Summary
Successful manufacture of specialized human cells requires process understanding of directed differentiation. Here, we apply high-dimensional Design of Experiments (HD-DoE) methodology to identify critical process parameters (CPPs) that govern neural territory patterning from pluripotency—the first stage toward specification of central nervous system (CNS) cell fates. Using computerized experimental design, 7 developmental signaling pathways were simultaneously perturbed in human pluripotent stem cell culture. Regionally specific genes spanning the anterior-posterior and dorsal-ventral axes of the developing embryo were measured after 3 days and mathematical models describing pathway control were developed using regression analysis. High-dimensional models revealed particular combinations of signaling inputs that induce expression profiles consistent with emerging CNS territories and defined CPPs for anterior and posterior neuroectoderm patterning. The results demonstrate the importance of combinatorial control during neural induction and challenge the use of generic neural induction strategies such as dual-SMAD inhibition, when seeking to specify particular lineages from pluripotency.
Subject areas: Neuroscience, Stem cells research, Developmental biology
Graphical abstract

Highlights
-
•
Mathematical models describe pathway control of neuroectoderm marker expression
-
•
Stage 1 media conditions optimized for regionally specific neuroectoderm in 3 days
-
•
Optimized conditions are more consistent than dual-SMADi across hiPSC lines
Neuroscience; Stem cells research; Developmental biology
Introduction
Degenerative diseases are often characterized by the loss of specific cell types. Although surgical replacement of lost cells is a logical and feasible potential treatment for such diseases, cell-based therapies demand access to large quantities of high quality pure populations of subtype-specific human cells. These can be generated by directing the differentiation of human pluripotent stem cells (hPSCs) through targeted activation or blockade of specific developmental signaling pathways. Because hPSCs can commit to any lineage and are inherently renewable, they are an ideal source of large quantities of human cells (Odorico et al., 2001).
Like pharmacological treatments, cell-based therapies must be held to rigorous standards that maximize efficacy and safety. Protocols for directing differentiation must therefore be decidedly robust, yielding high-purity subtype-specific cell populations. Currently, however, published protocols rarely surpass 50% purity in their final yield, require months of cell culture, and often exhibit high levels of batch variability (Arenas et al., 2015).
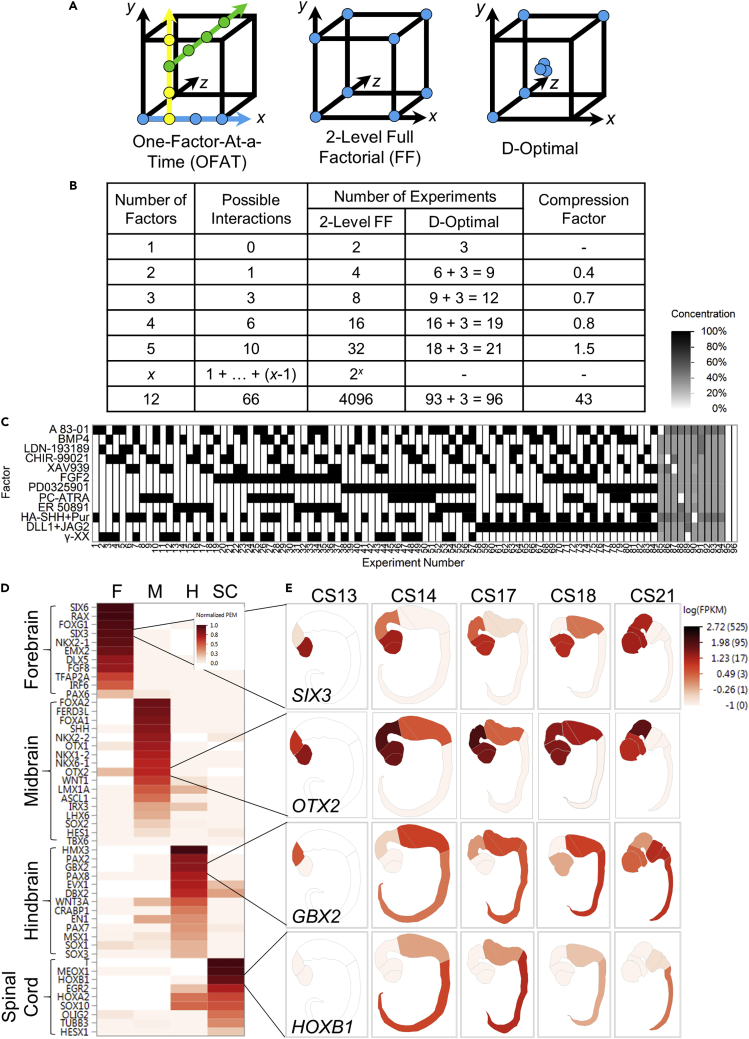
Improved differentiation efficiency can be achieved by more specifically targeting the precise developmental signals that induce cell fate conversions in the normal developing embryo (D'Amour et al., 2006; Tabar and Studer, 2014; Wichterle et al., 2002). However, determining how to effectively control commitment toward specific cell fates is enormously challenging because many signaling pathways operate simultaneously during development (Figure 1) and because those pathways interact heavily (Li and Elowitz, 2019). Although classical experimental designs, in which each pathway-modulating factor is tested independently (one-factor-at-a-time [OFAT], Figure 2A), provide essential understanding of pathway effects and mechanistic detail, they rarely assess interactions between more than two factors or pathways at a time. Since leveraging interactions is often paramount for highly efficient differentiation in vitro, improving protocol development demands a new approach.
Figure 1.
Proximity analysis of distinct signaling organizers impacting the emerging neuroectodermal territory
(A) Diagram of the Carnegie Stage (CS) 7 human embryo based on the 3D Atlas of Human Embryology (de Bakker et al., 2016).
(B) Regional expression of endogenous pathway activators and inhibitors in the gastrulating embryo. Expression patterns are based on mouse expression data from in situ hybridization and scRNA-Seq studies (Table S1). For all pathways except Retinoic Acid (RA), pathway activators were defined as protein products that have been shown to increase pathway activity, typically by binding and activating pathway receptors. Inhibitors were defined as endogenous protein products that have been shown to reduce pathway activity, typically by binding and inhibiting receptor activation or by binding and sequestering pathway activators. Because RA is not a protein but a metabolite, activators for this pathway include all-trans RA (ATRA) synthesis enzymes whereas inhibitors include enzymes that convert ATRA to other species. Regions marked by solid colors indicate areas of activator expression and regions marked with the outlined diamond pattern indicate areas of inhibitor expression. See also Table S1.
(C) Soluble factors used to modulate pathway activity in hPSC culture. The highest concentration tested for each factor is indicated in parentheses. Abbreviations: A, anterior; BMP, bone morphogenetic protein; D, dorsal; DLL1, delta-like protein 1; FGF, fibroblast growth factor ; L, left; γ-XX, γ-Secretase Inhibitor XX; JAG2, protein jagged-2; HA-SHH, high-activity sonic hedgehog; HH, hedgehog; P, posterior; PC-ATRA, photo-converted all-transretinoic acid; Pur, purmorphamine; R, right; RA, retinoic acid; V, ventral.
Figure 2.
High-Dimensional Design of Experiments (HD-DoE) exploration of the neural differentiation space
(A) Experimental design strategies for optimization. Circles indicate experimental conditions in a hypothetical 3-factor design space. A triple circle represents a single experimental condition, tested in triplicate. Each color represents a round of experimentation. The D-Optimal diagram is simplified to indicate that, at higher dimensions, only a fraction of the vertices are included in the design. (D-Optimal is not a practical choice for a 3-dimensional design, because the compression factor is < 1, as shown in B).
(B) The attributes of 2-level full factorial and D-optimal designs as the number of factors increases. The number of runs required for D-Optimal designs were determined by creating designs with the indicated number of variables in MODDE software; “+3” indicates the inclusion of a replicate center point.
(C) D-Optimal interaction screening design used to simultaneously test for effects of and interactions between 12 soluble pathway-modulating factors in 96 experiments. The design is constrained to exclude regions of the design space where agonist/antagonist pairs are both ≥50% of their maximum concentration. See also Table S2.
(D) Marker genes selected to assess neural patterning from pluripotency and their specificity within the human CNS at CS14. Specificity was calculated using Preferential Expression Measure (PEM, Kryuchkova-Mostacci and Robinson-Rechavi, 2017) and normalized to the highest PEM calculated among measured genes.
(E) Expression of selected genes across human early CNS development mapped onto corresponding diagrams adapted from the 3D Atlas of Human Embryology (de Bakker et al., 2016). Because no reconstruction is available for CS14, expression data for that stage is mapped onto the CS15 embryo. For CS13, data were available for forebrain and midbrain only.
We have previously developed a multi-stage small-molecule induction protocol for differentiation of hPSCs to pancreatic insulin-producing cells, utilizing a statistical methodology known as Design of Experiments (DoE), applied at high dimensions (HD-DoE, Bukys et al., 2020). HD-DoE utilizes deliberate experimental design and multivariate regression analysis to simultaneously vary a large number of pathway-modulating factors and assess their effects on a large number of marker genes. Resulting polynomial models describing pathway control of lineage-specific genes then identify factor combinations that reliably produce expression of desired markers.
Considering the broader utility of the HD-DoE approach and high demand for specific neuroectoderm-derived cell types, this study aims to develop unbiased Stage 1 protocols that begin to direct hPSCs toward regionally specific human neurons. Current neural induction protocols generally begin with a 5–7 days’ dual-SMAD inhibition treatment to induce neuroectoderm identity (Chambers et al., 2009; Galiakberova and Dashinimaev, 2020). However, in the developing mouse embryo, anterior/posterior (A/P) patterning occurs before neural induction. ATAC-seq analysis revealed differences in chromatin accessibility between anterior and posterior neural progenitors (Metzis et al., 2018) and single cell RNA sequencing (scRNA-Seq) between gastrulation and somitogenesis (E6.5 – E8.5) revealed an early split of neuroectoderm into distinct rostral and caudal populations (Pijuan-Sala et al., 2019). In fact, general neural markers Pax6 and Sox1 are not detected in developing neuroectoderm until after the onset of somitogenesis, when A/P-patterning has already been established (Callaerts et al., 1997; Wood and Episkopou, 1999).
Here, we demonstrate the use of HD-DoE to identify Stage 1 media conditions that specifically direct cells toward anterior and posterior neuroectoderm fates across hPSC lines in only 3 days. To determine initial combinatorial inputs underlying territory control in the developing neuroectoderm, we simultaneously varied 12 soluble pathway-modulating factors that control the following classical developmental signaling pathways: Activin/Nodal (SMAD2/3), bone morphogenetic protein (BMP; SMAD1/5/8), WNT, fibroblast growth factor (FGF), retinoic acid (RA), hedgehog (HH), and Notch. We assessed pathway effects on hPSC differentiation by measuring 53 lineage-specific marker genes after 3 days of treatment with DoE-generated combinations and concentrations of pathway-modulating factors. Models of expression for regionally specific neuroectoderm genes SIX3 and GBX2 were then optimized to develop specific 3-day anterior and posterior neuroectoderm differentiation protocols. The HD-DoE-derived protocols specifically directed differentiation and did so more consistently than a general neural induction strategy in all 4 human induced pluripotent stem cell (hiPSC) lines tested.
In addition to identifying robust protocols, HD-DoE-derived mathematical models comprehensively describe control of marker gene expression by tested factors, providing the opportunity to more generally explore signaling control in the human neuroectodermal fate space from pluripotency. Models estimate main effects and interactions between tested factors on each gene, provide response surface modeling for predictive analyses within the design space, and facilitate identification of phenotype-determining Critical Process Parameters (CPPs)—the factors that are most important for controlling the differentiation process—a necessary step toward industrial manufacture of specialized human cells for therapy. The results of the study were consistent with decades of developmental research, identifying BMP inhibition as critical to inducing neuroectoderm and WNT, FGF, and RA control as essential for A/P-patterning. In contrast to previous studies, our protocols did not require SMAD2/3 pathway inhibition for neuroectoderm induction when other pathways were controlled.
Results
Proximity analysis of distinct signaling organizers impacting the emerging neuroectodermal territory
To identify potentially critical signaling pathways for neuroectoderm formation and effectively control human cell fate from pluripotency in vitro, we first looked to the anatomical structure of the early human embryo. Human embryos at various stages of development have been sectioned, scanned, and digitally reconstructed in the 3D Atlas of Human Embryology (de Bakker et al., 2016). The earliest available reconstruction is Carnegie Stage 7 (CS7), which occurs between 15 and 17 days after conception (Figure 1A). Because hPSCs grown in conventional culture conditions are in a state of primed pluripotency, similar to post-implantation epiblast cells (Nakamura et al., 2016), we assume the epiblast cells at this stage are similar to hPSCs. Because of that, the signaling pathways that are regulated in this region are likely to be important for directing cell fate from pluripotency (Figure 1B).
At CS7, the embryo is implanted and the primitive node has appeared in the center of the epiblast layer. The epiblast is surrounded on the ventral side by the hypoblast and on the dorsal side by the amniotic cavity, which is bounded by amniotic ectoderm cells lining trophoblast-derived extraembryonic tissue (Figure 1A, Shahbazi, 2020). As gastrulation proceeds, the primitive streak forms in the posterior epiblast extending toward the node as cells migrate through the streak to the ventral side of the epiblast, creating the three germ layers: endoderm (ventral-most), mesoderm, and ectoderm (dorsal-most).
The early human embryo differs anatomically from the equivalent stage mouse embryo (∼E6.75): the mouse embryo—often called the egg cylinder—is a cup shape whereas the human embryo is a flat disc. In the egg cylinder, the hypoblast-derived visceral endoderm surrounds the entire embryo, directly lining the epiblast on its ventral side, whereas the trophectodoerm-derived extraembryonic ectoderm is positioned on the dorsal side of the epiblast, contacting it directly only along the perimeter (Weinberger et al., 2016).
