Abstract
Background
Glioma is the most common intracranial primary malignancy, characterized by abnormal signal transductions caused by transcriptional and post-transcriptional regulators. Studies show the palmitoylation of oncoproteins and tumor suppressors participate in cancer progression, while studies of protein S-palmitoyltransferases in glioma are limited. A systematic analysis of zinc finger DHHC-type palmitoyltransferases (ZDHHC) in glioma is still lacking.
Methods
A prognostic heatmap and Kaplan-Meier overall survival plot of 24 members of the ZDHHC family in pan-cancer created. The expression and prognostic significance of ZDHHC12 was analyzed by using Gene Expression Profiling Interactive Analysis (GEPIA) and PrognoScan. DBTRG and U251 cells with silenced ZDHHC12 expression were constructed and used for cell counting kit-8 (CCK-8), Transwell assay and wound healing assay in vitro.
Results
Here, we first conducted expression and prognostic analyses of 24 ZDHHCs from The Cancer Genome Atlas (TCGA), the Chinese Glioma Genome Atlas (CGGA), and other glioma datasets. We found ZDHHC12 to be the only unfavorable prognostic marker in glioma. The function of ZDHHC12 in glioma was then investigated with loss-of-function strategies and in vitro cell assays. Results showed that ZDHHC12 knockdown remarkably reduced the growth, migration, and invasion capabilities in DBTRG and U251 cell lines, suggesting that ZDHHC12 may contribute to malignant behavior in glioma cells. Finally, the molecular basis for ZDHHC12 expression in glioma was analyzed, and DNA hypomethylation was found to be responsible for increased ZDHHC12 mRNA expression and related prognoses.
Conclusions
ZDHHC12 positively promoted the proliferation and migration of glioma cells. Decreased DNA methylation may lead to increased ZDHHC12 expression in gliomas. This study may deepen the understanding of glioma progression and therapeutics.
Keywords: Glioma, ZDHHC12, prognosis, hypomethylation
Introduction
Gliomas are the most common intracranial primary malignancy, with glioblastoma (GBM) the most malignant form (1). Based on their histological properties, gliomas have been categorized into the World Health Organization (WHO) classifications of astrocytic, oligodendroglial, or ependymal tumors, which indicate different degrees of malignancy (2). Accumulating evidence shows that gliomas are genetic, epigenetic, and highly heterogeneous (3). Glioma development, such as apoptosis and immune evasion, is characterized by active and passive DNA demethylation processes, that promote certain apoptosis and epigenetic reprogramming of immune system participants (4). The abnormal signal transductions caused by transcriptional and post-transcriptional regulators underlie glioma pathogenesis, progression, and therapeutic responses (5,6). Although diagnostic (7,8) and therapeutic advances (e.g., surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy) (1,5,9) have been achieved, outcomes in patients with glioma still remain unsatisfactory. Therefore, it is necessary to identify novel genes and prognostic markers in glioma.
Protein S-palmitoylation is a reversible post-translational modification and one of the most common lipid modifications (10). Protein S-palmitoylation has been reported to play a role in protein stability, transport, and human disease processes (11,12). Protein palmitoylation is regulated by palmitoyl S-acyltransferases from the DHHC family, while the removal of palmitate is catalyzed by serine hydrolases, including acyl-protein thioesterases (APTs) (13). Studies have shown that the palmitoylation of oncoproteins and tumor suppressors participate in cancer progression (14-17). However, studies of protein palmitoylation in glioma are limited. In p53-mutated glioma, Chen et al. proved that zinc finger DHHC-type palmitoyltransferase 5 (ZDHHC5) acts as an oncogene via EZH2 palmitoylation (18). Zhang et el. recently found that DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis (19). In a study by Fan et al., ZDHHC15-mediated GP130 palmitoylation was found to be involved in the effect of local anesthetics on the repression of growth and self-renewal of glioblastoma stem cells (20). In another study, Fan et al. found that Smad3 palmitoylation by ZDHHC19 promoted the activation of transforming growth factor-beta (TGFβ) in the mesenchymal subtype of GBM (21). Differential expression, prognostic analysis and functional correlation analysis of 10 kinds of ZDHHC in renal clear cell carcinoma were systematically investigated (22). However, there is still a lack of systematic analyses of ZDHHCs in glioma.
ZDHHC12 is one of the protein palmitoylation regulated by palmitoyl s-acyltransferases from the DHHC family (12). Currently, there are few studies on the occurrence and progression of tumors related to ZDHHC12, which is why we chose ZDHHC12 as the main research object. This study analyzed the expression and prognostic of ZDHHCs in glioma and found ZDHHC12 to be the hub gene. The function of ZDHHC12 in glioma was then investigated using loss-of-function strategies and in vitro cell assays. Finally, the molecular basis for ZDHHC12 expression in glioma was analyzed, and DNA hypomethylation was found to be responsible for increased ZDHHC12 mRNA expression and related prognoses. Our research aimed to reveal the clinical significance and cellular function of ZDHHC12 in glioma were revealed for the first time, which could provide a theoretical basis for ZDHHC12 as a marker for early screening of glioma in the future, and might deepen the understanding of glioma progression and therapeutics. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-520/rc).
