Abstract
A clinical strain of Escherichia coli (Ec GCE) displayed resistance to cefoxitin, cefotetan, cefotaxime, and ceftazidime. Susceptibility was not restored by the addition of clavulanic acid. Two β-lactamases with apparent pIs of 5.4 and 6.4 were identified; the β-lactamase with a pI of 6.4 was transferred by conjugation and associated with a 40-kb plasmid. Analysis of the nucleotide sequence showed a new ampC β-lactamase gene that is closely related to those encoding the FOX-3, FOX-2, and FOX-1 β-lactamases but whose product has four novel amino acid mutations, at positions 11 (M→T), 43 (A→E), 233 (V→A), and 280 (Y→H). This first cephamycinase from Spain was named FOX-4.
Plasmid-mediated class A β-lactamases derived from the commonly found TEM and SHV enzymes have been extensively reported (9, 10). They confer resistance to extended-spectrum cephalosporins and aztreonam, but cephamycins are unaffected. The enzymes remain susceptible to inhibition with the addition of clavulanic acid.
Plasmid-mediated class C β-lactamases have been reported in the last decade for Klebsiella pneumoniae, Escherichia coli, Salmonella spp., and Proteus spp. (2, 3, 4, 7, 14, 24). These β-lactamases show sequence similarities to AmpC β-lactamases (1) of Enterobacter cloacae (ACT-1 and MIR-1), Citrobacter freundii (CMY-2, BIL-1, LAT-2, and LAT-1), and Pseudomonas aeruginosa (CMY-1, FOX-1, and MOX-1) and confer a similar resistance pattern to that of AmpC chromosomally mediated enzymes (8).
An E. coli strain (Ec GCE) that is resistant to a broad spectrum of β-lactam antibiotics, including cephamycins, was isolated recently at Hospital Insular in Las Palmas de Gran Canaria (Canary Islands, Spain) from a patient suffering from a urinary tract infection who was previously treated with cloxacillin, aztreonam, and cefotaxime for an abdominal surgical infection. Our results showed that this strain harbored a new plasmid-encoded cephamycin-hydrolyzing β-lactamase, the first plasmid-mediated cephamycinase isolated in Spain.
Susceptibility to β-lactams.
Antibiotic susceptibility patterns of the Ec GCE clinical strain, as well as its transconjugant and transformant, are shown in Table 1. MICs were determined by the agar dilution method as recommended by the National Committee for Clinical Laboratory Standards (18). Antibiotics were provided as powders by the corresponding manufacturers.
TABLE 1.
MICs of β-lactams for the Ec GCE clinical strain, E. coli MC4100(pGC1), E. coli TG1, and E. coli TG1(pGC-2)
| Antibiotica | MIC (μg/ml) for:
|
|||
|---|---|---|---|---|
| Ec GCE (produces TEM-1 + FOX-4) | MC4100 (pGC1) (produces FOX-4) | TG-1 | TG1 (pGC-2) (produces FOX-4) | |
| Ampicillin | >1,024 | 64 | 4 | 256 |
| Ampicillin + clavulanate | 64 | 64 | 4 | 256 |
| Ticarcillin | >1,024 | 64 | 4 | 512 |
| Temocillin | 4 | 2 | 2 | 16 |
| Cefoxitin | 512 | 256 | 4 | >512 |
| Cefoxitin + clavulanate | 512 | 256 | 4 | >512 |
| Cefotetan | 64 | 32 | 0.2 | 128 |
| Moxalactam | 8 | 4 | 0.06 | 32 |
| Moxalactam + clavulanate | 8 | 4 | 0.06 | 32 |
| Cefotaxime | 32 | 16 | 0.03 | 64 |
| Cefotaxime + clavulanate | 32 | 16 | 0.03 | 64 |
| Ceftazidime | 128 | 64 | 0.12 | >128 |
| Ceftazidime + clavulanate | 128 | 64 | 0.12 | >128 |
| Cefepime | 1 | 0.5 | 0.03 | 2 |
| Aztreonam | 16 | 8 | 0.03 | 64 |
| Imipenem | 0.12 | 0.25 | 0.12 | 0.5 |
| Meropenem | 0.03 | 0.03 | 0.03 | 0.12 |
Clavulanate was used at 4 μg/ml.
