Summary
Cells elaborate transcriptional programs in response to external signals. In the peripheral nerves, Schwann cells (SC) sort axons of given caliber and start the process of wrapping their membrane around them. We identify Actin-like protein 6a (ACTL6a), part of SWI/SNF chromatin remodeling complex, as critical for the integration of axonal caliber recognition with the transcriptional program of myelination. Nuclear levels of ACTL6A in SC are increased by contact with large caliber axons or nanofibers, and result in the eviction of repressive histone marks to facilitate myelination. Without Actl6a the SC are unable to coordinate caliber recognition and myelin production. Peripheral nerves in knockout mice display defective radial sorting, hypo-myelination of large caliber axons, and redundant myelin around small caliber axons, resulting in a clinical motor phenotype. Overall, this suggests that ACTL6A is a key component of the machinery integrating external signals for proper myelination of the peripheral nerve.
Subject areas: Biological sciences, Neuroscience, Molecular neuroscience, Cellular neuroscience
Graphical abstract
Highlights
-
•
ACTL6a levels in Schwann cells respond to stiffness and caliber of PLA nanofibers
-
•
ACTL6a integrates axonal caliber recognition signals with Schwann cells transcriptome
-
•
ACTL6a null mice have thin myelin on large axons and redundant myelin on small axons
-
•
Mice lacking ACTL6a in Schwann cells show severe clinical symptoms
Biological sciences; Neuroscience; Molecular neuroscience; Cellular neuroscience
Introduction
Cells are constantly exposed to a variety of physical and chemical stimuli in their local microenvironment. These different signals are integrated and transduced into transcriptional changes that ultimately generate a biological response. The transduction of chemical and physical signals has been studied extensively in stem cells, whose fate can be modulated by both soluble factors and matrix elasticity (Chaudhuri et al., 2016; Engler et al., 2006). Schwann cells in the peripheral nervous system (PNS) must similarly integrate and respond to a wide range of external signals. During development, myelination of peripheral nerve results from the response of Schwann cells to a diverse array of physical (e.g., axonal caliber recognition, tensile strain, extracellular matrix (ECM) stiffness), and chemical signals (e.g., neuregulins and ECM proteins) (Wilson et al., 2021).
Within the peripheral nerve, axons can be either myelinated (caliber greater or equal to 1μm diameter) and in close contact with myelinating Schwann cells or unmyelinated (small caliber) and organized in clusters, surrounded by the membrane of non-myelinating Schwann cells in histological structures called “Remak bundles”. This specialized cellular architecture of the peripheral nerve results from a sequence of events occurring during development, which includes radial sorting and myelination. During radial sorting, immature Schwann cells extend protrusions into axonal bundles and, after identifying those with a caliber greater or equal to 1μm, they establish a 1:1 relation and start an elaborate transcriptional program which eventually leads to formation of the specialized myelin membrane (Feltri et al., 2016). These two inter-related processes (i.e., radial sorting and myelination) require the integration of physical and chemical signals (Feltri et al., 2002; Pellegatta et al., 2013; Poitelon et al., 2016; Yu et al., 2005) and the responsible molecular components remain incompletely understood.
The recognition of large caliber axons has thus far been shown to be mediated by a chemical signal known as neuregulin-1 type III (NRG1-III). NRG1-III is a transmembrane protein that is present on axonal membranes and activates Schwann cell ErbB2/ErbB3 receptor complexes. The amount of NRG1-III present on the axonal surface has been shown to dictate myelination and the thickness of the myelin sheath (Michailov et al., 2004; Taveggia et al., 2005). NRG1-III therefore functions as a signal of axonal size with larger axons expressing higher levels. It remains unclear, however, whether Schwann cells are also able to “sense” axonal caliber independently of chemical signals. Schwann cells are mechano-sensitive and they respond to both tension and substrate stiffness, by modulating gene expression (Poitelon et al., 2016; Urbanski et al., 2016). The mechano-sensitivity of these cells has been mostly attributed to the HIPPO pathway and shown to require the presence of two co-activators: Yes-Associated Protein 1 (YAP1) and WW domain containing transcription regulator 1 (TAZ/WWTR1) (Poitelon et al., 2016). YAP and TAZ nuclear localization, in response to mechanical stimuli leads to their association with the transcription factor TEA Domain Transcription Factor 1 (TEAD1) and regulates gene expression (Dupont et al., 2011). Both YAP1 and TAZ were demonstrated to be required for radial sorting and myelination, but it is unclear if the same pathway may also be involved in sensing of axonal caliber, a process which is crucial for the development of the peripheral nerve.
Process extension during radial sorting and membrane wrapping during myelination require extensive morphological changes that are mediated by actin cytoskeleton remodeling (Jin et al., 2011; Montani et al., 2014; Nawaz et al., 2015; Novak et al., 2011; Sparrow et al., 2012; Zuchero et al., 2015). However, the potential role that nuclear actin and related proteins play in these processes, remains virtually unexplored. Actin-Like Protein 6a (ACTL6a) is a nuclear actin-related protein which is part of several ATP-dependent chromatin remodeling complexes, such as the SWItch/Sucrose Non-Fermentable (SWI/SNF) complexes. Among some of these complexes, one containing Brahma-related Gene 1 (BRG1) as the core ATPase, has been previously shown to be - essential in Schwann cells, for radial sorting and for the expression of the pro-myelinating transcription factors Pou3f1 and Egr2 (Limpert et al., 2013; Weider et al., 2012). ACTL6A facilitates BRG1 optimal ATPase activity and is also part of other SWI/SNF complexes (Szerlong et al., 2008; Zhao et al., 1998).
In this study we asked whether ACTL6a could integrate the recognition of axonal caliber - with the transcriptional program regulating radial sorting and myelination in the peripheral nerve.
Results
ACTL6a is highly expressed during developmental myelination in the sciatic nerve
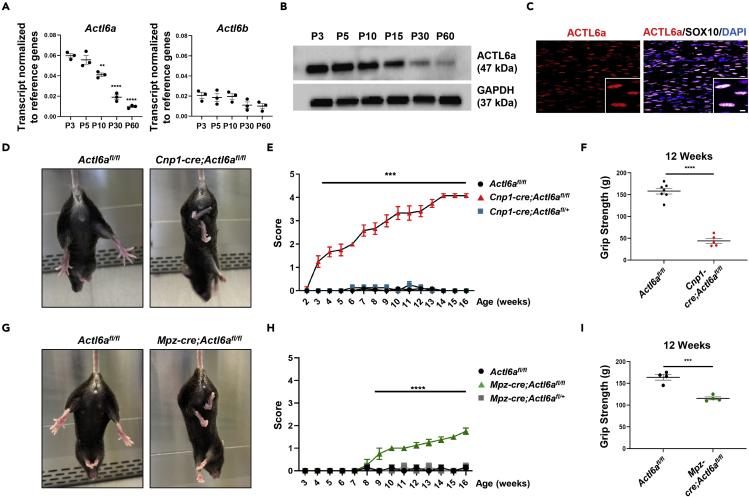
Previous studies on the role of SWI/SNF complexes in neural progenitors identified an important dynamic exchange of ACTL6a for its homolog ACTL6b during their differentiation into post-mitotic neurons (Lessard et al., 2007). However, a time course of the expression of Actl6a and Actl6b transcripts in the developing sciatic nerve, identified Actl6a as the only subunit (Figure 1A). This finding was consistent with several previous reports, including microarray data in Schwann cells (Verdier et al., 2012), RNA sequencing datasets in neural crest cells (Clements et al., 2017), and single cell RNAsequencing of Schwann cells in postnatal nerves (Gerber et al., 2021). Thus, Schwann cells did not display the subunit switch between Actl6a and Actl6b like neural progenitors. ACTL6A was the only subunit expressed in the developing sciatic nerve, with the highest levels detected postnatally (Figures 1B and S1A) during the temporal window characterized by the selection of axons of proper caliber and onset of myelination (Feltri et al., 2016). Immunohistochemical analysis of developing nerves further validated the expression of ACTL6a in the nuclei of Schwann cells, identified by immunoreactivity for the lineage marker SOX10 (Figure 1C). These data collectively suggested a prominent role for ACTL6a, and not ACTL6b, in peripheral myelination at a time consistent with the recognition of axonal caliber by Schwann cells and the integration with the complex transcriptional program of myelination.
Figure 1.
ACTL6a is detected in the developing sciatic nerve and its ablation in Schwann cells induces a progressive motor phenotype
(A) The scatterplot represents the average transcript levels ±SEM of Actl6a and Actl6b in sciatic nerves at postnatal day 3, 5, 10, 30 and 60 (n = 3 per age), normalized to the geometric mean of three reference genes (Gapdh, Wdr33, and 18s). Statistical analysis was performed using one-way ANOVA with multiple comparisons.
(B) Western blot of ACTL6a protein levels in the developing sciatic nerve at postnatal day 3, 5, 10, 15, 30 and 60. GAPDH levels were used as loading control.
(C) Confocal images of postnatal day 5 mouse sciatic nerve sections stained for ACTL6a (red) and SOX10 (white). DAPI (blue) as nuclear stain. Scale bar, 5μm.
(D) Tail suspension test showing abnormal hindlimb clasping reflex in Cnp1-cre;Actl6afl/fl mice, compared to controls.
(E) The curve represents the average clinical score ±SEM in mutantCnp1-cre;Actl6afl/f (n = 6), heterozygous Cnp1-cre;Actl6afl/+(n = 4), and wildtype Actl6afl/fl (n = 12) controls. Motor scoring was performed starting at P15 and conducted for 14 weeks, until mice were 16 weeks old. Scoring was as follows: 1 – hindlimb clasping, 2 – unable to hold on to inverted grid, 2.5 – can only grip upright grid with front paws, 3 – not able to hold on to upright grid, 3.5 no forelimb resistance when pulled across grid, 4 – hindlimb paralysis, 5 – dead. Statistical analysis was performed using two-way ANOVA.
(F) Average grip strength values ±SEM obtained in Cnp1-cre;Actl6afl/fl mice (n = 5) and control littermates (n = 7) at age 12 weeks. Statistical analysis was performed using Student’s t-test.
(G) Tail suspension test results in Mpz-cre;Actl6afl/fl mice and relative controls.
(H) Motor scoring represents the average clinical score ±SEM over time (from 2 to 16 weeks) for mutant Mpz-cre;Actl6afl/f (n = 4), compared to heterozygous Mpz-cre;Actl6afl/+(n = 3), and Actl6afl/fl (n = 4) controls. Statistical analysis was performed using two-way ANOVA.
(I) Average grip strength values ±SEM for Mpz-cre;Actl6afl/fl mice (n = 4) and control littermates (n = 4) at 12 weeks of age. Statistical analysis was performed using Student’s t-test. ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.001.
