Abstract
Large, observational genetic studies are commonly used to identify genetic factors associated with diseases and disease‐related traits. Such cohorts have not been commonly used to identify genetic predictors of drug dosing or concentrations, perhaps because of the heterogeneity in drug dosing and formulation, and the random timing of blood sampling. We hypothesized that large sample sizes relative to traditional pharmacokinetic studies would compensate for this variability and enable the identification of pharmacogenetic predictors of drug concentrations. We performed a cross‐sectional, proof‐of‐concept association study to replicate the well‐established association between metoprolol concentrations and CYP2D6 genotype‐inferred metabolizer phenotypes in participants from the Montreal Heart Institute Hospital Cohort undergoing metoprolol therapy. Plasma concentrations of metoprolol and α‐hydroxymetoprolol (α‐OH‐metoprolol) were measured in samples collected randomly regarding the previous metoprolol dose. A total of 999 individuals were included. The metoprolol daily dose ranged from 6.25 to 400 mg (mean 84.3 ± 57.1 mg). CYP2D6‐inferred phenotype was significantly associated with both metoprolol and α‐OH‐metoprolol in unadjusted and adjusted models (all p < 10−14). Models for metoprolol daily dose showed consistent results. Our study suggests that randomly drawn blood samples from biobanks can serve as a new approach to discover genetic associations related to drug concentrations and dosing, with potentially broader implications for genomewide association studies on the pharmacogenomics of drug metabolism.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Observational cohorts have been leveraged numerous times to identify genetic determinants of disease‐related traits. Whether biobanks can be utilized to identify pharmacogenomic (PGx) determinants affecting drug concentrations and other clinical parameters remains largely untested.
WHAT QUESTION DID THIS STUDY ADDRESS?
Using the Montreal Heart Institute (MHI) Hospital Cohort and its information comprised within the MHI Biobank, we investigated if pharmacogenetic associations could be observed with the level of statistical significance of genomewide association studies (GWAS). We sought to recreate a PGx association previously validated through traditional pharmacokinetic study design between metoprolol plasma concentrations and CYP2D6 genotype‐inferred phenotypes.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
With a sample of ~ 1000 patients, we demonstrate that single, random blood samplings can be used to identify PGx associations influencing average metoprolol plasma concentrations, along with those of its associated CYP2D6 metabolite. Even after correcting for cofactors through simple multivariable modeling, statistical significance of GWAS magnitude persists. Additional associations regarding drug response, such as average daily dosage and heart rate, can be detected, even when analyzing a single CYP2D6 polymorphism.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
This study suggests that biorepositories can be leveraged to discover and validate pharmacogenetic targets associated to variations in drug metabolism, and potentially clinical response. Biobanks represent a new resource by which PGx consortia could identify predictors of drug metabolism, overcoming methodological limitations encountered in traditional pharmacokinetic studies and GWAS.
INTRODUCTION
The discovery of pharmacogenomic (PGx) determinants influencing drug concentrations, dosing, and overall pharmacokinetics (PKs) is critical to the emergence of precision medicine. 1 Typical PGx‐PK studies generally involve small to medium sample sizes (n < 100) using “classic” PK study designs. 2 Although classic PK studies are indispensable to determine the PK parameters of a drug, such as its half‐life and clearance, they present several limitations to discover new PGx markers. First, these studies are generally limited to one drug and require multiple blood samples taken at specific intervals, which are not routinely performed outside this context. 3 Thus, costly investigations necessitating the recruitment of new patients are generally required for each study. Second, because they usually include only a few patients, they are underpowered to identify common genetic determinants that have a modest effect on a drug’s concentrations, or rare genetic factors that could have a major impact on dosing requirements.
Large, observational genetic studies have been used successfully to identify genetic factors associated with diseases and traits. 4 , 5 , 6 Most commonly, data are collected on multiple conditions or phenotypes and are being leveraged to conduct genetic association studies on multiple phenotypes. Because blood samples are frequently drawn at baseline in these studies, we hypothesized that such studies could also be used to identify new genetic predictors of drug concentrations. Although it could be argued that the random timing of blood sampling since the last dose taken, as well as the variability in drug dosing, formulation, and manufacturer could instill too much heterogeneity for such an approach, we hypothesized that using a large sample size would compensate for this variability.
The cytochrome P450 (CYP) 2D6 is involved in the hepatic metabolism of drugs. 7 Over 140 genetic variants of CYP2D6 have been reported and their impacts range from null to increased metabolic activity. 8 A harmonized classification of CYP2D6 genotype‐inferred metabolizing phenotypes based on their cumulative impact was recently brought forward by major PGx consortia. 8 Four phenotypes are established, including poor metabolizers (PMs), intermediate metabolizers (IMs), normal metabolizers (NMs), and ultrarapid metabolizers (UMs).
