Abstract
Background & Aims
Direct-acting antiviral (DAA) regimens provide a cure in >95% of patients with chronic HCV infection. However, in some patients in whom therapy fails, resistance-associated substitutions (RASs) can develop, limiting retreatment options and risking onward resistant virus transmission. In this study, we evaluated RAS prevalence and distribution, including novel NS5A RASs and clinical factors associated with RAS selection, among patients who experienced DAA treatment failure.
Methods
SHARED is an international consortium of clinicians and scientists studying HCV drug resistance. HCV sequence linked metadata from 3,355 patients were collected from 22 countries. NS3, NS5A, and NS5B RASs in virologic failures, including novel NS5A substitutions, were examined. Associations of clinical and demographic characteristics with RAS selection were investigated.
Results
The frequency of RASs increased from its natural prevalence following DAA exposure: 37% to 60% in NS3, 29% to 80% in NS5A, 15% to 22% in NS5B for sofosbuvir, and 24% to 37% in NS5B for dasabuvir. Among 730 virologic failures, most were treated with first-generation DAAs, 94% had drug resistance in ≥1 DAA class: 31% single-class resistance, 42% dual-class resistance (predominantly against protease and NS5A inhibitors), and 21% triple-class resistance. Distinct patterns containing ≥2 highly resistant RASs were common. New potential NS5A RASs and adaptive changes were identified in genotypes 1a, 3, and 4. Following DAA failure, RAS selection was more frequent in older people with cirrhosis and those infected with genotypes 1b and 4.
Conclusions
Drug resistance in HCV is frequent after DAA treatment failure. Previously unrecognized substitutions continue to emerge and remain uncharacterized.
Lay summary
Although direct-acting antiviral medications effectively cure hepatitis C in most patients, sometimes treatment selects for resistant viruses, causing antiviral drugs to be either ineffective or only partially effective. Multidrug resistance is common in patients for whom DAA treatment fails. Older patients and patients with advanced liver diseases are more likely to select drug-resistant viruses. Collective efforts from international communities and governments are needed to develop an optimal approach to managing drug resistance and preventing the transmission of resistant viruses.
Keywords: RAS, HCV, DAA, virologic failure, NS5A
Abbreviations: aOR, adjusted odds ratio; DAA, direct-acting antiviral; DCV, daclatasvir; DSV, dasabuvir; GT, genotype; LDV, ledipasvir; NS5AI, NS5A replication complex inhibitor; NI, nucleoside; NNI, non-nucleoside; OR, odds ratio; PI, NS3 protease inhibitor; PIB, pibrentasvir; RASs, resistance-associated substitutions; sFC, substitution frequency change; SHARED, The Surveillance of Hepatitis C Antiviral Resistance, Epidemiology and methoDologies; SOF, sofosbuvir; SVR, sustained virologic response; VEL, velpatasvir
Graphical abstract
Highlights
-
•
An international cohort of 3,355 patients with hepatitis C from 22 countries was evaluated for drug resistance following DAA therapy.
-
•
Nearly all patients harbored drug-resistant variants after treatment failure, with over 2/3 having resistance against ≥2 drug classes.
-
•
Pathways to drug resistance included enrichment of highly resistant variants and selection of multiple resistant variants.
-
•
Previously unrecognized variants in patients who failed NS5A inhibitors were identified at high frequencies.
-
•
Resistance selection was frequent in older people with cirrhosis and those infected with genotypes 1b and 4 following DAA failure.
Introduction
The discovery and licensing of direct-acting antiviral (DAA) agents ushered in a new era of hepatitis C treatment. To date, over 95% of DAA-treated patients have achieved cure, defined as a sustained virologic response (SVR).1 Currently, approved regimens for HCV contain a combination of DAAs, namely, NS3 protease inhibitors (PIs), NS5A replication complex inhibitors (NS5AIs), and NS5B polymerase nucleoside (NI) and non-nucleoside (NNI) inhibitors. NS5AIs (daclatasvir [DCV], ledipasvir [LDV], elbasvir, and ombitasvir), PIs (simeprevir, grazoprevir, asunaprevir, and paritaprevir/r), and NNI (dasabuvir [DSV]) are considered "first-generation" DAAs because their activities are restricted to specific genotypes (GTs). NS5AIs (pibrentasvir [PIB] and velpatasvir [VEL]), PIs (glecaprevir and voxilaprevir), and an NI (sofosbuvir [SOF]) are pan-genotypic and referred to as "second-generation" DAAs.
Despite the remarkable progress in therapeutics, the extreme genetic diversity of HCV and the inability to mount a protective immune response against it present significant barriers to vaccine development. Nevertheless, the success of these DAAs has rekindled optimism around the potential for global elimination of HCV, which will rely on the "Treatment as Prevention" approach in the absence of an effective vaccine.2 One potential barrier to the success of this approach is the selection of drug-resistant viruses. The occurrence of resistance-associated substitutions (RASs), either natural or during treatment, can limit the efficiency of treatment and its scale-up. Transmission of resistant variants, particularly among high-risk groups, is of potential concern.3
Paradoxically, the therapeutic success has made studying HCV drug resistance a formidable task. Existing knowledge of HCV drug resistance is often derived from small, regional, short-term clinical studies with limited power to draw generalizable conclusions given the genetic heterogeneity of HCV GTs and subtypes.4 Current treatment regimens combining highly potent DAAs further mask the negative impact of RASs, making it difficult to delineate the mechanism of virologic failure. In studies where HCV resistance was addressed, there have been no consistent criteria to define RASs and no standardization of analytical methods. As a result, although Treatment Guidelines have recommended resistance testing in a subgroup of patients for some regimens, there has been no clear interpretation of drug resistance results.1
The Surveillance of Hepatitis C Antiviral Resistance, Epidemiology and methoDologies, SHARED, is an international consortium which aims to better understand and avoid HCV drug resistance and transmission through the development, application, and sharing of HCV genomic data, methods, software, and technologies.5 SHARED has brought together clinicians, virologists, and researchers from 22 countries, including over 110 individual clinics, hospitals, and reference laboratories. The pooled database comprises HCV sequences linked with detailed patient information, disease characteristics, treatment history, and clinical outcomes. The comprehensive data collected and its diverse sources provide an excellent opportunity to conduct in-depth analyses, generating insights into HCV antiviral resistance that are not possible from individual studies.
This report aims to a) provide a global resource on the prevalence and characteristics of RASs across GTs in patients who failed DAA therapy, b) explore clinical characteristics associated with RAS selection and, c) present evidence for new potential RASs in the NS5A region of the HCV genome. These results will raise awareness of HCV drug resistance, identify unmet needs, and encourage further data sharing on RASs.
Patients and methods
SHARED database
The SHARED database contains multiple types of metadata linked with HCV sequences.5 Data were collected at each collaborator's site under protocols approved by local medical or human research ethics committees. De-identified data were sent through a secured file transfer protocol server to the coordinating center at the University of British Columbia, Canada. Data were curated, formatted, and stored in a relational database using MYSQL. The schema of the SHARED database can be found at https://hcvdb.med.ubc.ca. The SHARED study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in a priori approval by the University of British Columbia Research Ethics Board (H17-10589).
Sequence handling
Consensus nucleotide sequences for HCV NS3, NS5A, and NS5B generated by population sequencing or next-generation sequencing (NGS) were collected. The cut-offs for variant calling were set at 15–20% for population sequencing and 5% for NGS sequencing. Genotypes and subtypes were assigned by mapping the sequences with a panel of 217 reference sequences downloaded from the International Committee on Taxonomy of Viruses.4 Consensus nucleotide sequences from each sample were codon aligned to the respective GT-specific references (Table S1) using the NucAmino program adapted from HIV, which was designed to handle IUPAC ambiguity codons caused by multiple substitutions.6 Amino acid sequences generated from the NucAmino program were used for RAS evaluations. Amino acids that differed from those in the reference sequences were considered substitutions. In amino acid positions with a mixture of 3 or fewer substitutions, this mixture was parsed into individual amino acids proportionally, e.g., Q30R/I (n = 7) was counted as Q30R (n = 3.5) and Q30I (n = 3.5). Positions that contained >3 amino acids or a stop codon were considered unknown and excluded from the analysis.
Prevalence, class resistance, and patterns of resistance-associated substitutions
The prevalence and patterns of natural RASs were evaluated using the baseline HCV sequences from DAA-treated patients and those waiting for treatment. The prevalence and patterns of post-treatment NS3, NS5A, and NS5B RASs were examined in virologic failures treated with PI-, NS5AI-, and NI/NNI-containing regimens, respectively. Virologic failures were considered for DAA class resistance evaluation if sequences from NS3, NS5A, NS5B were available. Patients were stratified by GT, and those who had ≥1 GT-specific RAS variant per the 2020 EASL recommendations on the treatment of hepatitis C were considered as harboring RAS in the prevalence analysis (supplementary patients and methods – M1).1 The RAS pattern and class RAS analyses included all substitutions from all listed amino acid positions of the respective drug target genes in the recommendations. Computations were done in Python v3.7 and visualized using Matplotlib (https://www.python.org).
Potential NS5A substitutions associated with virologic failure
Amino acid sequences derived from NS5AI-naive and NS5AI-exposed individuals were stratified according to their GTs/subtypes. The analyses were restricted to the first 200 amino acids in GTs 1a, 1b, 3, and 4 because most sequences collected were limited to this region. Sequences from GTs 2, 5, and 6 were too few to warrant analysis. Two independent methods were used. In the first approach, the substitution frequency change (sFC) before and after treatment at each amino acid position was determined. Amino acids with an sFC greater than 2 standard deviations from the background drifts were considered potentially associated with selection pressure from treatment. In the second approach, a pairwise comparison, with one amino acid at a time, between the treated and untreated groups was conducted using Fisher's exact test. A Benjamini-Hochberg method was employed to determine the minimum alpha values for multiple comparisons.7 In this analysis, amino acid positions with a p value associated with a false discovery rate of 15% (q <0.15) were considered positions of interest. Specific amino acids within the positions of interest associated with drug selection were identified using odds ratios (supplementary patients and methods – M2).
Association of resistance-associated substitutions and clinical factors
We assessed factors associated with the presence of NS3 and NS5A RASs in sequences collected after treatment among people who failed PI and NS5A therapy, respectively, with separate models evaluating people receiving PI-based and NS5A-based therapies. Patients who had ≥1 RAS variant of the respective drug target gene per the 2020 EASL recommendations were considered as harboring RAS(s). Independent variables considered included sex, age, injection drug use, HIV coinfection, cirrhosis, DAA regimen, treatment history, and HCV genotype. The age of the cohort was divided into quartiles (<52, 52–58, 58–63, and ≥64). Participants with a history of injection drug use (past and recent) were categorized as people who inject drugs. The definition of cirrhosis was based on the classification used by each of the participating centers. Unadjusted logistic regression analyses were performed to estimate odds ratios (ORs) and corresponding 95% CIs to identify factors associated with the presence of RASs in those who experienced treatment failure. All included variables were considered for multivariate logistic regression models to generate adjusted odds ratios (aORs) except injection drug use due to incomplete data. Missing data was assessed as its own category for all hypothesized predictor variables with less than 90% data availability. The analysis was performed using Stata v14.0 (StataCorp, College Station, Texas). A p value <0.05 was considered statistically significant.
