Abstract
The transcriptional response to cellular stress relies upon trafficking of regulators of transcription between the nuclear and cytoplasmic compartments, which occurs through action of members of the importin (IPO) superfamily. As a result of stresses such as oxidative or osmotic stress, one consequence is that importins become mislocalised, leading to inhibition of conventional nuclear transport. Here, we examine IPO13, which has a number of nonconventional characteristics, in the context of cell stress. We used Next Generation RNA Sequencing using the Illumina platform to compare the transcriptomes of Wild-type (WT) and IPO13-Knockout (KO) mouse embryonic stem cells in the absence and presence of oxidative stress. Differences in the mRNA expression profiles were observed between the cell lines in the absence and in the presence of stress. This data will be a key resource to enable characterization of the contribution of nuclear transporter IPO13 to cellular transcription in the absence and presence of oxidative stress, as well as more broadly, in the study of stem cell biology and effect of stress on embryonic stem cell transcription.
Keywords: Embryonic stem cells, RNA sequencing, Oxidative stress, Nuclear transport
Specifications Table
| Subject | Omics:Transcriptomics |
| Specific subject area | Stem cell biology, Molecular biology, Stress biology. |
| Type of data | Table Chart Graph Figure |
| How the data were acquired | RNA sequencing of mouse embryonic stem cells with and without IPO13 knock out treated with hydrogen peroxide induced oxidative stress. |
| Data format | Raw Analyzed |
| Description of data collection |
|
| Data source location |
|
| Data accessibility | Repository name: Gene Expression Omnibus Data identification number: GSE108913 Direct URL to data: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108913 Repository name: Degust Direct URL to data: https://degust.erc.monash.edu/degust/compare.html?code=d0c8b150384a6f560dfc81cdf97cfaeb |
| Related research article | K.A. Gajewska, H. Lescesen, M. Ramialison, K.M. Wagstaff, D.A. Jans, Nuclear transporter Importin-13 plays a key role in the oxidative stress transcriptional response, Nature Communications. 12 (1) (2021) 5904. https://doi.org/10.1038/s41467-021-26125-x |
Value of the Data
-
•
The data here delineates the transcriptome of embryonic stem cells with and without exposure to oxidative stress and embryonic stem cells with a knock out of the nuclear transport protein IPO13, enabling identification of differentially regulated genes in the absence and presence of stress and thus shedding insight into the molecular basis of oxidative stress response in embryonic stem cells and the contribution of IPO13 thereto.
-
•
This data will be particularly useful to stem cell biology researchers, providing the transcriptomic basis to understand the oxidative stress response in stem cells.
-
•
Since IPO13 is a nuclear transport protein with critical roles in early embryonic development, the IPO13-dependent transcriptome will be invaluable to the biology of nuclear transport as well as the study of embryonic development and related diseases.
1. Data Description
The data provided here are the result of a transcriptomic approach to examine the impact of oxidative stress and the stress response in embryonic stem cells and the contribution of IPO13 therein. We provide the raw sequencing data (deposited at GEO) and the differential expression analysis (deposited at Degust) which contains all data, including all sub-significant data, that is associated with our related research article in which we had only reported genes significantly differentially expressed (≥log2-fold differentially expressed with an FDR cut off of 0.05) and thus provide additional insights not previously described.
2. Experimental Design, Materials and Methods
2.1. Cell culture
Wild-type (WT) and IPO13-Knock Out (KO) mouse embryonic stem cells (ESCs) used in this study have been characterized previously with cell culture conditions previously described [1]. Briefly, ESCs maintained in feeder-free culture conditions in Dulbecco's Modified Eagle Medium (DMEM) (12.5% FBS, 1x Glutamax, 0.1 mM NEAA, 0.1 mM β mercaptoethanol and 1000 U/ml LIF) at 37 °C on 0.1% gelatin-coated surfaces in 5% CO2 atmosphere in a humidified incubator. For passage, ESCs were trypsonised with TrypLE Select every 2 d. WT and KO ESC lysates were analyzed by Western blotting to confirm negligible expression of IPO13 in KO ESCs as previously described [1], with rabbit polyclonal anti-IPO13 (Protein Tech, Cat no.11696-2-AP) and mouse monoclonal anti-β-actin (CST, Cat no. 3700) antibodies (Supplementary Fig. 1).
