Abstract
Corynebacterium glutamicum has been successfully employed for the industrial production of amino acids and other bioproducts, partially due to its native ability to utilize a wide range of carbon substrates. We demonstrated C. glutamicum as an efficient microbial host for utilizing diverse carbon substrates present in biomass hydrolysates, such as glucose, arabinose, and xylose, in addition to its natural ability to assimilate lignin-derived aromatics. As a case study to demonstrate its bioproduction capabilities, L-lactate was chosen as the primary fermentation end product along with acetate and succinate. C. glutamicum was found to grow well in different aromatics (benzoic acid, cinnamic acid, vanillic acid, and p-coumaric acid) up to a concentration of 40 mM. Besides, 13C-fingerprinting confirmed that carbon from aromatics enter the primary metabolism via TCA cycle confirming the presence of β-ketoadipate pathway in C. glutamicum. 13C-fingerprinting in the presence of both glucose and aromatics also revealed coumarate to be the most preferred aromatic by C. glutamicum contributing 74 and 59% of its carbon for the synthesis of glutamate and aspartate respectively. 13C-fingerprinting also confirmed the activity of ortho-cleavage pathway, anaplerotic pathway, and cataplerotic pathways. Finally, the engineered C. glutamicum strain grew well in biomass hydrolysate containing pentose and hexose sugars and produced L-lactate at a concentration of 47.9 g/L and a yield of 0.639 g/g from sugars with simultaneous utilization of aromatics. Succinate and acetate co-products were produced at concentrations of 8.9 g/L and 3.2 g/L, respectively. Our findings open the door to valorize all the major carbon components of biomass hydrolysate by using C. glutamicum as a microbial host for biomanufacturing.
Keywords: lignocellulosic biomass hydrolysate, 13C-fingerprinting, lignin-derived aromatics, mixed-acid fermentation, L-lactate
Introduction
Research efforts are underway worldwide for production of biofuels and chemicals from various renewable resources as a sustainable alternative to fossil resources (Zhou et al., 2008; Zhou et al., 2011; Varman et al., 2013; Salvachua et al., 2015; Liu et al., 2017; Wu et al., 2017). Amongst the various renewable resources that are currently being investigated, lignocellulosic biomass represents the largest net resource with a worldwide annual production of 200 billion tons (Simmons et al., 2008; Tuck et al., 2012). Industrial biotechnology, wherein microbial strains are utilized for coupled bioconversion of various carbon substrates present in biomass, is expected to provide a scalable and cost-competitive production method for renewable fuels and chemicals (Sievert et al., 2017; Flores et al., 2020; Abdel-Rahman et al., 2021; Flores et al., 2021; Wu et al., 2021; Zan et al., 2021). However, several challenges still need to be overcome for efficient bioconversion of biomass to bioproducts. The first challenge is the structural heterogeneity of lignocellulosic biomass, resulting in hydrolysates that contain diverse substrates such as sugars, organic acids and aromatics as the main depolymerized components (Bruijnincx, 2014; Socha et al., 2014). Furthermore, aromatics derived from hydrolysis of lignocellulosic biomass can be inhibitory to the growth of common model microbial conversion hosts such as Escherichia coli and Saccharomyces cerevisiae (Fitzgerald et al., 2004; Adeboye et al., 2014; Varman et al., 2018). Although significant progress is underway for utilization of each of the potential fermentation substrates, the diversity of these substrates can result in spiralling costs for upstream separations, supplementation of fermentation medium, serial fermentation steps, and product purification. From a techno-economic perspective, bottlenecks associated with any one of these unit operations can tip the scales against economic viability for a promising biotechnology. Process integration provides a means for achieving dramatic improvements in reaction kinetics, yields, product titers, and separations, thereby minimizing cost and risk associated with biomanufacturing (Van Gerven and Stankiewicz, 2009). One-way bioprocess integration could be achieved for valorization of lignocellulosic biomass is by employing omnivorous microbial cell factories that can utilize diverse carbon substrates present in biomass hydrolysates.
Corynebacterium glutamicum is a fast growing, aerobic, and non-pathogenic Gram-positive soil bacterium exhibiting physiological traits that could make it an interesting alternative to these model microbes for the reasons discussed below. C. glutamicum naturally has a wide substrate range and has been further engineered to uptake other carbon substrates such as xylose and arabinose (Kawaguchi et al., 2008; Xia et al., 2016). Owing to the industrial importance of C. glutamicum, the genomes of several of its strains have been sequenced (Ikeda and Nakagawa, 2003; Kalinowski et al., 2003; Yukawa et al., 2007; Lv et al., 2011). The availability of genetic data for C. glutamicum led to the discovery of genetic clusters encoding aromatic degradation pathways with the aid of genomic bioinformatics. As C. glutamicum possesses catabolic pathways for the utilization of all major aromatic compounds present in biomass hydrolysates, it can be grown in the presence of lignin-derived aromatics (Shen et al., 2005; Sakai et al., 2007). Moreover, C. glutamicum is known to secrete L-lactate, succinate, and amino acids at higher concentrations as fermentation end products from glucose due to its robust glycolytic pathway (Wieschalka et al., 2012; Tsuge et al., 2019b). Because of these remarkable features, C. glutamicum was selected as a development target for investigation of microbial omnivory in bioprocessing.
Amongst the various lignocellulosic biomass available in United States, corn stover (the biomass residue left from harvest of the corn from Zea mays) represents both a cheap and an abundant biomass feedstock available for bioconversion. As found in other plant biomass, cellulose, hemicellulose, and lignin make up 70–90% of its biochemical constituents (Saha, 2003; Wang, 2007; Öhgren et al., 2007; Girio et al., 2010). Through mechanical, chemical or biochemical pretreatment, these biopolymers can be converted into the corresponding monomers, such as glucose, xylose, arabinose, and various aromatics (Chen et al., 2016). These monomers can in turn be used as carbon substrates in bioprocessing for conversion into fungible fuels and chemicals by a suitable biocatalyst like C. glutamicum.
Mixed-acid fermentation products containing lactate, succinate, and acetate have gained significant attention due to their potential application as platform chemicals for manufacturing of biobased chemicals and materials. Succinate has been identified as one of the top 12 building block chemicals by the U.S. Department of Energy, with various applications in the pharmaceutical, food, and agricultural indutries (Zheng et al., 2012; Meng et al., 2016; Flores et al., 2020). L-lactate, one of the two lactate optical isomers, is mainly used in the production of a bio-based polymer poly-L-lactic acid (PLLA). Stereochemistry of lactate has a significant effect on the material properties of polylactic acid (PLA). The two common types of polylactic acid are poly-L-lactic acid (PLLA) and poly-D-lactic acid (PDLA). Both types of lactic acids are semicrystalline polymers. However, the PDLA obtained from the racemic mixtures is amorphous (Casalini et al., 2019). PLLA is used as a packaging material for fresh fruit containers, drinking cups, lamination films, and other items that can replace petroleum derived plastics. In the present study, by using C. glutamicum as a microbial host we demonstrate bioproduction of L-lactate, succinate, and acetate from corn stover biomass hydrolysates with simultaneous utilization of sugars and aromatics (Figure 1). As aromatics metabolic pathways are not well understood in C. glutamicum, 13C-fingerprinting studies have been used to throw more light on the aromatics metabolic pathway and thereby, aid future engineering efforts.
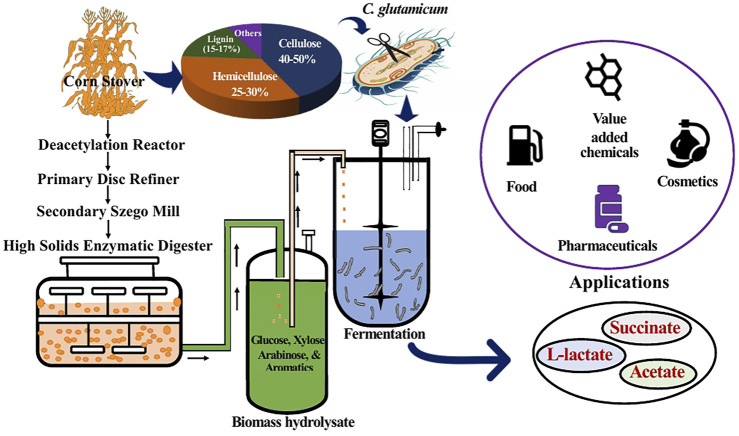
FIGURE 1.
Schematic representation of the overall process employed in this study. Corn stover was subjected to deacetylation and mechanical refining (DMR) to obtain liquid biomass hydrolysate. The hydrolysate constituting glucose, xylose, arabinose, and aromatics was used as fermentation feedstock for C. glutamicum to produce mixed acids having various applications.
Materials and Methods
Chemicals and Reagents
All organic solvents, glucose, xylose, arabinose, aromatics, and other chemicals used in this study were purchased from Sigma-Aldrich (St. Louis, MO, United States). 13C labeled glucose was purchased from Cambridge Isotope Laboratories (Tewksbury, MA, United States). The biomass hydrolysate used in this study was obtained from NREL and used without any further processing, unless otherwise stated. In brief, the biomass hydrolysate used in this study was obtained by dilute alkali-based Deacetylation and Mechanical Refining (DMR) pretreatment and enzymatic hydrolysis technology developed at NREL (Chen et al., 2016).
