Abstract
Background
Non-small cell lung cancer (NSCLC) frequently metastasizes to bone, leading to poor prognosis. Siglec15 has been identified as a newly discovered immune checkpoint and exists in a variety of tumors. However, the expression and function of Siglec15 in NSCLC and bone metastasis remains largely unclear.
Methods
Siglec15 expression in NSCLC and the correlation between Siglec15 expression and the clinicopathological factors of patients with NSCLC were analyzed using The Cancer Genome Atlas (TCGA) dataset. Correlation analysis between Siglec15 and bone metastasis-related genes expression was based on the Molecular Signatures Database (MSigDB). Western blotting and immunohistochemistry were applied to detect Siglec15 expression in NSCLC and spinal metastasis. Human A549 and mouse CMT167 cells were transfected with Siglec15 siRNA to investigate its biological functions in NSCLC proliferation, migration, and invasion. The immune-related signaling pathways and correlations between Siglec15 and tumor-infiltrating immune cells and different immune checkpoints in the NSCLC tumor microenvironment (TME) were analyzed using Estimating Relative Subsets of RNA Transcripts (CIBERSORT) and gene set enrichment analysis (GSEA). To demonstrate Siglec15 in NSCLC cell-mediated T cell suppression and investigate the potential mechanism of Siglec15 silencing in antitumor immunity, we used a T cell killing assay in vitro and the high‑throughput sequencing approach.
Results
Siglec15 expression was positively associated with the tumor stage and lymph node metastasis, and was markedly up-regulated in NSCLC bone metastasis. Functionally, Siglec15 knockdown inhibited the proliferation, migration, and invasion of NSCLC cells (A549 and CMT167 cell lines). A total of eight kinds of tumor-infiltrating immune cells were found to have a strong association with the Siglec15 expression in NSCLC cases. The expression of previously discovered immune checkpoints was higher in the high Siglec15 expression NSCLC group. Furthermore, an in vitro T cell killing assay showed that the down-regulation of Siglec15 in tumor cells could enhance the antitumor immune responses of CD8+ T cells. High‑throughput sequencing revealed the potential molecular mechanisms underlying the Siglec15-mediated immunosuppression effect of tumor cells on immune cells.
Conclusions
Siglec15 may be involved in the pathogenesis of spinal metastasis in NSCLC and provide a new potential therapeutic target for the treatment of NSCLC and bone metastasis.
Keywords: Non-small cell lung cancer (NSCLC), bone metastasis, Siglec15, immunosuppression, tumor progression
Introduction
Non-small cell lung cancer (NSCLC) is one of the most commonly diagnosed cancers worldwide (1). Invasion and distant metastasis are the leading cause of death in NSCLC cases, and bone is considered to be the main metastatic site of NSCLC (2). It has been reported that more than 70% of patients with advanced NSCLC suffer from bone metastasis, 80% of which are located in the spine (3). Spinal metastasis often leads to severe bone pain, pathological fracture of the vertebral body, spinal cord compression, and neurological dysfunctions such as incontinence and even paralysis (4). The treatment of this disease includes chemotherapy, radiotherapy, surgery, molecular targeted therapy, etc. Although the combination of these treatments can improve the quality of life of NSCLC spinal metastasis patients, these treatments often cannot significantly improve the overall survival of patients. Therefore, an in-depth understanding of the underlying mechanism of NSCLC spinal metastasis will be beneficial to the prevention and treatment of the disease (5).
Programmed cell death-ligand 1 (PD-L1) immune checkpoint blockade therapy is regarded as a successful normalization cancer immunotherapy (6). However, this treatment strategy is only applicable in less than 40% of solid tumors, indicating the presence of other important immune suppressors (7). A recent study published by the Wang et al. in Nature Medicine found that Siglec15, a member of the Sialic acid-binding immunoglobulin-like lectins, was a new immunosuppressive checkpoint that could suppress anti-tumor T cell responses in the tumor microenvironment (TME). Siglec15 expression was shown to be significantly higher in tumor cells and tumor-infiltrating myeloid cells in TME (8). Furthermore, they also found that Siglec15 expression is mutually exclusive to PD-L1, indicating that these two immune checkpoints may have different mechanisms for tumor immunosuppression. Siglec15 could favor M2-like TAM polarization and immunosuppressive microenvironment of pancreatic ductal adenocarcinoma, meanwhile high Siglec15+ TAM abundance significantly correlated with severe microvascular invasion, poor tumor differentiation (9). It has shown that Siglec15 plays an important role in tumor immune microenvironment and has the potential to be a vital immunotherapy-related target. Recently, it has been reported that Siglec15 participates in the development of different kinds of cancers. While in NSCLC, there is only one study has preliminarily investigated the expression of Siglec15, an emerging immune suppressor, in early-stage lung cancers and its relationship with the clinicopathological characteristics of NSCLC patients yet no potential mechanism has been deeply explored (10). Hence, the role and specific mechanism of Siglec15 in NSCLC, especially in NSCLC bone metastasis, still remains further investigated.
The aim of the present study was to investigate Siglec15 expression in clinical NSCLC spinal metastasis and primary NSCLC tissues, and to evaluate the biological functions of Siglec15 in NSCLC progression. Notably, we further explored the role and molecular mechanisms of Siglec15 in NSCLC immunosuppression. Our results suggested that Siglec15 could possibly serve as a biomarker and molecular target for treating NSCLC and bone metastasis. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-764/rc).
Methods
Clinical specimens
Samples of three NSCLC spinal metastasis tissues, three primary NSCLC tissues, and paired para-carcinoma tissues (distance >3 cm from the tumor tissue) were collected from patients who underwent surgical resection in Zhongshan Hospital, Fudan University between April 2020 and August 2020, and subsequently further confirmed by pathological results. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Zhongshan Hospital, Fudan University, Shanghai, China (approval No. Y2019-085) and informed consent was taken from all the patients.
Cell culture
Cell lines (A549 and CMT167) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco, USA) in humidified air with 5% carbon dioxide (CO2) at 37 ℃.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
The total ribonucleic acid (RNA) was extracted from tissues or cultured cells with TRIzol reagent (Invitrogen, USA), according to the manufacturer’s protocol. One microgram of total RNA was reverse transcribed using PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). Complementary DNA (cDNA) was used for subsequent qRT-PCR reactions (SYBR, TaKaRa, Japan) according to the manufacturer’s instructions. The results were normalized to the expression of β-actin. The expression levels of each target gene were determined according to the 2−ΔΔCt method, as previously described (11). The primer sequences were listed in the Table 1 below. The primers were synthesized by HuaGene Biotech Co., Ltd. (Shanghai, China).
Table 1. The primer sequences.
| Gene | Primer sequences |
|---|---|
| Mice Siglec15 | Forward: TCAGGCTCAGGAGTCCAATTAT |
| Reverse: GGTCTAGCCTGGTACTGTCCTTT | |
| Mice ACTB | Forward: GTGACGTTGACATCCGTAAAGA |
| Reverse: GCCGGACTCATCGTACTCC | |
| Human Siglec15 | Forward: CAGCCACCAACATCCATTTC |
| Reverse: CGCTCAAGCTAATGCGTGTA | |
| Human ACTB | Forward: GGCCAACCGCGAGAAGATGAC |
| Reverse: GGATAGCACAGCCTGGATAGCAAC |
Western blotting
Tissue or cell proteins were collected using radio-immunoprecipitation assay (RIPA) lysis buffer, and a bicinchoninic acid (BCA) assay was applied to quantify the concentration of proteins. After boiling, the same amount of protein was separated on sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% skim milk at room temperature and probed with primary antibodies for Siglec15 (Sigma Aldrich, USA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Beyotime, China) overnight at 4 ℃, followed by incubation with secondary antibodies at 25 ℃ for 2 h. The signals were visualized using a chemiluminescence system with the enhanced chemiluminescence (ECL) reagent (WBKlS0100, Millipore, USA).
