Abstract
COVID-19 outbreak revealed fundamental weaknesses of current diagnostic systems, particularly in prediction and subsequently prevention of pandemic infectious diseases (PIDs). Among PIDs detection methods, wastewater-based epidemiology (WBE) has been demonstrated to be a favorable mean for estimation of community-wide health. Besides, by going beyond purely sensing usages of WBE, it can be efficiently exploited in Healthcare 4.0/5.0 for surveillance, monitoring, control, and above all prediction and prevention, thereby, resulting in smart sensing and management of potential outbreaks/epidemics/pandemics. Herein, an overview of WBE sensors for PIDs is presented. The philosophy behind the smart diagnosis of PIDs using WBE with the help of digital technologies is then discussed, as well as their characteristics to be met. Analytical techniques that are pushing the frontiers of smart sensing and have a high potential to be used in the smart diagnosis of PIDs via WBE are surveyed. In this context, we underscore key challenges ahead and provide recommendations for implementing and moving faster toward smart diagnostics.
Keywords: Smart diagnostics, Emerging infectious diseases, SARS-CoV-2, Internet of things, Big data analysis, Artificial intelligence, Real-time connectivity
Graphical abstract
1. Introduction
Biosensors and bioanalytical devices have received significant and unprecedented achievements over the last decades; however, the occurrence of the present coronavirus disease 2019 (COVID-19) pandemic with serious global health and economic effects worldwide has revealed the fundamental weakness of current diagnostic systems, especially for efficient sensing, management, and prediction of epidemics/pandemics [1,2]. Particularly, its severity has increased due to new mutations and variants and caused health experts to re-evaluate the effectiveness of current strategies for pandemic management [3]. Indeed, the present pandemic, perhaps more than ever, revealed that our sensing/diagnostics systems demand an in-depth renovation and fast inevitable transition from pure diagnostics toward smart diagnostics to be able to efficiently cope with pandemics and prevent possible future epidemics/pandemics [4].
The advent of a smart diagnostic system and concept goes back to the work described by Chowdhury and Saha in 2006, which used fuzzy logic for diagnostic decision-making in renal therapeutic applications. The system could monitor and diagnose regularly and help the healthcare specialists for therapeutic decision-making and transferring some health-related parameters wirelessly [5]. Over the years, the idea of smart diagnostic has been included in the concept of smart city, which is a term used for technologies and concepts that are directed toward making cities technologically more advanced, socially inclusive, and greener [6]. Generally speaking, smart diagnostics refers to a diagnosis on account of smart data analyzed/resulting from the big data of diagnostic devices/tests through digital technologies including the Internet of things (IoT) [7], machine learning (ML), deep learning (DL), big data analytics (BDA), artificial neural networks (ANN), artificial intelligence (AI) [8], blockchain analysis (BA), system integration, augmented reality (AR), system integration, fog/cloud computing, cybersecurity, and smartphone [9]. Among various detection scenarios, which can be effectively utilized toward smart diagnostics of COVID-19, the detection of biomolecules forming part of the structure of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in sewage through the so-called wastewater-based epidemiology (WBE) has drawn lots of attention as a potent epidemiological tool for early monitoring and warning of the virus spreading in communities and consequently clarifying the pandemic status of SARS-CoV-2. In this arena, point-of-need biosensors provide self-testing criteria for rapid detection of suspicious individual cases of COVID-19 [10,11]; however, an approach such as WBE, which enables targeting large population and zonal analysis instead of individual tests, is more advantageous in terms of cost-effectiveness, speed, and regional healthcare [12]. Indeed the genetic material of viral RNA reproduced in infected individuals, with or without symptoms, will also go into the wastewater system/treatment plant; hence, analyzing wastewater pay a way for examining all people in an area even at an early stage [13]. Specifically, it has been proved that the SARS-CoV-2 RNA is shed in feces, saliva, and sputum, and these body fluids are often poured into wastewater systems. Since the symptoms of COVID-19 appear after a few days of infection or even in most cases are not observed, WBE can be employed as an early detection indicator and monitoring system to determine the occurrence of COVID-19 within a specific region [14] (Fig. 1 ).
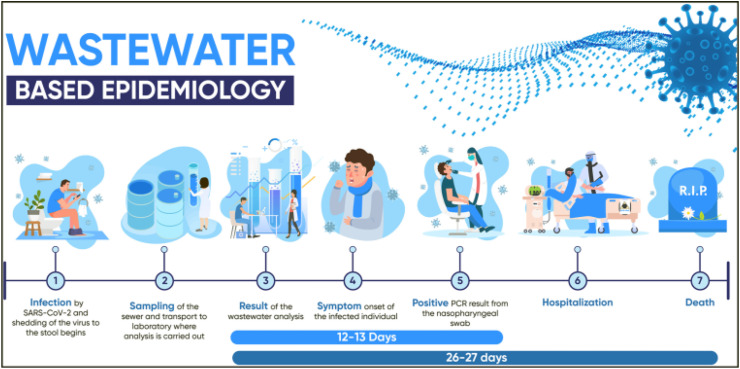
Fig. 1.
Schematic representation of wastewater-based epidemiology. A rise in the value of viral in the wastewater is measured to be about 12 days earlier than a rise in clinical PCR testing and about 26 days before an upturn in death numbers, reprinted with permission from Ref. [15].
Observations of COVID-19 genetic material in sewage have been reported in a variety of countries, including Australia [16], France [17], Italy [18], the Netherlands [19], Spain [20], the Czech Republic [21], Japan [22], Turkey [23], India [24], Pakistan [25], China [26] and the USA [27], which has been summarized in some reviews [14]. These findings demonstrate the presence of SARS-CoV-2 in sewage and hence provide a way for WBE. Due to the high importance of this concept, a plethora of reviews and perspectives have been reported on the WBE, providing technical frameworks [28], analysis, and quantification methods [[29], [30], [31], [32], [33]], highlighting its early warning capabilities to cope efficiently [[34], [35], [36], [37], [38], [39], [40], [41], [42]], demonstrating prospects [12,43] and challenges of using electrochemical immunosensors [44] and PCR-based methods [[45], [46], [47], [48], [49]]. Other studies have been devoted to the data-driven estimation of COVID-19 through WBE [50], the employment of artificial intelligence (AI) techniques [[51], [52], [53]], and fuzzy-Bayesian optimization modeling [54]. Some studies also examined the employment of sensor systems in wastewater plants to monitor their virus content [55] and highlighted the employment of digital technologies [56] and smart cities [57] in this area. Recently, we have also discussed and highlighted the features that ought to be addressed by COVID-19 diagnostic tests/devices to realize smart diagnostics of future epidemics/pandemics [58]. Nonetheless, there are still some unanswered questions in the utilization of WBE as a promising tool in combat with pandemics, particularly through their early prediction: “What criteria should be addressed by WBE sensors and how they can be efficiently exploited toward smart diagnostics of pandemic infectious diseases (PIDs)”?
Although WBE offers life-saving information related to infectious diseases; WBE sensors can only be exploited toward smart diagnostics of PIDs if they can be connected to each other and other systems via Internet/IoT as the heart of Healthcare 4.0 for sharing, analyzing, and eventually generating smart data in combination with digital technologies. Moreover, due to the ubiquitous character of COVID-19, real-time connected WBE sensors should meet other criteria to be globally exploited for smart PIDs diagnostics, even at sites far from centralized laboratories and resource-limited settings. In this context, we foresee that the realization of smart diagnostics will lead to resourceful strategies and life-saving smart decisions toward monitoring, control, treatment, surveillance, and management of pandemics during the pandemic occurrence period, and more importantly prediction and consequently on time prevention of the next possible outbreak/epidemic/pandemic scenarios in the future.
This review aims to illustrate the inevitability of fast transition from pure sensing platforms towards smart sensing systems and provide the possible application pathways for the employment of smart sensing devices for effective detection, monitoring, management, and diagnosis of PIDs via WBE. We outline an overview of WBE sensors for PIDs as well as the philosophy behind the smart diagnosis of PIDs using WBE. We also bring up the features and criteria that should be come across by WBE sensors to be employed for smart diagnosis of PIDs with the help of digital technologies and to defeat the existing pandemic and hinder the occurrence of the next possible outbreaks/epidemics/pandemics. Finally, the current challenges and our recommendations to move faster toward realizing the smart diagnosis of PIDs using WBE are highlighted.
2. Wastewater-based biosensors for pandemic infectious diseases monitoring
In this section, different types of employed wastewater-based biosensing systems for detection of COVID-19 or those of biosensors possessing the potential to be utilized in this area will be discussed and the respective analytical performances and figures of merit are presented. These biosensing platforms include polymerase chain reaction (PCR)-based assays, lab-on-a-chip (LOC) and microfluidic devices, μpaper-based analytical devices (μPADs), and lateral flow assays (LFAs).
