Abstract
Despite the initial benefit from treating ERBB2-positive breast cancer with tyrosine kinase inhibitor lapatinib, resistance develops inevitably. Since the expression of protein tyrosine phosphatase receptor-type O (PTPRO), a member of the R3 subfamily of receptor protein tyrosine phosphatases (PTPs), is inversely correlated with the aggressiveness of multiple malignancies, we decided to explore the correlation between PTPRO and lapatinib resistance in ERBB2-positive breast cancer. Results of immunohistochemical (IHC) staining and the correlation analysis between the expression levels of PTPRO and the clinicopathological parameters indicate that PTPRO is downregulated in cancer tissues as compared with normal tissues and negatively associated with differentiation, tumor size, tumor depth, as well as the expression of ERBB2 and Ki67. Results from Kaplan–Meier analyses indicate that lower expression of PTPRO is correlated with shorter relapse-free survival for patients with ERBB2-positive breast cancer, and multivariable Cox regression analysis found that PTPRO can potentially serve as an independent prognostic indicator for ERBB2-positive breast cancer. Results from both human breast cancer cells with PTPRO knockdown or overexpression and mouse embryonic fibroblasts (MEFs) which derived from Ptpro +/+ and Ptpro −/− mice with then stably transfected plasmid FUGW-Erbb2 consistently demonstrated the essentiality of PTPRO in the lapatinib-mediated anticancer process. Our findings suggest that PTPRO is not only able to serve as an independent prognostic indicator, but upregulating PTPRO can also reverse the lapatinib resistance of ERBB2-positive breast cancer.
Keywords: PTPRO, phosphatase, tyrosine kinase, ERBB2-positive breast cancer, prognosis, lapatinib resistance
Introduction
Breast cancer is by far the most common cancer and the leading cause of cancer-related mortality in women worldwide and therefore breast cancer presents a significant burden for public health (DeSantis et al., 2017; Bray et al., 2018). ERBB2, a member of the ERBB family of receptor tyrosine kinases (RTKs) (EGFR/ERBB1, ERBB2, ERBB3, and ERBB4), is overexpressed in approximately 15–20% of human breast cancers and its expression is highly associated with poorer overall survival (Nahta and Esteva, 2006; Murthy et al., 2016; Ethier et al., 2021). ERBB2 status is a determinant for stratifying invasive breast cancer concerning ERBB2-directed targeted therapy (Ahmed et al., 2015; Goutsouliak et al., 2020; Oh and Bang, 2020), including ERBB2-targeted monoclonal antibodies and small-molecule tyrosine kinase inhibitors such as lapatinib (Xia et al., 2005). Unfortunately, only about 24% of patients respond to lapatinib treatment and most of the responders become lapatinib resistant (D'Amato et al., 2015). Thus, understanding the molecular mechanisms of lapatinib resistance and developing new therapeutic strategies are urgently needed.
Protein tyrosine phosphatase receptor-type O (PTPRO), a member of the R3 subfamily of receptor protein tyrosine phosphatases (PTPs), has been found to correlate negatively with clinical aggressiveness in different malignancies (Motiwala et al., 2003; Motiwala et al., 2004; You et al., 2012; Hou et al., 2013; Huang et al., 2013; Dong et al., 2017a; Gan and Zhang, 2020). We and others have demonstrated that PTPRO is capable of dephosphorylating RTKs such as ERBB2 and EPH receptors (Shintani et al., 2006; Dong et al., 2017a) and repressing tumor growth (Motiwala et al., 2003; Motiwala et al., 2004; Dong et al., 2017a). In this study, we explored the correlation between PTPRO and lapatinib resistance in ERBB2-positive breast cancer. We found that 1) reduced levels of PTPRO correlate with poorer clinical outcomes; and 2) overexpressed PTPRO in lapatinib resistant ERBB2-positive breast cancer cells is capable of reversing lapatinib-resistance.
Materials and Methods
Patients and Tissue Samples
Surgically treated female breast cancer patients (n = 180) with confirmed pathology of invasive ductal carcinoma were included, and their clinicopathological characteristics are summarized in Table 1. Breast cancer tissues were obtained from the patients when undergoing surgical treatment at the Department of Surgery, Cancer Hospital affiliated to Shantou University Medical College, during the period from 2010 to 2013. All patients received primary treatment by surgery followed by adjuvant radiotherapy, chemotherapy, or hormone therapy. The mean age of the patients was 50 years (range = 19–78 years). The histological types and grades of the primary tumors were determined according to a system modified from the WHO classification. Tumor staging was defined according to the 7th Edition of the American Joint Committee on Cancer TNM Staging manual. For all cases, representative parts of the primary invasive ductal carcinoma, along with adjacent noncancerous tissue were frozen in liquid nitrogen-cooled isopentane at the time of diagnosis and stored at a temperature below -80°C until use. The rest of the tissue was fixed in buffered formalin, embedded in paraffin, and used for routine histology. Clinical research protocols of this study were reviewed and approved by the Ethics Committee of Shantou University Medical College (IRB serial number: # 04–070). Written informed consents were obtained from patients in accordance with principles expressed in the Declaration of Helsinki.
TABLE 1.
Relationship between PTPRO expression and clinicopathologic variables in tissue samples of breast cancer patients (n = 180).
| Variables | No. of samples | PTPRO expression | P | |
|---|---|---|---|---|
| Negative, no. (%) | Positive, no. (%) | |||
| All samples | 180 | 111 (61.7) | 69 (38.3) | — |
| Age (years) | ||||
| ≤50 | 105 | 64 (61.0) | 41 (39.0) | 0.816 |
| >50 | 75 | 47 (62.7) | 28 (37.3) | |
| Histologic differentiation | ||||
| Well | 26 | 10 (38.5) | 16 (61.5) | 0.031 |
| Moderate | 101 | 66 (65.3) | 35 (34.7) | |
| Poor | 53 | 35 (66.0) | 18 (34.0) | |
| Tumor size | ||||
| ≤2 | 31 | 13 (41.9) | 18 (58.1) | 0.013 |
| >2 | 149 | 98 (65.8) | 51 (34.2) | |
| Tumor depth | ||||
| T1/T2 | 138 | 77 (55.8) | 61 (44.2) | 0.003 |
| T3/T4 | 42 | 34 (81.0) | 8 (19.0) | |
| Lymph node metastasis | ||||
| N0 | 108 | 70 (64.8) | 38 (35.2) | 0.287 |
| N1 | 72 | 41 (56.9) | 31 (43.1) | |
| Stage | ||||
| I/II | 124 | 72 (58.1) | 52 (41.9) | 0.139 |
| III/IV | 56 | 39 (69.6) | 17 (30.4) | |
| ERBB2 expression | ||||
| Negative | 115 | 64 (55.7) | 51 (44.3) | 0.027 |
| Positive | 65 | 47 (72.3) | 18 (27.7) | |
| Ki67 expression | ||||
| <20 | 38 | 3 (7.89) | 35 (92.1) | 0.000 |
| ≥20 | 142 | 108 (76.1) | 34 (23.9) | |
Immunohistochemical Analysis
Four-μm-thick sections from representative tumor areas and normal mammary gland tissue from surgical specimen tissue blocks were processed according to standard immunohistochemistry (IHC) protocols (Dong et al., 2017b; Wang et al., 2019; Wang et al., 2020; Wang et al., 2021a) and stained with antibodies against PTPRO (Cat. No. HPA034525, Sigma-Aldrich, St. Louis, United States), and Ki67 (Cat. No. #9449, Cell Signaling Technology, Danvers, MA, United States). Sections incubated with immunoglobulin (Ig)G of appropriate species (rabbit or mouse) as the primary antibody were used as negative controls.