Immunohistochemistry
Immunohistochemical analysis was performed to measure the level of Siglec15. Briefly, the resected NSCLC spinal metastasis tissues were fixed in paraformaldehyde for 48 h and decalcified with 10% ethylenediaminetetraacetic acid (EDTA) for 8 weeks. Next, paraffin-embedded and formalin-fixed tissues were cut into 4 µm sections and incubated on slides. The endogenous peroxidase was blocked using 3% hydrogen peroxide (H2O2). Primary antibody against Siglec15 (Sigma Aldrich, USA) was applied to the slides, which were incubated at 4 ℃ overnight. After washing, the sections were incubated with the secondary antibodies. The sections were photographed using a microscope (DM 3000, Leica, Wetzlar, Germany).
Small interfering RNA (siRNA) transfection
siRNA was purchased from OBiO Technology Corp., Ltd. (Shanghai, China). The siRNA sequences were shown in the Table 2 below. Lipofectamine® 3000 reagent (Invitrogen, USA) was used for siRNA transfection according to the manufacturer’s instructions. Transfection occurred by adding 0.2 nmoL siRNA and 5 µL lipofectamine 3000 in six-well plates with cells at 50–60% confluence. After being transfected for 48 h, the knockdown efficiency of Siglec15 was assessed by western blotting and qRT-PCR.
Table 2. The siRNA sequences.
| siRNA | Sequence (5'-3') |
|---|---|
| siSiglec15-mus-#1 | CCUCAGCCAUCACCAUCUATT |
| siSiglec15-mus-#2 | GGCUCAGGAGUCCAAUUAUTT |
| siSiglec15-mus-#3 | UCGCUAUGAGAGUCGCCAUTT |
| scramble siRNA-mus | UUCUCCGAACGUGUCACGUTT |
| siSiglec15-homo-#1 | UCUCCCGACAGGCUCAUUUTT |
| siSiglec15-homo-#2 | GGAGAACUUGCUCAACACATT |
| siSiglec15-homo-#3 | AGGCCCAGGAGUCCAAUUATT |
| scramble siRNA-homo | CAGGAGUCCAAUUAUGAAATT |
Cell proliferation and plate colony formation assays
Cell proliferation was assessed using the cell counting kit-8 (CCK-8) assay. Transfected cells were seeded in a 96-well plate at 1.5×103 cells per well, and subsequently followed by incubation for 24, 48, 72, 96, and 120 h. Ten µL of CCK-8 solution (Beyotime, China) was added to each well and the cells were incubated at 37 ℃ for another 1 h in darkness. The absorbance value was detected at 450 nm (OD450 nm) using a microplate reader (Thermo Fisher Scientific, USA).
For plate colony formation assays, a total of 1×103 transfected cells were seeded in a six-well plate and cultured in DMEM containing 10% FBS for 1 week to allow for colony formation. Cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet (Beyotime, China) for 20 min. Cell colonies was captured using a digital camera following analysis with an inverted microscope (Olympus, Japan).
Transwell cell migration and invasion assays
Cell migration and invasion assays were evaluated using Transwell chambers (8-µm pore, Corning, USA) with or without Matrigel (BD Biosciences, USA). After cell transfection for 48 h, 5×104 transfected cells in serum-free medium were added to the upper chamber, and 600 µL of the corresponding medium containing 10% FBS was added into the lower chamber. After 24 h of 37 ℃ incubation, the remaining cells on the upper surface of the membrane were removed with a cotton swab and the migrated cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 20 min. The migrated cells in the chamber were counted at 10× magnification and photographed using an inverted microscope (Olympus, Japan).
Wound healing assays
Cell migration of the transfected cells was evaluated by wound healing assays. The cells were seeded in six-well plate and cultured in DMEM containing 10% FBS to 90% confluence. Wounds at the cell surface were created using 200 µL pipette tip, and the cells were washed three times using phosphate-buffered saline (PBS) to remove detached cells. After 24 h of 37 ℃ incubation with medium containing 1% FBS, images were taken using an inverted microscope (Olympus, Japan) at 4× magnification.
In vitro co-culture of CD8+ T cells together with tumor cells
EasySep Mouse CD8+ T Cell Isolation Kit (STEMCELL Technologies Inc., Canada) was used to isolate mouse CD8+ T cells from splenocytes according to the manufacturer’s protocol. CD8+ T cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 10% FBS and antibiotics. Isolated CD8+ T cells were activated using anti-mouse CD3 (2 µg/mL) and anti-mouse CD28 (1 µg/mL) for 2 days.
After CMT167 cells transfection for 48 h, 1×105 transfected cells were seeded into six-well plates per well and cultured at 37 ℃. After overnight incubation to allow tumor cell attachment, activated CD8+ T cells were added at the effector cell (CD8+ T cells) to target cell (tumor cells) (E/T) ratio of 10:1. After co-culturing for 48 h, the CD8+ T cells in supernatant were isolated and stained with Brilliant Violet 510™ anti-mouse CD8 and FITC anti-mouse Ki-67 (BioLegend, USA) to analyze T cell proliferation using a FACSAriaIII flow cytometer (BD Biosciences, USA). To analyze IFN -γ+ CD8+ T cells, T cells in the supernatant were collected, fixed, permeablized, and stained with Brilliant Violet 421™ anti-mouse IFN-γ and Brilliant Violet 510™ anti-mouse CD8 (BioLegend, USA), and assessed using a FACSAriaIII flow cytometer. Adherent tumor cells were isolated with trypsin. The Annexin V -FITC Apoptosis Detection Kit (Invitrogen, USA) was used to assess the percentage of apoptotic tumor cells.
Bioinformatics analysis of The Cancer Genome Atlas (TCGA) database
The normalized TCGA messenger RNA (mRNA) expression of Siglec15 and the corresponding clinical information of the patients with NSCLC were downloaded from TCGA (https://portal.gdc.cancer.gov/). Bioinformatics analysis of the NSCLC dataset from TCGA was performed as previously described (12).
Analysis of tumor-infiltrating immune cells
The Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithm, created by Newman et al., was used to assess the proportion of 22 types of tumor-infiltrating immune cells profile in all NSCLC cases (13). Samples with a result of P<0.05 in the CIBERSORT analysis were used for further analysis. These tumor-infiltrating immune cells included resting memory CD4+ T cells, activated memory CD4+ T cells, CD8+ T cells, naive CD4+ T cells, gamma delta T cells, follicular helper T cells, regulatory T cells, memory B cells, naive B cells, activated mast cells, resting mast cells, activated dendritic cells, resting dendritic cells, macrophages (M0, M1, and M2), eosinophils, monocytes, activated natural killer cells, resting natural killer cells, plasma cells, and neutrophils.
RNA sequencing
Six independent CD8+ T cell samples were collected (n=3 from the siSiglec15 group and normal control group) to obtain RNA samples. RNA extraction and RNA-seq were performed by OBiO Technology Corp., Ltd. (Shanghai, China). Differences in gene expression levels between two groups were analyzed using the Bioconductor package edgeR. |log2FC| ≥1 and P<0.05 were used as the cut-off criteria to identify significantly differentially-expressed genes (DEGs). Heatmaps of DEGs were drawn using the pheatmap package. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of DEGs were conducted using the R packages clusterProfiler, enrichplot, and ggplot2 to explore the biological functions and signaling pathways. Gene set enrichment analysis (GSEA) was performed using default parameters in the Molecular Signatures Data base version 4.1.
Statistical analysis
Quantitative variables were expressed as mean ± standard deviation. Statistically significant differences were determined by one-way analysis of variance (ANOVA) or the two-tailed Student’s t-test. The Kaplan-Meier method was used to estimate survival curves and the log-rank test was used to test the differences between groups. The experiments were replicated for 3 times. All statistical analyses were performed using GraphPad Prism version 8 (Graphpad Software, USA). A P value <0.05 was considered statistically significant.
