Abstract
Impressive achievements in clinical trials to treat hemophilia establish a milestone in the development of gene therapy. It highlights the significance of AAV-mediated gene delivery to liver. AAV5 is a unique serotype featured by low neutralizing antibody prevalence. Nevertheless, its liver infectivity is relatively weak. Consequently, it is vital to exploit novel AAV5 capsid mutants with robust liver tropism. To this aim, we performed AAV5-NNK library and barcode screening in mice, from which we identified one capsid variant, called AAVzk2. AAVzk2 displayed a similar yield but divergent post-translational modification sites compared with wild-type serotypes. Mice intravenously injected with AAVzk2 demonstrated a stronger liver transduction than AAV5, roughly comparable with AAV8 and AAV9, with undetectable transduction of other tissues or organs such as heart, lung, spleen, kidney, brain, and skeletal muscle, indicating a liver-specific tropism. Further studies showed a superior human hepatocellular transduction of AAVzk2 to AAV5, AAV8 and AAV9, whereas the seroreactivity of AAVzk2 was as low as AAV5. Overall, we provide a novel AAV serotype that facilitates a robust and specific liver gene delivery to a large population, especially those unable to be treated by AAV8 and AAV9.
Keywords: MT: delivery strategies, AAV5, directed evolution, library screening, liver tropism, low seroreactivity
Graphical abstract
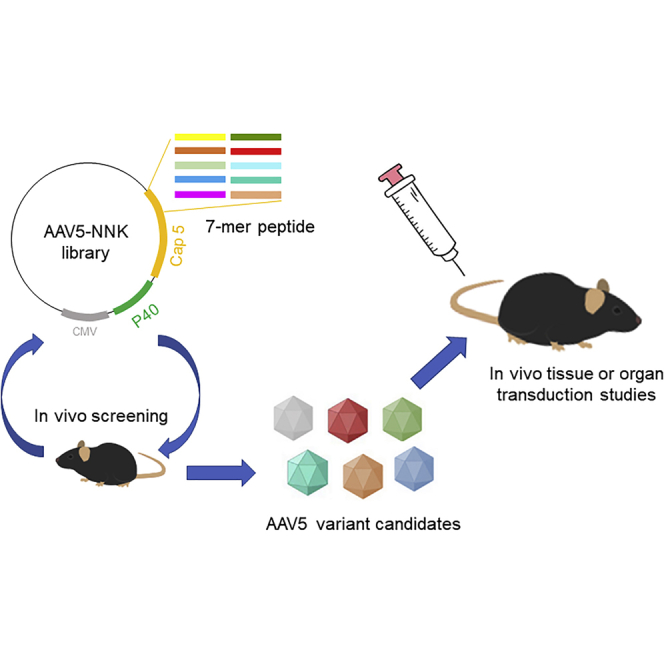
In this study, Wang et al. performed AAV capsid library screening in mice and identified a capsid variant, AAVzk2, which specifically transduces murine liver rather than other tissues or organs. Moreover, the seroreactivity of AAVzk2 was as low as AAV5, implicating its promising usage in a large population.
Introduction
Applications of adeno-associated virus (AAV) vectors in the treatment of liver diseases have drawn enormous attention in recent years, as featured by the impressive success in several clinical trials for hemophilia A and B.1, 2, 3, 4, 5 Fortunately, AAV administration at moderate doses is sufficient to restore blood coagulation safely in these cases.3, 4, 5 However, extensive usage of AAV beyond hemophilia may require more robust gene delivery into hepatocyte and less off-target tissue/organ transduction. Moreover, preexisting neutralizing antibodies (NAbs) against the AAV particles also set barriers to the application of gene therapy in a broader range of population.6, 7, 8 NAbs in the circulation are able to shield the receptor binding sites exposed on the capsid protein surface and nullify the therapeutic effect of the AAV vectors.9,10 Therefore, developing novel AAV serotypes with superior hepatocyte transduction efficiency is an interesting direction to take. During the past decade, many studies have tried various methods such as DNA shuffling, error-prone PCR, and directed point mutations to evolve natural capsid proteins.11, 12, 13, 14 Although improved liver transduction has been reported among their engineered AAV particles in primary human hepatocytes and human hepatocellular xenograft mouse models, preexisting NAbs still block the activity of these AAV serotypes.15 It is probably associated with the conserved epitope sequences from the natural AAV capsids, which is hard to eradicate.
AAV5 is a distinct serotype due to its relatively low amino acid identity compared with others, unique Rep protein cleavage site, divergent crystal structure, and its preference for the herpes simplex virus (HSV) as a helper in nature.16, 17, 18, 19, 20, 21 Interaction with 2,3-N-linked sialic acid containing glycans is a prerequisite for the entry of AAV5 into neuronal and airway epithelial cells, together with platelet-derived growth factor receptor (PDGFR) as a co-receptor.22, 23, 24 The top edge of AAV5 is the low preexisting NAbs levels among multiple groups of people, ranging from European to Asian and from healthy to hemophilia and HIV-infected patients.25, 26, 27, 28 In addition, in some clinical trials, sustained systemic Factor VIII levels were detected even in hemophilia patients with preexisting anti-AAV5 NAbs,29,30 implicating the potential ability of AAV5 to circumvent circulatory neutralizing factors, although the detailed mechanisms remain obscure. Therefore, despite the inferior transduction efficiency of murine and human hepatocytes to AAV3, AAV8, or AAV9,13,14 AAV5 is a promising backbone to exploit desired serotypes that are suitable for a large number of subjects. We previously combined error-prone PCR and shuffling to engineer AAV5.11 Whereas the directed evolution generated capsid mutants exhibiting profound transduction capability in Huh7 cells and human primary hepatocytes without impeding the advantage of mild seroreactivity, most of them displayed higher lung infectivity compared with AAV5, suggesting the lack of liver specificity.
Oligopeptide-insert library and barcode selections stand out with the advantage of high-throughput screening of more than 1 × 106 capsid variants both in vitro and in vivo. These methods have been successful in developing novel AAV serotypes with more splendid tissue and organ tropisms toward the brain, skeletal muscle, and retina as compared with wild-type AAVs.31, 32, 33, 34 To generate novel AAV5 capsid mutants with better liver specificity while retaining low seroreactivity and NAb prevalence, we tried the above methods with minor modifications. After 2 rounds of library screening and another single round of barcode selection in mice, we picked up a candidate with a 7-mer oligopeptide “FAPTPGP” after the amino acid Q574 on the variable region VIII of the AAV5 capsid (referred to as AAVzk2 hereafter), which was demonstrated to have no negative impact on virus production. However, mass spectrum analysis revealed significant differences in multiple post-translational modification (PTM) patterns between AAV5 and AAVzk2, implying distinct biological properties. Furthermore, the intravenous administration of the novel AAV vector in mice that expressed green fluorescent protein (GFP) enabled a robust and specific gene delivery in the liver, but not other tissues and organs such as the heart, lung, spleen, kidney, brain, and skeletal muscle. Gender differences had little effect on the biodistribution of AAVzk2. Additional study on human hepatocellular carcinoma and normal liver cell lines revealed a relatively higher transduction of the engineered serotype than AAV5, AAV8, and AAV9. Concomitantly, the prevalence of NAbs against the novel serotype was comparable to AAV5, significantly lower than AAV8 and AAV9, illustrating its potential usage in a large population group that is not able to be treated with AAV8 and AAV9. Overall, this engineered AAVzk2 possesses specific liver tropism with diminished off-target tissue/organ transduction and seroreactivity. It may pave the way for the extensive application of gene therapy in more liver diseases.
Results
Selection of liver tropic AAV5 capsid variant
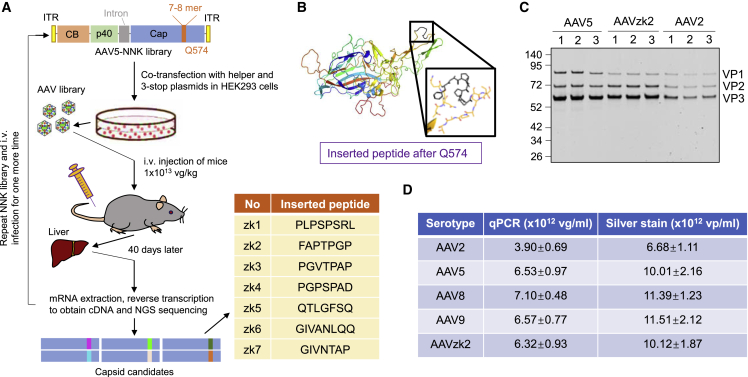
To develop novel AAV capsids, we generated an AAV5-NNK library by inserting the coding sequence of a 7- to 8-amino acid (aa) oligopeptide library after Q574 on the AAV5 capsid (Figure 1A), which has been reported as having an affinity with the sialic acid receptor binding pocket.23 We transformed TOP10 bacteria with the AAV5-NNK plasmid library, and approximately 5 × 106 distinct insert variants were estimated, which was deduced from the single clone sequencing result of 1/100 in volume of the transformed bacteria. The corresponding AAV library was produced by co-transfection of the AAV5-NNK library plasmid with AdHelper and 3-stop plasmid in a way similar to that of previous studies.31,32 We performed 2 rounds of in vivo screening and harvested the liver mRNA from the mice treated with the AAV5 variant library for next-generation sequencing (NGS) to obtain oligopeptide candidates. In the next step, each capsid candidate was distributed with a unique barcode in the cis-element plasmid encoding GFP as depicted in earlier studies33 and packaged into a distinct AAV serotype with GFP transgene and the barcode (Figure S1A). After the treatment of mice with these AAVs and quantification of the barcodes by qPCR, we found 1 variant, AAVzk2, that transduced murine liver impressively, with transgene mRNA levels more than 10-fold higher than the AAV5-treated group (Figure S1B). AAVzk2 harbors a 7-mer oligopeptide FAPTPGP inserted after Q574 of the AAV5 capsid. The inserted oligopeptide was expectedly exposed on the exterior region of loop 8 (Figure 1B), close to the variable region IV that contributes to the 3-fold axes protrusion.