Bacterial Strains and Plasmids
C. glutamicum ATCC 13032 was used as the microbial host in this work (Kalinowski et al., 2003). E. coli TOP10 was used as the host for all plasmids constructed in this study. Genetic manipulations were carried out by following standard procedures (Sambrook and Russell, 2001). The plasmids pEKEx-xylAxc-xylBec and pVWE1araBAD provided by Dr. Wendisch (Bielefeld University) were used for the metabolic engineering of C. glutamicum to enable xylose and arabinose utilization (Schneider et al., 2010; Meiswinkel et al., 2013). All the plasmids and bacterial strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strain | Description | Source |
|---|---|---|
| E. coli Top 10 | General cloning host | Stratagene, La Jolla, CA |
| C. glutamicum | Wild-type, ATCC 13032 | ATCC |
| SSL03 | C. glutamicum strain harboring pEKEx-xylAxc-xylBec | This study |
| SSL09 | SSL03 harboring pVWE1araBAD | This study |
| Plasmid | ||
| pEKEx-xylAxc-xylBec | pEKEx3 based recombinant plasmid harboring xylA, and xylB | Meiswinkel et al. (2013) |
| pVWE1araBAD | pVWEx1 based recombinant plasmid harboring araB, araA and araD | Schneider et al. (2010) |
Transformation
Transformation of C. glutamicum was conducted for construction of SSL03 and SSL09 strains as described previously (van der Rest et al., 1999; Eggeling and Bott, 2005) with minor modifications. Briefly, overnight cultures of C. glutamicum were inoculated into a 1L conical flask containing 100 ml of BHIS medium (brain-heart infusion medium containing 91 g/L sorbitol) and grown at 30°C until the optical density at 600 nm [OD600] reached between 0.9 and 1.0. The cells were washed twice with ice-cold 10% (v/v) glycerol. Finally, the cells were resuspended in 1 ml of 10% (v/v) glycerol. For each transformation, 100 μL of the cells was mixed with at least 100 ng of DNA. Electroporation was then performed in a 0.2 cm electroporation cuvette and electroporation was done with a gene pulser system (Bio-Rad Laboratories, Richmond, CA), operated at 25 uF, 200 Ω and 2.5 kV, 5 ms. The electroporated cells were immediately transferred to 1 ml of fresh BHIS medium and recovered at 30°C for 2 h. Subsequently, the cultures were then plated onto a BHIS agar plate containing appropriate antibiotics. Recombinant colonies usually appeared after 24 h of plating. Single colonies were propagated on a fresh brain heart infusion (BHI) liquid medium containing appropriate antibiotics to serve as inoculum for conducting fermentation experiments.
Media and Culture Conditions
E. coli strains were cultivated in LB medium supplemented with kanamycin (50 μg/ml), and/or spectinomycin (100 μg/ml) as required at 37°C in a shaker at 200 rpm or on solidified LB plates. C. glutamicum strains were cultured in BHI medium or BTM2 medium (modified from BT medium) as required (Kawaguchi et al., 2006). For L-lactate production, the cells were grown in BTM2 medium. The BTM2 medium composition is as follows (per liter): (NH4)2SO4 7 g, KH2PO4 0.5 g, K2HPO4 0.5 g, MgSO4.7H2O 0.5 g, urea 2 g, NaHCO3 8.4 g, 1 × vitamin solution, and 1 × trace metal solution. The 1,000x trace metal solution (per liter) contained FeSO4.7H2O 6 g, MnSO4.H2O 10 g, ZnSO4.7H2O 0.56 g, CuSO4.5H2O 0.2 g, and 1,000x vitamin solution (per liter) contained biotin 4 g and thiamine 4 g. In addition, the BTM2 medium was supplemented with 2% MOPS to provide additional buffering capacity during L-lactate production (Rouabhia et al., 2014). BHI agar plates were supplemented with kanamycin (50 μg/ml), and spectinomycin (200 μg/ml) as required to obtain colonies of the engineered strains at 30°C.
Single colonies were inoculated into 5 ml of BHI medium and cultured overnight in 50 ml culture tubes at 30°C and 250 rpm. For the fermentation experiments, 1% of the overnight seed culture was inoculated into BTM2 containing 4% glucose or aromatics (benzoic acid, cinnamic acid, vanillic acid, and p-coumaric acid) at varying concentrations and grown for 5 days at 30°C and 250 rpm.
Analytical Methods
Growth of C. glutamicum was monitored by measuring optical density of cultures at 600 nm with an Infinite 200 Pro plate reader (Tecan, Männedorf, Switzerland). Estimation of glucose, xylose, arabinose, acetate, L-lactate, and succinate concentration in culture broths were analyzed using HPLC. The supernatants were collected, filtered, and injected onto an ion exchange HPLC column at 70oC under an isocratic condition with a flow rate of 0.5 ml/min using 0.005 M sulfuric acid in water as solvent. Compounds were monitored by a refractive index detector in positive mode (kept at 40°C) and their concentrations were calculated by integration of peak areas and comparison to a calibration curve prepared from pure standards. Furthermore, enzyme based D (-)/L (+) lactic acid detection kit (R-biopharm) was used to verify the optical purity of L-lactate.
For aromatics analysis, supernatant liquid obtained from above was analyzed by an Agilent Technologies 1,260 Infinity II HPLC system equipped with an Agilent Eclipse Plus Phenyl-Hexyl column (250 mm length, 4.6 mm diameter, 5 µm particle size). The mobile phase gradient profile was as follows: 30% B (0 min; 0.5 ml/min), 80% B (12 min; 0.5 ml/min), 100% B (12.1 min; 0.5 ml/min), 100% B (12.6 min; 1 ml/min), 30% B (12.8 min; 1 ml/min), 30% B (15.6 min; 1 ml/min), where solvent A was 10 mM ammonium acetate in water and solvent B was 10 mM ammonium acetate in 90% acetonitrile. Sample injection volumes of 5 µL and a column temperature of 50°C were used. Prior to analysis, samples were filtered through 0.45 µm centrifuge filters. Compounds were monitored by a UV detector and their concentrations were calculated by integration of peak areas and comparison to a calibration curve prepared from pure standards.
Fermentation Culture Conditions
Batch fermentation experiments were conducted in 1 L bioreactors with 500 ml working volume (Bioflo 115, New Brunswick Scientific Inc., Enfield, CT, United States). C. glutamicum was grown in BHI broth until the OD600 value reached ∼3 and was used as inoculum. Batch fermentations were conducted by the addition of 1% (v/v) inoculum to the fermenter containing 500 ml of BTM2 medium with 40 g/L of glucose. pH was maintained at seven by adding 25% Ca(OH)2. Fed-batch fermentations experiment were conducted by the addition of glucose at 72 h. All other fermentation conditions were identical to the batch fermentation experiments. Aerobic fermentation was conducted by sparging the cultures with air at 1 vvm and by continuously stirring the culture at 300 rpm. Anaerobic conditions were maintained by growing the cells without sparging air or oxygen. It can be verified from Supplementary Figure S2 that growing the cultures without sparging air or oxygen allowed the dissolved oxygen (DO) levels to be maintained at 0% except for the initial 10–12 h period.
A batch fermentation experiment with DMR hydrolysate was conducted with C. glutamicum SSL09 in a 1-L fermenter with 500 ml working volume (Bioflo 115, New Brunswick Scientific Inc., Enfield, CT, United States). The other fermentation conditions were identical to the batch fermentation experiment, with pH maintained at seven by adding 25% Ca(OH)2. Similarly, a batch fermentation experiment was conducted with BTM2 medium supplemented with pure sugars i.e. 40 g/L glucose, 20 g/L xylose, and 10 g/L arabinose supplemented with kanamycin (50 μg/ml), and spectinomycin (200 μg/ml). The rest other fermentation conditions were identical to the batch fermentation experiment conducted above.
13C Labeling Studies to Decipher Carbon Contribution by Aromatics
As a first step, C. glutamicum seed cultures were grown in BHI medium as described previously. Cells were centrifuged and washed with minimal medium to avoid any carryover of carbon substrates and nutrients from BHI medium. For the labeling study, 50 µL of washed cells were inoculated into 5 ml of BTM2 medium containing 5 g/L [U-13C] glucose and/or different aromatics at 20 mM concentration. Likewise, 2.5 g/L [U-13C] xylose with 9.25 g/L BHI in BTM2 minimal medium was used to investigate the presence of catabolic pathways for metabolizing xylose in C. glutamicum. Bacterial cells were harvested at the end of third day and the biomass was hydrolyzed to obtain proteinogenic amino acids. The amino acids were derivatized with TBDMS (N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide, Sigma-Aldrich) by following previously reported protocols (You et al., 2012; Varman et al., 2014). The derivatized amino acids were analyzed for their mass isotopomer abundance by GC-MS, as described elsewhere (Xiao et al., 2012; You et al., 2012). The m/z ion [M-57]+ that corresponds to the entire amino acid was used to calculate the mass fraction of labeled and unlabeled amino acids. m0, m1,…., mn represents mass distribution vector, where m0 is mass fraction of amino acids with zero labeled carbon, m1 is mass fraction of amino acids with one labeled carbon, and so on. The m/z of [M-15]+ was used for leucine and isoleucine since their [M-57]+ overlaps with other mass peaks (Wahl et al., 2004). The natural abundance of isotopes, including 13C (1.13%), 18O (0.20%), 29Si (4.70%), and 30Si (3.09%) contributes noise to the mass isotopomer spectrum. This background noise was rectified in the calculation of the mass distribution vector using a published algorithm and the detailed correction protocol can be found elsewhere (Tang et al., 2009). 13C fraction (13C carbons/total carbons) for amino acids were calculated using following formula (Xiao et al., 2013): 13C , where “n” is maximum number of carbons in the amino acid, mk is mass fraction of amino acids with “k” number of labeled carbon.