Schwann cell-specific ablation of ACTL6a induces a phenotype characterized by motor impairment and altered developmental myelination of the peripheral nerve
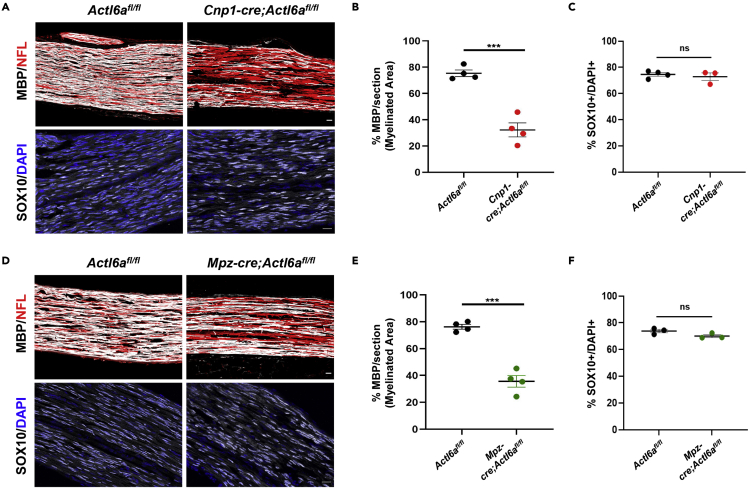
To investigate ACTL6a function in myelin formation, we generated a conditional mouse line by breeding Actl6a floxed mice (Actl6afl/fl), with loxP-sites flanking exons 4 and 5 of the gene (Krasteva et al., 2012), with a mouse line with the Cre recombinase knock-in within the Cnp1 promoter (Cnp1-cre) to target myelinating cells in the peripheral and central nervous system (Lappe-Siefke et al., 2003).Cnp1-cre;Actl6afl/fl mice showed a recombination efficiency of 76% +/− 0.06 (Figures S1B and S1C). They were born at expected Mendelian ratios and were indistinguishable from Actl6afl/fl littermates during the first few postnatal weeks. Over the course of development, however, mutant mice displayed abnormal hindlimb clasping upon tail suspension (Figure 1D), and developed a progressive motor phenotype, characterized by gradual loss of strength in hindlimbs and, to a lesser extent, also of the forelimbs. By age 8 weeks, mutant mice were unable to grasp onto an inverted grid. This loss of hindlimb strength progressively deteriorated into complete hindlimb paralysis by age 16 weeks (Figure 1E). To further characterize the motor deficit, we assessed the mice using a grip strength meter over the course of development. Grip strength of Cnp1-cre;Actl6afl/fl mice was significantly impaired at 12 weeks compared to control littermates (Figure 1F). The motor defects were highly suggestive of peripheral neuropathy, consequent to aberrant PNS myelination , since myelinationin the mutant spinal cord was indistinguishable from controls (data not shown). To further validate this interpretation, we generated a second mouse line, selectively targeting myelinating cells in the PNS, by crossing the Actl6afl/fl mice with the Mpz-cre line (Feltri et al., 1999) The recombination efficiency in this line was 66% +/− 0.03 (Figures S1D and S1E), and we detected a similar, albeit milder motor phenotype characterized by hindlimb clasping (Figures 1G and 1H) and subsequent reduction in grip strength at 12 weeks of age (Figure 1I). Overall, the similar clinical phenotype detected in the two lines of mouse mutants was highly suggestive of altered myelination in the developing peripheral nerves, an interpretation supported by the immunohistochemical data in adult sciatic nerves (Figures S2A and S2B). To determine whether the phenotype detected in adult mice was the result of aberrant development of the peripheral nerves, we conducted a similar analysis in neonatal sciatic nerves.Reduced MBP immunoreactivity was detected at postnatal day 5 (P5) in Cnp1-cre;Actl6afl/fl (Figures 2A and 2B) and Mpz-cre;Actl6afl/fl mice (Figures 2D and 2E), compared to controls.and it was not accompanied by decreased SOX10+ Schwann cell numbers(Figures 2C and 2F). Quantification of caspase-3+/SOX10+, to measure apoptosis and Ki67+/SOX10+ cells, to measure proliferation in Schwann cells, did not reveal significant differences between mutants and controls (Figures S2C and S2D). Thus, the phenotype detected in Actl6a mutants was consequent to an alteration of Schwann cell biological processes, rather than a putative reduction in their number secondary to an apoptotic or proliferative phenotype Figures S2.
Figure 2.
Impaired myelination in the neonatal sciatic nerve following ACTL6a ablation
(A) Confocal image of postnatal day 5 sciatic nerve sections from Actl6afl/fl controls and Cnp1-cre;Actl6afl/fl null mutants, stained with antibodies specific for neurofilament light chain (NFL, red) and MBP (white) in the upper panels, and for SOX10 (white) and DAPI (blue) counterstain in the lower panels sciatic nerves at P5.
(B) Quantification of the average MBP immunoreactive area ±SEM measured in at least two sections per mouse in Actl6afl/fl control (n = 4 mice) and Cnp1-cre;Actl6afl/fl (n = 4 mice) at postnatal day 5.
(C) Quantification of the average number of SOX10+ Schwann cells determined in at least 2 sections from each Actl6afl/fl control (n = 4 mice) and Cnp1-cre;Actl6afl/fl (n = 3) mice. Data represent the percentage relative to DAPI nuclei ±SEM.
(D) Confocal image of postnatal day 5 sciatic nerve sections from Actl6afl/fl control and Mpz-cre;Actl6afl/fl mice stained as described in (A).
(E) Quantification of the average MBP immunoreactive area ±SEM measured as described in (B) in controls (n = 4) and Mpz-cre;Actl6afl/fl mutants (n = 4 mice).
(F) Quantification of the average number of SOX10+ Schwann cells in at least 2 sections from Actl6afl/fl control (n = 4 mice) and Mpz-cre;Actl6afl/fl (n = 3) mice. Data represents the percentage relative to DAPI nuclei ±SEM. Student’s t-test. ∗∗∗p< 0.001. Scale bars in panels A and D, 20μm.
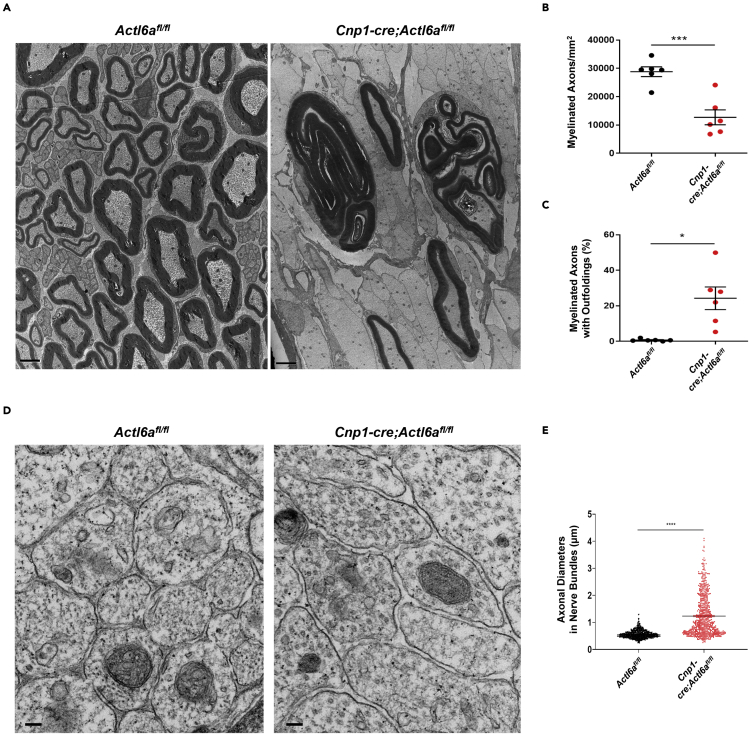
Consistent with the immunohistochemical data, ultrastructural analysis of adult P60 nerves revealed a ∼50% reduction of myelinated fibers in mutants, compared to controls (Figures 3A and 3B). Besides hypomyelination of large caliber axons (Figure S3A), we detected a higher number of smaller caliber axons with myelin outfoldings (Figures 3A and 3C), consistent with production of redundant myelin membranes (Figures 3A and S3B), a feature indicative of lack of coordination between axonal caliber and myelin synthesis. An additional feature of the mutant nerves was the presence of large caliber axons tightly associated in clusters (Figure 3A). Although control nerves displayed the characteristic “Remak bundles”, characterized by clusters of small caliber axons, the bundles in mutant nerves were characterized by the presence of large caliber axons (Figures 3D and 3E), and suggested that Schwann cells were unable to select and sort axons of caliber proper for myelination. Since aberrant sorting had been previously reported in laminin and integrin mutants (Feltri et al., 2002; Yang et al., 2005), we also analyzed the basal lamina integrity in the Actl6a mutants. Our analysis revealed an intact basal lamina in pro-myelinating Schwann cells and in myelin-forming Schwann cells (arrows in Figure S3C), while the basal lamina was discontinuous or even absent in immature mutant Schwann cells that were unable to sort (arrowhead in Figure S3D). Defective sorting and altered myelination were already detected in the neonatal mutant nerves (Figure S4), suggesting that hypomyelination and radial sorting defects began early in peripheral development and persisted in adulthood. Based on these results we suggest that the presence of ACTL6a is necessary for the ability of Schwann cells to recognize axonal caliber and integrate it with the transcriptional program allowing for proper sorting and coordinated myelin synthesis and wrapping.
Figure 3.
Impaired myelination in the adult sciatic nerve of Actl6a mutants
(A) Representative electron micrographs showing impaired myelination in Cnp1-cre;Actl6afl/fl sciatic nerves at P60 with fewer myelinated exons and myelin outfoldings. Scale bars, 2μm.
(B) Average numbers ±SEM of myelinated axons per square millimeter in sciatic nerve sections from adult Actl6afl/fl control (n = 6) and Cnp1-cre;Actl6afl/fl (n = 6) nerves.
(C) Quantification of axons with myelin outfoldings calculated as percentages of myelinated axons ±SEM in adult Actl6afl/fl control (n = 6) and Cnp1-cre;Actl6afl/fl (n = 6) nerves.
(D) Representative electron micrographs showing radial sorting defects in Cnp1-cre;Actl6afl/fl sciatic nerves at P60. Note the difference in caliber of the naked axons within bundles. Scale bars, 100nm.
(E) Quantification of the average diameter ±SEM of axons in bundles in control (n = 6) and mutant (n = 6) nerves. A total of 100 axons were measured per animal. Statistical analysis was performed using Student’s t-test ∗p< 0.05, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001.