Clavulanic acid did not act synergistically with ampicillin and cephalosporins on E. coli MC4100 and TG1 strains harboring pGC-1 and pGC-2 plasmids, respectively. A slight synergistic effect was observed only with ampicillin for the clinical strain Ec GCE, which produced a TEM-1 enzyme in addition to the AmpC β-lactamase. In addition, high levels of resistance to cefoxitin and cefotetan were observed, together with a moderate level of resistance to moxalactam. Higher MICs were obtained with the E. coli TG1 transformant harboring the pGC-2 plasmid, probably due to more copies of the ampC gene.
Isoelectric focusing.
Isoelectric focusing was performed in polyacrylamide gels containing Ampholine with a pH range of 3.5 to 9.5, as previously described (17). The clinical isolate produced two enzymes, one with a pI of 5.4 (TEM-1 type) and one with a pI of 6.4 (AmpC type).
Conjugation experiments.
In this study the E. coli MC4100 strain, with nalidixic acid and kanamycin resistances for markers, was used in the conjugation experiments and E. coli TG1 was used as the host for cloning the ampC gene. Plasmid pBGS18− (23), with a kanamycin resistance marker, was used for cloning the β-lactamase gene.
The Ec GCE clinical strain had two plasmids. Plasmid DNA from Ec GCE, which harbored the β-lactamase with a pI of 6.4, was transferred by conjugation into E. coli MC4100, with kanamycin (256 μg/ml), nalidixic acid (50 μg/ml), and ceftazidime (16 μg/ml) as selector antibiotics. A few transconjugants grew which harbored an identical plasmid of approximately 40 kb, named pGC-1. The other plasmid from the Ec GCE clinical strain carried the blaTEM-1 gene and would probably be transferred if an appropriate antibiotic (i.e., ampicillin) was used for selection. The presence of the blaTEM-1 gene in the Ec GCE strain was demonstrated by PCR assay with specific blaTEM primers C1 (5′-GGGAATTCTCGGGGAAATGTGCGCGGAAC) and C2 (5′-GGGATCCGAGTAAACTTGGTCTGACAG).
Cloning experiments and nucleotide sequencing.
Plasmid DNA was isolated by the alkaline lysis method (21) from the ceftazidime-resistant transconjugant that produced a single β-lactamase with a pI of 6.4. Plasmid DNA was digested with HindIII, and the resulting fragments were ligated to the plasmid pBGS18; which was previously digested with the same restriction enzyme. The ligation mixture was introduced into E. coli TG1 by transformation with CaCl2, and transformants were detected on Luria-Bertani agar plates supplemented with ceftazidime (4 μg/ml) and kanamycin (50 μg/ml). The resultant plasmid carrying the bla gene was named pGC-2. The molecular size of the insert was estimated by using restriction enzymes and electrophoresis on 1% agarose gels.
Double-stranded templates were subjected to nucleotide sequencing by the method of Sanger et al. (22). Sequencing was carried out with the Taq DyeDeoxiTerminator cycle sequencing kit and specific primers to the coding sequence, and the sequence was analyzed in an automatic DNA sequencer (377 Abi-Prism; Perkin-Elmer).