Results and Discussion
Bioreactor Studies for Mixed Acid Production From Glucose
Glucose is the primary carbon substrate for C. glutamicum, which is assimilated through central metabolism and leads to the formation of various fermentation end products such as L-lactate, succinate, and acetate (Inui et al., 2004; Tsuge et al., 2015). L-lactate is produced from pyruvate and succinate is produced in the TCA cycle, whereas acetate is formed via acetyl-CoA (Figure 2). Fermentations with 40 g/L of glucose in BTM2 medium were performed to evaluate the L-lactate fermentation ability of C. glutamicum under aerobic and anaerobic conditions. As shown in Figure 3A, after 120 h of fermentation, 16.2 g/L of L-lactate was produced under anaerobic conditions along with 0.9 g/L and 3.1 g/L of acetate and succinate, respectively. Aerobic fermentation produced 9.8 g/L of L-lactate at a yield of 0.265 g/g from glucose, in comparison to 0.42 g/g obtained in the anaerobic fermentation (Table 2 and Figure 3B. Under aerobic conditions, pyruvate (precursor for L-lactate) produced in the glycolytic pathway is further metabolized in the citric acid cycle, thereby reducing the availability of pyruvate for lactate production. In the absence of oxygen, pyruvate is instantaneously converted to L-lactate (Okino et al., 2008; Chai et al., 2016). As a result, higher yields and titers of L-lactate were observed under anaerobic conditions.
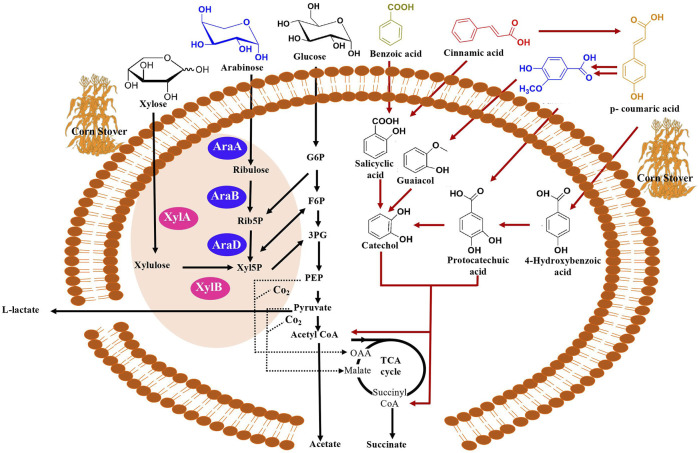
FIGURE 2.
Overview of natural and engineered metabolic pathways for the production of mixed acids (succinate, acetate and lactate) from diverse carbon substrates (xylose, arabinose, glucose, benzoic acid, cinnamic acid, vanillic acid, p-coumaric acid) in C. glutamicum. Assimilation of xylose to be facilitated by XylA (xylose isomerase) and XylB (xylulose kinase) denoted by pink in the figure and assimilation of arabinose facilitated by AraA (arabinose isomerase), AraB (ribolokinase) and AraD (ribulose 5-phosphate 4-epimerase) denoted by purple. Abbreviations in arabinose and xylose assimilation Rib5P (ribulose 5-phosphate), Xyl5P (D-xylulose 5-phosphate), G6P (glucose 6-phosphate), F6P (fructose 6-phosphate), 3PG (3-phosphoglycerate). Succinate is synthesized through the reductive branch of TCA cycle from pyruvate or PEP (phosphoenolpyruvate) under anaerobic conditions (Shirai et al., 2007; Thakker et al., 2012). Acetate is synthesized from acetyl-CoA and L-lactate is synthesized via pyruvate. Aromatic catabolic pathway represented by red arrows. Anaplerotic pathways denoted by dotted lines.
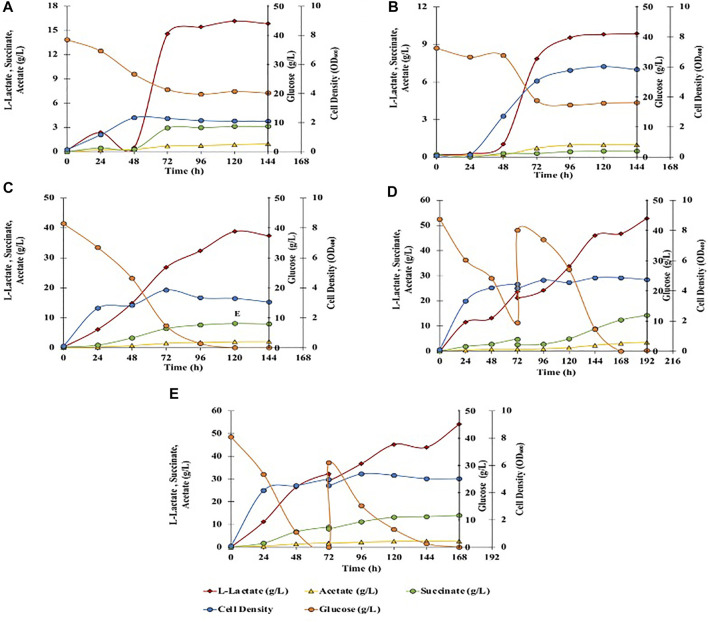
FIGURE 3.
Effect of different fermentation cultivation parameters on mixed-acids production by C. glutamicum. Data is shown for single fed batch fermentation. (A) Anaerobic, batch, no pH control, BTM2 supplemented with glucose; (B) Aerobic, batch, no pH control, BTM2 supplemented with glucose; (C) Anaerobic, batch, pH 7, BTM2 supplemented with glucose; (D) Anaerobic, fed-batch: 40 g/L (0 h) + 30 g/L (72 h), pH 7, BTM2; (E) Anaerobic with initial aeration for 16 h, fed-batch: 40 g/L (0 h) + 30 g/L (72 h), pH 7, BTM2.
TABLE 2.
Fermentation kinetics of different cultivation strategies employed for production of mixed acids.
| Batch | Initial glucose (g/L) | Initial xylose (g/L) | Initial arabinose (g/L) | % Glucose consumption | % Xylose consumption | % Arabinose consumption | Mixed acids titer | ||
|---|---|---|---|---|---|---|---|---|---|
| Acetate (g/L) | L- lactate (g/L) | Succinate (g/L) | |||||||
| I | 36.21 | 0 | 0 | 50 | − | − | 1 | 9.6 | 0.5 |
| II | 38.47 | 0 | 0 | 46 | − | − | 0.9 | 16.2 | 3.1 |
| III | 41.45 | 0 | 0 | 100 | − | − | 2.1 | 38.6 | 8.1 |
| IV | 43.79 | 0 | 0 | 100 | − | − | 3.6 | 52.8 | 14.3 |
| V | 40.45 | 0 | 0 | 100 | − | − | 2.7 | 54.1 | 14 |
| VI | 37.30 | 18.15 | 8.05 | 100 | 17% | 60% | 2 | 31.5 | 8.1 |
| VII | 45.13 | 29.91 | 0 | 100 | 15% | − | 3.2 | 47.9 | 8.9 |
Where BTM2 is modified BTM medium supplemented with sugar/s.
I- Aerobic, batch, BTM2 supplemented with glucose, no pH control, 120 h.
II- Anaerobic, batch, BTM2 supplemented with glucose, no pH control, 120 h.
III- Anaerobic, batch, BTM2 supplemented with glucose, pH 7, 120 h.
IV- Anaerobic, fed-batch: 40 g/L (0 h) + 30 g/L (72 h), BTM2 supplemented with glucose, pH 7, 192 h.
V- Anaerobic with initial aeration for 16 h, Fed-batch: 40 g/L (0 h) + 30 g/L (72 h), BTM2 supplemented with glucose, pH 7, 168 h.
VI- Anaerobic, batch; BTM2 supplemented with glucose, xylose and arabinose, pH 7, 168 h, SSL09.
VII- Anaerobic, batch; DMR hydrolysate, pH 7, 168 h, SSL09.
Energy metabolism in C. glutamicum gets impaired when the pH is maintained below six or above 8.5 owing to the non-generation of proton motive force (pmf) (Follmann et al., 2009). Therefore, it is crucial to maintain an optimal pH to maximize flux through the energy metabolism and to increase the product titer. Therefore, pH dependence of product formation was investigated at identical substrate loading to evaluate L-lactate production under chemo statically controlled neutral pH (pH 7). Ca(OH)2 has been effectively used as a low cost neutralizing agent in the production of L-lactate (Zhu et al., 2007; Nakano et al., 2012; Liu et al., 2014). Downstream processing for the recovery of L-lactate from calcium precipitate is also established, and generally performed with the addition of sulfuric acid. Therefore, pH was maintained with the addition of 25% Ca(OH)2. As shown in Figure 3C, 38.6 g/L of L-lactate was produced after 120 h with an increase in L-lactate yield to 0.932 g/g glucose. In addition, acetate and succinate were also produced in small amounts as byproducts at a titer of 2 g/L and 8.1 g/L, respectively. Downstream seperations of mixed acids can be successfully performed using the emulsion liquid membrane and adsorbent resins method as shown by Lee and Kim (2011) and Nam et al. (2012). Acetate and succinate along with L-lactate have extensive industrial applications and present work investigates C. glutamicum’s ability to produce organic acids from lignocellulosic substrates.
After establishing batch culture setup, further work focused on setting up a fed batch culture to check if fed batch culture will lead to higher mixed acid yields. A fed-batch fermentation was carried out with addition of 40 ml of 400 g/L glucose stock solution to the fermentation broth at 72 h, and fermentation was continued for another 120 h. In the end of the fermentation time course, 52.8 g/L of L-lactate (0.71 g/g glucose) was produced with 3.1 g/L of acetate and 12.4 g/L of succinate (Figure 3D). To further enhance the L-lactate production, fermentation was performed in high density cultures. Towards this end, aeration was performed for initial 16 h to enhance the cell growth at the beginning of the fermentation and to accumulate microbial biomass. Aeration was ceased and 40 ml of 400 g/L glucose was added at 72 h of fermentation. Under these conditions, 54.1 g/L of L-lactate was produced with 2.7 g/L of acetate and 14 g/L of succinate with corresponding yield of L-lactate 0.758 g/g, and productivity of 0.322 g/L/h (Figure 3E). No further improvements in L-lactate yield suggests the presence of pathway bottlenecks and can be resolved by established metabolic engineering approaches for lactate synthesis (Papagianni, 2012; Upadhyaya et al., 2014; Tsuge et al., 2015), which is not the primary focus of this study.
