Abstract
Current delivery strategies for cancer therapeutics commonly cause significant systemic side effects due to required high doses of therapeutic, inefficient cellular uptake of drug, and poor cell selectivity. Peptide-based delivery systems have shown the ability to alleviate these issues and can significantly enhance therapeutic loading, delivery, and cancer targetability. Peptide systems can be tailor-made for specific cancer applications. This review describes three peptide classes, targeting, cell penetrating, and fusogenic peptides, as stand-alone nanoparticle systems, conjugations to nanoparticle systems, or as the therapeutic modality. Peptide nanoparticle design, characteristics, and applications are discussed as well as peptide applications in the clinical space.
Keywords: Peptide delivery, Nanoparticle conjugation, Gene therapy, Drug therapy, Cancer
Graphical abstract
1. Introduction
Cancer is one of the leading causes of death globally and the second leading cause of death in the United States [1]. Current cancer treatment regimens often cause significant adverse side effects and fall short in effectively eliminating advanced disease [2]. Low therapeutic targeting ability, insufficient cellular uptake, and development of multidrug resistance are among the current treatment limitations [3]. These limitations can result in decreased efficacy and increased likelihood of metastatic disease. Numerous research endeavors have been initiated to improve personalized therapeutics that address the challenges of cancer treatment, including rapid growth, distant metastatic spread, dysregulation of oncogenes, and development of treatment resistance [4]. Exploiting some of these inherent characteristics of cancer to increase treatment efficacy and circumvent off-target adverse effects are key to developing better therapeutics and therapeutic delivery systems [4].
Because of the diverse functions that peptides offer, this nonviral carrier has rapidly gained exposure and popularity as a prospective delivery vehicle. Peptide-conjugated and peptide-based delivery systems are commonly comprised of peptides derived from viral proteins, which assist viruses in delivering their genome into host cells and are known to have high delivery efficiency [5]. Synthetic peptides that have been designed to optimize delivery properties, including efficient cargo complexation, cell targeting, stability in physiological conditions, and controlled cargo release are also commonly used [[6], [7], [8], [9], [10], [11]]. Combinations of the 20 naturally occurring amino acids provide a spectrum of characteristics, including vehicle conformation, charge, polarity, and hydrophobicity, all of which are vital to ensuring efficient drug loading and delivery to cancer tissue [6,12]. These delivery systems have been used to target tumor microenvironments (TME), enhance cellular uptake, disrupt lysosomal degradation pathways, and assist in controlled and sustained release of therapeutics [[4], [5], [6],13]. Additionally, the release of therapeutic cargo can be initiated by internal and external stimuli including pH, enzyme activity, redox potential, thermal shifts, and light [11,[14], [15], [16]]. Due to their versatility, these peptides, either natural or synthetic, contain specific biochemical properties that can be divided into three areas of classification: cell-penetrating, fusogenic, and targeting peptides. These classes of peptides have been implemented in several in vitro and in vivo applications as delivery systems for cancer therapy and as therapeutic modalities (peptide vaccines) in clinical trials [[17], [18], [19], [20]]. One such application that has garnered recent attention is the implementation of peptide systems as ‘shuttles’ to address the significant disadvantage in effectively treating malignant brain disease [21]. By implementing characteristics of fusogenic, cell-penetrating, and targeting peptides, delivery of therapeutics across the blood-brain barrier has been achieved via paracellular diffusion, adsorptive-mediated transcytosis, receptor-mediated transcytosis, and carrier-mediated transport [21,22]. Furthermore, some peptide shuttles have been advantageously linked to antibody Fc domains, enhancing the pharmacokinetic properties of the delivery system to better target and sustain delivery of therapeutics to hard-to-access areas of the brain that may contain malignant tumors [23].
Through the addition of functional peptides, nanoparticle delivery systems can become well-suited for systemic delivery with minimal immune response and efficient cargo loading [24]. Additionally, exploiting overexpressed cell surface receptors has been a focus in developing peptide-modified delivery systems for patient- and cell-specific medicine to reduce adverse systemic reactions [24]. Nonviral nanoparticle-based delivery systems primarily comprised of lipids and polymers have been well-studied in preclinical and clinical models [25]. Barriers to implementing lipid and polymer-based systems include particle aggregation, cytotoxicity, reduced efficacy at low doses, and low cell specificity [[25], [26], [27]]. Fortunately, many of these issues can be addressed through conjugation of natural or synthetically derived peptide systems [25].
While peptides have been used extensively as complementary components of nanoparticle delivery systems [28,29], using peptides directly as delivery systems has several advantages. Peptides yield a low immunogenic response because of their small size and ability to be tailor-made with minimal surface charge densities to minimize opsonization, avoiding downstream phagocytotic destruction when complexed with nanoparticles [30,31]. Additionally, peptide-based systems can accumulate quickly in the TME due to their size and utilization of the enhanced-permeability and retention effect. Peptides can also be designed to avoid clearance via the reticuloendothelial system through inclusion of hydrophilic residues that pull in water to shield the peptide from opsonization or for cell receptor specificity to achieve enhanced tissue targeting capabilities [8,32].
This review discusses peptide delivery systems and highlights applications of targeting, cell penetrating, and fusogenic peptides as delivery systems for cancer therapy. Peptide systems and peptide-conjugated nanoparticles will be discussed as carriers for chemo and gene therapeutics individually and as combination approaches. Characteristics of each subtype will be discussed as well as current research that exemplifies the advantages and disadvantages of each. Having a complete understanding of the benefits and pitfalls of each peptide subtype can assist in designing peptides as complementary components or direct delivery systems. Several efforts have been successful in identifying optimal peptide formulations to advance these systems to clinical trials, with some achieving phase III completion [33]. Special focus will be given to past and current clinical trials exploring peptide-based systems.
2. Targeting peptides and cancer specificity
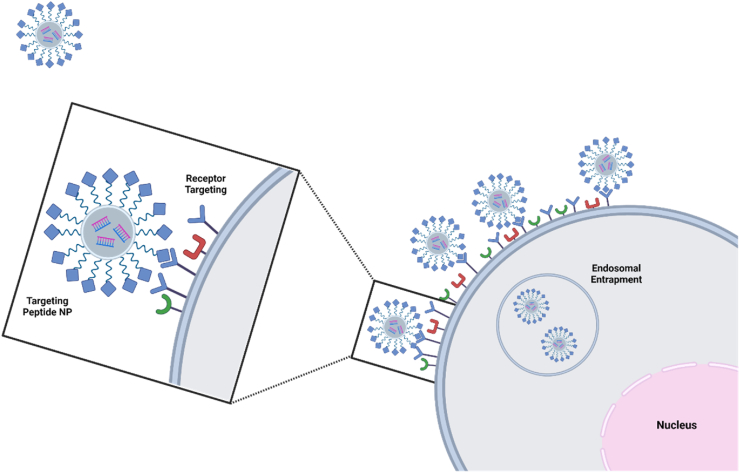
Integrating advanced targeted delivery systems can provide significant improvement of cellular uptake, and reduce off-target adverse effects, improving clinical translatability compared to non-targeted delivery systems, addressing a major barrier to therapeutic delivery [34]. Used as complementary moieties to nanoparticle systems, targeting peptides have demonstrated enhanced cell specificity and affinity for cancer cells with unique, overexpressed cell receptors, depicted in Fig. 1 [35]. Conjugation of a targeting peptide to a nanocarrier or drug increases precision for targeting cancerous cells and decreases off-target effects, reducing the quantity of drug that must be administered and enhancing drug concentration at target sites [36].
Fig. 1.
Targeting peptides bind to overexpressed cell receptors commonly seen in cancer cells. This binding can induce receptor-mediated endocytosis, enhancing peptide-cargo cell uptake. Created with Biorender.com.
Targeting peptides are largely used in cancer therapeutics to target various receptors present on cancer cells or tumors with little to no expression on healthy tissues but can also be implemented for specific delivery of therapeutics to endothelial cells, cardiac cells, and even chondrocytes to elicit cell differentiation and recovery after injury or due to degenerative disease [[37], [38], [39], [40]].
Before targeting peptides became more prevalent as active targeting moieties, monoclonal antibodies (mAb) were explored as targeting moieties, but many drawbacks were reported [37,41]. mAbs are large (molecular weight around 150 kDa), which can hinder infiltration of mAb-conjugated nanoparticles into tumors [41,42]. Another complication is the uptake of mAbs by the spleen, liver, and other organs of the reticuloendothelial system (RES) due to the presence of phagocytic cells that bind to these circulating antibody complexes [43]. Use of peptides as targeting entities can overcome these barriers because of their significantly lower molecular weight and decreased immunogenicity [43]. Additional advantages of using peptides as targeting agents include lower manufacturing costs, increased efficiency for infiltrating tumor masses, high stability, and high tunability [44,45]. Targeting peptides increase the specificity of drug delivery systems for targeting tumors and cancer cells, enhancing uptake of therapeutics in cancer cells and minimizing off target-effects. However, a disadvantage of targeting peptides is that nonspecific uptake is still possible, especially if the target is expressed in healthy cells.
2.1. Identification and synthesis of targeting peptides
Various techniques have been developed to identify and synthesize targeting peptides. One common technique is phage display, which utilizes bacteriophage libraries to screen for a peptide sequence that has the highest affinity for a given target [45]. Among bacteriophage libraries are Ph.D.™-7 and Ph.D.™-12 phage display peptide libraries. These libraries use M13 bacteriophages that express multiple copies of a randomized 7- or 12-amino acid peptide sequence on their surfaces [46,47]. Peptide phenotypes are altered by fusing genes that encode for peptides to genes that encode for phage surface expression [48]. These phages can be introduced in vivo; however in vitro panning has been proven to be more effective for identifying targeting peptides more quickly [49]. For in vitro panning, phages are incubated with target cells or target receptors; phages that do not bind to the target are washed away, while the bound phages are amplified using bacteria, completing one round of biopanning [50]. This procedure is repeated up to 3–5 times, narrowing the library to sequences with the highest affinity for the chosen cell receptor [50]. The peptides present on these phages, commonly 10–20 amino acids in length, can then be identified through DNA sequencing and enzyme-linked immunosorbent assays [50].
An alternative method for identifying targeting peptides is the one-bead one-compound (OBOC) combinational library [51]. This method utilizes resin beads that each display one peptide sequence [52]. The OBOC library is synthesized by adding peptide sequences to the bead through PEG linkers, then incubated with the target of interest [[53], [54], [55]]. Usually, this method is performed in vitro by incubating the beads with target cell receptors or cell lines [55]. The beads that bind to the target of interest are separated from the nonbinding beads and can be characterized through microsequencing [53,54]. An advantage of the OBOC method over phage display is that an OBOC library can consist of peptides that have not only l-amino acids, but also unnatural amino acids and d-amino acids [51].
Solid- or liquid-phase synthesis are used as the modern methods of fabrication for targeting peptides as well as for synthesizing most other peptide varieties. Solid-phase synthesis, the most commonly used method, was first reported in 1963 and is based upon the solubility of the group shielding the exposed carboxyl group [56]. If this protecting group is insoluble in the synthesis-reaction medium, solid-phase synthesis is performed. Briefly, solid-phase synthesis is achieved by attaching the first amino acid to a resin, commonly 1–2% divinylbenzene-cross-linked polystyrene, and the peptide is then elongated to its final product [56]. Numerous other resins can be utilized to immobilize the first amino acid of a peptide block and can be expanded via side chain, backbone, N-terminus, and C-terminus expansion. Advantageously, solid-phase synthesis can be used to create vast libraries of peptides with few steps and has been improved by incorporating multicomponent reactions, developing a final product from various points of synthesis [57]. However, solid-phase synthesis can have some difficulties producing sequences that rapidly aggregate into β-sheets, experience side-chain reactions, contain large portions of glycine-rich residues, or have amino acids with bulky side chains [[56], [57], [58]]. Liquid-phase synthesis was the first approach in synthesizing peptides, but many studies have investigated liquid-phase synthesis as an improvement on solid-phase in order to improve the scalability, reduce financial costs, and reduce amino acid and solvent waste [59]. Liquid-phase peptide synthesis is implemented for high volume industrial production but can achieve higher peptide purity as a trade-off for higher time consumption. However, many one-pot experiments to build liquid-phase synthesized peptides can encounter product and membrane instability as well as difficulty in synthesizing highly hydrophobic peptides [60].
Through methods including “click chemistry,” one-pot self-assembly, or other chemical conjugations, peptides can form robust three-dimensional structures or be complexed to other nanoparticle systems [61]. However, dating back to the 1980s, groups of peptides were collectively termed “difficult sequences” due to their high aggregation and low solubility, primarily due to excessive hydrophobic side chains [62]. The primary structure of the amino acid configuration governs the interactions for self-assembly through hydrogen bonding or π-π stacking and can lend to increased solubility and aggregation issues if the sequences have a high content of hydrophobic residues [61]. To address this, external factors, including environmental pH, temperature, ionic strength, and solvent can be manipulated to influence nanostructure formation and stability [63]. Another approach to improve stability and functionality is through chemical conjugation. For example, a pH-dependent one-pot approach utilizing a chemical linker composed of maleimide and iodoacetyl groups can be reacted with cysteine amino acid residues to facilitate heterodimerization of a peptide system, dramatically increasing peptide stability in solution [64]. Functionality can also be improved through chemical conjugation using “click chemistry.” Nucleic acids, including siRNAs, can be alkynyl modified at their 5’ ends for conjugation to an azido-bearing peptide, causing an increase in nanocomplex yield and purity [65]. Taken together, many strategies have been used to improve peptide stability and avoid aggregation in forming peptide-based or peptide-conjugated nanoparticles that enhance the usability of peptides for drug delivery. Through chemical ligation, solvents, internal peptide modifications, and external factors, including temperature, pressure, and agitation, peptide stability can be improved while reducing aggregation during particle formation [62,63].
2.2. Targeting receptors
Targeting peptides can bind to a variety of cell receptors that are either overexpressed by or unique to cancer cells, many of which are listed in Table 1. Targeting receptors can be sorted into three main groups: integrins, G-protein coupled receptors, and growth factor receptors. Integrins are transmembrane cell receptors that bind to proteins in the extracellular matrix [74]. Integrin αvβ3, which is often overexpressed in ovarian, prostate, and breast cancers, has been a popular integrin for target studies [75]. G-protein coupled receptors (GPCRs) are membrane receptors that transduce stimuli into various cellular activities by activating G proteins when they bind to ligands [76]. Luteinizing hormone-releasing hormone receptor is a GPCR which has been demonstrated to be overexpressed in a range of tumors including prostate, breast, and ovarian tumors [77]. Growth factor receptors (GFRs) are surface receptors that are activated by binding to growth factors and can cause cell proliferation [78]. A common targeting receptor in this category is epidermal growth factor (EGFR), which is found overexpressed on breast, lung, and colon cancer cells and tumors [69,79].
Table 1.
Targeting peptides conjugated to other nanoparticles or used alone to deliver chemo and gene therapeutics.
| Peptide | Target | Delivery System | Cell Type |
|---|---|---|---|
| Luteinizing-Hormone-Releasing Hormone (LHRH) | LHRH Receptor | Complexed with PEG and campothecin [66] | ovarian, breast, and prostate cancer cells |
| MQLPLAT | Fibroblast Growth Factor Receptor | M13 phage displaying MQLPLAT in vivo [67] | gastric cancer cells |
| T7 | Transferrin Receptor | Lipid nanoparticles conjugated with T7 loaded with antisense nucleotides in vitro [68] | lung cancer cells |
| MC11 | Fibroblast Growth Factor Receptor | MC11 was conjugated to branched PEI and PEG encapsulating plasmid DNA [28] | HepG2 |
| GE11 | Epidermal Growth Factor Receptor | Doxorubicin (DOX) loaded liposomes conjugated to GE11 [69] | lung cancer cells |
| KCCYSL | Human Epidermal Growth Factor Receptor 2 | KCCYSL conjugated toTGX-D1 chemotherapy [70] | prostate cancer cells |
| RGD peptide Derivative | αvβ3 integrin | Paclitaxel conjugated to RGD peptide [71] | breast cancer cells |
| AP Peptide | Interleukin-4 receptor | Polymeric micelles loaded with doxorubicin conjugated to AP peptide [72] | breast cancer cells |
| P1c | αvβ3 integrin | Liposomes loaded with doxorubicin conjugated to P1c [73] | glioblastoma cells |
2.3. Targeting peptides conjugated to therapeutics
Once targeting peptides are developed, they can be conjugated to nanoparticles that carry therapeutics or even to the therapeutic itself with the help of linkers [37,41]. Cleavable linkers are commonly used because of their ability to release a drug once it has reached its target site [37]. Cleavable linkers can be separated into two main groups: chemically cleavable or enzyme-cleavable [80]. Chemically cleavable linkers can be cleaved in acidic environments such as the lysosome or endosome since they are designed to be stable only at a pH of 7 [80]. Enzyme-cleavable linkers can be cleaved by proteases or other enzymes relevant to the tumor microenvironment [37,41]. Peptide hydrolysis, acid-base catalysis, or covalent restructuring of amide bonds are methods by which enzymatically cleaved linkers are broken after peptide-mediated delivery of nanocomplexes to the TME or after internalization in cancer cells, in the example of cathepsin-B cleavable linkers [[81], [82], [83]].