Results
Cohort characteristics
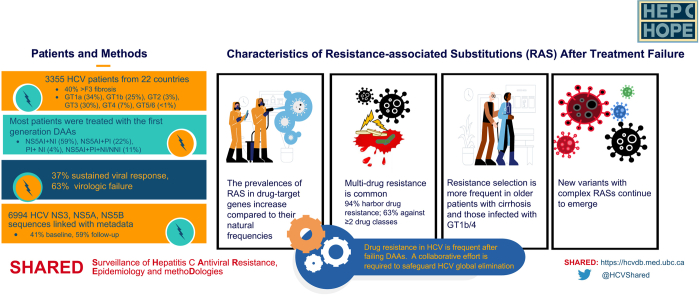
The study cohort (n = 3,355) comprised HCV sequences and metadata from the SHARED database collected from Argentina, Australia, Austria, Canada, France, Germany, Israel, Italy, Luxemburg, the Netherlands, Norway, New Zealand, Poland, Portugal, Romania, Russia, Slovenia, Spain, Switzerland, Sweden, Turkey, and the United States.5 Overall, 76% (2,483/3,273) of the population were male, and the median age was 57 years (interquartile range: 51–63). Most SHARED data came from reference laboratories and "real-world" clinics; therefore, not all records had the same metadata, especially the behavioral information. The available data are summarized in Table 1.
Table 1.
Cohort characteristics.
| Characteristics | Total1 |
|---|---|
| Number of patients, n | 3,355 |
| Sex, n (%) | 3,273 (98%) |
| Male, n (%) | 2,483 (76%) |
| Age, n (%) | 2,417 (72%) |
| Age in 2021, median (IQR) | 57 (51 - 63) |
| Ethnicity, n (%) | 652 (19%) |
| Caucasian, n (%) | 579 (89%) |
| Black, n (%) | 20 (3%) |
| Other, n (%) | 53 (8%) |
| Illicit drug use, n (%) | 746 (22%) |
| Injection drug use, n (%)2 | 488 (65%) |
| Non-injection drug use, n (%)2 | 219 (29%) |
| Sexual orientation, n (%) | 496 (15%) |
| Heterosexual, n (%) | 290 (58%) |
| Homosexual, n (%) | 187 (38%) |
| Bisexual, n (%) | 18 (4%) |
| Coinfection, n (%) | 2,030 (61%) |
| HIV-HCV, n (%)3 | 482 (24%) |
| HBV-HCV, n (%)3 | 73 (4%) |
| Fibrosis, n (%) | 1,418 (42%) |
| F0-F1, n (%) | 438 (31%) |
| F2, n (%) | 286 (20%) |
| F3, n (%) | 182 (13%) |
| F4, n (%) | 512 (36%) |
| Cirrhosis, n (%) | 2,139 (64%) |
| Yes, n (%) | 911 (43%) |
| Hepatocellular carcinoma, n (%) | 262 (8%) |
| Yes, n (%) | 26 (10%) |
| Genotype4, n (%) | 3,355 (100%) |
| GT1a, n (%) | 1,145 (34%) |
| GT1b, n (%) | 848 (25%) |
| GT1-other, n (%) | 27 (1%) |
| GT2, n (%) | 102 (3%) |
| GT3, n (%) | 999 (30%) |
| GT4, n (%) | 219 (7%) |
| GT5, n (%) | 1 (0.03%) |
| GT6, n (%) | 14 (0.4%) |
| DAA treatment5, n (%) | 3,353 (100%) |
| NS5AI + NI, n (%) | 1,990 (59%) |
| NS5AI + PI, n (%) | 722 (22%) |
| PI + NI, n (%) | 134 (4%) |
| NS5AI + PI + NI or NNI, n (%) | 373 (11%) |
| Other, n (%) | 134 (4%) |
| Response to DAA treatment, n (%) | 3,015 (90%) |
| Sustained viral response | 1,121 (37%) |
| Virologic failure | 1,894 (63%) |
| Treatment history6, n (%) | 2,260 (67%) |
| Treatment naïve, n (%) | 1,557 (69%) |
| Treatment experienced, n (%) | 703 (31%) |
| Prior PEG/RBV, n (%) | 357 (51%) |
| Prior DAA, n (%) | 107 (15%) |
| Unknown, n (%) | 239 (34%) |
| HCV sequences, n | 6,994 |
| NS3, n (%) | 1,872 (27%) |
| NS5A, n (%) | 3,367 (48%) |
| NS5B, n (%) | 1,755 (25%) |
DAA, direct-acting antiviral; GT, genotype; NI, nucleoside (sofosbuvir); NNI, non-nucleoside (dasabuvir); NS5AI, NS5A inhibitor; other, pegylated interferon +/- ribavirin +/- DAA including boceprevir, telaprevir; PEG, pegylated interferon; PI, protease inhibitor; RBV, ribavirin.
The number of patients with data available in the specified category
Injection drug use and non-injection drug use were not mutually exclusive; 189 (25%) participants engaged in both. Injection drug use and non-injection drug use referred to having a history of drug use, including past and recent.
HIV- and HBV-coinfection with HCV were not mutually exclusive; 14 (0.7%) participants were infected with HIV and HBV in addition to HCV.
Genotypes were derived from the HCV NS5A, NS3, or NS5B sequences. GT1-other included GT1c/d/e/g/h/i/j/k/l/n/o. Non-GT1 subtypes included GT2a/b/c/d/i/j/m/r/q/t/u, GT3a/b/g/h/i/k, GT4a/b/d/f/g/k/l/n/o/q/r/v, GT5a, GT6a/e/h/l/p/q/r/xe.
DAA treatment associated with the HCV sequence examined.
Treatment history at the time when DAA treatment was administered.
Clinically, 482 of 2,030 patients (24%) providing coinfection data were co-infected with HIV. Most had advanced liver diseases; over 40% had greater than F3 fibrosis score (694/1,418) or cirrhosis (911/2,139), and 10% (26/262) had hepatocellular carcinoma. Most patients were infected with HCV GT1 (2,020/3,355, 60%) and GT3 (999/3,355, 30%). A majority were treated with regimens containing an NS5AI: 59% (1,990/3,353) NS5AI+NI, 22% (722/3,353) NS5AI+PI, and 11% (373/3,353) NS5AI+PI+NI/NNI. A small group of patients (134/3,353, 4%) were prescribed a PI+NI combination. Ribavirin (RBV) was used in 22% of the DAA regimens.
Treatment response at the time of sample collection for sequencing was reported in 90% (3,015/3,353) of the DAA-treated patients; 37% (1,121/3,015) achieved SVR, and 63% (1,894/3,015) experienced virologic failure. Fig. S1 shows the distribution of DAA regimens administered among the virologic failures in this cohort. Among the DAA-treated patients, treatment history information was available in 2,260 patients, of whom 31% (703/2,260) had prior treatment experience, mainly with pegylated interferon and RBV (357/703, 51%).
A total of 6,994 HCV sequences, including NS3 (n = 1,872), NS5A (n = 3,367), and NS5B (n = 1,755), were available, and 59% of all sequences were collected at end-of-treatment or follow-up visits after treatment; the remaining 41% were collected before treatment.
Prevalence of resistance-associated substitutions in DAA-naïve and -experienced patients
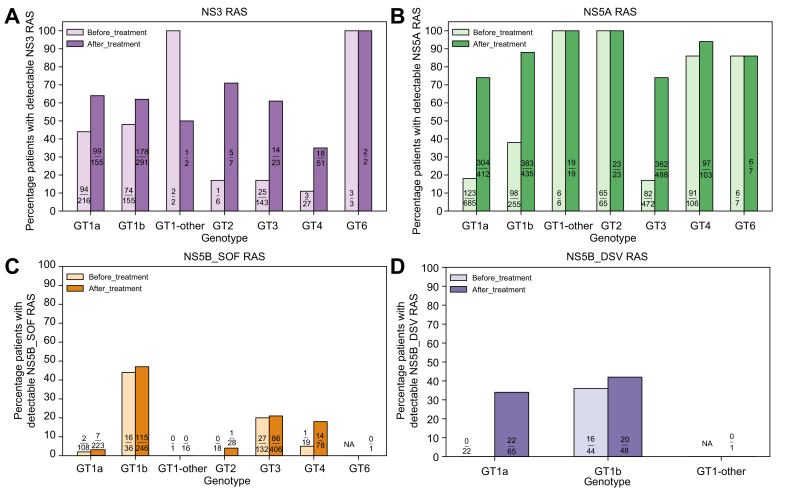
The natural RAS prevalence varied among GTs with an average frequency of 37% (202/552) in NS3, 29% (417/1,597) in NS5A, 15% (46/314) in NS5B_SOF, and 24% (16/66) in NS5B_DSV based on the RAS variants listed in the 2020 EASL recommendations (Fig. 1).1
Fig. 1.
Prevalence of resistance-associated substitutions in DAA-naïve and -experienced patients with HCV.
HCV sequences from virologic failures treated with combination regimens containing a PI (SIM, GZR, PAR/r, GLE, ASV, VOX, BOC, TVR), an NS5AI (LDV, DCV, EBR, VEL, OMB, PIB), or an NI (SOF)/NNI (DSV) were evaluated for the prevalence of RASs in NS3, NS5A, NS5B_SOF, and NS5B_DSV, respectively. HCV DAA-naïve patients were included to estimate the natural RAS prevalence. Patients who harbored ≥1 RAS variant listed in the 2020 EASL recommendations on treatment of hepatitis C were counted. ASV, asunaprevir; BOC, boceprevir; DAA, direct-acting antiviral; DCV, daclatasvir; DSV, dasabuvir; EBR, elbasvir; GLE, glecaprevir; GT, genotype; GT1-other, any GT1 subtypes except GT1a and GT1b; GZR, grazoprevir; LDV, ledipasvir; NA, not available; NI, nucleoside inhibitor; NNI, non-nucleoside inhibitor; NS5AI, NS5A inhibitor; OMB, ombitasvir; PAR/r, paritaprevir/ritonavir; PI, protease inhibitor; PIB, pibrentasvir; RAS(s), resistance-associated substitution(s); SIM, simeprevir; SOF, sofosbuvir; TVR, telaprevir; VEL, velpatasvir; VOX, voxilaprevir.
