Abstract
Neuroimmunology plays a critical role in our understanding of the pathophysiological processes that underlie a variety of diseases treated by neurosurgeons including degenerative disc disease (DDD), glioblastoma multiforme (GBM), aneurysmal subarachnoid hemorrhage (aSAH), and others. Compared to traditional methods in neuroimmunology which would study one pathway or gene at a time, emerging multiomics methodologies allow for holistic interrogation of multiple immune signaling pathways to test hypotheses and the effects of therapeutics at a systems level. In this review, we summarize key concepts for gathering and analyzing multiomics data so that neurosurgeons can contribute to the emerging field of systems neuroimmunology. We describe three use cases based on original research published from our groups and others which utilize transcriptomic, metabolomic, and proteomic analyses to study immune signaling pathways in DDD, aSAH, and GBM. Additionally, through our use cases, we share techniques for performing machine learning and network-based analyses to generate new clinical insights from multiomics data. We hope that neurosurgeons may use our review as a summary of common tools and principles in systems immunology, so that they may better engage in creating the immunotherapies of tomorrow.
Keywords: systems neuroimmunology, multiomics, transcriptomics, proteomics, metabolomics, degenerative disc disease, glioblastoma
1. Introduction
The cellular microenvironment of the nervous system is composed of a dynamic milieu of vascular, immune, and neural cell types which interact with one another in a way that produces complex physiological responses.1 Neuroimmune interactions in particular play a critical role in both normal physiology, as well as the pathophysiological processes that underlie cranial and spinal neurosurgical disease. In fact, the immune system has been found to have a key role in a variety of conditions treated by spine, tumor, and cerebrovascular neurosurgeons including (1) the immune-mediated progression of degenerative disc disease (DDD)2, (2) the susceptibility of brain tumors like glioblastoma (GBM)3–6 to novel immunotherapies, and (3) the response of brain tissue to extravasated blood products during aneurysmal subarachnoid hemorrhage (aSAH).7
Systems neuroimmunology is an emerging framework for interrogating neuroimmune interactions at a systems level, which can be used to study the immune processes that drive neurosurgical disease.8–10 Systems neuroimmunology leverages multiomics data from genomic, proteomic, metabolomic, and other sources in order to arrive at a systems level picture of which genes, proteins, and metabolites of the immune system are active during healthy and diseased states. The evolution of systems neuroimmunology into its modern form stems from advancements in molecular technologies, such as expression profiling, microchip array technology, immunological assay methods, and large-scale epitope screening.9 Neurosurgeons are particularly well poised to engage in systems neuroimmunology research, as their role is essential in gathering the tissue samples that generate multiomics data.