Results
Higher Siglec15 expression is associated with malignant progression and spinal metastasis in patients with NSCLC
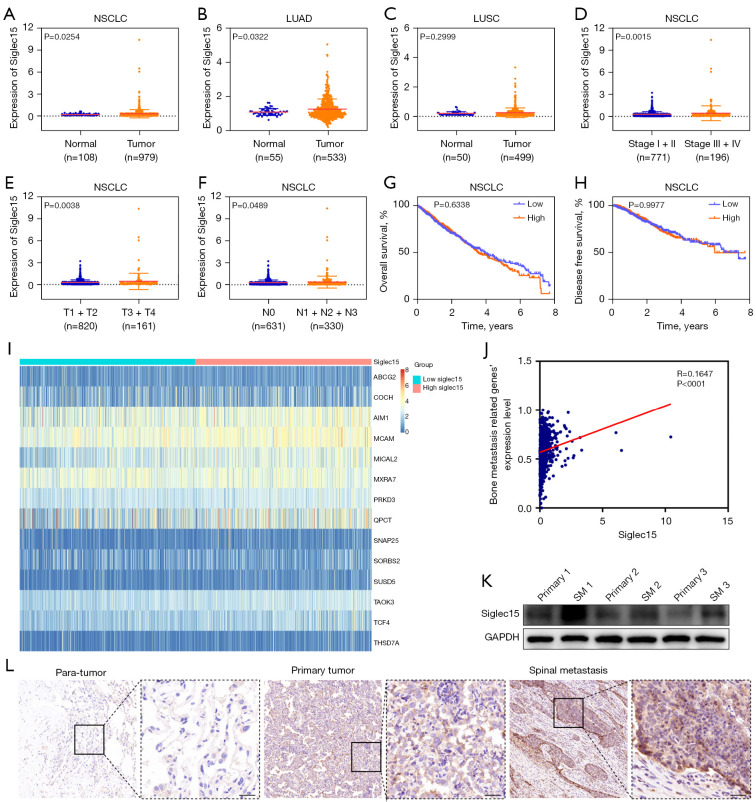
Siglec15 expression in NSCLC was analyzed using TCGA dataset. The results indicated that Siglec15 expression level in NSCLC was significantly higher than those in the corresponding normal tissues (Figure 1A). Moreover, further analysis demonstrated that Siglec15 expression was significantly higher in lung adenocarcinoma (LUAD) than normal lung tissues, while no significant Siglec15 overexpression was observed in lung squamous carcinoma (LUSC) compared with normal tissues (Figure 1B,1C). Next, we explored the correlation between Siglec15 expression and the clinicopathological factors of patients with NSCLC. In general, Siglec15 level was associated with the TNM stage. Patients with advanced tumor stages (III + IV), higher T stage (T3 + T4), or lymph node metastasis (N1 + N2 + N3) were associated with higher Siglec15 expression, suggesting that Siglec15 may promote the malignant progression of NSCLC (Figure 1D-1F). We also examined the relationship between Siglec15 expression and NSCLC patient survival, and found that high Siglec15 expression was not associated with either overall survival (Figure 1G) or disease-free survival (Figure 1H) in patients with NSCLC.
Figure 1.
Siglec15 was highly expressed in NSCLC and was correlated with spinal metastasis. (A) The expression profiles of Siglec15 mRNA in NSCLC (n=979) and normal lung tissues (n=108) based on TCGA database are represented. (B,C) Data collected from TCGA database showed the relative expression of Siglec15 mRNA in TCGA-LUAD (n=533) and TCGA-LUSC (n=499) tissues and normal lung tissues (n=55, n=50). (D-H) Siglec15 mRNA expression and the relationship between tumor stage, T stage, lymph node metastasis, overall survival, and disease-free survival in TCGA-NSCLC. (I) Heatmap of bone metastasis-related genes expression between the high- and low-Siglec15 expression groups. (J) Correlation analysis between Siglec15 expression and bone metastasis-related genes expression based on the MSigDB. (K) The levels of Siglec15 protein expression in human primary NSCLC tissues (primary) and NSCLC SM tissues were examined by western blotting. (L) Siglec15 protein expression in human normal lung and primary NSCLC, as well as NSCLC spinal metastasis tissues were examined by IHC. Scare bar =50 μm. NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma; LUSC, lung squamous carcinoma; MSigDB, Molecular Signatures Database; SM, spinal metastasis; IHC, immunohistochemistry.
As has been reported in previous studies, Siglec15 has immune-suppressive ability and is also an important modulator for osteoclast differentiation and bone resorption (8,14-16), and thus, Siglec15 may play a unique role in malignant tumor metastasis into the bone by mediating immune evasion and osteolytic bone destruction in the bone microenvironment. To explore this issue, bone metastasis-related genes were extracted from the metastasis gene signature, which were up-regulated genes during the metastasis of NSCLC tumors to bone sourced from the Molecular Signatures Database (MSigDB) (17). The results showed that Siglec15 expression was positively correlated with bone metastasis-related genes expression (Figure 1I,1J). Furthermore, human NSCLC spinal metastasis tissues were analyzed using western blotting, which showed significantly higher levels of Siglec15 expression than primary NSCLC tissues (Figure 1K). Immunohistochemistry (IHC) assay results also demonstrated that Siglec15 expression was increased in human NSCLC spinal metastasis tissues relative to the primary NSCLC tissues and paired para-carcinoma tissues (Figure 1L).
Knockdown efficiency of Siglec15 gene
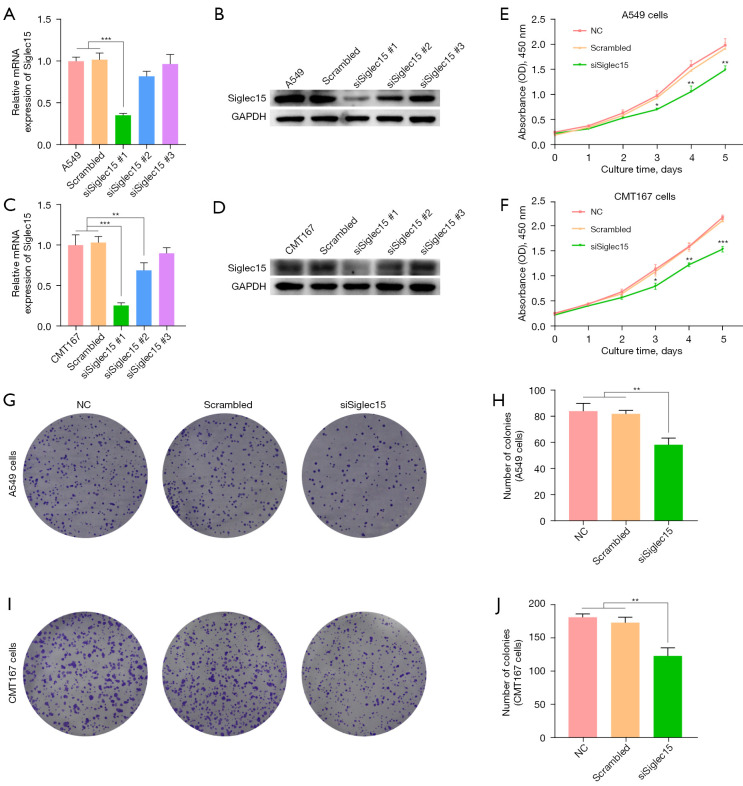
To confirm the RNAi-mediated silencing of Siglec15 in human A549 and mouse CMT167 cells, Siglec15 mRNA and protein expression were respectively quantified by RT-PCR and western blotting 48 h after transfection of Siglec15 siRNA (Figure 2A-2D). Three siRNAs targeting Siglec15 gene of A549 and CMT167 cells were respectively designed. As shown in Figure 2A, the expression of Siglec15 mRNA declined to 35.4% in the siSiglec15#1 transfected A549 cells, compared to the untransfected control A549 cells (P<0.001). There were no significant differences between the untransfected control, negative control scrambled siRNA transfected A549 cells, siSiglec15#2 transfected A549 cells, and siSiglec15#3 transfected A549 cells. Western blotting analysis confirmed the results of the RT-PCR analysis (Figure 2B). Similarly, siSiglec15#1 transfected CMT167 cells showed the best Siglec15 knockdown efficiency (Figure 2C,2D). There were no significant differences between the untransfected control and negative control scrambled siRNA transfected CMT167 cells. Therefore, siSiglec15#1 targeting A549 and CMT167 cells, respectively, were selected for use in subsequent experiments.
