Abstract
The cellular prion protein (PrPC) has a C-terminal globular domain and a disordered N-terminal region encompassing five octarepeats (ORs). Encounters between Cu(II) ions and four OR sites produce interchangeable binding geometries; however, the significance of Cu(II) binding to ORs in different combinations is unclear. To understand the impact of specific binding geometries, OR variants were designed that interact with multiple or single Cu(II) ions in specific locked coordinations. Unexpectedly, we found that one mutant produced detergent-insoluble, protease-resistant species in cells in the absence of exposure to the infectious prion protein isoform, scrapie-associated prion protein (PrPSc). Formation of these assemblies, visible as puncta, was reversible and dependent upon medium formulation. Cobalamin (Cbl), a dietary cofactor containing a corrin ring that coordinates a Co3+ ion, was identified as a key medium component, and its effect was validated by reconstitution experiments. Although we failed to find evidence that Cbl interacts with Cu-binding OR regions, we instead noted interactions of Cbl with the PrPC C-terminal domain. We found that some interactions occurred at a binding site of planar tetrapyrrole compounds on the isolated globular domain, but others did not, and N-terminal sequences additionally had a marked effect on their presence and position. Our studies define a conditional effect of Cbl wherein a mutant OR region can act in cis to destabilize a globular domain with a wild type sequence. The unexpected intersection between the properties of PrPSc's disordered region, Cbl, and conformational remodeling events may have implications for understanding sporadic prion disease that does not involve exposure to PrPSc.
Keywords: prion, oligomer, conformational change, glycoprotein, plasma membrane, cobalamin, vitamin B12, sporadic disease
Abbreviations: Al, aluminum; Ca, calcium; Cbl, cobalamin; CJD, Creutzfeldt–Jakob disease; Co, cobalt; Cr, chromium; Cu, copper; CV, coefficient of variation; DEPC, diethylpyrocarbonate; DMEM, Dulbecco's modified Eagle's medium; Fe, iron; GPI, glycosylphosphatidylinositol; GSS, Gerstmann–Sträussler syndrome; HMW, high–molecular weight; LLPS, liquid–liquid phase separation; MD, molecular dynamics; Mg, magnesium; Mn, manganese; mPrP, mouse PrP; MS, mass spectrometry; Ni, nickel; OR, octarepeat; PDB, Protein Data Bank; PK, proteinase K; PNGase F, peptide-N-glycosidase F; PrPC, cellular prion protein; PrPSc, scrapie-associated prior protein; recPrP, recombinant PrP; RIPA, radioimmunoprecipitation assay; Tg, transgenic; Zn, zinc
The cellular prion protein (PrPC) is a widely expressed, conserved glycoprotein; it serves as a precursor to disease-associated and misfolded isoforms, including a form designated scrapie-associated prion protein (PrPSc) found in the prototypical prion disease, scrapie. PrPC is attached to the outer leaflet of the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor (1) and is abundant in the central nervous system (2, 3). The C-terminal part of PrPC has a globular fold with three α-helices and a short β-sheet, whereas the N-terminal portion of the protein is natively disordered and contains five octarepeats (ORs) and a hydrophobic tract (4). Besides having dissimilar amino acid composition and folding properties, these two regions within the full-length PrPC holoprotein can be separated physically by physiological endoproteolysis events; α-cleavage in the vicinity of residue 110 gives rise to N1 and C1 fragments, whereas β-cleavage approximately 20 residues upstream of this site generates N2 and C2 fragments, with the latter fragment possibly subject to further cleavages (5, 6, 7, 8).
PrPSc is associated with infectivity in experimental scrapie disease and often has an intrinsic resistance to in vitro digestion with the broad-spectrum protease proteinase K (PK), with the coordinates of this resistant fragment, PrP 27–30, being similar to those of C2 PrP. Furthermore, PrP species with a similar electrophoretic mobility to PrP 27–30 exist in Creutzfeldt–Jakob disease (CJD) (9). However, closer chemical analyses indicate heterogeneities that add complexity to these overarching ideas; some misfolded PrP is protease sensitive, PK cleavage sites within the N terminus are different between prion diseases, and PrPSc forms differ in their chemical denaturation profiles (10, 11, 12). Indeed, these observations have been marshaled into the notion of a “cloud” of possible PrPSc forms (13). While considered to be natively disordered in solution, the N-terminal part of PrP has a role in familial prion diseases. Insertions and deletions in the OR are associated with heterogeneous clinical manifestations, such as Gerstmann–Sträussler syndrome (GSS), psychiatric disorders, and some forms of genetic CJD (see the study by Lloyd et al. (14)). Some binding partners have been identified for this region, with transition metals being notable (15, 16, 17, 18, 19, 20, 21, 22). Copper (Cu) ions produce metalated forms of PrP, binding via four different histidines located in the OR region (23, 24) and with a fifth site described involving histidines 95 and 110 (mouse PrP [mPrP] numbering scheme) (17, 25) (Fig. 1A). Interestingly, with increasing stoichiometries of Cu(II), the PrP OR region adopts different structures (18, 26) and interacts with a negatively charged area on the globular domain (27). As these metal-induced OR structures can interconvert in WT PrP, we created transgenic (Tg) mice encoding PrP with engineered OR region variants, which blocked interconversion to potentially delineate the biological properties associated with the different structures (18, 28). Briefly, the S1-PrP allele has additional prolines in histidine-containing ORs 1 and 3, PHGGGWGQ->PHGGGWPP; these substitutions hold the OR region in an extended conformation and emphasize a binding mode (component 1) where each OR can bind one Cu2+ ion. Conversely, the S3PrP allele is engineered to preclude component 1 binding that requires tetrahedral coordination of Cu2+ complexed with a water molecule; this is achieved by swapping out glycine and tryptophan residues needed for this binding geometry, PHGGGWGQ->PHGPGFGQ (Fig. 1B). While both S1 and S3 alleles supported prion replication in the brains of infected animals (28), we encountered unanticipated effects for S3-PrP when expressed in cells in the absence of prion infection. As described here, these effects favoring protein assembly could be traced to a cofactor, vitamin B12 (cobalamin [Cbl]) present in a medium formulation commonly used to enhance the success rate of de novo prion infections (29, 30, 31, 32, 33). Cbl comprises a cobalt(III) (Co(III)) ion located in the center of a complex coordinated structure; it is coordinated in a planar fashion by a corrin ring structure and in an axial fashion by a fifth nitrogen located within a dimethylbenzimidazole group (34). In studies herein, the impact of Cbls upon PrP assembly was documented by different end point measures and assessed versus known metal coordination structures and tetrapyrrole-binding sites.
Figure 1.
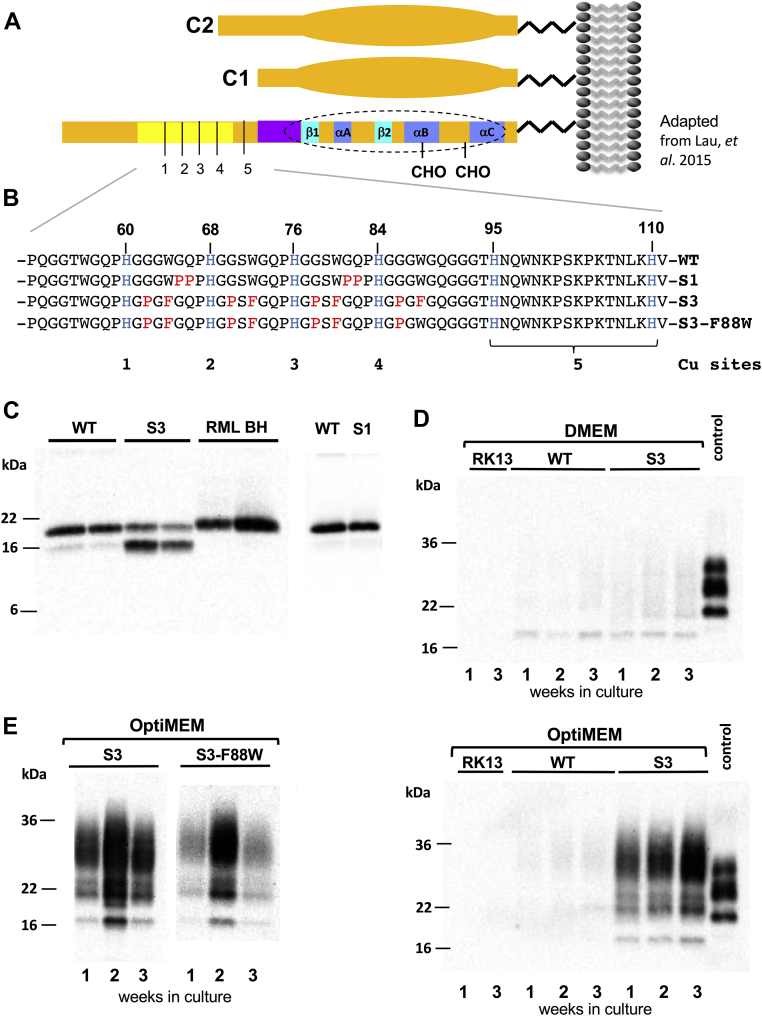
Spontaneous production of protease-resistant PrP.A, schematic of PrP and positions of C1 and C2 fragments. The globular domain is indicated by an extended oval and structures within this (dotted perimeter) include three α-helices and two β-strands, as well as N-glycosylation sites (“CHO”). The OR region is shaded yellow and 1 to 5 refer to Cu-binding sites. B, expanded view of the OR region showing amino acid replacements in the S1, S3, and S3-F88W alleles (modified from Lau et al., 2015 (28)). C, RK13 cells expressing WT-PrP, S3-PrP, or S1-PrP alleles were infected with the RML prion isolate and subjected to PNGase F and PK digestion (50 μg/ml). RML BH refers to biological replicate samples of mouse brain homogenate from infected animals. The origin and identity of the bands in S3 cells (lanes 3 and 4) is elaborated in Figure 3, D and E. D, protease digest comparison of stable clones of WT-PrP and S3-PrP and untransfected RK13 cells grown in two different media, DMEM and Opti-MEM. Cells were grown for 3 consecutive weeks. Cell extracts were treated with 50 μg/ml PK and probed with Sha31 antibody. As a comparison, a brain homogenate of a WT mouse infected with RML is presented (“control”). E, S3 and S3-F88W cells were grown for 3 consecutive weeks in Opti-MEM. Cell lysates were digested with PK as aforementioned. DMEM, Dulbecco's modified Eagle's medium; OR, octarepeat; PK, proteinase K; PNGase F, peptide-N-glycosidase F; PrP, prion protein.
