Abstract
Background
Allergic conjunctivitis (AC) is an inflammation caused by a hypersensitive immune reaction of conjunctiva to external allergens. The microRNA (miRNA) miR-146a has been reported to suppress the exacerbation of inflammation. However, the underlying influence and mechanism of miR-146a in AC has not been completely elucidated.
Methods
We first successfully established an AC mouse model and AC cell model. After each model was treated based on the experimental purposes, miR-146a, FOXP3, and homeodomain-interacting protein kinases 3 (HIPK3) expressions were estimated by reverse transcription quantitative polymerase chain reaction (RT-qPCR). The levels of immunoglobulin E (IgE), tumor necrosis factor-α (TNF-α), interleukin-10 (IL-10), interleukin-4 (IL-4), and transforming growth factor-β (TGF-β) were assessed using enzyme-linked immunosorbent assay (ELISA) kits; the related proteins were analyzed by western blot, immunofluorescence, or immunohistochemistry (IHC) assays; the interaction between miR-146a and HIPK3 were validated by a dual-luciferase reporter gene assay; and the inflammatory infiltration was certified by hematoxylin and eosin (H&E) staining.
Results
Our results indicated that miR-146a and FOXP3 were downregulated in AC model mice. Meanwhile, miR-146a overexpression could upregulate FOXP3 and inhibit inflammatory response in TGF-β-induced thymocytes. Besides, our results testified that HIPK3, as a target gene of miR-146a, could reverse miR-146a-mediated FOXP3 upregulation and inflammation inhibition. Moreover, we discovered that miR-146a could downregulate p-STAT3 by targeting HIPK3, and activation of STAT3 also could reverse miR-146a-mediated inflammation suppression in TGF-β-induced thymocytes. More importantly, miR-146a could ameliorate inflammatory infiltration and downregulate HIPK3 and p-STAT3 in AC model mice.
Conclusions
We demonstrated a possible protective mechanism by the miR-146a/HIPK3/STAT3 axis, by which decrease of miR-146a could aggravate the inflammation of AC.
Keywords: miR-146a, allergic conjunctivitis (AC), homeodomain-interacting protein kinases 3 (HIPK3), STAT3, FOXP3, inflammation
Introduction
Allergic conjunctivitis (AC) refers to a hypersensitive immune response of the conjunctiva to an external allergen (1). It is mainly divided into type I and type IV allergic reactions, of which type I allergic reaction is the predominant cause of AC (2). Eye-itching is the most common symptom of AC, with the majority of patients also experiencing tears, photophobia, burning sensations, and increased secretions, among others. Some patients may even experience corneal infection that threatens vision (3,4). Due to constant aggravation by air pollution and frequent applications of eye cosmetics and contact lenses, people are increasingly likely to be exposed to allergens (5). It has been reported outside of China that more than 25% of the population experiences immunoglobulin E (IgE)-mediated allergic reactions (6). At present, there are multiple clinical treatment methods for AC, among which, western medicine mostly employs antihistamines, short-term corticosteroids, mast cell stabilizers, and subcutaneous or sublingual immunotherapy (5,7,8). For patients with severe AC, immunosuppressants such as cyclosporine eye drops are also applied (9). However, these therapies are largely based on symptomatic relief. Besides, the duration of eye drops remaining active in the conjunctival sac is relatively short, and it is difficult to maintain their efficacy (10). When the hypersensitivity reaction is re-ignited, patient relapse can easily occur. Therefore, it is of vital clinical significance to explore the relevant mechanisms of AC and screen effective biomarkers and targeted drugs for the diagnosis and treatment of AC.
Homeodomain-interacting protein kinase 3 (HIPKs) are a family of cofactors that have different functions with homologous proteins, including HIPK1, HIPK2, HIPK3 and HIPK4, which regulate the expression of proteins in the nucleus, cytoplasm and membrane. These factors change the phosphorylation state of target proteins. Involved in cell proliferation, differentiation, apoptosis and other biological processes. HIPK3 plays a key role in the regulation of inflammation (11). However, the mechanism of HIPK3 in allergic conjunctivitis remains to be explored. STAT3 has been shown to be upregulated in allergic conjunctivitis and to play a role in promoting inflammation. At the same time, STAT3 is also a key transcription factor for Th17 cell differentiation (12). When STAT3 is phosphorylated, it enters the nucleus and functions as a transcription factor (13). Therefore, we hypothesized that HIPK3 regulates T cell differentiation by phosphorylating STAT3 and ultimately affects the inflammatory response of allergic conjunctivitis.
MicroRNAs (miRNAs) are a class of small molecular non-coding RNAs in eukaryotes with a length of about 22 nucleotides, which are mainly synthesized in the nucleus and cytoplasm (14). They can induce the degradation of target messenger RNA (mRNA) and block the translation of target genes by binding to the 3'-untranscribed region (3'-UTR) sequences of target mRNA in an incomplete complementary way (15). In this way, miRNAs can act as key regulators of various intracellular mechanisms and physiological processes, such as cell proliferation, cycle, apoptosis, metastasis, and so on (16,17). Recently, studies have reported that certain miRNAs, such as miR-19b and miR-146a, also play major roles in AC (12,18,19). In particular, miR-146a has been shown to improve AC, which might be relevant to thymic stromal lymphopoietin (TSLP) and CD4+CD25− T cells (18,19). Therefore, miR-146a might be used as a potential biomarker for AC. However, the specific mechanism of miR-146a in AC is not completely clear.
Through bioinformatics prediction, we discovered that HIPK3 may be the downstream target gene of miR-146a. As an intracellular serine/threonine protein kinase, HIPK3 negatively regulates cell apoptosis through phosphorylation of Jun and Runx2, and also enhances androgen receptor (AR)-mediated transcriptional activation (20). It has also been reported to accelerate the progression of multiple diseases, such as certain cancers (21,22), Huntington’s disease (23), and sepsis (11). Additionally, HIPK3 also could regulate inflammatory cytokines to affect the inflammatory response (11). However, it has not been clarified whether miR-146a could alter AC progression by regulating HIPK3.
Foxp3 is one of the key transcription factors controlling the development and function of Treg cells. Treg-like immunosuppression can occur after in vitro or in vivo induction of FOXP3 expression on naive T cells (24). In our study, we proposed that miR-146a, as a crucial link in AC, can alleviate AC. We further investigated the possible regulatory mechanism of miR-146a in AC progression. This study is the first to confirm the role of miR-146a in allergic conjunctivitis by targeting HIPK3, reducing the phosphorylation of STAT3, promoting expression of FOXP3 and inhibiting inflammation. The elucidation of the role and mechanism of miR-146a in AC might contribute to a deeper understanding of the AC process. The miR-146a/HIPK3 axis might provide a theoretical basis for the therapeutic target of AC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-982/rc)
Methods
Animal
Adult male C57BL/6 mice [specific-pathogen-free (SPF) grade] were purchased from Shenzhen Eye Hospital affiliated to Jinan University Experimental Animal Center (Shenzhen, China), all of which were housed at Shenzhen Eye Hospital Affiliated to Jinan University Animal Experimental Center. The feeding conditions involved a barrier system with 12 h light and dark cycle. Experiments were performed under a project license (Approval No. TOP-IACUC-2021-0106) granted by the Ethics Committee of Shenzhen Eye Hospital Affiliated to Jinan University, in compliance with guidelines of Jinan University for the care and use of animals.