Figure 2.
Down-regulated Siglec15 expression inhibited A549 and CMT167 cell proliferation abilities. (A,B) Validation of Siglec15 knockdown A549 cell at mRNA and protein levels after Siglec15-specific siRNAs transfection by RT-PCR and western blotting, respectively. (C,D) Validation of Siglec15 knockdown CMT167 cell at mRNA and protein levels after Siglec15-specific siRNAs transfection by RT-PCR and western blotting, respectively. (E,F) The proliferation of A549 and CMT167 cells was performed by CCK-8 assays after Siglec15-specific siRNAs transfection and negative control. (G-J) Colony formation assay in A549 and CMT167 cells and a corresponding number of colonies. Cells were stained with 0.1% crystal violet. *, P<0.05; **, P<0.01; ***, P<0.001. CCK-8, cell counting kit-8; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NC, normal control.
Knock-down of Siglec15 inhibited the proliferation of NSCLC cells
To detect the potential function of Siglec15 in NSCLC, we assessed cell proliferation using the CCK-8 and cell colony formation assays. As shown in Figure 2E,2F, we found that cell proliferation was significantly decreased in the siSiglec15 transfected A549 and CMT167 cells group (siSiglec15) compared to untransfected control group (NC) and negative control scrambled siRNA transfected cells group (Scrambled). Similarly, the colony formation assay indicated that down-regulation of Siglec15 expression significantly suppressed proliferation ability of A549 and CMT167 cells (Figure 2G-2J).
Knock-down of Siglec15 inhibited the migration and invasion of NSCLC cells
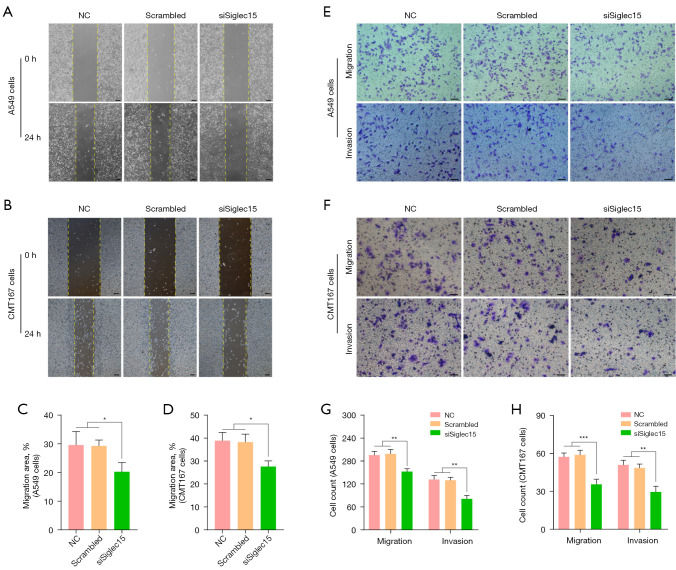
Next, we examined the cell migration and invasion ability in A549 and CMT167 cells with Siglec15 knockdown. According to the wound healing assays, the down-regulation of Siglec15 led to the inhibition of migration of A549 and CMT167 cells (Figure 3A-3D). We demonstrated the character of Siglec15 in NSCLC using Transwell migration and invasion assays, which are often used to evaluate tumor metastasis capacity. The results indicated that number of wandering cells of A549 and CMT167 cells, which shifted from upper chamber to the nether chamber, exhibited a different tendency (Figure 3E-3H). According to Transwell assays, the migration and invasion abilities of A549 and CMT167 cells were lower in the siSiglec15 group than in the NC and Scrambled groups (Figure 3E-3H).
Figure 3.
Down-regulation Siglec15 gene expression in A549 and CMT167 cells inhibited migration and invasion. (A-D) Representative images and statistical analysis of wound healing assay of A549 and CMT167 cells after Siglec15-specific siRNAs transfection and negative control (4× magnification), scare bar equals to 150 μm. (E-H) Representative images and statistical analysis of Transwell migration and invasion assays of A549 and CMT167 cells after Siglec15-specific siRNAs transfection and negative control (10× magnification). Cells were stained with 0.1% crystal violet. Scare bar equals to 100 μm. *, P<0.05; **, P<0.01; ***, P<0.001. NC, normal control.
Relationship between Siglec15 with the proportion of tumor-infiltrating immune cells
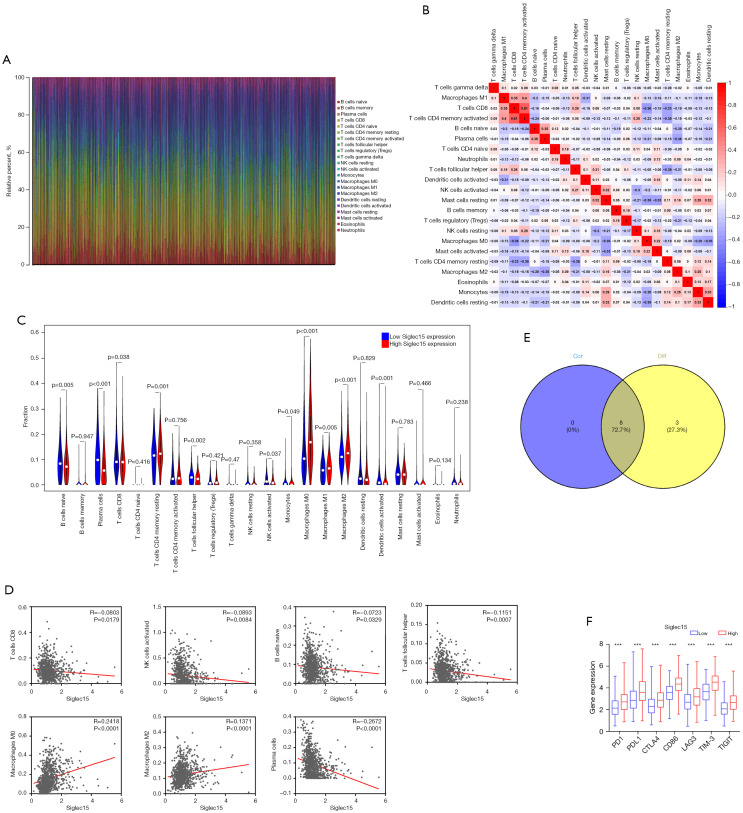
We applied the CIBERSORT method to further confirm the relationship between Siglec15 expression and the immune component, constructing 22 types of immune cell profiles in NSCLC cases and analyzing the proportion of tumor-infiltrating immune subtypes (Figure 4A,4B). Next, a total of eight kinds of tumor-infiltrating immune cells were found to have a strong association with the Siglec15 expression from the correlation and difference analyses (Figure 4C-4E, Table 3). The results revealed that five tumor-infiltrating immune cells had a negative relationship with Siglec15 expression, including CD8+ T cells, follicular helper T Cells, activated natural killer (NK) cells, plasma cells, and naive B cells. Macrophages (M0 and M2) and resting memory CD4+ T cells were positively correlated with Siglec15 expression. The above results confirmed that Siglec15 expression is associated with immunosuppression in NSCLC.
Figure 4.
