Summary
High-throughput antibody repertoire analysis via next-generation sequencing is a key method in understanding humoral immunity. Errors introduced during library preparation and sequencing can be corrected with molecular amplification fingerprinting using unique molecular identifiers. Here, we provide a step-by-step protocol for laboratory and bioinformatic workflows to perform sequencing in murine cells with isotype-specific primers, obtaining total and isotype-specific B cell receptor repertoires. This enables the examination of isotype-dependent immune responses and improves the understanding of underlying mechanisms in humoral immunity.
For complete details on the use and execution of this protocol, please refer to Khan et al. (2016).
Subject areas: Bioinformatics, Sequence analysis, Cell Biology, Cell isolation, Sequencing, Immunology, Molecular Biology
Graphical abstract

Highlights
-
•
Step-by-step protocol to perform isotype-specific BCR sequencing in murine cells
-
•
Correction of PCR and sequencing errors using unique molecular identifiers
-
•
Setup and use of bioinformatic tools for the analysis and QC of BCR repertoire data
High-throughput antibody repertoire analysis via next-generation sequencing is a key method in understanding humoral immunity. Errors introduced during library preparation and sequencing can be corrected with molecular amplification fingerprinting using unique molecular identifiers. Here, we provide a step-by-step protocol for laboratory and bioinformatic workflows to perform sequencing in murine cells with isotype-specific primers, obtaining total and isotype-specific B cell receptor repertoires. This enables the examination of isotype-dependent immune responses and improves the understanding of underlying mechanisms in humoral immunity.
Before you begin
Ensure that all reagents are available, and buffers have been prepared.
Before RNA isolation, properly clean all surfaces with RNase AWAY or another alternative (this includes for example bench, centrifuge, pipettors, pipette tip boxes or tube holder).
Note: It is recommended to work on a working space, which is used for RNA isolation only, to reduce contamination with RNases and DNA.
Prepare 70% ethanol for RNA isolation.
Note: As ethanol is hygroscopic fresh 70% ethanol should be prepared for optimal results. To obtain a proper solution of 70% ethanol measure 70 mL ethanol and 30 mL water separately and combine. Do not top off 70 mL of ethanol to 100 mL with water as this will generate lower percentage of ethanol.
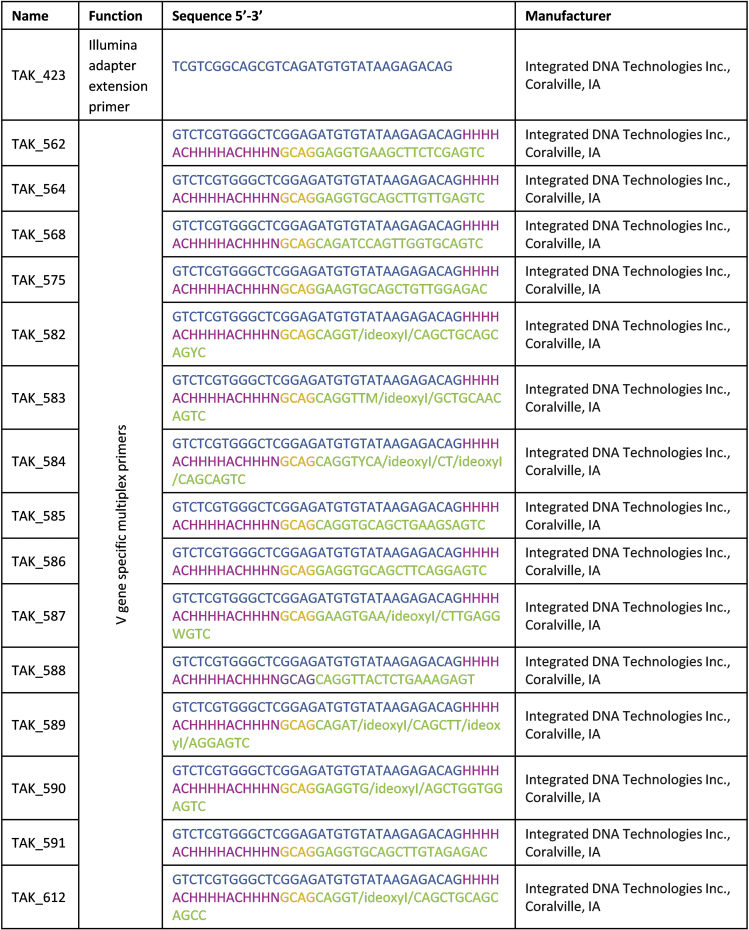
Synthesize primer sets according to Figures 3 and 6 as well as Tables S1 and S2.
CRITICAL: Make sure that primer sets contain real “H” and “N” nucleotides according to IUPAC (Cornish-Bowden, 1985), where “H” stands for everything except guanine and “N” for any base (not a gap).
Note: This nomenclature is accepted by Integrated DNA Technologies Inc., which we recommend for ordering of the primers.
Figure 3.
Primers used for cDNA Synthesis (Stock concentration: 100 μM)
Partial Illumina Adapter (blue), reverse unique molecular identifier (red), spacer (yellow) and Ig-specific sequences (black) are highlighted. Nucleotides according to IUPAC nomenclature (1985), where “H” stands for everything except guanine and “N” for any base (not a gap). See also Table S1.
Figure 6.
Primers used for Multiplex PCR (Stock concentration: 100 μM)
Partial Illumina Adapter (blue), forward unique molecular identifier (purple), spacer (yellow) and VDJ-specific sequence (green) are highlighted. Nucleotides according to IUPAC nomenclature (1985), where “H” stands for everything except guanine and “N” for any base (not a gap). See also Table S2.
Make sure that animal studies have been approved and are performed in accordance with relevant institutional and national guidelines and regulations. Exemplary graphs after data analysis (see Figure 10, Figure 11, Figure 12) show data of 18-week-old female and male BXSB mice treated with or without a B cell depleting anti-CD20-mIgG2c antibody performed in our working group by Werner et al. (2021). The animal study was reviewed and approved by government of Lower Franconia.
Figure 10.
Exemplary circos plots
VDJ recombination in (d) μ, (e) γ1/2 and (f) α heavy chains (HC) of anti-CD20 (αCD20) or isotype control antibody treated male (♂) autoimmune-prone BXSB mice at the age of 18 weeks was analyzed by high-throughput sequencing. Circos plots depicting the frequencies of VHDH, VHJH and DHJH usage and combinations of productive sequences from four individual mice are depicted. One independent experiment with a total of n = 4 mice.
Figure 11.
Exemplary quantification of V/D/J gene usage
Shown is the abundance (in percent of total reads) of the indicated V (IGHV), D (IGHD) and J (IGHJ) gene segments in (a) μ, (b) γ1/2 and (c) α heavy chains (HC) in splenic cells of male (♂) autoimmune-prone BXSB mice at 18 weeks of age, which were treated with an isotype control or CD20-specific (αCD20) antibody during the pre-phase of disease as determined by high-throughput RNA sequencing. Bars represent mean ± SD. One independent experiment with a total of n=4. Significant differences between groups were determined with an ordinary two-way ANOVA with Sidak’s multiple comparison test. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001.
Figure 12.
