Abstract
Background
High-throughput population screening for the novel coronavirus disease (COVID-19) is critical to controlling disease transmission. Convolutional neural networks (CNNs) are a cutting-edge technology in the field of computer vision and may prove more effective than humans in medical diagnosis based on computed tomography (CT) images. Chest CT images can show pulmonary abnormalities in patients with COVID-19.
Methods
In this study, CT image preprocessing are firstly performed using fuzzy c-means (FCM) algorithm to extracted the region of the pulmonary parenchyma. Through multiscale transformation, the preprocessed image is subjected to multi scale transformation and RGB (red, green, blue) space construction. After then, the performances of GoogLeNet and ResNet, as the most advanced CNN architectures, were compared in COVID-19 detection. In addition, transfer learning (TL) was employed to solve overfitting problems caused by limited CT samples. Finally, the performance of the models were evaluated and compared using the accuracy, recall rate, and F1 score.
Results
Our results showed that the ResNet-50 method based on TL (ResNet-50-TL) obtained the highest diagnostic accuracy, with a rate of 82.7% and a recall rate of 79.1% for COVID-19. These results showed that applying deep learning technology to COVID-19 screening based on chest CT images is a very promising approach. This study inspired us to work towards developing an automatic diagnostic system that can quickly and accurately screen large numbers of people with COVID-19.
Conclusions
We tested a deep learning algorithm to accurately detect COVID-19 and differentiate between healthy control samples, COVID-19 samples, and common pneumonia samples. We found that TL can significantly increase accuracy when the sample size is limited.
Keywords: Coronavirus disease 2019 (COVID-19), computed tomography (CT), convolutional neural network, transfer learning
Introduction
Since the outbreak of coronavirus disease 2019 (COVID-19) at the end of 2019, approximately 12 million people worldwide have been infected. Infection rates are still increasing at the time of writing by more than 100,000 people every day, causing a serious threat to human life and health. To control the spread of the virus, rapid screening of many suspected cases to implement appropriate isolation and treatment measures is vital (1,2). Viral nucleic acid laboratory testing is the gold standard for diagnosing suspected cases. However, this method is very time-consuming and has a high rate of false negatives. Therefore, there is an urgent need for a complementary diagnostic method to improve diagnostic efficiency and accuracy in suspected cases. Chest computed tomography (CT) findings have been an important reference and of clinical value in diagnosing COVID-19 (2,3).
With the rapid development of artificial intelligence technologies, computer-aided diagnosis techniques could help doctors quickly detect and screen lesions on CT images. Convolutional neural networks (CNNs) are a common deep learning technology and have been widely used in auxiliary diagnoses with medical imaging (4). Many scholars have studied CT image analysis and modeling methods based on deep learning technology to achieve intelligent diagnoses of COVID-19. However, due to the lack of training samples, this method often leads to the overfitting of the deep learning network model or non-convergence of the objective function, resulting in poor performance on independent test sets (5,6).
COVID-19 is a serious infectious disease discovered only in December 2019, and its CT findings, and performance and the application prospect of computer-aided technology are unknown to a large extent. Therefore, it may be an important challenge in the diagnosis of COVID-19 and the differentiation between COVID-19 and common pneumonia although these computer-aided diagnosis strategies and advanced deep learning algorithms are widely used and valuable in non-medical computer vision tasks. Consequently, it is an urgent task, at present, to develop more diagnostic tools, especially objective and efficient diagnostic tools.
In response to the above problem, this study proposed a CT-based intelligent diagnosis method combining transfer learning (TL) and CNN to accurately diagnose COVID-19 and successfully differentiate between healthy patients, those with COVID-19, and those with common pneumonia. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-534/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of Shenzhen Third People's hospital (No. 2020-258), and written informed consent was obtained from all the participants.
