Summary
Members of the genus Klebsiella have rapidly evolved within the past decade, generating organisms that simultaneously exhibit both multidrug resistance and hypervirulence (MDR-hv) phenotypes; such organisms are associated with severe hospital- and community-acquired infections. Carbapenem-resistant infections with unknown optimal treatment regime were of particular concern among the MDR-hv Klebsiella strains. Recent studies have revealed the molecular features and the mobile resistance elements they harbour, allowing identification of genetic loci responsible for transmission, stable inheritance, and expression of mobile resistance or virulence-encoding elements that confer the new phenotypic characteristics of MDR-hv Klebsiella spp. Here, we provide a comprehensive review on the taxonomic position, species composition and different phylotypes of Klebsiella spp., describing the diversity and worldwide distribution of the MDR-hv clones, the genetic mutation and horizontal gene transfer events that drive the evolution of such clones, and the potential impact of MDR-hv infections on human health.
Keywords: Klebsiella spp., Hypervirulence, Multidrug resistance, Convergence, Taxonomy, Evolution
Introduction
Various strains in the genus Klebsiella have evolved to become a major clinical and public health threat worldwide.1 Klebsiella spp. are opportunistic pathogens which are normally found in the flora of healthy individuals’ nose, throat, skin, and intestinal tract, but can also cause a range of infections, including pneumonia, soft tissue and surgical wound infections, urinary tract infections, bloodstream infections and sepsis.2 The Klebsiella genus comprises a wide diversity of species, including the Klebsiella pneumoniae species complex (KpSC) and several more genetically distant species.3 A large proportion of infections caused by Klebsiella spp. are due to two major pathotypes, namely the multidrug-resistant (MDR) and hypervirulent (hv) clones.4 Strains of the two branches were considered to be non-overlapping since they each exhibit different genetic backgrounds.4 However, Klebsiella spp. has demonstrated the ability to acquire genetic elements and mutations that confer antimicrobial resistance and/or virulence traits, leading to the ultimate emergence of the convergent clones, termed multidrug-resistant and hypervirulent (MDR-hv) Klebsiella spp.5,6 MDR-hv Klebsiella spp. are simultaneously hypervirulent and resistant to multiple antibiotics, and are known to be undergoing further evolution to produce phenotypically novel strains.6,7 A broad diversity of MDR-hv strains of Klebsiella spp. that evolved through diverse mechanisms has been reported across different continents in the world.8 The rise in number of severe infections and the increasing limitations in effective treatments rendered MDR-hv Klebsiella spp. real superbugs that pose serious challenges to public health.9 In this review, we provide an overview of the taxonomic position and species composition of Klebsiella spp. Based on the current information we classify different Klebsiella sp. clones of diverse phenotypes, explored the genetic diversity and worldwide distribution of the MDR-hv Klebsiella spp., describe the genetic mutation and horizontal gene transfer (HGT) events driving the evolution of such clones, and discuss the clinical impact of infections caused by MDR-hv strains.
Taxonomic position and species composition of the Klebsiella genus
The genus Klebsiella is a class of Gram-negative, encapsulated, nonmotile, rod-shaped and oxidase-negative bacteria.10 Strains of this genus were first isolated in the late 19th century and named by Trevisan (1885) to honor the German microbiologist Edwin Klebs (1834-1913).10 Klebsiella is classified under the Enterobacteriaceae family which contained a large array of biochemically distinct genus, including the model organism Escherichia coli and the notorious human pathogens Salmonella, Yersinia, Serratia, Enterobacter, Citrobacter, Kluyvera, Leclercia, Raoultella, Cronobacter, etc.3 The Klebsiella genus currently comprises a wide diversity of species, including species belonging to the K. pneumoniae species complex (KpSC) and other Klebsiella species (K. indica, K. terrigena, K. spallanzanii, K. huaxiensis, K. oxytoca, K. grimontii, K. pasteurii and K. michiganensis) that share an average of only 90% nucleotide identity with KpSC.3 KpSC has no formal taxonomic designation, and it commonly refers to closely related species that share 95%–96% average nucleotide identity with K. pneumoniae sensu stricto.3 At the time of writing, seven phylogroups that belong to KpSC have been classified, including K. pneumoniae (Kp1), K. quasipneumoniae subsp. quasipneumoniae (Kp2), K. variicola subsp. variicola (Kp3), K. quasipneumoniae subsp. similipneumoniae (Kp4), K. variicola subsp. tropica (Kp5), K. quasivariicola (Kp6) and K. africana (Kp7) .11 All taxa of the KpSC used to be misassigned by biochemical or proteomics assays as K. pneumoniae, since they possess overlapping features.12 Modern species identification methods rely largely on sequence-based classifiers or mass spectrometry (MALDI-TOF) platforms, both of which perform comparative analysis with reference genomes or strains, and require updated databases to reflect an accurate taxonomy.12,13 The multilocus sequence typing (MLST) scheme developed for typing K. pneumoniae isolates can be applied across the entire species complex.11 Closely related sequence types (ST) could be further designed as clonal complexes (CC) using eBURST (http://eburst.mlst.net/).14 The species K. terrigena, K. planticola and K. ornithinolytica have been transferred to the Raoultella genus based on the rpoB sequence.15