Metoprolol is a lipophilic β1‐adrenergic receptor antagonist used to treat various cardiovascular conditions, including angina, atrial fibrillation, and hypertension. It is primarily metabolized by CYP2D6 into α‐hydroxymetoprolol (α‐OH‐metoprolol), which exhibits ~ 10% of metoprolol’s potency. 9 Previous investigations using classic PK designs have consistently found that metoprolol concentrations decrease across all four CYP2D6 genotype‐inferred phenotypes, from PMs to UMs, and the opposite has been shown for α‐OH‐metoprolol. 2 , 3 , 10
In order to test our hypothesis that randomly collected blood samples collected in observational studies could be used to identify genetic markers of drug concentrations, we performed a proof‐of‐concept association study to replicate the well‐established association between metoprolol concentrations and CYP2D6 genotype‐inferred metabolizer phenotypes. We used plasma samples collected at random timepoints relative to the previous metoprolol dose in participants from the Montreal Heart Institute (MHI) Hospital Cohort. 11 , 12
METHODS
Study design
We performed a cross‐sectional study that included participants from the MHI Hospital Cohort taking metoprolol at the baseline visit. The methods of the MHI Hospital Cohort, including its sample collection protocol, have been reported elsewhere. 11 , 12
Study population
The study population included self‐reported “White” men and women aged greater than or equal to 18 years, who reported being treated with oral metoprolol and who had plasma collected according to the standardized plasma collection protocol of the MHI Biobank. We put no restriction regarding the dose or formulation used. In Canada, only metoprolol tartrate is available. The only exclusion criterion was a history of liver transplant, as the genotypes from the donor and the recipient could differ. 13 Given the exploratory nature of our approach, we sought to include a convenience sample of 1000 individuals.
Study end points
The primary objective of this study was to replicate the association between CYP2D6 genotype‐inferred phenotypes and the primary study end point of plasma concentrations of racemic metoprolol. Association among CYP2D6 phenotype and α‐OH‐metoprolol, metoprolol daily dosing, and resting heart rate were also investigated.
Measurement of metoprolol and α‐hydroxymetoprolol concentrations
The plasma samples of individuals included as part of this study was collected as previously described. 11 Briefly, blood samples were collected in ethylenediaminetetraacetic acid vacutainer tubes on ice. Plasma was obtained by centrifugation within 30 min (1900 g at 4°C for 15 min). The plasma was then transferred to 1‐ml microtubes, rapidly frozen at −21°C, and then stored at −80°C on the same day. Blood sampling was performed randomly regarding the intake of metoprolol or other concomitant medications, time of day, or food intake. Metoprolol and α‐OH‐metoprolol measurements were conducted in the bioanalytical laboratory of the Platform of Biopharmacy at Université de Montréal. The analyses were conducted blinded to any information related to the samples, including CYP2D6 genotypes and metoprolol doses. Lower limits of quantification (LLOQ) were set at 1 ng/ml for metoprolol and α‐OH‐metoprolol, with upper limits set at 1000 ng/ml. The complete bioanalytical method is described in the Supplementary Material section, along with a schematic representation of sample preparation.
Genotyping and CYP2D6 metabolism
DNA isolation was performed under GLP conditions using the Gentra Autopure LS system, as previously described. 10 , 14 The targeted regions of genomic DNA were amplified through polymerase chain reaction before conducting single‐base extension using either Agena’s iPLEX ADME PGx Pro Panel 1.0 or MHI ADME Panel V3.0 (Agena Bioscience), as previously described. 15 Subsequent conditioning of the extension conditions was made using Clean Resin before being dispensed on a SpectroChipII Array using a MassARRAY Nanodispenser (Agena Bioscience). Data acquisition was made on the MassARRAY Analyzer Compact (Agena Bioscience MALDI‐TOF mass spectrometer; Agena Bioscience). CYP2D6 variant alleles genotyped were *1, *2, *3, *4, *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, *19, *20, *29, *41, *69, and *109. CYP2D6 alleles were assigned according to the PharmVar core alleles (https://www.pharmvar.org/gene/CYP2D6). As for copy number variants (CNVs), seven probes were designed, with target sequences located on introns 4 and 6, and exon 9 of CYP2D6. For both variant alleles and CNVs, the different extended mass intensities were analyzed using the MassArray Typer software version 4.1, including analysis application Typer Analyzer version 4.1.0.83 with PGxReporter v3.51 software. Using allelic activity scores, genotypes, and CNVs, we categorized participants into PM, IM, NM, and UM phenotypes according to the most recent CYP2D6 classification guidelines. 8
Statistical analyses
We performed descriptive statistics of clinical and genetic variables for all included participants. Frequencies and proportions were reported for categorical variables, whereas means, SDs, and medians were used for continuous variables. Multiple linear regression analyses were conducted to investigate the relationship between CYP2D6 genotype‐inferred phenotypes and the study end points. To do so, we considered a crude model and different adjusted models with an increasing number of covariables for each analysis. Candidate adjustment variables were age, sex, metoprolol dose, and weight for models of analyte concentrations and metoprolol dosing, with the addition of a history of atrial fibrillation/flutter and the use of heart rate lowering drugs for the end point of heart rate. In addition, for every fully adjusted model, we considered concomitant uses of CYP2D6 inhibitors. The complete list of all medications taken by the cohort were screened to identify CYP2D6 inhibitors. We referred to the US Food and Drug Administration (FDA) Table of Inhibitors to provide an objective list of such medications to consider. 16 Furthermore, we also built models where patients’ phenotype was converted prior to multivariable adjustment by multiplying the patients’ CYP2D6 activity scores by factors of 0.5 and 0 for the concomitant intake of moderate and strong CYP2D6 inhibitors, respectively. 17 CYP2D6 genotype‐inferred phenotype was treated as an ordinal variable and coded as 0, 1, 2, or 3 for PM, IM, NM, and UM, respectively. Although inferred phenotypes are widely used in PK association studies, 18 , 19 , 20 as a sensitivity analysis, we also assessed a single key variant of CYP2D6*4, rs3892097, which is prevalent within populations of European descent. 21 When necessary, to satisfy the normality assumption, a regression model for each outcome and its natural logarithm transformation were fitted and the distributions of residuals were compared. In all analyses, the residuals of the models with transformed outcomes are more normal than the residuals of the models with original outcomes. Therefore, we kept the models with transformed outcomes. Samples with concentrations lower than the LLOQ were attributed a value of 0. Statistical tests were two‐tailed and a p value less than 0.05 was considered statistically significant. All analyses were carried using SAS version 9.4 (SAS Institute).