Analysis of the ACTL6a mutant nerve transcriptome provides mechanistic insights on its role in Schwann cells
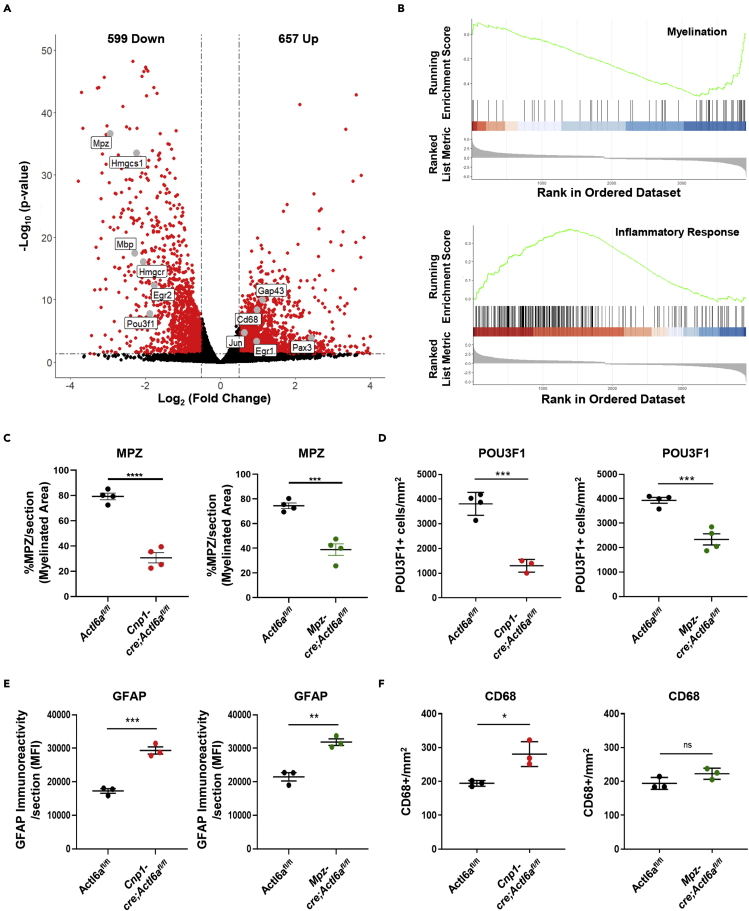
To begin characterizing the transcriptional consequences of Actl6a deletion in Schwann cells, we performed RNA-sequencing of P3 sciatic nerves in Cnp1-cre;Actl6afl/fl and compared it with that of Actl6afl/fl control mice. The analysis identified 1,256 dysregulated transcripts, of which 599 were significantly decreased (Table S1) and 657 were significantly increased (Table S2) in mutant nerves (Figures 4A and S5A). A Gene Set Enrichment Analysis (GSEA) of these transcripts (Figure 4B) identified decreased levels of myelination-related genes and increased levels of inflammation-related genes. More specifically, the transcript levels of myelin protein (e.g., Pmp22, Mbp, Mpz), pro-myelinating transcription factors (e.g., Egr2, Pou3f1) and cholesterol biosynthesis (e.g., Lss, Hmgcs1, Hmgcr, Dhcr24) encoding genes were all significantly decreased whereas the markers of pre-myelinating Schwann cells (e.g., Egr1, Gap-43, c-Jun, and GFAP) were increased in the P3 mutant nerves compared to control. These results suggested that the absence of Actl6a impacted the coordinated synthesis of pro-myelinating transcription factors myelin proteins and lipids. However, it is important to note that ACTL6A is part of the SWI/SNF complex, which contains BRG1 as the ATPase motor and previously reported to be critical for Schwann cell development (Limpert et al., 2013; Weider et al., 2012). Complexes containing BRG1 were previously reported in Schwann cells to bind to NF-κB and activate Sox10 expression (Limpert et al., 2013), but also bind to SOX10 and increase transcription of Pou3f1 and Egr2 (Weider et al., 2012) and bind to SOX10 and EGR2 to activate myelin gene expression in other cell types (Marathe et al., 2013). Importantly, the transcriptome of Actl6a mutant nerves was different from that of Brg1 mutant because transcript levels of genes increased in Brg1 null Schwann cells (e.g., Sox2), were decreased in Actl6a mutants. Validation of the transcriptome analysis was conducted by using immunohistochemistry which identifies a significant reduction in the levels of myelin protein zero (MPZ) immunoreactive nerve fibers in Actl6a mutants (Figure 4C). The levels of the pro-myelinating transcription factor POU3F1 were also reduced in mutant nerves compared to controls (Figure 4D) whereas markers of pre-myelinating stage, such as GFAP protein levels were increased (Figure 4E). Consistent with increased expression of transcripts involved in inflammatory response, the number of nerve macrophages (CD68+) was increased in the neonatal nerves of Cnp1-cre;Actl6afl/fl, although this was not observed in the Mpz-cre;Actl6afl/fmutant mice with a milder clinical phenotype (Figure 4F). Analysis of enriched transcription factor binding sites in genes with decreased expression in Actl6a mutant nerves, using the Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST) (Han et al., 2015), identified an enrichment for SOX10 and POU3F1 binding sites (data not shown), thereby suggesting an important functional relation between ACTL6A and these transcription factors.
Figure 4.
Dysregulated transcriptome in Actl6a mutant neonatal nerves
(A) Volcano plot depicting the log2 fold change plotted against –log10 p values of transcripts with differential expression (p,0.05 and FC < or > than 1) in postnatal day 3 nerves from Cnp1-cre;Actl6afl/fl (n = 3) compared to controls (n = 3). The number on top indicates the total transcripts.
(B) Gene set enrichment analysis (GSEA) of downregulated transcripts in Actl6a mutant nerves identifies myelination genes (GO:0042552) whereas GSEA of upregulated transcripts identifies inflammatory response genes (GO:0006954). Blue bars signify downregulated genes and red bars signify upregulated genes.
(C) Quantification of MPZ immunoreactivity in Actl6afl/fl control, Cnp1-cre;Actl6afl/fl, and Mpz-cre;Actl6afl/fl sciatic nerves at P5. The graph represents the average area of immunoreactivity ±SEM measured in at least 2 sections per mouse.
(D) Quantification of POU3F1+ cells per square millimeter in P5 nerve sections from mice of the indicated genotype. Data represent average number of POU3F1 immunoreactive cells ±SEM.
(E) Quantification of GFAP immunoreactivity expressed as mean fluorescence intensity (MFI) ± SEM in postnatal day 5 sciatic nerves in mice of the indicated genotype. GFAP immunoreactivity was quantified as percentages of the total section area from 2 sections per mouse and then averaged.
(F) Quantification of total number of macrophages (CD68+) per square millimeter in P5 nerve sections from mice of the indicated genotype. Data represent the average number ±SEM of CD68+ cells in at least 2 sections per animal. Statistical analysis was performed using Student’s t-test, ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001.
The TRRUST analysis also identified the presence of binding sites for the Sterol regulatory element binding transcription factor (SREBF) in the genes with decreased expression in mutant nerves (data not shown). Genes within the SREBF pathway were previously reported to be expressed at lower levels also in the peripheral nerves of mutants in the HIPPO mechano-transduction pathway (Mpz-cre;Tazfl/fl;Yapfl/+) (Poitelon et al., 2016). Therefore, we compared the transcriptome of Actl6a mutant nerves with that of P3 sciatic nerves Yap/Taz mutants. We detected a significant overlap (p = 8.9e-189), as one third of the transcripts were decreased in both Actl6a and Yap/Taz mutants neonatal nerves, compared to controls (Figure S5B). Common transcripts expressed at lower levels in mutants (Table S3) included several cholesterol biosynthesis genes (Hmgcr, Dhcr24, Lss), myelin genes (Mpz, Pmp22, Mbp), and extracellular matrix components (Col28a1, CollagenType XXVIII Alpha 1 Chain). This overlap suggested the existence of a synergism between ACTL6a-containing SWI/SNF complexes and the mechano-sensing YAP1/TAZ pathway, as also reported in cancer (Chang et al., 2018). However, the fact that the overlap was limited to 27% of the transcripts and that Pou3f1 expression was only affected in the Actl6a and not in the Yap1/Taz mutants, supported the interpretation that ACTL6a containing SWI/SNF complexes may be part of the HIPPO mechanotransduction pathway, but also exert a distinct function in Schwann cells.
Nuclear ACTL6a levels in schwann cells in response to mechanostimulation
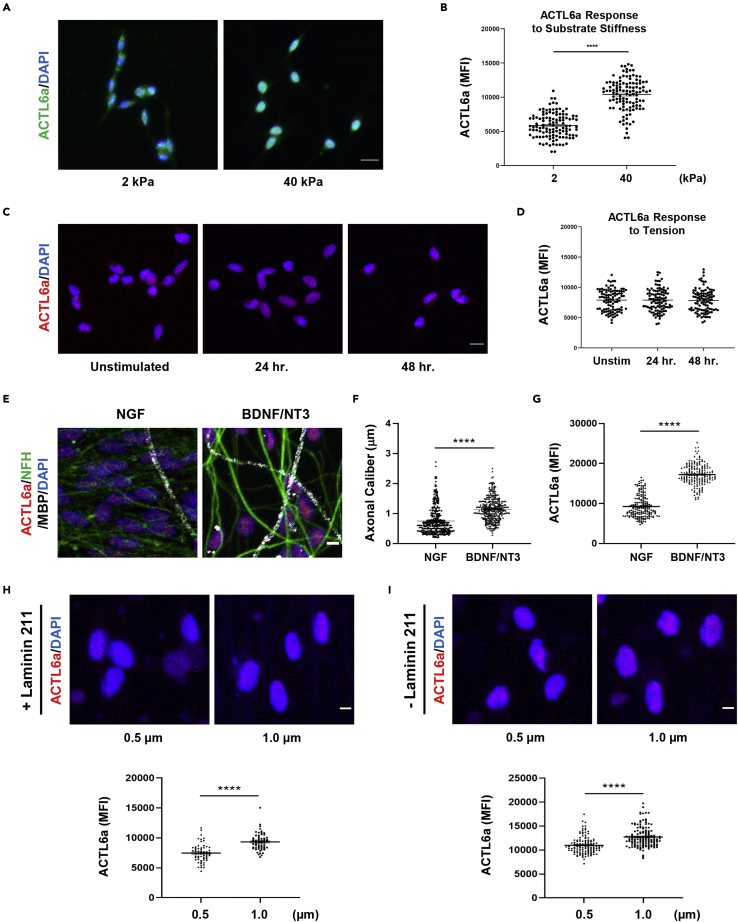
Based on the transcriptional overlap between Actl6a and Yap/Taz mutant nerves and the similar histological features of the peripheral nerve (i.e. hypomyelination and altered sorting ) we asked whether stiffness of growth substrate, a mechanical stimulus previously reported to affect nuclear localization of YAP in Schwann cells (Poitelon et al., 2016), also affected ACTL6A nuclear immunoreactivity. Indeed, Schwann cells cultured on stiffer (40 kPa) laminin 211-coated polyacrylamide hydrogels showed higher ACTL6A nuclear levels compared to softer (2 kPa) matrices (Figure 5A and 5B), and further supported the role of ACTL6A in the mechano-transduction pathway in Schwann cells. To further define whether higher ACTL6A nuclear immunoreactivity was achieved by any sort of mechano-stimulation, we cultured Schwann cells on 2D deformable silicone membrane coated with laminin 211 and subjected them to stretching for varying amounts of time. The levels of nuclear ACTL6A remained the same in the two conditions, thereby underscoring the specificity of the response to distinct physical signals (Figures 5C and 5D). The sorting phenotype detected in mice suggested that ACTL6a could act as potential axonal caliber recognition molecule. This question was initially approached by assessing ACTL6A nuclear levels in Schwann cells plated on dorsal root ganglia (DRGs) axons of different caliber, due to differential supplementation either with brain-derived neurotrophic factor (BDNF) and neurotrophin-3 (NT3), to promote survival of neurons with large caliber axons or with nerve growth factor (NGF) to promote survival of neurons with small caliber axons (Taveggia et al., 2005). The nuclear levels of ACTL6a were higher in Schwann cells co-cultured on the large caliber BDNF/NT3-supplemented DRG axons than in Schwann cells co-cultured on NGF supplemented DRG neurons (Figures 5E–5G). However, since distinct neurotrophins differentially affect the levels of NRG1 type III (Taveggia et al., 2005) we could not rule out the possibility that the increased levels of ACTL6A in Schwann cells co-cultured with neurons grown in BDNF and NT3 were related to neuregulin signaling.
Figure 5.
ACTL6a nuclear immunoreactivity is regulated by substrate stiffness and fiber caliber
(A) Confocal imaging of rat Schwann cells cultured on either soft (2 kPa) or stiff (40 kPa) laminin-coated polyacrylamide hydrogels and then stained for ACTL6a (green) and DAPI (blue) as nuclear counterstain.
(B) The scatterplot represents the mean fluorescence intensity (MFI) of nuclear ACTL6a in individual cells, measured in 100 nuclei for each condition. Average value is represented by a line ±SEM.
(C) Confocal imaging of rat Schwann cells exposed to 12% substrate elongation (tension) using the Flexcell FX-5000 Tension system for either 24 or 48 h and then stained for ACTL6a (red) and DAPI (blue) as nuclear counterstain.
(D) The scatter plot represents the individual determinations of fluorescence intensity, measured for 100 nuclei for each condition in three independent experiments. Statistical analysis was performed using one-way ANOVA with multiple comparisons.
(E) Immunofluorescent imaging of ACTl6a (red), neurofilament heavy chain (NFH, green), and MBP (white) in rat Schwann cells co-cultured with rat dorsal root ganglia (DRG) neurons grown either in 50 ng/mL NGF or in 25 ng/mL BDNF and 10 ng/mL NT3.
