Summary
Polygenic risk scores (PRS) measure genetic disease susceptibility by combining risk effects across the genome. For coronary artery disease (CAD), type 2 diabetes (T2D), and breast and prostate cancer, we performed cross-ancestry evaluation of genome-wide PRSs in six biobanks in Europe, the United States, and Asia. We studied transferability of these highly polygenic, genome-wide PRSs across global ancestries, within European populations with different health-care systems, and local population substructures in a population isolate. All four PRSs had similar accuracy across European and Asian populations, with poorer transferability in the smaller group of individuals of African ancestry. The PRSs had highly similar effect sizes in different populations of European ancestry, and in early- and late-settlement regions with different recent population bottlenecks in Finland. Comparing genome-wide PRSs to PRSs containing a smaller number of variants, the highly polygenic, genome-wide PRSs generally displayed higher effect sizes and better transferability across global ancestries. Our findings indicate that in the populations investigated, the current genome-wide polygenic scores for common diseases have potential for clinical utility within different health-care settings for individuals of European ancestry, but that the utility in individuals of African ancestry is currently much lower.
Keywords: polygenic risk score, health disparities, global health, ancestry, precision medicine, risk prediction, coronary artery disease, diabetes, breast cancer, prostate cancer
Highlights
-
•
An evaluation of cross-ancestry transferability of polygenic risk scores
-
•
Four common diseases in four global ancestry groups and across Europe were studied
-
•
PRS transferability was high across European ancestry and lowest for African ancestry
-
•
PRS transferability was good across population substructures in Finland
Combining six biobanks in Europe, the United States, and Asia, Mars et al. evaluated cross-ancestry transferability of polygenic risk scores for four common diseases: coronary artery disease, type 2 diabetes, and breast and prostate cancer. They observed good cross-ancestry transferability between individuals with different European ancestry, but poorer transferability in individuals of African, South Asian, and East Asian ancestry, which highlights the need for diversity in polygenic risk score development for clinical translation.
Introduction
Polygenic risk scores (PRSs) capture an individual’s genetic susceptibility to diseases by summarizing the estimated polygenic effects across the genome. PRSs have shown great promise for improving detection of high-risk individuals in many common complex diseases, such as cardiometabolic diseases and common cancers.1, 2, 3, 4 However, these studies have been heavily biased toward individuals of European ancestry and have provided limited understanding about the transferability of the PRSs across ancestries. This currently limits the potential clinical utility of the PRS and may lead to exacerbation of health disparities in implementation of the PRSs across different societies and health-care systems.5
We evaluated the variability of the PRS risk estimates across multiple populations and ancestry groups in four common complex diseases that have shown promise beyond routinely used clinical risk scores: coronary artery disease (CAD), type 2 diabetes (T2D), breast cancer, and prostate cancer.2,6, 7, 8, 9, 10 We combined genome-wide genotype data with disease endpoints for four ancestry groups across six biobanks covering one million individuals. We calculated genome-wide PRSs, obtaining input weights from genome-wide association studies (GWASs) published and made available by large disease genetics consortia.11, 12, 13, 14 These consortia GWASs and corresponding linkage disequilibrium (LD) reference panels consisted primarily of individuals of European ancestry, and they provided weights for genetic variants used for generating the PRS. This reflects the current reality where most PRSs are developed and tested in individuals of European ancestry. To extensively assess the impact of Eurocentric study biases on PRS portability, we performed a cross-ancestry evaluation of our genome-wide PRSs of on three levels: across global ancestries, across European populations, and locally within Finland, a European country with a well-known population substructure.15
Results
The descriptive statistics for the six biobank studies are shown in Table 1. These include BioBank Japan (n = 178,726), Estonian Biobank (n = 110,597), FinnGen (n = 258,402), The Trøndelag Health Study (HUNT, n = 69,422), Mass General Brigham (MGB) Biobank (n = 27,231), and UK Biobank (n = 358,922). The represented ancestries are European, South Asian, East Asian, and African ancestry. The total number of cases was 88,830 for CAD, 110,685 for T2D, 32,922 for breast cancer, and 26,700 for prostate cancer, and the mean age ranged from 43.4 years in Estonian Biobank to 63.1 in BioBank Japan. The proportion of women ranged from 46.3% in BioBank Japan to 67.3% in Estonian Biobank.
Table 1.
Study characteristics
| Biobank | Ancestry | Sample size | Age, mean (SD) | Women, % | CAD |
T2D |
Breast cancer |
Prostate cancer |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases N = 88,830 | AAO, mean (SD) | Cases N = 110,685 | AAO, mean (SD) | Cases, N = 32,922 | AAO, mean (SD) | Cases, N = 26,700 | AAO, mean (SD) | ||||||
| European ancestry (n = 807,793) | |||||||||||||
| Estonian Biobank | Estonia | EUR | 110,597 | 43.4 (16.0) | 67.3 | 5,064 | 67.5 (11.9) | 7,066 | 60.9 (12.7) | 1,379 | 59.5 (13.4) | 1,202 | 68.7 (9.2) |
| FinnGen | Finland | EUR | 258,402 | 60.3 (17.1)∗ | 56.5 | 25,706 | 64.9 (11.8) | 37,001 | 60.1 (11.9) | 11,573 | 59.0 (11.6) | 8,709 | 68.5 (8.1) |
| HUNT | Norway | EUR | 69,422 | 50.8 (17.0) | 53.0 | 6,594 | 69.0 (12.5) | 5,228 | 68.1 (13.4) | 1,731 | 61.7 (13.4) | 2,224 | 70.6 (9.2) |
| MGB Biobank | United States | EUR | 25,696 | 60.0 (16.5) | 53.1 | 3,206 | – | 5,182 | – | 1,513 | – | 1,593 | – |
| UK Biobank | United Kingdom | EUR | 343,676 | 56.9 (8.0) | 53.7 | 17,986 | 62.1 (8.9) | 13,616 | 54.6 (8.5) | 11,075 | 54.0 (8.1) | 7,429 | 59.7 (6.2) |
| Other ancestry (n = 195,507) | |||||||||||||
| BioBank Japan | Japan | EAS | 178,726 | 63.1 (14.0) | 46.3 | 29,080 | 61.7 | 40,121 | 56.2 | 5,316 | 56.1 | 5,192 | 71.1 |
| MGB Biobank | United States | AFR | 1,535 | 54.1 (16.3) | 61.4 | 285 | – | 660 | – | 64 | – | 80 | – |
| UK Biobank | United Kingdom | AFR | 7,618 | 51.9 (8.1) | 57.0 | 169 | 56.9 (10.3) | 691 | 50.2 (8.9) | 132 | 50.2 (9.1) | 199 | 57.4 (7.4) |
| UK Biobank | United Kingdom | SAS | 7,628 | 53.4 (8.5) | 46.1 | 740 | 58.6 (9.7) | 1,120 | 50.0 (8.7) | 139 | 51.2 (7.9) | 72 | 59.6 (7.2) |
EUR = European, EAS = East Asian, AFR = African (self-reported African/Caribbean in UK Biobank), SAS = South Asian, CAD = coronary artery disease, T2D = type 2 diabetes, AAO = age at onset, SD = standard deviation. Disease definitions are listed by cohort in STAR Methods. In HUNT, we show the age at baseline for those participating in either HUNT2 or HUNT3, and a mean of these baseline ages for individuals participating in both. ∗Age at the end of follow-up.
For each disease, our main PRSs were calculated with LDpred (>6 million variants in each PRS; Table S1), using weights from the largest published GWASs that do not contain data from the UK Biobank.11, 12, 13, 14 The PRSs were rescaled in each dataset and for each ancestry subset, to have mean 0 and standard deviation (SD) at 1. We then assessed the transferability of PRSs by comparing the odds ratio (OR) estimates between biobanks and ancestry groups on three levels of variation in ancestry: (1) across global ancestries, (2) across populations with European ancestry but with varying health-care systems, and (3) across subpopulations in Finland, a country with a nationwide uniform health-care system and a well-known early- and late-settlement division in population structure, with previous evidence of PRS stratification.16
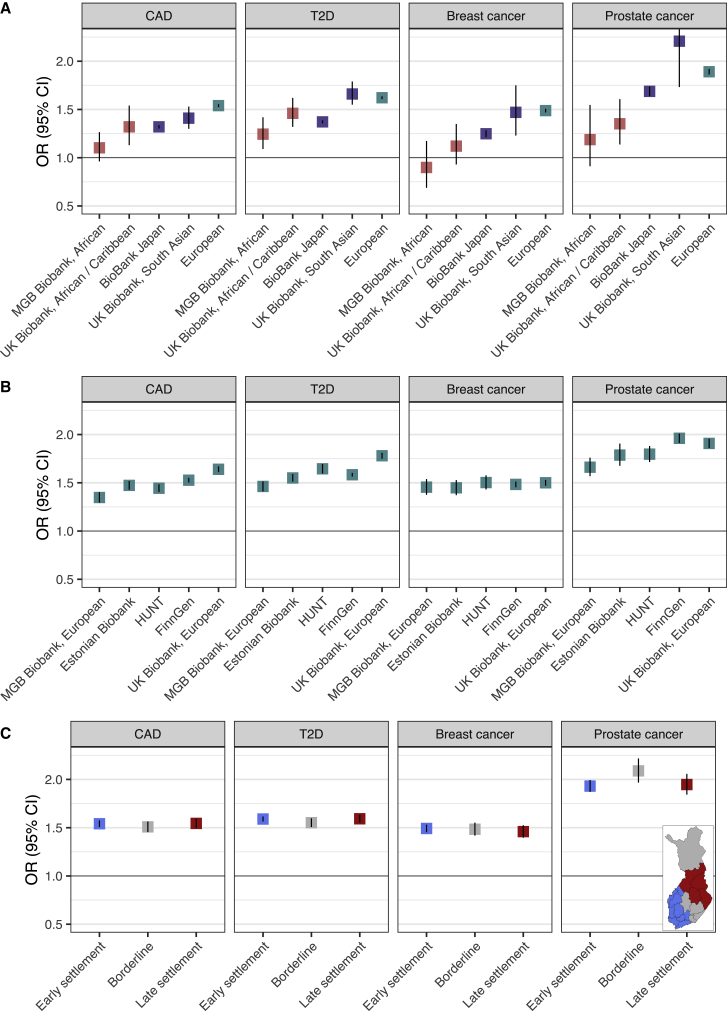
Figure 1A shows the ORs per SD increase in PRS across the three ancestry groups: European, South and East Asian, and African ancestries. The OR estimates ranged from 1.10 to 1.53 for CAD, from 1.24 to 1.66 for T2D, from 0.90 to 1.49 for breast cancer, and from 1.35 to 2.21 for prostate cancer (Table S2). For all four diseases, the effect sizes were lowest in individuals of African ancestry and highest in individuals of European ancestry, followed by individuals of South and East Asian ancestry with similar or slightly lower effect sizes. In breast cancer, we did not detect an association for women of African ancestry (OR 1.12, 95% CI 0.93–1.35 in UK Biobank, OR 0.90, 0.69–1.35 in MGB Biobank), but looking at the effects across different LDpred parameters for fraction of causal variants in UK Biobank (Figure S1), the PRS would be associated with OR 1.40 (1.13–1.72) in individuals of African ancestry, had the fraction been chosen based on individuals of African ancestry, instead of individuals of European ancestry. In other diseases, the choice of the fraction had only a fairly small effect.