Cleavable linkers can act as activators of prodrug conjugates, causing release of activated chemotherapeutics in their target site. Daunorubicin linked to leucine residues responsible for TME accumulation, retention, and linker cleavage, dissociated via aminopeptidase cleavage, was delivered to L1210 leukemia cells, and exhibited significantly higher TME accumulation and anticancer effects compared to free daunorubicin when delivered intravenously to subcutaneous tumor-bearing mice [84]. In another application, the doxorubicin (DOX) prodrug, R8DB, comprised of an octa-arginine peptide complexed to doxorubicin via a glycine-phenylalanine-leucine-glycine linker, enabled high levels of free doxorubicin delivery upon cleavage via cathepsin-B, a lysosomal protease, in ADR-Res drug-resistant ovarian cancer [85]. Most notable, was the ability to include a DOX quencher, allowing for drug tracking prior to release via cathepsin-B cleavage, a phenomenon not seen with free DOX [81,85]. DOX is a popular drug conjugate to evaluate linker efficacy, as chemically cleavable linkers have also been shown to enhance internalization and release of DOX in cancer. Modified human α-fetoprotein receptor-binding peptide containing a conjugate poly-glutamic acid linker bound to DOX increased solubility, tumor selectivity, and pH-dependent disulfide bond degradation between the peptide sequence and DOX conjugate within the lysosome in SKOV3 ovarian cancer and MCF7 breast cancer cells [86].
Many successful studies have included targeted peptide-drug systems conjugated via enzymatically or chemically cleavable linkers. An RGD peptide derivative conjugated to paclitaxel (PTX) via ester bond coupling has proven effective in targeting breast cancer cells with overexpression of αvβ3 integrin and delivering PTX following esterase cleavage in an acidic environment [71]. An in vivo study showed that the tumor uptake of tritium-labeled (3H) particles, 3H-RGD-PTX, in MDA-MB-435 tumors via αvβ3 integrin binding was higher at all time points in comparison to nontargeted 3H-PTX and 3H-PTX + RGD [71]. Tumor to muscle ratio was also higher for 3H-RGD-PTX at each recorded time point, indicating selective tumor targeting over muscle cells. 3H-RGD-PTX treatment also exhibited greater tumor growth inhibition in comparison to the free PTX controls [71]. Human epidermal growth factor receptor 2 (HER2) targeting peptide, KCCYSL, conjugated to TGX-D1, a chemotherapeutic, enhanced cell-specific binding and uptake into LNCaP prostate cancer cells [70]. The targeting peptide-drug conjugate, KCC-TGX, was compared to a cleavable linker-drug conjugate, containing the SSKYQ prostate specific antigen-cleavable peptide (Ac-SSKyQSL-TGX), the TGX-D1 intermediate (NH2-SL-TGX), and free TGX-D1 [70]. Cellular uptake studies showed that KCC-TGX had the highest uptake in LNCaP prostate cancer cells [70]. Furthermore, a competitive binding assay with preincubation of anti-HER2 ligand followed by delivery of KCC-TGX resulted in significantly lower cellular uptake, demonstrating that internalization of the drug was largely due to the HER2 targeting peptide [70]. These overexpressed cell receptors provide a point of entry for targeting peptide-based delivery systems and exhibit greatly improved targeting abilities that are further enhanced using cleavable linkers. With these attributes, drug payloads can be reduced, as off-target delivery becomes less of a barrier in addition to fewer possible systemic side effects.
2.4. Targeting peptides conjugated to nanocarriers
Although research on targeting peptides initially focused on direct conjugation of the peptide to a drug, most recent work focuses on conjugating peptides to nanocarriers such as micelles, polymers, or liposomes with proven abilities to effectively deliver not just monotherapies, but also types of combination therapy to cancer cells [37]. AP peptide (CRKRLDRN) is a targeting peptide that binds to interleukin-4 receptor and has been conjugated to pH-responsive polymeric micelles carrying doxorubicin [72]. DOX-loaded micelle uptake in MDA-MB-231 breast cancer tissue was enhanced at each time point for the micelles conjugated to the targeting peptide when compared to micelles alone when delivered intravenously [72]. As a result, 15 days after micelle-DOX injection, breast cancer tumors in mice injected with DOX-loaded targeting micelles were 18.6% or 57% the weight of the tumors in mice injected with saline or non-targeted micelles, respectively [72]. In another study, P1c peptide (CIRTPKISKPIKFELSG) targeting αvβ3 integrin was conjugated to doxorubicin-loaded liposomes [73]. P1c-conjugated liposomes had a significantly higher mean fluorescence intensity following incubation with αvβ3 positive U87MG glioblastoma cells compared to non-targeted liposomes [73]. A CCK-8 assay confirmed that P1c conjugated liposomes loaded with DOX mediated significantly higher cytotoxicity in U87MG tumors compared to nontargeted DOX-loaded liposomes, while the MCF-7 cell line, an integrin αvβ3-negative cell line, did not exhibit a significant difference in cytotoxicity between the targeted and nontargeted liposomes [73]. U87MG tumors were significantly smaller for mice that were treated with P1c-conjugated liposomes loaded with DOX compared to free DOX, nontargeting DOX-loaded liposomes, or saline [73].
Fibroblast growth factor receptor (FGFRs) has been shown to be overexpressed on cancer cells, and a targeting peptide, MC11, has been identified for receptor-specific binding with fibroblast growth factor [28,67]. A polyplex consisting of eight-armed PEG (EAP), PEG600, and MC11 was constructed and delivered to HepG2 hepatocellular carcinoma cells in vitro and displayed significantly higher luciferase gene transfection compared to the nontargeted polyplex [28]. Similarly, the T7 peptide was developed to specifically target transferrin receptor (TfR) in MCF-7breast cancer cells and was complexed with core-shell nanoparticles to form DSPE-PEG2000-T7 nanocomplexes [24]. DSPE-PEG2000-T7 nanoparticles displayed increased fluorescence in the cytoplasm of MCF-7 cells compared to nontargeted nanoparticles and also showed significantly EGFR protein expression when delivering short-interfering RNA (siRNA) targeting EGFR intravenously into mice bearing MCF-7 mammary tumors compared to nontargeted complex formulations [24].
The development of targeting regions on cancer therapeutics has been advantageous in delivering therapeutics to targeted sites on cancer cells. Targeting peptides are efficient for this goal due to their low molecular weight and abundant versatility, especially when compared to monoclonal antibodies. Use of targeting peptides to enhance current nanoparticle delivery systems or as additional moieties with other peptide systems to deliver cell-specific therapeutics may help to establish peptides as promising candidates for clinical translation.
3. Cell penetrating peptides
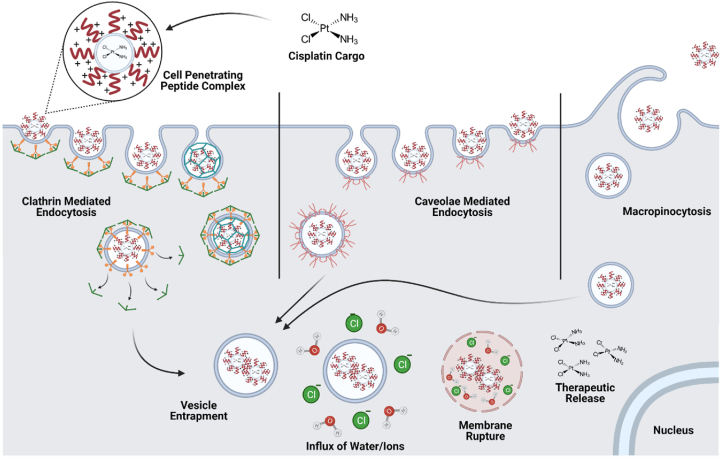
Efficient internalization of cargo at therapeutically effective concentrations is a major barrier to drug delivery. Limitations to nanoparticle-cargo cellular uptake can be attributed to particle size, polarized charge distribution, and net neutrality or negativity that negate cell membrane interactions [87]. Cell penetrating peptides (CPP), a well-studied peptide class, play a major role in increasing cellular uptake of nanoparticle delivery systems and can also influence endosomal escape. Physiochemical properties, including charge density, degree of hydrophobicity, and cell penetrating peptide concentration, create interactions with the cell membrane and activation of cellular internalization. The known methods of cellular internalization using cell penetrating peptide-conjugated nanoparticles and cell penetrating peptide complexes include inverted micelle internalization, direct translocation via membrane pore formation, and endocytotic mechanisms, such as micropinocytosis, clathrin-mediated, and caveolae-mediated endocytosis, illustrated in Fig. 2 [7,88]. Although each of these mechanisms have been demonstrated to play a role in cell penetrating peptide delivery, pinocytosis and clathrin-mediated endocytosis are the primary routes of internalization [89].
Fig. 2.
Cell penetrating peptides can be utilized to deliver siRNA and other cargo through electrostatic interactions with the positively charged amino acid residues. Cell penetrating peptides are primarily internalized via endocytotic mechanisms, including micropinocytosis, clathrin-mediated, and caveolae-mediated endocytosis. Under acidic conditions in the endosome, some cell penetrating peptides are protonated and can induce an influx of water and chloride ions into the endosome, resulting in increased internal pressure and eventual endocytotic rupture and release chemotherapeutics, allowing for subsequent RNAi activity. Created with BioRender.com.
3.1. Characteristics of cell penetrating peptides for internalization and endosome escape
Cell penetrating peptides are commonly less than 40 amino acids long and are often cationic or amphipathic in nature [33,89]. With these size and charge qualities, peptide nanocarriers enable small therapeutic cargo, including siRNAs microRNAs, oligonucleotides, and low-molecular weight chemotherapeutics, to traverse the cell membrane upon electrostatic interaction and/or hydrophobic internal interactions with cell penetrating peptides [90]. Many cell penetrating peptides can be designed from naturally occurring viral proteins that have the ability to efficiently complex with cargos. The first cell penetrating peptide was derived from the human immunodeficiency virus (HIV) [91,92]. The transactivator of transcription (TAT) protein domain is a major player in HIV-gene expression, has been chemically synthesized, and comprehensively characterized to describe its interaction with the cell membrane [91].
Synthetic variants of TAT have led to significant advances in nanoparticle-based delivery systems and have seen substantially increasing uses over years of drug delivery research [13,[90], [91], [92], [93], [94], [95], [96]]. Cell penetrating peptide sequences form specific secondary structures that have high affinity for the cell membrane; however, structures formed depend on total peptide charge and helical properties [33,89,91]. Increased peptide helical content has been shown to increase cellular uptake through aromatic tryptophan inclusions, while higher overall peptide charge can induce rapid oligonucleotide complexation and interaction with the negatively charged cell membrane [33,89,97].
Varying characteristics of cell penetrating peptide designs, including the charge density and amino acid chain length, can affect the method and extent of cellular uptake [98]. Antennapedia, a homeoprotein identified in Drosophila [97,99], R9, a nona-arginine cell penetrating peptide sequence, and TAT were investigated to determine their route of cellular internalization None of the peptides showed evidence of a single, isolated pathway; Macropinocytosis, clathrin-mediated endocytosis, and caveolae-mediated endocytosis were all observed [97]. However, the results revealed that endocytosis of the peptides was concentration dependent, with R9 and TAT requiring micromolar concentrations for rapid endocytosis and distribution [97]. R9 and TAT were also able to directly translocate into the cytoplasm of HeLa cells [97,98]. Another cell penetrating peptide, R8, an octo-arginine sequence, was delivered in CHO cells, and single-molecule spectroscopy was used to examine the system diffusion coefficient and residence time of the peptide within the cellular membrane [100]. Similar to other cell penetrating peptides, R8 exhibited heterogeneous uptake behavior with evidence of active endocytotic mechanisms and the capacity to translocate the cell membrane when delivered after CHO cell treatment with cytochalasin D, an actin filament disruptor [100]. Solid-state nuclear magnetic resonance (NMR) paramagnetic relaxation of penetratin was able to show direct insertion into single- and double-membrane Mn2+ vesicles at depths dependent on the concentration of peptide delivered, driven by cationic-anionic interactions [93,98]. The ability of cell penetrating peptides to use multiple routes of uptake can be advantageous for delivery of therapeutic cargo when concentration of delivered cargos is variable, spatially limited, or cells are receptor-inhibited through competitive binding [93,97,100].
Inclusion of amino acid structure modifications in cell penetrating peptides has been explored for increased surface charge, cargo affinity, and biocompatibility to promote efficient and stable cellular uptake [90,101]. To avoid low synthesis yield, low solubility, aggregation, and toxicity, modifications to the side chain of cell penetrating peptide transportan 10 (TP10), including stearylation, addition of novel amino acids, and site-specific hydrophilic inclusions, were implemented [[102], [103], [104]]. These design variants were complexed with antisense splice-correcting RNA oligonucleotide cargo and delivered in HeLa cells. Each peptide variant enabled higher levels of transfection and increased splice-correction in comparison to unmodified peptides and Lipofectamine controls [[102], [103], [104]]. Additional modifications, including amidation and inclusion of pyroglutamic acid residues at the N-terminus of cell penetrating peptide sequences, increased peptide half-lives and cellular uptake [105].
In addition to promoting cellular internalization, some cell penetrating peptides can also enable release of cargo from the endosome. Because cell penetrating peptides are primarily cationic, their main route of endosomal escape is through the proton-sponge effect [106,107]. The cationic charges associated with cell penetrating peptides and additional pH-responsive residue modifications become reactive to the acidic environment within the endosome and can absorb free protons, increasing the peptide charge [106]. This significant proton concentration causes an influx of extravesicular water and chloride ions, giving rise to a dramatic increase in osmotic pressure, resulting in destabilization of the endosomal membrane and eventual endosomal lysing [108]. Rupture of the endosome releases the internal components, including therapeutic cargo, into the cytosol. However, a drawback of cell penetrating peptides is that they have been known to increase nonspecific delivery, resulting in off-target effects, and increased cytotoxicity compared to other delivery systems due to highly acidic conditions created by a large number of positively charged residues [14,88,[109], [110], [111]].
3.2. Therapeutic applications
Two primary modes of incorporating cell penetrating peptides into therapeutics, including conjugation to nanoparticles or using cell penetrating peptides alone to deliver therapeutics, have been extensively studied since 1988 [112]. Cell penetrating peptides can be conjugated to nanoparticles via short sequences of peptide linkers to create additional functionality on polymeric nanoparticles, lipid nanoparticles, or drug therapies [90,113]. More specifically, conjugation of cell penetrating peptides with other nanoparticle systems can enhance chemotherapeutic delivery. Additionally, cell penetrating peptides alone have been used extensively for delivery of nucleic acids, due to the ease of electrostatic complexation of the cell penetrating peptide with its negatively charged cargo. Several therapeutic applications of cell penetrating peptides as either conjugates to nanoparticles or used alone to directly complex nucleic acids are listed in Table 2.
Table 2.
