Abstract
The COVID‐19 crisis and the development of the first approved mRNA vaccine have highlighted the power of RNA‐based therapeutic strategies for the development of new medicines. Aside from RNA‐vaccines, antisense oligonucleotides (ASOs) represent a new and very promising class of RNA‐targeted therapy. Few drugs have already received approval from the Food and Drug Administration. Here, we underscored why and how ASOs hold the potential to change the therapeutic landscape to beat SARS‐CoV‐2 viral infections.
This article is categorized under:
RNA Interactions with Proteins and Other Molecules > Small Molecule‐RNA Interactions
Keywords: antisense oligonucleotide, ASO, SARS‐CoV‐2, therapeutic option
Adapted from “Human Coronavirus Structure” and “Genome Organization of SARS‐CoV” (Acknowledgements: Glaunsinger Lab: Jessica M Tucker, Britt A Glaunsinger et al.) by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender-templates
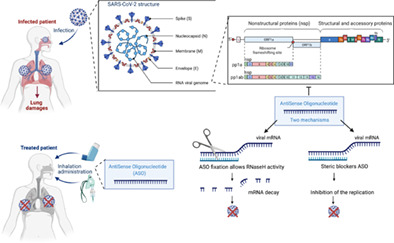
1. THE CORONAVIRUS DISEASE 2019 OUTBREAK
Over the last century, the world has been confronted with a number of viral epidemics: the 1918‐flu pandemic, severe acute respiratory syndrome (SARS, 2002–2004), the H1N1 flu (2009), Ebola (2014–2016), and now coronavirus disease 2019 (COVID‐19). COVID‐19 is a respiratory infectious disease caused by a novel coronavirus named severe acute respiratory syndrome corona virus 2 (SARS‐CoV‐2), initially identified in China. The COVID‐19 outbreak has been steadily advancing, country by country, and now affects the entire globe, despite never‐seen‐before worldwide control measures. In addition, despite the diverse array of governmental sanitary measures worldwide, many countries have experienced several deadly waves of the COVID‐19 epidemic. Massive testing campaigns to isolate infected individuals have been used to control the epidemic, but are not sufficient to eradicate the threat. In this unprecedented sanitary crisis, vaccination has arisen to be among the most effective approaches to protect the population.
In just 1 year, vaccines based on a never‐seen‐before technology have emerged. This expedited process has been the result of a major effort by many actors. Scientists have shared data to establish the genomic sequence of SARS‐CoV‐2, characterize its structure, and understand its infectious behavior. Such knowledge has been indispensable for development of the current vaccines. The vaccine technologies already existed. Adenovirus technologies have been built on decades of research and experience. The mRNA vaccines are based on a newer technology but are also backed by decades of research, the first product being developed in 1999 (Pardi et al., 2018). Administrative logistics were dramatically accelerated. It typically takes years of administrative work (submissions, answering questions from regulatory agencies) to conduct a clinical trial. Rolling reviews were instituted, in which pharmaceutical companies were able to provide regulatory agencies with data on safety and efficacy. In addition, volunteers massively signed up to participate in COVID‐19 vaccine clinical trials to confront the world crisis. Countries began manufacturing vaccines for use as soon as they had a product in clinical trials. Overall, these measures have led to the extremely rapid and successful development of highly efficient and safe vaccines. The Pfizer‐BioNTech mRNA vaccine was approved for emergency use by the FDA on December 11, 2020, the Moderna mRNA vaccine, 1 week later. These two‐approved mRNA vaccines were followed by adenovirus‐based vaccines. The AstraZeneca adenovirus‐based vaccine received conditional marketing‐authorization from the European Commission after evaluation by the European Medicine Agency and consultation with the EU Member States on January 29, 2021. Finally, the Johnson and Johnson laboratory developed the first single‐shot adenovirus‐based vaccine that was approved on February 27, 2021. All four vaccines target the spike protein.
2. HARD WORK AND LUCK PLAYED A PART, WITH THE CHOICE OF THE RIGHT VIRAL ANTIGENS
The SARS‐CoV‐2 RNA virus belongs to the β‐coronavirus subfamily, which includes SARS‐CoV and MERS‐CoV. It shares 89.1% homology with SARS‐CoV. It has a positive single‐stranded non‐segmented RNA genome, composed of 29,903 nucleotides (Genbank accessible number MN908947). Coronavirus (CoVs) are enveloped, and the surface of each virion is decorated with, among other features, characteristic glycoprotein spikes. The spike (S) protein mediates host cell attachment and is indispensable for viral entry. SARS‐CoV‐2 S proteins recognize the host ACE2 receptor, as that of SARS‐CoV, whereas the MERS‐CoV S protein recognizes the dipeptidyl 4‐receptor (Figure 1). The S protein is also involved in cell tropism and pathogenesis (Belouzard et al., 2012). Knowledge gained from the initial development of vaccines against SARS‐CoV and MERS identified the S protein as a potent target. In less than 16 months since the establishment of the SARS‐CoV‐2 genome sequence, more than 280 candidate vaccines have been developed, according to the World Health Organization. 1 Among them, mRNA vaccines have emerged as good candidates. Indeed, four vaccines have received approval (Chung et al., 2020). As mentioned above, mRNA‐vaccines represent a powerful alternative to conventional vaccine approaches. They are based on the use of a synthetic mRNA encoding viral antigens in human cells to trigger immune responses. They are highly potent (Lederer et al., 2020) and are able to be developed in record time, as seen with the COVID‐19 outbreak, at a lower cost than conventional vaccines.
FIGURE 1.
