Abstract
Kefir grains consist of complex symbiotic mixtures of bacteria and yeasts, and are reported to impart numerous health-boosting properties to milk and water kefir beverages. The objective of this work was to investigate the microbial communities in kefir grains, and explore the possibility of deriving useful probiotic strains from them. A total of 158 microbial strains, representing six fungal and 17 bacterial species, were isolated from milk and water kefir grains collected from a Singapore-based homebrewer. Based on 16S rRNA sequencing, isolated genera included Lactobacillus, Liquorilactobacillus, Lacticaseibacillus, Lentilactobacillus, Leuconostoc, Lactococcus, Acetobacter, Gluconobacter, Oenococcus, Clostridium, Zymomonas, Saccharomyces, Kluyveromyces, Pichia, Lachancea, Candida, and Brettanomyces. To characterize these isolates, a funnel approach, involving numerous phenotypic and genomic screening assays, was applied to identify kefir-derived microbial strains with the highest probiotic potential. Particular focus was placed on examining the pathogen inhibitory properties of kefir isolates toward enteric pathogens which pose a considerable global health burden. Enteric pathogens tested include species of Bacillus, Salmonella, Vibrio, Clostridium, Klebsiella, Escherichia, and Staphylococcus. Well diffusion assays were conducted to determine the propensity of kefir isolates to inhibit growth of enteric pathogens, and a competitive adhesion/exclusion assay was used to determine the ability of kefir isolates to out-compete or exclude attachment of enteric pathogens to Caco-2 cells. Seven bacterial strains of Lentilactobacillus hilgardii, Lacticaseibacillus paracasei, Liquorilactobacillus satsumensis, Lactobacillus helveticus, and Lentilactobacillus kefiri, were ultimately identified as potential probiotics, and combined to form a “kefir probiotics blend.” Desirable probiotic characteristics, including good survival in acid and bile environments, bile salt hydrolase activity, antioxidant activity, non-cytotoxicity and high adhesion to Caco-2 cells, and a lack of virulence or antimicrobial resistance genes. In addition, vitamin and γ-aminobutyric acid (GABA) synthesis genes, were identified in these kefir isolates. Overall, probiotic candidates derived in this study are well-characterized strains with a good safety profile which can serve as novel agents to combat enteric diseases. These kefir-derived probiotics also add diversity to the existing repertoire of probiotic strains, and may provide consumers with alternative product formats to attain the health benefits of kefir.
Keywords: kefir, probiotics, screening, pathogen inhibition, competitive, sequencing, safety, Singapore
Introduction
The human body is an ecosystem which is home to trillions of microbes. In the recent two decades, international collaborative projects such as the Human Microbiome Project (HMP) (Turnbaugh et al., 2007) and METAgenomics of the Human Intestinal Tract (MetaHIT) (Qin et al., 2010), have brought to light the intricate interactions between the human microbiome and our human health. Dysbiosis of the human gut microbiota, which indicates a state where microbiota richness and diversity is significantly deviated from a healthy control state, has been correlated with numerous gut diseases, and chronic health conditions such as obesity, type II diabetes, immune diseases and neurological disorders (Shreiner et al., 2015). The idea that perturbations of the human microbiome may cause disease sparked considerable interest and the question posed: Could there be healthy microbes supplemented to the human body that reverse the disease state—i.e., probiotics? Because of the promising health benefits of probiotics, interest in these “healthy microbes” has risen rapidly in recent years, with a market estimate projecting a compound annual growth rate (CAGR) of 7% through 2028 (Grand View Research, 2022). Existing commercial probiotic bacterial strains are mostly Lactobacilli,1 Streptococcus, Bacillus, Bifidobacterium, or Saccharomyces, and have been sourced from the native microbiota in human feces, human breast milk as well as from fermented food sources.
One crucial mechanism by which probiotics elicit health benefits is through inhibition of enteric bacterial pathogens. Enteric pathogens are disease-causing microorganisms which infect the gastrointestinal tract, typically leading to symptoms of diarrhea, nausea, abdominal pain, fever or in severe cases, death. Enteric bacterial pathogen-associated diseases are a significant global health burden; in 2010, based on a World Health Organization (WHO) report, enteric bacterial pathogens accounted for 350 million cases of illnesses, 187 thousand deaths and the loss of 5.7 million healthy years of life worldwide (WHO, 2022). Currently, antibiotics are employed as first-line therapy for enteric bacterial infections. However, there is a need to reduce antibiotics usage because of rising instances of antimicrobial resistance (AMR) and the spread of multi-drug resistant organisms. Such concerns have led to a 2019 report by the Centers for Disease Control and Prevention (CDC) declaring that humankind has entered the “post-antibiotic” era (Centers for Disease Control Prevention, 2019), calling for alternative solutions to target enteric bacterial infections. As such, probiotics are now being studied in greater detail for their potential to inhibit and treat enteric bacterial infections.
Kefir is an ancient fermented beverage well-known for its purported health-boosting properties. Originating from the Caucasus mountains, kefir is traditionally produced from kefir grains, which are three-dimensional cauliflower-like granules comprising a symbiotic mixture of bacteria, yeasts, and their extracellular polymeric substances. There are two types of kefir—milk kefir and water kefir, each of them being fermented by placing respective grains in milk or sugary water and incubating at ambient temperature. Bacterial and fungal genera which have been previously identified in kefir include lactobacilli, Lactococcus, Streptococcus, Leuconostoc, Acetobacter, Saccharomyces, Kluyveromyces, and Candida (Bourrie et al., 2016; Moretti et al., 2022). The microbial composition of milk and kefir grains are reported to differ slightly (Fiorda et al., 2017). Numerous scientific and clinical studies have taken closer examination of kefir’s nutritional and medicinal efficacies. Health-promoting qualities, such as antipathogenic effects, antioxidant properties, cholesterol assimilation, tumor suppression, and gastrointestinal immunity have been identified (Bourrie et al., 2016; Gut et al., 2021). For instance, several lactobacilli and yeast strains from kefir exhibited antipathogenic activities, inhibiting the growth of Salmonella typhimurium and enteropathogenic Escherichia coli O157:H7 (Santos et al., 2003; Menezes et al., 2020). Lactobacillus acidophilus, Lactiplantibacillus plantarum, and Lentilactobacillus kefiri strains from milk kefir were found to lower serum total cholesterol, low-density lipoprotein (LDL) and triglyceride levels in Sprague Dawley rats fed with a high cholesterol diet (Zheng et al., 2013). In a study by Vinderola et al. (2005), kefir containing viable microorganisms was found to be more effective in modulating the gut mucosal immune system than pasteurized kefir. These examples suggest that the health-promoting properties of the kefir beverage can be attributed to the properties of strains present within kefir. Herein, the motivation of this study is to identify beneficial probiotic strains from both milk and water kefir grains.
In this paper, microorganism from locally-sourced milk and water kefir grains were isolated and characterized. A series of phenotypic and genotypic screening assays were conducted to identify strains with the highest probiotic potential. These assays include agar well diffusion for detection of antipathogenic effects, acid and bile resistance, bile salt hydrolase (BSH) activity, antioxidant activity, non-cytotoxicity, and adhesion to Caco-2 cells. DNA extracted from the isolates of interest was subjected to whole genome sequencing (WGS). The seven best performing kefir-derived strains were combined in a “kefir probiotic blend” formulation, and a competitive adhesion/exclusion assay of kefir probiotics against enteric pathogens to Caco-2 cells was conducted. Tested human enteric pathogens include Bacillus cereus, Salmonella enterica, Vibrio parahaemolyticus, E. coli O157:H7, Klebsiella pneumoniae, Staphylococcus aureus, Clostridium difficile, and Clostridium perfringens. Overall, this study explores the potential of using kefir-sourced microbial strains as probiotics to inhibit enteric pathogens, and provides detailed in vitro characterization of their safety and other functional properties.
Materials and Methods
Materials
Milk and water kefir grains used in this study were active grains donated by a homebrewer in Singapore. Lacticaseibacillus rhamnosus GG (LGG) (i-Health Inc., Cromwell, CT, United States) and L. plantarum (wild type strain), were used as positive probiotic controls in various assays. E. coli K-12, provided by colleagues at SCELSE, was used as a negative control for the well diffusion assay. Enteric pathogens used in this study are detailed in Table 1. All enteric pathogens were purchased from ATCC, Manassas, VA, United States, with exception of K. pneumoniae and S. aureus which were provided by colleagues at SCELSE. Human intestinal Caco-2 cell lines HTB-37™ were purchased from ATCC, Manassas, VA, United States.
TABLE 1.
Growth conditions of pathogens.
| Pathogen species | Strain | Media | Temperature (°C) | Aerobic/anaerobic (OX/ANA) | Incubation duration (days) |
| Bacillus cereus | ATCC 11778 | NB | 30 | OX | 1 |
| Salmonella enterica | ATCC-BAA-190 | NB | 37 | OX | 1 |
| Vibrio parahaemolyticus | ATCC 17802 | NB w/3% NaCl | 37 | OX | 1 |
| Clostridium perfringens | ATCC 13124 | TSB | 37 | ANA | 1 |
| Clostridium difficile | ATCC 9689-FZ | TSB | 37 | ANA | 2 |
| Klebsiella pneumoniae | KP-1 | TSB | 37 | OX | 1 |
| Escherichia coli O157:H7 | ATCC 43888 | TSB | 37 | OX | 1 |
| Staphylococcus aureus | USA300 | TSB | 37 | OX | 1 |
Static conditions were used for incubation of all strains.
De Man, Rogosa, and Sharpe (MRS), Mannitol-yeast extract peptone (MYP), Yeast Extract-Peptone-Glycerol (YPG), Yeast Extract-Peptone-Dextrose (YPD), M17, and Nutrient broth/agar (NB/NA) media, were used for isolation of strains from the kefir granules. MRS, M17, and NB media were purchased from Thermo Fisher Scientific, Waltham. MA, United States. YPD was purchased from Sigma-Aldrich, St. Louis, MO, United States. MYP medium was prepared with 25 g/l mannitol, 5 g/l yeast extract, 3 g/l peptone (Sievers and Swings, 2005). YPG contained 10 g/l peptone, 5 g/l yeast extract, 0.01 g/l bromophenol blue. 150 μg/ml of cycloheximide were added to MRS, GM and NB media to inhibit yeast growth; 100 mg/l of chloramphenicol was added into YPG and YPD media to inhibit bacteria growth. Antibiotics were filtered and added into respective media after autoclaving. Tryptic soy (TS) medium, used for culturing of enteric pathogens, was purchased from Thermo Fisher Scientific, Waltham. MA, United States. Bacto agar was purchased from BD, Franklin Lakes, NJ, United States. Yeast extract, Yeast protein extract reagent (YPER), 2,2-diphenyl-1-picrylhydrazyl (DPPH), Dulbecco’s Modified Eagle Medium (DMEM), fetal bovine serum (FBS), trypsin-EDTA were purchased from Thermo Fisher Scientific, Waltham. MA, United States. Q5 High Fidelity PCR kit was purchased from New England Biolabs, Ipswich, MA, United States. The CytoTox 96® non-radioactive cytotoxicity kit was purchased from Promega, Madison, WI, United States. DNeasy Ultraclean Microbial Kit was purchased from QIAgen, Hilden, Germany. All other chemicals used in this experiment were purchased from Sigma Aldrich, St. Louis, MO, United States. Sterilization by autoclaving at 121°C for 15 min was done for every media, chemical solutions, or apparatus prior to use, where necessary.