Grouping of mice
We purchased miR-146a mimics from GenePharma (Suzhou, China). The AC mouse model was established by inducing irritation for 14 days. Mice were randomly divided into 3 groups including a sham group, AC group, and an AC+miR-146a mimics group (5 mice per group). Oligonucleotides were injected into the vitreous cavity of mice using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA).
Cell treatment
Thymocytes were extracted from the mice based on previous research. An AC cell model was also established in thymocytes through transforming growth factor-β (TGF-β) induction. We purchased HIPK3 small interfering RNAs (siRNAs) and STAT3 siRNAs from GenePharma (Suzhou, China) and vector and HIPK3-overexpressed plasmids from HanBio Biotechnology (Shanghai, China). We randomly divided TGF-β-induced thymocytes into groups, which included (I) NC, mimics, and Inhibitor group; (II) NC, mimics, HIPK3 siRNA, and mimics + HIPK3; and (III) NC, mimics, STAT3 siRNA, and mimics + STAT3. The TGF-β-induced thymocytes were transfected with these oligonucleotides and plasmids with Lipofectamine 3000.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay
Total RNAs were extracted from the conjunctival tissues of mice and thymocytes in each group by applying TRIzol reagent (Invitrogen). Then, reverse transcription was conducted to produce complementary DNAs (cDNAs) using PrimeScriptTM RT Reagent Kit (Takara, Tokyo, Japan). The levels of miR-146a, FOXP3, and HIPK3 were confirmed using SYBR Green qPCR master Mix (DBI Bioscience, Ludwigshafen, Germany). The data was also counted with 2−ΔΔCt method. The sequences of primers are shown in Table 1.
Table 1. Primer sequences used in this study.
| Gene | Sequence (5'- 3') |
|---|---|
| β-actin F | CATTGCTGACAGGATGCAGA |
| β-actin R | CTGCTGGAAGGTGGACAGTGA |
| Foxp3 F | ACCATTGGTTTACTCGCATGT |
| Foxp3 R | TCCACTCGCACAAAGCACTT |
| Hipk3 F | CAGCGATGCGGGTTAAAGC |
| Hipk3 R | TGGGTTTCCCATGTTGGTTTG |
| U6 F | CTCGCTTCGGCAGCACA |
| U6 R | AACGCTTCACGAATTTGCGT |
| All R | CTCAACTGGTGTCGTGGA |
| mmu-miR-146a-5p RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATGGGTT |
| mmu-miR-146a-5p F | ACACTCCAGCTGGGTGAGAACTGAATTCCA |
Enzyme-linked immunosorbent assay (ELISA)
The blood of mice was collected in a 2 mL EP tube by eyeball extraction. After 8 h, the serum was collected by centrifugation (2,000 ×g/min for 10 min). Then, the contents of IgE, tumor necrosis factor-α (TNF-α), interleukin-10 (IL-10), interleukin-4 (IL-4), and TGF-β in mouse serum or cell medium supernatant were determined in line with the instructions of each ELISA kit.
Western blot assay
The conjunctiva tissue was pulverized at low temperature with liquid nitrogen. Radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China) containing phenylmethylsulfonyl fluoride (PMSF; 1:100) was added into the serous fluid of conjunctival tissues and thymocytes in each group. After incubation and centrifugation, the supernatant was taken to obtain the total proteins. After quantification, 50 µg protein was subjected to electrophoresis on the sodium dodecyl sulfate (SDS)-polyacrylamide gel, and then transferred onto polyvinylidene difluoride (PVDF) membranes. Then, 5% skim milk was applied for sealing the membranes at 37 ℃ for 1 h. The diluent primary antibodies were adopted to incubate the membranes overnight at 4 ℃, followed by secondary antibodies for 2 h. The imprinting of target protein was visualized using chemiluminescence reagent (Millipore, Burlington, MA, USA).
Immunofluorescence assay
The treated thymocytes were washed, collected, fixed using 4% paraformaldehyde for 15 min, and addressed with 0.5%Triton X-100 in phosphate-buffered saline (PBS) for 20 min. After washing, thymocytes were incubated with normal goat serum for 30 min, primary antibodies including anti-HIPK3 and anti-STAT3 overnight at 4 ℃, and diluted fluorescent secondary antibody at 37 ℃ for 1 h. After incubation with 4’,6-diamidino-2-phenylindole (DAPI) in the dark for 5 min, the images were observed and collected under a fluorescence microscope.
Dual-luciferase reporter gene assay
In accordance with the predicted binding sites between miR-146a and HIPK3 promoter region, we successfully constructed the wild type (WT) and mutant (Mut) HIPK3 plasmids with pGL3-Basic vector. Then, miR-146a mimics and the corresponding plasmids were con-transfected into thymocytes using Lipofectamine 3000 for 48 h. Finally, the luciferase activity was determined using dual luciferase assay kit (Promega, Madison, WI, USA).
Hematoxylin and eosin (H&E) staining
The conjunctival tissue in each group was first fixed in 4% paraformaldehyde. Then, the tissues were treated with xylene and dehydrated using 100%, 95%, 90%, 80%, and 70% ethyl alcohol. After embedding in paraffin, the tissues were continuously cut into slices (about 4 µm). After dewaxing and hydration, the slices were processed with Harris hematoxylin (5 min), 1% hydrochloric acid alcohol (5 s), and 0.6% ammonia and eosin (2 min). After dehydration and transparency, the inflammatory infiltration was evaluated with a light microscope.
Immunohistochemistry (IHC) assay
The slices were prepared using xylene I, xylene II, 95%, 90%, 80%, 70% ethyl alcohol and distilled water, respectively. Then, the slices were treated with 3% hydrogen peroxide and ethylenediamine tetraacetic acid (EDTA). Subsequently, the slices were blocked and treated with anti-FOXP3 (Abcam, Cambridge, MA, USA) at 37 ℃ for 1 h and secondary antibody at 37 ℃ for 30 min. After processing with 3,3'-diaminobenzidine (DAB), the slices were stained with hematoxylin, 0.1% hydrochloric acid. Finally, the images were photographed under a light microscope.
Statistical analysis
The statistical software SPSS 20.0 (IBM Corp., SPSS, Inc., Chicago, IL, USA) was used to analyze the experimental data, which was expressed as mean ± SD. One-way analysis of variance (ANOVA) was adopted for the comparison among groups. A P value <0.05 indicated statistical significance.