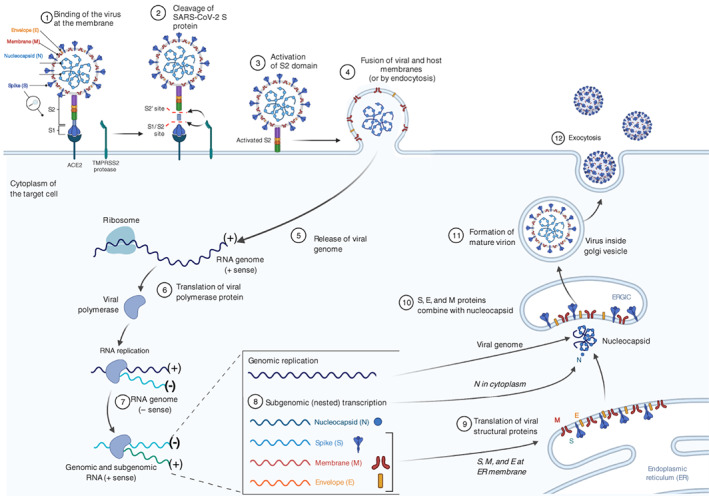
Life cycle of SARS‐CoV‐2. SARS‐CoV‐2 is an enveloped RNA virus. Its surface is decorated with the spike protein that is responsible for the recognition and entry of the virus into the cell. After entry of the virus into the cell, the ORF1a and ORF1b are translated by the ribosome of the host cell followed by the assembly of the replicase‐transcriptase complex. Then, the viral RNA is replicated to form a negative strand, ready for both replication of the viral genome and transcription of the subgenomic genome. Finally, the mature virions are formed and exit the cell by exocytosis. Adapted from “Human Coronavirus Structure” and “Coronavirus Replication Cycle” (Acknowledgements Dr. Benjamin Goldman‐Israelow) by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender‐templates
The mRNA vaccines manufactured by Pfizer‐BioNTech (BNT162b2) and Moderna (mRNA‐1273) encode the full‐length SARS‐CoV‐2 S glycoprotein stabilized in its prefusion conformation (Baden et al., 2021; Jackson et al., 2020; Polack et al., 2020). Two consecutive proline substitutions at amino acid positions 986 and 987 (at the top of the central helix in the S2 subunit) lock the protein in a stable conformation. The nucleoside‐modified mRNA is encapsulated in a lipid nanoparticle to ensure delivery and stability (Baden et al., 2021; Corbett et al., 2020). Both have shown high efficacy (Baden et al., 2021; Lederer et al., 2020; Polack et al., 2020). Several countries have relied on them, at least in part, to establish their immunization strategy. 2 Importantly, the development of these mRNA vaccine platforms to engineered mRNA sequences makes them very good candidates for adaption to potential variants (Jackson et al., 2020).
3. WHAT ABOUT THERAPEUTIC OPTIONS?
We have learned about the need for accurate viral‐detection methods and prophylactic methods, such as vaccination, to control the spread of the disease from previous pandemics, but what about therapeutic options? Even though vaccines appear to be the best weapon to protect the population, we need to have therapeutic options to combat the virus in the non‐vaccinated population, the vaccinated‐population with low immunity, and in cases of reinfection for systemic and local use. Therapeutics are also required to mitigate the spread of new SARS‐CoV‐2 variants, for which the vaccines would show only partial protection. This is vindicated by the easy and rapid viral transmission and the severe respiratory distress syndrome that requires intensive healthcare, leading to complete re‐organization of the healthcare system to the detriment of other patients (Petersen et al., 2020; Piroth et al., 2021).
No specific antiviral treatments against SARS‐CoV‐2 are currently available, despite the testing of a number of antiviral therapies in clinical trials (Lee et al., 2020). Antiviral drugs that have shown efficacy in the treatment of the coronavirus‐induced diseases SARS and MERS, or influenza viruses, have been tested (namely Remdesivir) (Fernandes et al., 2020). Remdesivir inhibits RNA‐dependent RNA polymerase (RdRp), preventing the self‐replication of viral RNA. Additional approaches have been considered, including corticosteroid therapy and treatment with angiotensin II among others (Li & De Clercq, 2020; Stasi et al., 2020). Therapeutic options rely on symptomatic and supportive care. Repurposing well‐known and widely available FDA‐approved molecules is an attractive option to address the lack of specific drugs and save time to combat the epidemic. It is, however, surprising that, in the era of deep sequencing and computational biology, we do not take more advantage of our knowledge of the sequence of SARS‐CoV‐2 to design a therapy that specifically targets the viral genome, as done to design an accurate vaccine. The development of RNA‐based therapies, such as antisense oligonucleotides (ASOs), warrants further exploration in the context of frontline therapeutics for the treatment of infected patients.
4. ASOs AS A PROMISING THERAPEUTIC OPTION
ASOs are single‐stranded nucleic acids that specifically bind to their cognate RNA‐target via Watson–Crick base pairing. Upon RNA‐binding ASO modulates the function of the targeted RNA. Their antisense mechanisms can be broadly classified according to their capacity to promote or not RNA‐degradation (Figure 2). RNA‐degradation may be elicited by the recruitment of endogenous enzyme such as the RNase H. Protein production is thereby canceled. ASO lacking RNase H‐dependent activity may act as steric blockers. As reported they can modulate RNA‐processing by interfering with the splicing machinery, promoting exon inclusion, exon skipping, or cryptic splice site. They can also induce translation arrest by masking the AUG translation initiation codon (reviewed in Geary et al., 2015; Quemener et al., 2020).
FIGURE 2.
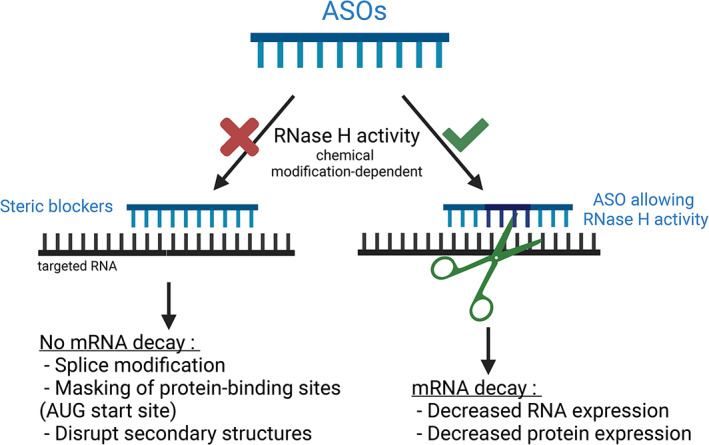
Mechanism of action of antisense oligonucleotides. ASOs are small single‐stranded nucleic acids that specifically bind to cognate RNA‐targets. They act through two main mechanisms, depending on their chemical backbone. First, ASOs that do not mediate RNase H activity, act as steric blockers disrupting the splicing machinery, Protein/RNA interactions and RNA secondary structures. Second, ASOs also known as GapmeRs recruit and induce RNase H cleavage of the targeted‐RNA. GapmeRs are typically composed of Lock Nucleic Acids (LNA) at both ends to provide high‐affinity base pairing, with a central block of unlocked bases to mediate RNase H activation. They decrease RNA and protein expression. Created with BioRender.com (2021)
Antisense mechanisms are dictated by the chemical modifications incorporated in the oligonucleotide structure (Figure 3). These chemical modifications have also been made to major potency, pharmacokinetics, and reduce toxicity. The most common modification is the replacement of one of the non‐bridging oxygen by a sulfur atom. Phosphorothioate (PS) backbone elicits RNase H cleavage and greatly increases resistance to nucleolytic degradation, conferring pharmacokinetic benefit. PS are commonly referred as first‐generation ASOs.
FIGURE 3.