Figure 1.
Directed evolution of AAV5 capsid and candidate selection
(A) Screening of AAV5 capsid mutants with liver tropism. The AAV5 capsid NNK library plasmid was transfected into HEK293 cells together with AdHelper and 3-stop plasmids. Mosaic AAVs in HEK293 medium and lysate were purified by iodixanol gradient ultracentrifugation and used to inject mice via tail vein with a dose of 1 × 1013 vg/kg. Liver mRNAs were harvested 40 days post-injection, reverse-transcribed to cDNA, and subjected to NGS sequencing after 2 rounds of selection. CB, hybrid CB promoter with CMV enhancer and β-actin promoter elements; p40, p40 promoter region in the AAV2 Rep sequence; ITR, inverted terminal repeats. Representative oligopeptide inserts are shown in the table. (B) PDB construction of the 3-dimensional (3D) structure of AAVzk2 VP1 protein by Chimera 1.15 (University of California, San Francisco). The inserted oligopeptide was highlighted and magnified in the black rectangle frame. (C) Silver staining of the indicated AAV particles produced by 2 rounds of iodixanol gradient ultracentrifugation, with each AAV serotype in 3 lanes. (D) Table illustrating that the indicated AAVs packaged from 2 × 108 cells (medium + lysates) were quantified by qPCR and silver staining. n = 3. vg, vector genome; vp, viral particle.
We then packaged a self-complementary GFP transgene cassette controlled by the chicken β-actin (CB) promoter (hybrid of cytomegalovirus [CMV] enhancer and the CB promoter, scCB-GFP hereafter) into AAV2, AAV5, AAV8, AAV9, and zk2 capsids. The corresponding AAV particles were purified by 2 rounds of iodixanol gradient ultracentrifugation. Silver staining showed clear VP1, VP2, and VP3 bands without impurities (Figure 1C), implicating the eligible quality of the purified virus. Quantification of both the vector genome (vg) and the viral particle (vp) revealed that the yield of AAVzk2 was not significantly lower than its parent serotype AAV5, even being similar to AAV8 and AAV9 (Figure 1D). The results indicate little effect of the inserted oligopeptide on viral production.
Discrepancies in post-translational modification sites between AAVzk2 and AAV5
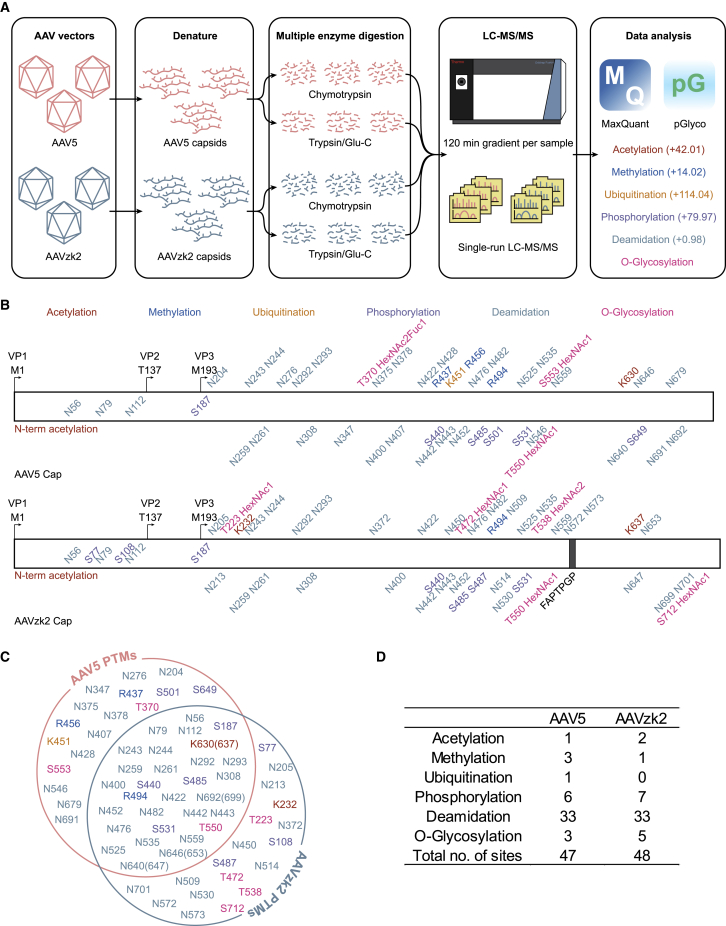
During vector production, AAV capsids acquire PTMs, which play a critical role in immunogenicity, targeting, functional activity, and stability. To determine whether the oligopeptide insertion could alter the capsid PTM patterns, we established a liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based workflow (Figure 2A). After digestion by chymotrypsin or trypsin/Glu-C, 6 types of PTMs, including acetylation, methylation, phosphorylation, ubiquitination, deamidation, and O-glycosylation, were analyzed by LC-MS/MS based on the b/y ion fragments. PTMs were distributed along the capsid proteins of the AAV5 and AAVzk2, and more PTMs were identified on the VP3 (Figure 2B). We identified protein N-terminal acetylation on both AAV5 and AAVzk2 capsids, which was similar to other AAV serotypes.35 In spite of a vast majority of conserved sites on both, we identified many unique PTMs only on AAV5 or AAVzk2 capsids individually, such as acetylation site K232 on AAVzk2 and methylation sites R437 and R456 on AAV5 (Figures 2C, S2A, and S2B). We also observed that residues S77, S108, and S487 were only phosphorylated on the AAVzk2. Residue K451 was only ubiquitinated on AAV5 capsids. Intriguingly, we failed to identify any ubiquitination site on the AAVzk2 capsids (Figures 2C, S2C, and S2D), which are widely known to degrade capsid proteins and impair transgene delivery. Furthermore, the most abundant PTM we characterized was asparagine deamidation, accounting for ∼70% of the total PTMs (33/47 for AAV5 and 33/48 for AAVzk2; Figures 2D and S2E). By means of pGlyo analysis, we found multiple O-glycosylation structures on the capsid sequences, featured by HexNAc2Fuc1 on the T370 of AAV5 that was not observed on AAVzk2 (Figures 2B, 2C, and S2F). To summarize, PTM patterns vary magnificently between AAV5 and AAVzk2, although only a 7-mer difference merges on their aa sequences.
Figure 2.
AAV5 and AAVzk2 capsids were post-translationally modified and exhibited differential PTM patterns
(A) A schematic workflow for mass spectrometry-based PTM analysis of AAV5 and AAVzk2 capsids. (B) PTM residue positions along the capsid sequences of the AAV5 and AAVzk2 from the N to the C terminus. Different colors represent different PTMs (red, acetylation; blue, methylation; orange, ubiquitination; purple, phosphorylation; light blue, deamidation; violet, O-glycosylation). The inserted sequence FAPTPGP in AAVzk2 capsid is highlighted in the gray box. (C) Common and unique capsid PTMs for AAV5 and AAVzk2 capsids. The color and its corresponding PTMs were the same as those in (B). (D) The number of the individual PTM site detected in AAV5 and AAVzk2 capsids.
Specific liver tropism of AAVzk2 in mice
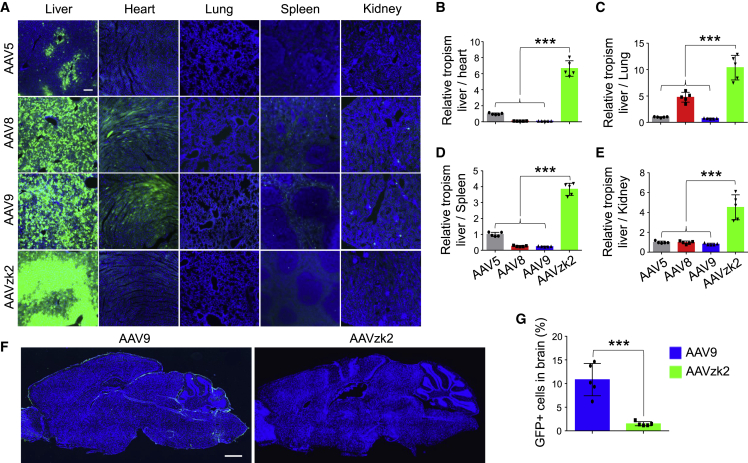
To study global transduction in vivo comprehensively, we treated male mice with the indicated AAV encoding GFP via tail vein injection at a dosage of 1 × 1013 vg/kg. Three weeks post-administration, we found that 92.25% hepatocytes were GFP positive in the AAVzk2-treated group, dramatically superior to the AAV5-treated liver—only 22.64% expressed GFP. This level was even comparable with the AAV8- (85.37%) and AAV9- (88.11%) treated groups (Figure 3A). Nevertheless, unlike AAV8 and AAV9 demonstrating splendid transduction in the heart and mild to moderate infectivity of the lung, spleen, and kidney, there were few GFP-positive cells in those tissues/organs of the AAVzk2-treated mice, implicating the specificity of liver transduction. Thus, we quantified the relative tropisms between the liver and the heart, lung, spleen, and kidney by calculating the ratios of GFP-positive cells in the liver versus other tissues/organs. Intriguingly, the relative tropisms of liver versus heart, lung, spleen, and kidney of AAVzk2 were 4- to 11-fold higher than AAV5, AAV8, and AAV9 (Figures 3B–3E). Furthermore, AAVzk2 exhibited little brain infectivity at dosages up to 5 × 1013 vg/kg (Figures 3F and 3G). A discrepancy has been observed between genders regarding AAV transduction of murine livers.36,37 The copy number of vgs delivered by AAV into male livers could be several-fold higher than those into female livers, probably associated with the androgen levels. To illustrate whether the liver transduction of AAVzk2 would be diverse in females, we intravenously injected the female mice with the indicated AAV particles and found that 68.4% ± 8.6% of the hepatocytes were transduced by AAVzk2 (Figure S3A). Although the level was lower than AAV8 (78.4% ± 3.6%) and AAV9 (82.6% ± 3.1%), few cells in the heart, lung, spleen, kidney, and brain were infected by AAVzk2 as opposed to AAV8 and AAV9 (Figures S3B–S3G), consolidating the specific liver transduction of AAVzk2 in females.
