Abstract
Machine learning techniques used in computer-aided medical image analysis usually suffer from the domain shift problem caused by different distributions between source/reference data and target data. As a promising solution, domain adaptation has attracted considerable attention in recent years. The aim of this paper is to survey the recent advances of domain adaptation methods in medical image analysis. We first present the motivation of introducing domain adaptation techniques to tackle domain heterogeneity issues for medical image analysis. Then we provide a review of recent domain adaptation models in various medical image analysis tasks. We categorize the existing methods into shallow and deep models, and each of them is further divided into supervised, semi-supervised and unsupervised methods. We also provide a brief summary of the benchmark medical image datasets that support current domain adaptation research. This survey will enable researchers to gain a better understanding of the current status, challenges and future directions of this energetic research field.
Index Terms—: Domain adaptation, domain shift, machine learning, deep learning, medical image analysis
I. Introduction
Machine learning has been widely used in medical image analysis, and typically assumes that the training dataset (source/reference domain) and test dataset (target domain) share the same data distribution [1]. However, this assumption is too strong and may not hold in practice. Previous studies have revealed that the test error generally increases in proportion to the distribution difference between training and test datasets [2], [3]. This is referred to as the “domain shift” problem [4]. Even in the deep learning era, deep Convolutional Neural Networks (CNNs) trained on large-scale image datasets may still suffer from domain shift [5]. Thus, how to handle domain shift is a crucial issue to effectively apply machine learning methods to medical image analysis.
Unlike natural image analysis with large-scale labeled datasets such as ImageNet [6], [7], in medical image analysis, a major challenge for medical image analysis is the lack of labeled data to construct reliable and robust models. Labeling medical images is generally expensive, time-consuming and tedious, requiring labor-intensive participation of physicians, radiologists and other experts. An intuitive solution is to reuse pre-trained models for some related domains [8]. However, the domain shift problem is still widespread among different medical image datasets due to different scanners, scanning parameters, and subject cohorts, etc. As a promising solution to tackle the domain shift/heterogeneity among medical image datasets, domain adaptation has attracted increasing attention in the field, aiming to minimize distribution gap among different but related domains, as shown in Fig. 1. Many researchers have engaged in leveraging domain adaptation methods to solve various tasks in medical image analysis.
Fig. 1.
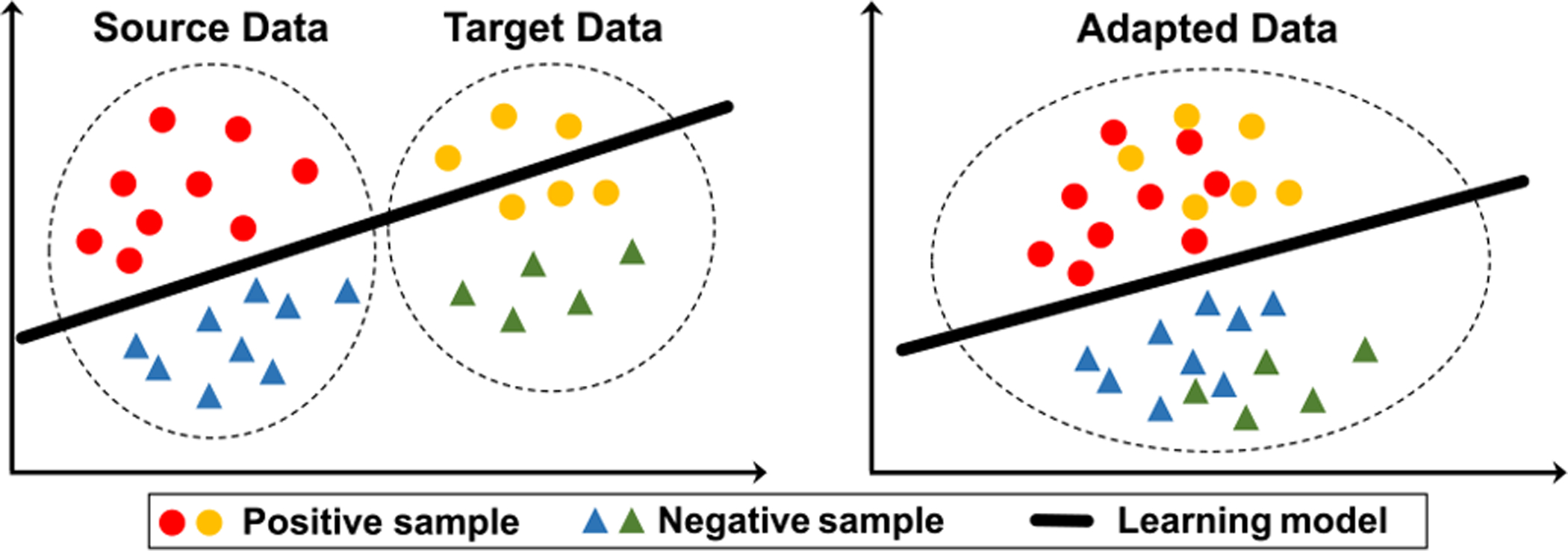
Illustration of source and target data with (left) original feature distributions, and (right) new feature distributions after domain adaptation, where domain adaptation techniques help to alleviate the “domain shift” problem [4] between source and target domains.
There have been a number of surveys on domain adaptation [9]–[15] and transfer learning [16]–[21] with natural images. However, there are only very limited reviews related to domain adaptation and their applications in medical image analysis which is a broad and important research area. Cheplygina et al. [22] provide a broad survey covering semi-supervised, multi-instance, and transfer learning for medical image analysis. Due to the wide scope, they only review general transfer learning methods in medical imaging applications without focusing on domain adaptation. Valverde et al. [23] conduct a survey on transfer learning in magnetic resonance (MR) brain imaging. Morid et al. [24] review transfer learning methods based on pre-training on ImageNet [6] that can benefit from knowledge from large-scale labeled natural images. Recent studies propose to design deep learning based domain adaptation models for medical image analysis, but many of them are not included in these review/survey papers.
In this paper, we review and discuss recent advances and challenges of domain adaptation for medical image analysis. We systematically summarize the existing methods according to their characteristics. Specifically, we categorize different methods into two groups: 1) shallow models, and 2) deep models. Each group is further divided into three categories, i.e., supervised, semi-supervised and unsupervised methods. To the best of our knowledge, this is among the first attempts to systematically review domain adaptation methods for analyzing multi-site multi-modality medical imaging data. Unlike previous review/survey papers that usually focus on a single modality or organ/object (e.g., brain MRI [23]), this survey reviews studies that cover multiple imaging modalities and organs/objects (e.g., brain, lung and heart). In addition, we also review the most advanced deep domain adaptation models recently used in medical image analysis.
The rest of this paper is organized as follows. We firstly introduce some background knowledge in Section II. In Sections III–IV, we review the recent advances of domain adaptation methods in medical image analysis. Challenges and future research directions are discussed in Section V. Finally, the conclusion is given in Section VI. A summary of benchmark medical image datasets used in domain adaptation problems can be found in Supplementary Materials.
II. Background
A. Domain Shift in Medical Image Analysis
For a machine learning model, domain shift [4], [26]–[28] refers to the change of data distribution between its training dataset (source/reference domain) and test dataset (target domain). The domain shift problem is very common in practical applications of various machine learning algorithms and may cause significant performance degradation. Especially for multi-center studies, domain shift is widespread among different imaging centers/sites that may use different scanners, scanning protocols, and subject populations, etc. Fig. 2 illustrates the problem of inter-center domain shift in terms of the intensity distribution of structural magnetic resonance imaging (MRI) from four independent sites (i.e., UCL, Montreal, Zurich and Vanderbilt) in the Gray Matter segmentation challenge [25], [29]. Fig. 3 illustrates the image-level distribution heterogeneity caused by different scanners [30]. From Figs. 2–3, one can observe that there are clear distribution shifts in these medical imaging data. However, many conventional machine learning methods ignore this problem, which would lead to performance degradation [27], [31]. Recently, domain adaptation has attracted increasing interests and attention of researchers, and become an important research topic in machine learning based medical image analysis [22], [32]–[34].
Fig. 2.
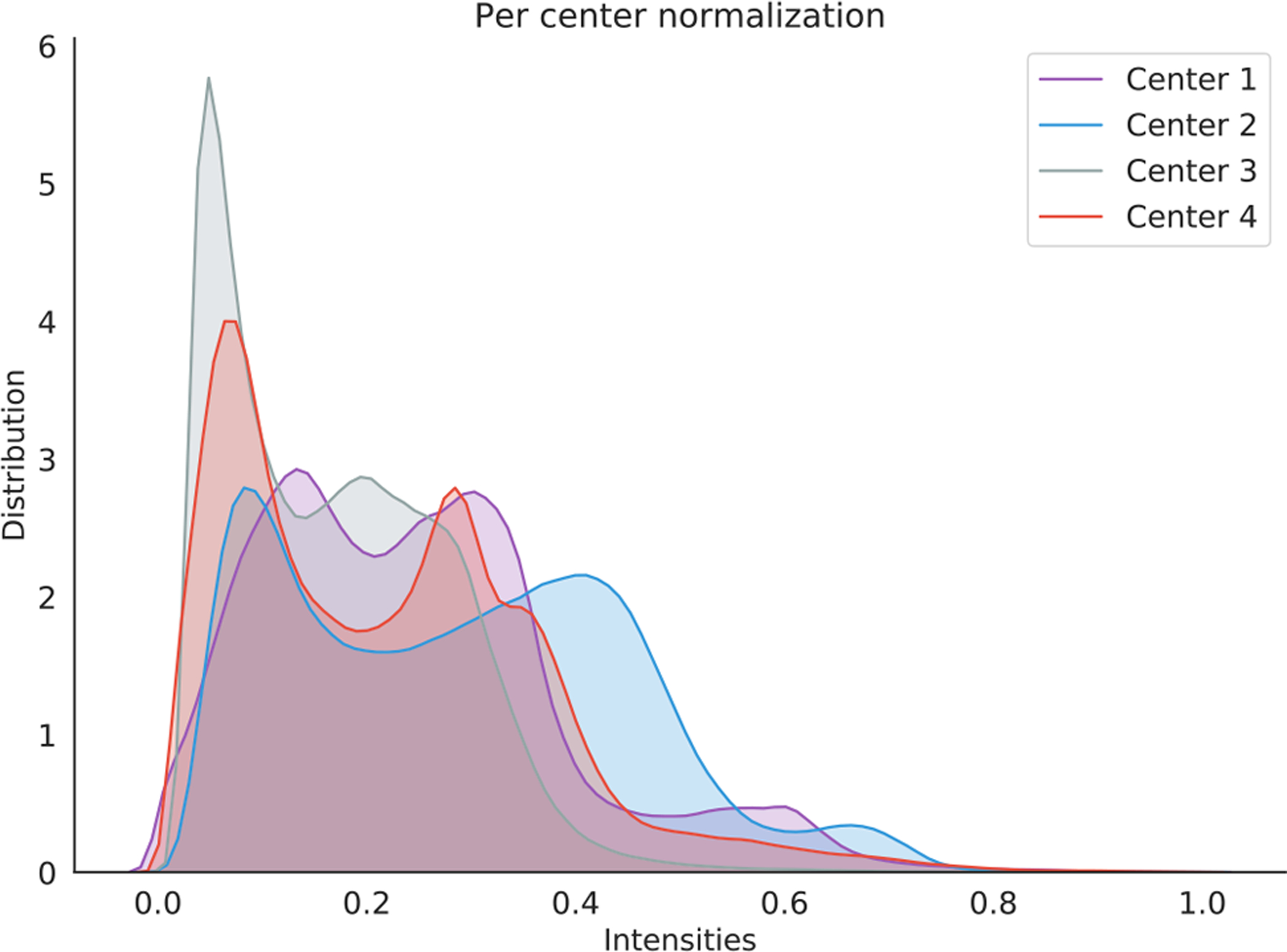
Intensity distribution of MRI axial-slice pixels from four different datasets (i.e., UCL, Montreal, Zurich, and Vanderbilt) that collected for gray matter segmentation. Intensity is normalized between 0 and 1 for each site. Image courtesy to C. Perone [25].
Fig. 3.