The percentage of positively stained cells was scored using the following scales: 0, no staining of cells in any field; 1, ≤10%; 2, 11–50%; 3, 51–75%; 4, >75%. The intensity of staining was scored using the following scales: 1+, weak staining; 2+, moderate staining; 3+, strong staining. Percentage (P) and intensity (I) of nuclear or cytoplasm or membrane expression were multiplied to generate a numerical score (S = P*I). The immunoreactivity was evaluated by two different pathologists with no prior knowledge of patient data. When the opinions of the two evaluators were different, the agreement was then compared and discussed.
Analysis of Breast Cancer Microarray Datasets
Raw data from 1,079 breast cancer gene expression profiles from 5 independent datasets (GSE3494, GSE7390, GSE6532, GSE1456, and GSE2034) were collected from NCBI Gene Expression Omnibus (GEO) (Pawitan et al., 2005; Wang et al., 2005; Sotiriou et al., 2006; Desmedt et al., 2007; Loi et al., 2007). Each dataset selected for this study was based on Affymetrix U133A, and all of the datasets contained clinical outcome data. The raw expression data were normalized by Robust multi-array average (RMA) (Irizarry et al., 2003) and compiled. The batch effect was removed by using Bayesian Factor Regression Modeling (BFRM) (Carvalho et al., 2008). Following this procedure, a normalized gene expression dataset compiling 1,079 breast cancer samples was generated.
Cell Culture
Human breast cancer cell lines were obtained from American Type Culture Collection (ATCC, USA). SKBR3 and BT474 cells were cultured in DMEM/F12 (GIBCO/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS). All cells were maintained at 37°C in an incubator containing 5% CO2. Primary mouse embryonic fibroblasts (MEFs) were derived from Ptpro +/+ and Ptpro −/− mice. In brief, uteri firstly isolated from 13.5-days-pregnant mice, then washed uteri with phosphate-buffered saline (PBS) and removed the head and visceral tissues. After washed with fresh PBS, the remaining bodies were minced using a pair of scissors, transferred into a 0.25% trypsin/EDTA solution (1 mL per embryo) at 37°C for 20 min. The trypsin was inactivated by DMEM with 10% FBS. The cells were cultured for 24 h at 37°C. Adherent cells were used as MEF cells.
Plasmid Constructs and Transfection
The full-length sequence encoding PTPRO was inserted into a pCR3.1 expression vector. Lentiviral pGIPZ shRNA vectors targeting human PTPRO (pGIPZ-shPTPRO, V2LHS_226,171) and non-targeting pGIPZ control vector (pGIPZ-shCtrl) were obtained from Open Biosystems (Huntsville, AL, United States). Transfection of plasmids was performed using Lipofectamine 3,000 (Cat. No. L3000015, Thermo Fisher Scientific) according to the manufacturer’s instructions. To overexpress mouse Erbb2 in Ptpro +/+ MEFs and Ptpro −/− MEFs, the plasmid FUGW-Erbb2 (a generous gift from Prof. Yi Li, Baylor College of Medicine) was transfected into the cells, which expresses mouse Erbb2 under control of a ubiquitin promoter, using the Calcium Phosphate Transfection Kit (Invitrogen) based on the manufacturer’s instructions.
Cell Viability Assay and Colony Formation Assay
The cell viability was examined by MTT assay. MTT and colony formation assays were performed as described previously (Feng et al., 2014; Gan et al., 2016). For MTT assay, cells were inoculated in a 96-well plate and incubated with 10-fold serial dilutions of lapatinib ranging from 0.001 to 10 μM for 72 h at 37°C. The optical density (OD) of each well was measured with a microplate reader set at 570 nm. Cell inhibition ratio (%) = [ODcontrol-ODlapatinib treatment]/ODcontrol ×100%.
For colony formation assay, cells were trypsinized and 1,000 viable cells (depending on the experiment) were inoculated into six-well plates (in triplicate). Cells were allowed to adhere and colonize for 2 weeks. To visualize colonies, media was removed and cells were fixed in 100% methanol for 15 min and stained with crystal violet (0.1%, Sigma) for 20 min.
Cell Death Assay
The cell death was confirmed by a photometric enzyme-immunoassay (Cat. No. 11774425001, cell death detection ELISAPLUS kit, Roche Diagnostics, Mannheim, Germany) according to manufacturer’s protocol as previously described (Chandrasekar et al., 2008). In brief, the cells (1×105 cells/mL) were placed in a streptavidin-coated microtiter plate with 10-fold serial dilutions of lapatinib ranging from 0.001 to 10 μM for 72 h at 37°C. After that, cells were incubated with a mixture of biotin-labeled monoclonal histone antibody and peroxidase-conjugated monoclonal DNA antibody at 37°C for 2 h. The amount of mono- and oligonucleosomes was measured as the absorbance at 405 nm with a microplate spectrophotometer (Bio-Rad, United States) after washing to remove unbound antibodies.
Reverse Transcription-Quantitative PCR and Western Blot
Total RNA was extracted from cells using TRIzol (Invitrogen). Four-μg RNA was reverse transcribed using M-MLV Reverse Transcriptase (Invitrogen) with oligo-(dT)20 primer (Invitrogen) according to the manufacturer’s instructions. Then equal amounts of cDNA were amplified using SYBR Green PCR amplification kit (Invitrogen) with the Applied Biosystems 7,500 Real-Time PCR system (Applied Biosystems, Foster City, CA, United States), as described previously (Zhang et al., 2013; Lin et al., 2019). The results were normalized to GAPDH expression. All reactions were run in triplicate. Western blot was performed as described previously (Du et al., 2019; Xiong et al., 2020; Wang et al., 2021b). Whole-cell extracts from breast cancer cell lines. The following antibodies were used: anti-PTPRO (Cat. No. 12161-1-AP; Proteintech Group Inc., Chicago, IL, United States); anti-β-actin (Cat. No. #4967, Cell Signaling Technology, Beverly, MA, United States). Specially, the membranes of PTPRO proteins were incubated with horseradish enzyme-labeled secondary antibody for 1 h at room temperature. An enhanced chemiluminescence (IBright FL1000, Life Technologies, United States) detection system was applied to detect protein blots.
Statistical Analysis
All statistical analyses were performed using the SPSS 19.0 statistical software package (SPSS Inc., IL, United States). The differences in PTPRO expression immunoreactivity score were compared by the Mann-Whitney U test. The statistical significance of differences was evaluated with a t-test, and means, standard error, and 95% confidence intervals were calculated using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA). Correlations between PTPRO expression and clinicopathological factors were investigated using the χ2 test. The correlation coefficient between variables was performed using Spearman’s rank test. Survival curves were generated using the Kaplan-Meier estimates, and the statistical significance of differences between curves was evaluated by the log-rank test. Furthermore, hazard ratios (HR) and 95% confidence intervals (CI), which were computed from univariate and multivariable Cox proportional hazards regression models (approximate proportionality was verified by visual examination of the Kaplan–Meier estimates), were used to assess associations between relapse-free survival and clinicopathological characteristics. A p-value of less than 0.05 was considered to be significant, and all tests were 2-sided.