Figure 1.
Effect sizes of polygenic risk scores (PRSs) across ancestries
(A) The results across ancestry groups, with “European” representing a pooled OR of effect sizes from (B).
(B) The results across different populations with European ancestry.
(C) The results across early- and late-settlement regions in Finland (FinnGen).
ORs with 95% CIs (CI) are shown for 1 SD increase in PRS. See Table 1 for respective number of cases and controls. The pooled OR (“European”) in (A) was obtained by random effects meta-analysis of effects shown in (B). In (C), out of 258,402 in FinnGen, 8,117 individuals were excluded, comprising 3,157 born abroad, 4,304 born in regions ceded to the Soviet Union, 182 born in Åland Islands, and 474 with missing data. Detailed information of the Finnish regions in (C) is provided in the description of FinnGen in STAR Methods. CAD = coronary artery disease, T2D = type 2 diabetes.
Figure 1B compares the effect sizes across different populations with European ancestry. Overall, the variation between estimates was much smaller in European ancestry samples, ranging from 1.35 to 1.64 for CAD, from 1.46 to 1.78 for T2D, from 1.45 to 1.50 for breast cancer, and from 1.66 to 1.96 for prostate cancer. For CAD and T2D, the estimates were highest in the UK Biobank and lowest in MGB Biobank. Breast cancer estimates were highly similar across all biobanks, and prostate cancer estimates were highest in Finns.
Figure 1C shows the estimates in early- and late-settlement regions in Finland. The effect sizes were highly consistent throughout the regions for all four diseases. The most similar effect sizes were again detected for breast cancer. The findings were highly similar also across a more detailed set of geographic regions (Figure S2).
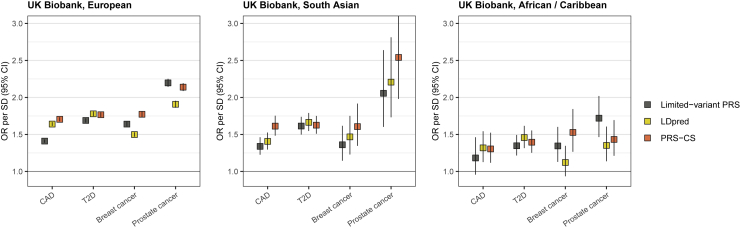
Lastly, we compared in UK Biobank the LDpred PRSs to two other types of PRSs generated primarily in individuals of European ancestry: (1) to previously published PRSs containing a smaller number of variants3,10,17,18 and (2) to genome-wide PRSs generated with PRS-CS, which restricts analyses to HapMap3 variants (Figure 2, Table S3). The highest effect size was observed in 2/4 diseases (European) and 3/4 diseases (South Asian) for PRS-CS. In T2D, the effect sizes were fairly similar across the three PRSs. In African/Caribbean ancestry, the best-performing PRS varied by disease: in CAD, the LDpred and PRS-CS had the highest and highly similar effects; in T2D, LDpred had the highest effect size, but the difference between the different PRSs was fairly small; in breast cancer, the PRS-CS PRS had the highest effect size, with a considerable drop (to 27% of the effect size) with the LDpred PRS and a moderate drop to 70% for the limited-variant PRS; in prostate cancer, the limited-variant PRS had the highest effect size, with considerable effect size drops with the other PRSs.
Figure 2.
Comparison of polygenic risk scores (PRSs) generated with different methods
The figure shows a comparison of three types of PRSs in UK Biobank: previously published PRSs using a smaller number of variants (“limited-variant PRS”),3,10,17,18 PRSs generated with LDpred, and PRSs generated with PRS-CS. ORs with 95% CI are shown across ancestries for 1 SD increase in the PRS. Detailed effect size comparisons are in Table S3. CAD = coronary artery disease, T2D = type 2 diabetes. Table 1 shows the respective number of cases and controls.
Looking at the transferability of the different CAD PRSs across ancestries in UK Biobank (Figure 2; Table S3), the best transferability was observed for the PRS-CS PRS (drop to 90% for South Asian ancestry, and to 56% for African/Caribbean ancestry, compared to European ancestry). For the T2D PRSs, the transferability between PRSs was highly similar (drops to 85%–91% for South Asian ancestry and to 58%–65% for African/Caribbean ancestry). For the breast cancer PRSs, the best transferability to South Asian ancestry was observed for the LDpred PRS (drop to 95%) and for the PRS-CS PRS (drop to 83%), with a drop to 62% for the limited-variant PRS. For the breast cancer PRSs, the best transferability to African/Caribbean ancestry was observed for the PRS-CS PRS (drop to 74%), followed by the limited-variant PRS (drop to 60%). For prostate cancer PRSs, all PRSs showed good transferability to South Asian ancestry, but the best transferability to African/Caribbean ancestry was observed for the limited-variant PRS.
Discussion
By combining data across six biobanks with one million samples, we show that in four major diseases with great public health impact and well-developed genome-wide PRSs—CAD, T2D, breast and prostate cancer—the scores transfer well across European and, to a lesser extent, South and East Asian populations. We also show that the PRSs transfer much more poorly to individuals of African ancestry. Within populations of European ancestry, we observed only small variability in risk estimates. Within Finland, a country with well-documented genetic differences between the early-settlement region in the South and West and the late-settlement region in the East and North, we observed essentially no variability in risk estimates.16
Several studies have looked at trans-ancestry performance of PRSs for common diseases, but the majority of such studies have used PRSs containing a small number of variants, consisting of approximately tens to a few hundred genetic variants.18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Contemporary PRSs have focused on liberalizing variant inclusion to build genome-wide PRSs, which typically contain hundreds of thousands to a few million variants.30, 31, 32, 33 But, only a few studies have assessed transferability of such PRSs across ancestries,34, 35, 36 with even fewer comparing these genome-wide PRSs to ones containing a smaller number of variants.31,34,37 To our knowledge, this is the largest study to date evaluating these genome-wide European ancestry PRSs across ancestries, with additional evaluation of effects across different cohorts of European ancestry, and within a country with well-known east-west differences. Our order of effect sizes by ancestry—largest in Europeans, followed by South and East Asians, with generally lowest effect sizes detected in Africans—are consistent with population history, and they are in line with the previous studies using a smaller number of variants, with further evidence from comparisons of prediction accuracy of anthropometric traits and lipid biomarkers.5,19,22,24,26,34,38,39
The genome-wide PRSs were also compared to the PRSs containing a smaller number of variants. In general, the genome-wide PRSs, particularly PRSs generated with PRS-CS, conferred the largest effect sizes. The limited-variant PRS in prostate cancer was an exception, but it is based on a twice as large and a diverse GWAS18 compared to the LDpred and PRS-CS PRSs for prostate cancer,13 which may explain why it performed best in individuals of European ancestry. Compared to the PRSs containing a smaller number of variants, the genome-wide PRSs showed generally better performance and higher transferability to individuals of South Asian and African ancestry.13,18 The main exception was African ancestry, where the prostate cancer PRS consisting of 269 variants outperformed the LDpred and PRS-CS PRSs. One reason for this may be that the GWAS underlying the 269 PRSs is highly diverse, containing multiple cohorts of individuals of African ancestry,18 whereas in the other PRSs across the diseases, the GWAS was primarily based on individuals of European ancestry. This finding further highlights the need for more diversity in genetic discovery studies and the need for research on optimizing trans-ancestry polygenic risk prediction.
Finland has two well-known genetic subpopulations, for which population stratification has been observed previously.16 Previous studies have shown geographical differences in allele frequencies of rare high-impact variants for recessive Mendelian diseases as well as for common diseases in Finland with well-documented genetic differences between early- and late-settlement regions.40,41 We therefore studied whether such gradients would impact the utility of PRSs. Despite these genetic substructures, our results showed highly similar effect sizes between early- and late-settlement regions, indicating that fine-scale population structures and recent genetic bottlenecks did not affect the transferability of the PRSs.
PRSs have been particularly promising for identifying individuals at risk for early-onset disease and for improving accuracy of risk estimation in individuals carrying mutations in high-impact disease-causing genes, such as known breast cancer susceptibility genes.2,6,42 There are two key steps in creating risk functions for PRS: (1) calculation of weighted sums of the genetic variants using effect sizes from an independent dataset and (2) estimating the predictive accuracy and the dose response between the PRS and the disease risk. Ancestry needs to be considered in both steps to allow for transferability of PRSs. Large-scale GWASs widely used for drawing weights for the variants are currently heavily biased toward individuals of European ancestry. This makes them less optimal for generating PRSs for individuals of other ancestries due to, for example, differing allele frequencies and genetic architectures across populations, as well as varying LD patterns.38 The PRS distribution in each ancestry group is also dependent on these same genetic factors and can therefore create considerable differences of the raw PRS distributions between the ancestry groups.43 The optimal way to adjust for these differences is to have a reference genome that correspond to the target ancestry group. In addition, the PRS distributions may differ due to methodological choices used for constructing the PRS,26 and it is likely that rescaling should be done only for similarly processed datasets, to reduce the influence of factors such as genotype quality control and technical artifacts.