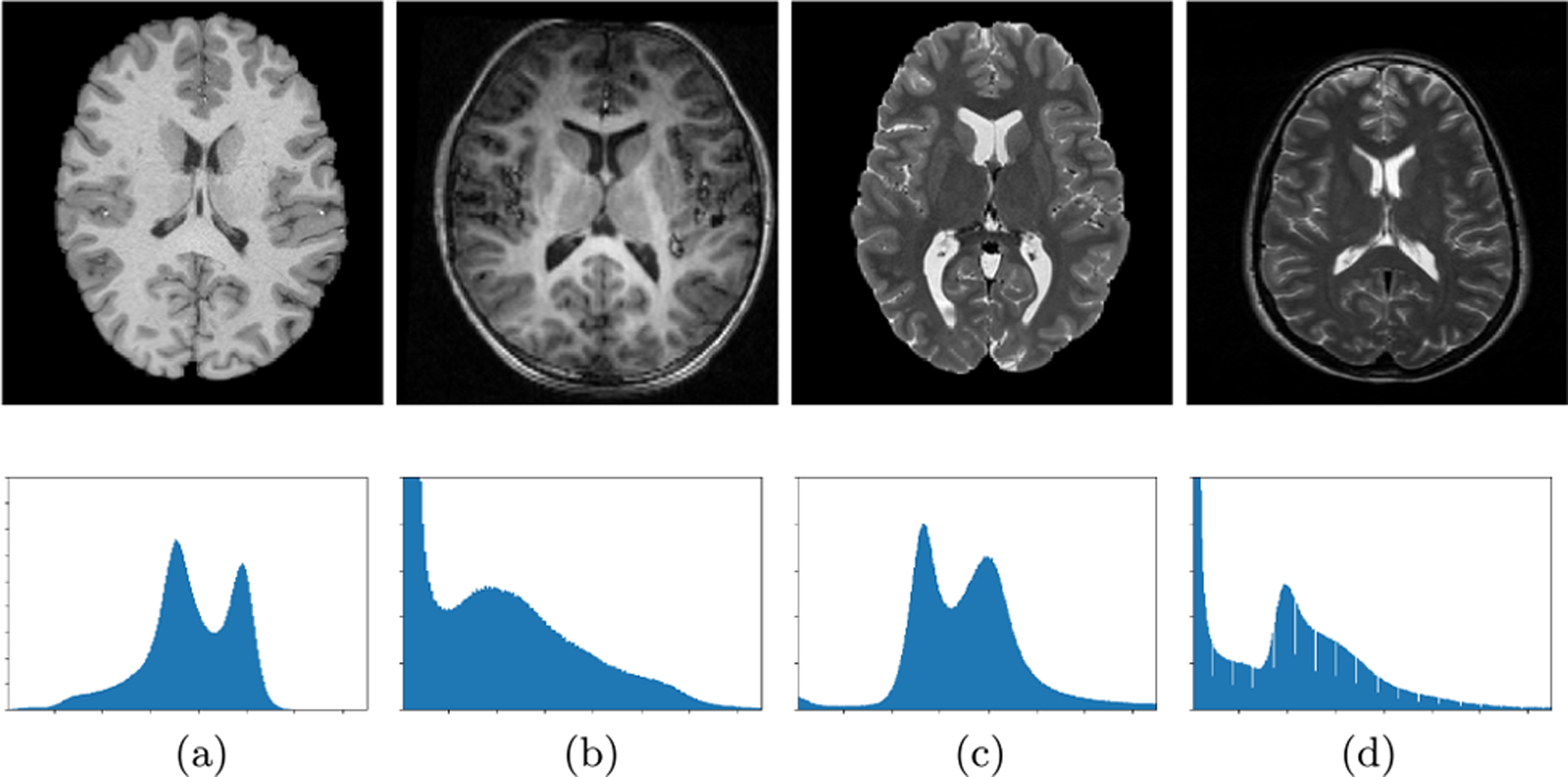
Image slices (top) and corresponding intensity distribution (bottom) of normalized T1-weighted (a), (b) and T2-weighted (c), (d) MRIs from different scanners. Image courtesy to N. Karani [30].
B. Domain Adaptation and Transfer Learning
This survey focuses on domain adaptation for medical image analysis. Since domain adaptation can be regarded as a special type of transfer learning, we first review their definitions to provide a clear understanding of their differences. In a typical transfer learning setting, there are two concepts: “domain” and “task” [16]–[18]. A domain relates to the feature space of a specific dataset and the marginal probability distribution of features. A task relates to the label space of a dataset and an objective predictive function. The goal of transfer learning is to transfer the knowledge learned from the task Ta on domain A to the task Tb on domain B. Note that either the domain and the task may change during the transfer learning process.
Domain adaptation, the focus of this survey, is a particular and popular type of transfer learning [9]–[13]. For domain adaptation, it is assumed that the domain feature spaces and tasks remain the same while the marginal distributions are different between the source and target domains (datasets). It can be mathematically depicted as follows. Let represent the joint feature space and the corresponding label space, respectively. A source domain S and a target domain T are defined on , with different distributions Ps and Pt, respectively. Suppose we have ns labeled samples (subjects) in the source domain, i.e., , and also have nt samples (with or without labels) in the target domain, i.e., . Then the goal of domain adaptation (DA) is to transfer knowledge learned from S to T to perform a specific task on T, and this task is shared by S and T.
C. Problem Settings of Domain Adaptation
In DA, the source domain and target domain share the same learning tasks. In practice, DA can be categorized into different groups according to different scenarios, constraints, and algorithms. In Fig. 4, we summarize different categories of DA methods for medical image analysis based on six problem settings, i.e., model type, label availability, modality difference, number of sources, and adaptation steps. Each group is not mutually exclusive to each other, and we employ the model type as the main criterion in this paper.
Fig. 4.
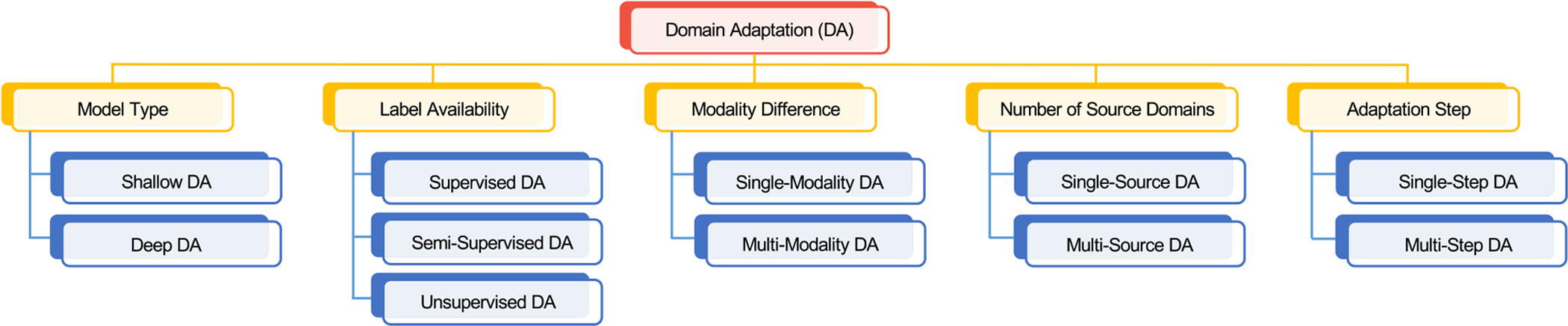
An overview of different categories of domain adaptation methods for medical image analysis.
1). Model Type:
Shallow DA & Deep DA. In terms of whether the learning model is shallow or deep, the DA methods can be divided into shallow DA and deep DA [9], [10], [13]. Shallow DA methods usually rely on human-engineered imaging features and conventional machine learning models. To avoid hand-crafted feature engineering, deep learning based domain adaptation methods have emerged and achieved state-of-the-art performance in many applications. Existing deep DA methods (especially those with CNN architectures) generally integrate feature learning and model training into end-to-end learning models, where the data adaptation is performed in a task-oriented manner.
2). Label Availability:
Supervised DA & Semi-Supervised DA & Unsupervised DA. In terms of label availability, existing DA methods can be divided into supervised DA, semi-supervised DA, and unsupervised DA [11], [13]. In supervised DA, a small number of labeled data in the target domain are available for model training. Since labeling/annotating medical images is usually time-consuming and tedious, there are often limited or even no labeled data in some sites/domains. To address this issue, semi-supervised and unsupervised DA methods have been developed. In semi-supervised DA, a small number of labeled data as well as redundant unlabeled data in the target domain are available in the training process. In unsupervised DA, only unlabeled target data are available for training the adaptation model. There is an extreme case that even unlabeled target data are not accessible during the training time. The learning model is only allowed to be trained on several related source domains to gain enough generalization ability for the target domain. This problem is referred to as domain generalization, while a few related research has been conducted for medical image analysis [35].
3). Modality Difference:
Single-Modality DA & Cross-Modality DA. In terms of modality difference, the existing DA methods can be divided into single-modality DA and cross-modality DA [9], [36], [37]. In single modality DA, the source and target domains share the same data modality. For example, the source domain consists of MRIs collected in vendor A, and the target domain contains MRIs acquired from vendor B [38]. With the development of imaging techniques, more multi-modality data (e.g., MRI, CT and PET) have been collected. To bridge the distribution gap between different imaging modalities, many cross-modality DA methods have emerged. In cross-modality DA, the modalities of source and target domains are different with various scanning technologies. For example, the source domain consists of MR images, whereas the target domain contains CT images [37].
4). Number of Sources:
Single-Source DA & Multi-Source DA. In terms of the number of source domains, the existing DA methods can be divided into single-source DA and multi-source DA approaches [12], [14], [15]. Single-source DA is usually based on the assumption that there is only one source domain, but this assumption is too strong and may not hold in practice. To tackle scenarios where training data may come from multiple sites/domains, multi-source DA has been proposed. Since there is also data heterogeneity among different source domains, multi-source DA is quite challenging. Most of the existing DA methods for medical image analysis fall into the single-source DA category.
5). Adaptation Step:
One-Step DA & Multi-Step DA. In terms of adaptation steps, existing methods can be divided into one-step and multi-step DA [10], [39], [40]. In one-step DA, adaption between source and target domains is accomplished in one step due to a relatively close relationship between them. For scenarios where data heterogeneity between source and target domains is significant (e.g., from ImageNet to medical image datasets), multi-step DA (also called distant/transitive DA) methods have been proposed, where intermediate domains are often introduced to bridge the distribution gap between source and target domains.
Since extensive domain adaptation methods have been proposed, it is typically suggested to fully consider the background and characteristics of specific problems when applying different adaptation methods to medical image analysis. When there are sufficient data and computation resources, deep domain adaptation methods tend to achieve better performance, compared with conventional machine learning methods. For medical image based learning tasks, domain adaptation can be performed at two levels, i.e., feature-level and image-level. Generally, feature-level methods are more suitable for classification or regression problems. When there are well-defined image features (e.g., tissue intensity), it is also a good choice to utilize feature-level methods for domain adaptation. Image-level adaptation methods, e.g., via generative adversarial networks (GANs), are often suitable for segmentation tasks to preserve more original structure information of pixels/voxels. For cross-modality tasks, GAN-based adaptation is also a good choice for bridging different data modalities.
III. Shallow Domain Adaptation Methods
In this section, we review shallow domain adaptation methods based on human-engineered features and conventional machine learning models for medical image analysis. We first introduce two commonly-used strategies in shallow DA methods: 1) instance weighting, and 2) feature transformation.
Instance weighting is one of the most popular strategies adopted by shallow DA methods for medical image analysis [41]–[45]. In this strategy, samples/instances in the source domain are assigned with different weights according to their relevance with target samples/instances. Generally, source instances that are more relevant to the target instances will be assigned larger weights. After instance weighting, a learning model (e.g., classifier or regressor) is trained on the re-weighted source samples, thus reducing domain shift between the source and target domains. Fig. 5 illustrates the effect of instance weighting strategy on the source and target domains. From Fig. 5(a)–(b), we can see that the domain difference between the source and target domain is large. After instance weighting, the domain difference is reduced based on those re-weighted source instances, as shown in Fig. 5(b)–(c).
Fig. 5.