Figure 3.
Remarkable murine liver infectivity of AAVzk2 in male mice
(A) Male C57BL/6 mice were injected via tail vein with the indicated AAVs that carry cis-element cassette encoding GFP at the dose of 1 × 1013 vg/kg, and liver, heart, lung, spleen, and kidney were subjected to GFP (green) and DAPI (blue) immunostaining 21 days post-viral injection. Scale bar, 200 μm. (B–E) Quantification of AAV tropism in liver relative to heart (B), lung (C), spleen (D), and kidney (E), which was referred to as the ratio of GFP-positive liver cells versus GFP-positive heart, lung, spleen, and kidney cells based on DAPI spots number. n = 5 mice per group. ∗∗∗p < 0.001, One-way ANOVA. (F) Male C57BL/6 mice were intravenously injected with 5 × 1013 vg/kg, the indicated AAVs encoding GFP, and whole-brain sagittal sections were stained for GFP (green) and DAPI (blue) 21 days post-viral injection. Scale bar, 1 mm. (G) Quantification of GFP-positive cells in the brain in (F), n = 5 mice per group. ∗∗∗p = 0.0003, unpaired t test.
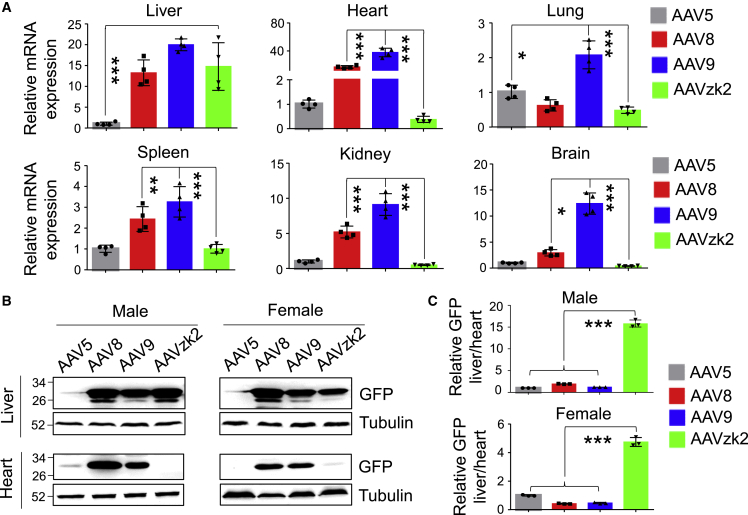
We also tested the GFP expression in the tissues and organs of the AAV-treated male mice. Compared with AAV5, the GFP mRNA levels were 14.7 ± 5.7-fold higher in AAVzk2, 13.3 ± 3.1-fold higher in AAV8 and 19.98 ± 1.4-fold higher in AAV9-treated livers (Figure 4A). In contrast, the GFP mRNA levels in the AAVzk2-treated heart, lung, spleen, kidney, and brain were similar to or less than AAV5, significantly lower than AAV8 and AAV9 groups. Moreover, a western blot showed similar GFP protein levels among AAV8-, AAV9-, and zk2-treated livers, whereas remarkable GFP expression was observed only in AAV8 and AAV9, but not in AAV5- and zk2-treated hearts (Figures 4B and 4C). The trend of biodistribution was recapitulated in female mice (Figure S4). Altogether, the results suggest a specific tropism of AAVzk2 toward the liver, but not the heart, lung, spleen, kidney, and brain for both genders.
Figure 4.
Selective GFP mRNA and protein expression in the livers of AAVzk2-treated mice
(A) Male C57BL/6 mice were injected via tail vein with the indicated AAV serotypes encapsulating GFP transgene at a dose of 1 × 1013 vg/kg, and liver, heart, lung, spleen, kidney, and brain were subjected to qRT-PCR to detect GFP expression, in which the GFP mRNA levels of the AAV8, AAV9, and AAVzk2 groups were normalized to the AAV5 group. n = 4 mice per group. (B) Western blot to detect GFP protein expression in the livers and hearts from the mice treated with the indicated AAVs. (C) Quantification of GFP protein levels in the liver relative to the heart. n = 4 male and 4 female mice per group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, One-way ANOVA.
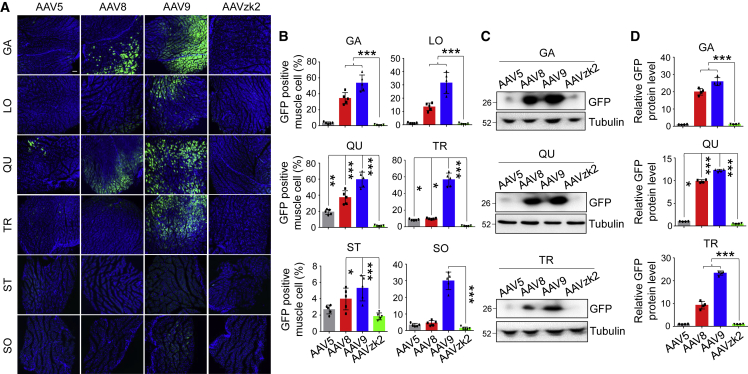
Dimmed skeletal muscle transduction of AAVzk2
Skeletal muscle is the largest tissue, accounting for 35%–40% of total body weight.38 Metabolic and functional issues of the muscle cells trigger multiple disorders such as muscular dystrophies and congenital myasthenic syndromes.39 The current application of AAV5 in the treatment of hemophilia has exhibited a relatively high dose, ranging from 2 × 1013 vg/kg to 6 × 1013 vg/kg,1, 2, 3 implying the potential risk of off-target gene delivery into muscle cells. To investigate whether AAVzk2 could exert any impact on skeletal muscles via systemic administration, we treated the mice with a relatively high dose (5 × 1013 vg/kg) of the indicated AAVs. Muscles from multiple regions of the body, including the hindlimb (gastrocnemius, quadriceps, soleus), the back (longissimus thoracis), the forelimb (triceps), and the neck (sternocleidomastoid), were stained with GFP. As shown in Figures 5A and 5B, AAV9 displayed the best transduction efficiency, with ∼60% of the gastrocnemius, quadriceps, and triceps muscle cells and 30%–40% soleus and longissimus thoracis muscle cells successfully expressing GFP; AAV8 ranked second, with ∼15%–40% gastrocnemius, quadriceps, and longissimus muscle cells effectively transduced. Even in AAV5-treated mice, the percentage of GFP-positive quadriceps muscle cells was 19.19%. In contrast, in all of the muscles of the AAVzk2-treated group, negligible numbers of muscle fibers were transduced. To further evaluate GFP expression, we performed western blot to test the GFP protein levels in gastrocnemius, quadriceps, and triceps, the three types of skeletal muscles that AAV transduced more efficiently (Figure 5A). Consistently, GFP protein bands were hardly visible in AAV5- and AAVzk2-treated gastrocnemius, quadriceps, and triceps, as opposed to the AAV8- and AAV9-treated muscles (Figures 5C and 5D), suggesting little GFP protein expression in the muscle cells faced with circulatory AAV5 and AAVzk2. These observations further consolidate the specific liver transduction property of AAVzk2.
Figure 5.
Negligible skeletal muscle cells transduced by AAVzk2 in vivo
(A) C57BL/6 mice of both genders were intravenously injected with 5 × 1013 vg/kg, the indicated AAV serotypes encapsulating the GFP transgene. Multiple skeletal muscles were subjected to immunostaining 21 days post-viral treatment. The types of muscles were as follows: GA, gastrocnemius; LO, longissimus thoracis; QU, quadriceps; TR, triceps; ST, sternocleidomastoid; SO, soleus. Scale bar, 200 μm. (B) Quantification of the muscle cells effectively transduced by AAV based on the percentage of GFP-positive muscle cells versus total muscle area. n = 5 mice (3 females and 2 males) per group, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, 1-way ANOVA. (C) Western blot to detect muscular GFP protein levels in the AAV-treated mice. (D) Quantification of the relative GFP protein level in (C). n = 4 mice per group. ∗p < 0.05, ∗∗∗p < 0.001, One-way ANOVA.