Several measures can be undertaken to improve the utility of PRSs across ancestries. Most importantly, we need better representation of different ancestries in GWASs.18,44, 45, 46, 47, 48 Of the four GWASs used for generating our genome-wide PRSs, the proportion of individuals of other than European ancestry was highest for CAD (23%), the majority of whom were of South or East Asian ancestry. In breast cancer, the proportion of individuals of East Asian ancestry was 11%, whereas the T2D and prostate cancer GWASs were limited to individuals of European ancestry. Similarly, we need strategies to account for genetic admixture, as well as careful alignment of the PRSs against adequate reference samples with respect to ancestry and, when relevant, with respect to relevant subpopulations.46,49, 50, 51 Several tools are currently being developed for improved trans-ancestry polygenic risk prediction,52,53 and the transferability could also be improved by leveraging information about functional annotations.54
Limitations of the study
This study should be interpreted in light of certain limitations. Despite the large number of individuals studied, the sample size in South Asian or African ancestries remained fairly small, particularly for analyses on breast and prostate cancer. While our comparisons show relatively small differences between cohorts with European ancestry, it may be that the risk estimates vary considerably between individuals due to, for example, admixed ancestry, and the role of admixture in this variability warrants further research. Differences in predictive performance and dose response can reflect true differences in genetic architecture, but the results can be affected by multiple other population and reference sample-related factors, such as age and sex distribution, disease definitions, sample ascertainment, as well as variation in environmental risk factors.55 This study involved biobanks with hospital-based ascertainment (BioBank Japan, MGB Biobank) and population-based ascertainment (Estonian Biobank, HUNT, UK Biobank), as well as a mixture of the two (FinnGen). Phenotyping differences between datasets existed, ranging from single ICD-based records to high-quality cancer and medication reimbursement registries. Despite the differences across countries, health systems, and biobank characteristics, we observed good transferability of all PRSs across similar populations. Our observations may help in defining the population and ancestry-specific reference samples for PRS calculation in the four diseases studied. Moreover, differences in risk between ancestries may arise from a range of factors, including socioeconomic and health-care system-related factors and differing levels of traditional disease risk factors.56, 57, 58 They may also reflect differing impacts of clinical risk factors: for instance, weight gain is considered particularly detrimental for risk of T2D in Asians.59
In conclusion, we observed good transferability of largely European ancestry-derived, genome-wide PRSs for CAD, T2D, breast and prostate cancer across biobanks of European and Asian ancestry, but not for individuals of African ancestry. The highly polygenic, genome-wide PRSs generally displayed better transferability across ancestries than PRSs containing a smaller number of variants. This large-scale study further emphasizes the pressing need for diversity in genetic studies and the need for population and ancestry-based reference samples. Without prioritizing diversity in PRS evaluations and translation efforts, widely adopting PRSs to clinical care may exacerbate health disparities, and efforts to overcome the lack of diversity have great potential to improve health outcomes across ancestries.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| The FinnGen data may be accessed through Finnish Biobanks’ FinBB portal | - | http://www.finnbb.fi |
| GWAS genotype data of BioBank Japan are available at the National Bioscience Database Center Human Database | Nagai et al., 2017 | Research ID: hum0014; https://humandbs.biosciencedbc.jp/en/hum0014-v21 |
| UK Biobank data are available through a procedure described at http://www.ukbiobank.ac.uk/using-the-resource/ | Bycroft et al., 2018 | http://www.ukbiobank.ac.uk/using-the-resource/ |
| The Trøndelag Health Study (HUNT). The HUNT data may be accessed by application to the HUNT Research Centre. | Krokstad et al., 2013 | https://www.ntnu.edu/hunt |
| De-identified data of the MGB Biobank that supports this study is available from the MGB Biobank portal. Restrictions apply to the availability of these data, which are available to MGB-affiliated researchers via a formal application. | Karlson et al., 2016 | https://biobank.partners.org/ |
| Estonian Biobank. Researchers interested in Estonian Biobank can request the access at https://www.geenivaramu.ee/en/access-biobank | Leitsalu et al., 2015 | https://www.geenivaramu.ee/en/access-biobank |
| PGS Catalog/LDpred polygenic risk scores | This paper |
https://www.pgscatalog.org/ PGS002241–PGS002244 |
| PGS Catalog/PRS-CS polygenic risk score for prostate cancer | This paper |
https://www.pgscatalog.org/ PGS002240 |
| PGS Catalog/PRS-CS polygenic risk score for breast cancer | Mars et al., 2020 |
https://www.pgscatalog.org/ PGS000335 |
| PGS Catalog/PRS-CS polygenic risk score for coronary artery disease and type 2 diabetes | Tamlander et al., 2022 |
https://www.pgscatalog.org/ PGS001780, PGS001781 |
| Software and algorithms | ||
| PRS-CS (version Sep 10, 2020) | Ge et al., 2019 | https://github.com/getian107/PRScs |
| PLINK v2.00a2.3LM | Chang et al., 2015 | https://www.cog-genomics.org/plink/2.0/ |
| STEROID 0.1 | - | https://genomics.ut.ee/en/tools/steroid |
| Eagle v2.3.5 | Loh P-R et al. 2016 | https://alkesgroup.broadinstitute.org/Eagle/ |
| R statistical programming v3.2.0 or later | - | https://www.r-project.org/ |
| LDpred v1.0.7 | Vilhjálmsson et al. 2015 | https://github.com/bvilhjal/ldpred |
| Other | ||
| PRS-CS pipeline | - | https://github.com/FINNGEN/CS-PRS-pipeline |
| Project code | This paper | https://doi.org/10.5281/zenodo.6203211 |
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Samuli Ripatti (samuli.ripatti@helsinki.fi).
Materials availability
This study did not generate new materials.
Experimental model and subject details
Each of the six studies had undergone the pre-processing, imputation and quality control steps according to local pipelines. All analyses were limited to adults (age ≥18).
BioBank Japan
BioBank Japan (BBJ) is a multi-institutional hospital-based biobank with DNA and serum samples from 12 medical institutions in Japan and approximately 200,000 participants.62 The individuals are mainly of Japanese ancestry, and all patients have a diagnosis of at least 1 of 47 diseases. The study participants have been followed up for their health record after an initial visit, collecting information on disease onset and cause of death. Each participant has provided written informed consent and the BBJ project was approved by the research ethics committees of the RIKEN Center for Integrative Medical Sciences and the Institute of Medical Sciences at the University of Tokyo.
All disease endpoints were defined based on physician’s diagnosis. The coronary artery disease (CAD) diagnosis comprises individuals diagnosed with myocardial infarction, stable angina, or unstable angina. Age at disease onset was available for a subset of individual: for 11,717 with CAD, for 30,475 with type 2 diabetes (T2D), for 4,962 with breast cancer and for 4,374 with prostate cancer. The detailed definitions can be found elsewhere.63 Age at diagnosis was retrieved from medical records.
We genotyped samples with either (i) the Illumina HumanOmniExpressExome BeadChip or (ii) a combination of the Illumina HumanOmniExpress and HumanExome BeadChips. We applied standard quality control criteria for both samples and variants as detailed elsewhere.64 We then prephased genotypes with Eagle65 and imputed dosages with Minimac3 using 1000 Genomes Project Phase 3 (version 5) data (n = 2,504) and Japanese whole-genome sequencing (WGS) data (n = 1,037) as a reference.64 The dataset uses genome build 37 (hg19). The polygenic risk score (PRS) calculation was performed with PLINK v2.00a2LM66 using genotype dosages.
Estonian Biobank
The Estonian Biobank is a population-based biobank of the Estonian Genome Center at the University of Tartu (EstBB).67 The biobank consists of Estonians (83%), Russians (14%), and other nationalities (3%). The genotypes have been linked to several national health records, including National Health Insurance Fund, hospital databases, prescription data, infraction registries, the cancer registry, and the Causes of Death registry. All biobank participants have signed a broad informed consent form. Analysis in the EstBB was carried out under ethics approval 1.1-12/624 from the Estonian Committee on Bioethics and Human Research.
The disease diagnoses were defined based on ICD-10 codes (International Classification of Diseases, 10th revision) as follows: for CAD, any of I21-I23 or Z95; for type 2 diabetes (T2D), any of E11, excluding gestational diabetes with E10; for breast cancer, any of C50; for prostate cancer, any of C61. Age at diagnosis was defined as the time in years from birth until the date of first record for each diagnosis.
All EstBB participants have been genotyped at the Core Genotyping Lab of the Institute of Genomics, University of Tartu, using Illumina GSAv1.0, GSAv2.0, and GSAv2.0_EST arrays. Samples were genotyped and PLINK format files were created using Illumina GenomeStudio v2.0.4. Individuals were excluded from the analysis if their call-rate was < 95% or if sex defined based on heterozygosity of X chromosome did not match sex in phenotype data. Before imputation, variants were filtered by call-rate < 95%, HWE p-value < 1e-4 (autosomal variants only), and minor allele frequency < 1%. Variant positions were updated to b37 (hg19) and all variants were changed to be from TOP strand using GSAMD-24v1-0_20011747_A1-b37.strand.RefAlt.zip files from https://www.well.ox.ac.uk/∼wrayner/strand/ webpage. Pre-phasing was done using Eagle v2.3 software22 (number of conditioning haplotypes Eagle2 uses when phasing each sample was set to: --Kpbwt=20000) and imputation was done using Beagle v.28Sep18.79323 with effective population size ne=20,000. Population specific imputation reference of 2297 WGS samples was used.68 The PRS calculation was performed with STEROID 0.1 (https://genomics.ut.ee/en/tools/steroid) using imputed genotype dosages.
FinnGen
FinnGen is a collection of prospective epidemiological and disease-based cohorts and hospital biobank samples, aiming for a collection of 500,000 genotype samples from Finnish individuals by 2023. The Data Freeze 6 consists of 258,402 adult individuals, with their genotypes linked to national health registries, including the national hospital discharge (available from 1968), death (1969–), cancer (1953–) and medication reimbursement (1964–) and purchase (1995–) registries. Information on region of birth was obtained from the Finnish Population Information System.