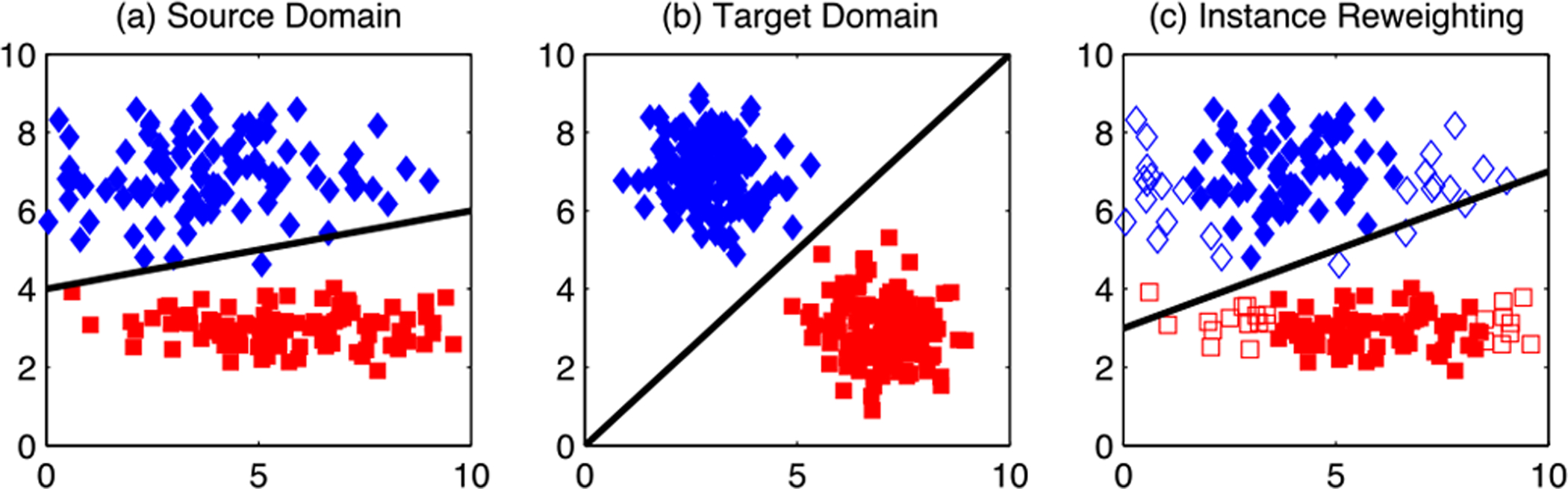
Illustration of instance weighting which can alleviate domain shift. (a) Source domain after feature matching (i.e., discovering a shared feature representation by jointly reducing the distribution difference and preserving the important properties of input data). (b) Target domain after feature matching. (c) Source domain after joint feature matching and instance weighting, with unfilled markers indicating irrelevant source instances that have smaller weights. Image courtesy to Long et al. [46].
Feature transformation strategy focuses on transforming source and target samples from their original feature spaces to a new shared feature representation space [47]–[49]. As shown in Fig. 6, the goal of feature transformation for DA is to construct a common/shared feature space for the source and target domains to reduce their distribution gap, based on various techniques such as low-rank representation [49]. Then, a learning model can be trained on the new feature space, which is less affected by the domain shift in the original feature space between the two domains.
Fig. 6.
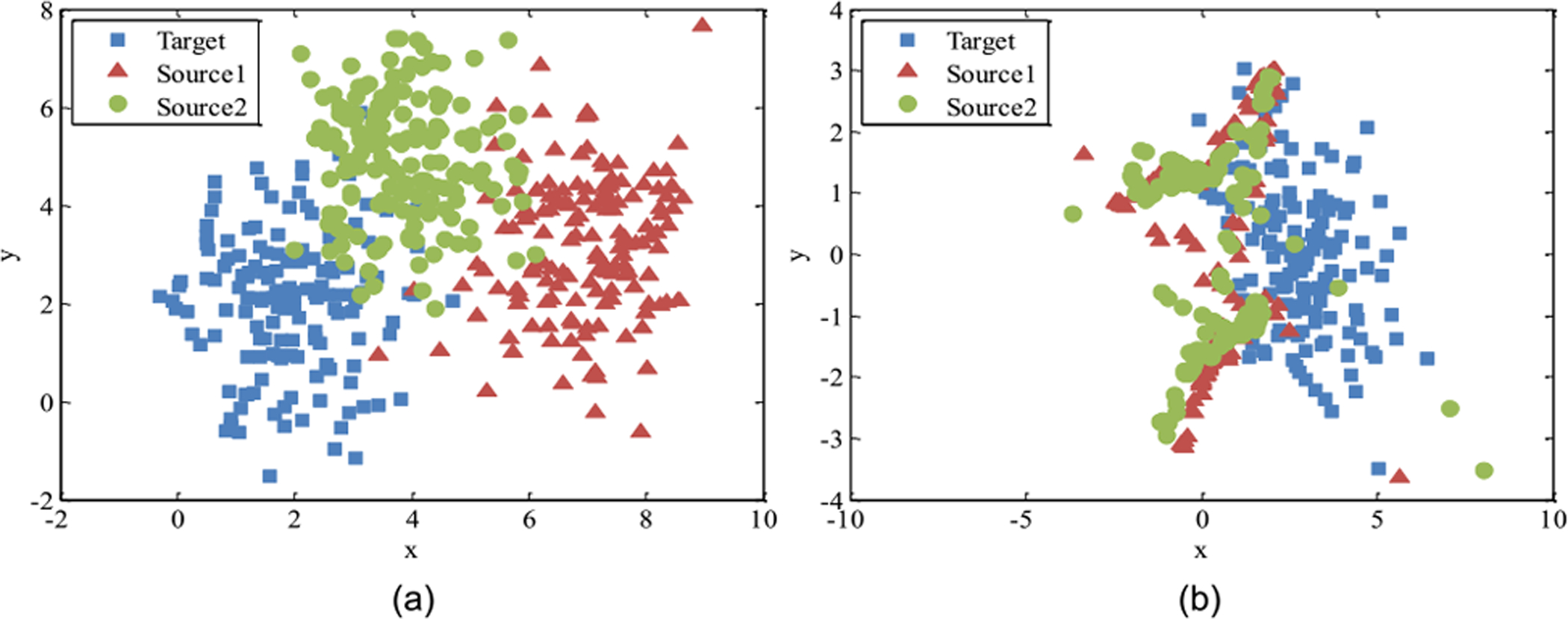
Illustration of feature transformation to reduce domain shift. Each color denotes a specific domain. (a) Data distributions of two source and one target domains before feature transformation, and (b) data distributions after feature transformation. Image courtesy to Wang et al. [49].
A. Supervised Shallow DA
Wachinger et al. [41] propose an instance weighting-based DA method for Alzheimer’s disease (AD) classification. Specifically, labeled target samples are used to estimate the target distribution. Then source samples are re-weighted by calculating the probability of source samples in the target domain. A multi-class logistic regression classifier is finally trained on the re-weighted samples and labeled target data (with volume, thickness, and anatomical shape features), and applied to the target domain. Experiments on the ADNI [50], AIBL [51] and CADDementia challenge datasets [52] show that the DA model can achieve better results than methods that only use data from either the source or target domain. Goetz et al. [42] propose an instance weighting-based DA method for brain tumor segmentation. To compute the weights of source samples, a domain classifier (logistic regression) is trained with paired samples from source and target domains. The output of the domain classifier (in terms of probability) is then used to calculate the adjusted weights for source samples. A random forest classifier is trained to perform segmentation. Experiments on the BraTS dataset [53] demonstrate the effectiveness of this method. Van Opbroek et al. [43] propose an instance weighting-based DA method for brain MRI segmentation. In their approach, each training image in the source domain is assigned a weight that can minimize the difference between the weighted probability density functions (PDFs) of voxels in the source and target data. The re-weighted training data are then used to train a classifier for segmentation. Experiments on three segmentation tasks (i.e., brain-tissue segmentation, skull stripping, and white-matter-lesion segmentation) demonstrate its effectiveness.
Becker et al. [47] propose a feature transformation-based DA method for microscopy image analysis. Specifically, they propose to learn a nonlinear mapping that can project samples in both domains to a shared discriminative latent feature space. Then, they develop a boosting classifier trained on the data in the transformed common space. In [54], [55], the authors propose a domain transfer support vector machine (DTSVM) for mild cognitive impairment (MCI) classification. Based on the close relationship of Alzheimer’s disease (AD) and MCI, DTSVM is trained on MCI subjects in the target domain and fine-tuned with an auxiliary AD dataset to enhance its generalization ability. They validate their method on ADNI and achieve good performance in MCI conversion prediction.
B. Semi-Supervised Shallow DA
Conjeti et al. [56] propose a two-step DA framework for ultrasound image classification. In the first step, they use principle component analysis (PCA) to transform the source and target domains to a common latent space, through which the global domain difference is minimized. In the second stage, a random forest classifier is trained on the transformed source domain, and then fine-tuned with a few labeled target data to further reduce the domain shift.
C. Unsupervised Shallow DA
Cheplygina et al. [44] employ the instance weighting strategy for lung disease classification. Specifically, Gaussian texture features are first extracted from all CT images, based on which a weighted logistic classifier is trained. Source samples that are similar to the target data are assigned high weights to reduce domain shift. Experiments on four chest CT image datasets show that this method can significantly improve the classification performance. Heimann et al. [45] use instance weighting-based DA strategy for ultrasound transducer localization in X-ray images. The instance weights are computed through a domain classifier (i.e., logistic regression). Based on Haar-like features of X-ray images, a cascade of tree-based classifiers is trained for ultrasound transducer localization. Li et al. [57] propose a subspace alignment DA method for AD classification using functional MRI (fMRI) data. They first conduct feature extraction and selection for source and target samples, followed by a modified subspace alignment strategy to align samples from both domains into a shared subspace. Finally, the aligned samples in the shared subspace are used as an integrated dataset to train a discriminant analysis classifier. This method is validated on both ADNI and a private fMRI datasets. Kamphenkel et al. [58] propose an unsupervised DA method for breast cancer classification based on diffusion-weighted MR images (DWI). They use the Diffusion Kurtosis Imaging (DKI) algorithm to transform the target data to the source domain without any label information of target data.
D. Multi-Source Shallow DA
Wang et al. [48] propose a multi-source DA framework for Autism Spectrum Disorder (ASD) classification with resting-state functional MRI (rs-fMRI) data. Each subject/sample is represented by both functional connectivity (FC) features in gray matter regions and functional correlation tensor features extracted from white matter regions. All subjects in each of the multiple source domains are transformed to the target domain via low-rank regularization and graph embedding. Then, a classifier is trained on the source samples (with transformed features). This method is validated on the ABIDE dataset [59]. Wang et al. [49] propose a low-rank representation based multi-source domain adaptation framework for ASD classification. They assume that multiple domains share an intrinsic latent data structure, and map multiple source data and target data into a common latent space via a low-rank representation to reduce the domain shift. Cheng et al. [60] propose a Multi-Domain Transfer Learning (MDTL) framework for the early diagnosis of AD and MCI. They first select a subset of discriminative features (i.e., gray matter tissue volume) by using training data from multiple auxiliary domains and target domain, and then construct a final classifier based on multiple classifiers learned from auxiliary domains.
IV. Deep Domain Adaptation Methods
Deep learning [61], [62] has greatly pushed forward the development of artificial intelligence and machine learning. Trained with large-scale labeled data in a full supervision manner, CNN has made breakthroughs in computer vision and medical image analysis [63]–[65].
A. Supervised Deep DA
With deep features (e.g., extracted by CNNs), several studies focus on using shallow DA models for medical image analysis. Kumar et al. [66] use ResNet as the feature extractor of mammographic images. Based on the CNN features, they evaluate three shallow domain adaptation methods, i.e., Transfer Component Analysis (TCA) [67], Correlation Alignment (CORAL) [68], and Balanced Distribution Adaptation (BDA) [69] for breast cancer classification, and provide some empirical results on the DDSM [70] and InBreast [71] datasets. Huang et al. [72] propose to use LeNet-5 to extract features of histological images from different domains for epithelium-stroma classification, and then project them into a subspace (via PCA) to align them for adaptation. Experiments on the NKI, VGH [73] and IHC [74] datasets verify its effectiveness.