The type species of the Klebsiella genus, K. pneumoniae, is highly prevalent in clinical collections, accounting for an isolation rate of approximately 85%.3 K. pneumoniae was classified as one of the ESKAPE organisms (Enterococcus faecium,Staphylococcus aureus,K. pneumoniae,Acinetobacter baumannii,Pseudomonas aeruginosa and Enterobacter species), which are well-known highly virulent and antimicrobial resistant clinical pathogens.16 Two subspecies of K. pneumoniae, namely K. pneumoniae subsp. ozaenae and K. pneumoniae subsp. rhinoscleromatis, which are associated with a specific disease syndrome (atrophic rhinitis and rhinoscleroma, respectively), and one monomorphic clone, have been reported.17 Both subspecies sit squarely within the general population of K. pneumoniae phylogenetically and are regarded as hypervirulent clones derived from K. pneumoniae.11 Species including K. variicola and K. oxytoca, which are associated with clinical infections, have also emerged, yet their virulence profiles has not been fully characterized.12,18,19 The genetic features and clinical relevance of K. pneumoniae virulence factors in distant species remain poorly understood.11
Defining classical, hypervirulent and MDR-hv Klebsiella spp
Klebsiella spp. have gained the ability to acquire external genetic materials that enable the organisms to undergo extensive evolution.6 Two pathotypes, classical and hypervirulent Klebsiella spp., are reported to be associated with infections.6 Currently, the majority of currently available data are concentrated on K. pneumoniae, which is also of highest clinical importance.3 Classical K. pneumoniae (cKP) strains have gained increasing notoriety owing to its propensity to accumulate mutations and acquire determinants, leading to the emergence of multiple, extensive, or pan-drug resistant clones.1,20 More than 100 acquired antimicrobial resistance genes have been identified in K. pneumoniae, which encodes different products that confer resistance towards distinct classes of antibiotics including β-lactams, aminoglycosides, quinolones, tigecycline and polymyxins.16,20 The majority of K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae worldwide belong to the notorious CC258 clone (including ST258, ST11, ST340, ST437 and ST512).21 Several other clonal groups (CG), including CG14/15, CG17/20, CG29, CG37, CG43, CG101, CG147, CG152, CG231, CG307 and CG490, are also globally distributed and associated with multidrug resistance.20,22
Hypervirulent K. pneumoniae (hvKp), initially reported in the mid-1980s from the Asian Pacific Rim, are increasingly reported worldwide.23 Despite the lack of 100% specific and sensitive marker for hvKp, several phenotypic and clinical features have defined this K. pneumoniae variant.24 The first is their ability to cause severe infections associated with high pathogenicity and mortality in both immunocompromised and healthy hosts, typically presenting as pyogenic liver abscesses.6,25 Also, hvKp has been observed to trigger diseases at unusual sites, including endophthalmitis, necrotizing fasciitis, central nervous diseases (meningitis, bacteremia), etc.26 A second trait is their propensity for metastatic spread to distinct sites, which is uncommon among enteric Gram-negative bacilli, including cKP.9,23, 24, 25 Furthermore, a large proportion of hvKp colonies present a hypermucoviscous phenotype on agar plates which could be semi-quantitatively defined by a “string test”.6 However, the degree of correlation between hypermucoviscosity, capsule gene expression and hypervirulence remains to be clarified.26 Clinical hvKp clones are known to be less diverse than MDR K. pneumoniae.22 The majority of reported hvKp clones belong to serotype K1 or K2, with CG23 being the dominant K1 hvKp clone, whereas several genetically unrelated groups (CG25, CG65, CG66, CG86 and CG380, etc.) constitute the K2 strains.3,27 To date, hvKp belonging to serotypes K5, K16, K20, K54, K57, and KN1 have also been documented.26 The hvKp strains rely on a battery of virulence factors for survival and infection, among which enhanced capsule production and the synthesis of siderophores are dominant.26 Evidence from previous studies highlighted the importance of pLVPK-like virulence plasmids in the expression of the hypermucoid phenotype and siderophore production among hvKp isolates, since they harbor virulence-associated determinants that encode the regulators of the mucoid phenotype (rmpADC and rmpA2), production of siderophores (iutAiucABCD encoding aerobactin and iroBCDN clusters encoding salmochelin), expression of the metabolite transporter PEG344 and ABC-type transporter (fepBC) and the regulatory system for iron uptake (fecIRA).28, 29, 30 Additionally, a “high pathogenicity island” (KPHP1208) harboring genes that encode colibactin, microcin E492, and the siderophore yersiniabactin is related to the hypervirulence phenotype.27 A few candidate biomarkers were proposed for differentiation between hvKp from cKP, among which the genes peg-344, iucA, iroB, and plasmid-borne rmpA and rmpA2, and quantitative siderophore production of ≥30 μg/mL demonstrated >0.95 diagnostic accuracy, whereas the string test exhibited an accuracy of only 0.90.31 Besides, a previous study has demonstrated that the murine model, but not the Galleria mellonella model, is appropriate for validating suspected hvKp strains.32
MDR K. pneumoniae and hvKp are generally associated with two distinct subsets of K. pneumoniae lineages distinguishable by the presence of acquired resistance genes and several key virulence loci.22 The two lineages were considered to be non-overlapping over a period of 30 years after the discovery of hvKp.33 Yet the last few years witnessed an increasing number of reports on the convergent K. pneumonia clones (MDR-hvKp) that are simultaneously multidrug resistant and hypervirulent.5,34 MDR-hvKp isolates are highly diverse in genetic backgrounds and exhibit diversified antimicrobial resistance profiles.8 Hypervirulent and MDR-hv Klebsiella spp. other than K. penumoniae, including K. quasipneumoniae subsp. similipneumoniae, K. variicola, etc., has been reported, posing a new challenge to public health.12,35,36
Worldwide distribution and genetic backgrounds of MDR-hv Klebsiella spp
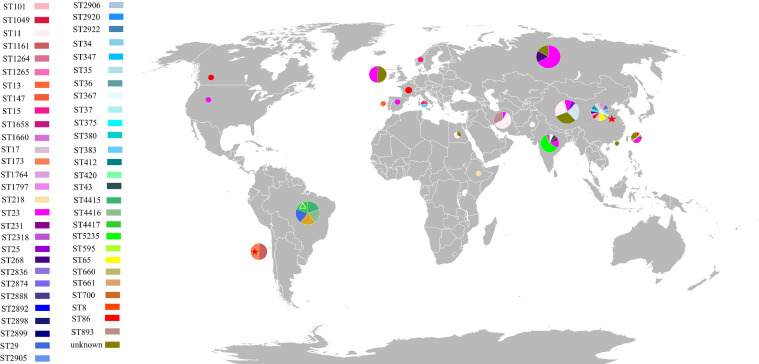