Exemplary quantification of CDR3 lengths and amino acid composition of CDR3 regions
Analysis of CDR3 regions of μ, γ1/2 and α heavy chains (HC) of male (♂) autoimmune-prone BXSB mice at 18 weeks of age, which were treated with an isotype control or CD20-specific (αCD20) antibody during the pre-phase of disease. (A) Individual CDR3 length distribution and (B) individual amino acids in the CDR3 region are shown for respective groups. Bars represent mean ± SD. One independent experiment with a total of n=4. Significant differences between isotype and αCD20 treated males for each CDR3 length or amino acid were determined with ordinary two-way ANOVA with Sidak’s multiple comparison test. ∗p<0.05; ∗∗p<0.01.
Preparation of buffers and solutions
Timing: 10 min
Freshly prepare working solutions of buffers used for RNA isolation according to the manufacturer’s instructions of RNeasy Plus Mini Kit from Qiagen (#74134).
CRITICAL: To obtain good RNA quality, RLT-Lysis buffer with beta-Mercaptoethanol should be prepared freshly at the day of RNA extraction.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 1× PBS, pH 7.4 | No recommended vendor | n/a |
| 10× PBS, pH 7.4 | No recommended vendor | n/a |
| Distilled water | No recommended vendor | n/a |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#M6250-100ML |
| Ethanol ≥99,8%, p.a. | Carl Roth | Cat#9065.4 |
| Agarose | Carl Roth | Cat#3810.3 |
| 6× DNA Loading Buffer | No recommended vendor | n/a |
| 100 bp DNA ladder | NIPPON genetics | Cat#MWD100 |
| Critical commercial assays | ||
| RNeasy Plus Mini Kit | QIAGEN | Cat#74134 |
| QIAshredder | QIAGEN | Cat#79656 |
| SCRIPTUM FIRST cDNA synthesis kit | Bio&SELL | Cat#BS48.100 |
| Agencourt AMPure XP | Beckman Coulter | Cat#A63881 |
| Buffer EB | QIAGEN | Cat#19086 |
| KAPA HIFI HotStart Uracil+ ReadyMix | KAPA Biosystems | Cat#KK2801 |
| Q5® High-Fidelity PCR Kit | New England Biolabs | Cat#E0555S |
| Nextera XT Index Kit v2 Set D | Illumina | Cat#FC-131-2004 |
| MiSeq Reagent Kit v3 (600-cycle) | Illumina | MS-102-3003 |
| Experimental models: Organisms/strains | ||
| Mus Musculus: C57BL/6 | The Jackson Laboratory | Cat#000664 |
| Oligonucleotides | ||
| TAK_402, IgG1, IgG2a, IgG2b, IgG2c primer: (Figure 3 and Table S1) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_403, IgG3 primer: (Figure 3 and Table S1) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_IgM, IgM primer: (Figure 3 and Table S1) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_IgA, IgA primer: (Figure 3 and Table S1) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_423, Illumina adapter extension primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_562, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_564, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_568, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_575, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_582, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_583, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_584, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_585, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_586, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_587, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_588, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_589, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_590, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_591, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| TAK_612, Multiplex primer: (Figure 6 and Table S2) | Integrated DNA Technologies Inc., Coralville, IA | n/a |
| Software and algorithms | ||
| ImageJ 1.53k | Wayne Rasband, NIH, USA | https://imagej.nih.gov/ij/ |
| MultiQC 1.9 | Ewels et al., 2016 | www.multiqc.info |
| FastQC v0.11.9 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| IMGT® (the international ImMunoGeneTics information system®) 3.5.28 | Alamyar et al., 2012 | www.imgt.org |
| ARGalaxy (Antigen Receptor Galaxy) | IJspeert et al., 2017 |
https://argalaxy.researchlumc.nl/ https://github.com/ErasmusMC-Bioinformatics/ARGalaxy-docker |
| Script for UMI error correction and isotype filtering (Github) | Winkler lab, FAU Erlangen | https://github.com/Simon-K-Schaefer/umify_iso |
| Script for UMI error correction and isotype filtering (Zenodo) | Winkler lab, FAU Erlangen | https://doi.org/10.5281/zenodo.6327191 |
| Immcantation framework 4.2 | Gupta et al. (2015) | https://immcantation.readthedocs.io/en/stable/ |
| Python 2.7.18 | Python Software Foundation | https://www.python.org/ |
| Python 3.8.5 | Python Software Foundation | https://www.python.org/ |
| Perl 5.30.0 | Perl Foundation | https://www.perlfoundation.org/ |
| Docker version 20.10.7 | Docker, Inc. | https://www.docker.com/ |
| Other | ||
| Dissection mat | No recommended vendor | n/a |
| Blunt forceps | No recommended vendor | n/a |
| Scissors | No recommended vendor | n/a |
| 40 μm and 70 μm cell strainer | No recommended vendor | n/a |
| 5 mL syringe | No recommended vendor | n/a |
| Hemocytometer or automated cell counter | No recommended vendor | n/a |
| NanoDrop™ 2000 | Thermo Fisher Scientific | ND-2000 |
| PCR machine | No recommended vendor | n/a |
| Gel chamber | No recommended vendor | n/a |
| Gel documentation system | No recommended vendor | n/a |
| Workflow was validated on a test system containing: Ubuntu 20.04.3 LTS with 128 GB RAM and an Intel Core i9-10980XE CPU | No recommended vendor | n/a |
Step-by-step method details
Spleen collection and preparation of erythrocyte-lysed single cell suspension
Timing: 30 min
The following steps describe the isolation of murine spleens with subsequent preparation of erythrocyte lysed single cell suspensions.
Note: Prepare all steps on ice or with appropriate cooling. The temperature for centrifugation should be 10°C as we noticed that centrifugation at the typically stated 4°C can lead to lower temperatures during the centrifugation process and thus freezing of the samples (especially when working with small volumes).
-
1.Spleen Collection.
-
a.Euthanize mice using an institutionally approved method (e.g., CO2 asphyxiation).
-
b.Place the mouse right side down on a dissection mat and spray fur with 70% ethanol to disinfect.
-
c.Using blunt forceps and scissors, make an incision in the skin on the left thorax just below the ribcage until the spleen is visible.
-
d.Carefully lift the spleen out of the abdomen with an additional pair of forceps and remove any non-splenic tissue as good as possible.
-
a.
Note: remaining non-splenic tissue can lead to increased clump formation and thus reduced cell counts.
-
2.Preparation of single cell suspension.
-
a.Place the spleen in a 70 μm cell strainer on top of a 50 mL conical tube.
-
b.Smash the spleen using a sterile plunger of a 5 mL syringe while rinsing the cell strainer with appropriate amount of ice cold 1× PBS.
-
c.Make sure to flush through all remaining cells in the cell strainer and attached to the plunger.
-
d.Discard the cell strainer, close the tube and keep on ice until further procedure.
-
a.
-
3.Hypotonic erythrocyte lysis.Note: This is a protocol for performing hypotonic erythrocyte lysis. Of course, other established protocols for red blood cell lysis can be used.
-
a.Centrifuge the sample at 400 × g for 5 min at 10°C.
-
b.Discard the supernatant and resuspend the pellet in 1 mL ice-cold 1× PBS.
-
c.Add 9 mL of distilled water and incubate for 20 s.
-
d.Stop lysis by adding 1 mL 10× PBS.
-
e.Pour cell suspension through 40 μm cell strainer to remove clumps that may have formed inter alia due to remaining non-splenic tissue.
-
f.Centrifuge the sample at 400 × g for 5 min at 10°C.
-
g.Discard the supernatant and resuspend in 10 mL ice-cold 1× PBS.