Results
MiR-146a and FOXP3 were lowly expressed in AC model mice
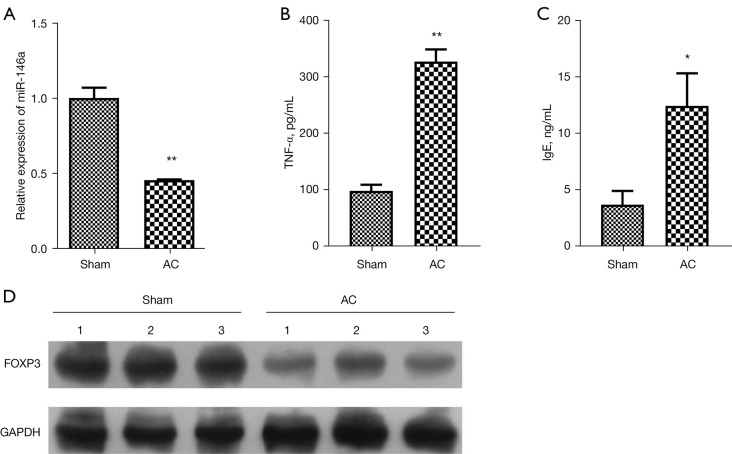
To verify the change in expression of miR-146a in AC, we first successfully established an AC mouse model. The RT-qPCR results showed that the level of miR-146a was notably decreased in the conjunctival tissues of AC model mice relative to that in sham mice (P<0.01, Figure 1A). The results of ELISA showed that the levels of TNF-α and IgE were obviously raised in the serum of AC model mice compared to that in sham mice (P<0.05, P<0.01, Figure 1B,1C). Moreover, western blot data revealed that the protein level of FOXP3 was also prominently diminished in the conjunctival tissues of AC model mice with respect to that in sham mice (Figure 1D). On the whole, these data showed that the expression changes of miR-146a and FOXP3 are relevant to AC process, and the downregulations of miR-146a and FOXP3 might contribute to the development of AC progression.
Figure 1.
MiR-146a and FOXP3 were lowly expressed in AC model mice. (A) RT-qPCR assay was used to confirm the expression of miR-146a in the conjunctival tissues of sham and AC model mice. (B) ELISA assay displayed the change in the serum level of TNF-α in sham and AC model mice. (C) IgE content in the serum of the sham and AC model mice was determined by ELISA assay. (D) Western blot assay was used to evaluate the change of FOXP3 expression in sham and AC model mice. *P<0.05; **P<0.01. AC, allergic conjunctivitis; RT-qPCR, reverse transcription quantitative polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; TNF-α, tumor necrosis factor-α; IgE, immunoglobulin E.
Overexpression of miR-146a dramatically suppressed inflammation in TGF-β-induced thymocytes
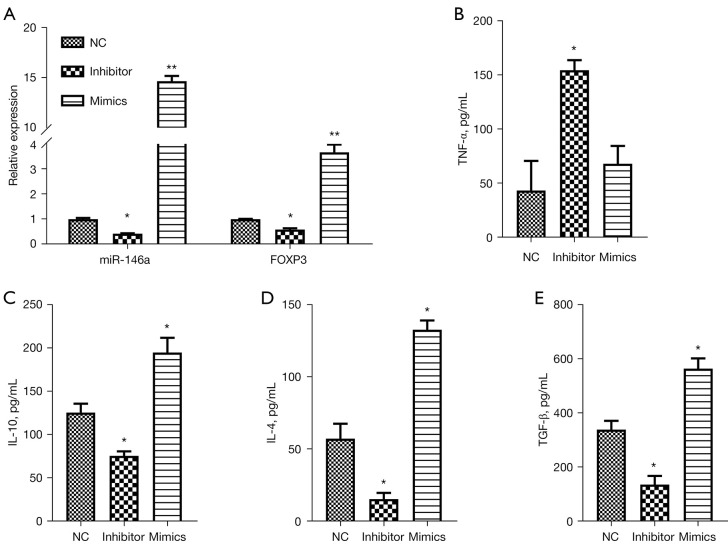
We further investigated the influence of miR-146a on the inflammatory process of AC in vitro. Thymocytes were extracted and cultured, and stimulated with TGF-β to establish an AC cell model. Then, TGF-β-induced thymocytes were transfected with miR-146a inhibitor, miR-146a mimics, or NC, respectively. The RT-qPCR results first displayed that inhibition of miR-146a markedly downregulated miR-146a and FOXP3, and overexpression of miR-146a dramatically upregulated miR-146a and FOXP3 in TGF-β-induced thymocytes (P<0.05, P<0.01, Figure 2A). Additionally, ELISA results showed that inhibition of miR-146a elevated the level of TNF-α and reduced the levels of IL-10, IL-4, and TGF-β, and overexpression of miR-146a signally increased the levels of IL-10, IL-4, and TGF-β in TGF-β-induced thymocytes (P<0.05, Figure 2B-2E). Consequently, these data revealed that aberrant expression of miR-146a was involved in the inflammation of TGF-β-induced thymocytes, and increase of miR-146a might relieve cellular inflammation in TGF-β-induced thymocytes.
Figure 2.
Overexpression of miR-146a dramatically suppressed inflammation in TGF-β-induced thymocytes. TGF-β induced thymocytes were transfected with miR-146a NC, miR-146a inhibitor, or miR-146a mimics, respectively. (A) Changes in miR-146a and FOXP3 expressions in thymocytes were detected by RT-qPCR assay in each group. (B-E) ELISA assay displayed the changes of TNF-α, IL-10, IL-4, TGF-β levels in the transfected thymocytes. *P<0.05; **P<0.01. NC, negative control; TNF-α, tumor necrosis factor-α; IL-10, interleukin 10; IL-4, interleukin-4; TGF-β, transforming growth factor-β.