Results
Low Levels of PTPRO Correlate With Tumor Progression and Poor Prognosis in Patients With ERBB2-Positive Breast Cancer
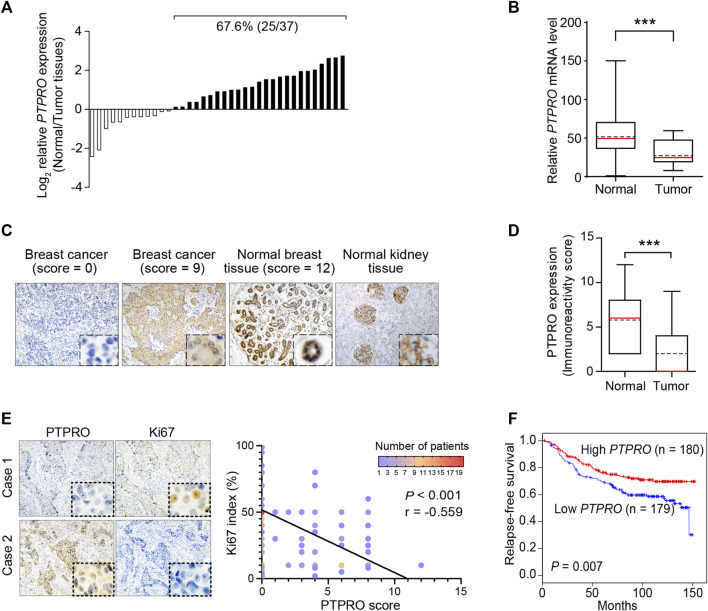
To determine if PTPRO plays any role in breast cancer, we first conducted reverse transcription-quantitative PCR (RT-qPCR) to estimate the mRNA levels of PTPRO in 37 breast tumor tissues and found that compared with their adjacent normal tissues, the mRNA level of PTPRO is significantly lower in 67.6% of cancer tissues (Figures 1A,B). We then conducted IHC analysis of PTPRO on a tissue microarray (TMA) containing 180 human breast cancer samples and 18 normal breast tissues. Figure 1C showed the representative photomicrographs with PTPRO IHC scores of 0, 9, and 12 along with normal human kidneys which expresses a high level of PTPRO as a positive reference (Dong et al., 2017a). Compared with that of the normal breast tissues, PTPRO IHC scores in breast cancer samples are significantly lower (p < 0.001) (Figure 1D). The association between the levels of PTPRO and the clinicopathological parameters in the cohort of 180 breast cancer specimens used in the TMA was also examined (Table 1). The levels of PTPRO in poorly and moderately differentiated cancer specimens were significantly lower than that of well-differentiated cancer (p = 0.031). In addition, the levels of PTPRO were consistently lower in larger (>2cm, p = 0.013), more invasive (T3+T4 vs. T1+T2, p = 0.003), and ERBB2-positive tumors (p = 0.027). Of note, the levels of PTPRO were inversely correlated with the levels of Ki67 (p < 0.001) and the representative cases shown in Figure 1E (Left panel). This finding is consistent with the relationship between PTPRO expression and the Ki67 index of the breast cancer specimens used in the TMA (r = - 0.559; p < 0.001) (Figure 1E, right panel). These data altogether suggest that reduced PTPRO expression is associated with a wide spectrum of clinicopathological parameters in breast cancer.
FIGURE 1.
Downregulation of PTPRO correlates with poor prognosis in ERBB2-positive breast cancer patients. (A) The mRNA level of PTPRO in cancer tissues and corresponding adjacent normal breast tissues was determined in 37 breast cancer patients. (B) Paired t-test revealed the significant alteration of PTPRO mRNA in tissue samples (***p < 0.001). (C) Representative images are shown as IHC scores (0–9); Normal kidney and breast tissues were positive references. (D) The immunoreactivity scores of PTPRO in normal breast tissues and breast cancer tissues were compared using the Mann-Whitney U test (***p < 0.001). (E) Images of IHC staining of PTPRO and Ki67 in serial sections of two representative human breast cancer specimens (left panel); Pearson’s correlation analysis of Ki67 index and PTPRO score in 180 human breast cancer specimens (right panel). (F) Relapse-free survival analysis in breast cancer patients according to PTPRO high and low expression levels (log-rank test). These data came from the publicly available cohorts (GSE3494, GSE7390, GSE6532, GSE1456, GSE2034; the top tertile (359 cases) of ERBB2 mRNA expression was assumed to be ERBB2-positive tumors).
Since PTPRO expression is closely related to ERBB2 status (Dong et al., 2017a), we thus further investigated the relationship between PTPRO expression and clinical outcomes in ERBB2-stratified breast cancer patients in a large dataset. In patients with lower levels of PTPRO, the relapse-free survival is significantly shorter than those with higher levels of PTPRO (p = 0.007) (Figure 1F). Multivariate analyses suggest that PTPRO can serve as an independent prognostic factor for patients with high levels of ERBB2 (HR = 0.512, 95% CI = 0.324 to 0.807, p = 0.004) (Table 2).
TABLE 2.
Univariate and multivariate Cox proportional hazards model predicting survival in ERBB2-positive breast cancer patients.
| Variables | Univariate analysis | P | Multivariate analysis | P |
|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||
| Age | ||||
| ≤50 vs. > 50 | 0.752 (0.479–1.182) | 0.217 | 0.785 (0.459–1.342) | 0.376 |
| Tumor depth | ||||
| T3/T4 vs. T1/T2 | 1.710 (1.028–2.845) | 0.039 | 1.735 (1.045–2.881) | 0.033 |
| Lymph node metastasis | ||||
| Positive vs. Negative | 2.079 (1.249–3.459) | 0.005 | 1.506 (1.195–3.138) | 0.007 |
| Histologic differentiation | ||||
| Poor/moderate vs. Well | 2.192 (1.311–3.665) | 0.003 | 2.301 (1.394–3.798) | 0.001 |
| PTPRO expression | ||||
| High vs. Low | 0.523 (0.330–0.828) | 0.006 | 0.512 (0.324–0.807) | 0.004 |
HR, hazard ratio; CI, confidence interval.
