Abstract
Several variants of multicolor single-molecule localization microscopy (SMLM) have been developed to resolve the spatial relationship of nanoscale structures in biological samples. The oligonucleotide-based SMLM approach “DNA-PAINT” robustly achieves nanometer localization precision and can be used to count binding sites within nanostructures. However, multicolor DNA-PAINT has primarily been realized by “Exchange-PAINT”, which requires sequential exchange of the imaging solution and thus leads to extended acquisition times. To alleviate the need for fluid exchange and to speed up the acquisition of current multichannel DNA-PAINT, we here present a novel approach that combines DNA-PAINT with simultaneous multicolor acquisition using spectral demixing (SD). By using newly designed probes and a novel multichannel registration procedure, we achieve simultaneous multicolor SD-DNA-PAINT with minimal crosstalk. We demonstrate high localization precision (3–6 nm) and multicolor registration of dual- and triple-color SD-DNA-PAINT by resolving patterns on DNA origami nanostructures and cellular structures.
Keywords: Super-resolution, single-molecule imaging, fluorescence microscopy, single-molecule localization microscopy, DNA-PAINT, multicolor, spectral demixing
Introduction
Single-molecule localization microscopy (SMLM) became a widely used method to investigate biological nanostructures with unprecedented spatial resolution in fluorescence-based light microscopy.1−3 To investigate the spatial relationships between different protein structures on the nanoscale, the precision and applicability of multicolor SMLM is of central importance. However, the development of robust multicolor SMLM approaches was challenged by several factors. These include a limited number of well-performing photoswitchable fluorophores, unwanted color channel crosstalk, chromatic aberrations, multicolor registration errors, long acquisition times, and an increasing complexity in the required hardware. Over the past decade, several optimized SMLM approaches that overcome at least some of these limitations have been developed.
Recently, DNA point accumulation for imaging in nanoscale topography (DNA-PAINT) has emerged as a promising SMLM method.4,5 DNA-PAINT uses transient binding of freely diffusing fluorescently labeled oligonucleotides (“imager strands”, or in short, “imagers”) to complementary DNA oligonucleotides (“docking strands”) that are linked to the target structure (i.e., antibody against protein of interest). The transient binding of the imagers produces the required on/off blinking for SMLM. In contrast to dSTORM,6 DNA-PAINT does not require a chemical switching buffer and is largely insensitive to photobleaching, as the imagers are constantly exchanged within the imaging volume. By taking advantage of bright organic fluorophores and the controlled binding kinetics of the imagers, DNA-PAINT can be used to determine the number of binding sites within a biological sample with high spatial resolution.7
Multicolor DNA-PAINT can be achieved by using multiple docking/imager strand pairs with orthogonal sequences. In the recently developed “Exchange-PAINT” method, multiplexing is realized by sequential fluid exchange of the imaging buffer that contains distinct imagers for each channel.4 This approach reaches very high localization precisions and is insensitive to chromatic errors by using the same dye in each acquisition round. Exchange-PAINT, however, relies on the complete exchange of the imagers containing buffer in between the acquisition phases of each channel. Each fluid exchange, including the washing steps, is prone to cause extended sample drift through thermal or pressure gradients and therefore requires precise drift-correction and multichannel registration. The combination of long exposure times that are needed to average out the noise from diffusing imagers and the sequential color channel acquisition leads to long total acquisition periods. By optimizing the DNA sequences and buffer conditions, the acquisition time was dramatically shortened.8,9 However, these improved multicolor DNA-PAINT approaches were still limited by the fluid exchange that was required for sequential acquisition.
Multicolor DNA-PAINT without fluid exchange has recently been achieved by modulation of the excitation frequency.10 However, this approach (fm-DNA-PAINT) requires a minimum of three rapidly triggered lasers and suffers from chromatic aberrations like in any other multicolor method with spectrally separate fluorophores. Compared to DNA-PAINT, fm-DNA-PAINT was limited to two color channels with a compromised spatial resolution. Another DNA-PAINT variant used three spectrally different dyes and different binding frequencies to achieve sequential multicolor DNA-PAINT without fluid exchange.11 This highly multiplexed approach massively speeds up fluorescence in situ hybridization, however, it relies on separable point-like objects and cannot resolve dense cellular structures.
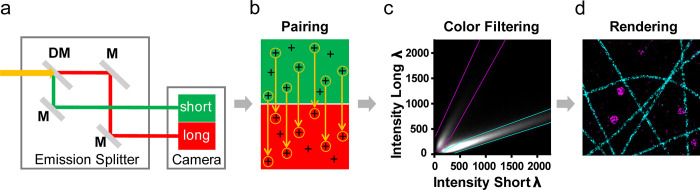
We recently developed a robust multicolor dSTORM approach that achieved high localization precision with fast, simultaneous, dual-color acquisition based on spectral demixing (SD).12−15 The SD-mode uses spectrally overlapping fluorophores excited by a single laser line and a simple dichroic-based emission splitter to image short and long wavelength components of the emission on two sides of the same camera (Figure 1a). By using a custom open-source software tool (SD-Mixer216), the single-molecule localizations are “paired”, while nonpaired localizations, from which a large part is random noise, are excluded (Figure 1b). Depending on their emission spectra, localizations from each dye display a spatially distinct population within the 2D intensity histogram of long and short channel intensity values (Figure 1c). By applying binary masks to exclude or include populations of the intensity distribution, the colors are assigned to each paired localization before multicolor rendering (Figure 1d). These color-separation masks are optimized for maximal inclusion of localizations and minimal crosstalk between the channels.