HIPK3 was a target gene of miR-146a
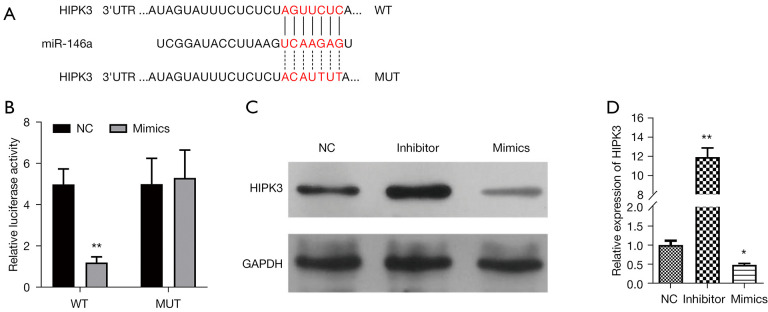
Meanwhile, we screened the potential target genes of miR-146a by bioinformatics prediction. Through analysis, we preliminarily identified HIPK3 as the downstream target gene of miR-146a. As shown in Figure 3, there is a binding site between miR-146a and HIPK3 3'UTR sequence, and the sequence after point mutation is not complementary to miR-146a (Figure 3A). Accidentally, the results from dual-luciferase reporter gene assay revealed that introduction of miR-146a mimics could result in a prominent decrease in the luciferase activity of WT-HIPK3, while miR-146a mimics cannot affect the luciferase activity of Mut-HIPK3 (P<0.01, Figure 3B). In addition, we discovered that compared with the NC group of TGF-β-induced thymocytes, the protein level of HIPK3 was heightened in the miR-146a inhibitor group, and significantly lowered in miR-146a mimics group (Figure 3C). The RT-qPCR data indicated that relative to NC-transfected TGF-β-induced thymocytes, HIPK3 was prominently upregulated in the miR-146a inhibitor group, and dramatically downregulated in the miR-146a mimics group (P<0.01, P<0.05, Figure 3D). Therefore, our results verified that HIPK3, as a target gene, could be notably downregulated by miR-146a in TGF-β-induced thymocytes.
Figure 3.
HIPK3 as a target gene of miR-146a. A The binding site between miR-146a and HIPK3 3’UTR. B. A dual-luciferase reporter gene assay was conducted to assess the interaction between miR-146a and HIPK3. (C) Western blotting analysis of HIPK3 expression in TGF-β-induced thymocytes transfected with NC, miR-146a inhibitor, or miR-146a mimics, respectively. (D) After transfection with miR-146a inhibitor or miR-146a mimics, the changed HIPK3 expression was monitored by RT-qPCR in TGF-β-induced thymocytes. *P<0.05, **P<0.01. NC, negative control; HIPK3, homeodomain-interacting protein kinases 3; TGF-β, transforming growth factor-β; RT-qPCR, reverse transcription quantitative polymerase chain reaction.
Overexpression of HIPK3 markedly attenuated miR-146a-mediated downregulation of p-STAT3 and suppression of inflammation in TGF-β-induced thymocytes
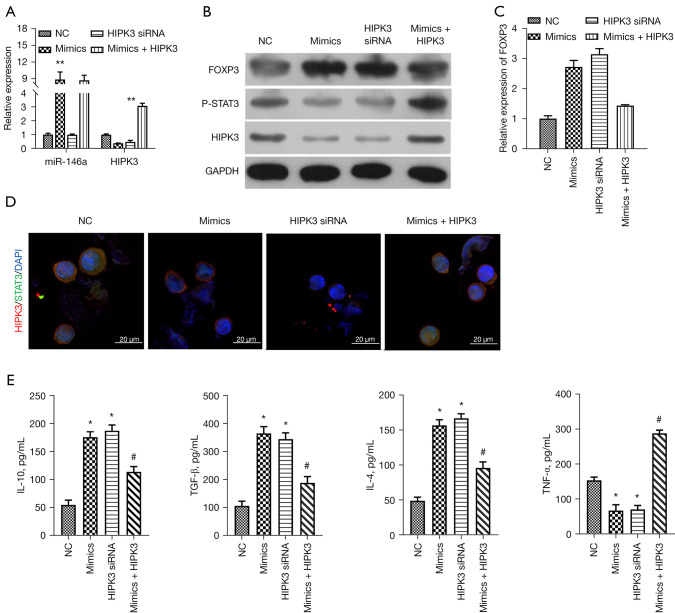
On account of the targeted down-regulation of miR-146a to HIPK3 in TGF-β-induced thymocytes, we further explored whether HIPK3 could participate in the inhibitory role of miR-146a on the inflammation of TGF-β-induced thymocytes. In this part of the experiment, an miR-146a mimic and HIPK3 siRNAs were transfected separately into TGF-β-induced thymocytes, while miR-146a mimics and HIPK3-overexpressed plasmids were co-transfected into TGF-β-induced thymocytes. The RT-qPCR data first showed that overexpression of miR-146a markedly upregulated miR-146a, while knockdown or overexpression of HIPK3 did not affect the expression of HIPK3 in TGF-β-induced thymocytes (P<0.01, Figure 4A). The data revealed that overexpression of miR-146a markedly downregulated HIPK3, while overexpression of HIPK3 could prominently reverse miR-146a overexpression-mediated downregulation of HIPK3 in TGF-β-induced thymocytes, and the silence alone of HIPK3 by siRNAs also dramatically downregulated HIPK3 (P<0.01, Figure 4A,4B). Thus, we identified the successful transfection of these oligonucleotides and HIPK3-overexpressed plasmids in TGF-β-induced thymocytes. Next, western blotting results also certified that upregulation of FOXP3 and downregulation of p-STAT3, which were mediated by miR-146a mimics, could be markedly diminished by HIPK3 overexpression, and single knockdown of HIPK3 also obviously strengthened FOXP3 expression and lowered p-STAT3 expression in TGF-β-induced thymocytes (Figure 4B). Similarly, qRT-qPCR results also revealed that the mRNA expression trend of FOXP3 was basically consistent with the protein expression trend in the western blot results in the TGF-β-induced thymocytes after the overexpression or knockdown of miR-146a and HIPK3 (Figure 4C). Meanwhile, immunofluorescence (IF) results also indicated the impacts of miR-146a and HIPK3 on the expression changes of HIPK3 and p-STAT3 in TGF-β-induced thymocytes, and we discovered that the expression trends of HIPK3 and p-STAT3 were also basically the same as those in western blotting assay in Figure 4B (Figure 4D). Furthermore, the ELISA data indicated that overexpression of HIPK3 also could markedly weaken the increases of IL-10, TGF-β, IL-4 and reduction of TNF-α, and the isolated silencing of HIPK3 also could significantly elevate IL-10, TGF-β, and IL-4 levels and lower TNF-α level as overexpression of miR-146a in TGF-β-induced thymocytes (P<0.05, Figure 4E). Thus, the current results certified that miR-146a overexpression can not only prevent inflammation and but also downregulate p-STAT3 by targeting HIPK3 in TGF-β-induced thymocytes.
Figure 4.
Overexpression of HIPK3 memorably attenuated miR-146a-mediated downregulation of p-STAT3 and suppression of inflammation in TGF-β-induced thymocytes. TGF-β-induced thymocytes were transfected with miR-146a mimics, HIPK3 siRNAs or MiR-146a mimics + HIPK3-overexpressed plasmids. (A) Changes of miR-146a and HIPK3 expressions in each group were evaluated via RT-qPCR assay. (B) Western blot assay was carried out to analyze the expression of HIPK3, FOXP3 and p-STAT3 in each group. (C) The relative expression of FOXP3 in each group was determined by RT-qPCR assay. (D) IF assay exhibited the co-localization and expression of HIPK3 and STAT3 in the processed thymocytes. Magnification, 400×, scale bar =20 µm. (E) After transfection, the levels of IL-10, TGF-β, IL-4, and TNF-α in thymocytes induced by TGF-β were detected by ELISA assay. *P<0.05, **P<0.01 vs. NC group; #P<0.05 vs. mimics group. NC, negative control; HIPK3, homeodomain-interacting protein kinases 3; TGF-β, transforming growth factor-β; siRNA, small interfering RNA; RT-qPCR, reverse transcription quantitative polymerase chain reaction; IF, immunofluorescence; IL-10, interleukin 10; IL-4, interleukin 4; TNF-α, tumor necrosis factor-α; ELISA, enzyme-linked immunosorbent assay.