Performance of CIBERSORT across immune cells in NSCLC samples. (A) Barplot showing the proportion of 22 types of immune cells in NSCLC tumor samples. (B) Heatmap shows the correlation between 22 kinds of immune cell proportions in TCGA datasets. Red represents a positive correlation and blue represents a negative correlation; the darker the color, the stronger the correlation. (C) Violin plot showing the ratio differentiation of 22 types of immune cells between NSCLC tumor samples with low or high Siglec15 expression relative to the median of Siglec15 expression level. (D) Scatter plot showing the correlation of eight kinds of infiltrated immune cells with the Siglec15 expression (P<0.05). (E) Venn plot displaying eight kinds of immune cells correlated with Siglec15 expression co-determined by difference and correlation tests displayed in the violin and scatter plots, respectively. (F) The results showed that the expression of previously discovered immune checkpoints was significantly higher in the high Siglec15 expression group. ***, P<0.001. CIBERSORT, Cell-type Identification by Estimating Relative Subsets of RNA Transcripts; NSCLC, non-small cell lung cancer; NK, natural killer cells; PD-1, programmed cell death 1; PD-L1, programmed cell death-ligand 1; TIM-3, T cell immunoglobulin mucin; TIGIT, T cell immunoreceptor with Ig and ITIM domains.
Table 3. Tumor-infiltrating immune cells co-determined by difference test and correlation test.
| Tumor infiltrating immune cells | Correlation test (P value) | Difference test (P value) |
|---|---|---|
| naive B cells | −0.072 (0.033) | 0.005 |
| Plasma cells | −0.267 (<0.001) | <0.001 |
| CD8+ T cells | −0.080 (0.018) | 0.038 |
| Resting memory CD4+ T cells | 0.093 (0.006) | 0.001 |
| Follicular helper T cells | −0.115 (<0.001) | 0.002 |
| Activated NK cells | −0.089 (0.008) | 0.037 |
| M0 macrophages | 0.242 (<0.001) | <0.001 |
| M2 macrophages | 0.137 (<0.001) | <0.001 |
NK, natural killer.
To evaluate the immunotherapy responses through Siglec15 expression in NSCLC, we explored the correlation of Siglec15 levels with other common immune checkpoints [programmed cell death 1 (PD-1), PD-L1, cytotoxic T lymphocyte antigen 4 (CTLA4), cluster of differentiation 86 (CD86), lymphocyte activation gene-3 (LAG3), T Cell Immunoglobulin Mucin 3 (TIM-3), and T Cell Immunoreceptor with Ig and ITIM Domains (TIGIT)]. The results showed that the high expression of previously discovered immune checkpoints was observed in the high Siglec15 expression NSCLC group, suggesting that patients with high Siglec15 expression tended to have a better immunotherapy response (Figure 4F).
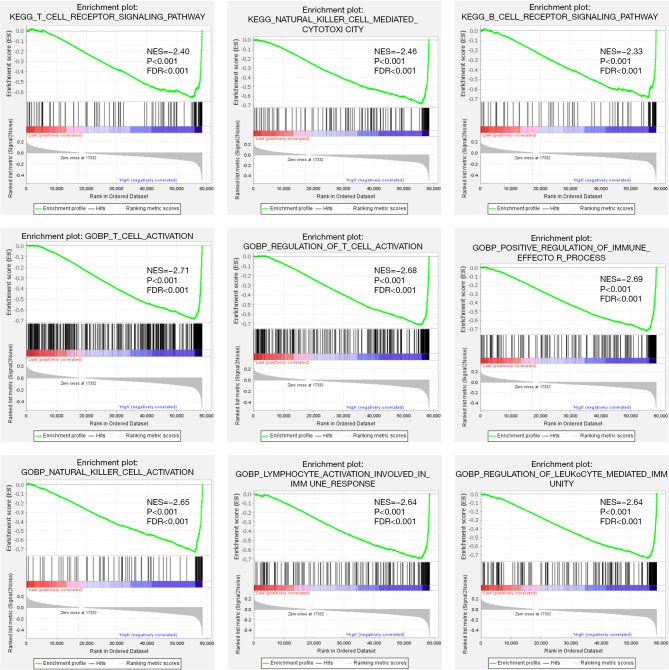
To further determine the potential mechanisms of Siglec15 in NSCLC, both KEGG and GO pathway analyses were performed using GSEA (Figure 5). The results of the KEGG pathway analysis showed that Siglec15 was significantly associated with the “T cell receptor signaling pathway”, “natural killer cell mediated cytotoxicity”, and the “B cell receptor signaling pathway”. GO biological processes (BPs) analysis further revealed that Siglec15 was significantly associated with “T cell activation”, “regulation of T cell activation”, “positive regulation of immune effector process”, “natural killer cell activation”, “lymphocyte activation involved in immune response”, and “regulation of leukocyte-mediated immunity”. These results confirmed that Siglec15 expression significantly influenced the immune activity in TME.
Figure 5.
GSEA of the Siglec15 gene in TCGA-NSCLC patients. GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology; NES, normalized enrichment score; FDR, false discovery rate; TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer.
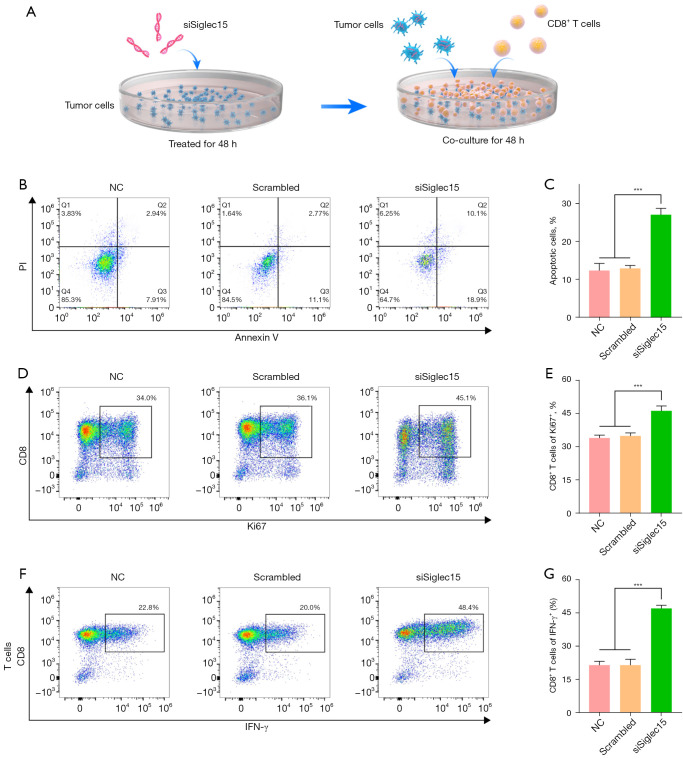
CD8+ T cell antitumor immune suppression is reversed by Siglec15 silencing
Here, we identified that Siglec15 was significantly associated with T cell activation in NSCLC. To demonstrate Siglec15 in NSCLC cell-mediated T cell suppression and investigate the potential mechanism of Siglec15 silencing in antitumor immunity, we used a T cell killing assay in vitro and the high‑throughput sequencing approach. The schedule of the T cell killing assay, which was performed with co-culture of CD8+ T cells and tumor cells, is described in Figure 6A. After Siglec15 silencing for 48 h, Siglec15-silenced CMT167 cells were co-cultured with activated CD8+ T cells (cytotoxic T cells). After being co-cultured with activated CD8+ T cells for 48 h, more apoptosis cells were found in siSiglec15 group compared with NC and Scrambled groups, suggesting that Siglec15 silencing on CMT167 cells successfully promotes T cell-mediated killing (Figure 6B,6C).
Figure 6.