Since the first report of MDR-hvKp in 2015 and the fatal outbreak of ST11 carbapenem-resistant hvKp (CR-hvKp) reported in 2017 in China, research on MDR-hv Klebsiella spp. has become a hot topic.5,37 To date, MDR-hv Klebsiella spp. have been reported across several continents including Asia (China (China mainland, Hong Kong and Taiwan), India, Iran), Europe (France, Norway, the United Kingdom, Russia), Africa (Egypt), South (Brazil) and North (Canada, the USA) America.8 Most MDR-hv Klebsiella spp.-related studies were from Asia, in particular China. Infections caused by such convergent clones could be dated back to as early as 2008, almost a decade before the first report of such clones, when ST2888 and ST1264 extended-spectrum-β-lactamase (ESBL)-producing hvKp strains were isolated from the elderly in China.38 Results from previous retrospective studies suggested MDR-hv Klebsiella spp. is increasing in China over the recent years, with their resistance phenotypes expanding from ESBL-producing to non-susceptibility to last-line antibiotics including carbapenems, tigecycline and colistin.38,39 The majority of MDR-hv Klebsiella spp. reported belonged to K. pneumoniae sensu stricto, which could potentially be associated with the inaccuracy in species identification. MDR-hvKp belonging to at least 51 diverse sequence types and 13 serotypes (K1, K2, K5, K16, K19, K20, K24, K47, K51, K54, K57, K62, K64, KL108) have been reported, with regional differences seen from the literature (Figure 1). Only a few cases of MDR-hvKp infections were reported from Europe, Africa, South and North America, most of which were ESBL- or carbapenemase-producing isolates belonging to the sequence and sero-types of ST15:K24, ST23:K1, ST29: K19, ST268:K20, ST661: KNA, ST4415:KNA, and ST4416:KNA (NA, not available).8 MDR-hvKp isolates circulating outside Asia was reported in as early as the year 2012, when an ESBL-producing ST86:K2 hvKP strain was isolated from an immunocompromised patient in France.40 In India, infection cases caused by MDR-hvKp producing ESBLs or encoding resistance to carbapenems or colistin were reported, including those belonging to ST11: KNA, ST23:K1, ST43:KNA, ST231:K51, ST2318:KNA and ST5235:KNA.41 Particularly, ST5235 hvKp isolates resistant to the last-line antibiotics carbapenems and colistin were isolated from a series of neonatal sepsis.42 Infections caused by ST11:KNA, ST893:KNA and ST23:K1 CR-hvKp were reported from Iran from 2012-2018.43,44 In Taiwan (China), ESBL-producing and/or tigeycline-resistant hvKp that belong to ST1049:K5, ST23:K1, ST380:K2, ST65:K2, ST660:K16, ST8:K1, and ST86:K2 were reported during the period 2015-2018.45,46 Mainland China has reported the most cases of MDR-hvKp infections, covering at least 38 diverse sequence types and 11 serotypes.5,34,47 ST11 and ST23 MDR-hvKp isolates were most frequently reported from China, which is consistent with the finding in a previous study that ST11 and ST23 accounted for the majority of MDR K. pneumoniae (MDR-KP, 51/66, ∼77.27%) and hvKp (16/23, ∼69.57%), respectively.48 Species other than K. pneumoniae were reported, including carbapenem-resistant and colistin-resistant ST595:K16 K. variicola strains isolated in 2015 and 2016 in China, respectively, and strains of ST4417:K47 K. quasipneumoniae subsp. similipneumoniae isolated in Brazil in 2016 that were resistant to various antibiotics (fluoroquinolones, β-lactams, tetracyclines, trimethoprim, aminoglycosides, sulfonamides, macrolides, and fosfomycin) (Supplementary Table S1).36,47,49 CR-hvKp isolates producing carbapenemases were of particular concern among all MDR-hvKp isolates. KPC-2 was the most frequently reported carbapenemase produced by CR-hvKp isolates, and the production of other carbapenemases including NDM (NDM-1, NDM-5, and NDM-7), IMP, SIM, VIM (VIM-1 and VIM-2), and OXA-48-like has also been described.8
Figure 1.
Worldwide distribution of MDR-hv Klebsiella spp. Different colours of sectors represent different sequence types of the MDR-hv Klebsiella spp. as shown in the left of the figure. Sectors with asterisk and triangle represent the existence of K. variicola and K. quasipneumoniae subsp. similipneumoniae, respectively, and sectors without symbols represent that of K. pneumoniae. All MDR-hv Klebsiella spp. reported up to February 20th, 2022, were included. Multidrug-resistant Klebsiella sp. strains were defined based on the resistance to at least one agent in three or more antimicrobial categories in addition to ampicillin. Pie graph areas are relatively proportional to the total number of MDR-hv strains reported in each country/region.
Mutation and horizontal gene transfer events driving the formation of MDR-hv Klebsiella spp
Klebsiella spp. have gained the capacity to acquire both drug resistance and hypervirulence phenotype through accumulation of mutations and/or horizontal gene transfer (HGT).50 MDR-hv Klebsiella spp. was commonly considered to have evolved as a result of acquisition of virulence determinants by MDR-KP or through development of MDR phenotypes by hvKp.8 Accumulation of mutations, including mutations in the quinolone resistance-determining region (QRDR mutations in gyrA and parC), porin gene mutations (ompK36) and mutations leading to the over-expression of efflux pumps (OEP mutations: acrAB, oqxAB, ramA and rarA), was reported among hypervirulent clones (Table 1 g,h,i), leading to the emergence of MDR-hv Klebsiella spp. These mutations confer resistance to a diverse range of antibiotics including quinolones, carbapenems and tigecyclines. Taiwan (China) has reported the most cases of resistance-associated mutations among MDR-hvKp, covering isolates of ST23:K1, ST86:K2, ST268:K20, ST307:K1 and ST1049:K5 which often contain QRDR and OEP mutations.46,51 An ST86:K2 hvKp isolate carrying the plasmid-borne blaCTX-M-15 gene from France has developed carbapenem resistance due to deletion of a 11 bp region in the ompK36 gene, which resulted in truncation of the outer membrane protein.52 In China, QRDR mutations were reported among ST23:K1 hvKp strains carrying plasmid-mediated quinolone resistance determinants.53
Table 1.