CAD was defined as A) any of I20–I25, I46, R96 or R98 (ICD-10), or 410–414 or 798 (ICD-9) as underlying or direct cause of death, or B) any of I20.0, I21–I22 (ICD-10) or 410, 4110 (ICD-9) as the main diagnosis at hospital discharge, or C) coronary bypass surgery or coronary angioplasty at hospital discharge or identified from the specific country-wide register of invasive cardiac procedures. T2D was defined as any of E11.[0-9] (ICD-10), 250.[0-8]A (ICD-9), or use of blood-glucose lowering drugs, and by excluding individuals with type 1 diabetes with E10.[0-9] (ICD-10), 250.[0-8]B (ICD-9) or with eligibility for special reimbursement for insulin with ICD-10 E10.[0-9]. Breast cancer cases were identified from the cancer registry with diagnosis C50 (International Classification of Diseases for Oncology, 3rd Edition; ICD-O-3), from the death registry with C50 (ICD-10) and 174 (ICD-9), and from the drug reimbursement registry by selecting individuals with a reimbursement code for C50 (ICD-10). Similarly, prostate cancer cases were identified from the cancer registry with diagnosis C61 (ICD-O-3), from the death registry with C61 (ICD-10) and 185 (ICD-9), and from the reimbursement registry with C61 (ICD-10). Age at diagnosis was defined as the date of first record for each diagnosis.
The early- and late-settlement analyses were based on information about birthplace. Early settlement comprised the regions Central Ostrobothnia, Ostrobothnia, South Ostrobothnia, Southwest Finland, Pirkanmaa, Uusimaa, Päijät-Häme, Satakunta, Kanta-Häme; late settlement contained the regions Kainuu, North Karelia, North Savo and North Ostrobothnia; the borderline area contained the regions South Savo, Central Finland, Lapland, Kymenlaakso, and South Karelia.
The samples are genotyped with Illumina and Affymetrix arrays (Illumina Inc., San Diego, and Thermo Fisher Scientific, Santa Clara, CA, USA). The genotypes have been imputed with using the SISu v3 population-specific reference panel developed from high-quality data for 3,775 high-coverage (25-30x) whole-genome sequencing in Finns. The detailed genotype imputation workflow can be found at https://dx.doi.org/10.17504/protocols.io.xbgfijw. The dataset uses genome build 38 (hg38). The PRS calculation was performed with PLINK v2.00a2.3LM.66
Patients and control subjects in FinnGen provided informed consent for biobank research, based on the Finnish Biobank Act. Alternatively, older research cohorts, collected prior the start of FinnGen (in August 2017), were collected based on study-specific consents and later transferred to the Finnish biobanks after approval by Valvira, the National Supervisory Authority for Welfare and Health. Recruitment protocols followed the biobank protocols approved by Valvira. The Ethics Review Board of the Hospital District of Helsinki and Uusimaa approved the FinnGen study protocol Nr HUS/990/2017. The FinnGen project is approved by the Finnish Institute for Health and Welfare (THL), approval number THL/2031/6.02.00/2017, amendments THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019), Digital and population data service agency VRK43431/2017-3, VRK/6909/2018-3, the Social Insurance Institution (KELA) KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019 and Statistics Finland TK-53-1041-17.
Following biobanks are acknowledged for collecting the FinnGen project samples: Auria Biobank (https://www.auria.fi/biopankki), THL Biobank (https://thl.fi/fi/web/thl-biopankki), Helsinki Biobank (https://www.terveyskyla.fi/helsinginbiopankki), Biobank Borealis of Northern Finland (https://www.oulu.fi/university/node/38474), Finnish Clinical Biobank Tampere (https://www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (https://ita-suomenbiopankki.fi), Central Finland Biobank (https://www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (https://www.veripalvelu.fi/verenluovutus/biopankkitoiminta) and Terveystalo Biobank (https://www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi).
The FinnGen project is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and by twelve industry partners (AbbVie Inc, AstraZeneca UK Ltd, Biogen MA Inc, Celgene Corporation, Celgene International II Sàrl, Genentech Inc, Merck Sharp & Dohme Corp, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc, and Novartis AG).
HUNT
The Trøndelag Health Study (HUNT) is a large population-based cohort from the county Nord-Trøndelag in Norway. All residents in the county, aged 20 years and older, have been invited to participate. Data was collected through three cross-sectional surveys, HUNT1 (1984-1986), HUNT2 (1995-1997) and HUNT3 (2006-2008), and has been described in detail previously,69 with the fourth survey recently completed (HUNT4, 2017-2019). DNA from whole blood was collected from HUNT2 and HUNT3, with genotypes available from 71,860 participants. Participation in the HUNT Study is based on informed consent and the study has been approved by the Data Inspectorate and the Regional Ethics Committee for Medical Research in Norway (REK: 2014/144, 2015/1205).
CAD was defined as A) any I20.0, I21, or I22 (ICD-10) or 410 or 411 (ICD-9) in the Hospital Registry, or B) any ICD-10 I21-5, I46, R96 or R98 in the Cause of Death Registry. T2D was defined as any E11 (ICD-10) in the Hospital Registry, breast cancer as any C50 in the Cancer Registry or the Hospital Registry, and prostate cancer as any C61 in the Cancer Registry or the Hospital Registry. Age used as a covariate was coded as birth year. Age given in the population overview was defined in two ways; The first as estimated age at first diagnosis occurrence. Age estimation was calculated by subtracting birthyear June 1st from date at first diagnosis occurrence. The second was estimated age at date of death. Age estimation was calculated by subtracting birthyear June 1st from date of death.
Imputation was performed on the 69,716 samples of recent European ancestry using Minimac3 (v2.0.1, http://genome.sph.umich.edu/wiki/Minimac3)70 with default settings (2.5 Mb reference-based chunking with 500kb windows) and a customized Haplotype Reference consortium release 1.1 (HRC v1.1) for autosomal variants and HRC v1.1 for chromosome X variants.71 The customized reference panel represented the merged panel of two reciprocally imputed reference panels: (1) 2,201 low-coverage whole-genome sequences samples from the HUNT study and (2) HRC v1.1 with 1,023 HUNT WGS samples removed before merging. We excluded imputed variants with Rsq < 0.3 resulting in over 24.9 million well-imputed variants. The dataset uses genome build 37 (hg19). The PRS calculation was performed with PLINK v2.00a2.3LM.66
The Trøndelag Health Study (HUNT) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology NTNU), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. The genotype quality control and imputation has been conducted by the K. G. Jebsen center for genetic epidemiology, Department of public health and nursing, Faculty of medicine and health sciences, Norwegian University of Science and Technology (NTNU).
MGB biobank
The Mass General Brigham (MGB) Biobank [https://biobank.partners.org] is a hospital-based research program launched in 2010 designed to empower genomic and translational research for human health. Participants are patients above age 18 who provided informed consent to join the biobank in the Mass General Brigham network (previously Partners HealthCare), including Massachusetts General Hospital, Brigham and Women’s Hospital, and other affiliated institutions. Sample recruitment of the MGB Biobank was approved by the Partners Human Research Committee (PHRC) (the Institutional Review Board). PHRC provides continued ethical and scientific oversight of the MGB activities.72 For each consented subject, a collection of blood samples is obtained (plasma, serum, and DNA), which are then linked to their clinical data in the electronic health records (EHR) as well as survey data on lifestyle, behavioral and environmental factors, and family history.72 To date, MGB Biobank has enrolled more than 120,000 participants and released genotyping array data for 36,424 subjects (December 2019). MGB investigators can access the de-identified datasets from the MGB Biobank under a Data Use Agreement (DUA) without additional study protocols.
The biobank samples are genotyped on Multi-Ethnic Global array (MEGA) from Illumina (Illumina Inc., San Diego, USA) and are released in several batches. We performed batch-specific genotype data QC to remove SNPs with genotype missing rate >0.05, samples with genotype missing rate >0.02, and SNPs with differential missing rate >0.01 between any two batches, after which different batches were merged for subsequent QC steps. As MGB Biobank included individuals from diverse populations, we inferred genetic ancestry of biobank participants using 1000 Genomes samples (1KG)73 as the population reference panel. Specifically, we computed principal components (PCs) for biobank samples and 1KG samples combined, and trained a Random Forest classifier to assign a “super population” label for biobank samples with a prediction probability ≥0.9 using the first 6 PCs of the 1KG samples as the training data. This resulted in 26,677 individuals classified as European (EUR), 1,607 as African (AFR), 1,840 as Admixed American (AMR), 504 as East Asian (EAS) and 297 as South Asian (SAS) ancestry. Within each ancestry, we removed samples with a mismatched reported and genetic sex, outliers of the absolute value of heterozygosity (>5SD from the mean), and one from each pair of related individuals (IBD >0.2); SNPs that showed significant batch associations at P < 1 × 10−4, with a missing rate > 0.02 or HWE test P < 1 × 10−10 were also discarded. Next, we used Michigan Imputation Server (Minimac4) to impute genotype dosages for biobank samples, with the Haplotype Reference Consortium (HRC) as the reference panel for EUR ancestry and 1KG phase3 AFR data as the reference for AFR samples. Lastly, we removed markers with imputation quality INFO score <0.8, minor allele frequency (MAF) <0.01, a significant deviation from HWE with P < 1 × 10−10, and missing rate >0.02. The dataset uses genome build 37 (hg19).
EUR and AFR ancestries were chosen for PRS analysis in the present study based on having >50 cases available for all four diseases. The disease diagnoses were the following ICD-10 diagnoses in the linked EHR data for biobank participants (with ICD-9 codes converted to ICD-10): for CAD, any of a I20.0, I21, or I22; for T2D, any of E11.[0-9]; for breast cancer, any of C50; for prostate cancer, any of C61. Age at disease onset was not available from the de-identified dataset. The PRS calculation was performed with PLINK2 using genotype dosages.
