Summary
The increased prevalence of obesity and metabolic diseases has heightened interest in adipose tissue biology and its potential as a therapeutic target. To better understand cellular heterogeneity and complexity of white adipose tissue (WAT), we employed cytometry by time-of-flight (CyTOF) to characterize immune and stromal cells in visceral and subcutaneous WAT depots under normal and high-fat diet feeding, by quantifying the expression levels of 32 surface marker proteins. We observed comparable proportions of immune cells in two WAT depots under steady state, but depot-distinct subtypes of adipose precursor cells (APC), suggesting differences in their adipogenic and fibrogenic potential. Furthermore, in addition to pro-inflammatory immune cell shifts, significant pro-fibrotic changes were observed in APCs under high-fat diet, suggesting that APCs are early responders to dietary challenges. We propose CyTOF as a complementary and alternative tool to current high-throughput single-cell transcriptomic analyses to better understand the function and plasticity of adipose tissue.
Subject areas: Immunology, Cell biology, Specialized functions of cells
Graphical abstract

Highlights
-
•
Application of CyTOF for cellular characterization in two adipose depots
-
•
Adipose depot-distinct APC subpopulations
-
•
APCs are early responders under obesogenic conditions to regulate WAT fibrosis
Immunology; Cell biology; Specialized functions of cells
Introduction
White adipose tissue (WAT) is a highly active metabolic and endocrine organ that plays a vital role in modulating systemic homeostasis and whole-body metabolism. WAT is widely distributed throughout the organism and largely categorized into subcutaneous WAT, which is located beneath the skin and visceral WAT, which accumulates around internal organs (Tandon et al., 2018). The regional distribution and functional differences between WAT depots have been associated with the development of metabolic syndromes and cardiovascular health. While the accumulation of visceral WAT is a strong predictor of insulin resistance and cardiovascular diseases, subcutaneous fat has been shown to have protective effects against metabolic dysfunction (Gabriely et al., 2002; Galassi et al., 2006; Manolopoulos et al., 2010; Tran et al., 2008). Previous murine studies have demonstrated that transplantation of inguinal WAT (IWAT), a major subcutaneous WAT depot, into the abdominal cavity of recipient mice leads to improvements in glucose tolerance and insulin sensitivity (Hocking et al., 2008; Tran et al., 2008). Intriguingly, these improvements were not observed upon transplantation of perigonadal WAT (PWAT), a visceral WAT depot, into the intra-abdominal or subcutaneous space, indicating inherent depot-specific functional characteristics of fat tissues. Additionally, while IWAT possesses stronger thermogenic capacity than PWAT in response to cold stimulation, PWAT is more sensitive to energy intake status, such as high-fat diet or fasting, than IWAT (Ding et al., 2016a; Lim et al., 2012; Marcelin et al., 2017). These results suggest that WAT depots possess unique and distinct functional properties that mediate differential responses to various physiological and environmental stimuli. However, the cause and underlying mechanism for the intrinsic differences between WAT depots are still unclear.
WAT is a heterogeneous organ comprised of diverse cell populations, including mature adipocytes, adipocyte precursor cells (APC) (also termed adipose stromal cells), vascular cells, and various immune cells. Dynamic interactions of these cell populations are essential for WAT remodeling and its functional adaptation under diverse metabolic and environmental challenges (Burl et al., 2018; Cao, 2007, 2010, 2013; Shao et al., 2021; Spallanzani et al., 2019). A number of studies have identified cellular heterogeneity and complexity of adipose stromal cells by employing single-cell transcriptomic analyses, such as single-cell and single-nuclei RNA sequencing (sc-RNAseq and sn-RNAseq, respectively). These subsets of stromal cells have unique roles in regulating tissue homeostasis, adipogenesis, and remodeling, and they play a critical role in regulating immune responses (Jimenez et al., 2021; Shao et al., 2021; Spallanzani et al., 2019). Recently, Merrick et al. identified a DPP4+ progenitor subpopulation as highly proliferative and multipotent stromal cells in murine IWAT that are resistant to adipogenic differentiation and are maintained by TGFβ signaling (Merrick et al., 2019). Furthermore, they identified cellular trajectories and lineage hierarchy in WAT, showing that DPP4+ progenitors give rise to ICAM1+ and CD142+ preadipocytes committed to the adipogenic lineage. Similarly, another study used sc-RNAseq to identify APCs and immune cells in IWAT and PWAT and showed that β3-adrenergic receptor activation promotes de novo adipogenesis in PWAT by triggering rapid proliferation of APCs, but not in IWAT (Burl et al., 2018). Notably, clinical studies have also identified transcriptional differences in adipose depots between lean and obese populations (Gerhard et al., 2014; Lefebvre et al., 1998; Modesitt et al., 2012; Passaro et al., 2017). A recent study identified cellular differences in subcutaneous and visceral WAT from bariatric surgery patients (with or without type 2 diabetes) using sc-RNAseq (Vijay et al., 2020). This study identified unique depot- and disease-specific APC populations that vary in their origins and mitochondrial activity. Collectively, these studies show that sc-RNAseq has emerged as a powerful tool to identify and characterize cellular heterogeneity of various WAT depots at the transcriptional level. However, few studies have characterized proteomic differences between IWAT and PWAT in multiple cellular subtypes.
High-dimensional mass cytometry (also known as cytometry by time-of-flight; CyTOF) has emerged as a powerful tool to investigate heterogeneous cell populations in tissues (Ajami et al., 2018; Korin et al., 2017; van Unen et al., 2016). The combination of over 50 surface and intracellular markers enables the identification and characterization of various cellular clusters with a high degree of reliability and consistency. While single-cell transcriptomic analyses reveal the diverse and complex nature of adipose cellular architecture, such methods do not provide information at the protein level which may provide additional insight into cellular function and composition and can reveal the lineage and maturation state of individual cells (Spitzer and Nolan, 2016). Conventional protein detection methods such as flow cytometry and immunohistochemistry are technically limited in the number of parameters that can be analyzed simultaneously due to spectral overlap. This is a major limitation in the ability of these tools to capture cellular heterogeneity and detect rare immune and stromal cell subsets within adipose tissue. Detecting an equivalent range of cell types would require multiple flow cytometry analyses, which may lead to batch-to-batch and experimental variability in results (Chuah and Chew, 2020). Meanwhile, CyTOF analysis employs antibodies tagged with heavy metal isotopes, allowing simultaneous assessment of more than 50 markers on single cells. As a result, CyTOF is frequently applied to conduct deep profiling of immune and stromal subsets in the tumor microenvironment and other disease-targeted tissues (Chew et al., 2017; Chuah and Chew, 2020). For example, peripheral blood immunophenotyping by CyTOF has provided insight into immune changes associated with autoimmune diseases (Cheung and Utz, 2011) and highlighted the differences between activated, bystander, and infected cells to elucidate viral disease pathogenesis and progression of diseases such as type 1 diabetes (Wiedeman et al., 2020). In clinical settings, understanding the differences in these populations can reveal predictive markers that may provide insight into disease progression, treatment response, and efficacy.
Evidently, CyTOF is a powerful tool for the identification and characterization of heterogeneous cell populations. As WAT is a key organ in regulating whole-body metabolism, a comprehensive cellular characterization of its depots will provide insight into the functional properties of the tissue. Although previous studies have demonstrated transcriptomic differences between the depots, heterogeneity of WAT depots has not been comprehensively analyzed based on cell surface marker expression using CyTOF. Our aim is to phenotype multiple cellular populations in murine PWAT and IWAT and their response to high-fat diet (HFD) feeding to highlight their proteomic differences.