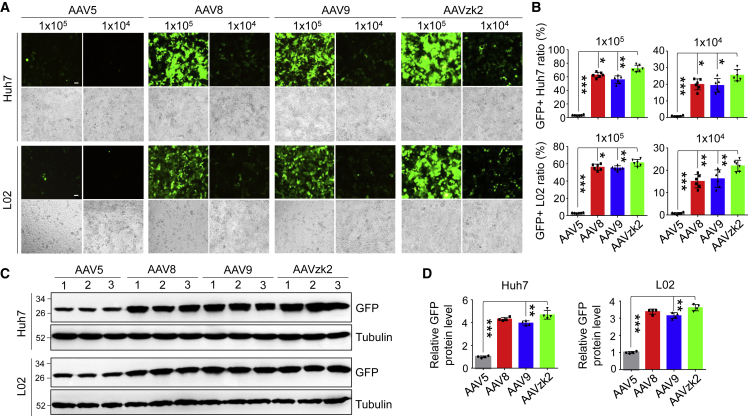
Robust transduction of human liver cell lines by AAVzk2
To further test the transduction efficiency of AAVzk2 in human liver cells, we selected a human hepatocellular carcinoma cell line, Huh7, and a normal liver cell line, L02, that have been frequently applied to studies on liver drug delivery.11,40, 41, 42, 43 As shown in Figures 6A and 6B, The GFP-positive cell ratio of the AAVzk2 group, which refers to the GFP signal area versus total cell area, was modestly more than the AAV8 and AAV9 groups, but dramatically (21.51-fold for Huh7 cells and 20.83-fold for L02 cells) higher than the AAV5-treated cells at the multiplicity of infection (MOI) of 1 × 105 vg/cell. When the MOI declined to 1 × 104 vg/cell, the percentages of Huh7 and L02 cells transduced by AAVzk2 were 25.51% and 21.98%, respectively, which is still significantly higher than the wild-type AAV groups (0.84% and 0.74% for AAV5-, 19.96% and 15.07% for AAV8-, 19.35% and 16.27% for AAV9-treated Huh7 and L02 cells). Similar results were observed at the protein level, while the total GFP protein delivered by AAVzk2 was 4.68- and 3.61-fold higher than AAV5 into Huh7 and L02 cells, respectively (Figures 6C and 6D). Furthermore, the cell binding and internalization assays revealed significantly more AAVzk2 viral particles bound to and internalized by the Huh7 cells than AAV5, AAV8, and AAV9 (Figures S5A and S5B), suggesting cellular mechanisms underlying the superior hepatocyte transduction of AAVzk2. To further investigate the physicochemical properties of AAVzk2, the purified viral particles underwent a gradual increase in temperature from 30°C to 100°C, when the ratio of fluorescent signals at 350 nm versus 330 nm was measured to reflect conformational changes and the denaturation of capsid proteins, an index of thermostability. F350/F330 displayed a sharp increase since 70°C for AAVz2, and the melting temperature (Tm) value was 75.39°C (Figure S5C), which was close to AAV8 and AAV9 (79.59°C and 77.9°C, respectively) rather than AAV5 (88.5°C), implicating a drastic alteration in thermostability stemming from oligopeptide insertion.
Figure 6.
Robust human hepatocellular transduction property of AAVzk2
(A) Human hepatocellular carcinoma cell Huh7 and normal liver cell L02 were infected by the indicated AAV particles encapsulating the GFP gene at a multiplicity of infection (MOI) of 1 × 105 and 1 × 104 vg/cell, respectively. Images were captured 48 h post-virus treatment. Upper panel, GFP signal; lower panel, bright field. Scale bar, 100 μm. (B) Quantification of GFP-positive cell percentage. The ratios of GFP signal area of Huh7 and L02 cells to the whole bright-field area were quantified at an MOI of 1 × 105 and 1 × 104 vg/cell, respectively. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. n = 6 wells of cells. One-way ANOVA. (C) Lysates of Huh7 and L02 cells treated with the indicated AAV particles at an MOI of 1 × 105 vg/cell were subjected to immunoblotting for GFP. Three lanes were performed for each AAV-treated group. (D) Quantification of relative GFP protein levels in (C). ∗∗p < 0.01, ∗∗∗p < 0.001. n = 4 wells of cells. One-way ANOVA.
Low seroreactivity retained by oligopeptide insertion
The preexisting NAbs in serum have elicited some major concerns regarding the deactivation of AAV vectors used for gene therapy. AAV5 stands out with the lowest circulatory NAb levels, with a prevalence of 3% in certain population groups.25 To investigate the effect of oligopeptide insertion on the capsid seroreactivity, we collected sera samples from 10 hemophilia B patients, a representative disease requiring liver gene delivery by AAV vectors. These sera samples were diluted at a 1:2 ratio and mixed with the indicated AAV vectors encoding luciferase, followed by the treatment of Huh7 cells with the virus-serum mixture. The highest serum dilution ratio that inhibits luciferase activity by at least 50% was defined as the NAbs titer. As shown in Table 1, we found that 4/10 of the patients demonstrated NAb titers higher than 1:4 against AAV8 and 9, which was determined as NAb positive. However, none of the patients were NAb positive against AAV5 and AAVzk2. Moreover, we also checked the NAb prevalence in 21 healthy rhesus monkeys. There were 3/21 monkeys NAbs positive against AAV5, 4/21 monkeys NAbs positive against AAV8, 9/21 monkeys NAbs positive against AAV9, and 1/21 monkey NAbs positive against AAVzk2 (Table S1). These results illustrate little effect of the inserted oligopeptide FAPTPGP on the capsid seroreactivity. AAVzk2 retains low NAb prevalence similar to AAV5.
Table 1.
Anti-AAV neutralizing antibody titers in hemophilia B patients’ sera
| Patient ID | Anti-AAV5 | Anti-AAV8 | Anti-AAV9 | Anti-AAVzk2 |
|---|---|---|---|---|
| 1 | <1:1 | 1:16 | 1:16 | 1:2 |
| 2 | <1:1 | <1:1 | <1:1 | <1:1 |
| 3 | <1:1 | <1:1 | <1:1 | <1:1 |
| 4 | <1:1 | 1:2 | <1:1 | <1:1 |
| 5 | <1:1 | <1:1 | <1:1 | <1:1 |
| 6 | <1:1 | 1:8 | 1:16 | <1:1 |
| 7 | <1:1 | <1:1 | <1:1 | <1:1 |
| 8 | 1:1 | 1:32 | 1:32 | 1:4 |
| 9 | <1:1 | 1:32 | 1:32 | 1:1 |
| 10 | <1:1 | <1:1 | <1:1 | <1:1 |
The samples with neutralizing antibody titers >1:4 (NAb positive) are indicated in boldface type.
Discussion
In this study, we performed library screening in mice and identified a novel AAV5 capsid mutant by inserting a 7-aa-oligopeptide FAPTPGP on the variable region VIII after Q574, called AAVzk2. The production of this engineered serotype in suspension HEK293 cells was comparable with wild-type AAV serotypes such as AAV2, AAV5, AAV8, and AAV9. Systemic administration revealed a robust liver transduction of AAVzk2 in both male and female mice, whereas its infectivities of heart, lung, spleen, kidney, brain, and skeletal muscle were rarely observed. In addition, the capability of AAVzk2 to transduce human hepatocellular cell lines overwhelmed AAV5, AAV8, and AAV9, further supported by better cell binding and uptake. Concomitantly, the seroreactivities of AAVzk2 in human and non-human primates were at low levels comparable with AAV5. These results implicate the potential application of AAVzk2 in a broad range of population groups to safely and effectively treat liver diseases, particularly for those patients unable to receive AAV8 or AAV9, probably due to high NAb titers. Moreover, the LC-MS/MS detected great divergency in acetylation, methylation, phosphorylation, ubiquitination, deamidation, and O-glycosylation sites on the individual capsid proteins between AAV5 and AAVzk2. The thermostability of AAVzk2 was also lower than that of AAV5. These data indicate that the PTM patterns and physicochemical properties of AAVzk2 differ from its parental AAV5.
There has been remarkable progress in inventing novel capsids that target the skeletal muscle, retina, and central nervous system exceptionally well in the insertion of peptides on the variable region VIII of many AAV serotypes, such as AAV2 and AAV9.31, 32, 33, 34,44,45 However, sporadic successes have been achieved in the evolution of the AAV5 capsid, probably due to the sequence and structure diversities between AAV5 and other serotypes.18,20,46 Khabou and colleagues tried to transfer the 7m8 oligopeptide, which enhanced the retinal transduction of AAV2 in several species, onto the AAV5 capsid after aa 575, yet they barely observed any improvement.47 Introducing the PHP.B peptide onto AAV5, together with the dipeptide flanking sequence Ala-Gln from AAV9, still failed to increase blood-brain barrier permeability.48 Furthermore, Arnold and colleagues incorporated an oligopeptide in AAV5 that could be biotinylated in producer cells.49 Although this modification could facilitate efficient viral purification by streptavidin beads and impressive transduction of HeLa and rat glioma BT4C cells, in vivo applications are still insufficient. Here, we conducted AAV5-directed evolution by in vivo NNK library and barcode screening in mice. We succeeded in obtaining the candidate oligopeptide FAPTPGP, which could be inserted into variable region VIII of AAV5 capsid sequence and was capable of improving hepatocyte transduction in vitro and in vivo (Figures 3, 4, and 6); this indicated the compatibility of certain foreign oligopeptides in AAV5 variable region VIII to induce some gain-of-function effect. The thermostability of AAVzk2 was lower than that of AAV5 (Tm = 75.39°C versus 88.5°C; Figure S5C), which may imply that AAVzk2 are more prone to uncoating and releasing of vector genome into the host cell nucleus. Binding and internalization of AAVzk2 by Huh7 cells were also significantly better than AAV5 (Figures S5A and S5B), suggesting different transmembrane co-receptors for AAVzk2. Altogether, our results illuminate a cunning direction for future study either to manipulate AAV5 for extensive utility or to delve into the nature of this unique serotype.
