Abstract
The mef(A) gene from a clinical isolate of Streptococcus pneumoniae exhibiting the M-type resistance to macrolides was found to be part of the 7,244-bp chromosomal element Tn1207.1, which contained 8 open reading frames. orf2 encodes a resolvase/invertase, and orf5 is a homolog of the macrolide-streptogramin B resistance gene msr(SA).
A new mechanism of resistance to macrolides based on an efflux system has recently emerged among clinical isolates of Streptococcus pyogenes and Streptococcus pneumoniae (35, 37). It is due to the presence of macrolide efflux (mef) genes conferring the M phenotype, which is characterized by resistance to macrolides and sensitivity to lincosamides and streptogramin B antibiotics (37). mef(A), originally described in S. pyogenes (8), and mef(E), originally described in S. pneumoniae (38), are 90% identical and were assigned to the same mef(A) class of macrolide resistance determinants (32). Genes of the mef(A) class (i) were found in different gram-positive genera including Corynebacterium, Enterococcus, Micrococcus, and Streptococcus, (ii) were never found associated with extrachromosomal plasmid DNA, and (iii) in some instances were shown to be transferred by conjugation (3–5, 10, 14, 15, 17, 26, 40). Both mef(E) and mef(A) have been reported to occur in pneumococci from Italy (19, 20, 25). Here we report the first characterization of a mef-carrying genetic element. The recently proposed nomenclature for macrolide resistance determinants (32) was adopted throughout this paper.
Transfer of mef(A).
To isolate and characterize the genetic element that carries the macrolide efflux gene of S. pneumoniae STR174b, mef(A) was transferred from STR174b to Rx1 (34), a rough (unencapsulated) laboratory strain of pneumococcus. Clinical strain STR174b, isolated from sputum and exhibiting the M-type resistance to macrolides, was used as donor in conjugation and transformation experiments (Table 1). The mef(A) gene could not be transferred from STR174b by conjugation (<3.4 × 10−8 transconjugants per donor), in filter matings performed as described by Smith and Guild (36), while successful transfer to Rx1 was accomplished by transformation (Table 1). Competent cells of Rx1 were prepared as previously described (28); transforming DNA of STR174b was added at 1 μg per ml of competent cells, and erythromycin-resistant transformants were isolated at a frequency of 4.7 × 10−4 per recipient (Table 1). MF4, a representative transformant, was kept for further analysis. MF4 showed the same resistance phenotype as the parent clinical strain, being resistant to erythromycin (MIC, 8 μg/ml) and sensitive to lincosamides and streptogramin B antibiotics. Again, mef(A) could not be transferred by conjugation in matings where MF4 was the donor and S. pneumoniae, S. pyogenes, and Streptococcus gordonii were used as recipients (Table 1).
TABLE 1.
Transfer of Tn1207.1
| Streptococcus pneumoniae donora | Recipientb | Frequencyc |
|---|---|---|
| Transformation | ||
| STR174b (clinical isolate) | S. pneumoniae Rx1 (34) | 4.7 × 10−4 |
| Conjugation | ||
| STR174b (clinical isolate) | S. pneumoniae DP1002 (34) | <3.4 × 10−8 |
| MF4 (transformant of Rx1, this work) | S. pneumoniae DP1002 (34) | <1.1 × 10−8 |
| MF4 (transformant of Rx1, this work) | S. pyogenes D471 (33) | <1.4 × 10−8 |
| MF4 (transformant of Rx1, this work) | S. gordonii GP204 (29, 30) | <1.6 × 10−8 |
The origin is shown in parentheses.
The reference(s) are shown in parentheses.
Conjugation frequency is expressed as CFU of transconjugants per CFU of donor, whereas transformation frequency is expressed as CFU of transformants per CFU of recipient.
Sequencing and sequence analysis.