Results
Cellular heterogeneity between visceral and subcutaneous WAT
We performed CyTOF to analyze the cellular composition of PWAT (i.e., visceral fat) and IWAT (i.e., subcutaneous fat) depots from 12-week-old C57Bl/6J male mice that were fed with normal diet (ND). The lymphoid cell-rich lymph nodes were removed from IWAT to solely assess the fat-resident immune and stromal cells. The PWAT and IWAT depots were pooled from 2–3 mice in each sample and were dissociated into single-cell suspensions using conventional collagenase digestion. The CyTOF antibody panel was assembled from 32 metal-tagged surface markers to identify hematopoietic, stromal, and vascular cells within WAT depots (Table S1). As shown in the workflow in Figure 1A, mass cytometry data were collected on the Helios instrument and all downstream analyses were performed on live singlets (which were pre-gated on the following channels: DNA1-191Ir, DNA2-193Ir, EQ-140Ce, Event-length, Center, Offset, and Cisplatin-195Pt). Two-dimensional biaxial gating was used to identify and quantify the CD45+ hematopoietic cells and PDGFRα+ stromal cells. We used two different CyTOF analysis tools available on Cytobank (www.cytobank.org): viSNE (visualization of t-distributed Stochastic Neighbor Embedding) and SPADE (Spanning-tree Progression Analysis of Density-normalized Events) algorithms to visualize and analyze the cellular composition of the two WAT depots—PWAT and IWAT. viSNE is a well established CyTOF analysis tool that captures the high-dimensional single-cell data into a spatial 2D representation. SPADE algorithm downsamples data and clusters cells with similar protein expression levels into a customizable hierarchy (i.e., number of nodes, downsampling percentage) (Figure 1A). Using the combination of these methods, we were able to quantify and visualize various cell types in the adipose depots based on clustering from 32 parameters (Tables S1 and S3). The median expression level of each of the 32 markers was quantified and summarized in a heatmap (Figure 1B). The cells were broadly grouped into immune cells (CD45+), which were further grouped into myeloid and lymphoid clusters, and lineage-negative PDGFRα+ stromal cells, which collectively comprise the majority of the SVF cells (Figures 1C and 1D). These immune and stromal cellular subtypes were clearly visualized as distinct clusters on viSNE (Figure 1E) and SPADE (Figure 1F) plots of visceral and subcutaneous adipose tissues under normal diet.
Figure 1.
Characterization of white adipose tissue immune and stromal cell populations by mass cytometry
(A) Schematic illustration of experimental design. Visceral (PWAT) and subcutaneous (IWAT) fat pads were isolated from 12-week-old male mice fed with either normal chow diet (ND) or 45% high-fat diet (HFD) for 4 weeks. WAT pooled from 2–3 mice was dissociated into a single cell suspension and stained with heavy metal-tagged antibodies. Live singlets were processed on Cytobank for further analysis.
(B) The median expression of 32 markers in WAT. Heatmap indicating the normalized transformed median intensity of each marker.
(C) Cellular composition of CD45+ hematopoietic, PDGFRα+ stromal, and CD45-/PDGFRα-cells in PWAT and IWAT.
(D) Immune cell composition of myeloid and lymphoid populations in PWAT and IWAT. ∗p < 0.05.
(E). Representative viSNE plots of ND PWAT and ND IWAT of the identified cellular populations.
(F) Representative SPADE plots of ND PWAT and ND IWAT, using automatic clustering by SPADE algorithm. Plots are displaying CD45 expression.
Characterization of adipose progenitor cells (APCs) in WAT depots
Earlier studies identified APCs in WAT as lineage-negative (CD45-/CD31-)/CD29+/CD34+/Sca-1+/PDGFRα+ cells (Rodeheffer et al., 2008). In our study, lineage-negative PDGFRα+ APCs accounted for approximately 44% and 33% of total SVF cells in lean PWAT and IWAT, respectively (Figure 1C). Merrick et al. identified DPP4+ APCs as highly proliferative multipotent cells exhibiting stem-like properties. DPP4-expressing progenitors were more prevalent in IWAT compared to PWAT, and their identity was maintained by TGFβ signaling (Merrick et al., 2019). These cells were shown to give rise to ICAM1+ or CD142+ pre-adipocytes, committed to the adipogenic lineage. Based on these studies, we used six different APC markers—DPP4, ICAM1, CD34, CD142, CD9, and Sca-1—in our CyTOF panel to identify previously defined subsets within PDGFRα+ stromal cells in IWAT and PWAT (Figure 2A). DPP4-expressing APCs were enriched in IWAT but were rare in PWAT of lean animals, with higher DPP4 median expression, confirming the presence of more stem-like progenitor populations in IWAT (Figure 2B). In contrast, the subpopulation of APCs expressing ICAM1 were primarily distinct from those expressing DPP4 and showed a trend of higher expression in PWAT compared to IWAT although the difference was not statistically significant (Figure 2C). Interestingly, while CD34, a conventional stem/progenitor marker, has often been suggested to be a general APC marker, we found that its expression was highest in DPP4-expressing cells (black arrows in SPADE plots in Figures 2B and 2D), further identifying these cells as more multipotent and stem-like in nature (Buffolo et al., 2019; Raajendiran et al., 2019). CD142 was first identified in a subpopulation of progenitor cells in mouse IWAT that was shown to inhibit adipogenic differentiation both in vivo and in vitro through paracrine mechanisms. These cells were consequently termed adipogenesis-regulatory cells (Aregs) (Schwalie et al., 2018). Contrary to this report, in the aforementioned study by Merrick et al., CD142+ cells derived from DPP4+ cells marked a population of preadipocytes that were fully adipogenic (Merrick et al., 2019). Therefore, these distinct APC clusters within the two adipose depots were further characterized and assessed, along with the expression of other conventional APC surface markers. As shown in Figure 2E, CD142 was broadly expressed across APCs but was largely absent in the cluster of DPP4-expressing cells (pink arrowheads in viSNE plots in Figures 2B and 2E). Furthermore, the median expression of CD142 was significantly higher in PWAT compared to IWAT (Figure 2E, violin plot). As expected, Sca-1 and CD9 were broadly expressed across various APC subsets in both depots (Figures 2F and 2G), confirming their use as APC surface markers. Interestingly, Sca-1 median expression was significantly higher in IWAT compared to PWAT, confirming the presence of a larger pool of stem-like progenitors in IWAT.
Figure 2.
Detection of depot-specific heterogeneity of adipose progenitor cell populations by mass cytometry
(A) Representative biaxial plots of adipocyte precursor cells (APC; PDGFRα+) and immune cells (CD45+). APCs were further analyzed with 6 additional markers including DPP4, ICAM1, CD142, CD9, CD34, and Sca-1. Representative viSNE and SPADE plots illustrating median expression of APC markers within PWAT and IWAT of ND-fed mice (B–G). B) DPP4, C) ICAM1, D) CD34, E) CD142, F) Sca-1, and G) CD9. ∗∗∗p < 0.001.
To further compare the inflammatory, fibrotic, and adipogenic characteristics of CD9low and CD9high APCs from PWAT and IWAT, we sorted CD9low and CD9high PDGFRα+ APCs from the two depots using FACS (Figure 3A, Table S2) and conducted qPCR analysis (Figures 3B-3F) to assess the expression of various surface markers and fibrosis and adipogenesis-related genes. First, we confirmed that the expression of CD9 is significantly higher in CD9high-sorted cells in both depots (Figure 3B). In alignment with our CyTOF data, DPP4 expression was much higher in CD9low cells from IWAT, compared to PWAT (Figure 3C). In contrast, the expression of CD142 was much lower in IWAT (Figure 3C). In agreement with previous studies, CD9high cells expressed significantly higher levels of fibrotic genes, particularly in PWAT (Figure 3D). Similarly, the expressions of adipogenic genes were significantly lower in CD9high APCs in both depots (Figure 3E). A recent study identified Cxcl14 as an anti-inflammatory factor secreted specifically from IWAT APCs (Nahmgoong et al., 2022). Indeed, we have identified that Cxcl14 expression was higher in IWAT (Figure 3F), suggesting that secreted factors from IWAT APCs may prevent adipose tissue fibrosis. Additionally, as shown in Figure 3G, CD9lowAPCs from PWAT also exhibited greater in vitro adipogenic potential than those from IWAT, confirming that APCs from PWAT are primed to mediate more active de novo adipogenesis than IWAT. Collectively, these data support our findings that APCs within discrete adipose depots have differing functional properties that are likely reflected in their unique expression pattern of surface markers which can be identified by CyTOF analysis.
Figure 3.
Characterization of CD9low and CD9high APC populations in PWAT and IWAT
(A) Schematic diagram of FACS sorting of CD9low and CD9high APCs from PWAT and IWAT for gene expression analysis and in vitro adipogenic differentiation and immunofluorescence staining.
(B) Relative gene expression of CD9 in sorted cells from two depots.
(C) Relative gene expression of various APC markers in sorted cells.
(D) Fibrotic gene signatures of sorted cells.
(E) Adipogenic gene signatures of sorted cells.
(F) Relative gene expression of Cxcl14 of sorted cells.