Cell-penetrating peptides conjugated to other nanoparticles or used alone for delivery of chemo and gene therapeutics.
|
Peptide Conjugates | ||
|
Peptide |
Sequence |
Application |
| TAT | GRKKRRQRRRPQ | TAT conjugated to paclitaxel-loaded poly (dl-lactide-co-glycolide), PLGA nanoparticles were delivered to mesenchymal stem cells in vivo [127] |
| R9 | (d)R9 | R9-conjugation to the cytotoxic caPeptide, was delivered to MDA-MB-436 triple-negative breast cancer cells in vitro and in vivo causing cell death [116] |
| Penetratin |
RQIKIWFQNRRMKWKK |
Penetratin conjugated to Grb7 targeting peptide displayed high levels of biotin and FITC fluorophore internalization in MDA-MB-231 breast cancer cells [128] |
|
Peptide Complexes | ||
|
Peptide |
Sequence |
Application |
| ARF (1-22) | MVRRFLVTLRIRRACGPPRVRV | ARF (1-22) delivered to MDA-MB-231 breast cancer cells displayed a dose-dependent decrease in cell viability [129] |
| 599 | GLFEAIEGFIENGWEGMID-GWYGGGGRRRRRRRRRK | Chimeric peptide-siCIP2A complex delivered to oral squamous cell carcinoma in vitro and in vivo [119,130] |
| NP1 | STR-H16R8 | NP1 complexed with siRNA enhanced cellular uptake in a tumor spheroid model and Bcl2 gene knockdown in human colon cancer cells HCT 116 [94] |
| Azurin-p28 | LSTAADMQGVVTDG MASGLDKDYLKPDD |
P28 exhibited preferential and temperature dependent entry into A549 lung cancer, DU145 prostate cancer, MCF-7 breast cancer, HCT116 colon cancer, HT1080 fibrosarcoma, and SKOV3 ovarian cancer cells, shown via P28 Alexa Fluor 568-labeling, compared to corresponding healthy CCD-13Lu lung, CRL11611 prostate, MCF-10A breast, CCD33Co colon, and HOSE6-3 ovarian cells [131] |
| PepFect-1 | KETWWETWWTEWSQPKKKRKV | PepFect-1 enhanced intracellular delivery of complexed proteins in human fibroblasts [[132], [133], [134]] |
| PepFect-2 | KETWFETWFTEWSQPKKKRKV | PepFect-2, and its derivatives, PepFect-20 through PepFect-47, enhanced uptake of HypNA-pPNA in HeLa cervical cancer cells [132,135] |
| PepFect-3 | KETWFETWFTEWSQPKKKRKV | PEGylated PepFect-3 effectively delivered DNA mimics to PC3 prostate adenocarcinoma cells in vivo [132,135] |
| PepFect-14 | AGYLLGKLLOOLAAAALOOLL | PepFect-14 complexed to splice-correcting oligonucleotides and ciproxifan modulated cell signaling pathways in HeLa cells [136] |
| Transportan 10 | AGYLLGKINLKALAALAKKIL | Transportan 10 improved the delivery and anticancer effects of cisplatin in HeLa cervical cancer and OS143B osteosarcoma cells [137] |
TAT has been used extensively in drug delivery systems as a conjugated moiety to nanoparticles including lipid, metallic, and microbubble-based therapeutic carriers [13,90,[92], [93], [94], [95], [96]]. TAT conjugated to lipid-based nanobubbles encapsulating siRNA was delivered to MDA-MB-231 triple negative breast cancer cells in vitro and in vivo [95]. Significantly enhanced internalization of siRNA was observed in cells treated with the cell penetrating peptide-complexed nanobubbles in comparison to cells treated with siRNA alone or nanobubbles without the cell penetrating peptide [95]. In another study, the selective cell penetrating peptide (SCPP) was conjugated to a polymersome (PS) nanoparticle encapsulating methotrexate disodium (MTX) [114]. The SCPP-PS-MTX nanoparticle was delivered to A549 human lung cancer cells in vivo and exhibited rapid penetration into cells and efficient cargo release in comparison to PS-MTX and free MTX [114]. Additionally, Gao et al. demonstrated that the arginine-leucine-tryptophan (RLW) cell penetrating peptide conjugated to poly(ethyleneglycol)-poly(ε-caprolactone) nanoparticles increased A549 lung cancer tumor spheroid penetration and delivery of docetaxel compared to octo-arginine-conjugated nanoparticles and docetaxel alone [115].
In addition to conjugation of cell penetrating peptides to other nanoparticle formulations, cell penetrating peptides can also be used as stand-alone delivery systems for enhanced nucleic acid delivery. Poly-arginine (pR) cell penetrating peptide sequences are extensively used in peptide delivery systems as complementary sequences for efficient cargo delivery and complexation with negatively charged nucleic acids [109,116,117]. NP1, a stearylated modification of a pR cell penetrating peptide, was directly complexed with siRNA and exhibited significantly higher levels of siRNA uptake in 2- and 3-dimensional models of HCT 116 human colon cancer cells in comparison to Lipofectamine-2000 [94]. Furthermore, NP1 complexed with Bcl2-siRNA (siBcl2) enhanced knockdown of Bcl-2 protein compared to delivery of siBcl2 alone and siBcl2 delivered via Lipofectamine 2000 [94]. Modifications to pR similar to NP1, in addition to the formulation of peptide-siRNA nanoparticles and cell penetrating peptide-conjugated nanoparticles, have exhibited increased levels of cellular internalization and subsequent gene knockdown when delivering siRNA to oral squamous cell carcinoma, breast cancer cells, and neuronal cells [[118], [119], [120]]. Lyp-1, a tumor-penetrating peptide sequence, complexed in tandem with numerous cell penetrating peptides, including pR variants, significantly increased cellular uptake of the delivery system and GFP silencing in HeLa cells compared to a lipofectamine control [117]. This study also demonstrated that pR cell penetrating peptide-mediated delivery was more effective in silencing GFP than both TAT and penetratin-complexed tandem systems.
Although cell penetrating peptides can enhance cellular uptake and therapeutic activity, their cytotoxicity has caused significant limitations in the clinical translatability of cell penetrating peptide systems both alone and when conjugated to nanoparticles [121]. Thus, it is important to evaluate and minimize cytotoxicity of cell penetrating peptides to avoid viability loss in off-target and healthy cells. Cationic cell penetrating peptides have been shown to elicit significantly less cytotoxicity in vitro in comparison to amphipathic cell penetrating peptides [[122], [123], [124]]. Transportan, an amphiphilic cell penetrating peptide, was shown to induce oxidative stress through a metabolic panel analysis in comparison to cationic cell penetrating peptides R9, MAP, and pTAT [125]. Additionally, the cargo attachment site has been shown to affect the toxicity profile. TP10 exhibited significant proliferation reduction in HeLa and CHO cells compared to penetratin and TAT; however, with orthogonal cargo complexation, a significant reduction in long-term toxicity was observed compared to N-terminal loading [126]. Selection and design of future cell penetrating peptides should include these considerations to yield low toxicity and thus improve the clinical translatability of these systems.
4. Fusogenic peptides
Another major barrier to nanocarrier-mediated delivery of therapeutics is their lack of endosomal escape, which causes degradation of both the delivery system and its therapeutic cargo. When a molecule is unable to readily pass through the cell membrane via diffusion, endocytosis allows uptake of the molecule into the cell. Through endocytosis, the cell internalizes a molecule by vesicle formation, and the molecule is sorted into the endosome, where it is recycled or trafficked to lysosomes for degradation [138]. Once in the endosome, molecules often lack the ability to escape into the cytoplasm where they can perform their desired function. Use of fusogenic peptides in delivery systems combats this issue by allowing cargo to escape from the endosome. Fusogenic peptides release their cargo through the fusion process. This process begins with the membrane and peptide in close proximity, followed by formation and expansion of an aqueous pore for a completed fusion reaction [139]. Within this process, the transition states typically consist of non-bilayer phospholipids and curved monolayers, both of which are unfavorable for membrane interaction due to electrostatic and steric barriers [140]. Therefore, fusogenic peptides fulfill the need to facilitate endosomal escape through a variety of mechanisms.
4.1. Characteristics of fusogenic peptides and mechanisms of membrane disruption
Though there are many fusogenic peptides, all share common characteristics that enable their functional ability for endosomal escape. A fusogenic sequence is typically 13–20 residues in length and is critical for the fusion process to be anchored into the membrane [141]. The fusion sequence is typically hydrophobic in nature and located at the N-terminus of the peptide sequence [142,143]. Typically, a mutation within this segment leads to a complete loss of fusogenic function [143], but specific residue additions at this location have been shown to increase fusogenicity [144]. The primary structure of the fusogenic peptide sequence is amphiphilic in nature, enabling the peptide to maintain a high affinity for lipid bilayers. Additionally, when forming amphiphilic helices, these sequences can create a hydrophobicity gradient, which plays a role in the peptide's endosomal escape ability through accelerating fusion [145].
Fusogenic peptides are pH dependent, membrane disrupting peptides. At a physiological pH, fusogenic peptides are inactive, but become protonated and undergo activation in an acidic environment, such as an endosome [146]. At an acidic pH, most fusogenic peptides undergo a conformational change to an α-helix secondary structure to fuse with and disrupt the endosomal membrane to allow for endosomal escape [143,147]. The “spring-loaded” nature of the conformational change of peptides usually happens in two major steps [147]. First, fusion peptides are released from the native complex to refold into long helical bundles, thus projecting the N-terminus toward the target membrane [147]. Next, the peptides form a six-helix bundle, composed of N-terminal helices and antiparallel C-terminal helices [147]. This typical post-fusion structure forces the peptide and membrane into close proximity and anchors the peptide into the membrane through the fusion peptide and transmembrane area [148]. These secondary structures are required for endosomal escape because the insertion of amide groups into membranes is unfavorable unless the amide groups are hydrogen bonded through protonation [149].
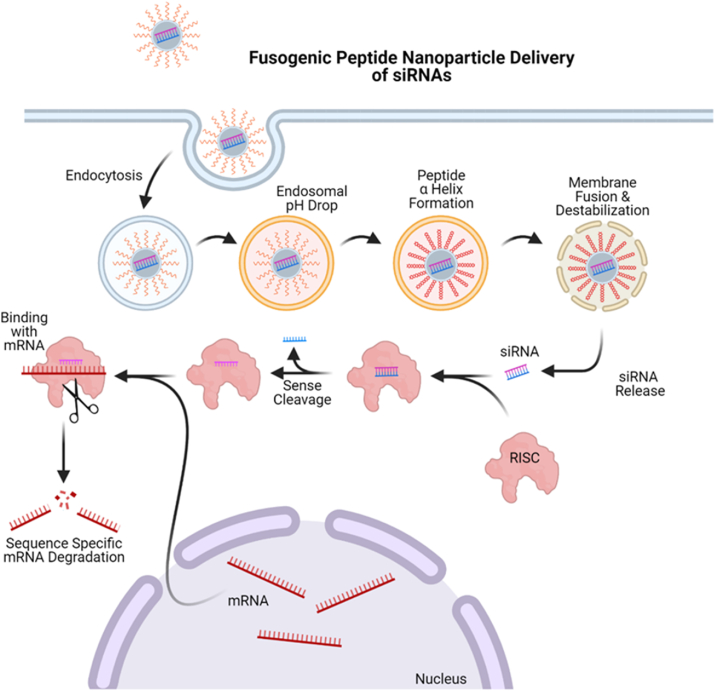
After fusogenic peptides initiate their secondary structure at an acidic pH, they utilize a variety of techniques to facilitate endosomal escape. Fusogenic peptides can cause curvature modulation through lowering of the bilayer hexagonal phase transition temperature to promote negative curvature [141]. This increase in negative curvature of contacting monolayers can form an intermediate membrane state in the process of fusion [141]. Another method is to disrupt the membrane through peptide helix insertion into the target endosomal membrane during the fusion process to perturb the regular packing of lipids [142]. The hydrophobic characteristics of a fusogenic sequence allow for membrane disruption in two ways. For two bilayers to merge, each monolayer must rupture and reform; hydrophobic peptides have the ability to lower membrane rupture tension, which is important for acceleration of fusion [145]. Hydrophobic peptides also form a hydrophobic gradient along the helical axis; the gradient determines the angle of insertion. Attenuated total reflectance-Fourier transform infrared experiments determined that the peptide inserts at an oblique angle to properly disrupt the target membrane [150]. Finally, through the energy associated with their conformational change, fusogenic peptides have the energy to drive the fusion process by lowering the activation energy for the unfavorable intermediates within the process [150]. Once the fusion process is complete, the membrane of the endosome has been disturbed, and the contents are released into the cytosol. Fig. 3 illustrates the process of a fusogenic peptide nanoparticle inducing endocytosis of siRNA cargo into a cell and causing endosomal escape, releasing the therapeutic cargo into the cytosol to achieve RNA interference (RNAi). Without this endosomal release, trapped siRNA cargo will be trafficked for lysosomal degradation, rendering the therapeutic ineffective. Upon release into the cell cytosol via fusogenic peptide mechanisms, siRNA can bind with the RNA-induced silencing complex and be trafficked for endonuclease argonaut-2 degradation of targeted mRNA responsible for oncogenesis.
Fig. 3.
Depiction of fusogenic peptide-based nanoparticles delivering bioactive siRNAs through pH-sensitive endosomal escape. Taken up through endocytosis, fusogenic peptides undergo a conformational change to adopt a helical secondary structure under acidic conditions within the endosome, resulting in fusion and disruption of the membrane to allow the release of complexed cargo into the cytosol for subsequent incorporation with RNAi machinery. Adapted from “siRNA Nanoparticle Delivery System,” by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates.
4.2. Subtypes of fusogenic peptides
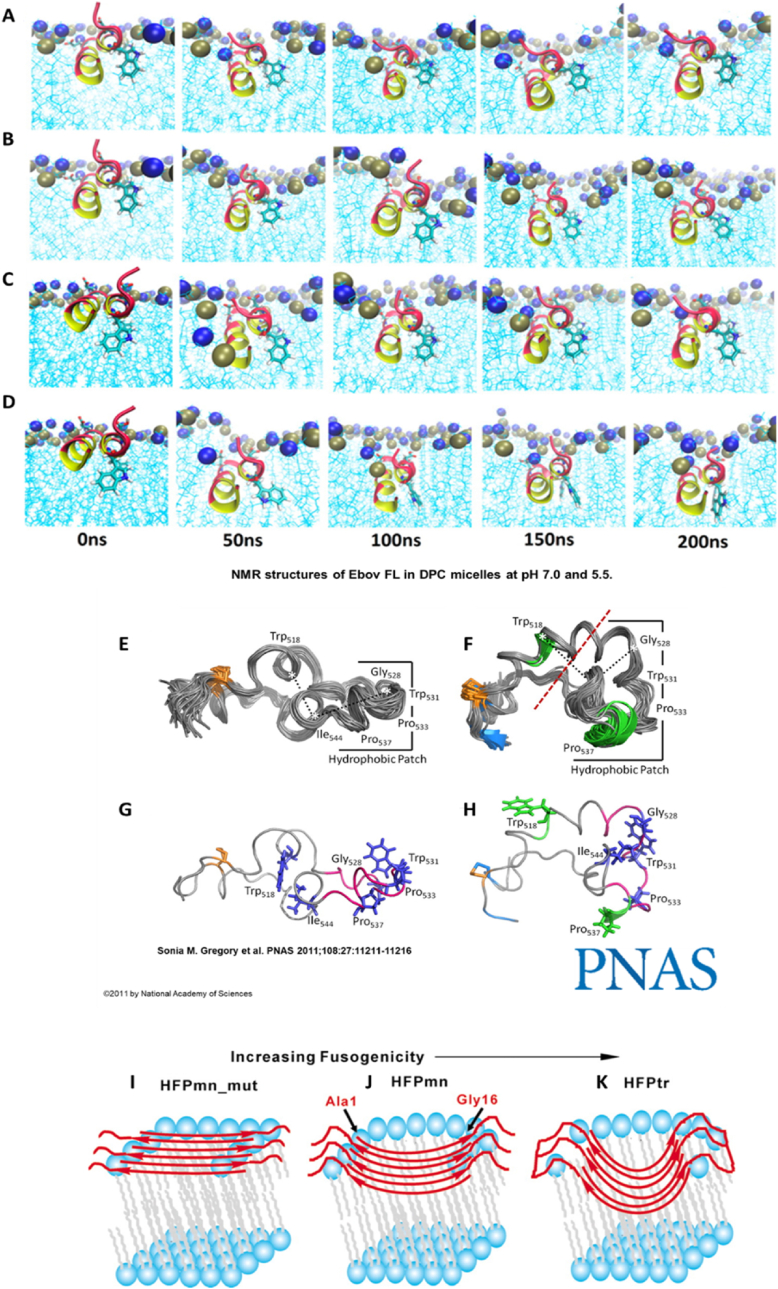
Three subtypes of fusogenic peptides exist due to variations in the secondary structure formed at acidic pH, including α-helical, β-sheet, internal fusogenic peptides, which are illustrated in Fig. 3. The conformational structure of a peptide can be evaluated by using circular dichroism, X-ray, or nuclear magnetic resonance. Most fusogenic peptides form an α-helix structure to promote endosomal escape at acidic pH, and is shown in molecular simulations in Fig. 4A–D, where the Ebola viral fusion sequence was inserted into to a dodecylphosphocholine (DPC) micelle as a model membrane. The most studied fusion protein is the glycoprotein hemagglutinin (HA) of the influenza virus. HA2, a synthetic peptide derived from HA, changes conformation to form an α-helix at the endosome's acidic pH and inserts itself into the membrane [6]. HA2 has functioned as an endolytic moiety on cell penetrating peptide and targeting peptide systems to deliver proteins into cancer cells [151,152]. Liou et al. demonstrated enhanced delivery and cytoplasmic localization of red fluorescent proteins (RFP) into A549 adenocarcinoma alveolar epithelial cells via HA2-functionalized cell penetrating peptide in comparison to delivery with nuclear localization signal-fused RFP [153]. Furthermore, HA2 conjugated to the TAT cell penetrating peptide exhibited higher cellular uptake and endosomal disruption capabilities in HeLa, HEK, 10T1/2, and HepG2 cells compared to cell penetrating peptide formulations of Transportan, octo-arginine (R8), penetratin, and TAT alone [154]. The polypeptide structure targeting the CXC chemokine receptor 4, T22-green fluorescent protein (GFP)-H6, with internal HA2 conjugation to form a tandem T22-GFP-HA2-H6 system, demonstrated the ability of the fusogenic HA2 to enhance endosomal lysis and nuclear accumulation of GFP into HeLa cells; however, these effects were concomitant with reduced targeting efficacy [155,156]. Though enhanced cell uptake and endosomal breakdown was achieved, the importance of peptide design considerations, specifically when conjugated in tandem, should not be overlooked as, interactions between the peptide regions can cause reduced efficacy of each peptide's specific function.