The RAS prevalence increased following treatment failure with an average frequency of 60% (317/531) in NS3, 80% (1,194/1,487) in NS5A, 22% (223/999) in NS5B_SOF, and 37% (42/114) in NS5B_DSV. The RAS prevalence varied among GTs in patients who failed NS5AI-, PI- or SOF-containing regimens: 35–100% in NS3, 74–100% in NS5A, and 0–47% in NS5B (NS5B_SOF), respectively (Fig. 1). About 34–42% of GT1 patients who failed DSV had detectable NS5B_DSV RAS. Compared to DAA-naïve patients, the frequencies of NS3 RASs following DAA exposure were significanlty higher in GTs 1a (64% vs. 44%, p <0.001), 1b (61% vs. 48%, p = 0.007), 3 (61% vs. 17%, p <0.001), and 4 (35% vs. 12%, p = 0.03) (Fig. 1A). Similarly, a notable difference of NS5A RAS prevalence was seen in GT1a (74% vs. 18%, p <0.001), 1b (88% vs. 38%, p <0.001), and 3 (74% vs. 17%, p <0.001) after NS5AI exposure (Fig. 1B). There was no major difference in pre- and post-treatment NS5B_SOF RAS frequencies (Fig. 1C). Interestingly, the NS5B_DSV RAS prevalence was 0% (0/22) pre-treatment vs. 34% (22/65) post-treatment in GT1a, but similar before (16/44, 36%) and after (20/48, 42%) treatment in GT1b (Fig. 1D).
The RAS variants' characteristics in different GTs are described in the supplementary results – R1. Additionally, Table S2 summarizes the newly observed RAS variants in the SHARED cohort that were not listed in the 2020 EASL recommendations.
Direct-acting antiviral class resistance in patients after first-line DAA therapies
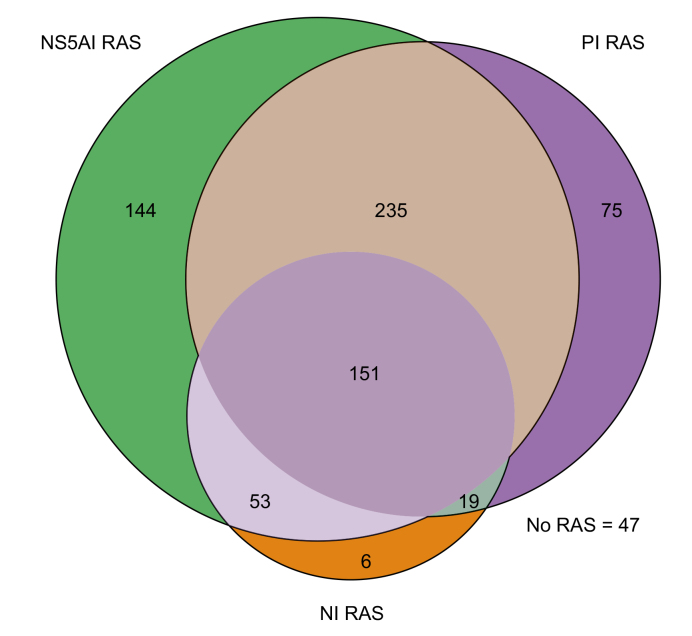
HCV DAAs appear to generate little or no cross-resistance to drugs from other classes. Rescue therapies switching to a different DAA class or adding a new DAA class not present in the previous regimen are practical approaches to treat patients who failed first-line therapies.8 Information on DAA class resistance following treatment failure can provide insights on retreatment strategies. To evaluate class resistance distribution following DAA failure, we included only virologic failures (n = 730) with available NS3, NS5A, and NS5B sequences to reduce sampling bias. This subgroup's patient characteristics and treatment regimen distribution were similar to all virologic failures (n = 1,894) in the SHARED cohort (Table S3); most were treated with the first-generation DAAs. Overall, 94% (683/730) of this subgroup selected drug resistance against ≥1 DAA class. Among them, 31% (225/730) had single-class resistance, 42% (307/730) harbored dual-class resistance, predominantly against PIs and NS5AIs, 21% (151/730) selected resistance against all 3 DAA classes, and 6% (47/730) did not have RAS (Fig. 2). The resistance profiles following NS5AI+PI, NS5AI+SOF, and PI+SOF failure are provided in Fig. S2A-C, which shows that resistance contributed by SOF was small compared to those contributed by PIs and NS5AIs. Nevertheless, 27% (23/85) and 31% (214/688) of patients harbored dual-class resistance after PI+SOF and NS5AI+SOF treatments, respectively. Strikingly, dual-class resistance in NS3 and NS5A increased to 62% (82/132) for those who failed NS5AI+PI combinations.
Fig. 2.
Micro-representation of direct-acting antiviral class resistance after failing first-line therapies.
A subset of virologic failures (n = 730) with sequences available from all 3 drug target genes (NS3, NS5A, and NS5B) were included for the analyses. Amino acid substitutions at all positions of the respective genes listed in the 2020 EASL recommendations on treatment of hepatitis C were included for the evaluation. PI RAS, NS5AI RAS, and NI RAS represent RASs selected in the HCV NS3, NS5A, and NS5B genes, respectively. Each circle represents the number of patients with detectable RAS in each drug class: purple, PI; green, NS5AI; and orange, NI. The size of the circle is proportional to the number of patients with detectable RASs. The intersecting regions represent dual- or triple-class resistance. NS5AI, NS5A inhibitor; NI, nucleoside inhibitor (sofosbuvir); PI, protease inhibitor; RASs, resistance-associated substitutions.
Patterns of resistance-associated substitutions in NS5A
There are over 3,300 NS5A sequences in the SHARED database, providing an opportunity to conduct an in-depth RAS characterization. Since longitudinal samples were not usually available from real-world clinics, we used the NS5A sequences collected before (n = 1,546) and after (n = 1,480) NS5AI treatment and compared the frequencies and RAS patterns to explore the resistance pathways. This RAS pattern analysis mainly focused on GT1a, 1b, 3, and 4; GT2 and GT6 could not be adequately evaluated due to the small sample size. The number of NS3 and NS5B sequences was too small to permit analysis with confidence after treatment and GT stratifications. For completeness, a summary of the key RAS patterns in NS3, NS5A, and NS5B for all GTs is listed in Tables S4–S7.
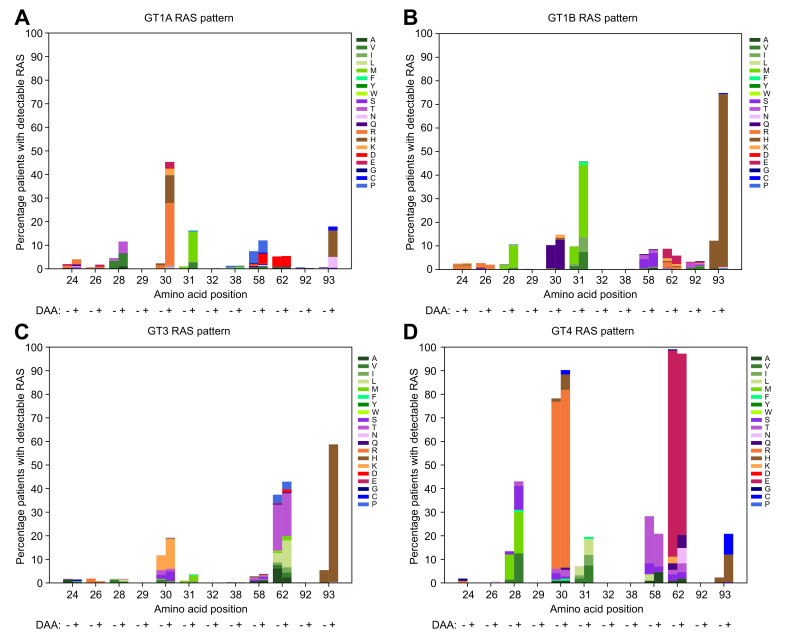
In GT1a, Q30R/H (163/424, 38%), followed by L31M (54/424, 13%) and Y93H/N (67/424, 16%), were most frequently detected in the samples collected after exposure to NS5AIs (Fig. 3A). The frequencies of RASs at these amino acids were low (<3%) at baseline but rose 18- to 20-fold after treatment (Fig. 3A). M28V/T, present in 4% (29/697) of the samples before treatment, had a modest 2-fold increase in frequency after treatment. M28T and M28V conferred up to 9,000- and 58-fold resistance in vitro, respectively, to the first-generation NS5AIs.9 Among the 424 GT1a virologic failures, 23% had no detectable RAS and 39% had single substitutions (Fig. 4). Q30R/H, L31M, and Y93H/N made up the majority of the viral variants with single substitutions (Table S4). In HCV replicons, these RASs displayed high resistance levels to most NS5AIs (100's- to 1,000's-fold) except PIB (1- to 7-fold).9,10
Fig. 3.
NS5A resistance patterns in different genotypes.
NS5A amino acid sequences obtained from patients treated with NS5A inhibitor-containing regimens before (DAA: -) and after (DAA: +) treatment were grouped according to their genotypes. The number of sequences before/after DAA treatment was 697/424 in GT1a, 262/451 in GT1b, 479/497 in GT3, 108/108 in GT4. The number of variant amino acids relative to the reference at each indicated position was enumerated and normalized to the total number of sequences in the respective genotypes. The amino acids are colored according to their side-chain biochemical properties: green = hydrophobic, purple = polar uncharged, orange = positively charged, red = negative charged, blue = special cases. The percentages of patients with detectable RASs in genotypes 1a, 1b, 3, and 4 are shown. The NS5AI-naïve sequences contained subtypes 3a/b/g/h and 4a/d/f/l/o/q/r/v. The NS5AI-exposed sequences contained subtypes 3a/b/g/h/k and 4a/b/d/f/g/k/n/o/q/r/v. DAA, direct-acting antiviral; GT, genotype; RASs, resistance-associated substitutions.
Fig. 4.
Number of NS5A substitutions after direct-acting antiviral failure in different genotypes.
NS5A sequences collected from patients at the follow-up visits after failing NS5AI-treatment were stratified according to their genotypes. There were 424 GT1a, 451 GT1b, 23 GT2, 497 GT3, 108 GT4, and 7 GT6. Amino acid variants relative to the genotype-specific reference strains were evaluated at positions 24, 26, 28, 29, 30, 31, 32, 38, 58, 62, 92, and 93 within NS5A. The number of detectable RASs for each sample was binned into the respective substitution subgroups. The number of patients in each substitution subgroup was summed and normalized to the total number in each genotype (percentage of patients, y-axis). GT, genotype; RASs, resistance-associated substitutions.
The signature substitutions for GT1b after NS5AI failure were L31M and Y93H in 31% and 73% of cases, respectively, out of 451 follow-up samples (Fig. 3B). These RASs existed at a moderate level (∼11%) at baseline but increased about 6-fold following drug selection. A third RAS, R30Q, was present in 10% (27/262) of patients before treatment and remained almost the same at 15% (67/451) after treatment. R30Q and L31M have low resistance (<3-fold) to NS5AIs. However, Y93H is highly resistant (30- to 1,325-fold) to all first-generation NS5AIs.9 In contrast to GT1a, in which most treatment failures had no or one substitution, 48% (214/451) and 18% (79/451) of the GT1b virologic failures had 2 or ≥3 substitutions, respectively (Fig. 4). Most double substitutions contained Y93H with L31M/V/I (Table S4).