Downregulated PTPRO Correlates to Lapatinib Resistance
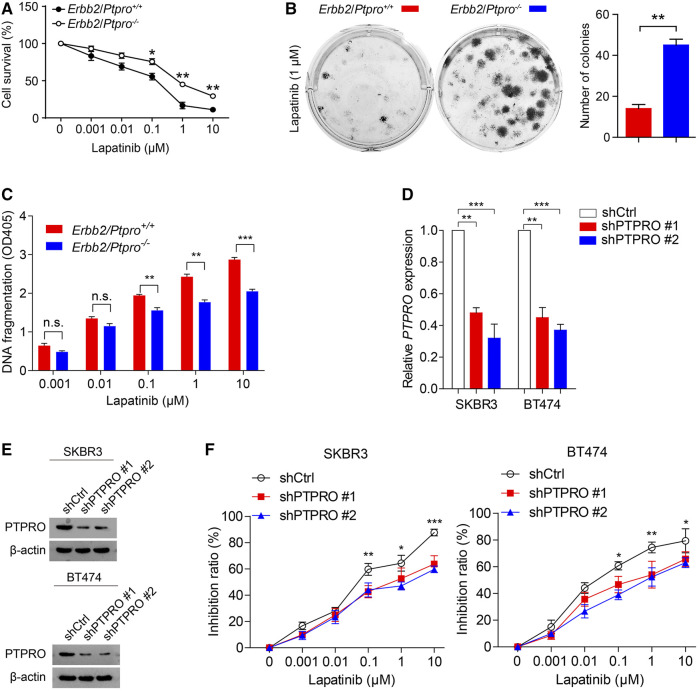
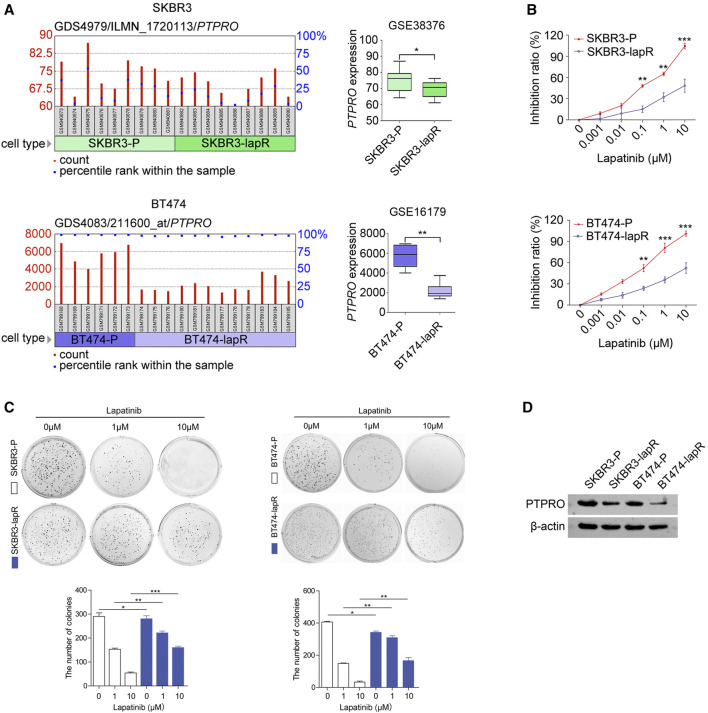
To investigate whether the levels of PTPRO affect lapatinib sensitivity, we generated mouse MEFs derived from two genotypic mice, Ptpro +/+ and Ptpro −/− , and stably expressed ERBB2 by transfecting the cells with the plasmid FUGW-Erbb2. When the cells were treated with either 1 μM or 10 μM lapatinib, the survival rate of Erbb2/Ptpro −/− MEFs is significantly higher than that of Erbb2/Ptpro +/+ MEFs (p < 0.01; Figure 2A). Colony formation assay also showed that more colonies were formed by Erbb2/Ptpro −/− MEFs than Erbb2/Ptpro +/+ MEFs in the presence of 1 μM lapatinib (p < 0.01; Figure 2B). Cell death assay suggested that Erbb2/Ptpro −/− MEFs can attenuate apoptotic cell death than Erbb2/Ptpro +/+ MEFs in the presence of 0.1–10 μM lapatinib (p < 0.01; Figure 2C). Furthermore, depletion of PTPRO from ERBB2-positive breast cancer cells SKBR3 and BT474 also reduced their lapatinib sensitivity (Figures 2D–F). These findings altogether suggest the importance of PTPRO in lapatinib sensitivity. Next, we analyzed two public available datasets GSE38376 (Komurov et al., 2012) and GSE16179 (Liu et al., 2009) and found that the PTPRO mRNA level is lower in lapatinib-resistant breast cancer cells (SKBR3-lapR and BT474-lapR) than that of their parental cells (SKBR3-P and BT474-P) (Figure 3A). We then established two lapatinib-resistant ERBB2-positive breast cancer cell lines (SKBR3-lapR and BT474-lapR) in our lab following the published protocol (Bianco et al., 2008). As expected, the SKBR3-lapR and BT474-lapR cells showed their resistance to the lapatinib treatment (Figures 3B,C). More importantly, we found that the levels of PTPRO in SKBR3-lapR and BT474-lapR are significantly lower than that of their parental cells (Figure 3D). These findings demonstrated that downregulated PTPRO is correlated with lapatinib resistance.
FIGURE 2.
PTPRO deficiency reduces lapatinib sensitivity in Ptpro −/− and littermate-control MEFs stably transfected with Erbb2 and ERBB2-positive breast cancer cells. (A) Erbb2/Ptpro +/+ MEFs and Erbb2/Ptpro −/− MEFs were plated in 96-well plates and treated with 10-fold serial dilutions of lapatinib ranging from 0.001 to 10 μM for 72 h. The percentage cell viability was evaluated by MTT assay. (B) Erbb2/Ptpro +/+ MEFs and Erbb2/Ptpro −/− MEFs were incubated with lapatinib (1 μM) for 2 weeks to allow colony formation (left panel) and the number of colonies was compared (right panel; **p < 0.01). (C) Erbb2/Ptpro +/+ MEFs and Erbb2/Ptpro −/− MEFs were incubated with lapatinib ranging from 0.001 to 10 μM for 72 h. Cell death was determined using a Cell Death detection ELISA and absorbance was measured at 405 nm. (D) RT-qPCR of PTPRO in SKBR3 and BT474 cells with PTPRO depleted. (E) The protein level of PTPRO in SKBR3 and BT474 cells with PTPRO knockdown were analyzed by western blot. (F) Cell inhibition ratio analysis of SKBR3 and BT474 cells with PTPRO depleted was evaluated by MTT assay, 72 h after treatment with 10-fold serial dilutions of lapatinib ranging from 0.001 to 10 μM. Data were shown as the means of three independent experiments or representative data. Error bars indicate SEM; n.s., not statistically significant; *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test or a one-way ANOVA with post hoc intergroup comparisons, where appropriate.
FIGURE 3.
PTPRO is downregulated in lapatinib-resistant ERBB2-positive breast cancer cells. (A) The mRNA level of PTPRO in the lapatinib resistant ERBB2-positive breast cancer cells, SKBR3-lapR and BT474-lapR, compared with their parental cells, SKBR3-P and BT474-P. The dataset GSE38376 and GSE16179 were get from GEO (https://www.ncbi.nlm.nih.gov/geo/). (B) Inhibition ratio analysis of SKBR3-lapR and SKBR3-P (upper panel), BT474-lapR and BT474-P (bottom panel) was evaluated by MTT assay after treatment with lapatinib. (C) Colony formation abilities of SKBR3-lapR and SKBR3-P (left panel), BT474-lapR and BT474-P (right panel) were evaluated after treatment with lapatinib, and the number of colonies was counted (bottom panel). (D) The PTPRO protein expression derived from lapatinib resistant ERBB2-positive cells and their parental cells were assessed by western blot analysis. Data were shown as the means of three independent experiments or representative data. Error bars indicate SEM. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t-test or one-way ANOVA with post hoc intergroup comparisons, where appropriate.