Results
Conditional production of protease-resistant S3-PrP species
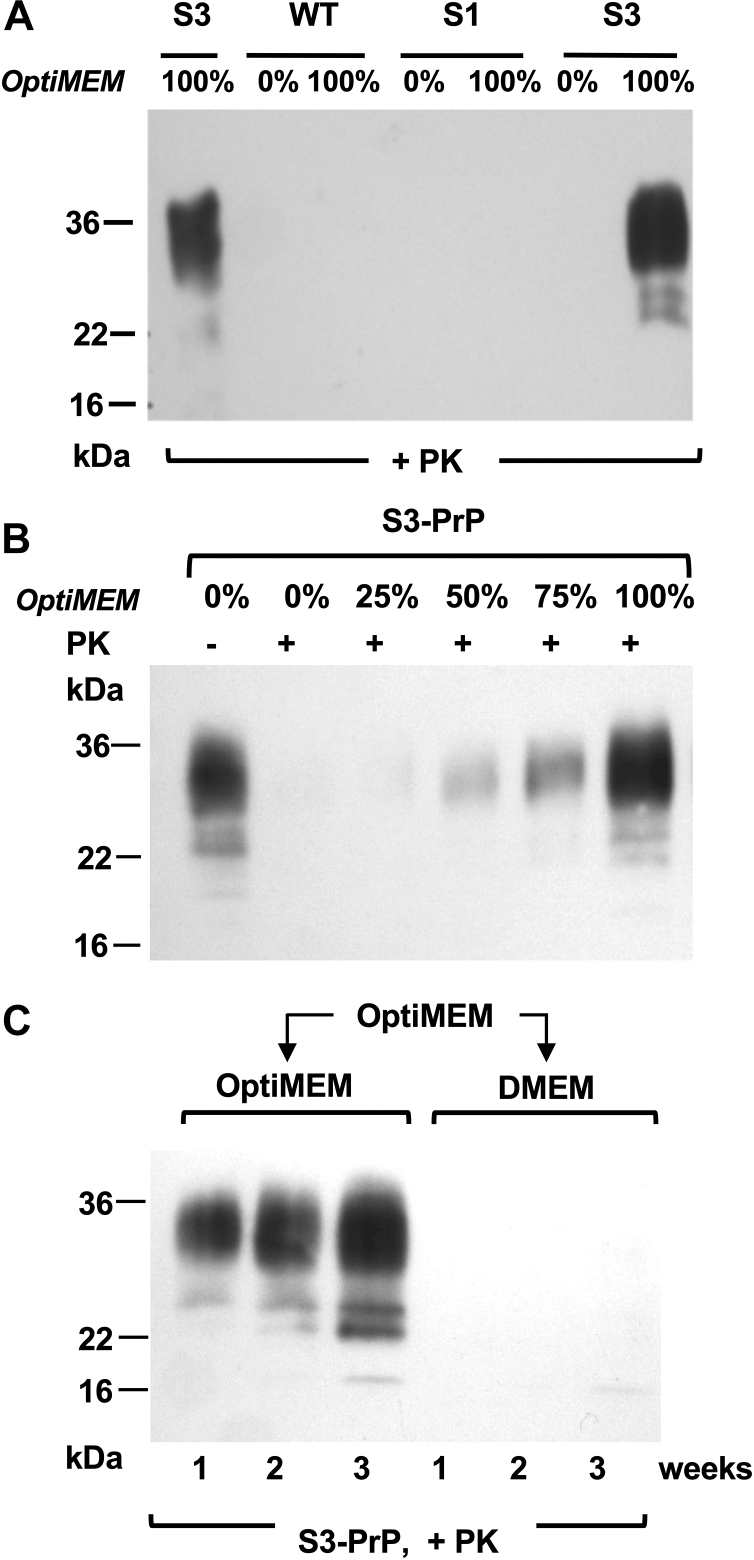
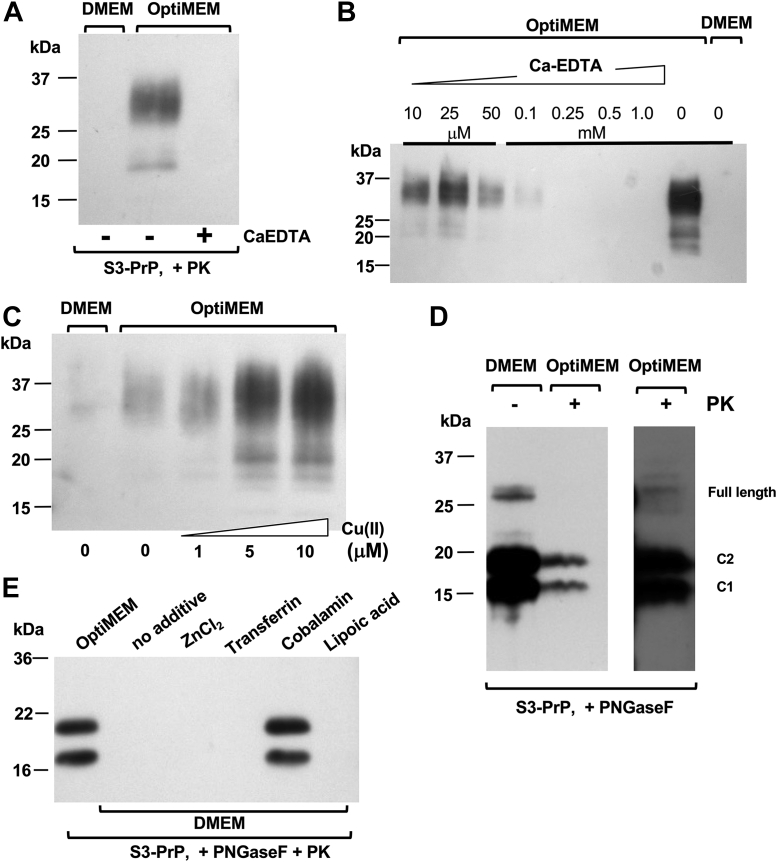
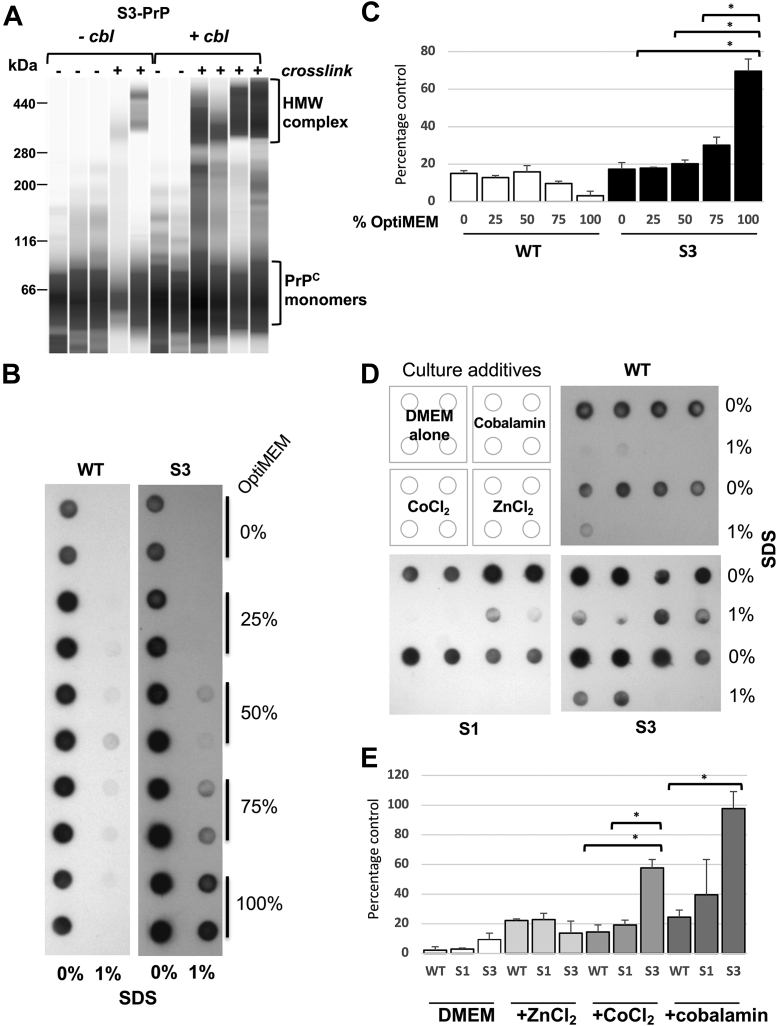
The S3-PrP allele (Fig. 1, A and B) expressed in the brains of uninfected Tg mice (TgS3.F88W) gives abundant production of a C2 fragment (28) with mobility similar to a PK-resistant fragment that increases during prion infections (35, 36, 37, 38, 39, 40). As the C2 fragment is also notable in uninfected RK13 cells expressing S3-PrP (“S3 cells”) (28), we first sought to understand how these cells respond to prion infection. When exposed to mouse-adapted scrapie prions (RML isolate), S3-expressing RK13 cells produced a two-band signature of deglycosylated PK-resistant PrP. This signature differed from cells expressing WT mPrP or expressing an alternative OR allelic form, S1-PrP, that was designed to have a different metal-binding geometry from S3-PrP (28) (Fig. 1, B and C), these producing signatures resembling PK-resistant PrP from the brain. This behavior of S3 cells proved to be unrelated to prion infection. Uninfected cells grown in standard Dulbecco's modified Eagle's medium (DMEM) and expressing WT-PrP or S3-PrP produced PK-sensitive PrP species (Fig. 1D, top panel) but within 1 week after change into Opti-MEM, S3-PrP-expressing cells—but not WT-PrP cells—produced PK-resistant material (50 μg/ml PK). By gel mobility, these species included full-length diglycosylated forms of PrP (Fig. 1D, bottom panel). A variant of S3-PrP called S3-F88W with a residue exchange in codon 88 also produced PK-resistant species after culture in Opti-MEM (Fig. 1E); the F88W substitution represents a change to the WT tryptophan residue at codon 88. Control analyses indicated that the S1-PrP OR variant also did not behave similarly to S3-PrP (Fig. 2A), whereas mixing DMEM with increasing proportions of Opti-MEM increased protease-resistant material from S3-PrP (Fig. 2B). This experiment, as well as the procedure for sample processing that includes a pellet-washing step and resuspension in a different buffer before the PK digestion, excludes trivial effects that could derive from inhibition of the in vitro PK digestion reaction (41). To study the kinetics of PK-resistant S3-PrP production, cells grown for several passages in Opti-MEM were split into either fresh Opti-MEM or DMEM; samples maintained in Opti-MEM remained PK resistant (Fig. 2C), whereas cells switched back to DMEM reverted to PK sensitivity within 1 week. Thus, PK resistance of S3-PrP is a conditional and reversible phenotype. As PrP binds Cu and zinc (Zn) (15, 23, 24, 42) and since S3-PrP has an OR region that facilitates “component 3” Cu binding (18, 28), we applied a broad-spectrum chelator, EDTA (used in calcium [Ca]–EDTA form (43)); this chelator diminished the amount of PK-resistant S3-PrP (Fig. 3, A and B), whereas addition of Cu ions had the reverse effect (Fig. 3C). Using peptide-N-glycosidase F (PNGase F) to remove N-linked sugars, PK-treated lysates from S3 cells grown in Opti-MEM revealed similar electrophoretic mobilities to those of PK-sensitive C1 and C2 PrP fragments of cells grown in DMEM (Fig. 3D). Given the chelator effects in Figure 2, we profiled the metal content of Opti-MEM and DMEM. We analyzed seven transition metals (chromium [Cr], iron [Fe], manganese [Mn], cobalt, nickel [Ni], Cu, and Zn) as well as the levels of magnesium (Mg), aluminum (Al), and Ca) (Table 1). The two media had similar content of Mg, Mn, Ni, and Cu. DMEM had twice as much Cr as Opti-MEM, whereas Opti-MEM had approximately twice the amount of Al, Ca, and Zn. Most notably, Opti-MEM had almost 50 times more Co than DMEM. With this information, we used reconstitution experiments to define the particular medium components relevant to producing PK resistance. Cells expressing S3-PrP were grown in DMEM spiked with either Zn chloride, transferrin (Fe), or Cbl (Co) to obtain levels similar to those present in Opti-MEM (Table 1). We also tested lipoic acid (cf. https://www.thermofisher.com/) in these analyses to make reconstituted versions of DMEM. While supplementation of DMEM with Cbl to a level of 0.85 μM produced the typical two-band pattern produced by S3-PrP grown in Opti-MEM, addition of Zn, Fe, or lipoic acid did not (Fig. 3E).
Figure 2.
Medium composition and conditional production of protease-resistant S3-PrP.A, S3, WT, and S1 cells were grown in DMEM (“O%”) or Opti-MEM for a period of 3 weeks. Cell lysates were digested with 50 μg/ml PK. Opti-MEM.B, as per panel A but with S3-PrP cells grown in different ratios of DMEM:Opti-MEM. C, S3 cells previously grown in Opti-MEM for several passages were grown for an additional 3 consecutive weeks in either Opti-MEM or DMEM. Cell extracts were digested with 50 μg/ml PK. DMEM, Dulbecco's modified Eagle's medium; PK, proteinase K; PrP, prion protein.
Figure 3.
Medium additives affecting the production of protease-resistant S3-PrP.A, S3 cells were grown in DMEM alone or in Opti-MEM with or without calcium (Ca)–EDTA. B, diminution of production of PK-resistant PrP following titrated addition of Ca–EDTA. Cell lysates were treated with 50 μg/ml PK. C, copper addition to S3-PrP cells propagated in Opti-MEM increased the yield of protease-resistant PrP. D, S3 cell extracts from cells grown in DMEM or Opti-MEM were digested (+) or not digested (−) with 50 μg/ml PK, and then samples were treated with PNGase F. The third lane represents a higher exposure of the second lane. E, S3 cells were grown in Opti-MEM, in DMEM alone, or in DMEM supplemented with 1.12 μM ZnCl2, 40 μM transferrin, 0.85 μM Cbl, or 40 μM lipoic acid. Cell lysates were treated with 50 μg/ml PK and PNGase F. Cbl, cobalamin; DMEM, Dulbecco's modified Eagle's medium; PK, proteinase K; PNGase F, peptide-N-glycosidase F; PrP, prion protein.
Table 1.
Metal composition comparison of DMEM and Opti-MEM determined by inductively coupled plasma MS
| Analyte | Mg | Al | Ca | Cr | Fe | Mn | Co | Ni | Cu | Zn |
|---|---|---|---|---|---|---|---|---|---|---|
| Detection limits | 0.002 | 0.0002 | 0.031 | 0.00005 | 0.0037 | 0.00003 | 0.00003 | 0.00006 | 0.00003 | 0.00008 |
| Units | ppm | ppm | ppm | ppm | ppm | ppm | ppm | ppm | ppm | ppm |
| Opti-MEM | 24.475 ± 3.09 | 0.004 ± 0.002 | 40.958 ± 5.27 | 0.060 ± 0.03 | 0.040 ± 0.00 | 0.001 ± 0.00 | 0.046 ± 0.01 | 0.001 ± 0.00 | 0.037 ± 0.02 | 0.075 ± 0.01 |
| DMEM | 19.580 ± 1.18 | 0.002 ± 0.00 | 71.99 ± 11.11 | 0.084 ± 0.04 | <DL | 0.001 ± 0.00 | 0.001 ± 0.00 | 0.002 ± 0.00 | 0.055 ± 0.02 | 0.025 ± 0.02 |
| DMEM + ZnCl2 | 17.407 | 0.006 | 72.361 | 0.041 | <DL | 0.001 | 0.000 | 0.002 | 0.027 | 0.108 |
| DMEM + transferrin | 19.006 | 0.004 | 71.206 | 0.04 | 0.026 | 0.002 | 0.000 | 0.002 | 0.027 | 0.034 |
| DMEM + cbl | 19.287 ± 0.29 | 0.002 | 61.032 | 0.056 | <DL | 0.001 | 0.035 ± 0.01 | 0.001 | 0.036 ± 0.01 | 0.023 ± 0.00 |
<DL, below detection limit.