Ethics statement
The MHI Hospital Cohort protocol has been approved by the institution’s Scientific and Ethics Committees. All participants provided a signed, informed consent prior to their inclusion in the MHI Hospital Cohort. This investigation was approved by the Cohort Management Committee, as well as the MHI’s Scientific and Ethics Committees.
RESULTS
Study population
A total of 1007 participants from the MHI Cohort met our inclusion and exclusion criteria. Of these, complete CYP2D6 genotyping was unsuccessful for seven patients, whereas plasma volume was insufficient at the time of analysis for one patient. Thus, 999 patients were included in the analyses. At baseline, patients presented characteristics consistent with a population with cardiovascular disorders treated with metoprolol (Table 1). Specifically, it was constituted mainly of males (73%), aged 66.5 ± 8.7 years with a weight of 84.4 ± 17.0 kg. As expected, patients were treated with multiple concomitant cardiovascular medications (Table 1). Regarding metoprolol, the mean daily dose was 84.3 ± 57.1 mg, with total daily doses ranging from 6.25 to 400 mg. Frequencies of CYP2D6 diplotypes, genotypes, and inferred phenotypes are listed in Table 1 and Supplementary Table S1. Of the 999 patients, phenotypes could not be determined in three patients because of triallelic genotyping results. In total, 4.4%, 34.3%, 54.4%, and 6.8% were PM, IM, NM, and UM, respectively. Only 36 patients (3.6%) used either one or more moderate and/or strong inhibitors, of which 33 were reclassified with lower phenotypic activity. Since considering the use of CYP2D6 inhibitors gave very similar results either as a covariable or as a phenoconversion factor, we present only the models where it was used as a covariable.
TABLE 1.
Baseline cohort characteristics
|
All a 999 (100%) |
PM 44 (4%) |
IM 342 (34%) |
NM 542 (54%) |
UM 68 (7%) |
|
|---|---|---|---|---|---|
| Socio‐demographic variables | |||||
| Age, n (%) | 66.5 ± 8.7 | 68.4 ± 7.7 | 66.3 ± 8.8 | 66.6 ± 8.6 | 65.7 ± 9.9 |
| Female, n (%) | 269 (27) | 11 (25) | 90 (26) | 142 (26) | 26 (38) |
| Self‐reported White ethnicity, n (%) | 999 (100) | 44 (100) | 342 (100) | 542 (100) | 68 (100) |
| Lifestyle and physical measure | |||||
| Smoking status, n (%) | |||||
| Never‐smoker | 278 (28) | 12 (27) | 89 (26) | 158 (29) | 19 (28) |
| Past‐smoker | 638 (64) | 29 (66) | 227 (66) | 336 (62) | 43 (63) |
| Current‐smoker | 83 (8) | 3 (7) | 26 (8) | 48 (9) | 6 (9) |
| Weight, kg | 84.4 ± 17.0 | 81.2 ± 13.9 | 85.0 ± 16.1 | 84.5 ± 17.9 | 82.1 ± 16.4 |
| BMI | 30.0 ± 5.4 | 30.0 ± 4.6 | 30.1 ± 5.2 | 30.0 ± 5.6 | 29.9 ± 5.4 |
| Cardiovascular chronic conditions at baseline | |||||
| Hypertension, n (%) | 784 (78) | 33 (75) | 280 (82) | 420 (77) | 50 (74) |
| Diabetes, n (%) | |||||
| Type 1 | 7 (1) | 0 (0) | 3 (1) | 4 (1) | 0 (0) |
| Type 2 | 296 (30) | 16 (36) | 113 (33) | 140 (26) | 26 (38) |
| Dyslipidemia, n (%) | 847 (85) | 41 (93) | 295 (86) | 455 (84) | 53 (78) |
| Myocardial infarction, n (%) | 428 (43) | 22 (50) | 143 (42) | 231 (43) | 31 (46) |
| Atrial fibrillation or flutter, n (%) | 358 (36) | 15 (34) | 120 (35) | 190 (35) | 31 (46) |
| Medications | |||||
| Aspirin, n (%) | 788 (79) | 35 (80) | 278 (81) | 422 (78) | 52 (76) |
| Other antiplatelet agents, n (%) | 149 (15) | 9 (20) | 56 (16) | 70 (13) | 13 (19) |
| ACE inhibitors, n (%) | 342 (34) | 16 (36) | 126 (37) | 182 (34) | 17 (25) |
| Angiotensin II receptor blockers, n (%) | 281 (28) | 13 (30) | 101 (30) | 149 (27) | 18 (26) |
| Calcium channel blockers, n (%) | 261 (26) | 13 (30) | 98 (29) | 133 (25) | 17 (25) |
| Warfarin, n (%) | 199 (20) | 13 (30) | 71 (21) | 100 (18) | 14 (21) |
| Novel oral anticoagulants, n (%) | 36 (4) | 1 (2) | 11 (3) | 20 (4) | 3 (4) |
| Digoxin, n (%) | 43 (4) | 2 (5) | 11 (3) | 25 (5) | 5 (7) |
| Amiodarone, n (%) | 36 (4) | 0 (0) | 16 (5) | 18 (3) | 2 (3) |
| Other antiarrhythmic agents, n (%) | 14 (1) | 0 (0) | 2 (1) | 10 (2) | 2 (3) |
| Diuretics, n (%) | 321 (32) | 18 (41) | 106 (31) | 178 (33) | 19 (28) |
| Statins, n (%) | 818 (82) | 37 (84) | 287 (84) | 438 (81) | 53 (78) |
