Abstract
Worldwide, breast cancer is the leading cause of cancer-related deaths in women. Breast cancer is a heterogeneous disease characterized by different clinical outcomes in terms of pathological features, response to therapies, and long-term patient survival. Thus, the heterogeneity found in this cancer led to the concept that breast cancer is not a single disease, being very heterogeneous both at the molecular and clinical level, and rather represents a group of distinct neoplastic diseases of the breast and its cells. Indubitably, in the past decades we witnessed a significant development of innovative therapeutic approaches, including targeted and immunotherapies, leading to impressive results in terms of increased survival for breast cancer patients. However, these multimodal treatments fail to prevent recurrence and metastasis. Therefore, it is urgent to improve our understanding of breast tumor and metastasis biology. Over the past few years, high-throughput “omics” technologies through the identification of novel biomarkers and molecular profiling have shown their great potential in generating new insights in the study of breast cancer, also improving diagnosis, prognosis and prediction of response to treatment. In this review, we discuss how the implementation of “omics” strategies and their integration may lead to a better comprehension of the mechanisms underlying breast cancer. In particular, with the aim to investigate the correlation between different “omics” datasets and to define the new important key pathway and upstream regulators in breast cancer, we applied a new integrative meta-analysis method to combine the results obtained from genomics, proteomics and metabolomics approaches in different revised studies.
Subject terms: Cancer genomics, Breast cancer
Introduction
Breast cancer (BC) is the most frequently diagnosed cancer and a major health issue in women, being the leading cause of cancer-related deaths among women, worldwide [1–3]. Even if 5–10% of BC cases are due to hereditary and genetic factors, non-hereditary factors have been described as the primarily responsible for differences in incidence between countries and ethnic groups [1]. BC is recognized as a heterogeneous disease, both at the molecular and clinical level. In this regard, at the beginning of the millennium, Perou and colleagues did a groundbreaking discovery in BC using DNA microarray. Their extensive gene expression profiling found variation in expression of 1753 genes in 84 experimental samples. They concluded that BC is not a uniform disease, instead it is composed of five distinct subtypes: luminal A, luminal B, basal-like, normal breast-like, and HER-2 enriched [2–5]. In fact, these subtypes of BC are also well known as “Perou’s molecular subtypes” [6]. Moreover, being a multifaceted disease, BC is characterized by intratumoral and intertumoral heterogeneity. Specifically, it is defined as intratumoral when cells within a tumor in a single patient are involved, while intertumoral when cells of the same subgroup of tumors in different patients are involved [2]. Thus, the heterogeneity found among BCs led to the concept that BC is not a single disease and rather represents a group of distinct neoplastic diseases of the breast and its cells [7]. Nowadays, therapeutic options for BC treatment include surgery, radiotherapy, chemotherapy, and targeted therapies [4, 8]. Despite recent and important advances in understanding BC biology, diagnosis and treatment, several significant clinical issues still remain unclear. In particular, these unmet clinical needs are related to prevention, diagnosis, tumor progression, treatment, therapeutic resistance and metastasis formation [3, 9]. In this context, modern systems biology based on “omics” approaches can potentially make a major contribution to overcome these problems. In fact, in the era of precision medicine, “omics” strategies and their integration in the study of BC may be considered as a new biomarker discovery tool, leading to novel biomarker molecules and molecular signature with a potential in clinical practice [9]. It is worth noting that a molecular profiling is involved in BC, as in any phenotypic alterations, and is recognizable on different levels: genome, transcriptome, proteome, and metabolome. At the beginning of the twenty-first century, revolutionary progress of high-yield and innovative technologies in nucleic acid sequencing and mass spectrometry (MS) have driven the advent of Genomics, Transcriptomics (functional genomics), Proteomics and finally of Metabolomics, leading to the “multi-omics” era [9–11]. Anyway, we should consider that, even if each “omics” approach is essential to systems biology, giving its contribution in a specific way to shape the biological phenotype under study, some are more mature than others [11]. These “omics” approaches are quite different from the conventional methods for the study of BC complex biology, mainly for the possibility of obtaining a huge number of molecular measurements within cells, a tissue or in biological fluids [12]. Once applied to a pathological condition of interest, they allow to obtain a snapshot of the underlying biology, with a resolution never achieved before. Interestingly, the application of each aforementioned “omics” technology and their integration by network science for studying BC gives the possibility of holistic investigation and contextually of a comprehensive pathophysiologic understanding of such a complex disease, with the promise of providing novel insights into precise diagnosis, potential therapeutic options and tailored treatment [11, 13, 14]. Thus, the main advantage of “omics” strategies is to bring out the omics-based molecular profiling with the clinical outcome under study. Actually, in the multi-omics context it has become evident that the use of integrative tools and of computational approaches is necessary [14] to deeply understand biological mechanisms from a system-wide perspective [15].
The present manuscript reviewed recent works in literature of the findings by omics-based studies in BC until 2020. In particular, we summarized and updated previously published literature on BC molecular candidates obtained by the application of three kinds of omics approaches, including genomics, proteomics, and metabolomics. In addition, we applied a network science paradigm performing a systematic integration of the molecular alterations at multiple levels including genome, proteome, and metabolome. In fact, elaborating heterogeneous-omics data sets has the potential to gain novel, mechanistically significant insights into the BC disease [14, 16].
Finally, we aimed to prove that the real challenge of multi-omics investigations lies in the integration of their frameworks and the cautious interpretation of the myriad of data in order to gain further insights on BC and to move toward P4 medicine (preventive, predictive, personalized, and participatory) [11, 14].
Genomics in BC
The history of BC genomics can be broadly divided into two categories, before next-generation sequencing (NGS), or pre-NGS, and after NGS, or post-NGS. Pre-NGS era is mainly characterized by studying of individual genes associated with BC. During the pre-NGS era, the hallmark genes such as BRCA1 and BRCA 2 were discovered. After the advent of NGS, the study of BC genomics boomed, and BC study was not limited to only few genes. A number of new genes and intergenic interactions were discovered during post-NGS era of BC. The genes associated with BC can be found on Supplementary Table S1. The genes are categorized in different groups, based on the type of alteration (i.e., mutation/polymorphism) or susceptibility of developing cancer (high/moderate/low penetrance). Gene expression or transcriptomic data are not included in this review.
Pre-NGS era of BC
Genomics is one of the important factors determining the outcome of BC. In the ’90 s, researchers observed that family history is the strongest single predictor of a woman’s chance of getting BC. After a long search, two genes, BRCA1, was discovered in 1994, and the second, BRCA2, in 1995, was found to be associated with the BC in women [17, 18]. Importantly, a meta-analysis shows that the mean cumulative BC risks at the age of 70 were 57% for BRCA1 and 49% for BRCA2 mutation carriers [19]. The search for other genes continues and several genes are known to be involved in somatic and inherited susceptibility to BC. Apart from BRCA1 and BRCA2 genes, which are used as the gold standard for genetic testing for BC, there are several other genes involved in varying extent in the susceptibility of BC. The pieces of evidence at that time made two things clear: first, BC is not a single disease, but instead, it is composed of a spectrum of tumor subtypes with distinct molecular, cellular and somatic changes [20]. Secondly, various rare genetic syndromes are linked with increased BC risk. For example, mutations that inactivate the Tp53 gene, which primarily causes Li–Fraumeni syndrome, are also associated with increased susceptibility to BC [21, 22]. The risk of developing BC before the age of 45 is 18-fold higher with the females affected with Tp53 mutation as compared to the general population [21]. Germline mutations in the Tp53 gene have been estimated to account for <1% of BC cases [23–25]. However, somatic mutations in the Tp53 gene are reported in 19–57% of human BCs [26–28]. Inactivation of just one allele for Tp53 gene may be sufficient for BC development [28]. Likewise, mutations in the ATM gene are responsible for Ataxia-telangiectasia (A-T) disease. Though A-T patients do not survive to an age at which BC generally occurs [29] A-T carriers (heterozygous for ATM mutations) appear to have an increased BC risk [30–33], with an estimated increased risk of 11% by the age of 50 and 30% by the age of 70 [34]. A study of 138 Austrian hereditary breast and ovarian cancer patients without BRCA1 and BRCA2 mutations showed functionally significant ATM germline mutations in at least 8.7% of the patients [35]. The penetrance for one of the mutations (L1420F) was estimated to be 85% at age 60. Renwick et al. sequenced ATM in 443 BRCA-negative cases from families with at least three BC-affected members and in 521 controls. Nine truncating and exon-skipping mutations were identified in cases, while only two were found in controls. All mutations found in cases were predicted to cause AT, and seven had been observed previously in AT cases [36]. Bernstein et al. performed an ATM mutation screen in 708 unilateral BC survivors who developed contralateral BC following radiotherapy and 1397 who did not. They found that women with AT-associated ATM mutations treated previously with radiation had a significantly greater risk of contralateral BC than unexposed women either with no mutation or unexposed women with the same mutation [37]. Similarly, a mutation in the PTEN gene leads to the Cowden syndrome in 80% of Cowden syndrome families [38, 39]. On the other hand, the same truncating PTEN mutations in Cowden syndrome families are associated with 25–50% lifetime BC risk in women [39–41]. Loss of heterozygosity (LOH) at the PTEN locus is found in 11–41% of sporadic BCs [42–45]. In one study (in 177 BC patients with a positive family history for BC and without BRCA1 and BRCA2 mutations), an association was found between a polymorphism in intron 4 of the PTEN gene and a lower age of diagnosis of BC (42.7 versus 48.1 years) [46]. Moreover, Peutz–Jeghers syndrome is an autosomal dominant disorder, caused by truncating germline mutations in the LKB1 gene [47, 48]. Patients with Peutz–Jeghers syndrome have an increased BC risk [47–49]. Another genetic disorder, Neurofibromatosis type 1 (NF1) is a common autosomal dominant disorder associated with an increased risk for neoplasms [50]. Women with NF1 develop BC at younger ages than the general population, with an average of <50 years old. Moreover, the risk of developing BC wit NF is 6.5-fold higher in women aged 30–39 years and 4.4 times higher among women aged 40–49 [51]. In patients with no BRCA1 or BRCA2 mutations, LOH in the NF1 region was found to be responsible for the onset of BC [52].