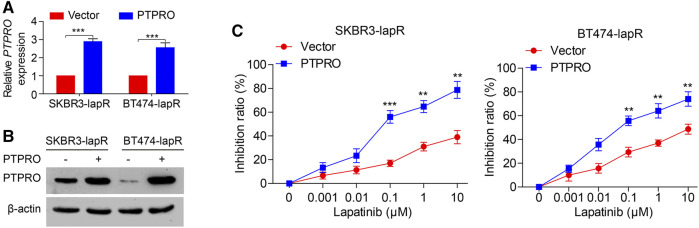
Overexpressed PTPRO is Capable of Reversing Lapatinib-Resistance in ERBB2-Positive Breast Cancer Cells
We next assessed the impact of PTPRO on the lapatinib resistance in ERBB2-positive breast cancer cells. Two established lapatinib resistant ERBB2-positive breast cancer cells, SKBR3-lapR and BT474-lapR, were transfected with the plasmid overexpressing PTPRO. The mRNA and protein levels of PTPRO were then estimated by RT-qPCR and Western blot (Figures 4A,B respectively). As shown in Figure 4C, cells were treated with 10-fold serial dilutions of lapatinib from 0.001 to 10 μM for 72 h, and the inhibition ratio exhibited a dose-dependent increase in the dose range of 0.1–10 μM. Of note, both SKBR3-lapR and BT474-lapR cells become more sensitive to lapatinib when PTPRO is overexpressed (Figure 4C). Altogether, our data strongly suggest that upregulated PTPRO in ERBB2-positive breast cancer cells can enhance lapatinib sensitivity.
FIGURE 4.
PTPRO sensitizes lapatinib in ERBB2-positive breast cancer cells. (A) The level of PTPRO mRNA in SKBR3-lapR and BT474-lapR cells with PTPRO overexpression were analyzed by RT-qPCR. (B) The protein level of PTPRO in SKBR3-lapR and BT474-lapR cells with PTPRO overexpression were analyzed by Western blot. (C) Cell inhibition ratio analysis of SKBR3-lapR and BT474-lapR cells with PTPRO overexpression was evaluated by MTT assay, 72 h after treatment with 10-fold serial dilutions of lapatinib ranging from 0.001 to 10 μM. Data were shown as the means of three independent experiments or representative data. Error bars indicate SEM. **p < 0.01, ***p < 0.001 by one-way ANOVA with post hoc intergroup comparisons, where appropriate.
Discussion
We found in this study that the expression level of PTPRO in breast cancer tissues is lower than that of the normal tissue. Further, the reduced expression level of PTPRO is associated with tumor size, poor histological differentiation, increased tumor depth, ERBB2, and Ki67 expression as well as shorter patient survival. It can also serve as an independent prognostic indicator for survival. In addition, PTPRO knockdown robustly reduced the sensitivity of the cancer cells to lapatinib. More importantly, overexpressed PTPRO is capable of reversing lapatinib resistance suggesting that upregulating PTPRO may be an effective strategy for overcoming lapatinib resistance.
The receptor tyrosine kinase ERBB2 is overexpressed in 15–20% of breast tumors and increased ERBB2 is capable of enhancing cancer cell proliferation and invasion, which is associated with a poor prognosis for overall survival (Murthy et al., 2016; Ethier et al., 2021). Therefore, targeting ERBB2 has been a strategy for drug development. ERBB2-targeted therapy drugs approved by the Food and Drug Administration (FDA), including the monoclonal antibodies trastuzumab and pertuzumab, the small molecule ERBB1/2 tyrosine kinase inhibitor (TKI) lapatinib, neratinib, tucatinib and the antibody-drug conjugate trastuzumab emtansine (TDM1), have significantly improved outcomes for patients with ERBB2-positive breast cancer (Xu et al., 2017). Lapatinib, an oral tyrosine kinase inhibitor that potently and specifically inhibits ERBB2, is now indeed a third-line therapy for ERBB2-positive disease (Vogel et al., 2010). However, lapatinib has been shown to improve progression-free survival (PFS) in patients with advanced/metastatic ERBB2-positive breast cancer (Geyer et al., 2006). Moreover, lapatinib is also approved for patients with ERBB2-positive advanced-stage breast cancer, and can show synergistic activity when used in combination with anti-ERBB2 antibodies such as trastuzumab (Guarneri et al., 2008; Medina and Goodin, 2008). A preclinical study shows that lapatinib inhibits the growth of trastuzumab-resistant ERBB2-positive breast cancer cells and increases apoptosis of anti-ERBB2 antibodies (O'Donovan et al., 2011). Evidences above suggest that lapatinib alone may be effective in treating trastuzumab-resistant ERBB2-positive patients (Blackwell et al., 2010). In addition, lapatinib is able to diffuse in the central nervous system (CNS) (Saleem et al., 2015), so it has the potential to improve the control of CNS disease compared to other monoclonal antibodies (Gril et al., 2008). Randomized trial has shown that lapatinib combined with chemotherapy reduces CNS involvement (Cetin et al., 2012). Of note, lapatinib appears to have a lower incidence of cardiotoxicity than trastuzumab, according to a comprehensive analysis of the clinical trials in metastatic breast cancer (MBC) treatment (Perez et al., 2008). Although lapatinib has many advantages such as we mentioned above, acquired resistance to lapatinib remains a major concern for its clinical utilization and lapatinib shares some of the mechanisms of resistance described for anti-ERBB2 targeted therapies, thus highlighting the importance of finding mechanisms of primary/acquired resistance.
As a member of the PTP family, PTPRO can serve as a tumor suppressor (Motiwala et al., 2003; Motiwala et al., 2004; You et al., 2012; Huang et al., 2013; Dong et al., 2017a). We have reported previously that PTPRO promoter hypermethylation is associated with poor survival for patients with ERBB2-positive breast cancer (Huang et al., 2013). In the current study, we showed that low expression of PTPRO is associated with poor prognosis and PTPRO could serve as an independent prognostic factor for relapse-free survival for patients with ERBB2-enriched breast cancer. Similar results have been reported in lung squamous cell carcinoma (Ming and Sun, 2017) and colorectal cancer (Asbagh et al., 2014).
Lapatinib resistance involves alterations of multiple signaling pathways including compensatory activation of RTKs, non-receptor tyrosine kinases (NRTKs), as well as the ERBB2-regulated downstream pathways (Shi et al., 2016). In addition, cross-talks among these pathways further exacerbated therapeutic resistance which makes current strategies preventing lapatinib resistance by targeting individual pathways far from ideal (Shi et al., 2016; Xuhong et al., 2019). To overcome this dilemma, we explored the possibility of preventing lapatinib resistance by focusing our attention on PTPRO, the upstream regulator of these pathways. It is known that PTPRO can dephosphorylate EGFR at Y845 and subsequently inhibit EGFR-mediated activation of SRC (Asbagh et al., 2014). PTPRO also can inactivate ERK/AKT pathways by dephosphorylating and repressing ERBB2 (Dong et al., 2017a). Therefore, our findings provide a novel therapeutic strategy to counteract lapatinib resistance. Based on the fact that PTPRO can sensitize cancer cells to different therapies (Ramaswamy et al., 2009; Asbagh et al., 2014), we overexpressed PTPRO in ERBB2-positive cancer cells and demonstrated that the overexpressed PTPRO is capable of reversing lapatinib-insensitivity. In our previous study, we explore the potentials of reexpressing PTPRO by using 5-azacytidine in ERBB2-positive breast cancer cells (Dong et al., 2017a). After treated with 5-azacytidine, cells decreased methylation leading to reexpression of PTPRO. In addition, plenty of methods currently have been reported to overexpress genes and proteins, such as lipid nanoparticles-based mRNA delivery strategy (LNPs) (Davies et al., 2021). Thus, it is also possible to overexpress PTPRO in vivo using LNP-formulated PTPRO mRNA. We will explore this in the further. Overall, through these methods above, we can overexpress PTPRO in breast cancer and overcome the resistance in patients.