(F) Distribution of axonal calibers of unmyelinated fibers in DRG explants supplemented with either NGF or BDNF/NT3. Axonal calibers were measured in NFH + fibers.
(G) The scatterplot represents the mean fluorescence intensity of ACTL6a in individual Schwann cells cultured on NGF-supplemented and BDNF/NT3-supplemented DRGs. The average value is represented by a line ±SEM.
(H) Immunofluorescence of ACTL6a (red) in rat Schwann cells cultured for 48 h on 0.5 and 1 μm diameter polylactic acid (PLA) nanofibers with or without laminin 211 coating.
(I) The scatterplot represents the mean fluorescence intensity (MFI) for ACTL6a measured in at least 60 nuclei in each condition and in experiments performed 3 times. The mean ± SEM are shown. Statistical analysis was performed by Student’s t-test. ∗∗∗∗p < 0.0001. Scale bars in panels A, C, E, H and I, 5μm.
To establish the relationship between ACTL6A levels and caliber recognition independently of NRG signaling, we then used a reductionist approach and cultured Schwann cells on polylactic acid (PLA) nanofibers of different caliber (0.5μm, 1μm), either coated with laminin 211 (Figure 5H) or uncoated (Figure 5I). The levels of ACTL6a nuclear immunoreactivity were significantly higher in Schwann cells cultured on large caliber (1μm) nanofibers than those cultured on nanofibers of smaller caliber (0.5μm), regardless of whether nanofibers were coated with laminin or poly-lysine (Figures 5H and 5I). This supported the relation between ACTL6A nuclear levels and the caliber of the nanofibers.
ACTL6a integrates axonal caliber recognition and regulation of gene expression during developmental myelination of peripheral nerves
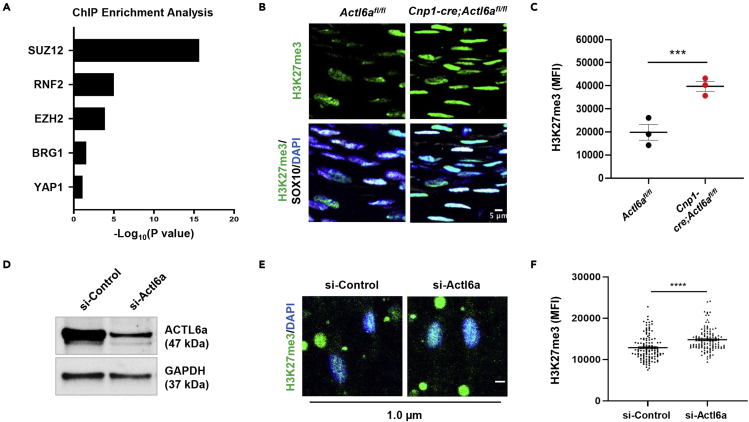
Insights regarding the molecular mechanism of ACTL6a function in Schwann cells were suggested by the results of the ChEA (Chip sequencing Enrichment Analysis), which identified within the list of transcripts decreased in the Actl6a mutant nerve, an enrichment for previously published ChIP Seq datasets for Polycomb Repressive Complex 2 (PRC2) subunits (SUZ12, EZH2, EED), PRC1 subunits (RNF2) and for BRG1 (Figure 6A). This was of extreme interest, as it had been previously reported that ACTL6A containing SWI/SNF complexes regulate gene expression through opposition of PRC2 complexes and eviction of its repressive trimethylation mark on lysine 27 in histone H3 (H3K27me3) (Ho et al., 2011; Kadoch et al., 2017; Kennison and Tamkun, 1988; Kia et al., 2008; Stanton et al., 2017). H3K27me3-mediated repression had been shown to play critical roles in repressing gene expression in Schwann cells (Ma et al., 2016) and H3K27me3 chromatin immunoprecipitation (ChIP)-sequencing studies revealed the importance of this mark for the repression of alternative lineages in adult sciatic nerves and neural crest cells (Ma et al., 2016; Rada-Iglesias et al., 2012). We therefore asked whether ACTL6A integrates mechanosensing to the coordinated production of myelin membrane by removal of the repressive H3K27me3 marks from genes regulating the myelination program.
Figure 6.
ACTL6a regulates developmental myelination by opposing H3K27me3-mediated gene repression
(A) Enrichment analysis of downregulated genes performed using Enrichr.
(B) Confocal images of neonatal nerves stained with antibodies specific for the repressive histone mark H3K27me3 (green) and for the SC marker SOX10 (white) cells.
(C) Average immunofluorescence intensity (MFI) ± SEM of H3K27me3 immunoreactivity measured in at least 150 nuclei in postnatal day 5 neonatal nerves from mutant Cnp1-cre;Actl6afl/fl (n = 3) and Actl6afl/fl controls (n = 3).
(D) Western blot analysis of protein extracts from cultured rat Schwann cells after siRNA-mediated silencing of ACTL6a. GAPDH used as loading control.
(E) Confocal images of rat Schwann cells in control conditions of after silencing of Actl6a, stained with antibodies for H3K27me3 (green) and DAPI (blue) as nuclear counterstain. Cells were cultured for 48 h on 1 μm diameter polylactic acid (PLA) nanofibers without laminin 211 coating before fixation and staining.
(F) Scatterplot represents the average immunofluorescence intensity ±SEM of H3K27me3 in Schwann cells with or without Actl6a silencing. Student’s t-test, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001. Scale bars in panels B and E, 5μm.
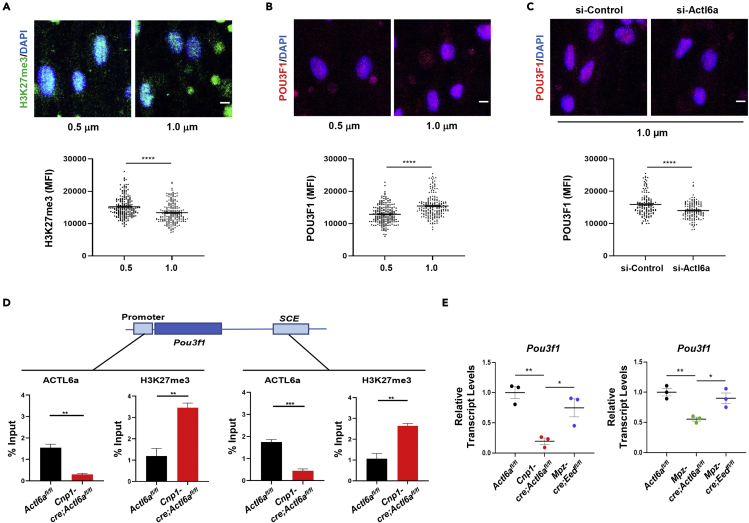
Consistent with a role for ACTL6a in evicting H3K27me3 repressive marks in Schwann cells, we detected increased global H3K27me3 immunoreactivity in SOX10+ cells in the mutant nerves compared to controls (Figures 6B and 6C). Higher nuclear levels of H3K27me3 were also detected in Actl6a silenced rat Schwann cells that were cultured on large caliber (1μm) nanofibers (Figure 6D–DF). The levels of H3K27me3 in Schwann cells cultured on nanofibers of different caliber were inversely proportional to the nuclear levels of ACTL6A, with higher levels of repressive histone marks in cells cultures on low caliber nanofibers and lower levels in those cells cultured on nanofibers with a diameter equivalent to that of myelinated nerve fibers (Figure 7A), which also showed higher nuclear levels of the pro-myelinating transcription factor POU3F1 (Figure 7B). The correlation between high nuclear levels of ACTL6A and POU3F1 in Schwann cells cultured on large caliber nanofibers was no longer detected when Actl6a was silenced (Figure 7C). To define the dynamic interplay between ACTL6A chromatin occupancy and the eviction of repressive H3K27me3 marks, we performed ChIP-qPCR of chromatin samples isolated from neonatal mutant (P3 Cnp1-cre;Actl6afl/fl) and control nerves. We used antibodies specific for ACTL6a and for H3K27me3 and primers directed at genomic locations containing genes with reduced transcript levels in mutant nerves, such as the Pou3f1 promoter and cell-specific enhancer region (SCE) approximately 10 kb downstream of the transcriptional start site. Activation of Pou3f1 expression, is driven by the activity of the distal SCE region (Ghazvini et al., 2002; Jagalur et al., 2011; Mandemakers et al., 2000) and regulated by SOX10 (Jagalur et al., 2011) and SOX10/BRG1 complexes (Weider et al., 2012). Consistent with the concept that ACTL6A containing SWI/SNF complexes render chromatin transcriptionally competent by removal of repressive histone marks, we noted that the absence of ACTL6a in mutant nerves resulted in the accumulation and persistence of the H3K27me3 repressive histone marks, compared to control nerves (Figure 7D). These results further consolidated the mechanistic interpretation of ACTL6a as modulator of Pou3f1 expression levels, due to eviction of H3K27me3, allowing key gene regulatory regions to become competent for transcription . Additional validation for this mechanism was provided by qRT-PCR analysis of nerves from mice lacking the enzyme EED, part of the PRC2 complex responsible for depositing the H3K27me3 repressive mark. The levels of Pou3f1 transcripts in Actl6a mutant nerves, characterized by increased H3K27me3 marks, were significantly lower than wildtype control, while the levels in the Mpz-cre;Eedfl/fl mutants, characterized by reduced H3K27me3 marks (Ma et al., 2015), were similar to control levels (Figure 7E). Because high levels of H3K27me3 (Cnp1-cre;Actl6afl/fl and Mpz-cre;Actl6afl/fl nerves) resulted in low Pou3f1 expression, whereas low H3K27me3 ( Mpz-cre;Eedfl/fl nerves) resulted in high Pou3f1 levels, we conclude that the eviction of H3K27me3 from key regulatory regions of target genes is a key mechanism by which ACTL6a coordinates axonal caliber recognition and a transcriptional program driving myelin synthesis in Schwann cells.
Figure 7.
ACTL6a-mediated eviction of H3K27me3 is a critical driver of Pou3f1 and myelin gene expression
(A and B) Confocal imaging and relative quantification of H3K27me3 (green) and POU3F1 (red) in rat Schwann cells cultured for 48 h on 0.5 and 1 μm diameter polylactic acid (PLA) nanofibers without laminin 211 coating. Scatterplot represents the average immunofluorescence intensity ±SEM of H3K27me3 or of Pou3f1 in individual cells. Average ±SEM is represented by a line.
(C) Confocal imaging and relative MFI quantification of POU3F1 (red) in rat Schwann cells after Actl6a silencing and cultured for 48 h on 1 μm diameter polylactic acid (PLA) nanofibers without laminin 211 coating. Scatterplot represents the average immunofluorescence intensity ±SEM of Pou3f1 in individual cells after Actl6a silencing. Average ±SEM is represented by a line. Panels A-D, statistical analysis was performed by Student’s t-test.
(D) ChIP-qPCR for ACTL6a and H3K27me3 in postnatal day3 Actl6afl/fl control vs. Cnp1-cre;Actl6afl/fl sciatic nerves. qPCR was performed with primers designed to target the promoter (positions −393 to −223 relative to transcriptional start site) and enhancer (SCE) (positions +10,043 to +10,288 relative to transcriptional start site) regions of the Pou3f1 gene loci (black bars). The results represent three independent experiments and are expressed as percentage of chromatin input ±SEM.
(E) Average Pou3f1 transcript levels ±SEM in postnatal day 3 Cnp1-cre;Actl6afl/fl (n = 3), Mpz-cre;Actl6afl/fl (n = 3) and Mpz-cre;Eedfl/fl (n = 3) nerves relative to wildtype controls (n = 3). Statistical analysis was performed using one-way ANOVA with multiple comparisons. Data represents mean ± SEM ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0001. Scale bars in panels A, B and C, 5μm.