Post-NGS era of BC
NGS of DNA introduced rapid and cost-efficient way to identify genes involved in BC [53]. NGS allows detecting multiple genetic alterations at the same time, using the same assay, leading to the concept of “multigene sequencing”. Based on the multigene sequencing, several other genes were found to be involved in the susceptibility of the BC. BRCA1-associated ring domain (BARD1), a direct interacting partner of BRCA1, is likely to be a low–moderate penetrance BC risk gene [54]. Loss-of-function (LoF) mutation in BRAD1 gene was present in 0.51% of BC patients. Also, BARD1-mutated BC patients showed a significantly younger mean age at first diagnosis (42.3 years, range 24–60 years) compared with the overall study sample (48.6 years, range 17–92 years) [55]. Similarly, germline LoF mutations in BRCA1 interacting protein C-terminal helicase 1 (BRIP1), which is a low penetrance gene, are associated to contribute to BC risk, particularly among patients who develop the disease at an early age [56]. Normal BRIP1 activity is required for DNA interstrand cross-link (ICL) repair and is thus central to the maintenance of genome stability. Next-generation sequencing of germline DNA in 2,160 early-onset BC and 1,199 patients with ovarian cancer revealed nearly 2% of patients carry a very rare missense variant in BRIP1, which is 3-fold higher than the frequency of all rare BRIP1 missense alleles reported in more than 60,000 individuals of the general population [56]. The study by Seal et al. sequenced the exons and exon-intron boundaries of BRIP1 in 1212 BC cases with a family history of the disease and no BRCA mutation and 2081 controls and found mutations in nine cases (0.74%) but only in two controls (0.10%) [57]. Similarly, mutated CHEK2 or CHEK2 pathogenic variant (PV) is a high penetrance BC gene [58]. Biallelic CHEK2 PV carriers have a higher risk for BC, are more likely to be diagnosed younger, and have multiple primary BCs compared to monoallelic carriers [58]. A population-based study found that deletion in CHEK2 (CHEK2*1100delC) is present at a frequency of 1.1% in controls, 5.1% in cases with a family history, and 13.5% in cases with a family history of male BC in a population with a positive history of BC but no BRCA mutation [59]. SMAD4 is another gene that gets inactivated in BC patients. SMAD4 is a common signal transducer in the bone morphogenetic protein (BMP)/transforming growth factor-β (TGF-β) signaling pathway, and functions as a transcription corepressor for human estrogen receptor α (ERα) [60]. SMAD4 is located on 18q21, a region frequently lost in BCs [61]. Inactivation or suppressed expression of TGF-β/SMAD4 signaling has been found to play an important role in BC development [61–63].
Besides the above-mentioned genes, NGS revealed that germline LoF mutations in PALB2 confers a predisposition to BC. PALB2 interacts with BRCA1 and BRCA2, and biallelic mutations in PALB2 (also known as FANCN), disrupts the Fanconi anemia–DNA repair pathway and increases BC predisposition [64, 65]. Furthermore, L35Pa, a pathogenic missense mutation in PALB2, abrogates the PALB2-BRCA1 interaction which may lead to failure in BC suppression [66]. In recent years, RAD51C and RAD51D are the other two genes that have been used in the screening of BC susceptibility [67, 68]. The estimated cumulative risks of developing BC to 80 years is 21% for RAD51C and 20% for RAD51D pathogenic variant carriers. BC risks for RAD51C and RAD51D pathogenic variants could be 44–46%, for carriers with two first-degree relatives diagnosed with BC [69]. Other notable genes that get mutated in BC are NBN and CDK12.
NBN gene mutation shows moderate to low penetrance [70]. Among NBN variants, a protein-truncating variant, c.657del5, is sufficiently common in some Eastern European populations to allow its evaluation in case-control studies. A meta-analysis of 10 studies reported strong evidence of an association with BC risk for this variant [34, 70].
Moreover, CDK12 (cyclin-dependent kinase 12), a low penetrance gene, is a regulatory kinase with evolutionarily conserved roles in modulating transcription elongation. In BCs, CDK12 is also frequently co-amplified with the HER2 oncogene [71]. CDK12 expression was found to be high in 21% of primary unselected BCs [72, 73]. Other low penetrance genes that are mutated in BC include MutYH, MSH2, CDKN2A and APC. The MutYH gene is involved in base excision repair. Carriers of variants in MutYH, although not very common, may have an increased risk of BC [74]. In a study in Italy by Rizzolo et al., biallelic MutYH pathogenic variants (p.Tyr179Cys/p.Arg241Trp) in one MBC patient with a phenotypic manifestation of adenomatous polyposis and Monoallelic pathogenic variants in 14 (2.5%) MBC patients were identified. Overall, the study suggests that MutYH pathogenic variants may have a role in MBC and, in particular, the p.Tyr179Cys variant may be a low/moderate penetrance risk allele for MBC [75]. On the other hand, Thibodeau et al. identified two patients with BC, each carrying a pathogenic germline MutYH variant with a somatic MutYH copy loss leading to the germline variant being homozygous in the tumor [76]. Regarding MSH2, a study showed that 1.1% woman with BC carries MSH2 mutation [77]. Moreover, another low penetrance gene CDKN2A mutation was identified (A148T variant) in 157 of 3,069 women with BC (5.1%) in a study in Poland. Their study shows that CDKN2A A148T variant seems to contribute to early-onset BC [78]. Furthermore, the adenomatous polyposis coli (APC) gene is a regulatory gene of the Wnt/β-catenin signaling pathway, which are independently involved in maintaining low levels of β-catenin in the cell. In an Indian study, a single nucleotide polymorphism (SNP), rs2229992 was identified in APC gene, with an increased risk of breast carcinogenesis in a BC and control population from eastern India [79].
Proteomics in BC
In recent years, omics approaches have emerged as a promising and extremely useful tool to reveal innovative molecular pathways as well as to identify and quantify the levels of molecules differentially expressed. In this scenario, mass spectrometry (MS) techniques have occupied an increasingly central position in the investigation of potential biomarkers, applied above all to complex and multifactorial pathologies, such as the study of cancer. To date, several studies based on the quantification of proteins are carried out with approaches based on the use of antibodies which are strongly linked to the availability, quantity, affinity and specificity [80]. Furthermore, as their use is inevitably linked to a starting hypothesis, this could hinder the study of the “neglected proteome” for the study of new potential biomarkers, or new biological pathways and functions related to BC [80, 81]. Proteomics can be defined as a high-throughput and large-scale study of proteins, investigating their classification, expression levels, properties and function [82]. Proteomic approaches based on MS techniques can be classified into two macro-groups: targeted and non-targeted proteomics. The main objective of the non-targeted applications is to cover almost complete proteomic knowledge, suitable for the application of the biomarkers discovery. The targeted approach, on the other hand, is more suitable for the validation of the results obtained from the first “discovery” approach and therefore for clinical applications. This fit-for-purpose approach has the aim of maximizing the coverage of potential objectives that can be assessed in the early stages for the discovery of biomarkers or therapeutic targets [80].
Proteomics features and studies in BC
In this literature review, to deepen proteomics features and studies in BC, we searched the PubMed site including quite recent scientific works approximately from 2010 to 2020, typing the keywords “Breast cancer proteomics Biomakers” and including in this study only the proteomics works followed by validation of the results obtained using different approaches. The research for BC is strongly aimed at the study of diagnosis, prognosis and disease course biomarkers in easily accessible biofluids, for this reason we listed in Supplementary Table S2 the candidate BC biomarkers found in serum [83–97], plasma [98–107], urine [108, 109], and Nipple Discharge biofluids [110].
In particular, human urine is considered one of the most interesting biofluids as it represents an excellent resource for the discovery of new biomarkers, with the advantage over tissue biopsy samples thanks to the ease and the less-invasive nature of the collection [111]. Furthermore, the high level of stability, the ease of sampling and an inactive and low complexity test matrix offer numerous potential advantages also compared to the use of other biofluids such as serum and plasma [112].
Interestingly, nipple secretion has also been proposed as a new clinical diagnostic technique and source of secreted proteomes that may reflect early pathological changes in the ductal-lobular epithelial microenvironment and could therefore provide specific BC biomarkers, while remaining an easily accessible and non-invasive source [110].
In recent years, much attention has been paid not only to the study of cell markers, but also especially to that of the “secretome”. “Secretome” is defined as the rich and complex set of molecules and proteins secreted by living cells and released from the surface. The need to develop increasingly effective cancer biomarkers has shifted the focus towards the study of tumor cell secretome as a means of identifying and characterizing diagnostic and prognostic markers and potential pharmacological and therapeutic targets, bearing in mind that secretome proteins carry out a key role in cell signaling, communication and migration [113].
Of note, recent technological developments in the field of proteomics have significantly stimulated and facilitated research in this direction. Hence, in this Review, the works involved in the research of protein biomarkers of BC through proteomics approaches on secretome and on extracellular matrices were collected, schematizing for each biomarker the clinical significance associated (Table S3).
As mentioned above, BC is considered a heterogeneous disease and the most common mistake is to treat BC as a single entity. Current insights from studies on intratumoral heterogeneity and cancer stem cells increase the possibility that multiple BC subtypes can coexist within a tumor and, therefore, the stratification of tumors is fundamental to obtain better clinical results [114].
In this work, we report a list of proteins considered Cell/Tissue Breast Cancer Biomarker, through proteomics studies and subsequent validation experiments (Table S4). Most of the research works cited in Table S4 are studies based on the research of protein biomarkers closely related to the onset of metastases or tumor growth and progression. Other proteins listed in the tables have been studied according to their correlation to epithelial–mesenchymal transition [115–117].
BC cell lines have been widely used for BC modeling, which includes a group of diseases with distinct phenotypic associations. The wide use of cell lines in biomarker research is due to its extremely homogeneous and potentially unlimited content for proteomics studies. Moreover, BC cell lines are also relatively easy to culture.
Numerous studies on protein biomarkers of prognosis, tumor growth and aggression have been conducted on various cellular subtypes of BC. Most of the proteomic studies reported on cellular models are exclusively conducted on Triple-negative (TN) BC (TNBC) tumor subtypes, any BC characterized by the lack of expression of estrogen and progesterone receptor, and of human epidermal growth factor receptor 2. In Table S4, asterisk (*) and hash (#) symbols are used to highlight the potential protein biomarkers found on the cell subtypes or on primary tumor tissues of BC patients, respectively.
As a group, TNBCs is viewed clinically as an aggressive subgroup of BC with a complex and heterogeneous genomic landscape, with an earlier age of presentation and requiring adjuvant chemotherapy to improve survival. The classification of TNBCs in subtypes on the basis of gene expression patterns can provide benefits from specific therapeutic agents [118].
In consideration of complex genomes, high levels of genetic instability, and a high degree of intertumor and intratumor heterogeneity, conventionally TNBCs are defined high-grade carcinomas.
Most TNBCs defined as high-grade tumors have an unfavorable prognosis. Anyway, a subset of TNBCs, comprising histologically low-grade lesions and therefore defined low-grade TNBCs, vastly differs from those high-grade TNBCs and has a favorable outcome. High-grade TNBC include carcinomas with apocrine differentiation, carcinomas with bone marrow characteristics and metaplastic breast carcinomas [118]. Furthermore, current studies suggest several subgroups of low-grade TN malignancies such as a subset of lesions that includes microglandular adenosis, atypical microglandular adenosis and acinic cell carcinoma. Low-grade variants of metaplastic breast carcinomas and solid papillary carcinoma with polarity reversal are additional rare special histological types of low-grade TNBC. The complexity and study of the various histological subtypes of TN disease should not be overlooked, as therapeutic approaches for rare low-grade TNBC subtypes are fundamentally different from those of high-grade TNBC [118].