(G) Representative images showing differential adipogenic potential in APCs sorted from IWAT and PWAT. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
Next, we investigated the expression patterns of these key APC markers in bulk RNA-sequencing analysis of human visceral and subcutaneous WAT depots obtained from the GTEX consortium (Consortium, 2020). Consistent with our findings in mice, expression of DPP4 and CD34 was higher in human subcutaneous WAT compared to visceral WAT, whereas ICAM1 and CD142 levels were higher in human visceral WAT (Figure S1). Taken as a whole, we have shown distinct clustering of APCs in IWAT and PWAT using CyTOF, and our data largely corroborate previous sc-RNAseq data from murine studies (Merrick et al., 2019; Schwalie et al., 2018) and bulk tissue RNA-sequencing data from human samples (Consortium, 2020).
HFD-induced changes in APC populations
Studies have addressed the dynamic changes that occur in APCs during obesity, but the functional role of each subpopulation in obesity-induced WAT remodeling is poorly understood. Recent studies showed that high expression of CD9 marked a subpopulation of progenitor cells that promoted obesity-induced fibrosis in both human and mouse visceral fat (Hepler et al., 2018; Marcelin et al., 2017). HFD-induced activation of PDGFRα promoted accumulation of these pro-fibrotic CD9high cells whereas CD9low pro-adipogenic counterparts were reduced under obese conditions. Similarly, CD9high PDGFRβ+ perivascular progenitors, termed fibro-inflammatory progenitors (FIPs), were described in mouse PWAT (Hepler et al., 2018). FIPs lack adipogenic capacity and mediate HFD-induced fibrotic phenotypes whereas their CD9low/PDGFRβ+ counterparts were highly adipogenic. At steady state, we observed that the proportion of CD9low APCs was greater in lean PWAT than IWAT (i.e., ND PWAT vs ND IWAT) (Figures 4A and 4B), suggesting that there is a larger pool of adipogenesis-prone progenitors in PWAT, which may explain their rapid expandability under diet challenges, such as HFD. In HFD-fed animals, the relative proportion of pro-fibrotic CD9high cells increased in PWAT but remained unchanged or slightly decreased in IWAT (Figures 4A and 4B). To determine how these changes are reflected in the alteration of other APC-specific cell surface markers, we characterized the CD9low (adipogenic) and CD9high (fibrogenic) APC subsets and assessed which subpopulations of APCs contribute to adipogenic and fibrogenic progenitor cell types in two depots. We first stratified CD9low and CD9high APCs and examined the expression levels of other markers (Figures 4C, 4D, S2A, and S2B). In alignment to our results above (Figures 2 and 3), DPP4+ cells were present in very low proportions within CD9low APCs in PWAT, while there was a large subset of DPP4+ cells in IWAT (Figure 4C, filled histograms of DPP4 panel). In contrast, most CD9low APCs in PWAT were highly expressing both ICAM1 and CD142 (Figure 4C, blue histograms). This suggests the heterogeneity of progenitor cells that contribute to adipogenesis between the two depots. During HFD feeding, WAT undergoes active adipogenesis to respond to excess nutrients. Thus, we assessed which APC subsets contribute to HFD-induced adipogenic remodeling, as shown in Figures 4C and 4D (unfilled histograms). CD9low APCs in PWAT (blue histograms) of HFD-fed mice expressed higher levels of ICAM1 and CD142 but reduced DPP4, compared to those in IWAT (Figures 4C and 4D), suggesting a more committed pro-adipogenic phenotype in response to nutrient excess. Further, 4 weeks of high-fat feeding led to increased expression of ICAM1 (presumably DPP4-) within the CD9low APC subset, indicating that these cells are likely responsible for HFD-induced early adipogenesis (Figure 4C, blue unfilled histogram in ICAM-1 panel). However, in IWAT, DPP4+and ICAM1 expression did not change by HFD within the CD9low APCs, suggesting that their adipogenic capacity may be limited and unaltered in response to short-term HFD feeding (Figure 4C, red histograms in DPP4, ICAM1, and CD142 panels). Although upregulation of CD9 expression has been associated with HFD-induced fibrogenesis in PWAT (Marcelin et al., 2017), the relationship between its expression level and tissue function in IWAT is not clear. Compared to PWAT, IWAT was reported to be relatively resistant to fibrotic changes in response to HFD feeding, and the ratio of CD9high to CD9low progenitors did not significantly change in obese conditions (Marcelin et al., 2017). In contrast to CD9low cells, CD9high progenitors did not show significant difference in DPP4 and ICAM1 expression between PWAT and IWAT in both ND and HFD conditions (Figures S2A and S2B). These data suggest that the CD9low progenitor cells in particular, may derive from different subpopulations within PWAT and IWAT, with differing function and response to metabolic challenge. Collectively, CyTOF analysis of a combination of APC markers can further elucidate the complexity of APC populations and provide insight into the differential response of PWAT and IWAT depots to HFD.
Figure 4.
HFD-mediated depot-specific changes in APC populations
(A) Biaxial plots displaying changes in CD9 expression in APCs from ND- and HFD-fed mice in PWAT and IWAT.
(B) Quantification of CD9low and CD9high APCs from ND and HFD in PWAT and IWAT.
Representative histograms (C) and violin plots (D) of APC-specific marker expression in CD9low populations of PWAT and IWAT. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, ∗∗∗∗p < 0.001.
Identification of immune cells in WAT depots by CyTOF
Adipose tissue-resident immune cells have been reported to play a key role in the maintenance of adipose tissue function and homeostasis, and the composition of immune cells determines its inflammatory state and metabolic function. Among the CD45+ immune cells, which represented 38% of live singlets in PWAT and 50% of live singlets in IWAT of mice on a normal diet (ND), we identified diverse lymphoid and myeloid cell populations (Figures 1C and 1D). Overall, the relative proportions of cellular subtypes within the lymphoid and myeloid fractions were similar between PWAT and IWAT depots at steady state, with myeloid cells comprising the greatest cellular proportion in both depots (Figure 1D). We chose cell surface markers that allow us to identify myeloid populations that have been reported to play a role in WAT homeostasis, including macrophages, which are the most prevalent immune cells in adipose tissue, dendritic cells (DCs), neutrophils, eosinophils, and monocytes (Figure 5A, Table S3). Overall, we did not observe drastic differences in most myeloid populations between the two depots, although there was a greater proportion of macrophages in IWAT, with no significant differences in M1 and M2 subsets. Dendritic cells, particularly DC-2 (characterized by high CD11b expression), were significantly enriched in PWAT, which may have implications in their role in inflammatory changes. Interestingly, monocytes appeared to be more prevalent in IWAT, although the biological implication is unclear. Moreover, T cells comprised the largest proportion of lymphoid cells (Figure 5B). The majority of T cells in both depots express TCRαβ chains, confirmed by co-expression of CD3ε and TCRβ (data not shown). The main T cell subsets identified in our panel included CD4+ T, CD8+ T, and regulatory T cells (Tregs) (Figure 5B, Table S3). Although the panel employed in the current study does not include Foxp3, a key transcription factor of Tregs, we were able to identify these cells with several surface markers (CD3+, CD4+, and CD25+) (Figure 5B, Tables S1 and S3). The remaining CD3ε+ cells may be γδ T cells, which have shown importance in modulating Tregs and adaptive thermogenesis but are present at lower levels in adipose depots at steady state (Kohlgruber et al., 2018). There were a few depot-specific differences in minor lymphoid cell populations but the biological significance of these differences at steady state is not clear (Figure 5B). Notably, there was a significantly higher proportion of ILC2 in PWAT compared to IWAT. This is consistent with previous reports, as visceral WAT (i.e., PWAT) progenitor cells produce IL-33 to promote ILC2 activity (Mahlakoiv et al., 2019).