Methods
Expression and prognosis analyses
A prognostic heatmap and Kaplan-Meier overall survival plot of 24 members of the ZDHHC family in pan-cancer were created, and the expression of ZDHHC12 in pan-cancer and glioma was investigated using GEPIA 2.0 (23). The expression and methylation of ZDHHC12 in the Chinese Glioma Genome Atlas (CGGA) samples (24) were analyzed using the CGGA website: http://cgga.org.cn/. The prognostic significance of ZDHHC12 was analyzed for other datasets using PrognoScan (25). The GSE4271 dataset (26-28) included 77 primary high-grade astrocytomas and 23 matched recurrences. The GSE4412 dataset included 85 gliomas from 74 patients, and platform were Affymetrix HG-U133A and HG-U133B arrays (29). This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
DBTRG and U251 glioma cell lines were preserved in our laboratory. Minimum essential medium-alpha (MEM) was used to culture cell lines with 10% fetal bovine serum (FBS) which is heat-inactivated, penicillin (100 IU/mL) and streptomycin (100 µg/mL) were used to prevent bacterial infection. Cells were cultured at 37 ℃ in humidified incubators, with 95% humidity and 5% carbon dioxide.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from the cells using a TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and then determined its concentration and purity. The total RNA was reverse transcribed into cDNA using a ReverTra Ace qPCR RT kit (Toyobo Life Science) following the manufacturer’s instructions. Primers were synthesized by Sangon Biotech Co. Ltd. (Shanghai, China).
Two-step quantitative polymerase chain reaction (qPCR) was performed, also using the ReverTra Ace qPCR RT kit (Toyobo Life Science), with GAPDH serving as the internal reference. The primers were used as following:
(F) 5'-GTGCTGACCTGGGGAATCAC-3' and (R) 5'-CTGCACATTCACGTAGCCA-3' for ZDHHC12; and (F) 5'-GGAGCGAGATCCCTCCAAAAT-3' and (R) 5'-GGCTGTTGTCATACTTCTCATGG-3' for GADPH.
Short hairpin RNAs targeting ZDHHC12 and cell transfection
To silence the expression of ZDHHC12 in glioma cells, 3 short hairpin (sh) RNAs were designed and synthesized by Sangon Biotech Co. Ltd. (Shanghai, China). The shRNA sequences were: shRNA-1: 5'- GGAATCACGCTGGTGCTCTTC-3'; shRNA-2: 5'-GAATCACGCTGGTGCTCTTCC-3' and shRNA-3: 5'- GCACGATACCGAGCTGCGGCA-3'. The shRNAs were cloned into PcDNA 3.1 and prepared for subsequent transfection. DBTRG and U251 cells were used for transfection at the exponential stage. 1×106 cells were cultured in 6-well plates before transfection (with 2 mL above-mentioned medium for 24 hours) until they were 90% confluent.
Cell counting kit-8 assay (CCK-8)
A CCK-8 assay was conducted as per our previous study (30). Briefly, cellular viability was determined using a CCK-8 (Beyotime Biotechnology). After transfection, DBTRG and U251 cells were plated into 96-well plates (at a density of 500 cells per well). Then, incubated 10 µL CCK-8 solution with the cell medium to each well for another 2 hours at 37 ℃ daily. Using Multiskan FC Microplate spectrophotometer to detect the absorbance of each well at 570 nm. Data were showed as mean ± standard deviation (SD) and comparisons were analyzed by 2-way repeated measure analysis of variance (RM ANOVA) using GraphPad Prism 8.0.
Wound-healing assay
A wound-healing assay was conducted as per our previous study (30). Briefly, DBTRG and U251 cells were seeded in a 6-well plate evenly with the cell concentration of 1×106 per well. To inhibit cell division, serum-free medium containing 1 mM mitomycin was used to replace the complete medium. When the cells covered 90% of the plate base, the plate was perpendicularly scratched with a 10 µL sterile pipette tip Then removed the culture medium in the plate and softly washed 3 times with phosphate-buffered saline (PBS). To make the visual field clear for photography phase, the cell debris was carefully rinsed off. A medium containing 1% fetal bovine serum (FBS) was added to the 6-well plate. Introduced 3 mm wound of each plate across the diameter.
Transwell migration assay
A transwell migration assay was conducted as per our previous study (30). Briefly, cells in the logarithmic growth phase were seeded in the upper transwell chamber insert (Corning, USA) at a density of 2×104 cells per well. To inhibit cell division, serum-free medium containing 1 mM mitomycin was used to replace the complete medium. A 24-well plate was used to place the chamber, where the upper chamber containing FBS-free medium and the lower chamber containing 10% FBS complete medium. Cells cultured for 24 hours. Then discarded the medium and stained the cells with 1% crystal violet solution in order to present the amount of migrated cells.
Statistical analysis
Comparisons were calculated using 2-sided, unpaired t-tests or 2-way ANOVAs in GraphPad Prism 8.0. and data were presented as means ± SDs. All assays were replicated 3 times. A P value of less than 0.05 was considered statistically significant.