However, BC cell lines are known to develop mutations during initial establishment and subsequent culture series [118]. In fact, BC cell lines are extremely useful, but often considered rough models for tumors of the same subtype.
The onset of metastasis is one of the most important factors causing the death of patients with BC. In fact, the detection of metastases from BC is an indication of tumor aggression, and if detected early, it should facilitate the correct management of the progression of BC. Therefore, it is very important to look for effective biomarkers for the metastasis and prognosis of BC.
Numerous studies focusing on the identification of metastasis-related factors, potentially used as prognostic markers related to tumor size, axillary lymph node status and histological grade / subtype, have been found [119–122]. The profiling of tumor tissue proteomics provides important information on the discovery of biomarkers [123, 124]. In Table S4 the protein markers obtained from studies conducted and validated in primary tumor tissues of BC patients have been highlighted with the symbol “#”.
In addition, in recent years many researchers have shifted their attention from the study of BC cell lines to cancer steam cells (CSCs). As a result, some protein biomarkers placed on the surface of the CSCs [125] or involved in self-renewal of CSCs [126] have been identified, indicated with the symbol “§“ in Table S4. CSCs are known to play an important role in the recurrence of cancer in almost 65% of cases [127, 128]. Unlike cancer cells, CSCs are quiescent, resulting resistant to anti-cancer drugs. Furthermore, after anti-cancer treatment, these cells can become active and multiply rapidly [129, 130].
For this reason, it has been necessary to develop specific CSC tracking techniques and markers in order to maximize the therapeutic effect of the treatment in cancer cells.
Metabolomics in BC
Metabolomics, one of the newest promising techniques in the “omics” field, allows the quantification of metabolites and/or the evaluation of their ratios in a biofluid, cell, tissue, organ or organism at a given state. As one of the most recent members of the omics family, there has been significant progress in metabolomics in the last decade, primarily driven by technological advances in MS. The metabolome is dynamic, so that metabolite levels and/or ratios can result altered in a pathological condition, thus highlighting abnormal metabolic functions, mainly in complex diseases as BC [11]. Moreover, variations in the metabolome may be the result of genetic, environmental factors, as well as exogenous and endogenous factors [131]. Nowadays, it has become clear that metabolomics, through the comprehensive and quantitative analysis of low-molecular-weight compounds in a system provides the clue to a phenotype, with the potential for a great clinical impact [12, 132, 133]. In fact, even a comprehensive understanding of the state of genes, transcripts, and proteins in a living system is not sufficient to reveal its phenotype [132, 133]. When combining metabolomics with genomics, possibly transcriptomics, and proteomics, a complete understanding of biological mechanisms from a system-wide perspective can be provided [7]. It is now well accepted the idea that metabolites represent the link between genotype and phenotype, and that the study of metabolome offers a significant advantage, allowing to highlight the end-point markers of biological events [133]. Unlike genomics and proteomics, metabolomics is able to provide evidence of end-point markers for diagnosis or evaluation of response to therapy [133]. In this view, the transcripts deriving from DNA are translated into proteins, enzymes necessary for the catalysis of metabolic intermediates [7]. In fact, metabolites, identified by metabolomics strategies either in a targeted or in unbiased manner, are downstream and thus are more sensitive signs of alterations in biological system [131, 134]. Anyway, we should recognize that metabolomics is still emerging with the potential to be deeply (highly) effective in the discovery of molecular candidates for cancer diagnosis, prognosis and treatment[135].
There are two main strategies for metabolic studies: targeted and untargeted analysis of endogenous and exogenous metabolites (<1500 Da) in biological samples at a given point of time. Targeted analysis aims to the quantitative measurement of predetermined compounds taking part in the same biochemical pathway. Thus, the metabolic profile characteristic of that sample might be altered as a result of a gene mutation, diet, drugs, or environmental factors [136]. Non-targeted metabolomics may be described as an open analysis not driven by any preliminary hypothesis for the comprehensive determination of all metabolites present in a sample, with the aim to define alterations in whole metabolome as metabolic fingerprinting characterizing the biological system under specific conditions [131, 136]. Even if metabolomics enables high-throughput analysis of different metabolic pathways and processes all at once, it should be emphasized that it is not yet possible to analyze the entire metabolome and that no single analytical platform can describe all the possible metabolites present in a complex sample, because of their chemical differences and concentration [11]. However, it should be remembered that a wide coverage of metabolism can be obtained by combining two high resolution analytical frameworks: MS, coupled with different separation techniques, and nuclear magnetic resonance (NMR) spectroscopy [137]. In both platforms for metabolomics investigations, after data acquisition, statistical analysis is crucial to give the right value and significance to the dataset previously obtained by the analytical tools [131]. NMR and MS are the most popular platforms for metabolomics and are complementary to each other, even if each approach has advantages and limitations [134]. NMR, the pioneering platform in metabolomics, requires no or low sample pretreatment and allows for reproducible, non-destructive and non-selective analysis, also enabling the simultaneous measurement of different classes of metabolites. Such approach generates a NMR spectrum providing structural information for metabolite identification. Anyway, it presents lower sensitivity if compared with MS [9, 131, 136]. MS is an increasingly used analytical tool for metabolomics applications aiming at the identification of potential biomarkers in different clinical fields. In general, direct-injection MS analysis allows to obtain metabolic profile or fingerprint, but this approach also has some limitations in terms of co-suppression and low ionization efficiency. For this reason, MS is often coupled to a separation technique, based on gas chromatography (GC-MS), liquid chromatography (LC-MS) or capillary electrophoresis. Briefly, GC-MS analysis are characterized by high specificity, sensitivity and accuracy, but they have limitations in mass range (mass-to-charge ratio, m/z 30–550) and in some requirements since the compounds of interest need to be volatile and also thermo stable. The overmentioned requirements are not necessary in LC-MS analysis, the most promising and widely used tool for metabolomics in clinical applications [131]. The growing use of LC-MS can be explained with its high-throughput, soft ionization, and with the possibility to cover a wide range of metabolites. The success and popularity of LC-MS-based metabolomic study is essentially due to the versatility dependent on the sample pretreatment, more simple and rapid in comparison to GC-MS technique, and to the variety of separation possibilities and mass analyzer [131, 138].
Metabolomics features and studies in BC
Bringing our attention back to BC and considering that cancer is a disease that contributes to alterations in cellular metabolism, metabolomics-based studies in the area of BC may be an useful tool for novel biomarker discovery, identification of the related disturbed pathway, early diagnosis, and the evaluation of treatments [131]. When mentioning the perturbed pathway and fighting BC, knowledge on metabolism is highly important [139]. In 1924, Otto Warburg put forward his metabolic hypothesis for cancer. In oncology, the term Warburg effect indicates cancer dependence on fermentative glycolysis, even when oxygen supply is adequate [140]. Therefore, in cancer tissues, the metabolic state often reflects hypoxic metabolism [139]. Metabolomics studies have also described an altered protein and lipid metabolism in cancer [141]. One hypothesis is that even small tumors influence the way metabolites are used in the whole organism. Several metabolic changes have been observed in the blood or urine which reflect one further step downstream in metabolic transformation. Samples for the metabolomic analysis of the BC include urine, serum, plasma, saliva, or tissue and, since metabolites are end products of cellular processes, their concentrations reflect the systems-level response of biological systems and may be valuable for diagnostic tests and therapeutic interventions.
Metabolomics is based on recently developed technologies that allow the quantitative investigation of a multitude of different metabolites. A comprehensive coverage of metabolism can be achieved only by a combination of analytical approaches. The most popular approaches for metabolomics involve GC-MS, LC-MS or NMR spectroscopy. MS-based approaches are typically more sensitive. NMR spectroscopy can be applied to intact tissue samples and even to observe metabolites in vivo, with the technology being referred to as magnetic resonance spectroscopy in the clinic. An improvement of NMR spectroscopic procedure is a technique called high resolution magic angle spinning (HR-MAS) NMR spectroscopy, which involves spinning of a biopsy sample at an angle to the magnetic field, to improve the spectrum resolution. GC-MS-based analyses of metabolic impact or changes in metabolism have a long history in BC research—for example, analysis of phospholipids [142], pharmacology (including tamoxifen metabolism) [143], exposure to xenobiotics [144], estrogen levels [145] or urinary metabolomic profiles [146, 147].
In this work, we report a list of metabolites considered Biofluids/Cell/Tissue Breast Cancer Biomarker, obtained using different approaches (Tables S5 and S6).
NMR studies of human BC samples [148] have found higher contents of Gly, Tau, Lact, and Succ, and lower levels of Gluc and inositol for tumors compared to noninvolved breast tissue. In addition, lipidomics studies have showed that the lipidomics profile correlates with cancer tissue and tumor grade. One of the other profound changes that accompany tumor proliferation is alteration in the proportion of choline-containing metabolites. A series of studies has provided a comprehensive picture of altered Cho metabolism in tissues, as shown in the table [149, 150]. Numerous studies have showed raised levels of choline and its phosphorylated metabolites in subjects with benign and malignant tumors only, and these metabolites have been used for classifying tumor types during the immortalization of cell lines and during apoptosis and necrosis. Choline, phosphocholine and glycerophosphocholine can be observed in clinical magnetic resonance spectroscopy. This has a considerable clinical potential, especially as the HR-NMR analysis is fast, relatively inexpensive, and nondestructive.
In recent years, much attention has been paid to the study of metabolite markers in biofluids.
Although both NMR and MS are commonly used for urine metabolomics, most BC studies were based on MS. Urine samples have the information that can discriminate between normal and BC groups. Urine BC study identified among metabolites as potential biomarkers, amino acids, organic acids, and nucleosides including dimethylarginine, tyrosine, phenylalanine, pantothenic acid, succinyladenosine, dimethylguanosine, apronal, threonyl carbamoyl adenosine, tryptophan, kynurenic acid, nico-tinuric acid, and indolelactic acid [151]. Homovanillate, 4-hydroxy-phenylacetate, 5-hydroxyindoleacetate and urea have been identified as biomarkers for BC from urine using GC-MS [146, 152].
The most easily identifiable clinical biomarkers are derived from blood. Therefore, it is an essential question whether the metabolic response observed in the blood is directly derived from tumor tissue, or whether it represents a more general response of the organism to the presence of a tumor. Several studies identified markers associated with amino acid metabolism, glycolysis, and fatty acid metabolism. Biomarkers reported for metastatic subjects include high values of Phe, Gluc, Pro, Lys, and N-acetyl-Cys [153, 154], and low values of lipids. The final products of β-oxidation (Acac and 3-HB) and lipid degradation (Gluc), N-acetyl glycoproteins (NAC 1 and 2), Pyr, Glut and mannose have been reported from the analysis of serum of early and advanced BC patients. Overall reoccurring marker metabolites include His, Pro, Phe, Glu, 3-HB, Lact, and lipids [155]. Thus, several critical pathways for the early diagnosis of BC have been discovered, including the metabolism of taurine and hypotaurin, and the metabolism of alanine, aspartate, and glutamate [153, 156, 157]. Wang et al. [158] used a dried blood spot approach for rapid BC detection. In this study, the target analytes were 23 aminoacids and 26 acylcarnitines, and based on the results piperamide, asparagine, proline, tetradecenoylcarnitine/palmitoylcarnitine, phenylalanine/tyrosine, and glycine/alanine could be used as potential biomarkers to diagnose BC.