| Oral hypoglycemic agents, n (%) | 255 (26) | 12 (27) | 100 (29) | 121 (22) | 21 (31) |
| Plasma concentrations | |||||
| Mean metoprolol plasma concentrations, ng/ml | 111 ± 141 | 270 ± 230 | 151 ± 153 | 101 ± 130 | 82.7 ± 117 |
| Mean α‐OH‐metoprolol plasma concentrations, ng/ml | 48.7 ± 52.5 | 2.4 ± 6.2 | 16.0 ± 15.9 | 52.2 ± 53.7 | 64.8 ± 50.8 |
| Daily metoprolol dose | |||||
| Mean daily dose, mg | 84.3 ± 57.1 | 81.0 ± 56.2 | 75.5 ± 51.8 | 88.6 ± 59.0 | 95.8 ± 61.5 |
| Daily metoprolol dose, by categories, n (%) | |||||
| ≤12.5 | 16 (2) | 2 (5) | 8 (2) | 6 (1) | 0 (0) |
| >12.5–25 | 144 (14) | 5 (11) | 65 (19) | 65 (12) | 8 (12) |
| >25–50 | 351 (35) | 17 (39) | 122 (36) | 189 (35) | 23 (34) |
| >50–100 | 318 (32) | 13 (30) | 107 (31) | 177 (33) | 21 (31) |
| >100–150 | 63 (6) | 1 (2) | 13 (4) | 44 (8) | 3 (4) |
| >150–200 | 91 (9) | 6 (14) | 22 (6) | 51 (9) | 12 (18) |
| >200 | 14 (1) | 0 (0) | 4 (1) | 9 (2) | 1 (1) |
| CYP2D6 inhibitors | |||||
| Moderate | 7 (0.7) | 0 (0) | 1 (0.3) | 5 (0.9) | 1 (1.5) |
| Strong | 29 (2.9) | 2 (4.6) | 13 (3.8) | 11 (2.0) | 3 (4.4) |
Values are presented as means ± SD unless otherwise specified.
Abbreviations: ACE, angiotensin‐converting enzyme; BMI, body mass index; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
Metabolizing phenotype could not be inferred in 3 participants due to triallelic variants.
CYP2D6 and plasma concentrations
The mean metoprolol and α‐OH‐metoprolol concentrations in the cohort were 111.0 ± 141.4 ng/ml (median 59.3) and 48.7 ± 52.5 ng/ml (median 35.1), respectively. Only 2% of patients presented metoprolol concentrations below the LLOQ, and 4% for α‐OH‐metoprolol. Those with values below the LLOQ for both analytes comprised 1% of the population, indicating that few patients had not taken metoprolol recently.
Metoprolol concentrations decreased with higher metabolizing capacity (Figure 1a). The association between CYP2D6 and metoprolol concentrations was highly significant across all models (all p < 10−14). Age, sex, metoprolol daily dose, and weight were significantly associated with metoprolol concentrations in the fully adjusted model (Table 2). We observed that the association between CYP2D6 genotype‐inferred phenotype and metoprolol concentrations increased when metoprolol daily dose was introduced in the multivariable model.
FIGURE 1.
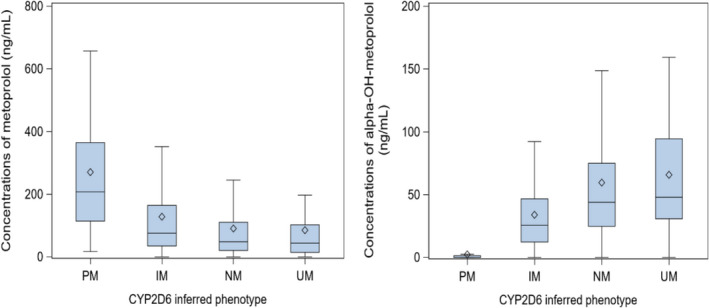
Plasma concentrations of metoprolol and α‐OH‐metoprolol. Concentrations of metoprolol (left) and α‐OH‐metoprolol (right) by CYP2D6‐inferred phenotype. Data presented as untransformed values. IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM: ultrarapid metabolizer. Central bar of the box: median; lower bar of the box: first quartile; upper bar of the box: third quartile; diamond: mean; bar below the box: minimum (excluding outliers); bar above the box: maximum (excluding outliers)
TABLE 2.
Association between metoprolol concentrations and CYP2D6 genotype‐inferred phenotypes
| Effect | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | |
| CYP2D6 inferred phenotype | −0.506 (0.062) | 1.4 × 10−15 | −0.511 (0.061) | 3.0 × 10−16 | −0.632 (0.051) | 7.0 × 10−33 | −0.634 (0.051) | 1.7 × 10−33 | −0.6306 (0.0503) | 1.5 × 10−33 |
| Age | ‐ | ‐ | 0.017 (0.005) | 0.0005 | 0.015 (0.004) | 0.0001 | 0.012 (0.004) | 0.0025 | 0.0121 (0.0040) | 0.0024 |
| Female sex | ‐ | ‐ | 0.398 (0.094) | 2.3 × 10−5 | 0.392 (0.077) | 4.6 × 10−7 | 0.298 (0.081) | 0.0002 | 0.2855 (0.0803) | 0.00040 |
| Metoprolol dose | ‐ | ‐ | ‐ | ‐ | 0.013 (0.001) | 7.7 × 10−85 | 0.013 (0.001) | 1.2 × 10−87 | 0.0133 (0.0006) | 5.5 × 10−89 |
| Weight | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | −0.008 (0.002) | 0.0002 | −0.0080 (0.0022) | 0.0002 |
| CYP2D6 inhibitors | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | 0.3851 (0.0978) | 0.00009 |
Model 1: crude model; model 2: model adjusted for age and sex; model 3: model adjusted for age, sex, and metoprolol dose; model 4: model adjusted for age, sex, metoprolol dose, and weight; model 5: model adjusted for age, sex, metoprolol dose, weight, and CYP2D6 inhibitors. Intercepts for model 1: 4.821, model 2: 4.012, model 3: 3.191, model 4: 4.003, and model 5: 3.670.