The nucleotide sequence of the DNA adjacent to mef(A) was determined in both STR174b and MF4, using templates obtained by ligation-mediated and/or inverse PCR (24, 31). Direct automated sequencing of long PCR fragments was performed as previously described (11, 12). The software BLAST (2) was used to conduct homology searches of the GenBank database and the microbial genome databases available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) and at the WIT Computational Biology Group at Argonne National Laboratory website (http://wit.mcs.anl.gov./WIT2/CGI). Sequence analysis was carried out at the Baylor College of Medicine Search Launcher server (http://dot.imgen.bcm.tmc.edu:9331/index.htlm). RNA structure prediction was done using RNA structure 2.5 (21).
Nucleotide sequence of the mef(A) element.
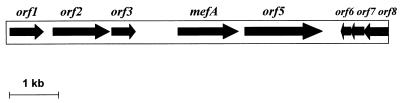
Both in STR174b and in MF4 the mef gene was shown to be 100% identical to the already described mef(A) gene (8) and was found to be part of a chromosomal element which was designated Tn1207.1. Tn1207.1 was found to be 7,244 bp in size (Fig. 1), and its nucleotide sequence was identical in both strains. Sequence data analysis showed the presence of 8 open reading frames (ORFs), of which the first 5 have the same direction of transcription, while orf6, orf7, and orf8 are oriented opposite to the others (Fig. 1). Between orf3 and mef(A) there is an intergenic region with a high potential for the formation of hairpins (21). Palindromes indicative of terminators can be detected downstream of orf1 and orf3. Terminal repeats could not be detected.
FIG. 1.
Structure of the chromosomal genetic element Tn1207.1, which is 7,244-bp long.
Chromosomal insertion site.
Tn1207.1 was found integrated at a specific site in the chromosome of S. pneumoniae, downstream of the A nucleotide in position 1676 of the celB gene (27). Both in clinical isolate STR174b and in transformant MF4, the integration of Tn1207.1 caused a 1,947-bp deletion in the pneumococcal genome (nucleotides 1393 to 3339 of contig 4139) (The Institute for Genomics Research [http://www.tigr.org]) involving the 3′ end of celB (nucleotides 2775 to 3339 of contig 4139) and the 5′ end of orf436 (nucleotides 1393 to 1784 of contig 4139) (WIT website [see above]).
ORFs of Tn1207.1.
To identify genes homologous to the ORFs of Tn1207.1, homology searches were done at a protein level, using the BLAST software (2). The E value, which is considered a very reliable statistical scoring measure (7), is reported for each homolog detected by the search (Table 2). No homolog was found for orf1 and orf3, while orf2 was found to be homologous to site-specific recombinases of genetic elements of gram-positive bacteria. These elements included chromosomal elements of Staphylococcus aureus (13) and Bacillus subtilis (39), a bacteriophage of Listeria monocytogenes (16), and a transposon of Clostridium perfringens (6) (Table 2). An additional ORF potentially involved in determining resistance to macrolides and streptogramin antibiotics was found adjacent to mef(A). orf5, whose coding sequence starts 119-bp downstream of the mef(A) stop codon (Fig. 1), was found to be homologous to msr(SA) (22) and vga(A) (1). msr(SA) confers resistance to macrolides and streptogramin B, and vga(A) confers resistance to streptogramin A antibiotics. Both genes are putative members of the ABC transporter superfamily and mediate resistance by encoding an antibiotic-specific efflux pump (32). orf6, orf7, and orf8 are homologous to 3 ORFs of the pneumococcal conjugative transposon Tn5252 (23). The 5′ end of orf8 appears truncated since its gene product is homologous to the C-terminal portion of UmuC, an UV-resistance protein encoded by Tn5252 (Table 2). No function is known for the gene products of orf6 and orf7 and the Tn5252 homologs. Even if no significant homology can be detected at the nucleotide level, orf6 to orf8 and their Tn5252 homologs show a similar organization characterized by overlapping ORFs. orf6 and orf7 overlapped by 14 bp, and orf7 and orf8 overlapped by 4 bp.
TABLE 2.