PTMs contribute vitally to AAV transduction, stability, and tissue tropism.50, 51, 52 For example, AAV3 S663V + T492V and AAV1 S669A mutations could enhance human and murine hepatocyte transduction, while AAV5 S485A mutation leads to a switch from liver to lung.14,53 Multiple tyrosine mutations on the AAV2 and AAV9 capsids boost gene delivery into the central nervous system and retina, which could barely be recapitulated in AAV8 mutants harboring similar mutations,54, 55, 56 implying differences in phosphorylation sites and functions among capsids. Ubiquitination also plays a critical role in virus uncoating and replication.57 Proteasome inhibition facilitates AAV2- and AAV5-mediated gene transfer in hepatocytes and airway epithelial cells, but not cardiac or skeletal muscles.50,58,59 Site-directed mutagenesis of potentially ubiquitinated lysine residues on the AAV2 capsid protein surface ameliorates liver transduction in vivo.60 Glycosylation of the capsid proteins has been reported to regulate the life cycle of many categories of viruses. Although the detailed functions of glycosylation on AAV capsid proteins remain largely elusive, the perturbation of glycosylation by either pharmacological ways or site-directed mutagenesis evidently altered the AAV2 transduction of HeLa, Huh7, and ARPE-19 cells.61
Our LC-MS/MS dataset showed that both AAV5 and AAVzk2 capsids were post-translationally modified, and the PTM patterns differed from one another (including acetylation, methylation, ubiquitination, phosphorylation, deamidation, and O-glycosylation; Figures 2 and S2). We observed that residues S77 and S108 were only phosphorylated on the N terminus of VP1 in AAVzk2; residue K451 was ubiquitinated only on the AAV5 capsid, while no ubiquitination pattern was detected on the AAVzk2 capsid. As to glycosylation sites, although common sites such as T550 were observed on both AAV5 and AAVzk2 capsids, residues T370 and S553 were glycosylated only on AAV5 capsids, and T223, T472, T538, and S712 glycosylations were solely present on AAVT36 capsids. Nevertheless, the most abundant PTM observed was deamidation, accounting for ∼70% of total PTMs (Figures 2C and 2D). It has been reported that multiple asparagine to aspartic acid mutations that mimic deamidation could affect the liver transduction efficiency of AAV8 in vitro and in vivo.62 Consequently, the large variety of different deamidation sites, together with other PTM patterns, between AAV5 and AAVzk2, may explain at least to some extent the improved liver tropism of AAVzk2. Although it is time intensive to fully figure out which distinct PTM or the combination of several on the AAVzk2 capsid contribute to the enhanced murine liver transduction efficiency, current PTM results may still provide valuable clues to illuminate the nature of AAV5, expand our knowledge about the field, and guide future studies.
Materials and methods
Reagents and antibodies and AAV serotypes
Unless otherwise indicated, the chemicals mentioned here were purchased from Sigma-Aldrich (St. Louis, MO, USA). The antibodies used here were as follows: GFP (11814460001, 1:2,000 for western blot and 1:500 for staining) from Sigma-Aldrich; β-tubulin (sc101527, 1:3,000 for western blot) from Santa Cruz Biotechnology (Dallas, TX, USA); Alexa Fluor 488 goat anti-rabbit immunoglobulin G (IgG), Alexa Fluor 488 goat anti-mouse IgG (1:1,000 for immunofluorescence), horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG, and goat anti-mouse IgG antibodies (1:4,000 for western blot) were as previously described.63,64 The accession numbers of the AAV sequences were from the NCBI database as follows: AAV2 (NC_001401.2), AAV5 (NC_006152.1), AAV8 (NC_006261.1), and AAV9 (AY530579.1).
AAV production and quantification
AAV particles were produced by means of triple-plasmid transfection in the suspension HEK293 cell line and purified by iodixanol gradient ultracentrifugation, as previously described.64, 65, 66, 67 Briefly, the transgene plasmid (scCB-GFP construct, as previously reported68), Rep/Cap plasmid and AdHelper plasmid were transfected at the concentrations of 1 μg/mL, 1 μg/mL, and 2 μg/mL, respectively, when the HEK293 cell density reached 5 × 106 cells/mL. Cell lysates plus medium were subjected to benzonase (E8263, Sigma-Aldrich) digestion 72 h post-transfection and then loaded onto iodixanol gradients, followed by ultracentrifugation at 68,000 rpm for 2 h. AAV samples were recovered from the 40% iodixanol layer and then resolved to another round of iodixanol gradient ultracentrifugation. Finally, the AAV particles were switched to phosphate-buffered saline (PBS) containing 0.001% poloxamer 188 via Amicon 15-100,000 molecular weight cutoff (MWCO) concentration unit to a final volume of ∼1 mL.
The purified AAV samples underwent DNase I (10104159001, Roche, Basel, Switzerland) digestion for 30 min at 37°C. Then, the vg was quantified by qPCR with the GFP primers (F: 5′-CGACCACTACCAGCAGAACACC-3′, R: 5′-CGAACTCCAGCAGGACCATG-3′) based on the standard curve made from a serial dilution of linearized CB-GFP plasmid samples. The amount of vp capsid protein was determined by the Pierce Silver Stain Kit (Cat#24612, Thermo Scientific, Waltham, MA, USA) based on the standard curve made from a serial dilution of standard bovine serum albumin (BSA) protein. Unless otherwise indicated, VP3 was quantified on behalf of all of the capsid proteins.
Systemic administration of AAV vectors in mice
C57BL/6 mice (000664, Jackson Laboratory, Bar Harbor, ME, USA) of both genders were housed in a room with a 12-h light/dark cycle and ad libitum access to water and rodent chow diet (Diet 7097, Harlan Teklad, Madison, USA). The animal protocols were approved by the institutional animal care and use committee (IACUC) of the Department of Ophthalmology, Daping Hospital, Chongqing, China. Mice 6–8 weeks old were injected via the tail vein with the indicated AAV vectors. Insulin syringes (REF328421, BD, Franklin Lakes, NJ, USA) were used for injection. Unless otherwise indicated, the mice were sacrificed 21 days after AAV treatment to collect tissues and organs for the experiments that followed.
Generation and screening of AAV5 capsid variant library
The capsid library screening process was generally based on previous studies.31, 32, 33 Briefly, the AAV5-NNK library plasmid contains part of a CB promoter (568-bp sequence of CMV enhancer and CB promoter) followed by the p40 region from Rep 2 to drive the expression of capsid variants. To construct the library, the inserted oligopeptide coding sequences were synthesized as NNK primers by GenScript (F: 5-GCGTACAACGTCGGCGGGCAG-3′, R: 5′-CGCGGGGGCAGTGGTGGANNKNNKNNKNNKNNKNNKNNKCTGCCCGCCGACGTTGTACGC-3′), amplified by PCR (F plus R primers) and ligated to the plasmid backbone by the Gibson assembly, which was digested in advance by restriction enzymes EcoRV and AfeI. To generate the 3-stop plasmid, 3 stop codons were introduced into the AAV5 capsid nucleotide sequence at 36 bp after VP1, 15 bp after VP2, and 42 bp after VP3 translation start codons, respectively, to annihilate the expression of the VP1, VP2, and VP3 proteins. Afterward, HEK293 cells were transfected with 16 μg AdHelper plasmid, 8 μg 3-stop plasmid, 8 μg pUC19 plasmid, and 10 ng AAV5-NNK library plasmid per 2 × 107 cells. The AAV library was then purified by iodixanol gradient ultracentrifugation 72 h post-transfection.
To screen the AAV5-NNK library for liver-tropic candidates, 3 mice were injected intravenously with 1 × 1013 vg/kg the purified AAV library. Forty days post-viral treatment, mRNAs in the livers were extracted by the RNAprep Pure Tissue Kit (DP431, TIANGEN BIOTECH, Beijing, China) and the TIANSeq mRNA Capture Kit (NR105, TIANGEN) and reverse-transcribed to cDNA. The sequences of AAV5 capsid variants in the cDNA were then amplified by PCR to produce a 160-bp band for NGS sequencing, which was performed and analyzed by GENEWIZ (Azenta Life Sciences, Suzhou, China). The data have been deposited on the NCBI Sequence Read Archive (SRA) (BioProject: PRJNA816319 and submission: SUB11193782). Two rounds of in vivo screening were conducted to obtain oligopeptide candidates inserted after Q574. Then, each capsid variant candidate was dispensed 1 barcode located in the scCB-GFP construct, between the GFP coding sequence and the bovine growth hormone (bGH) polyA signal. The scCB-GFP construct carrying individual barcodes was packaged into the corresponding capsid to form functional AAV particles and used to treat mice intravenously. The quantification of the vg product was actualized by barcode evaluation, as previously reported.33
Immunofluorescence
Mice were perfused with 20–25 mL PBS and then 20–25 mL 4% paraformaldehyde (PFA) solution. Tissues and organs were harvested for another 3 h fixation in 4% PFA solution, which was followed by dehydration in 30% sucrose solution at 4°C overnight. Tissue samples were sliced into 25-μm sections on a cryostat (HM550; Thermo Scientific) at −22°C, which were then blocked and penetrated with the blocking buffer (5% BSA, 2% Triton X-100, and 5% goat serum in PBS) for 1.5 h at room temperature. Afterward, they were incubated with primary antibodies in antibody dilution buffer (5% BSA, 0.5% Triton X-100, and 5% goat serum in PBS) at 4°C overnight. After washing with 0.5% Triton X-100 in PBS 3 times, 10 min each at room temperature (25°C), samples were incubated with Alexa Fluor-tagged secondary antibodies for 3 h at room temperature and mounted with Vectashield mounting medium. Images were collected with a Zeiss confocal laser scanning microscope (LSM 700) and analyzed by ImageJ (National Institutes of Health, Bethesda, MD, USA).