Discussion
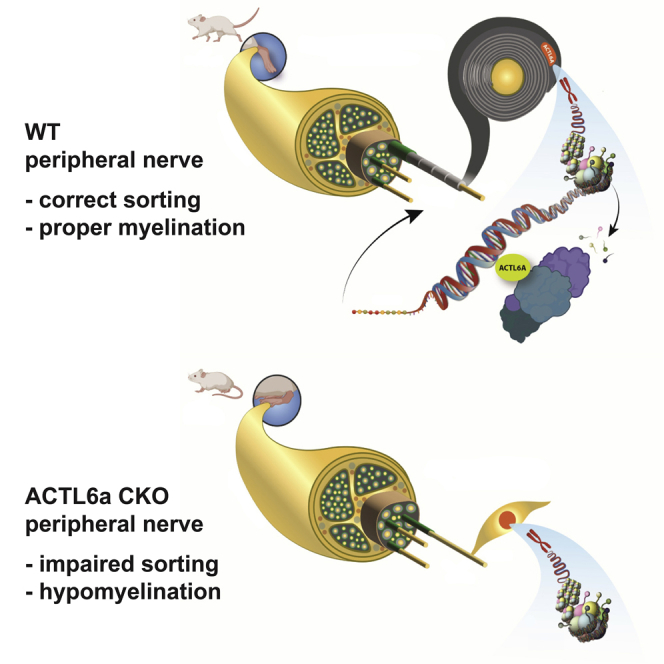
During development, Schwann cells transduce multiple signals into transcriptional programs that are responsible for coordinating proper radial sorting and myelination of axons with caliber greater or equal to 1μm. Overall, our data support ACTL6a as a mechanosensitive regulator of transcription in Schwann cells, that is required for proper radial sorting and developmental myelination. ACTL6a expression is higher in neonatal sciatic nerves, and in Schwann cells plated on large caliber axons or nano fibers of equivalent caliber, and its genetic ablation results in defective radial sorting and myelination. Thus, we identify ACTL6a, an actin-binding subunit and component of SWI/SNF complexes, as a critical molecule for the integration of physical and chemical signals in myelinating Schwann cells. Mammalian SWI/SNF complexes comprise a multitude of ATP-dependent chromatin remodelers that use energy to modify the interaction between histones and DNA and alter chromatin competence states. Complexes containing the BRG1 ATPase as enzymatic activity have been previously reported to regulate multiple steps of Schwann cell development. However, all previous studies focused on the interaction of BRG1 with specific transcription factors, leading to localized expression of specific genes. For instance, in response to neuregulin signaling, BRG1 was shown to complex with the p65 and p50 subunits of NF-κB and modulate Sox10 expression (Limpert et al., 2013). BRG1 was also shown to bind with SOX10 to activate myelin gene expression in Schwann cells (Weider et al., 2012). However, it is important to note that not all SWI/SNF complexes contain BRG1 and that the chromatin remodeling activity of the complex is the result of several other subunits, which are able to recognize and bind to chromatin domains and control gene expression. This study addresses the role of one subunit of a subset of SWI/SNF complexes, the actin-binding molecule ACTL6A, which has been reported to bind and regulate the enzymatic activity of BRG1 and also synergize with mechano-sensing pathways in cancer cells (Chang et al., 2018, 2021). Schwann cells are the myelin forming cells in the peripheral nerve, a structure which is highly susceptible to mechanical forces.
Previous studies in these cells had focused on the role of the transcriptional co-activators YAP1 and TAZ, in sensing the stiffness of substrates and lineage-specific ablation of these molecules in Schwann cells resulted in hypomyelination and defective radial sorting in the neonatal sciatic nerve (Poitelon et al., 2016). We report here that ACTL6A levels are also sensitive to stiffness and its ablation in Schwann cells results in a phenotype characterized by impaired radial sorting and hypomyelination of large caliber axon. However, ACTL6A nuclear levels in Schwann cells do not vary in response to physical forces, like tension or stretching, whereas they are very sensitive to the caliber of axons or of uncoated PLA nanofibers. Increased levels of nuclear ACTL6A were detected in Schwann cells cultured in the presence of fibers with a diameter equal or greater to 1μm, which represent the minimal size requirement for peripheral nerve myelination. The phenotype of the Actl6a mutants also differs from that of the Yap/Taz mutant mice in the presence of aberrant myelination of smaller caliber axons, with redundant myelin and membrane outfoldings. Together these results suggestthe existence of several signaling pathways in response to physical signals. Mechanotransduction regulates gene expression and we detected a significant overlap in the transcriptome of Schwann cells from Yap/Taz and Actl6a mutant nerves. For instance, in both mutants we detected downregulation of Col28a1, a collagen that is enriched in Schwann cell basal lamina (Veit et al., 2006) and also downregulation of the SREBF pathway (Poitelon et al., 2016), a critical transcription factor that activates expression of cholesterol and fatty acid synthesis in myelinating Schwann cells (Horton et al., 2002; Leblanc et al., 2005; Verheijen et al., 2003).
Mechanistically, however, YAP/TAZ are co-activators , binding to the transcription factor TEAD whereas ACTL6A is part of chromatin remodeling complexes, including the one containing BRG1 (Limpert et al., 2013; Weider et al., 2012). In Schwann cells, BRG1 was previously reported to bind several transcription factors, and activate Sox10 (Limpert et al., 2013), and Pou3f1 expression (Weider et al., 2012). Here, we show that ACTL6a can bind to the same genomic regulatory regions of the Pou3f1 gene locus (Weider et al., 2012), suggesting that Pou3f1 is regulated by the SWI/SNF complex containing both ACTL6A and BRG1. The phenotype of mice lacking Brg1 in Schwann cells shared several phenotypic similarities with Actl6a mutants such as hypomyelination, aberrant radial sorting and significant reduction in POU3F1+ cell numbers (Limpert et al., 2013; Weider et al., 2012). However, BRG1 is also part of other complexes which may activate the expression of Pou3f1 in response to NRG1-III dependent and independent signaling pathways (Leimeroth et al., 2002; Monk et al., 2009; Monuki et al., 1989), -while ACTL6a is able to remove the repressive histone marks from Pou3f1 regulatory regions, in response to axonal caliber recognition. In both cases, the reduced levels of expression of the pro-myelinating transcription factor Pou3f1, arrest the Schwann cell at an immature stage, characterized by the increased levels of Gap43, Egr1, cJun. Finally, the data on the declining levels of ACTL6A in the adult sciatic nerve, are also consistent with previous reports of enrichment of the repressive marks H3K27me3 on Pou3f1 (Ma et al., 2016) and further strengthen the conclusion that ACTL6A plays a central role in removing repressive histone marks from Pou3f1 gene regulatory elements to ensure that the chromatin is open and accessible to transcriptional activators.
However, it is important to underscore the fact that Pou3f1 is only one of several ACTL6A gene targets and therefore it is not surprising that the phenotype of Actl6a mutants is far more complex than the phenotype of mice with deletion of a single transcription factor. Besides hypomyelination of large caliber axons, the phenotype of Actl6a mutants show a paradoxical overproduction of myelin at small caliber axons, as indicated by redundant myelin and several outfoldings, collectively suggesting a role of ACTL6a in coordinating the caliber of the axon to the amount of myelin being produced by the Schwann cells. This phenotype is highly reminiscent of that observed in double transgenic mice with Schwann cell specific deletion of laminin 211 and overexpression of NRG1 type III, to mimic a dysregulated response of Schwann cells to opposing signals from the extracellular matrix and the axon (Ghidinelli et al., 2017). Similar to our mutant, the phenotype of thedouble transgenic mice was characterized by the presence of clusters of large naked axons with diameter >1μm and redundant myelin with outfoldings around small diameter axons (Ghidinelli et al., 2017). The authors concluded that Schwann cells in the double transgenic mice were unable to appropriately interpret and integrate the distinct signals. Mechanistically, the authors suggested that in immature Schwann cells, proper sorting depends on signals derived from the basal lamina, such as Laminin 211, which inhibits NRG1 function in order to prevent precocious myelination of inappropriate caliber axons, whereas in large caliber axons favors the fine-tuning of the myelin sheath components necessary for adequate wrapping. Because ACTL6a has several gene targets, including components of the basal lamina, based on the similarity of the Actl6a phenotype with that previously reported for the double transgenic (with dysregulated response of Schwann cells to opposing signals from the extracellular matrix and the axon), we suggest that both mutants may be characterized by a profound dysregulation between axonal size recognition and expression of genes related to myelination, basal lamina and lipid and myelin protein synthesis.
In summary, our data identify the chromatin remodeling complex containing ACTL6A in Schwann cells, as a point of signal convergence and transcriptional integration, functioning at multiple stages of peripheral nerve myelination. In immature Schwann cells during axonal sorting, in response to mechanical signals (such as axonal caliber and stiffness of the extracellular matrix), nuclear ACTL6A levels are increased and, together with other mechanosensitive complexes, such as TEAD/YAP1 complexes, facilitate the synthesis of basal lamina components that contribute to successful sorting. Once premyelinating cells have reached a 1:1 ratio with the axon, the production of myelin proteins and lipids needs to be coordinated by integrating chemical signals (such as Laminin 211 and NRG1 type III) and physical signals (axonal caliber recognition). The nuclear levels of ACTL6A respond to the size of the axonal caliber, and this leads to the removal of repressive histone marks at gene loci critical for the transcriptional program of myelination. The proper amount of myelin proteins and lipids is then carefully orchestrated in myelinating Schwann cells by the integration of chemical and physical signals. In the absence of ACTL6a, the expression of ECM components in immature Schwann cells is compromised, leading to discontinuity of the basal lamina and defective radial sorting. The levels of Pou3f1 in many cells are insufficient to initiate a myelination program, thereby leaving them at an immature stage. In the few cells which have been able to overcome these hurdles and successfully sort, the fine-tuning mechanism of membrane production in response to axonal caliber is impaired and this results in deficient myelin production for large caliber axons and redundant myelin for small caliber axons.
Future studies will be needed to further decipher the molecular nature of the mechanism of integration of chemical and physical signals in the PNS and understand whether similar or distinct mechanisms occur in the central nervous system.