Lapatinib is indicated to has good clinical activity, and acquired resistance to lapatinib remains a major concern for its clinical utilization. The efforts to study lapatinib resistance by us and others will offer the possibility of making lapatinib become a first-line drug in the further. At the same time, lapatinib treatment based on PTPRO level also reflects the significance of tumor precision treatment. Since the drugs such as trastuzumab, TDM1, neratinib and tucatinib also target ERBB2 like lapatinib, PTPRO may also reverse trastuzumab, TDM1, neratinib or tucatinib resistance. We will continue to explore and study this issue in the future. To our knowledge, this is the first to report the role of PTPRO for the response of breast cancers toward lapatinib, and upregulated PTPRO is capable of reversing lapatinib-insensitivity. However, the clinical significance of the overexpressed PTPRO needs to be validated in larger clinical cohorts.
Acknowledgments
We acknowledge members of HZ’s laboratory for the technical help and discussion.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by The Ethics Committee of Shantou University Medical College (IRB serial number: # 04–070). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HZ and HD conceived and designed the experiments, and interpreted data. LD, SC, WL, and CC performed the in vitro experiments. HD and ZY performed patient specimen analysis, bioinformatic assays, and data analysis. MS, RC, YP, DZ, and SG wrote the manuscript, contributed to the discussion, and reviewed the manuscript.
Funding
The work was supported by a grant in part by the National Natural Science Foundation of China (82072683, 81773087, 81071736, 81572876, and 30973508 to HZ, 81802404 to HD); Natural Science Foundation of Guangdong Province of China (2021A1515012522 and 9151018004000000 to HZ, 2021A1515011028 to HD); Science and Technology Planning Project of Guangdong Province of China (2019A030317024 to HZ); Special Project on the Integration of Industry, Education and Research of Guangdong Province (2011A090100024 to HZ); Jinan university Innovation and Entrepreneurship Fund for College Students (202010559081 and 202110559097 to HZ); Flagship specialty construction project General surgery (Funding no.: 711003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.838171/full#supplementary-material
References
- Ahmed S., Sami A., Xiang J. (2015). HER2-directed Therapy: Current Treatment Options for HER2-Positive Breast Cancer. Breast Cancer 22, 101–116. 10.1007/s12282-015-0587-x [DOI] [PubMed] [Google Scholar]
- Asbagh L. A., Vazquez I., Vecchione L., Budinska E., De Vriendt V., Baietti M. F., et al. (2014). The Tyrosine Phosphatase PTPRO Sensitizes colon Cancer Cells to Anti-EGFR Therapy through Activation of SRC-Mediated EGFR Signaling. Oncotarget 5, 10070–10083. 10.18632/oncotarget.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco R., Rosa R., Damiano V., Daniele G., Gelardi T., Garofalo S., et al. (2008). Vascular Endothelial Growth Factor Receptor-1 Contributes to Resistance to Anti-epidermal Growth Factor Receptor Drugs in Human Cancer Cells. Clin. Cancer Res. 14, 5069–5080. 10.1158/1078-0432.CCR-07-4905 [DOI] [PubMed] [Google Scholar]
- Blackwell K. L., Burstein H. J., Storniolo A. M., Rugo H., Sledge G., Koehler M., et al. (2010). Randomized Study of Lapatinib Alone or in Combination with Trastuzumab in Women with ErbB2-Positive, Trastuzumab-Refractory Metastatic Breast Cancer. J. Clin. Oncol. 28, 1124–1130. 10.1200/JCO.2008.21.4437 [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68, 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Carvalho C. M., Chang J., Lucas J. E., Nevins J. R., Wang Q., West M. (2008). High-Dimensional Sparse Factor Modeling: Applications in Gene Expression Genomics. J. Am. Stat. Assoc. 103, 1438–1456. 10.1198/016214508000000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin B., Benekli M., Oksuzoglu B., Koral L., Ulas A., Dane F., et al. (2012). Lapatinib Plus Capecitabine for Brain Metastases in Patients with Human Epidermal Growth Factor Receptor 2-positive Advanced Breast Cancer: a Review of the Anatolian Society of Medical Oncology (ASMO) Experience. Onkologie 35, 740–745. 10.1159/000345040 [DOI] [PubMed] [Google Scholar]
- Chandrasekar B., Boylston W. H., Venkatachalam K., Webster N. J., Prabhu S. D., Valente A. J. (2008). Adiponectin Blocks Interleukin-18-Mediated Endothelial Cell Death via APPL1-dependent AMP-Activated Protein Kinase (AMPK) Activation and IKK/NF-kappaB/PTEN Suppression. J. Biol. Chem. 283, 24889–24898. 10.1074/jbc.M804236200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amato V., Raimondo L., Formisano L., Giuliano M., De Placido S., Rosa R., et al. (2015). Mechanisms of Lapatinib Resistance in HER2-Driven Breast Cancer. Cancer Treat. Rev. 41, 877–883. 10.1016/j.ctrv.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Davies N., Hovdal D., Edmunds N., Nordberg P., Dahlén A., Dabkowska A., et al. (2021). Functionalized Lipid Nanoparticles for Subcutaneous Administration of mRNA to Achieve Systemic Exposures of a Therapeutic Protein. Mol. Ther. Nucleic Acids 24, 369–384. 10.1016/j.omtn.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis C. E., Ma J., Goding Sauer A., Newman L. A., Jemal A. (2017). Breast Cancer Statistics, 2017, Racial Disparity in Mortality by State. CA Cancer J. Clin. 67, 439–448. 10.3322/caac.21412 [DOI] [PubMed] [Google Scholar]
- Desmedt C., Piette F., Loi S., Wang Y., Lallemand F., Haibe-Kains B., et al. (2007). Strong Time Dependence of the 76-gene Prognostic Signature for Node-Negative Breast Cancer Patients in the TRANSBIG Multicenter Independent Validation Series. Clin. Cancer Res. 13, 3207–3214. 10.1158/1078-0432.CCR-06-2765 [DOI] [PubMed] [Google Scholar]
- Dong H., Ma L., Gan J., Lin W., Chen C., Yao Z., et al. (2017a). PTPRO Represses ERBB2-Driven Breast Oncogenesis by Dephosphorylation and Endosomal Internalization of ERBB2. Oncogene 36, 410–422. 10.1038/onc.2016.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Xu J., Li W., Gan J., Lin W., Ke J., et al. (2017b). Reciprocal Androgen Receptor/interleukin-6 Crosstalk Drives Oesophageal Carcinoma Progression and Contributes to Patient Prognosis. J. Pathol. 241, 448–462. 10.1002/path.4839 [DOI] [PubMed] [Google Scholar]