Another explored biological fluid is saliva and from metabolites identified, 3-methyl-pentanoic acid, 4-methyl-pentanoic acid, phenol, p-tert-butyl-phenol, acetic, propanoic, benzoic acids, 1,2-decanediol, 2-decanone, and decanal seem to be relevant for the discrimination of BC patients [159]. Another type of molecules, the polyamines, including N-acetylated forms, are associated with tumor growth due to their biosynthesis and accumulation [160].
In literature, the reports performed involving human cell lines focus mainly on diagnostic purposes. Finally, in the volatile composition (VOMs) of BC cell lines, 2-pentanone, 2-heptanone, 3-methyl-3-buten-1-ol, ethyl acetate, ethyl propanoate, and 2-methyl butanoate were detected only in cultured BC cell lines [161]. These VOMs are formed endogenously or obtained from exogenous sources (e.g., environmental, lifestyle, biological agents), and can be recognized as a useful tool to BC non-invasive diagnosis.
Data processing and elaboration
In this review, in order to combine the results from different “omics” studies, we introduced a new methodical framework by performing a meta-analysis through data “omics” integration.
In particular, we used Ingenuity Pathway Analysis software (IPA, Qiagen, Hilden, Germany) for “Core Analysis” to map statistically each gene or protein or metabolite for their functional annotation, such as network discovery, Upstream Regulator Analysis (URA) and downstream effects networks. Details of data processing and elaboration by IPA are fully described in Supplementary Materials.
“Omics” integration in BC
Following the revision of recent works in literature of the findings by omics-based studies in BC until 2020, we focalized our attention on BC molecular candidates obtained by the application of three specific omics approaches including genomics, proteomics and metabolomics, providing a systematic and detailed integration of the molecular alterations at multiple levels to better describe the pathological phenotype of such a complex disease.
We have discussed above each corresponding “omics” technique as used in the processing of biological data, starting from genomics, the oldest of the “omics” technologies, for DNA, and going on with proteomics for proteins and finally with metabolomics for metabolites [9]. Thus, the term “omics” means an approach capable of generating a complete data set of something measurable [133]. For sure, two are the most important tools which allowed “omics” approaches to reveal their great potential: NGS and MS [135]. Anyway, it should be recognized that statistical and bioinformatics tools are necessary for the processing of the large amount of data turning out by the use of such “omics” approaches [9].
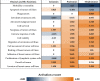
The meta-analysis we conducted on the IPA tool showed good agreement with the literature currently available especially considering the Proteomics and Metabolomics approaches in BC. As shown in Table 1 the “Disease and Functions” that the data uploaded on the IPA described are fully related to the topic and, above all, they showed functions up-regulated (in orange) and down-regulated (in blue) often in agreement considering the dataset of Proteomics and Metabolomics individually.
Table 1.
Disease and biofunctions resulted from the meta-analysis on IPA tool for Genomics, Proteomics, and Metabolomics single data sets, respectively.
 |
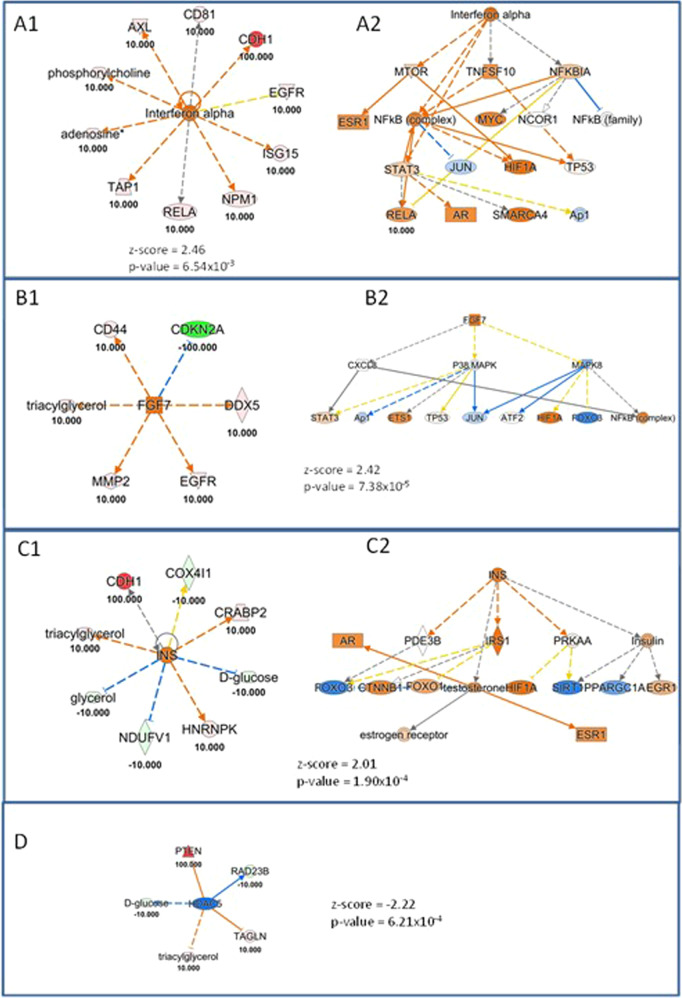
Moreover, we found upstream regulators resulting from first loading of the datasets individually and subsequently from the integration of protein and metabolic biomarker candidates much interesting. As shown by the Venn diagram in Fig. 1A, 17.6% of significantly regulated upstream are in common considering the Proteomics and Metabolomics datasets individually. When we put together all the data collected from the literature (protein and metabolic candidate biomarkers), 18 new upstream results were significant in the meta-analysis on IPA, demonstrating the enormous potential that an integrated omics approach can generate (Fig. 1B). Among these interesting upstream regulators (highlighted by data integration and listed in Table 2), we decided to discuss and deepen the most interesting ones: Histone deacetylase 5 (HDAC5), Insulin (INS), Fibroblast growth factor 7 (FGF7) and Interferon alpha. For each upstream regulator considered, we have plotted a network showing which genes, metabolites, and proteins of the loaded dataset were considered in identifying the aforementioned upstream (Fig. 2A1–C1, D). Interestingly, Fig. 2A2–C2 show the theoretical networks that the activation or inhibition of the aforesaid upstream would contribute within the biological system, as a sort of mechanistic prediction. In particular, a panel of 10 biomarker candidates including proteins and metabolites (Fig. 2A1) demonstrated an up-regulation of Interferon alpha, in agreement with the literature, especially in aggressive cell lines [162]. In fact, in agreement with literature data, increased levels of interferon alpha have been reported in inflammatory BC, the most aggressive and lethal subtype of BC. Another interesting upstream is FGF7 (Fig. 2B1, B2) which is also up-regulated and is known as a regulator involved in tumor growth and invasion not only in BC but also in breast and ovarian cancer [163, 164]. As reported in Fig. 2C1, the simultaneous modulation of some metabolites, such as D-Glucose, Glycerol and Triacylglycerols together with 5 proteins COX4I1, CRABP2, HNRNPK, NDUFV1, and CDH1 promotes an up-regulation of INS. INS is a known upstream regulator involved in tumorigenesis through a direct effect on epithelial tissues or indirectly by affecting the levels of other modulators, such as the family of insulin-like growth factor (IGF) receptors, sex hormones and adipokines [165]. This can also be highlighted from the mechanistic network in Fig. 2C2.
Fig. 1. Upstream Regulator Analysis Results.

A Venn diagram for significant upstream (both activated and inhibited) from the single “Core Analysis” using IPA tool based on Proteomics (in green), or Metabolomics (in light red). B Venn Diagram for significant upstream (both activated and inhibited) from integrating “Omics” approaches (in violet) vs the sum of the significant upstream obtained by each single approach (Proteomics + Metabolomics, in yellow).
Table 2.
List of significant upstream results only from the integration of Proteomics and Metabolomics data.
 |
The four upstream systems deemed most interesting and discussed in more detail are identified in bold.
Fig. 2. Upstream Regulator Analysis, based on “omics” integration using the Ingenuity Pathway Analysis software.
Orange and blue shapes represent predicted activation or inhibition, respectively. The predicted relationship between genes may lead to direct activation (orange solid lines) or direct inhibition (blue solid lines). Red and green color indicate genes, proteins, and metabolites increased and decreased in expression, respectively, while the numbers represent the measurements of their expression. A1 shows the proteins and metabolites of the loaded dataset involved in the activation of the upstream regulator Interferon alpha. A2 shows the mechanistic network, theoretically reconstructed that underlies the activation of the upstream Interferon alpha. B1 shows the proteins and metabolites of the loaded dataset involved in the activation of the upstream regulator Fibroblast growth factor 7 (FGF7). B2 shows the mechanistic network, theoretically reconstructed that underlies the activation of the upstream FGF7. C1 shows the proteins and metabolites of the loaded dataset involved in the activation of the upstream regulator Insulin (INS). C2 shows the mechanistic network, theoretically reconstructed that underlies the activation of the upstream INS. D shows the proteins and metabolites of the loaded dataset involved in the down-regulation of the upstream regulator Histone deacetylase 5 (HDAC5).
Finally, HDAC5 was significantly inhibited, as shown in Fig. 2 Panel D. Indeed, it is often down-regulated or eliminated in human cancers, such as prostate cancer [166]. Conversely, the elevated expression of SOX9 and HDAC5 is associated with lower survival rates in BC patients treated with tamoxifen. HDAC5 was widely expressed in human BC tissues and high HDAC5 expression was associated with a lower prognosis, while HDAC5 knockdown inhibited cell proliferation, migration, invasion and enhanced apoptosis [167].