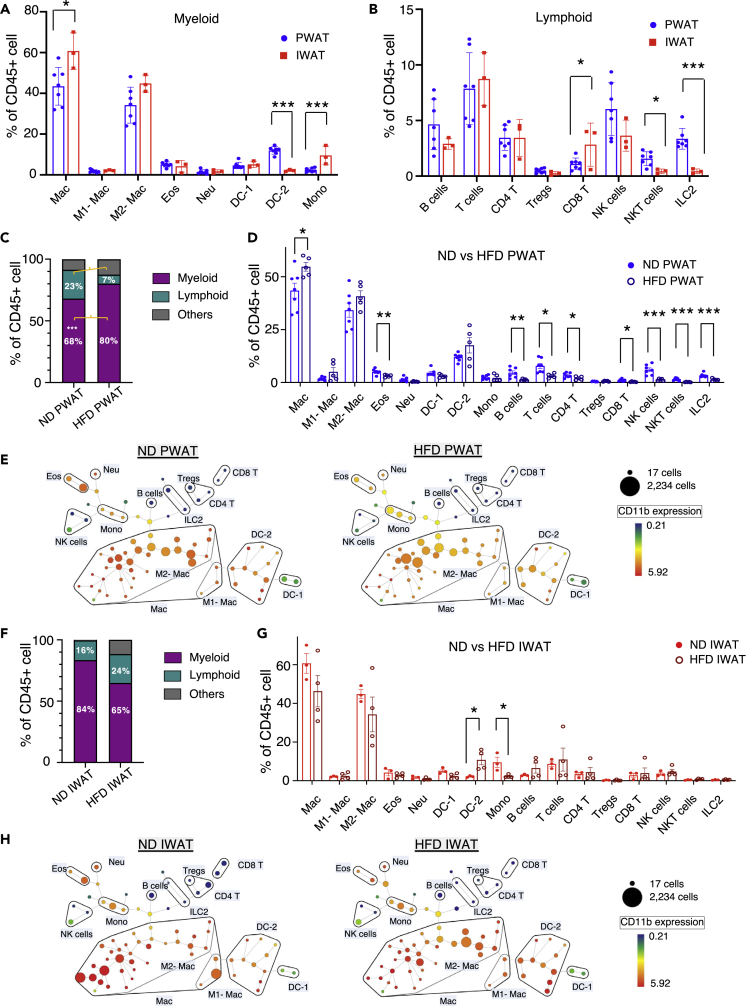
Figure 5.
Differential abundance of immune cell populations in PWAT and IWAT
Mass cytometry analysis of (A) myeloid and (B) lymphoid cell populations in PWAT and IWAT under normal diet (ND). High-fat diet (HFD)-induced alterations in immune cells (C and D) in PWAT and their SPADE plots (E). HFD-induced alteration in immune cells (F and G) in IWAT and their SPADE plots (H). Data are presented as mean ± SEM. n = 3–7. Illustrative SPADE plots of identified immune cell populations in (E) and (H), displaying CD11b+ expression. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
HFD-induced immune cell alterations in WAT depots
HFD-induced obesity results in a state of chronic inflammation. PWAT in particular has been reported to be more prone to obesity-induced inflammation whereas studies suggest that IWAT is relatively resistant to these changes (Marcelin et al., 2017). We observed significantly higher levels of total CD45+ immune cells by HFD in PWAT, but not IWAT (Figures S3A and S3B), likely through increased myeloid cell populations (Figure 5C), confirming that HFD leads to proliferation of tissue-resident subpopulations and infiltration of immune cells into adipose tissue (Liu et al., 2020; Lumeng et al., 2007; Weisberg et al., 2003). Studies using both flow cytometry and sc-RNAseq have identified molecular signatures of inflammatory myeloid cell populations present in obese tissue (Harasymowicz et al., 2021; Hill et al., 2018).
HFD leads to increased total macrophages and induces polarization of macrophages, resulting in increased CD11c+ M1 inflammatory macrophages with a relative decrease in anti-inflammatory CD206+ M2 macrophages (Russo and Lumeng, 2018). Recent studies identified a population of lipid-associated macrophages that highly express CD9 and are thought to play an important role in obesity-induced adipose tissue remodeling in both murine and human visceral fat (Harasymowicz et al., 2021; Jaitin et al., 2019). Stratification of macrophage populations based on expression of CD9 and Ly6C was reported to differentiate populations that contribute to adipogenesis and pro-inflammatory conditions (Hill et al., 2018). As shown in Figure S4, we were able to observe this reported heterogeneity in macrophages based on their expression of CD206, CD11c, CD9, and Ly6C. In alignment to previous reports, we observed that the total number of macrophages was significantly increased by HFD in PWAT (Figure 5D). Interestingly, we observed a mild increase of both M1 and M2 macrophage subsets, which could be attributed to the increase in pan-macrophages. As our HFD feeding regimen lasted for four weeks, this may be insufficient to observe significant changes in macrophage subtypes in WAT. We also identified two subsets of DCs (DC-1 and DC-2, Figure 1B), the second most prevalent immune cell population in adipose tissue, distinguished by the presence or absence of CD11b expression (Table S3). CD11b+ DCs, termed DC-2 in our analysis, have been reported to contribute to obesity-induced pathologies in adipose tissue (Macdougall et al., 2018). Concomitant with the increased proportion of immune cells and total macrophages observed selectively in PWAT, HFD feeding resulted in a moderate increase in pro-inflammatory DC-2, along with decreased type 2 innate immune cells (eosinophils, ILC2) and decreased NKT, which have been associated with HFD-induced reductions in adipose thermogenesis and obesity-induced chronic inflammation (Ding et al., 2016b) (Figure 5D). Interestingly, we observed an overall decrease in various lymphoid populations by HFD in PWAT, although increased B and T cell populations have been observed in obese adipose tissue (Misumi et al., 2019; Winer et al., 2011) (Figure 5D). This may be due to the significant increase in myeloid populations by HFD, resulting in decreased relative proportions of various lymphoid populations. In contrast, most of the immune populations in IWAT were not significantly changed by HFD, although we observed a significant increase in pro-inflammatory DC-2 (Figure 5G). It is likely that our current regimen of short-term HFD (4 weeks) may not be sufficient to induce robust and extensive remodeling of the adipose immune landscape but we observed a trend toward a proinflammatory phenotype in both fat depots (Harasymowicz et al., 2021; Jaitin et al., 2019). Furthermore, HFD-induced immune cell alterations were analyzed and visualized by SPADE algorithm, which displays cell number and marker expression in each immune cell population in PWAT (Figure 5E) and IWAT (Figure 5H). Collectively, these data show that deep immune profiling obtained by CyTOF offers comparable data to flow cytometry for the analysis of immune cells in adipose tissues. Importantly, our CyTOF antibody panel was able to distinguish myeloid and lymphoid cell populations that are known to be involved in adipose tissue function and inflammation.
Discussion
In this study, we used mass cytometry with a range of surface markers distinguished by other studies to analyze murine WAT at a single-cell resolution. We identified various immune and progenitor cell populations in WAT and compared their composition in visceral and subcutaneous depots. Moreover, we characterized their changes upon HFD, in order to gain insight into cellular changes in adipose tissues under obesogenic conditions. To date, the characterization of adipose immune populations at the protein level has depended heavily on flow cytometric immune profiling, which is limited by the number of parameters that can be analyzed simultaneously. Our findings show that extensive cellular profiling obtained with CyTOF offers reliable and comprehensive data for the analysis of adipose-resident immune and progenitor cell populations.
Characterization of stromal cell heterogeneity at single-cell resolution has allowed us to better understand the development and functional plasticity of WAT under physiological and pathological conditions. Previous studies have shown that HFD-induced de novo adipogenesis is primarily limited to PWAT, via activation of APCs leading to hyperplastic growth whereas IWAT depots display low rates of adipogenesis (Jeffery et al., 2015; Wang and Scherer, 2014). Marcelin et al. identified a CD9high subset of PDGFRα+ APCs that accumulated in obese PWAT, characterized by their loss of adipogenic and gain of fibrogenic phenotype, whereas the CD9low APCs had high adipogenic capacity. Our current study identified a greater proportion of CD9low APCs in PWAT than IWAT at steady state, suggesting a bigger pool of adipogenic APCs that can contribute to rapid adipogenesis and tissue expandability upon excess energy consumption. Moreover, we observed that HFD-induced loss of CD9low APC was exclusive to PWAT (Figures 4A and 4B). DPP4+ APCs, a multipotent, highly proliferative APC subpopulation maintained by TGFβ signaling, were reported to have low adipogenic potential but give rise to ICAM1+ and CD142+ preadipocytes, which are committed to the adipogenic lineage. Consistently, we found that the CD9low APCs in PWAT highly express ICAM1 with very low expression of DPP4 (Figure 4C). In contrast, in IWAT, DPP4 expression was high and ICAM1 was low in this same population, even under HFD conditions. While the functional significance of differential marker expression in APC subpopulations of IWAT and PWAT is not fully clear, our data suggest distinct origins of adipogenic progenitor cells in different fat depots. Notably, a previous study has demonstrated that APCs from subcutaneous and visceral adipose tissue depots display distinct adipogenic and fibrogenic programs (Shao et al., 2021). While visceral WAT contains distinct adipogenic and fibrotic PDGFRβ+ APC subpopulation, in subcutaneous WAT, these two processes appear to be co-enriched within the same APC subpopulation, which may explain depot-specific adipogenesis and pathological remodeling under HFD. Other depot-specific factors such as anatomical location of the APCs within the tissue and expression of cytokines and chemotactic factors may also confer unique properties and functions to each cell subpopulation (Jimenez et al., 2021; Kohlgruber et al., 2018; Maghsoudlou et al., 2019).