Fig. 4.
Fusogenic peptides can undergo three different conformational changes to cause endosomal membrane interactions. α-helices (A–D) [166] are the main structure formation upon protonation in an acidic environment and can exhibit time and pH-dependent fusion depth, as demonstrated through molecular simulations with dodecylphosphocholine (DPC) micelles. Internal loop (E–H) [164,165] and β-sheet (I–K) [167] structures have also been noted as major players in fusogenicity, dependent on the internal amino acid sequence, and have been described in viruses including Ebola, sarcoma, and the gp41 fusion domain of HIV (HFP). Panels A–D used with permission from Brice and Lazaridis [166], panels E–H with permission from Gregory et al. [165], and panels I–K with permission from Sackett and Shai [167]. Permission related to panel A–D should be directed to ACS and can be accessed at https://pubs.acs.org/doi/10.1021/jp409412g.
Though α-helices are the primary known structure of fusogenic peptides, they can also form β-sheets upon conformational change for membrane insertion through sequence editing or truncation, which has been observed using lipid membrane interactions, Fourier-transform infrared analysis, and solid-state nuclear magnetic resonance spectroscopy [[157], [158], [159], [160]]. Shown in the gp41 fusion domain of HIV (HFP), interactions of amphipathic repeats between individual fusogenic peptides can adopt an anti-parallel structure, illustrated in Fig. 4I–K, specifically when containing glycine residues, causing whole-sheet insertion of the fusogenic peptide supramolecular structure into the endosomal membrane [[158], [159], [160], [161]]. A third subtype of fusogenic peptides are known as internal fusogenic peptides, and do not form singular secondary structures [162,163]. Ebola and the sarcoma virus peptide are examples of internal fusogenic peptides containing subunits that respond to pH decrease through oligomerization, causing membrane fusion with evidence of α-helix structures, β-sheet conformations, and internal loop structures confirmed through nuclear magnetic resonance imaging, circular dichroism, and lipid mixing resonance [[162], [163], [164], [165]].
4.3. Therapeutic Application
Fusogenic peptide systems have shown great success in delivering gene and chemotherapies both in vitro and in vivo. Shown in Table 3, most fusogenic peptides have been implemented as conjugated moieties to other peptide or nanoparticle formulations. The exact number of fusogenic peptides that have been discovered is not entirely known. Derivations of natural viral fusion sequences, including the dimeric form of the influenza-derived fusogenic peptide INF-7 (diINF-7) and HA2, have almost endless permutations to evaluate optimum amphipathic qualities and endosomal membrane fusion ability. With each new fusogenic peptide evaluated, the window for clinical therapeutic applications grows wider through delivery of chemotherapies, immunotherapies, and gene therapies (see Table 4).
Table 3.
Fusogenic peptides conjugated to other nanoparticles or used alone for therapeutic delivery.
| Peptide | Sequence | Application |
|---|---|---|
| GALA | WEAALAEALAEALAEHLAEALAEALEALAA | GALA, complexed to a hepatitis B surface antigen bionanocapsule, delivered calcein to SKBR3 breast and HeLa cervical carcinoma cells [168] |
| KALA | WEAKLAKALAKALAKHLAKALAKALKACEA | A polyelectrolyte complex micelle displaying KALA delivered siRNA in breast cancer cells [169] |
| 599 | GLFEAIEGFIENGWEGMIDGWYGGGGRRRRRRRRRK | The 599 peptide enhanced siCIP2A bioactivity in CAL27 oral squamous cell carcinomas in vitro and in vivo [119,130] |
| HA2 | GDIMGEWGNEIFGAIAGFLG | HA2 complexed with TAT protein transduction domain enhanced internalization and macropinosome escape of enhanced green fluorescent protein gene in 3T3 fibroblasts, COS7 kidney cells, and CHO–K ovarian cells [170] |
| INF7 | GLFEAIEGFIENGWEGMIDGWYGC | INF7 enhances endosomal membrane interaction capabilities when exposed to the endosomal membrane following liposomal degradation shown through rhodamine signal in CV1 kidney cells [171] |
| H5WYG | GLFHAIAHFIHGGWHGLIHGWYG | H5WYG conjugated to polyethylene glycol (PEG)-tetraacrylate (PEG-TA), caused pH-dependent endosomal membrane interaction and release of antisense oligonucleotides in A549 non-small lung carcinoma and HeLa cervical carcinoma cells [172] |
| EALA | AALAEALAEALAEALAEALAEALAAAAGGC | EALA conjugated to a folate-PEG-PE liposome caused folate receptor-mediated endocytosis and endosomal escape of propidium iodide in KB cervical cancer cells [173] |
| SFP | FEAALAEALAEALA | Novel synthetic fusogenic peptide analyzed via circular dichroism, hemolysis, and lipid mixing enhanced membrane fusion and enhanced resonance energy transfer of 7-nitrobenz-2-oxa-l,3-diazol-4-yl and rhodamine fluorescence [174] |
| ccX31 | Undisclosed | ccX31 coiled-coil trimer enhances mixing with liposome bilayer for membrane destabilization and leakage of calcein out of the destabilized liposome [174] |
Table 4.
Peptides in clinical trials and their therapeutic mechanisms.
| Peptide | Therapeutic System | Disease Treated | Mechanism of Action |
|---|---|---|---|
| DPV1047 | SN38 conjugated to positively charged cell penetrating peptide to form DTS-108 | Colorectal Cancer | SN38 released upon cleavage of ester bond within blood via plasma esterases [195] |
| XG-102 | TAT (cell penetrating peptide) conjugated to dextrogyre peptide | Uveitis/ocular inflammation | Inhibition of the JNK pathway [[214], [215], [216]] |
| KAI-1678 | Synthetic cell penetrating peptide composed an inhibitor of epsilon PKC and carrier moiety | Postherpetic neuralgia | Inhibition of epsilon protein kinase C (εPKC) and isozyme-specific receptor for active C kinase (RACK) [217] |
| P28GST | Sole protein derived from schistosome helminth | Crohn's disease | Downregulating Th1/Th17 immune response to reduce intestinal inflammation [184] |
| p28 | cell penetrating peptide fragment of cupreodexin azurin | Solid tumors | Inhibits proteasomal degradation via HDM2-independent pathway [218] |
| UV1 | Three epitope peptides corresponding to reverse transcriptase subunit of telomerase | Metastatic hormone-naïve prostate cancer | Vaccine to induce immune response towards achieving tumor eradication via targeting of hTERT [204,208] |
| KIF20A-66 | HLA-A24-restricted epitope peptide | Pancreatic cancer | Vaccine targeting tumor-associated antigen kinesin family member 20A (KIF20A) [219] |
| NA-1 (Tat-NR2B9c) | cell penetrating peptide | Ischemic infarction | Disrupts protein–protein interactions of PSD-95, a postsynaptic scaffolding protein [220] |
One example of a gene therapy application for fusogenic peptides is in delivery of small interfering RNA (siRNA) to induce RNAi. Due to the large size and negative charge of siRNA, it lacks the ability to readily interact with the cell membrane for uptake. Fusogenic peptides can protect siRNA from degradation, facilitate cell membrane interactions, promote cellular uptake, and release siRNA into the cytosol [141]. This application is especially promising for cancer treatment to mediate cytosolic delivery of bioactive siRNAs for targeting and silencing oncogenes. In one study diINF-7 was evaluated for delivery of EGFR siRNA into human epidermoid carcinoma cells [175]. Electrostatic complexation of the diINF-7 peptide to EGFR-siRNA particles resulted in increased knockdown of EGFR in comparison to siRNA delivered alone, confirming the functionality of the fusogenic peptide [176].
The GALA peptide, a synthetic derivation of the amphipathic influenza virus peptide, HA2, mimics the fusogenic properties of the virus-derived peptide through pH-responsive conformational change, allowing insertion of the peptide into the endosomal membrane [177]. GALA was complexed to a bionanocapsule consisting of a hepatitis B surface antigen and a lipid bilayer. The complex was delivered to HER2-positive SKBR3 human breast carcinoma cells and HER2-negative HeLa human cervical carcinoma cells containing the green, fluorescent cargo calcein [168]. Cells treated with the GALA-functionalized nanoparticle displayed enhanced cytoplasmic localization of green fluorescence in comparison to non-GALA nanoparticles, indicating release of the fluorescent cargo from the endosomes into the cytosol. The non-GALA nanoparticles displayed colocalized green nanoparticle signal and Lysotracker® Red DND-99-stained endosomes, demonstrating endosomal entrapment [178].
Another application of fusogenic peptides is conjugation to other nanoparticles to improve efficacy of delivery systems. For example, shGALA, a shortened form of the GALA peptide, was conjugated to the liposomal delivery system polyethylene glycol-multifunctional envelope-type nano device (PEG-MEND) designed for siRNA delivery [179]. Cellular uptake studies comparing delivery of PEG-MEND to shGALA-conjugated PEG-MEND (shGALA-MEND) demonstrated that uptake efficiency of both liposomal systems into HT1080 fibrosarcoma cells was approximately 100% [179]. However, further studies revealed that only the shGALA-MEND delivery system allowed release of siRNA from the endosome into the cytosol [180]. Endosomal escape of siRNA was confirmed through RT-qPCR, revealing increased gene silencing mediated by the shGALA-MEND liposomes compared to PEG-MEND [177].
Fusogenic peptides could be highly beneficial for therapeutic delivery systems due to the peptides’ versatility in design and application. Encapsulation or conjugation of fusogenic peptides to other nanoparticle systems have consistently demonstrated enhanced cellular uptake efficacy and endolytic activity, with few drawbacks. The amphipathic nature of fusogenic peptides reduce cytotoxicity, enhance membrane affinity, and can assist in therapeutic loading [181]. One potential drawback of fusogenic peptides is that although they increase efficient release of therapeutics into the cell, they can also reduce cell targetability and cell uptake prior to endosomal encapsulation when delivered at lower doses [155].
5. Clinical implications and uses
While many preclinical studies have demonstrated the utility of peptides as a component of nanoparticle delivery systems, in most clinical applications, peptides are used as the therapeutic modality alone, as epitopes in vaccines, or as nanoparticle-therapeutic conjugates. Initial forms of therapeutics to involve peptides, with insulin being the most notable, were introduced in the 1920s [182]. Insulin was then followed by other common drugs, such as oxytocin, vasopressin, and calcitonin throughout the mid to late 20th century. As the genomic era emerged, more drug targets surfaced because of strategized therapeutic targeting. These strategies have led to recent developments in peptide therapeutics and can be applied to the peptide-conjugated nanoparticle strategies. More recently, a glutathione S-transferase recombinant peptide, P28GST, has been shown to reduce immune responses [183]. As a result, it was hypothesized that P28GST would reduce severity of intestinal inflammation in irritable bowel disease (IBD) patients. A phase 2a clinical study was completed in 2018 and showed promise after displaying decreased Crohn's disease activity index scores and blood calprotectin [184]. Ultimately, P28GST proved to be a safe therapeutic option. Other peptide therapeutics have also shown promise in recent developments [185]. While these therapies have highlighted the capabilities of peptides as therapies, they do not highlight the ability of peptides to serve as delivery systems in clinical applications.
The most commonly used drug delivery systems in clinical applications include polymers and liposomes [186,187]. Comparing the use of these systems to peptide applications, a significant gap exists in preclinical and clinical uses. Potential causes for the lack of clinical use may include the high cost of peptide research endeavors compared to other small molecules and suffering a long-time gap between research studies to mass production/commercialization [188]. The latter can be mostly attributed to difficulty in the upscaling of peptides due to rigor in the production process, such as peptides folding upon themselves during solid phase peptide synthesis [189,190]. When using peptides as or a part of a therapeutic, the likelihood of approval is 8% [191]. For peptide therapeutics approved from 2010 to 2017, the median development time was 9.4 years [192], contrasting with other molecule types that had a median of 8.1 years [192]. Increased development time may deter research in not only peptide therapeutics but also peptide nanocarriers. However, with a compound annual growth rate of 7.9% in global peptide therapeutics by 2027, there is promise for upcoming peptide-based research [193]. In addition, the proportion of conjugated peptides entering clinical development has increased over time, consisting of 30% of all peptides entering clinical trials since 2010 [194]. To continue the improvements of peptide inclusion in clinical work, extensive funding and research is necessary to translate peptide delivery systems from the bench to the bedside.
5.1. Peptide-prodrug conjugate
Pro-drugs using a peptide delivery system have been developed to circumvent the limitations in drug metabolism, such as activation and elimination [195]. Irinotecan is a pro-drug chemotherapeutic effective against colorectal cancer used in combination with 5-fluorouracil (5-FU) as of early 2000 [195,196]. It is limited by hepatic activation, resulting in variable effects from patient-to-patient [195,197]. To overcome this limitation, peptide-drug conjugate DTS-108 was developed, consisting of a chemotherapy drug, SN38, and an oligopeptide, DPV1047 [197]. DPV1047 is part of a family of cell-penetrating peptides named Vectocell® peptides (also referred to as Diatos peptide vectors, DPVs) and is derived from anti-DNA antibodies [198]. This cell penetrating peptide consists of 19 amino acids, with 35% representing basic amino acids, and is internalized independent of caveolar pathways [198]. Due to its insolubility, SN38 was linked to an oligopeptide, DPV1047, to form a water-soluble conjugate peptide: DTS-108 [195,196,199]. DTS-108 is cleaved by esterases in the blood via esterase-cleavable linkers on the peptide, resulting in non-hepatic release of SN38 [200]. This mechanism proves to be an advantage of peptides conjugates, given the efficacy of irinotecan is limited by hepatic activation. Preclinical studies showed that the release of SN38, antitumoral efficacy, and cytotoxicity of DTS-108 outperformed irinotecan [195]. Notably, systemic exposure of SN38 in the gut was decreased using the DPV1047 peptide, which could significantly decrease adverse side effects, as life threatening diarrhea is observed in some irinotecan-treated patients as a result of accumulation of SN38 from the pro-drug via bile excretion [195,201]. Ultimately, DTS-108 allowed for delivery of higher doses of SN38 without increased gastrointestinal toxicity. These characteristics were attributed to the SN38 oligopeptide, which served as the vector for efficient SN38 delivery in this model.
Higher levels of blood circulating SN38 observed with the use of the DTS-108 in comparison to irinotecan led to the first in-human clinical trial completed in 2016 [195,200]. This study revealed SN38 drug concentration levels in DTS-108 four times higher than irinotecan at similar dosage amounts [200]. While diarrhea and other gastrointestinal problems were observed in patients, they were less problematic and manageable with patients subjected to DTS-108 treatment in comparison to those historically treated with irinotecan [200]. The clinical trial investigators attributed this to the location of cleavage within the body (blood for DTS-108 and liver for irinotecan). Although tumor responses were not observed, disease stability was sustained for six or more cycles in several patients [200]. As a whole, the clinical study reinforced the observations seen in the preclinical models, including reduced toxic side-effects and overcoming pharmacokinetic barriers [195,200]. Moreover, this trial highlights the possibilities of peptides in increasing therapeutic effectiveness by circumventing the barriers imposed by natural human mechanisms.