Another research direction is to transfer models learned on the source domain onto the target domain with fine-tuning. Ghafoorian et al. [32] evaluate the impact of fine-tuning strategy on brain lesion segmentation, based on CNN models pre-trained on brain MRI scans. Their experimental results reveal that using only a small number of target training examples for fine-tuning can improve the transferability of models. They further evaluate the influence of both the size of the target training set and different network architectures on the adaptation performance. Based on similar findings, numerous methods have been proposed to leverage CNNs that are well pre-trained on ImageNet to tackle medical image analysis problems. Samala et al. [75] propose to first pre-train an AlexNet-like network on ImageNet, and then fine-tune it with regions-of-interest (ROI) from 2,454 mass lesions for breast cancer classification. Khan et al. [76] propose to pre-train a VGG network on ImageNet, and then use labeled MRI data to fine-tune it for Alzheimer’s disease (AD) classification. They propose to use image entropy to select the most informative training samples in the target domain, and verify their method on the ADNI database [50]. Similarly, Swati et al. [77] pre-train a VGG network on ImageNet and re-train the higher layers of the network with labeled MR images for brain tumor classification. Experiments are conducted on a publicly available CE-MRI dataset.1 Abbas et al. [78] employ the ImageNet to pre-train CNNs for chest X-ray classification. To deal with the irregularities in the datasets, they introduce class decomposition into the network learning process, by partitioning each class within the image dataset into k subsets and then assign new labels to the new set. Three different cohorts of chest X-ray images, histological images of human colorectal cancer, and digital mammograms are used for performance evaluation.
The above-mentioned methods employ the one-step DA strategy, i.e., directly transferring the pre-trained model to the target domain. To handle the problem when the target samples are too few to fine-tune a model, several studies propose to use intermediate domains to facilitate multi-step DA. Gu et al. [79] develop a two-step adaptation method for skin cancer classification. First, they fine-tune a ResNet on a relatively large medical image dataset (for skin cancer). Then, the network is trained on the target domain which is a relatively small medical image dataset. Their experimental results on the MoleMap dataset2 and the HAM10000 dataset3 show that the two-step adaptation method achieves better results than the directly transferring methods.
Although adopting pre-trained models on a dataset (e.g., ImageNet) is a popular way for supervised DA for medical image analysis, the 2D CNN structure may not be able to fully explore the rich information conveyed in 3D medical images. To this end, some researchers deliberately design task-specific 3D CNNs that are trained with medical images as the backbone to facilitate the subsequent data adaptation tasks. Hosseini-Asl et al. [80] design a 3D CNN for brain MR images classification. The network is pre-trained with MR images in the source domain. Then, its upper fully-connected layers are fine-tuned with samples in the target domain. Experiments on ADNI and CADDementia demonstrate its effectiveness. Valverde et al. [81] propose a similar 3D CNN for brain MR images segmentation. Instead of using the whole brain MRI, their network takes 3D image patches as input, while only partial fully-connected layers are fine-tuned using the target data. This method is evaluated on the ISBI2015 dataset [82] for MRI segmentation. Kaur et al. [83] propose to first pre-train a 3D U-Net on a source domain that has relevant diseases with a large number of samples, and then use a few labeled target data to fine-tune the network. Experiments on the BraTS dataset [53] show this strategy achieves a better performance than the network trained from scratch. Similar strategy is also used in [84] where a network is pre-trained using a large number of X-ray computed tomography (CT) and synthesized radial MRI datasets and then fine-tuned with only a few labeled target MRI scans. Zhu et al. [85] propose a boundary-weighted domain adaptive neural network for prostate segmentation. A domain feature discriminator is co-trained with the segmentation networks in an adversarial learning manner to reduce domain shift. Aiming to tackle the difficulty caused by unclear boundaries, they design a boundary-weighted loss and add it into the training process for DA and segmentation. The boundary contour needs to be extracted from the ground truth label to generate a boundary-weighted map as the supervision information to minimize the loss. Experiments on the PROMISE12 challenge dataset4 and BWH dataset [86] demonstrate its effectiveness. Bermúdez-Chacón et al. [87] design a two-stream U-Net for electron microscopy image segmentation. One stream uses source domain samples while the other uses target data. They utilize Maximum Mean Discrepancy (MMD) and correlation alignment as the domain regularization for DA. With a few labeled target data to fine-tune the model, their method achieves promising performance in comparison with several state-of-the-art ones on a private dataset. Laiz et al. [88] propose to use triplet loss for DA in endoscopy image classification. Each triplet consists of an anchor sample A from the source domain, a positive sample B from the target domain with the same label of A, and a negative sample C in the source domain. Through minimizing the triplet loss, their model can reduce the domain shift while keeping discrimination on different diseases.
B. Semi-Supervised Deep DA
Roels et al. [89] propose a semi-supervised DA method for electron microscopy image segmentation. They design a “Y-Net” with one feature encoder and two decoders. One decoder is used for segmentation, while a reconstruction decoder is designed to reconstruct images from both the source and target domains. The network is initially trained in an unsupervised manner. Then, the reconstruction decoder is discarded, and the whole network is fine-tuned with labeled target samples to make the model adapt to the target domain.
Madani et al. [90] propose a semi-supervised generative adversarial network (GAN) based DA framework for chest X-ray image classification. Different from conventional GAN, this model takes labeled source data, unlabeled target data and generated images as input. The discriminator performs three-category classification (i.e., normal, disease, or generated image). During training, unlabeled target data can be classified as any of those three classes, but can contribute to loss computation when they are classified as generated images. Through this way, both labeled and unlabeled data can be incorporated into a semi-supervised manner. Experiments on the NIH PLCO dataset [91] and NIH Chest X-Ray dataset [92] demonstrate its effectiveness.
C. Unsupervised Deep DA
Unsupervised deep domain adaptation has attracted increasing attention [11] in the field of medical image analysis, due to its advantage that does not require any labeled target data. We now introduce the existing unsupervised deep DA methods based on their specific strategies for knowledge transfer.
1). Feature Alignment:
This line of research aims to learn domain-invariant features across domains through specifically designed CNN models. Most of the models adopt a siamese architecture similar to the Domain Adversarial Neural Network (DANN) structure [93] as shown in Fig. 7. Kamnitsas et al. [34] propose a DANN-based multi-connected adversarial network for brain lesion segmentation. In their model, the domain discriminator is trained simultaneously with a segmentation network. In addition, the authors argue that only adapting the last layers of the segmentor is not ideal, thus the domain discriminator is connected at multiple layers of the network to make it less susceptible to image-quality variations between different domains. Javanmardi et al. [94] propose a DANN-based model for eye vasculture segmentation. A U-Net and a domain discriminator are co-trained during the training process. Experiments on the DRIVE [95] and STARE [96] datasets demonstrate its effectiveness. Their work won the first place in the optic disc (OD) and optic cup (OC) segmentation tasks in the MICCAI 2018 Retinal Fundus Glaucoma Challenge. Wang et al. [97] propose a DANN-based framework for fundus image segmentation. Besides segmentation and adversarial losses, they also introduce a smoothness loss to enforce the network to make homogeneous predictions in neighbor regions. Ren et al. [98] propose a DANN-based method for prostate histopathology classification. They introduce a Siamese structure in target domain to enforce patches from the same image to have the same classification score. Experiments on the Cancer Genome Atlas (TCGA)5 and a local dataset verify its effectiveness. Yang et al. [99] employ the DANN for lung texture classification, with experiments performed on the SPIROMICS dataset.6 Panfilov et al. [100] develop an adversarial learning based model for knee tissue segmentation. A U-Net-based segmentor and a domain discriminator with adversarial learning are co-trained for DA. Experiments on three knee MRI datasets verify its effectiveness. Zhang et al. [101] propose an adversarial learning based DA method for AD/MCI classification on ADNI. A classifier and a domain discriminator are co-trained to enhance the model’s transferability across different domains.
Fig. 7.
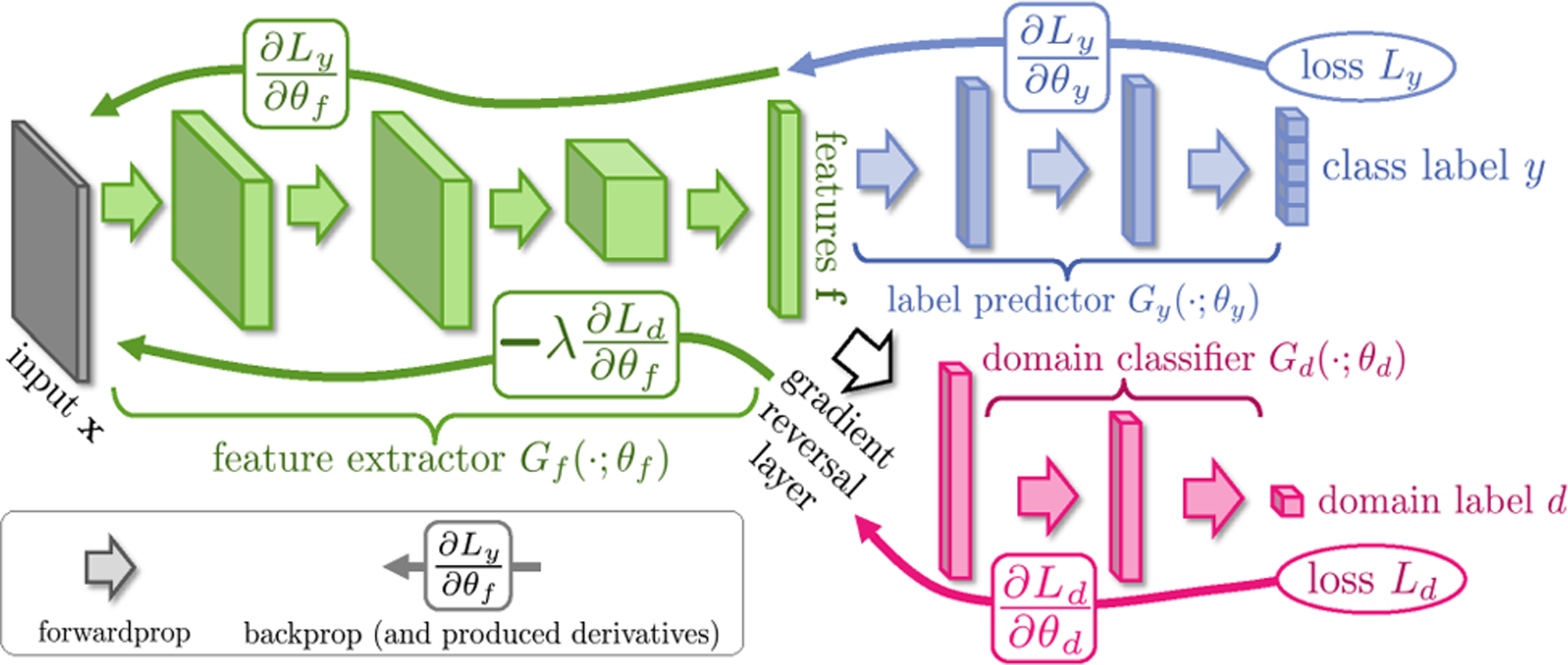
Illustration of Domain Adversarial Neural Network (DANN) framework. It is a classic and efficient DA model for domain-invariant feature learning through adversarial training. Image courtesy to Ganin et al. [93].
Different from the above-mentioned studies, Dou et al. [102] develop a cross-modality DA framework for cardiac MR and CT image segmentation, by only adapting low-level layers (with higher layers fixed) to reduce domain shift during training. They assume that the data shift between cross-modality domains mainly lies in low-level characteristics, and validate this assumption through experiments on the MM-WHS dataset [103]. Shen et al. [104] employ adversarial learning for fundus image quality assessment. Due to the demand for image assessment, they fix high-level weights during the adversarial training to focus on low-level feature adaptation. Yan et al. [105] propose an adversarial learning based DA method for MR image segmentation. A domain discriminator is co-trained with the segmentor to learn domain-invariant features for the task of segmentation. To enhance the model’s attention to edges, Canny edge detector is introduced into the adversarial learning process. Experiments on images from three independent MR vendors (Philips, Siemens, and GE) demonstrate the effectiveness. Shen et al. [106] propose an adversarial learning based method for mammogram detection which is an essential step in breast cancer diagnosis. A segmentor based on fully convolutional network (FCN) and a domain discriminator are co-trained for domain adaptation. Experiments on the public CBIS-DDSM [70], InBreast [71] and a self-collected dataset demonstrate its effectiveness. Yang et al. [107] propose an adversarial learning based DA method for lesion detection within the Faster RCNN framework [108]. Besides global feature alignment, they also extract ROI-based region proposals to facilitate local feature alignment.