Homologies of the ORFs of Tn1207.1
| ORFa | Homologous geneb | Originc | Proposed function of gene product | E value | Amino acid identity | Amino acid similarity |
|---|---|---|---|---|---|---|
| orf1 (218) | None | |||||
| orf2 (370) | ccrB (AB014438) | mec DNA (S. aureus) (13) | Site-specific recombinase | 4e-18 | 93/294 (31%) | 144/294 (48%) |
| cisA (M29040) | skin element (B. subtilis) (39) | Site-specific recombinase | 2e-14 | 79/304 (25%) | 139/304 (44%) | |
| int (AJ242593) | Bacteriophage A118 (L. monocytogenes) (16) | Integrase | 6e-12 | 59/196 (30%) | 104/196 (52%) | |
| tnpX (U15027) | Transposon Tn4451 (C. perfringens) (6) | Site-specific recombinase | 1e-11 | 78/312 (25%) | 132/312 (42%) | |
| orf3 (138) | None | |||||
| mefA (405) | mef(A) (U70055) | (S. pyogenes) (8) | Macrolide-efflux protein | 0 | 405/405 (100%) | |
| orf5 (487) | msr(SA) (AB016613) | Plasmid pEP2104 (S. aureus) (22) | Macrolide-resistance protein | 6e-91 | 180/471 (38%) | 285/471 (60%) |
| vga(A) (M90056) | Plasmid pIP524 (S. aureus) (1) | Streptogramin A-resistance protein | 2e-89 | 183/485 (37%) | 283/485 (57%) | |
| orf6 (99) | orf11 (L29324) | Conjugative transposon Tn5252 (S. pneumoniae) (23) | Unknown | 2e-22 | 45/76 (59%) | 61/76 (80%) |
| orf7 (122) | orf12 (L29324) | Conjugative transposon Tn5252 (S. pneumoniae) (23) | Unknown | 3e-06 | 30/96 (31%) | 46/96 (47%) |
| orf8 (117) | umuC (L29324) | Conjugative transposon Tn5252 (S. pneumoniae) (23) | UV resistance protein | 6e-36 | 70/116 (60%) | 89/116 (76%) |
The number of amino acids is shown in parentheses.
The accession number is shown in parentheses.
The number in parentheses is the reference.
Conclusions.
In this work we characterized a genetic element that carries the macrolide efflux gene mef(A) in a clinical isolate of S. pneumoniae exhibiting the M-type resistance to macrolides. mef(A) was found to be part of a chromosomal genetic element designated Tn1207.1. Tn1207.1 should be considered a defective transposon, since it terminates with a truncated ORF at the right side, while in different clinical isolates it is integrated at the same chromosomal site and has the same overall genetic organization but is larger in size at its right side (Santagati and colleagues, unpublished data). The presence of an ORF potentially encoding a site-specific recombinase of the resolvase/invertase family resembles clostridial transposons Tn4451, Tn4453, and Tn5397, in which the TnpX and TndX resolvases are involved in the transposon excision process (6, 9, 18, 41). Although it has been reported that mef genes from S. pneumoniae and group C streptococci can be transferred by conjugation (15, 17), Tn1207.1 appeared to be nonconjugative in nature.
Nucleotide sequence accession number.
The complete nucleotide sequence of Tn1207.1 was assigned GenBank accession no. AF227520.
Acknowledgments
This work was supported in part by grants from CNR (P. F. Biotecnologie) and MURST (Cofinanziamento 1999).
We thank Alessandra Zanchi for help in the initial phases of this work.