Western blot
Western blot was performed as previously reported.64,69 Briefly, cells and tissues were homogenized and lysed in the lysis buffer with ingredients as follows: 150 mM NaCl, 2.5 mM EDTA, 50 mM Tris-HCl (pH 7.4), 1% SDS, 20% glycerol, 1% PMSF, and 1% protease inhibitor cocktail (no. 04693159001, Sigma-Aldrich). The debris was removed by centrifugation at 12,000 × g for 10 min at 4°C, while supernatants (referred to as lysates) were collected and diluted with 4× loading buffer containing 20% Tris-HCl (pH, 8.8), 8% SDS, 8% β-mercaptoethanol, 0.04% bromophenol blue, and 40% glycerol, and boiled at 100°C for 10 min. Samples were resolved to SDS-PAGE running and transferred to a nitrocellulose membrane (cat. no. 1620112, Bio-Rad, Hercules, CA, USA). The membrane was blocked in 5% skim milk for 1 h and incubated in primary antibodies overnight at 4°C. After washing 3 times with 0.1% Tween 20 in PBS in a span of 10 min, membranes were incubated in HRP-conjugated secondary antibodies for 1 h at room temperature. Protein bands were visualized by the Pierce enhanced chemiluminescence kit.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay
Total messenger RNA (mRNA) was extracted from the indicated tissues/organs with Trizol (Invitrogen, Waltham, MA, USA) reagent and reverse -transcribed to cDNAs with GoScript reverse transcription kit (Promega, Madison, WI, USA). cDNAs were used as templates for qPCR in a 20-μL reaction system containing SYBR GreenER qPCR mix with gene-specific primers. The PCR reaction was initiated with a denaturation step at 95°C (3 min), followed by 40 cycles consisting of denaturation at 95°C (15 s), annealing, and extension at 60°C (60 s). The GFP primers were mentioned above and the internal control, Gapdh primers, were as follows: F: 5′-AAGGTCATCCCAGAGCTGAA-3′, R: 5′-CTGCTTCACCACCTTCTTGA-3′).
AAV neutralization assays
Human sera samples were collected from 10 hemophilia B patients at the Institute of Hematology & Blood Disease Hospital, Chinese Academy of Medical Sciences and Pekin Union Medical College (Tianjin, China), which was approved by the ethics review board. Sera samples from rhesus monkeys were obtained from Greentech Bioscience (Sichuan, China). NAb levels were detected following our previous protocols, with minor modifications.42,70 Briefly, the rationale of the ratio to mix AAV and sera (or sera dilutions) was grossly based on an intravenous administration at a dose of 1 × 1013 vg/kg, namely 2 × 108 vg vps per 1 μL serum. Huh7 cells were cultured at the density of 1 × 105 cells/well in a 48-well plate. A serial 2-fold dilution of sera samples of 5 μL in volume each were incubated with 1 × 109 vg AAV particles for 2 h at 4°C, followed by treatment of the Huh7 cells with the AAV-sera mixture. The activity of luciferase delivered by AAV was measured by the Wallac-1420 Victor 2 plate reader (PerkinElmer, Waltham, MA, USA) 48 h post-AAV treatment. NAb titers were defined as the largest number of dilution ratios able to reduce the luciferase activity by half compared with the AAV-PBS mixture group. A serum sample was considered NAb positive when the titer was higher than 1:4.
AAV cell binding and internalization assay
These 2 experiments were slightly modified from our previous paper.11 Briefly, for the binding assay, the indicated AAV vectors encapsulating dsCB-GFP transgene were incubated with pre-cooled Huh7 cells at 4°C for 30 min. After washing with PBS 3 times to rinse unbound AAV particles, the cells were harvested for DNA extraction; for the internalization assay, the AAV-treated cells were incubated at 37°C for 1 h to allow internalization of the bound virus by Huh7 cell. The cells were then digested by trypsin for 10 min at 37°C to remove the un-internalized AAV. After washing 3 times in PBS, the cells were subjected to DNA extraction. Viral genomic DNA was extracted by the DNeasy Blood & Tissue Kit (Cat#69506, QIAGEN, Hilden, Germany) and quantified by qPCR with the GAPDH gene copy number as the internal control. The relative vg numbers of the other AAV serotype groups were normalized to AAV5.
Preparation of AAV capsid samples for LC-MS/MS analysis
AAV capsid samples were lysed in SDT lysis buffer (4% SDS, 100 mM dithiothreitol [DTT], 100 mM Tris-HCl, pH 7.6), heated at 100°C for 5 min and centrifuged at 12,000 × g for 10 min. The supernatants were collected from which protein samples were precipitated with a 5-fold acidified acetone/ethanol buffer (ice-cold 50% acetone, 50% ethanol, 0.1% acetic acid) at −20°C overnight. Precipitated proteins were centrifuged at 16,000 × g for 25 min at 4°C and subsequently washed twice with ice-cold acetone. The extracted proteins were dissolved in 50 mM ammonium bicarbonate (ABC) buffer. After reduction with 10 mM DTT for 30 min at 56°C and alkylation with 20 mM iodoacetamide (IAA) for 30 min at room temperature in the dark, the AAV capsid samples were equally divided into 2 parts. One aliquot was treated with chymotrypsin (Promega) in an enzyme protein ratio of 1:50 (w/w) and incubated at 25°C for 18 h with an agitation speed of 1,200 rpm. The other aliquot was treated with trypsin and Glu-C (Promega) in an enzyme protein ratio of 1:50 (w/w) and incubated at 37°C for 18 h with an agitation speed of 1,200 rpm. The digested samples were subjected to vacuum centrifugation and the C18 solid-phase (3M Empore, Oxford, PA, USA) desalting process. The desalted peptides were evaporated using vacuum centrifugation again and stored at −80°C until further processing.
Nano-LC-MS/MS data acquisition and analysis
Data acquisition and analysis were technically supported by the Institutional Technology Service Center of the Shanghai Institute of Materia Medica. To obtain raw files for acetylation, methylation, ubiquitination, phosphorylation, and deamidation, the dried peptides (∼1 μg each sample) were resolved using 0.1% formic acid (FA) and separated on an analytical column (75 μm × 200 mm) packed with reverse-phase (RP) beads (3 μm ReproSil-Pur C18 beads, 120-Å pore size; Dr. Maisch GmbH, Ammerbuch, Germany) on a nanoflow HPLC Easy-nLC 1000 system (Thermo Scientific) with a 120-min gradient at a flow rate of 300 nL/min. Buffer A consisted of 0.1% (v/v) FA in water and buffer B consisted of 0.1% (v/v) FA in acetonitrile (ACN). The gradient was set as follows: 2%–5% B in 1 min, 5%–25% B in 100 min, 25%–35% B in 12 min, 35%–100% B in 2 min, and 100% B in 5 min. An Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) is integrated with the LC system. The spray voltage was set at 2,500 V in positive ion mode and the ion transfer tube temperature was set at 275°C. MS acquisition was programmed in data-dependent acquisition (DDA) mode, in which 1 full MS scan (350–1,500 m/z) was performed at 60,000 resolution, with a 4 × 105 automatic gain control (AGC) target and a 50-ms maximum injection time. Then, MS2 scans were generated by higher energy collisional dissociation (HCD) fragmentation in a 3-s cycle time and 45 s dynamic exclusion. MS/MS acquisition was performed at 15,000 resolution, with a 5 × 104 AGC target and a 100-ms maximum injection time. Precursor fragmentation was performed with a 1.6-m/z isolation window and 27% normalized collision energy (NCE). Precursors with charge 2–6 were selected for MS2 analysis.
To acquire raw files for O-glycosylation, the resolved peptides (∼1 μg each sample) were separated using an 80-min gradient at 300 nL/min. The RP chromatography column, buffer A, and buffer B were the same as above. The gradient was set as follows: 2% B in 5 min, 2%–35% B in 60 min, 35%–50% B in 7 min, 50%–90% B in 10 s, 90% B in 2 min and 50 s, and 90%–2% B in 5 min. An Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific) is integrated with the LC system. The spray voltage was set at 2,500 V in positive ion mode and the ion transfer tube temperature was set at 275°C. MS acquisition was programmed in DDA mode, in which 1 full MS scan (350–1,800 m/z) was first performed at 120,000 resolution, with a 4 × 105 AGC target and a 50-ms maximum injection time. Then, MS2 scans were generated in a 3-s cycle time and a 15-s dynamic exclusion. HCD was performed at 15,000 resolution, with a 100-ms maximum injection time. Precursor fragmentation was performed with a 1.6-m/z isolation window and 30% NCE. Precursors with charge 2–7 were selected for MS2 analysis. Electron-transfer/higher energy collision dissociation (EThcD) was performed after HCD, under the condition that one of the glyco-fingerprint ions (126.055, 129.1023, 138.0545, 144.07, 168.065, 186.0762, 204.0867, 274.0866, 292.103, and 366.1396) had to be present in the HCD spectrum with a 15-ppm mass accuracy. Calibrated charge-dependent electron transfer dissociation (ETD) parameters were used.
Raw files of acetylation, methylation, ubiquitination, phosphorylation, and deamidation were further analyzed by MaxQuant version 2.0.1.0 and O-GlcNAclytion raw files were analyzed by pGlyco, as previously described.71,72 All PTM mass spectra were manually validated and the data have been deposited to the integrated proteome resources (iProX) (https://www.iprox.cn/) via the PRIDE partner repository with the identifier IPX0003661000.