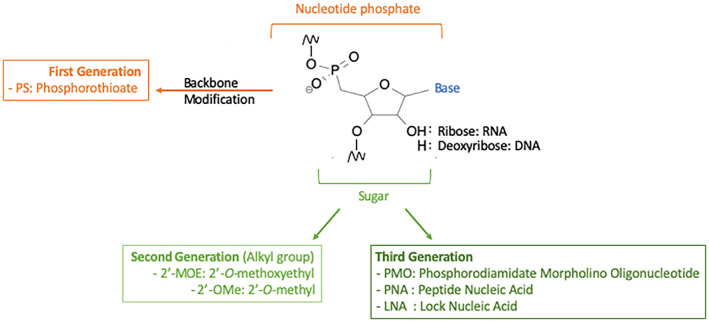
Chemical modification of ASO. Substitution of the non‐bridging oxygen atoms in the phosphodiester bond with a sulfur leads to phosphorothiote (PS) the most common first generation ASO. Second generation ASO include modifications at the 2′ position of the sugar moiety. The hydroxyl group can be replaced with a methyl group (2′‐O‐methyl or 2′‐OMe) or a methoxyethyl group (2′‐O‐methoxyethyl or 2′‐MOE). Third generation have various modifications: Locked nucleic acids (LNAs ‐ the 2′‐oxygen and the 4′‐carbon on the same ribose are connected through a methylene bridge); phosphrodiamidate morpholino oligomers (PMOs ‐ a morpholine moiety connected through methylene phosphorodiamidate replace the ribose); peptide nucleic acids (PNAs ‐ the deoxyribose phosphate is replaced by a speudopeptide backbone)
Second‐generation ASOs include modification of the 2′ position of the sugar moiety enhancing the drug‐like properties of the ASO. The introduction of an oxygened group leads to the formation of 2′‐O‐methoxyethyl (2′‐MOE) oligonucleotides and of 2′‐O‐methyl oligonucleotides. These second‐generation ASOs are the most advanced 2′‐modified series, leading to the development of drugs entering the clinic. Five FDA and or MEA approved drugs endorse 2′‐MOE modifications. These modifications do not support recruitment and cleavage by RNase H but offer the opportunity for the ASO to act as steric blockers.
Third‐generation ASOs have been developed to further improve nuclease resistance, increase binding affinity, and to enhance pharmacokinetics and biostability. Many modifications have been proposed however, peptide nucleic acid, phosphorodiamidate morpholino oligomer (PMO), and locked nucleic acid (LNA) are the three most studied third‐generation ASOs. LNAs are very promising candidates. The bicyclic modification of the sugar locks the conformation of the sugar improving drastically the binding affinity of the ASO. Thus, LNAs provide high‐affinity base‐pairing to complementary nucleic acid targets and confer resistance to nucleases. LNAs do not recruit RNase H.
Chemical modification that do not support recruitment and cleavage by RNase H acts as steric blockers. However, chimeric strategies have been developed to induce effective knockdown of mRNA through RNase H‐dependent activation. To this end, ASOs include a central block of unlocked bases sufficiently long to recruit and induce RNase H cleavage, flanked by locked base at both ends ensuring high‐affinity binding. Such ASOs, also known as GapmeRs, are designed to target mRNA and induce specific degradation through RNase H‐dependent activation, both in the nucleus and cytoplasm. The activity in the cytosol was found to be roughly comparable to that observed in the nucleus (Liang et al., 2017). Importantly, ASOs do not require vectorization to enter target cells, as they are internalized by micropinocytosis or gymnosis (Fazil et al., 2016; Ong et al., 2018).
The versatility of their mechanism of action and the flexibility of their design has led to the development of several FDA‐approved drugs for genetic and neurological disorders that lack therapeutic options (Rinaldi & Wood, 2017). This can be exemplified by RNase H‐competent ASO elaborated for the treatment of familial homozygous hypercholesterolemia, characterized by elevated concentration of atherogenic LDL particles carrying cholesterol (Mullick et al., 2011). Mipomersen also known as Kynamro® is a 20mer PS 2′‐MOE GapmeR ASO inhibiting the synthesis of ApoB100 the main protein at the surface of the LDL particles. Complementary binding of Mipomersen to the ApoB100 mRNA promotes its degradation by RNase H and progressive decrease of LDL‐cholesterol levels (Akdim et al., 2011; Ricotta & Frishman, 2012). Splice‐switching ASOs received FDA‐approval to treat Duchenne muscular dystrophy (DMD) a progressive and lethal genetic disorder caused by alteration of the dystrophin gene. Eteplirsen also known as Exondys51® is a 30‐nucleotide PMO ASO approved for the treatment of DMD in patients who have confirmed mutation in exon 51 (Lim et al., 2017; Nelson et al., 2009). Eteplirsen is a steric blocker directing the exclusion of the exon 51. By skipping exon 51, a truncated but partially functional dystrophin protein is produced shifting the severe DMD phenotype to that of the less severe phenotype of Becker muscular dystrophy, with functional albeit truncated versions of dystrophin. Eteplirsen is beneficial only for patient with amenable DMD mutation in exon 51 (Lim et al., 2017; Stein & Castanotto, 2017). Comparable exon‐skipping approaches have thus been developed to target the exon 53 (Golodirsen also known as Vyondys 53® in 2019) (Frank et al., 2020) and this year the exon 45 (Casimersen also known as Amondys 45®) demonstrating the capacity of those splice‐switch ASOM in reprogramming gene expression and mitigate the disease.
Remarkably, ASO may also promote exon‐retention. Nusinersen also known as Spiranza® is 18mer PS 2′‐MOE ASO indicated for the treatment of spinal muscular atrophy (SMA). SMA is caused by mutations in the survival of motor neuron 1 (SMN1) gene leading to SMN1 protein deficiency. Nusinersen acts to replace SMN1 protein deficit by increasing the splicing efficiency of the SMN2 pre‐mRNA a paralogous copy of the SMN1 gene. SMN2 differs from SMN1 by few nucleotides, which lessens the splice site of exon 7 leading to the removal of the exon 7 and the formation of a rapidly degraded SMN2 protein. Nusinersen is thus designed to bind a specific sequence in the intron 7 of the SMN2 pre‐mRNA to promote exon 7 retention and the production of a stable SMN2 protein (Biogen, 2019; Corey, 2017; Finkel et al., 2017). Recently, ASO development made a major breakthrough. In 14 months, a splice‐switch custom ASO, called Milasen, was designed for a never‐seen‐before mutation to halt the rare neurodegenerative disorder of a single patient. Knowledge of the genetic alteration led to the rational design, testing, and manufacture of the drug in record time (J. Kim et al., 2019).