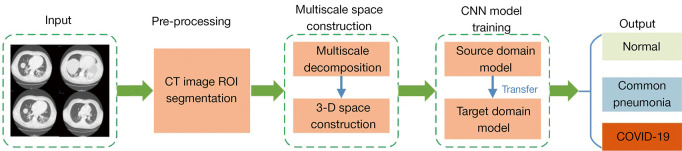
The study included 4 stages: CT image preprocessing, multiscale space construction, convolutional neural network training based on transfer learning, and CT image classification (Figure 1).
Figure 1.
Flow chart of the study. CT, computed tomography; ROI, region of interest; CNN, Convolutional neural network; COVID-19, novel coronavirus disease.
The data used in this study came from the Imaging Department of the Shenzhen Third People’s Hospital. The CT image data of 120 patients were collected, and 4,500 CT slices, including 1,500 COVID-19 slices, 1,500 common pneumonia slices, and 1,500 healthy sample slices, were selected for this study. All CT slice data were annotated by medical imaging experts. The annotation method involved the imaging experts selecting CT slices and marking them up with corresponding imaging changes from all the CT scan slices for healthy patients and those with COVID-19 or common pneumonia.
Preprocessing
Due to the limited number of training samples in this study, we found that if the CT image was directly input as the model, the image background interfered with the model training. Therefore, to improve the model’s accuracy, we first extracted the region of the pulmonary parenchyma from the CT images. The fuzzy c-means (FCM) algorithm is one of the most widely used clustering techniques. It introduces the membership function based on fuzzy theory and solves the defects of the hard segmentation method (7). The method is based on the following objective function:
| [1] |
where m is weighted index, xi is the ith pixel in the CT image, cj is the center of the jth category, and ui,j is the degree of membership of xi in the jth cluster. Fuzzy segmentation is achieved by iteratively updating the membership degree ui,j and cluster center cj to optimize the above objective function.
| [2] |
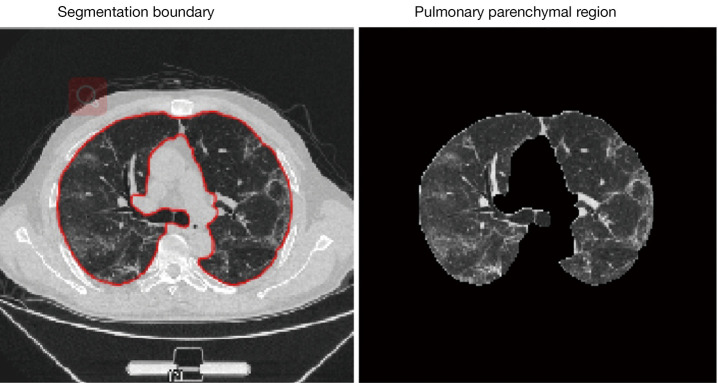
The termination condition of iteration is defined as , where k represents the number of iterations, and ε is a threshold of iteration stops. Segmentation results for the pulmonary parenchymal region of a CT image using this method are shown in Figure 2.
Figure 2.
Segmentation result from the algorithm.
Multi-scale space construction
Multiscale analysis is widely applied in medical image processing field. An image contains different frequency components that can be decomposed into different scales using a multi-resolution analysis method. Through multiscale transformation, an image can be analyzed from coarse-grained to fine-grained multi-scale (8). The Gaussian kernel is the only convolution kernel constructed in a multiscale space. Its essence is to obtain image sets I(x, y) with different degrees of blurring after image I(x, y) is convolved with Gaussian kernel functions G(x, y) at different scale factors σ. The specific formula is defined as follows:
| [3] |
| [4] |
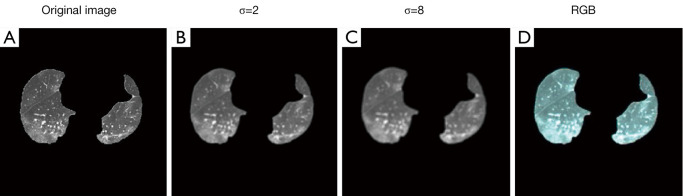
As shown in Figure 3, the original CT images were first converted to two scale Spaces (σ=2 and σ=8), (Figure 3A-3C). Then, we reconstructed the CT images into RGB (red, green, blue) 3-dimensional space using RGB image-space construction (Figure 3D). The RGB-space CT image integrated the image texture information of 3 scale-spaces through 3 independent channels, which was conducive to the CNN obtaining COVID-19 features from multiple perspectives.