The existing diagnostic systems for COVID-19 testing can be broadly categorized into two groups: genetic material and antigen detection for early identification of infected persons, and antibody detection in serum/blood samples for monitoring infected people during their treatment or past infections [59]. However, detection of genetic material is the only accepted method in studies regarding WBE; Hence, a plethora of approaches have been reported in genetic material detection of SARS-CoV-2 including reverse transcription-polymerase chain reaction (RT-PCR), clustered regularly interspaced short palindromic repeats (CRISPR), loop-mediated isothermal amplification (LAMP), digital PCR (dPCR) and nucleic acid sequence-based amplification (NASBA) [33]. Although these approaches often benefit from high sensitivity, reliability, laboratory availability, and high throughput capabilities, most of them suffer from the need for sophisticated instruments, centralized services, specialized users, and lack of point-of-use (POU) employment capabilities [59], which diminish their employment in massively accessible WBE utilization. In addition, qRT-PCR has been utilized as a golden approach in quantifying SARS-CoV-2 in sewage; however, due to the complexity of the matrix, the reported protocols are complicated and include several steps of sampling, storage, pre-concentration, RNA extraction, and finally measuring. At present, there are no consequences of targets endorsed for detecting SARS-CoV-2 in sewage [15]. Furthermore, various techniques have been utilized for concentrating viruses from wastewater samples such as employing electropositive/electronegative membranes, ultrafiltration, precipitation by polyethylene glycol, ultracentrifugation, and monolithic adsorption filtration columns [32]. Such different concentrating approaches certainly impact the accuracy of final results; hence, the necessity for standardization of detection approaches and the usage of standard strategies by all researchers to achieve reliable and comparable results are highly demanded [48,49]. In addition to these analytical complexities, other criteria such as the survivability of SARS-CoV-2 in various types of water matrices, the effect of water treatment processes on SARS-CoV-2, time of sampling, the storage and handling conditions of samples are all challenging factors that should be considered since they substantially affect outcomes of wastewater analysis [47]. The uncertainty of results is the biggest problem of WBE, as it is evident in the case of drugs of abuse [60]. The advent of new variants of SARS-CoV-2 makes it necessary to develop new and specific recognition elements for each type of virus, which further complicates the issue [61].
Interestingly, the basic working principle of PCR-based approaches can be integrated into LOC and microfluidic devices. Each LOC instrument usually contains reaction chambers and detectors connected by valves, mixers, and pumps. Hence, they are paving a way for automated virus detection without any need for laborious clinical protocols with considerably low mass-production costs [62]. These devices are broadly employed in clinical applications for viral detection of Ebola virus disease [63], dengue fever hepatitis [64], and human immunodeficiency virus (HIV) [65]. The existing COVID-19 pandemic has significantly accelerated the advance or adaptation of LOC platforms by various companies, which were previously used for diagnosis of infectious disease outbreaks, such as cobas® Liat® by Roche, ID NOW from Abbott Laboratories, Biofire® Filmarray® from BioMerieux, GeneXpert® developed by Cepheid™, and RTisochip® established by CapitalBio™. The most important advantage of these technologies is their employment by a non-expert user, which can be used for both home and clinical settings, and possibly in WBE. The use of LOC technology has been approved by the respective authorities in several countries, including the USA, China, and Japan, demonstrating its application value. These technologies aid simple, fast, sensitive, and on-site virus detection and can be utilized for WBE [32]. A multicenter study on the performance of cobas® Liat® in the quantification of SARS-CoV-2 showed highly accurate results within 20 min with a sensitivity of 100% and specificity of 98.6% that are in accordance with high-throughput laboratory molecular tests [66]. A systematic study of literature by Care et al. on sensitivity comparison of ID NOW and RT–PCR for detection of SARS-CoV-2 showed an overall sensitivity of 84% for ID NOW assay in an ambulatory population [67]. Liotti et al. compared the efficiency of Biofire® Filmarray® with that of Quanty COVID-19 assay. Compared to the reference method, the sensitivity and specificity for BioFire COVID-19 were 93.0%, 100.0%, respectively [68]. A meta-analysis approach by Lee and Song represented the pooled sensitivity and specificity of 99% and 97% for the Xpert® Xpress assay, and 79% and 100% for the ID NOW assay in detection of SARS-CoV-2, respectively [69]. A multi-index nucleic acid isothermal amplification analyzer (RTisochip™-W) involving a microfluidic chip to analyze 19 common respiratory viruses as well as SARS-CoV-2 was developed by Xing et al. [70]. They used NASBA that is mostly appropriate for the detection of single-stranded RNA. This approach needs fewer cycles than its conventional counterparts e.g. PCR or LAMP, and hence diminishes the time of assay and total error. Limit of detection (LOD) values of 50 copies/μL or lower were obtained for quantification of SARS-CoV-2 by RTisochip™-W system with a coincidence rate of 98.15%. Despite having outstanding features, these commercial LOC platforms still need connection to IoT gateways to be employed in smart sensing avenue. More recently, in this regard, an assimilated microfluidic chip was developed for multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in sewage (Fig. 2 ). The device includes on-chip nucleic acid extraction on a Flinders Technology Associates (FTA) membrane followed by isothermal amplification and finally colorimetric detection, which is processed timely by a smartphone needless to any complicated equipment. Within 1 h, sensitivities of 100 genome equivalent (GE)/mL and 500 colony-forming units (CFU)/mL were attained for SARS-CoV-2 and human enteric pathogens, respectively. Thanks to the wireless connection by smartphone, test results and locations as data, could be reported on a custom website enabling online infection source tracking [71]. However, there is still a necessity for availability of a database center to gather data, model and evaluate them for proper decision making and epidemic management. In addition, the smartphone-based quantification modalities suffer from camera-dependent results that can affect their performance and outputs [72,73].
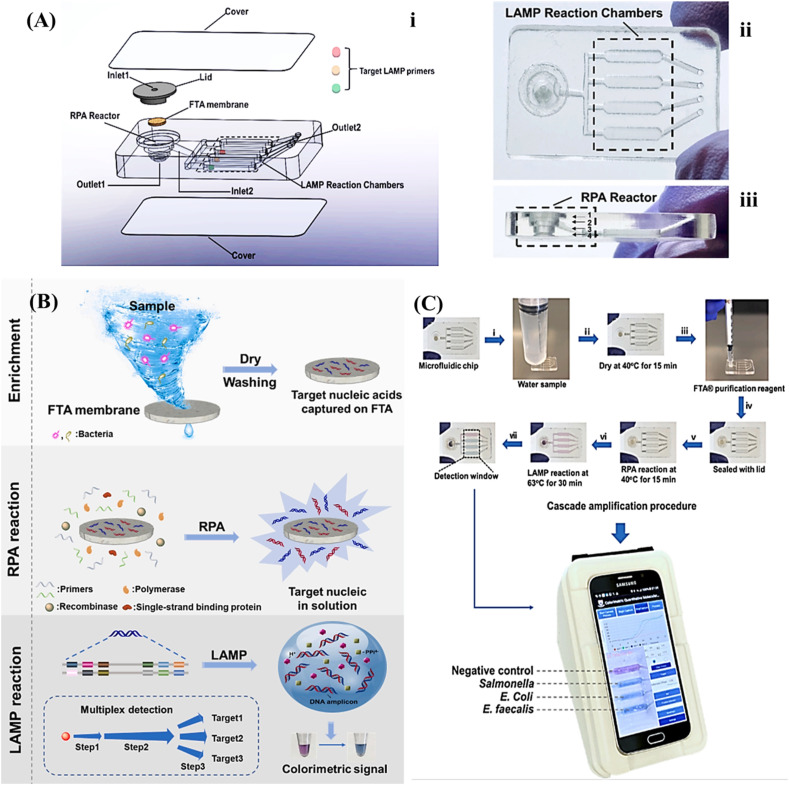
Fig. 2.
(A) Assimilated microfluidic chip for multiplexed colorimetric assay, i: exploded; ii: top; and iii: side views. (B) Working principle of the assay for the detection of different pathogens. (C) The procedure of multiplexed colorimetric detection, reprinted with permission from [71].
Recently, the combination of RT-LAMP and microfluidic tools for the quantification of SARS-CoV-2 in sewage has been reported. In this work, the products of RT-LAMP and PCR of wastewater samples were tested within a microfluidic device prepared from polydimethylsiloxane, in which the color changes from pink (negative sample) to yellow/orange (positive sample) can be observed easily by the naked eye. The results of RT-LAMP for wastewater samples were largely in accordance with those of RT-qPCR. They found that RNA extraction is necessary for successful analysis [74]. A miniaturized and integrated micro-capillary electrophoresis (μCE) system was also developed that can be manipulated by using a smartphone (Fig. 3 ). In the developed device, the smartphone not only powers the electrical section but is also used for detecting the fluorescence signal of amplified amplicons by RT-PCR for on-site quantification. With a weight of ∼1 kg, as a suitable device for point-of-care (POC) DNA testing, it could precisely analyze two genes of SARS-CoV-2 [75]; however, its utility for real sample analysis and especially in WBE should be evaluated.