Figure 1.
Spectral demixing (SD) DNA-PAINT principle and workflow. (a) Schematic beam path of the emission splitter installed between the microscope body (not displayed) and camera system (indicated). All dyes (here, dual-color example with ATTO-655/700) are excited by a single laser line (e.g., 647 nm), and the mixed emission (yellow) is split via a dichroic mirror (DM) and 100% mirrors (M) into “short” (green) and “long” (red) wavelength channels onto a single camera. (b) The localizations detected in both channels are subjected to a custom “pair-finding” algorithm that identifies corresponding localization pairs. Random (unpaired) localizations are excluded. (c) The channel-specific intensity values (short/long) of all localization pairs are plotted into a 2D intensity histogram. Color-specific masks (cyan/magenta dotted lines) are designed to minimize color crosstalk and maximize inclusion of localizations. Each included localization is assigned to a color channel according to the masks. (d) Example of a rendered dual-color SD-DNA-PAINT image of microtubules (cyan) and clathrin-coated vesicles (magenta).
To take advantage of the benefits of DNA-PAINT but to avoid the need of fluid exchange for multicolor imaging, we here present a novel approach that combines spectral demixing (SD) with DNA-PAINT. In “SD-DNA-PAINT”, multiple channels are acquired simultaneously not sequentially. Compared to Exchange-PAINT, SD-DNA-PAINT effectively speeds up the acquisition time by x-fold for an x-color experiment while keeping the benefits. By avoiding fluid exchange, sample drift and potential errors in multichannel registration are minimized. Using a new combination of spectrally overlapping dyes (ATTO-655/680/700), we achieved simultaneous triple-color SD-DNA-PAINT with maximal localization precision and minimal crosstalk. We demonstrate the feasibility of SD-DNA-PAINT by resolving known nanostructures within DNA origamis and cells.
Results and Discussion
To perform robust multicolor SD-DNA-PAINT, we first established a combination of organic dyes suited for this approach. In principle, the choice of dye is not limited to any specific spectral region. The far-red emitting ATTO-655 dye was successfully used to perform DNA-PAINT using a 647 nm laser.5 The use of far-red dyes offers the flexibility to add additional non-super-resolved channels with dyes excited in the visible range (405/488/561 nm) to the SD-based experiment.13 We therefore chose spectrally close and bright, stable dyes (ATTO-633/643/655/680/700) to be evaluated as potential multicolor SD-DNA-PAINT candidates. A detailed description of the dye selection is outlined in the Supporting Information (Figure S1, Text S1). In short, the evaluation criteria for the dyes included their spectral property (emission spectra at 647 nm excitation) and the final image quality (brightness, background).
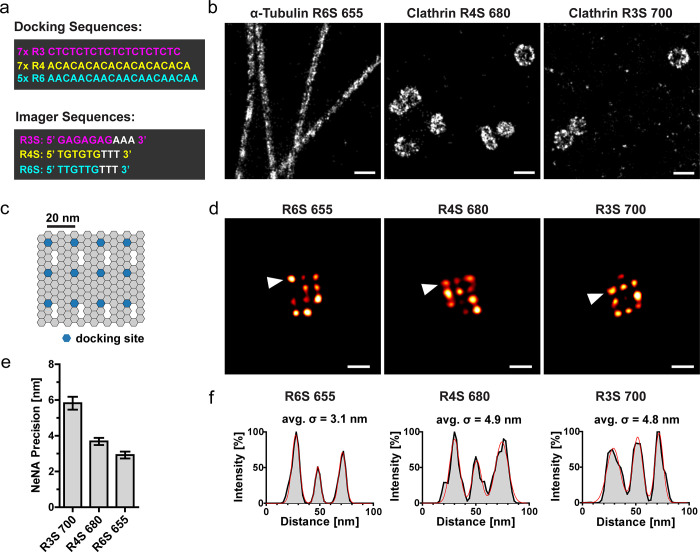
Based on these investigations, we chose ATTO-655, ATTO-680, and ATTO-700 as promising dye candidates for triple-color SD-DNA-PAINT. To allow speed-optimized and simultaneous imaging of three distinct dye-imager conjugates within the same buffer, we chose three concatenated docking sequences (R3/R4/R6) that were recently introduced.9 We then designed three compatible imager sequences with matching melting points and a short spacer between the dye and hybridization site to avoid guanine quenching of the ATTO-dyes (R3S/R4S/R6S, Figure 2a).24 To evaluate the image quality in SD-mode, we first used these optimized imagers to perform single-color DNA-PAINT experiments in SD-mode on COS-7 cells immunolabeled for α-tubulin or clathrin (Figure 2b). Image processing was done as described below (Figure S2a, without color-filtering).26−30 The nanostructures of microtubules and clathrin-coated vesicles were well-resolved.