Results
ZDHHC12 expression increased and might predict poor prognosis in glioma
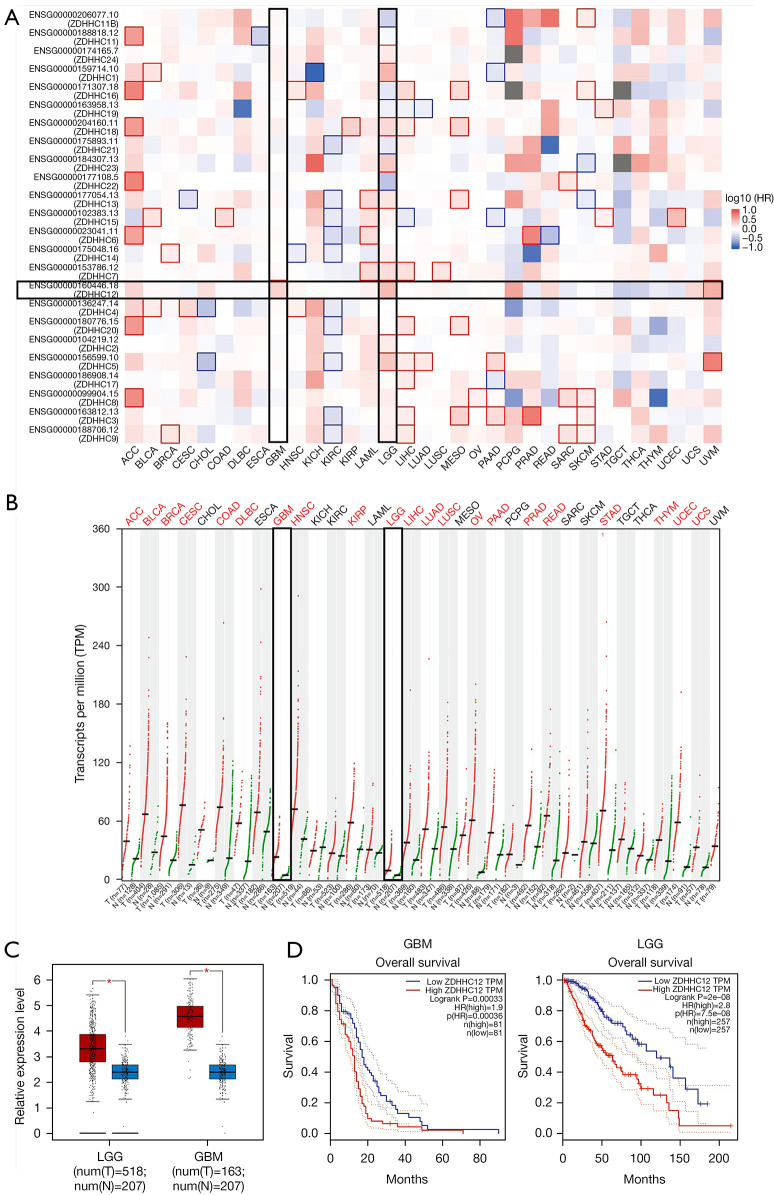
A pan-cancer prognosis heatmap was constructed using GEPIA 2.0 to investigate the prognostic significance of 24 members of the ZDHHC family in multiple cancer types included in The Cancer Genome Atlas (TCGA) database (23). Only ZDHHC12 showed significant performance as an unfavorable marker in glioblastoma (GBM) (Figure 1A). In low grade glioma (LGG), all 13 genes showed significant performance, with 9 genes (ZDHHC24, ZDHHC1, ZDHHC18, ZDHHC23, ZDHHC13, ZDHHC7, ZDHHC12, ZDHHC2, and ZDHHC5) showing unfavorable outcomes (hazard ratio >1, and P<0.05) and 4 genes (ZDHHC11B, ZDHHC16, ZDHHC21, and ZDHHC22) showing favorable outcomes (hazard ratio <1, and P<0.05). We chose ZDHHC12 for subsequent analysis, and its expression was compared in cancer and corresponding normal group (TCGA normal and GTEx data). Of the 33 cancer types analyzed, higher expression of ZDHHC12 was consistently observed in 21, including LGG and GBM, suggesting that ZDHHC12 may play an important role in multiple cancer types (Figure 1B). The differential expressions of ZDHHC12 in GBM and LGG can be seen in Figure 1C, while Figure 1D shows the Kaplan-Meier overall survival plots of ZDHHC12 in GBM and LGG.
Figure 1.
Analysis showing increased expression of ZDHHC12 as a hazardous marker in glioma. (A) Overall survival map of 24 members of the ZDHHC family. (B) Pan-cancer expression analysis showing upregulation of ZDHHC12 in multiple cancer types, including GBM and LGG. (C) Box plot showing significantly increased ZDHHC12 in GBM and LGG datasets. (D) Kaplan-Meier survival plot of ZDHHC12 in GBM and LGG. *, P<0.05. ZDHHC, zinc finger DHHC-type palmitoyltransferase; GBM, glioblastoma multiforme; LGG, low-grade glioma.