Figure 3.
Multi-scale transformation and RGB-space construction. RGB, red-green-blue.
Transfer learning
In recent years, CNN has been widely used in machine vision and speech recognition due to its powerful feature-extraction and induction capabilities (9,10). A basic CNN architecture usually consists of a convolutional layer, a pooling layer, and a fully connected layer. The convolutional layer consists of a series of convolution kernels used to calculate feature maps. This is followed by a pooling layer. Finally, the feature maps are connected to the fully connected layer, and the classification is realized using the softmax function. In the convolutional layer, the formula for calculating the jth feature map of the lth convolutional layer is
| [5] |
The most classic CNN architectures include AlexNet, VGGNet, GoogLeNet, and ResNet (11). GoogLeNet and ResNet have shown better performance in natural image classification tasks due to various technical improvements based on the former. The purpose of this study was to verify and analyze the performance of these 2 state-of-the-art CNN architectures on the COVID-19 CT image database. Considering the limitations of the patient sample size used in this study, training a CNN model with super-large parameters directly through random initialization could have easily led to overfitting or non-convergence. TL has been shown to improve the performance of training tasks with a small sample set by using the knowledge learned from classification tasks with large sample sets (12). Deep transfer learning (DTL) has been the most successful application of CNN for image classification of small sample sets (13).
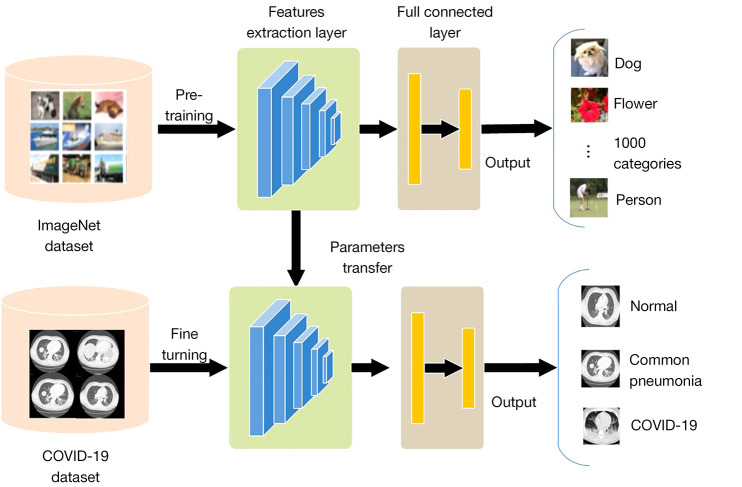
ImageNet is a large natural image database. In this study, we train the source domain model on ImageNet. Then, the parameters on the feature extraction module of the model were used to initialize the corresponding layer of the COVID-19 detection model. At the same time, the fully connected parameters were initialized randomly for COVID-19 detection, the output layer was adjusted to 3 outputs. Finally, the new model was fine-tuned and trained on the CT image database (Figure 4).
Figure 4.
The transfer learning (TL) process.
Although the data from ImageNet are very different from medical images, CNNs can still make model training of medical image recognition tasks more efficient by learning graphic primitives from large, well-annotated databases. In this paper, we refer to the models of random initialization (RI) as GoogLeNet-RI and ResNet-50-RI and the models of TL as GoogLeNet-TL and ResNet-50-TL. Since the CT images had a 512×512 resolution, and the input size for GoogLeNet and ResNet was 256×256, the CT images had to be down-sampled to 256×256.
