba H., Wada T., Fukagawa M. Relapse of minimal change disease following the Pfizer-BioNTech COVID-19 Vaccine. Am. J. Kidney Dis. 2021;78(3):469–470. doi: 10.1053/j.ajkd.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lebedev L., et al. Minimal change disease following the Pfizer-BioNTech COVID-19 Vaccine. Am. J. Kidney Dis. 2021;78(1):142–145. doi: 10.1053/j.ajkd.2021.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kervella D., et al. Minimal change disease relapse following SARS-CoV-2 mRNA vaccine. Kidney Int. 2021;100(2):457–458. doi: 10.1016/j.kint.2021.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perrin P., et al. Gross hematuria following SARS-CoV-2 vaccination in patients with IgA nephropathy. Kidney Int. 2021;100(2):466–468. doi: 10.1016/j.kint.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Holzworth A., et al. Minimal change disease following the moderna mRNA-1273 SARS-CoV-2 vaccine. Kidney Int. 2021;100(2):463–464. doi: 10.1016/j.kint.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sekar A., et al. ANCA glomerulonephritis after the moderna COVID-19 vaccination. Kidney Int. 2021;100(2):473–474. doi: 10.1016/j.kint.2021.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gillion V., et al. Granulomatous vasculitis after the AstraZeneca anti-SARS-CoV-2 vaccine. Kidney Int. 2021;100(3):706–707. doi: 10.1016/j.kint.2021.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Morlidge C., et al. Relapse of minimal change disease following the AstraZeneca COVID-19 vaccine. Kidney Int. 2021;100(2):459. doi: 10.1016/j.kint.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aydin M.F., et al. Relapse of primary membranous nephropathy after inactivated SARS-CoV-2 virus vaccination. Kidney Int. 2021;100(2):464–465. doi: 10.1016/j.kint.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Terracina K.A., Tan F.K. Flare of rheumatoid arthritis after COVID-19 vaccination. Lancet Rheuma. 2021;3(7):e469–e470. doi: 10.1016/S2665-9913(21)00108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baimukhamedov C. Arthritis of the left elbow joint after vaccination against SARS-CoV-2 infection. Int J. Rheum. Dis. 2021;24(9):1218–1220. doi: 10.1111/1756-185X.14202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baimukhamedov C., Makhmudov S., Botabekova A. Seropositive rheumatoid arthritis after vaccination against SARS-CoV-2 infection. Int J. Rheum. Dis. 2021 doi: 10.1111/1756-185X.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Othman M., et al. Adenovirus-induced thrombocytopenia: the role of von Willebrand factor and P-selectin in mediating accelerated platelet clearance. Blood. 2007;109(7):2832–2839. doi: 10.1182/blood-2006-06-032524. [DOI] [PubMed] [Google Scholar]
- 132.Tahir S., et al. Endothelial CD40 mediates microvascular von willebrand factor-dependent platelet adhesion inducing inflammatory venothrombosis in ADAMTS13 knockout mice. Thromb. Haemost. 2020;120(3):466–476. doi: 10.1055/s-0040-1702228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
No data was used for the research described in the article.