Fate maps from mouse embryos indicate that epiblast cells anterior to the node become ectoderm whereas cells posterior and lateral to the node primarily become endoderm and mesoderm, respectively (Lawson et al., 1991). Also lateral to the node are a mix of ectodermal and mesodermal fated cells, which have been defined as neuromesodermal progenitors (NMPs) and become either paraxial mesoderm or spinal cord (Tzouanacou et al., 2009). In this way, the future central nervous system (CNS; neuroectoderm) arises from anterior and lateral epiblast cells, as indicated in Figure 1A.
During CNS cell specification, three major processes occur overlapping in time and space: germ layer specification, ectoderm patterning, and neural plate patterning. Based mainly on rodent investigations, and other non-human vertebrate organisms, the signaling dynamics of these processes are well described and predicted to operate similarly in the human anatomical and genetic context. We focused our study on 7 signaling pathways known to be involved in early fate-defining processes (Figures 1B and 1C). Germ layer specification is controlled primarily by SMAD2/3, SMAD1/5/8, Wnt, and FGF signaling in the mouse embryo, where SMAD2/3 and SMAD1/5/8 signaling are required for mesendoderm formation, and FGF and WNT signaling are involved in mesoderm induction (Kiecker et al., 2016). Ectoderm patterning is controlled primarily by SMAD1/5/8, FGF, and WNT signals, all three of which interact to direct neural plate, neural crest, and non-neural ectoderm fates (Patthey and Gunhaga, 2014). Anterior-posterior patterning of the neural plate is attributed to WNT, RA, and FGF whereas dorsal-ventral patterning is directed by SMAD1/5/8 and HH signaling (Ozair et al., 2013; Tuazon and Mullins, 2015). We also considered Notch signaling, as it has been implicated in neuroectodermal commitment (Souilhol et al., 2015).
Examining the spatial distribution of signaling activators and inhibitors provides an anatomical representation of patterning inputs. Signaling patterns have been well described and illustrated in the gastrulating mouse embryo (Bardot and Hadjantonakis, 2020; Guzzetta et al., 2020; Kam et al., 2012; Przemeck et al., 2003; Tam and Loebel, 2007). However, the gastrulating human embryo differs anatomically from that of the mouse and gene expression data is not available for human embryos at this stage (CS7). To prioritize plausible critical combinatorial inputs for neuroectodermal patterning, we aimed to visualize signaling control from human pluripotency. To do so, we compiled available expression data from mouse gastrulation (TS9) and applied the resulting expression patterns to the previously described reconstruction of the CS7 human embryo (Figure 1B). Mouse expression data included in situ images of regionally-restricted developmental signaling pathway modulators cross-referenced with single-cell RNA sequencing data (scRNA-Seq, E6.75 unless otherwise noted, Pijuan-Sala et al., 2019) (Table S1, see legend for detailed analysis). Expression in the mouse visceral endoderm was overlaid on the human hypoblast, as it is hypoblast-derived and lines the ventral side of the embryo. Although there is no human equivalent of the mouse extraembryonic ectoderm, expression detected in this tissue was overlaid on the amnion layer to represent the trophoblast-derived extraembryonic tissue that surrounds the amniotic ectoderm on the dorsal side of the embryo and contacts the epiblast around its perimeter.
SMAD2/3-activating ligands (Nodal, Tgfb1, Gdf1) are expressed in the epiblast with a posterior bias and in the posterior region of the visceral endoderm (VE, corresponding to the human hypoblast). Secreted inhibitors are expressed in the anterior VE (AVE; Cer1, Lefty1) and throughout the epiblast (Lefty1/2), with high levels in the primitive streak.
SMAD1/5/8-activating ligands are expressed in posterior VE (Bmp2), posterior epiblast (Bmp2), around the node (Bmp7) and in the extraembryonic ectoderm (Bmp4, Bmp8b). Secreted inhibitors are expressed in the epiblast (Fst), with high levels in the primitive streak, and in AVE (Chrd, Nog). Intracellular inhibitors that target both SMAD2/3 and SMAD1/5/8 signaling pathways (Bambi, Smad7, Smad6) are also expressed in the epiblast.
WNT ligands are expressed in posterior regions of both epiblast and VE (Wnt3), and throughout the extraembryonic ectoderm (Wnt6, Wnt7b). Secreted inhibitors are expressed throughout the epiblast (Frzb) and in the AVE (Sfrp1, Sfrp2, Dkk1).
FGF ligands are expressed throughout the epiblast (Fgf5, 15), with some concentrated posteriorly (Fgf3, 4, 8, 10, 17), and in VE (Fgf5; Fgf8 restricted to AVE). Intracellular inhibitors that specifically regulate MAPK signaling are expressed throughout the epiblast (Il17rd, Spred1, Spred2, Spry4; Spry2 with anterior bias).
Aldh1a2, the enzyme primarily responsible for producing all-trans-retinoic acid (ATRA) in vivo, is detected in the epiblast starting around E7.0, as is its homolog, Aldh1a3, which performs the same function (Rhinn and Dollé, 2012). Meanwhile, Cyp26a1, which encodes an enzyme that converts ATRA to other RA species, is widely expressed in extraembryonic tissues, anterior primitive streak, and emerging mesoderm, but is notably absent from epiblast cells (Fujii et al., 1997; Pijuan-Sala et al., 2019).
Indian hedgehog (Ihh) is expressed in VE, but no inhibitors are strongly expressed at this stage.
Notch ligands (Dll1, Jag1, Jag2) are restricted to the epiblast layer whereas a secreted inhibitor (Dll3) is weakly expressed in extraembryonic ectoderm and epiblast, with higher levels in nascent mesoderm.
To replicate developmental signaling in vitro, we identified small molecule and recombinant protein pathway modulators that have previously been used to control pathway activity in hPSC culture (Figure 1C). We opted to test the 12 pathway modulators most strongly implicated in neural patterning from pluripotency. The maximum concentration for each factor was selected based on manufacturer provided ED50/IC50 values, typically in nanomolar ranges, and on previous HD-DoE experiments such that factors elicited a measurable response, but did not induce signs of toxicity.
High-Dimensional Design of Experiments (HD-DoE) exploration of the neural differentiation space
Optimization is commonly performed by sequential evaluation of candidate process parameters in a reductionist fashion (OFAT, Figure 2A). However, this approach yields only a small amount of information about how the system responds to perturbations and, in complex systems where factors interact, different starting conditions may result in different outcomes. Consequently, an OFAT approach is unlikely to identify a true optimum.
Full factorial (FF) designs are equipped to detect interactions between factors. For example, in a 2-level FF design, all possible combinations of factors are tested at 2 levels (low and high, Figure 2A). Two-level FF designs provide complete coverage of the design space and directly estimate all possible factor interactions, which can be highly useful for examining systems in fewer than 5 dimensions. However, as the number of factors increases, the number of experimental runs quickly becomes impractical and inefficient, growing exponentially with each additional dimension (Figure 2B).
To explore the neural differentiation space with high dimensionality in a manageable number of experiments, we employ a D-Optimal design approach (Figure 2A). D-optimal designs are fractional factorial designs, comprising a subset of full factorial runs that are computationally selected to sample the design space and maximize information about the system (Eriksson et al., 2000). In addition, because we do not want to consider conditions that include high levels of agonist/antagonist pairs (for example, both BMP4 and LDN-193189), we are exploring an irregular design space, which D-optimal designs are well suited to assess. DoE experiments also include center points tested in triplicate, which provide an estimate of pure error (i.e., reproducibility) and monitor response curvature (i.e., higher order factor effects). Multivariate regression analysis produces predictive mathematical models of response behavior within the design space. These models can then be interrogated to identify conditions that produce desired gene expression profiles and provide CPP analysis. Practically, a D-Optimal design can examine effects of 12 factors in only 96 experimental runs, compressing the corresponding full factorial design by a factor of 43, while testing for up to 66 factor interactions (Figure 2B).
We created a D-optimal interaction screening design, testing 12 factors at low (0) and high (maximum concentration) levels (as exemplified, Figures 1C and 2C; Table S2). Experiments numbered 1–84 represent vertices in the 12-dimensional design space. Experiments 85–94 represent various center-points, including one in triplicate (92, 93, 94). Experiments 95 and 96 provide a second set of replicates and a baseline measure of gene expression in basal media.
To assess regional identity of differentiating hPSCs after exposure to HD-DoE-defined experimental conditions, we selected genes known to mark specific regions and cell types in the developing vertebrate embryo (Figure 2D). Because expression data is not available for gastrulating human embryos (CS7-9), markers were selected based on their early regional expression in the gastrulating mouse embryo at corresponding stages (TS9-12, Mitiku and Baker, 2007). To ensure that the markers are also specifically expressed in developing human CNS, we examined their specificity within the developing CNS at the earliest available time point (Lindsay et al., 2016, CS13 – CS21; Figures 2D and 2E). Of the marker genes selected, those most specific for forebrain in the CS14 human embryo included SIX6, RAX, FOXG1 and SIX3. Midbrain-specific genes included FOXA2, FERD3L, FOXA1, and SHH. In certain cases, genes were expressed across multiple territories: although OTX2 is categorized as a midbrain-specific gene, it is also strongly expressed in forebrain. The genes most specific for hindbrain include HMX3, PAX2, GBX2, and PAX8, while T, MEOX1, HOXB1, and EGR2 were most specific for spinal cord. Pan-neural markers, like PAX6 and SOX1, whose expression has been targeted to develop widely used neural induction protocols (i.e., dual-SMAD inhibition, Chambers et al., 2009), are not regionally specific in the developing human CNS, but are expressed in all regions.
HD-DoE generates predictive models of marker gene control
HD-DoE-defined experimental conditions (Figure 2C) were applied to hPSCs with daily media exchange for 3 days. We measured marker gene (Figure 2D) expression after 3 days of treatment because, in dual-SMADi neural induction, general neuroectoderm marker SOX1 is detected as early as 3 days from pluripotency (Chambers et al., 2009). Before modeling, maker gene expression was normalized such that a value of 10,000 was equivalent to the average expression level of endogenous control genes in each sample (Figure 3A).
Figure 3.
HD-DoE generates predictive models of marker gene control
(A) Distribution of marker gene expression across experiments after 3 days exposure of H9 hPSCs to perturbation matrix (Figure 2C), with daily media exchange. Boxplots show median/IQR and outliers (>Q3 + 1.5∗IQR) are show as solid circles. Expression is normalized to endogenous genes (GAPDH, TBP, YWHAZ) such that the average expression of endogenous genes for a given treatment correspond to 10,000 and 0 indicates that the gene was not detected. The top panel is plotted on a log scale. See also Figure S1 and Table S2.
(B) Strength and directionality of linear relationships between factor concentration and marker gene expression. Tilde (∼) indicates data are transformed. See also Table S3, Figures S2, and S3.
(C) Measures of fit for partial least-squares regression (PLSR) models of gene control. R2 is the coefficient of determination, a measure of the variance in the response explained by the model, where R2 = 0.5 is a model with low significance. Q2 is expressed in the same units as R2, but represents the amount of variance predicted by the model, as calculated by cross-validation. Q2> 0.1 is a useful model and Q2> 0.5 is a good model. The difference between R2 and Q2 should be < 0.3 for a good model (reference line indicated). R2 is shown on a power of two scale for visual clarity. See also Table S4.
(D) Scaled and centered coefficients for the PLSR model of HOXA2∼ expression (=HOXA20.5). Error bars are 95% CIs. See also Tables S3, S5, and S6.
(E) Scaled and centered coefficients of the PLSR model of PAX6∼ expression (=PAX60.3). Error bars are 95% confidence intervals. See also Tables S3, S5, and S6. A83, A 83-01; BMP, BMP4; CHI, CHIR-99021; DLL, DLL1+JAG2; ER5, ER 50891; FGF, FGF2; LDN, LDN-193189; PD, PD0325901; RA, PC-ATRA; SHH, HA-SHH+Pur; XAV, XAV939.
Several marker genes were detected in all experimental conditions, with wide ranges of expression (Figure 3A). Of the marker genes measured, SOX2 was expressed at the highest level across the design space, with median (Mdn) normalized expression of 14152 (IQR 9546-22501) and was detected in all experimental conditions. CRABP1 (Mdn 6199, IQR 4344-9038), OTX2 (Mdn 3190, IQR 1875-7234), and TUBB3 (Mdn 2570, IQR 1713-3548) also tended to be expressed at high levels and were detected in all samples, whereas HES1 (Mdn 252, IQR 190-344), SOX3 (Mdn 160, IQR 55-269), and OLIG2 (Mdn 106, IQR 59-170) were detected at lower levels in all experimental conditions. This expression profile is generally consistent with differentiation toward ectoderm lineages because these genes are broadly expressed in the early mouse embryo and Sox2, Otx2, Tubb3 and Sox3 are specifically expressed at high levels in ectoderm-derived tissues by E8.0. Mesendoderm marker TBX6 was detected in all but one experiment (#13), at low levels (Mdn 62, IQR 46-75), indicating only limited off-target differentiation in the experiment.
Genes whose orthologues exhibit little or no expression in early neuroectoderm (e.g. E8.0 mouse embryo, Pijuan-Sala et al., 2019) are detected only at very low levels in a few experimental conditions. EMX2, EN1, FERD3L, KRT5, NKX2-1, and VSX2 are expressed later in development and were not detected at high levels in any experiments. Despite that, replicate reproducibility for these genes was high, and useful information about their pathway control was obtained. DBX2, LHX6, NKX2-2, and PROP1 were detected at low levels with low replicate reproducibility and were therefore excluded from further analyses.
The remaining marker genes ranged from undetectable to moderate or high levels across experiments. Genes with the widest ranges of expression include TFAP2A (range 13986), DLX5 (10140), SIX3 (9253), HOXB1 (8925), PAX6 (8772), and GBX2 (5942). Wide ranges of expression can indicate a high level of control by tested factors and, therefore, often produce highly useful regression models. Before further analysis, the distribution of each marker gene was independently analyzed and transformed, if necessary (indicated by ∼; see STAR Methods, Table S3 and Figure S1).