Gao et al. [109] propose an unsupervised method for classification of brain activity based on fMRI data from the Human Connectome Project (HCP) dataset [110]. They use the central moment discrepancy (CMD) which matches the higher-order central moments of the data distributions as the domain regularization loss to facilitate adaptation. They assume that the high-level features extracted by fully-connected layers have large domain shift, thus CMD is imposed on these layers to perform adaptation. Bateson et al. [111] propose an unsupervised constrained DA framework for disc MR image segmentation. They propose to use some useful prior knowledge that is invariant across domains as an inequality constraint, and impose such constraints on the predicted results of unlabeled target samples as the domain adaptation regularization. Mahapatra et al. [112] develop a deep DA approach for cross-modality image registration. To transfer knowledge across domains, they design a convolutional auto-encoder to learn latent feature representations of images, followed by a generator to synthesize the registered image.
2). Image Alignment:
Image-level alignment is also used for domain adaptation based on deep generative models, such as Generative Adversarial Network (GAN) [115]. The original GAN takes random noise as input. Zhu et al. [113] propose a cycle-consistent GAN (CycleGAN) model which can translate one image domain into another without the demand for paired training samples. The CycleGAN has been used for medical image synthesis (see Fig. 8), where an MR image is firstly mapped to the target domain (CT image) and then mapped back to the input domain. A cycle consistency loss is used to measure the difference between input image and reconstructed image. By minimizing this loss, the CycleGAN can realize image-to-image translation without paired training samples. Cai et al. [116] propose a CycleGAN-based framework for cross-modality segmentation, by introducing a shape-consistency loss during a cross-modality synthesis process to ensure consistent anatomical structures between source and target images. Experiments on self-collected MR/CT datasets and two public mammogram datasets demonstrate the effectiveness. Gadermayr et al. [117] leverage CycleGAN to facilitate image-image translation between different domains (digital pathology datasets), followed by a U-Net for kidney histology segmentation. Wollmann et al. [118] propose a CycleGAN-based DA method for breast cancer classification. They first use CycleGAN to transform whole-slide images (WSIs) of lymph nodes from a source domain (i.e., a medical center) to the target domain (i.e., another medical center). Then a densely connected deep neural network (DenseNet) is used for breast cancer classification. Image patches from regions-of-interest (ROIs) are used as the input to facilitate two-level classification. Experiments on the CAMELYON17 challenge dataset7 demonstrate the effectiveness. Manakov et al. [119] propose to leverage unsupervised DA to tackle retinal optical coherence tomography (OCT) image denoising problem. They treat image noises as domain shift between high and low noise domains. A CycleGAN-based DA mode is designed to learn a mapping between the source (i.e., high noise) and the target domain (i.e., low noise) on unpaired OCT images, thus achieving the goal of image denoising. Experiments on numerous in-house OCT images demonstrate the effectiveness of this method. Gholami et al. [120] propose to use CycleGAN to generate more training data for brain tumor segmentation. They first generate synthetic tumor-bearing MR images using their in-house simulation model, and then transform them to real MRIs using CycleGAN to augment training samples. They conduct experiment on the BraST [53] dataset and achieve good segmentation results for brain tumors. Jiang et al. [121], [122] propose to employ CycleGAN to generate MRIs from CT scans for lung tumor segmentation. The generated MRIs are helpful to segment tumors that are close to soft tissues. Zhang et al. [123] leverage CycleGAN for multi-organ segmentation in X-ray images. They first create Digitally Reconstructed Radiographs (DDRs) from labeled CT scans and train a segmentation model based on DDRs. Then, they use CycleGAN to map X-ray images (target domain) to DDRs (source domain) for segmentation via the trained network.
Fig. 8.
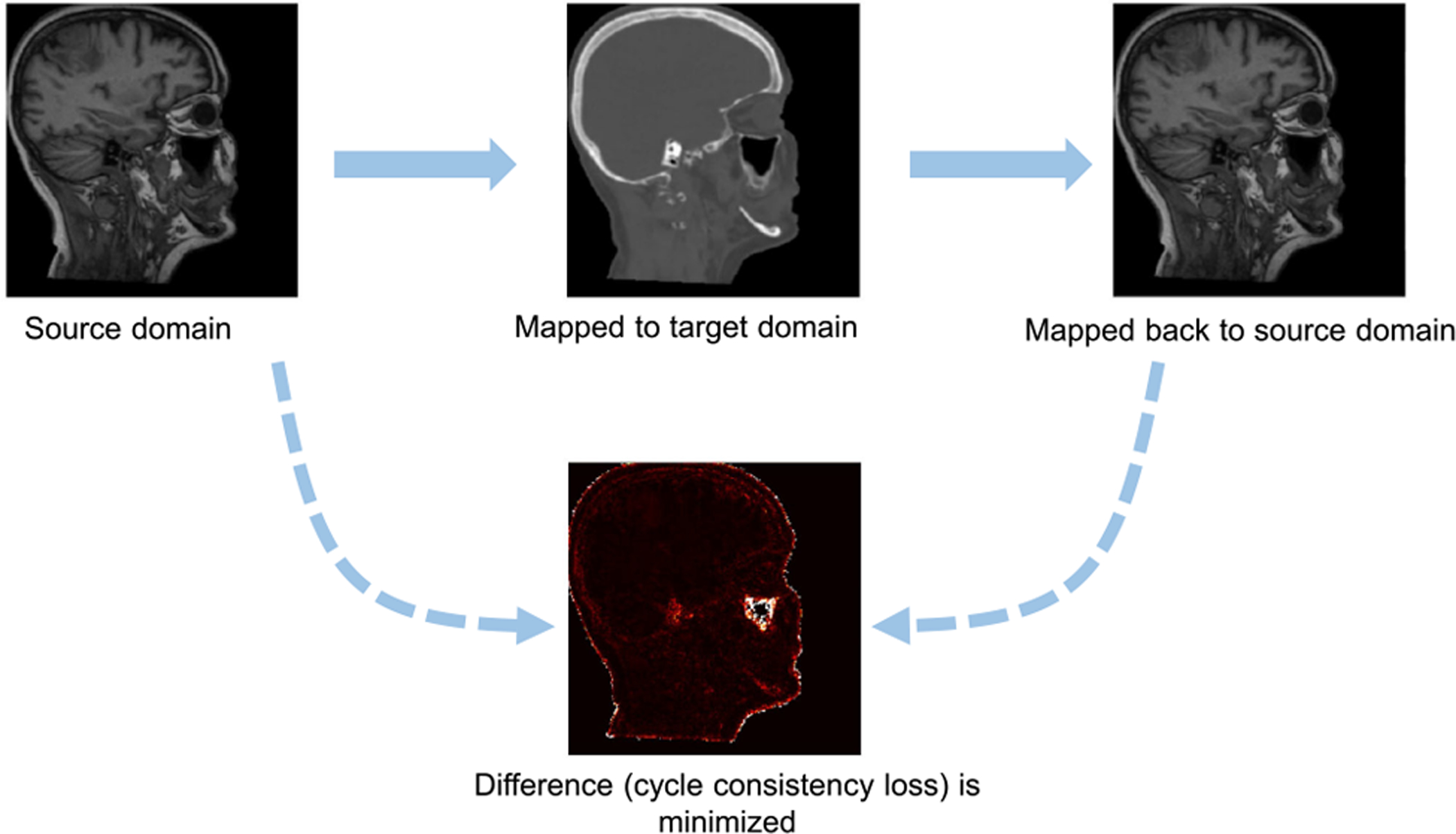
Image-to-image translation via cycle consistency loss (CycleGAN) [113], [114]. The source domain MR image is mapped to the target domain CT image, and then mapped back to the source domain. The difference between the input MR image and the reconstructed MR image is minimized.
Zhang et al. [124] propose a Noise Adaptation Generative Adversarial Network (NAGAN) for eye blood vessel segmentation, by formulating DA as a noise style transfer task. They use an image-to-image translation generator to map the target image to the source domain. Besides a normal discriminator which enforces the content similarity between generated images and real images, they also design a style discriminator which enforces the generated images to have the same noise patterns as those from the target domain. Experiments on the SINA [125] and a local dataset demonstrate the effectiveness. Mahmood et al. [126] propose a Reverse Domain Adaptation method based on GAN for endoscopy image analysis. This method reverses the flow and transforms real images to synthetic images, based on the insight that subjects with optical endoscopy images have patient-specific details and these details do not generalize across patients. Xing et al. [127], [128] leverage CycleGAN for cell detection across multi-modality microscopy images. Source images are firstly mapped to the target domain which are used to train a cell detector. Then the detector is used to generate pseudo-labels on target training data. The detector is further fine-tuned with labeled target data to boost learning performance.
3). Image+feature Alignment:
Chen et al. [37], [129] combine image alignment and feature alignment for cross-modality cardiac image segmentation. They firstly use CycleGAN to transform labeled source images into target-like images. Then, the synthesized target images and real target images are fed into a two-stream CNN with a domain discriminator which can further reduce the domain gap via adversarial learning. Experiments on the MM-WHS dataset [103] demonstrate its effectiveness. Yan et al. [38] propose a similar unsupervised framework for cross-vendor cardiac cine MRI segmentation. They first train a U-Net using labeled source samples, and then train a CycleGAN to facilitate image-level adaptation. In addition, they design a feature-level alignment mechanism by computing the Mean Square Error (MSE) of features between the original and translated images. Three major vendors including Philips, Siemens, and GE, are treated as three domains. For example, the Philips data are treated as the source domain, while the Siemens and GE data are defined as two target domains. The short-axis steady-state free precession (SSFP) cine MR images of 144 subjects acquired by those three vendors are used for performance evaluation.
4). Disentangled Representation:
Yang et al. [36] propose a cross-modality (between CT and MRI) DA method via disentangled representation for liver segmentation. Through disentangle presentation learning, images from each domain are embedded into two spaces, i.e., a shared domain-invariant content space and a domain-specific style space. Then, domain adaptation is performed in the domain-invariant space. Experiments on the LiTS benchmark dataset [130] and a local dataset demonstrate the effectiveness of this method.
5). Ensemble Learning:
Following [131], Perone et al. [25] propose a self-ensemble based DA method for medical image segmentation. A baseline CNN, called student network, takes labeled samples from the source domain, and makes its predictions after training with a task (segmentation) loss. Another network, called teacher network, gives its predictions using only unlabeled samples in the target domain. The teacher network is updated by the exponential moving average of the weights of the student network (a temporal ensemble strategy). During training, the domain shift is minimized by a consistency loss by comparing predictions from both the student and teacher networks. This method is evaluated on the SCGM challenge dataset [132], which is a multi-center multi-vendor dataset of spinal cord anatomical MR images from healthy subjects. Shanis et al. [133] employ a similar DA architecture for brain tumor segmentation. Besides a consistency loss that measures the difference between predictions of the teacher and student networks, they also utilize an adversarial loss to further improve adaptation performance. Effectiveness is validated with experiments on the BraTS dataset [53].
6). Soft Labels:
Bermúdez-Chacón et al. [134] propose an unsupervised DA method to reduce domain shift in electron microscopy images. Based on the observation that some ROIs in medical images still resemble each other across domains, the authors propose to use Normalized Cross Correlation (NCC) which computes the similarity of two structures from two different images (visual correspondence) to construct a consensus heatmap. The high-scoring regions of the heatmap are used as soft labels for the target domain, which are then used to train the target classifier for segmentation.
7). Feature Learning:
Ahn et al. [135], [136] introduce a convolutional auto-encoder to learn features for domain adaptation. Other than using labeled target data to fine-tune a pre-trained AlexNet, the authors propose to use a zero-bias convolutional auto-encoder as an adapter to transform the feature maps from the last convolution layer of AlexNet to relevant medical image features. The learned features can further be used for classification tasks.