Figure 2.
Image quality and localization precision. (a) Schematic of the oligonucleotide sequences of the docking strands (R3, R4, R6) and the optimized imager strands (R3S, R4S, R6S). (b) COS-7 cells were immunolabeled with primary antibodies against nanostructures (microtubules and clathrin-coated vesicles) and secondary nanobodies conjugated to unique docking strands. Samples were imaged in SD-mode without color-filtering, using the imagers coupled to the ATTO-dyes (R6S-655, R4S-680, R3S-700) as indicated. Recording modality: 20 000 frames for ATTO 680; 30 000 frames for ATTO (655/700), 100 ms exposure, 1 nM imager concentration. Selected regions show super-resolved nanostructures. Scale bar: 200 nm. (c) Schematic of the DNA origami structure with a 3 × 4 docking site arrangement with 20 nm grid distances. (d–f) Immobilized 20 nm DNA origamis with the docking sequences (R3, R4, R6) were imaged in SD-mode without color-filtering, using the optimized imager strands (R3S, R4S, R6S) coupled to the ATTO-dyes (655/680/700) as indicated. Recording modality: 20 000 frames, 100 ms exposure, 1 nM imager. (d) Representative SD-DNA-PAINT images of DNA origamis. Scale bar: 40 nm. (e) The NeNA precision was calculated on the same single-color data set as the images in (d). Mean ± SEM, images: 655 (n = 5), 680 (n = 4), 700 (n = 3). (f) Line profiles through the origamis (arrowheads in (d)) were fitted with a multi-Gaussian distribution (red line). The standard deviation of each single Gaussian was averaged (avg. σ).
To determine the localization precision and to clearly demonstrate the nanoscale resolution, we next performed single-color DNA-PAINT experiments using the optimized imagers on 20 nm DNA origami grids (Figure 2c).23 The imaging was done in SD-mode, and the data was processed as described below (Figure S2a, without color-filtering). The resulting images clearly show the resolved 20 nm patterns of the origami structures (Figure 2d). Next, we determined the experimental localization precision by performing a nearest neighbor analysis on subsequent localizations (NeNA precision).17,18,31−33 The values were between 3 and 6 nm for all used ATTO-dyes (Figure 2e), similar to previously achieved precisions using Cy3b-labeled imagers in Exchange-PAINT (5 nm9). To verify this precision, including all errors that could occur through data processing steps (e.g., bead-based drift-correction: Figure S2b,c), we performed intensity line scans through the origami spots. The standard deviation (σ) of the Gaussian fits along the line scans were between 3.1 and 4.8 nm, demonstrating that the NeNA precision (Figure 2e) is an accurate estimate for the experimentally achieved localization precision (Figure 2f).
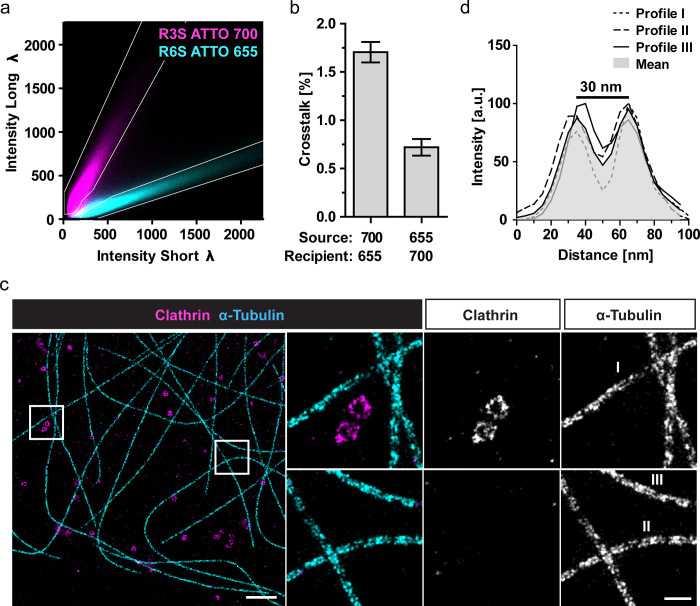
To perform dual-color SD-DNA-PAINT, we chose the ATTO-655/700-imagers as those showed spectrally clearly distinct intensity populations (Figure S1c,e). To design color-separation masks, we performed single-color SD-DNA-PAINT experiments on COS-7 cells immunolabeled for abundant cellular structures (microtubules, clathrin). We used secondary nanobodies coupled to the described docking strands (R3/R6) and acquired the data in SD-mode using the corresponding (optimized) imagers (R6S-655/R3S-700).25 The color-separation masks were designed to minimize the channel crosstalk while maximizing the inclusion of localizations (Figure 3a). For that, we quantified the crosstalk from the R6S-655 channel (source) into the R3S-700 channel (recipient) and vice versa based on the single-color ground truth data. The masks were designed based on merged single-color intensity histograms to tolerate a maximal color crosstalk of 2% between both channels (Figure 3b). These masks included more than 70% of all localizations for the reconstruction of the images (Figure S3a).