-
h.Count the cells using a hemocytometer or an automated cell counter.
-
i.Take out no more than 1 × 107 cells and transfer to a new tube. Remaining cells can be discarded or used for other applications.Note: For the RNeasy Plus Mini Kit from Qiagen the maximum number of cells is 1 × 107 cells. Here we used between 0.5 – 1 × 107 cells. When using other kits for RNA isolation cell numbers might need to be adapted according to the manufacturer’s instructions.
-
j.Centrifuge cells at 400 × g for 5 min at 10°C.
-
k.Carefully discard the supernatant completely.Note: Incomplete removal of the supernatant will inhibit lysis and dilute the lysate. These factors may reduce the RNA yield.
 Pause point: Proceed with RNA extraction following manufacturer’s protocol or snap freeze the cell pellet in liquid nitrogen before storage at −80°C.Note: We would recommend to immediately proceed with RNA isolation to yield high RNA amounts.
Pause point: Proceed with RNA extraction following manufacturer’s protocol or snap freeze the cell pellet in liquid nitrogen before storage at −80°C.Note: We would recommend to immediately proceed with RNA isolation to yield high RNA amounts.
-
a.
RNA extraction
Timing: 40 min
RNA extraction is performed using the RNeasy Plus Mini Kit from Qiagen (#74134) according to manufacturer’s instruction. However, other methods suitable for efficient RNA extraction can be used. Please refer to Figure 1 for a rapid overview of this step.
CRITICAL: Properly clean all surfaces with RNase AWAY or another alternative (this includes for example bench, centrifuge, pipettors, pipette tip boxes or tube holder).
Note: Freshly prepare Buffer RLT + beta-Mercaptoethanol (for example: Sigma-Aldrich; #M6250-100ML) according to manufacturer’s instruction for all samples processed on the same day. Thus, add 10 μL beta-Mercaptoethanol per 1 mL Buffer RLT.
-
4.
Flick the tube to thoroughly loosen the cell pellet, add 600 μL Buffer RLT + beta-Mercaptoethanol (10 μL beta-Mercaptoethanol per 1 mL Buffer RLT) and carefully pipette up and down to properly lyse the cells.
-
5.
Transfer the lysate into a QIAshredder spin column (Qiagen, #79656; not included in the RNeasy Plus Mini Kit from Qiagen) placed in a 2 mL collection tube and centrifuge for 2 min at 14,000 × g at room temperature (around 19°C–23°C).
-
6.
Transfer the homogenized lysate to a gDNA Eliminator spin column placed in a 2 mL collection tube. Centrifuge for 30 s at 14,000 × g at room temperature (around 19°C–23°C). Discard the column and save the flow-through.
-
7.
Add 600 μL of 70% ethanol to the flow-through and mix well by pipetting.
-
8.
Transfer up to 700 μL of the sample, including any precipitate that may have formed, to a RNeasy spin column placed in a 2 mL collection tube. Close the lid gently, and centrifuge for 15 s at 14,000 × g at room temperature (around 19°C–23°C). Discard the flow-through and repeat this step with the remaining flow-through (approximately 500 μL).
-
9.
Add 700 μL Buffer RW1 to the column and centrifuge for 15 s at 14,000 × g at room temperature (around 19°C–23°C) to wash the column. Discard the flow-through.
-
10.
Add 500 μL Buffer RPE working solution to the column and centrifuge for 14,000 × g at room temperature (around 19°C–23°C). Discard the flow-through.
Note: Buffer RPE is supplied as a concentrate. Ensure that 4 volumes of ethanol have been added before use, as indicated on the bottle. For example, for the RNeasy Plus Mini Kit (50) add 44 mL of ethanol to 11 mL of RPE stock solution.
-
11.
Add 500 μL Buffer RPE working solution to the column and centrifuge for 2 min at 14,000 × g at room temperature (around 19°C–23°C) to wash the column.
Optional: Place the column into a new 2 mL collection tube and centrifuge at 8,000 × g for 1 min at room temperature (around 19°C–23°C) to completely remove Buffer RPE.
-
12.
Place the RNeasy spin column into a new 1.5 mL collection tube. Add 30 μL RNase-free water directly to the middle of the spin column membrane without touching the surface and centrifuge for 1 min at 14,000 × g at room temperature (around 19°C–23°C) to elute the RNA.
-
13.
Determine the RNA concentration and check purity of each sample by NanoDrop™ 2000 spectrophotometer at 260 nm and 280 nm.
Note: NanoDrop can only give information on for example RNA quantity and salt or protein contamination. However, it does not tell anything on RNA integrity. Thus, if available, you could use an Agilent 2100 Bioanalyzer to get information on RNA quality. See troubleshooting problem 1 for examples of degraded and intact RNA analyzed by Agilent 2100 Bioanalyzer.
Pause point: RNA can be stored at −80°C until cDNA preparation. However, it is always recommended to process RNA as soon as possible to avoid degradation.
Figure 1.
Workflow for RNA extraction
Library preparation and synthesis
Timing: 6 days
Note: In contrast to Khan et al. (2016), who used MiSeq Illumina Primer, we adapted the protocol to the Nextera XT Index Kit v2 Set D from Illumina (#FC-131-2004).
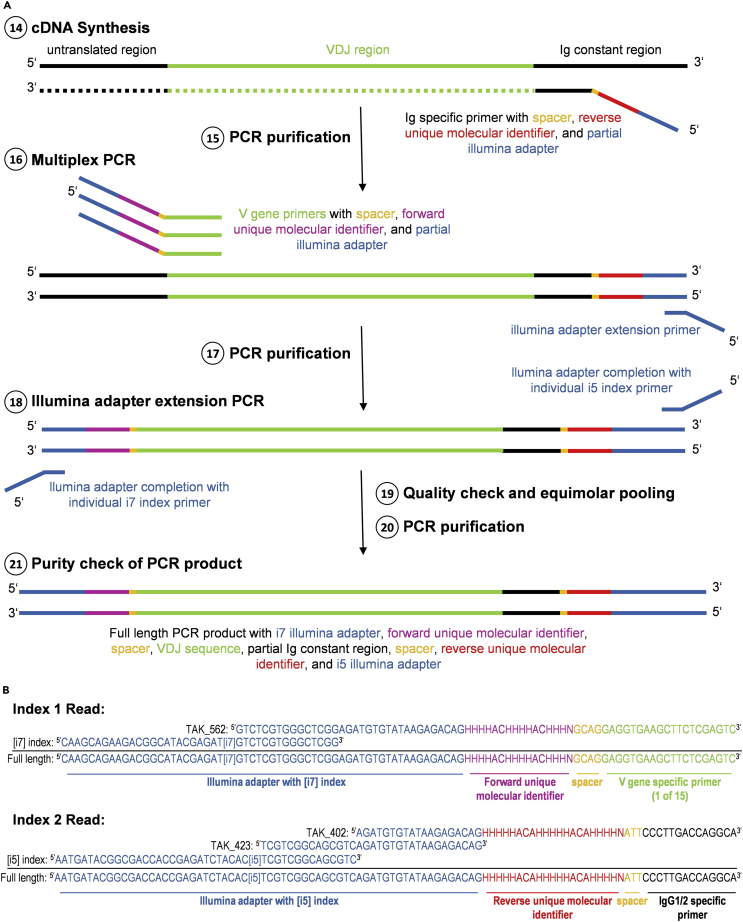
An overview of the different steps of library preparation and overlaps of the different primer sequences can be found in Figure 2.