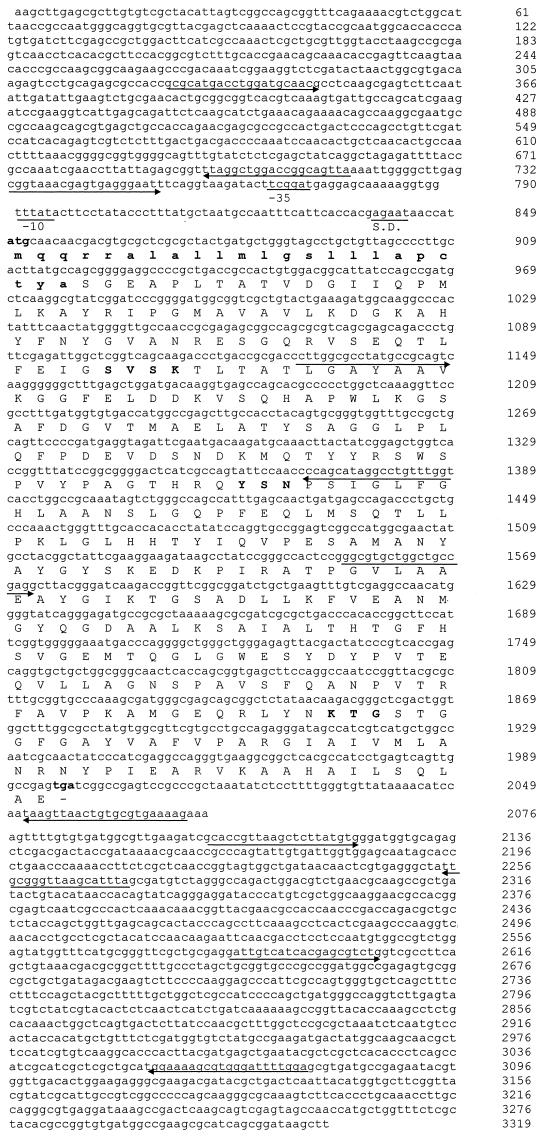
Several transformants harbored the recombinant pGC-2 plasmid, with an insert of about 3.3 kb. In isoelectric focusing, the pI 6.4 β-lactamase activity band from the E. coli transformant cofocused with the β-lactamase activity band from the Ec GCE clinical strain. Nucleotide sequencing of the 3.3 kb insert (Fig. 1) revealed, in addition to a new bla gene, the presence of ORF341 and a conserved integron sequence previously identified in the In7 integron (2, 19). This new bla gene was 1,149 bp long (Fig. 1), initiated with an ATG codon, and ended with a TGA codon (382 amino acids long). The initiation codon was preceded by a Shine-Dalgarno ribosome-binding sequence, GAGAA, and putative −10 and −35 promoter regions, TTTATA and TTCGGAT, respectively. EMBL and SwissProt database searches for this open reading frame revealed similarities with several class C chromosome- and plasmid-mediated β-lactamases. The new protein had the greatest homology with FOX-2, FOX-3, and FOX-1 β-lactamases (97.4, 96.9, and 95.3% homology), AER14 and CEPS β-lactamases (20, 25) from Aeromonas sobria (76 and 74% homology), and to a lesser extent, CMY-1 (5), MOX-1 (13), and AmpC from P. aeruginosa (73 to 55% homology) (15) and AmpC (6) from Acinetobacter baumannii (41.6% homology).
FIG. 1.
Nucleotide sequence of the 3.3-kb fragment. The deduced amino acid sequence of FOX-4 β-lactamase is shown in the line below the nucleotide triplets. The boldface ATG and TGA represent the initiation and termination codons, respectively. A putative Shine-Dalgarno (S.D.) ribosomal recognition site and −10 and −35 consensus sequences are indicated. The positions of the primers used to sequence the gene are indicated by arrows. The β-lactamase active site SVSK, the conserved triad KTG, and the typical class C motif YXN are presented in boldface. Nucleotides 1 to 64 and 2353 to 3319 correspond to ORF341, previously identified in the In7 integron. Nucleotides 65 to 808 and 2059 to 2352 correspond to a conserved region in the same In7 integron (EMBL database accession no. L06418).
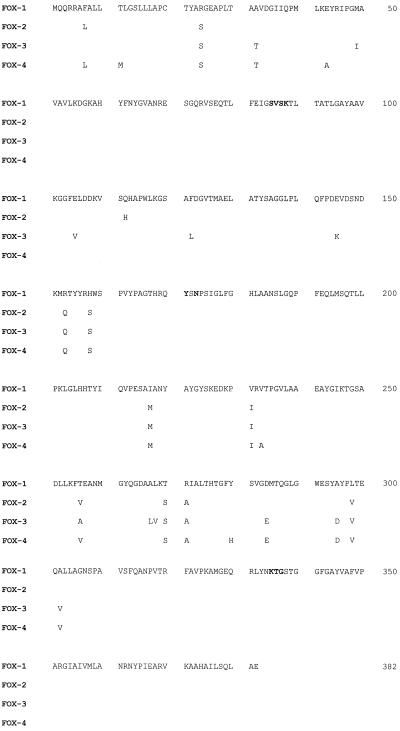
Multiple alignment was calculated with the Clustal V program (12). Figure 2 shows the amino acid replacements between FOX β-lactamases. In FOX-4, there are four novel mutations, at positions 11 (M→T), 43 (A→E), 233 (V→A), and 280 (Y→H). The other FOX-4 amino acid changes are found in positions previously described for FOX-2 and FOX-3 β-lactamases, although in some positions the amino acid changes were different from those previously reported.
FIG. 2.
Amino acid sequences of the FOX enzymes. Amino acid replacements are indicated. The β-lactamase active site SVSK, the conserved triad KTG, and the typical class C motif YXN are presented in boldface.
Determination of the β-lactamase kinetic constants Km, Vmax, and hydrolysis rate.