Mechanisms driving the convergence of multidrug resistance and hypervirulence in Klebsiella spp.
| Country/region | Year of isolation | ST/serotype | No. of plasmids | Plasmid replicons and plasmid-borne resistance genes | gene mutation and overexpression * | formation mechanism ⁎⁎ | Genetic marker | Reference |
|---|---|---|---|---|---|---|---|---|
| China mainland | 2014 | ST23/K1 | 1 | pvir-res: IncHI1B/FIB (blaCTX-M-24) | - | a | 8-bp TSD bordering IS903D⁎⁎⁎ | 59 |
| China mainland | 2013 | ST23/K1 | 1 | pvir-res: IncHI1B/FIB (blaKPC-2, dfrA14) | - | a | 8-bp TSD bordering IS26 | 58 |
| China mainland | 2015 | ST23/K1 | 2 |
pvir: IncHI1B/FIB; pres: IncHI5 (blaDHA-1, sul1, qnrB4) |
- | b | - | 69 |
| China mainland | 2018 | ST23/K1 | 3 |
pvir: IncHI1B/FIB; pres-A: IncA (blaVIM-1, dfrA14b, aacA4, aphA15, aacA4, aphA15, aadA1b, catB2); pres-B: IncFII (qnrS, blaCTX-M-15, blaTEM-1C) |
- | b | - | 70 |
| China mainland | 2017 | ST23/K1 | 7 |
pvir: IncHI1B; pres-A: IncHI1B/FIB (blaNDM-1, aac(6′)-Ib-cr, blaOXA-1, catB3, arr-3, sul1); pres-B: IncFII (blaCTX-M-14); pres-C: IncFIIk (aadA16, aph(3′)-Ia, qnrB2, qnrS1, aac(6′)-Ib-cr, mph(A), tet(A), dfrA27, sul1); others: IncX4, IncP, ColRNAI |
- | b | - | 71 |
| China mainland | 2014 | ST1265/K1 | 2 |
pvir: IncHI1B/FIB; pres: NA (blaKPC-2) |
- | b | - | 72 |
| China mainland | 2017 | ST23/K1 | NA⁎⁎⁎⁎⁎ | NA (qnrS1, acc(6′)-Ib-cr, blaCTX-M-14/15) | M: gyrA, parC | b+g | - | 53 |
| China mainland | 2015 | ST661/K1 | NA | NA (blaDHA-1, mcr-1) | - | b | - | 73 |
| China mainland | 2018 | ST29/K54 | NA |
pvir: IncHI1B/FIB; pres: IncX3 (blaNDM-5) |
- | b | - | 74 |
| China mainland | 2016 | ST35/KL108 ⁎⁎⁎⁎ | 3 |
pres: IncX3 (blaNDM-5); others: NA |
- | c+d | - | 64 |
| China mainland | 2017 | ST86/K2 | 2 |
pvir: IncHI1B; pres: IncX6 (blaKPC-2 and △blaTEM-1) |
- | b | - | 75 |
| China mainland | 2017 | ST15/K19 | 3-4 |
pvir: IncHI1B/FIB; pres-A: IncFIB/FII (blaOXA-1, tet(A), aph (3’)-Ia, mph(A), blaCTX-M-15, blaTEM-1B, blaSHV-11, mph(E), msr(E), armA, sul1, blaDHA-1, qnrB4, dfrA12, aadA2, sul1); pres-B: IncFII (blaKPC-2, aac(6′)-Ib-cr, aac(3)-IId, mdf(A), catB4); other: ColRNAI |
M: ramR, parC | e | homologous fragment (unknown size) | 60 |
| China mainland | 2016 | ST11/K64 | 4 |
pvir: IncHI1B/FIB; pres-A: IncFII (blaKPC-2, blaSHV-12, blaCTX-M-65, blaTEM-1); pres-B: IncFII (qnrS1, blaLAP-2, tet(A), catA2, sul2); other: IncFIA |
- | e | homologous fragment (241-bp) | 61 |
| China mainland | 2015 | ST595/K16 | 3 |
pvir: IncFIB; pres: IncX5 (blaKPC-2, blaTEM-1); other: IncFIB/IncFII |
- | f | homologous fragments (889-bp upstream and 1,246-bp downstream) | 62 |
| France | 2017 | ST86/K2 | NA |
pvir: IncHI1B/FIB; pres: IncL (blaOXA-48) |
- | c | - | 52 |
| France | 2017 | ST86/K2 | NA |
pvir: IncHI1B/FIB; pres: IncN (blaCTX-M-15) |
M: ompK36, ramR | c+h | - | 52 |
| India | 2018 | ST23/K1 | 7 |
pvir: IncFIB (catA1); pres: IncFIB (qnrB1, catB, aac(6’)-Ib3, rmtF, arr-2); others: IncA/C2, IncFIB, IncX3, ColRNAI, Col440II |
- | a+c | 10-bp TSD bordering IS5075 | 41 |
| India | 2017 | ST23/K1 | 8 |
pvir: IncFIB (catA1); pres-A: IncFIB(qnrB1, catB, aac(6’)-Ib3, rmtF, arr-2); pres-B: IncX3 (aac(6’)-Ib-cr, armA, mph(E), msr(E), sul1); pres-C: ColKP3 (blaOXA-232); others: IncA/C2, IncFIB, ColRNAI, Col440II |
- | a+c | 10-bp TSD bordering IS5075 | 41 |
| Iran | 2012 | ST23/K1 | NA |
pvir: NA; pres: IncN (aacA7, blaVIM-2, dhfrI) |
- | b | - | 44 |
| Norway | 2014 | ST15/K24 | 7 |
pvir: IncHI1B/FIB (blaCTX-M-15, aph(3’)-Ia, dfrA, sat2, blaSHV-5, sul1, aadA1); pres: IncFII (aacA4, blaOXA-1, blaTEM, cat, blaCTX-M-15); others: IncFIB, Col440I, ColpVC and 2 nontypable plasmids |
- | k | genes for conjugal transfer on fusion plasmid | 65 |
| Norway | 2015 | ST15/K24 | 4 | pvir: IncHI1B/FIB (aph(3’)-Ia, dfrA, sat2, blaSHV-5, sul1, aadA1); pres: IncFII (aacA4, blaOXA-1, blaTEM, cat, tet(A), blaCTX-M-15); others: IncFIB and a nontypable plasmid | - | k | genes for conjugal transfer on fusion plasmid | 65 |
| Taiwan, China | 2015-2018 | ST268/K20; ST307/K1; ST23/K1; ST86/K2; ST1049/K5 | NA | NA | ramA upstream alterations, etc. | i | - | 46 |
| Taiwan, China | 2013-2016 | ST23/K1 | NA | NA (blaDHA-1, qnrS) | M: oqxR, ramR | i+j | - | 51 |
| Taiwan, China | 2013-2016 | ST307/K1 | NA | NA (qnrB, qnrS, aac(6′)-Ib-cr) | M: ramR, gyrA, parC; O: AcrAB efflux pump and RamA |
g+i+j | - | 51 |
*M, mutation; O, overexpression. ⁎⁎ Formation mechanisms: a, acquisition of resistance gene by virulence plasmid via intermolecular transposition; b, acquisition of conjugative resistance plasmid; c, acquisition of resistance plasmid with its conjugal transferability unknown; d, chromosomal integration of ICE element carrying virulence factors; e, transfer of virulence plasmid by forming a cointegrate with a helper conjugative plasmid mediated by homologous recombination; f, acquisition of conjugative plasmid with virulence genes acquired by homologous recombination; g, mutations in quinolone resistance-determining region; h, porin mutation; i, overexpression of efflux pump; j, acquisition of resistance gene (unknown mechanism); k, acquisition of conjugative fusion plasmid with virulence and resistance genes (hypothetical). Among these mechanisms, g, h, and i are accumulation of mutations, and others are horizontal gene transfer. ⁎⁎⁎ TSD, target site duplication. ⁎⁎⁎⁎ KL108 is correspond to capsule loci defined on the basis of gene content, for which the corresponding serological capsule types are yet to be defined. ⁎⁎⁎⁎⁎ NA, not available. This table only covers strains with validated evolutionary mechanisms. Except the ST595:K16 isolate which belonged to K. variicola, all other strains belonged to K. pneumoniae.