To assess the overall factor effects on marker gene expression within the 12-dimensional design space, we examined the directionality and strength of linear relationships between individual factors and responses (Figure 3B). The strongest positive correlation coefficients were detected between PC-ATRA and HOXA2∼ (r = 0.93, 95% CI 0.89-0.95), HOXB1∼ (r = 0.82, 95% CI 0.74-0.88), and MEOX1∼ (r = 0.87, 95% CI 0.81-0.91), all of which are known RA responsive genes (Ishikawa and Ito, 2009; Kennedy et al., 2009; Ogura and Evans, 1995). The strongest negative correlation coefficient was detected between MEK inhibitor PD0325901 and GBX2∼ (r = −0.84, 95% CI -0.89 to -0.76), a known FGF responsive gene (Lin et al., 2005). Genes tended to have opposite responses to pathway agonists and antagonists (Figure S2A).
FGF and RA pathway modulators were most highly correlated with expression of marker genes (Figures S2B and S3). PD0325901 had the strongest overall effect on marker expression, with a median absolute correlation coefficient of 0.29 (IQR 0.16-0.44). PC-ATRA and FGF2 had median absolute correlation coefficient equaling 0.28 (IQR 0.14-0.36) and 0.21 (IQR 0.17-0.35), respectively. Furthermore, FGF2 and PC-ATRA together are strongly correlated with expression of a large number of marker genes, indicating strong additive effects between the two pathways (Figure S3).
SMAD2/3, HH, and Notch modulators were weakly correlated with marker gene expression, indicating that they had less of an overall effect on ectoderm differentiation from pluripotency. For instance, the ALK4/5/7 inhibitor (A 83-01) had a median absolute correlation of only 0.05 (IQR 0.03-0.08). Because hPSC maintenance media contained SMAD2/3 activators (Activin A or TGFβ1), it is possible that removal of the activating signal may have reduced pathway activity, such that additional inhibition during differentiation had little effect. HH activators (HA-SHH+Pur) and Notch activators (DLL1+JAG2) also had weak correlations with marker expression, both having median absolute correlations of 0.05 (IQR 0.02-0.11, 0.02-0.10, respectively). Although a correlation coefficient close to zero does not necessarily indicate that the factor has no effect on expression of a gene, it does indicate that the linear relationship is weak.
To detect individual factor and interaction effects on expression of marker genes and to predict expression across the design space, we used partial least squares regression (PLSR) modeling. To assess model strength, we consider two metrics of fit: the coefficient of determination (R2), which represents the amount of variance in the data explained by the model; and Q2, a similar calculation that represents the variation in the data predicted from the model by cross-validation (Figure 3C). High metrics of fit indicate that variation in the factors tested account for a large proportion of the variance in the data, suggesting that expression was well controlled by tested pathway-modulating factors. Variation that is not explained by the models may be because of the activity of uncontrolled pathways and models with low metrics of fit may benefit from further experimentation.
The strongest HD-DoE-generated model describes control of HOXA2∼ expression from pluripotency, explaining 95% of the observed variance and predicting 84% (Figure 3D, Tables S4, and S5). Main factor effects for each gene can be interpreted and compared by examining factor-specific regression coefficients, which are scaled and centered to correspond to the change in the response when the factor concentration increases from half-maximal to maximal and all other factors are half-maximal. For example, when all factors are present at their half-maximal concentrations, HOXA2∼ (=HOXA20.5, Table S3) equals 2.7 (95% CI 2.5-3.0), corresponding to normalized HOXA2 expression of 7.4 (95% CI 6.0-9.0). By far the largest predictor of HOXA2∼ was the concentration of PC-ATRA, with a regression coefficient of 2.22 ± 0.26 (p< 0.001). Thus, when PC-ATRA increases from its half-maximal concentration (1 μM) to its maximal concentration (2 μM), HOXA2∼ increases by 2.2 ± 0.26, resulting in a normalized HOXA2 expression value of 23 (95% CI 20-28) and amounting to an approximately 3-fold increase in normalized HOXA2 gene expression.
The model of PAX6∼ expression is also highly predictive (Figure 3E; Tables S4 and S5), with positive regression coefficients for LDN-193189 (1.02 ± 0.26, p< 0.001), PC-ATRA (1.00 ± 0.28, p< 0.001), PD0325901 (0.94 ± 0.27, p< 0.001) and negative coefficients for BMP4 (-0.52 ± 0.27, p< 0.001) and CHIR-99021 (-0.41 ± 0.27, p = 0.004). Because PAX6 was one of the first neural markers used to develop neural induction protocols (Chambers et al., 2009) it is not surprising that the pathways tested here—those known to be involved in neural induction—largely explain the control of PAX6 expression from pluripotency.
Overall, the experiment yielded 43 significant regression models containing a total of 429 significant model terms (160 main factor effects, 240 interaction effects, and 29 triple interaction effects; p< 0.05; Tables S5 and S6). The pathway interaction space was rich, with models averaging 3.7 main, 5.6 interaction, and 0.7 triple interaction terms detected per gene (p< 0.05). Including all regression terms, models ranged from 13 to 41 total terms, with a mean of 26.4 total terms per model.
Interpreting HD-DoE-derived PLSR models of regionally specific marker gene control
We focused the neural patterning analysis on 4 genes for which there is clear evidence of regional neural specificity in both human and mouse embryos, and which produced highly predictive PLSR models: SIX3∼ (R2 0.85, Q2 0.68), OTX2 (R2 0.86, Q2 0.67), GBX2∼ (R2 0.88, Q2 0.79), and HOXB1∼ (R2 0.83, Q2 0.60) (Figure 3C). By E8.0, mouse neuroectoderm has split into anterior and posterior populations, where Six3 (p = 0.03) and Otx2 (p<0.001) are more strongly expressed in anterior neuroectoderm than in other tissues. At the same time, Gbx2 and Hoxb1 are strongly expressed in posterior neuroectoderm, but not anterior neuroectoderm. Importantly, both posterior neuroectoderm markers are also expressed in mesodermal populations at this stage. To assess possible off-target mesodermal cell fates, we monitor expression of T, which is strongly expressed in mesoderm populations, but not in neuroectoderm at E8.0 (Pijuan-Sala et al., 2019).
SIX3 and OTX2 expression was confirmed by immunostaining for select HD-DoE-defined experimental conditions (Figures 4A–4D).
Figure 4.
Interpreting HD-DoE-derived PLSR models of gene control
(A and B) Expression of SIX3 (A) and OTX2 (B) across HD-DoE-defined experimental conditions (Figure 2C). Green points represent conditions that are validated by immunostaining in C and (D) Blue points represent replicates. See also Table S2.
(C and D) Representative immunofluorescence images of SIX3 (C) and OTX2 (D) after 3 days exposure to select experimental conditions. Nuclei are stained with DAPI. Scale bars = 250 μm.
(E) Coefficients of terms included in the PLSR model of SIX3∼ expression control. Regression coefficients are scaled and centered, representing the change in the response value when factors are varied from their half-maximal to maximal concentrations and all other factors are also half-maximal. Asterisks (∗) denote interaction terms between two or more factors. Error bars are 95% confidence intervals. See also Table S5, Figures S5, and S6.
(F) Interpretation of main factor effects on SIX3 expression, based on the PLSR model shown in (E). Green lines indicate a positive regression coefficient, purple lines indicate a negative regression coefficient. Arrows indicate activation and bar-headed lines indicate inhibition or repression. Solid lines, p< 0.05; dotted lines, p≥ 0.05. See also Figure S4.
(G) Interaction plots demonstrating effects of two significant two-factor interactions on SIX3 expression. See also Figures S5 and S6.
The main factor effects detected in the SIX3∼ model were consistent with the default model of differentiation, which postulates that the lack of signaling pathway activation leads to a default anterior neural fate (reviewed in Ozair et al., 2013, Figure 4F). Effects can be compared by examining their scaled and centered regression coefficients, where the value of the coefficient indicates the change in expression when the factor was increased from its half-maximal to maximal concentration, while other factors are half-maximal. RA, WNT, and FGF pathway-modulating factors were the strongest predictors of SIX3 expression (Figure 4E and Table S5). Increasing pathway activators PC-ATRA and CHIR-99021 from half-maximal to maximal concentrations reduced SIX3∼ (= SIX30.5) by 7.2 ± 1.0 (p< 0.001) and 5.9 ± 0.9 (p< 0.001) respectively. In contrast, doubling the concentration of MEK inhibitor PD0325901 increased SIX3∼ by 4.2 ± 1.0 (p< 0.001; Figure 4E). In addition, FGF2 (p< 0.001) and BMP4 (p = 0.005) reduced SIX3 expression, while LDN-193189 increased expression (p = 0.036).
Other regionally-specific neural patterning genes were also consistent with the default paradigm (Figure S4). Expression of a second anterior neuroectoderm marker, OTX2, was reduced with increasing BMP (p = 0.008), RA (p< 0.001), and FGF (p< 0.001) signaling and increased with higher PD0325901 concentration (p< 0.001, Figure S4). Conversely, the posterior neuroectoderm marker GBX2∼ had positive regression coefficients for FGF2 (p< 0.001), CHIR-99021 (p = 0.007), and LDN-193189 (p = 0.002), indicating that FGF and WNT signaling activation increased GBX2 expression. A second posterior marker, HOXB1∼, had a positive regression coefficient for PC-ATRA (p< 0.001), indicating that it was upregulated by increased RA signaling.
Additional PLSR regression terms account for interactions between factors—when the concentration of one factor influences the effect of another (Figure 4G). For instance, the model of SIX3∼ includes 7 significant interaction terms (p< 0.05) and 8 additional interaction terms that improve Q2 (Figures S5 and S6). The largest interaction term detected for SIX3∼ was between PC-ATRA and PD0325901 (Coeff.-4.4, 95% CI -6.4 to -2.5). As demonstrated in the interaction plot, in the absence of PC-ATRA (RA low), increasing PD0325901 concentration increases SIX3∼. However, when PC-ATRA is present at its maximum concentration (RA high) during differentiation, increasing PD0325901 has the opposite effect on SIX3∼. We also detected a synergistic interaction between LDN-193189 and PD0325901, such that SIX3∼ is responsive to increasing levels of LDN-193189 when PD0325901 is also present (PD high), but not when PD0325901 is absent (PD low). In addition, a triple interaction was detected between CHIR-99021, PC-ATRA, and PD0325901 (p = 0.006), such that the positive effect of PD0325901 on SIX3∼ was only observed when CHIR-99021 and PC-ATRA were omitted from media.
HD-DoE-derived PLSR models identify media conditions that direct differentiation specifically toward anterior and posterior neuroectoderm with built-in CPP analysis
High-dimensional predictive models of gene expression control allow identification of factor settings that optimize marker expression within the design space. Using PLSR models of SIX3∼ and GBX2∼ we identified factor conditions predicted to achieve high levels of SIX3 and GBX2 expression just 3 days from pluripotency (Figures 5A and S6).
Figure 5.
HD-DoE-derived PLSR models identify media conditions that direct differentiation specifically toward anterior and posterior neuroectoderm with built-in CPP analysis
(A) Pathway-modulating factor concentrations for a general neural induction strategy (dual-SMAD inhibition, dual-SMADi) compared to those optimized for early anterior (SIX3) and posterior (GBX2) neuroectoderm marker expression within the tested design space using PLSR models. Fill color of markers representing factor concentrations for optimized conditions correspond to their factor contribution (FC), a metric describing the predicted effect of altering the factor concentration by ±5%. A high FC indicates that a small change in factor concentration is likely to have a large effect on the response variable of interest. Maximum factor concentrations are defined in Figure 1C. See also Figures S7–S9.
(B) Developmental pathway control of regionally optimized neuroectoderm differentiation conditions compared to a dual-SMADi strategy.
(C) Representative images of marker expression in NCRM-1 hiPSCs at the start of differentiation (Day 0) and after 3 days of exposure to dual-SMADi or regionally-optimized neuroectoderm conditions. Scale bars = 250 μm. hiPSCs, human induced pluripotent stem cells; Opt, optimized; PLSR, partial least-squares regression. See Figure 1C for factor and pathway abbreviations. See also Figures S10A and S10B.
In addition to identifying differentiation recipes, optimization analysis includes factor contribution (FC) values for each recipe component, where high FC indicates that a small change in concentration will have a large effect on the desired result. For instance, high FCs for CHIR-99021 (26.7), PC-ATRA (20.6), and PD0325901 (16.5) in SIX3-optimized conditions identify these factors as CPPs, which must be well controlled in order to achieve maximal SIX3 expression. In other words, a small change in WNT agonist, RA agonist, or FGF antagonist concentrations will have large impacts on expression of SIX3. Again, this is consistent with previous developmental studies that have demonstrated that anterior neural identity cannot be achieved when posteriorizing RA, WNT, or FGF signals are activated (Figure 5B). On the other hand, GBX2-optimized conditions have high FCs for PC-ATRA (22.9), PD0325901 (21.6), and FGF2 (17.4), requiring high levels of RA and FGF activation to achieve maximal GBX2 expression (Figures 5A and 5B).
Having developed predictive models of control for a wide range of marker genes, we can assess predicted expression profiles of differentiating cells at any region within the design space with statistical confidence. The expression profile predicted for cells exposed to SIX3- and GBX2-optimized media conditions are similar to but more regionally specific than that predicted for a general neural induction strategy (dual-SMADi, Figure S10A). CRABP1, SOX2, and TUBB3, all of which are expressed in developing neuroectoderm, are expected at high levels in all three conditions. However, large differences are predicted in regionally-patterned neuroectodermal genes after exposure to regionally specific protocols. Expression of both GBX2 (54, 95% CI 23-127) and SIX3 (1035, 95% CI 628-1542) is predicted under dual-SMADi conditions, implying a mixture of anterior and posterior neuroectodermal cells. Conversely, under optimized conditions, the anterior and posterior marker genes are distinctly regulated. The GBX2 model predicts very high expression after exposure to GBX2-optimized conditions (10423, 95% CI 3591-30252), but low expression in SIX3-optimized conditions (4, 95% CI 2-11). Similarly, the SIX3∼ expression model predicts high expression after exposure to SIX3-optimized conditions (5600, 95% CI 4272-7108) and low expression in GBX2-optimized conditions (119, 95% CI 0.5-505).