5. WHAT DO WE KNOW ABOUT ASOs AS ANTIVIRAL DRUGS?
The first ASO used for a therapeutic strategy was, interestingly, designed to block the replication of Rous sarcoma virus and prevent in vitro oncogenic transformation of chicken fibroblast infected with Rous virus (Stephenson & Zamecnik, 1978). This pioneering proof of concept opened the way not only for the treatment of genetic disorders, as seen above, but also against viral infection. Remarkably, the first FDA approved ASO was designed to treat cytomegalovirus (CMV) retinitis. Formivirsen, a first‐generation ASO (PS backbone) was marketed by Ionis Pharmaceuticals and Norvatis Ophthalmics under the name of Vitravene®. This treatment was recommended for patients with acquired immunodeficiency syndrome, in whom CMV infection can lead to the progressive destruction of retinal cells. The designed‐ASO targeted specifically the viral mRNA encoding the major immediate early region (IE2) proteins, resulting in specific inhibition of IE2 expression and production of essential viral proteins. Intravitreous injections were performed giving very effective results. Unfortunately, the development of local adverse effects (increased intraocular pressure and local inflammation) led several years later (2006) to its withdrawn from the market (Geary et al., 2002; Roehr, 1998; Vitravene Study Group, 2002).
Other ASOs mediating RNase H activity have been shown to be potent antiviral agents. Javanbakht et al. showed that ASOs significantly reduce intracellular viral mRNA and viral antigen levels. Diminution of the production of hepatitis B surface antigen in cultured human hepatoma cell lines provided critical knowledge about the participation of this protein in the pathogenicity of HBV‐infection (Javanbakht et al., 2018).
ASOs acting through steric blockage of viral RNA regions, such as AUG start sites and critical regulatory motifs showed to be very effective. For example, ASOs have been designed to inhibit highly conserved Influenza A genes in vitro and in vivo (Gabriel et al., 2008; Kabanov et al., 1990). Targeting the AUG start site of the polymerase subunit genes (PB1, PB2, PB) and the nucleoprotein gene inhibited viral replication, significantly lowering the viral titer in the lungs of mice and protecting them from lethal infection. ASOs were shown to be specific and non‐toxic for cells. The combination of different ASOs slightly improved antiviral activity (Gabriel et al., 2008). In vivo work further underscored the relevance of using intranasal delivery to stop Influenza A infection (Lupfer et al., 2008).
Recently, the choice of ASOs has followed a new paradigm, in which they are designed on the basis of the modeled secondary structure of the genomic viral RNA. This approach was used to test ASOs that target segments 5 and 8 of Influenza A viral RNA. The most effective ASOs were shown to specifically bind to internal loops, supporting the notion that the effectiveness of ASOs is likely dependent on RNA structure and accessibility (Lenartowicz et al., 2016; Michalak et al., 2019). Importantly, chemical modifications of ASOs to improve their stability and binding affinity to their targets reduced their toxicity and increased their antiviral activity (Lenartowicz et al., 2016; Michalak et al., 2019).
6. ASO TECHNOLOGY HAS ALSO BEEN DEVELOPED AGAINST SARS‐CoV
The first ASOs reported against SARS‐CoV, the virus most closely related to SARS‐CoV‐2, targeted viral replicate polyprotein open reading frame 1a (ORF1a) and the transcription regulatory sequence located in the 5′‐UTR of the positive‐sense viral RNA genome. They showed high antiviral activity in vitro, underscoring the potential capacity of ASOs for antiviral treatment against coronavirus and, potentially, SARS‐CoV‐2 (Neuman et al., 2004; Neuman et al., 2005). Two types of ASOs were used where the sugar was replaced with a morpholine ring (PMOs [antisense morpholino oligomers] and P‐PMOs [peptide‐conjugated antisense morpholino oligomers]). These chemical modifications ensure steric blockage being generally used to block translation via translation arrest. P‐PMOs targeting the viral transcription start regulatory sequence in the 5′‐UTR were the most effective inhibitors tested; they disrupt RNA secondary structure critical for its function. This study acted as a proof of principal of the efficiency of P‐PMOs, a modified class of ASOs, as an anti‐viral treatment.
Three patents were concomitantly published. Stein et al. developed ASOs targeting the translational initiation start site (AUG) of the first open reading frame (ORF1) of the viral genome (217–245) (US20030224353). 3 Ionis Pharmaceutical developed an ASO to disrupt the pseudoknot in the frameshift site of the SARS‐CoV genomic RNA (WO2005023083). 4 This region is essential for the life cycle of the virus. It is required to produce the RdRp located downstream of the frameshift site and thus permit viral replication. AVI Biopharma, Inc (now called Sarepta Therapeutics) developed ASOs targeting the 40 bases at the 3′ terminal end of the negative strand of the viral RNA to disrupt the stem‐loop secondary structure in this region and inhibit replication. Indeed, the negative‐strand RNA is used as template for replication of multiple copies of positive‐strand RNA (genomic RNA), which are consequently incorporated into newly formed virions or transcribed and translated into viral structural proteins. As the stem‐loop motif is a highly conserved secondary structure, this targeting strategy was also proposed to treat infections by other ssRNA viruses, such as flavivirus, picoronavirus, tagovirus, and calcivirus (WO2005013905). 5
7. HOW COULD ASOs TARGET SARS‐CoV‐2?
Knowledge of the sequence and structure of the SARS‐CoV‐2 genome is required for the development of specific ASOs. As mentioned above, the SARS‐CoV‐2 genome is a single‐stranded non‐segmented RNA of 29,903 nucleotides. It is capped at the 5′ end, harbors a 3′ poly(A)‐tail, and contains two short UTR‐sequences. It encodes 14 ORFs, some which overlap. At the 5′ end, ORF1a and ORF1b encode the replicase polyprotein comprising approximately two‐thirds of the genome. In addition to these nonstructural proteins (nsps), the remaining genome contains nine small ORFs that encode the structural proteins spike (S), envelope (E), membrane (M), nucleocapsid (N), and several accessory proteins (D. Kim et al., 2020) (Figure 4). Understanding the mode of replication of the virus is also crucial to establishing accurate ASO strategies.
FIGURE 4.
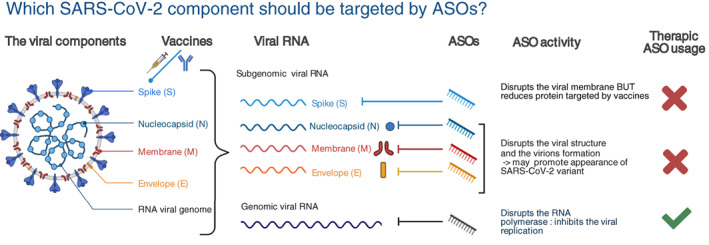
Which SARS‐CoV2 component should be targeted by ASOs? Schematic representation of the viral structure and components. Presently FDA approved vaccines mediate the production of antibodies against the spike protein that decorates the viral membrane. Viral RNAs comprise subgenomic and genomic entities. Subgenomic viral RNAs are translated to produce structural and accessory proteins. Genomic viral RNA supports RNA replication previous virion incorporation. Although, any viral sequences could be targeted with ASOs we recommend to target only genomic viral RNA and replication steps. Targeting, subgenomic RNA sequences may lower the efficacy of the present vaccines. Adapted from “Human Coronavirus Structure” and “Coronavirus Replication Cycle” (Acknowledgements: Dr. Benjamin Goldman‐Israelow) and “Genome Organization of SARS‐CoV” (Acknowledgements: Glaunsinger Lab: Jessica M Tucker, Britt A Glaunsinger et al.) by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender‐templates
After infection, the virus replicates its genome inside the host. Enzymes used for replication are encoded by the virus itself. Briefly, the 5′ cap and 3′ poly(A) tail allow direct translation of the nsps ORF1a and ORF1b, followed by assembly of the replicase‐transcriptase complex (RTC). The RTC drives both genome replication and discontinuous transcription of subgenomic mRNAs (sgRNAs). Discontinuous transcription is mediated by short AU‐rich transcription regulatory sequences (TRSs) located either immediately downstream of the 5′ leader (TRS‐L) or immediately upstream of each viral ORF (TRS‐B), except for ORF1a and ORF1b. The resulting sgRNAs are further translated to produce the structural and accessory proteins (Piyush et al., 2020).