2.2. Oxidative stress treatment and RNA isolation
Expanding on our related work [2], WT and KO ESCs were seeded into T25 flasks Fig. 1. Next day at 70–80% confluency, cells were either given fresh media or given media supplemented with hydrogen peroxide (H2O2) at 125 µM. After 1 h treatment, media was replaced allowing cells to recover for 2 h prior to RNA extraction. RNA was harvested from cell lines with the Isolate II RNA Mini Kit (Bioline) according to the manufacturer's instructions, and concentration estimated using the Qubit RNA HS Assay Kit on the Qubit Fluorometer (Invitrogen).
Fig. 1.
Schematic representation of experimental design and data analysis workflow.
2.3. Library preparation and RNA sequencing
RNA sequencing was performed at the Monash University Micromon Technology Research Platform using the Illumina HiSeq 2000 sequencer platform. mRNA samples were converted to Illumina sequencing libraries using Illumina's TruSeq Stranded mRNA Sample Prep Kit (Cat. # RS-122-2101). Briefly, 1 mg of total RNA from each sample was purified for polyA containing mRNA molecules using oligo (dT)-conjugated magnetic beads and fragmented. Next, first- and second- strand cDNA was synthesised and the blunt fragment ends 3′-adenylated. Indexing adapters were ligated to the ends of the cDNA and amplified by PCR to enrich DNA fragments bearing adapter sequences. Double stranded DNA was quantified using the Qubit DNA HS kit (Invitrogen, Carlsbad CA., USA). Libraries were then monitored for quality control using the Agilent Bioanalyzer 2100 microfluidics device, in conjunction with Agilent DNA HS kits and chemistry (Agilent Technologies, Waldbronn, Germany), enabling the size of the library to be estimated as well as assessed for contamination with unligated adapters.
Sample libraries, including each replicate (two of each of untreated WT ESC – WT-NS1 and WT-NS2, H2O2-treated WT ESC – WTST1 and WTST2, untreated KO ESC - KO-NS1 and KO-NS2, and H2O2-treated KO ESC – KO-ST1 and KO-ST2) were pooled in equimolar concentrations into 3 sequencing lanes of the Illumina HiSeq 2000 and underwent 101 bp paired end sequencing (Table 1). These methods are expanded versions of descriptions in our related work [2].
Table 1.
Samples in this study.
| Source | Protocol 1 | Protocol 2 | Samples | Protocol 3 | Protocol 4 | Data Citation |
|---|---|---|---|---|---|---|
| Wild-type ESC | No Treatment | Isolate II RNA Mini Kit |
WT-NS1 (Replicate 1) |
TruSeq Stranded mRNA | Illumina HiSeq 2000 |
Data Citation 1 GSM2915889 |
| Wild-type ESC | No Treatment | Isolate II RNA Mini Kit |
WT-NS2 (Replicate 2) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915890 |
| Wild-type ESC | H2O2 Treatment | Isolate II RNA Mini Kit |
WT-ST1 (Replicate 1) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915891 |
| Wild-type ESC | H2O2 Treatment | Isolate II RNA Mini Kit |
WT-ST2 (Replicate 2) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915892 |
| IPO13 Knock-Out ESC |
No Treatment | Isolate II RNA Mini Kit |
KO-NS1 (Replicate 1) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915893 |
| IPO13 Knock-Out ESC |
No Treatment | Isolate II RNA Mini Kit |
KO-NS2 (Replicate 2) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915894 |
| IPO13 Knock-Out ESC |
H2O2 Treatment | Isolate II RNA Mini Kit |
KO-ST1 (Replicate 1) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915895 |
| IPO13 Knock-Out ESC |
H2O2 Treatment | Isolate II RNA Mini Kit |
KO-ST2 (Replicate 2) |
TruSeq Stranded mRNA Prep Kit | Illumina HiSeq 2000 |
Data Citation 1 GSM2915896 |
2.4. Sequencing quality check and read alignment
Per-cycle base call (BCL) files were generated by the Illumina Real Time Analysis Software and converted to per-read FASTQ files for further analysis using bcl2fastq v1.8.4. This was followed by de-multiplexing by the software of samples that were multiplexed to run in 3 sequencing lanes. Adapter sequences were then trimmed by bcl2fastq and any adapter sequence detected masked in the resulting FASTQ file. After adapter removal, the quality of the paired-end sequence files was estimated using the FASTQC v0.11.5 analysis module. The sequence data were determined to be of high quality, as demonstrated by a representative result in Fig. 2, based on the distribution of quality score (Phred score) per base (plotting the phred score per base as box plots (Fig. 2a), the distribution of quality scores of the raw sequences (examining the distribution of average Phred scores of all sequences, Fig. 2b) and the GC content (%) distribution of the raw sequences [3]. The mean Phred score over all sequences was 27, with no sequences flagged as of poor quality. The distribution of the sequence GC content (%) was compared to the theoretical normal distribution (Fig. 2c). The curves overlapped to a considerable extent, confirming successful trimming of adapter sequence.