Additional anterior and posterior neuroectoderm markers—whose models were not involved in optimization of regionally-specific conditions—follow similar patterns. Strong OTX2 expression is predicted under SIX3-optimized conditions (10762, 95% CI 8697-12827), but not GBX2-optimized conditions (-1595, 95% CI -3663 to 473). Conversely, posterior marker HOXB1 expression is expected under GBX2-optimized conditions (939, 95% CI 455-7238), but not SIX3-optimized conditions (0.2, 95% CI -0.06 to 3). Anterior marker HESX1 and eye-field gene RAX are likely expressed strongly in SIX3-optimized conditions whereas posterior epiblast marker NKX1-2 is likely to be expressed in GBX2-optimized conditions. Note that mesodermal marker T is expected to remain at very low levels in all three conditions. These profiles are consistent with regionally specific neureoectoderm populations.
In fact, hiPSCs exposed to dual-SMADi for 3 days produced neuroectoderm populations with mixed anterior/posterior identity whereas regionally optimized conditions more specifically directed differentiation toward A/P-patterned neuroectodermal cell populations (Figures 5C and S10B). Two dual-SMADi approaches were assessed including factors at concentrations modeled in the HD-DoE experiment (LDN/A83) and a protocol described in literature (LDN/SB, Surmacz et al., 2012). In the NCRM-1 line, both dual-SMADi protocols resulted in upregulation of general neural marker SOX1 (Tukey’s HSD vs hiPSCs: LDN/A83 p< 0.001, LDN/SB p = 0.046) and posterior marker GBX2 (Tukey’s HSD versus hiPSCs: LDN/A83 p = 0.007, LDN/SB p = 0.033) after 3 days. LDN/A83 also upregulated anterior marker SIX3 (Tukey’s HSD versus iPSCs p = 0.002). In contrast, differentiation conditions optimized for anterior neural marker SIX3 yielded widespread SIX3 expression (Tukey’s HSD p< 0.05 versus all other groups) whereas those optimized for posterior neural marker GBX2 produced SIX3-negative populations (Tukey’s HSD p = 0.985 versus hiPSC) with widespread upregulation of GBX2 (Tukey’s HSD p< 0.05 versus all other groups). Both regionally optimized protocols also exhibited widespread SOX1 expression (Tukey’s HSD p< 0.001 versus hiPSCs for both conditions), indicating neuroectodermal commitment, and downregulation of OCT4, indicating exit from pluripotency.
HD-DoE-derived protocols specifically direct differentiation toward regional neuroectoderm populations across hiPSC lines
HD-DoE-derived protocols produced more consistent marker expression across cells lines compared to dual-SMADi (Figures 6, S10C, and Table S7). Three days of exposure to dual-SMADi yielded highly variable marker expression across hiPSC lines (Figures 6A and S10C). In addition, as observed in NCRM-1 cells, NCRM-2, -4, and -5 hiPSCs treated with dual-SMADi expressed both anterior (SIX3) and posterior (GBX2) markers of neuroectoderm.
Figure 6.
HD-DoE-derived protocols specifically direct differentiation toward regional neuroectoderm populations across hiPSC lines
(A–C) Marker expression after exposure of NCRM-1, -2, -4, and -5 hiPSC lines to (A) dual-SMADi (100 nM LDN-193189, 10 μM SB 431542), (B) SIX3-optimized conditions, and (C) GBX2-optimized conditions for 3 days. Scale bars = 250 μm. See also Figure S10 C and Table S7.
General neural marker SOX1 was detected at higher levels in cells exposed to regionally optimized conditions, compared to dual-SMADi. SOX1 expression was affected by differentiation condition and cell line (Two-way ANOVA, p< 0.001) but no interaction was detected between condition and cell line (F(5, 17) = 2.302, p = 0.091). SOX1 intensity was higher in cells treated with regionally optimized conditions compared to cells treated with LDN/SB (Table S7, Tukey’s HSD, SIX3-Opt diff = 12.932, 95% CI 5.093-20.772, p = 0.002; GBX2-Opt diff = 29.558, 95% CI 20.97-38.145, p< 0.001).
In addition, HD-DoE-derived protocols more specifically directed expression of regionally specific markers, splitting the anterior and posterior neuroectodermal fields across hiPSC lines. Anterior marker SIX3 is clearly upregulated in all cell lines under SIX3-optimized conditions (Figure 6B) and absent in all cell lines under GBX2-optimized conditions (Figure 6C). Expression of posterior marker GBX2 was affected by differentiation condition (Two-way ANOVA, p = 0.001) and cell line (p< 0.001) and an interaction between condition and cell line was detected (p = 0.002). GBX2 expression was lower in cells exposed to SIX3-optimized conditions compared to LDN/SB (Tukey’s HSD, diff = -4.280, 95% CI -8.488 to -0.073, p = 0.046) and GBX2 expression was higher in cells exposed to GBX2-optimized conditions compared to SIX3-optimized conditions (diff = 5.713, 95% CI 2.278-9.149, p = 0.001).
In summary, SIX3-optimized conditions induced SIX3 and SOX1 expression, consistent with anterior neuroectoderm, in all 4 hiPSC lines tested (Figure 6B) whereas GBX2-optimized conditions consistently induced GBX2, SOX1, and PAX6 in SIX3-negative populations, consistent with posterior neuroectoderm (Figure 6C).
CPPs for directing anterior/posterior neuroectoderm patterned from pluripotency include SMAD1/5/8, WNT, FGF, and RA pathway-modulating factors
Not only do predictive PLSR models of gene control identify conditions that produce highly reproducible, regionally specific cell types of interest, but they also provide deep in silico analysis of gene expression behavior across the design space. By visualizing model-predicted expression in four dimensions, we can quickly assess the relative importance of pathway-modulating factor concentrations in optimized conditions, providing deep understanding of system behavior (Figure 7). Other gene models can be examined in the same way, providing insight into how changes in each factor are likely to affect other important lineage-specific genes (Figure S11).
Figure 7.
CPPs for directing anterior/posterior neuroectoderm patterning from pluripotency include SMAD1/5/8, WNT, FGF, and RA pathway-modulating factors
(A and B) PLSR model-predicted behavior of (A) SIX3 expression around the SIX3-optimized set point and (B) GBX2 expression around the GBX2-optimized set point. Factors with the highest factor contributions are shown in the following order: outer x axis, outer y axis, x axis, y axis. All other factors are at their optimized concentrations, as indicated in the bottom-right corner of each panel. See also Figure S11.
Changes in SIX3-optimized CPP concentrations are expected to have detrimental effects on SIX3 expression (Figure 7A). The model of SIX3∼ expression predicts that adding half-maximal CHIR-99021 to SIX3-optimized differentiation media would reduce SIX3 expression by a factor of 2.5 to 2239 (95% CI 1493–3135) whereas adding half-maximal PC-ATRA is expected to reduce expression by a factor of 1.9 to 2875 (95% CI, 1936–3999). Conversely, omitting PD0325901 from differentiation media would reduce SIX3 expression by a factor of 3.4 to only 1667 (95% CI 1022–2468) whereas omitting LDN-193189 would reduce expression by a factor of 1.8 to 3114 (95% CI 2236–4138). Thus, the model indicates that SIX3 expression depends on preventing WNT and RA signaling, while maintaining inhibition of BMP receptors and MEK.
Predicted expression of OTX2 in the SIX3-optimized region of the design space reveals similar control by RA, SMAD1/5/8, and FGF signaling (Figure S11A). SIX3-optimized factor concentrations are also expected to produce high OTX2 expression, consistent with anterior neuroectoderm. Like SIX3, OTX2 was sensitive to RA signaling, with a predicted 28% reduction (to 7710, 95% CI 5431–9990) on addition of half-maximal PC-ATR. The model also predicts omission of LDN-193189 or PD0325901 would moderately reduce OTX2 expression. In contrast, addition of half-maximal CHIR-99021 is expected to increase OTX2 expression slightly, possibly moving the culture toward a more posterior mesencephalic identity.
RA, FGF, and SMAD1/5/8 pathway modulating factors are also CPPs for posterior neuroectoderm differentiation (Figure 7B). The PLSR model of GBX2 expression predicts that reducing the concentration of PC-ATRA to half-maximal (1μM) would reduce GBX2 expression by a factor of 5.3 to 1957 (95% CI 838–4570) whereas eliminating PC-ATRA altogether would reduce expression 28-fold to only 367 (95% CI 136–985). Omitting FGF2 would reduce expression almost 13-fold to 823 (95% CI 320–2120) whereas adding 100 nM MEK inhibitor PD0325901 would reduce expression by a factor of 23 to only 445 (95% CI 129–1531). Finally, omitting LDN-193189 would reduce GBX2 approximately 5-fold to 2021 (95% CI 709–5762). Thus, to achieve high GBX2 expression, it isimportant to provide RA and FGF activation while simultaneously inhibiting SMAD1/5/8 signaling.
The model of posterior neuroectoderm marker HOXB1, whose mouse orthologue is expressed at higher levels in mesoderm (E8.0), predicts moderate, but not maximal levels under GBX2-optimized conditions (939, 95% CI 122–7238, Figure S11B). Similar to GBX2, omission of PC-ATRA would drastically reduce HOXB1 expression to a normalized expression value of only 5 (95% CI 0.6-32). Unlike GBX2, however, omitting FGF2, omitting LDN-193189, or adding PD0325901 would likely increase HOXB1 expression.
Discussion
We have applied an HD-DoE approach to identify combinatorial signaling conditions that quickly and specifically direct expression of regionally-specific neuroectodermal genes from pluripotency. We previously used the method, which navigates a multidimensional factor space and optimizes conditions for differentiation toward specific cellular fates, to develop a small molecule induction protocol for pancreatic fate from pluripotency (Bukys et al., 2020). The method allows for optimized protocol development and critical process parameter identification, performed here for both anterior and posterior neuroectoderm, using known regionally restricted marker genes. With increasing availability of scRNA-Seq data from developing vertebrate embryos (Pijuan-Sala et al., 2019) and new lineage analysis techniques (Yao et al., 2017), improved identification of lineage-specific marker genes is resolving the fate space of the developing organism and may further improve HD-DoE-driven protocol development.
Directing differentiation toward neural fate from pluripotency almost invariably involves dual-SMAD inhibition (Galiakberova and Dashinimaev, 2020). This paradigmatic protocol was developed using 10 μM ALK5 receptor inhibitor (SB 431542) and 500 ng/mL recombinant Noggin protein (Chambers et al., 2009) and was later adapted using small molecule BMP receptor inhibitors (Morizane et al., 2011; Surmacz et al., 2012). Others have further tailored dual-SMAD inhibition to achieve regional patterning of neural progenitors by including control of FGF, WNT, RA, and SHH signaling from pluripotency (Kirkeby et al., 2012; Mariani et al., 2012; Reinhardt et al., 2013; Shi et al., 2012). However, these protocols typically require at least a week in culture to achieve expression of marker genes. They also require concentrations of pathway-modulating factors that are many fold greater than ED50/IC50 values, which could indicate that off-target signaling effects may be contributing to differentiation.
The results from this study are consistent with the current understanding of CNS patterning, while also providing additional knowledge of complex pathway interactions that must be understood and controlled in order to more specifically direct differentiation of hPSCs. Unlike other approaches, HD-DoE allows for direct comparison of many developmental signaling pathways simultaneously, strengthening our understanding of their importance in neural induction and patterning. For instance, our study reveals that ALK5 inhibition is less important than ALK2/3 inhibition for neuroectoderm marker expression, and that concurrent control of A/P patterning pathways is essential for imparting regional identity to the emerging neuroectoderm. Consequently, improved territory control is attained by providing BMP signaling inhibition, while also controlling signaling activity of RA, FGF, and WNT based on desired A/P identity to achieve rapid induction of regional neuroectodermal territory markers. The results extracted from this single HD-DoE experiment recapitulate decades of research on neural induction and A/P control of neural patterning (reviewed by Lupo et al., 2013).
Our previous work relating to induction of pancreatic fate revealed rapid fate conversion compared to previously published protocols. Here, we similarly demonstrated that neural territory specification is directly attainable within only 3 days of pluripotency. It is possible that epigenetic landscape changes, which are necessary for fate commitment, occur more effectively in hPSC culture when pathways are controlled simultaneously—as they are in the developing embryo—although further experimentation is needed to confirm whether this is the case. If so, the duration of differentiation protocols may be significantly shortened with more specific signaling control, reducing the total time, cost, and effort of manufacturing hPSC-derived cell therapies.
In contrast to traditional OFAT approaches, the HD-DoE method addresses variation across complex design spaces. HD-DoE simultaneously identifies both main factor effects and pathway interactions for a large number of responses, thereby explaining complex system behavior at an unprecedented level. Because developing organismal systems, like the human embryo, rely on combinatorial signaling to robustly produce a large number of diverse cell types, understanding signaling interactions is key to replicating development in vitro and underlies the success of the HD-DoE method. OFAT approaches rarely identify factor interactions with statistical confidence, but, as demonstrated here, they are extremely prevalent and important for effectively directing cell fate.
Importantly, HD-DoE-optimized conditions exhibited more consistent marker expression across cell lines compared to a typical neural induction approach. Strano et al. (2020) recently demonstrated that cell line-dependent differences in directed differentiation of hPSCs to cortical neurons could be attributed to differences in endogenous WNT signaling across cell lines and could be corrected by additional pathway control. Although further studies are needed to confirm whether this is the case, it is possible that providing differentiation media that simultaneously controls activity of many important signaling pathways, as we have done here, may help reduce cell-line variability often observed during hPSC differentiation. HD-DoE is an excellent tool to quickly identify the most important signaling pathways for particular markers and to identify permissive conditions that provide high-level pathway control.