Y93H emerged as the most commonly selected substitution in GT3, with a 10-fold increase in frequency following treatment failure (290/497, 58% in NS5AI-exposed vs. 27/479, 6% in NS5AI-naive) (Fig. 3D). Another RAS, A30K/S, which existed at a moderate level (40/479, 8%) at baseline, increased 2-fold after treatment. Both RASs have limited impact on PIB but are highly resistant to all other NS5AIs; 700- to 40,000-fold for Y93H and 50- to 177-fold for A30K.9 At virologic failure, 17% (85/497) of the patients had no RAS, 38% (189/497) had single substitutions, and 38% (189/497) had 2 substitutions (Fig. 4). Most of the samples with single substitutions were characterized by Y93H, whereas samples containing dual substitutions often presented as A30K/S + Y93H or S62T/L + Y93H (Table S4).
One distinguishing feature in the RAS pattern for GT4 virologic failures was the presence of multiple substitutions (Fig. 3D). Of the 108 follow-up samples, 102 (94%) contained ≥2 RASs (Fig. 4) (73% if position 62 was excluded). HCV variants containing L30R in combination with 1 to 3 substitutions from L28M/S/V, M31V/L/I, P58T, or Y93H/C were common (Table S4). L30R, mainly in GT4d as a natural polymorphism, remained constant before and after treatment (77/108, 71% vs. 82/108, 76%). In HCV GT4a replicons, L30R conferred 30- to 180-fold resistance to DCV and LDV and <2-fold to VEL and PIB.9
A sizable portion of samples contained substitutions at amino acids 58 and 62 with high frequencies before and after treatment (Fig. 3). These amino acids were polymorphic in natural isolates. The role of these 2 amino acids in drug resistance is unclear due to limited in vitro data; however, all available results to date showed minimal drug resistance except 58D/N.9
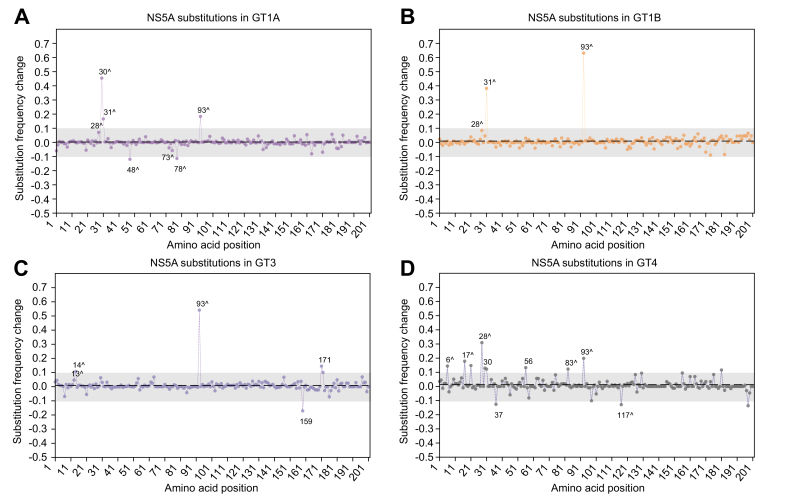
Novel substitutions potentially associated with virologic failure in NS5A
Using 2 independent approaches, we performed a systematic codon analysis across NS5A to identify novel substitutions potentially associated with virologic failure. Similar analyses were not conducted in NS3 and NS5B due to a limited number of sequences after GT stratification and variable sequence lengths. In the substitution frequency approach, the background genetic fluctuations (mean sFC ± standard deviation) were similar among the 4 GTs/subtypes examined: 0.003 ± 0.042 in GT1a, 0.009 ± 0.056 in GT1b, 0.009 ± 0.046 in GT3, and 0.012 ± 0.047 in GT4. Several amino acids (with sFC >±0.1) potentially associated with selection pressure from treatment were observed (Fig. 5A-D). Additional amino acids with a false discovery rate of 15% were identified in the pairwise comparison approach (Table 2). Known RASs (listed in the 2020 EASL recommendations and discussed earlier) at amino acids 28, 30, 31, 93 were confirmed by one or both methods (supplementary results – R2).
Fig. 5.
Substitution frequency change before and after treatment in NS5A.
The sFC relative to the reference amino acids before and after treatment was calculated independently at each of the first 200 amino acids within NS5A. The dotted line represents the mean sFC across the 200 amino acids in each genotype. The gray bar across the amino acids depicts a "genetic drift corridor" (2 SD from the mean sFC) to exclude variants with usual genetic drifts. Amino acid positions with a significant sFC before and after treatment are shown. A "positive" sFC represents an increase in the substitution levels (or decrease in the reference amino acid levels) after treatment; a "negative" sFC represents a decrease in the substitution levels (or increase in the reference amino acids) after treatment. Amino acid positions with a high-frequency change but without a specific substitution (i.e., only a mixture of substitutions) are not indicated. The amino acids of interest identified by the pairwise comparison are also noted (ˆ) for comparison. GT, genotype; sFC, substitution frequency change.
Table 2.
Amino acid substitutions in NS5A before and after treatment.
| Amino acid position | Substitution frequency change1 | FET p value | False discovery rate (q)2 | Amino acid3 | Odds ratio (95% CI)4 |
|---|---|---|---|---|---|
|
GT1a | |||||
| 28 | 0.07 | <0.0001ˆ,∗∗ | 0.003 | M28T, M28V | 2.6 (1.6–4.2) |
| 30 | 0.46# | <0.0001ˆ,∗∗ | <0.001 | Q30H, Q30K, Q30R,Q30E | 36.0 (20.9–62.3) |
| 31 | 0.17# | <0.0001ˆ,∗∗ | <0.001 | L31M, L31V | 18.1 (8.3–40.0) |
| 48 | -0.12# | <0.0001ˆ,∗∗ | 0.003 | R48K (decrease) | 0.5 (0.4–0.7) |
| 73 | -0.04 | 0.0007ˆ | 0.019 | R73K (decrease) | 0.3 (0.1–0.9) |
| 78 | -0.11# | <0.0001ˆ,∗∗ | 0.004 | R78K (decrease) | 0.6 (0.5–0.8) |
| 93 |
0.18# |
<0.0001ˆ,∗∗ |
<0.001 |
Y93C, Y93H, Y93N |
49.5 (15.5–158.3) |
|
GT1b | |||||
| 28 | 0.08 | <0.0001ˆ,∗∗ | 0.001 | L28M | 5.4 (2.1–13.9) |
| 31 | 0.38# | <0.0001ˆ,∗∗ | <0.001 | L31I, L31M, L31V | 7.4 (4.7–11.5) |
| 93 |
0.63# |
<0.0001ˆ,∗∗ |
<0.001 |
Y93H |
20.6 (13.4–31.7) |
|
GT3 | |||||
| 13 | 0.05 | 0.002ˆ | 0.114 | C13S | 3.3 (1.4–8.2) |
| 14 | 0.11# | <0.0001ˆ,∗∗ | 0.085 | S14T | 1.5 (1.2–2.0) |
| 93 | 0.54# | <0.0001ˆ,∗∗ | <0.001 | Y93H | 23.6 (15.3–36.2) |
| 159 | -0.17# | 0.02 | 0.207 | H159Q (decrease) | 0.3 (0.1–0.9) |
| 171 |
0.15# |
0.02 |
0.207 |
E171D |
13.9 (1.7–114.2) |
|
GT4 | |||||
| 6 | 0.14# | 0.004ˆ | 0.115 | W6R | 4.0 (1.3–12.7) |
| 17 | 0.18# | <0.0001ˆ,∗∗ | <0.001 | S17T | 34.3 (2.0–583.3) |
| 28 | 0.31# | <0.0001ˆ,∗∗ | <0.001 | L28S, L28V | 10.5 (3.0–36.1) |
| 30 | 0.13# | 0.001 | 0.241 | L30R | 2.8 (1.2–6.4) |
| 37 | -0.13# | 0.07 | 0.314 | L37F (decrease) | 0.5 (0.3–0.9) |
| 56 | 0.13# | 0.04 | 0.314 | T56R | 18.5 (1.1–325.4) |
| 83 | 0.12# | 0.0003ˆ | 0.015 | T83V | 12.3 (1.6–96.9) |
| 93 | 0.20# | <0.0001ˆ,∗∗ | 0.001 | Y93C, Y93H | 9.3 (2.7–32.0) |
| 117 | -0.13# | 0.0009ˆ | 0.031 | D117E (decrease) | 0.1 (0.02–0.5) |
FET, Fisher's exact test; GT, genotype.
Substitution frequency after treatment minus substitution frequency before treatment.
Acceptable false discovery rate was set at 15% (q-value ≤0.15)
Specific amino acid substitution observed within the amino acid position of interest. "Decrease" represents decreased substitution frequency, i.e., the level of wild-type reference amino acid increased.
Odds ratio for the amino acids at the positions of interest
Substitution frequency change >2 standard deviations from the mean frequency change across the first 200 amino acids in NS5A.
below Benjamin Hochberg critical value with 15% false discovery rate (q ≤0.15)
FET p value < Bonferroni adjusted alpha level of 0.00025.
In GT1a, previously unrecognized amino acid positions 48 (sFC = -0.12, p <0.0001), 73 (p = 0.0007), and 78 (sFC = -0.11, p <0.0001) had a significant frequency change following drug selection (Fig. 5A and Table 2). Within these positions, R48K (OR = 0.5, 95% CI 0.4–0.7), R73K (OR = 0.3, 95% CI 0.1–0.9), and R78K (OR = 0.6, 95% CI 0.5–0.8) were negatively selected after NS5AI exposure (i.e., the level of substitutions decreased and the level of wild-type increased); these were likely adaptive changes.
In GT3, several previously undescribed substitutions at amino acid positions 13 (p = 0.002), 14 (sFC = 0.11, p <0.0001), 159 (sFC = -0.17), and 171 (sFC = 0.15) were identified by one or both methods (Fig. 5C and Table 2). Specifically, C13S (OR = 3.3, 95% CI 1.4–8.2), S14T (OR = 1.5, 95% CI 1.2–2.0), and E171D (OR = 13.9, 95% CI 1.7–114.2) were more frequently detected in patients following NS5AI failure. In contrast, H159Q (OR = 0.3, 95% CI 0.1–0.9) was less frequently observed after treatment.
In GT4, several new amino acid positions were identified; these included positions 6 (sFC = 0.14, p = 0.004), 17 (sFC = 0.18, p <0.0001), 37 (sFC = -0.13), 56 (sFC = 0.13), 83 (sFC = 0.12, p = 0.0003), 117 (sFC = -0.13, p = 0.0009) (Fig. 5D and Table 2). The odds of having W6R (OR = 4.0, 95% CI 1.3–12.7), S17T (OR = 34.3, 95% CI 2.0–583.3), T56R (OR = 18.5, 95% CI 1.1–325.4), and T83V (OR = 12.3, 95% CI 1.6–96.9) were higher in patients after exposure to NS5AIs. In contrast, L37F (OR = 0.5, 95% CI 0.3–0.9) and D117E (OR = 0.1, 95% CI 0.02–0.5) were less frequently seen after treatment.
Factors associated with resistance-associated substitutions in virologic failure
To identify if clinical or demographic factors might affect RAS selection, we examined the presence of post-treatment NS3 and NS5A RASs in 531 PI- and 1,487 NS5AI-treated patients, respectively, who failed to achieve SVR.