Utilization of Aromatic Compounds in C. Glutamicum
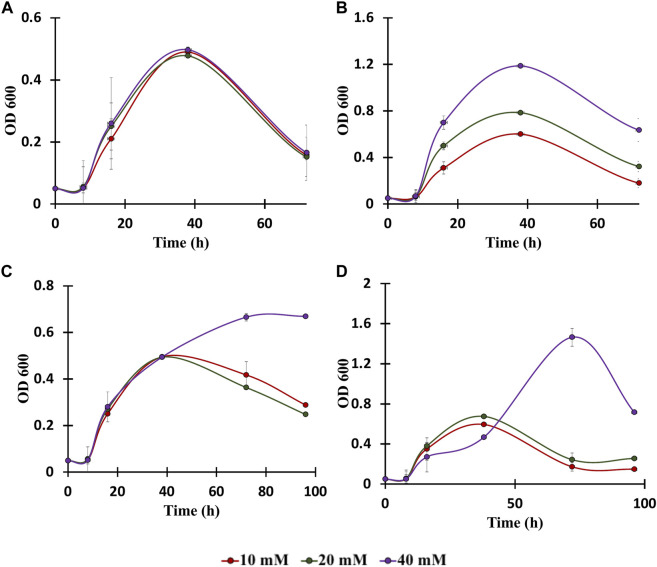
To date, numerous advances have been made for efficient bioconversion of monomers derived from cellulosic and hemicellulosic biomass components (Tsuge et al., 2019a). C. glutamicum can play a major role in bioconversion of lignin-derived aromatics since it is highly tolerant to the presence of aromatics and it can grow by utilizing aromatics as the only carbon substrate (Figure 2) (Shen et al., 2012). In this context, genomic and proteomic analysis confirm the presence of genes and proteins for the utilization of aromatics through various peripheral pathways as shown in Figure 2 (Shen et al., 2012). Although it is known that the aromatic metabolic pathways are present, growth of C. glutamicum in the presence of aromatics has not being well characterized and the upper limit of aromatics concentration at which C. glutamicum still grows is unknown (Ding et al., 2015; Kallscheuer et al., 2016). Therefore, to further understand the limits of aromatics utilization, C. glutamicum strains were cultured in BTM2 medium containing various lignin derived aromatics (vanillic acid, benzoic acid, cinnamic acid and p-coumaric acid), in varying concentrations (10 mM, 20 mM, and 40 mM). C. glutamicum showed two folds higher growth in the presence of benzoic acid and p-coumaric acid as substrates compared to vanillic acid and cinnamic acid (Figure 4). Cells showed growth up to 40 mM concentration for all the aromatics which indicates high tolerance as well as successful utilization of lignin derived aromatics by C. glutamicum. Previous studies on C. glutamicum have tested its growth at lower aromatics concentration of 5–10 mM (Ding et al., 2015; Kallscheuer et al., 2016), and therefore, the present findings further expand the aromatics concentration limit for C. glutamicum. This is important from the context of lignin valorization as biomass hydrolysates on an average contain higher (more than 10 mM) lignin derived aromatics (Linger et al., 2014; Rodriguez et al., 2017; Gomes and Rodrigues, 2019; Salvachúa et al., 2020). In comparison, other microbial hosts being studied for the bioconversion of aromatics such as Pseudomonas putida KT2440, P. putida mt-2, Rhodococcus jostii RHA1, and Sphingobium sp. SYK-6 have been tested at aromatic concentrations less than 10 mM (Chen et al., 2012; Varman et al., 2016; Ravi et al., 2017; Kohlstedt et al., 2018; Almqvist et al., 2021; Spence et al., 2021). This remarkable ability of C. glutamicum to naturally resist as well as assimilate high concentrations of aromatics makes it an ideal choice for the bioconversion of multiple substrates present in biomass hydrolysates.
FIGURE 4.
Growth assays of C. glutamicum in: (A) vanillic acid (10, 20, 40 mM), (B) Benzoic acid (10, 20, 40 mM), (C) Cinnamic acid (10, 20, 40 mM), and (D) p-coumaric acid (10, 20, 40 mM). The experiments were performed in biological triplicates. Data represents mean ± SD, n = 3.
13C-Fingerprinting to Decipher Pathways for the Utilization of Lignin-Derived Aromatics
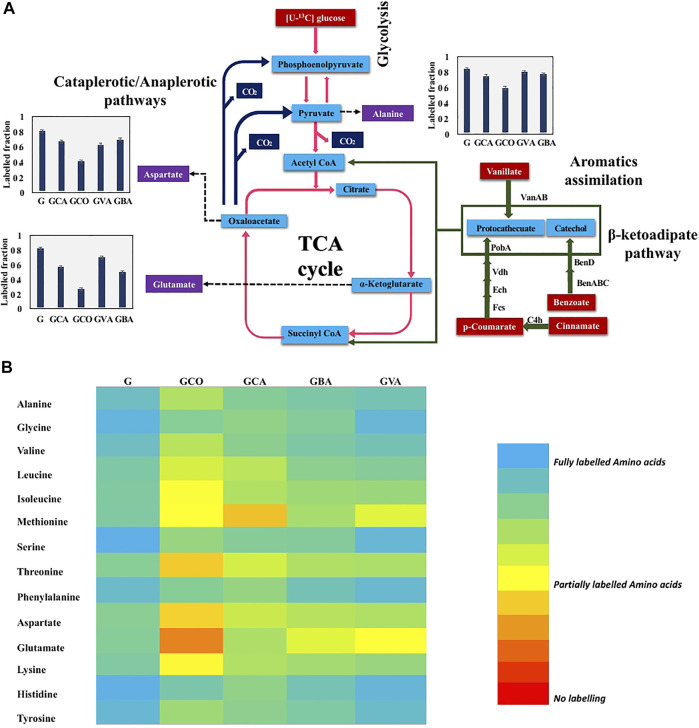
To further elucidate the aromatics metabolic pathways, 13C-fingerprinting was performed. Specifically, [U-13C] glucose was fed to C. glutamicum along with the following unlabeled aromatics: p-coumaric acid, cinnamic acid, benzoic acid, and cinnamic acid in four independent batch cultures. The 13C-labeling analysis revealed that in comparison to the cultures fed with only labeled glucose, the cultures that were fed with both labeled glucose and aromatics showed a reduction in the labeling fraction of the amino acids (Figure 5). Interestingly, a minor reduction in labeled fraction was observed for cultures that were fed with only [U-13C] glucose. This can be attributed to the unlabeled carbon that entered via CO2 through anaplerotic pathways (PEP → OAA, pyruvate→OAA) (Shirai et al., 2007). This in turn contributed to unlabeled carbon in amino acids derived from TCA cycle such as glutamate and aspartate. These results confirm that C. glutamicum is able to effectively metabolize the aromatics and incorporate the carbon from the aromatics into cell biomass components. Interestingly, amino acids synthesized from the TCA cycle showed the highest reduction in the labeled carbon fraction (Figure 5B). This finding confirms that the unlabeled carbon skeletons from aromatics entered the primary metabolism via succinyl-CoA and acetyl-CoA derived from the β-ketoadipate pathway, as shown in Figure 5A (Shen et al., 2012).
FIGURE 5.
13C-fingerprinting study for tracing aromatic assimilation. Fully labeled [U-13C] Glucose (5 g/L) in BTM 2 medium was used in culturing C. glutamicum strain along with 20 mM concentration of aromatics to study aromatic preference in C. glutamicum strain. (A) Lignin derived aromatics (p-coumaric acid, cinnamic acid, vanillic acid, and benzoic acid) assimilation determination by tracing reduction in labeled fraction in key amino acids (alanine, glutamic acid, and aspartic acid). Cinnamate to p-coumarate catalyzed by C4h (cinnamate 4-hydroxylase), p-coumarate to protocathecuate catalyzed by Fcs (feruloyl-CoA synthetase), Ech (Enoyl-CoA hydratase), Vdh (vanillin dehydrogenase), and PobA (p-hydroxybenzoate hydroxylase), vanillate to protocathecuate catalyzed by VanAB (vanillate monooxygenase), and benzoate to catechol catalyzed by BenABC (benzoate 1,2-dioxygenase), BenD (1,6-dihydroxycyclohexa-2,4-diene-1-carboxylate dehydrogenase). Labeled fraction in aspartate, alanine, and glutamate is shown in charts where G: [U-13C] Glucose (5 g/L) in BTM 2 medium batch; GCA: [U-13C] Glucose (5 g/L) + 20 mM cinnamic acid in BTM 2 medium batch; GCO: [U-13C] Glucose (5 g/L) + 20 mM p-coumaric acid in BTM 2 medium batch; GVA: [U-13C] Glucose (5 g/L) + 20 mM vanillic acid in BTM 2 medium batch; GBA: [U-13C] Glucose (5 g/L) + 20 mM benzoic acid in BTM 2 medium batch. The experiments were performed in biological triplicates. Data represents mean ± SD, n = 3. (B) Heatmap depicting variation in labeled fraction in different amino acids where Glucose only: [U-13C] Glucose (5 g/L) in BTM 2 medium batch; GCA: [U-13C] Glucose (5 g/L) + 20 mM cinnamic acid in BTM 2 medium batch; GCO: [U-13C] Glucose (5 g/L) + 20 mM p-coumaric acid in BTM 2 medium batch; GVA: [U-13C] Glucose (5 g/L) + 20 mM vanillic acid in BTM 2 medium batch; GBA: [U-13C] Glucose (5 g/L) + 20 mM benzoic acid in BTM 2 medium batch.