The HD-DoE approach has the potential to revolutionize hPSC differentiation protocol development—and other multifactorial biological optimization problems—by reducing the number of experiments necessary to deeply understand complex systems in high dimensions. While we tested known morphogen signaling inputs in this study, the approach can be applied to screen for effects of novel signaling pathways and pathway-modulating factors. It will also be highly useful for developing protocols for cell types whose differentiation control has not been well described or studied. In addition, once CPPs have been identified for particular marker genes and/or cell types, deeper DoE designs (i.e., those specifically devised for precise optimization and robustness testing) can be applied to further refine recipes, identify robust set points, and calculate process capability indices of complex media formulations, facilitating production of high-purity specific human cell populations on a large scale. By providing deep process understanding of developmental pathway control in differentiating hPSCs, the HD-DoE approach can aid in development of cell-based therapies and in vitro models for a wide variety of degenerative diseases.
Limitations of the study
Models were developed by measuring, modeling, and optimizing overall mRNA expression of marker genes in the culture. There may be differences in protein expression of markers and culture homogeneity that are not captured in the models of expression control presented here. Depending on the intended use of hPSC-derived cells, these may be important metrics for protocol development. Although these issues could not be addressed in this study, HD-DoE is well-suited to assess these needs with further experimentation. Protocols can be further optimized by modeling additional controllable variables (i.e., cell seeding density, treatment time, additional pathway-modulating factors) and by modeling different response metrics (i.e., % positive cells to optimize for homogeneity).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GBX2 | Thermo Fisher Scientific | Cat # PA5-66953; RRID:AB_2662957 |
| Mouse monoclonal anti-Oct-3/4 (C-10) | Santa Cruz Biotechnology | Cat# sc-5279; RRID:AB_628051 |
| Rabbit monoclonal anti-OTX2 (14H14L5) | Thermo Fisher Scientific | Cat# 701948; RRID: AB_2608961 |
| Rabbit polyclonal anti-Pax-6 | Covance | Cat# PRB-278P; RRID:AB_291612 |
| Mouse monoclonal anti-Six3 (A-1) | Santa Cruz Biotechnology | Cat# sc-398797 |
| Goat polyclonal anti-Sox-1 (C-20) | Santa Cruz Biotechnology | Cat# sc-17318; RRID:AB_2195365 |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Mouse IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 715-545-151; RRID:AB_2341099 |
| Alexa Fluor® 594 AffiniPure Donkey Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 711-585-152; RRID:AB_2340621 |
| Alexa Fluor® 647 AffiniPure Donkey Anti-Goat IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 705-605-147; RRID:AB_2340437 |
| Chemicals, peptides, and recombinant proteins | ||
| Vitronectin (VTN-N) Recombinant Human Protein, Truncated | Gibco | Cat# A14700 |
| Essential 8 (E8) Medium | Gibco | Cat# A1517001 |
| Essential 8 (E8) Flex Medium | Gibco | Cat# A2858501 |
| UltraPure™ 0.5M EDTA, pH 8.0 | Invitrogen | Cat# 15575020 |
| TrypLE Select Enzyme | Gibco | Cat# 12563029 |
| RevitaCell Supplement (100X) | Gibco | Cat# A2644501 |
| IMDM | Gibco | Cat# 12440053 |
| Ham’s F-12 Nutrient Mix | Gibco | Cat# 11765047 |
| Insulin, human | Roche | Cat# 11376497001 |
| Transferrin from human serum | Roche | Cat# 10652202001 |
| Chemically Defined Lipid Concentrate | Gibco | Cat# 11905031 |
| 1-Thioglycerol | Sigma-Aldrich | Cat# M6145; CAS: 96-27-5 |
| Poly(vinyl alcohol), 87–90% hydrolyzed | Sigma-Aldrich | Cat# P8136; CAS: 9002-89-5 |
| A 83-01 | Sigma-Aldrich | Cat# SML0788; CAS: 909910-43-6 |
| A 83-01 | Biogems | Cat# 9094360 |
| Animal-Free Recombinant Human BMP-4 (E.coli derived) | PeproTech | Cat# AF-120-05ET |
| LDN-193189 | Selleckchem | Cat# S2618; CAS: 1062368-24-4 |
| LDN-193189 | Biogems | Cat# 1066208 |
| CHIR-99021 (CT99021) HCl | Selleckchem | Cat# S2924; CAS: 1797989-42-4 |
| CHIR 99021 | Biogems | Cat# 2520691 |
| XAV939 | Sigma-Aldrich | Cat# X3004; CAS: 284028-89-3 |
| XAV939 | Biogems | Cat# 2848932 |
| bFGF Recombinant Human Protein | Gibco | Cat# 13256029 |
| Recombinant Human FGF-basic (154 a.a.) | PeproTech | Cat# 100-18B |
| PD0325901 (Mirdametinib) | Selleckchem | Cat# S1036; CAS: 391210-10-9 |
| All-trans Retinoic Acid | Sigma-Aldrich | Cat# R2625; CAS: 302-79-4 |
| ER 50891 | Tocris | Cat# 3823; CAS: 187400-85-7 |
| Recombinant Human Sonic Hedgehog/Shh Protein, High Activity | R&D Systems | Cat# 8908-SH |
| Purmorphamine | Stemcell Technologies | Cat# 72202; CAS: 483367-10-8 |
| Purmorphamine | Biogems | Cat# 4831086 |
| Recombinant Human DLL1 His-tag Protein, CF | R&D Systems | Cat# 1818-DL |
| Recombinant Human Jagged 2 Fc Chimera Protein, CF | R&D Systems | Cat# 1726-JG |
| γ-Secretase Inhibitor XX | Sigma-Aldrich | Cat# 565789; CAS: 209984-56-5 |
| SB 431542 | Biogems | Cat# 3014193 |
| Critical commercial assays | ||
| MagMAX™-96 Total RNA Isolation Kit | Invitrogen | Cat# AM1830 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| QuantStudio 12K Flex Real-Time PCR System with custom designed OpenArray plates | Applied Biosystems | |
| Experimental models: Cell lines | ||
| WA09 Human Embryonic Stem Cell Line | WiCell | RRID:CVCL_9773 |
| NCRM-1 | iXCells Biotechnologies | NHCDR Cat# ND50028; RRID:CVCL_1E71 |
| NCRM-4 | iXCells Biotechnologies | NHCDR Cat# ND50025; RRID:CVCL_1E74 |
| NCRM-5 | iXCells Biotechnologies | NHCDR Cat# ND50031; RRID:CVCL_1E75 |
| NCRM-2 | iXCells Biotechnologies | NHCDR Cat# ND50030; RRID:CVCL_1E72 |
| Software and algorithms | ||
| MODDE Pro v 12.0.0.3292 | Sartorius | https://www.sartorius.com/en/products/process-analytical-technology/data-analytics-software/doe-software/modde |
| JMP Pro v 14.2.0 | JMP Statistical Discovery LLC | https://www.jmp.com/en_us/software/predictive-analytics-software.html |
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| R v 4.1.1 for Windows | R Foundation for Statistical Computing | https://www.R-project.org |
| Other | ||
| E-MTAB-4840 data | Lindsay et al. (2016) | https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4840/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jan Jensen (jjensen@trailbio.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human embryonic stem cells
The WA09 (H9, RRID:CVCL_9773) female human embryonic stem cell (hESC) line was used to assess differentiation effects of factors listed in Figure 1C. Cells were grown at 37°C, in a humidified environment at 10% O2 and 5% CO2. Cells were maintained on tissue culture plates coated with 0.5 ug/cm2 Vitronectin (VTN-N; Gibco, A14700) in Essential 8 (E8) Medium (Gibco, A1517001) with daily media exchange, according to manufacturer instructions. Cells were passaged as colonies using 0.5 mM EDTA when they were approximately 80% confluent, at least every 5 days, and media was supplemented overnight with 1X RevitaCell (Gibco, A2644501) after passage. The cell line was authenticated by STR testing before use and karyotype analysis was performed at least every 10 passages (WiCell).
Human induced pluripotent stem cells
Two male (NCRM-1, RRID:CVCL_1E71; NCRM-5, RRID:CVCL_1E75) and two female (NCRM-2, RRID:CVCL_1E72; NCRM-4, RRID:CVCL_1E74) human induced pluripotent stem cell (hiPSC) lines were used to validate HD-DoE-derived protocols. Cells were grown at 37°C, in a humidified environment at atmospheric O2 and 5% CO2. Cells were maintained on tissue culture plates coated with 0.5 ug/cm2 Vitronectin (VTN-N; Gibco, A14700) in Essential 8 Flex (E8) Medium (Gibco, A2858501), according to manufacturer instructions. Cells were passaged as colonies using 0.5 mM EDTA when they were approximately 80% confluent, at least every 5 days, and media was supplemented overnight with 1X RevitaCell (Gibco, A2644501) after passage. Karyotype analysis was performed at least every 10 passages.
Method details
Expression of pathway modulators in the embryo
In order to model control of signaling pathway components from pluripotency in the human embryo, we used the 3D Atlas of Human Embryology (de Bakker et al., 2016) at the earliest epiblast-patterning time point (Carnegie Stage (CS) 7) and overlaid expression patterns from corresponding stages of mouse development. CS7 corresponds approximately to mouse Theiler stage (TS) 9, which begins around embryonic days (E) 6.5–6.75 (Otis and Brent, 1954). We compiled in situ hybridization data for all known endogenous ligands and inhibitors for the pathways examined at E6.5 and E6.75, beginning with data cataloged in the Mouse Genome Informatics Gene eXpression Database (MGI-GXD). When data was not available for a particular component at the appropriate stage, we searched PubMed for additional data. We also supplemented expression data using the recently published single-cell RNA sequencing dataset of the gastrulating mouse embryo at E6.75 (Pijuan-Sala et al., 2019). Genes whose expression patterns were included met the following conditions: 1) known or very likely to directly activate or inhibit pathway activity and 2) expressed in a regionally-restricted manner at E6.5–6.75. For secreted agonists and inhibitors, expression domains in both embryonic and extraembryonic tissues are depicted. For intracellular inhibitors, only embryonic expression was considered, as extraembryonic expression would be unlikely to directly influence signaling activity in embryonic cells.
Generating the HD-DoE design
The high-dimensional Design of Experiments (HD-DoE) design depicted in Figure 2C was generated using MODDE software (Sartorius). A D-Optimal interaction screening design was used, with linear constraints for opposing factors (defined as agonists and antagonists in Figure 1C), such that opposing factors were never tested together above half their maximum concentrations.
Preparing CDM2 basal differentiation medium
Chemically defined medium 2 (CDM2) was used as basal differentiation medium (Loh et al., 2014). To prepare CDM2, IMDM (Gibco, 12440053) and F12 (Gibco, 11765054) were mixed in equal proportions and supplemented with 0.7 μg/mL recombinant human insulin (Roche, 11376497001), 15 μg/mL transferrin from human serum (Roche, 10652202001), 1% chemically defined lipid concentrate (Gibco, 11905031), 450 μM 1-thioglycerol (Sigma-Aldrich, M6145), and 1 mg/mL poly(vinyl alcohol) (PVA; Sigma-Aldrich, P8136). PVA powder was solubilized in water at 50 mg/mL by heating to 85°C while stirring for up to 30 min until completely dissolved. Solution was cooled, filter-sterilized, and used within 6 months. CDM2 was sterilized using a 22 μm low protein-binding filter, stored at 4°C, and used within 2 weeks.
Differentiating hPSCs
For differentiation, hPSCs were plated as single cells and treated for 3 days with CDM2 basal medium supplemented with various combinations and concentrations of soluble pathway-modulating factors. When colonies were approximately 80% confluent, cells were collected and dissociated to single cell suspension using 1X TrypLE Select (Gibco, 12563029). Countess Automated Cell Counter (Invitrogen, 10227) was used to count cells in suspension and 0.4% Trypan blue stain (Invitrogen, T10282) to assess viability. hESCs were plated at 200,000 viable cells/cm2 and hiPSCs were plated at 100,000 cells/cm2 on 1 ug/cm2 VTN-N-coated 96-well plates in E8 medium supplemented overnight with 1X RevitaCell. Test factors (Figure 1C) were reconstituted and stored according to manufacturer instructions (key resources table). To create the perturbation matrix (PM) containing all experimental conditions defined in Figure 2C, factors were diluted in CDM2 basal medium and pipetted at appropriate concentrations by a Tecan Freedom Evo 150 liquid handling robot. Differentiation media was applied 36–48 h after single-cell plating, when cells were approximately 90% confluent. Media was exchanged every 24 h.
RNA extraction and cDNA synthesis
After 3 days of treatment with the PM, RNA was isolated from differentiated hESCs and cDNA was prepared for gene expression analysis. During RNA isolation, cells for each experimental condition were pooled from three identically-treated 96-well plates to ensure sufficient RNA collection. The MagMAX-96 Total RNA Isolation Kit (Invitrogen, AM1830) was used to isolate RNA and a Bio-Tek Epoch plate reader was used to assess the amount and purity of RNA collected for each condition. High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, 4368814) was used for cDNA synthesis.
Measuring gene expression
For each of the experimental conditions, expression of 56 carefully selected genes was measured using the QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems). OpenArray plates were custom designed to include fate-defining markers of germ layers (i.e. SOX1, T, MEOX1), general neuroectoderm (i.e. PAX6, SOX2), non-neural ectoderm (i.e. TFAP2A), and regionally-specific CNS (i.e. SIX3, GBX2), among others (Figure 2D and Table S3). In addition, 3 housekeeping genes were measured for normalization (GAPDH, YWHAZ, and TBP).
Immunofluorescence
Cells were fixed using 4% paraformaldehyde or 10% formalin at room temperature for 10 min. Samples were permeabilized and blocked using 0.2% Triton X-100 in 10% normal donkey serum with 0.2 M glycine for 1 h at room temperature. Antibodies were diluted in 0.1% Triton X-100, 1% normal donkey serum. Primary antibodies were incubated at 4°C overnight and secondary antibodies at room temperature for 1 h. Samples were subsequently incubated in 300 nM DAPI for 5 min at room temperature and stored in PBS. Immunofluorescent images were acquired using Keyence All-in-One Fluorescence Microscope BZ-X710.