In the unadjusted bivariate analyses, the presence of NS3 RASs in patients following PI-treatment failure was positively associated with cirrhosis (p = 0.001); and negatively associated with HCV GT4 infection (p <0.001) (Table 3). In multivariate logistic regression adjusted analysis, having cirrhosis (aOR 1.93, 95% CI 1.09–3.43; p = 0.02) was independently associated with increased NS3 RAS selection; while GT4 HCV infection (aOR 0.33, 95% CI 0.15–0.71; p = 0.005) was inversely associated with NS3 RASs compared to GT1a.
Table 3.
Factors associated with NS3 and NS5A resistance-associated substitutions in virologic failures.
| NS31 |
NS5A2 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAS present; n (%) n = 317 |
RAS absent; n (%) n = 214 |
Unadjusted OR | p value | Adjusted OR (95% CI) |
p value | RAS present; n (%) n = 1,194 |
RAS absent; n (%) n = 293 |
Unadjusted OR | p value | Adjusted OR (95% CI) |
p value | |
| Sex | ||||||||||||
| Female | 91 (58) | 65 (42) | 1.00 | – | 1.00 | – | 270 (88) | 38 (12) | 1.00 | – | 1.00 | |
| Male | 222 (60) | 148 (40) | 1.07 (0.73–1.57) | 0.72 | 1.25 (0.79–1.99) | 0.34 | 866 (78) | 238 (22) | 0.51 (0.35–0.74) | <0.001 | 0.71 (0.47–1.07) | 0.10 |
| Age (quartiles) | ||||||||||||
| <52 | 80 (57) | 61 (43) | 1.00 | – | 1.00 | – | 286 (76) | 92 (24) | 1.00 | – | 1.00 | |
| 52-58 | 59 (52) | 55 (48) | 0.82 (0.50–1.34) | 0.43 | 0.90 (0.31–1.54) | 0.71 | 300 (76) | 94 (24) | 1.03 (0.74–1.43) | 0.88 | 0.89 (0.62–1.29) | 0.54 |
| 58-63 | 79 (63) | 46 (37) | 1.31 (0.80–2.14) | 0.29 | 1.21 (0.71–2.06) | 0.49 | 228 (80) | 57 (20) | 1.29 (0.89–1.87) | 0.20 | 1.09 (0.72–1.67) | 0.68 |
| ≥ 64 | 70 (64) | 40 (36) | 1.33 (0.80–2.23) | 0.27 | 1.18 (0.65–2.14) | 0.60 | 316 (90) | 36 (10) | 2.82 (1.86–4.29) | <0.001 | 1.97 (1.21–3.19) | 0.01 |
| History of injecting drug use | ||||||||||||
| No | 43 (56) | 34 (44) | 1.00 | – | 64 (81) | 15 (19) | 1.00 | – | ||||
| Yes | 17 (61) | 11 (39) | 1.22 (0.51–2.95) | 0.66 | 56 (70) | 24 (30) | 0.55 (0.26–1.14) | 0.11 | ||||
| Unknown | 257 (60) | 169 (40) | 1.20 (0.74–1.96) | 0.46 | 1074 (81) | 254 (19) | 0.99 (0.56–1.77) | 0.98 | ||||
| HIV | ||||||||||||
| No | 204 (60) | 137 (40) | 1.00 | – | 1.00 | – | 507 (85) | 90 (15) | 1.00 | – | 1.00 | |
| Yes | 24 (48) | 22 (52) | 0.61 (0.32–1.16) | 0.13 | 0.72 (0.33–1.58) | 0.41 | 92 (69) | 42 (31) | 0.39 (0.25–0.60) | <0.001 | 0.60 (0.35–1.04) | 0.07 |
| Unknown | 93 (63) | 55 (37) | 1.14 (0.76–1.69) | 0.53 | 0.99 (0.44–2.23) | 0.98 | 595 (79) | 161 (21) | 0.66 (0.49–0.87) | 0.004 | 1.02 (0.48–2.18) | 0.96 |
| Cirrhosis | ||||||||||||
| No | 70 (49) | 72 (51) | 1.00 | – | 1.00 | – | 212 (75) | 71 (25) | 1.00 | – | 1.00 | |
| Yes | 129 (68) | 62 (32) | 2.14 (1.37–3.35) | 0.001 | 1.93 (1.09–3.43) | 0.02 | 520 (84) | 98 (16) | 1.78 (1.26–2.51) | 0.001 | 1.80 (1.18–2.76) | 0.01 |
| Unknown | 118 (60) | 80 (40) | 1.52 (0.98–2.34) | 0.06 | 1.79 (0.93–3.43) | 0.08 | 490 (84) | 96 (16) | 1.25 (0.89–1.74) | 0.19 | 1.50 (0.95–2.37) | 0.08 |
| DAA regimen | ||||||||||||
| NS5AI + PI + NI | 98 (59) | 69 (41) | 1.00 | – | 1.00 | – | 149 (84) | 29 (16) | 1.00 | – | 1.00 | |
| NS5AI + PI | 77 (55) | 62 (45) | 0.87 (0.55–1.38) | 0.56 | 1.34 (0.77–2.33) | 0.31 | 893 (79) | 242 (21) | 0.72 (0.47–1.10) | 0.13 | 0.70 (0.41–1.19) | 0.19 |
| PI + NI or NS5AI + NI | 92 (69) | 41 (31) | 1.58 (0.98–2.55) | 0.06 | 1.39 (0.74–2.62) | 0.30 | 147 (88) | 20 (12) | 1.43 (0.77–2.64) | 0.25 | 1.18 (0.58–2.38) | 0.65 |
| PI + PEG/RBV | 50 (54) | 42 (46) | 0.84 (0.50–1.40) | 0.50 | 1.07 (0.54–2.10) | 0.85 | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. |
| Treatment history | ||||||||||||
| Naïve | 267 (60) | 178 (40) | 1.00 | – | 1.00 | – | 994 (80) | 246 (20) | 1.00 | – | 1.00 | |
| Prior PEG/RBV | 36 (59) | 25 (41) | 0.96 (0.56–1.65) | 0.88 | 0.58 (0.29–1.15) | 0.12 | 140 (79) | 38 (21) | 0.91 (0.62–1.34) | 0.64 | 0.65 (0.40–1.06) | 0.08 |
| Prior DAA | 14 (56) | 11 (44) | 0.85 (0.38–1.91) | 0.69 | 0.62 (0.23–1.64) | 0.33 | 60 (87) | 9 (13) | 1.65 (0.81–3.37) | 0.17 | 1.40 (0.63–3.14) | 0.41 |
| Genotype | ||||||||||||
| 1a | 99 (64) | 56 (36) | 1.00 | – | 1.00 | – | 304 (74) | 108 (26) | 1.00 | – | 1.00 | |
| 1b | 178 (61) | 113 (39) | 0.89 (0.60–1.33) | 0.58 | 0.78 (0.45–1.35) | 0.40 | 383 (88) | 52 (12) | 2.62 (1.82–3.76) | <0.001 | 1.77 (1.15–2.75) | 0.01 |
| 1-other | 1 (50) | 1 (50) | 0.57 (0.03–9.22) | 0.69 | 0.56 (0.03–10.53) | 0.70 | 19 (100) | 0 (0) | n.a. | n.a. | n.a. | n.a. |
| 2 | 5 (71) | 2 (29) | 1.41 (0.27–7.53) | 0.69 | 1.44 (0.25–8.41) | 0.69 | 23 (100) | 0 (0) | n.a. | n.a. | n.a. | n.a. |
| 3 | 14 (61) | 9 (39) | 0.88 (0.36–2.16) | 0.78 | 1.19 (0.43–3.33) | 0.74 | 362 (74) | 126 (26) | 1.02 (0.76–1.38) | 0.89 | 0.83 (0.58–1.20) | 0.32 |
| 4 | 18 (35) | 33 (65) | 0.31 (0.16–0.60) | <0.001 | 0.33 (0.15–0.71) | 0.005 | 97 (94) | 6 (6) | 5.74 (2.45–13.28) | <0.001 | 10.20 (3.52–29.56)) | <0.001 |
| 6 | 2 (100) | 0 (0) | n.a. | n.a. | n.a. | n0.a0. | 6 (86) | 1 (14) | 2.13 (0.25–17.91) | 0.50 | 1.80 (0.20–16.48) | 0.60 |
Values in bold denote statistical significance. Odds ratio and logistic regression analysis were used to generate the results.
DAA, direct-acting antiviral; GT, genotype; 1-other; GT1 subtypes excluding GT1a and GT1b; n.a., not applicable; –, reference; NI, nucleoside inhibitor; NS5AI, NS5A inhibitor; OR, odds ratio; PEG, pegylated interferon; PI, protease inhibitor; RAS, resistance-associated substitution; RBV, ribavirin.
NS3 sequences from virologic failures treated with a PI-containing regimen.
NS5A sequences from virologic failures treated with an NS5AI-containing regimen.
In the unadjusted bivariate analyses, the presence of NS5A RASs in patients who failed NS5AI treatment were higher in people aged 64 and over (p <0.001), having cirrhosis (p <0.001), and being infected with GT1b (p <0.001) or GT4 (p <0.001) (Table 3). Whereas, male patients (p <0.001) and those who were co-infected with HIV were less likely to harbor NS5A RASs (p <0.001). In multivariate logistic regression adjusted analyses, older age (aOR 1.95, 95% CI 1.21–3.19; p = 0.01), cirrhosis (aOR 1.80, 95% CI 1.18–2.76; p = 0.01), GT1b (aOR 1.77, 95% CI 1.15–2.75; p = 0.01) and GT4 (aOR 10.20, 95% CI 3.52–29.56; p <0.001) infections (compared to GT1a infections) were positively and independently associated with the presence of NS5A RASs post-treatment.
Discussion
Despite the high efficacy of DAAs, our results confirm a high frequency of drug resistance in patients in whom treatment failed, mostly with first-generation DAAs. Although the current EASL recommendations focused primarily on the pan-genotypic DAAs and grazoprevir/elbasvir,1 many countries continue to use first-generation DAAs to reduce the treatment cost and maximize the number of patients eligible for publicly funded healthcare, so these drugs will remain relevant as part of the global attempt to eradicate HCV. Almost all patients in our cohort harbored drug-resistant variants after treatment failure, with over two-thirds having resistance to ≥2 DAA classes. Most patients were treated with NS5AIs, which have the lowest barrier to resistance among all DAA classes. Importantly NS5AI RASs tend to persist for a long time, presenting a challenge for retreatment.11 Transmission of resistant viruses further threatens global HCV elimination efforts. The spread of stable resistant variants is frequently observed in viral infections, as witnessed recently with SARS-CoV2.12 In the case of HCV, Q80K, a natural PI-resistant polymorphism, descended from a single lineage around the 1940s in the United States and expanded to 18–22% of the GT1a patients in Europe in 2014.13,14 HCV variants in NS5AI failures were characterized by complex RAS patterns interlaced with multiple substitutions with a high level of resistance; a critical question is whether these patients can be successfully re-treated with salvage therapies. SOF/VEL/VOX or 16–24 weeks of GLE/PIB + SOF and RBV are the regimens of choice (if available) to re-treat patients following DAA treatment failure.1,15 In people previously exposed to PIs and NS5AIs, 91–100% of the patients achieved SVR, although only a minority of patients had multi-class resistance in these studies.[16], [17], [18] Lower SVR rates were observed in GT3-infected patients with cirrhosis (69–92%), and GT1- and GT3-infected patients with prior SOF/VEL experience (78–85%).16,[19], [20], [21] In moderately resource-limiting settings where these new regimens may not be available, a resistance-guided retreatment approach using first-generation DAAs with a switch of the DAA drug class and additional RBV with or without extended duration is a workable option.8,22,23
Circulating HCV displays a high degree of genetic heterogeneity; variants with every possible individual substitution are generated daily.24 Under suboptimal drug concentrations, drug-resistant variants are rapidly enriched, leading to viral breakthrough or relapse. Based on resistance patterns before and after treatment, we postulate several pathways of RAS development. For example, a single highly resistant RAS with a low prevalence could be enriched during treatment leading to drug resistance (e.g., Q30R and Y93H/N in GT1a), or RASs with a moderate prevalence and resistance could persist at the same level (e.g., A30K/S in GT3) leading to the same outcome. Similarly, a baseline RAS with low resistance may acquire additional RASs as co-mutations (e.g., R30Q+Y93H in GT1b). Multiple low prevalence RASs at baseline may be sequentially/simultaneously enriched during treatment (e.g., 30R+31M/V+62E+93Y/C in GT4), leading to virologic failure.