Methods
Isolation of Microbes From Milk and Water Kefir
Milk and water kefir grains were ground with mortar and pestle, then serially diluted in phosphate buffered solution (PBS), before spreading on various media, including MRS, MYP, YPG, YPD, NA, and M17 agar (1.5% agar used). These media were selected in attempt to isolate of a variety of kefir microorganisms; MRS was used to isolate lactobacilli; MYP for acetic acid bacteria (Sievers and Swings, 2005); YPG and YPD for yeast (Martínez-Torres et al., 2017); NA as a general growth medium, and M17, for lactic streptococci (Terzaghi and Sandine, 1975). Plates were incubated at 30°C for 3–7 days at both aerobic (air) and anaerobic (4% H2, 5% CO2, 91% N2) conditions. Anaerobic handling of strains was done within the Bactron 300 anaerobic chamber (Sheldon Manufacturing, Cornelius, OR, United States). Representative colonies were selected for isolation and purified by streaking onto fresh agar plates. Purified colonies were then cultured in corresponding broths and aliquots were stored at –80°C with 20% (v/v) glycerol as cryoprotectant.
Species Identification by 16S rRNA and Internal Transcribed Spacer Sequencing
The 16S rRNA and internal transcribed spacer (ITS) segments were sequenced to determine the species identity of kefir-derived microbial strains. Individual isolates were cultured in respective broths and 100 μl of cell cultures were lysed by addition of 20 μl YPER and heating at 98°C for 5 min (Packeiser et al., 2013). The cell lysate was centrifuged at 14,000 × g for 10 min, and the supernatant was collected for polymerase chain reaction (PCR). Q5 High Fidelity PCR kit (New England Biolabs, MA, United States) was used with universal primers 27F (5′ AGAGTTTGATCMTGGCTCAG 3′), and 1492R (5′ TACGGYTACCTTGTTACGACTT 3′) to amplify 16S rRNA for bacteria. For yeast, primers used were ITS 1 (5′ TCCGTAGGTGAACCTGCGG 3′) and ITS 4 (5′ TCCTCCGCTTATTGATATGC 3′). The PCR reaction mix consisted of 10 μl 5X Q5 reaction buffer, 1 μl 10 mM dNTPs, 25 μM forward primer, 25 μM reverse primer, 0.5 μl Q5 High Fidelity DNA polymerase, 5 μl DNA template and 28.5 μl nuclease-free water, totaling 50 μl. PCR amplification was carried out with following parameters: 98°C for 3 min, 30 cycles of 98°C for 10 s, 57°C for 15 s, 72°C for 1 min, 72°C for 2 min and holding at 12°C. Quality of PCR products was checked by gel electrophoresis, performed at 100 V for 50 min with ethidium bromide (EtBr) staining for 30 min. Gels were viewed under UV light with Universal Hood II Gel Doc system (Bio-Rad Laboratories, Hercules, CA, United States) to obtain the band image. Samples that produced defined bands were sent to an external vendor (1st base, Singapore) for sequencing. Briefly, PCR products were purified using FavorPrep™ GEL/PCR Purification Kit (Favorgen Biotech Corporation, Pingtung, Taiwan). Purified DNA templates were then subjected to cycle sequencing following standard protocol from BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, United States). Products were purified by MagBio HighPrep™ DTR Clean-up System (Magbio Genomics, Gaithersburg, MD, United States) before loading into ABI 3730xl DNA Analyzer (Applied Biosystems, Waltham, MA, United States) for DNA Sanger Sequencing. Obtained nucleotide sequences were analyzed using the ApE plasmid editor software (Davis, 2022) and species assignment of kefir isolates was done using the National Centre for Biotechnology Information (NCBI) BLAST platform, based on the BLAST result which yielded highest total score.
Agar Well Diffusion Assay
Antimicrobial activities of isolates from kefir strains against pathogens were assessed using agar well diffusion (Ayala et al., 2019). Pathogen cultures were inoculated in respective media and growth conditions as described in Table 1, before adjustment to OD600 of 0.1 (except KP1, which was adjusted to 0.01), and spreading 100 μl onto the agar plates. Kefir isolates were cultured in MRS broth at 30°C, anaerobic conditions for 3 days. Thirty-six strains representing 23 species were selected for this assay. LGG, cultured in MRS at 37 °C, aerobic conditions for 24 h, was used as a positive probiotic control. Wells 6 mm in diameter were formed in the agar and filled with 50 μl of cultures (contains microbes) or culture supernatants (without microbes), with duplicates performed for each kefir isolate strain. Cell-free supernatants were obtained by centrifuging the incubated cultures at 10,000 × g for 5 min. Plates were incubated in growth conditions according to the growth requirements of each pathogen. Finally, plates were observed for the presence of inhibition zones around individual wells. Inhibition zones are clear sites that have no visible pathogen growth, indicating that the kefir isolates have successfully inhibited growth of the enteric pathogen.
Acid and Bile Resistance
The protocol of Chandel et al. (2019) was used to examine the acid and bile resistance of the kefir isolates to determine their survival capacity during exposure to acidic pH and bile typical of the human GIT. In this assay, MRS broths were adjusted to pH 2, pH 3, or with 0.3% (w/v) ox-bile added. Aliquots (100 μl) of cultures of the kefir isolate were added to 4.9 ml of each of the adjusted MRS broths in duplicates and incubated at 37°C for 2 h under 200 rpm shaking conditions in an Ecotron shaking incubator (Infors AG, Bottmingen, Switzerland). Ten-fold serial dilutions and drop plating (100 μl aliquots) was done to enumerate the colony-forming units (CFU/ml) before and after acid/bile exposure.
Bile Salt Hydrolase Activity Assay
Bile salt hydrolase activity is a desirable probiotic trait which has been postulated to increase survival and persistence of microbes in the intestinal tract, and has been reported to also induce cholesterol-lowering efficacies by controlling serum cholesterol levels (Begley et al., 2005). BSH activities of kefir isolates were evaluated as previously reported by Michael et al. (2016). Aliquots (5 μl) of the cultures of the kefir isolates were inoculated onto MRS agar containing 0.5% (w/v) taurodeoxycholate hydrate (TDC) in duplicates. Plates were incubated at 37°C under anaerobic conditions for 24 h. BSH activity was indicated by the development of a white precipitate on TDC-supplemented plates after 48 h. L. plantarum was used as a positive control in this assay (Michael et al., 2016).
Antioxidant Activity Assay
Antioxidant activities of kefir isolates were determined using the DPPH free radical assay (Kedare and Singh, 2011). Aliquots (0.5 ml) of cultures of kefir isolates were added to 3 ml of 0.05 mM DPPH in ethanol in duplicate. Controls were prepared by mixing MRS broth (0.5 ml) with 3 ml of absolute ethanol. The reaction mixture was then incubated in the dark at room temperature for 30 min. A color change from deep violet to light yellow could be observed if antioxidant activity was present. After incubation, the absorbance at 517 nm was measured with a spectrophotometer. The antioxidant activity percentage (AA%) was determined according to:
| (1) |
where Asample is the average of measured absorbance at 517 nm of the sample with DPPH added minus the absorbance of MRS broth without DPPH added, and Acontrol is the measured absorbance at 517 nm of MRS broth with DPPH added minus the absorbance of MRS broth without DPPH added.
Cytotoxicity of Kefir Isolates Using Caco-2
The in vitro cytotoxicity of kefir isolates was determined using Caco-2 cells. The Caco-2 cells were maintained in DMEM containing 20% (v/v) FBS, 100 units/ml of penicillin and 100 μg/ml of streptomycin, at 37°C in a humidified 5% CO2 atmosphere. Caco-2 cells were seeded at a concentration of 2.8 × 104 cells/cm2 in 24-well tissue culture plates and grown for 7 days until confluence was reached. For the last medium change, DMEM without antibiotics was used. Duplicate wells with confluent Caco-2 cells were inoculated with 0.5 ml of 1 × 107 CFU/ml (as determined by optical density from a standard culture of OD and CFU) kefir isolates, which were prepared by washing respective cultures of kefir isolates in PBS and resuspending in DMEM. Inoculated Caco-2 plates were then incubated for 24 h in 37°C, 5% CO2 conditions. Following incubation, Caco-2 cells were washed twice with PBS, and the number of Caco-2 cells that remained viable in each well was determined with the CytoTox 96® non-radioactive cytotoxicity kit, using a protocol from Van den Bossche et al. (2020) which reduces bacterial interference with lactate dehydrogenase (LDH) quantification. Caco-2 cells were completely lysed with 200 μl of 1× Lysis solution (from kit) and incubated at 37°C for 45 min. Efficient lysis of cells was further achieved through vigorous pipetting. Lysed Caco-2 cells were then centrifuged at 5,000 × g for 10 min, and 50 μl of the supernatant was added to 50 μl CytoTox 96® Reagent. Following a 30-min incubation at room temperature in dark conditions, 50 μl of Stop Solution (from kit) was added to each sample, and absorbance at 490 nm was measured in a microplate reader (Infinite M200, Tecan Group Ltd, Männedorf, Switzerland). Un-inoculated Caco-2 wells were used as maximum lysis controls, and 1× Lysis solution was used to determine the background absorbance. Background absorbance was subtracted from all measured absorbance values with Caco-2 cells. Percent cytotoxicity was calculated as follows:
Adhesion of Kefir Isolates to Caco-2
The ability of kefir isolates to adhere to an intestinal surface was determined by an in vitro adhesion assay using the human epithelial cell line Caco-2 (Ayala et al., 2019). Caco-2 cells were seeded at 2.8 × 104 cells/cm2 in 12-well tissue culture plates and the culture medium was changed daily for 21 days to allow growth into the late post-confluence stage. For the last medium change, DMEM without antibiotics was used. Duplicate confluent Caco-2 cell monolayers were inoculated with 1 ml of 1 × 108 CFU/ml kefir isolates, which were prepared by washing cultures of the kefir isolates in PBS and re-suspending in DMEM. Inoculated Caco-2 plates were then incubated for 2 h in 37°C, 5% CO2 conditions to allow for microbial attachment. Following incubation, non-attached or loosely adherent microbes were removed by washing Caco-2 monolayers three times with sterile PBS. To release adherent microbes, 200 μl of 0.25% (w/v) trypsin with 0.53 mM EDTA were added to each well, and incubated for 10 min in 37°C, 5% CO2 conditions. PBS (800 μl) was then added per well to dilute the trypsin-EDTA, and ten-fold serial dilutions and drop-plating was done to enumerate CFU of attached kefir microbes. Percentage adhesion was calculated by taking the ratio of attached microbes to the CFU of microbes added.