The substrate profile of the β-lactamase was determined with enzyme that was partially purified by G100 Sephadex (Pharmacia Fine Chemicals AB, Uppsala, Sweden), as previously described (6). Initial hydrolysis rates were monitored spectrophotometrically (UVIKON-930) at 25°C in 0.05 M phosphate buffer (pH 7.4) (14). Kinetic parameters were determined in duplicate experiments based on the initial steady-state rates at different substrate concentrations (Lineweaver-Burk transformation).
FOX-4 showed a hydrolytic profile similar to that expected for a class C molecular cephalosporinase (Table 2). The enzyme hydrolyzed cephaloridine with a Vmax 500-fold higher than that for ampicillin and only 30-fold higher than that for cefoxitin. This enzyme also showed moderate hydrolytic activity against cefotaxime, ceftazidime, and cefepime and very little hydrolytic activity against moxalactam and imipenem. A low Km value for cefotaxime (1.23 μM) was responsible for a high hydrolytic efficiency (Vmax/Km) for this antibiotic which was approximately equal to that of cephaloridine. Interestingly, a very high hydrolytic efficiency was obtained with cefoxitin, which is similar to results obtained with the FOX-1 β-lactamase (14). The hydrolysis rates for moxalactam and imipenem were very low, thus preventing the determination of reliable Km and Vmax values.
TABLE 2.
Kinetic parameters of FOX-4 β-lactamasea
| Antibiotic | Km (μM) | Relative Vmaxb (%) | Vmax/Km | Relative Vmax/Kmb (%) | Hydrolysis ratec (μmol/min/μl) | Relative hydrolysis rateb (%) |
|---|---|---|---|---|---|---|
| Cephaloridine | 1,400 | 100 | 357 | 100 | 92,900 | 100 |
| Ampicillin | 23.1 | 0.2 | 43.3 | 12.1 | 546 | 0.58 |
| Cefoxitin | 36.7 | 3.3 | 454 | 127.3 | 489 | 0.52 |
| Cefotaxime | 1.23 | 0.09 | 379 | 106 | 393 | 0.42 |
| Ceftazidime | 14.4 | 0.25 | 87 | 24.4 | 769 | 0.82 |
| Cefepime | 116 | 0.08 | 3.45 | 0.97 | 366 | 0.39 |
| Moxalactam | NDd | ND | ND | ND | 6.45 | 0.0069 |
| Imipenem | ND | ND | ND | ND | 40.8 | 0.043 |
Kinetic experiments were performed using a 9.5-mg/ml cellular extract with a specific activity of 232,937 μmol of nitrocefin/min/μg of protein.
Normalized with respect to cephaloridine (taken as 100%).
Hydrolysis rates were determined by using 100 μM concentrations of the indicated substrates.
ND, not done.
Comparative studies.
In recent years, ampC genes have been found in conjugative plasmids, mostly among K. pneumoniae and E. coli strains but also in Klebsiella oxytoca, Salmonella enterica serovar Enteritidis, and Proteus mirabilis (2, 3, 4, 7, 14, 24). The nucleotide and deduced amino acid sequences of the β-lactamase characterized here had very close homology with FOX-1, FOX-2, and FOX-3 β-lactamases (4, 14, 16). Moreover, the deduced peptide sequence of blaFOX-4 contained the common conserved motifs found in serine β-lactamases (11): the SXSK motif of the active site of AmpC, the typical class C motif YXN, and the KTG domain. In addition, kinetic experiments performed with the semipurified enzyme revealed typical cephalosporinase properties, especially a high hydrolytic efficiency of the enzyme for cefoxitin and cefotaxime. These biochemical results correlated well with the MICs of β-lactams for the clinical strain and its transconjugant and transformant. A high cefoxitin hydrolytic efficiency was also observed with the FOX-1 β-lactamase.
On the other hand, FOX-4 β-lactamase and all the FOX enzymes had the greatest homology to the ampC genes from A. sobria; therefore, the ampC β-lactamase genes from A. sobria may be ancestors of the FOX genes. However, it is possible that other ampC genes, from Enterobacteriaceae and other gram-negative rods, which have not yet been sequenced, may assume this role.
Recently, two new blaFOX genes have been identified (N. D. Hanson, P. Coudron, E. S. Moland, and C. C. Sanders, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1458, 1999; G. Jacoby, J. Tran, and M. Nato, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1481, 1999). Since we do not have enough experimental data to compare them, we named our enzyme FOX-4.
In summary, we report here the molecular characterization of a novel β-lactamase, FOX-4, from an E. coli clinical strain. To our knowledge, this is the first report of a cephamycinase in Spain.
Nucleotide sequence accession number.