Although the effect of baseline RASs on the overall DAA response is relatively small, reduced SVR rates in the presence of pre-treatment RASs have been described for GT1a and even more markedly for GT3 with cirrhosis.8,25 The resistance pathways discussed above further support the importance of baseline RASs on treatment outcomes. Emerging data from our analyses suggested that L30R in GT4d might present as a natural polymorphism that precipitates virologic failure in this subtype. Aside from the well-defined RASs, our exploratory studies have identified several new substitutions associated with virologic failure. Nevertheless, detailed reverse genetics and in vitro-in vivo correlations are required to establish the biological relevance of these substitutions. Lastly, individual polymorphisms that do not affect drug susceptibility can mount a severe resistance to DAAs when combined. For example, NS5A Q30R/K, L31M, and H58D individually do not affect PIB in HCV replicons, while Y93H only increases drug resistance by 2- to 7-fold. However, Q30R+L31M+H58D in GT1a or Q30K+Y93H in GT3a results in 1740- and 69-fold resistance, respectively, to PIB.9 In the SHARED dataset, >75% of the patients who failed glecaprevir/PIB or VEL/SOF/voxilaprevir had ≥2 RASs in NS5A; each has minimal resistance to NS5AIs, but their combined effect is largely unknown. Combination RASs should be considered and characterized when interpreting drug resistance results for the second-generation DAAs.
Our studies showed that cirrhosis and older age are associated with the presence of RASs in patients who failed DAA therapy, consistent with the observation of reduced SVR rates in patients with advanced liver diseases.26 Hepatic disease, notably cirrhosis, results in altered pharmacokinetics and pathophysiologic changes and can lead to treatment failure accompanied by RAS. However, long infection duration could also influence RAS selection. We have recently reported clock-like characteristics of the HCV genome in which intra-host viral genetic diversity increases over time.27 People with a longer duration of infection may have increased viral diversity shaped by immune selection, resulting in polymorphisms with increased fitness, including RASs and other compensatory substitutions. Indeed, a high prevalence of baseline NS5A RASs was reported in a cohort of HIV-HCV patients with advanced fibrosis and cirrhosis and older mono-infected patients in Poland.28 Although our database did not document the duration of infection, older patients and those with cirrhosis might have a long course of HCV infection. There is evidence that most people start risk behaviors in the second to third decade of life, and progression from acute hepatitis to cirrhosis usually takes at least 10-20 years after infection.29 The impacts of age, cirrhosis, and infection duration on viral diversity are important for future research.
There are many limitations and challenges. By nature of the real-world data collection, there is considerable variability and heterogeneity in data reporting, clinical interpretations, and length and location of HCV sequences, presenting structural and contextual challenges during data merging and analysis. This paper described HCV resistance mainly from the epidemic strains of GT1a/b, 3, and 4; resistance profiles from the other GT/subtypes are largely lacking. To comprehensively map global patterns of HCV resistance, we need more non-GT1/3 sequences from Africa, Asia, and South America. The current SHARED dataset is biased towards first-line DAAs; retreatment data, especially from the second-generation DAAs, are needed. The analyses and interpretations in this study also suffered from missing data (especially behavioral information), lack of longitudinal samples, and lack of in vitro data. These deficiencies underscore the pressing need to call for collective efforts across disciplines and organizations, including collaboration with health agencies to integrate epidemiological and genomic data. So far, SHARED participation and data contribution were entirely voluntary; support from government and funding agencies will provide the infrastructure required for data collection, knowledge dissemination, and in vitro characterization of RASs.
Nevertheless, the diverse collective expertise from our consortium has turned these challenges into an invaluable learning opportunity to set up a workable database and streamline future data collection. As a joint effort of clinicians, virologists and researchers, SHARED offers a unique opportunity to widen our knowledge of HCV drug resistance, improve resistance testing and interpretations, guide clinical management of HCV in the context of antiviral resistance, and provide a resource for collaborative research. This manuscript's pooled analyses, including large patient numbers and diverse clinical and academic settings, have adequately illustrated the current real-world HCV drug resistance landscape in patients treated with the first-generation DAAs. This new information can help refine future guidelines and generate a simple and actionable resistance algorithm to guide treatment decisions. The SHARED database is live, and as data accumulates, it will have sufficient power to address relatively rare events, such as RAS characteristics in "unusual" GTs/subtypes, novel RASs, and RAS characteristics following salvage regimens. We hope that the database can provide comprehensive surveillance of the global HCV resistance landscape and transmission networks in the future. SHARED is open to all scientists and clinicians who would like to contribute data, scientific expertise, or financial support for HCV drug resistance research. Interested researchers can contact the corresponding author or any SHARED member. For more information about SHARED, please visit https://hcvdb.med.ubc.ca.
Conclusions
Despite success in HCV therapeutics, drug resistance is almost inevitable in patients who fail to achieve SVR. In addition to providing the characteristics and clinical factors associated with RASs, this study highlights the complexity and multi-dimensional nature of drug resistance interpretation in HCV. There is room for improvement in the definition of RASs, as previously unknown mutations emerge and more epistatic interactions between co-occurring mutations are uncovered. A collaborative effort is required to keep HCV drug resistance in check and safeguard global HCV elimination.
Financial support
No financial support was received for the writing of this article. The initial SHARED development was funded in part by Merck and a User Partnership Program grant from Genome British Columbia to P.R.H and A.Y.M.H (UPP029). The Kirby Institute is funded by the Australian Government Department of Health and Ageing. The views expressed in this publication do not necessarily represent the position of the Australian (or any other) Government. C.R. and J.G. are supported by an NHMRC Investigator Grant (nos. 1173666, 1176131). An NHMRC Program Grant supported RAS testing and data collection by M.W.D. (1053206) and small grants from the Australian Centre for HIV and Hepatitis Virology Research, The University of Sydney, Western Sydney Local Health District Research Education Network, and the Robert W. Storr bequest to the Sydney Medical Foundation (University of Sydney). S.P. received personal fees from Gilead Sciences. Regarding the Italian data, the work was supported in part by the Italian Ministry of Instruction, University and Research (MIUR) (Bandiera InterOmics Protocollo PB05 1°), by the Italian Ministry of Health (RF-2016-02362422), and by Aviralia and Vironet C Foundations.
Authors' contributions
Concept and design: A.Y.M.H. Data collection: A.Y.M.H., M.W.D., F.G.G., F.C.S., S.P., J.D., C.S., J.G., C.R., J.L., M.K., H.K., J.M.P, S.F., M.S., M.P., M.S., J.Q. J.A.S., V.C. Data analyses: A.Y.M.H., P.R.H., E.C. Writing of article: A.Y.M.H., C.R. M.W.D., E.C. All authors contributed to the preparation and review of the manuscript.
Data availability statement
Aggregate and redacted data are available upon request. However, patient line data cannot be disclosed due to patient privacy and data sharing agreement conditions.
SHARED Collaborators
Marianne Martinello, The Kirby Institute, UNSW Sydney, Sydney, Australia
Gail Matthews, The Kirby Institute, UNSW Sydney, Sydney, Australia
Fay Fabián Fernando, Laboratorio CIBIC, Rosario, Argentina
Juan I. Esteban, Vall d'Hebron Institut de Recerca (VHIR), Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain and CIBEREHD
Beat Müllhaupt. University Hospital Zürich, Zürich, Switzerland.
Julian Schulze zur Wiesch. University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Peter Buggisch. Institute for Interdisciplinary Medicine IFI, Hamburg, Germany.
Christoph Neumann-Haefelin. University of Freiburg, Freiburg, Germany.
Thomas Berg. University Hospital Leipzig, Leipzig, Germany.
Christoph P. Berg. University of Tübingen, Tübingen, Germany.
Jörn M. Schattenberg. University Medical Center of the Johannes Gutenberg-University, Mainz, Germany.
Christophe Moreno. CUB Hôpital Erasme, Belgium.
Rudolf Stauber. Medical University of Graz, Graz, Austria.
Andrew Lloyd. The Kirby Institute, UNSW, Sydney, New South Wales, Australia.
Gregory Dore. The Kirby Institute, UNSW Sydney, Sydney, Australia.
Tanya Applegate. The Kirby Institute, UNSW Sydney, Sydney, Australia.
Juan Ignacio. Instituto de Salud Carlos III, Madrid, Spain.
Damir Garcia-Cehic. Vall d'Hebron Institut de Recerca (VHIR), Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain and CIBEREHD.
Josep Gregori. Vall d'Hebron Institut de Recerca (VHIR), Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain and CIBEREHD.
Francisco Rodriguez-Frias. Vall d'Hebron Institut de Recerca (VHIR), Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain and CIBEREHD.
Ariadna Rando. Vall d'Hebron Institut de Recerca (VHIR), Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain.
Yael Gozlan. Central Virology Laboratory, Ministry of Health, Sheba Medical Center, Ramat-Gan, Israel.
Mario Angelico. Hepatology Unit, University Hospital of Rome Tor Vergata, Rome, Italy.
Massimo Andreoni. Infectious Diseases, University Hospital of Rome Tor Vergata, Rome, Italy.
Sergio Babudieri. Medical, Surgical and Experimental Sciences, University of Sassari, Sassari, Italy.
Ada Bertoli. Department of Experimental Medicine, University of Rome Tor Vergata, Rome, Italy.
Valeria Cento. Department of Oncology and Hemato-oncology, Università degli Studi di Milano, Milan, Italy.
Nicola Coppola. University of Campania “L. Vanvitelli”, Napoli, Italy.
Antonio Craxì. Gastroenterology, “P. Giaccone” University Hospital, Palermo, Italy.
Stefania Paolucci. Molecular Virology Unit, Microbiology and Virology Department, IRCCS Policlinic Foundation San Matteo, Pavia, Italy.