Whole Genome Sequencing and Genotypic Characterization
Selected kefir isolates were genotypically characterized by WGS. Genomic DNA was extracted from kefir isolates using a DNeasy Ultraclean Microbial Kit, according to manufacturer’s instructions. The quality and concentration of extracted DNA concentration was checked via gel electrophoresis and a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham. MA, United States), respectively, prior to sequencing. Library preparation was performed using a TruSeq DNA HT Library Preparation Kit (Illumina, San Diego, CA, United States), and sequencing was done on the Illumina MiSeq platform with 300 base pairs (bp) paired end reads. Raw reads were cleaned using Trimmomatic version 0.39 (Bolger et al., 2014), and quality of reads was checked with FastQC (Babraham Bioinformatics, 2022). De novo genome assembly was done using SPAdes version 3.14.1 (Bankevich et al., 2012), and the quality of resultant assembled contigs was checked with the DDBJ Fast Annotation and Submission Tool (DFAST) (Tanizawa et al., 2016). DFAST was also used to confirm the species and taxonomy of kefir isolates via average nucleotide identity (ANI) analysis (Jain et al., 2018). Assembled contigs of kefir isolates Kef-w/m1-21 were submitted to GenBank under the BioProject ID PRJNA721546, with accession numbers JAGPZD000000000 to JAGPZX000000000. Functional gene annotation of assembled contigs was performed using the NCBI prokaryotic genome annotation pipeline (PGAP). Bacteriocins, virulence factors and AMR genes were identified using the BAGEL4 (Van Heel et al., 2018), Virulence Finder v2.0.3 (Joensen et al., 2014), and ResFinder 4.1 pipelines (Bortolaia et al., 2020) respectively. Plasmids were identified using PlasmidFinder 2.1 (Carattoli et al., 2014). Biogenic amines (BAs) and toxins production genes (including genes related to production of histidine decarboxylase, tyrosine decarboxylase, ornithine decarboxylase, agmatine dehydrolase, l-lysine decarboxylase, agmatine deiminase, hemolysin, cytotoxin, fengygin, surfactin, lychenisin, and lipopolysaccharides) were manually searched for among annotated genes of each kefir isolate. Vitamins and γ-aminobutyric acid (GABA) biosynthesis genes were also searched for manually. Taxonomic analysis was performed using the Type (Strain) Genome Server (TYGS), a free bioinformatics platform available under https://tygs.dsmz.de (Meier-Kolthoff and Göker, 2019). Briefly, the kefir isolate genomes were compared against all type strain genomes available in the TYGS database via the MASH algorithm to determine closely related type strains. These strains were compared pairwise to derive their intergenomic distances, then used to infer a balanced minimum evolution tree with branch support via FASTME 2.1.6.1 including SPR post-processing.
Preparation of Kefir “Probiotic Blend”
Based on the above phenotypic and genotypic characterization assays, seven best performing kefir bacterial strains were selected for combination in a multi-strain “kefir probiotic blend” (Table 2). The kefir probiotics blend was prepared by adjusting the cultures of the selected kefir isolates to indicated OD600 values and concentrating each of the isolates into the same volume by centrifugation and resuspending in PBS. Total CFU/ml of kefir probiotics blend was approximately 3 × 109 CFU/ml.
TABLE 2.
Composition of kefir “probiotics blend.”
| Kefir probiotic species | Strain | OD600 used in kefir probiotic blend | CFU in 1 ml of kefir probiotic blend | CFU ratio |
| Lentilactobacillus hilgardii | Kef-w9 | 5.2 | 6.03E+07 | 0.02 |
| Lacticaseibacillus paracasei | Kef-w14 | 6 | 8.57E+08 | 0.28 |
| Lacticaseibacillus paracasei | Kef-w19 | 6 | 8.25E+08 | 0.27 |
| Liquorilactobacillus satsumensis | Kef-w13 | 5.6 | 4.13E+08 | 0.13 |
| Liquorilactobacillus satsumensis | Kef-w11 | 5.6 | 5.56E+08 | 0.18 |
| Lactobacillus helveticus | Kef-m4 | 6 | 1.75E+08 | 0.06 |
| Lentilactobacillus kefiri | Kef-m15 | 2.5 | 2.06E+08 | 0.07 |
Competitive Adhesion/Exclusion of Enteric Pathogens by the Kefir Probiotic Blend
An important probiotic mechanism of action is through adhesion to host cells, which competitively blocks the adherence of, or excludes, pathogens from host cell binding sites (Monteagudo-Mera et al., 2019). The ability of kefir isolates to compete for adhesion sites with or exclude pathogens from Caco-2 epithelial cells, was tested using a modification of an established method (Candela et al., 2008; Inturri et al., 2016). Briefly, Caco-2 cells were seeded at a concentration of 2.8 × 104 cells/cm2 in 12-well tissue culture plates and grown for 21 days to allow growth into the late post-confluence stage. For the last medium change, DMEM without antibiotics was used. Cultures of the eight human enteric pathogens included in Table 1 and the kefir probiotics blend as indicated in Table 2 were used. For this assay, pathogens and kefir isolates were prepared by washing cultures in PBS and re-suspending in DMEM, to yield 1 × 108 CFU/ml respectively. Duplicate confluent Caco-2 cells were inoculated with either: (1) 1 ml of a mixture of kefir isolates and individual pathogens, and incubated for 2 h, for the competitive adhesion assay, or (2) 1 ml of kefir isolates, incubated for the first hour, prior to the removal of kefir isolates and the addition of 1 ml of individual pathogens and incubated for the next hour, for the competitive exclusion assay. Incubation was done under 37°C, 5% CO2 conditions to allow for probiotics/pathogens attachment. Following incubation, non-attached or loosely adherent bacteria were removed by washing Caco-2 monolayers three times with sterile PBS. To release adherent bacteria, 200 μl of trypsin-EDTA were added to each well, and incubated for 10 min in 37°C, 5% CO2 conditions. PBS (800 μl per well) was then added to dilute the trypsin-EDTA, and drop-plating of aliquots from ten-fold serial dilutions onto selective agars (e.g., NA or TS media were used to enumerate enteric pathogens, while MRS was used to enumerate kefir isolates) to enumerate attached probiotics/pathogens as CFU. The rationale of the competitive adhesion method is to determine if the kefir isolates could reduce pathogen adhesion to Caco-2 when added in combination, while competitive exclusion determines if pre-exposure to kefir microbes can reduce attachment of pathogens. The CFU data of attached probiotics/pathogens in the competitive adhesion and exclusion methods were hence compared to the adhesion data of individual pathogens to Caco-2 cells. Normalized adhesion was calculated as the percentage of pathogens adhered (competitive adhesion or competitive exclusion method) divided by the percentage of pathogens adhered when added alone.
Statistical Analysis
All experiments were performed at least in duplicate. Results for various assays were expressed as the mean of duplicates, with the relevant standard deviation data provided in Supplementary Table 1. Statistical evaluation was conducted for results of the competitive exclusion/adhesion assay, and one way analysis of variance and post-hoc Dunnett test was used. Letters on bars were based on *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns–no significant difference.
Results
List of Isolated Strains
In total, 158 strains representing 6 fungal and 17 bacterial species, were isolated from milk and water kefir (Table 3). Isolated genera include Lactobacillus, Liquorilactobacillus, Lacticaseibacillus, Lentilactobacillus, Leuconostoc, Lactococcus, Acetobacter, Gluconobacter, Oenococcus, Clostridium, Zymomonas, Saccharomyces, Kluyveromyces, Pichia, Lachancea, Candida, and Brettanomyces. Most species were isolated from milk or water kefir exclusively, with exception of Pichia fermentans, Saccharomyces cerevisiae, and Clostridium beijerinckii, which were isolated from both milk and water kefir.
TABLE 3.
List of microbial strains isolated from milk and water kefir.
| No. | Microorganism Species | Kingdom | Number of strains isolated | Strain names | Source | Isolation media | Previously isolated from kefir | Previously identified in kefir by metagenomic techniques |
| 1 | Liquorilactobacillus satsumensis | Bacteria | 20 | Kef-w1, Kef-w2, Kef-w11, Kef-w13, Kef-w18, Kef-w22-36 | Water Kefir | MRS, MYP, NA, YPD, M17 | Miguel et al., 2011; Zanirati et al., 2015; Gamba et al., 2019 | Kumar et al., 2021 |