5.2. Peptide epitopes
Peptides are most commonly used clinically as epitopes in subunit vaccines [202]. Subunit vaccines present antigens to the immune system using any molecule associated with the pathogen; in this case – peptides. Peptide vaccines are usually synthetic and serve as antigenic epitopes that will induce lymphocytic responses. These qualities allow peptide-vaccines to be highly versatile. However, they must be engineered with close attention to epitope structure and immunodominance to render an effective immune response. Successful peptide vaccines mimic the structures of naturally occurring antigens such as pathogens, which tend to cross-link B-cell receptors to promote antibody affinity maturation for increased immunogenicity [202]. Others may yield a T-cell by binding to T-cell receptors of class I or II major histocompatibility complexes of antigen presenting cells [202]. However, these require a free N-terminal amine group and an epitope peptide backbone that conforms to the class I or II major histocompatibility complex for successful recognition [202]. This alludes to the versatility in peptide-epitope structure engineering, which must be within fine detail to yield an effective immune response.
UV1 is a vaccine peptide designed based on epitope spreading, the formation of an immune response to epitopes that are distinct from those that cause disease, to target telomerase upregulated tumors [203]. Epitope spreading is common in vaccine development, leading to the diversification of the immune system to recognize multiple targets on a pathogen [203,204]. With the ability to modify peptides for targeting of certain antigens, epitope spreading can be used as an advantage in vaccine development. UV1 is composed of three telomerase reverse transcriptase (hTERT) peptides, known as GV1001. Previous work established that hTERT peptides elicit CD4+ T-cell reactivity [204]. hTERT, the catalytic subunit of telomerase, is an attractive antigen target since telomerase is often upregulated and contributes to proliferation of human tumors and cancer progression [[205], [206], [207]]. Upon vaccination with a peptide corresponding to hTERT, an immune response would be generated to eradicate tumors with upregulated telomerase activity. A phase I clinical trial using UV1 and granulocyte macrophage colony stimulating factor (GM-CSF) in metastatic hormone-naïve prostate cancer (mPC) demonstrated that 18 of 21 patients exhibited an immune response to the peptide [208]. When used as androgen deprivation therapy, 13 of 21 patients who received >10 UV1 vaccinations had decreased prostate-specific antigen (PSA) levels and 10 patients no longer had persisting tumors in the prostate gland [208]. However, the efficacy of the vaccine must be studied further, given moderate adverse side effects were observed in patients that could not be attributed specifically to UV1 or GM-CSF. Several other clinical trials have been conducted using an hTERT peptide and other adjuvants [[209], [210], [211], [212]]. More recently, the GV1001 peptide has been explored as a heat shock protein-mediated cell penetrating peptide [213]. These clinical studies highlight the versatility of peptides in bridging the gap between research and human application.
6. Conclusion
Peptide-based delivery systems have performed successfully in in vitro and in vivo studies, but few are translated to clinical studies, a gap that must be addressed further. In the vast majority of peptide-based therapies, peptides are used as the therapeutic system rather than as the delivery system. The success of peptide therapeutics in early clinical trials only helps to support the ongoing studies of peptide biocompatibility and efficacy when used as drug delivery methods. Nevertheless, peptide systems are largely investigatory and come with advantages and disadvantages that should be considered in weighing the applicability of their use.
Peptide systems can be advantageously selected and tailored for specific uses causing endosomal escape of therapeutic cargo, efficient cellular uptake, and cancer-specific cell targeting. Whether being delivered as a nanoparticle complex independently or as a conjugated moiety to other particle systems, peptide inclusions have shown tremendous promise in enhancing cancer therapeutic efficacy. When designed with proper considerations, peptide systems can effectively avoid the many threats to vehicle and therapeutic destruction prior to arrival at the desired therapeutic site. Though numerous advantages to using peptides as a delivery system have been discussed, there are disadvantages that must be addressed. Without the following considerations, utilization of some peptide complexes can hinder the capabilities of delivery systems and cause negative effects including peptidase degradation and local and systemic toxicity. While a large degree of peptide destruction can occur prior to arrival at a target cell via protease and peptidase activity, peptide systems can also be subject to degradation intracellularly without efficient endosomal escape and subsequent trafficking to the highly acidic lysosome. Still, the use of fusogenic, cell-penetrating, and targeting peptide systems in cancer drug delivery has shown great success in academic and preclinical studies, and the continued understanding of cancer biology, tumorigenesis, and the tumor microenvironment will continue to improve these methods of delivery.
Funding
This work was supported by the National Science Foundation EPSCoR program under NSF Award #OIA-1655740 and the National Science Foundation Faculty Early Career Development Program under NSF Award #204669. This work is also supported in-part by the Hollings Cancer Center Lowvelo Doctoral Fellowship Program. Any Opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect those of the National Science Foundation or the Hollings Cancer Center.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.He H., Sun L., Ye J., Liu E., Chen S., Liang Q., Shin M.C., Yang V.C. Enzyme-triggered, cell penetrating peptide-mediated delivery of anti-tumor agents. J. Contr. Release. 2016;240:67–76. doi: 10.1016/j.jconrel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 3.Zugazagoitia J., Guedes C., Ponce S., Ferrer I., Molina-Pinelo S., Paz-Ares L. Current challenges in cancer treatment. Clin. Therapeut. 2016;38:1551–1566. doi: 10.1016/j.clinthera.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Dissanayake S., Denny W.A., Gamage S., Sarojini V. Recent developments in anticancer drug delivery using cell penetrating and tumor targeting peptides. J. Contr. Release. 2017;250:62–76. doi: 10.1016/j.jconrel.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Wan Y., Dai W., Nevagi R.J., Toth I., Moyle P.M. Multifunctional peptide-lipid nanocomplexes for efficient targeted delivery of DNA and siRNA into breast cancer cells. Acta Biomater. 2017;59:257–268. doi: 10.1016/j.actbio.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 6.Tai W., Gao X. Functional peptides for siRNA delivery. Adv. Drug Deliv. Rev. 2017;110–111:157–168. doi: 10.1016/j.addr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings J.C., Zhang H., Jakymiw A. Peptide Carriers to the Rescue: Overcoming the Barriers to siRNA Delivery for Cancer Treatment. Transl. Res. 2019:92–104. doi: 10.1016/j.trsl.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander-Bryant A.A., Vander Berg-Foels W., Wen X. Bioengineering Strategies for Designing Targeted Cancer Therapies. Adv. Cancer Res. 2013:1–53. doi: 10.1038/mp.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boohaker R.J., Lee M.W., Vishnubholta P., Perez J.M., Khaled A.R. The use of therapeutic peptides to target and to kill cancer cells. Current Medicinal Chemsitry. 2012;19:3794–3804. doi: 10.2174/092986712801661004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varanko A., Saha S., Chilkoti A. 2020. Recent Trends in Protein and Peptide-Based Biomaterials for Advanced Drug Delivery. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y., Yang Z., Wang C., Yang T., Cai C., Zhao X., Yang L., Ding P. Exploring the role of peptides in polymer-based gene delivery. Acta Biomater. 2017;60:23–37. doi: 10.1016/j.actbio.2017.07.043. [DOI] [PubMed] [Google Scholar]
- 12.Fan T., Yu X., Shen B., Sun L. Peptide self-assembled nanostructures for drug delivery applications. J. Nanomater. 2017;2017 doi: 10.1155/2017/4562474. [DOI] [Google Scholar]
- 13.Apte A., Koren E., Koshkaryev A., Torchilin V.P. Doxorubicin in TAT peptide-modified multifunctional immunoliposomes demonstrates increased activity against both drug-sensitive and drug-resistant ovarian cancer models. Cancer Biol. Ther. 2014;15:69–80. doi: 10.4161/cbt.26609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Begum A.A., Wan Y., Toth I., Moyle P.M. Bombesin/oligoarginine fusion peptides for gastrin releasing peptide receptor (GRPR) targeted gene delivery. Bioorg. Med. Chem. 2018;26:516–526. doi: 10.1016/j.bmc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Al-Ahmady Z.S., Al-Jamal W.T., Bossche J.V., Bui T.T., Drake A.F., Mason A.J., Kostarelos K. Lipid-peptide vesicle nanoscale hybrids for triggered drug release by mild hyperthermia in vitro and in vivo. ACS Nano. 2012;6:9335–9346. doi: 10.1021/nn302148p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu N., Tan Y., Hu Y., Meng T., Wen L., Liu J., Cheng B., Yuan H., Huang X., Hu F. A54 peptide modified and redox-responsive glucolipid conjugate micelles for intracellular delivery of doxorubicin in hepatocarcinoma therapy. ACS Appl. Mater. Interfaces. 2016;8:33148–33156. doi: 10.1021/acsami.6b09333. [DOI] [PubMed] [Google Scholar]
- 17.Bezu L., Kepp O., Cerrato G., Pol J., Fucikova J., Spisek R., Zitvogel L., Kroemer G., Galluzzi L. Trial watch: peptide-based vaccines in anticancer therapy. OncoImmunology. 2018;7:1–15. doi: 10.1080/2162402X.2018.1511506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulsharova G., Lee M., Cheng F., Haque M., Choi H., Kim K., O'Brien W., Liu G.L. In vitro and in vivo imaging of peptide-encapsulated polymer nanoparticles for cancer biomarker activated drug delivery. IEEE Trans. NanoBioscience. 2013;12:304–310. doi: 10.1109/TNB.2013.2274781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L., Jing D., Jiang N., Rojalin T., Baehr C.M., Zhang D., Xiao W., Wu Y., Cong Z., Li J.J., Li Y., Wang L., Lam K.S. Transformable peptide nanoparticles arrest HER2 signalling and cause cancer cell death in vivo. Nat. Nanotechnol. 2020;15:145–153. doi: 10.1038/s41565-019-0626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira S., van Rooy I., Kranenburg O., Storm G., Schiffelers R.M. Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. Int. J. Pharm. 2007;331:211–214. doi: 10.1016/j.ijpharm.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 21.Sánchez-Navarro M., Giralt E., Teixidó M. Blood–brain barrier peptide shuttles. Curr. Opin. Chem. Biol. 2017;38:134–140. doi: 10.1016/j.cbpa.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 22.X. Peng, X. Liu, Y.-L. Liu, J.Y. Kim, Y.-I. Chen, P. Ang, A. Nguyen, J. Leal, H.-C. Yeh, D. Ghosh, Brain-penetrating Peptide Shuttles across the Blood-Brain Barrier and Extracellular-like Space, n.d. [DOI] [PubMed]
- 23.Cavaco M., Frutos S., Oliete P., Valle J., Andreu D., Castanho M.A.R.B., Vila-Perelló M., Neves V. Conjugation of a blood brain barrier peptide shuttle to an Fc domain for brain delivery of therapeutic biomolecules. ACS Med. Chem. Lett. 2021;12(11):1663–1668. doi: 10.1021/acsmedchemlett.1c00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M.Z., Pang W.H., Yang T., Wang J.C., Wei L., Qiu C., Wu Y.F., Liu W.Z., Wei W., Guo X.Y., Zhang Q. Systemic delivery of siRNA by T7 peptide modified core-shell nanoparticles for targeted therapy of breast cancer. Eur. J. Pharmaceut. Sci. 2016;92:39–48. doi: 10.1016/j.ejps.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Zununi Vahed S., Salehi R., Davaran S., Sharifi S. Liposome-based drug co-delivery systems in cancer cells, Materials. Science and Engineering C. 2017;71:1327–1341. doi: 10.1016/j.msec.2016.11.073. [DOI] [PubMed] [Google Scholar]
- 26.Akbarzadeh A., Rezaei-sadabady R., Davaran S., Joo S.W., Zarghami N. Liposome : classification , preparation , and applications. Nanoscale Res. Lett. 2013;8:1–9. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zielinska A., Carreiro F., Oliveira A., Neves A., Pires B., Venkatesh D., Durazzo A., Lucarini M., Eder P., Silva A., Santini A., Souto E. Polymeric nanoparticles: production, characterization, toxicology and ecotoxicology. Molecules. 2020;25:195–240. doi: 10.1201/9780429031236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D., Ping Y., Xu F., Yu H., Pan H., Huang H., Wang Q., Tang G., Li J. Construction of a star-shaped copolymer as a vector for FGF receptor-mediated gene delivery in vitro and in vivo. Biomacromolecules. 2010;11:2221–2229. doi: 10.1021/bm100141y. [DOI] [PubMed] [Google Scholar]
- 29.Cocco E., Deng Y., Shapiro E.M., Bortolomai I., Lopez S., Lin K., Bellone S., Cui J., Menderes G., Black J.D., Schwab C.L., Bonazzoli E., Yang F., Predolini F., Zammataro L., Altwerger G., de Haydu C., Clark M., Alvarenga J., Ratner E., Azodi M., Silasi D.-A., Schwartz P.E., Litkouhi B., Saltzman W.M., Santin A.D. Dual-targeting nanoparticles for in vivo delivery of suicide genes to chemotherapy-resistant ovarian cancer cells. Mol. Cancer Therapeut. 2017;16:323–333. doi: 10.1158/1535-7163.MCT-16-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie S. Understanding and overcoming major barriers in cancer nanomedicine. Nanomedicine. 2010;5:523–528. doi: 10.2217/nnm.10.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruno B.J., Miller G.D., Lim C.S. Basics and recent advances in peptide and protein drug delivery. Ther. Deliv. 2013;4:1443–1467. doi: 10.4155/tde.13.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dijkgraaf I., Kruijtzer J.A.W., Frielink C., Corstens F.H.M., Oyen W.J.G., Liskamp R.M.J., Boerman O.C. αvβ3 integrin-targeting of intraperitoneally growing tumors with a radiolabeled RGD peptide. Int. J. Cancer. 2007;120:605–610. doi: 10.1002/ijc.22297. [DOI] [PubMed] [Google Scholar]
- 33.Habault J., Poyet J.L. Recent advances in cell penetrating peptide-based anticancer therapies. Molecules. 2019;24:1–17. doi: 10.3390/molecules24050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristensen M., Birch D., Nielsen H.M. Applications and challenges for use of cell-penetrating peptides as delivery vectors for peptide and protein cargos. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vivès E., Schmidt J., Pèlegrin A. Cell-penetrating and cell-targeting peptides in drug delivery. Biochim. Biophys. Acta Rev. Canc. 2008:126–138. doi: 10.1016/j.bbcan.2008.03.001. 1786. [DOI] [PubMed] [Google Scholar]
- 36.Vinson V.K. Drug targeting. Sci. Signal. 2013;6:81–91. doi: 10.1126/scisignal.2004484. [DOI] [Google Scholar]
- 37.Mousavizadeh A., Jabbari A., Akrami M., Bardania H. Cell targeting peptides as smart ligands for targeting of therapeutic or diagnostic agents: a systematic review. Colloids Surf. B Biointerfaces. 2017;158:507–517. doi: 10.1016/j.colsurfb.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 38.White S.J., Nicklin S.A., Sawamura T., Baker A.H. Identification of peptides that target the endothelial cell–specific LOX-1 receptor. Hypertension. 2001;37:449–455. doi: 10.1161/01.hyp.37.2.449. [DOI] [PubMed] [Google Scholar]