One of the major advantages of ASOs is the possibility to target any conserved sequence as the positive or negative RNA strand, allowing the targeting of SARS‐CoV‐2 RNA at any step of the viral cycle life (Figure 5). As previously reported, ASOs should target the translational initiation start site (AUG) of the first open reading frame (ORF1) and/or secondary structures critical for translational processing to specifically mitigate translation of the replicase transcription complex. Thus, both steric blockers and RNase H‐dependent ASOs could be used to inhibit replication of the genomic RNA, which produces the intermediate negative‐sense RNA, as well as the positive‐sense RNA, which serves as the genomic‐RNA reservoir.
FIGURE 5.
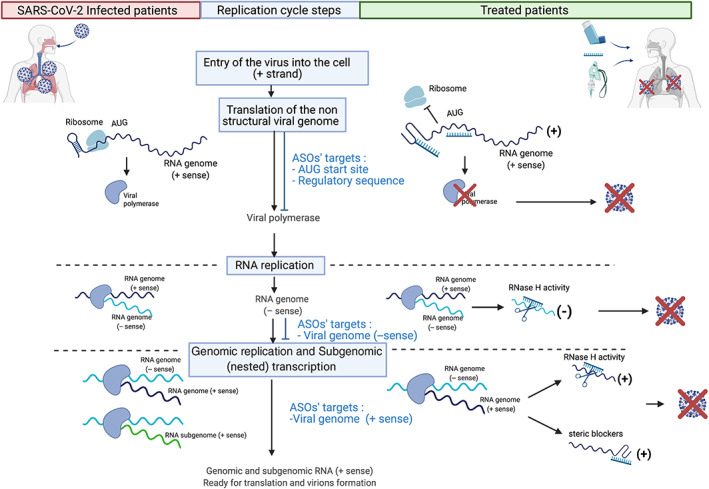
When and how targeting viral RNA with ASOs. Schematic representation of the genomic replication and the subgenomic (nested) transcription steps highlighting the RNA sequences to be targeted by ASOs using steric blockers and GapmeRs. Adapted from “Coronavirus Replication Cycle” (Acknowledgements: Dr. Benjamin Goldman‐Israelow) by BioRender.com (2021). Retrieved from https://app.biorender.com/biorender‐templates
The most elegant way to design ASO is to take advantage of the structure of the viral RNA‐sequence. This is what was recently proposed by Huston et al. (2021). They applied a novel long amplicon strategy to resolve the secondary structure of the SARS‐CoV‐2 genome at single resolution in infected cells. The RNA structure prediction revealed an elaborate genome architecture containing both unique and conserved well‐folded regions across beta‐coronaviruses. They tested their prediction, by developing steric blockers that disrupt two putative secondary structures within the genome (Regions 15 and 22). Both ASOs were designed with three consecutive LNAs at the 5′ and 3′ ends of each oligonucleotide, with stretches of unlocked bases within the oligonucleotide limited to three consecutive nucleotides to avoid RNase‐dependent activation. Both significantly inhibited viral growth, demonstrating that those two regions are critical for the life cycle of the virus and that these ASOs have antiviral potential (Huston et al., 2021). Further characterization of the structure of the SARS‐CoV‐2 RNA genome predicted the binding of 42 host proteins (Sun et al., 2021), underscoring potential vulnerability. In addition to targeting secondary structures, ASOs were used to disrupt predicted interactions between RNA‐binding proteins and SARS‐CoV‐2 RNA. The ASOs included 2'‐MOE and PS backbone modifications to enhance steric blocking activity. With this approach, they highlighted the important role played by host RNA binding proteins in the viral life cycle and identified druggable target regions (Sun et al., 2021).
In addition to steric blockers, RNase H‐dependent ASOs have been designed to target the highly conserved stem‐loop 2 motif (s2m) in the 3′‐UTR of the positive‐sense RNA. The ASOs were composed of LNA at both ends of the oligonucleotide, with a long stretch of unlocked nucleic acids to ensure RNase H recruitment and subsequent viral RNA cleavage. Interestingly, as RNase H is responsible for the cleavage of the targeted RNA in the RNA/ASO duplexes, the GapmeR remains intact and free to bind to other positive‐sense RNAs, while the viral RNA is cleaved. The GapmeRs were able to induce sequence‐specific RNA cleavage and inhibited SARS‐CoV‐2 replication in infected cells (Lulla et al., 2021). To our knowledge, this is the first experiment using GapmeRs to target a cytosolic RNA‐positive strand virus, showing the proof of concept of the efficacity of such a design and unlocking new therapeutic prospects. Also, as mentioned above, the knowledge of the RNA structure may help to the design of RNase H‐competent ASOs. By establishing the secondary structure maps of the full SARS‐CoV‐2 genome at the single‐base resolution, Manfredonia et al. identified a set of single stranded segments, with high sequence conservation. Such segments are likely to be persistently exposed representing accessible targets for RNase H‐competent ASO (Manfredonia et al., 2020).
Knowledge from previous ASO‐studies would predict the use of combinations of ASOs to maximize the impairment of vital SARS‐CoV‐2 functions and avoid the acquisition of resistant mutations. Such combinations could include steric blockers to inhibit translation of the replicase machinery (AUG start site) and replication process (secondary structure) and RNase H‐dependent ASOs to degrade the positive and negative‐strands of viral RNA. As the vaccines developed to provide protection against SARS‐CoV‐2 infection are directed against the S protein, the use of ASOs specifically targeting the S‐genes could potentially lower the efficacy of the vaccine or force the appearance of S‐variants, which would be counterproductive. A similar logic applies to ASOs targeting other components of the viral membrane. We thus recommend designing ASOs to target the replication steps to disrupt life‐dependent structural elements and induce genomic RNA degradation, acting on both the positive and negative strands. These complementary modes of action should ensure a strong anti‐viral effect (Figures 4 and 5).