| 2 | Lactobacillus kefiranofaciens | Bacteria | 4 | Kef-m3, Kef-m37-39 | Milk Kefir | MRS, MYP | Santos et al., 2003 | Nalbantoglu et al., 2014; Korsak et al., 2015; Chen et al., 2021 |
| 3 | Lactobacillus helveticus | Bacteria | 1 | Kef-m4 | Milk Kefir | MYP | Simova et al., 2002; Miguel et al., 2011 | Nalbantoglu et al., 2014; Chen et al., 2021 |
| 4 | Acetobacter persici | Bacteria | 4 | Kef-w5, Kef-w40-42 | Water Kefir | MYP | Brandt et al., 2017 | - |
| 5 | Leuconostoc mesenteroides | Bacteria | 5 | Kef-m6, Kef-m16, Kef-m43-45 | Milk Kefir | MRS, M17 | Pidoux, 1989; Gulitz et al., 2011; Zanirati et al., 2015 | Nalbantoglu et al., 2014; Korsak et al., 2015, Kazou et al., 2021 |
| 6 | Lacticaseibacillus paracasei | Bacteria | 12 | Kef-w7, Kef-w14, Kef-w17, Kef-w19-20, Kef-w46-52 | Water Kefir | MRS, NA, YPD, M17 | Santos et al., 2003; Miguel et al., 2011; Gamba et al., 2019; Talib et al., 2019 | Korsak et al., 2015; Verce et al., 2019; Chen et al., 2021 |
| 7 | Lentilactobacillus hilgardii | Bacteria | 3 | Kef-w8-10 | Water Kefir | MRS | Pidoux, 1989; Gamba et al., 2019 | Cao et al., 2019; Verce et al., 2019; Kumar et al., 2021 |
| 8 | Zymomonas mobilis | Bacteria | 10 | Kef-w12, Kef-w53-61 | Water Kefir | MRS, YPD, YPG | - | Cao et al., 2019 |
| 9 | Lentilactobacillus kefiri | Bacteria | 2 | Kef-m15, Kef-m62 | Milk Kefir | MRS, YPD | Gao et al., 2012; Zheng et al., 2013; Zanirati et al., 2015; Hurtado-Romero et al., 2021 | Nalbantoglu et al., 2014; Korsak et al., 2015; Kazou et al., 2021 |
| 10 | Liquorilactobacillus nagelii | Bacteria | 1 | Kef-w21 | Water Kefir | YPD | Gamba et al., 2019 | Verce et al., 2019 |
| 11 | Lactobacillus delbrueckii | Bacteria | 1 | Kef-m63 | Milk Kefir | MYP | Simova et al., 2002 | Kazou et al., 2021 |
| 12 | Lactococcus lactis | Bacteria | 31 | Kef-m64-93 | Milk Kefir | MRS, MYP, NA, YPD, M17 | Simova et al., 2002; Gao et al., 2012; Zanirati et al., 2015; Hurtado-Romero et al., 2021 | Nalbantoglu et al., 2014; Korsak et al., 2015 |
| 13 | Acetobacter fabarum | Bacteria | 7 | Kef-m94-100 | Milk Kefir | MRS, MYP, YPD | Gulitz et al., 2011; Gao et al., 2012 | - |
| 14 | Kluyveromyces marxianus | Fungi | 8 | Kef-m101-108 | Milk Kefir | MYP, YPG | Wyder et al., 1997; Simova et al., 2002; Gao et al., 2012; Hsu and Chou, 2021 | Chen et al., 2021; Kazou et al., 2021 |
| 15 | Acetobacter orientalis | Bacteria | 3 | Kef-m109-111 | Milk Kefir | MYP, YPD | Gulitz et al., 2011 | Korsak et al., 2015 |
| 16 | Pichia fermentans | Fungi | 3 | Kef-m112-113, Kef-w114 | Milk and Water Kefir | YPG | Miguel et al., 2011 | - |
| 17 | Saccharomyces cerevisiae | Fungi | 19 | Kef-m115, Kef-w116-125, Kef-m126-129, Kef-w130-133 | Milk and Water Kefir | YPD, YPG | Simova et al., 2002; Miguel et al., 2011; Gao et al., 2012; Gamba et al., 2019; Hsu and Chou, 2021 | Verce et al., 2019; Kazou et al., 2021 |
| 18 | Clostridium beijerinckii | Bacteria | 3 | Kef-w134, Kef-m135-136 | Milk and Water Kefir | MYP, YPD | - | Chen et al., 2021 |
| 19 | Lachancea fermentati | Fungi | 11 | Kef-w137-147 | Water Kefir | YPD, YPG | Gulitz et al., 2011 | Marsh et al., 2013 |
| 20 | Candida ethanolica | Fungi | 4 | Kef-w148-151 | Water Kefir | YPG | - | Sarikkha et al., 2015 |
| 21 | Gluconobacter oxydans | Bacteria | 1 | Kef-w152 | Water Kefir | YPD | Gamba et al., 2019 | Kumar et al., 2021 |
| 22 | Oenococcus oeni | Bacteria | 3 | Kef-w153-155 | Water Kefir | MYP | Zanirati et al., 2015 | Nalbantoglu et al., 2014; Kumar et al., 2021 |
| 23 | Brettanomyces anomalus | Fungi | 3 | Kef-w156-158 | Water Kefir | YPG | Wyder et al., 1997 | - |
Pathogen Inhibition Using Well Diffusion Assay
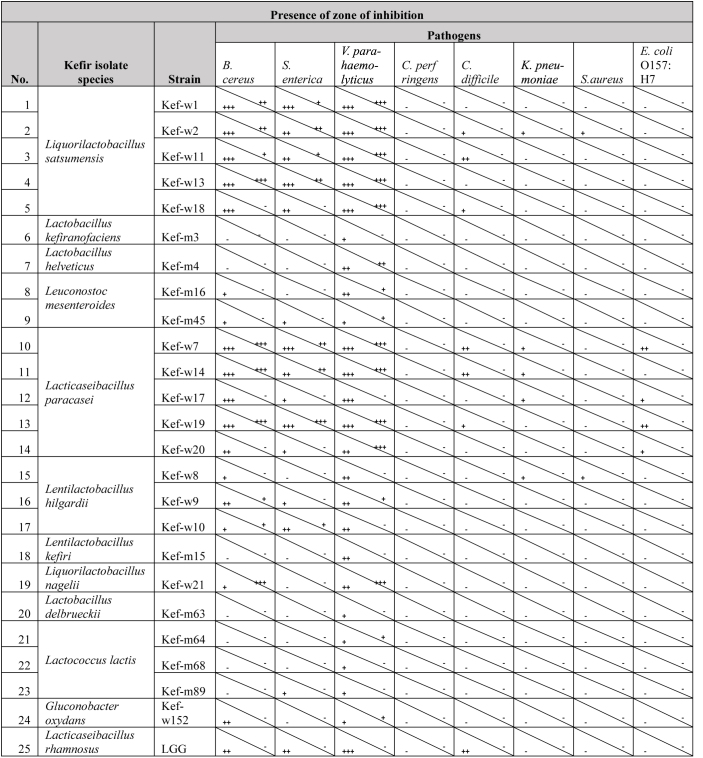
Amongst the 158 kefir-derived isolates, 23 strains representing the 23 distinct species isolated, together with 13 more strains selected to test for strain-specific activity, a total of 36 strains, were selected for a detailed screening. Of these 36 tested strains, 24 kefir-isolates showed inhibitory activities towards at least one of the tested pathogens (Table 4). Photos of agar wells with inhibition zones are included in Supplementary Figure 1. These kefir-isolates were from the Lactobacillus, Liquorilactobacillus, Lentilactobacillus, Lacticaseibacillus, Leuconostoc, Lactococcus, and Gluconobacter genera. None of the tested kefir-fungal strains showed antimicrobial properties. The growth of the common food borne pathogens V. parahaemolyticus, B. cereus, and S. enterica were most inhibited by multiple kefir isolates. Inhibition of C. difficile, a bacterium which causes diarrhea and colon inflammation in humans (Lessa et al., 2015), was observed for the isolates Lacticaseibacillus paracasei, Liquorilactobacillus satsumensis, and LGG. Enteropathogenic E. coli O157:H7, a particular virulent serotype of E. coli known to cause diarrhea, abdominal cramps and other complications (WHO, 2022), was found to be inhibited by several L. paracasei strains. K. pneumoniae and S. aureus were also inhibited by L. satsumensis, L. paracasei, and Liquorilactobacillus hilgardii kefir isolates. The kefir isolates inhibited both Gram-positive (B. cereus, C. difficile, S. aureus) and Gram-negative (V. parahaemolyticus, S. enterica, E. coli, K. pneumoniae) pathogens. Inhibitory mechanisms of kefir-isolates against these pathogens are most likely attributed to production of antimicrobial metabolites, such as organic acids, hydrogen peroxide and bacteriocins (Šušković et al., 2010). Notably, the pathogen inhibition capacity of several kefir isolates, particularly L. paracasei and L. satsumensis, outperformed the reference probiotic LGG. Comparing between different strains of the same species, some slight differences in inhibitory activities can be observed. Overall, L. satsumensis, Lactobacillus helveticus, Leuconostoc mesentorides, L. paracasei, L. hilgardii, L. kefiri, Liquorilactobacillus nagelii, L. paracasei, and Gluconobacter oxydans showed most promising antimicrobial efficacies. These promising strains were selected for further phenotypic screening.
TABLE 4.
Inhibitory activities of kefir isolates and Lacticaseibacillus rhamnosus GG (LGG) against human enteric pathogens.a
aThe bottom-left triangle indicates inhibition by kefir isolate cultures, while the top-right triangle indicates inhibition by the cell-free supernatant component. Inhibition zones of >4, 2–4, and <2 mm were reviewed as strong (+++), intermediate (++), and weak inhibition (+), respectively. Cells indicated with (-) are without any zone of inhibition observed. Only kefir isolates with inhibitory activities towards at least one tested pathogen are listed. Zones of inhibition were averaged from duplicate wells.
Phenotypic Characteristics of Kefir Isolates
From the well diffusion assay results, 24 kefir isolates were selected for further screening and results are presented in Table 5. For acid/bile tolerance, all selected kefir-isolates showed susceptibility to pH 2, similar to the control probiotic LGG. Twelve strains, namely L. satsumensis Kef-w1, Kef-w2, Kef-w13, L. helveticus Kef-m4, L. paracasei Kef-w7, Kef-w14, Kef-w17, Kef-w19, L. hilgardii Kef-w8-10, L. kefiri Kef-m15 showed the ability to survive 2-h exposure to pH 3 and 0.3% (w/v) ox-bile with <1 logCFU reduction, suggesting their intrinsic resistances to acid and bile exposure typical of the conditions in the human GIT. For BSH activity, seven kefir isolates of species L. hilgardii, L. kefiri, Lactobacillus delbrueckii, and Lactobacillus lactis demonstrated BSH activity. Antioxidant activities, ranging from 23 to 78%, were observed of the kefir isolates. Highest antioxidant activity was observed of L. paracasei Kef-w19, notably higher than that of the reference strain LGG (Mu et al., 2018). Adhesion of tested kefir isolates to Caco-2 cells were notably higher than that of LGG, measured at 0.9%. The adhesion of kefir isolates ranged from 1.2 to 18.1%, with L. satsumensis Kef-w11, L. hilgardii Kef-w8-10, L. kefiri Kef-m15, and L. nagelii Kef-w21 having adhesion percentages of more than 10%, indicative of good adhesion properties (Candela et al., 2005). All tested kefir isolates were found to be non-cytotoxic to Caco-2 cells, showing percentage cytotoxicities of <1.21%.
TABLE 5.