Giustino Parruti. Infectious Disease Unit, Pescara General Hospital, Pescara, Italy.
Caterina Pasquazzi. Infectious Diseases, Sant’Andrea Hospital – “La Sapienza”, Rome, Italy.
Carlo Federico Perno. Unit of Microbiology and Diagnostic Immunology, Bambino Gesù Children’s Hospital, IRCCS Rome, Italy.
Elisabetta Teti. Infectious Diseases, University Hospital of Rome Tor Vergata, Rome, Italy.
Vironet C. HCV Virology Italian Resistance Network Group, https://www.vironetc.org/ (18 November 2021).
Anders Lannergård. Uppsala University Hospital, Uppsala, Sweden.
Ann-Sofi Duberg. Örebro University, Örebro, Sweden.
Soo Aleman. Karolinska University Hospital, Stockholm, Sweden.
Tore Gutteberg. University Hospital of North Norway, Tromsø, Norway.
Alexandre Soulier. French National Agency for Research on AIDS and viral Hepatitis, Paris, France.
Aurélie Gourgeon. French National Agency for Research on AIDS and viral Hepatitis, Paris, France.
Stephane Chevaliez. French National Agency for Research on AIDS and viral Hepatitis, Paris, France.
Stanislas Pol. French National Agency for Research on AIDS and viral Hepatitis, Paris, France.
Fabrice Carrat. French National Agency for Research on AIDS and viral Hepatitis, Paris, France.
Dominique Salmon. Paris Descartes University, Paris, France.
Rolf Kaiser. University Hospital Cologne, Cologne, Germany.
Elena Knopes. University Hospital Cologne, Cologne, Germany.
Perpetua Gomes. Centro Hospitalar Lisboa Ocidental, Hospital Egas Moniz, Lisbon and CiiEM, Almada, Portugal.
Rob de Kneght. Erasmus University Medical Center, Rotterdam, Netherlands.
Bart Rijnders. Erasmus University Medical Center, Rotterdam, Netherlands.
Mario Poljak. University of Ljubljana, Slovenia.
Maja Lunar. University of Ljubljana, Slovenia.
Rafael Usubillaga. Paris Descartes University, Paris, France.
Carole Seguin_Devaux. Luxembourg Institute of Health, Luxembourg, Luxembourg.
Enoch Tay. Westmead Institute for Medical Research, New South Wales, Australia.
Caroline Wilson. Westmead Institute for Medical Research, New South Wales, Australia.
Dao Sen Wang. University of Sydney and WIMR, New South Wales, Australia.
Jacob George. University of Sydney and Westmead Hospital, New South Wales, Australia.
Jen Kok. NSW Health Pathology, ICPMR, Westmead, New South Wales, Australia.
Ana Belén Pérez. Instituto Maimónides de Investigación Biomédica de Córdoba (IMIBIC). Córdoba, Spain.
Natalia Chueca. University Hospital San Cecilio. Instituto de Investigacion Ibs.Granada, Granada, Spain.
Miguel García-Deltoro. Hospital General de Valencia, Valencia, Spain.
Ana María Martínez-Sapiña. Hosputal Miguel Servet, Zaragoza, Spain.
María Magdalena Lara-Pérez. Hospital Nuestra Señora de la Candelaria, Tenerife, Spain.
Silvia García-Bujalance. University Hospital La Paz, Madrid, Spain.
Teresa Aldámiz-Echevarría. Hospital Gregorio Marañón, Madrid, Spain.
Francisco Jesús Vera-Méndez. University Hospital Santa Lucía, Cartagena, Spain.
Juan Antonio Pineda. University Hospital Nuestra Señora de Valme, Sevilla, Spain.
Marta Casado. Complejo Hospitalario Torrecárdenas, Almería, Spain.
Juan Manuel Pascasio. University Hospital Virgen del Rocío, Sevilla, Spain.
Javier Salmerón. University Hospital San Cecilio Granada, Granada, Spain.
Juan Carlos Alados-Arboledas. University Hospital Jerez, Cadiz, Spain.
Antonio Poyato. University Hospital Reina Sofía, Córdoba, Spain.
Francisco Téllez. Hospital Puerto Real, Cádiz, Spain.
Antonio Rivero-Juárez. University Hospital Reina Sofía. IMIBIC. Universidad de Córdoba. Córdoba, Spain.
Dolores Merino. Complejo Hospitalario de Huelva, Huelva, Spain.
María Jesús Vivancos-Gallego. University Hospital Ramón y Cajal, Madrid, Spain.
José Miguel Rosales-Zábal. Hospital Costa del Sol, Marbella, Spain.
María Dolores Ocete. Hospital General de Valencia, Valencia, Spain.
Miguel Ángel Simón. Hospital Miguel Servet, Zaragoza, Spain.
Pilar Rincón. University Hospital Nuestra Señora de Valme, Sevilla, Spain.
Sergi Reus. Hospital General de Alicante, Alicante, Spain.
Alberto De la Iglesia. Complejo Hospitalario de Huelva, Huelva, Spain.
Isabel García-Arata. Hospital Universitario de Fuenlabrada, Fuenlabrada, Madrid, Spain.
Miguel Jiménez. Hospital Regional de Málaga, Málaga, Spain.
Fernando Jiménez. Complejo Hospitalario de Huelva, Huelva, Spain.
José Hernández-Quero. University Hospital San Cecilio Granada, Granada, Spain.
Carlos Galera. University Hospital Virgen de la Arrixaca, El Palmar, Murcia, Spain.
Mohamed Omar Balghata. Complejo Hospitalario de Jaén, Jaén, Spain.
Joaquín Primo. Hospital de Sagunto, Sagunto, Valencia, Spain.
Mar Masiá. Hospital General de Elche, Elche, Alicante, Spain.
Nuria Espinosa. University Hospital Virgen del Rocío, Sevilla, Spain.
Marcial Delgado. Hospital Regional de Málaga, Málaga, Spain.
Miguel Ángel von-Wichmann. University Hospital Donostia, Donostia, Spain.
Antonio Collado. Complejo Hospitalario Torrecárdenas, Almería, Spain.
Jesús Santos. University Hospital Virgen de la Victoria, Málaga, Spain.
Carlos Mínguez. Castellón II—Penitenciary Institution; Albocásser, Castellón de la Plana, Spain.
Felícitas Díaz-Flores. Hospital Universitario de Canarias, Santa Cruz de Tenerife, Canary Islands, Spain.
Elisa Fernández. Hospital de Poniente, El Ejido, Almería, Spain.
Enrique Bernal. Hospital General Reina Sofía, Murcia, Spain.
José De Juan. Penitenciary Institution, Córdoba, Spain.
José Joaquín Antón. Penitenciary Institution; Albolote, Granada, Spain.
Mónica Vélez. Hospital General de La Palma, Santa Cruz de Tenerife, Canary Islands, Spain.
Antonio Aguilera. Complejo Hospitalario Universitario de Santiago de Compostela, Santiago de Compostela, Spain.
Daniel Navarro. Complejo Hospitalario Universitario de Santiago de Compostela, Santiago de Compostela, Spain.
Juan Ignacio Arenas. University Hospital Donostia, San Sebastian, Spain.
Clotilde Fernández. University Hospital Puerta del Mar, Cádiz, Spain.
María Dolores Espinosa. University Hospital Virgen de las Nieves, Granada, Spain.
María José Ríos. University Hospital Virgen Macarena, Sevilla, Spain.
Roberto Alonso. University Hospital Gregorio Marañón, Madrid, Spain.
Carmen Hidalgo. University Hospital Virgen de las Nieves, Granada, Spain.
Rosario Hernández. University Hospital Torrevieja, Torrevieja, Alicante, Spain.
María Jesús Téllez. Hospital Clínico San Carlos, Madrid, Spain.
Francisco Javier Rodríguez. Hospital General Reina Sofía, Murcia, Spain.
Pedro Antequera. University Hospital José María Morales Meseguer, Murcia, Spain.
Cristina Delgado. Hospital Alto Guadalquivir, Andújar, Jaén, Spain.
Patricia Martín. Hospital de Denia, Denia, Alicante, Spain.
Javier Crespo. University Hospital Marqués de Valdecilla, Santander, Spain.
Berta Becerril. Hospital Punta de Europa, Algeciras,Cádiz, Spain.
Oscar Pérez. Hospital de Castellón de la Plana, Castellón de la Plana, Spain.
Antonio García-Herola. Hospital Marina Baixa, Vilajoyosa, Alicante, Spain.
José Montero. University Hospital Puerto Real, Puerto Real, Cádiz, Spain.
Carolina Freyre. University Hospital Puerto Real, Puerto Real, Cádiz, Spain.
Concepción Grau. Hospital Vega Baja, Orihuela, Alicante, Spain.
Joaquin Cabezas. Marqués de Valdecilla University Hospital, Santander, Spain.
Miguel Jimenez. Hospital Regional de Málaga, Málaga, Spain.
Manuel Alberto Macias Rodriguez. Hospital Universitario Puerta del Mar, Cádiz, Spain.
Cristina Quilez. Hospital Marina Baixa, Alicante, Spain.
Maria Rodriguez Pardo. Hospital Universitario Puerta del Mar, Cádiz, Spain.
Leopoldo Muñoz-Medina. University Hospital San Cecilio Granada, Granada, Spain.
Blanca Figueruela. Hospital Universitario Virgen de Valme, Sevilla, Spain.
Conflict of Interest
J.M.P. has been an advisor and/or speaker for AbbVie, Assembly Biosciences, Arbutus, Merck, Gilead, Regulus, and Memo Therapeutics. J.D. receives research support from Gilead. A.Y.M.H. is a consultant for Boston Pharmaceuticals. Outside the submitted work, J.G. reports grants and personal fees from AbbVie, Gilead Sciences, Merck, and Cepheid and grants from Hologic and Indivior. M.W.D has been an advisor and/or speaker for Gilead, AbbVie, and Merck and has received grants from Gilead and AbbVie. F.G.G. has been an advisor and/or speaker for AbbVie, Merck, and Gilead. S.F. has been an advisor and/or speaker for Abbott diagnostics, AbbVie, Gilead, and AB Science. F.C.S. has been an advisor and/or speaker for AbbVie, Merck, and Gilead and received grants from Merck and Gilead.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgments
We want to express our immense gratitude to our patients who participated in this project. This paper and the research behind it would not have been possible without the support from our contributing collaborators, staff, and research scientists, who have spent numerous hours organizing the data. Special thanks to Conan Woods, Yuan Ji, and Eric Newton for their contributions in developing the HCV database and HCV Nextstrain. We are grateful to Drs. Naveed Janjua and Mel Krajden for providing SHARED with the infrastructure and administrative support. This manuscript was written in memory of Dr. Charles Boucher. His dynamism, leadership, and unyielding enthusiasm in promoting virology education have inspired the SHARED community.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100462.
Contributor Information
Anita Y.M. Howe, Email: anita.howe@bccdc.ca.