Validating the expression and prognostic relevance of ZDHHC12 in other glioma datasets
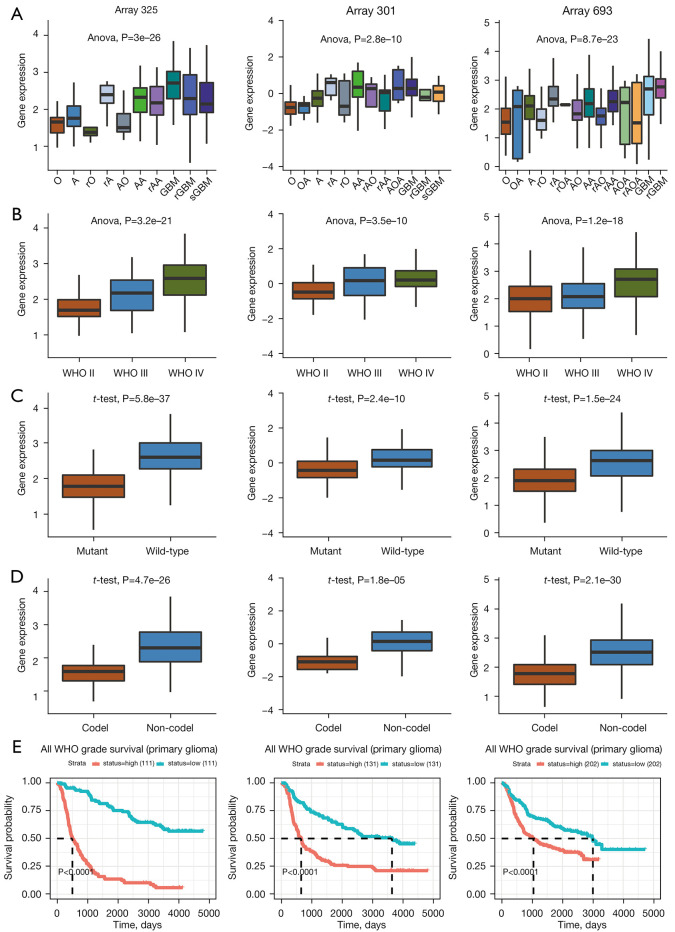
To further explore the clinical relevance of ZDHHC12 in glioma, we used data from the CGGA (24) and other datasets. In 3 of the CGGA datasets (array 325, array 301, and array 693), ZDHHC12 was found to be clinically relevant for histology types, WHO grades, isocitrate dehydrogenase (IDH) mutation, and 1p/19q co-deletion status. Higher expression was observed in GBM samples than in oligodendroglioma, astrocytoma, and other histological types of glioma (Figure 2A). ZDHHC12 showed consistent grade-dependent expression in groups classified according to the WHO grades (Figure 2B). For molecular subtypes, increased expression of ZDHHC12 was observed in IDH wild-type (IDH-WT) (Figure 2C) and non-1p/19q co-deletion (Figure 2D) glioma tissue. Finally, the Kaplan-Meier overall survival plots for the 3 datasets also supported TCGA datasets as unfavorable significant prognostic markers (P<0.0001) (Figure 2E).
Figure 2.
Expression and prognostic performance of ZDHHC12 in CGGA datasets. (A) Expression of ZDHHC12 in different histological types of glioma tissue. (B) Expression of ZDHHC12 in WHO grade II, III, and IV grades glioma tissue. (C) Abundance of ZDHHC12 in IDH mutant and wild-type glioma tissues. (D) ZDHHC12 expression in the 1p/19q co-deletion glioma tissue. (E) Kaplan-Meier survival plot of ZDHHC12 expression in patients with glioma. ZDHHC, zinc finger DHHC-type palmitoyltransferase; CGGA, the Chinese Glioma Gene Atlas; WHO, World Health Organization; IDH, isocitrate dehydrogenase.
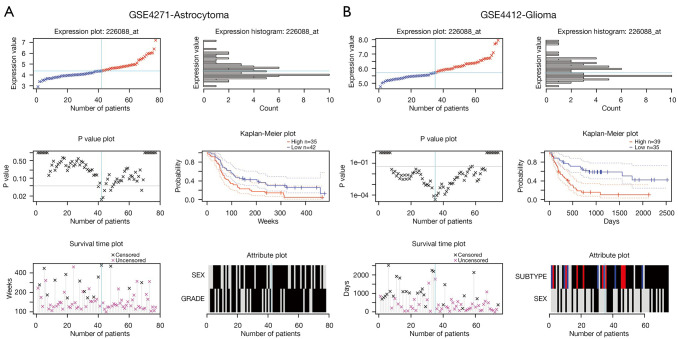
The prognostic significance of ZDHHC12 was then analyzed in other datasets using PrognoScan (25). ZDHHC12 showed prognostic significance in 2 datasets: GSE4271 (Figure 3A) and GSE4412 (Figure 3B). The GSE4271 dataset comprised high-grade astrocytoma samples, and GSE4412 included diffuse infiltrating glioma samples. Together, these results suggest that ZDHHC12 is upregulated, correlates with cancer progression, and can predict poor survival outcomes in glioma.