Phenotypic properties of selected kefir isolates, including results for acid and bile resistance, bile salt hydrolase (BSH) activity, antioxidant activity, adhesion, and cytotoxicity to Caco-2 cells.a
| No. | Kefir isolate species | Strain | Acid and bile resistance, Log10CFU surviving |
BSH act-ivity | Anti-oxidant activity (AA %) | % Adhered to Caco-2 (%) | Cyto-toxicity to Caco-2 (%) | |||
| Initial | After pH2 exposure | After pH3 exposure | After bile exposure | |||||||
| 1 | Kef-w1 | 7.55 | 0.00 | 7.54 | 6.70 | - | 56% | 7.6% | 0.53% | |
| 2 | Kef-w2 | 7.51 | 0.00 | 7.53 | 6.83 | - | 51% | 1.2% | –0.37% | |
| 3 | Liquorilactobacillus | Kef-w11 | 7.70 | 0.00 | 7.57 | 5.28 | - | 32% | 10.0% | –0.05% |
| 4 | satsumensis | Kef-w13 | 7.44 | 0.00 | 7.31 | 7.11 | - | 58% | 3.3% | –0.20% |
| 5 | Kef-w18 | 7.55 | 2.74 | 7.55 | 5.54 | - | 62% | 8.9% | 1.21% | |
| 6 | Lactobacillus kefiranofaciens | Kef-m3 | 6.58 | 3.22 | 4.11 | 6.42 | - | Nil | Nil | Nil |
| 7 | Lactobacillus helveticus | Kef-m4 | 7.37 | 3.35 | 7.24 | 6.58 | - | 60% | 1.2% | 0.81% |
| 8 | Leuconostoc mesenteroides | Kef-m16 | 6.10 | 2.74 | 3.59 | 5.82 | - | Nil | Nil | Nil |
| 9 | Kef-m45 | 6.90 | 0.00 | 2.74 | 6.44 | - | Nil | Nil | Nil | |
| 10 | Lacticaseibacillus paracasei | Kef-w7 | 8.02 | 0.00 | 7.74 | 8.21 | - | 56% | 4.0% | –0.66% |
| 11 | Kef-w14 | 6.32 | 3.22 | 6.36 | 5.85 | - | 68% | 4.8% | 0.23% | |
| 12 | Kef-w17 | 8.03 | 0.00 | 8.09 | 7.86 | - | 63% | 3.0% | 0.07% | |
| 13 | Kef-w19 | 7.49 | 0.00 | 7.61 | 7.43 | - | 78% | 5.4% | 0.03% | |
| 14 | Kef-w20 | 6.90 | 0.00 | 5.76 | 7.09 | - | Nil | Nil | Nil | |
| 15 | Lentilactobacillus hilgardii | Kef-w8 | 7.49 | 0.00 | 7.14 | 7.48 | - | 23% | 17.6% | –2.14% |
| 16 | Kef-w9 | 7.69 | 0.00 | 7.12 | 7.89 | + | 37% | 17.2% | –3.24% | |
| 17 | Kef-w10 | 7.64 | 0.00 | 7.02 | 7.48 | + | 37% | 13.3% | –1.62% | |
| 18 | Lentilactobacillus kefiri | Kef-m15 | 7.28 | 0.00 | 6.92 | 8.24 | + | 69% | 10.2% | 0.81% |
| 19 | Liquorilactobacillus nagelii | Kef-w21 | 7.58 | 0.00 | 7.29 | 5.35 | - | 72% | 18.1% | 1.11% |
| 20 | Lactobacillus delbrueckii | Kef-m63 | 6.70 | 0.00 | 2.74 | 6.43 | + | Nil | Nil | Nil |
| 21 | Lactococcus lactis | Kef-m64 | 6.49 | 0.00 | 0.00 | 6.43 | + | Nil | Nil | Nil |
| 22 | Kef-m68 | 7.14 | 0.00 | 0.00 | 6.86 | + | Nil | Nil | Nil | |
| 23 | Kef-m89 | 7.59 | 0.00 | 0.00 | 7.29 | + | Nil | Nil | Nil | |
| 24 | Gluconobacter oxydans | Kef-w152 | 5.86 | 0.00 | 4.91 | 5.60 | - | Nil | Nil | Nil |
| 25 | Lacticaseibacillus rhamnosus | LGG | 7.51 | 0.00 | 7.48 | 7.47 | - | 73% | 0.9% | 0.84% |
| 26 | Lactiplantibacillus plantarum | Wild Type | Nil | + | Nil | Nil | Nil | |||
aAll results were calculated as an average of duplicate experiments. Nil indicates that the kefir isolate was not studied for the particular assay.
Genotypic Characteristics of Kefir Isolates
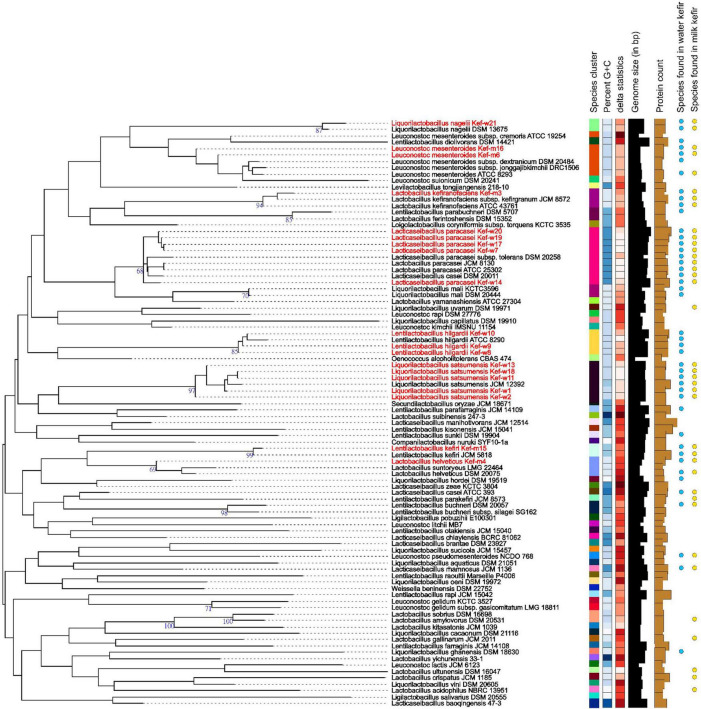
WGS was carried out for 15 selected kefir isolates, and the collated data of bacteriocins, virulence, antimicrobial resistance (AMR), biogenic amines (BAs), toxins, vitamins, γ-aminobutyric acid (GABA) genes and plasmids for each kefir isolate are presented in Table 6. Antibiotic resistance and virulence genes were absent for all kefir isolates. Plasmids, previously identified in L. plantarum (UniProt, 2022) and Lentilactobacillus buchneri (Liu et al., 2011), were found present in L. satsumensis Kef-w2, Kef-w13, L. helveticus Kef-m4 and L. kefiri Kef-m15. Genes encoding for production of bacteriocins, which are antimicrobial peptides produced by bacteria and serve as natural alternatives to antibiotics, were identified in L. satsumensis Kef-w13, L. helveticus Kef-m4, L. paracasei Kef-w7, Kef-w14, Kef-w17 and Kef-w19. The toxin protein, hemolysin III, a common hemolysin found in lactobacilli bacteria, was found in all kefir isolates. There are numerous reports about hemolysin III and its safety in lactobacilli, hence this toxin protein is not considered of significant concern (Senan et al., 2015; Surachat et al., 2017). L. nagelii Kef-w21 was found to harbor lipopolysaccharide (LPS) endotoxin proteins, which are potentially harmful as LPS can trigger intestinal inflammation and increased permeability. LPS are generally known to be present in Gram-negative bacteria, and the only Gram-positive bacteria so far known to contain an LPS is Listeria monocytogenes (Casey et al., 2014). To err on the safe side, L. nagelii Kef-w21 was excluded from the kefir probiotic blend. The production of BAs was investigated, as lactobacilli, particularly those involved in fermented foods, often produce them. Notably, synthesis genes associated with histamine and tyramine, two most concerning BAs which have higher incidences of intolerance upon ingestion (Durak-Dados et al., 2020), were found to be absent in the kefir isolates. Agmatine decarboxylase and ornithine decarboxylase genes, which are involved in the biosynthesis of putrescine, were identified in the kefir isolates. Since agmatine decarboxylase and ornithine decarboxylase genes have been previously been identified in many known probiotics and starter cultures (Costantini et al., 2013; Barbieri et al., 2019), this was not considered to be a major safety concern. All kefir isolates were found to contain genes for biosynthesis of riboflavin (vitamin B2), L. helveticus, L. paracasei, L. hilgardii were found to additionally contain cobalamin (vitamin B12) synthesis genes and L. hilgardii was found to also contain pyridoxal (vitamin B6) synthesis genes. Genes for biosynthesis of GABA, a neurotransmitter which reduces anxiety and stress (Komatsuzaki et al., 2005), was identified in L. hilgardii strains, consistent with other reports (Lee et al., 2013). Phylogenetic analysis revealed several highly similar strains amongst the kefir isolates (Figure 1). L. paracasei Kef-w7, Kef-17, Kef-w19, Kef-w20 were highly similar to each other while L. satsumensis Kef-w11 and Kef-w18 were similar to each other. Accordingly, only one isolate of very similar strains was selected for the kefir probiotic blend formulation. Ultimately, seven kefir isolates (Table 2) were selected to constitute the kefir probiotic blend based on the phenotypic and genotypic data analyses.
TABLE 6.
Genetic features of selected kefir isolates.
| No. | Kefir isolate species | Strain | AMR genes | Virulence genes | Plasmids | Bacteriocin genes | Toxin synthesis genes | BA synthesis genes | Vitamin synthesis genes | GABA syn-thesis genes |
| 1 | Kef-w1 | Absent | Absent | Absent | 0 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin | Absent | |
| 2 | Kef-w2 | Absent | Absent | CP-005948 | 0 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin | Absent | |
| 3 | Liquorilactobacillus satsumensis | Kef-w11 | Absent | Absent | Absent | 0 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin | Absent |
| 4 | Kef-w13 | Absent | Absent | CP-005948 | 1 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin | Absent | |
| 5 | Kef-w18 | Absent | Absent | Absent | 0 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin | Absent | |
| 6 | Lactobacillus helveticus | Kef-m4 | Absent | Absent | CP-002655, CP-002654 | 5 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin, Cobalamin | Absent |
| 7 | Lacticaseibacillus paracasei | Kef-w7 | Absent | Absent | Absent | 2 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin, Cobalamin | Absent |
| 8 | Kef-w14 | Absent | Absent | Absent | 4 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin, Cobalamin | Absent | |
| 9 | Kef-w17 | Absent | Absent | Absent | 2 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin, Cobalamin | Absent | |
| 10 | Kef-w19 | Absent | Absent | Absent | 2 | Hemolysin III protein | Ornithine decarb-oxylase | Riboflavin, Cobalamin | Absent | |
| 11 | Lentilactobacillus hilgardii | Kef-w8 | Absent | Absent | Absent | 0 | Hemolysin III protein | Agmatine deiminase | Riboflavin, Cobalamin, Pyridoxal | Present |
| 12 | Kef-w9 | Absent | Absent | Absent | 0 | Hemolysin III protein | Agmatine deiminase | Riboflavin, Cobalamin, Pyridoxal | Present | |
| 13 | Kef-w10 | Absent | Absent | Absent | 0 | Hemolysin III protein | Agmatine deiminase | Riboflavin, Cobalamin, Pyridoxal | Present | |
| 14 | Lentilactobacillus kefiri | Kef-m15 | Absent | Absent | CP-002654 | 0 | Hemolysin III protein | Agmatine deiminase | Riboflavin | Absent |
| 15 | Liquorilactobacillus nagelii | Kef-w21 | Absent | Absent | Absent | 0 | Hemolysin III protein, LPS protein | Ornithine decarb-oxylase | Riboflavin | Absent |
| 16 | Lacticaseibacillus rhamnosus | LGG | Absent | Absent | Absent | 1 | Absent | Ornithine decarb-oxylase | Riboflavin, Cobalamin | Absent |
FIGURE 1.
Phylogenetic tree of selected kefir isolates (highlighted in red) and other related strains. The branch lengths are scaled in terms of Genome BLAST Distance Phylogeny approach (GBDP) distance formula d5.