Assessing physical interactions between recombinant PrPs and Cbl
We next considered direct Cbl–PrP interactions. Given Cu(II) binding of the N-terminal region of PrPCs (23, 42, 44) and a PrP–Co2+ interaction reported for histidine 110 (45), we performed surface plasmon resonance experiments with tethered recombinant PrP (recPrP) and divalent cations presented in a mobile phase. As shown in Fig. S1, recWT-PrP and recS3-PrP responded to Cu (presented as Cu(glycine)2 chelate to mimic dietary presentation (46, 47) or with free Cu2+) with averaged KD values for chelated Cu of 101 and 78 nM, respectively, when fitted to a model with 1:1 binding stoichiometry. Cu presented in ionic form gave KD values of 65 and 100 nM for WT and S3 alleles (Table S1). These submicromolar binding constants are in broad agreement with the prior literature (18, 48, 49, 50). Unfortunately, use of Cbl in the surface plasmon resonance flow buffer produced sensorgrams with dramatic rise times, even in flow cells lacking recPrP, indicating a confounding interaction with the chip surface. Following an alternative analytical approach, it is known that imidazole groups of surface-exposed histidines react with diethylpyrocarbonate (DEPC) (51), and this reactivity, in the case of PrP, is reduced by preincubation with ionic Cu (17). This technique relies upon a 72.06 Da mass increase for each modified imidazole, measured by MALDI–TOF mass spectrometry (MS) and can be performed at neutral pH and physiological salt concentration; as previous experiments using recPrP 23–231 and recPrP 23–98 defined shielding of reactive His residues in the OR region (Cu sites 1–4), and at His 95, here we sought to assess shielding mediated by Cbl. WT mPrP 23–230, measured by MALDI–MS, had an observed mass of 23,054.7 Da ± 4.3 (n = 6) for WT PrP (predicted, 23,063.43 Da). WT mPrP 23–230 reacted with DEPC to give multiple adduct peaks with averaged increments of 70.46 ± 1.3 Da. These data (Table S2) define a strong effect deriving from four reactive histidines (peaks assigned with one to four modifications, 1m–4m) and further peaks of smaller signal intensity and higher charge-to-mass ratio indicating additional DEPC adducts, noting nine histidines in total for WT mPrP. Preincubation with Cu(glycine)2 increased the relative size of the “0m” peak (i.e., unmodified PrP) and peaks 2m–4m became less prominent (Fig. S2B versus Fig. S2A), indicating that the main effect of Cu(glycine)2 is to protect two histidines. Cbl alone had little effect (Fig. S2C), and a mixture of Cu(glycine)2 and Cbl produced a mass spectrum resembling the Cu(glycine)2 mass spectrum (Fig. S2D versus Fig. S2B). For S3-PrP, unmodified recS3-PrP 23–230 had a mass of 23060.9 Da ± 8.1 (n = 7), an observed mass increase over WT of 6.2 Da (predicted, 4.1 Da). Following reaction with DEPC, the relative yield of 2m–4m adducts was lower than that of WT-PrP (Fig. S3A versus Fig. S2A), suggesting an intrinsic difference between unmetalated S3-PrP and WT-PrP. Preincubation with Cu(glycine)2 altered the repertoire of S3 DEPC adducts (Fig. S3B versus Fig. S3A), though not as noticeably as for WT-PrP (Fig. S3B versus Fig. S2B). Preincubation of S3-PrP with Cbl before reaction with DEPC produced a different profile for a mass spectrum with four discernible adducts (Fig. S3C), an effect retained when Cu(glycine)2 was added along with Cbl (Fig. S3D). Overall, these in vitro analyses support conclusions that recWT-PrP and S3-PrP have high-affinity Cu-binding sites and that Cbl addition does not interfere with the ability of added Cu to protect prominent DEPC-reactive histidines from chemical modification (these mapped as lying within PrP 23–98 (17)). Consequently, we considered effects of Cbl upon other areas of PrP.
Molecular dynamics and docking analyses reveal allelic divergence in Cbl–PrP interactions
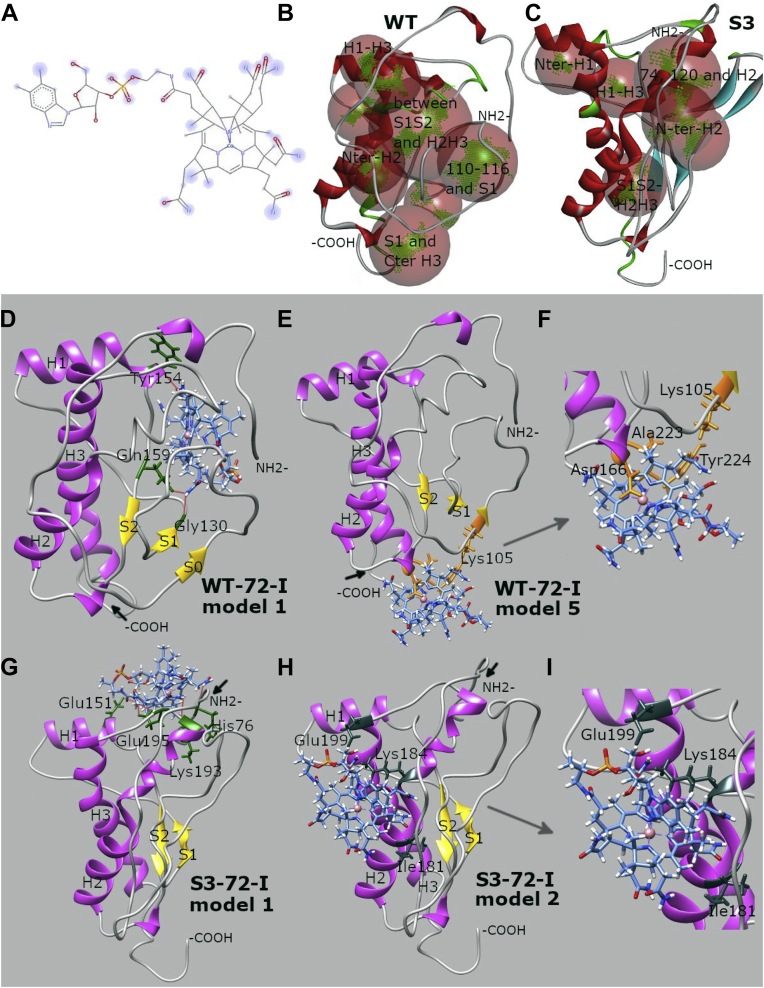
Cbl is a type of tetrapyrrole (52), containing a four-membered planar corrin ring as well as an axial benzamidazole linked via a ribonucleoside structure. Given that other tetrapyrroles interact with the PrPC globular domain (53), we considered analogous events for Cbl. Two sets of WT and S3 mPrP 72 to 231 conformations with different orientations of flexible N termini were obtained from different initial Protein Data Bank (PDB) structures and subjected to molecular dynamics (MD) simulations prior to docking (Fig. 4, A–F), whereas a third set of 119 to 231 conformations with truncated N termini was generated for control analyses (Table S3).
Figure 4.
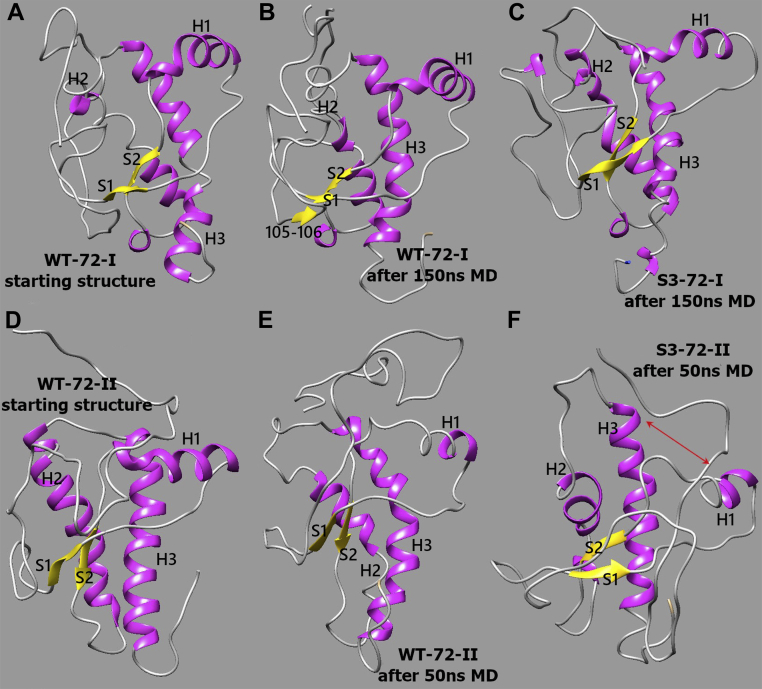
Comparison of mPrP 72–231 structures. Comparison of WT-72-II and S3-72-II mPrP 72–231 structures. A, original WT structure based on 2L39.pdb that was used to build WT-72-I and S3-72-I models. B, WT-72-I representative conformation after 150 ns MD simulations: α-helix H2 partially unfolded; β-sheet S1–S2 reduced to shorter β-strands; transient short β-strands appeared in the N terminus. C, S3-72-I representative conformation after 150 ns MD simulations: C-terminal regions of α-helices H2 and H3 substantially disrupted. D, original WT structure based on 2PRP.pdb that was used to build both WT-72-II and S3-72-II models. E, WT-72-II representative conformation after 50 ns MD simulations: C termini of helices partially unfolded. F, S3-72-II representative conformation after 50 ns MD simulations: region S1–H1–S2 and bundle H2–H3 shifted away from each other exposing the hydrophobic core between them; helix H1 considerably shifted toward C-terminal part of H3. MD, molecular dynamics; mPrP, mouse PrP.
Molecular dynamics
During MD simulations in a system with S3-72-I mPrP, we observed substantial disruption of α-helical structure (from 40.6% in the original conformation in 2L39.PDB to 31.1%), the formation of two stable short β-strands in the N terminus (total β content increased from 3.7% to 6.9%), and transient presence of an extended S1–S2 β-sheet. The structure of the C-terminal half of helix H2 was disrupted, and similar unfolding occurred in the C-terminal region of helix H3 (Fig. 4C). Changes were less notable for WT-72-I PrP (Fig. 4B); disruption of helix H2 was found (but not H3), only transient short β-strands appeared in the N terminus of WT-PrP, and β-sheet S1–S2 was reduced to shorter β-strands because of competitive interactions with the unstructured N-terminal tail. Similar changes in secondary structure were observed during MD simulations of systems mPrP WT-72-II and S3-72-II (Fig. 4, E and F): C termini of all three helices were disrupted, and total α-helical content was reduced from 39% to 25.8% in WT and 23.9% in S3. Formation of short β-strands in the N terminus was observed as well, with total β-content increasing from 3.8% to 8.1% in WT and 7.1% in S3. Interestingly, we observed a wide opening of the hydrophobic core between S1–H1–S2 and H2–H3 parts of PrP only in the case of the S3-72-II construct (Fig. 4F), a type of opening that has been associated with PrP misfolding (54). Furthermore, a new short β-strand appeared in position 194 of this construct, within a displaced loop LH2H3.
To summarize, changes in the residues involved in β-sheets and β-strands (mPrP numbering scheme) are as follows:
- Original WT/S3-72-I model
- β-strands: residues 78 and 81, β-sheets: residues 128 to 129 and 161 to 162; WT-72-I after 150 ns: β-sheets: residues 105 to 106, 128 to 129, and 161 to 162. S3-72-I after 150 ns: β-strands: 91, 106, 114, 124, β-sheets: 128 to 132 and 159 to 162.
- Original WT/S3-72-II model
- β-strands: residues 115, 121, and 124; β-sheets: residues 128 to 130 and 160 to 162; WT-72-II after 50 ns: β-strands: residues 87, 92, and 115, β-sheets: residues 119 to 120, 125 to 126, 128 to 130, and 160 to 162; S3-72-II after 50 ns: β-strands: 90, 107, 116, and 125 and β-sheets: 128 to 130 and 160 to 162.
Docking
Predicted docking sites are illustrated in Figures 5, S4 and S5. Binding sites in the WT-72-I and S3-72-I globular domains (domains with identical amino acid sequences) were predicted from receptor cavities but differed for the two alleles (Fig. 5, B and C). As seen in Fig. S4, positions of docking sites in 119 to 231 and 72 to 231 constructs are substantially different. While the top docking models against 72 to 231 constructs exhibit overlapping binding sites for Cbl and tetrapyrrole (Fig. S4, A and E), secondary binding sites for these two ring compounds are different (Fig. S4, B and F). For shorter 119 to 231 systems, the trend was the opposite: the top docking models tend to have different docking locations for Cbl and tetrapyrrole, whereas the locations overlap for secondary docking models (Fig. S4, C, D, G and H). Docking energies for tetrapyrrole are lower than Cbl docking energies (Table S4) because of the large size of Cbls. Both sets of WT-72-I/II and S3-72-I/II conformations, based on different structures, result in overlapping docking sites (Figs. S4 and S5), which have somewhat lower docking energy than their 119 to 231 counterparts.