Figure 3.
Dual-color SD-DNA-PAINT. (a,b) Single-color DNA-PAINT experiments in SD-mode using COS-7 cells immunolabeled for either clathrin (R3) or α-tubulin (R6). Recording modality: 20 000 frames, 100 ms exposure, 1 nM imager concentration. (a) Pseudocolor overlay of the 2D intensity histograms from separate single-color experiments (cyan: R6S-ATTO-655, magenta: R3S-ATTO-700, both: average of six experiments). Color-separation masks (white lines) were manually designed within the 2D intensity histogram based on the single-color experiments. (b) The crosstalk was calculated as the percentage of localizations from the source (i.e., ATTO-700) that were detected within the mask of the recipient channel (i.e., ATTO-655) based on the single-color ground truth and vice versa. Mean ± SEM, N = 4–5 images per channel. (c) COS-7 cells were immunolabeled for clathrin (R3) or α-tubulin (R6) simultaneously. Samples were imaged simultaneously in SD-mode (including color-filtering) using the imagers from (a). Representative dual-color SD-DNA-PAINT image of microtubules (R6S-655) and clathrin (R3S-700) with enlarged regions (white boxes) are shown. Scale bar: 1 μm, zoom: 200 nm. Both enlarged regions have the same contrast. Recording modality: 30 000 frames, 100 ms exposure, 1 nM total imager concentration. (d) Intensity line profiles across the indicated microtubules shown in (c). The mean profile shows a “valley-to-peak” intensity ratio of ∼50% (dotted red line).
Next, we performed a dual-color SD-DNA-PAINT experiment on immunolabeled cells, stained for microtubules and clathrin with both imagers (R6S-655/R3S-700) in the buffer (Figure 3c). The data was processed using the optimized color-separation masks as described above (Figure S2a). The resulting dual-color SD-DNA-PAINT images show microtubules and clathrin structures clearly separated with a very low crosstalk. The nanoscale structures of both tubular microtubules (double line) and spherical clathrin-coated vesicles (ring structure) are clearly resolved. The peak-to-peak distance of the intensity line profiles across the microtubules (30 nm, Figure 3d) is in agreement with the distances (∼29 nm) that have previously been calculated for similarly labeled microtubules.19
The steps used to process the imaging data for dual-color SD-DNA-PAINT include the localization of single-molecule emissions, the drift-correction, the pairing and grouping of the localizations, the color-filtering, and the rendering of the multicolor image (Figure S2a). Here, we would like to discuss the color-filtering step in more detail and highlight the implementation of a novel multichannel registration procedure to further improve the precision and image quality of any SD-based approach, including SD-DNA-PAINT or SD-dSTORM.
One seemingly critical point of the SD-mode was the exclusion of localizations through the color-filtering using the color-separation masks.12 In dual-color SD-DNA-PAINT, for example, about 30% (Figure S3a) of the initial localizations plotted within the 2D intensity histogram (Figure 3a) are excluded by the masks to keep the crosstalk below 2% (Figure 3b). This exclusion might be conceived as a disadvantage. However, most of the excluded localizations in fact had low-intensity values (Figure 3a) and were therefore less precise compared to the included localizations with high intensities. To demonstrate that, we reconstructed images from cellular nanostructures (Figure S2d) and 20 nm DNA origami grids (Figure S2e) using the included or excluded localizations of several dyes. Meanwhile, for some conditions, structures were still partially visible in the images rendered from the excluded localizations, these images were of much lower quality and contained grossly deteriorated nanostructures in comparison to the correctly filtered (“included”) images. The applied color-filtering procedure has a similar effect as a linear low-intensity threshold (Figure S2f); it reduces the number of low-quality localizations, and therefore enhances the quality of the super-resolved images (Figure S2d,e). The unique feature of the SD-mode is that the balance between the permitted level of crosstalk and the required density of localizations for structural analysis can be tuned by the color-filtering step.
In multicolor SMLM, the accuracy of color channel registration is of central importance for any experiment in which relative nanoscale distances between different objects are measured. In SD-mode, even mild flat-field distortions within the optical path (e.g., emission splitter) could contribute significantly to an error in registration of the short and long wavelength channels. Originally, potential errors in multichannel registration were prevented by reconstructing the final image using only the single-molecule localizations of one (i.e., short) wavelength channel.12,13 This resulted in a partial loss of the single-molecule emissions that were split into the long wavelength channel. Here, we optimized this approach by harvesting the single-molecule emissions from both channels to achieve the highest possible localization precision. To correct for nanoscale distortions between the split channels, we designed a custom registration and unwarping procedure that was implemented after the color-filtering step (Figure S2a). In this procedure, we used the entire population of paired localizations as intrinsic fiducial markers for correction of the local offset between the split channels.