In this article, we hope to provide neurosurgeons with a practical guide for how they may use systems neuroimmunology in their practice to advance our collective understanding of neuroimmune interactions in neurosurgical disease. We have selected three use cases featuring original research from our laboratories and others that apply systems neuroimmunology to the study of immune interactions in spinal and cranial neurosurgical disease. These use cases each respectively demonstrate how multiomics data including (1) transcriptomic, (2) metabolomic, and (3) proteomic data may be used to generate insights about how the immune system drives the progression in a broad variety of neurosurgical diseases including DDD, aSAH, and GBM (Figure 1).
Figure 1.
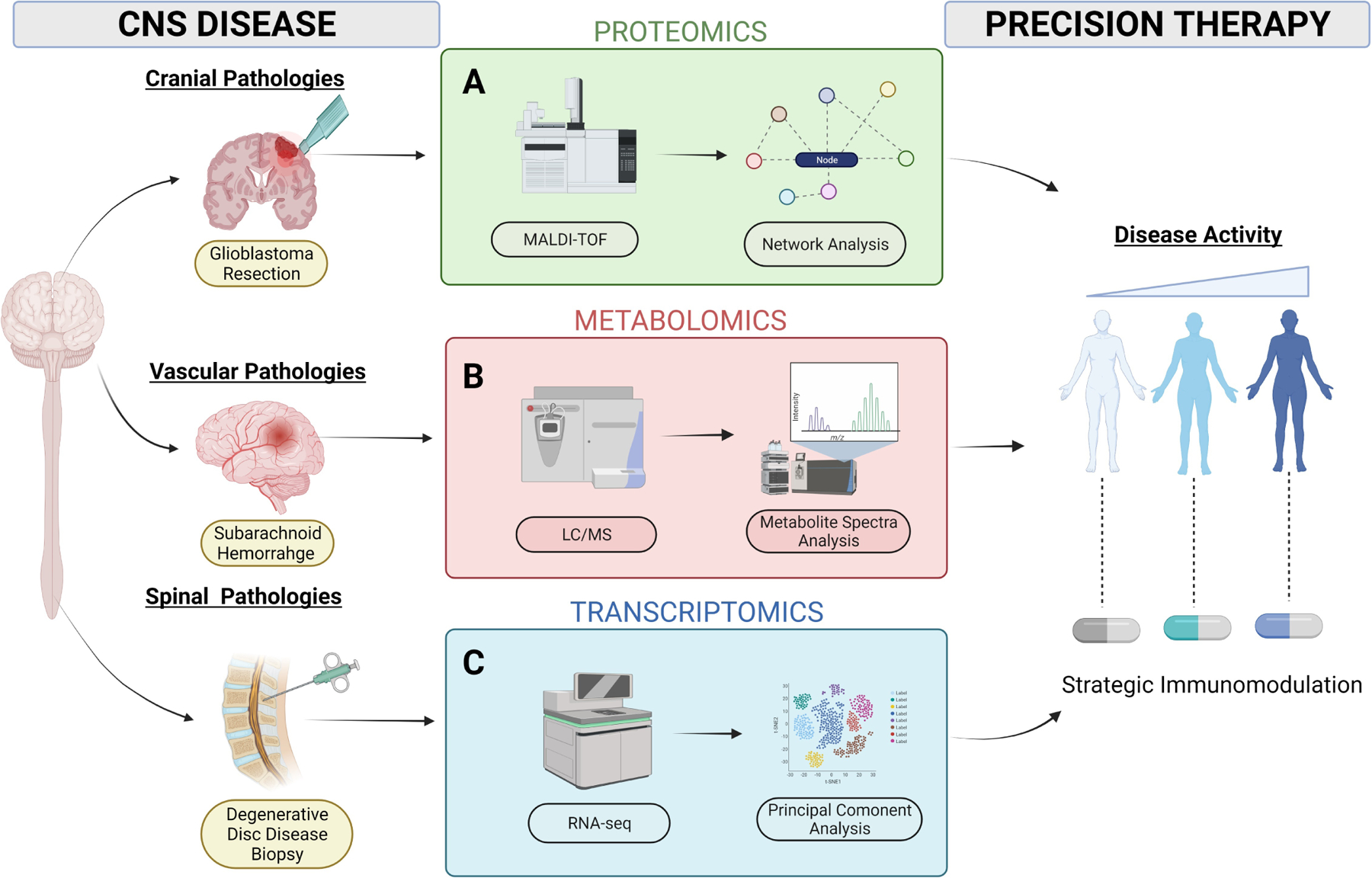
A summary of multiomics approaches to characterize neuroimmune interactions in degenerative disc disease and glioblastoma multiforme. (1A) Proteomics can be used to identify susceptibilities of GBM to immunotherapy based on protein-protein network analysis, (1B) Metabolomics can identify metabolites that modulate immune interactions after aSAH, and (1C) Transcriptomics can identify differentially expressed genes in immune signaling pathways that drive DDD progression. Figure 1 created using BioRender.com.
2. Multiomics technologies
Systems neuroimmunology has successfully matured as a discipline thanks to technological advances in (1) gathering multiomics data in a high-throughput fashion, as well as (2) robust analysis pipelines for identifying alterations in immune signaling. In this section, we present some general principles related to generating, analyzing, and interpreting multiomics data so that neurosurgeons can better engage in this line of research.
2.1. Generating multiomics data
Systems neuroimmunology as applied to the study of neurosurgical pathologies relies on robust multiomics datasets generated by carefully curating tissue samples from healthy and diseased subjects. Neurosurgeons are essential to the process of collecting multiomics data for systems neuroimmunology research because they are directly involved in procedures that result in the excision of diseased tissue. For example, as described in Use Case 1, a standard lumbar discectomy and fusion can result in the excision of inflamed disc tissue which can be used for systems neuroimmunology research. Several enabling technologies such as microarrays, RNA sequencing, and mass spectrometry then allow for multiomics data to be gathered from direct tissue biopsies, cerebrospinal fluid, or blood (Table 1).
Table 1.
A summary of multiomics studies including data source, enabling technologies, and insights created for neuroimmunologys.
| Study | -Omic Approach | Data Source | Enabling Technologies | Application in Neuroimmunology |
|---|---|---|---|---|
| Costantino et al, 201739 | Genomics | Genes | DNA microarray | Genome-wide association study from 906 subjects, including 486 with spondyloarthritis (SpA), revealed an association of SpA with MAPK14 |
| Bydon et al, 20202 | Transcriptomics | RNA transcripts | RNA sequencing (RNA-seq) | Transcriptomics data from intervertebral disc samples showed immune signaling pathways were upregulated in DDD |
| Ma et al, 20214 | Proteomics | Proteins | Matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) | A computational analysis of protein-protein interactions in GBM cell lines identified opportunities to strategically improve immunotherapy design |
| Valentin-Guillama et al, 20185 | Metabolomics | Small molecules and metabolites | Mass spectrometry, high performance liquid chromatography (HPLC) | Metabolomic data from gp120 treated glioma cells shows that HIV-1 can promote proliferation and activation of glycolysis, resulting in increased protein synthesis |
Generating genomic and transcriptomic data.