Statistical analysis
The performance of models were evaluated using the accuracy, recall, F1 score.
Results
This study evaluated and compared the performance of 4 CNN models: GoogLeNet-RI and ResNet-50-RI, which were randomly initialized from scratch training on the COVID-19 CT database; and GoogLeNet-TL and ResNet-50-TL, which used transfer learning to obtain initialization parameters from the ImageNet training model.
We used 5-fold cross-validation to evaluate COVID-19 classification tasks at the CT slice level as this validation method reflects clinical performance more effectively than the leave-one-out validation method. Performance evaluation indicators were accuracy, recall, and the F1 score. The performances of the 4 models are shown in Table 1 for comparison.
Table 1. Performance comparison between the 4 methods.
| Algorithm | Accuracy (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|
| GoogLeNet-RI | 62.7 | 61.2 | 60.3 |
| GoogLeNet-TL | 80.7 | 73.1 | 76.5 |
| ResNet-50-RI | 65.3 | 62.5 | 63.2 |
| ResNet-50-TL | 82.7 | 77.9 | 79.1 |
RI, random initialization; TL, transfer learning.
Our results showed that the GoogLeNet-TL and ResNet-50-TL models obtained accuracy rates of 80.7% and 82.7%, respectively, showing significantly better accuracy than the GoogLeNet-RI (62.7%) and ResNet-50-RI models (63.5%). These results showed that using the TL method for training with the COVID-19 database led to better model performance.
The confusion matrix of the ResNet-50-TL classification of COVID-19 CT images is shown in Figure 5. We found that the correct recognition rate of the model for the healthy control samples reached 92.1%, with only 4.7% misidentified as common pneumonia and 3.2% as COVID-19. The accurate identification rates for common pneumonia and COVID-19 samples were 77.5% and 79.1%, respectively. Among the incorrectly identified samples, 13.2% of the samples for common pneumonia were identified as COVID-19 and 9.3% as healthy samples, while 14.7% of the COVID-19 samples were identified as common pneumonia, and 6.2% as healthy samples.
Figure 5.
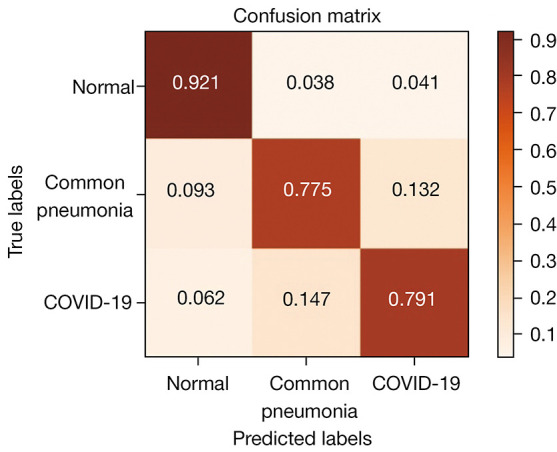
Confusion matrix of the ResNet-50-TL training model for COVID-19 CT image classification results. TL, transfer learning; COVID-19, novel coronavirus disease; CT, computed tomography.
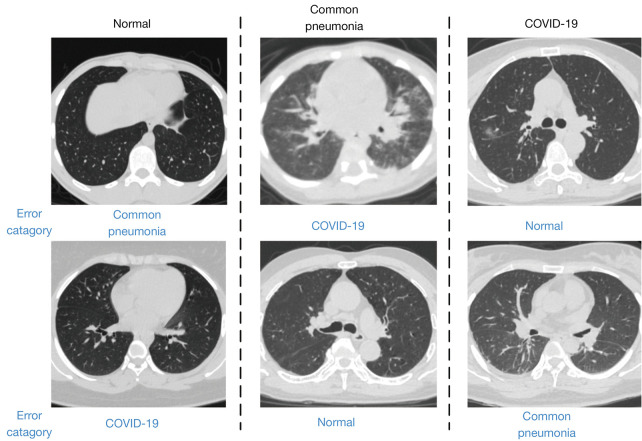
Some of the most typical misclassified images are shown in Figure 6. The CT images of patients with mild COVID-19 were more likely to be mistakenly identified as common pneumonia or healthy samples, and patients with severe pneumonia were likely to be misidentified as having COVID-19.