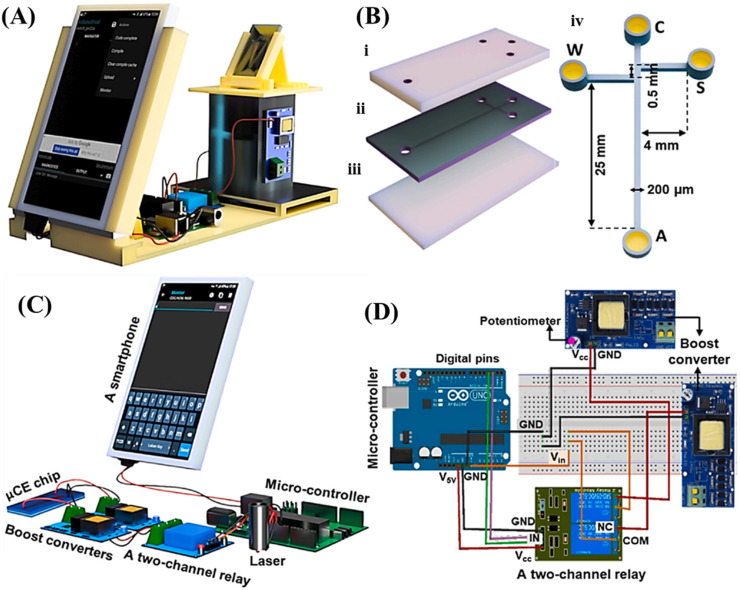
Fig. 3.
Scheme of the integrated portable μCE system. (A) smartphone containing system. (B) The assembly of the chip containing a polymethyl methacrylate (PMMA) layer for each of (i) patterning reservoirs, (ii) patterning a typical sample-stacking channel, and (iii) as a base. The dimension of the μ-channels was shown in (iv). (C) The schemes of individual components and their configuration within system. A smartphone is connected to a micro-controller, to supply electronic power for system. (D) The detailed circuit connection among the micro-controller, reprinted with permission from Ref. [75].
As the simplest yet most efficient approach, the possibility of μPADs was also tested in this arena. The μPADs consist of different printed sections that integrate all processes required for nucleic acid testing including extraction, purification, concentration, elution, amplification, and visual detection into an inexpensive paper assembly [76]. These tests rival PCR-based laboratory techniques in terms of multiplexity and sensitivity and enable high-quality and fast detection of viruses. Recently, the quantification of malaria from whole blood has been demonstrated using a multiplexed paper-based device in a rural region in Uganda. Offering a sensitive and multiplexed nucleic acid sequences analysis of pathogens within just 50 min. It showed a rapid and precise diagnosis for malaria compared with PCR [77]. Another advantage of μPADs is their easy coupling with various detection systems, especially electrochemical approaches to enhance sensitivity. By printing the electrodes on folded paper, a quantitative paper-based electrochemical sensor chip genetic material has been reported elsewhere [78] to facilitate the digital detection of SARS-CoV-2. As seen in Fig. 4 , this biosensor utilizes gold nanoparticles capped with antisense oligonucleotides highly specific to viral nucleocapsid phosphoprotein (N-gene) and immobilized on paper. In less than 5 min, the sensor showed a LOD of 6.9 copies/μL, without the need for any further amplification, with almost 100% accuracy, sensitivity, and specificity for real sample analysis. Such strategies have the potential to be employed in the smart detection of viruses’ genomes for WBE in the case of adding the capability of data sharing.
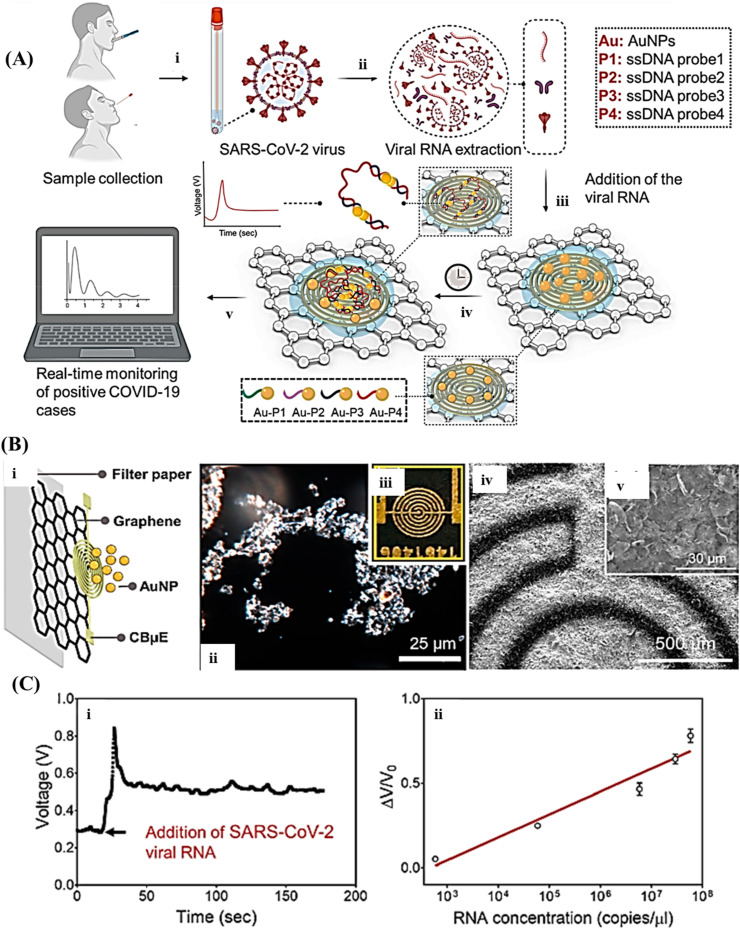
Fig. 4.
(A) An scheme representing the working principle of the COVID-19 electrochemical μPAD in which in step i: the samples are got from the nasal swab or saliva; step ii: the viral SARS-CoV-2 RNA is extracted; step iii: the viral RNA is added on top of the graphene-ssDNA-AuNP platform; step iv: incubated for 5 min; and step v: the digital electrochemical output is noted. (B) Illustrating the layer-by-layer building of the sensor (i) Zoom-in image. (ii) Dark-field view of the graphene film. (iii) Photograph of the graphene sensor. (iv, v) SEM images of the graphene-based platform. (C) Final signal, (i) as a function of time with the addition of SARS-CoV-2 viral RNA loads (5.85 × 104 copies/μL). (ii) The standard curve, reprinted with permission from Ref. [78].
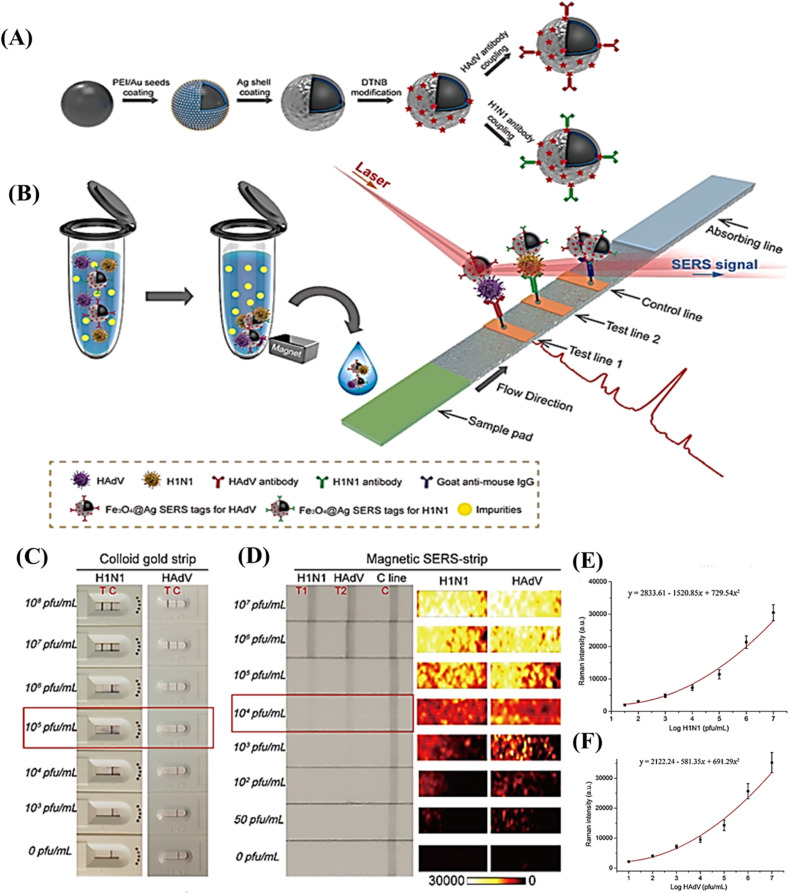
LFAs, as a mature technology in the POC field, are simple and economical devices. These rapid test devices can be easily coupled with quantification detectors and data sharing tools [79,80], and a variety of nanotags have been employed to enhance their sensitivities [81]. Since the desired target for virus detection in WBE is to quantify the genetic material of the virus, the oligonucleotide-based LFA or even lateral flow immunoassay (LFIA) could be used in this field. However, the amplification of genetic material before testing by LFA, the signal amplification within the test strip, or the use of highly sensitive detection approaches is crucial for quantification purposes. For instance, concurrent detection of influenza A H1N1 virus and human adenovirus (HAdV) was attained via a surface-enhanced Raman scattering-based lateral flow immunoassay (SERS-based LFIA) strip using Fe3O4@Ag nanoparticles as an enrichment agent and reporting tag (Fig. 5 ). The direct analysis of complex matrices can be achieved via this strip without any primary pretreatment. Some features such as ease to operate, rapidness, stable signals, and high throughput make it the desired approach for point-of-need analysis, possibly in WBE [82]. Another research group has reported the combination of an easy-to-perform isothermal and non-enzymatic signal boosting system named catalytic hairpin assembly reaction with an LFIA in the quantification of SARS-CoV-2 within swab samples. This method does not require RNA isolation, PCR amplification, and complicated analysis and benefits from a low LOD of 2000 copies/mL with 100% positive and negative predictive agreements [83]. A microfluidic integrated lateral flow assay that combines the reverse transcription recombinase polymerase amplification (RT-RPA) and a quantification dipstick into a single microfluidic chip (MI–IF–RPA) was settled for fast and highly sensitive detection of SARS-CoV-2. This device needs just conventional nucleic acid extraction and loading followed by incubation to yield results in approximately 30 min with a LOD of 1 copy/μL. The evaluation of chip performance by clinically approved cases of COVID-19 exhibited sensitivity and specificity values of 97% and 100%, respectively, which are highly comparable to RT-PCR assay [84].