Ubiquitous reduction in labeled fraction was observed in all four aromatics used in the present study. Amongst the various aromatics that were tested, the highest reduction in carbon labelling was observed with p-coumaric acid followed by cinnamic acid, benzoic acid and least in vanillic acid (Figure 5B). Specifically, labeling of glutamic acid and aspartic acid significantly reduced by 67 and 48%, respectively. However, alanine labeling was reduced by only 24%, which is strong evidence that carbon is entering the TCA cycle without being converted into pyruvate directly. This also suggest that p-coumaric acid is the aromatic preferred by C. glutamicum, which agrees with the higher growth rate observed in p-coumaric acid fed cultures compared to the other aromatics (Figure 5D). Interestingly, the p-coumaric acid catabolic pathway involves a greater number of steps compared to the catabolic pathways of vanillic acid or benzoic acid. One potential explanation can be that C. glutamicum has highly active enzymes in the coumarate catabolic pathway, resulting in better uptake and breakdown of coumarate. Another possibility is that the coumarate catabolic pathway is upregulated under these conditions compared to the other aromatics. Furthermore, these results are congruent with the growth assay results reported in Figure 4, which showed highest growth in the presence of p-coumaric acid among all the other aromatics that were tested. These results can be leveraged for the further engineering of C. glutamicum as we aspire to increase the catabolism of all aromatics simultaneously for increased production of aromatics derived fuels and chemicals. The p-coumaric acid catabolic pathway is a three-step process leading to the formation of protocatechuate as an intermediate. Protocatechuate is also an intermediate for both vanillate and cinnamate catabolic pathways. In general, protocatechuate degradation occurs via three different pathways known as the ortho, meta, and para cleavage pathways. Meta and para cleavage pathways provide pyruvate as an intermediate prior to the carbon entering the TCA cycle. Alanine labelling in an organism can be used to confirm the presence of these pathways, since alanine is directly biosynthesized from pyruvate. From Figure 5, it can be observed that aspartate and glutamate had higher reduction in labelling as compared to alanine which demonstrates that carbon enters the TCA cycle without being diverted through pyruvate. Therefore, this work confirms that the ortho-cleavage protocatechuate pathway is active in C. glutamicum, in agreement with the previous genomic studies (Shen et al., 2012; Kallscheuer et al., 2016). Furthermore, the 10–15% reduction in labeling fraction of alanine indicate that cataplerotic pathways are also active in C. glutamicum.
C. glutamicum has long been used for the production of amino acids, including glutamic acid and lysine. The present study shows that glutamic acid is highly labeled compared to aspartic acid, both derived from TCA cycle intermediates. This trend was consistent in all of the labeling studies that were performed with the various aromatics. A likely explanation for this observation is the presence of anaplerotic pathways, through which labeled carbon from pyruvate and phosphoenolpyruvate enters oxaloacetate to produce aspartic acid (Varman et al., 2016). Also, as C. glutamicum is a natural producer of glutamic acid, the flux from α-ketoglutarate to glutamate may be more robust, thereby restricting the unlabeled carbon flow from α-ketoglutarate to oxaloacetate. Overall, the 13C analysis performed improves our understanding of aromatic catabolism in C. glutamicum, and provides strong evidence that aromatics present in lignocellulosic biomass hydrolysates can be used as carbon substrates for the bioproduction of L-lactate using this chassis.
Mixed-Acid Production From Corn Stover Hydrolysate
In the above sections, we have established fermentation of mixed acids in the presence of glucose as well as gained a better understanding of the aromatics metabolism in C. glutamicum. As a next step conducting fermentation with actual biomass hydrolysates containing diverse substrates will establish C. glutamicum as a omnivorous host for bioproduction. However, several of the previous studies have reported that C. glutamicum lacks the ability to catabolize xylose. On the other hand, whole genome sequencing has confirmed the presence of xylB that encodes for xylulokinase, but its function was never studied (Kawaguchi et al., 2006). This led us to wonder if the other genes of the xylose pathway are present in C. glutamicum but are not yet known. Therefore, to investigate the presence of xylose pathway in C. glutamicum 13C isotopomer analysis was performed by feeding the cells with [U-13C] xylose. We observed that there was no change in MDV levels for cells grown with labeled xylose in comparison to cells that were grown without labeled xylose (Supplementary Figure S3). The absence of labelling conclusively proves that C. glutamicum lacks the ability to metabolize xylose under the conditions tested in the laboratory. Therefore, it was essential to impart C. glutamicum the ability to grown on pentose sugars. Towards this, the genes xylA, xylB of xylose pathway, and the genes araA, araB, araD of arabinose pathway were transformed into the C. glutamicum ATCC 13032 strain resulting into the strain SSL09 (Schneider et al., 2010; Meiswinkel et al., 2013). Also, to evaluate the growth of C. glutamicum in the presence of biomass hydrolysates, the wild-type strain was grown in the presence of varying volume ratios of the hydrolysate. Biomass hydrolysates contain chemicals like furfurals and hydroxymethylfurfurals (HMF) which can be inhibitory to growth of bacterial cells (Tsuge et al., 2014; Chen et al., 2016; Kuhn et al., 2020). However, DMR hydrolysate has been processed to remove these inhibitors (Chen et al., 2016). C. glutamicum was found to be tolerant to the presence of aromatics and other unknown inhibitory chemicals present in corn stover biomass hydrolysates (DMR) (Chen et al., 2016), growing well even in the presence of 80% hydrolysates (Supplementary Figure S1). However, the maximum growth rate was observed in the presence of 20% (vol/vol) of biomass hydrolysate, therefore this hydrolysate concentration was chosen for further experiments.
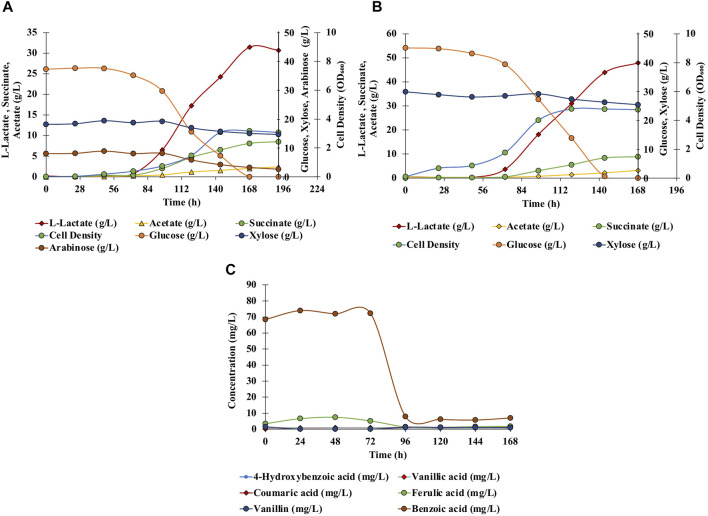
Anaerobic fermentation experiments were carried out using wild-type C. glutamicum and engineered SSL09 strains in BTM2 medium supplemented with glucose, xylose, and arabinose as carbon substrates. As seen in Figure 6A, complete utilization of glucose was observed in 168 h followed by utilization of arabinose and xylose. Delayed utilization of xylose and arabinose can be associated with the most common catabolite repression phenomenon observed in microorganisms in the presence of multiple carbon substrates (Dien et al., 2002; Abdel-Rahman et al., 2015; Flores et al., 2020). Although utilization of pentose sugars happened only after 90 h, other works have shown that through further engineering, C. glutamicum can be made to utilize both pentose and hexose sugars simultaneously (Sasaki et al., 2016). Lactate (31.5 g/L) along with acetate (2 g/L) and succinate (8.1 g/L) were produced at 168 h. Figure 6B shows the fermentation profile using DMR corn stover hydrolysate as a fermentation medium. Lactate 47.9 g/L along with acetate 3.2 g/L and succinate 8.9 g/L were produced with complete utilization of glucose in 144 h. Utilization of xylose (17%), arabinose (60%), glucose (100%) lead to production of acetate (2 g/L), L-lactate (31.5 g/L) and succinate (8.1 g/L) in SSL09 strain this shows active utilization of pentose and hexose sugar utilization for production of mixed acids. At the end of the fermentation time course, the yield of lactate from sugars available in DMR hydrolysate was 0.639 g/g of the total, and productivity was 0.285 g/L/h. Furthermore, C. glutamicum showed good growth in DMR hydrolysate and lead to reduction in aromatics levels (Figure 6C). In this fermentation trial, it was observed that the strain completely consumed the aromatics ferulic acid, vanillin, and benzoic acid which were present in DMR hydrolysate (Figure 6C). Although, we have demonstrated the production of mixed acids from 20% biomass hydrolysate, using high biomass concentration as feedstock is preferred from an industrial standpoint. However, the DMR hydrolysate used in this study contains very high total sugar concentration, 230 g/L (Chen et al., 2016). Growing C. glutamicum at high sugar concentrations would require adaptive laboratory evolution of the strain. Therefore, future works should focus on utilizing adaptive laboratory evolution to improve the tolerance of C. glutamicum at high sugar concentrations and further optimization of bioprocess conditions to grow the cells at very high hydrolysate levels. Overall, this work demonstrates the first application of C. glutamicum to utilize all the diverse carbon substrates present in lignocellulosic biomass for bioproduction of value-added chemicals.
FIGURE 6.