- 39.Zahid M., Feldman K., Garcia-Borrero G., Feinstein T., Pogodzinski N., Xu X., Turko R., Czachowski M., Wu Y., Mason N., Lo C. Cardiac targeting peptide, a novel cardiac vector:studies in bio-distribution, imaging Application,and mechanism of transduction. Biomolecules. 2018;8 doi: 10.3390/biom8040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue S., Zhou X., Sang W., Wang C., Lu H., Xu Y., Zhong Y., Zhu L., He C., Ma J. Cartilage-targeting peptide-modified dual-drug delivery nanoplatform with NIR laser response for osteoarthritis therapy. Bioactive Materials. 2021;6:2372–2389. doi: 10.1016/j.bioactmat.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shadidi M., Sioud M. Selective targeting of cancer cells using synthetic peptides. Drug Resist. Updates. 2003;6:363–371. doi: 10.1016/j.drup.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 42.a J.C., Jr., York N., Science G. The structure of a typical antibody molecule. Immunobiology. 2001:1–10. [Google Scholar]
- 43.Aina O.H., Sroka T.C., Chen M.L., Lam K.S. Therapeutic cancer targeting peptides. Biopolymers - Peptide Science Section. 2002;66:184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- 44.Ladner R.C., Sato A.K., Gorzelany J., De Souza M. Phage display-derived peptides as therapeutic alternatives to antibodies. Drug Discov. Today. 2004;9(12):525–529. doi: 10.1016/S1359-6446(04)03104-6. [DOI] [PubMed] [Google Scholar]
- 45.Le Joncour V., Laakkonen P. Seek & Destroy, use of targeting peptides for cancer detection and drug delivery. Bioorg. Med. Chem. 2018:2797–2806. doi: 10.1016/j.bmc.2017.08.052. [DOI] [PubMed] [Google Scholar]
- 46.Sidhu S.S. Phage display in pharmaceutical biotechnology. Curr. Opin. Biotechnol. 2000;11:610–616. doi: 10.1016/S0958-1669(00)00152-X. [DOI] [PubMed] [Google Scholar]
- 47.Turnbough C.L. Discovery of phage display peptide ligands for species-specific detection of Bacillus spores. J. Microbiol. Methods. 2003;53:263–271. doi: 10.1016/S0167-7012(03)00030-7. [DOI] [PubMed] [Google Scholar]
- 48.Sidhu S.S. Engineering M13 for phage display. Biomol. Eng. 2001;18:57–63. doi: 10.1016/S1389-0344(01)00087-9. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L., Yin G., Yan D., Wei Y., Ma C., Huang Z., Liao X., Yao Y., Chen X., Hao B. In vitro screening of ovarian tumor specific peptides from a phage display peptide library. Biotechnol. Lett. 2011;33:1729–1735. doi: 10.1007/s10529-011-0634-4. [DOI] [PubMed] [Google Scholar]
- 50.Wang L., Hu Y., Li W., Wang F., Lu X., Han X., Lv J., Chen J. Identification of a peptide specifically targeting ovarian cancer by the screening of a phage display peptide library. Oncol. Lett. 2016;11:4022–4026. doi: 10.3892/ol.2016.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aina O.H., Liu R., Sutcliffe J.L., Marik J., Pan C.X., Lam K.S. From combinatorial chemistry to cancer-targeting peptides. Mol. Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 52.Lam K.S., Salmon S.E., Hersh E.M., Hruby V.J., Kazmierski W.M., Knapp R.J. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 53.Yao N., Xiao W., Wang X., Marik J., Park S.H., Takada Y., Lam K.S. Discovery of targeting ligands for breast cancer cells using the one-bead one-compound combinatorial method. J. Med. Chem. 2009;52:126–133. doi: 10.1021/jm801062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng L., Liu R., Marik J., Wang X., Takada Y., Lam K.S. Combinatorial chemistry identifies high-affinity peptidomimetics against α4β1 integrin for in vivo tumor imaging. Nat. Chem. Biol. 2006;2:381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 55.Liu R., Li X., Xiao W., Lam K.S. Tumor-targeting peptides from combinatorial libraries. Adv. Drug Deliv. Rev. 2017;110–111:13–37. doi: 10.1016/j.addr.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaradat D.M.M. Thirteen decades of peptide synthesis: key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation. Amino Acids. 2018;50:39–68. doi: 10.1007/s00726-017-2516-0. [DOI] [PubMed] [Google Scholar]
- 57.Basso A., Banfi L., Riva R., Piaggio P., Guanti G. Solid-phase synthesis of modified oligopeptides via Passerini multicomponent reaction. Tetrahedron Lett. 2003;44:2367–2370. doi: 10.1016/S0040-4039(03)00238-7. [DOI] [Google Scholar]
- 58.Katritzky A.R., Haase D.N., Johnsons J.v., Chung A. Benzotriazole-assisted solid-phase assembly of Leu-Enkephalin, amyloid β segment 34-42, and other “difficult” peptide sequences. J. Org. Chem. 2009;74:2028–2032. doi: 10.1021/jo8026214. [DOI] [PubMed] [Google Scholar]
- 59.Okada Y., Takasawa R., Kubo D., Iwanaga N., Fujita S., Suzuki K., Suzuki H., Kamiya H., Chiba K. 2019. Improved Tag-Assisted Liquid-phase Peptide Synthesis: Application to the Synthesis of the Bradykinin Receptor Antagonist Icatibant Acetate. [DOI] [Google Scholar]
- 60.Yeo J., Peeva L., Chung S., Gaffney P., Kim D., Luciani C., Tsukanov S., Seibert K., Kopach M., Albericio F., Livingston A. Liquid phase peptide synthesis via one-pot nanostar sieving (PEPSTAR) Angew. Chem. Int. Ed. 2021;60:7786–7795. doi: 10.1002/anie.202014445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Hest J.C.M. Peptide conjugates for biological applications. Bioconjugate Chem. 2017;28:689–690. doi: 10.1021/acs.bioconjchem.7b00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mueller L.K., Baumruck A.C., Zhdanova H., Tietze A.A. Challenges and perspectives in chemical synthesis of highly hydrophobic peptides. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D'Orlyé F., Trapiella-Alfonso L., Lescot C., Pinvidic M., Doan B.T., Varenne A. Synthesis, characterization and evaluation of peptide nanostructures for biomedical applications. Molecules. 2021;26 doi: 10.3390/molecules26154587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He R., Mowery S.A., Chabenne J., Finan B., Mayer J.P., DiMarchi R.D. A facile procedure for one-pot stable conjugation of two proglucagon cysteine-containing peptide analogs. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.693958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gandioso A., Massaguer A., Villegas N., Salvans C., Sánchez D., Brun-Heath I., Marchán V., Orozco M., Terrazas M. Efficient siRNA-peptide conjugation for specific targeted delivery into tumor cells. Chem. Commun. 2017;53:2870–2873. doi: 10.1039/c6cc10287e. [DOI] [PubMed] [Google Scholar]
- 66.Dharap S.S., Wang Y., Chandna P., Khandare J.J., Qiu B., Gunaseelan S., Sinko P.J., Stein S., Farmanfarmaian A., Minko T. vol. 102. 2005. Tumor-specific targeting of an anticancer drug delivery system by LHRH peptide; pp. 12962–12967. (Proceedings of the National Academy of Sciences of the United States of America). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maruta F., Parker A.L., Fisher K.D., Hallissey M.T., Ismail T., Rowlands D.C., Chandler L.A., Kerr D.J., Seymour L.W. Identification of FGF receptor-binding peptides for cancer gene therapy. Cancer Gene Ther. 2002;9:543–552. doi: 10.1038/sj.cgt.7700470. [DOI] [PubMed] [Google Scholar]
- 68.Cheng X., Yu D., Cheng G., Yung B.C., Liu Y., Li H., Kang C., Fang X., Tian S., Zhou X., Liu Q., Lee R.J. T7 peptide-conjugated lipid nanoparticles for dual modulation of bcl-2 and akt-1 in lung and cervical carcinomas. Mol. Pharm. 2018;15:4722–4732. doi: 10.1021/acs.molpharmaceut.8b00696. [DOI] [PubMed] [Google Scholar]
- 69.Song S., Liu D., Peng J., Sun Y., Li Z., Gu J.-R., Xu Y. Peptide ligand-mediated liposome distribution and targeting to EGFR expressing tumor in vivo. Int. J. Pharm. 2008;363:155–161. doi: 10.1016/j.ijpharm.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 70.Tai W., Shukla R.S., Qin B., Li B., Cheng K. Development of a peptide-drug conjugate for prostate cancer therapy. Mol. Pharm. 2011;8:901–912. doi: 10.1021/mp200007b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao Q., Li Z.B., Chen K., Wu Z., He L., Neamati N., Chen X. Evaluation of biodistribution and anti-tumor effect of a dimeric RGD peptide-paclitaxel conjugate in mice with breast cancer. Eur. J. Nucl. Med. Mol. Imag. 2008;35:1489–1498. doi: 10.1007/s00259-008-0744-y. [DOI] [PubMed] [Google Scholar]
- 72.Wu X.L., Kim J.H., Koo H., Bae S.M., Shin H., Kim M.S., Lee B.H., Park R.W., Kim I.S., Choi K., Kwon I.C., Kim K., Lee D.S. Tumor-targeting peptide conjugated pH-responsive micelles as a potential drug carrier for cancer therapy. Bioconjugate Chem. 2010;21:208–213. doi: 10.1021/bc9005283. [DOI] [PubMed] [Google Scholar]
- 73.Xu W., Yan X., Liu N., Wu G. P1c peptide decorated liposome targeting αvβ3-expressing tumor cells in vitro and in vivo. RSC Adv. 2018;8:25575–25583. doi: 10.1039/c8ra05014g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park Y.K., Goda Y. Integrins in synapse regulation. Nat. Rev. Neurosci. 2016;17:745–756. doi: 10.1038/nrn.2016.138. [DOI] [PubMed] [Google Scholar]
- 75.Liu Z., Wang F., Chen X. Integrin targeted delivery of chemotherapeutics. Theranostics. 2012;1:201–210. doi: 10.7150/thno/v01p0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X.E., He Y., de Waal P.W., Gao X., Kang Y., Van Eps N., Yin Y., Pal K., Goswami D., White T.A., Barty A., Latorraca N.R., Chapman H.N., Hubbell W.L., Dror R.O., Stevens R.C., Cherezov V., Gurevich V.V., Griffin P.R., Ernst O.P., Melcher K., Xu H.E. Identification of phosphorylation codes for arrestin recruitment by G protein-coupled receptors. Cell. 2017;170:457–469. doi: 10.1016/j.cell.2017.07.002. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Accardo A., Aloj L., Aurilio M., Morelli G., Tesauro D. Receptor binding peptides for target-selective delivery of nanoparticles encapsulated drugs. Int. J. Nanomed. 2014;9:1537–1557. doi: 10.2147/IJN.S53593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mcinnes C., Sykes B.D. 1998. Growth Factor Receptors : Structure , Mechanism , and. [DOI] [PubMed] [Google Scholar]
- 79.Verbeek B.S., Adriaansen-Slot S.S., Vroom T.M., Beckers T., Rijksen G. Overexpression of EGFR and c-erbB2 causes enhanced cell migration in human breast cancer cells and NIH3T3 fibroblasts. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1998;425:145–150. doi: 10.1016/S0014-5793(98)00224-5. [DOI] [PubMed] [Google Scholar]
- 80.Bargh J.D., Isidro-Llobet A., Parker J.S., Spring D.R. Cleavable linkers in antibody-drug conjugates. Chem. Soc. Rev. 2019;48:4361–4374. doi: 10.1039/c8cs00676h. [DOI] [PubMed] [Google Scholar]
- 81.Poreba M. vol. 287. 2020. pp. 1936–1969. (Protease-activated Prodrugs : Strategies , Challenges , and Future Directions). [DOI] [PubMed] [Google Scholar]
- 82.Teich N., Bödeker H., Keim V. Cathepsin B cleavage of the trypsinogen activation peptide. BMC Gastroenterol. 2002;2:4–7. doi: 10.1186/1471-230X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dubowchik G.M., F I R.A. vol. 8. 1998. pp. 3341–3346. (Cathepsin B-Sensitive Dipeptide Prodrugs A Model Study of Structural Requirements for Efficient Release of Doxorubicin). [DOI] [PubMed] [Google Scholar]
- 84.Baurain R., Masquelier M., Campeneere D.D., Trouet A. 1980. Amino Acid and Dipeptide Derivatives of Daunorubicin . 2 . Cellular Pharmacology and Antitumor Activity on L1210 Leukemic Cells in Vitro and in Vivo; pp. 1171–1174. [DOI] [PubMed] [Google Scholar]
- 85.Lock L.L., Tang Z., Keith D., Reyes C., Cui H. 2015. Enzyme-Specific Doxorubicin Drug Beacon as Drug-Resistant Theranostic Molecular Probes; pp. 8–11. [DOI] [PubMed] [Google Scholar]
- 86.Mollaev M., Gorokhovets N., Nikolskaya E., Faustova M., Zabolotsky A., Zhunina O. Type of pH sensitive linker reveals different time-dependent intracellular localization , in vitro and in vivo efficiency in alpha-fetoprotein receptor targeted doxorubicin conjugate. Int. J. Pharm. 2019;559:138–146. doi: 10.1016/j.ijpharm.2018.12.073. [DOI] [PubMed] [Google Scholar]
- 87.Foroozandeh P., Aziz A. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res. Lett. 2018;13 doi: 10.1186/s11671-018-2728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jones S.W., Christison R., Bundell K., Voyce C.J., Brockbank S.M.V., Newham P., Lindsay M.A. Characterisation of cell-penetrating peptide-mediated peptide delivery. Br. J. Pharmacol. 2005;145:1093–1102. doi: 10.1038/sj.bjp.0706279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McClorey G., Banerjee S. Cell-penetrating peptides to enhance delivery of oligonucleotide-based therapeutics. Biomedicines. 2018;6:1–15. doi: 10.3390/biomedicines6020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Copolovici D.M., Langel K., Eriste E., Langel Ü. Cell-penetrating peptides: design, synthesis, and applications. ACS Nano. 2014;8:1972–1994. doi: 10.1021/nn4057269. [DOI] [PubMed] [Google Scholar]
- 91.Green M., Loewenstein P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus Tat trans-activator protein. Cell. 1988;55:1179–1188. doi: 10.1016/j.scitotenv.2011.11.081. [DOI] [PubMed] [Google Scholar]
- 92.Endoh T., Sisido M., Ohtsuki T. Cellular siRNA delivery mediated by a cell-permeant RNA-binding protein and photoinduced RNA interference. Bioconjugate Chem. 2008;19:1017–1024. doi: 10.1021/bc800020n. [DOI] [PubMed] [Google Scholar]
- 93.Su Y., Mani R., Hong M. Asymmetric insertion of membrane proteins in lipid bilayers by solid-state NMR paramagnetic relaxation enhancement: a cell-penetrating peptide example. J. Am. Chem. Soc. 2008;130:8856–8864. doi: 10.1021/ja802383t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Al-Husaini K., Elkamel E., Han X., Chen P. Therapeutic potential of a cell penetrating peptide (CPP, NP1) mediated siRNA delivery: evidence in 3D spheroids of colon cancer cells. Can. J. Chem. Eng. 2020;98:1240–1254. doi: 10.1002/cjce.23743. [DOI] [Google Scholar]
- 95.Jing H., Cheng W., Li S., Wu B., Leng X., Xu S., Tian J. Novel cell-penetrating peptide-loaded nanobubbles synergized with ultrasound irradiation enhance EGFR siRNA delivery for triple negative Breast cancer therapy. Colloids Surf. B Biointerfaces. 2016;146:387–395. doi: 10.1016/j.colsurfb.2016.06.037. [DOI] [PubMed] [Google Scholar]
- 96.Taha A.A., AL-Jawad S.M.H., AL-Barram L.F.A. Improvement of cancer therapy by TAT peptide conjugated gold nanoparticles. J. Cluster Sci. 2019;30:403–414. doi: 10.1007/s10876-019-01497-9. [DOI] [Google Scholar]
- 97.Duchardt F., Fotin-Mleczek M., Schwarz H., Fischer R., Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 98.Fonseca S.B., Pereira M.P., Kelley S.O. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv. Drug Deliv. Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 99.Bloch-Gallego E., Le Roux I., Joliot A.H., Volovitch M., Henderson C.E., Prochiantz A. Antennapedia homeobox peptide enhances growth and branching of embryonic chicken motoneurons in vitro. JCB (J. Cell Biol.) 1993;120:485–492. doi: 10.1083/jcb.120.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee H.L., Dubikovskaya E.A., Hwang H., Semyonov A.N., Wang H., Jones L.R., Twieg R.J., Moerner W.E., Wender P.A. Single-molecule motions of oligoarginine transporter conjugates on the plasma membrane of Chinese Hamster ovary cells. J. Am. Chem. Soc. 2008;130:9364–9370. doi: 10.1021/ja710798b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Farrera-Sinfreu J., Giralt E., Castel S., Albericio F., Royo M. Cell-penetrating cis-γ-amino-L-proline-derived peptides. J. Am. Chem. Soc. 2005;127:9459–9468. doi: 10.1021/ja051648k. [DOI] [PubMed] [Google Scholar]
- 102.Oskolkov N., Arukuusk P., Copolovici D.M., Lindberg S., Margus H., Padari K., Pooga M., Langel Ü. NickFects, phosphorylated derivatives of transportan 10 for cellular delivery of oligonucleotides. Int. J. Pept. Res. Therapeut. 2011;17:147–157. doi: 10.1007/s10989-011-9252-1. [DOI] [Google Scholar]