D. Multi-Target Deep DA
Orbes-Arteaga et al. [137] propose an unsupervised DA framework with two target domains for brain lesion segmentation. Their network is first trained with labeled source data. During the adaptation phase, labeled source data and the paired unlabeled data from the two target domains are fed into the DA model. An adversarial loss is imposed to reduce domain differences. To minimize the output probability distribution on the target data, a consistency loss is imposed on the paired target data. Experiments are performed on the MICCAI 2017 WMH Challenge [138] and two independent datasets. Karani et al. [30] propose a life-long multi-domain adaptation framework for brain structure segmentation. In their method, batch normalization plays the key role in adaptation, as suggested in [139]. They first train a network on d source domains with shared convolutional filters but different batch normalization (BN) parameters, and each domain corresponds to a specific set of BN parameters. By fine-tuning the BN parameters with a few labeled data in each new target domain, the model can then be adapted to those multi-target domains. This method is evaluated on 4 publicly available datasets, including HCP [110], ADNI [50], ABIDE [59], and IXI.8
V. Discussion
A. Challenges of Data Adaptation for Medical Image Analysis
1). 3D/4D Volumetric Representation:
Medical images are generally high-dimensional and stored in 3D or 4D formats (e.g., with the temporal dimension). Especially for time-series data, e.g., functional MRI, there are a series of 3D volumes for each subject, and each 3D volume is composed of hundreds of 2D image slices. These slices usually contain rich structural context information that is significant for representing medical images. It is challenging to design advanced domain adaptation models to effectively capture the 3D or 4D structural information convened in medical images.
2). Limited Training Data:
The existing medical image datasets usually contain a limited number of samples, ranging from a few hundred [51] to several hundred thousand [140]. Also, labeled medical images are generally even fewer, since labeling medical images is a time-consuming and expensive task which demands for the participation of medical experts. Even though one can adapt/transfer models pre-trained on large-scale ImageNet, the existing off-the-shelf deep models may not be well adapted to medical images, since they are designed for 2D image analysis and have a huge number of parameters at the higher layers for classification of a large number of categories (e.g., >1, 000). Raghu et al. [33] conduct an empirical study on two large-scale medical imaging datasets, and find that transferring standard CNN models pre-trained on ImageNet to medical imaging analysis does not benefit performance a lot. The problem of limited data has posed a great challenge to effective training of domain adaptation models, especially for deep learning based ones.
3). Inter-Modality Heterogeneity:
Various digital imaging techniques have been developed to generate heterogeneous visual representations of each subject, such as CT, structural MRI, function MRI, and positron emission tomography (PET). While these multi-modality data provide complementary information, the inter-modality heterogeneity brings many challenges for domain adaptation [37]. For instance, a pair of structural MRI and PET scans from the same subject contains large inter-modality discrepancy [141]. The large inter-modality difference brings many difficulties actually for efficient knowledge transfer between different domains.
B. Future Research Trends
1). Task-Specific 3D/4D Models for Domain Adaptation:
Compared with 2D CNNs pre-trained on ImageNet, 3D models are usually more powerful to explore medical image features and yield better learning performance [142]. For more high-dimensional medical images such as fMRI that involves temporal information, a 4D CNN has been introduced in the literature [143]. Besides, medical images usually contain redundant or noisy regions, while task-specific ROIs can help filter out these redundant/noisy regions. For instance, brain structural changes caused by AD usually locate in a limited number of ROIs in structural MRIs, such as hippocampus [144] and ventricles [145]. However, effectively defining these task-specific ROIs in medical images has always been an open issue. Currently, there are very few works on developing 3D/4D DA models with task-specific ROI definitions for medical image analysis, and we believe this is a promising future direction.
2). Unsupervised Domain Adaptation:
The lack of labeled data is one of the most significant challenges for medical image research. To tackle this problem, many recent studies tend to avoid using labeled target data for model fine-tuning, by using various unsupervised domain adaptation methods [11], [12] for medical image analysis. Also, completely avoiding any target data (even those unlabeled ones) for model training is an interesting research topic. Thus, research on domain generalization [35], [146]–[148] and zero-shot learning [149]–[151] will be welcome in the future.
3). Multi-Modality Domain Adaptation:
To make use of complementary but heterogeneous multi-modality neuroimaging data, it is desired to develop domain adaptation models that are trained on one modality and can be well generalized to another modality. For more challenging problems where each domain contains multiple modalities (e.g., MRI and CT), it is meaningful to consider both inter-modality heterogeneity and inter-domain difference when designing domain adaptation models. Several techniques including CycleGAN [37], [114] and disentangle learning [36], [141] have been introduced into this emerging area, while further exploration of multi-modality DA is required for medical image analysis.
4). Multi-Source/Multi-Target Domain Adaptation:
Existing DA methods usually focus on single-source domain adaptation, i.e., training a model on one source domain, but there may be multiple source domains (e.g., multiple imaging centers) in real-world applications. Multi-source domain adaptation [14], [15], [152]–[154], aiming to utilize training data from multi-source domains to improve models’ transferability on the target domain, is of great clinical significance. It is also practical to transfer a model to multiple target domains, i.e., multi-target DA. Currently, very limited works have been done on multi-source/multi-target DA in medical image analysis, so there is still a lot of room for future research.
5). Source-Free Domain Adaptation:
Existing domain adaptation methods usually require full access to source and target data. However, for privacy protection considerations, it is not always allowed to share medical image data between different sites/domains. Source-free domain adaptation, an emerging topic that avoids potential violation of data privacy policies, has drawn increasing attention recently [155]–[158]. It is interesting to develop source-free domain adaptation methods to handle multi-site/domain medical image data in accordance with corresponding data privacy policies.
VI. Conclusion
In this paper, we provide a survey of recent advances in domain adaptation for medical image analysis. We categorize existing methods into shallow and deep models, and each category is further divided into supervised, semi-supervised and unsupervised methods. We also introduce existing benchmark medical image datasets used for domain adaptation, summarize potential challenges and discuss future research directions.
Supplementary Material
Acknowledgment
We thank Dr. Dinggang Shen for his valuable suggestions on this paper.
This work was supported in part by NIH under Grant AG041721.
Footnotes
This article has supplementary downloadable material available at https://doi.org/10.1109/TBME.2021.3117407, provided by the authors.
Contributor Information
Hao Guan, Department of Radiology and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, USA..
Mingxia Liu, Department of Radiology, and Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA.
References
- [1].Valiant LG, “A theory of the learnable,” Commun. ACM, vol. 27, no. 11, pp. 1134–1142, 1984. [Google Scholar]
- [2].Ben-David S et al. , “Analysis of representations for domain adaptation,” Adv. Neural Inf. Process. Syst, vol. 19, pp. 137–144, 2007. [Google Scholar]
- [3].Torralba A and Efros AA, “Unbiased look at dataset bias,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2011, pp. 1521–1528. [Google Scholar]
- [4].Quiñonero-Candela J et al. , Dataset Shift in Machine Learning. Cambridge, MA, USA: MIT Press, pp. 1–229, 2009. [Google Scholar]
- [5].Donahue J et al. , “DeCAF: A deep convolutional activation feature for generic visual recognition,” in Proc. Int. Conf. Mach. Learn., 2014, pp. 647–655. [Google Scholar]
- [6].Russakovsky O et al. , “ImageNet large scale visual recognition challenge,” Int. J. Comput. Vis, vol. 115, no. 3, pp. 211–252, 2015. [Google Scholar]
- [7].Mahajan D et al. , “Exploring the limits of weakly supervised pretraining,” in Proc. Eur. Conf. Comput. Vis., 2018, pp. 181–196. [Google Scholar]
- [8].Ronneberger O et al. , “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2015, pp. 234–241. [Google Scholar]
- [9].Csurka G, “A comprehensive survey on domain adaptation for visual applications,” in Proc. Domain Adapt. Comput. Vis. Appl., 2017, pp. 1–35. [Google Scholar]
- [10].Wang M and Deng W, “Deep visual domain adaptation: A survey,” Neurocomputing, vol. 312, pp. 135–153, 2018. [Google Scholar]
- [11].Wilson G and Cook DJ, “A survey of unsupervised deep domain adaptation,” ACM Trans. Intell. Syst. Technol, vol. 11, no. 5, pp. 1–46, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kouw WM and Loog M, “A review of domain adaptation without target labels,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 43, no. 3, pp. 766–785, Mar. 2021. [DOI] [PubMed] [Google Scholar]
- [13].Patel VM et al. , “Visual domain adaptation: A survey of recent advances,” IEEE Signal Process. Mag, vol. 32, no. 3, pp. 53–69, May 2015. [Google Scholar]
- [14].Sun S et al. , “A survey of multi-source domain adaptation,” Inf. Fusion, vol. 24, pp. 84–92, 2015. [Google Scholar]
- [15].Zhao S et al. , “Multi-source domain adaptation in the deep learning era: A systematic survey,” 2020, arXiv:2002.12169.
- [16].Pan SJ and Yang Q, “A survey on transfer learning,” IEEE Trans. Knowl. Data Eng, vol. 22, no. 10, pp. 1345–1359, Oct. 2010. [Google Scholar]
- [17].Shao L, Zhu F, and Li X, “Transfer learning for visual categorization: A survey,” IEEE Trans. Neural Netw. Learn. Syst, vol. 26, no. 5, pp. 1019–1034, May 2015. [DOI] [PubMed] [Google Scholar]
- [18].Zhang J et al. , “Recent advances in transfer learning for cross-dataset visual recognition: A problem-oriented perspective,” ACM Comput. Surv, vol. 52, no. 1, pp. 1–38, 2019. [Google Scholar]
- [19].Tan C et al. , “A survey on deep transfer learning,” in Proc. Int. Conf. Artif. Neural Netw., 2018, pp. 270–279. [Google Scholar]
- [20].Zhuang F et al. , “A comprehensive survey on transfer learning,” Proc. IEEE, vol. 109, no. 1, pp. 43–76, Jan. 2021. [Google Scholar]
- [21].Zhang L and Gao X, “Transfer adaptation learning: A decade survey,” 2019, arXiv:1903.04687. [DOI] [PubMed]
- [22].Cheplygina V et al. , “Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis,” Med. Image Anal, vol. 54, pp. 280–296, 2019. [DOI] [PubMed] [Google Scholar]
- [23].Valverde JM et al. , “Transfer learning in magnetic resonance brain imaging: A systematic review,” J. Imag, vol. 7, no. 4, pp. 1–21, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Morid MA et al. , “A scoping review of transfer learning research on medical image analysis using ImageNet,” Comput. Biol. Med, 2020, Art. no. 104115. [DOI] [PubMed] [Google Scholar]
- [25].Perone CS et al. , “Unsupervised domain adaptation for medical imaging segmentation with self-ensembling,” NeuroImage, vol. 194, pp. 1–11, 2019. [DOI] [PubMed] [Google Scholar]
- [26].Moreno-Torres JG et al. , “A unifying view on dataset shift in classification,” Pattern Recognit, vol. 45, no. 1, pp. 521–530, 2012. [Google Scholar]
- [27].Pooch EH et al. , “Can we trust deep learning models diagnosis? The impact of domain shift in chest radiograph classification,” 2019, arXiv:1909.01940.