-
14.
First-strand cDNA synthesis (day 1; Timing: 2 h).
cDNA synthesis was performed using the SCRIPTUM FIRST cDNA synthesis kit from Bio&Sell (#BS 48.100) using immunoglobulin specific primers (see Figure 3 and Table S1) according to manufacturer’s instructions. By including IgM and IgA specific primer additionally to the primers used by Khan et al. (2016) discrimination of IgG1/2, IgG3, IgM and IgA VDJ recombination is possible. CRITICAL: Perform all steps on ice.
CRITICAL: Perform all steps on ice.-
a.Mix appropriate amounts of the four cDNA primers TAK_402, TAK_403, TAK_IgM and TAK_IgA (see Figure 3 and Table S1) and dilute 1:5 with nuclease-free water. Each Primer has a final concentration of 5 μM which taken together results in a final concentration of 20 μM for the primer mix. For example, for 100 μL of cDNA primer mix take 5 μL of each primer (TAK_402, TAK_403, TAK_IgM and TAK_IgA; stock concentration: 100 μM) and add 80 μL of nuclease-free water to receive a 20 μM stock concentration of cDNA primer mix.Note: cDNA primer mix can be stored at −20°C.
-
b.Prepare reverse transcription reaction mix as in Table 1 for each sample in a nuclease-free microcentrifuge tube, mix properly and keep on ice before use.Note: It is recommended to mix RNase-free water, total RNA and primers first before adding the other components.
-
c.Incubate mixture at 42°C for 10 min first and then at 50°C for 60 min.
-
d.Short-spin reaction tubes.
 Pause point: cDNA can be stored at −20°C until further use.
Pause point: cDNA can be stored at −20°C until further use.
-
a.
-
15.
PCR purification (day 1; Timing: 30 min).
SPRI paramagnetic bead cleanup was performed using Agencourt AMPure XP from Beckman and Coulter (#A63881) using manufacturer’s instruction with minor adaptions. Please refer to Figure 4 for a rapid overview of this step.Note: Purification steps are recommended to remove contaminants like dNTPs, salts, primers, or primer dimers that might interfere with further procedure.-
a.If cDNA has been stored at −20°C thaw samples.
-
b.Carefully resuspend AMPure XP by shaking the bottle to obtain a homogeneous suspension.
-
c.Add 20 μL of AMPure XP per 20 μL of sample and mix thoroughly by pipetting up and down (do not vortex).
-
d.Incubate 5 min at room temperature (around 19°C–23°C).
-
e.Place 1.5 mL tube in a magnetic bead separator and incubate for 2 min at room temperature (around 19°C–23°C) until solution turns clear and beads are stuck to the side close to the magnet.
- f.
-
g.Add 200 μL freshly prepared 70% ethanol on the beads in the tube and incubate for 30 s at room temperature (around 19°C–23°C).
-
h.Remove the supernatant completely and repeat ethanol-washing step to remove remaining contaminants.Note: A dry time of a couple of minutes under the fume hood is optional to ensure that all traces of ethanol are removed. For example, we dried the samples between 5–10 min until ethanol was completely evaporated. Since drying times may vary between experiments samples should be monitored closely to avoid overdrying of the beads.
-
i.Remove the tube from the magnetic bead separator and add 40 μL elution buffer (Buffer EB from Qiagen, #19086).
-
j.Resuspend by pipetting up and down 10 times.
-
k.Incubate 2 min at room temperature (around 19°C–23°C).
-
l.Place 1.5 mL tube in a magnetic bead separator and incubate for 1 min at room temperature (around 19°C–23°C) until beads are separated from the solution.
-
m.Transfer the eluate to a new tube.
 Pause point: cDNA can be stored at −20°C until further use.
Pause point: cDNA can be stored at −20°C until further use.
-
a.
-
16.
Multiplex PCR (day 2; Timing: 1 h).
This step was performed using the KAPA HIFI HotStart Uracil+ ReadyMix from KAPA Biosystems (#KK2801) with minor adaptions to manufacturer`s instructions. CRITICAL: KAPA HiFi HotStart Uracil+ reactions must be set up on ice since the high proofreading activity of the enzyme will result in rapid primer degradation at room temperature (around 19°C–23°C).
CRITICAL: KAPA HiFi HotStart Uracil+ reactions must be set up on ice since the high proofreading activity of the enzyme will result in rapid primer degradation at room temperature (around 19°C–23°C). CRITICAL: KAPA HiFi HotStart Uracil+ ReadyMix contains isostabilizers and may not freeze solidly, even when stored at −15°C to −25°C. This will not affect the shelf life of the product. Nevertheless, always ensure that the product has been fully thawed and mixed before use.
CRITICAL: KAPA HiFi HotStart Uracil+ ReadyMix contains isostabilizers and may not freeze solidly, even when stored at −15°C to −25°C. This will not affect the shelf life of the product. Nevertheless, always ensure that the product has been fully thawed and mixed before use.-
a.Prepare appropriate amount of Multiplex Primer Mix containing the 15 primers TAK_562, TAK_564, TAK_568, TAK_575, TAK_582 - TAK_591, TAK_612 (see Figure 6 and Table S2), dilute 1:8 with nuclease-free water and keep on ice before use. Each Primer has a final concentration of 0.833 μM which taken together results in a final concentration of 12.5 μM for the primer mix. For example, for 120 μL of Multiplex Primer Mix take 1 μL of each primer (TAK_562, TAK_564, TAK_568, TAK_575, TAK_582 - TAK_591, TAK_612; stock concentration: 100 μM) and add 105 μL of nuclease-free water to receive a 12.5 μM stock concentration of Multiplex Primer Mix.
-
b.Dilute Illumina adapter extension primer (TAK_423; see Figure 6 and Table S2) 1:8 with nuclease-free water. For example, for 120 μL of diluted Illumina adapter extension primer take 15 μL of Illumina adapter extension primer (TAK_423; stock concentration: 100 μM) and add 105 μL of nuclease-free water to receive a 12.5 μM stock concentration.Note:Multiplex Primer Mix and diluted Illumina adapter extension primer can be stored at −20°C.
-
c.Prepare Multiplex PCR mix for each sample as in Table 2, mix properly and keep on ice before use.
-
d.Spin down and incubate the reaction in a thermocycler with the Multiplex PCR program (see Table 3).
-
a.
-
17.
PCR purification (day 2; Timing: 1 h).
Figure 2.
Overview of library preparation
Shown are (A) the different steps of library preparation with color coding of the individual parts of the primers as well as (B) exemplary sequences of primer compositions for index read 1 and 2.
Table 1.
Reverse transcription reaction mix
| Component | Stock concentration | Final concentration | Amount per sample |
|---|---|---|---|
| RNase-free water | n/a | n/a | To 20 μL |
| Total RNA | n/a | n/a | 1 μga (Volume may vary among samples) |
| cDNA Primer Mix | 20 μM | 20 pmol | 1 μL |
| SCRIPTUM FIRST RT buffer | 5× | 1× | 4 μL |
| dNTP Mix | 10 mM per dNTP | 500 nM per dNTP | 1 μL |
| DTT | 100 mM | 5 mM | 1 μL |
| RNase inhibitor | 40 units/μL | 40 units | 1 μL |
| SCRIPTUM FIRST Reverse Transcriptase |
200 units/μL | 100 units | 0.5 μL |
| Total | n/a | n/a | 20 μL |
According to the manufacturer’s protocol, the amount of starting material can vary from 1 pg - 5 μg of total RNA. Therefore, also lower RNA amounts are sufficient. For example, we have successfully used about 0.1–0.3 μg of RNA.