Figure 5.
Docking analysis of PrP–Cbl interactions.A, chemical structure of Cbl with predicted docking sites highlighted in blue. B and C, binding sites in WT and S3 PrP structures (translucent red spheres with areas for compound centering highlighted in green) predicted from receptor cavities by the Biovia Discovery Studio Visualizer. D–I, predicted docking sites of Cbls: WT mPrP 72-I, docking model 1 (D); WT mPrP 72–231-I, docking model 5 (E), and the corresponding close-up view (F); S3 mPrP 72 to 231-I, docking model 1 (G); S3-72-I, docking model 2 (H), and the corresponding close-up view (I). D–I, α-helices of PrP are indicated in purple, β-strands in yellow, and random coils and turns in gray; PrP residues involved in hydrogen bonds are colored green; N and C termini for simplicity are presented as -NH2 and -COOH with the positions of occluded termini indicated by short black arrows; and atom/bonds of Cbl are colored blue with the phosphate group depicted in orange. E and F, in addition illustrate overlapping relationships of Cbl-binding sites with tetrapyrrole binding (globular domain contact residues for tetrapyrrole in orange). H and I, show relationships with Zn-loaded OR region (globular domain contact residues in slate gray) (55, 58), see also Table S5. Cbl, cobalamine; PrP, prion protein; mPrP, mouse prion protein; OR, octarepeat.
In predicted docking models, there was an inward orientation of the corrin ring of Cbl (i.e., with the benzimidazole/ribonucleoside axial structure pointing away from the globular domain) in 8 of 10 models for WT-72-I and 6/10 models for S3-72-I PrP. Only one WT model exhibited side docking of Cbl, and the remaining models exhibited inward orientation of the phosphate moiety side of Cbl (Table S4). The number of Cbl contacts with PrP differed for the two allelic forms, with an average number of three bonds per model for WT-72-I PrP versus four per model for S3-72-I PrP (Table S5). For WT-72-I PrP, most hydrogen bonds resulted from docking over the β-sheet region with involvement of the C terminus of helix H3, whereas for S3-72-I PrP, most hydrogen bonds were formed with helix H2 and the N terminus of H3 (Fig. 5, D and E versus Fig. 5, G and H; Table S5). Differences were also noted for the involvement of N-terminal residues in Cbl docking—W72, H84, N96, K100, and K105 for WT-PrP versus H76, P83, G87, F88, G91, G93, and T94 for S3-PrP.
We next compared these Cbl docking sites to a tetrapyrrole-binding site mapped to WT PrP. This tetrapyrrole-binding site corresponds to helix H3 and loop LS2H2 as established by solution NMR (55) and with overlapping, supportive findings from docking calculations used previously where we detected docking of tetrapyrrole to the site in the system WT-119-I, model 6 (Fig. S4D), and docking of Cbl to the site in the systems WT-119-I, model 2 (Fig. S4D) and WT-72-I, model 5 (Fig. S4B). Importantly, we did not observe docking to tetrapyrrole site for any of S3 systems. An S3-119-I model 1 had Fe(III)TMPyP docked to C terminus of H3, but those interactions did not include the S2–S1 bundle.
In order to make the tetrapyrrole site open for docking, we used a complementary set of WT-72-II and S3-72-II conformations based on the 2PRP structure wherein the 89 to 120 region is shifted away from the site and interacting with the beta-sheet bundle close to helix H1 (Fig. 4D). The representative conformations that we extracted from 50 ns MD trajectories did indeed have an open tetrapyrrole site (Fig. 4, E and F). Several models from conformation WT-72-II exhibited docking in slightly shifted locations closer to the S1S2 bundle (Fig. S5A). Nevertheless, docking to that site was not observed for the S3 allele. Because Autodock Chimera produces only top 10 docking models that correspond to the lowest docking energies, one can hypothesize that the mere presence of PrP residues 72 to 120 creates highly competitive docking sites involving N terminus, substantially reducing the probability of docking to the tetrapyrrole site defined by NMR analyses.
One of 10 top docking models for Cbl for WT-72-I mPrP exhibited almost the same site as published for Fe(III)TMPyP (model 5; Fig. 5F). Differences included N-terminal capping residue G63 (noting a shorter PrP construct, human PrP 90–232, in Ref. (55) and that G63 is G62 in mPrP) and slight differences in binding interfaces: the tetrapyrrole ring faces the helices H3–H2 of PrPs directly, whereas the corrin ring of Cbl is slightly twisted. In both cases, functional groups at the perimeter of the planar ring structures were involved in the binding. Turning to mPrP S3-72-I, overlaps with the tetrapyrrole-binding sites were not observed amongst the top 10 Cbl docking models. We extended this analysis to consider docking to the globular domain region implicated in cis interactions with a metalated OR region (Fig. 5, H and I, Table S5); these cis relationships were elucidated by others for the Zn2+-loaded OR region and extended using Cd2+ as a surrogate for Zn prototype, defining a key role for E199 (56, 57, 58). Our analyses revealed that only Cbl binding model 2 for S3-72-I PrP overlapped the cis interaction site for a metalated OR region (Fig. 5, H and I).
In sum, MD analyses revealed that (i) the tetrapyrrole site defined previously using the globular domain alone (55) is detected by docking to WT constructs, (ii) the globular domain alone behaves differently by docking and MD when extended by an N-terminal tail, (iii) cis effects are seen with the longer constructs, and (iv) allelic effects are represented by some divergent performances of WT and S3 for docking of tetrapyrrole versus Cbl (in addition to some overlapping properties). Furthermore, we noted two remarkable and unanticipated effects wherein (v) an N-terminal extension caused the published tetrapyrrole site to swap the other side of the globular domain, and (vi) S3 with an N terminus analyzed by MD is prone to open up the hydrophobic core between S1–H1–S2 and H2–H3.
Formation of high–molecular weight PrPC assemblies
Noting that mammalian PrPC differs from recPrP by having N-linked sugars and a GPI anchor, we used formaldehyde crosslinking to assess formation of macromolecular assemblies in PrP-expressing cells (59). Diffuse signals indicative of glycosylated PrPC monomers were detected in all samples, irrespective of culture condition or crosslinking (Fig. 6A). A mild diminution of these low–molecular weight signals and an increase in high–molecular weight (HMW) signals was noted for S3 cells grown in DMEM supplemented with Cbl and crosslinked; thus, these experiments indicate a latent propensity for HMW complex formation modulated by medium composition.
Figure 6.
Effect of Cbl on PrP assemblies and detergent insolubility.A, cells expressing S3-PrP were grown in DMEM with or without Cbl. Cells were left untreated (−) or crosslinked in situ with 2.5% formaldehyde for 15 min at room temperature. Immunoblot detection by capillary-based Western analysis of PrP was performed from crude cellular extracts using Sha31 antibody. Diglycosylated monomeric forms of PrP with a mass of ∼35 kDa run with an apparent size of 66 kDa on this capillary-based Western analysis system (as shown previously (109)) (lower bracket). In the presence of Cbl, there was increased representation of formation of high–molecular weight (HMW) assemblies (higher bracket) with an apparent mobility of 300 to 400 kDa. B, WT-PrP and S3-PrP cells were grown for 1 week in DMEM containing increasing concentrations of Opti-MEM. Cell lysates of WT-PrP, S1-PrP, and S3-PrP generated with RIPA buffer were treated or not treated with 1% SDS (final concentration). Products were analyzed by filter-trap assay. The proteins retained by the cellulose acetate were detected by incubation with Sha31 antibody. C, quantification of A. D, WT, S1, and S3 cells were grown for 1 week in DMEM containing various additives: 0.85 μM Cbl, 0.85 μM CoCl2, and 1.12 μM ZnCl2. Cell lysates were processed as in B. E, quantification of data presented in D. Cbl, cobalamin; DMEM, Dulbecco's modified Eagle's medium; PrP, prion protein; RIPA, radioimmunoprecipitation assay buffer.
PrP aggregates frequently feature in prion diseases, and these have altered solubility properties (60), suggesting the basis for resistance to protease digestion noted previously (Fig. 1, C and D). Accordingly, cells expressing WT-PrP and S3-PrP were grown in DMEM or Opti-MEM before using a filter-trap assay for SDS-insoluble aggregates (61). A further control in these analyses was furnished by the S1 form of PrP. Using 1% SDS, detergent-resistant aggregates were apparent for S3-PrP grown in increasing fractional concentrations of Opti-MEM, but not for other alleles (Fig. 6), and this was confirmed by Western blot analysis (Fig. S6). In a mixing experiment, as DMEM was progressively replaced with Opti-MEM, immunoblot signals for aggregated PrP increased (Fig. 6, B and C), whereas WT-PrP did not form SDS-insoluble species under any conditions. We also tested how specific medium components affected aggregate formation (Fig. 6, D and E). Regardless of the culture medium used, all PrP immunoreactivities for WT-PrP and S1-PrP became soluble with 1% SDS. In contrast, S3-PrP formed significantly more SDS-resistant signal in the presence of Cbl (Fig. 6E). Ionic Zn, in all cases, had no effect, but a weak effect (versus Cbl) was scored for ionic Co with S3-PrP; whether this outcome reflected a contribution from binding at PrP site 5 (45) is currently unclear. Parallel lysates from RK13 cells that did not express mPrP did not show signals (Fig. S6C), excluding any contribution of non-PrP proteins to immunoreactive signals seen in these dot-blot assays.
Clustered cellular PrP assemblies
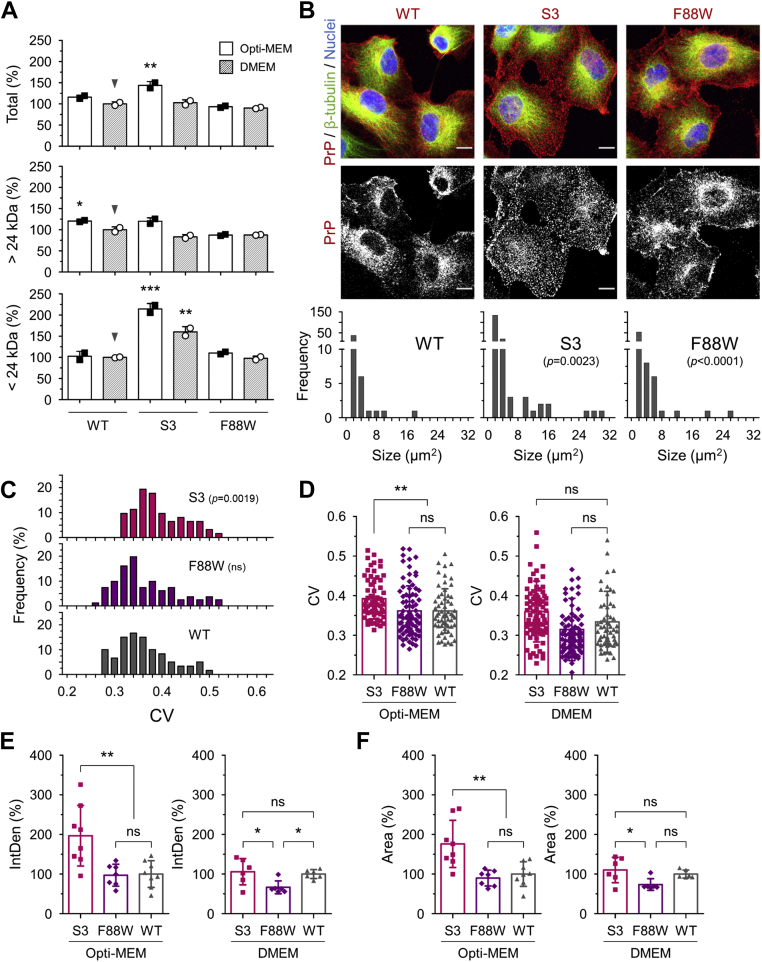
Next, cellular localization of WT PrPC and allelic variants (S3, S3-F88W, or S1) with or without medium additives was examined in stable cell lines. Equivalent expression of the S3 and F88W allelic forms versus WT-PrP in cell lines grown in DMEM was established (Figs. 7A and S7), considering either total PrP or PrP species of >24 kDa (to offset enhanced production of protease-sensitive C2 PrP by S3 and F88W allelic forms but not by WT PrP; (28), this article). Also, considering cells cultured in Opti-MEM, S3 and WT did not significantly differ with respect to species >24 kDa. A trend for higher levels of total PrP for the S3 PrP allele produced by cells cultured in Opti-MEM was attributed to <24 kDa species that include C2 fragments.
Figure 7.