Quantification and statistical analysis
Statistical tests, dispersion and precision measures, and p-values are embedded in Results text, while sample numbers (n) can be found in figure legends and supplemental tables. A significance level of 0.05 was used throughout.
Specificity of cell fate markers in human CNS
RNA-seq expression data from human embryos was downloaded from the Human Developmental Biology Resource (HDBR, E-MTAB-4840) (Lindsay et al., 2016). Expression was mapped to CNS regions of the 3D atlas human embryo models at the appropriate stages for CS13 - CS21 to visualize expression and specificity over time (Figure 2E). Specificity across the 4 CNS tissues was calculated for CS14 using Preferential Expression Measure (PEM) (Kryuchkova-Mostacci and Robinson-Rechavi, 2017) and normalized to the largest PEM of the genes measured. Genes were sorted into groups based on which tissues had the highest specificity score and organized in descending order of specificity by group (Figure 2D).
Gene expression analysis
The ΔΔCτ method was used to quantify relative gene expression. CRT, ΔCRT mean, amplification score, and Cq confidence values were exported from Expression Suite software for each treatment/gene combination. Expression values were normalized and transformed such that 0 indicates that the gene was not detected in the sample and 10000 corresponds to the mean expression of the endogenous control genes in that sample.
Two assays were omitted due to loading error (#73 DLX5 and FERD3L). Amplification plots for all assays with amplification score <1.24 and/or Cq confidence <0.8 were visually inspected and 16 assays and all assays for experiment #40 were omitted due to poor amplification.
Regression modeling
Partial Least Squares Regression (PLSR) was used to identify factors and interactions affecting marker gene expression and to predict expression across the design space.
Genes that exhibited high variability between replicates (DBX2, LHX6, NKX2-2, and PROP1) were excluded from analysis due to low reproducibility. EGR2, EVX1, and PAX2 had high variability between replicate experiments 95 and 96, but low variability between replicates in experiments 92, 93, and 94. As experiment 95/96 contained no factors, differentiation was less controlled, cells detached more easily, and RNA quantity was lower, all of which may have led to higher variability in that condition. Since the other set of replicates had low variability, experiment number 96 (which had a larger residual than 95 for all genes) was excluded for those genes.
PLSR models were refined using the Auto Tune feature in MODDE, which removes non-significant terms one at a time and checks for an increase in the predictive ability (Q2) of the model. If Q2 increases, the non-significant term is left out of the model and the next term is tested. After model tuning, all possible triple interaction terms were added manually and Auto Tune was applied again.
After initial model refinement, ANOVA was used to determine whether model fit was appropriate (Table S4). If the ANOVA lack of fit test provided evidence for lack of fit, the model was reset and response data were transformed. Transformations were selected based upon improved correlation of the standardized residuals normal probability plot, while ensuring reproducibility of replicates remained high after transformation. The model refinement process described above was repeated for transformed responses and models were re-assessed for lack of fit. Further adjustments to transformations were applied as necessary.
EMX2, KRT5, NKX2-1, and SHH were undetectable in all replicate experiments, precluding those models from ANOVA lack of fit testing (pure error = 0). These genes were log-transformed if and only if transformation improved correlation of the standardized residuals normal probability plot and were modeled as described above.
Transformation of SIX3 is illustrated in Figure S1 and all transformations are listed in Table S3. Models that had evidence for lack of fit after transformation and remodeling (EN1, FOXA2, LMX1A, MEOX1, NKX6-1, and PAX8) were excluded from predictive analyses.
Image quantification
Images were quantified using ImageJ v1.53o. DAPI images were used to create masks of regions containing cells and mean fluorescence intensity of marker proteins was quantified across biological replicates in masked regions. All images that were compared were treated identically. ANOVA with Tukey’s test was used to test for differences between groups using the stats package in R.
Data visualization
Mapping RNA-seq expression data onto the human embryo was done using JMP Pro v14.2.0. Graphs were created in JMP Pro v14.2.0, MODDE Pro v12.0.0.3292, and R v4.1.1.
Acknowledgments
Funding for this study was provided by the Cleveland Clinic and the Ohio Third Frontier grant number IPP 12-258. Support for the project was also provided by Trailhead Biosystems Inc., and ARMI (Advanced Regenerative Manufacturing Institute) through the “Biomanufacturing of the Neuroectodermal Fate Space” project, T0042.
Author contributions
Conceptualization, K.E.S and J.J.; Methodology, K.E.S, K.G., D.T., M.A.B., and J.J.; Validation, K.E.S.; Formal Analysis, K.E.S and A.M.; Investigation, K.E.S. and K.G.; Resources, J.J.; Writing – Original Draft, K.E.S.; Writing – Review & Editing, K.E.S and J.J.; Visualization, K.E.S.; Supervision, J.J.; Funding Acquisition, D.T. and J.J.
Declaration of interests
J. Jensen is founder and shareholder of Trailhead Biosystems Inc., Cleveland, OH, USA. D.T. and M.A.B. are shareholders of Trailhead Biosystems Inc.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104133.
Supporting citations
The following references appear in the supplemental information: Albano et al., 1994; Balasubramanian and Zhang, 2016; Becker et al., 1997; Belo et al., 1997; Bettenhausen et al., 1995; Böttcher and Niehrs, 2005; Bouillet et al., 1996; Boylan and Gudas, 1992; Conlon et al., 1994; Crossley and Martin, 1995; Cruciat and Niehrs, 2013; Dunwoodie et al., 1997; Finley et al., 2003; Haub and Goldfarb, 1991; Hébert et al., 1991; Hedger et al., 2011; Kemp et al., 2005; Kimura et al., 2001; Kispert et al., 1996; Lin et al., 2002; Maruoka et al., 1998; Niederreither et al., 1997; Niswander and Martin, 1992; Norris et al., 2002; Onichtchouk et al., 1999; Perea-Gómez et al., 1999; Shen, 2007; Solloway and Robertson, 1999; Wall et al., 2000; Ying and Zhao, 2001; Zakin et al., 2000; Zinski et al., 2018.
Supplemental information
SMAD2/3-activating ligands are expressed throughout the epiblast and in the posterior region of the visceral endoderm (VE). The classical developmental SMAD2/3-activating ligand, Nodal, is expressed throughout the epiblast and VE at E6.5 but very quickly becomes posteriorly restricted in both tissues (Conlon et al., 1994). In Figure 1B, the mouse VE corresponds to the human hypoblast, lining the ventral side of the embryonic layer. Tgfb1 expression is detected in the epiblast, concentrated in emerging mesoderm (Pijuan-Sala et al., 2019). Gdf1, is expressed ubiquitously in the epiblast (Wall et al., 2000), while Gdf3 and Tgfb2 are also detected in some epiblast cells (Pijuan-Sala et al., 2019) at E6.75.
Secreted inhibitors of the SMAD2/3 pathway are expressed in the anterior VE (AVE) and throughout the epiblast, with particularly high levels in the primitive streak. Cer1, Lefty1, and Lefty2 bind directly to Nodal, preventing it from interacting with its receptor (Zinski et al., 2018). Cer1 is expressed in the AVE (Belo et al., 1997) and in cells of the anterior primitive streak (Pijuan-Sala et al., 2019). Although Xenoups Cer1 inhibits SMAD2/3-activating, SMAD1/5/8-activating, and WNT ligands, mouse Cer1 is thought to primarily inhibit SMAD2/3-activating ligands (Cruciat and Niehrs, 2013; Shen, 2007). Lefty1 is co-expressed with Cer1 in AVE, and extends posteriorly toward the node (Norris et al., 2002). Lefty2 is strongly expressed in the middle of the primitive streak and in emerging mesoderm cells (Perea-Gómez et al., 1999). Both Lefty1 and Lefty2 are also expressed at low levels in epiblast cells (Pijuan-Sala et al., 2019).
In addition, intracellular inhibitors that target both SMAD2/3 and SMAD1/⅝ signaling pathways are expressed in the epiblast. Bambi, a TGFβ/BMP pseudoreceptor that binds pathway receptors and prevents their activation (Onichtchouk et al., 1999) is broadly expressed in the epiblast at E6.75, with particularly high levels in emerging mesoderm cells. Intracellular inhibitors Smad7 and, to a lesser extent Smad6, are also detected in the epiblast (Pijuan-Sala et al., 2019).
SMAD1/5/8-activating ligands are expressed in different regions in all three layers of the gastrulating mouse embryo. Bmp2 is expressed in the posterior-most region of the epiblast and VE (Pijuan-Sala et al., 2019; Ying and Zhao, 2001). Bmp7 is expressed in and around the node with a posterior bias (Solloway and Robertson, 1999). Bmp4 and Bmp8b are expressed strongly throughout the extraembryonic ectoderm, corresponding to the future amnion (Norris et al., 2002; Ying and Zhao, 2001). Bmp8a, Bmp7, and Gdf6 have also been detected in some extraembryonic ectoderm cells (Pijuan-Sala et al., 2019).
Secreted SMAD1/5/8 inhibitors are expressed in a pattern similar to SMAD2/3 inhibitors: throughout the epiblast, with particularly high levels in the primitive streak, and in AVE. Fst, whose product binds and neutralizes BMP ligands (Hedger et al., 2011) is expressed throughout the primitive streak (Albano et al., 1994). In situ data is unavailable for Chrd and Nog at this stage, so the spatial distribution of these inhibitors is not entirely clear, but both have been detected in AVE cells and in epiblast, including primitive streak cells. Additional secreted inhibitors, including Grem1, Nbl1, and Dand5, are also detected at low levels in some epiblast cells (Pijuan-Sala et al., 2019).
Wnt ligands are expressed in posterior regions of both epiblast and visceral endoderm, as well as throughout the extraembryonic ectoderm. Wnt3 is broadly expressed in the posterior epiblast (Kimura et al., 2001), where Wnt2, Wnt2b, Wnt3a, Wnt5a, Wnt5b, Wnt8a (Bouillet et al., 1996), and Wnt11 (Kispert et al., 1996) are also expressed at varying degrees in the primitive streak and emerging mesoderm (Pijuan-Sala et al., 2019). Wnt3 is also broadly expressed in posterior visceral endoderm (Kimura et al., 2001), where Wnt5a, Wnt9b and Wnt11 have been detected (Pijuan-Sala et al., 2019). Finally, Wnt6 and Wnt7b are strongly expressed in the extraembryonic ectoderm, where Wnt10a has also been detected (Pijuan-Sala et al., 2019) (Kemp et al., 2005).
Like SMAD2/3 and SMAD1/5/8 inhibitors, secreted Wnt inhibitors are expressed throughout the epiblast and in the AVE. Sfrps and Fzrb antagonize Wnt signaling by interact directly with Wnt ligands. Sfrp1 and Sfrp2 are detected at low levels throughout the epiblast and Frzb is expressed strongly in mesoderm and primitive streak cells and at lower levels in AVE (Pijuan-Sala et al., 2019). Sfrp1 (Kemp et al., 2005) and Sfrp5 (Finley et al., 2003) are strongly expressed in the anterior half of the visceral endoderm. And the product of Dkk1, which binds and inhibits the WNT co-receptor LRP6, is strongly expressed in the anterior-most region of the AVE (Zakin et al., 2000).
FGF ligands are expressed throughout the epiblast and visceral endoderm, while inhibitors are intracellular and expressed throughout the epiblast. Fgf5 (Hébert et al., 1991) and Fgf15 (Zakin et al., 2000) are detected throughout the epiblast at E6.5 and E6.75 and low levels of Fgf9 and Fgf18 have also been detected in epiblast cells (Pijuan-Sala et al., 2019). Embryonic expression of other ligands is restricted to the posterior region of the epiblast, including Fgf3, Fgf4 (Niswander and Martin, 1992), Fgf8 (Crossley and Martin, 1995), Fgf10 (Pijuan-Sala et al., 2019), and Fgf17 (Maruoka et al., 1998). Extraembryonic FGF expression is detected in the VE, where Fgf8 is restricted to AVE (Crossley and Martin, 1995), Fgf5 is expressed throughout the VE (Haub and Goldfarb, 1991), and Fgf10 has also been detected (Pijuan-Sala et al., 2019). Known FGF-signaling inhibitors are membrane bound proteins that regulate MAPK activation intracellularly (reviewed in Balasubramanian and Zhang, 2016; Böttcher and Niehrs, 2005). For this reason, we only considered embryonic expression of these genes. Il17rd (Lin et al., 2002), Spred1, Spred2, and Spry4 are expressed throughout the epiblast, where some Spry3 has also been detected (Pijuan-Sala et al., 2019). Spry2 is widely expressed in the embryo, with higher expression detected in the anterior half (Lin et al., 2002) and Spred3 and Spry1 are also expressed in the epiblast with an anterior bias (Pijuan-Sala et al., 2019).
Low levels of ATRA are likely produced in the epiblast and extraembryonic ectoderm at this stage. The primary enzyme thought to be responsible for producing all-trans-retinoic acid in vivo, Aldh1a2, is not yet expressed at this stage of development (Niederreither et al., 1997). However, expression is detected in the epiblast by the next stage, at E7.0. Additionally, its homolog Aldh1a3, which performs the same function (Rhinn and Dollé, 2012), has been detected in the epiblast and, at lower levels, in extraembryonic ectoderm by sc-RNA seq as early as E6.5 (Pijuan-Sala et al., 2019). Importantly, despite low expression of the ATRA synthesis enzyme, epiblast cells appear prepared to both produce and rapidly respond to the ATRA signal, as soon as the synthesis enzyme is present. This is evidenced by widespread embryonic expression of essential pathway components. For instance, synthesis enzymes and binding proteins specific to retinal, the precursor of ATRA and the substrate of the Aldh1a enzymes, are expressed in the epiblast at this stage, as are all of the RA receptor genes (Pijuan-Sala et al., 2019). Additionally, Crabp2, which produces a protein that binds available ATRA and translocates to the nucleus, so that the signaling molecule can interact with its receptors (Kam et al., 2012), is widely expressed in the epiblast at E6.5 and E6.75.