Figure 3.
Prognostic significance of ZDHHC12 in the GSE4271 (A) and GSE4412 (B) datasets. ZDHHC, zinc finger DHHC-type palmitoyltransferase.
Silencing ZDHHC12 repressed cancer growth, migration, and invasion in glioma
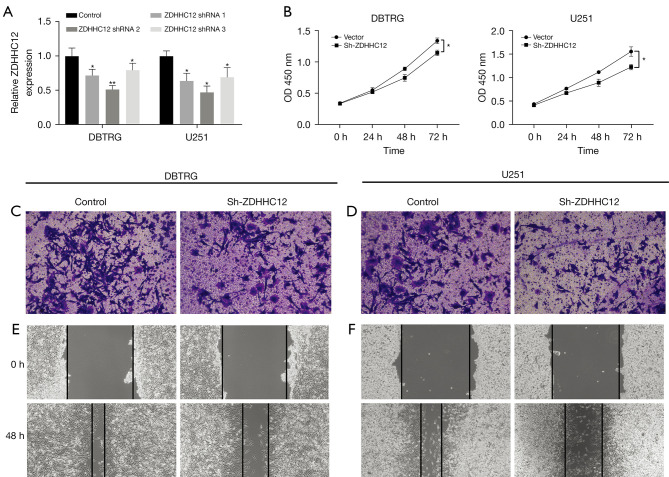
To further explore the function of ZDHHC12 in glioma, we designed 3 shRNAs to intervene in ZDHHC12 expression in DBTRG and U251 glioma cells. First, the knockdown efficiency was examined using qRT-PCR, and shRNA2 showed the overall best silencing, with a capability of more than 50% (Figure 4A). Therefore, we chose shRNA2 for the following phenotype assays. A CCK8 assay was conducted to study both cell lines with decreased ZDHHC12 expression. Both cell lines exhibited repressed growth capability in the ZDHHC12 silenced group at 72 hours (Figure 4B), suggesting that ZDHHC12 may contribute to glioma cell proliferation. Next, the function of ZDHHC12 on glioma cells was evaluated through transwell and wound healing assays for both cell lines. Compared to cells without ZDHCC silencing, ZDHHC12-silenced cells had significantly fewer trans-membrane cells at 48 hours in both cell lines (Figure 4C,4D, P<0.01). Finally, wound healing assays were conducted to analyze the migration potential of ZDHHC12 silenced cells. ZDHHC12 silenced cells showed a wider gap at 48 hours (Figure 4E,4F), implying that silencing ZDHHC12 may also reduce metastasis-related malignant behavior in glioma.
Figure 4.
Decreased ZDHHC12 attenuates the proliferation, invasion and migration in glioma cells. (A) The expression of ZDHHC12 was silenced in DBTRG and U251 glioma cells. (B) ZDHHC12 silencing repressed cell growth in DBTRG and U251 glioma cells. (C,D) Transwell assays showing that ZDHHC12 knockdown cells exhibited reduced invasion potential. The cells were stained with 1% crystal violet solution to observe the number of migrated cells. Magnification of image, 100×. (E,F) Wound healing assays demonstrating deceased migration ability in ZDHHC12 silenced glioma cells. ZDHHC, zinc finger DHHC-type palmitoyltransferase. *, P<0.05; **, P<0.01. Magnification of images, 40×.
ZDHHC12 hypomethylation to predict prognosis in glioma
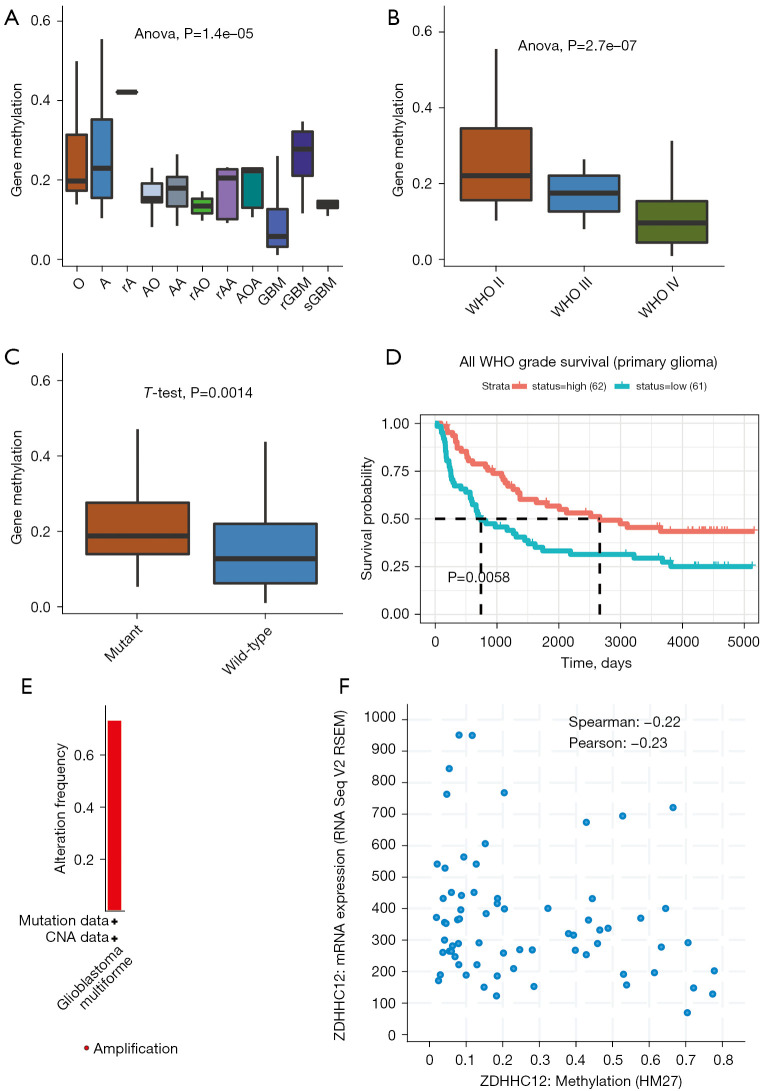
Finally, to investigate the molecular basis for increased ZDHHC12 expression in glioma, we analyzed the methylation levels in the CGGA database samples. Interestingly, in direct contrast to ZDHHC12 mRNA expression trends, the methylation level of ZDHHC12 decreased in GBM samples, compared with the other glioma histology types, such as oligodendroglioma and astrocytoma, among others (Figure 5A). Similarly, ZDHHC12 methylation also decreased in advanced WHO grades (II and III) (Figure 5B) and IDH-WT groups (Figure 5C), suggesting that the methylation score of ZDHHC12 is correlated with glioma progression. Next, by using the methylation level as a marker, we examined the prognostic significance of ZDHHC12 in glioma. Notably, glioma patients with higher ZDHHC12 methylation had a worse overall survival time (Figure 5D), implying that ZDHHC12 methylation may serve as an unfavorable biomarker. Finally, as ZDHHC12 exhibited low frequencies of genomic amplifications and deletions in TCGA glioma datasets (Figure 5E), we analyzed the relationship between ZDHHC12 methylation and mRNA levels. Moderate negative correlation (Spearman’s =−0.22, and Pearson’s =−0.23) was observed between methylation and mRNA expression, suggesting that increased mRNA expression of ZDHHC12 may be attributable to ZDHHC12 hypomethylation (Figure 5F).