Genomic and transcriptomic data can be gathered from high throughput technologies that quickly survey global gene expression patterns. Through microarray technology, global gene expression in tissue samples can be measured in parallel by the thousands of genes and quantified to deduce signaling pathways and regulatory networks.11 The emergence of single cell RNA sequencing (scRNA-seq) technologies allows transcriptomic data to be gathered from individual cells.12,13 RNA-seq requires single-cell suspensions to be obtained, which is typically achieved with high yield through mechanical disaggregation and enzymatic dissociation. Several technologies, such as flow-cytometry, can be used to then isolate individual cells. After isolation, cells are lysed to extract RNA which is subsequently amplified by PCR.12 It is important to assess the quality of the extracted RNA by measuring its integrity number (RIN). Given that RNA molecules are susceptible to degradation, the RIN is an important measure of RNA integrity which can be used for quality control.14 Before sequencing can be performed, the RNA must be converted into double stranded complementary DNA (cDNA), which can be achieved through poly-T oligo-attached magnetic beads. Sequencing can then be conducted, with reads mapped to a reference transcriptome or genome, and quantified to gene counts per gene or transcript.15 An analysis of differential gene expression (DGE) enables quantification through computational statistical models to distinguish overlapping transcripts between samples. In Use Case 1, we discuss our study that identified molecular regulators associated with DDD in an effort to identify promising candidates for therapeutic targets.12,16
Generating proteomic and metabolomic data.
Mass spectrometry is the foundational technology for generating proteomic data and allows for characterization of proteins through analyzing their mass-to-charge ratio.17 Studying proteins in this fashion is a formidable task, due to the fact that genes can generate multiple proteins through sequence polymorphisms, alternative splicing, and post-transcriptional modifications. These challenges have been overcome with several technological advances.17 For example, matrix-assisted laser desorption/ionization time of flight (MALDI-TOF), allows for the visualization of proteins in addition to its various proteoforms.18 To use this technology, a matrix is used to coat the tissue under study, which aids in desorption and ionization of endogenous biomolecules during laser irradiation. Then, individual mass spectra are collected to generate signal intensity maps and ion images across the sample area. As thousands of these ion images are created through a single MALDI-TOF experiment, researchers can use top-down protein identification methods to gain molecular contexts of the tissue sample under study.18 In Use Case 2, we discuss how mass spectrometry is used to generate metabolomic data from tumor cell lines.
2.2. Analyzing multiomics data
Broadly, algorithms for analyzing multiomics data can be subdivided into clustering and network-based approaches.19
Clustering analysis.
Clustering algorithms are a type of unsupervised machine learning algorithm that can infer groups of similar samples based on a distance metric using a weighted sum of expression values in multiomics datasets derived from different samples.20 Samples which have a distance apart from each other less than a given threshold are considered similar and are clustered into the same group. Common clustering algorithms used in neuroimmunology research include t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP).21 In Figure 2A, we demonstrate an example of hierarchical clustering that was used in Use Case 1 to assess tissue-to-tissue and patient-to-patient variation in musculoskeletal tissue samples from patients with DDD.
Figure 2.
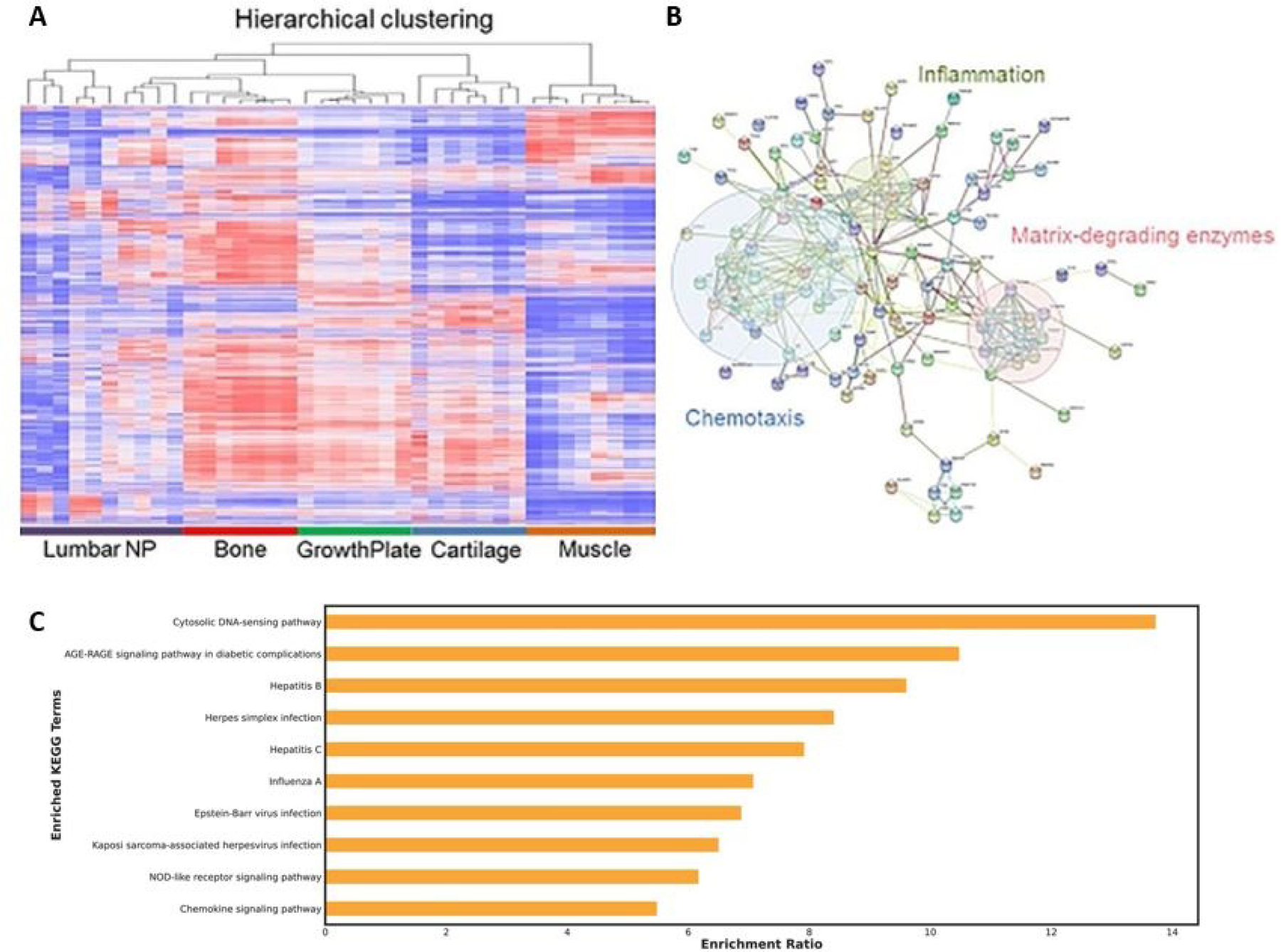