S3-PrP formed clusters under Opti-MEM culture conditions. RK13 cells expressing WT-PrP (WT), S3-PrP (S3), and F88W-PrP (F88W) were cultured in Opti-MEM or DMEM. A, the cells were harvested at 10 days of culture, and the expression levels of PrP were determined by Western blot analysis. Amounts of PrP immunoreactive species >24 kDa and <24 kDa are presented (also see Fig. S7). B, upper panel, representative images of each cell line grown in Opti-MEM (top and middle). Lower panel, frequency of clustered PrP particles with size ranging from 1 to 30 μm2 (bottom). PrP in red; β-tubulin in green; and nuclei in blue. The scale bars represent 10 μm. C, degree of clustered PrP was quantified by the coefficient of variation (CV) from regions exhibiting above threshold PrP signals. N = 60, 62, and 87 for WT, S3, and F88W, respectively. A total of 24 different sites (160 μm2 for each) were analyzed. Data were presented as percent frequency of the CVs. D, mean comparisons of CVs from each cell line cultured in Opti-MEM (left, the same data shown in C) and DMEM (right, n = 60, 82, and 85 for WT, S3, and F88W, respectively). A total of 18 different sites, 160 μm2 for each, were analyzed). E, integrated PrP signal densities (IntDen). F, sizes of individual cells (area) were analyzed from a total of 594 cells grown in DMEM and Opti-MEM. N = 154, 194, and 246 for WT, S3, and F88W, respectively. Error bars represent SD. ∗p < 0.05 and ∗∗p < 0.01 by one-way ANOVA with Tukey’s multiple comparison test (A, D, E, and F) and unpaired t test (C). The equality of variance was analyzed by F statistics (B). CV, coefficient of variation; DMEM, Dulbecco's modified Eagle's medium; PrP, prion protein.
Noting these comments about expression levels for cells grown in DMEM or Opti-MEM, all three allelic forms of PrP gave signals at the cell periphery, as well as yielding some internal immunoreactivity. However, processing cell monolayers with guanidine thiocyanate, which denatures proteins to reveal epitopes and renders cells more permeable to antibody entry, revealed puncta of immunostaining, these being most apparent for S3-PrP cells grown in Opti-MEM versus corresponding WT-PrP and S1-PrP controls (Fig. S8). While some signals corresponded to filamentous processes extending between adjacent cells, puncta were especially notable for S3-PrP versus a WT-PrP control, indicating these forms of PrP were clustered into macromolecular complexes. Production of puncta was also noted for S3-F88W PrP (Fig. 7B). Quantitative analyses confirmed this relationship for frequency of occurrence versus allelic type regarding numerical aspects of morphology, S3-PrP differed from the two other alleles with respect to area, integrated pixel intensity, and size distribution (Fig. 7, C–F). In these analyses, PrP signal intensities were measured from all the pixels across regions of interest, and coefficients of variations (CVs) were calculated. In the presence of Opti-MEM, clustered S3-PrP appeared as red puncta signals, whereas under other conditions, PrP signals were more uniformly distributed. Nonuniformity was quantified by computing the pixel CVs across the entire regions of interests above threshold PrP signals. In sum, for cells grown in Opti-MEM, S3-PrP expressed at a similar level to WT-PrP had a heightened tendency to form clustered assemblies, in line with data presented in Figure 6. In several respects, the S3-PrPF88W allele had a performance intermediate between that of S3- and WT-PrP.
Discussion
Conformers, cofactors, and a conditional PrP allele
While an intersection between Cbl and prion biology has been considered previously, these earlier studies considered effects upon PrP mRNA levels deriving from an endogenous Prnp locus (62). Here, we demonstrate an alternative form of PrPC (S3) with effects impinging upon a globular domain with a sequence identical to that of the WT protein; this phenomenon manifests as a conditional effect, observed most strongly for the combination of an S3-PrP allele and growth in a cell culture medium such as Opti-MEM containing Cbl. Although S3-PrP exhibits a striking overproduction of C2 fragment in RK13 cells and brain, this property exists irrespective of culture conditions and is inferred to reflect vulnerability to a membrane-anchored protease (28). Instead, conditional phenotypic hallmarks noted for S3-PrP include protease resistance, detergent insolubility, and a predisposition to form HMW assemblies; these phenotypes were documented by covalent crosslinking and formation of puncta seen by immunostaining of permeabilized cells.
S3-PrP (and S3-F88W-PrP) contains OR region amino acid substitutions that involve extra proline and phenylalanine residues, but the properties of these alleles cannot be a simple consequence of altered solubility, which might in any event be considered a constitutive property. The intrinsic hydrophobicity of the amino acid changes in the S3 versus WT OR region does include four changes of tryptophan to phenylalanine and increasing hydrophobicity, yet four other changes of glycine to proline reduce hydrophobicity (63), and in practice, robust production of monomeric glycosylated forms of PrP was documented for all alleles in this study (Figs. 1, 2 and 5). Also speaking to a lack of constitutive propensity for aggregation, the abundant C-terminal protease-resistant PrP fragments that can be generated by S3-PrP alleles (alongside full-length PrP) do not encompass the mutant OR region that define this allele; by this criterion, these protease-resistant fragments differ from the 8 and 11 kDa species commonly seen in GSS disease brain material that do include OR-derived sequences (64, 65, 66, 67, 68).
From the studies here, production of assemblies of alternative forms is accentuated in media containing Cbl. For S3-PrP, upon cell lysis with detergent, these clustered assemblies are rendered insoluble and include PK-resistant forms with electrophoretic mobilities similar to full-length glycosylated PrP as well as glycosylated forms of C1 and C2. PK-resistant forms of C1 PrP are unusual but were found in two cattle in Switzerland surveilled for bovine spongiform encephalopathy (69). After PNGase F treatment, which involves a further denaturation step, C2 and C1 fragments predominate with a putative ∼26 kDa full-length species visible only in extended exposures. As other studies have inferred interactions between a (methionine-bearing) PrP N-terminal 22 to 31 peptide and a peptide spanning residues 218 to 225 (57) and as a 23 to 41 peptide was found in formic acid–extracted PrP from GSS disease (65), it is possible that reciprocal products of C1 and C2 cleavage (often referred to as N1 and N2) are present in the initial detergent insoluble complexes formed by S3-PrP but are depleted by subsequent sample processing steps.
Some findings reported here bear a resemblance to unusual forms of WT-PrP reported in normal human brains lacking prion infectivity (70). Not least amongst these, the form of WT-PrP identified by Yuan et al. (70) was termed “iPrP”; it comprised 5 to 25% of net PrP and was characterized by more than one PK-resistant fragment (20, 18–19, and 7 kDa, after PNGase F treatment) and the presence of HMW aggregates. More generally, in disease states, conformational change transforms a cellular protein naturally expressed in the brain into a pathological isoform; this transformation involves remodeling PrPCs “three-helix bundle” globular domain to form new structures enriched in β-sheet (71, 72, 73, 74, 75, 76). Sporadic disease is the most common epidemiological manifestation in humans (77, 78), and its origins do not appear to lie in cryptic infection events or environmental exposure to toxins (79); sporadic CJD is defined de facto by the lack of missense mutations in the C-terminal domain of PrPC that is converted to a protease-resistant form. While in vitro studies have produced misfolded PrPC or recPrP by use of low pH, chemical denaturants, or mechanical energy (80, 81, 82, 83), the natural conformational repertoire of mature N-glycosylated GPI-anchored PrPC is less explored. Along the same lines, while there are empirically deduced small molecule additives that help cell-free amplification reactions to generate misfolded PrP from native substrate (83, 84), dietary components defined as authentic cofactors (because of their fixed stoichiometry with apoproteins) are of interest since they could lie in the hinterland between physiology and pathophysiology that must encompass the origins of sporadic disease.
Mechanistic origin of conditional PrP assemblies
Effects upon PrP described here have an allelic hierarchy—that is, most noted for S3-PrP, least for WT-PrP, S3.F88W often intermediate—yet are independent of (similar) expression levels for the highest molecular weight forms of mature PrP, and prompt the question of mechanism. As revealed by readouts of solubility and protease resistance monitored with C-terminal antibodies, how does the variant S3 OR region work in cis in cells to reconfigure PrP with a WT globular domain? One starting point is to consider PrP metallochemistry. Many studies have documented multiple binding sites for Cu and/or Zn in the OR region of PrPs (19, 23, 42, 56, 85, 86, 87, 88, 89) and, concerning WT-PrP, there are studies wherein Cu might favor multimerization (48, 90, 91). Furthermore, the pathogenic effects of extra ORs in certain PrP alleles have led to consideration of metal-binding geometries of the OR region and how these might affect the globular domain, culminating in the definition of cis interactions with a negatively charged pocket (56, 92). Yet, although recPrP is described as binding ionic Co (5, 45, 93) and in silico studies suggest that Co might induce a bigger distortion of PrP OR region compared with other metals (94), we found no compelling evidence for direct interactions between Cbl (or, where studied, Co ions) and OR histidines; this is in agreement with studies of N-acetylated OR region peptides failing to reveal novel X-ray signatures after the addition of Co (95). Nor did we find evidence by MD for effects upon globular domain cis interaction site with a metalated OR region.
Another starting point to deduce mechanism concerns PrP–tetrapyrrole interactions. Tetrapyrroles have been associated with antipathogenic effects: attenuation of infectivity or reduced production of PK-resistant PrP or reduced toxic effects of misfolded PrP accumulation (53, 96, 97, 98). The effects of Cbl described here, that is, generating detergent-insoluble protease-resistant forms of PrP, can be considered to be propathogenic. Cbl contains a heterocyclic four-membered corrin ring augmented by an axial dibenzylimidazole group, whereas tetrapyrroles—which include porphyrins and phthalocyanines—are solely planar. With these observations in mind, a simple explanation for the reciprocal effects of tetrapyrrole and Cbl effects on PrPC conformation is via alternative interactions with the known globular domain tetrapyrrole-binding site. However, as deduced from in silico analyses, this explanation is only partly correct, and it is also incomplete insofar as it does not encompass other types of interactions. One reason for the discrepancies is that the globular domain tetrapyrrole-binding site only comes to the fore with truncated constructs (in our case, starting at residue 119 and, for solution NMR studies, starting at residue 91) (55). With longer WT constructs starting at residue 72, binding of Fe(III)TMPyP to this site was not prominent in docking analyses, and a shift to alternative sites was complete for analyses of the S3 allele. Remarkably, Fe(III)TMPyP binding could switch to a new site on the other side of the globular domain with long S3 constructs. Overall, lack of concordance between Fe(III)TMPyP and Cbl-binding sites was most apparent for docking analyses with the S3-72-1 construct and less apparent for WT-72-1. Allelic effects were also noted for MD, where destabilization of the hydrophobic core of PrP was apparent for S3-72-1 construct (as apo-PrP in the absence of Fe(III)TMPyP or Cbl) and less apparent for WT-72-1.
In terms of equating in vitro/in silico results with in vivo results, these structural biology effects and their allelic hierarchy tally with endpoints scored in PrP-expressing RK13 cells, specifically the heightened formation of PrP immunoreactive puncta and detergent-insoluble complexes in S3 cells grown in Cbl-containing cell medium. A recent and perhaps parallel avenue of work on PrP concerns liquid–liquid phase separation (LLPS) effects, often studied with recombinant protein presented on glass surfaces (99, 100, 101). Notably, the PrP N-terminal region—the site of the S3 allele amino acid substitutions—has been deduced as being important for phase separation effects (100, 101), a result in accordance with possession of two polybasic regions (101), low sequence complexity, and regular positioning of aromatic residues as per the “sticker and spacer” model of phase transitions (102, 103). It is also striking that the PrP paralog protein shadoo has an N-terminal low-complexity region, in this case an RGG repeat domain (104, 105), with this class of proteins being well accepted as undergoing LLPS (106). Noting that protein droplets may birth fibrillar aggregates (107, 108), it is possible that phase separation within clusters of PrP molecules might elaborate and emphasize conformational changes inferred for individual molecules. While technologies to authenticate LLPS phenomena in living cells have lagged behind those used for recombinant proteins (106), future effort in this area might be rewarding to understand how the N-terminal sequences of PrPC might coerce a WT globular domain to undergo a fundamental conformation reorganization, as is commonly seen in sporadic prion diseases.
Experimental procedures
Cell culture
Rabbit kidney epithelial (RK13) cells stably expressing WT-PrP, S1-PrP, and S3-PrP have been described previously (28). Chemical additives for DMEM were from Sigma: we supplemented the cells with 0.05 ppm of Co, which correspond to 0.85 μM Cbl and Co chloride; 0.07 ppm of Zn, which correspond to 1.12 μM Zn chloride, and 40 μM lipoic acid (Sigma). Opti-MEM (Gibco) was supplemented with 500 μM Ca–EDTA, 100 μM Cu sulphate, or 100 μM Zn sulphate.
Western blot, PK proteolysis, and deglycosylation
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl [pH 7.4], 1% NP-40, 0.5% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA prior to protein analysis); other aspects of these methods have been described previously (28).
Aggregation and filter-trap assay
About 100 μg of cell lysate was diluted in 100 μl of RIPA buffer with or without 1% SDS (final concentration). A cellulose acetate membrane (OE66; Whatman) was equilibrated in Tris-buffered saline for 10 min, followed by the recommended assembly of the 96-well dot blot apparatus (Bio-Rad). The samples were vacuum filtered through the membrane, and the samples were rinsed twice in Tris-buffered saline. Then, the membrane was treated as outlined for Western blots.
Formaldehyde crosslinking
S3 cells were grown for 1 week in the presence/absence of Cbl (0.85 μM). After two washes in PBS, cells were crosslinked at room temperature in 2.5% formaldehyde in PBS for 15 min. The reaction was quenched with 125 mM glycine for 10 min. After two further washes in PBS, cells were lysed in RIPA buffer, with lysates then analyzed by capillary Western, as previously described (109) but using 60 to 440 kDa range capillaries.