Fig. 2.
Quality assessment of raw FASTQ sequence data for paired-end reads of one sample (read 1 of sample WT NS). (a) Box plots of the distribution of per-base quality score; (b) Distribution of quality scores of all sequences; (c) The distribution of GC content (%) over all sequences compared to the theoretical normal distribution. All figures were generated using the FASTQC v0.11.5 program.
Reads were then aligned to the mouse genome, release mm10, using STAR v2.3.1 in combination with SAMtools [4]. The aligned reads were sorted and indexed using BEDTools [5], with key outputs of the alignment shown in Table 2. Here we estimate that the quality of sequencing is considerably high with the uniquely mapped reads rate on average around 85% and the mismatch rate per base at only approx. 0.25%. We could confirm that fragmentation and PCR amplification during library preparation was of a high standard by looking at the consistency in the size of the average insert across the samples (∼199 bp) [6]. Thus, it was possible to be confident of unambiguous read alignment to the genome, together with greater accuracy in quantification of genes with low expression levels, given the high sequencing read depths in the datasets (between 79 and 100 million) and the fact that paired end sequencing was used [7,8].
Table 2.
Details of key QC metrics of RNA-seq library after alignment with STAR v.2.3.1.
| Sample | Number of Input Reads | Average Mapped Length | Uniquely Mapped Reads (%) | Mismatch Rate Per Base (%) |
|---|---|---|---|---|
| WTWT-NS1 WT-NS2 |
91,612,455 94,329,655 |
198.9 199.01 |
89.03 89.56 |
0.25 0.25 |
| WTWT-ST1 + H2O2WT-ST2 |
90,926,005 92,600,914 |
198.76 199.26 |
88.75 89.26 |
0.25 0.25 |
| KOKO-NS1 KO-NS2 |
97,557,544 79,090,958 |
198.69 199.03 |
89.42 88.87 |
0.25 0.26 |
| KOKO-ST1 + H2O2KO-ST2 |
85,788,342 80,659,051 |
199.27 199.02 |
88.38 88.98 |
0.26 0.25 |
2.5. Read counts, normalization, and differential gene expression
The number of aligned reads that overlap with each gene present in the alignment was counted to estimate gene expression using HTSeq-count [9] in union mode. Ambiguous reads that aligned to more than one location were not counted. Both reads of a pair had to overlap exons within the same gene for a read to count towards it. Across all samples 84% of aligned read pairs were exonic, with only 1% found to be ambiguous.
Differential gene expression among cell lines and treatments was calculated following Voom/Limma [10] using Degust v0.20 with the false discovery rate (FDR) cut-off of 0.05. The following are an expanded description of our related work [2]. Read counts that were obtained from HTSeq-count were analysed by normal linear modeling with Voom/Limma and normalization criteria included normalization for sequencing depth and to reduce composition bias. To this end, the reads for each gene were normalized to the library size generated by a sample to produce counts per million (cpm). We were interested in relative changes in expression between conditions rather than absolute expression and thus log-cpm values between samples were generated by Voom/Limma using the Limma empirical Bayes analysis pipeline. Log-cpm values between samples can be interpreted as log-fold-changes of expression.