Statistical analysis
Data were analyzed via GraphPad Prism 7 (GraphPad, San Diego, CA, USA) by unpaired t test and 1-way ANOVA (with Tukey’s multiple comparison test). Unless otherwise indicated, the data were shown as means ± SDs. Statistical difference was considered when p < 0.05. The sample size (n) was based on the literature.11,12,45,73
Acknowledgments
We greatly appreciate Dr. Wanxiang Jiang (Greentech Bioscience) for generous advice on and help with non-human primate experiments; Dr. Hu Zhou, Department of Analytical Chemistry and CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, for mass spectrum analysis; and Dr. Lei Zhang, Institute of Hematology & Blood Disease Hospital, Chinese Academy of Medical Sciences & Pekin Union Medical College, for providing hemophilia B patient sera samples. This study was supported by the National Natural Science Foundation of China (grant no. 31901052), the Basic Research and Scientific Frontier Foundation of Chongqing (cstc2019jcyj-msxmX0010), and Shanghai Municipal Science and Technology Major Project.
Author contributions
X.X. and K.Z. designed the project and drafted the manuscript. H.H., C.Y., and K.Z. constructed the AAV5 capsid NNK library. K.Z., Y.W., C.Y., H.H., C.C., M.Y., F.L., K.C.W., X.W., X.Z., and F.Z. performed the experiments and analyzed the data. S.L. and Z.D. provided the platforms for the rodent and non-human primate experiments and AAV production, respectively.
Declaration of interests
X.X., K.Z., C.Y., H.H., and Z.D. are co-inventors of a patent about the novel AAV serotype generated in this paper.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.03.017.
Contributor Information
Kai Zhao, Email: zhao-kai@beliefbiomed.com.
Xiao Xiao, Email: xiaoxiao@ecust.edu.cn.
Supplemental information
References
- 1.Leebeek F.W.G., Miesbach W. Gene therapy for hemophilia: a review on clinical benefit, limitations, and remaining issues. Blood. 2021;138:923–931. doi: 10.1182/blood.2019003777. [DOI] [PubMed] [Google Scholar]
- 2.Miesbach W., Meijer K., Coppens M., Kampmann P., Klamroth R., Schutgens R., Tangelder M., Castaman G., Schwable J., Bonig H., et al. Gene therapy with adeno-associated virus vector 5-human factor IX in adults with hemophilia B. Blood. 2018;131:1022–1031. doi: 10.1182/blood-2017-09-804419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangarajan S., Walsh L., Lester W., Perry D., Madan B., Laffan M., Yu H., Vettermann C., Pierce G.F., Wong W.Y., et al. AAV5-Factor VIII gene transfer in severe hemophilia A. N. Engl. J. Med. 2017;377:2519–2530. doi: 10.1056/NEJMoa1708483. [DOI] [PubMed] [Google Scholar]
- 4.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D., et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erles K., Sebokova P., Schlehofer J.R. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV) J. Med. Virol. 1999;59:406–411. doi: 10.1002/(sici)1096-9071(199911)59:3<406::aid-jmv22>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 7.Hurlbut G.D., Ziegler R.J., Nietupski J.B., Foley J.W., Woodworth L.A., Meyers E., Bercury S.D., Pande N.N., Souza D.W., Bree M.P., et al. Preexisting immunity and low expression in primates highlight translational challenges for liver-directed AAV8-mediated gene therapy. Mol. Ther. J. Am. Soc. Gene Ther. 2010;18:1983–1994. doi: 10.1038/mt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Calcedo R., Bell P., Lin J., Grant R.L., Siegel D.L., Wilson J.M. Impact of pre-existing immunity on gene transfer to nonhuman primate liver with adeno-associated virus 8 vectors. Hum. Gene Ther. 2011;22:1389–1401. doi: 10.1089/hum.2011.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapti K., Grimm D. Adeno-associated viruses (AAV) and host immunity - a race between the hare and the hedgehog. Front. Immunol. 2021;12:753467. doi: 10.3389/fimmu.2021.753467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 11.Qian R., Xiao B., Li J., Xiao X. Directed evolution of AAV serotype 5 for increased hepatocyte transduction and retained low humoral seroreactivity. Mol. Ther. Methods Clin. Dev. 2021;20:122–132. doi: 10.1016/j.omtm.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisowski L., Dane A.P., Chu K., Zhang Y., Cunningham S.C., Wilson E.M., Nygaard S., Grompe M., Alexander I.E., Kay M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A., et al. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. J. Am. Soc. Gene Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vercauteren K., Hoffman B.E., Zolotukhin I., Keeler G.D., Xiao J.W., Basner-Tschakarjan E., High K.A., Ertl H.C., Rice C.M., Srivastava A., et al. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol. Ther. J. Am. Soc. Gene Ther. 2016;24:1042–1049. doi: 10.1038/mt.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perocheau D.P., Cunningham S., Lee J., Antinao Diaz J., Waddington S.N., Gilmour K., Eaglestone S., Lisowski L., Thrasher A.J., Alexander I.E., et al. Age-Related seroprevalence of antibodies against AAV-LK03 in a UK population cohort. Hum. Gene Ther. 2019;30:79–87. doi: 10.1089/hum.2018.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiorini J.A., Afione S., Kotin R.M. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J. Virol. 1999;73:4293–4298. doi: 10.1128/jvi.73.5.4293-4298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiorini J.A., Kim F., Yang L., Kotin R.M. Cloning and characterization of adeno-associated virus type 5. J. Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walters R.W., Agbandje-McKenna M., Bowman V.D., Moninger T.O., Olson N.H., Seiler M., Chiorini J.A., Baker T.S., Zabner J. Structure of adeno-associated virus serotype 5. J. Virol. 2004;78:3361–3371. doi: 10.1128/JVI.78.7.3361-3371.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hida K., Won S.Y., Di Pasquale G., Hanes J., Chiorini J.A., Ostermeier M. Sites in the AAV5 capsid tolerant to deletions and tandem duplications. Arch. Biochem. Biophys. 2010;496:1–8. doi: 10.1016/j.abb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Govindasamy L., DiMattia M.A., Gurda B.L., Halder S., McKenna R., Chiorini J.A., Muzyczka N., Zolotukhin S., Agbandje-McKenna M. Structural insights into adeno-associated virus serotype 5. J. Virol. 2013;87:11187–11199. doi: 10.1128/JVI.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bantel-Schaal U., Delius H., Schmidt R., zur Hausen H. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J. Virol. 1999;73:939–947. doi: 10.1128/jvi.73.2.939-947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seiler M.P., Miller A.D., Zabner J., Halbert C.L. Adeno-associated virus types 5 and 6 use distinct receptors for cell entry. Hum. Gene Ther. 2006;17:10–19. doi: 10.1089/hum.2006.17.10. [DOI] [PubMed] [Google Scholar]
- 23.Afione S., DiMattia M.A., Halder S., Di Pasquale G., Agbandje-McKenna M., Chiorini J.A. Identification and mutagenesis of the adeno-associated virus 5 sialic acid binding region. J. Virol. 2015;89:1660–1672. doi: 10.1128/JVI.02503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Pasquale G., Davidson B.L., Stein C.S., Martins I., Scudiero D., Monks A., Chiorini J.A. Identification of PDGFR as a receptor for AAV-5 transduction. Nat. Med. 2003;9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 25.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 26.Liu Q., Huang W., Zhang H., Wang Y., Zhao J., Song A., Xie H., Zhao C., Gao D., Wang Y. Neutralizing antibodies against AAV2, AAV5 and AAV8 in healthy and HIV-1-infected subjects in China: implications for gene therapy using AAV vectors. Gene Ther. 2014;21:732–738. doi: 10.1038/gt.2014.47. [DOI] [PubMed] [Google Scholar]
- 27.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanford S., Pink R., Creagh D., Clark A., Lowe G., Curry N., Pasi J., Perry D., Fong S., Hayes G., et al. Adenovirus-associated antibodies in UK cohort of hemophilia patients: a seroprevalence study of the presence of adenovirus-associated virus vector-serotypes AAV5 and AAV8 neutralizing activity and antibodies in patients with hemophilia A. Res. Pract. Thromb. Haemostatsis. 2019;3:261–267. doi: 10.1002/rth2.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasi K.J., Rangarajan S., Mitchell N., Lester W., Symington E., Madan B., Laffan M., Russell C.B., Li M., Pierce G.F., et al. Multiyear follow-up of AAV5-hFVIII-SQ gene therapy for hemophilia A. N. Engl. J. Med. 2020;382:29–40. doi: 10.1056/NEJMoa1908490. [DOI] [PubMed] [Google Scholar]
- 30.Majowicz A., Nijmeijer B., Lampen M.H., Spronck L., de Haan M., Petry H., van Deventer S.J., Meyer C., Tangelder M., Ferreira V. Therapeutic hFIX activity achieved after single AAV5-hFIX treatment in hemophilia B patients and NHPs with pre-existing anti-AAV5 NABs. Mol. Ther. Methods Clin. Dev. 2019;14:27–36. doi: 10.1016/j.omtm.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonnenmacher M., Wang W., Child M.A., Ren X.Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol. Ther. Methods Clin. Dev. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabebordbar M., Lagerborg K.A., Stanton A., King E.M., Ye S., Tellez L., Krunnfusz A., Tavakoli S., Widrick J.J., Messemer K.A., et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184:4919–4938.e22. doi: 10.1016/j.cell.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinmann J., Weis S., Sippel J., Tulalamba W., Remes A., El Andari J., Herrmann A.K., Pham Q.H., Borowski C., Hille S., et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 2020;11:5432. doi: 10.1038/s41467-020-19230-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalkara D., Byrne L.C., Klimczak R.R., Visel M., Yin L., Merigan W.H., Flannery J.G., Schaffer D.V. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl Med. 2013;5:189ra176. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 35.Jin X., Liu L., Nass S., O'Riordan C., Pastor E., Zhang X.K. Direct liquid chromatography/mass spectrometry analysis for complete characterization of recombinant adeno-associated virus capsid proteins. Hum. Gene Ther. Methods. 2017;28:255–267. doi: 10.1089/hgtb.2016.178. [DOI] [PubMed] [Google Scholar]