Figure 4.
Workflow for PCR purification
Figure 5.
Exemplary pictures of PCR purification steps with AMPure XP beads
(A–C) (A) PCR product is (B) mixed thoroughly with AMPure XP beads and incubated for 5 min at room temperature (around 19°C–23°C) until (C) solution turns clear and beads are stuck to the side close to the magnet.
(D) Afterwards supernatant is carefully removed without disturbing the beads.
Table 2.
Multiplex PCR mix
| Component | Stock concentration | Final concentration | Amount per sample |
|---|---|---|---|
| Template DNA (purified in step 15) | n/a | n/a | 10.5 μL |
| KAPA HiFi HotStart Uracil+ ReadyMix |
2× | 1× | 12.5 μL |
| Diluted Illumina adapter extension primer (TAK_423) | 12.5 μM | 500 nM | 1 μL |
| Multiplex Primer Mix | 12.5 μM | 500 nM | 1 μL |
| Total | n/a | n/a | 25 μL |
Table 3.
Multiplex PCR program
| Step | Temperature | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 95°C | 2 min | 1 |
| Denaturation | 98°C | 20 s | 9× |
| Annealing | 60°C | 45 s | |
| Extension | 72°C | 1 min 10 s | |
| Final extension | 72°C | 5 min | 1 |
Perform PCR purification according to step 15 using 0.65 μL AMPure XP beads per 1 μL of sample. Thus, add 16.25 μL AMPure XP beads per 25 μL of sample Repeat this step by adding 26 μL AMPure XP beads per 40 μL of eluted sample.
Pause point: PCR products can be stored at −20°C until further use.
-
18.
Illumina adapter extension PCR (day 3; Timing: 2 h).
This step was performed using the Q5® from New England Biolabs (#E0555S) with minor adaptions to manufacturer`s instructions and i5 and i7 primers of Nextera XT Index Kit v2 Set D from Illumina (#FC-131-2004). CRITICAL: Perform all steps on ice.
CRITICAL: Perform all steps on ice.-
a.Prepare Illumina PCR mix with individual i5 (S513, S515-S518, S520-S522) and i7 (N716, N718-N724, N726-N729) primers for each sample as in Tables 4 and 5, mix properly and keep on ice before use. An overview of the sequences of i5 and i7 Illumina primers can be found in Table 6.Note: Each sample gets a unique combination of i5 and i7 Illumina primer so that it can be demultiplexed upon sequencing (see Table 4).
-
b.Spin down and incubate the reaction in a thermocycler with the Illumina PCR program (see Table 7).
 Pause point: Samples can be stored at 2°C–8°C for up to 2 days. For longer storage, please store at −20°C.
Pause point: Samples can be stored at 2°C–8°C for up to 2 days. For longer storage, please store at −20°C.
-
a.
-
19.Quality check and equimolar pooling (day 3; Timing: 2 h).
-
a.Prepare 1.5% agarose gel.
-
b.Load 10 μL sample/lane with 2 μL 6× DNA loading buffer as well as 4 μL 100 bp DNA ladder on the gel.
-
c.Run at 120 V in an appropriate gel chamber for approximately 30 min-1 h.Check quality of PCR product using a gel documentation system. The expected size of the PCR product is ∼600 bp. See Figure 7 for exemplary gel picture of PCR products.Note: If your samples don’t show a clear DNA band, but instead degradation or overamplification check template DNA or adapt PCR protocol (see troubleshooting problem 3).
-
d.Determine intensity of each band of the PCR product using ImageJ or other image processing programs.
-
e.Pool samples equimolar according to the intensity of the band of the PCR product.Note: If you have access to an Agilent Technology 2100 Bioanalyzer or Qubit fluorometer, bead cleanup every sample separately using Agencourt AMPure XP (see step 15) and quantify the samples for equimolar pooling (this will lead to a more accurate pool of barcoded libraries in equimolar ratios ensuring an even read distribution for all samples).
 Pause point: PCR products can be stored at −20°C until further use.
Pause point: PCR products can be stored at −20°C until further use.
-
a.
-
20.
PCR purification (day 4; Timing: 1 h).
Table 4.
Index primer combinations for 96 libraries
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N716 | N718 | N719 | V720 | N721 | N722 | N723 | N724 | N726 | N727 | N728 | N729 | ||
| A | S513 | ||||||||||||
| B | S515 | ||||||||||||
| C | S516 | ||||||||||||
| D | S517 | ||||||||||||
| E | S518 | ||||||||||||
| F | S520 | ||||||||||||
| G | S521 | ||||||||||||
| H | S522 |
Table 5.
Illumina PCR mix
| Component | Stock concentration | Final concentration | Amount per sample |
|---|---|---|---|
| Nuclease-free water | n/a | n/a | 4.5 μL |
| Template DNA (purified in step 17) | n/a | n/a | 3 μL |
| Q5® | 2× | 1× | 12.5 μL |
| i5 Illumina Primer | 10 μM | 1 μM | 2.5 μL |
| i7 Illumina Primer | 10 μM | 1 μM | 2.5 μL |
| Total | n/a | n/a | 25 μL |
Table 6.
Primer sequences of i5 and i7 index primers
| Index name | Primer sequence 5′-3′ | |
|---|---|---|
| i5 | S513 | AATGATACGGCGACCACCGAGATCTACAC[TCGACTAG]TCGTCGGCAGCGTC |
| S515 | AATGATACGGCGACCACCGAGATCTACAC[TTCTAGCT]TCGTCGGCAGCGTC | |
| S516 | AATGATACGGCGACCACCGAGATCTACAC[CCTAGAGT]TCGTCGGCAGCGTC | |
| S517 | AATGATACGGCGACCACCGAGATCTACAC[GCGTAAGA]TCGTCGGCAGCGTC | |
| S518 | AATGATACGGCGACCACCGAGATCTACAC[CTATTAAG]TCGTCGGCAGCGTC | |
| S520 | AATGATACGGCGACCACCGAGATCTACAC[AAGGCTAT]TCGTCGGCAGCGTC | |
| S521 | AATGATACGGCGACCACCGAGATCTACAC[GAGCCTTA]TCGTCGGCAGCGTC | |
| S522 | AATGATACGGCGACCACCGAGATCTACAC[TTATGCGA]TCGTCGGCAGCGTC | |
| i7 | N716 | CAAGCAGAAGACGGCATACGAGAT[TAGCGAGT]GTCTCGTGGGCTCGG |
| N718 | CAAGCAGAAGACGGCATACGAGAT[GTAGCTCC]GTCTCGTGGGCTCGG | |
| N719 | CAAGCAGAAGACGGCATACGAGAT[TACTACGC]GTCTCGTGGGCTCGG | |
| V720 | CAAGCAGAAGACGGCATACGAGAT[AGGCTCCG]GTCTCGTGGGCTCGG | |
| N721 | CAAGCAGAAGACGGCATACGAGAT[GCAGCGTA]GTCTCGTGGGCTCGG | |
| N722 | CAAGCAGAAGACGGCATACGAGAT[CTGCGCAT]GTCTCGTGGGCTCGG | |
| N723 | CAAGCAGAAGACGGCATACGAGAT[GAGCGCTA]GTCTCGTGGGCTCGG | |
| N724 | CAAGCAGAAGACGGCATACGAGAT[CGCTCAGT]GTCTCGTGGGCTCGG | |
| N726 | CAAGCAGAAGACGGCATACGAGAT[GTCTTAGG]GTCTCGTGGGCTCGG | |
| N727 | CAAGCAGAAGACGGCATACGAGAT[ACTGATCG]GTCTCGTGGGCTCGG | |
| N728 | CAAGCAGAAGACGGCATACGAGAT[TAGCTGCA]GTCTCGTGGGCTCGG | |
| N729 | CAAGCAGAAGACGGCATACGAGAT[GACGTCGA]GTCTCGTGGGCTCGG | |
Sequences of i5 and i7 bases are bold and in square brackets.