UK biobank
UK Biobank is a prospective cohort study comprising approximately 500,000 individuals from across the United Kingdom, aged between 40 and 69 at recruitment. The cohort contains deep phenotyping, including biological measurements, lifestyle factors, and clinically relevant blood biomarkers. Although most individuals in the cohort are of European ancestry, over 20,000 individuals have a self-reported ethnic background originating outside Europe. The dataset has been imputed using the merged UK10K and 1000 Genomes (phase 3) reference panels.74 Details on the cohort, as well as data generation and imputation have been previously described.75 The dataset uses genome build 37 (hg19). The PRS calculation was performed with PLINK v2.00a2.3LM.66
We thank all participants in the UK Biobank study. This research was conducted using the UK Biobank Resource under Application Number 22627. UK Biobank has obtained ethics approval from the North-West Multi-centre Research Ethics Committee (approval number: 11/NW/0382) that covers analysis of data by approved researchers. UK Biobank obtained informed consent from all participants.
CAD was defined as A) any of I20–I25, I46, or R96 (ICD-10) as the primary or secondary cause of death (from data fields 40001 and 40002, age from data field 40007), B) any of I20.0, I21–I22 (ICD-10) or 410, 4110 (ICD-9) in the hospital inpatient records (from data fields 41270 and 41271, age defined based on data fields 41280 and 41281), or C) any coronary revascularization procedure (OPCS-4, variable 41272, codes K40, K41, K42, K43, K44, K45, K46, K49, K501, and K75, and age defined based on data field 41282; OPSC-3, data field 41273, code 3043, age defined based on data field 41283; self-reported operations, data field 20004, codes 1070 and 1095, age defined based on data field 20010).
T2D was defined as A) diabetes diagnosed by doctor (data field 2443, age from data field 2976) excluding individuals with age at diagnosis under 18, and individuals with type 1 diabetes by ICD-10 diagnosis E10 (from data field 41270), or B) ICD-10 E11 as the primary or secondary cause of death (from data fields 40001 and 40002, age from data field 40007). Breast cancer was defined as A) ICD-10 C50 in the Cancer register (data field 40006, age at diagnosis from data field 40008), B) C50 (ICD-10) or 174 (ICD-9) in the hospital inpatient records (from data fields 41270 and 41271, age defined based on data fields 41280 and 41281), or C) C50 (ICD-10) as the primary or secondary cause of death (from data fields 40001 and 40002, age from data field 40007). Prostate cancer was defined as A) ICD-10 C61 in the Cancer register (data field 40006, age at diagnosis from data field 40008), B) C61 (ICD-10) in the hospital inpatient records (from data field 41270, age defined based on data field 41280), or C) C61 (ICD-10) as the primary or secondary cause of death (from data fields 40001 and 40002, age from data field 40007).
White British individuals within the UK Biobank represented European ancestry, with all European-ancestry pairs unrelated to KING's kinship value 0.0442. South Asian ancestry was defined based on self-report (data field 21000) of being Indian, Pakistani, or Bangladeshi (codes 3001, 3002, 3003). Black / Caribbean ancestry was similarly defined based on self-report of being Caribbean, African, or any other Black background (codes 4001, 4002, 4003). These two non-European ancestry groups where chosen based on having >50 cases available for analysis for all four diseases.
Method details
Polygenic risk scores
The PRSs were derived with LDpred,31 a software that weights the single-nucleotide polymorphisms in GWAS summary statistics by their effect sizes by accounting for linkage disequilibrium (LD) between markers. The input weights were obtained from the largest available disease consortia GWAS (Table S4).11, 12, 13, 14 The LD reference panel consisted of 503 European individuals from 1000 Genomes phase 3.73 Out of 10 candidate PRSs concerning the LDpred default parameters for the fraction of causal variants, the PRSs with the best discriminative capacity (measured with maximum area under the receiver-operator curve, AUC) were chosen based on an earlier FinnGen data freeze (DF4) with 176,899 individuals. The PRSs were then calculated over autosomal chromosomes as the weighted sum of effect alleles. The number of variants used for each LDpred PRS are shown in Table S1. The number of variants available for PRS calculation (e.g. due to being polymorphic in the population) was lowest in BioBank Japan (67.1%-67.5%) and in individuals of African ancestry in MGB Biobank (75.9%-77.3%), with amount for the rest ranging from 89.9% to 100%. To perform the analysis in a setting as similar as possible to clinical use cases, where variant optimization cannot always be done for the derivation and test sets, we did not seek to optimize variant overlap between datasets. Some of our datasets had small overlap with the GWASs used for building the PRSs. These overlapping proportions were 5.9% for CAD and 7.5% for T2D in Estonian Biobank and 2.0% in FinnGen for CAD, which may result in slight overestimation of effects within Estonian biobank and FinnGen.
In UK Biobank, the LDpred PRSs were compared to two other types of PRSs generated mostly in individuals of European ancestry: 1) to previously published PRSs containing a smaller number of variants (PGS Catalog IDs PGS000012, PGS000020, PGS000004, PGS000662)3,10,17,18 and 2) to genome-wide PRSs generated with PRS-CS. In the smaller PRSs, the number of variants in the final score in UK Biobank (out of the variants in the original score) was 48,523/49,310 for CAD, 7,491/7,502 for T2D, 306/313 for breast cancer, and 267/269 for prostate cancer. PRS-CS uses HapMap3 variants when inferring posterior effect sizes,32 and we used 1000 Genomes Project European sample (N = 503) as the external LD reference panel, using autosomes.73 The PRS-CS scores were generated with the PRS-CS-auto approach in the FinnGen dataset, using the same GWASs used for generating the LDpred PRSs. The number of variants in UK Biobank (out of the variants in the original PRS-CS score) was 1,087,714/1,090,048 for CAD, 1,089,342/1,091,673 for T2D, 1,077,906/1,079,089 for breast cancer, and 1,089,645/1,092,093 for prostate cancer. When comparing decreases in effect sizes between different PRSs and across ancestries, the decreases were calculated from regression estimates (log odds).
Quantification and statistical analysis
All sample sizes are shown in Tables 1 and Table S2. In each study, each PRS was scaled to zero mean and unit variance by ancestry. In analyses by settlement in FinnGen, the scaling was done in the full FinnGen dataset. The odds ratio for risk of disease by one SD increase for the PRS was assessed using a logistic regression model (Figures 1, 2, S1, and S2; Tables S2 and S3). In all models, the covariates were age (age at baseline, at the end of follow-up, or birth year; depending on biobank) sex (for CHD and T2D), batch or genotyping array (when available), and the first 10 principal components of ancestry. Incident and prevalent cases were considered jointly. For statistical analyses, each biobank used R (version 3.2.0 or later). ORs by ancestry were pooled by random effects meta-analysis with function metagen() in R package meta (Figure 1, Table S2). All tests were two-tailed. P-value for heterogeneity was calculated based on Cochran’s heterogeneity statistic (Table S2).
Consortia