Abbreviation: SE, standard error.
Concentrations of α‐OH‐metoprolol increased with higher CYP2D6 metabolizing capacity (Figure 1b). The association between CYP2D6‐inferred phenotype and α‐OH‐metoprolol remained highly significant in the fully adjusted model (all p < 10−57). Age, sex, metoprolol daily dose, but not weight, were also significantly associated with α‐OH‐metoprolol concentrations (Table 3). Finally, the concomitant use of CYP2D6 inhibitors was significantly associated with both metoprolol and α‐OH‐metoprolol concentrations.
TABLE 3.
Association between α‐OH‐metoprolol concentrations and CYP2D6 genotype‐inferred phenotype
| Effect | Model 1 | Model 2 | Model 3 | Model 4 | Model 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | |
| CYP2D6 inferred phenotype | 0.877 (0.051) | 2.3 × 10−58 | 0.876 (0.050) | 1.4 × 10−59 | 0.790 (0.044) | 3.1 × 10−63 | 0.789 (0.044) | 3.5 × 10−63 | 0.7851 (0.0432) | 6.8 × 10−64 |
| Age | ‐ | ‐ | 0.018 (0.004) | 6.4 × 10−6 | 0.016 (0.003) | 1.4 × 10−6 | 0.015 (0.003) | 1.7 × 10−5 | 0.0150 (0.0034) | 0.00001 |
| Female sex | ‐ | ‐ | 0.224 (0.077) | 0.0035 | 0.214 (0.066) | 0.0013 | 0.175 (0.070) | 0.0124 | 0.1879 (0.0690) | 0.00656 |
| Metoprolol dose | ‐ | ‐ | ‐ | ‐ | 0.009 (0.001) | 1.4 × 10−64 | 0.010 (0.001) | 3.6 × 10−65 | 0.0096 (0.0005) | 4.4 × 10−66 |
| Weight | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | −0.003 (0.002) | 0.0709 | −0.0035 (0.0018) | 0.06045 |
| CYP2D6 inhibitors | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | −0.4219 (0.0840) | 6.0 × 10−7 |
Model 1: crude model; model 2: model adjusted for age and sex; model 3: model adjusted for age, sex and metoprolol dose; model 4: model adjusted for age, sex, metoprolol dose, and weight; and model 5: model adjusted for age, sex, metoprolol dose, weight, and CYP2D6 inhibitors. Intercepts for model 1: 1.936, model 2: 0.929, model 3: 0.348, model 4: 0.689, and model 5: 0.554.
Abbreviation: SE, standard error.
CYP2D6 and daily metoprolol dosing
We observed a significant, yet more modest, association between CYP2D6‐inferred phenotype and metoprolol daily dose (all p < 0.0003). Other than CYP2D6‐inferred phenotype, only weight was associated with metoprolol daily dose in the fully adjusted model (Table 4).
TABLE 4.
Association between CYP2D6 genotype‐inferred phenotype and metoprolol daily dosing
| Effect | Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | Estimate (SE) | p value | |
| CYP2D6 inferred phenotype | 0.115 (0.031) | 0.0002 | 0.114 (0.031) | 0.0002 | 0.114 (0.031) | 0.0002 | 0.1141 (0.0307) | 0.00021 |
| Age | ‐ | ‐ | 0.002 (0.002) | 0.4619 | 0.004 (0.002) | 0.0860 | 0.0042 (0.0024) | 0.08601 |
| Female sex | ‐ | ‐ | 0.025 (0.047) | 0.6008 | 0.094 (0.049) | 0.0565 | 0.0948 (0.0492) | 0.05413 |
| Weight | ‐ | ‐ | ‐ | ‐ | 0.006 (0.001) | 6.9 × 10−6 | 0.0059 (0.0013) | 0.00001 |
| CYP2D6 inhibitors | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | −0.0336 (0.0599) | 0.57479 |
Model 1: crude model; model 2: model adjusted for age and sex; model 3: model adjusted for age, sex, and weight, and model 4: model adjusted for age, sex, weight, and CYP2D6 inhibitors. Intercepts for model 1: 4.041, model 2: 3.942, model 3: 3.333, and model 4: 3.242.
Abbreviation: SE, standard error.
CYP2D6 and heart rate
Consistent with the observed decrease in metoprolol concentrations with greater metabolic capacity, we observed increasing heart rate with increases in CYP2D6 inferred metabolizing capacity (Figure 2). This association was significant in the crude and the adjusted models (all p < 0.0004; Supplementary Table S2).
FIGURE 2.