Competitive Adhesion/Exclusion
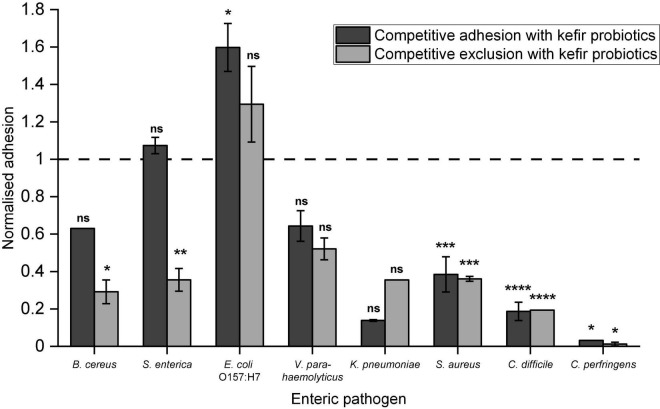
The kefir probiotics blend demonstrated reduction in pathogen adhesion to Caco-2 cells for seven of the eight tested human enteric pathogens, except E. coli O157:H7 (Figure 2). Statistically significant reduction in pathogen adhesion was observed for B. cereus, S. enterica, S. aureus, C. difficile, and C. perfringens. Highest reduction in pathogen adhesion was observed for C. perfringens, in which <0.05%, indicating around 3.5 log CFU reduction, of C. perfringens cells adhered when kefir probiotics were administered in combination. Notably, a significant reduction in pathogen adhesion (>50%) was also observed for S. aureus and C. difficile. In comparison with LGG (Supplementary Figure 2), the kefir probiotics blend showed greater inhibition of adhesion of V. parahaemolyticus and C. perfringens, and similar inhibition of adhesion of the other enteric pathogens. Competitive exclusion appeared to reduce pathogen adhesion more effectively than competitive adhesion, especially for pathogens B. cereus and S. enterica. This suggests that pre-consumption of kefir probiotics may prophylactically protect epithelial cells against pathogen adhesion better than consumption at the time of exposure to the pathogen.
FIGURE 2.
Ability of kefir probiotics to: (a) compete with enteric pathogens for adhesion sites (referred to as competitive exclusion) or (b) exclude adhesion of enteric pathogens when introduced before exposure to the pathogen (referred to as competitive exclusion). Normalized adhesion of 1 indicates that the number of pathogens adhered in competitive adhesion/exclusion with kefir probiotics, was the same as when the pathogens were added alone. Data are presented as an average of duplicates with standard deviation bars. Letters on bars were determined by Dunnett test based on a pairwise-comparison with pathogen-only treatment (refer to Supplementary Table 2 for statistical data). Letters on bars were based on *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns – no significant difference.
Discussion
Bacteria and fungi were isolated from locally-sourced milk and water kefir grains. Isolates were identified to the species level. This process yielded 158 isolates representing 23 microbial species, indicating the wide diversity of culturable microbes from both milk and water kefir. Most of these species have been previously reported to be isolated from kefir, while several were only identified in kefir by culture independent techniques. Notably, Zymomonas mobilis, C. beijerinckii, and Candida ethanolica have not been previously isolated from kefir, but have been purified from beer, soil, vinegar, and apple surfaces, respectively (Richards and Corbey, 1974; Bevardi et al., 2013; Little et al., 2015; Xing et al., 2018; Lynch et al., 2021). While most of these species have been identified in other kefir from different geographical origins, it is worth noting that the microbial species derived in this study were not completely identical to any prior publications. This may be due to differing culture techniques used for the isolation of the microbes from the granules. For instance, Miguel et al. (2011) used only MRS and malt yeast glucose peptone media to isolate bacteria and yeasts, respectively, under aerobic conditions, while in this study, MRS, MYC, NA and M17 media were used for bacteria isolation, YPG and YPD for yeasts isolation, under both aerobic and anaerobic conditions. Inherent differences in microbiota composition in kefir grains of different origins also explains the unique microbiota profile observed in this study. Microbial compositions within kefir grains are known to change between geographical regions, owing to differences in temperature, humidity, altitude conditions, water quality, type of milk/sugar available and hygiene conditions (Zhong et al., 2016). Singapore has a tropical climate, with year-round temperatures ranging from 24 to 32°C. As such, “room temperature” here is considerably higher and more consistent than other climates, which may give rise to microbiota differences in the complex community of the kefir granules. Findings in this study can prove valuable for comparison with studies of the diversity of the kefir microbiota from different geographic origins. Overall, the initial library of 158 strains provided a substantial sample size for this study to proceed, and various assays were conducted to identify the most promising probiotic candidates.
This study adopted a funnel approach to identify kefir-derived microbial strains with highest probiotic potential. Through a series of screening assays, including antimicrobial well diffusion, acid and bile resistance, BSH activity, antioxidant activity, cytotoxicity, adhesion to Caco-2 and whole genome sequencing (WGS), the seven best performing kefir isolates (Table 2) were identified from the initial repertoire of 158. These seven kefir isolates showed good survival [i.e., <2 log(CFU) reduction] in pH 3 acid and bile solutions, non-cytotoxicity and good adhesion to Caco-2, suggesting their suitability for use as oral probiotics. Whole genome analysis revealed an absence of AMR genes and virulence genes, corroborating the safety of these potential probiotic strains. Other qualities identified in the kefir isolates, including BSH activity, high antioxidant activity, presence of vitamins and GABA synthesis genes, add potential functionality of these strains in for general well-being, cholesterol management and mental health aspects. Since these kefir isolates are lactobacilli, and are also isolated from a fermented food source with a safe history of use, these can likely be considered Generally Regarded as Safe (GRAS) by the United States Food and Drug Administration (FDA) (U.S. Food & Drug Administration, 2022), or have Qualified Presumption of Safety (QPS) status granted by the European Food Safety Authority (EFSA) (Koutsoumanis et al., 2019), facilitating their use as potentially novel probiotic agents.
These kefir isolates also demonstrated potent antimicrobial effects against the enteric pathogens B. cereus, S. enterica, V. parahaemolyticus, E. coli O157:H7, and C. difficile, as demonstrated using the well diffusion assay. Generally, larger zones of inhibition were observed for the wells containing cultures of the kefir isolates, as compared to the cell-free supernatant of the same isolates (Table 4). This suggests that the activity of live cells contributed to the anti-pathogenic property of the kefir isolates, possibly due to additional production of antimicrobial compounds during the period of incubation. From WGS results, bacteriocin genes were identified in various kefir isolate strains, suggesting that bacteriocins production may have been the reason for strong pathogen inhibitory activity demonstrated by these kefir isolates. The seven kefir isolates applied in combination showed the ability to outcompete pathogens B. cereus, S. enterica, V. parahaemolyticus, K. pneumoniae, S. aureus, C. difficile, and C. perfringens for epithelial cell adhesion sites and can potentially reduce pathogenesis of these pathogens. It is known that adhesion to intestinal lining plays an important role in pathogenesis of the tested pathogen species (McKee and O’Brien, 1995; Maroncle et al., 2002; Haque et al., 2004; Ritchie et al., 2012; Misawa et al., 2015; Redondo et al., 2015; Tsilia et al., 2015; Anonye et al., 2019). Some of the reduction in pathogen adhesion may also be attributed to the growth inhibitory effects noted for the kefir isolates. As described, enteric bacterial diseases pose a significant global health burden and are an urgent problem to be tackled (WHO, 2022). In particular, non-typhoidal S. enterica and enteropathogenic E. coli, both which were found to be inhibited by kefir isolates, accounted for >50% of enteric pathogen associated deaths (WHO, 2022). Other tested enteric pathogens, including B. cereus [causes 63,400 foodborne disease cases per year in United States alone (Carroll et al., 2019)], V. parahaemolyticus [leading cause of seafood-associated infections in United States and Japan (Todd, 2014)], C. perfringens [second most frequent cause of bacterial foodborne outbreaks in United States (Todd, 2014)] and C. difficile [caused an estimated half a million infections and 29 000 deaths in 2012 in United States (Lessa et al., 2015)], also pose major health concerns and urgently require treatment and prophylactic solutions. The demonstrated impact of the kefir isolates against these pathogens suggests that kefir-derived probiotics are alternative, non-antibiotic, naturally derived candidates for prophylactic and therapeutic applications targeting these enteric pathogens. Further studies will be required to validate the effectiveness of kefir probiotics against enteric bacterial diseases, including examining the effects of kefir probiotics on the intestinal epithelial barrier, investigating the in vivo persistence of kefir probiotics in human gut and in vivo inhibition of the pathogens in model systems.
The findings from this study can be used to develop alternative means to reap the health benefits of kefir. Kefir is a well-known superfood with numerous purported beneficial qualities, including antipathogenic effects, antioxidant properties, cholesterol assimilation, tumor suppression, and gastrointestinal immunity (Bourrie et al., 2016). Currently, consumption of kefir is primarily via drinking the kefir beverage itself, while kefir-derived supplements and functional food products have yet to commercialize extensively. In this study, probiotic candidates isolated from kefir were found to exhibit several of the beneficial properties associated with the kefir beverage itself, such as enteric pathogen inhibition effects, antioxidant properties and some potential for cholesterol management. The probiotic isolates from kefir were also shown to be safe and could be consistently produced through batch fermentations. This suggests a potential avenue for kefir probiotics to be supplemented directly in other product formats to consumers, yet allowing them to receive the desired health benefits. Some of these formats include dietary supplements, such as capsules, tablets, or sachets, or within functional foods and beverages to enhance their nutritional profile. Kefir probiotics may also be encapsulated in polymeric matrices, to imbue additional functional qualities such as protection from the rigors of the GIT and shelf-life stability. These alternative product formats may provide consumers with a greater variety and convenience to attain the health benefits of kefir, thereby facilitating consumers to incorporate diverse microbiome-modulating products into their diet regime.
Conclusion
Microbes were isolated from Singapore-sourced kefir grains, and through a series of screening assays, seven bacterial isolates with the highest probiotic potential were identified. These kefir isolates demonstrate desirable probiotic characteristics, including good survival in acid and bile environments, BSH activity, antioxidant activity, non-cytotoxicity, and high adhesion to Caco-2 cells, lack of virulence or AMR genes. Notably, kefir isolates also demonstrate antimicrobial activity against enteric pathogens in the well diffusion assay, and showed the capacity to out-compete or exclude the attachment of enteric pathogens to Caco-2 cells. Overall, these kefir isolates represent novel probiotic candidates which can mitigate the burden of enteric pathogen associated diseases and benefit human health. Further studies via in vitro and in vivo models and clinical studies are required to ascertain the safety and efficacy of these kefir-derived probiotics.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The link to the datasets can be found at doi: 10.21979/N9/9TG8JH.
Author Contributions
LT planned and conducted all experiments and analyses and also drafted the manuscript. NN conducted the isolation and species identification of microbes from milk and water kefir. YT assisted in the phenotypic screening assays. CT and SL conceived the project direction. CT, PC, and SL provided advice and guidance on this project and also revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
LT was a recipient of the Nanyang President Graduate Scholarship from Nanyang Technological University, Singapore. We are grateful to SCELSE for the use of their facilities to conduct experiments in this project. We acknowledge Kimberly Kline’s lab at SCELSE for provision of multiple bacterial strains. We also acknowledge SCELSE’s sequencing facility for performing Illumina sequencing.
Footnotes
Note that lactobacilli is a generic term to designate all organisms that were classified as Lactobacillaceae until 2020 (Zheng et al., 2020).