Fig. 5.
(A) Synthetic procedure for antibody-immobilized Fe3O4@Ag magnetic particles and (B) Scheme of the magnetic-SERS strip for quantifying two respiratory viruses, (C) Photos of commercial colloidal gold-based strips for H1N1 and HAdV detection. (D) Photos and SERS mapping images of detection zones of magnetic SERS strips at increasing concentrations of H1N1 and HAdV. (E) Calibration graphs of H1N1 and HAdV based on averaged SERS spectrum of two test zones at different values of H1N1 and HAdV, reprinted with permission from [82].
A survey of some examples for each biosensing strategy for genome-based quantification of SARS-CoV-2 is given in Table 1 . As seen, the three strategies including LOC, μPAD, and LFA show desired characteristics, especially in terms of sensitivity and specificity compared to the gold standard method qRT-PCR. Furthermore, the assay time for these biosensing schemes, as the most important characteristic for on-time monitoring, is usually below 1 h. Additionally, most of them provide POU detection with no need for huge sample handling and processing steps; hence, they can be used as portable biosensors in WBE.
Table 1.
A survey of some reported biosensing strategies for genome-based quantification of SARS-CoV-2.
| Biosensing strategy |
LODa |
Sensitivity (%) |
Specificity (%) |
Assay time |
User |
Assay cost (USD) |
Sample |
Pretreatment |
Operating environment |
Equipment |
Ref. |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT-qPCR | 1000 TUa/mL | 100 100 |
99.26 99.83 |
2–3 h | Trained technician | NR.a | Swab Sputum |
RNA extraction and purification | Specialized laboratories | Precision instruments, non-portable | [85] | |
| LOC |
cobas® Liat® | 0.009 (target: ORF1a/b), 0.003 (target: E gene), TCID 50/ml |
100 | 98.6 | 20 min | Trained technician | NR. | Swab | Purification, Amplification | POC | POU, portable | [66] |
| ID NOW | 3900-20,000 gene copies/mL | 84 | 100 | ≤13 min | NR. | Nasal swab | Purification, Amplification | [67] | ||||
| Biofire® Filmarray® | 3.30 × 102 RNA copies/mL | 93 | 100 | 45 min | NR. | Nasal/oropharyngeal | Purification, Amplification | [68] | ||||
| Xpert® Xpress | 0.0200 PFU/mL | 99 | 97 | 45 min | NR. | upper respiratory specimens | processing, extraction, amplification |
[69] | ||||
| RTisochipTM-W | 50 copies/μL | NR. | NR. | ≤90 min | NR. | Swab | processing, extraction, amplification |
[70] | ||||
| integrated microfluidic chip | 100 GEa/mL 10 GE/mL | NR. | NR. | 1 h | Needless to professional technician |
∼2 | River water | Treatment free | On-site | [71] | ||
| integrated RT-LAMP microfluidic chip | NR. | 85.7 | NR. | ∼40 min | NR. | Wastewater | RNA extraction, Amplification | [74] | ||||
| Smartphone-μCE |
1 copy number/μL |
NR. |
NR. |
6 min |
NR. |
RT-PCR amplicons |
NR. |
[75] |
||||
| μPAD |
6.9 copies/μL |
100 |
100 |
5 min |
Needless to a professional technician |
NR. |
Swab |
RNA extraction |
[78] |
|||
| Multiplex RT-LAMP-LFA | 12 copies/reaction | 100 | 100 | 1 h | Needless to professional technician | ∼7.5 | Swab | RNA extraction | [86] | |||
| CHA-LFIA | 2000 copies/mL | 100 | 100 | <90 min | NR. | Nasopharyngeal | Pretreatment, signal amplification | [83] | ||||
| MI-lF-RPA | 1 copy/μL | 97 | 100 | 30 min | NR. | Swab | With extraction and amplification | [84] | ||||
| RT-LAMP-LFA | 2 copies/μL | 100 | 100 | 40 min | NR. | Swab | Extraction free | [87] | ||||
| Fluorescent LFA | 500 copies/mL | 87.69 91.94 |
95.01 92.41 |
<1 h | ∼2 | Swab Sputum |
Extraction free, amplification free | [85] | ||||
LOD: limit of detection, TU: transduction units, NR: not reported, GE: genome equivalents, TCID: tissue culture infective doses.
3. Toward smart diagnosis of pandemic infectious diseases using wastewater-based epidemiology
Despite the significant developments and fascinating features of WBE biosensors for PIDs targeting and early warning and monitoring of virus spreading in communities, we believe that only those WBE sensors meeting the criteria discussed below can play a critical role in the smart diagnosis, management, prediction, and prevention of future outbreaks/epidemics/pandemics.
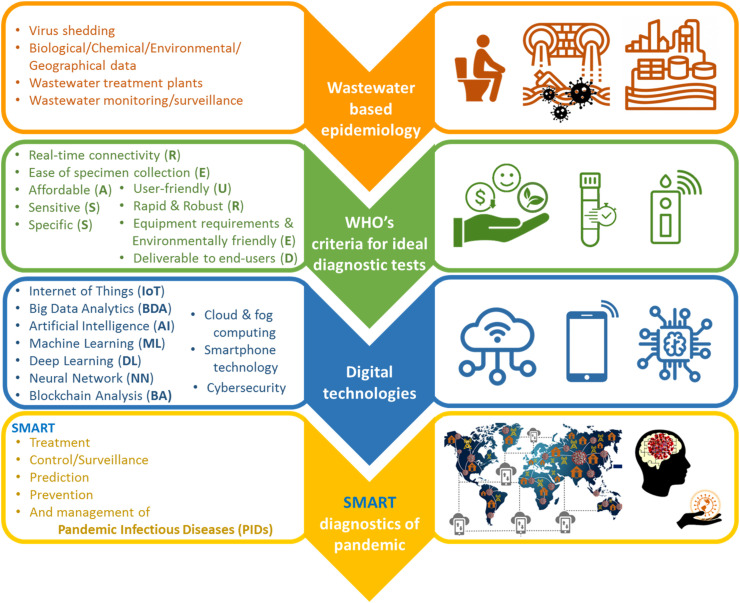
Indeed, as shown in Fig. 6 , we believe that wastewater epidemiological data obtained with WBE sensors that meet the following criteria, along with other related biological/chemical/environmental/geographical data, and with the help of digital technologies can finally lead to smart diagnostics of PIDs. One of the main requirements in the design and fabrication of smart WBE sensors for PIDs is that they should be able to connect to each other as well as other smart systems, and finally, end-users/decision-makers via the Internet as the most ideal candidate for inter/intra-connectivity, while having high sensitivity and specificity to avoid false-negatives and false-positives, respectively. This real-time connectivity through the IoT concept and sharing, analyzing, and optimizing the data with the assistance of new digital technologies such as ML, DL, BDA, ANN, AI, BA, and AR will lead to the generation of smart data as a required data for smart decisions. The IoT as the heart of Industry/Analytics/Healthcare 4.0 is a digital technology for connecting things/devices and data exchanging with other smart devices/systems via the Internet; as a result, those “things/devices” can be sensible, reportable, controllable, manageable, and smart (generally speaking) [88]. Indeed, IoT is considered the backbone of smart manufacturing and cyber-physical evolution in the Industry 4.0 era. Real-time connectivity is crucial for point of need/use analysis of samples and data transferring to the management center and receiving feedback for continuous real-time data gathering. The usage of WBE sensors with real-time connectivity capabilities will lead to timely testing and action to feedback from decision-makers, remote monitoring of conditions and hence management of the wastewater treatment processes, monitoring of disease speared, and subsequently its better control. Besides, it provides early warning that can be used for isolation of infected regions, delivery of required drugs, and society/health services, which will efficiently break the transmission chain of the disease.
Fig. 6.
Schematic representation of realizing smart diagnosis of PIDs using WBE, the corresponding ideal detection tests/devices, and digital technologies.
Hereof, smartphone technology can play a noteworthy and unique role in smart diagnostics owing to its fascinating and extraordinary features as an efficient universal tool and easy-to-use miniaturized computer. Besides, real-time connectivity with several wireless connectivity and data sharing/transferring modes can be achieved via smartphone. Interestingly, nowadays more than 90% of the world’s population is using smartphones contributing to about 6.8 billion Internet users [89,90]. Furthermore, the current developments in 4/5 G internet connectivity and data sharing, online cloud/fog services, near-field communication (NFC), wireless, Bluetooth connection, and powerful processors allow for the facile connection among decentralized devices/things and data-centers for smart services. Meanwhile, to reach the Internet of Medical Things (IoMT) [91] and the Internet of Analytical Things (IoAT), smartphones via interoperability can act as IoT gateways [92,93] and become cornerstone mediators for smart diagnostics. As a result, coupling/integrating the envisaged WBE sensors for PIDs to smartphones can be considered as an efficient strategy for meeting the real-time connectivity and some of the other criteria they need toward smart diagnostics of PIDs.