Examples of different approaches to analyzing multiomics data. (2A) Clustering methods can be used to assess patient-to-patient and tissue-to-tissue variation in musculoskeletal tissue samples from patients with DDD, (2B) Network methods can be used to assess immune signaling pathways that are active in patients with DDD, (2C) Overrepresentation analyses can be used to detect signatures of resistance to OV therapy in GBM. Figure 2A and 2B used with permission form Bydon et al, 20202.
Network-based analysis.
Network-based approaches for analyzing multiomics datasets seek to identify dependent relationships and interactions between genes, RNA transcripts, and proteins. Such algorithms use metrics like pairwise correlation, mutual information, or weighted co-expression in order to assess genes that are closely dependent on one another in order to create a network.22 Some common network-based algorithms include NetDecoder to derive context specificity, along with visualization packages Cytoscape and NetworkX.21–23 In Figure 2B, we show how a network-based algorithm was used to infer immune signaling pathways that are active in patients with DDD from Use Case 1.
For neurosurgeons first getting started with using such analytic approaches, there are several public multiomics data sources that may be applied to neuroimmunology (Table 2). We advise as a best practice to integrate different omics data sources which can serve to strengthen associations and reduce noise. In particular, similarity network fusion (SNF) is a commonly used method for integrating different omics data sources.23
Table 2.
Resources for public multiomic datasets.
| Study | Data Source | Multiomics Type | Description |
|---|---|---|---|
| Edgar et al, 200240 | Gene Expression Omnibus (GEO) | Genomics | A public curated repository of microarray experiment data |
| Darmanis et al, 201741 | GBMseq | Transcriptomics | A public repository of scRNA-seq on 3,589 cells from GBM samples in a cohort of 4 patients |
| Xie et al, 201542 | HGCC | Transcriptomics | An open resource useful to both basic and translational GBM research, containing a biobank of 48 GBM cell (GC) lines and an associated database containing high-resolution molecular data |
| Samaras et al, 202043 | ProteomicsDB | Proteomics | A public, curated repository of proteomics datasets from a variety of host organisms |
| Joshi, 201944 | TcellSubC | Proteomics | A public proteomic dataset of 6,572 proteins in the human CD4+ T cells proteome |
| Sud et al, 201645 | Metabolomics Workbench | Metabolomics | A public international repository for metabolomics data |
2.3. Generating new insights from multiomics data
Clinical insight generation is a critical component of multiomics pipelines which neurosurgeons can actively participate in. Often, the networks derived from the network-based analyses we described above can yield insights in and of themselves. For example, in Use Case 3, we describe how performing a network analysis on GBM multiomics data elucidated mechanisms of resistance to oncolytic viral therapies. Additionally, we describe how the same network analysis allowed us to postulate measles virus as a more effective oncolytic viral vector compared to herpes simplex virus for GBM with CCN1 resistance signatures (Figure 2C).
Additionally, once an interaction network has been derived from multiomics data, novel clinical insights can be gathered through performing in silico simulations of network behavior. Network simulator tools like BioNSi can estimate the behaviors of genes and proteins in a network once certain interactions are deleted or added.24
3. Systems neuroimmunology and degenerative disc disease (DDD)
3.1. Immunology review in DDD
DDD is a degenerative process affecting the mechanical integrity of intervertebral discs (IVDs) in the spine which are composed of an inner nucleus pulposus (NP), outer annulus fibrosus (AF), and the bordering cartilaginous end plate (EP).25 DDD is thought to be an age-related phenomenon that includes inflammation and cell apoptosis leading to decreased proteoglycan synthesis with resultant disc dehydration and loss of disc height. Multiomics can facilitate translational research to identify key genetic networks with sufficiently high levels of expression to make practical targets for genetic molecular therapy directed at ameliorating DDD.