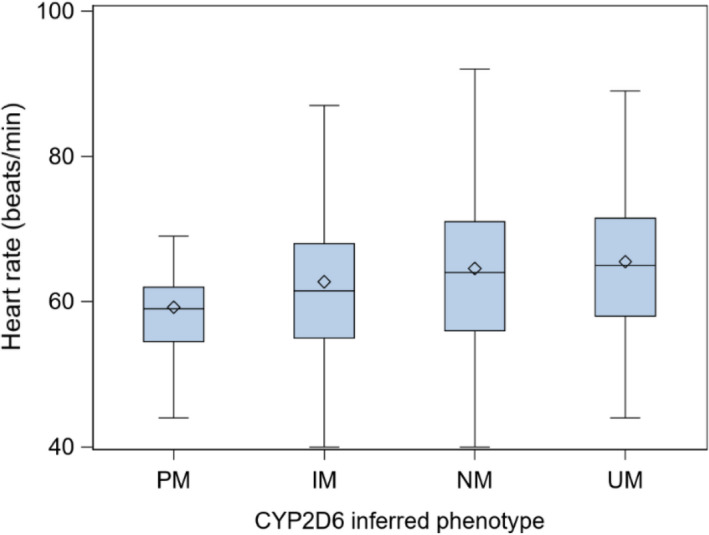
Resting heart rate across CYP2D6 genotype‐inferred phenotypes. Data presented as untransformed values. IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer. Central bar of the box: median; lower bar of the box: first quartile; upper bar of the box: third quartile; diamond: mean; bar below the box: minimum (excluding outliers); bar above the box: maximum (excluding outliers)
Sensitivity analyses: single‐variant models
For sensitivity analyses, we also investigated whether similar associations could be obtained using a single variant, rs3892097, which presented an allelic frequency of 15.94% in our cohort. As was found for the genotype‐inferred phenotype classification, in all unadjusted and adjusted models, the presence of the rs3892097 variant was significantly associated with both plasma concentration levels of metoprolol (all p < 10−7) and α‐OH‐metoprolol (all p < 10−47; Supplementary Tables S3 and S4). For the secondary end points of daily metoprolol heart rate and dosing, again, rs3892097 was associated with these end points, albeit more modestly in all models (Tables S5 and S6).
DISCUSSION
As part of this study, we provide proof to the concept that using randomly collected plasma samples as part of genetic cohort studies can be leveraged to identify predictors of drug concentrations. We observed highly significant associations between CYP2D6‐inferred metabolizer phenotype, as well as the rs3892097 variant, and plasma concentrations of metoprolol and α‐OH‐metoprolol measured in samples randomly drawn since the previous metoprolol dose at the baseline visit of the MHI Hospital Cohort. Our investigation provides consistent results in the magnitude of these inferred phenotypes with those from a previous meta‐analysis assessing the effect of CYP2D6‐inferred metabolizer phenotype on the variability in PK profiles of metoprolol. 2 Thus, the results of this study support the concept that random blood samples collected as part of large observational studies can be leveraged to conduct PGx association studies in unselected patients treated with a broad range of doses of a given medication to identify predictors of drug concentrations.
The most immediate implication of our study is the possibility that large observational genetic cohorts could be leveraged to identify PGx markers related to drug dosing requirements and drug concentrations, either as a single variant or using a complete phenotype classification, with the caveat that plasma or serum samples and information regarding drug dosing have also been collected. Our approach may facilitate the emergence of collaborative, multicohort, international consortia to conduct exploratory genomewide association studies (GWAS) of drug concentrations, beyond studies that initially focus on one or a few selected drugs investigated as part of a clinical study or drug monitoring programs. Such collaborations already represent a hallmark of genomic discovery for complex traits and diseases. 4 , 5 , 6 The cumulative experience from investigations of common diseases has shown that initial single‐cohort investigations generally allow the discovery of common variants with large effects that explain only a small proportion of genetic factors in complex diseases. For example, initial GWAS of hypertension that included only thousands of individuals led to few genetic discoveries, 22 whereas recent initiatives have included an excess of one million participants and reported over 900 loci associated with hypertension. 23 Based on the experience and results of the aforementioned GWAS of complex diseases, 4 , 5 , 6 , 23 large prospective PGx endeavors using biorepositories not initially designed to study drug PKs, in addition to aforementioned existing resources, could provide sample sizes required for the discovery of new and unsuspected genes or signaling pathways. 24 , 25 , 26 , 27 Moreover, such collaborative efforts could also incorporate clinical therapeutic drug monitoring PGx studies. 28 , 29
A noteworthy observation is that CYP2D6 phenotypes were more significantly associated to concentration levels of metoprolol and α‐OH‐metoprolol compared to metoprolol dosing. This is partly because doses used in the clinic are dependent on the prescriber’s preferences. 30 Therefore, drug doses by themselves may not be a suitable proxy to evaluate genetic determinants of dosing requirements or PK parameters and show that drug concentration measurements are critical. This is emphasized by drugs with only a single dosing regimen in clinical practice, such as clopidogrel, despite the presence of interindividual variability and genetic determinants of PK profiles and efficacy. 31 , 32
Interestingly, our models highlighted increases in resting heart rate relative associated higher CYP2D6 metabolic capacity. It is important to highlight that this observation should be made with caution. Due to the cross‐sectional nature of this analysis, it does not in fact reflect “response” to metoprolol. Thus, despite this observation in previous smaller prospective studies, 33 bias and chance cannot be excluded. Given the association between higher heart rate and worse outcomes in cardiovascular disease, 34 studies investigating the impact of CYP2D6 metabolizer status on the clinical outcomes of patients undergoing metoprolol therapy appear warranted.