REFERENCES
- 1.Allignet J, Loncle V, Sohl N. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene. 1992;117:45–51. doi: 10.1016/0378-1119(92)90488-b. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arpin C, Canron M H, Maugein J, Quentin C. Incidence of mefA and mefE genes in viridans group streptococci. Antimicrob Agents Chemother. 1999;43:2335–2336. doi: 10.1128/aac.43.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arpin C, Canron M H, Nourry P, Quentin C. Emergence of mefA and mefE genes in beta-haemolytic streptococci and pneumococci in France. J Antimicrob Chem. 1999;44:133–134. doi: 10.1093/jac/44.1.133. [DOI] [PubMed] [Google Scholar]
- 5.Arpin C, Daube H, Tessier F, Quentin C. Presence of mefA and mefE genes in Streptococcus agalactiae. Antimicrob Agents Chemother. 1999;43:944–946. doi: 10.1128/aac.43.4.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannam T L, Crellin P K, Rood J I. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site-specific recombinase excises a circular transposon molecule. Mol Microbiol. 1995;16:535–551. doi: 10.1111/j.1365-2958.1995.tb02417.x. [DOI] [PubMed] [Google Scholar]
- 7.Brenner S E, Chothia C, Hubbard T J P. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proc Natl Acad Sci USA. 1998;95:6073–6078. doi: 10.1073/pnas.95.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clancy J, Petitpas J, Dib-Hajj F, Yuan W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 9.Crellin P K, Rood J I. The resolvase/invertase domain of the site-specific recombinase TnpX is functional and recognizes a target sequence that resembles the junction of the circular form of the Clostridium perfringens transposon Tn4451. J Bacteriol. 1997;179:5148–5156. doi: 10.1128/jb.179.16.5148-5156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovanetti E, Montanari M P, Mingoia M, Varaldo P E. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob Agents Chemother. 1999;43:1935–1940. doi: 10.1128/aac.43.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iannelli F, Giunti L, Pozzi G. Direct sequencing of long PCR fragments. Mol Biotechnol. 1998;10:183–185. doi: 10.1007/BF02760864. [DOI] [PubMed] [Google Scholar]
- 12.Iannelli F, Pearce B J, Pozzi G. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol. 1999;81:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kataja J, Houvinen P, Skurnik M, Seppala H. Erythromycin resistance genes in group A streptococci in Finland. The Finnish study group for antimicrobial resistance. Antimicrob Agents Chemother. 1999;43:48–52. doi: 10.1128/aac.43.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kataja J, Seppala H, Skurnik M, Sarkkinen H, Huovinen P. Different erythromycin resistance mechanism in group C and group G streptococci. Antimicrob Agents Chemother. 1998;42:1493–1494. doi: 10.1128/aac.42.6.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loessner M J, Wendlinger G, Scherer S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence for conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol. 1995;16:1231–1241. doi: 10.1111/j.1365-2958.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 17.Luna V A, Coates P, Eady E A, Cove J H, Nguyen T T, Roberts M C. A variety of gram-positive bacteria carry mobile mef genes. J Antimicrob Chem. 1999;44:19–25. doi: 10.1093/jac/44.1.19. [DOI] [PubMed] [Google Scholar]
- 18.Lyras D, Storie C, Huggins A S, Crellin P K, Bannam T L, Rood J I. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob Agents Chemother. 1998;23:784–786. doi: 10.1128/aac.42.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchese A, Ramirez M, Schito G C, Tomasz A. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J Clin Microbiol. 1998;36:2944–2949. doi: 10.1128/jcm.36.10.2944-2949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchese A, Tonoli E, Debbia E A, Schito G C. Macrolide resistance mechanisms and expression of phenotypes among Streptococcus pneumoniae circulating in Italy. J Antimicrob Chem. 1999;44:461–464. doi: 10.1093/jac/44.4.461. [DOI] [PubMed] [Google Scholar]
- 21.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka M, Janosi K, Endou K, Nakajima K. Cloning and sequences of inducible and constitutive macrolide resistance genes in Staphylococcus aureus that correspond to an ABC transporter. FEMS Microbiol Lett. 1999;181:91–100. doi: 10.1111/j.1574-6968.1999.tb08830.x. [DOI] [PubMed] [Google Scholar]