In addition to this registration procedure, we used the intensity values of the localization in the short and long wavelength channels to calculate an intensity-weighted localization of each localization pair. This intensity-weighted localization correction ensured that we take advantage of the entire detected emission from each single-molecule event to determine its precise localization. Compared to using only one of the channels (i.e., short), this novel intensity-weighted multichannel registration procedure significantly improved the image quality on cellular nanostructures (Figure S2d) and 20 nm DNA origami grids (Figure S2e). Importantly, the localization precision measured from DNA origamis was significantly improved using this procedure. For ATTO-700, which has the weakest intensities in the short wavelength channel, the NeNA precision was almost doubled from 11 to 6 nm (Figure S2g). Importantly, this intensity-weighted multichannel registration procedure uses the localizations of the structure under investigation as fiducial markers rather than exogenously added fiducials (e.g., nanogold). That way, the structural signal governs the corrections rather than the signal from regions that are normally excluded from the final image.
As discussed earlier, DNA-PAINT achieves higher localization precisions compared to dSTORM approaches, because it offers a higher signal-to-noise ratio for the single-molecule emissions. To assess the difference in performance between dSTORM and DNA-PAINT using SD-mode, we directly compared the recently established multicolor SD-dSTORM12,13 with SD-DNA-PAINT. Since SD-dSTORM was so far limited to two colors, we directly compared it to dual-color SD-DNA-PAINT using the same microscope system. For SD-dSTORM, the emission splitter was equipped with a distinct dichroic mirror for optimal color separation of the specific dyes. Again, we used COS-7 cells immunolabeled for microtubules and clathrin with method-specific secondary antibodies. For direct comparison of the image quality, we used the same laser power settings and image processing steps as described above for SD-DNA-PAINT (Figure S2a). SD-DNA-PAINT achieved a significantly higher localization precision compared to SD-dSTORM (Figures 2e, S4c). In line with that, the cellular nanostructures in the SD-dSTORM images (double line of microtubules, ring structure of clathrin-coated vesicles, Figure S4a) were not as clearly resolved as in the SD-DNA-PAINT images (Figure 3c). The intensity line profiles across the microtubules showed more prominent “valley-to-peak” ratios for SD-DNA-PAINT (Figure 3d) compared to SD-dSTORM (Figure S4b). Similar to SD-DNA-PAINT, SD-dSTORM also gains precision by implementing the novel intensity-weighting procedure (Figure S4c).
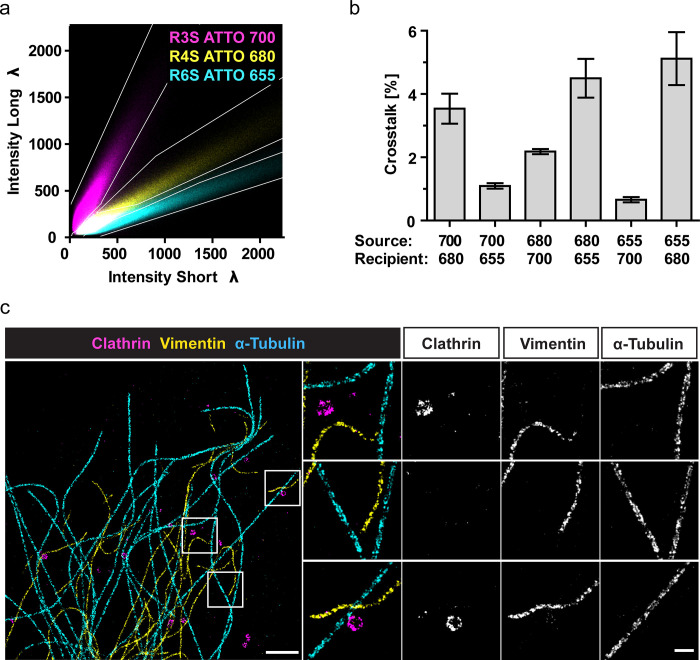
Next, we aimed at performing triple-color SD-DNA-PAINT using the spectrally suited triplet of ATTO-dyes (655/680/700) conjugated to distinct imagers (R6S/R4S/R3S). To optimize the color-separation masks for triple-color, we first performed single-color SD-DNA-PAINT experiments as described above. The emission spectrum of ATTO-680 peaks in between the spectra of ATTO-655 and ATTO-700 (Figure S1a,b). The 2D intensity histogram of the overlaid single-color experiments clearly shows three distinct populations of localizations with an overlap of ATTO-680 with ATTO-655 and ATTO-700 at low intensities (Figures 4a; S1c–e). In analogy to the dual-color masks, the triple-color-separation masks were designed to minimize color crosstalk while maximizing the inclusion of localizations. Despite the spectral overlap of ATTO-680 with both ATTO-655 and ATTO-700, up to 70% of the localizations could be included (Figure S3b) while tolerating a maximal crosstalk of 5% (Figure 4b). After optimizing the triple-color-separation masks, we performed triple-color SD-DNA-PAINT on cells immunolabeled for three cellular nanostructures (microtubules, clathrin-coated vesicles, vimentin filaments) with all three imagers (R6S-655/R4S-680/R3S-700) in the imaging buffer. The images were processed as described above using the optimized triple-color-separation masks. The resulting SD-DNA-PAINT images demonstrate that the nanoscale structures of all three biological structures are clearly resolved with minimal crosstalk using the triple-dye combination ATTO-655/680/700 (Figure 4c). The low crosstalk in triple-color SD-DNA-PAINT was verified using DNA origami grids with distinct docking strands (Figure S3c).