Table 7.
Illumina PCR program
| Step | Temperature | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 94°C | 3 min | 1 |
| Denaturation | 94°C | 60 s | 23× |
| Annealing | 55°C | 60 s | |
| Extension | 72°C | 30 s | |
| Final extension | 72°C | 10 min | 1 |
Figure 7.
Exemplary picture of PCR products
Displayed are four exemplary PCR products (#1 - #4) after Illumina PCR run on a 1.5% agarose gel showing specific bands at ∼600 bp and some smaller bands of primer, primer dimers etc. below 100 bp which will be removed after PCR purification with AMPure XP beads.
Perform PCR purification according to step 15 using 0.8 μL AMPure XP beads per 1 μL of pooled samples. As the volume of your pooled sample varies depending on the amount of sample used for pooling, no precise volume of AMPure XP beads can be specified here.
CRITICAL: Elute with 40 μL water instead of elution buffer as given in step 15!
-
21.Purity check of PCR product (day 4; Timing: 1 h).
-
a.Prepare 1.5% agarose gel.
-
b.Load 5 μL of pooled sample with 1 μL 6× DNA loading buffer as well as 4 μL 100 bp DNA ladder on the gel.
-
c.Run at 120 V in an appropriate gel chamber for approximately 30 min-1 h.
-
d.Check quality of PCR product using a gel documentation system. The expected size of the PCR product is ∼600 bp. See Figure 8 for exemplary picture of purified pooled PCR product.Note: If your samples still contain small bands repeat PCR purification (see troubleshooting problem 4).
 Pause point: Pooled sample can be stored at −20°C until further use.
Pause point: Pooled sample can be stored at −20°C until further use.
-
a.
-
22.
Illumina Miseq Sequencing (day 5–6; Timing: 2 d).
Figure 8.
Pooled and purified PCR product
Sequence the libraries on an Illumina MiSeq platform using a pair of overlapping paired-end reads (2 × 300 bp) with 20% PhiX. For a high-resolution repertoire analysis aim for 0.5–1 Mio raw reads per sample. Use the MiSeq Reagent Kit v3 (MS-102-3003 600-cycle) according to the Illumina protocol for Miseq.
Note: 20% PhiX was used according to Illumina recommendations to prevent sequencing error due to low sample complexity in antibody constant regions. In an additional step we used PhiX content after sequencing for a sample independent quality control for the Illumina workflow (see data processing step 24).
Note: It might be possible that problems with the Illumina MiSeq run are occurring (see troubleshooting problem 5 and problem 6). To enable a rerun, excess sample material should be stored in aliquots to avoid freeze thaw cycles.
Data processing
Timing: depends on sample number/size and computational power. Time-consuming steps are paired-end merging (takes some min/sample) the IMGT HighV-QUEST web analysis (takes some hours/days for all samples depending on server workload) and the ARGalaxy pipeline (takes some min/sample).
The following steps include the analysis from the Illumina output after demultiplexing (FASTQ) to the result visualizations as well as necessary quality control steps to ensure a successful workflow. Additionally to Khan et al. (2016), we used the IgG1/2, IgG3, IgM and IgA specific primer sequences which were used for cDNA synthesis to discriminate between those isotypes.
All workflows were evaluated for Ubuntu 20.04.3 LTS with 128 GB RAM and an Intel Core i9-10980XE CPU. General dependencies are Python2, Python3 and Perl5. All code depicted in boxes is entered via command line input.
Note: A modern Desktop PC running Linux is sufficient for most of the workflow. The external QC for Phage DNA as described here can require more RAM than most consumer systems provide (>90 GB).
-
23.Internal quality control and QC software setup.
-
a.FastQC analysis as standard QC for the Illumina workflow:Input: FASTQ files obtained after demultiplexing from the standard illumina workflow.Imports Phred Quality Score from FASTQ files and provides information about:Basic StatisticsPer base sequence qualityPer tile sequence qualityPer sequence quality scoresPer base sequence contentPer sequence GC contentPer base N contentSequence Length DistributionSequence Duplication LevelsOverrepresented sequencesAdapter ContentOutput: HTML files and archives containing QC graphics.#The working directory must be set to the location of FASTQ #files>sudo apt-get update -y #Software update>sudo apt-get install -y fastqc #one-time download>fastqc ∗.fastq
-
b.Aggregate all FastQC reports with MultiQC into one single summary HTML file:Input: MultiQC should be run in the same folder as the FastQC output files.Output: HTML files containing summary information about several FastQC results.#The working directory must be the location of the FastQC #output>pip install multiqc #One-time download>multiqc.Note: Compared to other NGS data sets the FastQC quality metrics of immune repertoire reads show a lower quality phred score at higher position (end of sequence). This is expected behavior and corrected by paired-end merging and UMI quality filtering. See Figure 9.
-
a.
Figure 9.
Exemplary FastQC pictures
Exemplary FastQC per base sequence quality scores (Phred Scores) for (A) read 1, (B) read 2 and (C) after paired-end merging.
-
24.
External QC with PhiX Phage DNA.
CRITICAL: For this analysis the undetermined reads output files from Illumina demultiplexing are required. This step is only recommended if PhiX was added to samples before sequencing.
Blast analysis with undetermined reads against the reference phage genome.
Input: Demultiplexing Undetermined Output Files:
Undetermined_R1.fasta
Undetermined_R2.fasta
Output: phiX-report_R1.csv
phiX-report_R2.csv
>sudo apt-get install ncbi-blast+ #one-time download
>blastn -subject Undetermined_R1.fasta -query phi-x174.fasta -
outfmt 7 > phiX-report_R1.csv
>blastn -subject Undetermined_R2.fasta -query phi-x174.fasta -
outfmt 7 > phiX-report_R2.csv
Note: This step as described here requires a lot of RAM (>90 GB depending on repertoire size). With the creation of a blast database or subsampling the input FASTA file this demand can be reduced.
CRITICAL: An alignment length for the first hundred reads shorter than max read length (4th column in phiX-report.scv) indicates a problem with the Illumina process. If PhiX aligned reads are full length but sample reads are shortened a problem with sample preparation is indicated (see troubleshooting problem 5 and problem 6).
-
25.
Merge paired-end reads.
Paired-end sequence merging is done via the presto command line tool (Vander Heiden et al., 2014) from the immcantation framework (Gupta et al., 2015).