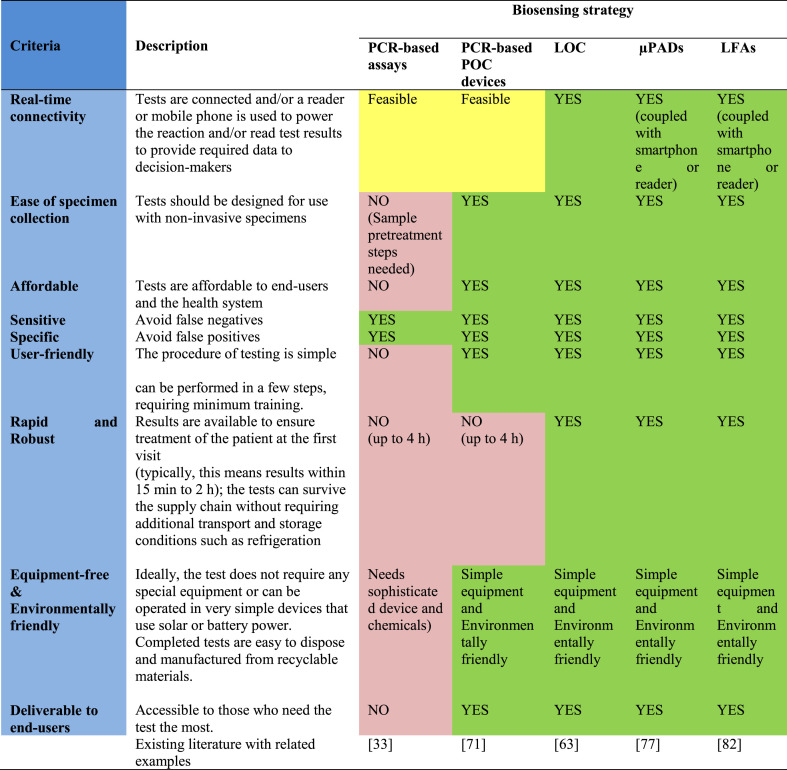
Furthermore, owing to the ubiquitous character of PIDs, WBE sensors should meet other criteria, in addition to real-time connectivity, high sensitivity, and specificity, to be globally utilized toward smart diagnostics of PIDs, even at resource-limited settings, the developing world, and generally sites far from well-equipped centralized laboratories as affordable and user/environmentally-friendly sensors. At this point, it is worth discussing those criteria proposed by the World Health Organization in 2018 to achieve the ideal diagnostic devices/tests. These criteria suggest that the devices/tests should be real-time connected (R), easy-to-sample collection, environmentally friendly (E), affordable (A), sensitive (S), specific (S), user-friendly (U), rapid and robust (R), equipment-free (E) and deliverable to end-users (D), that is REASSURED [94]. An evaluation of such criteria for the reported/high-potential wastewater-based biosensing systems toward the smart diagnosis of PIDs is displayed in Table 2 . In the case of real-time connectivity, only conventional PCR-based assays and their LOC counterparts cannot provide the desired results. In addition, POC PCR-based instruments still suffer from a time-consuming analysis process. Ease of specimen collection, user-friendly, rapidness and robustness, and equipment-free and environmentally friendly characteristics are also not satisfied by conventional PCR-based approaches. Three other proposed strategies, LOC, μPADs, and LFAs seem to be ideal candidates for smart monitoring of WBE because they could satisfy all the desired characteristics as detailed in Table 2.
Table 2.
The evaluation of COVID-19 diagnostic tests toward smart diagnostics of PIDs using WBE based on REASSURED criteria.
In addition to considering the WHO’s REASSURED criteria in the design and fabrication of WBE sensors for PIDs, the design of smart system architectures which can efficaciously integrate the digital technologies within a straightforward platform toward smart diagnostics of PIDs, is of great importance. Such applicable roadmaps with unified strategies derived from Industry 4.0/Healthcare 4.0 principles will pave the way to fulfill smart diagnostics of PIDs using the data obtained from the WBE sensors.
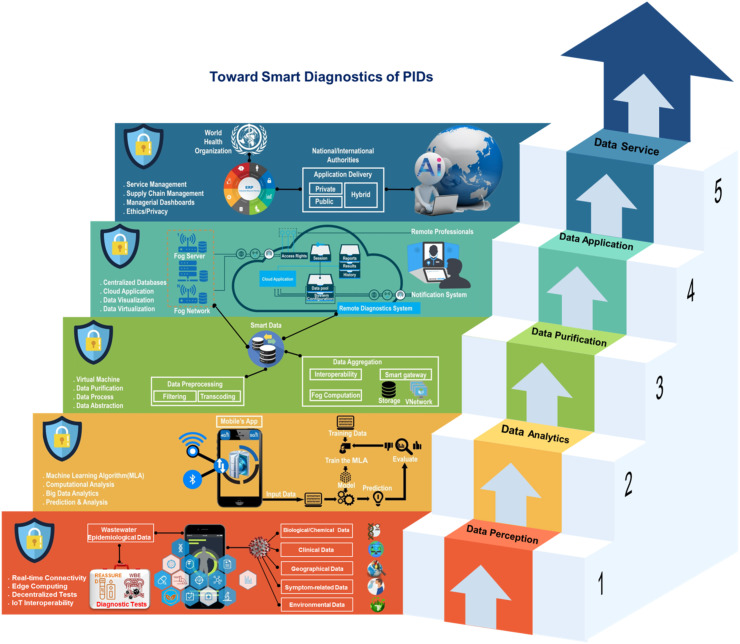
Hereof, we have proposed an IoT-Fog-Cloud model with joined approaches for data transmission/collection/interpretation using smartphones as IoT portals for smart diagnostics of PIDs using wastewater epidemiological data obtained with REASSURED-based diagnostic devices/tests, other related data, and with the help of digital technologies (See Fig. 7 ). The recommended model includes the interactions and integrations of cloud and fog, edge, and SEA (Service Endpoint Agent) computing technologies at regional, local, and device levels, respectively [95]. In the proposed roadmap design, a tailored Industry 4.0 unified 5C (Connection, Conversion, Cyber, Cognition, Configure)-level architecture [96] was adopted. The self-developed architecture includes five major inter-related (either within- or between-mediating) layers with the presupposition of security using Blockchain technology for all layers. Data aggregation and processing layers were designed to be served as mediating layers for generating smart data and additional adeptness for Enterprise Resource Planning (ERP) and smart cloud services management based on an AI. The proposed IoT-Fog-Cloud model with details of each layer toward smart diagnostics of PIDs based on WBE is depicted in Fig. 7. As seen, it follows the philosophy of knowledge pyramid (Data-Analysis-Knowledge-Wisdom) mixed with IoT-based architecture (Things-Gateway-Cloud), data from all REASSURED-based tests/devices via smartphone crowdsourcing with the help of cloud and fog servers are collected. Herein, the WBE data that include the main biological information related to the absence/presence of the virus, some geographical data, in combination with environmental factors that may affect results are transferred to the data center to make the first raw data layer. The simultaneous modeling and analysis of biological data with environmental data remedy the issue of complexity in the analysis step. In the following, the data center can even integrate diagnostics, clinical, and symptom-relayed data in this layer. The resultant multi-dimensional data are analyzed with the help of new digital technologies such as ML, DL, BDA methods in the second layer. Through some analytics procedures, including data processing and data aggregation in layer three, they will be classified, purified, filtered, and transcoded and yield smart data for further smart solutions and applications such as early prediction, warning, diagnosis, and treatment of the infectious diseases and timely isolation/quarantine of infected regions; thereby, controlling and breaking the transmission chain of the infectious diseases, before they become epidemics/pandemics. Such smart data generated via different types of analysis and filtering in layers two and three will be used for not only smart services such as Internet of Medical Things (IoMT), e-Diagnostics, and telemedicine but also smart management of the pandemics via a unified ERP used by WHO or any national or international authorities. Such a knowledge management system will create pandemic wisdom and that is the philosophy behind our proposed model.
Fig. 7.
Proposed IoT-Fog-Cloud model for smart diagnostics of PIDs using WBE with approaches for data gathering, transmission, and analysis to incorporate new digital technologies within a straightforward platform.