- [28].Stacke K et al. , “A closer look at domain shift for deep learning in histopathology,” 2019, arXiv:1909.11575. [DOI] [PubMed]
- [29].Prados F et al. , “Spinal cord grey matter segmentation challenge,” NeuroImage, vol. 152, pp. 312–329, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Karani N et al. , “A lifelong learning approach to brain MR segmentation across scanners and protocols,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2018, pp. 476–484. [Google Scholar]
- [31].AlBadawy EA et al. , “Deep learning for segmentation of brain tumors: Impact of crossinstitutional training and testing,” Med. Phys, vol. 45, no. 3, pp. 1150–1158, 2018. [DOI] [PubMed] [Google Scholar]
- [32].Ghafoorian M et al. , “Transfer learning for domain adaptation in MRI: Application in brain lesion segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2017, pp. 516–524. [Google Scholar]
- [33].Raghu M et al. , “Transfusion: Understanding transfer learning for medical imaging,” in Proc. Adv. Neural Inf. Process. Syst., 2019, pp. 3347–3357. [Google Scholar]
- [34].Kamnitsas K et al. , “Unsupervised domain adaptation in brain lesion segmentation with adversarial networks,” in Proc. Int. Conf. Inf. Process. Med. Imag., 2017, pp. 597–609. [Google Scholar]
- [35].Zhang L et al. , “Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation,” IEEE Trans. Med. Imag, vol. 39, no. 7, pp. 2531–2540, Jul. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yang J et al. , “Unsupervised domain adaptation via disentangled representations: Application to cross-modality liver segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2019, pp. 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen C et al. , “Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation,” IEEE Trans. Med. Imag, vol. 39, no. 7, pp. 2494–2505, Jul. 2020. [DOI] [PubMed] [Google Scholar]
- [38].Yan W et al. , “The domain shift problem of medical image segmentation and vendor-adaptation by Unet-GAN,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2019, pp. 623–631. [Google Scholar]
- [39].Tan B et al. , “Transitive transfer learning,” in Proc. 21th ACM SIGKDD Int. Conf. Knowl. Discov. Data Mining, 2015, pp. 1155–1164. [Google Scholar]
- [40].Tan B et al. , “Distant domain transfer learning,” in Proc. AAAI Conf. Artif. Intell., 2017, vol. 31, no. 1, pp. 2604–2610. [Google Scholar]
- [41].Wachinger C and Reuter M, “Domain adaptation for Alzheimer’s disease diagnostics,” NeuroImage, vol. 139, pp. 470–479, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Goetz M et al. , “DALSA: Domain adaptation for supervised learning from sparsely annotated MR images,” IEEE Trans. Med. Imag, vol. 35, no. 1, pp. 184–196, Jan. 2016. [DOI] [PubMed] [Google Scholar]
- [43].van Opbroek A et al. , “Weighting training images by maximizing distribution similarity for supervised segmentation across scanners,” Med. Image Anal, vol. 24, no. 1, pp. 245–254, 2015. [DOI] [PubMed] [Google Scholar]
- [44].Cheplygina V et al. , “Transfer learning for multicenter classification of chronic obstructive pulmonary disease,” IEEE J. Biomed. Health Informat, vol. 22, no. 5, pp. 1486–1496, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Heimann T et al. , “Learning without labeling: Domain adaptation for ultrasound transducer localization,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2013, pp. 49–56. [DOI] [PubMed] [Google Scholar]
- [46].Long M et al. , “Transfer joint matching for unsupervised domain adaptation,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2014, pp. 1410–1417. [Google Scholar]
- [47].Becker C, Christoudias CM, and Fua P, “Domain adaptation for microscopy imaging,” IEEE Trans. Med. Imag, vol. 34, no. 5, pp. 1125–1139, May 2015. [DOI] [PubMed] [Google Scholar]
- [48].Wang J et al. , “Multi-class ASD classification based on functional connectivity and functional correlation tensor via multi-source domain adaptation and multi-view sparse representation,” IEEE Trans. Med. Imag, vol. 39, no. 10, pp. 3137–3147, Oct. 2020. [DOI] [PubMed] [Google Scholar]
- [49].Wang M et al. , “Identifying autism spectrum disorder with multi-site fMRI via low-rank domain adaptation,” IEEE Trans. Med. Imag, vol. 39, no. 3, pp. 644–655, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jack CR Jr et al. , “The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods,” J. Magn. Reson. Imag, vol. 27, no. 4, pp. 685–691, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ellis KA et al. , “The Australian Imaging, Biomarkers and lifestyle (AIBL) study of aging: Methodology and baseline characteristics of 1112 individuals recruited for a longitudinal study of Alzheimer’s disease,” Int. Psychogeriatrics, vol. 21, no. 4, pp. 672–687, 2009. [DOI] [PubMed] [Google Scholar]
- [52].Bron EE et al. , “Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: The CADDementia challenge,” NeuroImage, vol. 111, pp. 562–579, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Menze BH et al. , “The multimodal brain tumor image segmentation benchmark (BRATS),” IEEE Trans. Med. Imag, vol. 34, no. 10, pp. 1993–2024, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cheng B et al. , “Domain transfer learning for MCI conversion prediction,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2012, pp. 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cheng B et al. , “Domain transfer learning for MCI conversion prediction,” IEEE Trans. Biomed. Eng, vol. 62, no. 7, pp. 1805–1817, Jul. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Conjeti S et al. , “Supervised domain adaptation of decision forests: Transfer of models trained in vitro for in vivo intravascular ultrasound tissue characterization,” Med. Image Anal, vol. 32, pp. 1–17, 2016. [DOI] [PubMed] [Google Scholar]
- [57].Li W et al. , “Detecting Alzheimer’s disease on small dataset: A knowledge transfer perspective,” IEEE J. Biomed. Health Informat, vol. 23, no. 3, pp. 1234–1242, May 2019. [DOI] [PubMed] [Google Scholar]
- [58].Kamphenkel J et al. , “Domain adaptation for deviating acquisition protocols in CNN-based lesion classification on diffusion-weighted MR images,” in Proc. Image Anal. Moving Organ, Breast, and Thoracic Images, 2018, pp. 73–80. [Google Scholar]
- [59].Di Martino A et al. , “The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism,” Mol. Psychiatry, vol. 19, no. 6, pp. 659–667, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cheng B et al. , “Multi-domain transfer learning for early diagnosis of Alzheimer’s disease,” Neuroinformatics, vol. 15, no. 2, pp. 115–132, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Goodfellow I et al. , Deep Learning. Cambridge, MA, USA: MIT Press, 2016, pp. 1–802. [Google Scholar]
- [62].Khan A et al. , “A survey of the recent architectures of deep convolutional neural networks,” Artif. Intell. Rev, vol. 53, no. 8, pp. 5455–5516, 2020. [Google Scholar]
- [63].Shen D et al. , “Deep learning in medical image analysis,” Annu. Rev. Biomed. Eng, vol. 19, pp. 221–248, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Litjens G et al. , “A survey on deep learning in medical image analysis,” Med. Image Anal, vol. 42, pp. 60–88, 2017. [DOI] [PubMed] [Google Scholar]
- [65].Mazurowski MA et al. , “Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI,” J. Magn. Reson. Imag, vol. 49, no. 4, pp. 939–954, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kumar D et al. , “Cross-database mammographic image analysis through unsupervised domain adaptation,” in Proc. IEEE Int. Conf. Big Data, 2017, pp. 4035–4042. [Google Scholar]
- [67].Pan SJ et al. , “Domain adaptation via transfer component analysis,” IEEE Trans. Neural Netw, vol. 22, no. 2, pp. 199–210, Feb. 2011. [DOI] [PubMed] [Google Scholar]
- [68].Sun B et al. , “Return of frustratingly easy domain adaptation,” in Proc. AAAI Conf. Artif. Intell, 2016, vol. 30, no. 1, pp. 2058–2065. [Google Scholar]
- [69].Wang J et al. , “Balanced distribution adaptation for transfer learning,” in Proc. IEEE Int. Conf. Data Mining, 2017, pp. 1129–1134. [Google Scholar]
- [70].Lee RS et al. , “A curated mammography data set for use in computer-aided detection and diagnosis research,” Sci. Data, vol. 4, no. 1, pp. 1–9, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Moreira IC et al. , “INbreast: Toward a full-field digital mammographic database,” Academic Radiol, vol. 19, no. 2, pp. 236–248, 2012. [DOI] [PubMed] [Google Scholar]
- [72].Huang Y et al. , “Epithelium-stroma classification via convolutional neural networks and unsupervised domain adaptation in histopathological images,” IEEE J. Biomed. Health Informat, vol. 21, no. 6, pp. 1625–1632, Nov. 2017. [DOI] [PubMed] [Google Scholar]
- [73].Beck AH et al. , “Systematic analysis of breast cancer morphology uncovers stromal features associated with survival,” Sci. Transl. Med, vol. 3, no. 108, pp. 108–113, 2011. [DOI] [PubMed] [Google Scholar]
- [74].Linder N et al. , “Identification of tumor epithelium and stroma in tissue microarrays using texture analysis,” Diagn. Pathol, vol. 7, no. 22, pp. 1–11, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Samala RK et al. , “Cross-domain and multi-task transfer learning of deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis,” Med. Imag.: Comput.-Aided Diagnosis, vol. 10575, no. 2, pp. 1–8, 2018. [Google Scholar]
- [76].Khan NM et al. , “Transfer learning with intelligent training data selection for prediction of Alzheimer’s disease,” IEEE Access, vol. 7, pp. 72726–72735, 2019. [Google Scholar]
- [77].Swati ZNK et al. , “Brain tumor classification for MR images using transfer learning and fine-tuning,” Comput. Med. Imag. Graph, vol. 75, pp. 34–46, 2019. [DOI] [PubMed] [Google Scholar]
- [78].Abbas A et al. , “DeTrac: Transfer learning of class decomposed medical images in convolutional neural networks,” IEEE Access, vol. 8, pp. 74901–74913, 2020. [Google Scholar]
- [79].Gu Y et al. , “Progressive transfer learning and adversarial domain adaptation for cross-domain skin disease classification,” IEEE J. Biomed. Health Informat, vol. 24, no. 5, pp. 1379–1393, May 2020. [DOI] [PubMed] [Google Scholar]
- [80].Hosseini-Asl E et al. , “Alzheimer’s disease diagnostics by adaptation of 3D convolutional network,” in Proc. IEEE Int. Conf. Image Process., 2016, pp. 126–130. [Google Scholar]
- [81].Valverde S et al. , “One-shot domain adaptation in multiple sclerosis lesion segmentation using convolutional neural networks,” NeuroImage: Clin, vol. 21, 2019, Art. no. 101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Carass A et al. , “Longitudinal multiple sclerosis lesion segmentation: Resource and challenge,” NeuroImage, vol. 148, pp. 77–102, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kaur B et al. , “Improving pathological structure segmentation via transfer learning across diseases,” in Proc. Domain Adapt. Representation Transfer Med. Image Learn. Less Labels Imperfect Data Springer, 2019, pp. 90–98. [Google Scholar]
- [84].Han Y et al. , “Deep learning with domain adaptation for accelerated projection-reconstruction MR,” Magn. Reson. Med, vol. 80, no. 3, pp. 1189–1205, 2018. [DOI] [PubMed] [Google Scholar]
- [85].Zhu Q, Du B, and Yan P, “Boundary-weighted domain adaptive neural network for prostate MR image segmentation,” IEEE Trans. Med. Imag, vol. 39, no. 3, pp. 753–763, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Fedorov A et al. , “An annotated test-retest collection of prostate multi-parametric MRI,” Sci. Data, vol. 5, no. 1, pp. 1–14, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Bermúdez-Chacón R et al. , “A domain-adaptive two-stream U-Net for electron microscopy image segmentation,” in Proc. 15th IEEE Int. Symp. Biomed. Imag., 2018, pp. 400–404. [Google Scholar]
- [88].Laiz P et al. , “Using the triplet loss for domain adaptation in WCE,” in Proc. IEEE Int. Conf. Comput. Vis. Workshops, 2019, pp. 1–7. [Google Scholar]
- [89].Roels J et al. , “Domain adaptive segmentation in volume electron microscopy imaging,” in Proc. 16th IEEE Int. Symp. Biomed. Imag., 2019, pp. 1519–1522. [Google Scholar]
- [90].Madani A et al. , “Semi-supervised learning with generative adversarial networks for chest X-ray classification with ability of data domain adaptation,” in Proc. 15th IEEE Int. Symp. Biomed. Imag., 2018, pp. 1038–1042. [Google Scholar]
- [91].Oken MM et al. , “Screening by chest radiograph and lung cancer mortality: The prostate, lung, colorectal, and ovarian (PLCO) randomized trial,” JAMA, vol. 306, no. 17, pp. 1865–1873, 2011. [DOI] [PubMed] [Google Scholar]
- [92].Demner-Fushman D et al. , “Preparing a collection of radiology examinations for distribution and retrieval,” J. Amer. Med. Informat. Assoc, vol. 23, no. 2, pp. 304–310, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ganin Y et al. , “Domain-adversarial training of neural networks,” J. Mach. Learn. Res, vol. 17, no. 1, pp. 2096–2030, 2016. [Google Scholar]