Funding
We would like to acknowledge the financial support from the Singapore Centre for Environmental Life Sciences Engineering (SCELSE) (MOE/RCE: M4330019.C70), Ministry of Education AcRF-Tier 1 grant [RG19/18 & RT08/19 (S)], Singapore Food Agency (SFS_RND_SUFP_001_06), and the Singapore National Biofilm Consortium (SNBC/2021/SF2/P04).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.857720/full#supplementary-material
References
- Anonye B. O., Hassall J., Patient J., Detamornrat U., Aladdad A. M., Schüller S., et al. (2019). Probing Clostridium difficile infection in complex human gut cellular models. Front. Microbiol. 10:879. 10.3389/fmicb.2019.00879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala D. I., Cook P. W., Franco J. G., Bugarel M., Kottapalli K. R., Loneragan G. H., et al. (2019). A systematic approach to identify and characterize the effectiveness and safety of novel probiotic strains to control foodborne pathogens. Front. Microbiol. 10:1108. 10.3389/fmicb.2019.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babraham Bioinformatics (2022). FastQC A Quality Control Tool for High Throughput Sequence Data [Internet]. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed February 14, 2022). [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/CMB.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri F., Montanari C., Gardini F., Tabanelli G. (2019). Biogenic amine production by lactic acid bacteria: a review. Foods 8:17. 10.3390/FOODS8010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley M., Gahan C. G. M., Hill C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29 625–651. 10.1016/j.femsre.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Bevardi M., Frece J., Mesarek D., Bošnir J., Mrvèić J., Delaš F., et al. (2013). Antifungal and antipatulin activity of Gluconobacter oxydans isolated from apple surface. Arh. Hig. Rada Toksikol. 64 279–284. 10.2478/10004-1254-64-2013-2308 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/BIOINFORMATICS/BTU170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R. S., Ruppe E., Roberts M. C., Schwarz S., Cattoir V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75 3491–3500. 10.1093/JAC/DKAA345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourrie B. C. T., Willing B. P., Cotter P. D. (2016). The microbiota and health promoting characteristics of the fermented beverage kefir. Front. Microbiol. 7:647. 10.3389/fmicb.2016.00647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt J. U., Jakob F., Geissler A. J., Behr J., Vogel R. F. (2017). Multiple genome sequences of heteropolysaccharide-forming acetic acid bacteria. Genome Announc. 5:e00185-17. 10.1128/GENOMEA.00185-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M., Perna F., Carnevali P., Vitali B., Ciati R., Gionchetti P., et al. (2008). Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 125 286–292. 10.1016/j.ijfoodmicro.2008.04.012 [DOI] [PubMed] [Google Scholar]
- Candela M., Seibold G., Vitali B., Lachenmaier S., Eikmanns B. J., Brigidi P. (2005). Real-time PCR quantification of bacterial adhesion to Caco-2 cells: competition between bifidobacteria and enteropathogens. Res. Microbiol. 156 887–895. 10.1016/J.RESMIC.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Cao C., Hou Q., Hui W., Kwok L., Zhang H., Zhang W. (2019). Assessment of the microbial diversity of Chinese Tianshan tibicos by single molecule, real-time sequencing technology. Food Sci. Biotechnol. 28 139–145. 10.1007/S10068-018-0460-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Zankari E., Garciá-Fernández A., Larsen M. V., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58 3895–3903. 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll L. M., Wiedmann M., Mukherjee M., Nicholas D. C., Mingle L. A., Dumas N. B., et al. (2019). Characterization of emetic and diarrheal Bacillus cereus strains from a 2016 foodborne outbreak using whole-genome sequencing: addressing the microbiological, epidemiological, and bioinformatic challenges. Front. Microbiol. 10:144. 10.3389/fmicb.2019.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey A., Fox E. M., Schmitz-Esser S., Coffey A., McAuliffe O., Jordan K. (2014). Transcriptome analysis of Listeria monocytogenes exposed to biocide stress reveals a multi-system response involving cell wall synthesis, sugar uptake, and motility. Front. Microbiol. 5:68. 10.3389/FMICB.2014.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2019). Antibiotic Resistance Threats in the United States, 1–113. Available online at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed February 14, 2022). [Google Scholar]
- Chandel D., Sharma M., Chawla V., Sachdeva N., Shukla G. (2019). Isolation, characterization and identification of antigenotoxic and anticancerous indigenous probiotics and their prophylactic potential in experimental colon carcinogenesis. Sci. Rep. 9:14769. 10.1038/s41598-019-51361-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Liu T., Ye T., Yang X., Xue Y., Shen Y., et al. (2021). Effect of lactic acid bacteria and yeasts on the structure and fermentation properties of Tibetan kefir grains. Int. Dairy J. 114:104943. 10.1016/J.IDAIRYJ.2020.104943 [DOI] [Google Scholar]
- Costantini A., Pietroniro R., Doria F., Pessione E., Garcia-Moruno E. (2013). Putrescine production from different amino acid precursors by lactic acid bacteria from wine and cider. Int. J. Food Microbiol. 165 11–17. 10.1016/J.IJFOODMICRO.2013.04.011 [DOI] [PubMed] [Google Scholar]
- Davis M. W. (2022). ApE – A Plasmid Editor [Internet]. Available online at: https://jorgensen.biology.utah.edu/wayned/ape/ (accessed February 14, 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak-Dados A., Michalski M., Osek J. (2020). Histamine and other biogenic amines in food. J. Vet. Res. 64 281–288. 10.2478/JVETRES-2020-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorda F. A., de Melo Pereira G. V., Thomaz-Soccol V., Rakshit S. K., Pagnoncelli M. G. B., Vandenberghe L. P. S., et al. (2017). Microbiological, biochemical, and functional aspects of sugary kefir fermentation – a review. Food Microbiol. 66 86–95. 10.1016/j.fm.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Gamba R. R., Yamamoto S., Sasaki T., Michihata T., Mahmoud A.-H., Koyanagi T., et al. (2019). Microbiological and functional characterization of kefir grown in different sugar solutions. Food Sci. Technol. Res. 25 303–312. 10.3136/fstr.25.303 [DOI] [Google Scholar]
- Gao J., Gu F., Abdella N. H., Ruan H., He G. (2012). Investigation on culturable microflora in Tibetan kefir grains from different areas of China. J. Food Sci. 77 M425–M433. 10.1111/J.1750-3841.2012.02805.X [DOI] [PubMed] [Google Scholar]
- Grand View Research (2022). Probiotics Market Size & Share Analysis Report, 2021-2028. Available online at: https://www.grandviewresearch.com/industry-analysis/probiotics-market (accessed February 14, 2022). [Google Scholar]
- Gulitz A., Stadie J., Wenning M., Ehrmann M. A., Vogel R. F. (2011). The microbial diversity of water kefir. Int. J. Food Microbiol. 151 284–288. 10.1016/J.IJFOODMICRO.2011.09.016 [DOI] [PubMed] [Google Scholar]
- Gut A. M., Vasiljevic T., Yeager T., Donkor O. N. (2021). Kefir characteristics and antibacterial properties – potential applications in control of enteric bacterial infection. Int. Dairy J. 118:105021. 10.1016/j.idairyj.2021.105021 [DOI] [Google Scholar]
- Haque A., Bowe F., Fitzhenry R. J., Frankel G., Thomson M., Heuschkel R., et al. (2004). Early interactions of Salmonella enterica serovar typhimurium with human small intestinal epithelial explants. Gut 53 1424–1430. 10.1136/gut.2003.037382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. A., Chou J. Y. (2021). Yeasts in fermented food and kefir: in vitro characterization of probiotic traits. J. Anim. Plant Sci. 31 567–582. 10.36899/JAPS.2021.2.0245 [DOI] [Google Scholar]
- Hurtado-Romero A., Del Toro-Barbosa M., Gradilla-Hernández M. S., Garcia-Amezquita L. E., García-Cayuela T. (2021). Probiotic properties, prebiotic fermentability, and GABA-producing capacity of microorganisms isolated from Mexican milk kefir grains: a clustering evaluation for functional dairy food applications. Foods 10:2275. 10.3390/FOODS10102275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inturri R., Stivala A., Furneri P. M., Blandino G. (2016). Growth and adhesion to HT-29 cells inhibition of Gram-negatives by Bifidobacterium longum BB536 & Lactobacillus rhamnosus HN001 alone and in combination. Eur. Rev. Med. Pharmacol. Sci. 20 4943–4949. [PubMed] [Google Scholar]
- Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T., Aluru S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensen K. G., Scheutz F., Lund O., Hasman H., Kaas R. S., Nielsen E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52 1501–1510. 10.1128/JCM.03617-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazou M., Grafakou A., Tsakalidou E., Georgalaki M. (2021). Zooming into the microbiota of home-made and industrial kefir produced in greece using classical microbiological and amplicon-based metagenomics analyses. Front. Microbiol. 12:621069. 10.3389/FMICB.2021.621069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedare S. B., Singh R. P. (2011). Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 48 412–422. 10.1007/s13197-011-0251-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuzaki N., Shima J., Kawamoto S., Momose H., Kimura T. (2005). Production of γ-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 22 497–504. 10.1016/j.fm.2005.01.002 [DOI] [Google Scholar]
- Korsak N., Taminiau B., Leclercq M., Nezer C., Crevecoeur S., Ferauche C., et al. (2015). Short communication: evaluation of the microbiota of kefir samples using metagenetic analysis targeting the 16S and 26S ribosomal DNA fragments. J. Dairy Sci. 98 3684–3689. 10.3168/jds.2014-9065 [DOI] [PubMed] [Google Scholar]
- Koutsoumanis K., Allende A., Alvarez-Ordóñez A., Bolton D., Bover-Cid S., Chemaly M., et al. (2019). Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 10: suitability of taxonomic units notified to EFSA until March 2019. EFSA J. 19:e06689. 10.2903/j.efsa.2019.5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. R., Yeap S. K., Mohamad N. E., Abdullah J. O., Masarudin M. J., Khalid M., et al. (2021). Metagenomic and phytochemical analyses of kefir water and its subchronic toxicity study in BALB/c mice. BMC Complement. Med. Ther. 21:183. 10.1186/s12906-021-03358-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S., Song T. Y., Kong W. S., Yoon M. H. (2013). Characterization of γ-aminobutyric acid (GABA) produced by a lactic acid bacterium from button mushroom bed. J. Mushrooms 11 181–186. 10.14480/jm.2013.11.4.181 [DOI] [Google Scholar]