Limitations of the study
A potential caveat of this study is that we relied on RNA-Seq analysis to define the overall effect of ACTL6a on the Schwann cell transcriptome; however, a ChIP-seq analysis would be needed to define the exact genome-wide distribution of ACTL6a in chromatin isolated from Schwann cells at distinct stages of development. It would be of great interest to compare and contrast with the results of BRG1 Chip-Seq in the same samples to define the degree of overlap between the distributions of the two SWI-SNF complex components. Finally, it would be important to define the role of ACTL6A in the integration of ECM and neuregulin signaling, possibly creating existent transgenic mouse lines or designing new ones.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-ACTL6a (IHC, ICC, WB) | Abcam | Cat#ab3882;RRID:AB 304125 |
| Rabbit polyclonal anti-H3K27me3 (IHC, ICC) | Millipore | Cat#07-449;RRID:AB 310624 |
| Goat polyclonal anti-SOX10 (IHC) | Santa Cruz | Cat#sc17342xRRID: N/A |
| Rabbit monoclonal anti-SOX10 (IHC) | Cell Signaling | Cat#89356;RRID:AB 2792980 |
| Mouse monoclonal anti-NfL (IHC) | Abcam | Cat#ab7255;RRID:AB 305803 |
| Rat monoclonal anti-MBP (IHC, ICC) | Millipore | Cat#MAB386;RRID:AB 94975 |
| Chicken polyclonal anti-MPZ (IHC) | Aves Labs | Cat#PZO;RRID:AB 2313561 |
| Rabbit polyclonal anti-POU3F1 (IHC) | Gift from Dr. Dies Meijer | N/A |
| Rabbit polyclonal anti-EGR2 (IHC) | Covance | Cat#pRB-236P;RRID:AB_291594 |
| Mouse monoclonal anti-GFAP (IHC) | BioLegend | Cat#644702;RRID:AB 2294566 |
| Rabbit polyclonal anti-CD68 (IHC) | Abcam | Cat#ab125212;RRID:AB 10975465 |
| Rabbit polyclonal anti-Caspase3 (IHC) | Cell Signaling | Cat#9661;RRID:AB 2341188 |
| Rabbit monoclonal anti-Ki67 (IHC) | Abcam | Cat#ab21700;RRID:AB 446486 |
| Mouse monoclonal anti-GAPDH (WB) | Abcam | Cat#ab8245;RRID:AB 2107448 |
| Goat anti-rabbit Alexa 546 (IHC, ICC) | Invitrogen | Cat#A11035;RRID:AB 143051 |
| Donkey anti-rabbit Alexa 594 (IHC) | Invitrogen | Cat#A21207;RRID:AB 141637 |
| Goat anti-mouse Alexa 488 (IHC, ICC) | Invitrogen | Cat#A21042;RRID:AB 141357 |
| Goat anti-rat Alexa 647 (IHC, ICC) | Life Technologies | Cat#A21247;RRID:AB 141778 |
| Goat anti-chicken Alexa 488 (IHC) | Life Technologies | Cat#A11039;RRID:AB 142924 |
| Donkey anti-goat Alexa 488 (IHC) | Abcam | Cat#ab150129;RRID:AB 2687506 |
| Mouse anti-rabbit IgG, light chain specific (WB) | Jackson ImmunoResearch | Cat#211-032-171;RRID:AB 2339149 |
| Goat anti-mouse IgG, light chain specific (WB) | Jackson ImmunoResearch | Cat#115-035-174;RRID:AB 2338512 |
| Chemicals, peptides, and recombinant proteins | ||
| MISSION siRNA Transfection Reagent | Sigma | Cat# S1452 |
| Deposited data | ||
| RNA-seq analysis data | This paper | GSE179340 |
| Experimental models: Organisms/strains | ||
| Mouse: Cnp1-cre | Gift from Dr. Klaus A Nave | N/A |
| Mouse: Mpz-cre | Gift from Dr. Laura Feltri | N/A |
| Mouse: ACTL6afl/fl | Gift from Dr. Gerald Crabtree | N/A |
| Oligonucleotides | ||
| MISSION siRNA-Actl6a | Sigma | siRNA ID; SASI_Rn02_00204786 |
| MISSION siRNA Universal Negative Control | Sigma | Cat# SIC001 |
| BaseScope Target Probe | Advanced Cell Diagnostics | N/A; Customized |
| 18s Forward: AGT CCC TGC CCT TTG TAC ACA18s Reverse: GAT CCG AGG GCC TCA CTA AAC | Scaglione et al. (2018) | N/A |
| Actl6b Forward: AAA TGC CCA ATG GCT ACA ACActl6b Reverse: GGC GAA TGT CAA TGT CAC AC | This paper | N/A |
| Actl6a Forward: GGA TAG AGC CAC CAA TCC ATActl6a Reverse: TCT CTA CGG CAG TGT GAT CG | This paper | N/A |
| Egr2 Forward: CAG TGA CTG CCA CCC CTT ATEgr2 Reverse: TAC TTT CGA GGT CGC AGG AT | He et al. (2010) | N/A |
| Wdr33 Forward: CAG GCA CAT AAG GAG GCG ATTWdr33 Reverse: CCG TCA TCA GAG CAT GTA GCA | This paper | N/A |
| Pou3f1 Forward: CAA GCA GTT CAA GCA ACG ACPou3f1 Reverse: TGG TCT GCG AGA ACA CGT TA | He et al. (2010) | N/A |
| Gapdh Forward: CAA GGT CAT CCC AGA GCT GAAGapdh Reverse: CAG ATC CAC GAC GGA CAC | This paper | N/A |
|
Pou3f1 Promoter (ChIP-qPCR)Forward: GGAAATTGAGCTTGTGTG GAAReverse: GCCCGCGTACACATTCACGC |
Weider et al. (2012) | N/A |
| Pou3f1 SCE (ChIP-qPCR)Forward: CCCCTGACACAAACAATReverse: GCAGGACAATAGCTGCA | Weider et al. (2012) | N/A |
| Software and algorithms | ||
| ImageJ-Fiji | NIH Image https://imagej.net/software/fiji/ |
RRID:SCR_002285 |
| GraphPad Prism 9 | GraphPad Software http://www.graphpad.com/ |
RRID:SCR_002798 |
| R Project for Statistical Computing | R Software http://www.r-project.org/ |
RRID:SCR_001905 |
| Other | ||
| Poly-L-Lactic Acid nanofiber inserts | Nanofiber Solutions | N/A; Customized |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Patrizia Casaccia (pcasaccia@gc.cuny.edu).
Materials availability
This study did not generate any new unique reagents. RNA-Seq data have been deposited into suitable repositories.
Experimental model and subject details
Animals
6-8 weeks-old male and female mice were used for breeders. All breeding and experiments were performed according to approved protocols by the Institutional Animal Care and Use Committee (IACUC) at Mount Sinai Medical Center, at the Advanced Science Research Center (ASRC) of the Graduate Center of The City University of New York (CUNY). Mice from either sex were used and mutants were checked for survival and weight every day from birth to weaning. For conditional knockout mouse lines, we crossed and bred the ACTL6afl/fl line (Gift from Dr. Gerald Crabtree) with either the Cnp1-cre (Gift from Dr. Klaus Nave) or the Mpz-cre (Gift from Dr. Laura Feltri) lines. Male and female mice at postnatal day 3 (P3), 5 (P5), 7 (P7), 10 (P10), 15 (P15), 30 (P30), 60 (P60) were used for sample collection. Motor scoring was conducted on mice starting at P15 (2 weeks) until age 16 weeks. Sample size (n) is mentioned in each figure legend.
Method details
Motor scoring
Motor scoring was performed on mice starting at P15 (2 weeks) until age 16 weeks, using the following scoring system: 1 – hindlimb clasping, 2 – unable to hold on to inverted grid, 2.5 – can only grip upright grid with front paws, 3 – not able to hold on to upright grid, 3.5 – no forelimb resistance when pulled across grid, 4 – hindlimb paralysis, 5 – dead. For grip strength measurements, mice were loaded onto a Bioseb GT3 grip strength meter and pulled in a horizontal direction until forelimbs were released from the meter. For each mouse, 3 measurements were taken and then averaged.
Cell culture and in vitro studies
Schwann cells (SC) were isolated from P2 rat sciatic nerves and expanded in vitro for 3 weeks in medium composed of MEM, 10% FBS and 2 mM L-glutamine (M media), supplemented with 4 μM forskolin (Sigma) and 5 ng/mL of the EGF domain of rhNRG-1-β1 (R&D Systems) (M+ media). Cells were maintained at 37°C in a 5% CO2 incubator with fresh medium change every 2-3 days.
For nanofiber experiments, electrospun poly-L-lactic acid (PLA) nanofiber inserts (NanoAlignedTM) were provided by Nanofiber Solutions, Inc. with diameters of either 0.5 or 1 μm. Before the cell culturing, nanofiber inserts were loaded in 12-well cell culture plates and coated with poly-D-lysine (PDL, 0.1 mg/mL) and laminin 211 (10 mg/mL). Rat Schwann cells were seeded at a density of 3 × 105 cells/well (insert) and cultured in M media for 48 h, then processed for immunocytochemistry.
For substrate stiffness experiments, polyacrylamide (PA) hydrogels were produced using the following protocol. 18-mm round coverslips were coated with 3-aminopropyltriethoxysilane and then incubated with 0.5% glutaraldehyde (ACROS organics, 233,280,250) for 30 min. PA gels of differing rigidities were then produced using differing mixtures of Acrylamide:Bis-acrylamide (7.5:0.05% and 8:0.48% were used to create 2 and 40 kPa PA gel substrates, respectively) and coated with PDL (0.01 mg/mL) and laminin 211 (10 μg/mL). Rat Schwann cells were then seeded at a density of 3 × 105 cells/well. For stretching experiments, six well BioFlex culture plates (Flexcell) were coated with PDL (0.01 mg/mL) and laminin 211 (10 μg/mL). Rat Schwann cells were then seeded at a density of 9 × 105 cells/well and cultured with M media. 12% static tension was then applied for up to 48 h using the Flexcell FX-5000 Tension system.
For co-culture experiments, DRG neurons were isolated from E16 rat spinal cords and either trypsinized or directly plated as explants on collagen-coated coverslips (BD Biosciences). Cultures were maintained in neurobasal medium (2% B27 supplement, 2 mM L-glutamine, 0.4% glucose) supplemented with either 50 ng/mL NGF (Peprotech) or 25 ng/mL BDNF (Peprotech) and 10 ng/mL NT3 (Peprotech). Nonneuronal cells were removed by feeding the cultures with NB medium containing 5-fluorodeoxyuridine and uridine. Myelinating cocultures were established by seeding purified DRG neuron cultures with 100,000 SCs in C medium (MEM, 10% FBS, 2 mM L-glutamine, 0.4% glucose) supplemented with either 50 ng/mL NGF (Peprotech) or 25 ng/mL BDNF (Peprotech) and 10 ng/mL NT3 (Peprotech). After 3 days, cocultures were changed to medium supplemented with 50 μg/mL ascorbic acid (Sigma-Aldrich) and further cultured for 18 days.
siRNA transfection
Rat Schwann cells were transiently transfected for 6 h with either siRNA targeting Actl6a or non-targeting control at a final concentration of 25 nM using MISSION siRNA Transfection Reagent (Sigma). After 48 h of knockdown, cells were harvested and protein extracted and processed for western blot analysis to verify knockdown efficiency. For the nanofiber experiment, cells were first seeded on 12-well cell culture plates containing PDL-coated nanofiber inserts, followed by transfection as described. After 48 h of knockdown, cells were prosessed for immunocytochemical analysis.
Western blot
Protein lysates from mouse sciatic nerves and rat Schwann cells were separated by precast 8-16% Bis-Tris gradient gels (GenScript). After electrophoresis, proteins were transferred onto PVDF membrane (Millipore), followed by 1 h blocking in blocking buffer (5% milk, 0.1% TBS-Tween) at room temperature, then incubated overnight at 4°C with primary antibodies in blocking buffer with 0.02 Sodium Azide. After washing with 0.1% TBS-Tween, membranes were incubated with secondary light-chain specific antibodies in blocking buffer for 2 h at room temperature, followed by washing. Immunoreactivity of target protein was detected using an ECL reagent (Amersham) with ChemiDoc Imaging System (Bio-Rad). GAPDH was used as loading control for normalization. Primary and secondary antibodies used are listed in the key resources table.
Immunohistochemistry
Sciatic nerves were fixed in 4% PFA and embedded in optimal cutting temperature (OCT) compound. 12μm thick longitudinal sections were cut and stored at −80°C. Before staining, tissue sections were permeabilized and blocked by incubation with PGBA (0.1 M pH 7.4, 0.1% Gelatin porcine type A, 1% BSA, and 0.002% Sodium Azide), 10% normal goat or donkey serum and 0.1% Triton X-100 at room temperature for 1 h. Sections were incubated overnight at 4°C with primary antibodies and then Alexa Fluor secondary antibodies for 2 h at room temperature. Primary and secondary antibodies used are in key resources table. Stained tissue was cover-slipped in DAPI Fluoromount G mounting medium (Thermo Fisher). Confocal images were captured using the Zeiss LSM-800 system and quantification was performed using ImageJ.