The nucleotide sequence of the blaFOX-4 gene has been given the EMBL database accession no. AJ277535.
Acknowledgments
We thank L. De Rafael for his critical comments, Dolores Malpica for technical assistance, and Reyes Garcia for secretarial assistance.
REFERENCES
- 1.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond B. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 2.Barnaud G, Arlet G, Verdet C, Gaillot O, Lagrange P H, Philippon A. Salmonella enteritidis: AmpC plasmid-mediated inducible β-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob Agents Chemother. 1998;42:2352–2358. doi: 10.1128/aac.42.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Stemplinger I, Jungwirth R, Giamarellou H. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob Agents Chemother. 1996;40:221–224. doi: 10.1128/aac.40.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Wagner S, Jungwirth R, Schneider I, Meyer D. A novel class C β-lactamase (FOX-2) in Escherichia coli conferring resistance to cephamycins. Antimicrob Agents Chemother. 1997;41:2041–2046. doi: 10.1128/aac.41.9.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhem R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi: 10.1128/aac.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou G, Martínez-Beltrán J. Cloning, nucleotide sequencing, and analysis of the gene encoding an AmpC β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 2000;44:428–432. doi: 10.1128/aac.44.2.428-432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford P A, Urban C, Mariano N, Projan S J, Rahal J J, Bush K. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob Agents Chemother. 1997;41:563–569. doi: 10.1128/aac.41.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chanal C, Sirot D, Romaszko J P, Bret L, Sirot J. Survey of prevalence of extended spectrum beta-lactamases among Enterobacteriaceae. J Antimicrob Chemother. 1996;38:127–132. doi: 10.1093/jac/38.1.127. [DOI] [PubMed] [Google Scholar]
- 10.De Champs C, Sirot D, Chanal C, Poupart M C, Dumas M P, Sirot J. Concomitant dissemination of three extended-spectrum beta-lactamases among different Enterobacteriaceae isolated in a French hospital. J Antimicrob Chemother. 1991;27:441–457. doi: 10.1093/jac/27.4.441. [DOI] [PubMed] [Google Scholar]
- 11.Ghuysen J M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 12.Higgins D G, Bleasby A J, Fuchs R. Clustal V: improved software for multiple alignments. CABIOS. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 13.Horii T, Arakawa Y, Ohta M, Ichiyama S, Wacharotayankun R, Kato N. Plasmid-mediated AmpC-type β-lactamase isolated from Klebsiella pneumoniae confers resistance to broad-spectrum β-lactams, including moxalactam. Antimicrob Agents Chemother. 1993;37:984–990. doi: 10.1128/aac.37.5.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiza M G, Perez-Diaz J C, Ayala J, Casellas J M, Martinez-Beltran J, Bush K, Baquero F. Gene sequence and biochemical characterization of FOX-1 from Klebsiella pneumoniae, a new AmpC-type plasmid-mediated β-lactamase with two molecular variants. Antimicrob Agents Chemother. 1994;38:2150–2157. doi: 10.1128/aac.38.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodge J M, Minchin S D, Piddock L, Busby J W. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem J. 1990;272:627–631. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchese A, Arlet G, Schito G C, Lagrange P H, Philippon A. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob Agents Chemother. 1998;42:464–467. doi: 10.1128/aac.42.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 19.Parsons Y, Hall R M, Stokes H W. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid pDGO100. Antimicrob Agents Chemother. 1991;35:2436–2439. doi: 10.1128/aac.35.11.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen B A, Keeney D, Yang Y, Bush K. Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob Agents Chemother. 1994;38:2078–2085. doi: 10.1128/aac.38.9.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt B G, Hedge P J, Heesen T S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 24.Verdet C, Arlet G, Ben-Redjeb S, Ben-Hassen A, Lagrange P H, Philippon A. Characterisation of CMY-4, an AmpC-type plasmid-mediated β-lactamase in a Tunisian clinical isolate of Proteus mirabilis. FEMS Microbiol Lett. 1998;169:235–240. doi: 10.1111/j.1574-6968.1998.tb13323.x. [DOI] [PubMed] [Google Scholar]
- 25.Walsh T, Hall L, MacGowan A P, Bennett P M. Sequence analysis of two chromosomally mediated inducible beta-lactamases from Aeromonas sobria, strain 163a, one a class D penicillinase, the other an AmpC cephalosporinase. J Antimicrob Chemother. 1995;36:41–52. doi: 10.1093/jac/36.1.41. [DOI] [PubMed] [Google Scholar]