Figure 5.
Elevated DNA methylation may contribute to increased expression of ZDHHC12 in glioma. (A) The DNA methylation level of ZDHHC12 significantly decreased in different histological types of glioma tissue (CGGA datasets). (B) The DNA methylation level of ZDHHC12 significantly decreased in glioma tissues with a higher WHO grade (CGGA datasets). (C) The DNA methylation level of ZDHHC12 significantly decreased in IDH wild-type glioma tissues (CGGA datasets). (D) Kaplan-Merrier survival plot of ZDHHC12 methylation level in primary glioma (CGGA datasets). (E) Low amplification was observed in glioma samples (TCGA dataset). (F) Significant negative correlation was observed between ZDHHC12 methylation and expression level (TCGA dataset). ZDHHC, zinc finger DHHC-type palmitoyltransferase; CGGA, Chinese Glioma Genome Atlas; TCGA, the Cancer Genome Atlas, WHO, World Health Organization; IDH, isocitrate dehydrogenase.
In conclusion, we first identified ZDHHC12 as an unfavorable prognostic marker in glioma, and loss-of-function assays suggest that ZDHHC12 may promote cancer cell growth and migration. Decreased DNA methylation may lead to elevated expression of ZDHHC12 in glioma.
Discussion
Protein palmitoylation levels are controlled by members of the DHHC family of protein palmitoyl transferases (PATs) that play oncogenic and tumor-suppressing roles in cancer (12,16,31-33). Therefore, targeting S-palmitoylation or the enzymes responsible for palmitoylation dynamics might be a therapeutic strategy for certain cancers (14,34-37). The role of the ZDHHC family in cancer has been widely reported. For example, ZDHHC5 has been found to regulate cancer stem cells and associated with malignant prognosis in lung adenocarcinoma (38), ZDHHC9 has been found to induce and stabilize PD-L1 protein, then drive cancer cells from immune surveillance in breast cancer (39), and the melanocortin-1 receptor palmitoylated by ZDHHC13 has been found to promote pigmentation and protect against melanoma (17).
In this study, a comprehensive analysis of ZDHHC12 in pan-cancer found that ZDHHC12 was upregulated and could predict poor prognosis in GBM and LGG. Further cellular assays showed that ZDHHC12 promoted glioma tumorigenesis. In previous studies, the role of ZDHHC12 has been shown to be implicated in neurological disorders like Huntington’s disease, Alzheimer’s disease, and schizophrenia (40). In human primary osteoblasts, it has also been found to be involved in 1,25 (OH)D, leading to improved viability and function (41). Furthermore, the knockdown of ZDHHC12 in ovarian cancer has been shown to significantly inhibit CLDN3 accurate membrane localization, protein stability, and ovarian cancer cell tumorigenesis (42). To our knowledge, the present study is the first to reveal the clinical significance and cellular function of ZDHHC12 in glioma.