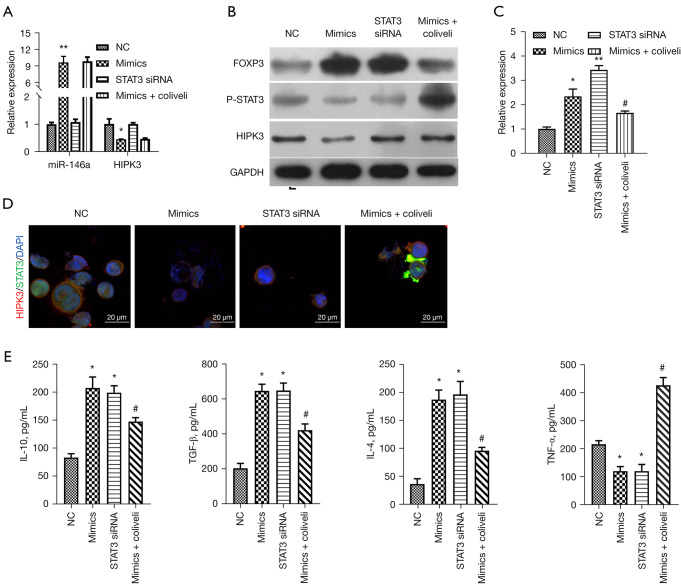
Activation of STAT3 prominently reversed miR-146a-mediated inhibition of p-STAT3 and inflammation in TGF-β-induced thymocytes
Based on our previous conclusion that the miR-146a/HIPK3 axis can regulate the expression of p-STAT3 in TGF-β-induced thymocytes, we further explored whether the STAT3 pathway is necessary for miR-146a to play its role in TGF-β-induced thymocytes. We separately transfected miR-146a mimics and STAT3 siRNAs into TGF-β-induced thymocytes, and after miR-146a overexpression, TGF-β-induced thymocytes were treated with STAT3 activator (colivelin). The results first signified that miR-146a overexpression memorably upregulated miR-146a and downregulated HIPK3, silence of STAT3 had no effect on the expressions of miR-146a and HIPK3 (P<0.05, P<0.01, Figure 5A,5B). We also discovered that miR-146a overexpression or STAT3 silencing significantly boosted FOXP3 expression and restrained p-STAT3 expression in TGF-β-induced thymocytes, while upregulation of FOXP3 and downregulation of p-STAT3 mediated by miR-146a overexpression could be notably reversed by colivelin (P<0.05, P<0.01, Figure 5C). Meanwhile, IF results also revealed that miR-146a overexpression markedly lowered the expressions of HIPK3 and p-STAT3, silence of STAT3 only reduced the expression of p-STAT3, and colivelin also only reversed the downregulation of p-STAT3 mediated by miR-146a overexpression in TGF-β-induced thymocytes (Figure 5D). Moreover, the results of ELISA showed that miR-146a overexpression or STAT3 silencing dramatically increased the levels of IL-10, TGF-β, and IL-4 and decreased the level of TNF-α, while the changes in IL-10, TGF-β, IL-4, and TNF-α levels mediated by miR-146a overexpression could also be prominently attenuated by colivelin in TGF-β-induced thymocytes (P<0.05, Figure 5E). Overall, these results supported that that STAT3 pathway is located downstream of HIPK3, and miR-146a also can suppress inflammation by regulating STAT3 pathway in TGF-β-induced thymocytes.
Figure 5.
Activation of STAT3 prominently reversed miR-146a-mediated inhibition of p-STAT3 and inflammation in TGF-β-induced thymocytes. TGF-β-induced thymocytes were transfected or treated with NC, miR-146a mimics, STAT3 siRNAs or miR-146a mimics + colivelin. (A) The relative expressions of miR-146a and HIPK3 were examined by RT-qPCR in the treated thymocytes. (B) Western blot analysis of expression of HIPK3, FOXP3 and p-STAT3 in each group. (C) Expression level of FOXP3 was assessed by RT-qPCR. (D) The co-localization and expression of HIPK3 and STAT3 were examined through IF assay. Magnification, 400×, scale bar =20 µm. (E) ELISA assay was carried out to evaluate the levels of IL-10, TGF-β, IL-4, and TNF-α in each group of thymocytes. *P<0.05, **P<0.01 vs. NC group; #P<0.05 vs. mimics group. NC, negative control; siRNA, small interfering RNA; HIPK3, homeodomain-interacting protein kinases 3; IL-10, interleukin 10; TGF-β, transforming growth factor-β; IF, immunofluorescence; IL-4, interleukin 4; TNF-α, tumor necrosis factor-α; RT-qPCR, reverse transcription quantitative polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay.