Immunofluorescence analysis of cell cultures
Cells were fixed in 4% paraformaldehyde and 4% sucrose and permeabilized or not for 10 min with 0.2% Triton X-100. After being blocked for 30 min in PBS containing 3% bovine serum albumin and 0.3 M glycine, the cells were incubated overnight with anti-PrP SAF83 antibody (Cayman; 1:2000 dilution) or Sha31 (Spi-Bio) and anti-β-tubulin (Novus Biologicals; 1:2000 dilution), followed by incubation with fluorescently conjugated secondary antibodies (Invitrogen). For the resistance to denaturation, before incubation with antibodies, the cells were treated for 2 h at 4 °C with 3 M guanidine thiocyanate. Cells were scanned with a NanoZoomer 2.0RS scanner and analyzed using NanoZoomer digital pathology software (Hamamatsu Photonics). For confocal analyses, cells were plated on poly-d-lysine (Sigma)-coated microscope cover glasses (Thermo Fisher Scientific). Cell images were acquired with the laser scanning confocal microscope, ZEN Digital Imaging for LSM 700 (Zeiss), and analyzed using Zen 2010b SP1 imaging software (Zeiss) and ImageJ (https://imagej.nih.gov/ij/). For measuring PrP signals, an identical threshold was applied to all images. Then intensities of PrP signals and clustered particles were measured using ImageJ software.
Immunocytochemistry
Cells were plated on poly-d-lysine (Sigma)-coated microscope cover glasses (Thermo Fisher Scientific). For immunocytochemistry, cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min and immersed in guanidinium thiocyanate for epitope retrieval. Then the cells were blocked with 1% bovine serum albumin in PBS with 0.1% Tween-20 for 30 min and probed with the first antibodies at 4 °C overnight: anti-PrP monoclonal antibody, Sha31 (Spi-Bio), and anti-β-tubulin polyclonal antibody (Novus). To visualize the target molecules, cells were then incubated with Alexa Fluor 594-conjugated or 488-conjugated secondary antibodies (Invitrogen). Counterstaining for nuclei was performed with Hoechst 33342 (Invitrogen). Cell images were acquired with the laser scanning confocal microscope, ZEN Digital Imaging for LSM 700 (Zeiss), and analyzed using Zen 2010b SP1 imaging software (Zeiss) and ImageJ. For measuring PrP signals, an identical threshold was applied to all images. Then intensities of PrP signals and clustered particles were measured using ImageJ software.
Metal analysis of cell media
Estimation of the media metal content was performed using inductively coupled plasma MS using a PerkinElmer Elan 6000. About 0.1 ml HNO3, 0.1 ml internal standards (In, Bi, and Sc) and 8.8 ml deionized water were added and mixed per 1 ml of medium to be analyzed. A nebulizer flow rate of 1 ml/min was used. For sample measurements, 35 sweeps/reading, one reading/replicate, and three replicates were used. Dwell times were 10 ms for Na, Al, K, Cu, Zn, and Sr, and the others were 20 ms, with the exception of Se (150 ms); the integration time was the dwell time multiplied by the number sweeps. Final results are presented as the average of three replicates. Instrument conditions were as per the manufacturer’s instructions: inductively coupled plasma RF power was 1300 W; dual detector mode; blank subtractions after internal standard correction. Measurement units were counts per second; auto lens on; four point calibration curves (0, 0.25, 0.50, and 1.00 ppm for Na, Ca, Mg, Fe, K, and P; 0, 0.005, 0.010, and 0.020 ppm for other elements. Bi, Sc, and In were used as internal standards.
Production of recombinant proteins
PJex-WT-PrP plasmid was a generous gift from Dr Glenn Millhauser (the University of California Santa Cruz) and was reengineered to make an S3 equivalent allele using Escherichia coli BL21(DE3)pLysS (Invitrogen). Recombinant proteins have been produced as previously described (110).
In silico modeling and preparation of WT and S3 structures for simulations
The coordinates of two solution-state NMR structures (mPrP 118–231 determined at 37 °C and pH of 4.5 [PDB entry: 2L39, model 1 (111)] and Syrian hamster prion protein 89 to 230 [mouse residue numbering] determined at 30 °C and pH of 5.2 [PDB entry: 2PRP, superseded by 1B10, model 6 (112)]) were used to build mPrP models denoted as WT-72-I and WT-72-II, respectively (Table S3). To obtain model WT-72-I, a sequence mPrP 72 to −117 was added to the former structure 2L39 using the Biovia Discovery Studio Visualizer (Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 2017: Dassault Systèmes, 2016). The 2PRP structure was used specifically to obtain a WT-72-II conformation without direct interactions between regions 72 and 120 and the C-terminal part of helix H3. To achieve this, we added a sequence mPrP 72 to 88 and made residue substitutions (i.e., hamster PrP -> mPrP) in the 89 to 230 part using the Biovia Discovery Studio Viewer. Mutations required to obtain the corresponding S3-72-I and S3-72-II constructs (28) were as described previously (68). Cbl coordinates were uploaded from the PDB (entry: 2BB5). After adding hydrogens where necessary, all the systems were prepared for simulations. Systems were solvated in 155 mM NaCl to approximate the composition of the cell medium.
MD simulations, docking, and analysis tools
The WT-72-I, WT-72-II, S3-72-I, and S3-72-II constructs (prepared as per Table S3) were subjected to minimizations, equilibrations, and production MD simulations in the Gromacs 5.0.7 package, as outlined previously (68). A 40 ns long equilibration/production step (“NPT”) was executed to make the 72 to 120 part of the construct more compact. The production simulations were conducted at a 310 K temperature and at a pressure of 1 atm with isotropic pressure coupling (NPT ensemble) with an 1 fs time step during 150 ns for WT-72-I and S3-72-I systems and during 50 ns for WT-72-II and S3-72-II systems. From the corresponding MD trajectories, representative snapshots were obtained using the clustering application in Gromacs. Representative conformations obtained from the four production MD trajectories were used as templates for subsequent ligand–protein docking. In addition, two truncated models WT-119-I and S3-119-I were prepared from the representative WT-72-I and S3-72-I conformations, respectively (Table S3). The ligand–protein docking was performed with Autodock Vina software implemented in the UCSF Chimera package (113, 114). Docking scores in Autodock represent the global minimum in the interaction energy between the ligand and the protein, which are obtained via an exploration of the system's available degrees of freedom. Rapid grid-based energy evaluation and torsional scanning were performed using the Lamarckian Genetic Algorithm and empirical free-energy scoring function, taking into account contributions from dispersion/repulsion, hydrogen bonding, electrostatics, and desolvation. The top 10 docking models were chosen for subsequent structural analysis. Table S4 lists the 10 top docking models for systems WT-72-I, S3-72-I, WT-119-I, and S3-119-I.
Data analysis and statistics
The number of biological and technical replicates of compared groups were represented in corresponding figure legends. Sample size (n) indicates biological replicates unless otherwise stated. For the most statistical analysis, comparisons of means were performed using the unpaired two-tailed Student's t test. The CV of PrP signals and the integrated PrP signal densities were compared using ANOVA with post hoc Tukey’s multiple comparison test. The equality of variance in particle distributions was analyzed by F statistics. Statistical analyses of all data were performed using PRISM, version 5 software (GraphPad Software, Inc).
Data availability
All data generated or analyzed during this study are included in this published article and its supporting information files.
Supporting information
This article contains supporting information including references (115, 116, 117, 118).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Jack Moore and Olivier Julien, Department of Biochemistry, University of Alberta, for help with MALDI analysis and Ghazaleh Eskandari-Sedighi for the Cu–glycine2 chelate. We are indebted to Glenn Millhauser for gifts of mouse PrP23 to 230 expression plasmids, for S3 recombinant protein, and for discussions. We thank Brian Sykes for discussions on binding pockets. This work was supported by the Canadian Institutes of Health Research (grant nos.: MOP123525 and PS165980) and the University Hospital Foundation (grant no.: SFR1404). Instrumentation was supported by the Canada Foundation for Innovation (grant no.: NIF21633) and the Alberta Synergies in Alzheimer’s and Related Disorders (SynAD) program, which is funded by the Alzheimer Society of Alberta and Northwest Territories through the “Hope for Tomorrow” program and by the University Hospital Foundation.
Author contributions
N. D., A. L., S. G. K., and D. W. methodology; N. D., A. L., H. W., D. W., D. G. P., M. S., and L. D. formal analysis; N. D., A. L., I. V., S. G. K., A. R. C., S. W., C. S., and L. D. investigation; N. D., A. L., H. W., and D. W. writing–original draft.
Funding and additional information
A. L. was funded by studentships from the National Sciences and Engineering Research Council, Alberta Innovates Health Solutions, and the University of Alberta, whereas D. W. was supported by the Canada Research Chairs and Canada Excellence Research Chairs programs, respectively. L. D. and M. S. were supported by the Alberta Prion Research Institute (grant no.: 20170016).
Edited by Paul Fraser
Footnotes
Present address for Agnes Lau: Tanz Centre for Research in Neurodegenerative Diseases, Krembil Discovery Tower, University of Toronto, Canada.
Supporting information
References
- 1.Stahl N., Borchelt D.R., Hsiao K., Prusiner S.B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 2.Benvegnu S., Poggiolini I., Legname G. Neurodevelopmental expression and localization of the cellular prion protein in the central nervous system of the mouse. J. Comp. Neurol. 2010;518:1879–1891. doi: 10.1002/cne.22357. [DOI] [PubMed] [Google Scholar]
- 3.Castle A.R., Gill A.C. Physiological functions of the cellular prion protein. Front. Mol. Biosci. 2017;4:19. doi: 10.3389/fmolb.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazan J.F., Fletterick R.J., McKinley M.P., Prusiner S.B. Predicted secondary structure and membrane topology of the scrapie prion protein. Protein Eng. 1987;1:125–135. doi: 10.1093/protein/1.2.125. [DOI] [PubMed] [Google Scholar]
- 5.Pan K.M., Stahl N., Prusiner S.B. Purification and properties of the cellular prion protein from Syrian hamster brain. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris D.A., Huber M.T., van Dijken P., Shyng S.L., Chait B.T., Wang R. Processing of a cellular prion protein: Identification of N- and C-terminal cleavage sites. Biochemistry. 1993;32:1009–1016. doi: 10.1021/bi00055a003. [DOI] [PubMed] [Google Scholar]
- 7.Chen S.G., Teplow D.B., Parchi P., Teller J.K., Gambetti P., Autilio-Gambetti L. Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 8.Liang J., Kong Q. alpha-Cleavage of cellular prion protein. Prion. 2012;6:453–460. doi: 10.4161/pri.22511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bockman J.M., Kingsbury D.T., McKinley M.P., Bendheim P.E., Prusiner S.B. Creutzfeldt-Jakob disease prion proteins in human brains. N. Engl. J. Med. 1985;312:73–78. doi: 10.1056/NEJM198501103120202. [DOI] [PubMed] [Google Scholar]
- 10.Safar J., Roller P.P., Gajdusek D.C., Gibbs C.J., Jr. Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J. Biol. Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 11.Wadsworth J.D., Hill A.F., Beck J.A., Collinge J. Molecular and clinical classification of human prion disease. Br. Med. Bull. 2003;66:241–254. doi: 10.1093/bmb/66.1.241. [DOI] [PubMed] [Google Scholar]
- 12.Cescatti M., Saverioni D., Capellari S., Tagliavini F., Kitamoto T., Ironside J., Giese A., Parchi P. Analysis of conformational stability of abnormal prion protein aggregates across the spectrum of Creutzfeldt-Jakob disease prions. J. Virol. 2016;90:6244–6254. doi: 10.1128/JVI.00144-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collinge J., Clarke A.R. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd S., Mead S., Collinge J. Genetics of prion disease. Top. Curr. Chem. 2011;305:1–22. doi: 10.1007/128_2011_157. [DOI] [PubMed] [Google Scholar]
- 15.Aronoff-Spencer E., Burns C.S., Avdievich N.I., Gerfen G.J., Peisach J., Antholine W.E., Ball H.L., Cohen F.E., Prusiner S.B., Millhauser G.L. Identification of the Cu2+ binding sites in the N-terminal domain of the prion protein by EPR and CD spectroscopy. Biochemistry. 2000;39:13760–13771. doi: 10.1021/bi001472t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns C.S., Aronoff-Spencer E., Dunham C.M., Lario P., Avdievich N.I., Antholine W.E., Olmstead M.M., Vrielink A., Gerfen G.J., Peisach J., Scott W.G., Millhauser G.L. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41:3991–4001. doi: 10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin K., Yang Y., Mastrangelo P., Westaway D. Mapping Cu(II) binding sites in prion proteins by diethyl pyrocarbonate modification and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometric footprinting. J. Biol. Chem. 2002;277:1981–1990. doi: 10.1074/jbc.M108744200. [DOI] [PubMed] [Google Scholar]