- Du L., Wang L., Gan J., Yao Z., Lin W., Li J., et al. (2019). MTA3 Represses Cancer Stemness by Targeting the SOX2OT/SOX2 Axis. iScience 22, 353–368. 10.1016/j.isci.2019.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethier J.-L., Desautels D., Robinson A., Amir E., Kong W., Booth C. M. (2021). Practice Patterns and Outcomes of Novel Targeted Agents for the Treatment of ERBB2-Positive Metastatic Breast Cancer. JAMA Oncol. 7, e212140. 10.1001/jamaoncol.2021.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Ke C., Tang Q., Dong H., Zheng X., Lin W., et al. (2014). Metformin Promotes Autophagy and Apoptosis in Esophageal Squamous Cell Carcinoma by Downregulating Stat3 Signaling. Cell Death Dis 5, e1088. 10.1038/cddis.2014.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J., Ke X., Jiang J., Dong H., Yao Z., Lin Y., et al. (2016). Growth Hormone-Releasing Hormone Receptor Antagonists Inhibit Human Gastric Cancer through Downregulation of PAK1-Stat3/nf-Κb Signaling. Proc. Natl. Acad. Sci. U S A. 113, 14745–14750. 10.1073/pnas.1618582114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J., Zhang H. (2020). PTPRO Predicts Patient Prognosis and Correlates with Immune Infiltrates in Human clear Cell Renal Cell Carcinoma. Transl Cancer Res. 9, 4800, 4810-+. doi: 10.21037/tcr-19-2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer C. E., Forster J., Lindquist D., Chan S., Romieu C. G., Pienkowski T., et al. (2006). Lapatinib Plus Capecitabine for HER2-Positive Advanced Breast Cancer. N. Engl. J. Med. 355, 2733–2743. 10.1056/NEJMoa064320 [DOI] [PubMed] [Google Scholar]
- Goutsouliak K., Veeraraghavan J., Sethunath V., De Angelis C., Osborne C. K., Rimawi M. F., et al. (2020). Towards Personalized Treatment for Early Stage HER2-Positive Breast Cancer. Nat. Rev. Clin. Oncol. 17, 233–250. 10.1038/s41571-019-0299-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gril B., Palmieri D., Bronder J. L., Herring J. M., Vega-Valle E., Feigenbaum L., et al. (2008). Effect of Lapatinib on the Outgrowth of Metastatic Breast Cancer Cells to the Brain. J. Natl. Cancer Inst. 100, 1092–1103. 10.1093/jnci/djn216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarneri V., Frassoldati A., Piacentini F., Jovic G., Giovannelli S., Oliva C., et al. (2008). Preoperative Chemotherapy Plus Lapatinib or Trastuzumab or Both in HER2-Positive Operable Breast Cancer (CHERLOB Trial). Clin. Breast Cancer 8, 192–194. 10.3816/CBC.2008.n.022 [DOI] [PubMed] [Google Scholar]
- Hou J., Xu J., Jiang R., Wang Y., Chen C., Deng L., et al. (2013). Estrogen-sensitive PTPRO Expression Represses Hepatocellular Carcinoma Progression by Control of STAT3. Hepatology 57, 678–688. 10.1002/hep.25980 [DOI] [PubMed] [Google Scholar]
- Huang Y. T., Li F. F., Ke C., Li Z., Li Z. T., Zou X. F., et al. (2013). PTPRO Promoter Methylation Is Predictive of Poorer Outcome for HER2-Positive Breast Cancer: Indication for Personalized Therapy. J. Transl Med. 11, 245. 10.1186/1479-5876-11-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., Scherf U., et al. (2003). Exploration, Normalization, and Summaries of High Density Oligonucleotide Array Probe Level Data. Biostatistics 4, 249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Komurov K., Tseng J. T., Muller M., Seviour E. G., Moss T. J., Yang L., et al. (2012). The Glucose-Deprivation Network Counteracts Lapatinib-Induced Toxicity in Resistant ErbB2-Positive Breast Cancer Cells. Mol. Syst. Biol. 8, 596. 10.1038/msb.2012.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Dong H., Deng W., Lin W., Li K., Xiong X., et al. (2019). Evaluation of Salivary Exosomal Chimeric GOLM1-NAA35 RNA as a Potential Biomarker in Esophageal Carcinoma. Clin. Cancer Res. 25, 3035–3045. 10.1158/1078-0432.CCR-18-3169 [DOI] [PubMed] [Google Scholar]
- Liu L., Greger J., Shi H., Liu Y., Greshock J., Annan R., et al. (2009). Novel Mechanism of Lapatinib Resistance in HER2-Positive Breast Tumor Cells: Activation of AXL. Cancer Res. 69, 6871–6878. 10.1158/0008-5472.CAN-08-4490 [DOI] [PubMed] [Google Scholar]
- Loi S., Haibe-Kains B., Desmedt C., Lallemand F., Tutt A. M., Gillet C., et al. (2007). Definition of Clinically Distinct Molecular Subtypes in Estrogen Receptor-Positive Breast Carcinomas through Genomic Grade. J. Clin. Oncol. 25, 1239–1246. 10.1200/JCO.2006.07.1522 [DOI] [PubMed] [Google Scholar]
- Medina P. J., Goodin S. (2008). Lapatinib: a Dual Inhibitor of Human Epidermal Growth Factor Receptor Tyrosine Kinases. Clin. Ther. 30, 1426–1447. 10.1016/j.clinthera.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Ming F., Sun Q. (2017). Epigenetically Silenced PTPRO Functions as a Prognostic Marker and Tumor Suppressor in Human Lung Squamous Cell Carcinoma. Mol. Med. Rep. 16, 746–754. 10.3892/mmr.2017.6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala T., Ghoshal K., Das A., Majumder S., Weichenhan D., Wu Y. Z., et al. (2003). Suppression of the Protein Tyrosine Phosphatase Receptor Type O Gene (PTPRO) by Methylation in Hepatocellular Carcinomas. Oncogene 22, 6319–6331. 10.1038/sj.onc.1206750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiwala T., Kutay H., Ghoshal K., Bai S., Seimiya H., Tsuruo T., et al. (2004). Protein Tyrosine Phosphatase Receptor-type O (PTPRO) Exhibits Characteristics of a Candidate Tumor Suppressor in Human Lung Cancer. Proc. Natl. Acad. Sci. U S A. 101, 13844–13849. 10.1073/pnas.0405451101 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Murthy P., Kidwell K. M., Schott A. F., Merajver S. D., Griggs J. J., Smerage J. D., et al. (2016). Clinical Predictors of Long-Term Survival in HER2-Positive Metastatic Breast Cancer. Breast Cancer Res. Treat. 155, 589–595. 10.1007/s10549-016-3705-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahta R., Esteva F. J. (2006). HER2 Therapy: Molecular Mechanisms of Trastuzumab Resistance. Breast Cancer Res. 8, 215. 10.1186/bcr1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan N., Byrne A. T., O'Connor A. E., McGee S., Gallagher W. M., Crown J. (2011). Synergistic Interaction between Trastuzumab and EGFR/HER-2 Tyrosine Kinase Inhibitors in HER-2 Positive Breast Cancer Cells. Invest. New Drugs 29, 752–759. 10.1007/s10637-010-9415-5 [DOI] [PubMed] [Google Scholar]