- [94].Javanmardi M and Tasdizen T, “Domain adaptation for biomedical image segmentation using adversarial training,” in Proc. 15th IEEE Int. Symp. Biomed. Imag., 2018, pp. 554–558. [Google Scholar]
- [95].Staal J et al. , “Ridge-based vessel segmentation in color images of the retina,” IEEE Trans. Med. Imag, vol. 23, no. 4, pp. 501–509, Apr. 2004. [DOI] [PubMed] [Google Scholar]
- [96].Hoover A et al. , “Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response,” IEEE Trans. Med. Imag, vol. 19, no. 3, pp. 203–210, Mar. 2000. [DOI] [PubMed] [Google Scholar]
- [97].Wang S et al. , “Patch-based output space adversarial learning for joint optic disc and cup segmentation,” IEEE Trans. Med. Imag, vol. 38, no. 11, pp. 2485–2495, Feb. 2019. [DOI] [PubMed] [Google Scholar]
- [98].Ren J et al. , “Adversarial domain adaptation for classification of prostate histopathology whole-slide images,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2018, pp. 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Yang J et al. , “Unsupervised domain adaption with adversarial learning (UDAA) for emphysema subtyping on cardiac CT scans: The mesa study,” in Proc. 16th IEEE Int. Symp. Biomed. Imag., 2019, pp. 289–293. [Google Scholar]
- [100].Panfilov E et al. , “Improving robustness of deep learning based knee MRI segmentation: Mixup and adversarial domain adaptation,” in Proc. IEEE Int. Conf. Comput. Vis. Workshops, 2019, pp. 1–10. [Google Scholar]
- [101].Zhang J et al. , “Unsupervised conditional consensus adversarial network for brain disease identification with structural MRI,” in Proc. Int. Workshop Mach. Learn. Med. Imag., 2019, pp. 391–399. [Google Scholar]
- [102].Dou Q et al. , “PnP-AdaNet: Plug-and-play adversarial domain adaptation network at unpaired cross-modality cardiac segmentation,” IEEE Access, vol. 7, pp. 99065–99076, 2019. [Google Scholar]
- [103].Zhuang X and Shen J, “Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI preparing a collection of radiology examinations for distribution and retrieval,” Med. Image Anal, vol. 31, pp. 77–87, 2016. [DOI] [PubMed] [Google Scholar]
- [104].Shen Y et al. , “Domain-invariant interpretable fundus image quality assessment,” Med. Image Anal, vol. 61, 2020, Art. no. 101654. [DOI] [PubMed] [Google Scholar]
- [105].Yan W et al. , “Edge-guided output adaptor: Highly efficient adaptation module for cross-vendor medical image segmentation,” IEEE Signal Process. Lett, vol. 26, no. 11, pp. 1593–1597, Nov. 2019. [Google Scholar]
- [106].Shen R et al. , “Unsupervised domain adaptation with adversarial learning for mass detection in mammogram,” Neurocomputing, vol. 393, pp. 27–37, 2020. [Google Scholar]
- [107].Yang S et al. , “Unsupervised domain adaptation for cross-device OCT lesion detection via learning adaptive features,” in Proc. 17th IEEE Int. Symp. Biomed. Imag., 2020, pp. 1570–1573. [Google Scholar]
- [108].Ren S et al. , “Faster R-CNN: Towards real-time object detection with region proposal networks,” in Proc. Adv. Neural Inf. Process. Syst., 2015, pp. 91–99. [DOI] [PubMed] [Google Scholar]
- [109].Gao Y et al. , “Decoding brain states from fMRI signals by using unsupervised domain adaptation,” IEEE J. Biomed. Health Informat, vol. 24, no. 6, pp. 1677–1685, Jun. 2020. [DOI] [PubMed] [Google Scholar]
- [110].Van Essen DC et al. , “The WU-Minn human connectome project: An overview,” NeuroImage, vol. 80, pp. 62–79, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bateson M et al. , “Constrained domain adaptation for segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2019, pp. 326–334. [Google Scholar]
- [112].Mahapatra D and Ge Z, “Training data independent image registration using generative adversarial networks and domain adaptation,” Pattern Recognit., vol. 100, 2020, Art. no. 107109. [Google Scholar]
- [113].Zhu JY et al. , “Unpaired image-to-image translation using cycle-consistent adversarial networks,” in Proc. IEEE Int. Conf. Comput. Vis., 2017, pp. 2223–2232. [Google Scholar]
- [114].Wolterink JM et al. , “Deep MR to CT synthesis using unpaired data,” in Proc. Int. Workshop Simul. Synth. in Med. Imag., 2017, pp. 14–23. [Google Scholar]
- [115].Goodfellow I et al. , “Generative adversarial nets,” in Proc. Adv. Neural Inf. Process. Syst., 2014, pp. 1–9. [Google Scholar]
- [116].Cai J et al. , “Towards cross-modal organ translation and segmentation: A cycle-and shape-consistent generative adversarial network,” Med. Image Anal, vol. 52, pp. 174–184, 2019. [DOI] [PubMed] [Google Scholar]
- [117].Gadermayr M et al. , “Generative adversarial networks for facilitating stain-independent supervised and unsupervised segmentation: A study on kidney histology,” IEEE Trans. Med. Imag, vol. 38, no. 10, pp. 2293–2302, Oct. 2019. [DOI] [PubMed] [Google Scholar]
- [118].Wollmann T et al. , “Adversarial domain adaptation to improve automatic breast cancer grading in lymph nodes,” in Proc. 15th IEEE Int. Symp. Biomed. Imag., 2018, pp. 582–585. [Google Scholar]
- [119].Manakov I et al. , “Noise as domain shift: Denoising medical images by unpaired image translation,” in Proc. Domain Adapt. Representation Transfer Med. Image Learn. Less Labels Imperfect Data Springer, 2019, pp. 3–10. [Google Scholar]
- [120].Gholami A et al. , “A novel domain adaptation framework for medical image segmentation,” in Proc. Int. MICCAI Brainlesion Workshop Springer, 2018, pp. 289–298. [Google Scholar]
- [121].Jiang J et al. , “Integrating cross-modality hallucinated MRI with CT to aid mediastinal lung tumor segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2019, pp. 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jiang J et al. , “Tumor-aware, adversarial domain adaptation from CT to MRI for lung cancer segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2018, pp. 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhang Y et al. , “Task driven generative modeling for unsupervised domain adaptation: Application to X-ray image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2018, pp. 599–607. [Google Scholar]
- [124].Zhang T et al. , “Noise adaptation generative adversarial network for medical image analysis,” IEEE Trans. Med. Imag, vol. 39, no. 4, pp. 1149–1159, Apr. 2020. [DOI] [PubMed] [Google Scholar]
- [125].Hoover AD et al. , “Validated automatic segmentation of AMD pathology including drusen and geographic atrophy in SD-OCT images,” Invest. Ophthalmol. Vis. Sci, vol. 53, no. 1, pp. 53–61, 2012. [DOI] [PubMed] [Google Scholar]
- [126].Mahmood F, Chen R, and Durr NJ, “Unsupervised reverse domain adaptation for synthetic medical images via adversarial training,” IEEE Trans. Med. Imag, vol. 37, no. 12, pp. 2572–2581, Dec. 2018. [DOI] [PubMed] [Google Scholar]
- [127].Xing F et al. , “Adversarial domain adaptation and pseudo-labeling for cross-modality microscopy image quantification,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2019, pp. 740–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xing F et al. , “Bidirectional mapping-based domain adaptation for nucleus detection in cross-modality microscopy images,” IEEE Trans. Med. Imag, vol. 40, no. 10, pp. 2880–2896, Oct. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chen C et al. , “Synergistic image and feature adaptation: Towards cross-modality domain adaptation for medical image segmentation,” in Proc. AAAI Conf. Artif. Intell, vol. 33, 2019, pp. 865–872. [Google Scholar]
- [130].Bilic P et al. , “The liver tumor segmentation benchmark (LiTS),” 2019, arXiv:1901.04056. [DOI] [PMC free article] [PubMed]
- [131].French G et al. , “Self-ensembling for visual domain adaptation,” in Proc. Int. Conf. Learn. Representations, 2018, pp. 1–20. [Google Scholar]
- [132].Prados F et al. , “Spinal cord grey matter segmentation challenge,” NeuroImage, vol. 152, pp. 312–329, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Shanis Z et al. , “Intramodality domain adaptation using self ensembling and adversarial training,” in Proc. Domain Adapt. Representation Transfer Med. Image Learn. Less Labels Imperfect Data, 2019, pp. 28–36. [Google Scholar]
- [134].Bermúdez-Chacón R et al. , “Visual correspondences for unsupervised domain adaptation on electron microscopy images,” IEEE Trans. Med. Imag, vol. 39, no. 4, pp. 1256–1267, Apr. 2020. [DOI] [PubMed] [Google Scholar]
- [135].Ahn E et al. , “Unsupervised deep transfer feature learning for medical image classification,” in Proc. 16th IEEE Int. Symp. Biomed. Imag., 2019, pp. 1915–1918. [Google Scholar]
- [136].Ahn E et al. , “Unsupervised domain adaptation to classify medical images using zero-bias convolutional auto-encoders and context-based feature augmentation,” IEEE Trans. Med. Imag, vol. 39, no. 7, pp. 2385–2394, Jul. 2020. [DOI] [PubMed] [Google Scholar]
- [137].Orbes-Arteaga M et al. , “Multi-domain adaptation in brain MRI through paired consistency and adversarial learning,” in Domain Adapt. Representation Transfer Med. Image Learn. Less Labels Imperfect Data, 2019, pp. 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Kuijf HJ et al. , “Standardized assessment of automatic segmentation of white matter hyperintensities and results of the WMH segmentation challenge,” IEEE Trans. Med. Imag, vol. 38, no. 11, pp. 2556–2568, Nov. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Li Y et al. , “Revisiting batch normalization for practical domain adaptation,” in Proc. Int. Conf. Learn. Representations Workshop, 2017, pp. 1–12. [Google Scholar]
- [140].Wang X et al. , “ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2017, pp. 2097–2106. [Google Scholar]
- [141].Yang E et al. , “Deep disentangled hashing with momentum triplets for neuroimage search,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2020, pp. 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Singh SP et al. , “3D deep learning on medical images: A review,” Sensors, vol. 20, no. 18, pp. 1–24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Zhao Y et al. , “Modeling 4D fMRI data via spatio-temporal convolutional neural networks (ST-CNN),” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2018, pp. 181–189. [Google Scholar]
- [144].Mu Y and Gage FH, “Adult hippocampal neurogenesis and its role in Alzheimer’s disease,” Mol. Neurodegener, vol. 6, no. 1, pp. 1–9, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Ott BR et al. , “Brain ventricular volume and cerebrospinal fluid biomarkers of Alzheimer’s disease,” J. Alzheimer’s Dis, vol. 20, no. 2, pp. 647–657, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Gan C et al. , “Learning attributes equals multi-source domain generalization,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2016, pp. 87–97. [Google Scholar]
- [147].Ghifary M et al. , “Scatter component analysis: A unified framework for domain adaptation and domain generalization,” IEEE Trans. Pattern Anal. Mach. Intell, vol. 39, no. 7, pp. 1414–1430, Jul. 2017. [DOI] [PubMed] [Google Scholar]
- [148].Li H et al. , “Domain generalization with adversarial feature learning,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2018, pp. 5400–5409. [Google Scholar]
- [149].Wang W et al. , “A survey of zero-shot learning: Settings, methods, and applications,” ACM Trans. Intell. Syst. Technol, vol. 10, no. 2, pp. 1–37, 2019. [Google Scholar]
- [150].Kodirov E et al. , “Unsupervised domain adaptation for zero-shot learning,” in Proc. IEEE Int. Conf. Comput. Vis., 2015, pp. 2452–2460. [Google Scholar]
- [151].Palatucci M et al. , “Zero-shot learning with semantic output codes,” in Proc. Adv. Neural Inf. Process. Syst., 2009, pp. 1410–1418. [Google Scholar]
- [152].Zhao S et al. , “Multi-source distilling domain adaptation,” in Proc. AAAI Conf. Artif. Intell., 2020, pp. 12975–12983. [Google Scholar]
- [153].Zhao H et al. , “Adversarial multiple source domain adaptation,” in Proc. Adv. Neural Inf. Process. Syst., 2018, pp. 8559–8570. [Google Scholar]
- [154].Guan H et al. , “Multi-source domain adaptation via optimal transport for brain dementia identification,” in Proc. IEEE 18th Int. Symp. Biomed. Imag., 2021, pp. 1514–1517. [Google Scholar]
- [155].Bateson M et al. , “Source-relaxed domain adaptation for image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.- Assist. Intervention, 2020, pp. 490–499. [Google Scholar]
- [156].Liang J et al. , “Do we really need to access the source data? source hypothesis transfer for unsupervised domain adaptation,” in Proc. Int. Conf. Mach. Learn., 2020, pp. 6028–6039. [Google Scholar]
- [157].Kundu JN et al. , “Universal source-free domain adaptation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., 2020, pp. 4544–4553. [Google Scholar]
- [158].Liu Y et al. , “Source-free domain adaptation for semantic segmentation,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., 2021, pp. 1215–1224. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