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (PFA) for 15 min at room temperature and non-specific binding was blocked by incubation in PGBA, 10% normal goat serum, and 0.1% Triton X-100 at room temperature for 1 h. Cells were then incubated with primary antibodies overnight at 4°C and then fluorescent-dye-conjugated secondary antibodies for 2 h at room temperature. Antibodies used are in key resources table. Stained cells were the cover-slipped in DAPI Fluormount G mounting medium (Thermo Fisher). Confocal images were captured using the Zeiss LSM-800 system. Quantification of the immunofluorescent intensity was performed on captured images, using ImageJ.
In situ hybridization with basescope red
In situ hybridization combining immunohistochemistry was performed using BaseScope probe and co-detection reagents from Advanced Cell Diagnostics in accordance with guidelines provided by the manufacturer. Sections were first fixed in chilled 4% paraformaldehyde for 15 min at 4°C, dehydrated in increasing gradients of ethanol baths and left to air dry for 5 min. Endogenous peroxidase activity was quenched with hydrogen peroxide reagent for 10 min, followed by co-detection antigen retrieval for 5 min in boiling buffer. Immunohistochemistry was performed afterward using SOX10 (Cell Signaling, #89356) to detect Schwann cells, followed by protease digestion for 30 min at 40°C. BaseScope probe, BA-Mn-Actl6a-1zz-st-C1 targeting exon junction between exon 4 and 5 of Actl6a, was then hybridized for 2 h at 40°C in a humidity-controlled oven (HybEZ II, ACDbio) before signal amplification steps with series of proprietary AMP reagents. The signal of target RNA was visualized through probe-specific horseradish-peroxidase-based detection by signal amplification with BaseScop Red solution. Slides were then counterstained with DAPI, coverslipped with Prolong Gold Antifade (Thermofisher), and we proceeded for imaging analysis with Zeiss LSM-800 system.
Electron microscopy
For electron microscopy, the EM fixative used consisted of 4% glutaraldehyde and 4% PFA in PBS containing 0.4 mM CaCl2. For EM of neonatal nerves, P7 mice were immersion fixed in EM fixative and then post-fixed in the same solution at 4°C for 2 weeks. For EM of adult nerves, 8 week animals were perfused with EM fixative and then post-fixed in the same solution at 4°C for 2 weeks. Neonatal and adult sciatic nerves were then dissected out for sectioning following post-fixation protocol. Cross sections of sciatic nerves were sliced at 1 mm thickness and treated with 2% osmium tetroxide overnight followed by epoxy resin embedding. The nerve sections were then sectioned at 1 μm and stained with toluidine blue. Ultrathin sections of the nerves were cut onto copper grides and stained with uranyl acetate. Imaging was performed with a Hitachi H-600 transmission electron microscope. G-ratios were calculated by dividing the diameter of the axon by the diameter of the entire myelinated fiber. Radius (R) measurements used for G ratio calculations were obtained from Area (A) measurements using the following formula: R = √(A/π) as is used in other publications (Dillenburg et al., 2018; Kaiser et al., 2021).
RNA isolation, qRT-PCR, and RNA-seq
RNA was extracted from sciatic nerves using TRIzol (Invitrogen) and the RNeasy Mini Kit (Qiagen, 74,106) with on-column DNase treatment. RNA purity was assessed by measuring the A260/A280 ratio using a NanoDrop, and RNA quality checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). For quantitative real-time PCR (qRT-PCR), RNA was reverse-transcribed with qScript cDNA Supermix (Quantabio, 95,047). qRT-PCR reactions were run in triplicates using PerfeCTa SYBR GREEN FastMix, ROX reagent (Quantabio, 95,072) at the ASRC Epigenetic Core facility. Expression levels for each transcript were normalized to the geometric mean of three housekeeping genes: Gapdh, 18S, and Wdr33. After normalization to housekeeping genes, CT values of technical triplicates were average for each biological replicate. Data was presented as either relative to either transcript levels in controls or transcript levels of housekeeping genes. Primer sequences used are listed in Table S2.
For RNA-sequencing, approximately 150 ng of total RNA per sample was used for library construction and sequenced using the Illumina HiSeq 4000 instrument. Raw reads were trimmed and filtered using sickle (v0.7) and remaining reads aligned to the mm10 reference genome with the subjunc aligner from the subread package (v2.0). Resulting BAM files were used for gene counts against the ENSEMBL GRCm38 v95 annotation with the featureCounts tool also from the subread package. Counts included only unique aligned fragments falling on exons. Gene count matrices were moved into an R environment where differential expression analysis and enrichment analysis was done. Differential Expression was performed with DESeq2 (v1.3). Enrichment analysis was performed with the clusterProfiler package (v3.16). All downstream analysis of differentially expressed genes were filtered by an adjusted pvalue ≤ 0.05. Gene ontology analysis was performed using Enrichr (Kuleshov et al., 2016) and geneset enrichment analysis was performed against the KEGG database. The overall mapped reads and sample names are listed in Table S6. Data deposited in GEO: accession number GSE179340.
Chromatin immunoprecipitation
Sciatic nerves were harvested from P3 Cnp1-cre;Actl6afl/fl and Actl6afl/fl nerves and flash frozen. Nerves were minced and cross-linked with 1% formaldehyde for 7 min. Cross-linked nerves were then sonicated into 200-500 bp fragments using a Bioruptor (Diagenode). Chromatin was then immunoprecipitated using either 2 ug anti-H3K27me3 anti-H3K27me3 antibody (Diagenode, rabbit polyclonal c15410195) or 5 μg anti-ACTL6a antibody (Abcam, rabbit polyclonal ab3882) and Dynabeads (Invitrogen). 5 μg Rabbit IgG was used as a negative control. Following reverse cross-linking, DNA was purified using phenol:chloroform:isoamyl alcohol (Invitrogen) extraction and ethanol precipitation. DNA purified from unprecipitated chromatin was used as input. qPCR was performed using PerfeCTa SYBR Green FastMix, ROX (Quanta) at the Epigenetics Core at the ASRC. Primers used to target Pou3f1 are listed in Table S4. Abundance of target genome DNA was calculated as the percentage of input. 3 biological replicates were performed for each experiment. For each biological replicate, nerves from 7-10 mice were pooled together. 3 technical replicates were performed for each biological replicate.
Quantification and statistical analysis
All images were analyzed using FIJI-Image J and all graphs and statistical tests were carried out using GraphPad Prism. Student’s t-test was used to compare 2 datasets. 2-way ANOVA was used to compare the motor scoring datasets. 1-way ANOVA was used to compare qRT-PCR measurements of Actl6a and Pou3f1 transcript levels. For all graphs, error bars are mean ±SEM. For image quantifications, n= 3-6 mice were examined (2 images were analyzed and averaged per mouse, for each staining). Significance levels are indicated as ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001. All statistical details for each graph can be found in the figure legends. The pvalue of the overlap between genes with decreased expression in the Actl6a mutants (compared to controls) and YAP/TAZ downregulated genes (Poitelon et al., 2016) was calculated using two-tailed Fisher tests in R.
Acknowledgments
We would like to thank Mr. Ben Inbar and Ms. Juanita Merrill with help with genotyping and animal husbandry. The majority of this work was supported by a grant from the National Institutes of Health NINDS-R35NS111604 (to PC) and a predoctoral fellowship F31NS110166-02 (to ET). Experiments on developmental expression, responsiveness to stiffness and Yap/Taz mice were supported by grant R01NS45630 (to LF). The work on cocultures with DRG neurons (CMV) was supported by NIH- SC1NS000001 and Research Centers in Minority Institutions Programgrant from the National Center for Research Resources (G12 RR003037). The data on repressive histones and nerves from mice with mutations in the PRC2 complex were supported by grant R01 NS100510 (to JS). The Actl6a floxed mice and all discussion related to eviction of repressive marks were supported by HHMI and NIH grants NIH5R01CA163915 and NIHR01MH126720 (to GRC).
Author contributions
HJP and ET equally contributed to this manuscript by performing the majority of the experiments reported in this manuscript. ET conducted initial data analysis, wrote the initial draft of the text and figures. HJP worked with PC on the revised text and reorganizing all the figures. JP contributed the clinical scoring scale and participated to the initial phases of the project. DH conducted the analysis of the RNA-Seq data, organized some of the figure panels and was responsible for uploading the data to GEO. MW and LF contributed data on the expression in the developing nerve and the data relative to stiffness and to the YAP/TAZ transcriptomics, AA performed the co-culture experiments with DRG neurons. CVM-V supervised AA and provided the rat Schwann cells. JM provided the nerves from EED mice and relative transcriptomics. CVM-V, GRC, MLF and JS provided edits to the paper, contributed methods, reagents and scientific discussions. PC conceived the project, coordinated and supervised the work, reorganized the figures with HJP and ET, wrote the final version of the manuscript
Declaration of interests
The authors declare no competing interests
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104132.
Supplemental information
and Tables S3 and S4
Data and code availability
-
•
RNA-Seq data have been deposited at the Gene Expression Omnibus database (GEO) with number GSE179340 . The data are publicly available as of March 8th 2022.
-
•
This study does not report any original code.