The members of the FinnGen Consortium are Aarno Palotie, Mark Daly, Bridget Riley-Gills, Howard Jacob, Dirk Paul, Athena Matakidou, Adam Platt, Heiko Runz, Sally John, George Okafo, Nathan Lawless, Robert Plenge, Joseph Maranville, Mark McCarthy, Julie Hunkapiller, Margaret G. Ehm, Kirsi Auro, Simonne Longerich, Caroline Fox, Anders Mälarstig, Katherine Klinger, Deepak Raipal, Eric Green, Robert Graham, Robert Yang, Chris O'Donnell, Tomi P. Mäkelä, Jaakko Kaprio, Petri Virolainen, Antti Hakanen, Terhi Kilpi, Markus Perola, Jukka Partanen, Anne Pitkäranta, Juhani Junttila, Raisa Serpi, Tarja Laitinen, Veli-Matti Kosma, Jari Laukkanen, Marco Hautalahti, Outi Tuovila, Raimo Pakkanen, Jeffrey Waring, Bridget Riley-Gillis, Fedik Rahimov, Ioanna Tachmazidou, Chia-Yen Chen, Heiko Runz, Zhihao Ding, Marc Jung, Shameek Biswas, Rion Pendergrass, Julie Hunkapiller, Margaret G. Ehm, David Pulford, Neha Raghavan, Adriana Huertas-Vazquez, Jae-Hoon Sul, Anders Mälarstig, Xinli Hu, Katherine Klinger, Robert Graham, Eric Green, Sahar Mozaffari, Dawn Waterworth, Nicole Renaud, Ma'en Obeidat, Samuli Ripatti, Johanna Schleutker, Markus Perola, Mikko Arvas, Olli Carpén, Reetta Hinttala, Johannes Kettunen, Arto Mannermaa, Katriina Aalto-Setälä, Mika Kähönen, Jari Laukkanen, Johanna Mäkelä, Reetta Kälviäinen, Valtteri Julkunen, Hilkka Soininen, Anne Remes, Mikko Hiltunen, Jukka Peltola, Minna Raivio, Pentti Tienari, Juha Rinne, Roosa Kallionpää, Juulia Partanen, Ali Abbasi, Adam Ziemann, Nizar Smaoui, Anne Lehtonen, Susan Eaton, Heiko Runz, Sanni Lahdenperä, Shameek Biswas, Julie Hunkapiller, Natalie Bowers, Edmond Teng, Rion Pendergrass, Fanli Xu, David Pulford, Kirsi Auro, Laura Addis, John Eicher, Qingqin S Li, Karen He, Ekaterina Khramtsova, Neha Raghavan, Martti Färkkilä, Jukka Koskela, Sampsa Pikkarainen, Airi Jussila, Katri Kaukinen, Timo Blomster, Mikko Kiviniemi, Markku Voutilainen, Mark Daly, Ali Abbasi, Jeffrey Waring, Nizar Smaoui, Fedik Rahimov, Anne Lehtonen, Tim Lu, Natalie Bowers, Rion Pendergrass, Linda McCarthy, Amy Hart, Meijian Guan, Jason Miller, Kirsi Kalpala, Melissa Miller, Xinli Hu, Kari Eklund, Antti Palomäki, Pia Isomäki, Laura Pirilä, Oili Kaipiainen-Seppänen, Johanna Huhtakangas, Nina Mars, Ali Abbasi, Jeffrey Waring, Fedik Rahimov, Apinya Lertratanakul, Nizar Smaoui, Anne Lehtonen, David Close, Marla Hochfeld, Natalie Bowers, Rion Pendergrass, Jorge Esparza Gordillo, Kirsi Auro, Dawn Waterworth, Fabiana Farias, Kirsi Kalpala, Nan Bing, Xinli Hu, Tarja Laitinen, Margit Pelkonen, Paula Kauppi, Hannu Kankaanranta, Terttu Harju, Riitta Lahesmaa, Nizar Smaoui, Alex Mackay, Glenda Lassi, Susan Eaton, Hubert Chen, Rion Pendergrass, Natalie Bowers, Joanna Betts, Kirsi Auro, Rajashree Mishra, Majd Mouded, Debby Ngo, Teemu Niiranen, Felix Vaura, Veikko Salomaa, Kaj Metsärinne, Jenni Aittokallio, Mika Kähönen, Jussi Hernesniemi, Daniel Gordin, Juha Sinisalo, Marja-Riitta Taskinen, Tiinamaija Tuomi, Timo Hiltunen, Jari Laukkanen, Amanda Elliott, Mary Pat Reeve, Sanni Ruotsalainen, Benjamin Challis, Dirk Paul, Julie Hunkapiller, Natalie Bowers, Rion Pendergrass, Audrey Chu, Kirsi Auro, Dermot Reilly, Mike Mendelson, Jaakko Parkkinen, Melissa Miller, Tuomo Meretoja, Heikki Joensuu, Olli Carpén, Johanna Mattson, Eveliina Salminen, Annika Auranen, Peeter Karihtala, Päivi Auvinen, Klaus Elenius, Johanna Schleutker, Esa Pitkänen, Nina Mars, Mark Daly, Relja Popovic, Jeffrey Waring, Bridget Riley-Gillis, Anne Lehtonen, Jennifer Schutzman, Julie Hunkapiller, Natalie Bowers, Rion Pendergrass, Diptee Kulkarni, Kirsi Auro, Alessandro Porello, Andrey Loboda, Heli Lehtonen, Stefan McDonough, Sauli Vuoti, Kai Kaarniranta, Joni A Turunen, Terhi Ollila, Hannu Uusitalo, Juha Karjalainen, Esa Pitkänen, Mengzhen Liu, Heiko Runz, Stephanie Loomis, Erich Strauss, Natalie Bowers, Hao Chen, Rion Pendergrass, Kaisa Tasanen, Laura Huilaja, Katariina Hannula-Jouppi, Teea Salmi, Sirkku Peltonen, Leena Koulu, Nizar Smaoui, Fedik Rahimov, Anne Lehtonen, David Choy, Rion Pendergrass, Dawn Waterworth, Kirsi Kalpala, Ying Wu, Pirkko Pussinen, Aino Salminen, Tuula Salo, David Rice, Pekka Nieminen, Ulla Palotie, Maria Siponen, Liisa Suominen, Päivi Mäntylä, Ulvi Gursoy, Vuokko Anttonen, Kirsi Sipilä, Rion Pendergrass, Hannele Laivuori, Venla Kurra, Laura Kotaniemi-Talonen, Oskari Heikinheimo, Ilkka Kalliala, Lauri Aaltonen, Varpu Jokimaa, Johannes Kettunen, Marja Vääräsmäki, Outi Uimari, Laure Morin-Papunen, Maarit Niinimäki, Terhi Piltonen, Katja Kivinen, Elisabeth Widen, Taru Tukiainen, Mary Pat Reeve, Mark Daly, Niko Välimäki, Eija Laakkonen, Jaakko Tyrmi, Heidi Silven, Eeva Sliz, Riikka Arffman, Susanna Savukoski, Triin Laisk, Natalia Pujol, Mengzhen Liu, Bridget Riley-Gillis, Rion Pendergrass, Janet Kumar, Kirsi Auro, Iiris Hovatta, Chia-Yen Chen, Erkki Isometsä, Kumar Veerapen, Hanna Ollila, Jaana Suvisaari, Thomas Damm Als, Antti Mäkitie, Argyro Bizaki-Vallaskangas, Sanna Toppila-Salmi, Tytti Willberg, Elmo Saarentaus, Antti Aarnisalo, Eveliina Salminen, Elisa Rahikkala, Johannes Kettunen, Kristiina Aittomäki, Fredrik Åberg, Mitja Kurki, Samuli Ripatti, Mark Daly, Juha Karjalainen, Aki Havulinna, Juha Mehtonen, Priit Palta, Shabbeer Hassan, Pietro Della Briotta Parolo, Wei Zhou, Mutaamba Maasha, Kumar Veerapen, Shabbeer Hassan, Susanna Lemmelä, Manuel Rivas, Mari E. Niemi, Aarno Palotie, Aoxing Liu, Arto Lehisto, Andrea Ganna, Vincent Llorens, Hannele Laivuori, Taru Tukiainen, Mary Pat Reeve, Henrike Heyne, Nina Mars, Joel Rämö, Elmo Saarentaus, Hanna Ollila, Rodos Rodosthenous, Satu Strausz, Tuula Palotie, Kimmo Palin, Javier Garcia-Tabuenca, Harri Siirtola, Tuomo Kiiskinen, Jiwoo Lee, Kristin Tsuo, Amanda Elliott, Kati Kristiansson, Mikko Arvas, Kati Hyvärinen, Jarmo Ritari, Olli Carpén, Johannes Kettunen, Katri Pylkäs, Eeva Sliz, Minna Karjalainen, Tuomo Mantere, Eeva Kangasniemi, Sami Heikkinen, Arto Mannermaa, Eija Laakkonen, Nina Pitkänen, Samuel Lessard, Clément Chatelain, Perttu Terho, Sirpa Soini, Jukka Partanen, Eero Punkka, Raisa Serpi, Sanna Siltanen, Veli-Matti Kosma, Teijo Kuopio, Anu Jalanko, Huei-Yi Shen, Risto Kajanne, Mervi Aavikko, Mitja Kurki, Juha Karjalainen, Pietro Della Briotta Parolo, Arto Lehisto, Juha Mehtonen, Wei Zhou, Masahiro Kanai, Mutaamba Maasha, Kumar Veerapen, Hannele Laivuori, Aki Havulinna, Susanna Lemmelä, Tuomo Kiiskinen, L. Elisa Lahtela, Mari Kaunisto, Elina Kilpeläinen, Timo P. Sipilä, Oluwaseun Alexander Dada, Awaisa Ghazal, Anastasia Kytölä, Rigbe Weldatsadik, Kati Donner, Timo P. Sipilä, Anu Loukola, Päivi Laiho, Tuuli Sistonen, Essi Kaiharju, Markku Laukkanen, Elina Järvensivu, Sini Lähteenmäki, Lotta Männikkö, Regis Wong, Auli Toivola, Minna Brunfeldt, Hannele Mattsson, Kati Kristiansson, Susanna Lemmelä, Sami Koskelainen, Tero Hiekkalinna, Teemu Paajanen, Priit Palta, Kalle Pärn, Mart Kals, Shuang Luo, Vishal Sinha, Tarja Laitinen, Mary Pat Reeve, Marianna Niemi, Kumar Veerapen, Harri Siirtola, Javier Gracia-Tabuenca, Mika Helminen, Tiina Luukkaala, Iida Vähätalo, Jyrki Pitkänen, Marco Hautalahti, Johanna Mäkelä, Sarah Smith, and Tom Southerington.
Acknowledgments
We would like to thank Julius Anckar, Ulla Tuomainen, and Anne Carson for management assistance. This work was supported by the Academy of Finland (grant number 331671 to N.M., grant number 285380 to S.R., 128650 to A.P., 288509 to M.P., and 323116 to A.G.); Academy of Finland Center of Excellence in Complex Disease Genetics (grant number 312062 to S.R., 312074 to A.P.,312076 to M.P.); European Union’s Horizon 2020 research and innovation program under grant agreement No 101016775; University of Helsinki HiLIFE Fellow grants 2017-2020 (to S.R.); Sigrid Jusélius Foundation (to S.R., A.P., and M.P.); National Institutes of Health (grant number K99MH117229 to A.M.); Estonian Research Council grant PUT (PRG687 to K.L and R.M). The research in BioBank Japan has been supported by JSPS KAKENHI (19H01021, 20K21834); AMED (JP21km0405211, JP21ek0109413, JP21gm4010006, JP21km0405217, JP21ek0410075); JST Moonshot R&D (JPMJMS2021); Takeda Science Foundation. In HUNT, the genotyping was financed by the National Institute of health (NIH), University of Michigan, The Norwegian Research Council, and Central Norway Regional Health Authority and the Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology (NTNU). G.C.F. is funded by the Faculty of Medicine and Health Sciences at NTNU and Central Norway Regional Health Authority. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
S.R. and N.M. conceived and designed the study. N.M., S.K., Y-C.A.F., M.K., K.L., L.F.T., and A.H.S. carried out the statistical and computational analyses with advice from S.R. and A.M. Quality control of the data was carried out by N.M., Y-C.A.F., M.K., K.L., L.F.T., and A.H.S. All authors provided critical inputs to interpretation of the data. The manuscript was written and revised by N.M. and S.R. with comments from all of the co-authors. All co-authors have approved the final version of the manuscript.
Declaration of interests
A.P. is a member of the Pfizer Genetics Scientific Advisory Panel. J.W.S is an unpaid member of the Bipolar/Depression Research Community Advisory Panel of 23andMe, a member of the Leon Levy Foundation Neuroscience Advisory Board, and received an honorarium for an internal seminar at Biogen, Inc. He is principal investigator of a collaborative study of the genetics of depression and bipolar disorder sponsored by 23andMe for which 23andMe provides analysis time as in-kind support but no payments. B.M.N. is a member of the Deep Genomics Scientific Advisory Board and serves as a consultant for the Camp4 Therapeutics Corporation, Takeda Pharmaceutical, and Biogen. The remaining authors declare no conflict of interests.
Published: April 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2022.100118.