Conclusion
As well know, adding “omics” to a molecular term connotes a comprehensive evaluation of a set of molecules, and thus multi-omics approaches through high-throughput technologies give the possibility to understand the flow of information underlying a disease, from the original cause of disease to the functional consequences [168], also leading to a crucial change in clinical research [169]. Importantly, if the analysis of data from a single omics technology is limited to correlations and mainly reflects reactive processes rather than causative ones, the integration of data from multi-omics approaches is often applied to explain potential causative changes that lead to disease, or the therapeutic targets. When applying a multi-omics strategy to a disease, it is important to consider the nature of the disorder: simple disease or complex disease. In fact, the etiology of a multifaceted disease as BC is much more complicated and is not focused on a single specific factor but rather on a combination of different factors [169]. In this review, we applied a new integrative meta-analysis method to combine the results obtained from different revised studies. Our meta-analysis proved to be a powerful tool not only to investigate and summarize the correlation between different “omics” datasets, but also to distinguish and highlight new important key pathways and upstream regulators related to BC. Hopefully, the results obtained by our speculation suggest that an in-depth description of the pathological phenotype in BC could be only reached by a proper integration of the large number of biological components, their complex interactions, and their relationships with environment. Therefore, in a systems biology view, data integration is necessary for the comprehensive understanding of the wide dataset arising from multi-omics approaches in the study of a complex disease, as BC. In fact, it is important to emphasized that BC, as each biological phenomenon, is characterized by interdependent layers of biological features. While a single omics approach can catch only a slice of the complex pathological system, multi-omics integration offers an unprecedented opportunity that is the possibility of capturing a deeper and more complete description of the pathological phenomena under study, with translation into clinically relevant information [11, 14, 170]. In conclusion, in comparison to traditional analysis focused on single biological layer, integrative and holistic multi-omics approaches, despite being complicated by the high dimensionality and heterogeneity of the data and the lack of universal analysis protocols, represent new opportunities for studying complex diseases in a more comprehensive way [171–173].
Supplementary information
Author contributions
CR developed the concept and flow of the review article, wrote and revised the manuscript. IC performed the literature review for the Proteomics section, as well as the data processing and integration. MCC performed the data processing and integration. AC performed the literature review for the Metabolomics section. PU performed the literature review for the Genomics section. GS, IA, and PDB assisted with revisions of the manuscript. LS edited the manuscript and supervised the study. VDL supervised the study, edited and final approval of the manuscript, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Fondazione AIRC (Italian Association for Cancer Research, Grant/Award Number: IG‐20043, 25696).
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data and information related to this paper may be requested from the authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41389-022-00393-8.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Jeibouei S, Akbari ME, Kalbasi A, Aref AR, Ajoudanian M, Rezvani A, et al. Personalized medicine in breast cancer: pharmacogenomics approaches. Pharmacogenomics Pers Med. 2019;12:59–73. doi: 10.2147/PGPM.S167886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polyak K. Breast cancer: origins and evolution. J Clin Investig. 2007;117:3155–63. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons J, Francavilla C. ‘Omics approaches to explore the breast cancer landscape. Front Cell Dev Biol. 2019;7:395. doi: 10.3389/fcell.2019.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 6.Cadenas C. Prognostic signatures of breast cancer: Perou’s molecular subtypes and Schmidt’s metagenes. EXCLI J. 2012;11:204–7. [PMC free article] [PubMed] [Google Scholar]
- 7.Dwivedi S, Purohit P, Misra R, Lingeswaran M, Vishnoi JR, Pareek P, et al. Single cell omics of breast cancer: an update on characterization and diagnosis. Indian J Clin Biochem. 2019;34:3–18. doi: 10.1007/s12291-019-0811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masoud V, Pages G. Targeted therapies in breast cancer: New challenges to fight against resistance. World J Clin Oncol. 2017;8:120–34. doi: 10.5306/wjco.v8.i2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wood SL, Westbrook JA, Brown JE. Omic-profiling in breast cancer metastasis to bone: implications for mechanisms, biomarkers and treatment. Cancer Treat Rev. 2014;40:139–52.. doi: 10.1016/j.ctrv.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 10.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Karnebeek CDM, Wortmann SB, Tarailo-Graovac M, Langeveld M, Ferreira CR, van de Kamp JM, et al. The role of the clinician in the multi-omics era: are you ready? J Inherit Metab Dis. 2018;41:571–82.. doi: 10.1007/s10545-017-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rai V, Mukherjee R, Ghosh AK, Routray A, Chakraborty C. “Omics” in oral cancer: New approaches for biomarker discovery. Arch Oral Biol. 2018;87:15–34. doi: 10.1016/j.archoralbio.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Benson M. Clinical implications of omics and systems medicine: focus on predictive and individualized treatment. J Intern Med. 2016;279:229–40. doi: 10.1111/joim.12412. [DOI] [PubMed] [Google Scholar]
- 14.de Anda-Jauregui G, Hernandez-Lemus E. Computational oncology in the multi-omics era: state of the art. Front Oncol. 2020;10:423. doi: 10.3389/fonc.2020.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song YN, Zhang GB, Zhang YY, Su SB. Clinical applications of omics technologies on ZHENG differentiation research in traditional Chinese medicine. Evid Based Complement Altern Med. 2013;2013:989618. doi: 10.1155/2013/989618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knox SS. From ‘omics’ to complex disease: a systems biology approach to gene-environment interactions in cancer. Cancer Cell Int. 2010;10:11. doi: 10.1186/1475-2867-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowan E, Poll A, Narod SA. A prospective study of breast cancer risk in relatives of BRCA1/BRCA2 mutation carriers. J Med Genet. 2007;44:e89. doi: 10.1136/jmg.2007.051631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somasundaram K. BRCA1 and BRCA1 genes and inherited breast and/or ovarian cancer: benefits of genetic testing. Indian J Surg Oncol. 2010;1:245–9. doi: 10.1007/s13193-011-0049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–33.. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50.. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garber JE, Goldstein AM, Kantor AF, Dreyfus MG, Fraumeni JF, Jr, Li FP. Follow-up study of twenty-four families with Li-Fraumeni syndrome. Cancer Res. 1991;51:6094–7. [PubMed] [Google Scholar]
- 22.Harris CC, Hollstein M. Clinical implications of the p53 tumor-suppressor gene. N Engl J Med. 1993;329:1318–27. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 23.Borresen AL, Andersen TI, Garber J, Barbier-Piraux N, Thorlacius S, Eyfjord J, et al. Screening for germ line TP53 mutations in breast cancer patients. Cancer Res. 1992;52:3234–6. [PubMed] [Google Scholar]
- 24.Prosser J, Elder PA, Condie A, MacFadyen I, Steel CM, Evans HJ. Mutations in p53 do not account for heritable breast cancer: a study in five affected families. Br J Cancer. 1991;63:181–4. doi: 10.1038/bjc.1991.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosser J, Porter D, Coles C, Condie A, Thompson AM, Chetty U, et al. Constitutional p53 mutation in a non-Li-Fraumeni cancer family. Br J Cancer. 1992;65:527–8. doi: 10.1038/bjc.1992.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coles C, Condie A, Chetty U, Steel CM, Evans HJ, Prosser J. p53 mutations in breast cancer. Cancer Res. 1992;52:5291–8. [PubMed] [Google Scholar]
- 27.Osborne RJ, Merlo GR, Mitsudomi T, Venesio T, Liscia DS, Cappa AP, et al. Mutations in the p53 gene in primary human breast cancers. Cancer Res. 1991;51:6194–8. [PubMed] [Google Scholar]
- 28.Thompson AM, Anderson TJ, Condie A, Prosser J, Chetty U, Carter DC, et al. p53 allele losses, mutations and expression in breast cancer and their relationship to clinico-pathological parameters. Int J Cancer. 1992;50:528–32. doi: 10.1002/ijc.2910500405. [DOI] [PubMed] [Google Scholar]
- 29.Swift M, Reitnauer PJ, Morrell D, Chase CL. Breast and other cancers in families with ataxia-telangiectasia. N Engl J Med. 1987;316:1289–94.. doi: 10.1056/NEJM198705213162101. [DOI] [PubMed] [Google Scholar]
- 30.Borresen AL, Andersen TI, Tretli S, Heiberg A, Moller P. Breast cancer and other cancers in Norwegian families with ataxia-telangiectasia. Genes Chromosomes Cancer. 1990;2:339–40. doi: 10.1002/gcc.2870020412. [DOI] [PubMed] [Google Scholar]
- 31.Pippard EC, Hall AJ, Barker DJ, Bridges BA. Cancer in homozygotes and heterozygotes of ataxia-telangiectasia and xeroderma pigmentosum in Britain. Cancer Res. 1988;48:2929–32. [PubMed] [Google Scholar]
- 32.Swift M, Morrell D, Massey RB, Chase CL. Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med. 1991;325:1831–6. doi: 10.1056/NEJM199112263252602. [DOI] [PubMed] [Google Scholar]
- 33.Morrell D, Chase CL, Swift M. Cancers in 44 families with ataxia-telangiectasia. Cancer Genet Cytogenet. 1990;50:119–23. doi: 10.1016/0165-4608(90)90245-6. [DOI] [PubMed] [Google Scholar]
- 34.Easton DF. The inherited component of cancer. Br Med Bull. 1994;50:527–35. doi: 10.1093/oxfordjournals.bmb.a072908. [DOI] [PubMed] [Google Scholar]
- 35.Thorstenson YR, Shen P, Tusher VG, Wayne TL, Davis RW, Chu G, et al. Global analysis of ATM polymorphism reveals significant functional constraint. Am J Hum Genet. 2001;69:396–412. doi: 10.1086/321296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renwick A, Thompson D, Seal S, Kelly P, Chagtai T, Ahmed M, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein JL, Haile RW, Stovall M, Boice JD, Jr, Shore RE, Langholz B, et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102:475–83.. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–7. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 39.Marsh DJ, Coulon V, Lunetta KL, Rocca-Serra P, Dahia PL, Zheng Z, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet. 1998;7:507–15. doi: 10.1093/hmg/7.3.507. [DOI] [PubMed] [Google Scholar]