Knock-down of Siglec15 reverses tumor-induced immunosuppression in vitro. (A) The schedule of the T cell killing assay. (B,C) Apoptosis analysis of CMT167 cells co-cultured with activated CD8+ T cells by flow cytometry. (D,E) Proliferation analysis on CD8+ T cells co-cultured with CMT167 cells by Ki-67 flow cytometric analysis. (F,G) Expression analysis of IFN-γ in CD8+ T cells co-cultured with CMT167 cells by flow cytometry. ***, P<0.001. NC, normal control; IFN-γ, interferon-γ.
To study the effect of tumor cells on immune cells, another important indicator of T-cell activation is their proliferative capacity. Activated CD8+ T cells proliferation was measured by Ki-67 flow cytometric analysis. We found that the percentage of proliferation was up to 46.1% for T cells co-cultured with Siglec15-silenced CMT167 cells, which was significantly higher than that of the NC group (33.9%) and the Scrambled group (34.8%) (Figure 6D,6E). This result demonstrated that eliminating the Siglec15-mediated immunosuppressive nature of cancer cells was able to enhance the proliferation of T cells. IFN-γ is a key cytokine produced by activated T cells and plays an important role in anti-tumor responses (18). In this study, after being co-cultured with CMT167 cells, significantly higher IFN-γ-positive T cells (46.9%) were found in the co-culture system compared with NC and Scrambled groups (Figure 6F,6G). These results suggested that Siglec15-mediated immune evasion can be blocked by knock-down of Siglec15.
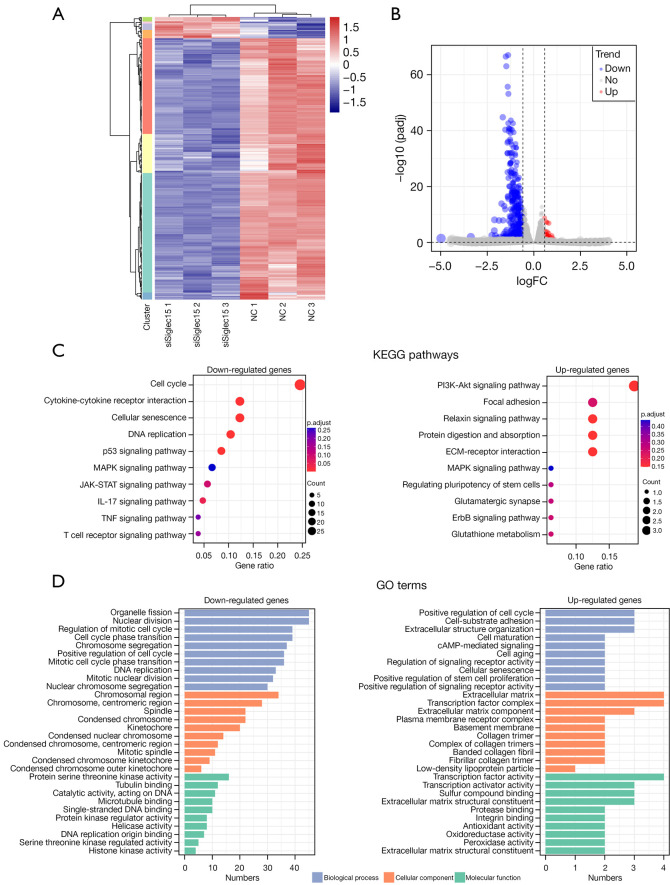
To further understand the molecular mechanisms underlying the Siglec15-mediated immunosuppression effect of tumor cells on immune cells, activated CD8+ T cells co-cultured with CMT167 cells for 48 h were subjected to RNA-Seq analysis (siSiglec15 group and NC group). Hierarchical clustering analysis and volcano plots were performed for the visualization of differential expression, showing that siSiglec15 treatment group and NC group could be obviously distinguished (Figure 7A,7B). A total of 440 genes were identified as DEGs, in which 33 genes were up-regulated and 407 genes were down-regulated (Figure 7B). In order to reveal potential mechanisms of Siglec15-mediated immunosuppression effect, we investigated the DEG-related pathways and performed enrichment analyses to identify the related pathways. KEGG analyses indicated that the down-regulated DEGs were significantly involved in KEGG pathways related to the cell cycle, cytokine-cytokine receptor interaction, cellular senescence, and the p53 signaling pathway (Figure 7C). The most significantly up-regulated DEGs KEGG pathway was the PI3K-Akt signaling pathway (Figure 7C). GO enrichment analysis showed that the DEGs were mainly associated with BPs such as regulation of the cell cycle (Figure 7D).
Figure 7.
mRNA expression profile changes in Siglec15-mediated immunosuppression effect of tumor cells on immune cells and function enrichment. (A) Heat map mRNA transcripts showing hierarchical clustering of altered mRNA transcripts in the siSiglec15 group vs. NC group. Red represents the up-regulated genes, while blue represents the down-regulated genes. (B) Volcano plot indicating up-and down-regulated mRNA transcripts in the siSiglec15 group vs. NC group. (C) KEGG pathway enrichment analysis of DEGs. The size of the point indicates the number of DEGs enriched in the pathway and the different colors represent the significance level of the enrichment result. (D) GO enrichment analysis of DEGs. The x-axis represents the number of DEGs and the y-axis represents the enriched GO terms. NC, normal control; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially-expressed genes; GO, Gene Ontology.
Discussion
Tumor metastasis is the main cause of poor prognosis and cancer-related mortality in NSCLC patients (19). It involves a series of steps, such as tumor cell invasion and migration, intravasation, circulation, extravasation, colonization in distant organs, and micrometastasis formation (20). Since bone is the most common metastasis site of NSCLC, inhibiting bone metastasis is considered to improve the survival rate of NSCLC patients (21). Recently, Wang et al. identified Siglec15 as a novel immune checkpoint responsible for inhibiting the anti-tumor immune response in the TME (8). In the current study, we comprehensively investigated the expression and function of Siglec15 in NSCLC and bone metastasis. In summary, our main findings were as follows: (I) Siglec15 expression was higher in NSCLC spinal metastasis compared to primary NSCLC and paired para-carcinoma tissues; (II) down-regulation of Siglec15 could impair NSCLC cell proliferation, migration, and invasion; (III) the expression of Siglec15 was negatively correlated with the tumor-infiltrating levels of CD8+ T cells, follicular helper T cells, activated NK cells, and plasma cells, while positively correlated with M0 and M2 macrophages in the NSCLC TME; (IV) the expression of common inhibitory checkpoint molecules was higher in the Siglec15 high expression NSCLC group; (V) down-regulation of Siglec15 in tumor cells could enhance the antitumor immune responses of CD8+ T cells in vitro; and (VI) the potential molecular mechanisms of the Siglec15-mediated immunosuppression effect of tumor cells on immune cells were mainly involved in cell cycle, cytokine-cytokine receptor interaction, cellular senescence, the p53 signaling pathway, and the PI3K-Akt signaling pathway.
Previous studies have shown that Siglec15 is essential for osteoclast differentiation, and that Siglec15 depletion could inhibit bone resorption (14-16,22,23). Interestingly, most NSCLC bone metastases were osteolytic bone metastases that involved tumor cell progression and hyperactive osteoclasts (24). Since Siglec15 has the unique function of mediating tumor cells immune escape and osteoclast differentiation, we hypothesized that Siglec15 may play an important role in NSCLC bone metastasis. Many previous studies also have demonstrated that Siglec15 is a tumor-promoting gene that is overexpressed in many primary malignancies, including osteosarcoma, kidney cancer, and liver cancer (25-27). Yet, the expression of Siglec15 in malignant tumor bone metastasis remains unclear. In this study, we found that Siglec15 expression in NSCLC was significantly higher compared to normal tissues. Moreover, patients with advanced tumor stages, higher T stage, or lymph node metastasis were associated with higher Siglec15 expression. These results indicated that Siglec15 may promote the malignant progression of NSCLC. More importantly, we firstly report that Siglec15 was correlated with bone metastasis-related genes, and was overexpressed in NSCLC spinal metastasis tissues compared to primary NSCLC tissues and paired para-carcinoma. These results suggest that Siglec15 is closely related to bone metastasis in NSCLC.