HGT is mainly mediated by several well-recognized mechanisms including transduction, transformation and conjugation, among which conjugation is considered the most significant.54,55 Acquired resistance and virulence-associated genes transferred among bacteria via mobile genetic elements include those located in conjugative and mobilizable plasmids, integrative conjugative elements (ICE), integrons, insertion sequences and transposons.56 Acquisition of resistance genes by hvKp through HGT involves mechanisms including resistance gene capture by virulence plasmid via intermolecular replicative transposition, acquisition of conjugative resistance plasmid, acquisition of resistance plasmid with unknown mechanism of conjugal transfer and acquisition of resistance genes through other unknown mechanisms (Table 1 a,b,c,j). HGT of resistance genes was reported from China, France, India and Iran, and involved hvKp clones ST23:K1, ST29:K54, ST35:KL108, ST86:K2, ST307:K1, ST661:K1 and ST1265:K1.8 Acquisition of resistance plasmids was the most commonly reported mechanism, followed by acquisition of resistance genes. Intermolecular transposition was an important mechanism responsible for integration of antibiotic resistance genes carried by the virulence plasmids. Mobile elements that mediate this transposition target the related hot spots on the virulence plasmids, leading to the integration of resistance genes and a duplicated hot spot sequence termed target site duplications (TSDs).57 The blaCTX-M-24, blaKPC-2 and dfrA14, and catA1 genes were integrated into the virulence backbones in ST23:K1 hvKp isolates upon mobilization of insertion sequences IS903D, IS26 and IS5075, generating 8-bp (GCACAGAGA), 8-bp (CTAAAATT) and 10-bp (TACCGGGAAG) TSD sequences bordering the mobile elements, respectively.41,58,59
The acquisition of virulence-associated genes by MDR Klebsiella spp. involves three main mechanisms: (i) obtaining a virulence plasmid by forming a cointegrate with a helper conjugative plasmid mediated by homologous recombination, (ii) acquisition of conjugative plasmid that contains virulence genes by homologous recombination, and (iii) chromosomal integration of ICE elements carrying the virulence factors (Table 1 d,e,f). Homologous recombination therefore plays a pivotal role in dissemination of hypervirulence-associated genes. An ST15:K19 MDR-hvKp and an ST11:K64 CR-hvKp strain with a homologous fragment of unknown size and 241-bp, respectively, were both found to have evolved through the first mechanism.60,61 An ST595:K16 CR-hv K. variicola strain which contained a 889-bp upstream and a 1,246-bp downstream homologous fragment, evolved from the second mechanisms.62 Strains that evolved from the third mechanism, which involves chromosomal integration of genetic elements harboring virulence-associated factors have also been reported.63 In addition, an ST35:KL108 CR-hvKp isolate was reported to have evolved through acquisition of a blaNDM-5-carrying plasmid and integration of a ∼76 Kb ICEKp1 element with the iroBCDN and rmpA genes in the chromosome.64 Another potential mechanism of evolution for MDR-hv Klebsiella spp. is the acquisition of a conjugative fusion plasmid which contains both virulence and resistance genes; yet reports of this mechanism are scarce and not supported by experimentally validated data (Table 1> k).65
The evolution of MDR-hv Klebsiella spp. is complicated and involves the aforementioned or other unknown mechanisms. For example, evolutionary events including simultaneous acquisition of resistance genes or plasmids and development of resistance-associated mutations, as well as acquisition of resistance genes or plasmids and chromosomal integration of ICE element, have been reported (Table 1). The highly variable genetic features of Klebsiella spp. rendered it able to rapidly adapt to diverse environmental niches.
Clinical implications of MDR-hv Klebsiella infections
Management of MDR-hv Klebsiella sp. infections requires both active antibiotic therapy and adequate infection control.66 No clinical trials assessing the ideal antibiotics for treating MDR-hv Klebsiella spp. infections has been conducted, hence empiric antibiotic therapy should take into account both the local antimicrobial resistance patterns and infection sites.66 A previous comparative study on evolutionary genomics suggested that MDR Klebsiella sp. clones pose the greatest risk, since they are more likely to acquire virulence genes than hypervirulent clones are to acquire resistance genes.22 Klebsiella spp. are intrinsically resistant to penicillin by producing different types of β-lactamases, such as SHV in K. pneumoniae, OKP in K. quasipneumoniae and LEN in K. variicola.2 The acquisition of antimicrobial resistance genes and accumulation of resistance-associated mutations rendered MDR Klebsiella spp. unresponsive to antimicrobial drugs.1 Infections caused by CR isolates are of key concern among all MDR Klebsiella strains, for which optimal treatment remains unavailable.67 According to a recent retrospective study, ceftazidime-avibactam is superior to other treatment regimens in treatment of CR K. pneumoniae bacteremia.68 Alternative antibiotic treatment options to consider include eravacycline, plazomicin, colistin, tigecycline, cefiderocol, meropenem/vaborbactam and imipenem/relebactam.5,66 These antibiotics have not been systematically evaluated for their efficacy against MDR-hv Klebsiella sp., thus they should only be consumed upon advice by clinical microbiologists. Besides, MDR-hv Klebsiella spp. infections should be controlled in a timely manner, since their hypervirulent nature and ability to undergo metastatic transfer would cause extensive damage in various vital organs.