SHARED Collaborators:
Marianne Martinello, Gail Matthews, Fay Fabián Fernando, Juan I. Esteban, Beat Müllhaupt, Julian Schulze zur Wiesch, Peter Buggisch, Christoph Neumann-Haefelin, Thomas Berg, Christoph P. Berg, Jörn M. Schattenberg, Christophe Moreno, Rudolf Stauber, Andrew Lloyd, Gregory Dore, Tanya Applegate, Juan Ignacio, Damir Garcia-Cehic, Josep Gregori, Francisco Rodriguez-Frias, Ariadna Rando, Yael Gozlan, Mario Angelico, Massimo Andreoni, Sergio Babudieri, Ada Bertoli, Valeria Cento, Nicola Coppola, Antonio Craxì, Stefania Paolucci, Giustino Parruti, Caterina Pasquazzi, Carlo Federico Perno, Elisabetta Teti, C. Vironet, Anders Lannergård, Ann-Sofi Duberg, Soo Aleman, Tore Gutteberg, Alexandre Soulier, Aurélie Gourgeon, Stephane Chevaliez, Stanislas Pol, Fabrice Carrat, Dominique Salmon, Rolf Kaiser, Elena Knopes, Perpetua Gomes, Rob de Kneght, Bart Rijnders, Mario Poljak, Maja Lunar, Rafael Usubillaga, Carole Seguin_Devaux, Enoch Tay, Caroline Wilson, Dao Sen Wang, Jacob George, Jen Kok, Ana Belén Pérez, Natalia Chueca, Miguel García-Deltoro, Ana María Martínez-Sapiña, María Magdalena Lara-Pérez, Silvia García-Bujalance, Teresa Aldámiz-Echevarría, Francisco Jesús Vera-Méndez, Juan Antonio Pineda, Marta Casado, Juan Manuel Pascasio, Javier Salmerón, Juan Carlos Alados-Arboledas, Antonio Poyato, Francisco Téllez, Antonio Rivero-Juárez, Dolores Merino, María Jesús Vivancos-Gallego, José Miguel Rosales-Zábal, María Dolores Ocete, Miguel Ángel Simón, Pilar Rincón, Sergi Reus, Alberto De la Iglesia, Isabel García-Arata, Miguel Jiménez, Fernando Jiménez, José Hernández-Quero, Carlos Galera, Mohamed Omar Balghata, Joaquín Primo, Mar Masiá, Nuria Espinosa, Marcial Delgado, Miguel Ángel von-Wichmann, Antonio Collado, Jesús Santos, Carlos Mínguez, Felícitas Díaz-Flores, Elisa Fernández, Enrique Bernal, José De Juan, José Joaquín Antón, Mónica Vélez, Antonio Aguilera, Daniel Navarro, Juan Ignacio Arenas, Clotilde Fernández, María Dolores Espinosa, María José Ríos, Roberto Alonso, Carmen Hidalgo, Rosario Hernández, María Jesús Téllez, Francisco Javier Rodríguez, Pedro Antequera, Cristina Delgado, Patricia Martín, Javier Crespo, Berta Becerril, Oscar Pérez, Antonio García-Herola, José Montero, Carolina Freyre, Concepción Grau, Joaquin Cabezas, Miguel Jimenez, Manuel Alberto Macias Rodriguez, Cristina Quilez, Maria Rodriguez Pardo, Leopoldo Muñoz-Medina, and Blanca Figueruela
Supplementary data
The following are the supplementary data to this article:
References
- 1.Pawlotsky J.-M., Negro F., Aghemo A., Berenguer M., Dalgard O., Dusheiko G., et al. EASL recommendations on treatment of hepatitis C: final update of the series. J Hepatol. 2020;73:1170–1218. doi: 10.1016/j.jhep.2020.08.018. [DOI] [PubMed] [Google Scholar]
- 2.Kåberg M., Weiland O. Hepatitis C elimination - macro-elimination. Liver Int. 2020;40(Suppl 1):61–66. doi: 10.1111/liv.14352. [DOI] [PubMed] [Google Scholar]
- 3.Popping S., Verwijs R., Cuypers L., Claassen M.A., van den Berk G.E., De Weggheleire A., et al. Transmission of NS5A-inhibitor resistance-associated substitutions among men who have sex with men recently infected with hepatitis C virus genotype 1a. Clin Infect Dis. 2020;71:e215–e217. doi: 10.1093/cid/ciaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Committee on Taxonomy of Viruses - HCV Classification . 2017. HCV Classification.https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification In. [Google Scholar]
- 5.Howe A.Y.M., Ceccherini-Silberstein F., Dietz J., Popping S., Grebely J., Rodrigo C., et al. SHARED: an international collaboration to unravel hepatitis C resistance. Viruses. 2021;13:1580. doi: 10.3390/v13081580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzou P.L., Huang X., Shafer R.W. NucAmino: a nucleotide to amino acid alignment optimized for virus gene sequences. BMC Bioinformatics. 2017;18:138. doi: 10.1186/s12859-017-1555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 1995;57:289–300. [Google Scholar]
- 8.Sarrazin C. Treatment failure with DAA therapy: importance of resistance. J Hepatol. 2021 doi: 10.1016/j.jhep.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Sorbo M.C., Cento V., Di Maio V.C., Howe A.Y.M., Garcia F., Perno C.F., et al. Hepatitis C virus drug resistance associated substitutions and their clinical relevance: update 2018. Drug Resist Updates. 2018 doi: 10.1016/j.drup.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Ng T.I., Krishnan P., Pilot-Matias T., Kati W., Schnell G., Beyer J., et al. In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS5A inhibitor pibrentasvir. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02558-16. e02558-02516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyles D., Mangia A., Cheng W., Shafran S., Schwabe C., Ouyang W., et al. Long-term persistence of HCV NS5A resistance-associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir Ther. 2018;23:229–238. doi: 10.3851/IMP3181. [DOI] [PubMed] [Google Scholar]
- 12.GISAID . Nextstrain; 2021, December 1. Genomic epidemiology of novel coronavirus - Global subsampling.https://nextstrain.org/ncov/gisaid/global [Google Scholar]
- 13.McCloskey R.M., Liang R.H., Joy J.B., Krajden M., Montaner J.S.G., Harrigan P.R., et al. Global origin and transmission of hepatitis C virus NS3 Q80 K polymorphism. J Infect Dis. 2014 doi: 10.1093/infdis/jiu613. jiu613. [DOI] [PubMed] [Google Scholar]
- 14.Sarrazin C., Lathouwers E., Peeters M., Daems B., Buelens A., Witek J., et al. Prevalence of the hepatitis C virus NS3 polymorphism Q80K in genotype 1 patients in the European region. Antivir Res. 2015;116:10–16. doi: 10.1016/j.antiviral.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 15.AASLD-IDSA . 2021. Recommendations for testing, managing and treating hepatitis C virus.http://www.hcvguidelines.org [DOI] [PubMed] [Google Scholar]
- 16.Degasperi E., Spinetti A., Lombardi A., Landonio S., Rossi M.C., Pasulo L., et al. Real-life effectiveness and safety of sofosbuvir/velpatasvir/voxilaprevir in hepatitis C patients with previous DAA failure. J Hepatol. 2019;71:1106–1115. doi: 10.1016/j.jhep.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Sarrazin C., Cooper C.L., Manns M.P., Reddy K.R., Kowdley K.V., Roberts S.K., et al. No impact of resistance-associated substitutions on the efficacy of sofosbuvir, velpatasvir, and voxilaprevir for 12 weeks in HCV DAA-experienced patients. J Hepatol. 2018;69:1221–1230. doi: 10.1016/j.jhep.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Saxena V., McKinney J., Chamberland S.L., Catalli L., Seo S.I., Ready J.B., et al. AASLD - The Liver Meeting (online) November 12 - 15. 2021. Excellent efficacy and safety of sofosbuvir, glecaprevir, pibrentasvir and ribavirin for retreatment of chronic hepatitis C after sofosbuvir, velpatasvir and voxilaprevir failure. [Google Scholar]
- 19.Belperio P.S., Shahoumian T.A., Loomis T.P., Backus L.I. Real-world effectiveness of sofosbuvir/velpatasvir/voxilaprevir in 573 direct-acting antiviral experienced hepatitis C patients. J Viral Hepat. 2019;26:980–990. doi: 10.1111/jvh.13115. [DOI] [PubMed] [Google Scholar]
- 20.Llaneras J., Riveiro-Barciela M., Lens S., Diago M., Cachero A., García-Samaniego J., et al. Effectiveness and safety of sofosbuvir/velpatasvir/voxilaprevir in patients with chronic hepatitis C previously treated with DAAs. J Hepatol. 2019;71:666–672. doi: 10.1016/j.jhep.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Smith D.A., Bradshaw D., Mbisa J., Manso C., Bibby D., Singer J., et al. Real-world retreatment of HCV-infected patients with prior failure to direct acting antiviral therapy using sofosbuvir, velpatasvir and voxilaprevir. J Hepatol. 2020;73:S336. [Google Scholar]
- 22.Pérez A.B., Chueca N., García-Deltoro M., Martínez-Sapiña A.M., Lara-Pérez M.M., García-Bujalance S., et al. High efficacy of resistance-guided retreatment of HCV patients failing NS5A inhibitors in the real world. J Hepatol. 2019;71:876–888. doi: 10.1016/j.jhep.2019.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Dietz J., Spengler U., Müllhaupt B., Schulze Zur Wiesch J., Piecha F., Mauss S., et al. Efficacy of retreatment after failed direct-acting antiviral therapy in patients with HCV genotype 1-3 infections. Clin Gastroenterol Hepatol. 2021;19:195–198.e192. doi: 10.1016/j.cgh.2019.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Ribeiro R.M., Li H., Wang S., Stoddard M.B., Learn G.H., Korber B.T., et al. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarrazin C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol. 2016;64:486–504. doi: 10.1016/j.jhep.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Miotto N., Mendes L.C., Zanaga L.P., Lazarini M.S.K., Goncales E.S.L., Pedro M.N., et al. All-oral direct antiviral treatment for hepatitis C chronic infection in a real-life cohort: the role of cirrhosis and comorbidities in treatment response. PLoS One. 2018;13 doi: 10.1371/journal.pone.0199941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montoya V., Howe A.Y.M., Dong W.Y., Dong W., Brumme C.J., Olmstead A.D., et al. Intra-host evolutionary dynamics of the hepatitis C virus among people who inject drugs. Scientific Rep. 2021;11:9986. doi: 10.1038/s41598-021-88132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parczewski M., Kordek J., Janczewska E., Pisula A., Łojewski W., Wawrzynowicz-Syczewska M., et al. Hepatitis C virus (HCV) genotype 1 NS5A resistance-associated variants are associated with advanced liver fibrosis independently of HCV-transmission clusters. Clin Microbiol Infect. 2019;25:513. e511–513. e516. doi: 10.1016/j.cmi.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigo C., Eltahla A.A., Bull R.A., Grebely J., Dore G.J., Applegate T., et al. Historical trends in the hepatitis C virus epidemics in North America and Australia. The J Infect Dis. 2016;214:1383–1389. doi: 10.1093/infdis/jiw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Aggregate and redacted data are available upon request. However, patient line data cannot be disclosed due to patient privacy and data sharing agreement conditions.