Figure 6.
Examples of misclassified samples. COVID-19, novel coronavirus disease.
In addition, we found that because the ResNet-50 architecture had a higher number of layers [50] than GoogLeNet [22], ResNet-50 showed stronger feature expression capabilities without fitting and thereby achieved a better classification performance.
Discussion
The CNN tool has been successfully tested and applied in clinical works. Several studies also reported the federated learning framework (14,15), which will open up new opportunities for global collaboration especially during this pandemic as this approach can avoids issues associated with data sharing ownership and privacy breaches. It will allow the rapid development of this model and improve its generalizability.
This study has made several contributions to the field. First, by introducing the TL method, the overfitting problems associated with CNN models for small sample sets were resolved. Experiments showed that the performance of the CNN model significantly improved after the introduction of TL. Secondly, we also introduced multi-scale analysis technology to map the original CT image to 3 different scale-spaces and then extracted deep multi-scale features through the CNN architecture, which effectively improved the ability to distinguish between common pneumonia and COVID-19 imaging features. In this way, the diagnostic accuracy of the models was further improved, and false positive and false negative rates were reduced.
The current gold standard diagnostic test for COVID-19 using polymerase chain reaction (PCR) has several limitations. It is resource intensive and time consuming. Its accuracy also depends on patient’s clinical condition, sampling technique and test kits used. The reported positive rate was around 60–70% and false negatives were possible. It also does not provide any information about patient’s disease severity. On the contrary, AI based CNN could provide more accurate and efficient tools during pandemic.
This study introduced TL into CNN model training to improve the accuracy of COVID-19 detection. The results showed that the ResNet-50-TL model achieved the best accuracy, recall, and F1 score. These results suggest that the ResNet-50-TL model has a strong ability to distinguish between healthy samples and the 2 types of disease samples. However, the model’s ability to distinguish between COVID-19 and common pneumonia CT images was lower than its ability to differentiate between diseased and healthy samples. This may be because the similarity between the CT images of the 2 disease samples was significantly higher than the similarity between the healthy and disease samples.
This study had certain limitations, primarily due to the relatively small training sample set, which only contained healthy, common pneumonia, and COVID-19 samples. However, under clinical conditions, the types of diseases of the samples would show greater diversity and concurrency. Therefore, the probability of model error detection would be higher when identifying such samples. From the perspective of machine learning theory, the most direct and effective way to address the problems associated with a small training set would be to increase the sample size of the training set and expand the sample diversity. In addition, radiologists could also perform more precise calibration on the lesion area of the CT image to achieve classification modeling based on the CT lesion area, ultimately improving the performance of the model. This will be the focus of future research and more in-depth exploration.
In this study, COVID-19 CT images were classified and recognized using CT image segmentation of the pulmonary parenchymal region, multi-scale CT space construction, and DTL methods. The performances of GoogLeNet and ResNet CNN architecture in the classification of COVID-19 CT image samples were evaluated. Our experiments showed that the ResNet-50 CNN architecture using TL achieved a COVID-19 recall rate of 79.1%, and an overall accuracy for the 3 sample classifications (healthy, common pneumonia, and COVID-19) of 82.7%. These results are of practical clinical significance for advancing the rapid detection of COVID-19. To further improve the performance of the model, future research will aim to increase the training sample size and integrate expert samples to accurately calibrate the lesion area.