- 40.Eng C. Genetics of Cowden syndrome: through the looking glass of oncology. Int J Oncol. 1998;12:701–10. doi: 10.3892/ijo.12.3.701. [DOI] [PubMed] [Google Scholar]
- 41.Starink TM, van der Veen JP, Arwert F, de Waal LP, de Lange GG, Gille JJ, et al. The Cowden syndrome: a clinical and genetic study in 21 patients. Clin Genet. 1986;29:222–33. doi: 10.1111/j.1399-0004.1986.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 42.Feilotter HE, Coulon V, McVeigh JL, Boag AH, Dorion-Bonnet F, Duboue B, et al. Analysis of the 10q23 chromosomal region and the PTEN gene in human sporadic breast carcinoma. Br J Cancer. 1999;79:718–23.. doi: 10.1038/sj.bjc.6690115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freihoff D, Kempe A, Beste B, Wappenschmidt B, Kreyer E, Hayashi Y, et al. Exclusion of a major role for the PTEN tumour-suppressor gene in breast carcinomas. Br J Cancer. 1999;79:754–8. doi: 10.1038/sj.bjc.6690121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol. 1999;155:1253–60.. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh B, Ittmann MM, Krolewski JJ. Sporadic breast cancers exhibit loss of heterozygosity on chromosome segment 10q23 close to the Cowden disease locus. Genes Chromosomes Cancer. 1998;21:166–71. doi: 10.1002/(SICI)1098-2264(199802)21:2<166::AID-GCC13>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 46.Carroll BT, Couch FJ, Rebbeck TR, Weber BL. Polymorphisms in PTEN in breast cancer families. J Med Genet. 1999;36:94–6. [PMC free article] [PubMed] [Google Scholar]
- 47.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–7. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 48.Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- 49.Boardman LA, Thibodeau SN, Schaid DJ, Lindor NM, McDonnell SK, Burgart LJ, et al. Increased risk for cancer in patients with the Peutz-Jeghers syndrome. Ann Intern Med. 1998;128:896–9. doi: 10.7326/0003-4819-128-11-199806010-00004. [DOI] [PubMed] [Google Scholar]
- 50.Madanikia SA, Bergner A, Ye X, Blakeley JO. Increased risk of breast cancer in women with NF1. Am J Med Genet A. 2012;158A:3056–60. doi: 10.1002/ajmg.a.35550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seminog OO, Goldacre MJ. Age-specific risk of breast cancer in women with neurofibromatosis type 1. Br J Cancer. 2015;112:1546–8. doi: 10.1038/bjc.2015.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guran S, Safali M. A case of neurofibromatosis and breast cancer: loss of heterozygosity of NF1 in breast cancer. Cancer Genet Cytogenet. 2005;156:86–8. doi: 10.1016/j.cancergencyto.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 53.Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, et al. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J Clin Oncol. 2015;33:3660–7. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 54.Alenezi WM, Fierheller CT, Recio N, Tonin PN. Literature review of BARD1 as a cancer predisposing gene with a focus on breast and ovarian cancers. Genes. 2020;11:856. [DOI] [PMC free article] [PubMed]
- 55.Irminger-Finger I, Soriano JV, Vaudan G, Montesano R, Sappino AP. In vitro repression of Brca1-associated RING domain gene, Bard1, induces phenotypic changes in mammary epithelial cells. J Cell Biol. 1998;143:1329–39. doi: 10.1083/jcb.143.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moyer CL, Ivanovich J, Gillespie JL, Doberstein R, Radke MR, Richardson ME, et al. Rare BRIP1 missense alleles confer risk for ovarian and breast cancer. Cancer Res. 2020;80:857–67.. doi: 10.1158/0008-5472.CAN-19-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41.. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 58.Rainville I, Hatcher S, Rosenthal E, Larson K, Bernhisel R, Meek S, et al. High risk of breast cancer in women with biallelic pathogenic variants in CHEK2. Breast Cancer Res Treat. 2020;180:503–9. doi: 10.1007/s10549-020-05543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2022;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 60.Wu L, Wu Y, Gathings B, Wan M, Li X, Grizzle W, et al. Smad4 as a transcription corepressor for estrogen receptor alpha. J Biol Chem. 2003;278:15192–200. doi: 10.1074/jbc.M212332200. [DOI] [PubMed] [Google Scholar]
- 61.Zhong D, Morikawa A, Guo L, Colpaert C, Xiong L, Nassar A, et al. Homozygous deletion of SMAD4 in breast cancer cell lines and invasive ductal carcinomas. Cancer Biol Ther. 2006;5:601–7. doi: 10.4161/cbt.5.6.2660. [DOI] [PubMed] [Google Scholar]
- 62.Deckers M, van Dinther M, Buijs J, Que I, Lowik C, van der Pluijm G, et al. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–9. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 63.Stuelten CH, Buck MB, Dippon J, Roberts AB, Fritz P, Knabbe C. Smad4-expression is decreased in breast cancer tissues: a retrospective study. BMC Cancer. 2006;6:25. doi: 10.1186/1471-2407-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–8. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foo TK, Tischkowitz M, Simhadri S, Boshari T, Zayed N, Burke KA, et al. Compromised BRCA1-PALB2 interaction is associated with breast cancer risk. Oncogene. 2017;36:4161–70.. doi: 10.1038/onc.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bagherzadeh M, Szymiczek A, Donenberg T, Butler R, Hurley J, Narod SA, et al. Association of RAD51C germline mutations with breast cancer among Bahamians. Breast Cancer Res Treat. 2020;184:649–51.. doi: 10.1007/s10549-020-05872-3. [DOI] [PubMed] [Google Scholar]
- 68.Konstanta I, Fostira F, Apostolou P, Stratikos E, Kalfakakou D, Pampanos A, et al. Contribution of RAD51D germline mutations in breast and ovarian cancer in Greece. J Hum Genet. 2018;63:1149–58.. doi: 10.1038/s10038-018-0498-8. [DOI] [PubMed] [Google Scholar]
- 69.Yang X, Song H, Leslie G, Engel C, Hahnen E, Auber B, et al. Ovarian and breast cancer risks associated with pathogenic variants in RAD51C and RAD51D. J Natl Cancer Inst. 2020;112:1242–50.. doi: 10.1093/jnci/djaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang G, Zeng Y, Liu Z, Wei W. Significant association between Nijmegen breakage syndrome 1 657del5 polymorphism and breast cancer risk. Tumour Biol. 2013;34:2753–7. doi: 10.1007/s13277-013-0830-z. [DOI] [PubMed] [Google Scholar]
- 71.Tien JF, Mazloomian A, Cheng SG, Hughes CS, Chow CCT, Canapi LT, et al. CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 2017;45:6698–716.. doi: 10.1093/nar/gkx187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naidoo K, Wai PT, Maguire SL, Daley F, Haider S, Kriplani D, et al. Evaluation of CDK12 protein expression as a potential novel biomarker for DNA damage response-targeted therapies in breast cancer. Mol Cancer Ther. 2018;17:306–15.. doi: 10.1158/1535-7163.MCT-17-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pharoah PD, Guilford P, Caldas C, International Gastric Cancer Linkage Consortium. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121:1348–53. doi: 10.1053/gast.2001.29611. [DOI] [PubMed] [Google Scholar]
- 74.Rennert G, Lejbkowicz F, Cohen I, Pinchev M, Rennert HS, Barnett-Griness O. MutYH mutation carriers have increased breast cancer risk. Cancer. 2012;118:1989–93. doi: 10.1002/cncr.26506. [DOI] [PubMed] [Google Scholar]
- 75.Rizzolo P, Silvestri V, Bucalo A, Zelli V, Valentini V, Catucci I, et al. Contribution of MUTYH variants to male breast cancer risk: results from a multicenter study in Italy. Front Oncol. 2018;8:583. doi: 10.3389/fonc.2018.00583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thibodeau ML, Zhao EY, Reisle C, Ch’ng C, Wong HL, Shen Y, et al. Base excision repair deficiency signatures implicate germline and somatic MUTYH aberrations in pancreatic ductal adenocarcinoma and breast cancer oncogenesis. Cold Spring Harb Mol Case Stud. 2019;5:a003681. [DOI] [PMC free article] [PubMed]
- 77.Buys SS, Sandbach JF, Gammon A, Patel G, Kidd J, Brown KL, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–30.. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 78.Debniak T, Cybulski C, Gorski B, Huzarski T, Byrski T, Gronwald J, et al. CDKN2A-positive breast cancers in young women from Poland. Breast Cancer Res Treat. 2007;103:355–9. doi: 10.1007/s10549-006-9382-x. [DOI] [PubMed] [Google Scholar]
- 79.Mukherjee N, Bhattacharya N, Sinha S, Alam N, Chakravarti R, Roychoudhury S, et al. Association of APC and MCC polymorphisms with increased breast cancer risk in an Indian population. Int J Biol Markers. 2011;26:43–9. doi: 10.5301/JBM.2011.6266. [DOI] [PubMed] [Google Scholar]
- 80.Sobsey CA, Ibrahim S, Richard VR, Gaspar V, Mitsa G, Lacasse V, et al. Targeted and untargeted proteomics approaches in biomarker development. Proteomics. 2020;20:e1900029. doi: 10.1002/pmic.201900029. [DOI] [PubMed] [Google Scholar]
- 81.Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, Yu FH. Too many roads not taken. Nature. 2011;470:163–5. doi: 10.1038/470163a. [DOI] [PubMed] [Google Scholar]
- 82.Chae YK, Gonzalez-Angulo AM. Implications of functional proteomics in breast cancer. Oncologist. 2014;19:328–35. doi: 10.1634/theoncologist.2013-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bera A, Russ E, Manoharan MS, Eidelman O, Eklund M, Hueman M, et al. Proteomic analysis of inflammatory biomarkers associated with breast cancer recurrence. Mil Med. 2020;185:669–75.. doi: 10.1093/milmed/usz254. [DOI] [PubMed] [Google Scholar]
- 84.Boccardo F, Rubagotti A, Nuzzo PV, Argellati F, Savarino G, Romano P, et al. Matrix-assisted laser desorption/ionisation (MALDI) TOF analysis identifies serum angiotensin II concentrations as a strong predictor of all-cause and breast cancer (BCa)-specific mortality following breast surgery. Int J Cancer. 2015;137:2394–402.. doi: 10.1002/ijc.29609. [DOI] [PubMed] [Google Scholar]
- 85.Chung L, Moore K, Phillips L, Boyle FM, Marsh DJ, Baxter RC. Novel serum protein biomarker panel revealed by mass spectrometry and its prognostic value in breast cancer. Breast Cancer Res. 2014;16:R63. doi: 10.1186/bcr3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fan Y, Wang J, Yang Y, Liu Q, Fan Y, Yu J, et al. Detection and identification of potential biomarkers of breast cancer. J Cancer Res Clin Oncol. 2010;136:1243–54.. doi: 10.1007/s00432-010-0775-1. [DOI] [PubMed] [Google Scholar]