If the formulation of a combination of ASOs proves to be clinically efficient against COVID‐19, it would be possible to prepare combinations of ASOs that match the sequence of variants responsible for any new outbreak. Given the flexibility of design and the facilities to synthesize ASOs, this strategy could also provide the ability to counter viral evolution and the appearance of important new mutations, as currently, during the course of an epidemic (Callaway, 2021; Kirby, 2021). Combinations of ASOs could be used to target several variants at once by acting on specific conserved and variant‐dependent regions. Importantly, the route of administration could be adapted to maximize the therapeutic index depending on the pathogenicity of the virus. ASOs could be delivered by inhalation, as reported for cystic fibrosis (Crosby et al., 2017). This route of administration is expected to locally decrease the local viral load, leading subsequently to the diminution of the spreading of viral particles. Overall, these recent advances underscore the importance of fostering the development and engineering of ASO technology as frontline antiviral agents.
The COVID‐19 outbreak shows our vulnerability to viral infection and, given that history repeats itself, we need to learn from this pandemic and be prepared to fast‐track the production of ready‐to‐use antiviral‐ASO therapeutics, as we did to produce RNA‐vaccines.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Anaïs Quemener: Conceptualization (equal); writing – original draft (equal); writing – review and editing (equal). Marie‐Dominique Galibert: Conceptualization (equal); writing – original draft (equal); writing – review and editing (lead).
RELATED WIREs ARTICLE
The powerful world of antisense oligonucleotides: From bench to bedside.
Quemener, A. M. , & Galibert, M.‐D. (2022). Antisense oligonucleotide: A promising therapeutic option to beat COVID‐19. Wiley Interdisciplinary Reviews: RNA, 13(4), e1703. 10.1002/wrna.1703
Edited by: Jeff Wilusz, Editor‐in‐Chief
Endnotes
European Centre for Disease Prevention and Control. Overview of the implementation of COVID‐19 vaccination strategies and vaccine deployment plans in the EU/EEA—February 1, 2021. ECDC: Stockholm; 2021. https://www.ecdc.europa.eu/en/publications-data/overview-implementation-covid-19-vaccination-strategies-and-vaccine-deployment#no-link
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- Akdim, F. , Tribble, D. L. , Flaim, J. D. , Yu, R. , Su, J. , Geary, R. S. , Baker, B. F. , Fuhr, R. , Wedel, M. K. , & Kastelein, J. J. P. (2011). Efficacy of apolipoprotein B synthesis inhibition in subjects with mild‐to‐moderate hyperlipidaemia. European Heart Journal, 32(21), 2650–2659. 10.1093/eurheartj/ehr148 [DOI] [PubMed] [Google Scholar]
- Baden, L. R. , El Sahly, H. M. , Essink, B. , Kotloff, K. , Frey, S. , Novak, R. , Diemert, D. , Spector, S. A. , Rouphael, N. , Creech, C. B. , McGettigan, J. , Khetan, S. , Segall, N. , Solis, J. , Brosz, A. , Fierro, C. , Schwartz, H. , Neuzil, K. , Corey, L. , … Zaks, T. (2021). Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. New England Journal of Medicine, 384, 403–416. 10.1056/nejmoa2035389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard, S. , Millet, J. K. , Licitra, B. N. , & Whittaker, G. R. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4, 1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biogen . (2019, April 1). Nusinersen. Australian Prescriber, 42, 75–76. 10.18773/austprescr.2019.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, E. (2021). Fast‐spreading COVID variant can elude immune responses. Nature, 589(7843), 500–501. 10.1038/d41586-021-00121-z [DOI] [PubMed] [Google Scholar]
- Chung, Y. H. , Beiss, V. , Fiering, S. N. , & Steinmetz, N. F. (2020, October 27). Covid‐19 vaccine frontrunners and their nanotechnology design. ACS Nano, 14, 12522–12537. 10.1021/acsnano.0c07197 [DOI] [PubMed] [Google Scholar]
- Corbett, K. S. , Edwards, D. K. , Leist, S. R. , Abiona, O. M. , Boyoglu‐Barnum, S. , Gillespie, R. A. , Himansu, S. , Schäfer, A. , Ziwawo, C. T. , DiPiazza, A. T. , Dinnon, K. H. , Elbashir, S. M. , Shaw, C. A. , Woods, A. , Fritch, E. J. , Martinez, D. R. , Bock, K. W. , Minai, M. , Nagata, B. M. , … Graham, B. S. (2020). SARS‐CoV‐2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature, 586(7830), 567–571. 10.1038/s41586-020-2622-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey, D. R. (2017). Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nature Neuroscience, 20(4), 497–499. 10.1038/nn.4508 [DOI] [PubMed] [Google Scholar]
- Crosby, J. R. , Zhao, C. , Jiang, C. , Bai, D. , Katz, M. , Greenlee, S. , Kawabe, H. , McCaleb, M. , Rotin, D. , Guo, S. , & Monia, B. P. (2017). Inhaled ENaC antisense oligonucleotide ameliorates cystic fibrosis‐like lung disease in mice. Journal of Cystic Fibrosis, 16(6), 671–680. 10.1016/j.jcf.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Fazil, M. H. U. T. , Ong, S. T. , Chalasani, M. L. S. , Low, J. H. , Kizhakeyil, A. , Mamidi, A. , Lim, C. F. H. , Wright, G. D. , Lakshminarayanan, R. , Kelleher, D. , & Verma, N. K. (2016). GapmeR cellular internalization by macropinocytosis induces sequence‐specific gene silencing in human primary T‐cells. Scientific Reports, 6, 37721. 10.1038/srep37721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, A. C. L. , Vale, A. J. M. , Guzen, F. P. , Pinheiro, F. I. , Cobucci, R. N. , & de Azevedo, E. P. (2020, September 15). Therapeutic options against the new coronavirus: Updated clinical and laboratory evidences. Frontiers in Medicine, 7, 546. 10.3389/fmed.2020.00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, R. S. , Mercuri, E. , Darras, B. T. , Connolly, A. M. , Kuntz, N. L. , Kirschner, J. , Chiriboga, C. A. , Saito, K. , Servais, L. , Tizzano, E. , Topaloglu, H. , Tulinius, M. , Montes, J. , Glanzman, A. M. , Bishop, K. , Zhong, Z. J. , Gheuens, S. , Bennett, C. F. , Schneider, E. , … de Vivo, D. C. (2017). Nusinersen versus sham control in infantile‐onset spinal muscular atrophy. New England Journal of Medicine, 377(18), 1723–1732. 10.1056/NEJMoa1702752 [DOI] [PubMed] [Google Scholar]