- Oh D. Y., Bang Y. J. (2020). HER2-targeted Therapies - a Role beyond Breast Cancer. Nat. Rev. Clin. Oncol. 17, 33–48. 10.1038/s41571-019-0268-3 [DOI] [PubMed] [Google Scholar]
- Pawitan Y., Bjöhle J., Amler L., Borg A. L., Egyhazi S., Hall P., et al. (2005). Gene Expression Profiling Spares Early Breast Cancer Patients from Adjuvant Therapy: Derived and Validated in Two Population-Based Cohorts. Breast Cancer Res. 7, R953–R964. 10.1186/bcr1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E. A., Koehler M., Byrne J., Preston A. J., Rappold E., Ewer M. S. (2008). Cardiac Safety of Lapatinib: Pooled Analysis of 3689 Patients Enrolled in Clinical Trials. Mayo Clin. Proc. 83, 679–686. 10.4065/83.6.679 [DOI] [PubMed] [Google Scholar]
- Ramaswamy B., Majumder S., Roy S., Ghoshal K., Kutay H., Datta J., et al. (2009). Estrogen-mediated Suppression of the Gene Encoding Protein Tyrosine Phosphatase PTPRO in Human Breast Cancer: Mechanism and Role in Tamoxifen Sensitivity. Mol. Endocrinol. 23, 176–187. 10.1210/me.2008-0211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem A., Searle G. E., Kenny L. M., Huiban M., Kozlowski K., Waldman A. D., et al. (2015). Lapatinib Access into normal Brain and Brain Metastases in Patients with Her-2 Overexpressing Breast Cancer. EJNMMI Res. 5, 30. 10.1186/s13550-015-0103-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhang W., Zhi Q., Jiang M. (2016). Lapatinib Resistance in HER2+ Cancers: Latest Findings and New Concepts on Molecular Mechanisms. Tumour Biol. 37, 15411–15431. 10.1007/s13277-016-5467-2 [DOI] [PubMed] [Google Scholar]
- Shintani T., Ihara M., Sakuta H., Takahashi H., Watakabe I., Noda M. (2006). Eph Receptors Are Negatively Controlled by Protein Tyrosine Phosphatase Receptor Type O. Nat. Neurosci. 9, 761–769. 10.1038/nn1697 [DOI] [PubMed] [Google Scholar]
- Sotiriou C., Wirapati P., Loi S., Harris A., Fox S., Smeds J., et al. (2006). Gene Expression Profiling in Breast Cancer: Understanding the Molecular Basis of Histologic Grade to Improve Prognosis. J. Natl. Cancer Inst. 98, 262–272. 10.1093/jnci/djj052 [DOI] [PubMed] [Google Scholar]
- Vogel C., Chan A., Gril B., Kim S. B., Kurebayashi J., Liu L., et al. (2010). Management of ErbB2-Positive Breast Cancer: Insights from Preclinical and Clinical Studies with Lapatinib. Jpn. J. Clin. Oncol. 40, 999–1013. 10.1093/jjco/hyq084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Du L., Xiong X., Lin Y., Zhu J., Yao Z., et al. (2021a). Repurposing Dextromethorphan and Metformin for Treating Nicotine-Induced Cancer by Directly Targeting CHRNA7 to Inhibit JAK2/STAT3/SOX2 Signaling. Oncogene 40, 1974–1987. 10.1038/s41388-021-01682-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Li K., Lin X., Yao Z., Wang S., Xiong X., et al. (2019). Metformin Induces Human Esophageal Carcinoma Cell Pyroptosis by Targeting the miR-497/PELP1 axis. Cancer Lett. 450, 22–31. 10.1016/j.canlet.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Wang L., Xiong X., Yao Z., Zhu J., Lin Y., Lin W., et al. (2021b). Chimeric RNA ASTN2-PAPPAas Aggravates Tumor Progression and Metastasis in Human Esophageal Cancer. Cancer Lett. 501, 1–11. 10.1016/j.canlet.2020.10.052 [DOI] [PubMed] [Google Scholar]
- Wang S., Lin Y., Xiong X., Wang L., Guo Y., Chen Y., et al. (2020). Low-Dose Metformin Reprograms the Tumor Immune Microenvironment in Human Esophageal Cancer: Results of a Phase II Clinical Trial. Clin. Cancer Res. 26, 4921–4932. 10.1158/1078-0432.CCR-20-0113 [DOI] [PubMed] [Google Scholar]
- Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., et al. (2005). Gene-expression Profiles to Predict Distant Metastasis of Lymph-Node-Negative Primary Breast Cancer. Lancet 365, 671–679. 10.1016/S0140-6736(05)17947-1 [DOI] [PubMed] [Google Scholar]
- Xia W., Gerard C. M., Liu L., Baudson N. M., Ory T. L., Spector N. L. (2005). Combining Lapatinib (GW572016), a Small Molecule Inhibitor of ErbB1 and ErbB2 Tyrosine Kinases, with Therapeutic Anti-ErbB2 Antibodies Enhances Apoptosis of ErbB2-Overexpressing Breast Cancer Cells. Oncogene 24, 6213–6221. 10.1038/sj.onc.1208774 [DOI] [PubMed] [Google Scholar]
- Xiong X., Ke X., Wang L., Yao Z., Guo Y., Zhang X., et al. (2020). Splice Variant of Growth Hormone-Releasing Hormone Receptor Drives Esophageal Squamous Cell Carcinoma Conferring a Therapeutic Target. Proc. Natl. Acad. Sci. U S A. 117, 6726–6732. 10.1073/pnas.1913433117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., De Angelis C., Burke K. A., Nardone A., Hu H., Qin L., et al. (2017). HER2 Reactivation through Acquisition of the HER2 L755S Mutation as a Mechanism of Acquired Resistance to HER2-Targeted Therapy in HER2+ Breast Cancer. Clin. Cancer Res. 23, 5123–5134. 10.1158/1078-0432.CCR-16-2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuhong J. C., Qi X. W., Zhang Y., Jiang J. (2019). Mechanism, Safety and Efficacy of Three Tyrosine Kinase Inhibitors Lapatinib, Neratinib and Pyrotinib in HER2-Positive Breast Cancer. Am. J. Cancer Res. 9, 2103–2119. [PMC free article] [PubMed] [Google Scholar]
- You Y. J., Chen Y. P., Zheng X. X., Meltzer S. J., Zhang H. (2012). Aberrant Methylation of the PTPRO Gene in Peripheral Blood as a Potential Biomarker in Esophageal Squamous Cell Carcinoma Patients. Cancer Lett. 315, 138–144. 10.1016/j.canlet.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lin W., Kannan K., Luo L., Li J., Chao P. W., et al. (2013). Aberrant Chimeric RNA GOLM1-MAK10 Encoding a Secreted Fusion Protein as a Molecular Signature for Human Esophageal Squamous Cell Carcinoma. Oncotarget 4, 2135–2143. 10.18632/oncotarget.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.