Parameters:
AssemblePairs.py align
--1 FASTQ file containing reads R1
--2 FASTQ file containing reads R2
--coord illumina Illumina (if Illumina formatted FASTQ)
--rc tail selects read to reverse complement before merging (paired-end)
--outname output file name
Input: FASTQ file containing reads R1
FASTQ file containing reads R2
Output: M1_assemble-pass.fastq
FilterSeq.py quality
-s merged FASTQ file from AssemblePairs
--q 20: reads with a phred score < 20 after merging are filtered out
-- outname output file name
--log log file name
Input: M1_assemble-pass.fastq
Output: M1_quality-pass.fastq
> pip3 install presto –user #One-time download to install
> AssemblePairs.py align -1 SampleX_1.fastq -2 SampleX
_2.fastq --coord illumina --rc tail --outname M1
> FilterSeq.py quality -s M1_assemble-pass.fastq -q 20 --
outname M1 --log FS.log
Note: QC for merged reads in FastQC format can be repeated with FastQC and MultiQC as described in internal quality control and QC software setup.
-
26.
File Conversion.
Convert merged FASTQ files to the FASTA format with Linux build-in commands. The FASTQ header with “@” is substituted for a FASTA header starting with “>”. Non-sequence information (QC scores) is removed in compliance with the FASTA format. This is required for UMI (unique molecular identifier) error correction and IMGT HighV-QUEST compatibility.
Input: M1_quality-pass.fastq
Output: M1_quality-pass.fasta
>sed -n '1∼4s/ˆ@/>/p;2∼4p' M1_quality-pass.fastq > M1_quality-pass.fasta
Note: The same results can be achieved by forcing presto to output FASTA files with the flag --fasta. This is not recommended as it will prevent QC after merging.
-
27.
UMI (unique molecular identifier) error correction and isotype filtering.
CRITICAL: The script only works with the primer design described in this protocol.
UMI correction is achieved with a perl script available at GitHub (Schaefer, 2022a) and Zenodo (Schaefer, 2022b).
PCR and sequencing errors are corrected by UMI pattern matching and selecting reads containing valid UMIs. Reads with matching UMIs are corrected to the most abundant read per UMI. To exclude quantitative bias introduced during Multiplex PCR reads are normalized to unique mRNA sequences. Reads shorter than 50 nucleotides are filtered for additional quality control. For problems during this step see troubleshooting problem 7.
Input: M1_quality-pass.fasta
Output: The script creates a folder containing a log file with primer and UMI information as well as new FASTA files with corrected sequences for all reads and primer defined isotypes.
>perl umify_normalized_iso.pl M1_quality-pass.fasta
Note: Depending on the workflow all reads or specific isotypes can be selected for further analysis.
-
28.
IMGT analysis/annotation.
UMI corrected FASTA files are uploaded to the IMGT HighV-QUEST web service for further analysis. The service takes a FASTA file and provides an archive file (.txz) containing immune receptor annotation and mutational analysis for the immune repertoire as tabular files.
Upload: http://www.imgt.org/HighV-QUEST/home.action
Input: Sample.fasta (repertoire or isotype specific)
Output: Archive file Sample.txz containing the following tabular files:
Summary (always provided in a spreadsheet and in a zip archive)
IMGT-gapped-nt-sequences
nt-sequences
IMGT-gapped-AA-sequences
AA-sequences
Junction
V-REGION-mutation-and-AA-change-table
V-REGION-nt-mutation-statistics
V-REGION-AA-change-statistics
V-REGION-mutation-hotspots
Parameters (always provided in a spreadsheet and in a zip archive)
scFv (for advanced functionalities, analysis of single chain fragment variable (scFv), only)
Note: IMGT HighV-QUEST has no batch mode. For huge collections of samples, we recommend the immcantation framework (see Alternatives).
-
29.
ARGalaxy analysis.
IMGT archives are further processed with the ARGalaxy pipeline or as docker image for local use.
Antigen Receptor Galaxy (ARGalaxy) is an online tool for the analysis and visualization of immune receptor sequencing data. The tool contains among basic utilities to load and concatenate IMGT archive files pipelines to create summary statistics and publication-ready graphics. The tool includes a pipeline to analyze immune repertoires (immune repertoire pipeline) and an additional pipeline to analyze class switch recombination and somatic hypermutation data (CSR and SHM pipeline).
CRITICAL: ARGalaxy depends on the output format of IMGT HighV-QUEST for full functionality. If the annotation was conducted with the immcantation framework or other tools, we recommend continuing with alternative workflows (see Alternatives).
Note: Docker download page: https://docs.docker.com/engine/install/ubuntu/
Input: Sample.txz archive obtained from IMGT HighV-QUEST
Upload Data: Get Data
+Upload File from your computer
Analysis: ARGalaxy Pipelines
+Immune Repertoire pipeline
+SHM & CSR pipeline
Output: The data is available inside the Galaxy GUI or can be downloaded as archive containing HTML summaries, summary tables and picture files.
>docker pull quay.io/erasmusmc_bioinformatics/argalaxy
>docker run -p 8080:80
quay.io/erasmusmc_bioinformatics/argalaxy
#Visit localhost:8080 in a webbrowser to start using
#ARGalaxy.
#If you don't want to lose your data when restarting the
#Docker image, add the -v flag to create a local mountpoint:
>docker run -p 8080:80 -v /path/to/local/dir/:/export/
argalaxy:latest
The graphical interface of locally deployed ARGalaxy servers can be accessed via any browser at: localhost:8080.
CRITICAL: The ID field in the Immune Repertoire pipeline is used as description in output graphics and should be named accordingly.
Alternatives: An alternative analysis method to the IMGT/ARGalaxy workflow is the immcantation framework (Gupta et al., 2015), which we recommend using as docker image.
Expected outcomes
Using the RNeasy Plus Mini Kit you should expect to receive 5–20 μg RNA out of 1 × 107 spleen cells. However, total RNA amounts can heavily depend on tissue handling, RNA working routine and individual mouse strains (Sellers et al., 2012).
ARGalaxy and IMGT/HighV-QUEST creates a multitude of different types of analysis for each individual sample. For example, VD, VJ and DJ pairings are depicted as circos plots. Furthermore, individual V/D/J gene frequencies, amino acid compositions and CDR3 lengths are visualized and quantified for each sample. Summary data is available and can be further analyzed via graphing and statistics software like GraphPad Prism. Exemplary presentation formats are depicted in Figure 10 (ARGalaxy graphic) and Figure 10, Figure 11, Figure 12 and Figure 10, Figure 11, Figure 12 (Statistical analysis with GraphPad Prism from IMGT/HighV-QUEST data) from (Werner et al., 2021). Further example graphics can be found in the original publication of ARGalaxy (IJspeert et al., 2017).
Limitations
This protocol outlines the analysis of murine B cell receptor repertoires using molecular amplification fingerprinting according to Khan et al. (2016), where they could dramatically reduce biases and errors introduced during library preparation and sequencing. We further refined this method, by utilizing additional immunoglobulin-specific primers and later separation according to the individual sequences, enabling us to distinguish between different isotypes and subclasses. However, due to sequence identities of IgG molecules we can only differentiate between IgG1/2 and IgG3 VH regions but not between IgG1 and IgG2a/b. As last-mentioned IgG subclasses differ particularly in functions like antibody dependent cell-mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) or binding to Fc-gamma receptors some IgG-specific information might not be elucidated.
This protocol is limited to murine B cell receptor repertoires. However, it can be adapted to investigate other parts of immune receptors such as variable light chains and T cell receptors or to analyze other species. Therefore, primer sequences need to be adjusted accordingly.
Troubleshooting
Problem 1
Step 13: NanoDrop can only give information on for example RNA quantity and salt and/or protein contamination. However, it does not tell anything on RNA integrity. For example, your RNA can be degraded, which would not be visible with a NanoDrop. However, RNA integrity can be analyzed by Agilent 2100 Bioanalyzer. See Figure 13 for examples of different RNA qualities analyzed by an Agilent 2100 Bioanalyzer.