Several studies in the neuroimmunology literature have elucidated the immune pathways that drive the progression of DDD. Initially, cells from the NP and AF secrete proinflammatory molecules that recruit immune cells to the IVD.26 Cytokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-1, IL-6, and IL-17 increase in concentration in the IVD, resulting in an inflammatory microenvironment.26 Such an environment reduces the structural integrity of the NP and AF, leading to herniation and annular tears. The degenerated IVDs subsequently amplify the inflammatory cascade through further production of TNF-α, IL-1, IL-6, IL-17, interferon (IFN)-γ, and inflammatory mediators such as nitric oxide and prostaglandin E2.27,28
It is imperative that neurosurgeons seeking therapeutic targets for DDD have a thorough understanding of the immunopathology for spine degeneration. In our first use case, we present work from our group that uses transcriptomic analysis to gain insight into DDD pathogenesis.
3.2. Use Case 1: Transcriptomics to characterize inflammatory pathways in DDD
Our group has published a study that aimed to identify immune signaling pathways distinct to DDD progression.2 DDD may lead to spinal stenosis, disc herniation (DH), and/or degenerative spondylolisthesis (DS) which may express as radiculopathy or myelopathy in the patient. To further enhance the understanding of DH and DS pathogenesis, the transcriptome data of IVD samples from lumbar degenerated discs were analyzed. Disc tissues were harvested intraoperatively and subsequently underwent RNA-seq.2
A total of 33 adult patients undergoing lumbar discectomy to treat DDD were enrolled in our study. During lumbar discectomy, NP samples from 33 adult patients were carefully dissected in the operating room by the primary surgeon. The harvested samples were then frozen in liquid nitrogen and stored until ready for processing. Of the 33 samples, only a total of 10 samples met our threshold RIN necessary for next generation sequencing.
The NP tissue biopsies were subsequently ground into powder and homogenized, then quantified with a spectrophotometer. Next-generation RNA sequencing was then performed to assess differential gene expression. Computational analysis compared NP RNA-seq to bone, cartilage, growth plate, and muscle tissue.2
When we compared transcriptomic data from NP samples compared to other musculoskeletal tissues in patients with DDD, we were able to identify several immune signaling pathways that were upregulated. We found 702 genes demonstrating significant upregulation and 3,734 genes with significant downregulation when comparing the NP samples to other musculoskeletal tissues. To better understand the unique regulatory pathway for DH and DS, the differential gene expression profile revealed 1514 upregulated and 281 downregulated mRNA. Furthermore, STRING analysis demonstrated differentially expressed genes for each gene ontology (GO) term from different types of networks with distinct cellular functions for each biomedical condition. Specifically, we found several differentially expressed genes in chemotactic signaling (CXCL10, CXCL11, IL1RL2, and IL-6) and matrix-degrading pathways (MMP16, ADAMTSL1, 5, 8, 12, and 15). Neurosurgeons can leverage these identified gene regulatory networks, or conduct similar transcriptomic analysis, to strategically engineer novel pharmacological approaches for the treatment of lumbar DDD and other degenerative diseases.
4. Systems neuroimmunology and aneursymal subarachnoid hemorrhage
4.1. Immunology review in aSAH
Extravasated blood products that are released after intracerebral hemorrhage (ICH) or aneurysmal subarachnoid hemorrhage (aSAH) are known triggers of inflammation in the brain.29,30 While primary injury, which occurs within hours after ICH, is related to the mechanical distortion of brain tissue due to hematoma expansion, secondary injury, which occurs on a much longer time scale after initial insult, is often mediated by immune signaling. Red blood cell lysis occurs approximately 24 hours after initial ICH and has been demonstrated to activate resident microglia and astrocytes. These neuroglia in turn release cytokines such as TNF-α and IL-1β that recruit circulating immune cells, including neutrophils and macrophages.29 In aSAH, the pathway towards neutrophil recruitment is similar and cytokines like IL-6, IL-1α, IL-1β, IL-8, and TNF-α have been implicated as contributors to cerebral vasospasm.30
High-throughput methods from multiomics are enabling researchers to interrogate changes in the cerebrospinal fluid of patients with aSAH and ICH. Here we summarize recent work from Koch et al.7 that used metabolomics and machine learning to study biomarkers in the cerebrospinal fluid (CSF) of patients with aSAH.
4.2. Use Case 2: Metabolomics to evaluate outcomes after aneurysmal subarachnoid hemorrhage
In their study, Koch et al. used metabolomic data derived from the CSF samples of 81 patients with aSAH to identify key metabolites in an immune signaling pathway that were predictive of poor outcomes after aSAH. CSF samples were obtained from unclamped external ventricular drains at three different time points (0–5 days, 6–10 days, and 11–15 days after admission). CSF samples were additionally obtained in a control cohort of 16 patients with nonruptured cerebral aneurysms either through lumbar drain or lumbar puncture.