Outstanding questions
Klebsiella, a notorious bacterial pathogen, comprises a wide diversity of species. It has evolved into hypervirulent and/or multidrug resistant strains over the past few decades. The convergent clone, MDR-hv Klebsiella sp., has disseminated worldwide, posing a significant threat on human health. Such strains evolved through acquisition of virulence genes by MDR clones or through development of MDR phenotypes by hv clones. The evolutionary events driving the emergence of MDR-hv Klebsiella sp. isolates is complicated, which predominantly involve accumulation of mutations and horizontal gene transfer. No optimal treatment is currently available for infections caused by MDR-hv Klebsiella sp. More work needs to be done to better understand this ‘superbug’ and help design feasible approaches to eradicate or halt further evolution of the existing strains of MDR-hv Klebsiella sp., including setting up a standard pipeline for accurate and rapid species identification, devising methods for rapid detection of capsule gene expression, hypermucoviscosity and hypervirulence, worldwide surveillance and molecular typing, and design of optimal treatment regime for a diverse range of MDR-hv Klebsiella spp.
Search strategy and selection criteria
Articles for this review were identified using PubMed, Google Scholar, and references from relevant articles using the search terms: ‘hypervirulence’ or ‘hypervirulent’ or ‘hypermucoviscous’ and ‘Klebsiella’, or ‘resistance’ or ‘resistant’ and ‘Klebsiella’. All papers reporting the case of multidrug resistant and hypervirulent Klebsiella sp. up to May 31st, 2021, were included in the MDR-hv Klebsiella spp. section and only the most impactful papers in other sections were considered.
Declaration of interests
All authors declare that they have no competing interests.
Acknowledgments
Contributors
DN drafted the manuscript, XY, EWCC and RZ edited the manuscript, SC edited the manuscript and supervised the project. All authors read and approved the final version of the manuscript. All figures are original creations for this manuscript, and no additional permissions are required for inclusion into the manuscript.
Acknowledgments
The research activities lead by SC are made possible by the following funders who had no role in writing this manuscript: Guangdong Major Project of Basic and Applied Basic Research (2020B0301030005). The funding sources played no part in the conceptualisation, drafting, editing, or reviewing of this manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103998.
Appendix. Supplementary materials
References
- 1.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences. 2015;112(27):E3574–E3E81. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyres KL, Lam MM, Holt KE. Population genomics of Klebsiella pneumoniae. Nature Reviews Microbiology. 2020;18(6):344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Kreiswirth BN. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. The Lancet infectious diseases. 2018;18(1):2–3. doi: 10.1016/S1473-3099(17)30517-0. [DOI] [PubMed] [Google Scholar]
- 5.Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. The Lancet infectious diseases. 2018;18(1):37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 6.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clinical microbiology reviews. 2019;32(3):e00001–e00019. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong N, Yang X, Zhang R, Chan EW-C, Chen S. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerging microbes & infections. 2018;7(1):1–8. doi: 10.1038/s41426-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang M, Kong X, Hao J, Liu J. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Frontiers in Microbiology. 2020;11:2774. doi: 10.3389/fmicb.2020.581543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert review of anti-infective therapy. 2019;17(2):71–73. doi: 10.1080/14787210.2019.1555470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brisse S, Grimont F, Grimont PAD. In: The Prokaryotes: A Handbook on the Biology of Bacteria Volume 6: Proteobacteria: Gamma Subclass. Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. Springer New York; New York, NY: 2006. The Genus Klebsiella; pp. 159–196. [Google Scholar]
- 11.RRW Margaret M.C.Lam, Watts Stephen C., Cerdeira Louise T., Wyres Kelly L., Kathryn E. Holt A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nature communications. 2021;12(1):1–16. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerging microbes & infections. 2019;8(1):973–988. doi: 10.1080/22221751.2019.1634981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues C, Passet V, Rakotondrasoa A, Brisse S. Identification of Klebsiella pneumoniae, Klebsiella quasipneumoniae, Klebsiella variicola and related phylogroups by MALDI-TOF mass spectrometry. Frontiers in microbiology. 2018;9:3000. doi: 10.3389/fmicb.2018.03000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. Journal of bacteriology. 2004;186(5):1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez J, Martínez L, Rosenblueth M, Silva J, Martínez-Romero E. How are gene sequences analyses modifying bacterial taxonomy? The case of Klebsiella. International Microbiology. 2004;7(4):261–268. [PubMed] [Google Scholar]
- 16.Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS microbiology reviews. 2017;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 17.Brisse S, Fevre C, Passet V, et al. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PloS one. 2009;4(3):e4982. doi: 10.1371/journal.pone.0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh L, Cariappa M, Kaur M. Klebsiella oxytoca: An emerging pathogen? Medical journal armed forces india. 2016;72:S59–S61. doi: 10.1016/j.mjafi.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neog N, Phukan U, Puzari M, Sharma M, Chetia P. Klebsiella oxytoca and Emerging Nosocomial Infections. Current Microbiology. 2021:1–9. doi: 10.1007/s00284-021-02402-2. [DOI] [PubMed] [Google Scholar]