- Frank, D. E. , Schnell, F. J. , Akana, C. , El‐Husayni, S. H. , Desjardins, C. A. , Morgan, J. , Charleston, J. S. , Sardone, V. , Domingos, J. , Dickson, G. , Straub, V. , Guglieri, M. , Mercuri, E. , Servais, L. , Muntoni, F. , & SKIP‐NMD Study Group . (2020). Increased dystrophin production with golodirsen in patients with Duchenne muscular dystrophy. Neurology, 94(21), e2270–e2282. 10.1212/WNL.0000000000009233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, G. , Nordmann, A. , Stein, D. A. , Iversen, P. L. , & Klenk, H. D. (2008). Morpholino oligomers targeting the PB1 and NP genes enhance the survival of mice infected with highly pathogenic influenza A H7N7 virus. Journal of General Virology, 89(4), 939–948. 10.1099/vir.0.83449-0 [DOI] [PubMed] [Google Scholar]
- Geary, R. S. , Henry, S. P. , & Grillone, L. R. (2002). Fomivirsen. Clinical Pharmacokinetics, 41(4), 255–260. 10.2165/00003088-200241040-00002 [DOI] [PubMed] [Google Scholar]
- Geary, R. S. , Norris, D. , Yu, R. , & Bennett, C. F. (2015, June 29). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Advanced Drug Delivery Reviews, 87, 46–51. 10.1016/j.addr.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Huston, N. C. , Wan, H. , Strine, M. S. , de Cesaris Araujo Tavares, R. , Wilen, C. B. , & Pyle, A. M. (2021). Comprehensive in vivo secondary structure of the SARS‐CoV‐2 genome reveals novel regulatory motifs and mechanisms. Molecular Cell, 81(3), 584–598.e5. 10.1016/j.molcel.2020.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, N. A. C. , Kester, K. E. , Casimiro, D. , Gurunathan, S. , & DeRosa, F. (2020, December 1). The promise of mRNA vaccines: A biotech and industrial perspective. Npj Vaccines, 5, 1–6. 10.1038/s41541-020-0159-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht, H. , Mueller, H. , Walther, J. , Zhou, X. , Lopez, A. , Pattupara, T. , Blaising, J. , Pedersen, L. , Albæk, N. , Jackerott, M. , Shi, T. , Ploix, C. , Driessen, W. , Persson, R. , Ravn, J. , Young, J. A. T. , & Ottosen, S. (2018). Liver‐targeted anti‐HBV single‐stranded oligonucleotides with locked nucleic acid potently reduce HBV gene expression in vivo. Molecular Therapy – Nucleic Acids, 11, 441–454. 10.1016/j.omtn.2018.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabanov, A. V. , Vinogradov, S. V. , Ovcharenko, A. V. , Krivonos, A. V. , Melik‐Nubarov, N. S. , Kiselev, V. I. , & Severin, E. S. (1990). A new class of antivirals: Antisense oligonucleotides combined with a hydrophobic substituent effectively inhibit influenza virus reproduction and synthesis of virus‐specific proteins in MDCK cells. FEBS Letters, 259(2), 327–330. 10.1016/0014-5793(90)80039-L [DOI] [PubMed] [Google Scholar]
- Kim, D. , Lee, J. Y. , Yang, J. S. , Kim, J. W. , Kim, V. N. , & Chang, H. (2020). The architecture of SARS‐CoV‐2 transcriptome. Cell, 181(4), 914–921.e10. 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Hu, C. , Moufawad el Achkar, C. , Black, L. E. , Douville, J. , Larson, A. , Pendergast, M. K. , Goldkind, S. F. , Lee, E. A. , Kuniholm, A. , Soucy, A. , Vaze, J. , Belur, N. R. , Fredriksen, K. , Stojkovska, I. , Tsytsykova, A. , Armant, M. , DiDonato, R. L. , Choi, J. , … Yu, T. W. (2019). Patient‐customized oligonucleotide therapy for a rare genetic disease. New England Journal of Medicine, 381(17), 1644–1652. 10.1056/NEJMoa1813279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby, T. (2021). New variant of SARS‐CoV‐2 in UK causes surge of COVID‐19. The Lancet. Respiratory Medicine, 9(2), e20–e21. 10.1016/s2213-2600(21)00005-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer, K. , Castaño, D. , Gómez Atria, D. , Oguin, T. H. , Wang, S. , Manzoni, T. B. , Muramatsu, H. , Hogan, M. J. , Amanat, F. , Cherubin, P. , Lundgreen, K. A. , Tam, Y. K. , Fan, S. H. Y. , Eisenlohr, L. C. , Maillard, I. , Weissman, D. , Bates, P. , Krammer, F. , Sempowski, G. D. , … Locci, M. (2020). SARS‐CoV‐2 mRNA vaccines Foster potent antigen‐specific germinal center responses associated with neutralizing antibody generation. Immunity, 53(6), 1281–1295.e5. 10.1016/j.immuni.2020.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, N. , Ison, M. , & Dunning, J. (2020, November 7). Early triple antiviral therapy for COVID‐19. The Lancet, 396, 1487–1488. 10.1016/S0140-6736(20)32274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowicz, E. , Nogales, A. , Kierzek, E. , Kierzek, R. , Martínez‐Sobrido, L. , & Turner, D. H. (2016). Antisense oligonucleotides targeting influenza A segment 8 genomic RNA inhibit viral replication. Nucleic Acid Therapeutics, 26(5), 277–285. 10.1089/nat.2016.0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , & De Clercq, E. (2020, March 1). Therapeutic options for the 2019 novel coronavirus (2019‐nCoV). Nature Reviews. Drug Discovery, 19, 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Liang, X. H. , Sun, H. , Nichols, J. G. , & Crooke, S. T. (2017). RNase H1‐dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Molecular Therapy, 25(9), 2075–2092. 10.1016/j.ymthe.2017.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K. R. Q. , Maruyama, R. , & Yokota, T. (2017, February 28). Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Design, Development and Therapy, 11, 533–545. 10.2147/DDDT.S97635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla, V. , Wandel, M. P. , Bandyra, K. J. , Ulferts, R. , Wu, M. , Dendooven, T. , Yang, X. , Doyle, N. , Oerum, S. , Beale, R. , O'Rourke, S. M. , Randow, F. , Maier, H. J. , Scott, W. , Ding, Y. , Firth, A. E. , Bloznelyte, K. , & Luisi, B. F. (2021). Targeting the conserved stem loop 2 motif in the SARS‐CoV‐2 genome. Journal of Virology, 95, e0066321. 10.1128/jvi.00663-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer, C. , Stein, D. A. , Mourich, D. V. , Tepper, S. E. , Iversen, P. L. , & Pastey, M. (2008). Inhibition of influenza A H3N8 virus infections in mice by morpholino oligomers. Archives of Virology, 153(5), 929–937. 10.1007/s00705-008-0067-0 [DOI] [PubMed] [Google Scholar]