- 87.Fernandez-Grijalva AL, Aguilar-Lemarroy A, Jave-Suarez LF, Gutierrez-Ortega A, Godinez-Melgoza PA, Herrera-Rodriguez SE, et al. Alpha 2HS-glycoprotein, a tumor-associated antigen (TAA) detected in Mexican patients with early-stage breast cancer. J Proteomics. 2015;112:301–12. doi: 10.1016/j.jprot.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 88.Fry SA, Sinclair J, Timms JF, Leathem AJ, Dwek MV. A targeted glycoproteomic approach identifies cadherin-5 as a novel biomarker of metastatic breast cancer. Cancer Lett. 2013;328:335–44. doi: 10.1016/j.canlet.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 89.Fu-Jun L, Shao-Hua J, Xiao-Fang S. Differential proteomic analysis of pathway biomarkers in human breast cancer by integrated bioinformatics. Oncol Lett. 2012;4:1097–103.. doi: 10.3892/ol.2012.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He W, Tong Y, Wang Y, Liu J, Luo G, Wu J, et al. Serum soluble CD14 is a potential prognostic indicator of recurrence of human breast invasive ductal carcinoma with Her2-enriched subtype. PLoS ONE. 2013;8:e75366. doi: 10.1371/journal.pone.0075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hyung SW, Lee MY, Yu JH, Shin B, Jung HJ, Park JM, et al. A serum protein profile predictive of the resistance to neoadjuvant chemotherapy in advanced breast cancers. Mol Cell Proteomics. 2011;10:M111.011023. [DOI] [PMC free article] [PubMed]
- 92.Lacombe J, Mange A, Jarlier M, Bascoul-Mollevi C, Rouanet P, Lamy PJ, et al. Identification and validation of new autoantibodies for the diagnosis of DCIS and node negative early-stage breast cancers. Int J Cancer. 2013;132:1105–13. doi: 10.1002/ijc.27766. [DOI] [PubMed] [Google Scholar]
- 93.Lee CS, Taib NA, Ashrafzadeh A, Fadzli F, Harun F, Rahmat K, et al. Unmasking heavily O-glycosylated serum proteins using perchloric acid: identification of serum proteoglycan 4 and protease C1 inhibitor as molecular indicators for screening of breast cancer. PLoS ONE. 2016;11:e0149551. doi: 10.1371/journal.pone.0149551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lopez-Arias E, Aguilar-Lemarroy A, Felipe Jave-Suarez L, Morgan-Villela G, Mariscal-Ramirez I, Martinez-Velazquez M, et al. Alpha 1-antitrypsin: a novel tumor-associated antigen identified in patients with early-stage breast cancer. Electrophoresis. 2012;33:2130–7. doi: 10.1002/elps.201100491. [DOI] [PubMed] [Google Scholar]
- 95.Valo I, Raro P, Boissard A, Maarouf A, Jezequel P, Verriele V, et al. OLFM4 expression in ductal carcinoma in situ and in invasive breast cancer cohorts by a SWATH-based proteomic approach. Proteomics. 2019;19:e1800446. doi: 10.1002/pmic.201800446. [DOI] [PubMed] [Google Scholar]
- 96.Zeidan B, Jackson TR, Larkin SE, Cutress RI, Coulton GR, Ashton-Key M, et al. Annexin A3 is a mammary marker and a potential neoplastic breast cell therapeutic target. Oncotarget. 2015;6:21421–7. doi: 10.18632/oncotarget.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeidan B, Manousopoulou A, Garay-Baquero DJ, White CH, Larkin SET, Potter KN, et al. Increased circulating resistin levels in early-onset breast cancer patients of normal body mass index correlate with lymph node negative involvement and longer disease free survival: a multi-center POSH cohort serum proteomics study. Breast Cancer Res. 2018;20:19. doi: 10.1186/s13058-018-0938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brauer HA, D’Arcy M, Libby TE, Thompson HJ, Yasui YY, Hamajima N, et al. Dermcidin expression is associated with disease progression and survival among breast cancer patients. Breast Cancer Res Treat. 2014;144:299–306. doi: 10.1007/s10549-014-2880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Correa S, Panis C, Binato R, Herrera AC, Pizzatti L, Abdelhay E. Identifying potential markers in breast cancer subtypes using plasma label-free proteomics. J Proteomics. 2017;151:33–42. doi: 10.1016/j.jprot.2016.07.030. [DOI] [PubMed] [Google Scholar]
- 100.Lee HB, Kang UB, Moon HG, Lee J, Lee KM, Yi M, et al. Development and validation of a novel plasma protein signature for breast cancer diagnosis by using multiple reaction monitoring-based mass spectrometry. Anticancer Res. 2015;35:6271–9. [PubMed] [Google Scholar]
- 101.Lobo MD, Moreno FB, Souza GH, Verde SM, Moreira RA, Monteiro-Moreira AC. Label-free proteome analysis of plasma from patients with breast cancer: stage-specific protein expression. Front Oncol. 2017;7:14. doi: 10.3389/fonc.2017.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Orlandi R, De Bortoli M, Ciniselli CM, Vaghi E, Caccia D, Garrisi V, et al. Hepcidin and ferritin blood level as noninvasive tools for predicting breast cancer. Ann Oncol. 2014;25:352–7. doi: 10.1093/annonc/mdt490. [DOI] [PubMed] [Google Scholar]
- 103.Panis C, Pizzatti L, Herrera AC, Cecchini R, Abdelhay E. Putative circulating markers of the early and advanced stages of breast cancer identified by high-resolution label-free proteomics. Cancer Lett. 2013;330:57–66. doi: 10.1016/j.canlet.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 104.Pitteri SJ, Amon LM, Busald Buson T, Zhang Y, Johnson MM, Chin A, et al. Detection of elevated plasma levels of epidermal growth factor receptor before breast cancer diagnosis among hormone therapy users. Cancer Res. 2010;70:8598–606.. doi: 10.1158/0008-5472.CAN-10-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scumaci D, Tamme L, Fiumara CV, Pappaianni G, Concolino A, Leone E, et al. Plasma proteomic profiling in hereditary breast cancer reveals a BRCA1-specific signature: diagnostic and functional implications. PLoS ONE. 2015;10:e0129762. doi: 10.1371/journal.pone.0129762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shao B, Tian Z, Ding H, Wang Q, Song G, Di L, et al. Plasma CAMK2A predicts chemotherapy resistance in metastatic triple negative breast cancer. Int J Clin Exp Pathol. 2018;11:650–63. [PMC free article] [PubMed] [Google Scholar]
- 107.Suh EJ, Kabir MH, Kang UB, Lee JW, Yu J, Noh DY, et al. Comparative profiling of plasma proteome from breast cancer patients reveals thrombospondin-1 and BRWD3 as serological biomarkers. Exp Mol Med. 2012;44:36–44. doi: 10.3858/emm.2012.44.1.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beretov J, Wasinger VC, Millar EK, Schwartz P, Graham PH, Li Y. Proteomic analysis of urine to identify breast cancer biomarker candidates using a label-free LC-MS/MS approach. PLoS ONE. 2015;10:e0141876. doi: 10.1371/journal.pone.0141876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gajbhiye A, Dabhi R, Taunk K, Vannuruswamy G, RoyChoudhury S, Adhav R, et al. Urinary proteome alterations in HER2 enriched breast cancer revealed by multipronged quantitative proteomics. Proteomics. 2016;16:2403–18. doi: 10.1002/pmic.201600015. [DOI] [PubMed] [Google Scholar]
- 110.Kurono S, Kaneko Y, Matsuura N, Oishi H, Noguchi S, Kim SJ, et al. Identification of potential breast cancer markers in nipple discharge by protein profile analysis using two-dimensional nano-liquid chromatography/nanoelectrospray ionization-mass spectrometry. Proteomics Clin Appl. 2016;10:605–13. doi: 10.1002/prca.201500016. [DOI] [PubMed] [Google Scholar]
- 111.Beretov J, Wasinger VC, Graham PH, Millar EK, Kearsley JH, Li Y. Proteomics for breast cancer urine biomarkers. Adv Clin Chem. 2014;63:123–67.. doi: 10.1016/B978-0-12-800094-6.00004-2. [DOI] [PubMed] [Google Scholar]
- 112.Nolen BM, Lokshin AE. -The advancement of biomarker-based diagnostic tools for ovarian, breast, and pancreatic cancer through the use of urine as an analytical biofluid. Int J Biol Markers. 2011;26:141–52. doi: 10.5301/JBM.2011.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J Proteomics. 2010;73:2291–305.. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 114.Yeo SK, Guan JL. Breast cancer: multiple subtypes within a tumor? Trends Cancer. 2017;3:753–60.. doi: 10.1016/j.trecan.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johansson HJ, Sanchez BC, Forshed J, Stal O, Fohlin H, Lewensohn R, et al. Proteomics profiling identify CAPS as a potential predictive marker of tamoxifen resistance in estrogen receptor positive breast cancer. Clin Proteomics. 2015;12:8. doi: 10.1186/s12014-015-9080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Silvestrini VC, Thome CH, Albuquerque D, de Souza Palma C, Ferreira GA, Lanfredi GP, et al. Proteomics analysis reveals the role of ubiquitin specific protease (USP47) in epithelial to mesenchymal transition (EMT) induced by TGFbeta2 in breast cells. J Proteomics. 2020;219:103734. doi: 10.1016/j.jprot.2020.103734. [DOI] [PubMed] [Google Scholar]
- 117.Uretmen Kagiali ZC, Sanal E, Karayel O, Polat AN, Saatci O, Ersan PG, et al. Systems-level analysis reveals multiple modulators of epithelial-mesenchymal transition and identifies DNAJB4 and CD81 as novel metastasis inducers in breast cancer. Mol Cell Proteomics. 2019;18:1756–71.. doi: 10.1074/mcp.RA119.001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Geyer FC, Pareja F, Weigelt B, Rakha E, Ellis IO, Schnitt SJ, et al. The spectrum of triple-negative breast disease: high- and low-grade lesions. Am J Pathol. 2017;187:2139–51.. doi: 10.1016/j.ajpath.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cawthorn TR, Moreno JC, Dharsee M, Tran-Thanh D, Ackloo S, Zhu PH, et al. Proteomic analyses reveal high expression of decorin and endoplasmin (HSP90B1) are associated with breast cancer metastasis and decreased survival. PLoS ONE. 2012;7:e30992. doi: 10.1371/journal.pone.0030992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dvorakova M, Jerabkova J, Prochazkova I, Lenco J, Nenutil R, Bouchal P. Transgelin is upregulated in stromal cells of lymph node positive breast cancer. J Proteomics. 2016;132:103–11. doi: 10.1016/j.jprot.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 121.Prochazkova I, Lenco J, Fucikova A, Dresler J, Capkova L, Hrstka R, et al. Targeted proteomics driven verification of biomarker candidates associated with breast cancer aggressiveness. Biochim Biophys Acta Proteins Proteomics. 2017;1865:488–98.. doi: 10.1016/j.bbapap.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 122.Zeng L, Deng X, Zhong J, Yuan L, Tao X, Zhang S, et al. Prognostic value of biomarkers EpCAM and alphaB-crystallin associated with lymphatic metastasis in breast cancer by iTRAQ analysis. BMC Cancer. 2019;19:831. doi: 10.1186/s12885-019-6016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brown JE, Westbrook JA, Wood SL. Dedicator of cytokinesis 4: a potential prognostic and predictive biomarker within the metastatic spread of breast cancer to bone. Cancer Informatics. 2019;18:1176935119866842. doi: 10.1177/1176935119866842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoneten KK, Kasap M, Akpinar G, Gunes A, Gurel B, Utkan NZ. Comparative proteome analysis of breast cancer tissues highlights the importance of glycerol-3-phosphate dehydrogenase 1 and monoacylglycerol lipase in breast cancer metabolism. Cancer Genomics Proteomics. 2019;16:377–97. doi: 10.21873/cgp.20143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Koh EY, You JE, Jung SH, Kim PH. Biological functions and identification of novel biomarker expressed on the surface of breast cancer-derived cancer stem cells via proteomic analysis. Mol Cells. 2020;43:384–96.. doi: 10.14348/molcells.2020.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kanojia D, Zhou W, Zhang J, Jie C, Lo PK, Wang Q, et al. Proteomic profiling of cancer stem cells derived from primary tumors of HER2/Neu transgenic mice. Proteomics. 2012;12:3407–15.. doi: 10.1002/pmic.201200103. [DOI] [PubMed] [Google Scholar]