Figure 4.
Triple-color SD-DNA-PAINT. (a,b) Single-color DNA-PAINT experiments were performed in SD-mode using COS-7 cells immunolabeled either for clathrin (R3S/R4S) or α-tubulin (R6S). Recording modality: 20 000 frames, 100 ms exposure, 1 nM imager concentration. (a) Pseudocolor overlay of the 2D intensity histograms from separate single-color experiments (cyan/magenta/yellow: R6S-655/R3S-700/R4S-680). Color-separation masks (white lines) were manually designed based on the single-color signals to minimize the crosstalk between the channels while maximizing the included localizations. Histograms from 5–6 images were averaged per channel. (b) The crosstalk was calculated for all dye combinations as the percentage of localizations from the source that are detected within the mask of the recipient channel based on the single-color ground truth. The triple-color masks were designed to tolerate a crosstalk below 5%. Mean ± SEM, N = 4–5 images per channel. Data in (a,b) were partially reused from Figure 3. (c) COS-7 cells immunolabeled for clathrin, vimentin, and α-tubulin were imaged in SD-mode, using the imagers from (a). Representative triple-color SD-DNA-PAINT image and indicated subregions (white boxes) of microtubules (R6S-655), vimentin (R4S-680), and clathrin (R3S-700) are shown. Scale bar: 1 μm, zoom: 200 nm. All three regions have the same contrast. Recording modality: 30 000 frames, 100 ms exposure, 1 nM total imager concentration.
Compared to the original SD-dSTORM approach,12 our novel intensity-weighted multichannel registration procedure is not intrinsically free from chromatic distortions; instead, these errors are corrected. To verify the high accuracy of this procedure in triple-color SD-DNA-PAINT mode, we imaged 20 nm DNA origami grids containing a single docking strand (R6) with three compatible imagers (R6S-ATTO-655/680/700) in the imaging buffer (Figure S5a). If the registration is precise, all three color channels should show a high degree of overlap. Indeed, the resulting triple-color overlay shows a clear colocalization of the docking site spots in all three channels. To quantify the accuracy of multichannel registration, we performed a nearest neighbor (NN) distance analysis between each of the channels on the origami data set (Figure S5c,d). The peaks of the multichannel NN distance distributions were significantly above the control for a random distribution and in the range of 1–3 nm for all comparative color channel combinations. This demonstrates the high accuracy of our custom-designed multichannel registration procedure that corrects for flat-field distortions of the emission splitter with an error that is below the localization precision of single-color images.
Conclusion
In summary, we presented a novel approach that combines DNA-PAINT with simultaneous multichannel acquisition using spectral demixing. SD-DNA-PAINT alleviates the need for sequential fluid exchange and therefore avoids all associated disadvantages, including the extended acquisition times. SD-DNA-PAINT speeds up the acquisition time of current multichannel DNA-PAINT while taking advantage of all the benefits of DNA-PAINT. To realize SD-DNA-PAINT, we selected a novel combination of spectrally overlapping dyes (ATTO-655/680/700) and designed DNA-PAINT probes for optimal color separation and resolution. Using these probes, we achieved simultaneous multicolor DNA-PAINT with minimal crosstalk and excellent localization precision (3–6 nm, Figure 2e). We demonstrated the ability of SD-DNA-PAINT to resolve 20 nm DNA origami grids and cellular nanostructures.
The presented SD-DNA-PAINT approach offers up to three color channels. Including more colors was limited by the partial overlap of the dye-specific intensity populations within the histogram that is used for color separation (Figure 4a). The laser power density of the SMLM system used here was limited to a maximum of 3 kW/cm2, while other applications (e.g., ultrahigh-resolution DNA-PAINT8) used higher power densities (>4 kW/cm2). We propose that by using higher laser powers, the intensity populations within the histogram could be shifted toward higher values and thus create sufficient space for additional dye channels.
A large number of researchers, including ourselves, routinely used variants of multicolor dSTORM to resolve cellular nanostructures.13,15,20−22 Compared to SD-dSTORM, SD-DNA-PAINT offers higher localization precisions and a third color channel. In analogy to qPAINT,7 the controlled binding kinetics of SD-DNA-PAINT could be used to count binding sites in biological structures. We therefore believe that SD-DNA-PAINT will become a powerful method to perform robust, fast, and quantitative multicolor SMLM in cases where simultaneous acquisition of up to three color channels is desired.
Acknowledgments
We thank the staff of the Advanced Medical Bioimaging Core Facility, Charité-Universitätsmedizin, Berlin, Germany (AMBIO), for support in sample preparation and usage of microscope systems. We thank Dr. Hylkje Geertsema (TU Delft) from the laboratory of Prof. Helge Ewers (FU Berlin) for initial discussions about DNA-PAINT probes.