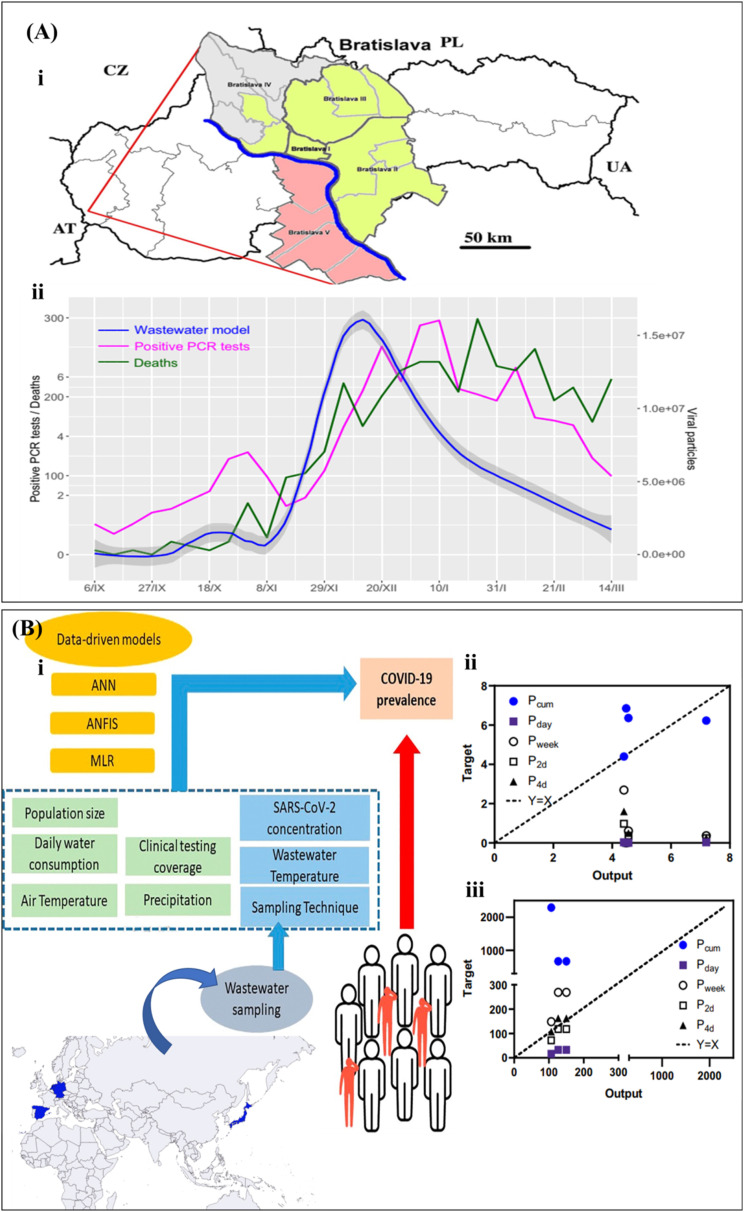
Although a complete smart sensing platform for WBE has not been reported yet, there are some recent reports on analyzing WBE data using digital technologies, which can be further applied for the generation of smart data toward smart diagnostics of PIDs. For example, a mathematical correlation between RT-qPCR tests of SARS-CoV-2 (viral particles) in wastewater samples and death cases or positive PCR-tests in Bratislava-Slovakia has been described by Krivoňáková et al. [15]. The established model was able to forecast the virus loads on the health system up to two weeks earlier (Fig. 8 A). The researchers pointed out that since every type of wastewater has its specific features (temperature, pH, (bio)chemical constituents, etc.), it is required to consider the effects of these factors in other monitored localities models. To push forward the WBE, Li et al. [50] used data-driven models for the meta-analysis of WBE datasets, in which three different models, multiple linear regression (MLR), ANN, and adaptive neuro-fuzzy inference system (ANFIS) were used for modeling a multi-national (Australia, Germany, Netherlands, Japan and Spain) WBE dataset. The inputs for models were the clinical results, the concentration of SARS-CoV-2 RNA detected in wastewater, temperature of wastewater and air, population, daily water intake, sampling and precipitation technique, where the prevalence of virus was chosen as the only output (Fig. 8B–i). In general, ANN and ANFIS models revealed better accuracy and robustness compared with MLR. Two environmental conditions, air, and wastewater temperature showed a detrimental effect on the prevalence approximation by these models, confirming their critical role in making uncertainties in conventional approaches. The ANN model foreshowed the prevalence of COVID-19 at the early phase and predicted the forthcoming new cases within 2–4 days at the later phase of the outbreak (Fig. 8B ii, iii). So, the data-driven methods could model several factors as input data with no need to control the sampling criteria, the property which reduces the difficulty and uncertainties in analysis and comparison of results for multi-national applications. The resultant models are more robust and provide more accurate and reliable data for WBE. This property eliminates the complexity of fixing conditions at constant values in the data gathering step. In another study, the use of ML and evolutionary computing methods in modeling of COVID-19 spreading dataset has been investigated [52]. The Long Short-Term Memory (LSTM) is the most universally employed algorithm in the COVID-19 spread once the regression is performed from time-series data, while Multilayer Perceptron (MLP) is used once the data are arranged as a regression dataset. The study revealed the highly prospective usage of AI-based algorithms for modeling and prediction of epidemics/pandemics in the future. In summary, the inclusion of all determinant factors in the sampling step along with diagnostic data as input factors for modeling could result in more reliable outputs in this arena.
Fig. 8.
(A) WBE for a local area via simple linear modeling of the qPCR test. (I) Geographical map of Slovakia and Bratislava with the districts. (II) the positive RT-qPCR tests and death cases vs. viral particles in wastewater in Bratislava. (B) WBE for a multi-national area via data-driven methods. (I) several sample-describing factors along with diagnostic test results are used as input data for modeling. Comparison of predictions from ANN model and prevalence determined by cumulative cases (Pcum), daily new cases (Pday), weekly new cases (Pweek), and upcoming new cases in the following 2 or 4 days (P2d, P4d) for the (II) early phase of an outbreak and (II) later phase of an outbreak, reprinted with permission from Refs. [15,50].
Unlike the COVID-19 diagnosis, the availability of datasets such as the COVID-19 WBE Collective (https://www.covid19wbec.org/) provides a way to empower collaborations on a worldwide scale for WBE of SARS CoV-2. With more than 500 collaborators worldwide, the website involves a map demonstrating their cooperating groups and also the locations of more than 50 societies analyzing their wastewater, more than 400 sampling sites, and more than 150 universities testing wastewater [97]. The availability of such datasets and collaborations makes it easy to develop a more generalized smart sensing system for global management and prediction of PIDs via WBE.
Finally, the integration of WBE data along with local clinical data can further increase the usefulness of WBE in the prediction and prevention of COVID-19 [59,98], which necessities ensure data security without any privacy violation [99,100], to reach more robust models for improved management. Eventually, the use of smartphone-connected REASSURED-based WBE diagnostic tests/devices can be considered as a superior strategy towards smart diagnostics, monitoring, and management of PIDs, owing to their capability to share the complementary data related to health, symptoms, and other useful and crucial information along with WBE-based PIDs diagnostic data. Blockchain combined sensing devices can be considered as safe platforms for real-time connectivity and data sharing of both personalized diagnostics and WBE. Blockchain technology enables a net of unknown members to agree on pooled data, with no implications on the centralized confirmation or a right-hand third party. The principles of decentralization make it appropriate for the management of data, consequently building a decentralized 'trustless' network lacking storage at an exact center. Herein, persons would own their medicinal records with no worries about misuse of their information. This approach may provide a new level of trust for worldwide cooperation to combat PIDs [101].
4. Conclusions
In summary, to achieve success in the use of WBE for managing COVID-19 and any other potential pandemics, the necessity to develop smart sensing platforms is crucial. While the complexity of measuring conditions makes it vital to follow the same protocols for regional analysis of wastewater in conventional PCR based approaches, the smart sensory platforms can provide rapid and online multi-dimensional data gathering and sharing via the internet to managing centers, enabling fast analysis of data via artificial intelligence routes for efficient management of pandemics. On the other hand, for realizing smart diagnostics via WBE to fight against COVID-19, the development of efficient REASSURED based sensory platforms along with online connection to the data center with an adaptive model that can powerfully incorporate all novel digital technologies into a single platform is required. The advent of new variants of SARS-CoV-2 complicates the sensing but could be overcome by utilizing new and specific DNA-based recognition elements. Such replacement of primers is also a necessity for PCR-based detection approaches. Besides, the predictive power of WBE supported by smart sensing could be improved using an innovative platform to continuously monitor and detect genetic material alerting on the presence of an unknown/emerging infectious disease agent as well as possible mutations directly in wastewater, which is a major challenge.
Besides the advantageous features of smart WBE diagnostics, there are some challenges mostly in the data-transferring area that should be resolved to overcome any outcoming epidemic/pandemics via the smart sensory strategies. Such issues include (i) the worldwide availability of 4/5 G Internet, which is a crucial necessity especially in times of pandemics, (ii) the availability of a sufficient number of high-speed fog and cloud servers to avoid network traffic issues, (iii) the creation of a decentralized "trustless' ecosystem such as Blockchain to be combined with sensing devices for safe real-time connectivity and data sharing, and finally (ix) collaborative smart system architecture for data collection, transmission, and interpretation by assembling digital technologies within a sole platform. Finally, WBE-related smart sensors can play an important role in the development of smart cities, homes, farming, etc.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
T.N. and H.G. acknowledge support from the Chemistry and Chemical Engineering Research Center of Iran (Tehran, Iran). E.M.-N. acknowledges financial support from CONACYT (Mexico, Granto No. 312271).