Several steps were taken to prepare CSF samples for metabolomic analysis. Samples were centrifuged to pellet out cellular material and the resulting supernatant was aliquoted and treated with cetonitrile/methanol (3:1) with citrulline-d8 in order to prepare the samples for processing through a high-performance liquid chromatography (HPLC) system. Mass spectrometry was then used to quantitatively measure the concentrations of a total of 138 metabolites.
To process their metabolomic data, Koch et al. used machine learning models to identify which metabolites were most strongly associated with modified Rankin scale (mRS) scores at discharge and at 90 days after discharge. They identified that the concentrations of key metabolites that alter the nitric oxide signaling pathway including symmetric dimethylarginine (SDMA), dimethylguanidine valeric acid (DMVA), and ornithine are associated with poor mRS at discharge and at 90 days.
The methodology from Koch et al. provides an example of how multiomics can shed light into how key metabolites that regulate immune signaling pathways can also shape outcomes after neurosurgical disease. Their findings provide key biomarkers for predicting outcomes, as well as point to potential drug targets for modulating the immune response after aSAH. Neurosurgeons may use a similar multiomics workflow as Koch et al. to study immune signaling pathways that are upregulated in the CSF samples of patients with epilepsy, spinal cord injury, and other neurosurgical diseases.
5. Systems neuroimmunology and glioblastoma multiforme
5.1. Immunology review in GBM
GBM is an aggressive brain tumor characterized by significant cellular and molecular heterogeneity, as well as complex interactions with the host immune system.31 Recent work characterizing the GBM microenvironment has led to the discovery of functionally distinct immune cells in tumors including tumor-infiltrating dendritic cells, regulatory T cells, cytotoxic T cells and tumor associated macrophages which each uniquely shape response to immunotherapies.32 For example, regulatory T cells recruited by GBM cells can promote tumorigenesis by suppressing the cytotoxic effect of tumor-infiltrating dendritic cells and cytotoxic T cells.33 Given how complex these interactions between immune cells and GBM cells are, there has been an increasing appreciation of the need for systems-based approaches to develop new immunotherapies for GBM.34
Herein, we present our second use case based on original work that uses systems neuroimmunology approaches to better understand how the immune system interacts with GBM. Use Case 3, discusses work done by our team to determine potential therapeutic targets for HSV-1 derived oncolytic viruses (OVs) to treat GBM based on a proteomic analysis. This use case highlights how proteomic data can generate insights about GBM neuroimmunology.
5.2. Use Case 3: Protein-protein interaction analysis to identify tumor susceptibility to immunotherapies
Immunotherapies use the host immune system to target GBM cells and are broadly divided into two categories: passive and active.37 Passive immunotherapies, such as monoclonal antibodies, require continuous administration over time as they produce short-lived, yet specific, immune responses. On the other hand, active immunotherapies, including OVs, are a direct stimulation of an immune response with long-term effects. OV immunotherapy for cancer dates back to the 1990s, when herpes simplex virus 1 (HSV-1) was engineered to selectively replicate and target tumor cells. Since then, active immunotherapies have made significant strides and are the focal point of many clinical trials.37
Our team has previously published a study3 that aimed to further understand HSV-1 OV resistance by investigating the role of cellular communication network factor 1 (CCN1) on GBM intracellular state. Found in most GBM microenvironments, CCN1 expression has shown to predict resistance to OV. This work builds on a study by Haseley et al.38, which found that CCN1 binds and activates cell surface integrin α6β1, promoting an antiviral and protumoral state. GBM with high concentrations of CCN1 (CCN1high) have demonstrated worse rates of overall and progression-free survival.3
To further understand the downstream effect of CCN1, transcriptomic data of LN229 GBM cell lines were mapped to known protein-protein interactions (PPIs) in the iRefIndex database. These PPIs were synthesized into networks and analyzed in NetDecoder to compare CCN1high to CCN1low GBM phenotypes to elucidate critical differences between the two states. High impact genes, network routers, key targets, and CCN1-specific edges were identified through information flow analysis. We further compared these cell states through network and motif modeling, overrepresentation analysis, and assessment of gene dependencies.3
After studying differential edge flows, we identified 39 nodes and 12 binary edges that may potentially determine susceptibility of CCN1high GBM to OV. Furthermore, CCN1high states were shown to exploit IDH1 and TP53 and increase dependency on RPL6, HUWE1, and COPS5. Our findings were reproduced in 65 other GBM cell lines and 174 clinical GBM patient sample datasets for validation.3
Our study identified novel pathways, proteins, and interactions critical to CCN1high GBM phenotype. Conducting a generalized network model and systems level analysis allowed us to identify several innate immune pathways in GBM that CCN1 leverages to disrupt HSV-1 OV immunotherapy. Additionally, we found opportunities to strategically improve HSV-1 OV design. For example, because measles virus is not present in the resistance signatures we identified through our overexpression analysis (Figure 2C), we postulated that measles virus may be a better vector than HSV-1 for GBM treatment.3 In a similar vein, other neurosurgeons can use this approach to design more effective immunotherapies for GBMs and other CNS malignancies.