Our finding that short-term HFD feeding increases the proportion of CD9high pro-fibrogenic APCs suggests that APCs are highly sensitive to energy fluctuations and rapidly respond to accommodate excess nutrient availability and obesogenic growth. The dynamic changes in APCs, particularly in PWAT, suggest that remodeling of the APCs may precede the remodeling of the immune cell compartment to a pro-inflammatory microenvironment during obesity. It is likely that in our study, 4 weeks of HFD was insufficient to observe a significant shift to an inflammatory milieu. One study tracked changes to the immune cell compartment of PWAT during several timepoints of HFD feeding and observed that dramatic remodeling of CD45+ immune populations occurred between 6 and 12 weeks on HFD compared to mice fed with ND (Jaitin et al., 2019). Similarly, others reported few HFD-induced changes in adipose lymphoid populations between 5 and 8 weeks of HFD whereas significant changes occurred after 8 weeks (Kohlgruber et al., 2018).
In the current study, CyTOF allowed us to simultaneously assess changes in multiple cell populations at the protein level, providing more robust information on the cellular composition of WAT, which would not have been possible with conventional flow cytometry, given the limitation in the number of markers that can be used. More recently, analysis of stromal and immune cell heterogeneity using sc-RNAseq has largely advanced the field of adipose tissue biology, through the discovery of new progenitor cell subpopulations and identification of a spectrum of macrophage subtypes. Sc-RNAseq captures a wider range of cellular profiles in an unbiased manner and pathway analyses can provide robust information on cellular patterns and biological changes that drive specific phenotypes. Thus, ultimately, we suggest the use of multiple single-cell level approaches to fully understand the functional role of heterogenous adipose cellular populations and immunoregulation of the tissue. Similar to flow cytometry, a limitation of CyTOF is that the markers of interest are pre-selected by the experimenter, and it requires adequate understanding of the tissue to select the most informative markers in identifying key cell populations. Hence, sc-RNAseq serves as a better tool for the discovery of new cell types and target molecular pathways. However, due to the massive quantity of information generated by sc-RNAseq, proper quality control and filtering cells with insufficient reads and high mitochondrial genes are required. Additionally, depending on the method of tissue dissociation and single cell isolation, various cellular stress response pathways could be significantly altered in the cellular transcriptome (O'Flanagan et al., 2019). Thus, CyTOF may provide a more robust deep immune and stromal profiling of multiple major cellular subsets within a sample at the protein level which directly mediates cellular processes (Spitzer and Nolan, 2016). One study used both RNA-seq-based analysis and CyTOF to assess heterogeneous cellular components of the tumor microenvironment in patients with gastric cancer. While sc-RNAseq identified new cell types not covered by the selected CyTOF markers, CyTOF was a better tool in detecting targeted immune cell subsets (Kashima et al., 2021). Therefore, the information obtained by combining protein- and sequencing-based technologies, can be applied to adipose tissue biology to offer deeper cellular phenotyping of heterogeneous stromal and immune cell populations at high resolution (Landhuis, 2018). Another limitation of CyTOF is that further sorting of specific cell populations is not possible as the cells are ionized during acquisition. Thus, for further analyses, populations of interest can be sorted and analyzed by flow cytometry. Furthermore, high-dimensional CyTOF analysis can be used as a prognostic tool in clinical settings to identify predictive biomarkers of disease and immune response (Lingblom et al., 2018; Subrahmanyam and Maecker, 2017). The identification of depot- and diet-associated alterations in immune and stromal composition provides opportunities to determine cellular parameters that correlate with prevalence of metabolic diseases and predict therapeutic response to dietary treatments. Moreover, our human data revealed that human adipose tissues exhibit similar regional variations as seen in mice. Thus, CyTOF can be also applied to the investigation of human WAT and its response to various nutritional and environmental stimuli which can be used in clinical settings.
Limitations of the study
The current study has employed CyTOF to assess various surface marker proteins on immune and stromal cells in adipose tissues. However, intracellular markers have not been assessed, which may shed more light on the cellular functionality as well as secretory and inflammatory properties of various cell types. In addition, the animals used in this study were all males. Previous studies have identified sex-dependent depot differences and tissue remodeling in response to high-fat diet. Thus, employing female animals may provide insight into sex-distinct adipogenesis and obesogenic WAT growth. Lastly, we were unable to detect a distinct cluster of APCs that expresses PDGFRβ. Although previous studies have identified PDGFRβ+ perivascular APCs, particularly in visceral WAT, there was an absence of cells that express a discernable level of PDGFRβ in our samples. This could be due to the discrepancy in signal intensity of PDGFRβ antibodies that are tagged with fluorochromes and heavy metals, or its transcript level may not correlate with surface protein level. To address this issue, lineage-tracing models or fluorescence-tagged mouse models may allow clear detection and isolation of PDGFRβ-expressing stromal cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD45 (30-F11) – 89Y | Fluidigm | Cat #3089005B; RRID:AB_2651152 |
| Anti-mouse Ly6C (HK1.4) | Biolegend | Cat #128016; RRID:AB_1732076 |
| Anti-mouse CD44 (IM7) | Biolegend | Cat #103002; RRID:AB_312953 |
| Anti-mouse CD11 c (N418) | Biolegend | Cat #117302; RRID:AB_313771 |
| Anti-mouse SiglecF (E50-2440) | BD Biosciences | Cat #552125; RRID:AB_394340 |
| Anti-mouse MHC2 (M5/114.15.2) | Biolegend | Cat #107614; RRID:AB_313329 |
| Anti-mouse CD11b (M1/70) | Biolegend | Cat #101202; RRID:AB_312785 |
| Anti-mouse CD19 (1D3) | BD Biosciences | Cat #550284; RRID:AB_393579 |
| Anti-mouse CD24 (M1/69; Maxpar-ready) | Biolegend | Cat #101829; RRID:AB_2563732 |
| Anti-mouse CD31 (390; Maxpar-ready) | Biolegend | Cat #102425; RRID:AB_2563741 |
| Anti-mouse DPP4 (H194-112) | Biolegend | Cat #137802; RRID:AB_2093730 |
| Anti-mouse ICAM1 (YN1/1.7.4) | Biolegend | Cat #116102; RRID:AB_313693 |
| Anti-mouse c-Kit (2B8; Maxpar-ready) | Biolegend | Cat #105829; RRID:AB_2563710 |
| Anti-mouse PDGFRα (APA5) | ThermoFisher | Cat #14-1401-82; RRID:AB_467491 |
| Anti-mouse CD64 (X54-5/7.1) | Biolegend | Cat #139302; RRID:AB_10613107 |
| Anti-mouse CD142 (AF3178) | R&D | Cat #AF3178; RRID:AB_2278143 |
| Anti-mouse CD25 (3C7) | Biolegend | Cat #101902; RRID:AB_312845 |
| Anti-mouse CD8b (H35-17.2) | ThermoFisher | Cat #14-0083-82; RRID:AB_657757 |
| Anti-mouse Sca1 (E13-161.7) | Biolegend | Cat #122502; RRID:AB_756187 |
| Anti-mouse CD4 (RM4-5; Maxpar-ready) | Biolegend | Cat #100561; RRID:AB_2562762 |
| Anti-mouse CD9 (EM-04) | Novus | Cat #NBP1-44876; RRID:AB_10008106 |
| Anti-mouse Ly6G (1A8) | Biolegend | Cat #127602; RRID:AB_1089180 |
| Anti-mouse KLRG1 (2F1) | BD Biosciences | Cat #562190: RRID:AB_11154418 |
| Anti-mouse TCRβ (H57-597) | Biolegend | Cat #109202; RRID:AB_313425 |
| Anti-mouse NK1.1 (PK136) | Biolegend | Cat #108702; RRID:AB_313389 |
| Anti-mouse CD29 (HMβ1-1) | Biolegend | Cat #102202; RRID:AB_312879 |
| Anti-mouse CD206 (C068C2) | Biolegend | Cat #141702; RRID:AB_10900233 |
| Anti-mouse CD34 (RAM34) | ThermoFisher | Cat #14-0341-82; RRID:AB_467210 |
| Anti-mouse CD3 (145-2C11; Maxpar-ready) | Biolegend | Cat #100345; RRID:AB_2563748 |
| Anti-mouse CD127 (A7R34; Maxpar-ready) | Biolegend | Cat #135029; RRID:AB_2563716 |
| Anti-mouse B220 (RA3-6B2) | Biolegend | Cat #103202; RRID:AB_312987 |