- 18.Chattopadhyay M., Walter E.D., Newell D.J., Jackson P.J., Aronoff-Spencer E., Peisach J., Gerfen G.J., Bennett B., Antholine W.E., Millhauser G.L. The octarepeat domain of the prion protein binds Cu(II) with three distinct coordination modes at pH 7.4. J. Am. Chem. Soc. 2005;127:12647–12656. doi: 10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson G.S., Murray I., Hosszu L.L., Gibbs N., Waltho J.P., Clarke A.R., Collinge J. Location and properties of metal-binding sites on the human prion protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8531–8535. doi: 10.1073/pnas.151038498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter E.D., Stevens D.J., Visconte M.P., Millhauser G.L. The prion protein is a combined zinc and copper binding protein: Zn(2+) alters the distribution of Cu(2+) coordination modes. J. Am. Chem. Soc. 2007;129:15440–15441. doi: 10.1021/ja077146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brazier M.W., Davies P., Player E., Marken F., Viles J.H., Brown D.R. Manganese binding to the prion protein. J. Biol. Chem. 2008;283:12831–12839. doi: 10.1074/jbc.M709820200. [DOI] [PubMed] [Google Scholar]
- 22.Beland M., Roucou X. The prion protein unstructured N-terminal region is a broad-spectrum molecular sensor with diverse and contrasting potential functions. J. Neurochem. 2012;120:853–868. doi: 10.1111/j.1471-4159.2011.07613.x. [DOI] [PubMed] [Google Scholar]
- 23.Brown D.R., Qin K., Herms J.W., Madlung A., Manson J., Strome R., Fraser P.E., Kruck T., von Bohlen A., Schulz-Schaeffer W., Giese A., Westaway D., Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 24.Stöckel J., Safar J., Wallace A.C., Cohen F.E., Prusiner S.B. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37:7185–7193. doi: 10.1021/bi972827k. [DOI] [PubMed] [Google Scholar]
- 25.Hasnain S.S., Murphy L.M., Strange R.W., Grossmann J.G., Clarke A.R., Jackson G.S., Collinge J. XAFS study of the high-affinity copper-binding site of human PrP(91-231) and its low-resolution structure in solution. J. Mol. Biol. 2001;311:467–473. doi: 10.1006/jmbi.2001.4795. [DOI] [PubMed] [Google Scholar]
- 26.Liu L., Jiang D., McDonald A., Hao Y., Millhauser G.L., Zhou F. Copper redox Cycling in the prion protein depends critically on binding mode. J. Am. Chem. Soc. 2011;133:12229–12237. doi: 10.1021/ja2045259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez J., Sanchez R., Castellanos M., Makarava N., Aguzzi A., Baskakov I.V., Gasset M. PrP charge structure encodes interdomain interactions. Sci. Rep. 2015;5:13623. doi: 10.1038/srep13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau A., McDonald A., Daude N., Mays C.E., Walter E.D., Aglietti R., Mercer R.C., Wohlgemuth S., van der Merwe J., Yang J., Gapeshina H., Kim C., Grams J., Shi B., Wille H., et al. Octarepeat region flexibility impacts prion function, endoproteolysis and disease manifestation. EMBO Mol. Med. 2015;7:339–356. doi: 10.15252/emmm.201404588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borchelt D.R., Scott M., Taraboulos A., Stahl N., Prusiner S.B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott M.R., Köhler R., Foster D., Prusiner S.B. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesebro B., Wehrly K., Caughey B., Nishio J., Ernst D., Race R. Foreign PrP expression and scrapie infection in tissue culture cell lines. Dev. Biol. Stand. 1993;80:131–140. [PubMed] [Google Scholar]
- 32.Priola S.A., Caughey B., Raymond G.J., Chesebro B. Prion protein and the scrapie agent: In vitro studies in infected neuroblastoma cells. Infect. Agents Dis. 1994;3:54–58. [PubMed] [Google Scholar]
- 33.Nishida N., Harris D.A., Vilette D., Laude H., Frobert Y., Grassi J., Casanova D., Milhavet O., Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodgkin D.C., Kamper J., Mackay M., Pickworth J., Trueblood K.N., White J.G. Structure of vitamin B12. Nature. 1956;178:64–66. doi: 10.1038/178064a0. [DOI] [PubMed] [Google Scholar]
- 35.Parchi P., Giese A., Capellari S., Brown P., Schulz-Schaeffer W., Windl O., Zerr I., Budka H., Kopp N., Piccardo P., Poser S., Rojiani A., Streichemberger N., Julien J., Vital C., et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann. Neurol. 1999;46:224–233. [PubMed] [Google Scholar]
- 36.Puoti G., Giaccone G., Rossi G., Canciani B., Bugiani O., Tagliavini F. Sporadic Creutzfeldt-jakob disease: Co-occurrence of different types of PrP(Sc) in the same brain. Neurology. 1999;53:2173–2176. doi: 10.1212/wnl.53.9.2173. [DOI] [PubMed] [Google Scholar]
- 37.Schoch G., Seeger H., Bogousslavsky J., Tolnay M., Janzer R.C., Aguzzi A., Glatzel M. Analysis of prion strains by PrPSc profiling in sporadic Creutzfeldt-Jakob disease. PLoS Med. 2006;3:e14. doi: 10.1371/journal.pmed.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uro-Coste E., Cassard H., Simon S., Lugan S., Bilheude J.M., Perret-Liaudet A., Ironside J.W., Haik S., Basset-Leobon C., Lacroux C., Peoch K., Streichenberger N., Langeveld J., Head M.W., Grassi J., et al. Beyond PrP type 1/type 2 dichotomy in Creutzfeldt-jakob disease. PLoS Pathog. 2008;4 [PubMed] [Google Scholar]
- 39.Cali I., Castellani R., Alshekhlee A., Cohen Y., Blevins J., Yuan J., Langeveld J.P., Parchi P., Safar J.G., Zou W.Q., Gambetti P. Co-existence of scrapie prion protein types 1 and 2 in sporadic Creutzfeldt-jakob disease: Its effect on the phenotype and prion-type characteristics. Brain. 2009;132:2643–2658. doi: 10.1093/brain/awp196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mays C.E., Kim C., Haldiman T., van der Merwe J., Lau A., Yang J., Grams J., Di Bari M.A., Nonno R., Telling G.C., Kong Q., Langeveld J., McKenzie D., Westaway D., Safar J.G. Prion disease tempo determined by host-dependent substrate reduction. J. Clin. Invest. 2014;124:847–858. doi: 10.1172/JCI72241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stone L.A., Jackson G.S., Collinge J., Wadsworth J.D., Clarke A.R. Inhibition of proteinase K activity by copper(II) ions. Biochemistry. 2007;46:245–252. doi: 10.1021/bi061646s. [DOI] [PubMed] [Google Scholar]
- 42.Hornshaw M.P., McDermott J.R., Candy J.M. Copper binding to the N-terminal tandem repeat region of mammalian and avian prion protein. Biochem. Biophys. Res. Comm. 1995;207:621–629. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 43.Koh J.Y., Suh S.W., Gwag B.J., He Y.Y., Hsu C.Y., Choi D.W. The role of zinc in selective neuronal death after transient global cerebral ischemia. Science. 1996;272:1013–1016. doi: 10.1126/science.272.5264.1013. [DOI] [PubMed] [Google Scholar]
- 44.Salzano G., Giachin G., Legname G. Structural consequences of copper binding to the prion protein. Cells. 2019;8:770. doi: 10.3390/cells8080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treiber C., Thompsett A.R., Pipkorn R., Brown D.R., Multhaup G. Real-time kinetics of discontinuous and highly conformational metal-ion binding sites of prion protein. J. Biol. Inorg. Chem. 2007;12:711–720. doi: 10.1007/s00775-007-0220-3. [DOI] [PubMed] [Google Scholar]
- 46.Viles J.H., Cohen F.E., Prusiner S.B., Goodin D.B., Wright P.E., Dyson H.J. Copper binding to the prion protein: Structural implications of four identical cooperative binding sites. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2042–2047. doi: 10.1073/pnas.96.5.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garnett A.P., Viles J.H. Copper binding to the octarepeats of the prion protein. Affinity, specificity, folding, and cooperativity: Insights from circular dichroism. J. Biol. Chem. 2003;278:6795–6802. doi: 10.1074/jbc.M209280200. [DOI] [PubMed] [Google Scholar]
- 48.Thompsett A.R., Abdelraheim S.R., Daniels M., Brown D.R. High affinity binding between copper and full-length prion protein identified by two different techniques. J. Biol. Chem. 2005;280:42750–42758. doi: 10.1074/jbc.M506521200. [DOI] [PubMed] [Google Scholar]
- 49.Wells M.A., Jackson G.S., Jones S., Hosszu L.L., Craven C.J., Clarke A.R., Collinge J., Waltho J.P. A reassessment of copper(II) binding in the full-length prion protein. Biochem. J. 2006;399:435–444. doi: 10.1042/BJ20060458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter E.D., Stevens D.J., Spevacek A.R., Visconte M.P., Dei Rossi A., Millhauser G.L. Copper binding extrinsic to the octarepeat region in the prion protein. Curr. Protein Pept. Sci. 2009;10:529–535. doi: 10.2174/138920309789352056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miles E.W. In: Hirs C.H.W., Timasheff S.N., editors. Vol. 47. Academic Press; NY: 1977. Modification of the histidyl residues in proteins by diethylpyrocarbonate; pp. 431–442. (Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- 52.Bryant D.A., Hunter C.N., Warren M.J. Biosynthesis of the modified tetrapyrroles-the pigments of life. J. Biol. Chem. 2020;295:6888–6925. doi: 10.1074/jbc.REV120.006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caughey W.S., Priola S.A., Kocisko D.A., Raymond L.D., Ward A., Caughey B. Cyclic tetrapyrrole sulfonation, metals, and oligomerization in anti-prion activity. Antimicrob. Agents Chemother. 2007;51:3887–3894. doi: 10.1128/AAC.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrec J., Tavernelli I., Rothlisberger U. Two misfolding routes for the prion protein around pH 4.5. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicoll A.J., Trevitt C.R., Tattum M.H., Risse E., Quarterman E., Ibarra A.A., Wright C., Jackson G.S., Sessions R.B., Farrow M., Waltho J.P., Clarke A.R., Collinge J. Pharmacological chaperone for the structured domain of human prion protein. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17610–17615. doi: 10.1073/pnas.1009062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans E.G., Pushie M.J., Markham K.A., Lee H.W., Millhauser G.L. Interaction between prion protein's copper-bound octarepeat domain and a charged C-terminal pocket suggests a mechanism for N-terminal regulation. Structure. 2016;24:1057–1067. doi: 10.1016/j.str.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonald A.J., Leon D.R., Markham K.A., Wu B., Heckendorf C.F., Schilling K., Showalter H.D., Andrews P.C., McComb M.E., Pushie M.J., Costello C.E., Millhauser G.L., Harris D.A. Altered domain structure of the prion protein caused by Cu(2+) binding and functionally relevant mutations: Analysis by Cross-linking, MS/MS, and NMR. Structure. 2019;27:907–922. doi: 10.1016/j.str.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Markham K.A., Roseman G.P., Linsley R.B., Lee H.W., Millhauser G.L. Molecular features of the Zn(2+) binding site in the prion protein probed by (113)Cd NMR. Biophys. J. 2019;116:610–620. doi: 10.1016/j.bpj.2019.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmitt-Ulms G., Legname G., Baldwin M.A., Ball H.L., Bradon N., Bosque P.J., Crossin K.L., Edelman G.M., DeArmond S.J., Cohen F.E., Prusiner S.B. Binding of neural cell adhesion molecules (N-CAMs) to the cellular prion protein. J. Mol. Biol. 2001;314:1209–1225. doi: 10.1006/jmbi.2000.5183. [DOI] [PubMed] [Google Scholar]
- 60.Pedersen J.T., Heegaard N.H. Analysis of protein aggregation in neurodegenerative disease. Anal. Chem. 2013;85:4215–4227. doi: 10.1021/ac400023c. [DOI] [PubMed] [Google Scholar]
- 61.Xu G., Gonzales V., Borchelt D.R. Rapid detection of protein aggregates in the brains of Alzheimer patients and transgenic mouse models of amyloidosis. Alzheimer Dis. Assoc. Disord. 2002;16:191–195. doi: 10.1097/00002093-200207000-00010. [DOI] [PubMed] [Google Scholar]
- 62.Scalabrino G., Veber D., Mutti E., Calligaro A., Milani S., Tredici G. Cobalamin (vitamin B(12)) regulation of PrP(C), PrP(C)-mRNA and copper levels in rat central nervous system. Exp. Neurol. 2012;233:380–390. doi: 10.1016/j.expneurol.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 63.Biro J.C. Amino acid size, charge, hydropathy indices and matrices for protein structure analysis. Theor. Biol. Med. Model. 2006;3:15. doi: 10.1186/1742-4682-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piccardo P., Seiler C., Dlouhy S.R., Young K., Farlow M.R., Prelli F., Frangione B., Bugiani O., Tagliavini F., Ghetti B. Proteinase-K-Resistant prion protein isoforms in gerstmann-straussler-scheinker disease (Indiana kindred) J. Neuropath. Exp. Neur. 1996;55:1157–1163. doi: 10.1097/00005072-199611000-00007. [DOI] [PubMed] [Google Scholar]
- 65.Tagliavini F., Lievens P.M., Tranchant C., Warter J.M., Mohr M., Giaccone G., Perini F., Rossi G., Salmona M., Piccardo P., Ghetti B., Beavis R.C., Bugiani O., Frangione B., Prelli F. A 7-kDa prion protein (PrP) fragment, an integral component of the PrP region required for infectivity, is the major amyloid protein in Gerstmann-Straussler-Scheinker disease A117V. J. Biol. Chem. 2001;276:6009–6015. doi: 10.1074/jbc.M007062200. [DOI] [PubMed] [Google Scholar]