Normalization ensured that the distribution of gene expression in each of the samples was comparable across the whole experiment (Fig. 3a). The Euclidean distances between each of the sample pairs were calculated from the log-cpm values of all 23,420 genes and approximated in a Multi Dimensional Scaling (MDS) plot (Fig. 3b). In the largest two dimensions, close clustering of replicate samples is seen, indicating the high reproducibility of the gene expression profile of treated and untreated cell lines. Furthermore, the plot highlights the significant changes in the gene expression profiles between WT and KO cell lines, as well as between cell lines, in the presence or absence of oxidative stress. Differentially expressed genes were identified by comparing normalized data from the untreated control of each cell line to the corresponding treated sample. Volcano plots indicate the magnitude and the confidence of gene expression regulation attributable to the lack of IPO13 in the KO ESCs (Fig. 3c), and in response to oxidative stress treatment in WT (Fig. 3d) and KO ESCs (Fig. 3e).
Fig. 3.
Exploratory analysis of RNA-seq data. (a). Boxplots of log-cpm values showing expression distributions for normalized data. (b). Multi-dimensional scaling (MDS) plot summarizing gene expression profile differences between Wild-type (“WT”) and IPO13-Knock Out (“KO”) ESCs for control (“NS”) and oxidative stress-treated (“ST”) conditions without FDR or log fold change cut-off. (c–e). Volcano plots comparing the KO ESC to WT ESC (c) or oxidative stress treatment in WT ESC (d) or KO ESC (e) to untreated cells. The x-axis represents differential expression of genes in the KO ESC relative to WT ESC (c) and in the oxidative stress-treated ESC relative to untreated (d,e) expressed as log base 2-fold changes (logFC). The y-axis represents the significance level of the differential expression, expressed as -log base 10 false discovery rate (-log10FDR). Genes indicated in yellow on the plot are of a log2-fold or greater change in expression and FDR = 0.05. Genes with highest logFC and -log10FDR are labelled in blue.
2.6. Data records
Raw FASTQ files for the RNA-seq libraries have been deposited to the NCBI Gene Expression Omnibus (GEO) with accession number GSE108913 (Data Citation 1). RNA-seq DEG data can be visualised at https://degust.erc.monash.edu/degust/compare.html?code=d0c8b150384a6f560dfc81cdf97cfaeb
2.7. Technical validation
To confirm the reliability of the dataset and the analysis performed, we performed RT-qPCR (Fig. 4) of six genes which the Voom/Limma software analysis indicates with a high confidence are either up- or down- regulated in response to stress treatment across the two cell lines, or non-responsive; Chac1, Ikbke, Lef1, Cyp2s1, Sdha and Tbp. RNA that was isolated from untreated and treated (125 µM H2O2 for 1 h followed by 2 h recovery) WT and KO ESC lines for RNA-Seq was reverse transcribed by first strand cDNA synthesis using Isolate II RNA Mini Kit (Bioline) according to the manufacturer's instructions, using 500 ng of total RNA and Superscript III Reverse Transcriptase/random hexamers (Invitrogen). cDNA was then amplified with the SensiMix SYBR Master Mix (Bioline Reagents, London, UK) including about 10 ng cDNA in each reaction. Quantitative RT-PCR was carried out in triplicate using the C1000 Touch Thermal Cycler (Biorad) using an annealing temperature of 62 °C. Primer sequences are detailed in Table 3. The comparative threshold cycle (Ct) method was used for the calculation of mRNA levels as previously [11], normalizing to the internal reference genes Sdha and Tbp to compare the change in mRNA levels of selected genes between the untreated and the 1 h H2O2 treated, followed by 2 h recovery in WT or KO ESC. Statistical analyses were performed by calculating the p-value using a two-tailed Student's t-test, assuming equal variances between two groups. A confidence level of 95% (p < 0.05) was considered a statistically significant difference.
Fig. 4.