Effect of different fermentation cultivation parameters on mixed-acids production by C. glutamicum. Data is shown for single fed batch fermentation. (A) Anaerobic, batch, pH 7, BTM2 supplemented with glucose, xylose, and arabinose; (B) Anaerobic, batch, pH 7, DMR hydrolysate; (C) Aromatics utilization: anaerobic, batch, pH 7, DMR hydrolysate.
conclusion
The omnivorous strain C. glutamicum ATCC 13032 was investigated for production of mixed acids from sugars and aromatics present in lignocellulosic biomass. Fermentative processes in a variety of cultivation modes, including an aerobic cell growth and anaerobic fermentation phase, batch, and fed-batch mode, using BTM2 medium with different carbon substrates, and DMR corn stover hydrolysate were evaluated for production of mixed acids from pure sugars as well as biomass hydrolysate. In all cultivation modes, L-lactate was produced in significantly high titers with acetate and succinate as co-products. C. glutamicum exhibited a maximum productivity of 0.322 g/L/h towards L-lactate with 54.1 g/L when grown aerobically for the first 16 h followed by anaerobic fermentation. Also, utilization of pentose and hexose sugars as well as aromatics present in corn stover hydrolysate for mixed acid production was demonstrated. Furthermore, C. glutamicum showed high tolerance and utilization of different aromatics commonly present in lignocellulosic hydrolysate, including benzoic acid, cinnamic acid, vanillic acid, and p-coumaric acid. 13C-fingerprinting confirmed the roles of the ortho-cleavage pathway, anaplerotic pathway, and cataplerotic pathways for utilization of aromatics in the TCA cycle, with p-coumaric acid as the preferred substrate amongst aromatics studied. These findings suggest C. glutamicum is a versatile omnivorous host for valorization of lignocellulosic feedstocks.
Acknowledgments
We thank Prof. Wendisch for generously donating arabinose and xylose utilization plasmids. Samples of DMR hydrolysate were provided by Richard Elender of the National Renewable Energy Laboratory through collaboration with the DOE Agile BioFoundry.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AV, RD, AM, SS, and AMJ conceived and designed this study. AM, SS, AMJ performed growth assays, aromatic assimilation, labelling studies, amino acid analysis and hydrolysate fermentation studies. ZW and ABJ assisted in wet lab experiments involving C. glutamicum cultures as well as glucose, xylose and arabinose fermentations. AV, RD, AM, SS, AG, JG, AJ performed data analysis, wrote and revised the manuscript.
Funding
AMV acknowledges start-up funds from the School for Engineering of Matter, Transport and Energy at Arizona State University, and the awards #2184871 and #1871463 from Sandia National Laboratories. This research was also supported in parts by the BioEnergy Technology Office (BETO), United States Department of Energy, through support for RWD, under agreement #26336.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.827386/full#supplementary-material
References
- Abdel-Rahman M. A., Hassan S. E., Alrefaey H. M. A., Elsakhawy T. (2021). Efficient Co-utilization of Biomass-Derived Mixed Sugars for Lactic Acid Production by Bacillus Coagulans Azu-10. Fermentation 7, 7010028. 10.3390/fermentation7010028 [DOI] [Google Scholar]
- Abdel-Rahman M. A., Xiao Y., Tashiro Y., Wang Y., Zendo T., Sakai K., et al. (2015). Fed-batch Fermentation for Enhanced Lactic Acid Production from Glucose/xylose Mixture without Carbon Catabolite Repression. J. Biosci. Bioeng. 119, 153–158. 10.1016/j.jbiosc.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Adeboye P. T., Bettiga M., Olsson L. (2014). The Chemical Nature of Phenolic Compounds Determines Their Toxicity and Induces Distinct Physiological Responses in Saccharomyces cerevisiae in Lignocellulose Hydrolysates. AMB Expr. 4, 46. 10.1186/s13568-014-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almqvist H., Veras H., Li K., Garcia Hidalgo J., Hulteberg C., Gorwa-Grauslund M., et al. (2021). Muconic Acid Production Using Engineered Pseudomonas Putida KT2440 and a Guaiacol-Rich Fraction Derived from Kraft Lignin. ACS Sustain. Chem. Eng. 9, 8097–8106. 10.1021/acssuschemeng.1c00933 [DOI] [Google Scholar]
- Bruijnincx P. C. A., Weckhuysen B. M. (2014). Lignin up for Break-Down. Nat. Chem 6, 1035–1036. 10.1038/nchem.2120 [DOI] [PubMed] [Google Scholar]
- Casalini T., Rossi F., Castrovinci A., Perale G. (2019). A Perspective on Polylactic Acid-Based Polymers Use for Nanoparticles Synthesis and Applications. Front. Bioeng. Biotechnol. 7, 259. 10.3389/fbioe.2019.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai X., Shang X., Zhang Y., Liu S., Liang Y., Zhang Y., et al. (2016). A Novel Pyruvate Kinase and its Application in Lactic Acid Production under Oxygen Deprivation in Corynebacterium Glutamicum. BMC Biotechnol. 16, 79. 10.1186/s12896-016-0313-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-P., Chow M., Liu C.-C., Lau A., Liu J., Eltis L. D. (2012). Vanillin Catabolism in Rhodococcus Jostii RHA1. Appl. Environ. Microbiol. 78, 586–588. 10.1128/aem.06876-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Kuhn E., Jennings E. W., Nelson R., Tao L., Zhang M., et al. (2016). DMR (Deacetylation and Mechanical Refining) Processing of Corn stover Achieves High Monomeric Sugar Concentrations (230 G L−1) during Enzymatic Hydrolysis and High Ethanol Concentrations (>10% V/v) during Fermentation without Hydrolysate Purification or Concentration. Energ. Environ. Sci. 9, 1237–1245. [Google Scholar]
- Dien B. S., Nichols N. N., Bothast R. J. (2002). Fermentation of Sugar Mixtures Using Escherichia coli Catabolite Repression Mutants Engineered for Production of L -lactic Acid. J. Ind. Microbiol. Biotechnol. 29, 221–227. 10.1038/sj.jim.7000299 [DOI] [PubMed] [Google Scholar]
- Ding W., Si M., Zhang W., Zhang Y., Chen C., Zhang L., et al. (2015). Functional Characterization of a Vanillin Dehydrogenase in Corynebacterium Glutamicum. Sci. Rep. 5, 8044. 10.1038/srep08044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling L., Bott M. (2005). Handbook of Corynebacterium Glutamicum. Boca Raton, Florida, USA: CRC Press. [Google Scholar]
- Fitzgerald D. J., Stratford M., Gasson M. J., Ueckert J., Bos A., Narbad A. (2004). Mode of Antimicrobial Action of Vanillin against Escherichia coli, Lactobacillus Plantarum and Listeria Innocua. J. Appl. Microbiol. 97, 104–113. 10.1111/j.1365-2672.2004.02275.x [DOI] [PubMed] [Google Scholar]
- Flores A. D., Choi H. G., Martinez R., Onyeabor M., Ayla E. Z., Godar A., et al. (2020). Catabolic Division of Labor Enhances Production of D-Lactate and Succinate from Glucose-Xylose Mixtures in Engineered Escherichia coli Co-culture Systems. Front. Bioeng. Biotechnol. 8, 329. 10.3389/fbioe.2020.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores A. D., Holland S. C., Mhatre A., Sarnaik A. P., Godar A., Onyeabor M., et al. (2021). A Coculture-Coproduction System Designed for Enhanced Carbon Conservation through Inter-strain CO2 Recycling. Metab. Eng. 67, 387–395. 10.1016/j.ymben.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Follmann M., Ochrombel I., Krämer R., Trötschel C., Poetsch A., Rückert C., et al. (2009). Functional Genomics of pH Homeostasis in Corynebacterium Glutamicum Revealed Novel Links between pH Response, Oxidative Stress, Iron Homeostasis and Methionine Synthesis. BMC Genomics 10, 621. 10.1186/1471-2164-10-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gírio F. M., Fonseca C., Carvalheiro F., Duarte L. C., Marques S., Bogel-Łukasik R. (2010). Hemicelluloses for Fuel Ethanol: A Review. Bioresour. Technol. 101, 4775–4800. 10.1016/j.biortech.2010.01.088 [DOI] [PubMed] [Google Scholar]
- Gomes E. D., Rodrigues A. E. (2019). Lignin Biorefinery: Separation of Vanillin, Vanillic Acid and Acetovanillone by Adsorption. Separat. Purif. Technol. 216, 92–101. 10.1016/j.seppur.2019.01.071 [DOI] [Google Scholar]
- Ikeda M., Nakagawa S. (2003). The Corynebacterium Glutamicum Genome: Features and Impacts on Biotechnological Processes. Appl. Microbiol. Biotechnol. 62, 99–109. 10.1007/s00253-003-1328-1 [DOI] [PubMed] [Google Scholar]
- Inui M., Murakami S., Okino S., Kawaguchi H., Vertès A. A., Yukawa H. (2004). Metabolic Analysis of Corynebacterium Glutamicum during Lactate and Succinate Productions under Oxygen Deprivation Conditions. J. Mol. Microbiol. Biotechnol. 7, 182–196. 10.1159/000079827 [DOI] [PubMed] [Google Scholar]
- Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., et al. (2003). The Complete Corynebacterium Glutamicum ATCC 13032 Genome Sequence and its Impact on the Production of L-Aspartate-Derived Amino Acids and Vitamins. J. Biotechnol. 104, 5–25. 10.1016/s0168-1656(03)00154-8 [DOI] [PubMed] [Google Scholar]
- Kallscheuer N., Vogt M., Kappelmann J., Krumbach K., Noack S., Bott M., et al. (2016). Identification of the Phd Gene Cluster Responsible for Phenylpropanoid Utilization in Corynebacterium Glutamicum. Appl. Microbiol. Biotechnol. 100, 1871. 10.1007/s00253-015-7165-1 [DOI] [PubMed] [Google Scholar]
- Kawaguchi H., Sasaki M., Vertès A. A., Inui M., Yukawa H. (2008). Engineering of an L-Arabinose Metabolic Pathway in Corynebacterium Glutamicum. Appl. Microbiol. Biotechnol. 77, 1053–1062. 10.1007/s00253-007-1244-x [DOI] [PubMed] [Google Scholar]
- Kawaguchi H., Vertès A. A., Okino S., Inui M., Yukawa H. (2006). Engineering of a Xylose Metabolic Pathway in Corynebacterium Glutamicum. Appl. Environ. Microbiol. 72, 3418–3428. 10.1128/aem.72.5.3418-3428.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstedt M., Starck S., Barton N., Stolzenberger J., Selzer M., Mehlmann K., et al. (2018). From Lignin to Nylon: Cascaded Chemical and Biochemical Conversion Using Metabolically Engineered Pseudomonas Putida. Metab. Eng. 47, 279–293. 10.1016/j.ymben.2018.03.003 [DOI] [PubMed] [Google Scholar]
- Kuhn E. M., Chen X., Tucker M. P. (2020). Deacetylation and Mechanical Refining (DMR) and Deacetylation and Dilute Acid (DDA) Pretreatment of Corn Stover, Switchgrass, and a 50:50 Corn Stover/Switchgrass Blend. ACS Sustain. Chem. Eng. 8, 6734–6743. 10.1021/acssuschemeng.0c00894 [DOI] [Google Scholar]
- Lee S. C., Kim H. C. (2011). Batch and Continuous Separation of Acetic Acid from Succinic Acid in a Feed Solution with High Concentrations of Carboxylic Acids by Emulsion Liquid Membranes. J. Membr. Sci. 367, 190–196. 10.1016/j.memsci.2010.10.057 [DOI] [Google Scholar]
- Linger J. G., Vardon D. R., Guarnieri M. T., Karp E. M., Hunsinger G. B., Franden M. A., et al. (2014). Lignin Valorization through Integrated Biological Funneling and Chemical Catalysis. Proc. Natl. Acad. Sci. U.S.A. 111, 12013–12018. 10.1073/pnas.1410657111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Wu W., Tran-Gyamfi M. B., Jaryenneh J. D., Zhuang X., Davis R. W. (2017). Bioconversion of Distillers' Grains Hydrolysates to Advanced Biofuels by an Escherichia coli Co-culture. Microb. Cel Fact 16, 192. 10.1186/s12934-017-0804-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gao W., Zhao X., Wang J., Garza E., Manow R., et al. (2014). Pilot Scale Demonstration of D -lactic Acid Fermentation Facilitated by Ca(OH) 2 Using a Metabolically Engineered Escherichia coli . Bioresour. Technol. 169, 559–565. 10.1016/j.biortech.2014.06.056 [DOI] [PubMed] [Google Scholar]
- Lv Y., Wu Z., Han S., Lin Y., Zheng S. (2011). Genome Sequence of Corynebacterium Glutamicum S9114, a Strain for Industrial Production of Glutamate. J. Bacteriol. 193, 6096–6097. 10.1128/jb.06074-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiswinkel T. M., Gopinath V., Lindner S. N., Nampoothiri K. M., Wendisch V. F. (2013). Accelerated Pentose Utilization byCorynebacterium Glutamicumfor Accelerated Production of Lysine, Glutamate, Ornithine and Putrescine. Microb. Biotechnol. 6, 131–140. 10.1111/1751-7915.12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Wang B., Liu D., Chen T., Wang Z., Zhao X. (2016). High-yield Anaerobic Succinate Production by Strategically Regulating Multiple Metabolic Pathways Based on Stoichiometric Maximum in Escherichia coli . Microb. Cel Fact 15, 141. 10.1186/s12934-016-0536-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S., Ugwu C. U., Tokiwa Y. (2012). Efficient Production of D-(−)-Lactic Acid from Broken rice by Lactobacillus Delbrueckii Using Ca(OH)2 as a Neutralizing Agent. Bioresour. Technol. 104, 791–794. 10.1016/j.biortech.2011.10.017 [DOI] [PubMed] [Google Scholar]
- Nam H.-G., Lim G.-W., Mun S. (2012). Separation of Acetic Acid, Formic Acid, Succinic Acid, and Lactic Acid Using Adsorbent Resin. J. Chem. Eng. Data 57, 2102–2108. 10.1021/je201065u [DOI] [Google Scholar]
- Ohgren K., Bura R., Saddler J., Zacchi G. (2007). Effect of Hemicellulose and Lignin Removal on Enzymatic Hydrolysis of Steam Pretreated Corn stover. Bioresour. Technol. 98, 2503–2510. 10.1016/j.biortech.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Okino S., Suda M., Fujikura K., Inui M., Yukawa H. (2008). Production of D-Lactic Acid by Corynebacterium Glutamicum under Oxygen Deprivation. Appl. Microbiol. Biotechnol. 78, 449–454. 10.1007/s00253-007-1336-7 [DOI] [PubMed] [Google Scholar]
- Papagianni M. (2012). Metabolic Engineering of Lactic Acid Bacteria for the Production of Industrially Important Compounds. Comput. Struct. Biotechnol. J. 3, e201210003. 10.5936/csbj.201210003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi K., García-Hidalgo J., Gorwa-Grauslund M. F., Lidén G. (2017). Conversion of Lignin Model Compounds by Pseudomonas Putida KT2440 and Isolates from Compost. Appl. Microbiol. Biotechnol. 101, 5059. 10.1007/s00253-017-8211-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Salvachúa D., Katahira R., Black B. A., Cleveland N. S., Reed M., et al. (2017). Base-Catalyzed Depolymerization of Solid Lignin-Rich Streams Enables Microbial Conversion. ACS Sustain. Chem. Eng. 5, 8171–8180. 10.1021/acssuschemeng.7b01818 [DOI] [Google Scholar]
- Rouabhia M., Asselin J., Tazi N., Messaddeq Y., Levinson D., Zhang Z. (2014). Production of Biocompatible and Antimicrobial Bacterial Cellulose Polymers Functionalized by RGDC Grafting Groups and Gentamicin. ACS Appl. Mater. Inter. 6, 1439–1446. 10.1021/am4027983 [DOI] [PubMed] [Google Scholar]
- Saha B. C. (2003). Hemicellulose Bioconversion. J. Ind. Microbiol. Biotechnol. 30, 279–291. 10.1007/s10295-003-0049-x [DOI] [PubMed] [Google Scholar]
- Sakai S., Tsuchida Y., Okino S., Ichihashi O., Kawaguchi H., Watanabe T., et al. (2007). Effect of Lignocellulose-Derived Inhibitors on Growth of and Ethanol Production by Growth-Arrested Corynebacterium Glutamicum R. Appl. Environ. Microbiol. 73, 2349–2353. 10.1128/aem.02880-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvachúa D., Rydzak T., Auwae R., De Capite A., Black B. A., Bouvier J. T., et al. (2020). Metabolic Engineering of Pseudomonas Putida for Increased Polyhydroxyalkanoate Production from Lignin. Microb. Biotechnol. 13, 813–298. 10.1111/1751-7915.13547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvachúa D., Karp E. M., Nimlos C. T., Vardon D. R., Beckham G. T. (2015). Towards Lignin Consolidated Bioprocessing: Simultaneous Lignin Depolymerization and Product Generation by Bacteria. Green. Chem. 17, 4951–4967. 10.1039/c5gc01165e [DOI] [Google Scholar]
- Sambrook J., Russell D. (2001). Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Sasaki M., Jojima T., Kawaguchi H., Inui M., Yukawa H. (2016). Engineering of Pentose Transport in Corynebacterium Glutamicum to Improve Simultaneous Utilization of Mixed Sugars. Appl. Microbiol. Biotechnol. 85, 105. 10.1007/s00253-009-2065-x [DOI] [PubMed] [Google Scholar]
- Schneider J., Niermann K., Wendisch V. F. (2010). Production of the Amino Acids L-Glutamate, L-Lysine, L-Ornithine and L-Arginine from Arabinose by Recombinant Corynebacterium Glutamicum. J. Biotechnol. 154, 191. 10.1016/j.jbiotec.2010.07.009 [DOI] [PubMed] [Google Scholar]
- Shen X.-H., Huang Y., Liu S.-J. (2005). Genomic Analysis and Identification of Catabolic Pathways for Aromatic Compounds in Corynebacterium Glutamicum. Microb. Environ. 20, 160–167. 10.1264/jsme2.20.160 [DOI] [Google Scholar]
- Shen X.-H., Zhou N.-Y., Liu S.-J. (2012). Degradation and Assimilation of Aromatic Compounds by Corynebacterium Glutamicum: Another Potential for Applications for This Bacterium? Appl. Microbiol. Biotechnol. 95, 77–89. 10.1007/s00253-012-4139-4 [DOI] [PubMed] [Google Scholar]