- 127.Rodini CO, Lopes NM, Lara VS, Mackenzie IC. Oral cancer stem cells - properties and consequences. J Appl Oral Sci. 2017;25:708–15.. doi: 10.1590/1678-7757-2016-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Song M, Giovannucci EL. Cancer risk: many factors contribute. Science. 2015;347:728–9. doi: 10.1126/science.aaa6094. [DOI] [PubMed] [Google Scholar]
- 129.Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 130.Zhu P, Fan Z. Cancer stem cells and tumorigenesis. Biophys Rep. 2018;4:178–88.. doi: 10.1007/s41048-018-0062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Silva C, Perestrelo R, Silva P, Tomas H, Camara JS. Breast cancer metabolomics: from analytical platforms to multivariate data analysis. a review. Metabolites. 2019;9:102. [DOI] [PMC free article] [PubMed]
- 132.Del Boccio P, Rossi C, di Ioia M, Cicalini I, Sacchetta P, Pieragostino D. Integration of metabolomics and proteomics in multiple sclerosis: from biomarkers discovery to personalized medicine. Proteomics Clin Appl. 2016;10:470–84. doi: 10.1002/prca.201500083. [DOI] [PubMed] [Google Scholar]
- 133.Goldsmith P, Fenton H, Morris-Stiff G, Ahmad N, Fisher J, Prasad KR. Metabonomics: a useful tool for the future surgeon. J Surg Res. 2010;160:122–32.. doi: 10.1016/j.jss.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 134.Hadi NI, Jamal Q. “OMIC” tumor markers for breast cancer: a review. Pak J Med Sci. 2015;31:1256–62.. doi: 10.12669/pjms.315.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chakraborty S, Hosen MI, Ahmed M, Shekhar HU. Onco-multi-OMICS approach: a new frontier in cancer research. Biomed Res Int. 2018;2018:9836256. doi: 10.1155/2018/9836256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bujak R, Struck-Lewicka W, Markuszewski MJ, Kaliszan R. Metabolomics for laboratory diagnostics. J Pharmacol Biomed Anal. 2015;113:108–20. doi: 10.1016/j.jpba.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 137.Zhang AH, Sun H, Qiu S, Wang XJ. Metabolomics in noninvasive breast cancer. Clin Chim Acta. 2013;424:3–7. doi: 10.1016/j.cca.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 138.Zhou B, Xiao JF, Tuli L, Ressom HW. LC-MS-based metabolomics. Mol Biosyst. 2012;8:470–81. doi: 10.1039/C1MB05350G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Menendez JA, Joven J, Cufi S, Corominas-Faja B, Oliveras-Ferraros C, Cuyas E, et al. The Warburg effect version 2.0: metabolic reprogramming of cancer stem cells. Cell Cycle. 2013;12:1166–79.. doi: 10.4161/cc.24479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M, et al. Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis. 2014;5:e1336. doi: 10.1038/cddis.2014.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551–61.. doi: 10.1038/nrc1390. [DOI] [PubMed] [Google Scholar]
- 142.Brochez V, Van Heuverswyn D, Diniz JA, De Potter CR, Van, den Eeckhout EG. Cellular uptake and retention measurements of alkylphosphocholines in the SK-BR-3 breast cancer and Molt-4 leukemia cell line using capillary gas chromatography. Lipids. 1999;34:511–6. doi: 10.1007/s11745-999-0392-6. [DOI] [PubMed] [Google Scholar]
- 143.Daniel CP, Gaskell SJ, Bishop H, Nicholson RI. Determination of tamoxifen and an hydroxylated metabolite in plasma from patients with advanced breast cancer using gas chromatography-mass spectrometry. J Endocrinol. 1979;83:401–8. doi: 10.1677/joe.0.0830401. [DOI] [PubMed] [Google Scholar]
- 144.Wolff MS, Toniolo PG, Lee EW, Rivera M, Dubin N. Blood levels of organochlorine residues and risk of breast cancer. J Natl Cancer Inst. 1993;85:648–52. doi: 10.1093/jnci/85.8.648. [DOI] [PubMed] [Google Scholar]
- 145.Ingram D, Sanders K, Kolybaba M, Lopez D. Case-control study of phyto-oestrogens and breast cancer. Lancet. 1997;350:990–4. doi: 10.1016/S0140-6736(97)01339-1. [DOI] [PubMed] [Google Scholar]
- 146.Nam H, Chung BC, Kim Y, Lee K, Lee D. Combining tissue transcriptomics and urine metabolomics for breast cancer biomarker identification. Bioinformatics. 2009;25:3151–7. doi: 10.1093/bioinformatics/btp558. [DOI] [PubMed] [Google Scholar]
- 147.Woo HM, Kim KM, Choi MH, Jung BH, Lee J, Kong G, et al. Mass spectrometry based metabolomic approaches in urinary biomarker study of women’s cancers. Clin Chim Acta. 2009;400:63–9. doi: 10.1016/j.cca.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 148.Giskeodegard GF, Lundgren S, Sitter B, Fjosne HE, Postma G, Buydens LM, et al. Lactate and glycine-potential MR biomarkers of prognosis in estrogen receptor-positive breast cancers. NMR Biomed. 2012;25:1271–9. doi: 10.1002/nbm.2798. [DOI] [PubMed] [Google Scholar]
- 149.Bathen TF, Geurts B, Sitter B, Fjosne HE, Lundgren S, Buydens LM, et al. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS ONE. 2013;8:e61578. doi: 10.1371/journal.pone.0061578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mimmi MC, Picotti P, Corazza A, Betto E, Pucillo CE, Cesaratto L, et al. High-performance metabolic marker assessment in breast cancer tissue by mass spectrometry. Clin Chem Lab Med. 2011;49:317–24. doi: 10.1515/CCLM.2011.060. [DOI] [PubMed] [Google Scholar]
- 151.Chen Y, Zhang R, Song Y, He J, Sun J, Bai J, et al. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst. 2009;134:2003–11. doi: 10.1039/b907243h. [DOI] [PubMed] [Google Scholar]
- 152.Cala M, Aldana J, Sanchez J, Guio J, Meesters RJW. Urinary metabolite and lipid alterations in Colombian Hispanic women with breast cancer: a pilot study. J Pharmacol Biomed Anal. 2018;152:234–41.. doi: 10.1016/j.jpba.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 153.Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, et al. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70:8309–18. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Oakman C, Tenori L, Claudino WM, Cappadona S, Nepi S, Battaglia A, et al. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol. 2011;22:1295–301.. doi: 10.1093/annonc/mdq606. [DOI] [PubMed] [Google Scholar]
- 155.Slupsky CM, Steed H, Wells TH, Dabbs K, Schepansky A, Capstick V, et al. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin Cancer Res. 2010;16:5835–41. doi: 10.1158/1078-0432.CCR-10-1434. [DOI] [PubMed] [Google Scholar]
- 156.Budczies J, Denkert C, Muller BM, Brockmoller SF, Klauschen F, Gyorffy B, et al. Remodeling of central metabolism in invasive breast cancer compared to normal breast tissue - a GC-TOFMS based metabolomics study. BMC Genomics. 2012;13:334. doi: 10.1186/1471-2164-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jove M, Collado R, Quiles JL, Ramirez-Tortosa MC, Sol J, Ruiz-Sanjuan M, et al. A plasma metabolomic signature discloses human breast cancer. Oncotarget. 2017;8:19522–33.. doi: 10.18632/oncotarget.14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang Q, Sun T, Cao Y, Gao P, Dong J, Fang Y, et al. A dried blood spot mass spectrometry metabolomic approach for rapid breast cancer detection. Oncotargets Ther. 2016;9:1389–98. doi: 10.2147/OTT.S95862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cavaco C, Pereira JAM, Taunk K, Taware R, Rapole S, Nagarajaram H, et al. Screening of salivary volatiles for putative breast cancer discrimination: an exploratory study involving geographically distant populations. Anal Bioanal Chem. 2018;410:4459–68.. doi: 10.1007/s00216-018-1103-x. [DOI] [PubMed] [Google Scholar]
- 160.Takayama T, Tsutsui H, Shimizu I, Toyama T, Yoshimoto N, Endo Y, et al. Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin Chim Acta. 2016;452:18–26. doi: 10.1016/j.cca.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 161.Silva CL, Perestrelo R, Silva P, Tomas H, Camara JS. Volatile metabolomic signature of human breast cancer cell lines. Sci Rep. 2017;7:43969. doi: 10.1038/srep43969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Provance OK, Lewis-Wambi J. Deciphering the role of interferon alpha signaling and microenvironment crosstalk in inflammatory breast cancer. Breast Cancer Res. 2019;21:59. doi: 10.1186/s13058-019-1140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Eiro N, Cid S, Fraile M, Cabrera JR, Gonzalez LO, Vizoso FJ. Analysis of the gene expression profile of stromal pro-tumor factors in cancer-associated fibroblasts from luminal breast carcinomas. Diagnostics. 2020;10:865. [DOI] [PMC free article] [PubMed]
- 164.Zhu Y, Yang L, Chong QY, Yan H, Zhang W, Qian W, et al. Long noncoding RNA Linc00460 promotes breast cancer progression by regulating the miR-489-5p/FGF7/AKT axis. Cancer Manag Res. 2019;11:5983–6001. doi: 10.2147/CMAR.S207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Biello F, Platini F, D’Avanzo F, Cattrini C, Mennitto A, Genestroni S, et al. Insulin/IGF axis in breast cancer: clinical evidence and translational insights. Biomolecules. 2021;11:125. [DOI] [PMC free article] [PubMed]
- 166.Zhou Y, Jin X, Ma J, Ding D, Huang Z, Sheng H, et al. HDAC5 loss impairs RB repression of pro-oncogenic genes and confers CDK4/6 inhibitor resistance in cancer. Cancer Res. 2021;81:1486–99.. doi: 10.1158/0008-5472.CAN-20-2828. [DOI] [PubMed] [Google Scholar]
- 167.Li A, Liu Z, Li M, Zhou S, Xu Y, Xiao Y, et al. HDAC5, a potential therapeutic target and prognostic biomarker, promotes proliferation, invasion and migration in human breast cancer. Oncotarget. 2016;7:37966–78.. doi: 10.18632/oncotarget.9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen YX, Rong Y, Jiang F, Chen JB, Duan YY, Dong SS, et al. An integrative multi-omics network-based approach identifies key regulators for breast cancer. Comput Struct Biotechnol J. 2020;18:2826–35.. doi: 10.1016/j.csbj.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Karahalil B. Overview of systems biology and omics technologies. Curr Med Chem. 2016;23:4221–30.. doi: 10.2174/0929867323666160926150617. [DOI] [PubMed] [Google Scholar]
- 172.Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinformatics. 2018;19:1370–81.. doi: 10.1093/bib/bbx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Worheide MA, Krumsiek J, Kastenmuller G, Arnold M. Multi-omics integration in biomedical research - a metabolomics-centric review. Anal Chim Acta. 2021;1141:144–62.. doi: 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data and information related to this paper may be requested from the authors.