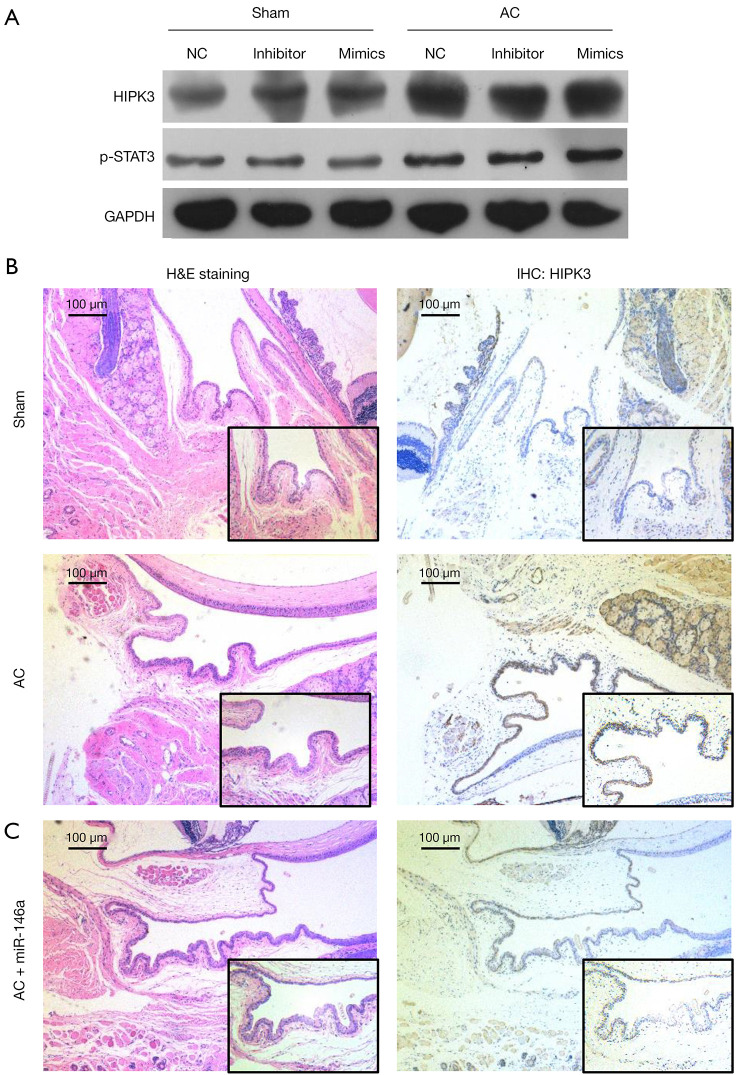
Increase of miR-146a markedly downregulated HIPK3, and ameliorated inflammatory infiltration in AC model mice
Furthermore, we finally confirmed the role and mechanism of miR-146a the inflammatory infiltration in AC model mice. Western blotting results revealed that HIPK3 and p-STAT3 were significantly upregulated in the conjunctival tissues of AC model mice versus that in sham mice (Figure 6A). Then, H&E staining results exhibited that in the sham group, the conjunctival epithelial cells were uniform in size and orderly in order with low inflammatory infiltration; in the AC group, conjunctival epithelial cells showed different sizes and shapes, accompanied by different degrees of cell infiltration; and overexpression of miR-146a notably prevented the pathological changes of the conjunctiva (Figure 6B). Besides, IHC results indicated that the protein level of FOXP3 was markedly lowered in the conjunctival tissues of AC model mice relative to that in sham mice, and overexpression of miR-146a markedly elevated FOXP3 expression in the conjunctival tissues of AC model mice (Figure 6C). In summary, these findings disclosed that overexpression of miR-146a could notably ameliorate inflammatory infiltration of AC model mice.
Figure 6.
Increase of miR-146a markedly downregulated HIPK3 and ameliorated inflammatory infiltration in AC model mice. The AC model mice were established using the irritability and addressed with miR-146a mimics. (A) Western blot analysis was applied to determine the relative expressions of HIPK3 and p-STAT3 in the treated sham and AC model mice. (B) The inflammatory infiltration was evaluated using H&E staining. Magnification, 200×, scale bar =50 µm. (C) The expression of HIPK3 was monitored by applying IHC assay. Magnification, 200×, scale bar =50 µm. AC, allergic conjunctivitis; HIPK3, homeodomain-interacting protein kinases 3; H&E, hematoxylin and eosin; IHC, immunohistochemical.
Discussion
In ophthalmology AC is a common disease, which involves a variety of cells and molecules during the pathological injury process (1). The pathogenesis of AC is type I hypersensitivity mediated by allergen-specific IgE. The ocular surface allergen binds to IgE on the FcεRI of conjunctival mast cell membrane, which immediately initiates the early phase reaction (EPR) of AC (25). The main characteristics of EPR are vasodilation, increased vascular permeability, and pruritus. The later phase reaction (LPR) can occur after 4–6 h and is characterized by an infiltration of multiple inflammatory cells, especially eosinophils (26). Antigen-specific T cells can initiate eosinophils to infiltrate the conjunctiva, resulting in tissue damage (27). The epithelial cells and fibroblasts of conjunctiva and cornea also accelerate inflammation and tissue remodeling by inducing the secretion and expression of cytokines (such as TNF-α, IL-10, IL-4, and TGF-β), chemokines, adhesion molecules, and so on (5,28,29). However, the mechanism of miRNA in AC is rarely studied.
Regulatory T (Treg) cells are a class of T-lymphocyte subsets expressing FOXP3, CD4, CD25, CTLA-4, and other surface molecules, which are characterized by immune response incapacity and immunosuppression (30,31). Inflammation involving T Helper 2 (Th2) is a marker of AC (32). It has been reported that mutations or deletions in the FOXP3 gene can cause a variety of autoimmune diseases, such as type 1 diabetes, thyroiditis, severe allergies, and inflammatory bowel disease (33-35). In our study, we discovered that miR-146a and FOXP3 were lowly expressed in AC model mice, and increase of miR-146a could prominently prevent the inflammatory process by altering the levels of TNF-α, IL-10, IL-4, TGF-β, and FOXP3 in TGF-β-induced thymocytes. Research has shown that TGF-β is a multifunctional cytokine that can regulate the growth and differentiation of diverse cells and the surrounding stromal environment, and plays major roles in the regulation of inflammation and tissue repair (36-38). Besides, TGF-β has also been applied in several studies to construct AC cell models (39,40). Therefore, our results revealed that high expression of miR-146a could suppress inflammation in AC, which is broadly consistent with a previous report (19). However, the mechanism of miR-146a in AC is not entirely clear.
Some data have implied that miRNAs are involved in 30–50% of gene expression regulation (41). Therefore, the determination of miRNA targets has gradually become a crucial link in the study of miRNA function. In our preliminary experiment, we predicted that HIPK3 had binding sites with miR-146a through bioinformatics. We speculated that HIPK3 might be a target gene of miR-146a. The HIPKs are a cofactor family that interact differently with homologous proteins, mainly including HIPK1, HIPK2, HIPK3, and HIPK4. They can participate in cell proliferation, differentiation, apoptosis, and other biological processes by changing the phosphorylation status of target proteins (42). Among them, HIPK3, as a known insulin secretion regulator, is relevant to multiple inflammation-related diseases, including sepsis (11) and diabetes (43). In this study, through the verification of dual-luciferase reporter gene assay, we verified that overexpression of miR-146a could prominently decrease the luciferase activity of WT-HIPK3, suggesting that miR-146a can target HIPK3. Besides, we showed that miR-146a could prevent the inflammatory process of TGF-β-induced thymocytes by targeting HIPK3. Thus, we suggested that miR-146a/HIPK3 axis is critical in the development of AC. Moreover, our data indicated that miR-146a also could downregulate p-STAT3 through HIPK3 in TGF-β-induced thymocytes, indicating that the STAT3 pathway might be required for the miR-146a/HIPK3 axis in AC.