-
•
All data reported in this study and any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
References
- Chang C.Y., Shipony Z., Lin S.G., Kuo A., Xiong X., Loh K.M., Greenleaf W.J., Crabtree G.R. Increased ACTL6A occupancy within mSWI/SNF chromatin remodelers drives human squamous cell carcinoma. Mol. Cell. 2021;81:4964–4978.e4968. doi: 10.1016/j.molcel.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Azzolin L., Di Biagio D., Zanconato F., Battilana G., Lucon Xiccato R., Aragona M., Giulitti S., Panciera T., Gandin A., et al. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018;563:265–269. doi: 10.1038/s41586-018-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S.A., Weaver J.C., Huebsch N., Lee H.P., Lippens E., Duda G.N., Mooney D.J. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements M.P., Byrne E., Camarillo Guerrero L.F., Cattin A.L., Zakka L., Ashraf A., Burden J.J., Khadayate S., Lloyd A.C., Marguerat S., Parrinello S. The wound microenvironment reprograms Schwann cells to invasive mesenchymal-like cells to drive peripheral nerve regeneration. Neuron. 2017;96:98–114.e117. doi: 10.1016/j.neuron.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillenburg A., Ireland G., Holloway R.K., Davies C.L., Evans F.L., Swire M., Bechler M.E., Soong D., Yuen T.J., Su G.H., et al. Activin receptors regulate the oligodendrocyte lineage in health and disease. Acta neuropathologica. 2018;135:887–906. doi: 10.1007/s00401-018-1813-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Feltri M.L., D'Antonio M., Previtali S., Fasolini M., Messing A., Wrabetz L. P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. N. Y. Acad. Sci. 1999;883:116–123. [PubMed] [Google Scholar]
- Feltri M.L., Graus Porta D., Previtali S.C., Nodari A., Migliavacca B., Cassetti A., Littlewood-Evans A., Reichardt L.F., Messing A., Quattrini A., et al. Conditional disruption of beta 1 integrin in Schwann cells impedes interactions with axons. J. Cell Biol. 2002;156:199–209. doi: 10.1083/jcb.200109021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltri M.L., Poitelon Y., Previtali S.C. How Schwann cells sort axons: new concepts. The Neuroscientist. 2016;22:252–265. doi: 10.1177/1073858415572361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber D., Pereira J.A., Gerber J., Tan G., Dimitrieva S., Yángüez E., Suter U. Transcriptional profiling of mouse peripheral nerves to the single-cell level to build a sciatic nerve ATlas (SNAT) eLife. 2021;10:e58591. doi: 10.7554/eLife.58591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazvini M., Mandemakers W., Jaegle M., Piirsoo M., Driegen S., Koutsourakis M., Smit X., Grosveld F., Meijer D. A cell type-specific allele of the POU gene Oct-6 reveals Schwann cell autonomous function in nerve development and regeneration. EMBO J. 2002;21:4612–4620. doi: 10.1093/emboj/cdf475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghidinelli M., Poitelon Y., Shin Y.K., Ameroso D., Williamson C., Ferri C., Pellegatta M., Espino K., Mogha A., Monk K., et al. Laminin 211 inhibits protein kinase A in Schwann cells to modulate neuregulin 1 type III-driven myelination. PLoS Biol. 2017;15:e2001408. doi: 10.1371/journal.pbio.2001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Shim H., Shin D., Shim J.E., Ko Y., Shin J., Kim H., Cho A., Kim E., Lee T., et al. TRRUST: a reference database of human transcriptional regulatory interactions. Sci. Rep. 2015;5:11432. doi: 10.1038/srep11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Kim J.Y., Dupree J., Tewari A., Melendez-Vasquez C., Svaren J., Casaccia P. Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat. Neurosci. 2010;13:1472–1480. doi: 10.1038/nn.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., Miller E.L., Ronan J.L., Ho W.Q., Jothi R., Crabtree G.R. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/jci15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagalur N.B., Ghazvini M., Mandemakers W., Driegen S., Maas A., Jones E.A., Jaegle M., Grosveld F., Svaren J., Meijer D. Functional dissection of the Oct6 Schwann cell enhancer reveals an essential role for dimeric Sox10 binding. J. Neurosci. 2011;31:8585–8594. doi: 10.1523/jneurosci.0659-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Dong B., Georgiou J., Jiang Q., Zhang J., Bharioke A., Qiu F., Lommel S., Feltri M.L., Wrabetz L., et al. N-WASp is required for Schwann cell cytoskeletal dynamics, normal myelin gene expression and peripheral nerve myelination. Development. 2011;138:1329–1337. doi: 10.1242/dev.058677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C., Williams R.T., Calarco J.P., Miller E.L., Weber C.M., Braun S.M., Pulice J.L., Chory E.J., Crabtree G.R. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2017;49:213–222. doi: 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T., Allen H.M., Kwon O., Barak B., Wang J., He Z., Jiang M., Feng G. MyelTracer: a semi-automated software for myelin g-ratio quantification. eNeuro. 2021;8 doi: 10.1523/eneuro.0558-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison J.A., Tamkun J.W. Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. U S A. 1988;85:8136–8140. doi: 10.1073/pnas.85.21.8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia S.K., Gorski M.M., Giannakopoulos S., Verrijzer C.P. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 2008;28:3457–3464. doi: 10.1128/mcb.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva V., Buscarlet M., Diaz-Tellez A., Bernard M.A., Crabtree G.R., Lessard J.A. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood. 2012;120:4720–4732. doi: 10.1182/blood-2012-04-427047. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C., Goebbels S., Gravel M., Nicksch E., Lee J., Braun P.E., Griffiths I.R., Nave K.A. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat.Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Leblanc S.E., Srinivasan R., Ferri C., Mager G.M., Gillian-Daniel A.L., Wrabetz L., Svaren J. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J.Neurochem. 2005;93:737–748. doi: 10.1111/j.1471-4159.2005.03056.x. [DOI] [PubMed] [Google Scholar]
- Leimeroth R., Lobsiger C., Lüssi A., Taylor V., Suter U., Sommer L. Membrane-bound neuregulin1 type III actively promotes Schwann cell differentiation of multipotent Progenitor cells. Dev. Biol. 2002;246:245–258. doi: 10.1006/dbio.2002.0670. [DOI] [PubMed] [Google Scholar]
- Lessard J., Wu J.I., Ranish J.A., Wan M., Winslow M.M., Staahl B.T., Wu H., Aebersold R., Graef I.A., Crabtree G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert A.S., Bai S., Narayan M., Wu J., Yoon S.O., Carter B.D., Lu Q.R. NF-κB forms a complex with the chromatin remodeler BRG1 to regulate Schwann cell differentiation. J. Neurosci. 2013;33:2388–2397. doi: 10.1523/jneurosci.3223-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K.H., Hung H.A., Srinivasan R., Xie H., Orkin S.H., Svaren J. Regulation of peripheral nerve myelin maintenance by gene repression through polycomb repressive complex 2. J. Neurosci. 2015;35:8640–8652. doi: 10.1523/jneurosci.2257-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K.H., Hung H.A., Svaren J. Epigenomic regulation of Schwann cell reprogramming in peripheral nerve injury. J. Neurosci. 2016;36:9135–9147. doi: 10.1523/jneurosci.1370-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandemakers W., Zwart R., Jaegle M., Walbeehm E., Visser P., Grosveld F., Meijer D. A distal Schwann cell-specific enhancer mediates axonal regulation of the Oct-6 transcription factor during peripheral nerve development and regeneration. EMBO J. 2000;19:2992–3003. doi: 10.1093/emboj/19.12.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe H.G., Mehta G., Zhang X., Datar I., Mehrotra A., Yeung K.C., de la Serna I.L. SWI/SNF enzymes promote SOX10- mediated activation of myelin gene expression. PLoS One. 2013;8:e69037. doi: 10.1371/journal.pone.0069037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailov G.V., Sereda M.W., Brinkmann B.G., Fischer T.M., Haug B., Birchmeier C., Role L., Lai C., Schwab M.H., Nave K.A. Axonal neuregulin-1 regulates myelin sheath thickness. Science. 2004;304:700–703. doi: 10.1126/science.1095862. [DOI] [PubMed] [Google Scholar]
- Monk K.R., Naylor S.G., Glenn T.D., Mercurio S., Perlin J.R., Dominguez C., Moens C.B., Talbot W.S. A G protein-coupled receptor is essential for Schwann cells to initiate myelination. Science. 2009;325:1402–1405. doi: 10.1126/science.1173474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montani L., Buerki-Thurnherr T., de Faria J.P., Pereira J.A., Dias N.G., Fernandes R., Gonçalves A.F., Braun A., Benninger Y., Böttcher R.T., et al. Profilin 1 is required for peripheral nervous system myelination. Development. 2014;141:1553–1561. doi: 10.1242/dev.101840. [DOI] [PubMed] [Google Scholar]
- Monuki E.S., Weinmaster G., Kuhn R., Lemke G. SCIP: a glial POU domain gene regulated by cyclic AMP. Neuron. 1989;3:783–793. doi: 10.1016/0896-6273(89)90247-x. [DOI] [PubMed] [Google Scholar]
- Nawaz S., Sánchez P., Schmitt S., Snaidero N., Mitkovski M., Velte C., Brückner B.R., Alexopoulos I., Czopka T., Jung S.Y., et al. Actin filament turnover drives leading edge growth during myelin sheath formation in the central nervous system. Dev. Cell. 2015;34:139–151. doi: 10.1016/j.devcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak N., Bar V., Sabanay H., Frechter S., Jaegle M., Snapper S.B., Meijer D., Peles E. N-WASP is required for membrane wrapping and myelination by Schwann cells. J. Cell Biol. 2011;192:243–250. doi: 10.1083/jcb.201010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegatta M., De Arcangelis A., D'Urso A., Nodari A., Zambroni D., Ghidinelli M., Matafora V., Williamson C., Georges-Labouesse E., Kreidberg J., et al. α6β1 and α7β1 integrins are required in Schwann cells to sort axons. J. Neurosci. 2013;33:17995–18007. doi: 10.1523/jneurosci.3179-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poitelon Y., Lopez-Anido C., Catignas K., Berti C., Palmisano M., Williamson C., Ameroso D., Abiko K., Hwang Y., Gregorieff A., et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat.Neurosci. 2016;19:879–887. doi: 10.1038/nn.4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A., Bajpai R., Prescott S., Brugmann S.A., Swigut T., Wysocka J. Epigenomic annotation of enhancers predicts transcriptional regulators of human neural crest. Cell Stem Cell. 2012;11:633–648. doi: 10.1016/j.stem.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglione A., Patzig J., Liang J., Frawley R., Bok J., Mela A., Yattah C., Zhang J., Teo S.X., Zhou T., et al. PRMT5-mediated regulation of developmental myelination. Nat. Commun. 2018;9:2840. doi: 10.1038/s41467-018-04863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow N., Manetti M.E., Bott M., Fabianac T., Petrilli A., Bates M.L., Bunge M.B., Lambert S., Fernandez-Valle C. The actin-severing protein cofilin is downstream of neuregulin signaling and is essential for Schwann cell myelination. J. Neurosci. 2012;32:5284–5297. doi: 10.1523/jneurosci.6207-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton B.Z., Hodges C., Calarco J.P., Braun S.M., Ku W.L., Kadoch C., Zhao K., Crabtree G.R. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 2017;49:282–288. doi: 10.1038/ng.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlong H., Hinata K., Viswanathan R., Erdjument-Bromage H., Tempst P., Cairns B.R. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C., Zanazzi G., Petrylak A., Yano H., Rosenbluth J., Einheber S., Xu X., Esper R.M., Loeb J.A., Shrager P., et al. Neuregulin-1 type III determines the ensheathment fate of axons. Neuron. 2005;47:681–694. doi: 10.1016/j.neuron.2005.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanski M.M., Kingsbury L., Moussouros D., Kassim I., Mehjabeen S., Paknejad N., Melendez-Vasquez C.V. Myelinating glia differentiation is regulated by extracellular matrix elasticity. Sci. Rep. 2016;6:33751. doi: 10.1038/srep33751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit G., Kobbe B., Keene D.R., Paulsson M., Koch M., Wagener R. Collagen XXVIII, a novel von Willebrand factor A domain-containing protein with many imperfections in the collagenous domain. J. Biol. Chem. 2006;281:3494–3504. doi: 10.1074/jbc.M509333200. [DOI] [PubMed] [Google Scholar]
- Verdier V., Csárdi G., de Preux-Charles A.S., Médard J.J., Smit A.B., Verheijen M.H., Bergmann S., Chrast R. Aging of myelinating glial cells predominantly affects lipid metabolism and immune response pathways. Glia. 2012;60:751–760. doi: 10.1002/glia.22305. [DOI] [PubMed] [Google Scholar]
- Verheijen M.H., Chrast R., Burrola P., Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M., Küspert M., Bischof M., Vogl M.R., Hornig J., Loy K., Kosian T., Müller J., Hillgärtner S., Tamm E.R., et al. Chromatin-remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev. Cell. 2012;23:193–201. doi: 10.1016/j.devcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Wilson E.R., Della-Flora Nunes G., Weaver M.R., Frick L.R., Feltri M.L. Schwann cell interactions during the development of the peripheral nervous system. Dev. Neurobiol. 2021;81:464–489. doi: 10.1002/dneu.22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Bierman J., Tarumi Y.S., Zhong Y.P., Rangwala R., Proctor T.M., Miyagoe-Suzuki Y., Takeda S., Miner J.H., Sherman L.S., et al. Coordinate control of axon defasciculation and myelination by laminin-2 and -8. J. Cell Biol. 2005;168:655–666. doi: 10.1083/jcb.200411158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.M., Feltri M.L., Wrabetz L., Strickland S., Chen Z.L. Schwann cell-specific ablation of laminin gamma1 causes apoptosis and prevents proliferation. J. Neurosci. 2005;25:4463–4472. doi: 10.1523/jneurosci.5032-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Wang W., Rando O.J., Xue Y., Swiderek K., Kuo A., Crabtree G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- Zuchero J.B., Fu M.M., Sloan S.A., Ibrahim A., Olson A., Zaremba A., Dugas J.C., Wienbar S., Caprariello A.V., Kantor C., et al. CNS myelin wrapping is driven by actin disassembly. Dev. Cell. 2015;34:152–167. doi: 10.1016/j.devcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and Tables S3 and S4
Data Availability Statement
-
•
RNA-Seq data have been deposited at the Gene Expression Omnibus database (GEO) with number GSE179340 . The data are publicly available as of March 8th 2022.
-
•
This study does not report any original code.
-
•
All data reported in this study and any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.