Another important finding in this study was the hypomethylation of ZDHHC12 in glioma. Gene expression is governed by multiple processes on transcriptional and post-transcriptional levels. Given that the genomic alteration frequencies of ZDHHC12 in glioma were low, we investigated the epigenetic elements influencing increased ZDHHC12 abundance in high-grade glioma. Notably, decreased ZDHHC12 methylation was found in advanced glioma groups and IDH WT samples, suggesting that hypomethylation in the ZDHHC12 gene may occur prior to increased ZDHHC12 expression, providing an early screening marker for glioma. In addition, the methylation levels of ZDHHC12 were found to be relevant in predicting glioma survival outcomes and times, which may also benefit patients. The exact regulatory mechanism of DNA methylation with mRNA expression still needs to be studied to explore DNA elements and transcriptional regulons within.
In conclusion, although in vitro cellular assays proved that ZDHHC12 has a role in glioma tumorigenesis and progression, further research is required, including the in vivo study of ZDHHC12 using gain-of-function technologies to assess rescue strategies. Also, the exact substrate of ZDHHC12 in glioma is not clear, even though claudin 3 (CLDN3) emerged as the target of CLDN3 in ovarian cancer (42). Finally, more in-house glioma samples and related clinical data will need to be collected in the future to assess the performance of ZDHHC12 in glioma diagnosis and prognosis.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Fujian Province (No. 2019J01500).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-520/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-520/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-520/coif). The authors have no conflicts of interest to declare.
References
- 1.Wang H, Xu T, Huang Q, et al. Immunotherapy for Malignant Glioma: Current Status and Future Directions. Trends Pharmacol Sci 2020;41:123-38. 10.1016/j.tips.2019.12.003 [DOI] [PubMed] [Google Scholar]
- 2.Weller M, Wick W, Aldape K, et al. Glioma. Nat Rev Dis Primers 2015;1:15017. 10.1038/nrdp.2015.17 [DOI] [PubMed] [Google Scholar]
- 3.Nicholson JG, Fine HA. Diffuse Glioma Heterogeneity and Its Therapeutic Implications. Cancer Discov 2021;11:575-90. 10.1158/2159-8290.CD-20-1474 [DOI] [PubMed] [Google Scholar]
- 4.Dai C, Celestino JC, Okada Y, et al. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev 2001;15:1913-25. 10.1101/gad.903001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomiyama A, Ichimura K. Signal transduction pathways and resistance to targeted therapies in glioma. Semin Cancer Biol 2019;58:118-29. 10.1016/j.semcancer.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 6.Chaligne R, Gaiti F, Silverbush D, et al. Epigenetic encoding, heritability and plasticity of glioma transcriptional cell states. Nat Genet 2021;53:1469-79. 10.1038/s41588-021-00927-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weller M, van den Bent M, Preusser M, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 2021;18:170-86. 10.1038/s41571-020-00447-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sturm D, Pfister SM, Jones DTW. Pediatric Gliomas: Current Concepts on Diagnosis, Biology, and Clinical Management. J Clin Oncol 2017;35:2370-7. 10.1200/JCO.2017.73.0242 [DOI] [PubMed] [Google Scholar]
- 9.Meel MH, Kaspers GJL, Hulleman E. Preclinical therapeutic targets in diffuse midline glioma. Drug Resist Updat 2019;44:15-25. 10.1016/j.drup.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 10.Rowland EA, Snowden CK, Cristea IM. Protein lipoylation: an evolutionarily conserved metabolic regulator of health and disease. Curr Opin Chem Biol 2018;42:76-85. 10.1016/j.cbpa.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R, Sahu M, Srivastava D, et al. Post-translational modifications: Regulators of neurodegenerative proteinopathies. Ageing Res Rev 2021;68:101336. 10.1016/j.arr.2021.101336 [DOI] [PubMed] [Google Scholar]
- 12.Jin J, Zhi X, Wang X, et al. Protein palmitoylation and its pathophysiological relevance. J Cell Physiol 2021;236:3220-33. 10.1002/jcp.30122 [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Hannoush RN. A Decade of Click Chemistry in Protein Palmitoylation: Impact on Discovery and New Biology. Cell Chem Biol 2018;25:236-46. 10.1016/j.chembiol.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 14.Ko PJ, Dixon SJ. Protein palmitoylation and cancer. EMBO Rep 2018;19:e46666. 10.15252/embr.201846666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu J, Sun Y, Chen B, et al. Fatty acids and cancer-amplified ZDHHC19 promote STAT3 activation through S-palmitoylation. Nature 2019;573:139-43. 10.1038/s41586-019-1511-x [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Du W, Hua F, Li X, et al. Loss of Optineurin Drives Cancer Immune Evasion via Palmitoylation-Dependent IFNGR1 Lysosomal Sorting and Degradation. Cancer Discov 2021;11:1826-43. 10.1158/2159-8290.CD-20-1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Zhu B, Yin C, et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature 2017;549:399-403. 