The STAT family, as transcription factors, can regulate the expression of genes related to cell proliferation, survival, and angiogenesis, as well as the genes downstream of immune and inflammatory responses (44,45). As a member of the STAT family, STAT3 can be activated by a variety of cytokines, including IL-6 and IL-23 (46). It also can regulate the biological behavior of immune cells by mediating the extracellular signals of inflammatory mediators (47). It has been reported that the STAT3 pathway can significantly affect the maturation and activation of dendritic cells in allergic diseases (48), and STAT3 is also required for the development of Th2 cells and production of cytokines (49). Therefore, we further speculated that the STAT3 pathway might also have a significant effect in the regulation of miR-146a/HIPK3 axis-mediated AC progression. Our current research also supported that activation of STAT3 could reverse miR-146a-mediated inflammation inhibition in TGF-β-induced thymocytes, suggesting that miR-146a could also suppress the inflammatory process of TGF-β-induced thymocytes by regulating the phosphorylation of STAT3. Besides, we also demonstrated that HIPK3 is located upstream of the STAT3 pathway, because STAT3 silencing did not affect the expression of HIPK3, while HIPK3 silencing down-regulated p-STAT3.
Conclusions
Our current study certified that miR-146a was downregulated in AC, and increase of miR-146a could markedly alleviate the inflammatory response of AC. Besides, HIPK3, as a target gene of miR-146a, also could be involved in the inhibitory effect of miR-146a on AC inflammation. More importantly, phosphorylation of STAT3 also played a critical role in the inflammatory response of AC. Overexpression of miR-146a could attenuate the inflammatory response by down-regulating HIPK3 to restrain STAT3 phosphorylation in AC (Figure 7). Therefore, we demonstrated that the miR-146a/HIPK3/STAT3 axis might be therapeutic targets for relieving inflammation of AC.
Figure 7.
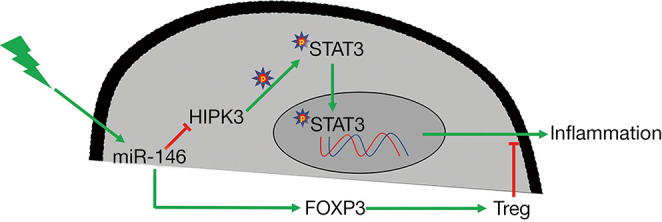
Graphical summary of this study. HIPK3, homeodomain-interacting protein kinases 3.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This research was supported by Shenzhen Fund for Guangdong Provincial High-level Clinical Key Specialties (SZGSP014) and the National Natural Science Foundation of China (81870626).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Shenzhen Eye Hospital Affiliated to Jinan University (Approval No. TOP-IACUC-2021-0106), in compliance with guidelines of Jinan University for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-982/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-982/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-982/coif). The authors have no conflicts of interest to declare.
(English Language Editor: J. Jones)
References
- 1.Baab S, Le PH, Kinzer EE. Allergic Conjunctivitis. StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2021, StatPearls Publishing LLC., 2021. [Google Scholar]
- 2.Sacchetti M, Abicca I, Bruscolini A, et al. Allergic conjunctivitis: current concepts on pathogenesis and management. J Biol Regul Homeost Agents 2018;32:49-60. [PubMed] [Google Scholar]
- 3.Miyazaki D, Fukagawa K, Okamoto S, et al. Epidemiological aspects of allergic conjunctivitis. Allergol Int 2020;69:487-95. 10.1016/j.alit.2020.06.004 [DOI] [PubMed] [Google Scholar]
- 4.Kuruvilla M, Kalangara J, Lee FEE. Neuropathic Pain and Itch Mechanisms Underlying Allergic Conjunctivitis. J Investig Allergol Clin Immunol 2019;29:349-56. 10.18176/jiaci.0320 [DOI] [PubMed] [Google Scholar]
- 5.Berger WE, Granet DB, Kabat AG. Diagnosis and management of allergic conjunctivitis in pediatric patients. Allergy Asthma Proc 2017;38:16-27. 10.2500/aap.2017.38.4003 [DOI] [PubMed] [Google Scholar]
- 6.Ding Y, Li C, Zhang Y, et al. Quercetin as a Lyn kinase inhibitor inhibits IgE-mediated allergic conjunctivitis. Food Chem Toxicol 2020;135:110924. 10.1016/j.fct.2019.110924 [DOI] [PubMed] [Google Scholar]
- 7.Fauquert JL. Diagnosing and managing allergic conjunctivitis in childhood: The allergist's perspective. Pediatr Allergy Immunol 2019;30:405-14. 10.1111/pai.13035 [DOI] [PubMed] [Google Scholar]
- 8.Elieh Ali Komi D, Rambasek T, Bielory L. Clinical implications of mast cell involvement in allergic conjunctivitis. Allergy 2018;73:528-39. 10.1111/all.13334 [DOI] [PubMed] [Google Scholar]
- 9.Vazirani J, Shukla S, Chhawchharia R, et al. Allergic conjunctivitis in children: current understanding and future perspectives. Curr Opin Allergy Clin Immunol 2020;20:507-15. 10.1097/ACI.0000000000000675 [DOI] [PubMed] [Google Scholar]
- 10.Zi Y, Deng Y, Ji M, et al. The effectiveness of olopatadine hydrochloride eye drops for allergic conjunctivitis: Protocol for a systematic review. Medicine (Baltimore) 2020;99:e18618. 10.1097/MD.0000000000018618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Hou Q, Ma Y, et al. HIPK3 Mediates Inflammatory Cytokines and Oxidative Stress Markers in Monocytes in a Rat Model of Sepsis Through the JNK/c-Jun Signaling Pathway. Inflammation 2020;43:1127-42. 10.1007/s10753-020-01200-5 [DOI] [PubMed] [Google Scholar]
- 12.Guo C, Liu J, Hao P, et al. The Potential Inhibitory Effects of miR-19b on Ocular Inflammation are Mediated Upstream of the JAK/STAT Pathway in a Murine Model of Allergic Conjunctivitis. Invest Ophthalmol Vis Sci 2020;61:8. 10.1167/iovs.61.3.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Zhou L, Xu Y, et al. A STAT3 palmitoylation cycle promotes TH17 differentiation and colitis. Nature 2020;586:434-9. 10.1038/s41586-020-2799-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018;141:1202-7. 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goeman F, Strano S, Blandino G. MicroRNAs as Key Effectors in the p53 Network. Int Rev Cell Mol Biol 2017;333:51-90. 10.1016/bs.ircmb.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 16.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol 2019;234:5451-65. 10.1002/jcp.27486 [DOI] [PubMed] [Google Scholar]
- 17.Biswas S. MicroRNAs as Therapeutic Agents: The Future of the Battle Against Cancer. Curr Top Med Chem 2018;18:2544-54. 10.2174/1568026619666181120121830 [DOI] [PubMed] [Google Scholar]