Abbreviations
- 5C
Connection, Conversion, Cyber, Cognition, Configure
- AI
artificial intelligence
- ANFIS
adaptive neuro-fuzzy inference system
- ANN
artificial neural networks
- AR
augmented reality
- BA
blockchain analysis
- BDA
big data analytics
- CFU/mL
colony-forming units/mL
- COVID-19
coronavirus disease 2019
- CRISPR
clustered regularly interspaced short palindromic repeats
- DL
deep learning
- dPCR
digital PCR
- ERP
Enterprise Resource Planning
- GE/mL
genome equivalent/mL
- HAdV
human adenovirus
- HIV
human immunodeficiency virus
- μCE
integrated micro-capillary electrophoresis
- IoAT
Internet of Analytical Things
- IoMT
Internet of Medical Things
- IoT
Internet of things
- LAMP
loop-mediated isothermal amplification
- LFA
Lateral flow assay
- LFIA
Lateral flow immunoassay
- ML
machine learning
- MLP
Multilayer Perceptron
- MLR
multiple linear regression
- NASBA
nucleic acid sequence-based amplification
- NFC
near-field communication
- PIDs
pandemic infectious diseases
- POC
point-of-care
- POU
point-of-use
- PMMA
poly methyl methacrylate
- RT-PCR
reverse transcription-polymerase chain reaction
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SEA
Service Endpoint Agent
- SERS-based LFIA
surface-enhanced Raman scattering-based lateral flow immunoassay
- SWVs
square-wave voltammograms
References
- 1.Callaway E. Time to use the p-word? Coronavirus enters dangerous new phase. Nature. 2020 doi: 10.1038/d41586-020-00551-1. [DOI] [PubMed] [Google Scholar]
- 2.Syal K. J. Med. Virol. 2021;93:1837. doi: 10.1002/jmv.26673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiwari S., Juneja S., Ghosal A., Bandara N., Khan R., Wallen S., Ramakrishna S., Kaushik A. Curr. Opin. Biomed. Eng. 2021 doi: 10.1016/j.cobme.2021.100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mujawar M.A., Gohel H., Bhardwaj S.K., Srinivasan S., Hickman N., Kaushik A. Mater. Today Chem. 2020;17 doi: 10.1016/j.mtchem.2020.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury S.R., Saha H. IETE Tech. Rev. 2006;23:297. [Google Scholar]
- 6.Hancke G.P., Hancke G.P., Jr. Sensors. 2013;13:393. [Google Scholar]
- 7.Jain S., Nehra M., Kumar R., Dilbaghi N., Hu T.Y., Kumar S., Kaushik A., Li C.-z. Biosens. Bioelectron. 2021 doi: 10.1016/j.bios.2021.113074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushik A.K., Dhau J.S., Gohel H., Mishra Y.K., Kateb B., Kim N.-Y., Goswami D.Y. ACS Appl. Bio Mater. 2020;3:7306. doi: 10.1021/acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 9.Ghazal T.M., Hasan M.K., Alshurideh M.T., Alzoubi H.M., Ahmad M., Akbar S.S., Al Kurdi B., Akour I.A. Future Internet. 2021;13:218. [Google Scholar]
- 10.Sharma P.K., Kim E.-S., Mishra S., Ganbold E., Seong R.-S., Kaushik A.K., Kim N.-Y. ACS Sens. 2021;6:3468. doi: 10.1021/acssensors.1c01437. [DOI] [PubMed] [Google Scholar]
- 11.Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. Biosens. Bioelectron. 2021;177 doi: 10.1016/j.bios.2021.112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao K., Zhang H., Pan Y., Yang Z. Water Res. 2020 doi: 10.1016/j.watres.2020.116787. [DOI] [PubMed] [Google Scholar]
- 13.Vitale D., Suárez-Varela M.M., Picó Y. Curr. Opin. Environ. Sci. Health. 2021 [Google Scholar]
- 14.Ali W., Zhang H., Wang Z., Chang C., Javed A., Ali K., Du W., Niazi N.K., Mao K., Yang Z. J. Hazard Mater. 2021 doi: 10.1016/j.jhazmat.2021.125439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krivoňáková N., Šoltýsová A., Tamáš M., Takáč Z., Krahulec J., Ficek A., Gál M., Gall M., Fehér M., Krivjanská A. Sci. Rep. 2021;11:1. doi: 10.1038/s41598-021-98653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. medRxiv. 2020 http://conacyt.repositorioinstitucional.mx/jspui/handle/1000/2983 [Google Scholar]
- 18.La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Environ. Sci. Technol. Lett. 2020;7:511. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 20.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Int. J. Environ. Res. Publ. Health. 2020;17:5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haramoto E., Malla B., Thakali O., Kitajima M. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First data-set on SARS-CoV-2 detection for Istanbul wastewaters in Turkey. medRxiv. 2020 doi: 10.1101/2020.05.03.20089417. [DOI] [Google Scholar]
- 24.Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharif S., Ikram A., Khurshid A., Salman M., Mehmood N., Arshad Y., Ahmad J., Angez M., Alam M.M., Rehman L. Detection of SARs-CoV-2 in wastewater, using the existing environmental surveillance network: an epidemiological gateway to an early warning for COVID-19 in communities. medRxiv. 2020 doi: 10.1101/2020.06.03.20121426. [DOI] [Google Scholar]
- 26.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C. J. Med. Virol. 2020;92:833. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 27.Wu Q., Liu W.-T. Water Res. 2009;43:1101. doi: 10.1016/j.watres.2008.11.039. [DOI] [PubMed] [Google Scholar]
- 28.Wu J., Wang Z., Lin Y., Zhang L., Chen J., Li P., Liu W., Wang Y., Yao C., Yang K. Sci. Total Environ. 2021;791 doi: 10.1016/j.scitotenv.2021.148271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philo S.E., Keim E.K., Swanstrom R., Ong A.Q., Burnor E.A., Kossik A.L., Harrison J.C., Demeke B.A., Zhou N.A., Beck N.K. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kweinor Tetteh E., Opoku Amankwa M., Armah E.K., Rathilal S. Water. 2020;12:2680. [Google Scholar]
- 31.Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Jr., Campiglia P., Belgiorno V., Korshin G., Naddeo V. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barceló D. Case Stud. Chem. Environ. Eng. 2020;2 doi: 10.1016/j.cscee.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alygizakis N., Markou A.N., Rousis N.I., Galani A., Avgeris M., Adamopoulos P.G., Scorilas A., Lianidou E.S., Paraskevis D., Tsiodras S. Trac. Trends Anal. Chem. 2020 doi: 10.1016/j.trac.2020.116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackuľak T., Gál M., Špalková V., Fehér M., Briestenská K., Mikušová M., Tomčíková K., Tamáš M., Butor Škulcová A. Int. J. Environ. Res. Publ. Health. 2021;18:5629. doi: 10.3390/ijerph18115629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar M., Joshi M., Patel A.K., Joshi C.G. Environ. Res. 2021;196 doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhama K., Patel S.K., Yatoo M.I., Tiwari R., Sharun K., Dhama J., Natesan S., Malik Y.S., Singh K.P., Harapan H. J. Environ. Manag. 2021;280:111825. doi: 10.1016/j.jenvman.2020.111825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims N., Kasprzyk-Hordern B. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. 2020. Current Opinion in Environmental Science & Health. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamouda M., Mustafa F., Maraqa M., Rizvi T., Hassan A.A. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adelodun B., Ajibade F.O., Ibrahim R.G., Bakare H.O., Choi K.-S. Sci. Total Environ. 2020;742 doi: 10.1016/j.scitotenv.2020.140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Zahedi, P. Monis, D. Deere, U. Ryan, Parasitol. Res. 1. [DOI] [PMC free article] [PubMed]
- 43.Hui Q., Pan Y., Yang Z. Case Stud. Chem. Environ. Eng. 2020;2 doi: 10.1016/j.cscee.2020.100064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu D., Zhu D.Z., Gan H., Yao Z., Fu Q., Zhang X.J. Sci. Total Environ. 2021 [Google Scholar]
- 45.Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Environ. Sci. Technol. 2021;55:3514. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- 46.Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuschi C., Pu H., Negri M., Colwell R., Chen J. Snowballing transmission of COVID-19 (SARS-CoV-2) through wastewater: Any sustainable preventive measures to curtail the scourge in low-income countries? ACS Es&t Water. 2021;742:140680. doi: 10.1016/j.scitotenv.2020.140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. J. Environ. Chem. Eng. 2020;8:104306. doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu D., Huang Z., Luo J., Zhang X., Sha S. Sci. Total Environ. 2020;747 doi: 10.1016/j.scitotenv.2020.141245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., Kulandaivelu J., Zhang S., Shi J., Sivakumar M., Mueller J., Luby S., Ahmed W., Coin L., Jiang G. 2021. Science of the Total Environment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salem H.S.A., Shams M.Y., Hassanien A.E., Nosair A.M. Springer; 2021. COVID-19 and Water Resources Nexus: Potential Routes for Virus Spread and Management Using Artificial Intelligence Techniques; p. 19. [Google Scholar]