Supplemental information
Data and code availability
-
•
The FinnGen data can be accessed through the Fingenious® services (https://site.fingenious.fi/en/) managed by FINBB. GWAS genotype data of BioBank Japan are available at the National Bioscience Database Center Human Database (research ID: hum0014; https://humandbs.biosciencedbc.jp/en/hum0014-v21). UK Biobank data are available through a procedure described at http://www.ukbiobank.ac.uk/using-the-resource/.The HUNT data may be accessed by application to the HUNT Research Centre (https://www.ntnu.edu/hunt). Researchers interested in Estonian Biobank can request the access at https://www.geenivaramu.ee/en/access-biobank. De-identified data of the MGB Biobank that supports this study is available from the MGB Biobank portal (https://biobank.partners.org/). Restrictions apply to the availability of these data, which are available to MGB-affiliated researchers via a formal application. Weights for the LDpred PRSs are available at PGS Catalog (pgs-info@ebi.ac.uk) with PGS IDs PGS002241–PGS002244, and weights for the PRS-CS PRSs with PGS001780–PGS001781,60 PGS000335,61 and PGS002240.
-
•
Original code generated within this project has been deposited at Zenodo and is publicly available. DOIs are listed in the key resources table.
-
•
Any additional information is available from the lead contact upon request.
References
- 1.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mars N., Koskela J.T., Ripatti P., Kiiskinen T.T.J., Havulinna A.S., Lindbohm J.V., Ahola-Olli A., Kurki M., Karjalainen J., Palta P., et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med. 2020;26:549–557. doi: 10.1038/s41591-020-0800-0. [DOI] [PubMed] [Google Scholar]
- 3.Mavaddat N., Michailidou K., Dennis J., Lush M., Fachal L., Lee A., Tyrer J.P., Chen T.H., Wang Q., Bolla M.K., et al. Polygenic risk scores for prediction of breast cancer and breast cancer subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seibert T.M., Fan C.C., Wang Y., Zuber V., Karunamuni R., Parsons J.K., Eeles R.A., Easton D.F., Kote-Jarai Z., Al Olama A.A., et al. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ. 2018;360:j5757. doi: 10.1136/bmj.j5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee A., Mavaddat N., Wilcox A.N., Cunningham A.P., Carver T., Hartley S., Babb de Villiers C., Izquierdo A., Simard J., Schmidt M.K., et al. BOADICEA: a comprehensive breast cancer risk prediction model incorporating genetic and nongenetic risk factors. Genet. Med. 2019;21:1708–1718. doi: 10.1038/s41436-018-0406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inouye M., Abraham G., Nelson C.P., Wood A.M., Sweeting M.J., Dudbridge F., Lai F.Y., Kaptoge S., Brozynska M., Wang T., et al. Genomic risk prediction of coronary artery disease in 480,000 adults: implications for primary prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindy G., Aragam Krishna G., Ng K., Chaffin M., Lotta Luca A., Baras A., Drake I., Orho-Melander M., Melander O., Kathiresan S., Khera Amit V. Genome-wide polygenic score, clinical risk factors, and long-term trajectories of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2020;40:2738–2746. doi: 10.1161/ATVBAHA.120.314856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanes T., Young M.A., Meiser B., James P.A. Clinical applications of polygenic breast cancer risk: a critical review and perspectives of an emerging field. Breast Cancer Res. 2020;22:21. doi: 10.1186/s13058-020-01260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lall K., Magi R., Morris A., Metspalu A., Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet. Med. 2017;19:322–329. doi: 10.1038/gim.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michailidou K., Lindstrom S., Dennis J., Beesley J., Hui S., Kar S., Lemacon A., Soucy P., Glubb D., Rostamianfar A., et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher F.R., Al Olama A.A., Berndt S.I., Benlloch S., Ahmed M., Saunders E.J., Dadaev T., Leongamornlert D., Anokian E., Cieza-Borrella C., et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 2018;50:928–936. doi: 10.1038/s41588-018-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott R.A., Scott L.J., Magi R., Marullo L., Gaulton K.J., Kaakinen M., Pervjakova N., Pers T.H., Johnson A.D., Eicher J.D., et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerminen S., Havulinna A.S., Hellenthal G., Martin A.R., Sarin A.P., Perola M., Palotie A., Salomaa V., Daly M.J., Ripatti S., Pirinen M. Fine-scale genetic structure in Finland. G3 (Bethesda) 2017;7:3459–3468. doi: 10.1534/g3.117.300217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerminen S., Martin A.R., Koskela J., Ruotsalainen S.E., Havulinna A.S., Surakka I., Palotie A., Perola M., Salomaa V., Daly M.J., et al. Geographic variation and bias in the polygenic scores of complex diseases and traits in Finland. Am. J. Hum. Genet. 2019;104:1169–1181. doi: 10.1016/j.ajhg.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham G., Havulinna A.S., Bhalala O.G., Byars S.G., De Livera A.M., Yetukuri L., Tikkanen E., Perola M., Schunkert H., Sijbrands E.J., et al. Genomic prediction of coronary heart disease. Eur. Heart J. 2016;37:3267–3278. doi: 10.1093/eurheartj/ehw450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conti D.V., Darst B.F., Moss L.C., Saunders E.J., Sheng X., Chou A., Schumacher F.R., Olama A.A.A., Benlloch S., Dadaev T., et al. Trans-ancestry genome-wide association meta-analysis of prostate cancer identifies new susceptibility loci and informs genetic risk prediction. Nat. Genet. 2021;53:65–75. doi: 10.1038/s41588-020-00748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho W.K., Tan M.M., Mavaddat N., Tai M.C., Mariapun S., Li J., Ho P.J., Dennis J., Tyrer J.P., Bolla M.K., et al. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat. Commun. 2020;11:3833. doi: 10.1038/s41467-020-17680-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shieh Y., Fejerman L., Lott P.C., Marker K., Sawyer S.D., Hu D., Huntsman S., Torres J., Echeverry M., Bohorquez M.E., et al. A polygenic risk score for breast cancer in US Latinas and Latin American women. J. Natl. Cancer Inst. 2020;112:590–598. doi: 10.1093/jnci/djz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polfus L.M., Darst B.F., Highland H., Sheng X., Ng M.C.Y., Below J.E., Petty L., Bien S., Sim X., Wang W., et al. Genetic discovery and risk characterization in type 2 diabetes across diverse populations. Hum. Genet. Genom. Adv. 2021;2:100029. doi: 10.1016/j.xhgg.2021.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Z., Gao G., Adedokun B., Ahearn T., Lunetta K.L., Zirpoli G., Troester M.A., Ruiz-Narvaez E.A., Haddad S.A., Pal Choudhury P., et al. Evaluating polygenic risk scores for breast cancer in women of African ancestry. J. Natl. Cancer Inst. 2021;113:1168–1176. doi: 10.1093/jnci/djab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Z., Lubmawa A., Gundell S., Wan P., Nalukenge C., Muwanga P., Lutalo M., Nansereko D., Ndaruhutse O., Katuku M., et al. Genetic risk of prostate cancer in Ugandan men. Prostate. 2018;78:370–376. doi: 10.1002/pros.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekoru K., Adeyemo A.A., Chen G., Doumatey A.P., Zhou J., Bentley A.R., Shriner D., Rotimi C.N. Genetic risk scores for cardiometabolic traits in sub-Saharan African populations. Int. J. Epidemiol. 2021;50:1283–1296. doi: 10.1093/ije/dyab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iribarren C., Lu M., Jorgenson E., Martinez M., Lluis-Ganella C., Subirana I., Salas E., Elosua R. Weighted multi-marker genetic risk scores for incident coronary heart disease among individuals of African, Latino and East-Asian ancestry. Sci. Rep. 2018;8:6853. doi: 10.1038/s41598-018-25128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R., Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi Q., Stilp A.M., Sofer T., Moon J.Y., Hidalgo B., Szpiro A.A., Wang T., Ng M.C.Y., Guo X., MEta-analysis of type 2 DIabetes in African Americans (MEDIA) Consortium. Chen Y.I., et al. Genetics of type 2 diabetes in U.S. Hispanic/Latino individuals: results from the Hispanic Community health study/study of Latinos (HCHS/SOL) Diabetes. 2017;66:1419–1425. doi: 10.2337/db16-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chande A.T., Rishishwar L., Conley A.B., Valderrama-Aguirre A., Medina-Rivas M.A., Jordan I.K. Ancestry effects on type 2 diabetes genetic risk inference in Hispanic/Latino populations. BMC Med. Genet. 2020;21:132. doi: 10.1186/s12881-020-01068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen W., Shu X.O., Guo X., Cai Q., Long J., Bolla M.K., Michailidou K., Dennis J., Wang Q., Gao Y.T., et al. Prediction of breast cancer risk based on common genetic variants in women of East Asian ancestry. Breast Cancer Res. 2016;18:124. doi: 10.1186/s13058-016-0786-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mak T.S.H., Porsch R.M., Choi S.W., Zhou X., Sham P.C. Polygenic scores via penalized regression on summary statistics. Genet. Epidemiol. 2017;41:469–480. doi: 10.1002/gepi.22050. [DOI] [PubMed] [Google Scholar]
- 31.Vilhjalmsson B.J., Yang J., Finucane H.K., Gusev A., Lindstrom S., Ripke S., Genovese G., Loh P.R., Bhatia G., Do R., et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge T., Chen C.Y., Ni Y., Feng Y.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prive F., Arbel J., Vilhjalmsson B.J. LDpred2: better, faster, stronger. Bioinformatics. 2020;36:5424–5431. doi: 10.1093/bioinformatics/btaa1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dikilitas O., Schaid D.J., Kosel M.L., Carroll R.J., Chute C.G., Denny J.A., Fedotov A., Feng Q., Hakonarson H., Jarvik G.P., et al. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am. J. Hum. Genet. 2020;106:707–716. doi: 10.1016/j.ajhg.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahed A.C., Aragam K.G., Hindy G., Chen Y.I., Chaudhary K., Dobbyn A., Krumholz H.M., Sheu W.H.H., Rich S.S., Rotter J.I., et al. Transethnic transferability of a genome-wide polygenic score for coronary artery disease. Circ. Genom. Precis. Med. 2020;14:e003092. doi: 10.1161/CIRCGEN.120.003092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M., Menon R., Mishra S., Patel A.P., Chaffin M., Tanneeru D., Deshmukh M., Mathew O., Apte S., Devanboo C.S., et al. Validation of a genome-wide polygenic score for coronary artery disease in South Asians. J. Am. Coll. Cardiol. 2020;76:703–714. doi: 10.1016/j.jacc.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamri A., Mao S., Desai D., Gupta M., Pare G., Anand S.S. Fine-tuning of genome-wide polygenic risk scores and prediction of gestational diabetes in South Asian women. Sci. Rep. 2020;10:8941. doi: 10.1038/s41598-020-65360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Guo J., Ni G., Yang J., Visscher P.M., Yengo L. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat. Commun. 2020;11:3865. doi: 10.1038/s41467-020-17719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuchenbaecker K., Telkar N., Reiker T., Walters R.G., Lin K., Eriksson A., Gurdasani D., Gilly A., Southam L., Tsafantakis E., et al. The transferability of lipid loci across African, Asian and European cohorts. Nat. Commun. 2019;10:4330. doi: 10.1038/s41467-019-12026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norio R. Finnish Disease Heritage I: characteristics, causes, background. Hum. Genet. 2003;112:441–456. doi: 10.1007/s00439-002-0875-3. [DOI] [PubMed] [Google Scholar]