- 18.Sun W, Sheng Y, Chen J, et al. Down-Regulation of miR-146a Expression Induces Allergic Conjunctivitis in Mice by Increasing TSLP Level. Med Sci Monit 2015;21:2000-7. 10.12659/MSM.894563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Yin X, Yi J, et al. MiR-146a overexpression effectively improves experimental allergic conjunctivitis through regulating CD4+CD25-T cells. Biomed Pharmacother 2017;94:937-43. 10.1016/j.biopha.2017.07.157 [DOI] [PubMed] [Google Scholar]
- 20.Sierra OL, Towler DA. Runx2 trans-activation mediated by the MSX2-interacting nuclear target requires homeodomain interacting protein kinase-3. Mol Endocrinol 2010;24:1478-97. 10.1210/me.2010-0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ba Y, Liu Y, Li C, et al. HIPK3 Promotes Growth and Metastasis of Esophageal Squamous Cell Carcinoma via Regulation of miR-599/c-MYC Axis. Onco Targets Ther 2020;13:1967-78. 10.2147/OTT.S217087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao W, Wang T, Ye Y, et al. Identification of HIPK3 as a potential biomarker and an inhibitor of clear cell renal cell carcinoma. Aging (Albany NY) 2021;13:3536-53. 10.18632/aging.202294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Sun X, Lu B. HIPK3 modulates autophagy and HTT protein levels in neuronal and mouse models of Huntington disease. Autophagy 2018;14:169-70. 10.1080/15548627.2017.1393130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y, Yang C, Pan F. Post-Translational Regulations of Foxp3 in Treg Cells and Their Therapeutic Applications. Front Immunol 2021;12:626172. 10.3389/fimmu.2021.626172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azari AA, Arabi A. Conjunctivitis: A Systematic Review. J Ophthalmic Vis Res 2020;15:372-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shii D, Nakagawa S, Yoshimi M, et al. Inhibitory effects of cyclosporine a eye drops on symptoms in late phase and delayed-type reactions in allergic conjunctivitis models. Biol Pharm Bull 2010;33:1314-8. 10.1248/bpb.33.1314 [DOI] [PubMed] [Google Scholar]
- 27.Erdinest N, London N, Solomon A. Chemokines in allergic conjunctivitis. Curr Opin Allergy Clin Immunol 2020;20:516-27. 10.1097/ACI.0000000000000676 [DOI] [PubMed] [Google Scholar]
- 28.Masrur A, Adnan M, Khan FA, et al. Association of Severity of Allergic Conjunctivitis with Skin Prick Test. J Coll Physicians Surg Pak 2020;30:1166-9. 10.29271/jcpsp.2020.11.1166 [DOI] [PubMed] [Google Scholar]
- 29.Wei CC, Kung YJ, Chen CS, et al. Allergic Conjunctivitis-induced Retinal Inflammation Promotes Myopia Progression. EBioMedicine 2018;28:274-86. 10.1016/j.ebiom.2018.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Göschl L, Scheinecker C, Bonelli M. Treg cells in autoimmunity: from identification to Treg-based therapies. Semin Immunopathol 2019;41:301-14. 10.1007/s00281-019-00741-8 [DOI] [PubMed] [Google Scholar]
- 31.Zhang BH, Li L, Ji J, et al. Prognostic value of Foxp3+ regulatory T cells in endometrial cancer: a meta-analysis. Eur J Gynaecol Oncol 2020;41:339-42. 10.31083/j.ejgo.2020.03.5181 [DOI] [Google Scholar]
- 32.Chen X, Deng R, Chi W, et al. IL-27 signaling deficiency develops Th17-enhanced Th2-dominant inflammation in murine allergic conjunctivitis model. Allergy 2019;74:910-21. 10.1111/all.13691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat Immunol 2018;19:665-73. 10.1038/s41590-018-0120-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr A, Atif M, Balderas R, et al. The role of FOXP3+ regulatory T cells in human autoimmune and inflammatory diseases. Clin Exp Immunol 2019;197:24-35. 10.1111/cei.13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao JH, Cheng M, Tang JP, et al. Foxp3, Regulatory T Cell, and Autoimmune Diseases. Inflammation 2017;40:328-39. 10.1007/s10753-016-0470-8 [DOI] [PubMed] [Google Scholar]
- 36.Fabregat I, Moreno-Càceres J, Sánchez A, et al. TGF-β signalling and liver disease. FEBS J 2016;283:2219-32. 10.1111/febs.13665 [DOI] [PubMed] [Google Scholar]
- 37.Goumans MJ, Ten Dijke P. TGF-β Signaling in Control of Cardiovascular Function. Cold Spring Harb Perspect Biol 2018;10:a022210. 10.1101/cshperspect.a022210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol 2016;12:325-38. 10.1038/nrneph.2016.48 [DOI] [PubMed] [Google Scholar]
- 39.Unsal AIA, Kocaturk T, Gunel C, et al. Effect of Pycnogenol® on an experimental rat model of allergic conjunctivitis. Graefes Arch Clin Exp Ophthalmol 2018;256:1299-304. 10.1007/s00417-018-3988-7 [DOI] [PubMed] [Google Scholar]
- 40.Yan A, Luo G, Zhou Z, et al. Tear osteopontin level and its relationship with local Th1/Th2/Th17/Treg cytokines in children with allergic conjunctivitis. Allergol Immunopathol (Madr) 2018;46:144-8. 10.1016/j.aller.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 41.Correia de Sousa M, Gjorgjieva M, Dolicka D, et al. Deciphering miRNAs' Action through miRNA Editing. Int J Mol Sci 2019;20:6249. 10.3390/ijms20246249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conte A, Pierantoni GM. Update on the Regulation of HIPK1, HIPK2 and HIPK3 Protein Kinases by microRNAs. Microrna 2018;7:178-86. 10.2174/2211536607666180525102330 [DOI] [PubMed] [Google Scholar]
- 43.Shojima N, Hara K, Fujita H, et al. Depletion of homeodomain-interacting protein kinase 3 impairs insulin secretion and glucose tolerance in mice. Diabetologia 2012;55:3318-30. 10.1007/s00125-012-2711-1 [DOI] [PubMed] [Google Scholar]
- 44.Dong Z, Chen Y, Yang C, et al. STAT gene family mRNA expression and prognostic value in hepatocellular carcinoma. Onco Targets Ther 2019;12:7175-91. 10.2147/OTT.S202122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verhoeven Y, Tilborghs S, Jacobs J, et al. The potential and controversy of targeting STAT family members in cancer. Semin Cancer Biol 2020;60:41-56. 10.1016/j.semcancer.2019.10.002 [DOI] [PubMed] [Google Scholar]
- 46.Hillmer EJ, Zhang H, Li HS, et al. STAT3 signaling in immunity. Cytokine Growth Factor Rev 2016;31:1-15. 10.1016/j.cytogfr.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarino AV, Kanno Y, O'Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol 2017;18:374-84. 10.1038/ni.3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell BD, Kitajima M, Larson RP, et al. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat Immunol 2013;14:364-71. 10.1038/ni.2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stritesky GL, Muthukrishnan R, Sehra S, et al. The transcription factor STAT3 is required for T helper 2 cell development. Immunity 2011;34:39-49. 10.1016/j.immuni.2010.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as