- 23.Munoz-Najar U, Vijayakumar M N. An operon that confers UV resistance by evoking the SOS mutagenic response in streptococcal conjugative transposon Tn5252. J Bacteriol. 1999;181:2782–2788. doi: 10.1128/jb.181.9.2782-2788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman H, Gerber A S, Hartl D L. Genetic application of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oster P, Zanchi A, Cresti S, Lattanzi M, Montagnani F, Cellesi C, Rossolini G M. Patterns of macrolide resistance determinants among community-acquired Streptococcus pneumoniae isolates over a 5-year period of decreased macrolide susceptibility rates. Antimicrob Agents Chemother. 1999;43:2510–2512. doi: 10.1128/aac.43.10.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Trallero E, Urbieta M, Montes M, Ayestaran I, Marimon J M. Emergence of Streptococcus pyogenes strains resistant to erythromycin in Gipuzkoa, Spain. Eur J Clin Microbiol. 1998;17:25–31. doi: 10.1007/BF01584359. [DOI] [PubMed] [Google Scholar]
- 27.Pestova E V, Morrison D A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pozzi G, Masala L, Iannelli F, Manganelli R, Havarstein L S, Piccoli L, Simon D, Morrison D A. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of peptide pheromone. J Bacteriol. 1996;178:6087–6090. doi: 10.1128/jb.178.20.6087-6090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pozzi G, Musmanno R A, Lievens P M J, Oggioni M R, Plevani P, Manganelli R. Method and parameters for genetic transformation of Streptococcus sanguis Challis. Res Microbiol. 1990;141:659–670. doi: 10.1016/0923-2508(90)90060-4. [DOI] [PubMed] [Google Scholar]
- 30.Pozzi G, Musmanno R A, Renzoni E A, Oggioni M R, Cusi M G. Host-vector system for integration of recombinant DNA into chromosomes of transformable and nontransformable streptococci. J Bacteriol. 1988;170:1969–1972. doi: 10.1128/jb.170.4.1969-1972.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prod'hom G, Lagier B, Pelicic V, Hance A J, Gicquel B, Guilhot C. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol Lett. 1998;158:75–81. doi: 10.1111/j.1574-6968.1998.tb12803.x. [DOI] [PubMed] [Google Scholar]
- 32.Roberts M C, Sutcliffe J, Courvalin P, Jensen L B, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott J R, Fischetti V A. Expression of streptococcal M protein in Escherichia coli. Science. 1983;221:758–760. doi: 10.1126/science.6192499. [DOI] [PubMed] [Google Scholar]
- 34.Shoemaker N B, Guild W R. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol Gen Genet. 1974;128:283–290. doi: 10.1007/BF00268516. [DOI] [PubMed] [Google Scholar]
- 35.Shortridge V D, Flamm R K, Ramer N, Beyer J, Tanaka S K. Novel mechanism of macrolide resistance in Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 1996;26:73–78. doi: 10.1016/s0732-8893(96)00183-6. [DOI] [PubMed] [Google Scholar]
- 36.Smith M D, Guild W R. Improved method for conjugative transfer by filter mating of Streptococcus pneumoniae. J Bacteriol. 1980;144:457–459. doi: 10.1128/jb.144.1.457-459.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutcliffe J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tait-Kamradt A, Clancy J, Cronan M, Dib-Hajj F, Wondrack L, Yuan W, Sutcliffe J. mefE is necessary for erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:2251–2255. doi: 10.1128/aac.41.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takemaru K, Mizuno M, Sato T, Takeuchi M, Kobayashi Y. Complete nucleotide sequence of a skin element excised by DNA rearrangement during sporulation in Bacillus subtilis. Microbiology. 1995;141:323–327. doi: 10.1099/13500872-141-2-323. [DOI] [PubMed] [Google Scholar]
- 40.Valisena S, Falci C, Mazzariol A, Cornaglia G, Cocuzza C E, Nicoletti P, Rescaldani R, Fontana R. Molecular typing of erythromycin-resistant Streptococcus pyogenes strains with the M phenotype isolated in Italy. Eur J Clin Microbiol. 1999;18:260–264. doi: 10.1007/s100960050274. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Robert A P, Lyras D, Rood J I, Wilks M, Mullany P. Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase, TndX. J Bacteriol. 2000;182:3775–3783. doi: 10.1128/jb.182.13.3775-3783.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]