10.1038/nature23887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Ma H, Wang Z, et al. EZH2 Palmitoylation Mediated by ZDHHC5 in p53-Mutant Glioma Drives Malignant Development and Progression. Cancer Res 2017;77:4998-5010. 10.1158/0008-5472.CAN-17-1139 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Li X, Yang F, et al. DHHC9-mediated GLUT1 S-palmitoylation promotes glioblastoma glycolysis and tumorigenesis. Nat Commun 2021;12:5872. 10.1038/s41467-021-26180-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan X, Yang H, Zhao C, et al. Local anesthetics impair the growth and self-renewal of glioblastoma stem cells by inhibiting ZDHHC15-mediated GP130 palmitoylation. Stem Cell Res Ther 2021;12:107. 10.1186/s13287-021-02175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan X, Fan J, Yang H, et al. Heterogeneity of subsets in glioblastoma mediated by Smad3 palmitoylation. Oncogenesis 2021;10:72. 10.1038/s41389-021-00361-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Liu C, Xiao M, et al. Bioinformatics Analysis of the Prognostic and Biological Significance of ZDHHC-Protein Acyltransferases in Kidney Renal Clear Cell Carcinoma. Front Oncol 2020;10:565414. 10.3389/fonc.2020.565414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. 10.1093/nar/gkz430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z, Zhang KN, Wang Q, et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genomics Proteomics Bioinformatics 2021;19:1-12. 10.1016/j.gpb.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuno H, Kitada K, Nakai K, et al. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2009;2:18. 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9:157-73. 10.1016/j.ccr.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 27.Costa BM, Smith JS, Chen Y, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res 2010;70:453-62. 10.1158/0008-5472.CAN-09-2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin K, Taylor JR, Jr, Wu TD, et al. TMEFF2 is a PDGF-AA binding protein with methylation-associated gene silencing in multiple cancer types including glioma. PLoS One 2011;6:e18608. 10.1371/journal.pone.0018608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res 2004;64:6503-10. 10.1158/0008-5472.CAN-04-0452 [DOI] [PubMed] [Google Scholar]
- 30.Wei Shen S, Lin K, et al. NPC2 as a Prognostic Biomarker for Glioblastoma Based on Integrated Bioinformatics Analysis and Cytological Experiments. Front Genet 2021;12:611442. 10.3389/fgene.2021.611442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser NJ, Howie J, Wypijewski KJ, et al. Therapeutic targeting of protein S-acylation for the treatment of disease. Biochem Soc Trans 2020;48:281-90. 10.1042/BST20190707 [DOI] [PubMed] [Google Scholar]
- 32.Resh MD. Palmitoylation of proteins in cancer. Biochem Soc Trans 2017;45:409-16. 10.1042/BST20160233 [DOI] [PubMed] [Google Scholar]
- 33.Anderson AM, Ragan MA. Palmitoylation: a protein S-acylation with implications for breast cancer. NPJ Breast Cancer 2016;2:16028. 10.1038/npjbcancer.2016.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin DTS, Davis NG, Conibear E. Targeting the Ras palmitoylation/depalmitoylation cycle in cancer. Biochem Soc Trans 2017;45:913-21. 10.1042/BST20160303 [DOI] [PubMed] [Google Scholar]
- 35.Fhu CW, Ali A. Protein Lipidation by Palmitoylation and Myristoylation in Cancer. Front Cell Dev Biol 2021;9:673647. 10.3389/fcell.2021.673647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan M, Song ZH, Ying MD, et al. N-myristoylation: from cell biology to translational medicine. Acta Pharmacol Sin 2020;41:1005-15. 10.1038/s41401-020-0388-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang WH, Yuan T, Qian MJ, et al. Post-translational modification of KRAS: potential targets for cancer therapy. Acta Pharmacol Sin 2021;42:1201-11. 10.1038/s41401-020-00542-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Li F, Fu K, et al. Potential Role of S-Palmitoylation in Cancer Stem Cells of Lung Adenocarcinoma. Front Cell Dev Biol 2021;9:734897. 10.3389/fcell.2021.734897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Hsu JM, Sun L, et al. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res 2019;29:83-6. 10.1038/s41422-018-0124-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young FB, Butland SL, Sanders SS, et al. Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases. Prog Neurobiol 2012;97:220-38. 10.1016/j.pneurobio.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 41.Al Saedi A, Myers DE, Stupka N, et al. 1,25(OH)2D3 ameliorates palmitate-induced lipotoxicity in human primary osteoblasts leading to improved viability and function. Bone 2020;141:115672. 10.1016/j.bone.2020.115672 [DOI] [PubMed] [Google Scholar]
- 42.Yuan M, Chen X, Sun Y, et al. ZDHHC12-mediated claudin-3 S-palmitoylation determines ovarian cancer progression. Acta Pharm Sin B 2020;10:1426-39. 10.1016/j.apsb.2020.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as