Figure 13.
RNA samples of different quality
Shown are examples of (A) strongly degraded, (B) partially degraded and (C) intact RNA analyzed by an Agilent 2100 Bioanalyzer.
Potential solution
As an RNase free environment is essential when working with RNA samples it is recommended to work on a working space, which is used for RNA isolation only. Furthermore, all surfaces (this includes for example bench, centrifuge, pipettors, pipette tip boxes or tube holder) need to be cleaned with RNase AWAY or other sufficient reagents.
Moreover, we observed degraded RNA and low RNA content, when Buffer RLT + beta-Mercaptoethanol was kept for a couple of weeks as suggested by the manufacturer’s instruction. Therefore, always freshly prepare Buffer RLT + beta-Mercaptoethanol on the day of experiment.
Problem 2
Steps 15, 17, and 20: AMPure XP beads slid down during removal of the supernatant and are soaked into the pipette tip.
Potential solution
Resuspend the beads in the remaining supernatant and put the tube back in the magnetic bead separator. Incubate for another 2 min at room temperature (around 19°C–23°C) until solution turns clear and beads are stuck to the side close to the magnet again and carefully remove the supernatant once more.
Problem 3
Step 19: PCR products show overamplification and unspecific bands (Figure 14A) in contrast to a concise PCR product (Figure 14B).
Figure 14.
Exemplary gel pictures of PCR products
Example of (A) overamplified compared to (B) specific PCR products.
Potential solution
Check template DNA or adapt Multiplex PCR protocol (e.g., change number of cycles or incubation times).
Problem 4
Step 21: It can be possible that primer dimers are not completely removed during PCR purification using AMPure XP beads after cDNA synthesis. Therefore, they get amplified during multiplex and Illumina PCR leading to small DNA fragments of ∼200 bp of size (see Figure 15).
Figure 15.
Example of PCR product containing amplified primer dimers
Potential solution
To remove these small DNA fragments, you can repeat the PCR purification (see step 15) using 1.2 μL AMPure XP beads per 1 μL of sample.
Problem 5
Step 24: If PhiX aligned reads are full length but sample reads are shortened a problem with sample preparation is indicated.
Potential solution
Review PCR analysis if there are any abnormalities with your PCR products.
Problem 6
Step 24: In Case of PhiX aligned reads shorter than max read length a problem with the Illumina process is indicated.
Potential solution
Rerun Illumina sequencing. To enable the rerun, excess sample material should be stored in aliquots to avoid freeze thaw cycles.
Problem 7
Step 27: Unsuccessful UMI error correction can be caused by wrongly trimmed sequences. Reads should not contain Illumina adapters and reads have to start and end with the UMI sequence. Furthermore, false labeling of read 1 and 2 can cause the correction to fail.
Potential solution
If the problem is caused by wrongly trimmed sequences, trim/retrim raw Illumina output. If false labeling is responsible for unsuccessful UMI error correction repeat the step “Merge paired-end reads” (step 25) and exchange sample order in the call of “AssemblePairs.py”.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas H. Winkler (thomas.winkler@fau.de). For technical support regarding sample preparation and data processing please contact Anja Werner (anja.werner@fau.de) or Simon Schäfer (simon.schaefer@fau.de).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
We thank Dr. Arif Bülent Ekici, Department of Human Genetics, Erlangen, for performing the Illumina MiSeq run. This work was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation, Research Training Group GRK1660 – Projektnummer 161220270 to F.N. and T.H.W., TRR130 – Projektnummer 215346292 to T.H.W. and F.N., and FOR2953 – Projektnummer 409784463 and FOR2886 – Projektnummer 405969122 to F.N.). We acknowledge financial support by Deutsche Forschungsgemeinschaft and Friedrich-Alexander-Universität Erlangen-Nürnberg within the funding programme “Open Access Publication Funding."
Author contributions
N.G. adapted and modified the protocol from the original publication by Khan et al. (2016). A.W. performed experiments and S.S. implemented and adapted the codes for data analysis. A.W. and S.S. wrote the manuscript with the help of all authors. T.H.W. and F.N. acquired funding and resources and provided discussion of the results. T.H.W. supervised the study.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103076.
Contributor Information
Anja Werner, Email: anja.werner@fau.de.
Simon Schäfer, Email: simon.schaefer@fau.de.
Thomas H. Winkler, Email: thomas.winkler@fau.de.
Supplemental information
Data and code availability
The accession number for codes needed for data analysis (perl script for UMI correction) reported in this paper is GitHub: https://github.com/Simon-K-Schaefer/umify_iso or Zenodo: https://doi.org/10.5281/zenodo.6327191. Original data for creating Figure 10, Figure 11, Figure 12 have been deposited to Mendeley Data: https://doi.org/10.17632/smtxktdtfc.1.
References
- Alamyar E., Duroux P., Lefranc M.P., Giudicelli V. IMGT((R)) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A., Nomenclature Committee of the International Union of Biochemistry (NC-IUB) Nomenclature for incompletely specified bases in nucleic acid sequences. Recommendations 1984. Eur. J. Biochem. 1985;150:1–5. doi: 10.1111/j.1432-1033.1985.tb08977.x. [DOI] [PubMed] [Google Scholar]
- Ewels P., Magnusson M., Lundin S., Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJspeert H., Van Schouwenburg P.A., Van Zessen D., Pico-Knijnenburg I., Stubbs A.P., Van Der Burg M. Antigen receptor galaxy: a user-friendly, web-based tool for analysis and visualization of T and B cell receptor repertoire data. J. Immunol. 2017;198:4156–4165. doi: 10.4049/jimmunol.1601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T.A., Friedensohn S., Gorter De Vries A.R., Straszewski J., Ruscheweyh H.J., Reddy S.T. Accurate and predictive antibody repertoire profiling by molecular amplification fingerprinting. Sci. Adv. 2016;2:e1501371. doi: 10.1126/sciadv.1501371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer S.K. GitHub; 2022. Simon-K-Schaefer/umify_iso: Umify_iso (V1.3)_GitHub. [Google Scholar]
- Schaefer S.K. Zenodo; 2022. Simon-K-Schaefer/umify_iso: Umify_iso (V1.3)_Zenodo. [Google Scholar]
- Sellers R.S., Clifford C.B., Treuting P.M., Brayton C. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet. Pathol. 2012;49:32–43. doi: 10.1177/0300985811429314. [DOI] [PubMed] [Google Scholar]
- Vander Heiden J.A., Yaari G., Uduman M., Stern J.N., O'connor K.C., Hafler D.A., Vigneault F., Kleinstein S.H. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A., Schafer S., Zaytseva O., Albert H., Lux A., Kristic J., Pezer M., Lauc G., Winkler T., Nimmerjahn F. Targeting B cells in the pre-phase of systemic autoimmunity globally interferes with autoimmune pathology. iScience. 2021;24:103076. doi: 10.1016/j.isci.2021.103076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for codes needed for data analysis (perl script for UMI correction) reported in this paper is GitHub: https://github.com/Simon-K-Schaefer/umify_iso or Zenodo: https://doi.org/10.5281/zenodo.6327191. Original data for creating Figure 10, Figure 11, Figure 12 have been deposited to Mendeley Data: https://doi.org/10.17632/smtxktdtfc.1.