RT-qPCR validation of selected genes showing IPO13-dependent stress responses. WT and KO ESCs were treated without and with 125 μM H2O2 for 1 h followed by 2 h recovery prior to RNA extraction. Transcript levels were assessed using SensiMix SYBR Green (Bioline Reagents, London, UK) on a C1000 Touch Thermal Cycler (Biorad). Results for gene expression were normalized to the expression of housekeeping genes Sdha and Tbp. Results represent the mean ± SEM (n = 4, where 2 cDNA samples were measured in duplicate), p values indicate statistical differences and NS indicates not significant as determined by students t-test.
Table 3.
Oligonucleotides used for qPCR analysis.
| Primer Name | Sequence |
|---|---|
| Chac1 | Forward - CTGTGGATTTTCGGGTACGG Reverse - CCCCTATGGAAGGTGTCTCC |
| Cyp2s1 | Forward - TGGCACAGGAGAAACAAGAC Reverse - CCAGCAAACAGCAGGTATGT |
| Ikbke | Forward - GCGGAGGCTGAATCACCAG Reverse - GAAAGCCCGAACGTGTTCTCA |
| Lef1 | Forward - GAGCACTTTTCTCCGGGATC Reverse - GGGATTTCAGGAGCTGGAG |
| Sdha | Forward - GCTCCTGCCTCTGTGGTTGA Reverse - AGCAACACCGATGAGCCTG |
| Tbp | Forward - GAAGAACAATCCAGACTAGCAGCA Reverse - CCTTATAGGGAACTTCACATCACAG |
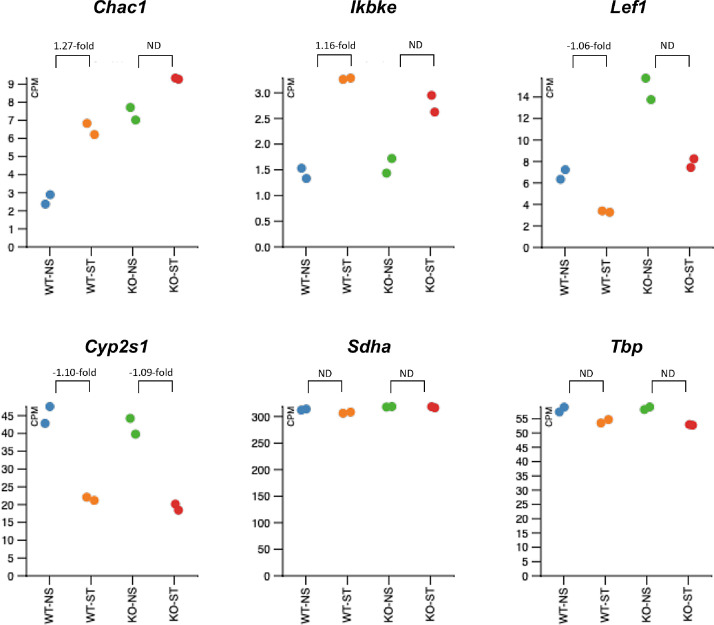
The corresponding differential expression values for each of these genes in response to oxidative stress in either cell line as identified by RNA Seq are presented in Fig. 5. The genes were chosen to identify DEGs exclusive to the treated WT cell line to reflect the role of and dependency on IPO13 in the transcriptional response to oxidative stress in ESC. Genes independent of IPO13 for differential expression under oxidative stress or that do not respond to oxidative stress are also represented. The qPCR validation of these genes shows that Chac1, Ikbke and Lef1 are indeed dependent on IPO13 for differential expression under oxidative stress (Fig. 4). Chac1 and Ikbke are both upregulated under conditions of stress in the WT ESCs. However, there is no significant change in expression of Chac1 and Ikbke in stressed KO ESCs compared to the untreated cells. In contrast, Lef1 is significantly downregulated by stress in WT ESCs, but not in KO cells. Cyp2s1 is down-regulated in response to stress independent of IPO13, while Sdha and Tbp show no change in expression across both cell lines and under all conditions, consistent with their house-keeping gene roles.
Fig. 5.
Counts per million and fold changes in expression values (obtained from Limma/Voom analysis of RNA-Seq data) of the selected genes relative to the untreated corresponding cell line. Abbreviations; ND, no differential expression. Log-2-fold values denote up- and down-regulated genes respectively in response to stress in the WT or KO ESCs.