The expression pattern of ATRA inhibitors provides further evidence that RA may be important for differentiation from pluripotency in vivo. Cyp26a1, which encodes an enzyme that converts ATRA to other RA species, is widely expressed in extraembryonic tissues, anterior primitive streak, and emerging mesoderm, but notably absent from epiblast cells (Fujii et al., 1997; Pijuan-Sala et al., 2019). This suggests that ATRA levels in the embryo are tightly controlled, but also that some signaling activity may be essential for embryonic development from pluripotency, particularly in future neuroectoderm. Additionally, ATRA binding protein Crabp1 may act as an intracellular inhibitor (Boylan and Gudas, 1992) and is expressed in the epiblast layer (Pijuan-Sala et al., 2019).
Hedgehog signaling protein Indian hedgehog (Ihh) is expressed in the visceral endoderm at E6.5 (Becker et al., 1997). No inhibitors are strongly expressed at this stage.
Finally, Notch ligands are restricted to the epiblast layer, while an inhibitor is expressed in both epiblast and extraembryonic ectoderm. Activating ligands include Dll1, which is expressed in nascent mesoderm cells (Bettenhausen et al., 1995), Jag1, expressed in nascent mesoderm and primitive streak cells, and Jag2, found in some epiblast cells (Pijuan-Sala et al., 2019). Dll3, the secreted inhibitor, is weakly expressed throughout the epiblast with higher levels in nascent mesoderm (Dunwoodie et al., 1997) and is also detected at low levels in extraembryonic ectoderm (Pijuan-Sala et al., 2019).
Excluded values are indicated by gray background
Data and code availability
-
•
The published article includes all datasets generated during this study (Tables S2 and S7). Original/source data for Figure 2D in the paper is publicly available at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4840/(E-MTAB-4840, Lindsay et al., 2016). RT-PCR and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Albano R.M., Arkell R., Beddington R.S., Smith J.C. Expression of inhibin subunits and follistatin during postimplantation mouse development: decidual expression of activin and expression of follistatin in primitive streak, somites and hindbrain. Development. 1994;120:803–813. doi: 10.1242/dev.120.4.803. [DOI] [PubMed] [Google Scholar]
- Arenas E., Denham M., Villaescusa J.C. How to make a midbrain dopaminergic neuron. Development. 2015;142:1918–1936. doi: 10.1242/dev.097394. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Zhang X. Mechanisms of FGF gradient formation during embryogenesis. Semin. Cell Dev. Biol. 2016;53:94–100. doi: 10.1016/j.semcdb.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot E.S., Hadjantonakis A.K. Mouse gastrulation: coordination of tissue patterning, specification and diversification of cell fate. Mech. Dev. 2020;163:103617. doi: 10.1016/j.mod.2020.103617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S., Wang Z.J., Massey H., Arauz A., Labosky P., Hammerschmidt M., St-Jacques B., Bumcrot D., McMahon A., Grabel L. A role for Indian hedgehog in extraembryonic endoderm differentiation in F9 cells and the early mouse embryo. Dev. Biol. 1997;187:298–310. doi: 10.1006/dbio.1997.8616. [DOI] [PubMed] [Google Scholar]
- Belo J.A., Bouwmeester T., Leyns L., Kertesz N., Gallo M., Follettie M., De Robertis E.M. Cerberus-like is a secreted factor with neutralizing activity expressed in the anterior primitive endoderm of the mouse gastrula. Mech. Dev. 1997;68:45–57. doi: 10.1016/s0925-4773(97)00125-1. [DOI] [PubMed] [Google Scholar]
- Bettenhausen B., Hrabĕ de Angelis M., Simon D., Guénet J.L., Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- Böttcher R.T., Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr. Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- Bouillet P., Oulad-Abdelghani M., Ward S.J., Bronner S., Chambon P., Dollé P. A new mouse member of the Wnt gene family, mWnt-8, is expressed during early embryogenesis and is ectopically induced by retinoic acid. Mech. Dev. 1996;58:141–152. doi: 10.1016/s0925-4773(96)00569-2. [DOI] [PubMed] [Google Scholar]
- Boylan J.F., Gudas L.J. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J. Biol. Chem. 1992;267:21486–21491. [PubMed] [Google Scholar]
- Bukys M.A., Mihas A., Finney K., Sears K., Trivedi D., Wang Y., Oberholzer J., Jensen J. High-dimensional design-of-experiments extracts small-molecule-only induction conditions for dorsal pancreatic endoderm from pluripotency. iScience. 2020;23:101346. doi: 10.1016/j.isci.2020.101346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaerts P., Halder G., Gehring W.J. PAX-6 in development and evolution. Annu. Rev. Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon F.L., Lyons K.M., Takaesu N., Barth K.S., Kispert A., Herrmann B., Robertson E.J. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Crossley P.H., Martin G.R. The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development. 1995;121:439–451. doi: 10.1242/dev.121.2.439. [DOI] [PubMed] [Google Scholar]
- Cruciat C.-M., Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 2013;5:a015081. doi: 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- de Bakker B.S., de Jong K.H., Hagoort J., de Bree K., Besselink C.T., de Kanter F.E., Veldhuis T., Bais B., Schildmeijer R., Ruijter J.M., et al. An interactive three-dimensional digital atlas and quantitative database of human development. Science. 2016;354:aag0053. doi: 10.1126/science.aag0053. [DOI] [PubMed] [Google Scholar]
- Dunwoodie S.L., Henrique D., Harrison S.M., Beddington R.S. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Eriksson L., Johansson E., Kettaneh-Wold N., WikstrÄom C., Wold S. Umetrics; 2000. Design of Experiments: Principles and Applications. [Google Scholar]
- Finley K.R., Tennessen J., Shawlot W. The mouse secreted frizzled-related protein 5 gene is expressed in the anterior visceral endoderm and foregut endoderm during early post-implantation development. Gene Expr. Patterns. 2003;3:681–684. doi: 10.1016/s1567-133x(03)00091-7. [DOI] [PubMed] [Google Scholar]
- Fujii H., Sato T., Kaneko S., Gotoh O., Fujii-Kuriyama Y., Osawa K., Kato S., Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 1997;16:4163–4173. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiakberova A.A., Dashinimaev E.B. Neural stem cells and methods for their generation from induced pluripotent stem cells. Front. Cell Dev. Biol. 2020;8:815. doi: 10.3389/fcell.2020.00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzetta A., Koska M., Rowton M., Sullivan K.R., Jacobs-Li J., Kweon J., Hidalgo H., Eckart H., Hoffmann A.D., Back R., et al. Hedgehog-FGF signaling axis patterns anterior mesoderm during gastrulation. Proc. Natl. Acad. Sci. U S A. 2020;117:15712–15723. doi: 10.1073/pnas.1914167117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haub O., Goldfarb M. Expression of the fibroblast growth factor-5 gene in the mouse embryo. Development. 1991;112:397–406. doi: 10.1242/dev.112.2.397. [DOI] [PubMed] [Google Scholar]
- Hébert J.M., Boyle M., Martin G.R. mRNA localization studies suggest that murine FGF-5 plays a role in gastrulation. Development. 1991;112:407–415. doi: 10.1242/dev.112.2.407. [DOI] [PubMed] [Google Scholar]
- Hedger M.P., Winnall W.R., Phillips D.J., de Kretser D.M. The regulation and functions of activin and follistatin in inflammation and immunity. Vitam. Horm. 2011;85:255–297. doi: 10.1016/B978-0-12-385961-7.00013-5. [DOI] [PubMed] [Google Scholar]
- Ishikawa S., Ito K. Plasticity and regulatory mechanisms of Hox gene expression in mouse neural crest cells. Cell Tissue Res. 2009;337:381–391. doi: 10.1007/s00441-009-0827-5. [DOI] [PubMed] [Google Scholar]
- Kam R.K.T., Deng Y., Chen Y., Zhao H. Retinoic acid synthesis and functions in early embryonic development. Cell Biosci. 2012;2:11. doi: 10.1186/2045-3701-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp C., Willems E., Abdo S., Lambiv L., Leyns L. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev. Dyn. 2005;233:1064–1075. doi: 10.1002/dvdy.20408. [DOI] [PubMed] [Google Scholar]
- Kennedy K.A.M., Porter T., Mehta V., Ryan S.D., Price F., Peshdary V., Karamboulas C., Savage J., Drysdale T.A., Li S.-C., et al. Retinoic acid enhances skeletal muscle progenitor formation and bypasses inhibition by bone morphogenetic protein 4 but not dominant negative beta-catenin. BMC Biol. 2009;7:67. doi: 10.1186/1741-7007-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C., Bates T., Bell E. Molecular specification of germ layers in vertebrate embryos. Cell. Mol. Life Sci. 2016;73:923–947. doi: 10.1007/s00018-015-2092-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura C., Shen M.M., Takeda N., Aizawa S., Matsuo I. Complementary functions of Otx2 and Cripto in initial patterning of mouse epiblast. Dev. Biol. 2001;235:12–32. doi: 10.1006/dbio.2001.0289. [DOI] [PubMed] [Google Scholar]
- Kirkeby A., Grealish S., Wolf D.A., Nelander J., Wood J., Lundblad M., Lindvall O., Parmar M. Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep. 2012;1:703–714. doi: 10.1016/j.celrep.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Kispert A., Vainio S., Shen L., Rowitch D.H., McMahon A.P. Proteoglycans are required for maintenance of Wnt-11 expression in the ureter tips. Development. 1996;122:3627–3637. doi: 10.1242/dev.122.11.3627. [DOI] [PubMed] [Google Scholar]
- Kryuchkova-Mostacci N., Robinson-Rechavi M. A benchmark of gene expression tissue-specificity metrics. Brief Bioinform. 2017;18:205–214. doi: 10.1093/bib/bbw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson K.A., Meneses J.J., Pedersen R.A. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- Li P., Elowitz M.B. Communication codes in developmental signaling pathways. Development. 2019;146:dev170977. doi: 10.1242/dev.170977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Fürthauer M., Thisse B., Thisse C., Jing N., Ang S.-L. Cloning of the mouse Sef gene and comparative analysis of its expression with Fgf8 and Spry2 during embryogenesis. Mech. Dev. 2002;113:163–168. doi: 10.1016/s0925-4773(02)00018-7. [DOI] [PubMed] [Google Scholar]
- Lin W., Jing N., Basson M.A., Dierich A., Licht J., Ang S.-L. Synergistic activity of Sef and Sprouty proteins in regulating the expression of Gbx2 in the mid-hindbrain region. Genesis. 2005;41:110–115. doi: 10.1002/gene.20103. [DOI] [PubMed] [Google Scholar]
- Lindsay S.J., Xu Y., Lisgo S.N., Harkin L.F., Copp A.J., Gerrelli D., Clowry G.J., Talbot A., Keogh M.J., Coxhead J., et al. HDBR expression: a unique resource for global and individual gene expression studies during early human brain development. Front. Neuroanat. 2016;10:86. doi: 10.3389/fnana.2016.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Ang L.T., Zhang J., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y., Nichane M., et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo G., Novorol C., Smith J.R., Vallier L., Miranda E., Alexander M., Biagioni S., Pedersen R.A., Harris W.A. Multiple roles of Activin/Nodal, bone morphogenetic protein, fibroblast growth factor and Wnt/β-catenin signalling in the anterior neural patterning of adherent human embryonic stem cell cultures. Open Biol. 2013;3:120167. doi: 10.1098/rsob.120167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Simonini M.V., Palejev D., Tomasini L., Coppola G., Szekely A.M., Horvath T.L., Vaccarino F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruoka Y., Ohbayashi N., Hoshikawa M., Itoh N., Hogan B.L., Furuta Y. Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mech. Dev. 1998;74:175–177. doi: 10.1016/s0925-4773(98)00061-6. [DOI] [PubMed] [Google Scholar]
- Metzis V., Steinhauser S., Pakanavicius E., Gouti M., Stamataki D., Ivanovitch K., Watson T., Rayon T., Mousavy Gharavy S.N., Lovell-Badge R., et al. Nervous system regionalization entails axial allocation before neural differentiation. Cell. 2018;175:1105–1118.e17. doi: 10.1016/j.cell.2018.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitiku N., Baker J.C. Genomic analysis of gastrulation and organogenesis in the mouse. Dev. Cell. 2007;13:897–907. doi: 10.1016/j.devcel.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Morizane A., Doi D., Kikuchi T., Nishimura K., Takahashi J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J. Neurosci. Res. 2011;89:117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Okamoto I., Sasaki K., Yabuta Y., Iwatani C., Tsuchiya H., Seita Y., Nakamura S., Yamamoto T., Saitou M. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. 2016;537:57–62. doi: 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- Niederreither K., McCaffery P., Dräger U.C., Chambon P., Dollé P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- Niswander L., Martin G.R. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- Norris D.P., Brennan J., Bikoff E.K., Robertson E.J. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- Odorico J.S., Kaufman D.S., Thomson J.A. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- Ogura T., Evans R.M. A retinoic acid-triggered cascade of HOXB1 gene activation. Proc. Natl. Acad. Sci. U S A. 1995;92:387–391. doi: 10.1073/pnas.92.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onichtchouk D., Chen Y.G., Dosch R., Gawantka V., Delius H., Massagué J., Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- Otis E.M., Brent R. Equivalent ages in mouse and human embryos. Anat. Rec. 1954;120:33–63. doi: 10.1002/ar.1091200104. [DOI] [PubMed] [Google Scholar]
- Ozair M.Z., Kintner C., Brivanlou A.H. Neural induction and early patterning in vertebrates. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:479–498. doi: 10.1002/wdev.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C., Gunhaga L. Signaling pathways regulating ectodermal cell fate choices. Exp. Cell Res. 2014;321:11–16. doi: 10.1016/j.yexcr.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Perea-Gómez A., Shawlot W., Sasaki H., Behringer R.R., Ang S. HNF3beta and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo. Development. 1999;126:4499–4511. doi: 10.1242/dev.126.20.4499. [DOI] [PubMed] [Google Scholar]