Next, a series of in vitro experiments were performed to explore the functions of Siglec15 in NSCLC. In the present study, we found that down-regulation of Siglec15 could impair the proliferation and aggressiveness of NSCLC cells. Similar results have been reported in other tumors (26-27). Fan et al. demonstrated that Siglec15 silencing suppresses the proliferation, migration, and invasion of osteosarcoma cells and, conversely, that overexpression of Siglec15 could promote the growth, migration, and invasion of osteosarcoma cells (27). Liu et al. showed that Siglec15 promoted the migration of hepatoma cells by regulating CD44 (26). Therefore, Siglec15 might play a pivotal role in NSCLC progression and metastasis.
Previous studies have also reported that the immune component in TME was closely related to the invasion and metastasis of tumors (28,29). However, the correlation between Siglec15 and the immune component in NSCLC remains undefined. Therefore, we further explored the relationship between Siglec15 expression and tumor-infiltrating immune cells in the TME using CIBERSORT and GSEA analysis. The results showed that the expression of Siglec15 was negatively correlated with the infiltration levels of CD8+ T cells, follicular helper T cells, activated NK cells, and plasma cells, and was positively correlated with M0 and M2 macrophages. Tumor-infiltrating CD8+ T cells, follicular helper T cells, NK cells, and plasma cells are important components of antitumor immunity (30-32). M2 macrophages reportedly contribute to tumor progression and immune suppression (33). Given the high expression of Siglec15 in NSCLC and NSCLC spinal metastasis, Siglec15 may create an immunosuppressive TME by regulating the type of tumor-infiltrating immune cells. Immune checkpoint therapy to improve T cell activity has demonstrated encouraging progress in the treatment of many human cancers (34,35), especially malignant and chemotherapy‐resistant cancers (36). However, previous studies have suggested that single immune checkpoint therapy produces limited antitumor responses in cancer patients (37,38). Therefore, dual-immune checkpoint inhibitor combination therapy is an alternative approach to overcome this limitation (39). PD-L1 is a classical immune checkpoint, while Siglec15 may show a complementary expression profile to PD-L1, which possibly means Siglec15 may be independent of classical PD-1/PD-L1 pathway further to perform functions in cancers. In other words, Siglec15 may provide a new therapeutic strategy for patients resistant to anti-PD therapy (40). In this study, we found that the expression of common immune checkpoints was higher in the high Siglec15 expression group, suggesting Siglec15 may be combined with other immune checkpoints to suppress the antitumor immune response in NSCLC patients. Thus, for NSCLC patients with high expression of Siglec15, immune checkpoint combination therapy may achieve better efficacy.
The tumor immune response is an important process in the occurrence and development of malignant tumors (41,42). The malignant biological behavior of tumors is further accelerated without immune monitoring, thereby promoting tumor proliferation, invasion, and metastasis (43). Therefore, understanding the tumor cell immune escape mechanism is crucial for tumor treatment, especially immunotherapy. Previous study has shown that in osteosarcoma cancer cells, Siglec15 leads to tumor progression via the DUSP1/MAPK pathway (27). In the TME, tumor-associated macrophages can produce transforming growth factor-β (TGF-β), which is a major immunosuppressive cytokine in the advanced stage of cancer. Siglec15 can enhance TGF-β secretion by coupling tumor cell recognition with the DAP12-Syk signal transduction pathway (44).
Regardless, the mechanism through which Siglec15 regulates the functions of CD8+ T cells remains unclear. In the present study, the down-regulation of Siglec15 successfully reversed the immunosuppression of NSCLC cells and improved T cell anti-tumor immunity. More importantly, we explored the molecular mechanisms underlying the Siglec15-mediated immunosuppression effect of tumor cells on immune cells by high‑throughput sequencing. We found that the underlying molecular mechanisms were significantly involved in the cell cycle, cytokine-cytokine receptor interaction, cellular senescence, the p53 signaling pathway, and the PI3K-Akt signaling pathway. These results will be confirmed in our future investigations.
Conclusions
In conclusion, our research showed that Siglec15 was significantly up-regulated in NSCLC spinal metastasis compared with primary NSCLC, and exerted an oncogenic function in NSCLC progression, including tumor cell proliferation, migration, and invasion. Furthermore, we explored the relationship between Siglec15 expression and tumor-infiltrating immune cells, as well as different immune checkpoints, offering novel insights into the immunosuppressive effect of siglec15 in the TME. Lastly, we found that the down-regulation of Siglec15 successfully reversed the tumor immunosuppression in vitro, and explored its molecular mechanisms by high‑throughput sequencing. However, some limitations to this study remain, as the potential molecular mechanisms and associated proteins were not verified experimentally. Therefore, further studies have been planned in our future work. Collectively, our findings indicate that Siglec15 was significantly correlated with NSCLC spinal metastasis and could be a promising new target for the treatment of NSCLC and spinal metastasis.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 81972508 and 82172738); Natural Science Foundation of Shanghai (No. 21ZR1412300); Technology Innovation Action Plan of Shanghai Science and Technology Commission (No. 21S11902700); Shanghai “Rising Stars of Medical Talent” Youth Development Program [Youth Medical Talents-Specialist Program (2020)087]; Shanghai Talent Development Fund (No. 2020067); Shanghai Orthopaedic Clinical Medicine Research Center (No. 21MC1930100); and the Construction of Orthopaedic Specialist Alliance for Clinical Ability Promotion and Enhancement (No. SHDC22021308).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Zhongshan Hospital, Fudan University, Shanghai, China (approval No. Y2019‑085) and informed consent was taken from all the patients.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-764/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-764/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-764/coif). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Liu Y, Fan J, Xu T, et al. Extracellular vesicle tetraspanin-8 level predicts distant metastasis in non-small cell lung cancer after concurrent chemoradiation. Sci Adv 2020;6:eaaz6162. 10.1126/sciadv.aaz6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang K, Jiang L, Hu A, et al. Vertebral-specific activation of the CX3CL1/ICAM-1 signaling network mediates non-small-cell lung cancer spinal metastasis by engaging tumor cell-vertebral bone marrow endothelial cell interactions. Theranostics 2021;11:4770-89. 10.7150/thno.54235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang Y, Qu J, Wu J, et al. Effect of Surgery on Quality of Life of Patients with Spinal Metastasis from Non-Small-Cell Lung Cancer. J Bone Joint Surg Am 2016;98:396-402. 10.2106/JBJS.O.00629 [DOI] [PubMed] [Google Scholar]