- 20.Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends in microbiology. 2016;24(12):944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Dong N, Zhang R, Liu L, et al. Genome analysis of clinical multilocus sequence Type 11 Klebsiella pneumoniae from China. Microbial genomics. 2018;4(2) doi: 10.1099/mgen.0.000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyres KL, Wick RR, Judd LM, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS genetics. 2019;15(4) doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shon AS, Russo TA. Hypervirulent Klebsiella pneumoniae: the next superbug? Future microbiology. 2012;7(6):669–671. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- 24.Pomakova D, Hsiao C, Beanan J, et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. European journal of clinical microbiology & infectious diseases. 2012;31(6):981–989. doi: 10.1007/s10096-011-1396-6. [DOI] [PubMed] [Google Scholar]
- 25.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J, Wang T, Chen L, Du H. Virulence factors in hypervirulent Klebsiella pneumoniae. Frontiers in Microbiology. 2021;12:734. doi: 10.3389/fmicb.2021.642484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Struve C, Roe CC, Stegger M, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio. 2015;6(4):e00630. doi: 10.1128/mBio.00630-15. -15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y-T, Chang H-Y, Lai Y-C, Pan C-C, Tsai S-F, Peng H-L. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Walker KA, Miner TA, Palacios M, et al. A Klebsiella pneumoniae regulatory mutant has reduced capsule expression but retains hypermucoviscosity. MBio. 2019;10(2) doi: 10.1128/mBio.00089-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker KA, Treat LP, Sepúlveda VE, Miller VL. The small protein RmpD drives hypermucoviscosity in Klebsiella pneumoniae. Mbio. 2020;11(5):e01750. doi: 10.1128/mBio.01750-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Russo TA, Olson R, Fang C-T, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. Journal of clinical microbiology. 2018;56(9):e00776. doi: 10.1128/JCM.00776-18. -18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo TA, MacDonald U. The Galleria mellonella infection model does not accurately differentiate between hypervirulent and classical Klebsiella pneumoniae. Msphere. 2020;5(1):e00850. doi: 10.1128/mSphere.00850-19. -19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hennequin C, Robin F. Correlation between antimicrobial resistance and virulence in Klebsiella pneumoniae. European Journal of Clinical Microbiology & Infectious Diseases. 2016;35(3):333–341. doi: 10.1007/s10096-015-2559-7. [DOI] [PubMed] [Google Scholar]
- 34.Wyres KL, Nguyen TN, Lam MM, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome medicine. 2020;12(1):1–16. doi: 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shankar C, Nabarro LE, Muthuirulandi Sethuvel DP, et al. Draft genome of a hypervirulent Klebsiella quasipneumoniae subsp. similipneumoniae with novel sequence type ST2320 isolated from a chronic liver disease patient. J Glob Antimicrob Resist. 2017;9:30–31. doi: 10.1016/j.jgar.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Wai-Chi Chan E, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019 doi: 10.1038/s41564-019-0566-7. [DOI] [PubMed] [Google Scholar]
- 37.Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. International journal of infectious diseases. 2015;37:107–112. doi: 10.1016/j.ijid.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 38.Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Annals of clinical microbiology and antimicrobials. 2019;18(1):1–11. doi: 10.1186/s12941-018-0302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Sun G, Yu Y, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clinical infectious diseases. 2014;58(2):225–232. doi: 10.1093/cid/cit675. [DOI] [PubMed] [Google Scholar]
- 40.Surgers L, Boyd A, Girard P-M, Arlet G, Decré D. ESBL-producing strain of hypervirulent Klebsiella pneumoniae K2, France. Emerging infectious diseases. 2016;22(9):1687. doi: 10.3201/eid2209.160681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shankar C, Jacob JJ, Vasudevan K, et al. Emergence of Multidrug Resistant Hypervirulent ST23 Klebsiella pneumoniae: Multidrug Resistant Plasmid Acquisition Drives Evolution. Frontiers in cellular and infection microbiology. 2020;10:719. doi: 10.3389/fcimb.2020.575289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerjee T, Wangkheimayum J, Sharma S, Kumar A, Bhattacharjee A. Extensively Drug-Resistant Hypervirulent Klebsiella pneumoniae From a Series of Neonatal Sepsis in a Tertiary Care Hospital, India. Frontiers in medicine. 2021;8:186. doi: 10.3389/fmed.2021.645955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solgi H, Shahcheraghi F, Bolourchi N, Ahmadi A. Molecular characterization of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae ST11 harbouring blaNDM-1 and blaOXA-48 carbapenemases in Iran. Microbial Pathogenesis. 2020;149 doi: 10.1016/j.micpath.2020.104507. [DOI] [PubMed] [Google Scholar]
- 44.Tabrizi AMA, Badmasti F, Shahcheraghi F, Azizi O. Outbreak of hypervirulent Klebsiella pneumoniae harbouring blaVIM-2 among mechanically-ventilated drug-poisoning patients with high mortality rate in Iran. Journal of global antimicrobial resistance. 2018;15:93–98. doi: 10.1016/j.jgar.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 45.Lin Y-T, Cheng Y-H, Chuang C, et al. Molecular and clinical characterization of multidrug-resistant and hypervirulent Klebsiella pneumoniae strains from liver abscess in Taiwan. Antimicrobial agents and chemotherapy. 2020;64(5) doi: 10.1128/AAC.00174-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y-H, Huang T-W, Juan C-H, et al. Tigecycline-non-susceptible hypervirulent Klebsiella pneumoniae strains in Taiwan. Journal of Antimicrobial Chemotherapy. 2020;75(2):309–317. doi: 10.1093/jac/dkz450. [DOI] [PubMed] [Google Scholar]
- 47.Furlan JPR, Gallo IFL, de Campos TA, Stehling EG. Genomic Characterization of a Multidrug-Resistant and Hypermucoviscous/Hypervirulent Klebsiella quasipneumoniae subsp. similipneumoniae ST4417 Isolated from a Sewage Treatment Plant. Microbial Drug Resistance. 2020;26(11):1321–1325. doi: 10.1089/mdr.2019.0417. [DOI] [PubMed] [Google Scholar]
- 48.Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Frontiers in microbiology. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Y, Feng Y, McNally A, Zong Z. Occurrence of colistin-resistant hypervirulent Klebsiella variicola. Journal of Antimicrobial Chemotherapy. 2018;73(11):3001–3004. doi: 10.1093/jac/dky301. [DOI] [PubMed] [Google Scholar]