- 103.Mäe M., EL Andaloussi S., Lundin P., Oskolkov N., Johansson H.J., Guterstam P., Langel Ü. A stearylated CPP for delivery of splice correcting oligonucleotides using a non-covalent co-incubation strategy. J. Contr. Release. 2009;134:221–227. doi: 10.1016/j.jconrel.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 104.Bodor N., Tóth-Sarudy É., Holm T., Pallagi I., Vass E., Buchwald P., Langel Ü. Novel, cell-penetrating molecular transporters with flexible backbones and permanently charged side-chains. J. Pharm. Pharmacol. 2007;59:1065–1076. doi: 10.1211/jpp.59.8.0003. [DOI] [PubMed] [Google Scholar]
- 105.Journal A.I., Soleymani-goloujeh M., Nokhodchi A., Niazi M., Najafi S., Shahbazi-mojarrad J., Zarghami N., Zakeri P., Mohammadi A., Karimi M., Valizadeh H. Effects of N-terminal and C-terminal modification on cytotoxicity and cellular uptake of amphiphilic cell penetrating peptides, Artificial Cells. Nanomedicine, and Biotechnology. 2018;46:S91–S103. doi: 10.1080/21691401.2017.1414823. [DOI] [PubMed] [Google Scholar]
- 106.Freeman E.C., Weiland L.M., Meng W.S. Modeling the proton sponge hypothesis: examining proton sponge effectiveness for enhancing intracellular gene delivery through multiscale modeling. J. Biomater. Sci. Polym. Ed. 2013;24:398–416. doi: 10.1080/09205063.2012.690282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bus T., Traeger A., Schubert U.S. The great escape: how cationic polyplexes overcome the endosomal barrier. J. Mater. Chem. B. 2018;6:6904–6918. doi: 10.1039/c8tb00967h. [DOI] [PubMed] [Google Scholar]
- 108.Meng Z., Luan L., Kang Z., Feng S., Meng Q., Liu K. Histidine-enriched multifunctional peptide vectors with enhanced cellular uptake and endosomal escape for gene delivery. J. Mater. Chem. B. 2017;5:74–84. doi: 10.1039/c6tb02862d. [DOI] [PubMed] [Google Scholar]
- 109.Liu Y., Lu Z., Mei L., Yu Q., Tai X., Wang Y., Shi K., Zhang Z., He Q. Tandem peptide based on structural modification of poly-arginine for enhancing tumor targeting efficiency and therapeutic effect. ACS Appl. Mater. Interfaces. 2017;9:2083–2092. doi: 10.1021/acsami.6b12611. [DOI] [PubMed] [Google Scholar]
- 110.Vives E. Present and future of cell-penetrating peptide mediated delivery systems: “Is the Trojan horse too wild to go only to Troy? J. Contr. Release. 2005;109:77–85. doi: 10.1016/j.jconrel.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 111.Svensen N., Walton J., Bradley M. Peptides for cell-selective drug delivery. Trends Pharmacol. Sci. 2012;33:186–192. doi: 10.1016/j.tips.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 112.Heitz F., Morris M.C., Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Langel Ü. Springer US; 2015. Cell-Penetrating Peptides: Methods and Protocols. [Google Scholar]
- 114.Yang W., Xia Y., Fang Y., Meng F., Zhang J., Cheng R., Deng C., Zhong Z. Selective cell penetrating peptide-functionalized polymersomes mediate efficient and targeted delivery of methotrexate disodium to human lung cancer in vivo. Advanced Healthcare Materials. 2018;7:1–8. doi: 10.1002/adhm.201701135. [DOI] [PubMed] [Google Scholar]
- 115.Gao H., Zhang Q., Yang Y., Jiang X., He Q. Tumor homing cell penetrating peptide decorated nanoparticles used for enhancing tumor targeting delivery and therapy. Int. J. Pharm. 2015;478:240–250. doi: 10.1016/j.ijpharm.2014.11.029. [DOI] [PubMed] [Google Scholar]
- 116.Smith S.J., Gu L., Phipps E.A., Dobrolecki L.E., Mabrey K.S., Gulley P., Dillehay K.L., Dong Z., Fields G.B., Chen Y.R., Ann D., Hickey R.J., Malkas L.H. A peptide mimicking a region in proliferating cell nuclear antigen specific to key protein interactions is cytotoxic to breast cancer. Mol. Pharmacol. 2015;87:263–276. doi: 10.1124/mol.114.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ren Y., Cheung H.W., Von Maltzhan G., Agrawal A., Glenn S., Weir B.A., Boehm J.S., Tamayo P., Karst A.M., Joyce F., Hirsch M.S., Mesirov J.P., Drapkin R., Root D.E., Lo J., Fogal V., Ruoslahti E., Hahn W.C., Sangeeta N. 2013. Targeted Tumor-Penetrating siRNA Nanocomplexes for Credentialing the Ovarian Cancer Target ID4; p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shukla R.S., Qin B., Cheng K. Peptides used in the delivery of small noncoding RNA. Mol. Pharm. 2014;11:3395–3408. doi: 10.1021/mp500426r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alexander-Bryant A.A., Dumitriu A., Attaway C.C., Yu H., Jakymiw A. Fusogenic-oligoarginine peptide-mediated silencing of the CIP2A oncogene suppresses oral cancer tumor growth in vivo. J. Contr. Release. 2015;218:72–81. doi: 10.1016/j.jconrel.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao Z.X., Gao S.Y., Wang J.C., Chen C.J., Zhao E.Y., Hou W.J., Feng Q., Gao L.Y., Liu X.Y., Zhang L.R., Zhang Q. Self-assembly nanomicelles based on cationic mPEG-PLA-b-Polyarginine(R15) triblock copolymer for siRNA delivery. Biomaterials. 2012;33:6793–6807. doi: 10.1016/j.biomaterials.2012.05.067. [DOI] [PubMed] [Google Scholar]
- 121.Shin M.C., Zhang J., Min K.A., Byun Y., David A.E., He H., Yang V.C. vol. 102. National Institutes of Health; 2015. p. 1. (Cell-penetrating Peptides: Achievements and Challenges in Application for Cancer Treatment). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dinca A., Chien W.M., Chin M.T. Intracellular delivery of proteins with cell-penetrating peptides for therapeutic uses in human disease. Int. J. Mol. Sci. 2016;17 doi: 10.3390/ijms17020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saar K., Lindgren M., Hansen M., Eiríksdóttir E., Jiang Y., Rosenthal-Aizman K., Sassian M., Langel Ü. Cell-penetrating peptides: a comparative membrane toxicity study. Anal. Biochem. 2005;345:55–65. doi: 10.1016/j.ab.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 124.El-Andaloussi S., Järver P., Johansson H.J., Langel Ü. Cargo-dependent cytotoxicity and delivery efficacy of cell-penetrating peptides: a comparative study. Biochem. J. 2007;407:285–292. doi: 10.1042/BJ20070507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kilk K., Mahlapuu R., Soomets U., Langel Ü. Analysis of in vitro toxicity of five cell-penetrating peptides by metabolic profiling. Toxicology. 2009;265:87–95. doi: 10.1016/j.tox.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 126.El-Andaloussi S., Järver P., Johansson H.J., Langel Ü. Cargo-dependent cytotoxicity and delivery efficacy of cell-penetrating peptides: a comparative study. Biochem. J. 2007;407:285–292. doi: 10.1042/BJ20070507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moku G., Layek B., Trautman L., Putnam S., Panyam J., Prabha S. Improving payload capacity and anti-tumor efficacy of mesenchymal stem cells using tat peptide functionalized polymeric nanoparticles. Cancers. 2019;11 doi: 10.3390/cancers11040491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Watson G.M., Kulkarni K., Brandt R., Del Borgo M.P., Aguilar M.I., Wilce J.A. Shortened penetratin cell-penetrating peptide is insufficient for cytosolic delivery of a Grb7 targeting peptide. ACS Omega. 2017;2:670–677. doi: 10.1021/acsomega.6b00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Johansson H.J., El-Andaloussi S., Holm T., Mäe M., Jänes J., Maimets T., Langel Ü. Characterization of a novel cytotoxic cell-penetrating peptide derived from p14ARF protein. Mol. Ther. 2008;16:115–123. doi: 10.1038/sj.mt.6300346. [DOI] [PubMed] [Google Scholar]
- 130.Cantini L., Attaway C.C., Butler B., Andino L.M., Sokolosky M.L., Jakymiw A. Fusogenic-Oligoarginine peptide-mediated delivery of siRNAs targeting the CIP2A oncogene into oral cancer cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taylor B.N., Mehta R.R., Yamada T., Lekmine F., Christov K., Chakrabarty A.M., Green A., Bratescu L., Shilkaitis A., Beattie C.W., Das Gupta T.K. Noncationic peptides obtained from azurin preferentially enter cancer cells. Cancer Res. 2009;69:537–546. doi: 10.1158/0008-5472.CAN-08-2932. [DOI] [PubMed] [Google Scholar]
- 132.Tripathi P.P., Arami H., Banga I., Gupta J., Gandhi S. Cell penetrating peptides in preclinical and clinical cancer diagnosis and therapy. Oncotarget. 2018;9:37252–37267. doi: 10.18632/oncotarget.26442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Margus H., Padari K., Pooga M. Cell-penetrating peptides as versatile vehicles for oligonucleotide delivery. Mol. Ther. 2012;20:525–533. doi: 10.1038/mt.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morris M.C., Depollier J., Heitz F., Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells: application to the delivery of antibodies and therapeutic proteins. Cell Biology, Four-Volume Set. 2006;4:13–18. doi: 10.1016/B978-012164730-8/50187-8. [DOI] [Google Scholar]
- 135.Morris M.C., Gros E., Aldrian-Herrada G., Choob M., Archdeacon J., Heitz F., Divita G. A non-covalent peptide-based carrier for in vivo delivery of DNA mimics. Nucleic Acids Res. 2007;35 doi: 10.1093/nar/gkm053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gestin M., Helmfors H., Falato L., Lorenzon N., Michalakis F.I., Langel Ü. Effect of small molecule signaling in PepFect14 transfection. PLoS One. 2020;15:1–21. doi: 10.1371/journal.pone.0228189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Izabela R., Jarosław R., Magdalena A., Piotr R., Ivan K. Transportan 10 improves the anticancer activity of cisplatin. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2016;389:485–497. doi: 10.1007/s00210-016-1219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schmid S.L., Sorkin A., Zerial M., Lewis W.H. Endocytosis : Past , Present , and Future. 2014;3:1–9. doi: 10.1101/cshperspect.a022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lau W.L., Ege D.S., Lear J.D., Hammer D.A., DeGrado W.F. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lau W.L., Ege D.S., Lear J.D., Hammer D.A., DeGrado W.F. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Epand R.M. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta Biomembr. 2003;1614:116–121. doi: 10.1016/S0005-2736(03)00169-X. [DOI] [PubMed] [Google Scholar]
- 142.Ishiguro R., Matsumoto M., Takahashi S. Interaction of fusogenic synthetic peptide with phospholipid bilayers: orientation of the peptide α-helix and binding isotherm. Biochemistry. 1996;35:4976–4983. doi: 10.1021/bi952547+. [DOI] [PubMed] [Google Scholar]
- 143.Carr C.M., Chaudhry C., Kim P.S. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Chemtracts. 1999;12:158–164. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Galdiero S., Falanga A., Vitiello M., Raiola L., Russo L., Pedone C., Isernia C., Galdiero M. The presence of a single N-terminal histidine residue enhances the fusogenic properties of a membranotropic peptide derived from herpes simplex virus type 1 glycoprotein H. J. Biol. Chem. 2010;285:17123–17136. doi: 10.1074/jbc.M110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Decout A., Labeur C., Vanloo B., Goethals M., Vandekerckhove J., Brasseur R., Rosseneu M. Contribution of the hydrophobicity gradient to the secondary structure and activity of fusogenic peptides. Mol. Membr. Biol. 1999;16:237–246. doi: 10.1080/096876899294553. [DOI] [PubMed] [Google Scholar]
- 146.Schmid S.L., Sorkin A., Zerial M., Lewis W.H. vol. 3. 2014. pp. 1–9. (Endocytosis : Past , Present , and Future). [Google Scholar]
- 147.Carr C.M., Kim P.S. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-W. [DOI] [PubMed] [Google Scholar]
- 148.Gruenke J.A., Armstrong R.T., Newcomb W.W., Brown J.C., White J.M. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J. Virol. 2002;76:4456–4466. doi: 10.1128/jvi.76.9.4456-4466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.M I., R J. Common properties of fusion peptides from diverse systems. Biosci. Rep. 2000;20:483–500. doi: 10.1023/A:1010454803579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Charloteaux B., Lorin A., Brasseur R., Lins L. The “tilted peptide theory” links membrane insertion properties and fusogenicity of viral fusion peptides. Protein Pept. Lett. 2010;16:718–725. doi: 10.2174/092986609788681724. [DOI] [PubMed] [Google Scholar]
- 151.Liou J., Revon B., Martin A.L., Huang Y., Chiang H., Lee H. Protein transduction in human cells is enhanced by cell-penetrating peptides fused with an endosomolytic HA2 sequence. Peptides. 2012;37:273–284. doi: 10.1016/j.peptides.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van den Berg A., Dowdy S.F. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011;22:888–893. doi: 10.1016/j.copbio.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 153.Liou J., Revon B., Martin A.L., Huang Y., Chiang H., Lee H. Protein transduction in human cells is enhanced by cell-penetrating peptides fused with an endosomolytic HA2 sequence. Peptides. 2012;37:273–284. doi: 10.1016/j.peptides.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Patel S.G., Sayers E.J., He L., Narayan R., Williams T.L., Mills E.M., Allemann R.K., Luk L.Y.P., Jones A.T., Tsai Y.H. Cell-penetrating peptide sequence and modification dependent uptake and subcellular distribution of green florescent protein in different cell lines. Sci. Rep. 2019;9:1–9. doi: 10.1038/s41598-019-42456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sanchez-Garcia L., Serna N., Mattanovich M., Cazzanelli P., Sanchez-Chardi A., Conchillo-Sole O., Cortes F., Daura X., Unzueta U., Mangues R., Villaverde A., Vazquez E. The fusogenic peptide HA2 impairs selectivity of CXCR4-targeted protein nanoparticles. Chem. Commun. 2017;53:4565–4568. doi: 10.1039/c6cc09900a. [DOI] [PubMed] [Google Scholar]
- 156.Brock D.J., Kondow-mcconaghy H.M., Hager E.C., Pellois P. Endosomal escape and cytosolic penetration of macromolecules mediated by synthetic delivery agents. Bioconjugate Chem. 2019;30:293–304. doi: 10.1021/acs.bioconjchem.8b00799.Endosomal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sackett K., Shai Y. The HIV fusion peptide adopts intermolecular parallel β-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: a 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 158.Yang J., Gabrys C.M., Weliky D.P. Solid-state nuclear magnetic resonance evidence for an extended β strand conformation of the membrane-bound HIV-1 fusion peptide. Biochemistry. 2001;40:8126–8137. doi: 10.1021/bi0100283. [DOI] [PubMed] [Google Scholar]
- 159.Pereira F.B., Goñi F.M., Muga A., Nieva J.L. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: dose and sequence effects. Biophys. J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang J., Prorok M., Castellino F.J., Weliky D.P. Oligomeric β-structure of the membrane-bound HIV-1 fusion peptide formed from soluble monomers. Biophys. J. 2004;87:1951–1963. doi: 10.1529/biophysj.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sackett K., Shai Y. The HIV fusion peptide adopts intermolecular parallel β-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: a 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 162.Babel A.R., Bruce J., Young J.A.T. The hr1 and fusion peptide regions of the subgroup B avian sarcoma and leukosis virus envelope glycoprotein influence low pH-dependent membrane fusion. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cheng S.F., Wu C.W., Kantchev E.A.B., Chang D.K. Structure and membrane interaction of the internal fusion peptide of avian sarcoma leukosis virus. Eur. J. Biochem. 2004;271:4725–4736. doi: 10.1111/j.1432-1033.2004.04436.x. [DOI] [PubMed] [Google Scholar]