6. Conclusions
Recent evidence has demonstrated that neuroimmune mediated processes have an essential role in both the normal physiology of the nervous system, as well as the pathophysiological processes that drive neurosurgical disease. The study of the multiome, which integrates genomic, transcriptomic, proteomic, and other omics data sources, is driving new discoveries in neuroimmunology. Compared to traditional approaches in neuroimmunology, multiomics methods allow for holistic interrogation of multiple immune signaling pathways at a systems level. In this review, we described three use cases where multiomics approaches were used to investigate the neuroimmunology of different conditions treated by neurosurgeons including DDD, GBM, and aSAH. We hope that the methods outlined here may catalyze further research using systems immunology and empower neurosurgeons to generate future discoveries in neuroimmunology.
Acknowledgements:
Figure 1 created using BioRender.com
Funding:
D.D.M. was supported by an individual fellowship from the National Cancer Institute (F30 CA250122), an institutional training grant from the National Institute of General Medical Sciences (T32 GM65841), the Mayo Clinic Medical Scientist Training Program, and the Mayo Clinic Center for Regenerative Medicine. A.Q.H. was supported by research grants from the National Institute of Health (R01 CA216855R01 CA195503, R01 CA200399, R01 CA216855, R33 CA240181)
Abbreviations:
- AF
outer annulus fibrosus
- cDNA
complementary DNA
- CCN1
cellular communication network factor
- DDD
degenerative disc disease
- DH
disc herniation
- DGE
differential gene expression
- DS
degenerative spondylolisthesis
- EP
end plate
- GBM
glioblastoma
- GEO
Gene Expression Omnibus
- GO
gene ontology
- HIV-1
human immunodeficiency virus 1
- HPLC
high performance liquid chromatography
- HSV-1
herpes simplex virus 1
- IFN
interferon
- IL
interleukin
- IPA
Ingenuity Pathway Analysis
- IVD
intervertebral disc
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- NP
nucleus pulposus
- OV
oncolytic virus
- PPI
protein-protein interaction
- RIN
RNA integrity number
- RNA-seq
RNA sequencing
- scRNA-seq
single cell RNA sequencing
- SNF
similarity network fusion
- SpA
spondyloarthritis
- TNF
tumor necrosis factor
- t-SNE
t-distributed stochastic neighbor embedding
- UMAP
uniform manifold approximation and projection
- aSAH
aneurysmal subarachnoid hemorrhage
Footnotes
Disclosures: No conflicts of interest to disclose
References
- 1.Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. 2015;7(1):a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bydon M, Moinuddin FM, Yolcu YU, et al. Lumbar intervertebral disc mRNA sequencing identifies the regulatory pathway in patients with disc herniation and spondylolisthesis. Gene. 2020;750:144634. [DOI] [PubMed] [Google Scholar]
- 3.Monie DD, Correia C, Zhang C, Ung CY, Vile RG, Li H. Modular network mechanism of CCN1-associated resistance to HSV-1-derived oncolytic immunovirotherapies for glioblastomas. Sci Rep. 2021;11(1):11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma T, Hu C, Lal B, et al. Reprogramming Transcription Factors Oct4 and Sox2 Induce a BRD-Dependent Immunosuppressive Transcriptome in GBM-Propagating Cells. Cancer Res. 2021;81(9):2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valentín-Guillama G, López S, Kucheryavykh YV, et al. HIV-1 Envelope Protein gp120 Promotes Proliferation and the Activation of Glycolysis in Glioma Cell. Cancers. 2018;10(9). doi: 10.3390/cancers10090301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Kharboosh R, ReFaey K, Lara-Velazquez M, Grewal SS, Imitola J, Quiñones-Hinojosa A. Inflammatory Mediators in Glioma Microenvironment Play a Dual Role in Gliomagenesis and Mesenchymal Stem Cell Homing: Implication for Cellular Therapy. Mayo Clin Proc Innov Qual Outcomes. 2020;4(4):443–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koch M, Acharjee A, Ament Z, et al. Machine Learning-Driven Metabolomic Evaluation of Cerebrospinal Fluid: Insights Into Poor Outcomes After Aneurysmal Subarachnoid Hemorrhage. Neurosurgery. 2021;88(5):1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis MM, Tato CM, Furman D. Systems immunology: just getting started. Nat Immunol. 2017;18(7):725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khan-Farooqi HR, Prins RM, Liau LM. Tumor immunology, immunomics and targeted immunotherapy for central nervous system malignancies. Neurol Res. 2005;27(7):692–702. [DOI] [PubMed] [Google Scholar]
- 10.Villani A-C, Sarkizova S, Hacohen N. Systems Immunology: Learning the Rules of the Immune System. Annu Rev Immunol. 2018;36:813–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braga-Neto UM, Marques ETA Jr. From functional genomics to functional immunomics: new challenges, old problems, big rewards. PLoS Comput Biol. 2006;2(7):e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20(11):631–656. [DOI] [PubMed] [Google Scholar]
- 13.Liang Q, Dharmat R, Owen L, et al. Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat Commun. 2019;10(1):5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owens NDL, De Domenico E, Gilchrist MJ. An RNA-Seq Protocol for Differential Expression Analysis. Cold Spring Harb Protoc. 2019;2019(6). doi: 10.1101/pdb.prot098368 [DOI] [PubMed] [Google Scholar]