Strengths and limitations
A strength of our investigation is that we validated our concept using two PGx associations between CYP2D6 and two analytes of metoprolol metabolism which have been repeatedly validated. Moreover, the associations observed regarding the plasma concentrations of both analytes were consistent with those previously reported. 2
Another strength from this study is that it was conducted in a “real‐life,” polymedicated population presenting multiple morbidities. This represents a strength for two reasons. As most previous studies investigating the association between CYP2D6 and metoprolol’s PKs focused exclusively on healthy individuals, the current study validates that it can be extended to “real‐life” populations. Ultimately, these are the populations in which PGx and precision medicine are expected to improve drug efficacy and safety. Regarding the concept and hypothesis tested here, this makes our findings more widely generalizable and applicable to other unselected populations which constitute most large cohorts used in genetic research.
One limitation is that the exact formulation of metoprolol was not routinely collected. Despite this limitation, which would have added heterogeneity to our analyses, we were able to convincingly validate the drug‐gene pair association. Furthermore, a caveat to our finding is that, considering that metoprolol’s half‐life ranges between 3 and 7 h from NM to PM, 2 whether our approach can be extended to drugs with shorter half‐lives warrants confirmation. Finally, the usefulness to identify rare factors, genetic or other, that can influence the concentrations of metoprolol requires further studies. Expanding the size of collaborative PGx investigations to sufficient sizes to explore the contribution of rare variants, as well as sex‐specific PGx associations, could help explain part of the “missing heritability” in PGx.
In conclusion, our results provide supporting evidence that randomly drawn blood samples from biorepositories could be leveraged to identify genetic determinants associated with drug concentrations and dosing. New methods or uses for already existing databases may be exploited to discover PGx associations related to drug metabolism and response without the limitations associated with current approaches like GWAS and traditional PK studies.
CONFLICT OF INTEREST
S.D. reports grants outside the submitted work from Pfizer, AstraZeneca, and RMS/Dalcor. A patent pertaining to pharmacogenomics‐guided CETP inhibition was granted and J.C.T. and M.P.D. are mentioned as authors. J.C.T. and M.P.D. have a minor equity interest in DalCor. J.C.T. has received research grants from Amarin, AstraZeneca, DalCor, Esperion, Ionis, Sanofi and Servier; honoraria from DalCor, Pfizer, Sanofi, and Servier. M.P.D. has received honoraria from Dalcor, GSK and Illumina, and has research support (access to samples and data) from AstraZeneca, Pfizer, Servier, Sanofi, and GlaxoSmithKline. All other authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
M.M., E.O., M.J.G., and S.D. wrote the manuscript. M.M., G.L., M.J., E.O., I.M., M.P.D., J.C.T. and S.D. designed the research. M.M., E.O., M.J.G., D.B., and J.C.T. performed the research. M.M., E.O., I.M., M.P.D., and S.D. analyzed the data. G.L. and D.B. contributed new reagents/analytical tools.
Supporting information
Figure S1
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Supplementary Material
ACKNOWLEDGEMENTS
None.
Meloche M, Leclair G, Jutras M, et al. Leveraging large observational studies to discover genetic determinants of drug concentrations: A proof‐of‐concept study. Clin Transl Sci. 2022;15:1063‐1073. doi: 10.1111/cts.13230
Funding information
This work was supported by the Montreal Heart Institute Foundation, the Université de Montréal Beaulieu‐Saucier Chair in Pharmacogenomics (S.D.), the Molson Foundation (S.D.), the Université de Montréal Faculty of Pharmacy (M.M.), and the Université de Montréal department of Higher Education and Postdoctoral Studies (M.M.)
REFERENCES
- 1. Abdullah‐Koolmees H, van Keulen AM, Nijenhuis M, Deneer VHM. Pharmacogenetics guidelines: overview and comparison of the DPWG, CPIC, CPNDS, and RNPGx guidelines. Front Pharmacol. 2021;11:595219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blake CM, Kharasch ED, Schwab M, Nagele P. A meta‐analysis of CYP2D6 metabolizer phenotype and metoprolol pharmacokinetics. Clin Pharmacol Ther. 2013;94(3):394‐399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hamelin BA, Bouayad A, Methot J, et al. Significant interaction between the nonprescription antihistamine diphenhydramine and the CYP2D6 substrate metoprolol in healthy men with high or low CYP2D6 activity. Clin Pharmacol Ther. 2000;67(5):466‐477. [DOI] [PubMed] [Google Scholar]
- 4. Auer PL, Teumer A, Schick U, et al. Rare and low‐frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat Genet. 2014;46(6):629‐634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ehret GB, Ferreira T, Chasman DI, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48(10):1171‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao W, Rasheed A, Tikkanen E, et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet. 2017;49(10):1450‐1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103‐141. [DOI] [PubMed] [Google Scholar]
- 8. Caudle KE, Sangkuhl K, Whirl‐Carrillo M, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the clinical pharmacogenetics implementation consortium and Dutch Pharmacogenetics Working Group. Clin Transl Sci. 2020;13(1):116‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laurent‐Kenesi MA, Funck‐Brentano C, Poirier JM, Decolin D, Jaillon P. Influence of CYP2D6‐dependent metabolism on the steady‐state pharmacokinetics and pharmacodynamics of metoprolol and nicardipine, alone and in combination. Br J Clin Pharmacol. 1993;36(6):531‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Batty JA, Hall AS, White HL, et al. An investigation of CYP2D6 genotype and response to metoprolol CR/XL during dose titration in patients with heart failure: a MERIT‐HF substudy. Clin Pharmacol Ther. 2014;95(3):321‐330. [DOI] [PubMed] [Google Scholar]
- 11. Turcot V, Brunet J, Daneault C, Tardif JC, Des Rosiers C, Lettre G. Validation of fatty acid intakes estimated by a food frequency questionnaire using erythrocyte fatty acid profiling in the Montreal Heart Institute Biobank. J Hum Nutr Diet. 2015;28(6):646‐658. [DOI] [PubMed] [Google Scholar]
- 12. Chami N, Chen MH, Slater AJ, et al. Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am J Hum Genet. 2016;99(1):8‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coller JK, Ramachandran J, John L, Tuke J, Wigg A, Doogue M. The impact of liver transplant recipient and donor genetic variability on tacrolimus exposure and transplant outcome. Br J Clin Pharmacol. 2019;85(9):2170‐2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mottet F, Vardeny O, de Denus S. Pharmacogenomics of heart failure: a systematic review. Pharmacogenomics. 2016;17(16):1817‐1858. [DOI] [PubMed] [Google Scholar]
- 15. de Denus S, Rouleau JL, Mann DL, et al. CYP3A4 genotype is associated with sildenafil concentrations in patients with heart failure with preserved ejection fraction. Pharmacogenomics J. 2018;18(2):232‐237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. US Food and Drug Administration . Drug development and drug interactions: table of substrates, inhibitors and inducers. Available: https://www.fda.gov/drugs/drug‐interactions‐labeling/drug‐development‐and‐drug‐interactions‐table‐substrates‐inhibitors‐and‐inducers. Last updated March 6, 2020. Accessed on September 9, 2021.