- 4.Wang HL, Wang HR, Liang Y, et al. Hsa_circ_0006571 promotes spinal metastasis through sponging microRNA-138 to regulate sirtuin 1 expression in lung adenocarcinoma. Transl Lung Cancer Res 2020;9:2411-27. 10.21037/tlcr-20-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ban J, Fock V, Aryee DNT, et al. Mechanisms, Diagnosis and Treatment of Bone Metastases. Cells 2021;10:2944. 10.3390/cells10112944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kraehenbuehl L, Weng CH, Eghbali S, et al. Enhancing immunotherapy in cancer by targeting emerging immunomodulatory pathways. Nat Rev Clin Oncol 2022;19:37-50. 10.1038/s41571-021-00552-7 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Chen L. Classification of Advanced Human Cancers Based on Tumor Immunity in the MicroEnvironment (TIME) for Cancer Immunotherapy. JAMA Oncol 2016;2:1403-4. 10.1001/jamaoncol.2016.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Sun J, Liu LN, et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat Med 2019;25:656-66. 10.1038/s41591-019-0374-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li TJ, Jin KZ, Li H, et al. SIGLEC15 amplifies immunosuppressive properties of tumor-associated macrophages in pancreatic cancer. Cancer Lett 2022;530:142-55. 10.1016/j.canlet.2022.01.026 [DOI] [PubMed] [Google Scholar]
- 10.Hao JQ, Nong JY, Zhao D, et al. The significance of Siglec-15 expression in resectable non-small cell lung cancer. Neoplasma 2020;67:1214-22. [DOI] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Shi M, Yang N, et al. GPR115 Contributes to Lung Adenocarcinoma Metastasis Associated With LAMC2 and Predicts a Poor Prognosis. Front Oncol 2020;10:577530. 10.3389/fonc.2020.577530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu T, Takahata M, Kameda Y, et al. Sialic acid-binding immunoglobulin-like lectin 15 (Siglec-15) mediates periarticular bone loss, but not joint destruction, in murine antigen-induced arthritis. Bone 2015;79:65-70. 10.1016/j.bone.2015.05.029 [DOI] [PubMed] [Google Scholar]
- 15.Ishida-Kitagawa N, Tanaka K, Bao X, et al. Siglec-15 protein regulates formation of functional osteoclasts in concert with DNAX-activating protein of 12 kDa (DAP12). J Biol Chem 2012;287:17493-502. 10.1074/jbc.M111.324194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiruma Y, Hirai T, Tsuda E. Siglec-15, a member of the sialic acid-binding lectin, is a novel regulator for osteoclast differentiation. Biochem Biophys Res Commun 2011;409:424-9. 10.1016/j.bbrc.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 17.Vicent S, Luis-Ravelo D, Antón I, et al. A novel lung cancer signature mediates metastatic bone colonization by a dual mechanism. Cancer Res 2008;68:2275-85. 10.1158/0008-5472.CAN-07-6493 [DOI] [PubMed] [Google Scholar]
- 18.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. 10.1172/JCI91190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer 2019;18:179. 10.1186/s12943-019-1111-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan X, Banerjee P, Liu X, et al. Transcriptional control of a collagen deposition and adhesion process that promotes lung adenocarcinoma growth and metastasis. JCI Insight 2022;7:153948. 10.1172/jci.insight.153948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han L, Huang Z, Liu Y, et al. MicroRNA-106a regulates autophagy-related cell death and EMT by targeting TP53INP1 in lung cancer with bone metastasis. Cell Death Dis 2021;12:1037. 10.1038/s41419-021-04324-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang FB, Chen W, Wang L, et al. The diverse functions of Siglec-15 in bone remodeling and antitumor responses. Pharmacol Res 2020;155:104728. 10.1016/j.phrs.2020.104728 [DOI] [PubMed] [Google Scholar]
- 23.Sato D, Takahata M, Ota M, et al. Siglec-15-targeting therapy increases bone mass in rats without impairing skeletal growth. Bone 2018;116:172-80. 10.1016/j.bone.2018.07.026 [DOI] [PubMed] [Google Scholar]
- 24.Wu S, Pan Y, Mao Y, et al. Current progress and mechanisms of bone metastasis in lung cancer: a narrative review. Transl Lung Cancer Res 2021;10:439-51. 10.21037/tlcr-20-835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang WB, Qin CP, Du YQ, et al. Siglec-15 promotes progression of clear renal cell carcinoma. Chin Med J (Engl) 2021;134:2635-7. 10.1097/CM9.0000000000001752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Ji Z, Wu B, et al. Siglec-15 promotes the migration of liver cancer cells by repressing lysosomal degradation of CD44. FEBS Lett 2021;595:2290-302. 10.1002/1873-3468.14169 [DOI] [PubMed] [Google Scholar]
- 27.Fan MK, Zhang GC, Chen W, et al. Siglec-15 Promotes Tumor Progression in Osteosarcoma via DUSP1/MAPK Pathway. Front Oncol 2021;11:710689. 10.3389/fonc.2021.710689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J, Huang J, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int 2019;19:264. 10.1186/s12935-019-0986-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu F, Shen J, Xu S. Integrated Bioinformatical Analysis Identifies GIMAP4 as an Immune-Related Prognostic Biomarker Associated With Remodeling in Cervical Cancer Tumor Microenvironment. Front Cell Dev Biol 2021;9:637400. 10.3389/fcell.2021.637400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 31.Shi W, Dong L, Sun Q, et al. Follicular helper T cells promote the effector functions of CD8+ T cells via the provision of IL-21, which is downregulated due to PD-1/PD-L1-mediated suppression in colorectal cancer. Exp Cell Res 2018;372:35-42. 10.1016/j.yexcr.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 32.Kroeger DR, Milne K, Nelson BH. Tumor-Infiltrating Plasma Cells Are Associated with Tertiary Lymphoid Structures, Cytolytic T-Cell Responses, and Superior Prognosis in Ovarian Cancer. Clin Cancer Res 2016;22:3005-15. 10.1158/1078-0432.CCR-15-2762 [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Geng X, Hou J, et al. New insights into M1/M2 macrophages: key modulators in cancer progression. Cancer Cell Int 2021;21:389. 10.1186/s12935-021-02089-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geva R, Voskoboynik M, Dobrenkov K, et al. First-in-human phase 1 study of MK-1248, an anti-glucocorticoid-induced tumor necrosis factor receptor agonist monoclonal antibody, as monotherapy or with pembrolizumab in patients with advanced solid tumors. Cancer 2020;126:4926-35. 10.1002/cncr.33133 [DOI] [PubMed] [Google Scholar]
- 35.Li B, Chan HL, Chen P. Immune Checkpoint Inhibitors: Basics and Challenges. Curr Med Chem 2019;26:3009-25. 10.2174/0929867324666170804143706 [DOI] [PubMed] [Google Scholar]
- 36.Galluzzi L, Humeau J, Buqué A, et al. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol 2020;17:725-41. 10.1038/s41571-020-0413-z [DOI] [PubMed] [Google Scholar]
- 37.Adams S, Schmid P, Rugo HS, et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol 2019;30:397-404. 10.1093/annonc/mdy517 [DOI] [PubMed] [Google Scholar]
- 38.Shen X, Huang S, Xiao H, et al. Efficacy and safety of PD-1/PD-L1 plus CTLA-4 antibodies ± other therapies in lung cancer: a systematic review and meta-analysis. Eur J Hosp Pharm 2021. [Epub ahead of print]. doi: . 10.1136/ejhpharm-2021-002803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang C, Zhang L, Xu X, et al. Engineering a Smart Agent for Enhanced Immunotherapy Effect by Simultaneously Blocking PD-L1 and CTLA-4. Adv Sci (Weinh) 2021;8:e2102500. 10.1002/advs.202102500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J, Lu Q, Sanmamed MF, et al. Siglec-15 as an Emerging Target for Next-generation Cancer Immunotherapy. Clin Cancer Res 2021;27:680-8. 10.1158/1078-0432.CCR-19-2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerezo M, Robert C, Liu L, et al. The Role of mRNA Translational Control in Tumor Immune Escape and Immunotherapy Resistance. Cancer Res 2021;81:5596-604. 10.1158/0008-5472.CAN-21-1466 [DOI] [PubMed] [Google Scholar]
- 42.Galassi C, Musella M, Manduca N, et al. The Immune Privilege of Cancer Stem Cells: A Key to Understanding Tumor Immune Escape and Therapy Failure. Cells 2021;10:2361. 10.3390/cells10092361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian Y, Zhai X, Yan W, et al. Clinical outcomes of immune checkpoint blockades and the underlying immune escape mechanisms in squamous and adenocarcinoma NSCLC. Cancer Med 2021;10:3-14. 10.1002/cam4.3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takamiya R, Ohtsubo K, Takamatsu S, et al. The interaction between Siglec-15 and tumor-associated sialyl-Tn antigen enhances TGF-β secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology 2013;23:178-87. 10.1093/glycob/cws139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as