- 50.Barlow M. Springer; 2009. What antimicrobial resistance has taught us about horizontal gene transfer. Horizontal Gene Transfer; pp. 397–411. [DOI] [PubMed] [Google Scholar]
- 51.Lin Y-T, Cheng Y-H, Juan C-H, et al. High mortality among patients infected with hypervirulent antimicrobial-resistant capsular type K1 Klebsiella pneumoniae strains in Taiwan. International journal of antimicrobial agents. 2018;52(2):251–257. doi: 10.1016/j.ijantimicag.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Beyrouthy R, Dalmasso G, Birer A, Robin F, Bonnet R. Carbapenem Resistance Conferred by OXA-48 in K2-ST86 Hypervirulent Klebsiella pneumoniae, France. Emerging infectious diseases. 2020;26(7):1529. doi: 10.3201/eid2607.191490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Du F-l, Xiang T-x, et al. High prevalence of plasmid-mediated quinolone resistance determinants among serotype K1 hypervirulent Klebsiella pneumoniae isolates in China. Microbial Drug Resistance. 2019;25(5):681–689. doi: 10.1089/mdr.2018.0173. [DOI] [PubMed] [Google Scholar]
- 54.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nature Reviews Genetics. 2015;16(8):472. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 55.Holmes AH, Moore LS, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. The Lancet. 2016;387(10014):176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 56.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clinical microbiology reviews. 2018;31(4):e00088. doi: 10.1128/CMR.00088-17. -17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He S, Hickman AB, Varani AM, et al. Insertion sequence IS 26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. MBio. 2015;6(3):e00762. doi: 10.1128/mBio.00762-15. -15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong N, Lin D, Zhang R, Chan EW-C, Chen S. Carriage of bla KPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. Journal of Antimicrobial Chemotherapy. 2018;73(12):3317–3321. doi: 10.1093/jac/dky358. [DOI] [PubMed] [Google Scholar]
- 59.Shen D, Ma G, Li C, et al. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a rare blaCTX-M-24-harboring virulence plasmid. Antimicrobial agents and chemotherapy. 2019;63(3) doi: 10.1128/AAC.02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li R, Cheng J, Dong H, et al. Emergence of a novel conjugative hybrid virulence multidrug-resistant plasmid in extensively drug-resistant Klebsiella pneumoniae ST15. International journal of antimicrobial agents. 2020;55(6) doi: 10.1016/j.ijantimicag.2020.105952. [DOI] [PubMed] [Google Scholar]
- 61.Xie M, Chen K, Ye L, et al. Conjugation of virulence plasmid in Clinical Klebsiella pneumoniae strains through formation of a fusion plasmid. Advanced biosystems. 2020;4(4) doi: 10.1002/adbi.201900239. [DOI] [PubMed] [Google Scholar]
- 62.Yang X, Chan EW-C, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nature microbiology. 2019;4(12):2039–2043. doi: 10.1038/s41564-019-0566-7. [DOI] [PubMed] [Google Scholar]
- 63.Lam MM, Wyres KL, Duchêne S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nature communications. 2018;9(1):1–10. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen Z, Gao Q, Qin J, Liu Y, Li M. Emergence of an NDM-5-producing hypervirulent Klebsiella pneumoniae sequence type 35 strain with chromosomal integration of an integrative and conjugative element, ICEKp1. Antimicrobial agents and chemotherapy. 2020;64(1):e01675. doi: 10.1128/AAC.01675-19. -19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lam MM, Wyres KL, Wick RR, et al. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. Journal of Antimicrobial Chemotherapy. 2019;74(5):1218–1222. doi: 10.1093/jac/dkz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choby J, Howard-Anderson J, Weiss D. Hypervirulent Klebsiella pneumoniae–clinical and molecular perspectives. Journal of internal medicine. 2020;287(3):283–300. doi: 10.1111/joim.13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ernst CM, Braxton JR, Rodriguez-Osorio CA, et al. Adaptive evolution of virulence and persistence in carbapenem-resistant Klebsiella pneumoniae. Nature medicine. 2020;26(5):705–711. doi: 10.1038/s41591-020-0825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shields RK, Nguyen MH, Chen L, et al. Ceftazidime-avibactam is superior to other treatment regimens against carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrobial agents and chemotherapy. 2017;61(8):e00883. doi: 10.1128/AAC.00883-17. -17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie Y, Tian L, Li G, et al. Emergence of the third-generation cephalosporin-resistant hypervirulent Klebsiella pneumoniae due to the acquisition of a self-transferable bla DHA-1-carrying plasmid by an ST23 strain. Virulence. 2018;9(1):838–844. doi: 10.1080/21505594.2018.1456229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dong N, Sun Q, Huang Y, et al. Evolution of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae by acquisition of blaVIM-1-bearing plasmid. Antimicrobial agents and chemotherapy. 2019;63(9):e01056. doi: 10.1128/AAC.01056-19. -19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu B-T, Su W-Q. Whole genome sequencing of NDM-1-producing serotype K1 ST23 hypervirulent Klebsiella pneumoniae in China. Journal of medical microbiology. 2019;68(6):866–873. doi: 10.1099/jmm.0.000996. [DOI] [PubMed] [Google Scholar]
- 72.Li C, Ma G, Yang T, et al. A rare carbapenem-resistant hypervirulent K1/ST1265 Klebsiella pneumoniae with an untypeable blaKPC-harboured conjugative plasmid. Journal of global antimicrobial resistance. 2020;22:426–433. doi: 10.1016/j.jgar.2020.04.009. [DOI] [PubMed] [Google Scholar]
- 73.Gu D-x, Huang Y-l, Ma J-h, et al. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrobial agents and chemotherapy. 2016;60(8):5099. doi: 10.1128/AAC.00476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan Y, Li Y, Wang G, et al. bla NDM-5 carried by a hypervirulent Klebsiella pneumoniae with sequence type 29. Antimicrobial Resistance & Infection Control. 2019;8(1):1–9. doi: 10.1186/s13756-019-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu Z, Chu W, Li X, et al. Genomic Features and Virulence Characteristics of a Community-Acquired Bloodstream Infection-Causing Hypervirulent Klebsiella pneumoniae ST86 Strain Harboring KPC-2-Encoding IncX6 Plasmid. Microbial Drug Resistance. 2021;27(3):360–368. doi: 10.1089/mdr.2019.0394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.