| Anti-mouse PDGFRα-PE (APA5) | Biolegend | Cat #135906; RRID:AB_1953269 |
| Anti-mouse CD31-Pacific Blue (390) | Biolegend | Cat #102422; RRID:AB_10612926 |
| Anti-mouse CD45-PECy7 (104) | BD Biosciences | Cat #560696; RRID:AB_1727494 |
| Anti-mouse CD9-PE/Dazzle 594 (MZ3) | Biolegend | Cat #124822; RRID:AB_2800602 |
| CD16/CD32 Fc Block (2.4G2) | BD Biosciences | Cat #553142; RRID:AB_394657 |
| Chemical, peptides, and recombinant proteins | ||
| Type II Collagenase | Worthington Biochem | Cat #LS004176 |
| Critical commercial assays | ||
| Fixable Viability Stain 510 | BD Biosciences | Cat #564406 |
| HCS LipidTox Red | ThermoFisher | Cat #H34476 |
| DAPI | BD Sciences | Cat #564907 |
| RNeasy Micro Kit | Qiagen | Cat #74004 |
| Red Blood Cell Lysis Buffer | Sigma | Cat #R7757 |
| Cell-ID Intercalator | Fluidigm | Cat #201192A |
| TRIzol | ThermoFisher | Cat #15596026 |
| M-MLV Reverse Transcriptase | ThermoFisher | Cat #28025013 |
| Deposited data | ||
| Genotype-Tissue Expression Project | GTEx Consortium, 2013 | https://www.gtexportal.org/home/ dbGAP Study Accession phs000424.v8.p2 |
| Software | ||
| GraphPad Prism 9.0 | GraphPad Software, CA, USA | graphpad.com |
| Cytobank | Beckman Coulter Life Sciences, IN, USA | mybeckman.ca/flow-cytometry |
| FCS Express 7.0 | DeNovo Software, CA, USA | denovosoftware.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hoon-Ki Sung (hoon-ki.sung@sickkids.ca).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
All animal experimental protocols were approved by the Animal Care Committee of The Centre for Phenogenomics (TCP) conformed to the standards of the Canadian Council on Animal Care. The investigators were not blinded to allocation during experiments. The animals were housed in a specific pathogen-free facility in ventilated cages with controlled environmental settings (22°C±1, 30–60% humidity), 12-h light/dark cycles, and free access to water. C57Bl/6J mice were bred from TCP in-house mouselines (#000664). Male mice at 12 weeks of age were used for the study. Half the cohort was fed with normal chow, and half were fed with 45% high-fat diet for 4 weeks.
Method details
Adipose tissue stromal cell isolation
Visceral and subcutaneous white adipose tissues, pooled from 3 mice per sample, (∼1 g) was dissected, finely minced, and digested with Type II collagenase in a 37°C incubator for 30–45 min. Digested tissue was neutralized with DMEM/F12 media supplemented with 10% FBS and filtered through a 100 μm cell strainer. The flow-through was centrifuged for 5 min at 300 g. After aspirating the supernatant, red blood cell lysis buffer was added to the cell pellet and incubated in room temperature for 1–2 min. The lysis buffer was neutralized with DMEM/F12 media (with 10% FBS) and centrifuged for 5 min at 300 g. The cell pellets were further processed for CyTOF or for FACS sorting of APC populations.
CyTOF (mass cytometry) staining and data processing
The cells were washed in cell staining medium (CSM) and pelleted by centrifuging for 5 min at 300 g. Fc receptors were blocked by suspending the cell pellet in 25 μL of purified anti-mouse CD16/CD32 for 10′ at RT. Metal-tagged antibodies specific for surface markers were diluted to 2X the desired final concentration in CSM. Clone and vendor information for the antibodies is provided in Table S1. An equal volume of the antibody cocktail was added to the cells without washing out Fc block, and the cells were incubated for 30′ at RT. Cells were washed with 2 mL of CSM, and pellet was collected by centrifugation for 5′ at 300 g. After washing with 2 mL PBS, cells were re-suspended in 100 μL PBS and 100 μL of 2X cisplatin viability stain for 5′ at RT and quenched by adding 2 mL CSM. Cells were washed by 3 mL of CSM and collected by centrifugation for 5′ at 600 g. Cells were stained with 1 mL of 100 nM Cell-ID Iridium intercalator overnight at 4°C. Next day, the cells were washed once with CSM and once with PBS. Cell pellets were re-suspended in Maxpar Cell Acquisition Solution containing EQ normalization beads (#201237, Fluidigm), according to the manufacturer’s protocol. Data were collected by Helios instrument and converted to FCS files. The FCS files were normalized using the global EQ bead passport value by the Helios software. Cytobank Enterprise (Beckman Coulter) was used to generate viSNE and SPADE plots as well as biaxial plots. The histograms were generated using FCS Express 7.
FACS sorting of CD9high and CD9 low APCs
For flow cytometry, SVF cells were washed once in PBS by centrifugation at 300 g for 5 min. After removal of the supernatant, cells were incubated with anti-mouse antibodies recognizing CD45, CD31, PDGFRα, CD9 and Fixable Viability Stain 510 (BD) for identification of dead cells (Table S2). Cells were washed with FACS staining buffer by centrifugation at 300 g for 5 min and the cell pellets were resuspended in FACS staining buffer. CD9high and CD9low APC populations were sorted and collected into FBS using a Sony SH800 cell sorter. Immediately after sorting, the collected cells were washed in FACS staining buffer and plated for cell differentiation experiments or lysed in Trizol for gene expression analyses.
Real-time PCR analysis of stromal cells
Cells sorted by FACS were lysed in Trizol and total RNA was extracted using RNeasy Micro Kit (Qiagen). Complementary DNA was synthesized from 100–500 ng of RNA using M-MLV reverse transcriptase (Invitrogen). Gene expression assay was conducted using SYBR green methods on Quantstudio 5 (Applied Biosystems), and relative CT values were normalized by 36B4 gene. Primer sequences are available upon request.
In vitro adipogenic differentiation and immunofluorescent staining
Sorted CD9high and CD9low APCs from IWAT and PWAT were seeded in 96- and 24-well plates, respectively, in high-glucose DMEM (Gibco) supplemented with 10% FBS, penicillin-streptomycin, gentamicin, sodium pyruvate and MEM non-essential amino acid solution. Once the cells reached confluence, differentiation was induced through the addition of medium containing 3-isobutyl-1-methylxanthine (0.5 mM), dexamethasone (1uM) and insulin (5ug/mL). After 2 days, the medium was changed to medium containing insulin only (5ug/mL). Culture medium was changed every 2 days and replaced with medium containing insulin until the end of the study. On day 7 of culture, cells were washed with PBS and fixed with 4% PFA for 30 min. After washing 3X with PBS, cells were stained with HCS LipidTox Red (Thermo Fisher) and DAPI. Final washes were done with PBS 3X and images were obtained using a NIKON A1R confocal microscope.
Human adipose tissue – GTEx differential gene expression analysis
Bulk RNA sequencing data was accessed from the Genotype-Tissue Expression (GTEx) (Consortium, 2020) project data release version 8 from https://gtexportal.org/home/datasets. Tissue sample extraction, sequencing (Illumina TruSeq RNA protocol) and data normalization and preprocessing (Broad Institute’s Picard pipeline) are described in detail in GTEx Consortium et al. (2020). Expression data were filtered to include samples taken from visceral omental fat (n = 541) and subcutaneous adipose tissue (n = 663). Linear mixed effects models were used to assess group differences in gene expression between visceral and subcutaneous tissue. Expression values, expressed in transcripts per million (TPM), were log10 (TPM +1) transformed for normality. Fixed effects included age, sex and other potential covariates known to impact RNA seq data (Consortium, 2015) (Hardy scale death classification, ischemic time for sample, RNA quality score, collection center, and expression batch ID). Subject identifier was used as a random effect to account for samples taken from the same donor. Linear mixed effects modeling was conducted in R using the ‘lme4’ and ‘lmerTest’ packages (Bates et al., 2007; Kuznetsova et al., 2017).
Quantification and statistical analysis
Statistics
All results are presented as mean ± standard errors of mean. Statistically significant differences among groups were determined by two-tailed unpaired Student’s t-test and one-way ANOVA followed by Sidak’s ad-hoc multiple comparisons test using GraphPad Prism, version 9.