- 66.Liberski P.P. Gerstmann-Straussler-Scheinker disease. Adv. Exp. Med. Biol. 2012;724:128–137. doi: 10.1007/978-1-4614-0653-2_10. [DOI] [PubMed] [Google Scholar]
- 67.Pirisinu L., Di Bari M.A., D'Agostino C., Marcon S., Riccardi G., Poleggi A., Cohen M.L., Appleby B.S., Gambetti P., Ghetti B., Agrimi U., Nonno R. Gerstmann-Straussler-Scheinker disease subtypes efficiently transmit in bank voles as genuine prion diseases. Sci. Rep. 2016;6:20443. doi: 10.1038/srep20443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mercer R.C.C., Daude N., Dorosh L., Fu Z.L., Mays C.E., Gapeshina H., Wohlgemuth S.L., Acevedo-Morantes C.Y., Yang J., Cashman N.R., Coulthart M.B., Pearson D.M., Joseph J.T., Wille H., Safar J.G., et al. A novel Gerstmann-Straussler-Scheinker disease mutation defines a precursor for amyloidogenic 8 kDa PrP fragments and reveals N-terminal structural changes shared by other GSS alleles. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serra F., Muller J., Gray J., Luthi R., Dudas S., Czub S., Seuberlich T. PrP-C1 fragment in cattle brains reveals features of the transmissible spongiform encephalopathy associated PrPsc. Brain Res. 2017;1659:19–28. doi: 10.1016/j.brainres.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 70.Yuan J., Xiao X., McGeehan J., Dong Z., Cali I., Fujioka H., Kong Q., Kneale G., Gambetti P., Zou W.Q. Insoluble aggregates and protease-resistant conformers of prion protein in uninfected human brains. J. Biol. Chem. 2006;281:34848–34858. doi: 10.1074/jbc.M602238200. [DOI] [PubMed] [Google Scholar]
- 71.Pan K.M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R.J., Cohen F.E., Prusiner S.B. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Caughey B.W., Dong A., Bhat K.S., Ernst D., Hayes S.F., Caughey W.S. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 73.Requena J.R., Wille H. The structure of the infectious prion protein: Experimental data and molecular models. Prion. 2014;8:60–66. doi: 10.4161/pri.28368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wille H., Requena J.R. The structure of PrP(Sc) prions. Pathogens. 2018;7:20. doi: 10.3390/pathogens7010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spagnolli G., Rigoli M., Orioli S., Sevillano A.M., Faccioli P., Wille H., Biasini E., Requena J.R. Full atomistic model of prion structure and conversion. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kraus A., Hoyt F., Schwartz C.L., Hansen B., Artikis E., Hughson A.G., Raymond G.J., Race B., Baron G.S., Caughey B. High-resolution structure and strain comparison of infectious mammalian prions. Mol. Cell. 2021;81:4540–4551.e6. doi: 10.1016/j.molcel.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Parchi P., Saverioni D. Molecular pathology, classification, and diagnosis of sporadic human prion disease variants. Folia Neuropathol. 2012;50:20–45. [PubMed] [Google Scholar]
- 78.Ironside J.W., Ritchie D.L., Head M.W. Prion diseases. Handb. Clin. Neurol. 2017;145:393–403. doi: 10.1016/B978-0-12-802395-2.00028-6. [DOI] [PubMed] [Google Scholar]
- 79.Will R.G., Ironside J.W. Sporadic and infectious human prion diseases. Cold Spring Harb. Perspect. Med. 2017;7 doi: 10.1101/cshperspect.a024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown D.R., Hafiz F., Glasssmith L.L., Wong B.S., Jones I.M., Clive C., Haswell S.J. Consequences of manganese replacement of copper for prion protein function and proteinase resistance. EMBO J. 2000;19:1180–1186. doi: 10.1093/emboj/19.6.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li X.L., Dong C.F., Wang G.R., Zhou R.M., Shi Q., Tian C., Gao C., Mei G.Y., Chen C., Xu K., Han J., Dong X.P. Manganese-induced changes of the biochemical characteristics of the recombinant wild-type and mutant PrPs. Med. Microbiol. Immunol. 2009;198:239–245. doi: 10.1007/s00430-009-0120-y. [DOI] [PubMed] [Google Scholar]
- 82.Legname G., Baskakov I.V., Nguyen H.O., Riesner D., Cohen F.E., DeArmond S.J., Prusiner S.B. Synthetic mammalian prions. Science. 2004;305:673–676. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 83.Wang F., Wang X., Yuan C.G., Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science. 2010;327:1132–1135. doi: 10.1126/science.1183748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Supattapone S. Phosphatidylethanolamine as a prion cofactor: Potential implications for disease pathogenesis. Prion. 2013;6:417–419. doi: 10.4161/pri.21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burns C.S., Aronoff-Spencer E., Legname G., Prusiner S.B., Antholine W.E., Gerfen G.J., Peisach J., Millhauser G.L. Copper coordination in the full-length, recombinant prion protein. Biochemistry. 2003;42:6794–6803. doi: 10.1021/bi027138+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Millhauser G.L. Copper binding in the prion protein. Acc. Chem. Res. 2004;37:79–85. doi: 10.1021/ar0301678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leach S.P., Salman M.D., Hamar D. Trace elements and prion diseases: A review of the interactions of copper, manganese and zinc with the prion protein. Anim. Health Res. Rev. 2006;7:97–105. doi: 10.1017/S1466252307001181. [DOI] [PubMed] [Google Scholar]
- 88.Choi C.J., Kanthasamy A., Anantharam V., Kanthasamy A.G. Interaction of metals with prion protein: Possible role of divalent cations in the pathogenesis of prion diseases. Neurotoxicology. 2006;27:777–787. doi: 10.1016/j.neuro.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 89.McDonald A., Pushie M.J., Millhauser G.L., George G.N. New insights into metal interactions with the prion protein: EXAFS analysis and structure calculations of copper binding to a single octarepeat from the prion protein. J. Phys. Chem. B. 2013;117:13822–13841. doi: 10.1021/jp408239h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qin K., Yang D.S., Yang Y., Chishti M.A., Meng L.J., Kretzschmar H.A., Yip C.M., Fraser P.E., Westaway D. Copper(II)-induced conformational changes and protease resistance in recombinant and cellular PrP. Effect of protein age and deamidation. J. Biol. Chem. 2000;275:19121–19131. doi: 10.1074/jbc.275.25.19121. [DOI] [PubMed] [Google Scholar]
- 91.Quaglio E., Chiesa R., Harris D.A. Copper converts the cellular prion protein into a protease-resistant species that is distinct from the scrapie isoform. J. Biol. Chem. 2001;276:11432–11438. doi: 10.1074/jbc.M009666200. [DOI] [PubMed] [Google Scholar]
- 92.Stevens D.J., Walter E.D., Rodriguez A., Draper D., Davies P., Brown D.R., Millhauser G.L. Early onset prion disease from octarepeat expansion correlates with copper binding properties. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pergami P., Jaffe H., Safar J. Semipreparative chromatographic method to purify the normal cellular isoform of the prion protein in nondenatured form. Anal. Biochem. 1996;236:63–73. doi: 10.1006/abio.1996.0132. [DOI] [PubMed] [Google Scholar]
- 94.Pandey K.K., Snyder J.P., Liotta D.C., Musaev D.G. Computational studies of transition metal selectivity of octapeptide repeat region of prion protein (PrP) J. Phys. Chem. B. 2010;114:1127–1135. doi: 10.1021/jp909945e. [DOI] [PubMed] [Google Scholar]
- 95.Pushie M.J., Ross A.R., Vogel H.J. Mass spectrometric determination of the coordination geometry of potential copper(II) surrogates for the mammalian prion protein octarepeat region. Anal. Chem. 2007;79:5659–5667. doi: 10.1021/ac070312l. [DOI] [PubMed] [Google Scholar]
- 96.Caughey W.S., Raymond L.D., Horiuchi M., Caughey B. Inhibition of protease-resistant prion protein formation by porphyrins and phthalocyanines. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mays C.E., Joy S., Li L., Yu L., Genovesi S., West F.G., Westaway D. Prion inhibition with multivalent PrP(Sc) binding compounds. Biomaterials. 2012;33:6808–6822. doi: 10.1016/j.biomaterials.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 98.Massignan T., Cimini S., Stincardini C., Cerovic M., Vanni I., Elezgarai S.R., Moreno J., Stravalaci M., Negro A., Sangiovanni V., Restelli E., Riccardi G., Gobbi M., Castilla J., Borsello T., et al. A cationic tetrapyrrole inhibits toxic activities of the cellular prion protein. Sci. Rep. 2016;6:23180. doi: 10.1038/srep23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matos C.O., Passos Y.M., do Amaral M.J., Macedo B., Tempone M.H., Bezerra O.C.L., Moraes M.O., Almeida M.S., Weber G., Missailidis S., Silva J.L., Uversky V.N., Pinheiro A.S., Cordeiro Y. Liquid-liquid phase separation and fibrillation of the prion protein modulated by a high-affinity DNA aptamer. FASEB J. 2020;34:365–385. doi: 10.1096/fj.201901897R. [DOI] [PubMed] [Google Scholar]
- 100.Tange H., Ishibashi D., Nakagaki T., Taguchi Y., Kamatari Y.O., Ozawa H., Nishida N. Liquid-liquid phase separation of full-length prion protein initiates conformational conversion in vitro. J. Biol. Chem. 2021;296:100367. doi: 10.1016/j.jbc.2021.100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kamps J., Lin Y.H., Oliva R., Bader V., Winter R., Winklhofer K.F., Tatzelt J. The N-terminal domain of the prion protein is required and sufficient for liquid-liquid phase separation: A crucial role of the abeta-binding domain. J. Biol. Chem. 2021;297:100860. doi: 10.1016/j.jbc.2021.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choi J.M., Holehouse A.S., Pappu R.V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020;49:107–133. doi: 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hastings R.L., Boeynaems S. Designer Condensates: A toolkit for the biomolecular architect. J. Mol. Biol. 2021;433:166837. doi: 10.1016/j.jmb.2021.166837. [DOI] [PubMed] [Google Scholar]
- 104.Corley S.M., Gready J.E. Identification of the RGG box motif in shadoo: RNA-binding and signaling roles? Bioinform Biol. Insights. 2008;2:383–400. doi: 10.4137/bbi.s1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lau A., Mays C.E., Genovesi S., Westaway D. RGG repeats of PrP-like shadoo protein bind nucleic acids. Biochemistry. 2012;51:9029–9031. doi: 10.1021/bi301395w. [DOI] [PubMed] [Google Scholar]
- 106.Alberti S., Gladfelter A., Mittag T. Considerations and Challenges in studying liquid-liquid phase separation and biomolecular Condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boyko S., Surewicz K., Surewicz W.K. Regulatory mechanisms of tau protein fibrillation under the conditions of liquid-liquid phase separation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:31882–31890. doi: 10.1073/pnas.2012460117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kang S.G., Han Z.Z., Daude N., McNamara E., Wohlgemuth S., Molina-Porcel L., Safar J.G., Mok S.A., Westaway D. Pathologic tau conformer ensembles induce dynamic, liquid-liquid phase separation events at the nuclear envelope. BMC Biol. 2021;19:199. doi: 10.1186/s12915-021-01132-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Castle A.R., Daude N., Gilch S., Westaway D. Application of high-throughput, capillary-based Western analysis to modulated cleavage of the cellular prion protein. J. Biol. Chem. 2019;294:2642–2650. doi: 10.1074/jbc.RA118.006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Daude N., Ng V., Watts J.C., Genovesi S., Glaves J.P., Wohlgemuth S., Schmitt-Ulms G., Young H., McLaurin J., Fraser P.E., Westaway D. Wild-type Shadoo proteins convert to amyloid-like forms under native conditions. J. Neurochem. 2010;113:92–104. doi: 10.1111/j.1471-4159.2010.06575.x. [DOI] [PubMed] [Google Scholar]
- 111.Damberger F.F., Christen B., Perez D.R., Hornemann S., Wuthrich K. Cellular prion protein conformation and function. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17308–17313. doi: 10.1073/pnas.1106325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.James T.L., Liu H., Ulyanov N.B., Farr-Jones S., Zhang H., Donne D.G., Kaneko K., Groth D., Mehlhorn I., Prusiner S.B., Cohen F.E. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 114.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wuerges J., Garau G., Geremia S., Fedosov S.N., Petersen T.E., Randaccio L. Structural basis for mammalian vitamin B12 transport by transcobalamin. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4386–4391. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hastings J., Owen G., Dekker A., Ennis M., Kale N., Muthukrishnan V., Turner S., Swainston N., Mendes P., Steinbeck C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44:D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trzesowska-Kruszynska A. Copper complex of glycine Schiff base: In situ ligand synthesis, structure, spectral, and thermal properties. J. Mol. Struct. 2012;1017:72–78. [Google Scholar]
- 118.Hamada Y.Z., Makoni N., Hamada H. Cu2+ complexes with the simplest amino acid glycine (Gly) J. Nanomed. Res. 2017;5 00123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supporting information files.