Glossary
Abbreviations
- SMLM
single-molecule localization microscopy
- STORM
stochastic optical reconstruction microscopy
- SD
spectral demixing
- PAINT
points accumulation in nanoscale topography
- fm-DNA-PAINT
frequency multiplexing DNA-PAINT
- NeNA
nearest-neighbor-based analysis
- NN
nearest neighbor
- AB
antibody
- NB
nanobody
- F(ab’)1
antigen binding fragment
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c04520.
Supplementary Data: SD-DNA-PAINT filter set (file type: PNG); SD-dSTORM filter set (file type: PNG); SDmixer configuration file (file type: TXT); ThunderSTORM configuration file for SD-DNA-PAINT (file type: TXT); ThunderSTORM configuration file for SD-dSTORM (file type: TXT) (ZIP)
Supporting Text: Detailed experimental methods and materials; Supporting Text to S1: Dye selection for SD-DNA-PAINT. Supporting Figures: Supporting Figure S1: Dye selection; Supporting Figure S2: Image processing; Supporting Figure S3: Color-filtering; Supporting Figure S4: SD-dSTORM; Supporting Figure S5: Multichannel SD-DNA-PAINT registration accuracy. Supplementary Tables: Supplementary Table 1: Imager sequences; Supplementary Table 2: Docking sequences; Supplementary Table 3: Experimental settings (PDF)
Author Contributions
J.S. and N.G. conceived the scientific concept and designed the experiments. N.G. performed the experiments, designed the software tools, and analyzed the data. J.S. supervised the study and together with N.G. wrote the manuscript. S.S. and R.J. contributed DNA-PAINT reagents and expertise in how to perform experiments. R.J. supervised S.S. and gave advice on the structure and content of the manuscript. All authors contributed to writing of the manuscript.
This work has been supported in part by the German Research Foundation (DFG) through the SFB958 project Z02 to J.S. (grant agreement number SFB958/Z02) and in part by the European Research Council through an ERC Starting Grant (MolMap; grant agreement number 680241) and the Max Planck Society.
The authors declare no competing financial interest.
Supplementary Material
References
- Sahl S. J.; Hell S. W.; Jakobs S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18 (11), 685–701. 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Sauer M.; Heilemann M. Single-Molecule Localization Microscopy in Eukaryotes. Chem. Rev. 2017, 117 (11), 7478–7509. 10.1021/acs.chemrev.6b00667. [DOI] [PubMed] [Google Scholar]
- Sigal Y. M.; Zhou R.; Zhuang X. Visualizing and discovering cellular structures with super-resolution microscopy. Science 2018, 361 (6405), 880–887. 10.1126/science.aau1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11 (3), 313–8. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Steinhauer C.; Scheible M.; Kuzyk A.; Tinnefeld P.; Simmel F. C. Single-molecule kinetics and super-resolution microscopy by fluorescence imaging of transient binding on DNA origami. Nano Lett. 2010, 10 (11), 4756–61. 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- Heilemann M.; van de Linde S.; Schuttpelz M.; Kasper R.; Seefeldt B.; Mukherjee A.; Tinnefeld P.; Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem., Int. Ed. Engl. 2008, 47 (33), 6172–6. 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Dai M.; Woehrstein J. B.; Agasti S. S.; Feiger Z.; Rodal A.; Yin P. Quantitative super-resolution imaging with qPAINT. Nat. Methods 2016, 13 (5), 439–42. 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueder F.; Stein J.; Stehr F.; Auer A.; Sperl B.; Strauss M. T.; Schwille P.; Jungmann R. An order of magnitude faster DNA-PAINT imaging by optimized sequence design and buffer conditions. Nat. Methods 2019, 16 (11), 1101–1104. 10.1038/s41592-019-0584-7. [DOI] [PubMed] [Google Scholar]
- Strauss S.; Jungmann R. Up to 100-fold speed-up and multiplexing in optimized DNA-PAINT. Nat. Methods 2020, 17 (8), 789–791. 10.1038/s41592-020-0869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Garcia P. A.; Garbacik E. T.; Otterstrom J. J.; Garcia-Parajo M. F.; Lakadamyali M. Excitation-multiplexed multicolor superresolution imaging with fm-STORM and fm-DNA-PAINT. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (51), 12991–12996. 10.1073/pnas.1804725115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade O. K.; Woehrstein J. B.; Nickels P. C.; Strauss S.; Stehr F.; Stein J.; Schueder F.; Strauss M. T.; Ganji M.; Schnitzbauer J.; Grabmayr H.; Yin P.; Schwille P.; Jungmann R. 124-Color Super-resolution Imaging by Engineering DNA-PAINT Blinking Kinetics. Nano Lett. 2019, 19 (4), 2641–2646. 10.1021/acs.nanolett.9b00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe A.; Haucke V.; Sigrist S. J.; Heilemann M.; Schmoranzer J. Multi-colour direct STORM with red emitting carbocyanines. Biol. Cell 2012, 104 (4), 229–37. 10.1111/boc.201100011. [DOI] [PubMed] [Google Scholar]