- 16.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312(5771):212–217. [DOI] [PubMed] [Google Scholar]
- 18.Ryan DJ, Spraggins JM, Caprioli RM. Protein identification strategies in MALDI imaging mass spectrometry: a brief review. Curr Opin Chem Biol. 2019;48:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform Biol Insights. 2020;14:1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rappoport N, Shamir R. Multi-omic and multi-view clustering algorithms: review and cancer benchmark. Nucleic Acids Res. 2018;46(20):10546–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun. 2019;10(1):5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform. 2018;19(6):1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B, Mezlini AM, Demir F, et al. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11(3):333–337. [DOI] [PubMed] [Google Scholar]
- 24.Rubinstein A, Bracha N, Rudner L, Zucker N, Sloin HE, Chor B. BioNSi: A Discrete Biological Network Simulator Tool. J Proteome Res. 2016;15(8):2871–2880. [DOI] [PubMed] [Google Scholar]
- 25.Leone G, Torricelli P, Chiumiento A, Facchini A, Barbucci R. Amidic alginate hydrogel for nucleus pulposus replacement. J Biomed Mater Res A. 2008;84(2):391–401. [DOI] [PubMed] [Google Scholar]
- 26.Livshits G, Kalinkovich A. Hierarchical, imbalanced pro-inflammatory cytokine networks govern the pathogenesis of chronic arthropathies. Osteoarthritis Cartilage. 2018;26(1):7–17. [DOI] [PubMed] [Google Scholar]
- 27.Navone SE, Marfia G, Giannoni A, et al. Inflammatory mediators and signalling pathways controlling intervertebral disc degeneration. Histol Histopathol. 2017;32(6):523–542. [DOI] [PubMed] [Google Scholar]
- 28.Weber KT, Alipui DO, Sison CP, et al. Serum levels of the proinflammatory cytokine interleukin-6 vary based on diagnoses in individuals with lumbar intervertebral disc diseases. Arthritis Res Ther. 2016;18:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Wang Y, Wang J, Anne Stetler R, Yang Q-W. Inflammation in intracerebral hemorrhage: from mechanisms to clinical translation. Prog Neurobiol. 2014;115:25–44. [DOI] [PubMed] [Google Scholar]
- 30.Schneider UC, Xu R, Vajkoczy P. Inflammatory Events Following Subarachnoid Hemorrhage (SAH). Curr Neuropharmacol. 2018;16(9):1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Sun D, Chen Y-J, et al. Cell lineage-based stratification for glioblastoma. Cancer Cell. 2020;38(3):366–379.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pombo Antunes AR, Scheyltjens I, Duerinck J, Neyns B, Movahedi K, Van Ginderachter JA. Understanding the glioblastoma immune microenvironment as basis for the development of new immunotherapeutic strategies. Elife. 2020;9. doi: 10.7554/eLife.52176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeCordova S, Shastri A, Tsolaki AG, et al. Molecular Heterogeneity and Immunosuppressive Microenvironment in Glioblastoma. Front Immunol. 2020;11:1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monie DD, Bhandarkar AR, Parney IF, et al. Synthetic and systems biology principles in the design of programmable oncolytic virus immunotherapies for glioblastoma. Neurosurg Focus. 2021;50(2):E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall JR, Short SC. Management of glioblastoma multiforme in HIV patients: a case series and review of published studies. Clin Oncol. 2009;21(8):591–597. [DOI] [PubMed] [Google Scholar]
- 36.Yang J-M, Schiapparelli P, Nguyen H-N, et al. Characterization of PTEN mutations in brain cancer reveals that pten mono-ubiquitination promotes protein stability and nuclear localization. Oncogene. 2017;36(26):3673–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbott M, Ustoyev Y. Cancer and the Immune System: The History and Background of Immunotherapy. Semin Oncol Nurs. 2019;35(5):150923. [DOI] [PubMed] [Google Scholar]
- 38.Haseley A, Boone S, Wojton J, et al. Extracellular matrix protein CCN1 limits oncolytic efficacy in glioma. Cancer Res. 2012;72(6):1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costantino F, Talpin A, Said-Nahal R, et al. A family-based genome-wide association study reveals an association of spondyloarthritis with MAPK14. Ann Rheum Dis. 2017;76(1):310–314. [DOI] [PubMed] [Google Scholar]
- 40.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darmanis S, Sloan SA, Croote D, et al. Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma. Cell Rep. 2017;21(5):1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Y, Bergström T, Jiang Y, et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine. 2015;2(10):1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt T, Samaras P, Frejno M, et al. ProteomicsDB. Nucleic Acids Res. 2018;46(D1):D1271–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joshi RN, Stadler C, Lehmann R, et al. TcellSubC: An Atlas of the Subcellular Proteome of Human T Cells. Frontiers in Immunology. 2019;10. doi: 10.3389/fimmu.2019.02708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sud M, Fahy E, Cotter D, et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44(D1):D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]