Conclusions
We tested a deep learning algorithm to detect and classify COVID-19 accurately and differentiate between COVID-19, healthy, and common pneumonia samples. TL significantly increased diagnostic accuracy in a limited sample size.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of Shenzhen Third People's hospital (No. 2020-258), and written informed consent was obtained from all the participants.
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-534/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-534/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-534/coif). The authors have no conflicts of interest to declare.
(English Language Editor: L. Roberts)
References
- 1.Zhang J, Xie Y, Li Y, et al. COVID-19 Screening on Chest X-ray Images Using Deep Learning based Anomaly Detection. arXiv, 2020.
- 2.Hasan AM, Aljawad MM, Jalab HA, et al. Classification of Covid-19 Coronavirus, Pneumonia and Healthy Lungs in CT Scans Using Q-Deformed Entropy and Deep Learning Features. Entropy 2020;22:517. 10.3390/e22050517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong Q, Li Z, Shen XY, et al. CT imaging features of patients with different clinical types of COVID-19. Journal of Zhejiang University (Medical Sciences) 2020;49:198-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanagar SB, Al-Ehaideb A, Maganur PC, et al. Developments, application, and performance of artificial intelligence in dentistry - A systematic review. J Dent Sci 2021;16:508-22. 10.1016/j.jds.2020.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho T, Rezende ERSD, Alves MTP, et al. Exposing Computer Generated Images by Eye's Region Classification via Transfer Learning of VGG19 CNN[C]// 16th IEEE International Conference On Machine Learning And Applications. IEEE, 2017. [Google Scholar]
- 6.Deepak S, Ameer P M. Brain tumor classification using deep CNN features via transfer learning. Computers in Biology and Medicine 2019;111:103345. 10.1016/j.compbiomed.2019.103345 [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Jiang H, Gao M, et al. An Assisted Diagnosis System for Detection of Early Pulmonary Nodule in Computed Tomography Images. Journal of Medical Systems 2017;41:1-9. 10.1007/s10916-016-0669-0 [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Shi K. Multi-scale Attention Convolutional Neural Network for time series classification. Neural Netw 2021;136:126-40. 10.1016/j.neunet.2021.01.001 [DOI] [PubMed] [Google Scholar]
- 9.Yao G, Lei T, Zhong J. A Review of Convolutional-Neural-Network-Based Action Recognition. Pattern Recognition Letters 2018;118:14-22. 10.1016/j.patrec.2018.05.018 [DOI] [Google Scholar]
- 10.Zhou FY, Jin LP, Dong J. Review of Convolutional Neural Network. Chinese Journal of Computers 2017;40:1-23. [Google Scholar]
- 11.Dhillon A, Verma GK. Convolutional neural network: a review of models, methodologies and applications to object detection. Progress in Artificial Intelligence 2020:1-28. doi: 10.1007/s13748-019-00203-0. 10.1007/s13748-019-00203-0 [DOI] [Google Scholar]
- 12.Liu XP, Luan XD, Xie YX, et al. Transfer Learning Research and Algorithm Review. Journal of Changsha University 2018;32:28-31. [Google Scholar]
- 13.Zhang Y, Gao X, He L, et al. Objective Video Quality Assessment Combining Transfer Learning With CNN. IEEE Transactions on Neural Networks and Learning Systems, 2019:1-15. . 10.1109/TNNLS.2018.2890310 [DOI] [PubMed] [Google Scholar]
- 14.Aruru M, Truong HA, Clark S. Pharmacy Emergency Preparedness and Response (PEPR): a proposed framework for expanding pharmacy professionals' roles and contributions to emergency preparedness and response during the COVID-19 pandemic and beyond. Res Social Adm Pharm 2021;17:1967-77. 10.1016/j.sapharm.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng D, Lan X, Yao MM, et al. Federated learning: a collaborative effort to achieve better medical imaging models for individual sites that have small labelled datasets. Quant Imaging Med Surg 2021;11:852-7. 10.21037/qims-20-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as