Acknowledgments
We thank Miki Gams from Dr. Cynthia Guidos’ laboratory and Tina Chen and Joe Cozzarin from the Center for Advanced Single Cell Analysis at the Hospital for Sick Children Research Institute for their considerable technical contribution, including optimization of staining and panel design. For this research, H.-K.S. is supported by grants from Canadian Institutes of Health Research (CIHR, PJT-162083), Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2016-06610), and Sun Life Financial New Investigator Award of Banting & Best Diabetes Centre (BBDC) of University of Toronto. H.-K.S and J.-R. K. are supported by Korea-Canada Research Fund (2019K1A3A1A74107385) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT. S.Y.P. is supported by the Medical Research Center Program (2015R1A5A2009124) through the National Research Foundation of Korea (NRF). J.H.L. is supported by Doctoral Program Postgraduate Scholarship (PGS-D) from NSERC. N.V. is supported by Novo Nordisk Studentship from BBDC. N.T. is supported by SickKids Restracomp Scholarship.
Author contributions
J.H.L., K.E, J.-R.K, and H.-K.S. conceived, designed, and performed the research. Y.P. performed human GTEx adipose tissue transcriptome analysis. N.V. and N.T. analyzed the median expression level of the 32 markers and summarized in a heatmap and performed the research. All authors discussed the results and agreed to the final version of the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104166.
Contributor Information
Jae-Ryong Kim, Email: kimjr@ynu.ac.kr.
Hoon-Ki Sung, Email: hoon-ki.sung@sickkids.ca.
Supplemental information
Data and code availability
-
•
This paper analyzes existing, publicly available data from the Genotype-Tissue Expression (GTEx) project data release version 8 from https://gtexportal.org/home/datasets. The accession number for the dataset is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information needed to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ajami B., Samusik N., Wieghofer P., Ho P.P., Crotti A., Bjornson Z., Prinz M., Fantl W.J., Nolan G.P., Steinman L. Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci. 2018;21:541–551. doi: 10.1038/s41593-018-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Sarkar D., Bates M.D. The lme4 package. R package. https://cran.r-project.org/web/packages/lme4/index.html
- Buffolo M., Pires K.M., Ferhat M., Ilkun O., Makaju A., Achenbach A., Bowman F., Atkinson D.L., Holland W.L., Amri E.Z., et al. Identification of a paracrine signaling mechanism linking CD34(high) progenitors to the regulation of visceral fat expansion and remodeling. Cell Rep. 2019;29:270–282. e275. doi: 10.1016/j.celrep.2019.08.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burl R.B., Ramseyer V.D., Rondini E.A., Pique-Regi R., Lee Y.H., Granneman J.G. Deconstructing adipogenesis induced by beta3-adrenergic receptor activation with single-cell expression profiling. Cell Metab. 2018;28:300–309. e304. doi: 10.1016/j.cmet.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. Angiogenesis modulates adipogenesis and obesity. J. Clin. Invest. 2007;117:2362–2368. doi: 10.1172/JCI32239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. Adipose tissue angiogenesis as a therapeutic target for obesity and metabolic diseases. Nat. Rev. Drug Discov. 2010;9:107–115. doi: 10.1038/nrd3055. [DOI] [PubMed] [Google Scholar]
- Cao Y. Angiogenesis and vascular functions in modulation of obesity, adipose metabolism, and insulin sensitivity. Cell Metab. 2013;18:478–489. doi: 10.1016/j.cmet.2013.08.008. [DOI] [PubMed] [Google Scholar]
- Cheung R.K., Utz P.J. Screening: CyTOF-the next generation of cell detection. Nat. Rev. Rheumatol. 2011;7:502–503. doi: 10.1038/nrrheum.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew V., Lai L., Pan L., Lim C.J., Li J., Ong R., Chua C., Leong J.Y., Lim K.H., Toh H.C., et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc. Natl. Acad. Sci. U S A. 2017;114:E5900–E5909. doi: 10.1073/pnas.1706559114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuah S., Chew V. High-dimensional immune-profiling in cancer: implications for immunotherapy. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2019-000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G.T. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H., Zheng S., Garcia-Ruiz D., Hou D., Wei Z., Liao Z., Li L., Zhang Y., Han X., Zen K., et al. Fasting induces a subcutaneous-to-visceral fat switch mediated by microRNA-149-3p and suppression of PRDM16. Nat. Commun. 2016;7:11533. doi: 10.1038/ncomms11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Luo Y., Zhang X., Zheng H., Yang X., Yang X., Liu M. IL-33-driven ILC2/eosinophil axis in fat is induced by sympathetic tone and suppressed by obesity. J. Endocrinol. 2016;231:35–48. doi: 10.1530/JOE-16-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely I., Ma X.H., Yang X.M., Atzmon G., Rajala M.W., Berg A.H., Scherer P., Rossetti L., Barzilai N. Removal of visceral fat prevents insulin resistance and glucose intolerance of aging: an adipokine-mediated process? Diabetes. 2002;51:2951–2958. doi: 10.2337/diabetes.51.10.2951. [DOI] [PubMed] [Google Scholar]
- Galassi A., Reynolds K., He J. Metabolic syndrome and risk of cardiovascular disease: a meta-analysis. Am. J. Med. 2006;119:812–819. doi: 10.1016/j.amjmed.2006.02.031. [DOI] [PubMed] [Google Scholar]
- Gerhard G.S., Styer A.M., Strodel W.E., Roesch S.L., Yavorek A., Carey D.J., Wood G.C., Petrick A.T., Gabrielsen J., Ibele A., et al. Gene expression profiling in subcutaneous, visceral and epigastric adipose tissues of patients with extreme obesity. Int. J. Obes. 2014;38:371–378. doi: 10.1038/ijo.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harasymowicz N.S., Rashidi N., Savadipour A., Wu C.L., Tang R., Bramley J., Buchser W., Guilak F. Single-cell RNA sequencing reveals the induction of novel myeloid and myeloid-associated cell populations in visceral fat with long-term obesity. FASEB J. 2021;35 doi: 10.1096/fj.202001970R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler C., Shan B., Zhang Q., Henry G.H., Shao M., Vishvanath L., Ghaben A.L., Mobley A.B., Strand D., Hon G.C., Gupta R.K. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018;7 doi: 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D.A., Lim H.W., Kim Y.H., Ho W.Y., Foong Y.H., Nelson V.L., Nguyen H.C.B., Chegireddy K., Kim J., Habertheuer A., et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl. Acad. Sci. U S A. 2018;115:E5096–E5105. doi: 10.1073/pnas.1802611115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking S.L., Chisholm D.J., James D.E. Studies of regional adipose transplantation reveal a unique and beneficial interaction between subcutaneous adipose tissue and the intra-abdominal compartment. Diabetologia. 2008;51:900–902. doi: 10.1007/s00125-008-0969-0. [DOI] [PubMed] [Google Scholar]
- Jaitin D.A., Adlung L., Thaiss C.A., Weiner A., Li B., Descamps H., Lundgren P., Bleriot C., Liu Z., Deczkowska A., et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178:686–698. e614. doi: 10.1016/j.cell.2019.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E., Church C.D., Holtrup B., Colman L., Rodeheffer M.S. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol. 2015;17:376–385. doi: 10.1038/ncb3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M.T., Michieletto M.F., Henao-Mejia J. A new perspective on mesenchymal-immune interactions in adipose tissue. Trends Immunol. 2021;42:375–388. doi: 10.1016/j.it.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima Y., Togashi Y., Fukuoka S., Kamada T., Irie T., Suzuki A., Nakamura Y., Shitara K., Minamide T., Yoshida T., et al. Potentiality of multiple modalities for single-cell analyses to evaluate the tumor microenvironment in clinical specimens. Sci. Rep. 2021;11:341. doi: 10.1038/s41598-020-79385-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlgruber A.C., Gal-Oz S.T., LaMarche N.M., Shimazaki M., Duquette D., Koay H.F., Nguyen H.N., Mina A.I., Paras T., Tavakkoli A., et al. Gammadelta T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol. 2018;19:464–474. doi: 10.1038/s41590-018-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korin B., Ben-Shaanan T.L., Schiller M., Dubovik T., Azulay-Debby H., Boshnak N.T., Koren T., Rolls A. High-dimensional, single-cell characterization of the brain's immune compartment. Nat. Neurosci. 2017;20:1300–1309. doi: 10.1038/nn.4610. [DOI] [PubMed] [Google Scholar]