- 165.Gregory S.M., Harada E., Liang B., Delos S.E., White J.M., Tamm L.K. vol. 108. 2011. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2; pp. 11211–11216. (Proceedings of the National Academy of Sciences of the United States of America). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brice A.R., Lazaridis T. Structure and dynamics of a fusion peptide helical hairpin on the membrane surface: comparison of molecular simulations and NMR. J. Phys. Chem. B. 2014;118:4461–4470. doi: 10.1021/jp409412g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sackett K., Shai Y. The HIV fusion peptide adopts intermolecular parallel β-sheet structure in membranes when stabilized by the adjacent N-terminal heptad repeat: a 13C FTIR study. J. Mol. Biol. 2005;350:790–805. doi: 10.1016/j.jmb.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 168.Nishimura Y., Takeda K., Ezawa R., Ishii J., Ogino C., Kondo A. A display of pH-sensitive fusogenic GALA peptide facilitates endosomal escape from a Bio-nanocapsule via an endocytic uptake pathway. J. Nanobiotechnol. 2014;12:11. doi: 10.1186/1477-3155-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Choi S.W., Lee S.H., Mok H., Park T.G. Multifunctional siRNA delivery system: polyelectrolyte complex micelles of six-arm PEG conjugate of siRNA and cell penetrating peptide with crosslinked fusogenic peptide. Biotechnol. Prog. 2010;26:57–63. doi: 10.1002/btpr.310. [DOI] [PubMed] [Google Scholar]
- 170.Wadia J.S., Stan R.V., Dowdy S.F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 171.Burks S.R., Legenzov E.A., Martin E.W., Li C., Lu W., Kao J.P.Y. Co-encapsulating the fusogenic peptide INF7 and molecular imaging probes in liposomes increases intracellular signal and probe retention. PLoS One. 2015;10:1–17. doi: 10.1371/journal.pone.0120982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Freulon I., Roche A., Monsigny M., Mayer R. Delivery of oligonucleotides into mammalian cells by anionic peptides: a comparison between monomeric and dimeric peptides. Peptides. 2001;679:671–679. doi: 10.1042/0264-6021:3540671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Vogel K., Wang S., Lee R.J., Chmielewski J., Low P.S. Peptide-mediated release of folate-targeted liposome contents from endosomal compartments. J. Am. Chem. Soc. 1995;118:4976–4983. doi: 10.1021/ja952725m. [DOI] [Google Scholar]
- 174.Lau W.L., Ege D.S., Lear J.D., Hammer D.A., DeGrado W.F. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 2004;86:272–284. doi: 10.1016/S0006-3495(04)74103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Oliveira S., van Rooy I., Kranenburg O., Storm G., Schiffelers R.M. Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. Int. J. Pharm. 2007;331:211–214. doi: 10.1016/j.ijpharm.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 176.Oliveira S., van Rooy I., Kranenburg O., Storm G., Schiffelers R.M. Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. Int. J. Pharm. 2007;331:211–214. doi: 10.1016/j.ijpharm.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 177.Evans J.C., Malhotra M., Sweeney K., Darcy R., Nelson C.C., Hollier B.G., O'Driscoll C.M. Folate-targeted amphiphilic cyclodextrin nanoparticles incorporating a fusogenic peptide deliver therapeutic siRNA and inhibit the invasive capacity of 3D prostate cancer tumours. Int. J. Pharm. 2017;532:511–518. doi: 10.1016/j.ijpharm.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 178.Nishimura Y., Takeda K., Ezawa R., Ishii J., Ogino C., Kondo A. A display of pH-sensitive fusogenic GALA peptide facilitates endosomal escape from a Bio-nanocapsule via an endocytic uptake pathway. J. Nanobiotechnol. 2014;12:11. doi: 10.1186/1477-3155-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sakurai Y., Hatakeyama H., Sato Y., Akita H., Takayama K., Kobayashi S., Futaki S., Harashima H. Endosomal escape and the knockdown efficiency of liposomal-siRNA by the fusogenic peptide shGALA. Biomaterials. 2011;32:5733–5742. doi: 10.1016/j.biomaterials.2011.04.047. [DOI] [PubMed] [Google Scholar]
- 180.Evans J.C., Malhotra M., Sweeney K., Darcy R., Nelson C.C., Hollier B.G., O'Driscoll C.M. Folate-targeted amphiphilic cyclodextrin nanoparticles incorporating a fusogenic peptide deliver therapeutic siRNA and inhibit the invasive capacity of 3D prostate cancer tumours. Int. J. Pharm. 2017;532:511–518. doi: 10.1016/j.ijpharm.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 181.Yang J., Bahreman A., Daudey G., Bussmann J., Olsthoorn R.C.L., Kros A. Drug delivery via cell membrane fusion using lipopeptide modified liposomes. ACS Cent. Sci. 2016;2:621–630. doi: 10.1021/acscentsci.6b00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gerstein H.C., Rutty C.J. Insulin therapy: the discovery that shaped a century. Can. J. Diabetes. 2021:1–6. doi: 10.1016/j.jcjd.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 183.Driss V., El Nady M., Delbeke M., Rousseaux C., Dubuquoy C., Sarazin A., Gatault S., Dendooven A., Riveau G., Colombel J.F., Desreumaux P., Dubuquoy L., Capron M. The schistosome glutathione S-transferase P28GST, a unique helminth protein, prevents intestinal inflammation in experimental colitis through a Th2-type response with mucosal eosinophils. Mucosal Immunol. 2016;9:322–335. doi: 10.1038/mi.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Capron M., Béghin L., Leclercq C., Labreuche J., Dendooven A., Standaert A., Delbeke M., Porcherie A., Nachury M., Boruchowicz A., Dupas J.-L., Fumery M., Paupard T., Catteau S., Deplanque D., Colombel J.-F., Desreumaux P. Safety of P28GST, a protein derived from a schistosome helminth parasite, in patients with Crohn's disease: a pilot study (ACROHNEM) J. Clin. Med. 2020;9 doi: 10.3390/jcm9010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Melmed S., Popovic V., Bidlingmaier M., Mercado M., van der Lely A.J., Biermasz N., Bolanowski M., Coculescu M., Schopohl J., Racz K., Glaser B., Goth M., Greenman Y., Trainer P., Mezosi E., Shimon I., Giustina A., Korbonits M., Bronstein M.D., Kleinberg D., Teichman S., Gliko-Kabir I., Mamluk R., Haviv A., Strasburger C. Safety and efficacy of oral octreotide in acromegaly: results of a multicenter phase III trial. J. Clin. Endocrinol. Metabol. 2015;100:1699–1708. doi: 10.1210/jc.2014-4113. [DOI] [PubMed] [Google Scholar]
- 186.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 187.Anselmo A.C., Mitragotri S. An overview of clinical and commercial impact of drug delivery systems. J. Contr. Release. 2014;190:15–28. doi: 10.1016/j.jconrel.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Muttenthaler M., King G.F., Adams D.J., Alewood P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021;20:309–325. doi: 10.1038/s41573-020-00135-8. [DOI] [PubMed] [Google Scholar]
- 189.Glaser V. Reducing the cost of peptide synthesis. Genetic Engineering & Biotechnology News. 2013;33:32–34. doi: 10.1089/gen.33.13.18. [DOI] [Google Scholar]
- 190.Bray B.L. Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discov. 2003;2:587–593. doi: 10.1038/nrd1133. [DOI] [PubMed] [Google Scholar]
- 191.Q.A. BIO, Informa Pharma Intelligence . 2021. Clinical Development Success Rates and Contributing Factors 2011-2020. [Google Scholar]
- 192.C. International, CMR International 2015 Pharmaceutical R&D Factbook. 2015. [Google Scholar]
- 193.Transparency Market Research . 2020. Peptide Therapeutics Market. [Google Scholar]
- 194.Lau J.L., Dunn M.K. Therapeutic peptides: historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018;26:2700–2707. doi: 10.1016/j.bmc.2017.06.052. [DOI] [PubMed] [Google Scholar]
- 195.Meyer-Losic F., Nicolazzi C., Quinonero J., Ribes F., Michel M., Dubois V., de Coupade C., Boukaissi M., Chéné A.-S., Tranchant I., Arranz V., Zoubaa I., Fruchart J.-S., Ravel D., Kearsey J. DTS-108, A novel peptidic prodrug of SN38: <em>In vivo</em> efficacy and toxicokinetic studies. Clin. Cancer Res. 2008;14 doi: 10.1158/1078-0432.CCR-07-4580. 2145 LP – 2153. [DOI] [PubMed] [Google Scholar]
- 196.Hartmann J.T., Lipp H.-P. Camptothecin and podophyllotoxin derivatives. Drug Saf. 2006;29:209–230. doi: 10.2165/00002018-200629030-00005. [DOI] [PubMed] [Google Scholar]
- 197.Smith N.F., Figg W.D., Sparreboom A. Pharmacogenetics of irinotecan metabolism and transport: an update. Toxicol. Vitro. 2006;20:163–175. doi: 10.1016/j.tiv.2005.06.045. [DOI] [PubMed] [Google Scholar]
- 198.De Coupade C., Fittipaldi A., Chagnas V., Michel M., Carlier S., Tasciotti E., Darmon A., Ravel D., Kearsey J., Giacca M., Cailler F. Novel human-derived cell-penetrating peptides for specific subcellular delivery of therapeutic biomolecules. Biochem. J. 2005;390:407–418. doi: 10.1042/BJ20050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Senter P.D., Beam K.S., Mixan B., Wahl A.F. Identification and activities of human carboxylesterases for the activation of CPT-11, a clinically approved anticancer drug. Bioconjugate Chem. 2001;12:1074–1080. doi: 10.1021/bc0155420. [DOI] [PubMed] [Google Scholar]
- 200.Coriat R., Faivre S.J., Mir O., Dreyer C., Ropert S., Bouattour M., Desjardins R., Goldwasser F., Raymond E. Pharmacokinetics and safety of DTS-108, a human oligopeptide bound to SN-38 with an esterase-sensitive cross-linker in patients with advanced malignancies: a Phase I study. Int. J. Nanomed. 2016;11:6207–6216. doi: 10.2147/IJN.S110274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Takasuna K., Hagiwara T., Watanabe K., Onose S., Yoshida S., Kumazawa E., Nagai E., Kamataki T. Optimal antidiarrhea treatment for antitumor agent irinotecan hydrochloride (CPT-11)-induced delayed diarrhea. Cancer Chemother. Pharmacol. 2006;58:494–503. doi: 10.1007/s00280-006-0187-8. [DOI] [PubMed] [Google Scholar]
- 202.Malonis R.J., Lai J.R., Vergnolle O. Peptide-based vaccines: current progress and future challenges. Chem. Rev. 2020;120:3210–3229. doi: 10.1021/acs.chemrev.9b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Powell A.M., Black M.M. Epitope spreading: protection from pathogens, but propagation of autoimmunity? Clin. Exp. Dermatol. 2001;26:427–433. doi: 10.1046/j.1365-2230.2001.00852.x. [DOI] [PubMed] [Google Scholar]
- 204.Inderberg-Suso E.-M., Trachsel S., Lislerud K., Rasmussen A.-M., Gaudernack G. Widespread CD4+ T-cell reactivity to novel hTERT epitopes following vaccination of cancer patients with a single hTERT peptide GV1001. OncoImmunology. 2012;1:670–686. doi: 10.4161/onci.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266 doi: 10.1126/science.7605428. 2011 LP – 2015. [DOI] [PubMed] [Google Scholar]
- 206.Counter C.M., Meyerson M., Eaton E.N., Ellisen L.W., Caddle S.D., Haber D.A., Weinberg R.A. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 207.Kirkpatrick K.L., Mokbel K. The significance of human telomerase reverse transcriptase (hTERT) in cancer. Eur. J. Surg. Oncol. 2001;27:754–760. doi: 10.1053/EJSO.2001.1151. [DOI] [PubMed] [Google Scholar]
- 208.Lilleby W., Gaudernack G., Brunsvig P.F., Vlatkovic L., Schulz M., Mills K., Hole K.H., Inderberg E.M. Phase I/IIa clinical trial of a novel hTERT peptide vaccine in men with metastatic hormone-naive prostate cancer, Cancer Immunology. Immunotherapy. 2017;66:891–901. doi: 10.1007/s00262-017-1994-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hunger R.E., Kernland Lang K., Markowski C.J., Trachsel S., Møller M., Eriksen J.A., Rasmussen A.-M., Braathen L.R., Gaudernack G. Vaccination of patients with cutaneous melanoma with telomerase-specific peptides, Cancer Immunology. Immunotherapy. 2011;60:1553. doi: 10.1007/s00262-011-1061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brunsvig P.F., Aamdal S., Gjertsen M.K., Kvalheim G., Markowski-Grimsrud C.J., Sve I., Dyrhaug M., Trachsel S., Møller M., Eriksen J.A., Gaudernack G. Telomerase peptide vaccination: a phase I/II study in patients with non-small cell lung cancer, Cancer Immunology. Immunotherapy. 2006;55:1553–1564. doi: 10.1007/s00262-006-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kyte J.A., Gaudernack G., Dueland S., Trachsel S., Julsrud L., Aamdal S. Telomerase peptide vaccination combined with temozolomide: a clinical trial in stage IV melanoma patients. Clin. Cancer Res. 2011;17 doi: 10.1158/1078-0432.CCR-11-0184. 4568 LP – 4580. [DOI] [PubMed] [Google Scholar]
- 212.Bernhardt S.L., Gjertsen M.K., Trachsel S., Møller M., Eriksen J.A., Meo M., Buanes T., Gaudernack G. Telomerase peptide vaccination of patients with non-resectable pancreatic cancer: a dose escalating phase I/II study. Br. J. Cancer. 2006;95:1474–1482. doi: 10.1038/sj.bjc.6603437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kim H., Seo E.-H., Lee S.-H., Kim B.-J. The telomerase-derived anticancer peptide vaccine GV1001 as an extracellular heat shock protein-mediated cell-penetrating peptide. Int. J. Mol. Sci. 2016;17:2054. doi: 10.3390/ijms17122054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Deloche C., Lopez-Lazaro L., Mouz S., Perino J., Abadie C., Combette J.-M. XG-102 administered to healthy male volunteers as a single intravenous infusion: a randomized, double-blind, placebo-controlled, dose-escalating study. Pharmacology Research & Perspectives. 2014;2 doi: 10.1002/prp2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.El Zaoui I., Touchard E., Berdugo M., Abadie C., Kowalczuk L., Deloche C., Zhao M., Naud M.-C., Combette J.-M., Behar-Cohen F. Subconjunctival injection of XG-102, a c-jun N-terminal kinase inhibitor peptide, in the treatment of endotoxin-induced uveitis in rats. J. Ocul. Pharmacol. Therapeut. 2014;31:17–24. doi: 10.1089/jop.2014.0019. [DOI] [PubMed] [Google Scholar]
- 216.Touchard E., Omri S., Naud M.-C., Berdugo M., Deloche C., Abadie C., Jonet L., Jeanny J.-C., Crisanti P., de Kozak Y., Combette J.-M., Behar-Cohen F. A peptide inhibitor of c-jun N-terminal kinase for the treatment of endotoxin-induced uveitis. Investig. Ophthalmol. Vis. Sci. 2010;51:4683–4693. doi: 10.1167/iovs.09-4733. [DOI] [PubMed] [Google Scholar]
- 217.Cousins M.J., Pickthorn K., Huang S., Critchley L., Bell G. The safety and efficacy of KAI-1678— an inhibitor of epsilon protein kinase C (εPKC)—versus lidocaine and placebo for the treatment of postherpetic neuralgia: a crossover study design. Pain Med. 2013;14:533–540. doi: 10.1111/pme.12058. [DOI] [PubMed] [Google Scholar]
- 218.Warso M.A., Richards J.M., Mehta D., Christov K., Schaeffer C., Rae Bressler L., Yamada T., Majumdar D., Kennedy S.A., Beattie C.W., Das Gupta T.K. A first-in-class, first-in-human, phase I trial of p28, a non-HDM2-mediated peptide inhibitor of p53 ubiquitination in patients with advanced solid tumours. Br. J. Cancer. 2013;108:1061–1070. doi: 10.1038/bjc.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Asahara S., Takeda K., Yamao K., Maguchi H., Yamaue H. Phase I/II clinical trial using HLA-A24-restricted peptide vaccine derived from KIF20A for patients with advanced pancreatic cancer. J. Transl. Med. 2013;11:291. doi: 10.1186/1479-5876-11-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hill M.D., Martin R.H., Mikulis D., Wong J.H., Silver F.L., terBrugge K.G., Milot G., Clark W.M., MacDonald R.L., Kelly M.E., Boulton M., Fleetwood I., McDougall C., Gunnarsson T., Chow M., Lum C., Dodd R., Poublanc J., Krings T., Demchuk A.M., Goyal M., Anderson R., Bishop J., Garman D., Tymianski M. Safety and efficacy of NA-1 in patients with iatrogenic stroke after endovascular aneurysm repair (ENACT): a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2012;11:942–950. doi: 10.1016/S1474-4422(12)70225-9. [DOI] [PubMed] [Google Scholar]