- Shirai T., Fujimura K., Furusawa C., Nagahisa K., Shioya S., Shimizu H. (2007). Study on Roles of Anaplerotic Pathways in Glutamate Overproduction of Corynebacterium Glutamicum by Metabolic Flux Analysis. Microb. Cel Fact 6, 19. 10.1186/1475-2859-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert C., Nieves L. M., Panyon L. A., Loeffler T., Morris C., Cartwright R. A., et al. (2017). Experimental Evolution Reveals an Effective Avenue to Release Catabolite Repression via Mutations in XylR. Proc. Natl. Acad. Sci. USA 114, 7349–7354. 10.1073/pnas.1700345114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons B. A., Loque D., Blanch H. W. (2008). Next-generation Biomass Feedstocks for Biofuel Production. Genome Biol. 9, 242. 10.1186/gb-2008-9-12-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socha A. M., Parthasarathi R., Shi J., Pattathil S., Whyte D., Bergeron M., et al. (2014). Efficient Biomass Pretreatment Using Ionic Liquids Derived from Lignin and Hemicellulose. Proc. Natl. Acad. Sci. U S A. 111, E3587–E3595. 10.1073/pnas.1405685111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence E. M., Calvo-Bado L., Mines P., Bugg T. D. H. (2021). Metabolic Engineering of Rhodococcus Jostii RHA1 for Production of Pyridine-Dicarboxylic Acids from Lignin. Microb. Cel Fact 20, 15. 10.1186/s12934-020-01504-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. J., Shui W., Myers S., Feng X., Bertozzi C., Keasling J. D. (2009). Central Metabolism in Mycobacterium Smegmatis during the Transition from O2-Rich to O2-Poor Conditions as Studied by Isotopomer-Assisted Metabolite Analysis. Biotechnol. Lett. 31, 1233–1240. 10.1007/s10529-009-9991-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker C., Martínez I., San K.-Y., Bennett G. N. (2012). Succinate Production in Escherichia coli . Biotechnol. J. 7, 213–224. 10.1002/biot.201100061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge Y., Hasunuma T., Kondo A. (2015). Recent Advances in the Metabolic Engineering of Corynebacterium Glutamicum for the Production of Lactate and Succinate from Renewable Resources. J. Ind. Microbiol. Biotechnol. 42, 375–389. 10.1007/s10295-014-1538-9 [DOI] [PubMed] [Google Scholar]
- Tsuge Y., Hori Y., Kudou M., Ishii J., Hasunuma T., Kondo A. (2014). Detoxification of Furfural in Corynebacterium Glutamicum under Aerobic and Anaerobic Conditions. Appl. Microbiol. Biotechnol. 98, 8675. 10.1007/s00253-014-5924-z [DOI] [PubMed] [Google Scholar]
- Tsuge Y., Kato N., Yamamoto S., Suda M., Inui M. (2019a). Enhanced Production of D-Lactate from Mixed Sugars in Corynebacterium Glutamicum by Overexpression of Glycolytic Genes Encoding Phosphofructokinase and Triosephosphate Isomerase. J. Biosci. Bioeng. 127, 288–293. 10.1016/j.jbiosc.2018.08.002 [DOI] [PubMed] [Google Scholar]
- Tsuge Y., Kato N., Yamamoto S., Suda M., Jojima T., Inui M. a.-O. (2019b). Metabolic Engineering of Corynebacterium Glutamicum for Hyperproduction of Polymer-Grade L- and D-Lactic Acid. Appl. Microbiol. Biotechnol. 103, 3381. 10.1007/s00253-019-09737-8 [DOI] [PubMed] [Google Scholar]
- Tuck C. O., Pérez E., Horváth I. T., Sheldon R. A., Poliakoff M. (2012). Valorization of Biomass: Deriving More Value from Waste. Science 337, 695–699. 10.1126/science.1218930 [DOI] [PubMed] [Google Scholar]
- Upadhyaya B. P., Deveaux L. C., Christopher L. P. (2014). Metabolic Engineering as a Tool for Enhanced Lactic Acid Production. Trends Biotechnol. 32, 637. 10.1016/j.tibtech.2014.10.005 [DOI] [PubMed] [Google Scholar]
- Van Der Rest M. E., Lange C., Molenaar D. (1999). A Heat Shock Following Electroporation Induces Highly Efficient Transformation of Corynebacterium Glutamicum with Xenogeneic Plasmid DNA. Appl. Microbiol. Biotechnol. 52, 541–545. 10.1007/s002530051557 [DOI] [PubMed] [Google Scholar]
- Van Gerven T., Stankiewicz A. (2009). Structure, Energy, Synergy, Time-The Fundamentals of Process Intensification. Ind. Eng. Chem. Res. 48, 2465–2474. 10.1021/ie801501y [DOI] [Google Scholar]
- Varman A. M., Yu Y., You L., Tang Y. J. (2013). Photoautotrophic Production of D-Lactic Acid in an Engineered Cyanobacterium. Microb. Cel Fact 12, 117–118. 10.1186/1475-2859-12-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varman A. M., Follenfant R., Liu F., Davis R. W., Lin Y. K., Singh S. (2018). Hybrid Phenolic-Inducible Promoters towards Construction of Self-Inducible Systems for Microbial Lignin Valorization. Biotechnol. Biofuels 11, 182. 10.1186/s13068-018-1179-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varman A. M., He L., Follenfant R., Wu W., Wemmer S., Wrobel S. A., et al. (2016). Decoding How a Soil Bacterium Extracts Building Blocks and Metabolic Energy from Ligninolysis Provides Road Map for Lignin Valorization. Proc. Natl. Acad. Sci. USA 113, E5802–E5811. 10.1073/pnas.1606043113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varman A. M., He L., You L., Hollinshead W., Tang Y. J. (2014). Elucidation of Intrinsic Biosynthesis Yields Using 13C-Based Metabolism Analysis. Microb. Cel Fact 13, 42. 10.1186/1475-2859-13-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl S. A., Dauner M., Wiechert W. (2004). New Tools for Mass Isotopomer Data Evaluation in13C Flux Analysis: Mass Isotope Correction, Data Consistency Checking, and Precursor Relationships. Biotechnol. Bioeng. 85, 259–268. 10.1002/bit.10909 [DOI] [PubMed] [Google Scholar]
- Wang P. (2007). “Nonconventional Biocatalysis for Production of Chemicals and Polymers from Biomass,” in Bioprocessing for Value-Added Products from Renewable Resources. Editor Yang S.-T. (Amsterdam: Elsevier; ), 325–350. 10.1016/b978-044452114-9/50013-x [DOI] [Google Scholar]
- Wieschalka S., Blombach B., Eikmanns B. J. (2012). Engineering Corynebacterium Glutamicum for the Production of Pyruvate. Appl. Microbiol. Biotechnol. 94, 449–459. 10.1007/s00253-011-3843-9 [DOI] [PubMed] [Google Scholar]
- Wu M., Zhao X., Shen Y., Shi Z., Li G., Ma T. (2021). Efficient Simultaneous Utilization of Glucose and Xylose from Corn Straw by Sphingomonas Sanxanigenens NX02 to Produce Microbial Exopolysaccharide. Bioresour. Technol. 319, 124126. 10.1016/j.biortech.2020.124126 [DOI] [PubMed] [Google Scholar]
- Wu W., Dutta T., Varman A. M., Eudes A., Manalansan B., Loqué D., et al. (2017). Lignin Valorization: Two Hybrid Biochemical Routes for the Conversion of Polymeric Lignin into Value-Added Chemicals. Sci. Rep. 7, 8420. 10.1038/s41598-017-07895-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q., Chen Z., Shao Y., Gong X., Wang H., Liu X., et al. (2016). Direct Hydrodeoxygenation of Raw Woody Biomass into Liquid Alkanes. Nat. Commun. 7, 11162. 10.1038/ncomms11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Feng X., Varman A. M., He L., Yu H., Tang Y. J. (2012). Kinetic Modeling and Isotopic Investigation of Isobutanol Fermentation by Two Engineered escherichia Coli Strains. Ind. Eng. Chem. Res. 51, 15855–15863. 10.1021/ie202936t [DOI] [Google Scholar]
- Xiao Y., Ruan Z., Liu Z., Wu S. G., Varman A. M., Liu Y., et al. (2013). Engineering Escherichia coli to Convert Acetic Acid to Free Fatty Acids. Biochem. Eng. J. 76, 60–69. 10.1016/j.bej.2013.04.013 [DOI] [Google Scholar]
- You L., Page L., Feng X., Berla B., Pakrasi H. B., Tang Y. J. (2012). Metabolic Pathway Confirmation and Discovery through 13C-Labeling of Proteinogenic Amino Acids. J. Vis. Exp. 59, e3583. 10.3791/3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa H., Omumasaba C. A., Nonaka H., Kós P., Okai N., Suzuki N., et al. (2007). Comparative Analysis of the Corynebacterium Glutamicum Group and Complete Genome Sequence of Strain R. Microbiology 153, 1042–1058. 10.1099/mic.0.2006/003657-0 [DOI] [PubMed] [Google Scholar]
- Zan X., Sun J., Chu L., Cui F., Huo S., Song Y., et al. (2021). Improved Glucose and Xylose Co-utilization by Overexpression of Xylose Isomerase And/or Xylulokinase Genes in Oleaginous Fungus Mucor Circinelloides. Appl. Microbiol. Biotechnol. 105, 5565–5575. 10.1007/s00253-021-11392-x [DOI] [PubMed] [Google Scholar]
- Zheng Z., Chen T., Zhao M., Wang Z., Zhao X. (2012). Engineering Escherichia coli for Succinate Production from Hemicellulose via Consolidated Bioprocessing. Microb. Cel Fact 11, 37–11. 10.1186/1475-2859-11-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C.-H., Beltramini J. N., Fan Y.-X., Lu G. Q. (2008). Chemoselective Catalytic Conversion of Glycerol as a Biorenewable Source to Valuable Commodity Chemicals. Chem. Soc. Rev. 37, 527–549. 10.1039/b707343g [DOI] [PubMed] [Google Scholar]
- Zhou C.-H., Xia X., Lin C.-X., Tong D.-S., Beltramini J. (2011). Catalytic Conversion of Lignocellulosic Biomass to fine Chemicals and Fuels. Chem. Soc. Rev. 40, 5588–5617. 10.1039/c1cs15124j [DOI] [PubMed] [Google Scholar]
- Zhu Y., Eiteman M. A., Dewitt K., Altman E. (2007). Homolactate Fermentation by Metabolically Engineered Escherichia coli Strains. Appl. Environ. Microbiol. 73, 456–464. 10.1128/aem.02022-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.