- 41.Martin A.R., Karczewski K.J., Kerminen S., Kurki M.I., Sarin A.P., Artomov M., Eriksson J.G., Esko T., Genovese G., Havulinna A.S., et al. Haplotype sharing provides insights into fine-scale population history and disease in Finland. Am. J. Hum. Genet. 2018;102:760–775. doi: 10.1016/j.ajhg.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahed A.C., Wang M., Homburger J.R., Patel A.P., Bick A.G., Neben C.L., Lai C., Brockman D., Philippakis A., Ellinor P.T., et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 2020;11:3635. doi: 10.1038/s41467-020-17374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirugo G., Williams S.M., Tishkoff S.A. The missing diversity in human genetic studies. Cell. 2019;177:1080. doi: 10.1016/j.cell.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 45.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L., et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyama S., Ito K., Terao C., Akiyama M., Horikoshi M., Momozawa Y., Matsunaga H., Ieki H., Ozaki K., Onouchi Y., et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 2020;52:1169–1177. doi: 10.1038/s41588-020-0705-3. [DOI] [PubMed] [Google Scholar]
- 47.Marquez-Luna C., Loh P.R., South Asian Type 2 Diabetes (SAT2D) Consortium. SIGMA Type 2 Diabetes Consortium. Price A.L. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol. 2017;41:811–823. doi: 10.1002/gepi.22083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gettler K., Levantovsky R., Moscati A., Giri M., Wu Y., Hsu N.Y., Chuang L.S., Sazonovs A., Venkateswaran S., Korie U., et al. Common and rare variant prediction and penetrance of IBD in a large, multi-ethnic, health system-based biobank cohort. Gastroenterology. 2020;160:1546–1557. doi: 10.1053/j.gastro.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakaue S., Hirata J., Kanai M., Suzuki K., Akiyama M., Lai Too C., Arayssi T., Hammoudeh M., Al Emadi S., Masri B.K., et al. Dimensionality reduction reveals fine-scale structure in the Japanese population with consequences for polygenic risk prediction. Nat. Commun. 2020;11:1569. doi: 10.1038/s41467-020-15194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marnetto D., Parna K., Lall K., Molinaro L., Montinaro F., Haller T., Metspalu M., Magi R., Fischer K., Pagani L. Ancestry deconvolution and partial polygenic score can improve susceptibility predictions in recently admixed individuals. Nat. Commun. 2020;11:1628. doi: 10.1038/s41467-020-15464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bitarello B.D., Mathieson I. Polygenic scores for height in admixed populations. G3 (Bethesda) 2020;10:4027–4036. doi: 10.1534/g3.120.401658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan Y., Anne Feng Y.-C., Chen C.-Y., Lam M., Stanley Global Asia I., Sawa A., Martin A.R., Qin S., Huang H., Ge T. Improving polygenic prediction in ancestrally diverse populations. medRxiv. 2021 doi: 10.1101/2020.12.27.20248738. Preprint at. [DOI] [Google Scholar]
- 53.Weissbrod O., Kanai M., Shi H., Gazal S., Peyrot W., Khera A., Okada Y., The Biobank Japan, Project. Martin A., Finucane H., Price A.L. Leveraging fine-mapping and non-European training data to improve trans-ethnic polygenic risk scores. medRxiv. 2021 doi: 10.1101/2021.01.19.21249483. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amariuta T., Ishigaki K., Sugishita H., Ohta T., Koido M., Dey K.K., Matsuda K., Murakami Y., Price A.L., Kawakami E., et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat. Genet. 2020;52:1346–1354. doi: 10.1038/s41588-020-00740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mostafavi H., Harpak A., Agarwal I., Conley D., Pritchard J.K., Przeworski M. Variable prediction accuracy of polygenic scores within an ancestry group. Elife. 2020;9:e48376. doi: 10.7554/eLife.48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gathani T., Ali R., Balkwill A., Green J., Reeves G., Beral V., Moser K.A., Million Women Study Collaborators Ethnic differences in breast cancer incidence in England are due to differences in known risk factors for the disease: prospective study. Br. J. Cancer. 2014;110:224–229. doi: 10.1038/bjc.2013.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiscella K., Sanders M.R. Racial and ethnic disparities in the quality of health care. Annu. Rev. Publ. Health. 2016;37:375–394. doi: 10.1146/annurev-publhealth-032315-021439. [DOI] [PubMed] [Google Scholar]
- 58.Carnethon M.R., Pu J., Howard G., Albert M.A., Anderson C.A.M., Bertoni A.G., Mujahid M.S., Palaniappan L., Taylor H.A., Jr., Willis M., et al. Cardiovascular health in African Americans: a scientific statement from the American heart association. Circulation. 2017;136:e393–e423. doi: 10.1161/CIR.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 59.Shai I., Jiang R., Manson J.E., Stampfer M.J., Willett W.C., Colditz G.A., Hu F.B. Ethnicity, obesity, and risk of type 2 diabetes in women: a 20-year follow-up study. Diabetes Care. 2006;29:1585–1590. doi: 10.2337/dc06-0057. [DOI] [PubMed] [Google Scholar]
- 60.Tamlander M., Mars N., Pirinen M., Finn G., Widén E., Ripatti S. Integration of questionnaire-based risk factors improves polygenic risk scores for human coronary heart disease and type 2 diabetes. Commun. Biol. 2022;5:158. doi: 10.1038/s42003-021-02996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mars N., Widén E., Kerminen S., Meretoja T., Pirinen M., della Briotta Parolo P., Palta P., Havulinna A., Elliott A., Shcherban A., et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat. Commun. 2020;11:6383. doi: 10.1038/s41467-020-19966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagai A., Hirata M., Kamatani Y., Muto K., Matsuda K., Kiyohara Y., Ninomiya T., Tamakoshi A., Yamagata Z., Mushiroda T., et al. Overview of the BioBank Japan project: study design and profile. J. Epidemiol. 2017;27:S2–S8. doi: 10.1016/j.je.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishigaki K., Akiyama M., Kanai M., Takahashi A., Kawakami E., Sugishita H., Sakaue S., Matoba N., Low S.-K., Okada Y., et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 2020;52:669–679. doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akiyama M., Ishigaki K., Sakaue S., Momozawa Y., Horikoshi M., Hirata M., Matsuda K., Ikegawa S., Takahashi A., Kanai M., et al. Characterizing rare and low-frequency height-associated variants in the Japanese population. Nat. Commun. 2019;10:4393. doi: 10.1038/s41467-019-12276-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loh P.R., Danecek P., Palamara P.F., Fuchsberger C., Reshef Y.A., Finucane H.K., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leitsalu L., Haller T., Esko T., Tammesoo M.L., Alavere H., Snieder H., Perola M., Ng P.C., Magi R., Milani L., et al. Cohort profile: Estonian biobank of the Estonian genome center, University of Tartu. Int. J. Epidemiol. 2015;44:1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 68.Mitt M., Kals M., Parn K., Gabriel S.B., Lander E.S., Palotie A., Ripatti S., Morris A.P., Metspalu A., Esko T., et al. Improved imputation accuracy of rare and low-frequency variants using population-specific high-coverage WGS-based imputation reference panel. Eur. J. Hum. Genet. 2017;25:869–876. doi: 10.1038/ejhg.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krokstad S., Langhammer A., Hveem K., Holmen T.L., Midthjell K., Stene T.R., Bratberg G., Heggland J., Holmen J. Cohort profile: the HUNT study, Norway. Int. J. Epidemiol. 2013;42:968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 70.Das S., Forer L., Schonherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karlson E.W., Boutin N.T., Hoffnagle A.G., Allen N.L. Building the partners HealthCare biobank at partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. J. Personalized Med. 2016;6:2. doi: 10.3390/jpm6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genomes Project Consortium. Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang J., Howie B., McCarthy S., Memari Y., Walter K., Min J.L., Danecek P., Malerba G., Trabetti E., Zheng H.F., et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat. Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The FinnGen data can be accessed through the Fingenious® services (https://site.fingenious.fi/en/) managed by FINBB. GWAS genotype data of BioBank Japan are available at the National Bioscience Database Center Human Database (research ID: hum0014; https://humandbs.biosciencedbc.jp/en/hum0014-v21). UK Biobank data are available through a procedure described at http://www.ukbiobank.ac.uk/using-the-resource/.The HUNT data may be accessed by application to the HUNT Research Centre (https://www.ntnu.edu/hunt). Researchers interested in Estonian Biobank can request the access at https://www.geenivaramu.ee/en/access-biobank. De-identified data of the MGB Biobank that supports this study is available from the MGB Biobank portal (https://biobank.partners.org/). Restrictions apply to the availability of these data, which are available to MGB-affiliated researchers via a formal application. Weights for the LDpred PRSs are available at PGS Catalog (pgs-info@ebi.ac.uk) with PGS IDs PGS002241–PGS002244, and weights for the PRS-CS PRSs with PGS001780–PGS001781,60 PGS000335,61 and PGS002240.
-
•
Original code generated within this project has been deposited at Zenodo and is publicly available. DOIs are listed in the key resources table.
-
•
Any additional information is available from the lead contact upon request.