- Pijuan-Sala B., Griffiths J.A., Guibentif C., Hiscock T.W., Jawaid W., Calero-Nieto F.J., Mulas C., Ibarra-Soria X., Tyser R.C.V., Ho D.L.L., et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature. 2019;566:490–495. doi: 10.1038/s41586-019-0933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck G.K., Heinzmann U., Beckers J., Hrabé de Angelis M. Node and midline defects are associated with left-right development in Delta1 mutant embryos. Development. 2003;130:3–13. doi: 10.1242/dev.00176. [DOI] [PubMed] [Google Scholar]
- Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Höing S., Moritz S., Parga J.A., Wagner L., Bruder J.M., et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 2013;8:e59252. doi: 10.1371/journal.pone.0059252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinn M., Dollé P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N. Mechanisms of human embryo development: from cell fate to tissue shape and back. Development. 2020;147:dev190629. doi: 10.1242/dev.190629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M.M. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Smith J., Robinson H.P.C., Livesey F.J. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15:477–486.S1. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloway M.J., Robertson E.J. Early embryonic lethality in Bmp5;Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development. 1999;126:1753–1768. doi: 10.1242/dev.126.8.1753. [DOI] [PubMed] [Google Scholar]
- Souilhol C., Perea-Gomez A., Camus A., Beck-Cormier S., Vandormael-Pournin S., Escande M., Collignon J., Cohen-Tannoudji M. NOTCH activation interferes with cell fate specification in the gastrulating mouse embryo. Development. 2015;142:3649–3660. doi: 10.1242/dev.121145. [DOI] [PubMed] [Google Scholar]
- Strano A., Tuck E., Stubbs V.E., Livesey F.J. Variable outcomes in neural differentiation of human PSCs arise from intrinsic differences in developmental signaling pathways. Cell Rep. 2020;31:107732. doi: 10.1016/j.celrep.2020.107732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmacz B., Fox H., Gutteridge A., Fish P., Lubitz S., Whiting P. Directing differentiation of human embryonic stem cells toward anterior neural ectoderm using small molecules. Stem Cells. 2012;30:1875–1884. doi: 10.1002/stem.1166. [DOI] [PubMed] [Google Scholar]
- Tabar V., Studer L. Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nat. Rev. Genet. 2014;15:82–92. doi: 10.1038/nrg3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P.P., Loebel D.A. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tuazon F.B., Mullins M.C. Temporally coordinated signals progressively pattern the anteroposterior and dorsoventral body axes. Semin. Cell Dev. Biol. 2015;42:118–133. doi: 10.1016/j.semcdb.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanacou E., Wegener A., Wymeersch F.J., Wilson V., Nicolas J.-F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell. 2009;17:365–376. doi: 10.1016/j.devcel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Wall N.A., Craig E.J., Labosky P.A., Kessler D.S. Mesendoderm induction and reversal of left-right pattern by mouse Gdf1, a Vg1-related gene. Dev. Biol. 2000;227:495–509. doi: 10.1006/dbio.2000.9926. [DOI] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N., Hanna J.H. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Wichterle H., Lieberam I., Porter J.A., Jessell T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Wood H.B., Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech. Dev. 1999;86:197–201. doi: 10.1016/s0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Yao Z., Mich J.K., Ku S., Menon V., Krostag A.-R., Martinez R.A., Furchtgott L., Mulholland H., Bort S., Fuqua M.A., et al. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell. 2017;20:120–134. doi: 10.1016/j.stem.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y., Zhao G.Q. Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev. Biol. 2001;232:484–492. doi: 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- Zakin L., Reversade B., Virlon B., Rusniok C., Glaser P., Elalouf J.M., Brulet P. Gene expression profiles in normal and Otx2-/- early gastrulating mouse embryos. Proc. Natl. Acad. Sci. U S A. 2000;97:14388–14393. doi: 10.1073/pnas.011513398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinski J., Tajer B., Mullins M.C. TGF-beta family signaling in early vertebrate development. Cold Spring Harb. Perspect. Biol. 2018;10:a033274. doi: 10.1101/cshperspect.a033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SMAD2/3-activating ligands are expressed throughout the epiblast and in the posterior region of the visceral endoderm (VE). The classical developmental SMAD2/3-activating ligand, Nodal, is expressed throughout the epiblast and VE at E6.5 but very quickly becomes posteriorly restricted in both tissues (Conlon et al., 1994). In Figure 1B, the mouse VE corresponds to the human hypoblast, lining the ventral side of the embryonic layer. Tgfb1 expression is detected in the epiblast, concentrated in emerging mesoderm (Pijuan-Sala et al., 2019). Gdf1, is expressed ubiquitously in the epiblast (Wall et al., 2000), while Gdf3 and Tgfb2 are also detected in some epiblast cells (Pijuan-Sala et al., 2019) at E6.75.
Secreted inhibitors of the SMAD2/3 pathway are expressed in the anterior VE (AVE) and throughout the epiblast, with particularly high levels in the primitive streak. Cer1, Lefty1, and Lefty2 bind directly to Nodal, preventing it from interacting with its receptor (Zinski et al., 2018). Cer1 is expressed in the AVE (Belo et al., 1997) and in cells of the anterior primitive streak (Pijuan-Sala et al., 2019). Although Xenoups Cer1 inhibits SMAD2/3-activating, SMAD1/5/8-activating, and WNT ligands, mouse Cer1 is thought to primarily inhibit SMAD2/3-activating ligands (Cruciat and Niehrs, 2013; Shen, 2007). Lefty1 is co-expressed with Cer1 in AVE, and extends posteriorly toward the node (Norris et al., 2002). Lefty2 is strongly expressed in the middle of the primitive streak and in emerging mesoderm cells (Perea-Gómez et al., 1999). Both Lefty1 and Lefty2 are also expressed at low levels in epiblast cells (Pijuan-Sala et al., 2019).
In addition, intracellular inhibitors that target both SMAD2/3 and SMAD1/⅝ signaling pathways are expressed in the epiblast. Bambi, a TGFβ/BMP pseudoreceptor that binds pathway receptors and prevents their activation (Onichtchouk et al., 1999) is broadly expressed in the epiblast at E6.75, with particularly high levels in emerging mesoderm cells. Intracellular inhibitors Smad7 and, to a lesser extent Smad6, are also detected in the epiblast (Pijuan-Sala et al., 2019).
SMAD1/5/8-activating ligands are expressed in different regions in all three layers of the gastrulating mouse embryo. Bmp2 is expressed in the posterior-most region of the epiblast and VE (Pijuan-Sala et al., 2019; Ying and Zhao, 2001). Bmp7 is expressed in and around the node with a posterior bias (Solloway and Robertson, 1999). Bmp4 and Bmp8b are expressed strongly throughout the extraembryonic ectoderm, corresponding to the future amnion (Norris et al., 2002; Ying and Zhao, 2001). Bmp8a, Bmp7, and Gdf6 have also been detected in some extraembryonic ectoderm cells (Pijuan-Sala et al., 2019).
Secreted SMAD1/5/8 inhibitors are expressed in a pattern similar to SMAD2/3 inhibitors: throughout the epiblast, with particularly high levels in the primitive streak, and in AVE. Fst, whose product binds and neutralizes BMP ligands (Hedger et al., 2011) is expressed throughout the primitive streak (Albano et al., 1994). In situ data is unavailable for Chrd and Nog at this stage, so the spatial distribution of these inhibitors is not entirely clear, but both have been detected in AVE cells and in epiblast, including primitive streak cells. Additional secreted inhibitors, including Grem1, Nbl1, and Dand5, are also detected at low levels in some epiblast cells (Pijuan-Sala et al., 2019).
Wnt ligands are expressed in posterior regions of both epiblast and visceral endoderm, as well as throughout the extraembryonic ectoderm. Wnt3 is broadly expressed in the posterior epiblast (Kimura et al., 2001), where Wnt2, Wnt2b, Wnt3a, Wnt5a, Wnt5b, Wnt8a (Bouillet et al., 1996), and Wnt11 (Kispert et al., 1996) are also expressed at varying degrees in the primitive streak and emerging mesoderm (Pijuan-Sala et al., 2019). Wnt3 is also broadly expressed in posterior visceral endoderm (Kimura et al., 2001), where Wnt5a, Wnt9b and Wnt11 have been detected (Pijuan-Sala et al., 2019). Finally, Wnt6 and Wnt7b are strongly expressed in the extraembryonic ectoderm, where Wnt10a has also been detected (Pijuan-Sala et al., 2019) (Kemp et al., 2005).
Like SMAD2/3 and SMAD1/5/8 inhibitors, secreted Wnt inhibitors are expressed throughout the epiblast and in the AVE. Sfrps and Fzrb antagonize Wnt signaling by interact directly with Wnt ligands. Sfrp1 and Sfrp2 are detected at low levels throughout the epiblast and Frzb is expressed strongly in mesoderm and primitive streak cells and at lower levels in AVE (Pijuan-Sala et al., 2019). Sfrp1 (Kemp et al., 2005) and Sfrp5 (Finley et al., 2003) are strongly expressed in the anterior half of the visceral endoderm. And the product of Dkk1, which binds and inhibits the WNT co-receptor LRP6, is strongly expressed in the anterior-most region of the AVE (Zakin et al., 2000).
FGF ligands are expressed throughout the epiblast and visceral endoderm, while inhibitors are intracellular and expressed throughout the epiblast. Fgf5 (Hébert et al., 1991) and Fgf15 (Zakin et al., 2000) are detected throughout the epiblast at E6.5 and E6.75 and low levels of Fgf9 and Fgf18 have also been detected in epiblast cells (Pijuan-Sala et al., 2019). Embryonic expression of other ligands is restricted to the posterior region of the epiblast, including Fgf3, Fgf4 (Niswander and Martin, 1992), Fgf8 (Crossley and Martin, 1995), Fgf10 (Pijuan-Sala et al., 2019), and Fgf17 (Maruoka et al., 1998). Extraembryonic FGF expression is detected in the VE, where Fgf8 is restricted to AVE (Crossley and Martin, 1995), Fgf5 is expressed throughout the VE (Haub and Goldfarb, 1991), and Fgf10 has also been detected (Pijuan-Sala et al., 2019). Known FGF-signaling inhibitors are membrane bound proteins that regulate MAPK activation intracellularly (reviewed in Balasubramanian and Zhang, 2016; Böttcher and Niehrs, 2005). For this reason, we only considered embryonic expression of these genes. Il17rd (Lin et al., 2002), Spred1, Spred2, and Spry4 are expressed throughout the epiblast, where some Spry3 has also been detected (Pijuan-Sala et al., 2019). Spry2 is widely expressed in the embryo, with higher expression detected in the anterior half (Lin et al., 2002) and Spred3 and Spry1 are also expressed in the epiblast with an anterior bias (Pijuan-Sala et al., 2019).
Low levels of ATRA are likely produced in the epiblast and extraembryonic ectoderm at this stage. The primary enzyme thought to be responsible for producing all-trans-retinoic acid in vivo, Aldh1a2, is not yet expressed at this stage of development (Niederreither et al., 1997). However, expression is detected in the epiblast by the next stage, at E7.0. Additionally, its homolog Aldh1a3, which performs the same function (Rhinn and Dollé, 2012), has been detected in the epiblast and, at lower levels, in extraembryonic ectoderm by sc-RNA seq as early as E6.5 (Pijuan-Sala et al., 2019). Importantly, despite low expression of the ATRA synthesis enzyme, epiblast cells appear prepared to both produce and rapidly respond to the ATRA signal, as soon as the synthesis enzyme is present. This is evidenced by widespread embryonic expression of essential pathway components. For instance, synthesis enzymes and binding proteins specific to retinal, the precursor of ATRA and the substrate of the Aldh1a enzymes, are expressed in the epiblast at this stage, as are all of the RA receptor genes (Pijuan-Sala et al., 2019). Additionally, Crabp2, which produces a protein that binds available ATRA and translocates to the nucleus, so that the signaling molecule can interact with its receptors (Kam et al., 2012), is widely expressed in the epiblast at E6.5 and E6.75.
The expression pattern of ATRA inhibitors provides further evidence that RA may be important for differentiation from pluripotency in vivo. Cyp26a1, which encodes an enzyme that converts ATRA to other RA species, is widely expressed in extraembryonic tissues, anterior primitive streak, and emerging mesoderm, but notably absent from epiblast cells (Fujii et al., 1997; Pijuan-Sala et al., 2019). This suggests that ATRA levels in the embryo are tightly controlled, but also that some signaling activity may be essential for embryonic development from pluripotency, particularly in future neuroectoderm. Additionally, ATRA binding protein Crabp1 may act as an intracellular inhibitor (Boylan and Gudas, 1992) and is expressed in the epiblast layer (Pijuan-Sala et al., 2019).
Hedgehog signaling protein Indian hedgehog (Ihh) is expressed in the visceral endoderm at E6.5 (Becker et al., 1997). No inhibitors are strongly expressed at this stage.
Finally, Notch ligands are restricted to the epiblast layer, while an inhibitor is expressed in both epiblast and extraembryonic ectoderm. Activating ligands include Dll1, which is expressed in nascent mesoderm cells (Bettenhausen et al., 1995), Jag1, expressed in nascent mesoderm and primitive streak cells, and Jag2, found in some epiblast cells (Pijuan-Sala et al., 2019). Dll3, the secreted inhibitor, is weakly expressed throughout the epiblast with higher levels in nascent mesoderm (Dunwoodie et al., 1997) and is also detected at low levels in extraembryonic ectoderm (Pijuan-Sala et al., 2019).
Excluded values are indicated by gray background
Data Availability Statement
-
•
The published article includes all datasets generated during this study (Tables S2 and S7). Original/source data for Figure 2D in the paper is publicly available at https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-4840/(E-MTAB-4840, Lindsay et al., 2016). RT-PCR and microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.