- Lessa F. C., Mu Y., Bamberg W. M., Beldavs Z. G., Dumyati G. K., Dunn J. R., et al. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372 825–834. 10.1056/nejmoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little G. T., Winzer K., Minton N. P. (2015). Genome sequence of the solvent-producing Clostridium beijerinckii strain 59B, isolated from Staffordshire garden soil. Genome Announc. 3:e00108-15. 10.1128/GENOMEA.00108-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Leathers T. D., Copeland A., Chertkov O., Goodwin L., Mills D. A. (2011). Complete genome sequence of Lactobacillus buchneri NRRL B-30929, a novel strain from a commercial ethanol plant. J. Bacteriol. 193 4019–4020. 10.1128/JB.05180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch K. M., Wilkinson S., Daenen L., Arendt E. K. (2021). An update on water kefir: microbiology, composition and production. Int. J. Food Microbiol. 345:109128. 10.1016/J.IJFOODMICRO.2021.109128 [DOI] [PubMed] [Google Scholar]
- Maroncle N., Balestrino D., Rich C., Forestier C. (2002). Identification of Klebsiella pneumoniae genes involved in intestinal colonization and adhesion using signature-tagged mutagenesis. Infect. Immun. 70 4729–4734. 10.1128/IAI.70.8.4729-4734.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh A. J., O’Sullivan O., Hill C., Ross R. P., Cotter P. D. (2013). Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol. Lett. 348 79–85. 10.1111/1574-6968.12248 [DOI] [PubMed] [Google Scholar]
- Martínez-Torres A., Gutiérrez-Ambrocio S., Heredia-del-Orbe P., Villa-Tanaca L., Hernández-Rodríguez C. (2017). Inferring the role of microorganisms in water kefir fermentations. Int. J. Food Sci. Technol. 52 559–571. 10.1111/ijfs.13312 [DOI] [Google Scholar]
- McKee M. L., O’Brien A. D. (1995). Investigation of enterohemorrhagic Escherichia coli O157:H7 adherence characteristics and invasion potential reveals a new attachment pattern shared by intestinal E. coli. Infect. Immun. 63 2070–2074. 10.1128/iai.63.5.2070-2074.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Göker M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. 10.1038/s41467-019-10210-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes A. G. T., Melo D. S., Ramos C. L., Moreira S. I., Alves E., Schwan R. F. (2020). Yeasts isolated from Brazilian fermented foods in the protection against infection by pathogenic food bacteria. Microb. Pathog. 140:103969. 10.1016/j.micpath.2020.103969 [DOI] [PubMed] [Google Scholar]
- Michael D. R., Moss J. W. E., Lama Calvente D., Garaiova I., Plummer S. F., Ramji D. P. (2016). Lactobacillus plantarum CUL66 can impact cholesterol homeostasis in Caco-2 enterocytes. Benef. Microbes 7 443–451. 10.3920/BM2015.0146 [DOI] [PubMed] [Google Scholar]
- Miguel M. G. C. P., Cardoso P. G., Magalhães K. T., Schwan R. F. (2011). Profile of microbial communities present in tibico (sugary kefir) grains from different Brazilian States. World J. Microbiol. Biotechnol. 27 1875–1884. 10.1007/s11274-010-0646-6 [DOI] [Google Scholar]
- Misawa Y., Kelley K. A., Wang X., Wang L., Park W. B., Birtel J., et al. (2015). Staphylococcus aureus colonization of the mouse gastrointestinal tract is modulated by wall teichoic acid, capsule, and surface proteins. PLoS Pathog. 11:e1005061. 10.1371/JOURNAL.PPAT.1005061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo-Mera A., Rastall R. A., Gibson G. R., Charalampopoulos D., Chatzifragkou A. (2019). Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 103:6463. 10.1007/S00253-019-09978-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A. F., Moure M. C., Florencia Q., Fiorella E., Simonelli N., Medrano M., et al. (2022). Water kefir, a fermented beverage containing probiotic microorganisms: from ancient and artisanal manufacture to industrialized and regulated commercialization. Future Foods 5:100123. 10.1016/j.fufo.2022.100123 [DOI] [Google Scholar]
- Mu G., Gao Y., Tuo Y., Li H., Zhang Y., Qian F., et al. (2018). Assessing and comparing antioxidant activities of lactobacilli strains by using different chemical and cellular antioxidant methods. J. Dairy Sci. 101 10792–10806. 10.3168/JDS.2018-14989 [DOI] [PubMed] [Google Scholar]
- Nalbantoglu U., Cakar A., Dogan H., Abaci N., Ustek D., Sayood K., et al. (2014). Metagenomic analysis of the microbial community in kefir grains. Food Microbiol. 41 42–51. 10.1016/j.fm.2014.01.014 [DOI] [PubMed] [Google Scholar]
- Packeiser H., Lim C., Balagurunathan B., Wu J., Zhao H. (2013). An extremely simple and effective colony PCR procedure for bacteria, yeasts, and microalgae. Appl. Biochem. Biotechnol. 169 695–700. 10.1007/s12010-012-0043-8 [DOI] [PubMed] [Google Scholar]
- Pidoux M. (1989). The microbial flora of sugary kefir grain (the ginger beer plant): biosynthesis of the grain from Lactobacillus hilgardii producing a polysaccharide gel. MIRCEN J. Appl. Microbiol. Biotechnol. 5 223–238. 10.1007/BF01741847 [DOI] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo L. M., Carrasco J. M. D., Redondo E. A., Delgado F., Miyakawa M. E. F. (2015). Clostridium perfringens type E virulence traits involved in gut colonization. PLoS One 10:e0121305. 10.1371/JOURNAL.PONE.0121305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M., Corbey D. A. (1974). Isolation of Zymomonas mobilis from primed beer. J. Inst. Brew. 80 241–244. 10.1002/j.2050-0416.1974.tb03612.x [DOI] [Google Scholar]
- Ritchie J. M., Rui H., Zhou X., Iida T., Kodoma T., Ito S., et al. (2012). Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 8:e1002593. 10.1371/JOURNAL.PPAT.1002593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A., San Mauro M., Sanchez A., Torres J. M., Marquina D. (2003). The antimicrobial properties of different strains of Lactobacillus spp. isolated from kefir. Syst. Appl. Microbiol. 26 434–437. 10.1078/072320203322497464 [DOI] [PubMed] [Google Scholar]
- Sarikkha P., Nitisoravut R., Poljungreed I. (2015). Identification of bacteria and yeast communities in a Thai sugary kefir by polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) analyses. J. Ind. Technol. 11 26–39. [Google Scholar]
- Senan S., Prajapati J. B., Joshi C. G. (2015). Feasibility of genome-wide screening for biosafety assessment of probiotics: a case study of Lactobacillus helveticus MTCC 5463. Probiotics Antimicrob. Proteins 7 249–258. 10.1007/s12602-015-9199-1 [DOI] [PubMed] [Google Scholar]
- Shreiner A. B., Kao J. Y., Young V. B. (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31 69–75. 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers M., Swings J. (2005). Bergey’s Manual® of Systematic Bacteriology, Vol. 2. Boston, MA: Springer US, 41–95. [Google Scholar]
- Simova E., Beshkova D., Angelov A., Hristozova T., Frengova G., Spasov Z. (2002). Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J. Ind. Microbiol. Biotechnol. 28 1–6. 10.1038/sj/jim/7000186 [DOI] [PubMed] [Google Scholar]
- Surachat K., Sangket U., Deachamag P., Chotigeat W. (2017). In silico analysis of protein toxin and bacteriocins from Lactobacillus paracasei SD1 genome and available online databases. PLoS One 12:e0183548. 10.1371/JOURNAL.PONE.0183548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šušković J., Kos B., Beganović J., Pavunc A. L., Habjaniè K., Matoć S. (2010). Antimicrobial activity – the most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 48 296–307. [Google Scholar]
- Talib N., Mohamad N. E., Yeap S. K., Hussin Y., Mubin Aziz M. N., Masarudin M. J., et al. (2019). Isolation and characterization of Lactobacillus spp. from kefir samples in Malaysia. Molecules 24:2606. 10.3390/MOLECULES24142606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa Y., Fujisawa T., Kaminuma E., Nakamura Y., Arita M. (2016). DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 35 173–184. 10.12938/bmfh.16-003l [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. (1975). Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29 807–813. 10.1128/am.29.6.807-813.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd E. C. D. (2014). “Foodborne diseases: overview of biological hazards and foodborne diseases,” in Encyclopedia of Food Safety, ed. Motarjemi Y. (Cambridge, MA: Academic Press; ), 221–242. [Google Scholar]
- Tsilia V., Uyttendaele M., Kerckhof F. M., Rajkovic A., Heyndrickx M., Van De Wiele T. (2015). Bacillus cereus adhesion to simulated intestinal mucus is determined by its growth on mucin, rather than intestinal environmental parameters. Foodborne Pathog. Dis. 12 904–913. 10.1089/fpd.2014.1926 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449 804–810. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food & Drug Administration (2022). GRAS Notices [Internet]. Available online at: https://www.accessdata.fda.gov/scripts/fdcc/?set=GRASNotices (accessed February 14, 2022). [Google Scholar]
- UniProt (2022). Lactobacillus plantarum subsp. Plantarum P-8. Available from: https://www.uniprot.org/taxonomy/767468 (accessed February 14, 2022). [Google Scholar]
- Van den Bossche S., Vandeplassche E., Ostyn L., Coenye T., Crabbé A. (2020). Bacterial interference with lactate dehydrogenase assay leads to an underestimation of cytotoxicity. Front. Cell. Infect. Microbiol. 10:494. 10.3389/fcimb.2020.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel A. J., De Jong A., Song C., Viel J. H., Kok J., Kuipers O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46 W278–W281. 10.1093/nar/gky383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verce M., De Vuyst L., Weckx S. (2019). Shotgun metagenomics of a water kefir fermentation ecosystem reveals a novel Oenococcus species. Front. Microbiol. 10:479. 10.3389/fmicb.2019.00479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinderola C. G., Duarte J., Thangavel D., Perdigón G., Farnworth E., Matar C. (2005). Immunomodulating capacity of kefir. J. Dairy Res. 72 195–202. 10.1017/S0022029905000828 [DOI] [PubMed] [Google Scholar]
- WHO (2022). WHO Estimates of the Global Burden of Foodborne Diseases: Executive Summary [Internet]. WHO Executive Summary. 2007. Available online at: https://apps.who.int/iris/bitstream/handle/10665/199350/?sequence=1 (accessed February 14, 2022). [Google Scholar]
- Wyder M. T., Spillmann H., Puhan Z. (1997). Investigation of the yeast flora in dairy products: a case study of kefyr. Food Technol. Biotechnol. 35 299–304. [Google Scholar]
- Xing X., Wang Y., Huo N., Wang R. (2018). Candida ethanolica strain Y18 enhances aroma of Shanxi aged-vinegar. Food Sci. Technol Res. 24 1069–1081. 10.3136/FSTR.24.1069 [DOI] [Google Scholar]
- Zanirati D. F., Abatemarco M., Sandes S. H. C., Nicoli J. R., Nunes ÁC., Neumann E. (2015). Selection of lactic acid bacteria from Brazilian kefir grains for potential use as starter or probiotic cultures. Anaerobe 32 70–76. 10.1016/j.anaerobe.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Zheng J., Wittouck S., Salvetti E., Franz C. M. A. P., Harris H. M. B., Mattarelli P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70 2782–2858. 10.1099/ijsem.0.004107 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Lu Y., Wang J., Yang L., Pan C., Huang Y. (2013). Probiotic properties of Lactobacillus strains isolated from Tibetan kefir grains. PLoS One 8:e69868. 10.1371/journal.pone.0069868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Hou Q., Kwok L., Yu Z., Zheng Y., Sun Z., et al. (2016). Bacterial microbiota compositions of naturally fermented milk are shaped by both geographic origin and sample type. J. Dairy Sci. 99 7832–7841. 10.3168/jds.2015-10825 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The link to the datasets can be found at doi: 10.21979/N9/9TG8JH.