Ethics Statement
There is no conflict of interest. This data is available in the public domain. This work did not involve human subjects, animal experiments or data collected from social media.
CRediT authorship contribution statement
Katarzyna A. Gajewska: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing, Methodology, Visualization. Mirana Ramialison: Conceptualization, Formal analysis. Kylie M. Wagstaff: Conceptualization, Formal analysis. David A. Jans: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We extend our gratitude to Stuart Archer from the Monash Bioinformatics Platform for advice on RNA-seq and thank Monash Micromon Sequencing Facility for their support in performing the sequencing. We acknowledge the financial support of the National Health and Medical Research Council Australia (fellowship APP1002486/APP1103050), the National Breast Cancer Foundation (fellowship CDA-17-007), National Health and Medical Research Council/Heart Foundation Career Development Fellowship (1049980) and MNHS Platform Access Grant (PAG15-0016). K. A. G. acknowledges the scholarship support by Monash University (MBio Discovery Scholarship). The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government. This research was supported by use of the Nectar Research Cloud, a collaborative Australian research platform supported by the National Collaborative Research Infrastructure Strategy (NCRIS).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2022.108099.
Appendix. Supplementary materials
Supplementary Fig. 1. IPO13 expression in WT and KO ESCs. Total WT and KO ESC extracts were subjected to Western analysis using rabbit-anti-IPO13 with mouse-anti-β-actin as a loading control with blots imaged using the ChemiDoc Gel Imaging System (Biorad).
Data Availability
Wild-type and IPO13-Knock Out (+/- Oxidative Stress) RNA-Sequencing (Original data) (DEGUST).
Transcriptomic profiling of untreated and oxidative stress treated Wild-type and IPO13-/- mouse embryonic stem cells to study the role of IPO13 in transcriptional response to stress (Original data) (NCBI Gene Expression Omnibus).
References
- 1.Fatima S., Wagstaff K.M., Lieu K.G., Davies R.G., Tanaka S.S., Yamaguchi Y.L., Loveland K.L., Tam P.P., Jans D.A. Interactome of the inhibitory isoform of the nuclear transporter Importin 13. Biochim. Biophys. Acta. 2017;1864(3):546–561. doi: 10.1016/j.bbamcr.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Gajewska K.A., Lescesen H., Ramialison M., Wagstaff K.M., Jans D.A. Nuclear transporter Importin-13 plays a key role in the oxidative stress transcriptional response. Nat. Commun. 2021;12(1):5904. doi: 10.1038/s41467-021-26125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang L., Zhang X., Zhang G., Pang C.P., Leung Y.F., Zhang M., Zhong W. Expression profiling of the retina of pde6c, a zebrafish model of retinal degeneration. Sci. Data. 2017;4 doi: 10.1038/sdata.2017.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paul J.A., Aich A., Abrahante J.E., Wang Y., LaRue R.S., Rathe S.K., Kalland K., Mittal A., Jha R., Peng F., Largaespada D.A., Bagchi A., Gupta K. Transcriptomic analysis of gene signatures associated with sickle pain. Sci. Data. 2017;4 doi: 10.1038/sdata.2017.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conesa A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A A., Szczesniak M.W., Gaffney D.J., Elo L.L., Zhang X., Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15(2):121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 9.Anders S., Pyl P.T., Huber W. HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law C.W., Chen Y., Shi W., Smyth G.K. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dussault A.A., Pouliot M. Rapid and simple comparison of messenger RNA levels using real-time PCR. Biol. Proced. Online. 2006;8:1–10. doi: 10.1251/bpo114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. IPO13 expression in WT and KO ESCs. Total WT and KO ESC extracts were subjected to Western analysis using rabbit-anti-IPO13 with mouse-anti-β-actin as a loading control with blots imaged using the ChemiDoc Gel Imaging System (Biorad).
Data Availability Statement
Wild-type and IPO13-Knock Out (+/- Oxidative Stress) RNA-Sequencing (Original data) (DEGUST).
Transcriptomic profiling of untreated and oxidative stress treated Wild-type and IPO13-/- mouse embryonic stem cells to study the role of IPO13 in transcriptional response to stress (Original data) (NCBI Gene Expression Omnibus).