- Manfredonia, I. , Nithin, C. , Ponce‐Salvatierra, A. , Ghosh, P. , Wirecki, T. K. , Marinus, T. , Ogando, N. S. , Snijder, E. J. , van Hemert, M. J. , Bujnicki, J. M. , & Incarnato, D. (2020). Genome‐wide mapping of SARS‐CoV‐2 RNA structures identifies therapeutically‐relevant elements. Nucleic Acids Research, 48(22), 12436–12452. 10.1093/nar/gkaa1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak, P. , Soszynska‐Jozwiak, M. , Biala, E. , Moss, W. N. , Kesy, J. , Szutkowska, B. , Lenartowicz, E. , Kierzek, R. , & Kierzek, E. (2019). Secondary structure of the segment 5 genomic RNA of influenza A virus and its application for designing antisense oligonucleotides. Scientific Reports, 9(1), 1–16. 10.1038/s41598-019-40443-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullick, A. E. , Fu, W. , Graham, M. J. , Lee, R. G. , Witchell, D. , Bell, T. A. , Whipple, C. P. , & Crooke, R. M. (2011). Antisense oligonucleotide reduction of apoB‐ameliorated atherosclerosis in LDL receptor‐deficient mice. Journal of Lipid Research, 52(5), 885–896. 10.1194/jlr.M011791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S. F. , Crosbie, R. H. , Miceli, M. C. , & Spencer, M. J. (2009, October). Emerging genetic therapies to treat Duchenne muscular dystrophy. Current Opinion in Neurology, 22, 532–538. 10.1097/WCO.0b013e32832fd487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, B. W. , Stein, D. A. , Kroeker, A. D. , Churchill, M. J. , Kim, A. M. , Kuhn, P. , Dawson, P. , Moulton, H. M. , Bestwick, R. K. , Iversen, P. L. , & Buchmeier, M. J. (2005). Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. Journal of Virology, 79(15), 9665–9676. 10.1128/jvi.79.15.9665-9676.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, B. W. , Stein, D. A. , Kroeker, A. D. , Paulino, A. D. , Moulton, H. M. , Iversen, P. L. , & Buchmeier, M. J. (2004). Antisense morpholino‐oligomers directed against the 5′ end of the genome inhibit coronavirus proliferation and growth. Journal of Virology, 78(11), 5891–5899. 10.1128/jvi.78.11.5891-5899.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, S. T. , Chalasani, M. L. S. , Fazil, M. H. U. T. , Prasannan, P. , Kizhakeyil, A. , Wright, G. D. , Kelleher, D. , & Verma, N. K. (2018). Centrosome‐ and golgi‐localized protein kinase N‐associated protein serves as a docking platform for protein kinase A signaling and microtubule nucleation in migrating T‐cells. Frontiers in Immunology, 9(March), 397. 10.3389/fimmu.2018.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi, N. , Hogan, M. J. , Porter, F. W. , & Weissman, D. (2018, March 28). mRNA vaccines – A new era in vaccinology. Nature Reviews. Drug Discovery, 17, 261–279. 10.1038/nrd.2017.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, E. , Koopmans, M. , Go, U. , Hamer, D. H. , Petrosillo, N. , Castelli, F. , Storgaard, M. , Al Khalili, S. , & Simonsen, L. (2020, September 1). Comparing SARS‐CoV‐2 with SARS‐CoV and influenza pandemics. The Lancet. Infectious Diseases, 20, e238–e244. 10.1016/S1473-3099(20)30484-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piroth, L. , Cottenet, J. , Mariet, A. S. , Bonniaud, P. , Blot, M. , Tubert‐Bitter, P. , & Quantin, C. (2021). Comparison of the characteristics, morbidity, and mortality of COVID‐19 and seasonal influenza: A nationwide, population‐based retrospective cohort study. The Lancet. Respiratory Medicine, 9(3), 251–259. 10.1016/S2213-2600(20)30527-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyush, R. , Rajarshi, K. , Chatterjee, A. , Khan, R. , & Ray, S. (2020, September 1). Nucleic acid‐based therapy for coronavirus disease 2019. Heliyon, 6, e05007. 10.1016/j.heliyon.2020.e05007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack, F. P. , Thomas, S. J. , Kitchin, N. , Absalon, J. , Gurtman, A. , Lockhart, S. , Perez, J. L. , Pérez Marc, G. , Moreira, E. D. , Zerbini, C. , Bailey, R. , Swanson, K. A. , Roychoudhury, S. , Koury, K. , Li, P. , Kalina, W. V. , Cooper, D. , Frenck, R. W., Jr. , Hammitt, L. L. , … Gruber, W. C. (2020). Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. New England Journal of Medicine, 383(27), 2603–2615. 10.1056/nejmoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemener, A. M. , Bachelot, L. , Forestier, A. , Donnou‐Fournet, E. , Gilot, D. , & Galibert, M.‐D. (2020). The powerful world of antisense oligonucleotides: From bench to bedside. WIREs RNA, 11(5), e1594. 10.1002/wrna.1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricotta, D. N. , & Frishman, W. (2012). Mipomersen: a safe and effective antisense therapy adjunct to statins in patients with hypercholesterolemia. Cardiology in Review, 20(2), 90–95. 10.1097/CRD.0b013e31823424be [DOI] [PubMed] [Google Scholar]
- Rinaldi, C. , & Wood, M. J. A. (2017). Antisense oligonucleotides: The next frontier for treatment of neurological disorders. Nature Reviews. Neurology, 14(1), 9–21. 10.1038/nrneurol.2017.148 [DOI] [PubMed] [Google Scholar]
- Roehr, B. (1998). Fomivirsen approved for CMV retinitis. Journal of the International Association of Physicians in AIDS Care, 4(10), 14–16. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11365956 [PubMed] [Google Scholar]
- Stasi, C. , Fallani, S. , Voller, F. , & Silvestri, C. (2020). Treatment for COVID‐19: An overview. European Journal of Pharmacology, 889, 173644. 10.1016/j.ejphar.2020.173644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, C. A. , & Castanotto, D. (2017). FDA‐approved oligonucleotide therapies in 2017. Molecular Therapy, 25(5), 1069–1075. 10.1016/j.ymthe.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson, M. L. , & Zamecnik, P. C. (1978). Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proceedings of the National Academy of Sciences of the United States of America, 75(1), 285–288. 10.1073/PNAS.75.1.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Li, P. , Ju, X. , Rao, J. , Huang, W. , Ren, L. , Zhang, Q. C. , Xiong, T. , Xu, K. , Zhou, X. , Gong, M. , Miska, E. , Ding, Q. , Wang, J. , & Zhang, Q. C. (2021). In vivo structural characterization of the SARS‐CoV‐2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell, 184(7), 1865–1883.e20. 10.1016/j.cell.2021.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitravene Study Group . (2002). A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with AIDS. American Journal of Ophthalmology, 133(4), 467–474. 10.1016/s0002-9394(02)01327-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