- 52.Musulin J., Baressi Šegota S., Štifanić D., Lorencin I., Anđelić N., Šušteršič T., Blagojević A., Filipović N., Ćabov T., Markova-Car E. Int. J. Environ. Res. Publ. Health. 2021;18:4287. doi: 10.3390/ijerph18084287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doorn N. Sci. Total Environ. 2021;755 doi: 10.1016/j.scitotenv.2020.142561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rallapalli S., Aggarwal S., Singh A.P. Sci. Total Environ. 2021;778 doi: 10.1016/j.scitotenv.2021.146294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nourinejad M., Berman O., Larson R.C. PLoS One. 2021;16 doi: 10.1371/journal.pone.0248893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rahman A., Belia E., Kirim G., Hasan M., Borzooei S., Santoro D., Johnson B. 2021. Water Environment Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karn A.L., Pandya S., Mehbodniya A., Arslan F., Sharma D.K., Phasinam K., Aftab M.N., Rajan R., Bommisetti R.K., Sengan S. Soft Comput. 2021:1. [Google Scholar]
- 58.Hosseinifard M., Naghdi T., Morales-Narváez E., Golmohammadi H. Front. Bioeng. Biotechnol. 2021;9:510. doi: 10.3389/fbioe.2021.637203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morales-Narváez E., Dincer C. Biosens. Bioelectron. 2020;163 doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorenzo M., Picó Y. Curr. Opin. Environ. Sci. Health. 2019;9:77. [Google Scholar]
- 61.Mostafavi E., Dubey A.K., Teodori L., Ramakrishna S., Kaushik A. Med. Comm. 2022;3:e119. doi: 10.1002/mco2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhuang J., Yin J., Lv S., Wang B., Mu Y. Biosens. Bioelectron. 2020 doi: 10.1016/j.bios.2020.112291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Du K., Cai H., Park M., Wall T., Stott M., Alfson K., Griffiths A., Carrion R., Patterson J., Hawkins A. Biosens. Bioelectron. 2017;91:489. doi: 10.1016/j.bios.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoo H.J., Baek C., Lee M.-H., Min J. Analyst. 2020;145:2405. doi: 10.1039/c9an02435b. [DOI] [PubMed] [Google Scholar]
- 65.Phillips E.A., Moehling T.J., Ejendal K.F., Hoilett O.S., Byers K.M., Basing L.A., Jankowski L.A., Bennett J.B., Lin L.-K., Stanciu L.A. Lab Chip. 2019;19:3375. doi: 10.1039/c9lc00506d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hansen G., Marino J., Wang Z.-X., Beavis K.G., Rodrigo J., Labog K., Westblade L.F., Jin R., Love N., Ding K. J. Clin. Microbiol. 2021;59 doi: 10.1128/JCM.02811-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tu Y.-P., Iqbal J., O'Leary T. Elife. 2021;10 doi: 10.7554/eLife.65726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liotti F.M., Menchinelli G., Marchetti S., Morandotti G.A., Sanguinetti M., Posteraro B., Cattani P. Clin. Microbiol. Infect. 2020;26:1699. doi: 10.1016/j.cmi.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J., Song J.U. J. Med. Virol. 2021;93:4523. doi: 10.1002/jmv.26994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xing W., Liu Y., Wang H., Li S., Lin Y., Chen L., Zhao Y., Chao S., Huang X., Ge S. A high-throughput, multi-index isothermal amplification platform for rapid detection of 19 types of common respiratory viruses including SARS-CoV-2. Engineering. 2020;6(10):1130–1140. doi: 10.1016/j.eng.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin K., Ding X., Xu Z., Li Z., Wang X., Zhao H., Otis C., Li B., Liu C. Sensor. Actuator. B Chem. 2021 doi: 10.1016/j.snb.2021.130242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahmoudi T., Pourhassan-Moghaddam M., Shirdel B., Baradaran B., Morales-Narváez E., Golmohammadi H. Biosensors. 2021;11:392. doi: 10.3390/bios11100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahmoudi T., Tazehkand A.P., Pourhassan-Moghaddam M., Alizadeh-Ghodsi M., Ding L., Baradaran B., Bazaz S.R., Jin D., Warkiani M.E. Microchem. J. 2020;154 [Google Scholar]
- 74.Donia A., Shahid M.F., Ahmad A., Javed A., Nawaz M., Yaqub T., Bokhari H. Integration of RT-LAMP and Microfluidic Technology for Detection of SARS-CoV-2 in Wastewater as an Advanced Point-of-care Platform. bioRxiv. 2021 doi: 10.1101/2021.08.18.456880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen V.D., Nguyen H.Q., Bui K.H., Ko Y.S., Park B.J., Seo T.S. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng W., Wang K., Xu H., Zheng C., Cao B., Qin Q., Jin Q., Cui D. Anal. Bioanal. Chem. 2021:1. doi: 10.1007/s00216-021-03213-x. [DOI] [PubMed] [Google Scholar]
- 77.Reboud J., Xu G., Garrett A., Adriko M., Yang Z., Tukahebwa E.M., Rowell C., Cooper J.M. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:4834. doi: 10.1073/pnas.1812296116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alafeef M., Dighe K., Moitra P., Pan D. ACS Nano. 2020;14 doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahmoudi T., de la Guardia M., Shirdel B., Mokhtarzadeh A., Baradarn B. Trac. Trends Anal. Chem. 2019;116:13–30. doi: 10.1016/j.trac.2019.04.016. [DOI] [Google Scholar]
- 80.Mahmoudi T., de la Guardia M., Baradaran B. Trac. Trends Anal. Chem. 2020 [Google Scholar]
- 81.Mahmoudi T., Pourhassan-Moghaddam M., Shirdel B., Baradaran B., Morales-Narváez E., Golmohammadi H. (Nano) tag–antibody conjugates in rapid tests. J. Mater. Chem. B. 2021;9(27):5414–5438. doi: 10.1039/D1TB00571E. [DOI] [PubMed] [Google Scholar]
- 82.Wang C., Wang C., Wang X., Wang K., Zhu Y., Rong Z., Wang W., Xiao R., Wang S. ACS Appl. Mater. Interfaces. 2019;11 doi: 10.1021/acsami.9b03920. [DOI] [PubMed] [Google Scholar]
- 83.Zou M., Su F., Zhang R., Jiang X., Xiao H., Yan X., Yang C., Fan X., Wu G. Sensor. Actuator. B Chem. 2021;342 doi: 10.1016/j.snb.2021.129899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu D., Shen H., Zhang Y., Shen D., Zhu M., Song Y., Zhu Z., Yang C. Lab Chip. 2021;21 doi: 10.1039/d0lc01222j. 2019. [DOI] [PubMed] [Google Scholar]
- 85.Wang D., He S., Wang X., Yan Y., Liu J., Wu S., Liu S., Lei Y., Chen M., Li L. Nat. Biomed. Eng. 2020;4:1150. doi: 10.1038/s41551-020-00655-z. [DOI] [PubMed] [Google Scholar]
- 86.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang C., Zheng T., Wang H., Chen W., Huang X., Liang J., Qiu L., Han D., Tan W. Anal. Chem. 2021;93:3325. doi: 10.1021/acs.analchem.0c05059. [DOI] [PubMed] [Google Scholar]
- 88.Javaid M., Khan I.H. J. Oral Biol. Craniofac. Res. 2021;11:209. doi: 10.1016/j.jobcr.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vashist S.K., Luong J.H. Springer; 2019. Smartphone-based Point-Of-Care Technologies for Mobile Healthcare; p. 27. [Google Scholar]
- 90.Xu X., Akay A., Wei H., Wang S., Pingguan-Murphy B., Erlandsson B.-E., Li X., Lee W., Hu J., Wang L. Proc. IEEE. 2015;103:236. [Google Scholar]
- 91.Islam S.R., Kwak D., Kabir M.H., Hossain M., Kwak K.-S. IEEE Access. 2015;3:678. [Google Scholar]
- 92.Aloi G., Caliciuri G., Fortino G., Gravina R., Pace P., Russo W., Savaglio C. J. Netw. Comput. Appl. 2017;81:74. [Google Scholar]
- 93.Mayer M., Baeumner A.J. Chem. Rev. 2019;119:7996. doi: 10.1021/acs.chemrev.8b00719. [DOI] [PubMed] [Google Scholar]
- 94.Land K.J., Boeras D.I., Chen X.-S., Ramsay A.R., Peeling R.W. Nat. Microbiol. 2019;4:46. doi: 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang Y. Nat. Electron. 2019;2:4. [Google Scholar]
- 96.Lee J., Bagheri B., Kao H.-A. Manuf. Lett. 2015;3:18. [Google Scholar]
- 97.A. Bivins, D. North, A. Ahmad, W. Ahmed, E. Alm, F. Been, P. Bhattacharya, L. Bijlsma, A.B. Boehm, J. Brown, (2020).
- 98.Chamola V., Hassija V., Gupta V., Guizani M. IEEE Access. 2020;8 doi: 10.1109/JIOT.2020.3044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ienca M., Vayena E. Nat. Med. 2020;26:463. doi: 10.1038/s41591-020-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morley J., Cowls J., Taddeo M., Floridi L. Nature Publishing Group; 2020. Ethical Guidelines for COVID-19 Tracing Apps. [DOI] [PubMed] [Google Scholar]
- 101.Altay A., Learney R., Güder F., Dincer C. Sensors in blockchain. Trends Biotechnol. 2021;40(2):141–144. doi: 10.1016/j.tibtech.2021.04.011. [DOI] [PubMed] [Google Scholar]