- Kuznetsova A., Brockhoff P.B., Christensen R.H.B. lmerTest package: Tests in linear mixed effects models. J. Stat. Softw. 2017;82:1–26. doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- Landhuis E. Single-cell approaches to immune profiling, 2018. Nature. 2018;557:595–597. doi: 10.1038/d41586-018-05214-w. [DOI] [PubMed] [Google Scholar]
- Lefebvre A.M., Laville M., Vega N., Riou J.P., van Gaal L., Auwerx J., Vidal H. Depot-specific differences in adipose tissue gene expression in lean and obese subjects. Diabetes. 1998;47:98–103. doi: 10.2337/diab.47.1.98. [DOI] [PubMed] [Google Scholar]
- Lim S., Honek J., Xue Y., Seki T., Cao Z., Andersson P., Yang X., Hosaka K., Cao Y. Cold-induced activation of brown adipose tissue and adipose angiogenesis in mice. Nat. Protoc. 2012;7:606–615. doi: 10.1038/nprot.2012.013. [DOI] [PubMed] [Google Scholar]
- Lingblom C.M.D., Kowli S., Swaminathan N., Maecker H.T., Lambert S.L. Baseline immune profile by CyTOF can predict response to an investigational adjuvanted vaccine in elderly adults. J. Transl. Med. 2018;16:153. doi: 10.1186/s12967-018-1528-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Lu X., Li X., Du P., Qin G. High-fat diet triggers obesity-related early infiltration of macrophages into adipose tissue and transient reduction of blood monocyte count. Mol. Immunol. 2020;117:139–146. doi: 10.1016/j.molimm.2019.11.002. [DOI] [PubMed] [Google Scholar]
- Lumeng C.N., Deyoung S.M., Bodzin J.L., Saltiel A.R. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56:16–23. doi: 10.2337/db06-1076. [DOI] [PubMed] [Google Scholar]
- Macdougall C.E., Wood E.G., Loschko J., Scagliotti V., Cassidy F.C., Robinson M.E., Feldhahn N., Castellano L., Voisin M.B., Marelli-Berg F., et al. Visceral adipose tissue immune homeostasis is regulated by the crosstalk between adipocytes and dendritic cell subsets. Cell Metab. 2018;27:588–601. e584. doi: 10.1016/j.cmet.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghsoudlou S., Yu Z.M., Beyene J., McDonald S.D. Phenotypic classification of preterm birth among nulliparous women: a population-based cohort study. J. Obstet. Gynaecol. Can. 2019;41:1423–1432. e1429. doi: 10.1016/j.jogc.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Mahlakoiv T., Flamar A.L., Johnston L.K., Moriyama S., Putzel G.G., Bryce P.J., Artis D. Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aax0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolopoulos K.N., Karpe F., Frayn K.N. Gluteofemoral body fat as a determinant of metabolic health. Int. J. Obes. 2010;34:949–959. doi: 10.1038/ijo.2009.286. [DOI] [PubMed] [Google Scholar]
- Marcelin G., Ferreira A., Liu Y., Atlan M., Aron-Wisnewsky J., Pelloux V., Botbol Y., Ambrosini M., Fradet M., Rouault C., et al. A PDGFRalpha-mediated switch toward CD9(high) adipocyte progenitors controls obesity-induced adipose tissue fibrosis. Cell Metab. 2017;25:673–685. doi: 10.1016/j.cmet.2017.01.010. [DOI] [PubMed] [Google Scholar]
- Merrick D., Sakers A., Irgebay Z., Okada C., Calvert C., Morley M.P., Percec I., Seale P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364 doi: 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misumi I., Starmer J., Uchimura T., Beck M.A., Magnuson T., Whitmire J.K. Obesity expands a distinct population of T cells in adipose tissue and increases vulnerability to infection. Cell Rep. 2019;27:514–524. e515. doi: 10.1016/j.celrep.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesitt S.C., Hsu J.Y., Chowbina S.R., Lawrence R.T., Hoehn K.L. Not all fat is equal: differential gene expression and potential therapeutic targets in subcutaneous adipose, visceral adipose, and endometrium of obese women with and without endometrial cancer. Int. J. Gynecol. Cancer. 2012;22:732–741. doi: 10.1097/IGC.0b013e3182510496. [DOI] [PubMed] [Google Scholar]
- Nahmgoong H., Jeon Y.G., Park E.S., Choi Y.H., Han S.M., Park J., Ji Y., Sohn J.H., Han J.S., Kim Y.Y., et al. Distinct properties of adipose stem cell subpopulations determine fat depot-specific characteristics. Cell Metab. 2022;34:458–472.e6. doi: 10.1016/j.cmet.2021.11.014. [DOI] [PubMed] [Google Scholar]
- O'Flanagan C.H., Campbell K.R., Zhang A.W., Kabeer F., Lim J.L.P., Biele J., Eirew P., Lai D., McPherson A., Kong E., et al. Dissociation of solid tumor tissues with cold active protease for single-cell RNA-seq minimizes conserved collagenase-associated stress responses. Genome Biol. 2019;20:210. doi: 10.1186/s13059-019-1830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro A., Miselli M.A., Sanz J.M., Dalla Nora E., Morieri M.L., Colonna R., Pisot R., Zuliani G. Gene expression regional differences in human subcutaneous adipose tissue. BMC Genomics. 2017;18:202. doi: 10.1186/s12864-017-3564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raajendiran A., Ooi G., Bayliss J., O'Brien P.E., Schittenhelm R.B., Clark A.K., Taylor R.A., Rodeheffer M.S., Burton P.R., Watt M.J. Identification of metabolically distinct adipocyte progenitor cells in human adipose tissues. Cell Rep. 2019;27:1528–1540. e1527. doi: 10.1016/j.celrep.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Rodeheffer M.S., Birsoy K., Friedman J.M. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Russo L., Lumeng C.N. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018;155:407–417. doi: 10.1111/imm.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalie P.C., Dong H., Zachara M., Russeil J., Alpern D., Akchiche N., Caprara C., Sun W., Schlaudraff K.U., Soldati G., et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559:103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- Shao M., Hepler C., Zhang Q., Shan B., Vishvanath L., Henry G.H., Zhao S., An Y.A., Wu Y., Strand D.W., Gupta R.K. Pathologic HIF1alpha signaling drives adipose progenitor dysfunction in obesity. Cell Stem Cell. 2021;28:685–701. e687. doi: 10.1016/j.stem.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spallanzani R.G., Zemmour D., Xiao T., Jayewickreme T., Li C., Bryce P.J., Benoist C., Mathis D. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aaw3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M.H., Nolan G.P. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam P.B., Maecker H.T. CyTOF measurement of immunocompetence across major immune cell types. Curr. Protoc. Cytom. 2017;82:9.54.1–9.54.12. doi: 10.1002/cpcy.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon P., Wafer R., Minchin J.E.N. Adipose morphology and metabolic disease. J. Exp. Biol. 2018;221 doi: 10.1242/jeb.164970. [DOI] [PubMed] [Google Scholar]
- Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Unen V., Li N., Molendijk I., Temurhan M., Hollt T., van der Meulen-de Jong A.E., Verspaget H.W., Mearin M.L., Mulder C.J., van Bergen J., et al. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity. 2016;44:1227–1239. doi: 10.1016/j.immuni.2016.04.014. [DOI] [PubMed] [Google Scholar]
- Vijay J., Gauthier M.F., Biswell R.L., Louiselle D.A., Johnston J.J., Cheung W.A., Belden B., Pramatarova A., Biertho L., Gibson M., et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab. 2020;2:97–109. doi: 10.1038/s42255-019-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.A., Scherer P.E. The AdipoChaser mouse: a model tracking adipogenesis in vivo. Adipocyte. 2014;3:146–150. doi: 10.4161/adip.27656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., McCann D., Desai M., Rosenbaum M., Leibel R.L., Ferrante A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedeman A.E., Muir V.S., Rosasco M.G., DeBerg H.A., Presnell S., Haas B., Dufort M.J., Speake C., Greenbaum C.J., Serti E., et al. Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression. J. Clin. Invest. 2020;130:480–490. doi: 10.1172/JCI126595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer D.A., Winer S., Shen L., Wadia P.P., Yantha J., Paltser G., Tsui H., Wu P., Davidson M.G., Alonso M.N., et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat. Med. 2011;17:610–617. doi: 10.1038/nm.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data from the Genotype-Tissue Expression (GTEx) project data release version 8 from https://gtexportal.org/home/datasets. The accession number for the dataset is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information needed to reanalyze the data reported in this paper is available from the lead contact upon request.