- Lehmann M.; Lichtner G.; Klenz H.; Schmoranzer J. Novel organic dyes for multicolor localization-based super-resolution microscopy. J. Biophotonics 2016, 9 (1–2), 161–70. 10.1002/jbio.201500119. [DOI] [PubMed] [Google Scholar]
- Lampe A.; Tadeus G.; Schmoranzer J. Spectral demixing avoids registration errors and reduces noise in multicolor localization-based super-resolution microscopy. Methods Appl. Fluoresc 2015, 3 (3), 034006. 10.1088/2050-6120/3/3/034006. [DOI] [PubMed] [Google Scholar]
- Lehmann M.; Gottschalk B.; Puchkov D.; Schmieder P.; Schwagerus S.; Hackenberger C. P.; Haucke V.; Schmoranzer J. Multicolor Caged dSTORM Resolves the Ultrastructure of Synaptic Vesicles in the Brain. Angew. Chem., Int. Ed. Engl. 2015, 54 (45), 13230–5. 10.1002/anie.201505138. [DOI] [PubMed] [Google Scholar]
- Tadeus G.; Lampe A.; Schmoranzer J. SDmixer-a versatile software tool for spectral demixing of multicolor single molecule localization data. Methods Appl. Fluoresc 2015, 3 (3), 037001. 10.1088/2050-6120/3/3/037001. [DOI] [PubMed] [Google Scholar]
- Endesfelder U.; Malkusch S.; Fricke F.; Heilemann M. A simple method to estimate the average localization precision of a single-molecule localization microscopy experiment. Histochem Cell Biol. 2014, 141 (6), 629–38. 10.1007/s00418-014-1192-3. [DOI] [PubMed] [Google Scholar]
- Schnitzbauer J.; Strauss M. T.; Schlichthaerle T.; Schueder F.; Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc 2017, 12 (6), 1198–1228. 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- Pengo T.; Olivier N.; Manley S., Away from resolution, assessing the information content of super-resolution images. arXiv, 2015. https://arxiv.org/abs/1501.05807v1 (accessed 22 Apr 2021).
- Schmerl B.; Gimber N.; Kuropka B.; Rentsch J.; Kunde S.-A.; Ewers H.; Freund C.; Schmoranzer J.; Rademacher N.; Shoichet S. A.. The postsynaptic MAGUK scaffold protein MPP2 organises a distinct interactome that incorporates GABAA receptors at the periphery of excitatory synapses. bioRxiv, 2020. https://www.biorxiv.org/content/10.1101/2020.05.29.123034v1 (accessed 14 Jan 2021).
- Schoneberg J.; Lehmann M.; Ullrich A.; Posor Y.; Lo W. T.; Lichtner G.; Schmoranzer J.; Haucke V.; Noe F. Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 2017, 8, 15873. 10.1038/ncomms15873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimber N.; Tadeus G.; Maritzen T.; Schmoranzer J.; Haucke V. Diffusional spread and confinement of newly exocytosed synaptic vesicle proteins. Nat. Commun. 2015, 6, 8392. 10.1038/ncomms9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadeh J. N.; Steenberg C. D.; Bois J. S.; Wolfe B. R.; Pierce M. B.; Khan A. R.; Dirks R. M.; Pierce N. A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32 (1), 170–3. 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- Knemeyer J. P.; Marme N.; Sauer M. Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem. 2000, 72 (16), 3717–24. 10.1021/ac000024o. [DOI] [PubMed] [Google Scholar]
- Sograte-Idrissi S.; Oleksiievets N.; Isbaner S.; Eggert-Martinez M.; Enderlein J.; Tsukanov R.; Opazo F. Nanobody Detection of Standard Fluorescent Proteins Enables Multi-Target DNA-PAINT with High Resolution and Minimal Displacement Errors. Cells 2019, 8 (1), 48. 10.3390/cells8010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9 (7), 671–5. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovesny M.; Krizek P.; Borkovec J.; Svindrych Z.; Hagen G. M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 2014, 30 (16), 2389–90. 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeus G.SDmixer2. https://github.com/gtadeus/sdmixer2 (accessed 01 Sept 2020).
- Gimber N.ThunderSTORM-SDmixer Converter. https://github.com/ngimber/Converter_ThunderSTORM_SDmixer/releases/tag/v1.0.0.
- Gimber N.SD-DNA-PAINT. https://github.com/ngimber/SD-DNA-PAINT/releases/tag/v1.0.1. [DOI] [PMC free article] [PubMed]
- Levet F.; Hosy E.; Kechkar A.; Butler C.; Beghin A.; Choquet D.; Sibarita J. B. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 2015, 12 (11), 1065–71. 10.1038/nmeth.3579. [DOI] [PubMed] [Google Scholar]
- Gimber N.Voronoi Tesselation for Single-Molecule Localization Microscopy. https://github.com/ngimber/SMLM_VoronoiTesselation/releases/tag/v1.0.0.
- Gimber N.Nearest Neighbor Analysis for Single-Molecule Localization Microscopy. https://github.com/ngimber/SMLM_NearestNeighbor/tree/v1.0.0.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.