- 36.Guenzel A.J., Collard R., Kraus J.P., Matern D., Barry M.A. Long-term sex-biased correction of circulating propionic acidemia disease markers by adeno-associated virus vectors. Hum. Gene Ther. 2015;26:153–160. doi: 10.1089/hum.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davidoff A.M., Ng C.Y., Zhou J., Spence Y., Nathwani A.C. Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood. 2003;102:480–488. doi: 10.1182/blood-2002-09-2889. [DOI] [PubMed] [Google Scholar]
- 38.Chal J., Pourquie O. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017;144:2104–2122. doi: 10.1242/dev.151035. [DOI] [PubMed] [Google Scholar]
- 39.Cohen S., Nathan J.A., Goldberg A.L. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015;14:58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 40.Feng L., Wang S., Chen F., Zhang C., Wang Q., Zhao Y., Zhang Z. Hepatic knockdown of endothelin type A receptor (ETAR) ameliorates hepatic insulin resistance and hyperglycemia through suppressing p66Shc-mediated mitochondrial fragmentation in high-fat diet-fed mice. Diabetes, Metab. Syndrome Obes. 2021;14:963–981. doi: 10.2147/DMSO.S299570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng J., Liu Y., Zhang M., Liu F., Ma L., Yu C.Y., Wei H. One-pot fabrication of dual-redox sensitive, stabilized supramolecular nanocontainers for potential programmable drug release using a multifunctional cyclodextrin unit. J. Control. Release. 2021;334:290–302. doi: 10.1016/j.jconrel.2021.04.027. [DOI] [PubMed] [Google Scholar]
- 42.Zhang F., Yan X., Li M., Hua B., Xiao X., Monahan P.E., Sun J. Exploring the potential feasibility of intra-articular adeno-associated virus-mediated gene therapy for hemophilia arthropathy. Hum. Gene Ther. 2020;31:448–458. doi: 10.1089/hum.2019.355. [DOI] [PubMed] [Google Scholar]
- 43.Gao Y.Y., Chen H., Zhou Y.Y., Wang L.T., Hou Y., Xia X.H., Ding Y. Intraorgan targeting of gold conjugates for precise liver cancer treatment. ACS Appl. Mater. Inter. 2017;9:31458–31468. doi: 10.1021/acsami.7b08969. [DOI] [PubMed] [Google Scholar]
- 44.Pavlou M., Schon C., Occelli L.M., Rossi A., Meumann N., Boyd R.F., Bartoe J.T., Siedlecki J., Gerhardt M.J., Babutzka S., et al. Novel AAV capsids for intravitreal gene therapy of photoreceptor disorders. EMBO Mol. Med. 2021;13:e13392. doi: 10.15252/emmm.202013392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buning H., Srivastava A. Capsid modifications for targeting and improving the efficacy of AAV vectors. Mol. Ther. Methods Clin. Dev. 2019;12:248–265. doi: 10.1016/j.omtm.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khabou H., Desrosiers M., Winckler C., Fouquet S., Auregan G., Bemelmans A.P., Sahel J.A., Dalkara D. Insight into the mechanisms of enhanced retinal transduction by the engineered AAV2 capsid variant -7m8. Biotechnol. Bioeng. 2016;113:2712–2724. doi: 10.1002/bit.26031. [DOI] [PubMed] [Google Scholar]
- 48.Pietersz K.L., Plessis F.D., Pouw S.M., Liefhebber J.M., van Deventer S.J., Martens G.J.M., Konstantinova P.S., Blits B. PhP.B enhanced adeno-associated virus mediated-expression following systemic delivery or direct brain administration. Front. Bioeng. Biotechnol. 2021;9:679483. doi: 10.3389/fbioe.2021.679483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnold G.S., Sasser A.K., Stachler M.D., Bartlett J.S. Metabolic biotinylation provides a unique platform for the purification and targeting of multiple AAV vector serotypes. Mol. Ther. J. Am. Soc. Gene Ther. 2006;14:97–106. doi: 10.1016/j.ymthe.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 50.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Jr., Weigel-Van Aken K.A., et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. U S A. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Q., Njenga R., Leuchs B., Chiocca S., Kleinschmidt J., Muller M. SUMOylation targets adeno-associated virus capsids but mainly restricts transduction by cellular mechanisms. J. Virol. 2020;94 doi: 10.1128/JVI.00871-20. e00871–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurya S., Mary B., Jayandharan G.R. Rational engineering and preclinical evaluation of neddylation and SUMOylation site modified adeno-associated virus vectors in murine models of hemophilia B and leber congenital amaurosis. Hum. Gene Ther. 2019;30:1461–1476. doi: 10.1089/hum.2019.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sen D., Balakrishnan B., Gabriel N., Agrawal P., Roshini V., Samuel R., Srivastava A., Jayandharan G.R. Improved adeno-associated virus (AAV) serotype 1 and 5 vectors for gene therapy. Sci. Rep. 2013;3:1832. doi: 10.1038/srep01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kay C.N., Ryals R.C., Aslanidi G.V., Min S.H., Ruan Q., Sun J., Dyka F.M., Kasuga D., Ayala A.E., Van Vliet K., et al. Targeting photoreceptors via intravitreal delivery using novel, capsid-mutated AAV vectors. PLoS One. 2013;8:e62097. doi: 10.1371/journal.pone.0062097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dalkara D., Byrne L.C., Lee T., Hoffmann N.V., Schaffer D.V., Flannery J.G. Enhanced gene delivery to the neonatal retina through systemic administration of tyrosine-mutated AAV9. Gene Ther. 2012;19:176–181. doi: 10.1038/gt.2011.163. [DOI] [PubMed] [Google Scholar]
- 56.Kanaan N.M., Sellnow R.C., Boye S.L., Coberly B., Bennett A., Agbandje-McKenna M., Sortwell C.E., Hauswirth W.W., Boye S.E., Manfredsson F.P. Rationally engineered AAV capsids improve transduction and volumetric spread in the CNS. Mol. Ther. Nucleic Acids. 2017;8:184–197. doi: 10.1016/j.omtn.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu H., Jan Fada B. Specificity in ubiquitination triggered by virus infection. Int. J. Mol. Sci. 2020;21:4088. doi: 10.3390/ijms21114088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan Z., Zak R., Luxton G.W., Ritchie T.C., Bantel-Schaal U., Engelhardt J.F. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duan D., Yue Y., Yan Z., Yang J., Engelhardt J.F. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li B., Ma W., Ling C., Van Vliet K., Huang L.Y., Agbandje-McKenna M., Srivastava A., Aslanidi G.V. Site-directed mutagenesis of surface-exposed lysine residues leads to improved transduction by AAV2, but not AAV8, vectors in murine hepatocytes in vivo. Hum. Gene Ther. Methods. 2015;26:211–220. doi: 10.1089/hgtb.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mary B., Maurya S., Kumar M., Bammidi S., Kumar V., Jayandharan G.R. Molecular engineering of adeno-associated virus capsid improves its therapeutic gene transfer in murine models of hemophilia and retinal degeneration. Mol. Pharm. 2019;16:4738–4750. doi: 10.1021/acs.molpharmaceut.9b00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giles A.R., Sims J.J., Turner K.B., Govindasamy L., Alvira M.R., Lock M., Wilson J.M. Deamidation of amino acids on the surface of adeno-associated virus capsids leads to charge heterogeneity and altered vector function. Mol. Ther. J. Am. Soc. Gene Ther. 2018;26:2848–2862. doi: 10.1016/j.ymthe.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao K., Shen C., Lu Y., Huang Z., Li L., Rand C.D., Pan J., Sun X.D., Tan Z., Wang H., et al. Muscle yap is a regulator of neuromuscular junction formation and regeneration. J. Neurosci. 2017;37:3465–3477. doi: 10.1523/JNEUROSCI.2934-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao K., Shen C., Li L., Wu H., Xing G., Dong Z., Jing H., Chen W., Zhang H., Tan Z., et al. Sarcoglycan alpha mitigates neuromuscular junction decline in aged mice by stabilizing LRP4. J. Neurosci. 2018;38:8860–8873. doi: 10.1523/JNEUROSCI.0860-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grieger J.C., Soltys S.M., Samulski R.J. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol. Ther. J. Am. Soc. Gene Ther. 2016;24:287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rabinowitz J.E., Rolling F., Li C., Conrath H., Xiao W., Xiao X., Samulski R.J. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z., Ma H.I., Li J., Sun L., Zhang J., Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- 69.Shen C., Lu Y., Zhang B., Figueiredo D., Bean J., Jung J., Wu H., Barik A., Yin D.M., Xiong W.C., et al. Antibodies against low-density lipoprotein receptor-related protein 4 induce myasthenia gravis. J. Clin. Invest. 2013;123:5190–5202. doi: 10.1172/JCI66039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun J., Hua B., Chen X., Samulski R.J., Li C. Gene delivery of activated factor VII using alternative adeno-associated virus serotype improves hemostasis in hemophiliac mice with FVIII inhibitors and adeno-associated virus neutralizing antibodies. Hum. Gene Ther. 2017;28:654–666. doi: 10.1089/hum.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu M.Q., Zeng W.F., Fang P., Cao W.Q., Liu C., Yan G.Q., Zhang Y., Peng C., Wu J.Q., Zhang X.J., et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017;8:438. doi: 10.1038/s41467-017-00535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 73.Yang L., Jiang J., Drouin L.M., Agbandje-McKenna M., Chen C., Qiao C., Pu D., Hu X., Wang D.Z., Li J., et al. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. U S A. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.