- 17. Cicali EJ, Elchynski AL, Cook KJ, et al. How to integrate CYP2D6 phenoconversion into clinical pharmacogenetics: a tutorial. Clin Pharmacol Ther. 2021;110(3):677‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rau T, Heide R, Bergmann K, et al. Effect of the CYP2D6 genotype on metoprolol metabolism persists during long‐term treatment. Pharmacogenetics. 2002;12(6):465‐472. [DOI] [PubMed] [Google Scholar]
- 19. Kirchheiner J, Heesch C, Bauer S, et al. Impact of the ultrarapid metabolizer genotype of cytochrome P450 2D6 on metoprolol pharmacokinetics and pharmacodynamics. Clin Pharmacol Ther. 2004;76(4):302‐312. [DOI] [PubMed] [Google Scholar]
- 20. Sharma A, Pibarot P, Pilote S, et al. Modulation of metoprolol pharmacokinetics and hemodynamics by diphenhydramine coadministration during exercise testing in healthy premenopausal women. J Pharmacol Exp Ther. 2005;313(3):1172‐1181. [DOI] [PubMed] [Google Scholar]
- 21. Gaedigk A, Sangkuhl K, Whirl‐Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19(1):69‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wellcome Trust Case Control Consortium . Genome‐wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evangelou E, Warren HR, Mosen‐Ansorena D, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412‐1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelson MR, Bacanu SA, Mosteller M, et al. Genome‐wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J. 2009;9(1):23‐33. [DOI] [PubMed] [Google Scholar]
- 25. Takeuchi F, McGinnis R, Bourgeois S, et al. A genome‐wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5(3):e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giacomini KM, Yee SW, Mushiroda T, Weinshilboum RM, Ratain MJ, Kubo M. Genome‐wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov. 2017;16(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh S, El Rouby N, McDonough CW, et al. Genomic association analysis reveals variants associated with blood pressure response to beta‐blockers in European Americans. Clin Transl Sci. 2019;12(5):497‐504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jukić MM, Haslemo T, Molden E, Ingelman‐Sundberg M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a retrospective study based on 2,087 patients. Am J Psychiatry. 2018;175(5):463‐470. [DOI] [PubMed] [Google Scholar]
- 29. Jukic MM, Smith RL, Haslemo T, Molden E, Ingelman‐Sundberg M. Effect of CYP2D6 genotype on exposure and efficacy of risperidone and aripiprazole: a retrospective, cohort study. Lancet Psychiatry. 2019;6(5):418‐426. [DOI] [PubMed] [Google Scholar]
- 30. Andrade JG, Krahn AD, Skanes AC, Purdham D, Ciaccia A, Connors S. Values and preferences of physicians and patients with nonvalvular atrial fibrillation who receive oral anticoagulation therapy for stroke prevention. Can J Cardiol. 2016;32(6):747‐753. [DOI] [PubMed] [Google Scholar]
- 31. Mega JL, Close SL, Wiviott SD, et al. Cytochrome p‐450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354‐362. [DOI] [PubMed] [Google Scholar]
- 32. Claassens DMF, Vos GJA, Bergmeijer TO, et al. A genotype‐guided strategy for oral P2Y12 inhibitors in primary PCI. N Engl J Med. 2019;381(17):1621‐1631. [DOI] [PubMed] [Google Scholar]
- 33. Meloche M, Khazaka M, Kassem I, Barhdadi A, Dubé M‐P, de Denus S. CYP2D6 polymorphism and its impact on the clinical response to metoprolol: a systematic review and meta‐analysis. Br J Clin Pharmacol. 2020;86(6):1015‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steinberg BA, Kim S, Thomas L, et al. Increased heart rate is associated with higher mortality in patients with atrial fibrillation (AF): results from the outcomes registry for better informed treatment of AF (ORBIT‐AF). J Am Heart Assoc. 2015;4(9):e002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Table S2
Table S3
Table S4
Table S5
Table S6
Supplementary Material


