Abstract
The COVID‐19 infections have posed an unprecedented global health emergency, with nearly three million deaths to date, and have caused substantial economic loss globally. Hence, an urgent exploration of effective and safe diagnostic/therapeutic approaches for minimizing the threat of this highly pathogenic coronavirus infection is needed. As an alternative to conventional diagnosis and antiviral agents, nanomaterials have a great potential to cope with the current or even future health emergency situation with a wide range of applications. Fundamentally, nanomaterials are physically and chemically tunable and can be employed for the next generation nanomaterial‐based detection of viral antigens and host antibodies in body fluids as antiviral agents, nanovaccine, suppressant of cytokine storm, nanocarrier for efficient delivery of antiviral drugs at infection site or inside the host cells, and can also be a significant tool for better understanding of the gut microbiome and SARS‐CoV‐2 interaction. The applicability of nanomaterial‐based therapeutic options to cope with the current and possible future pandemic is discussed here.
Keywords: antiviral drugs, biosensors, drug delivery, nanoformulations, nanotechnology, vaccines
COVID‐19 pandemic is posing a threat to humanity. The tunable physicochemical properties and wide applications of nanomaterial are providing new avenues to cope with the COVID‐19 pandemic. The biomedical applications of inherent properties and enormous potential of nanomaterials as a diagnostic tool to antiviral drugs for the cure of SARS‐CoV‐2 infection with related safety issues are focused here.
1. Introduction
A novel coronavirus (SARS‐CoV‐2) causing pneumonia was traced and identified in China in December 2019. Based on the phylogenetic and taxonomical finding on February 11, the coronavirus (COV) study group (CGS) of the International Committee on Virus Taxonomy (ICTV) designated the virus as SARS‐CoV‐2.[ 1 ] The same day, the director general of the World Health Organization (WHO) designated the disease caused by SARS‐CoV‐2 “coronavirus disease 2019” (COVID‐19). The WHO declared COVID‐19 a pandemic On March 11, 2020. Globally as of June 2021, 179 686 071 confirmed cases of COVID‐19, including 3 899 172 deaths, reported by WHO.[ 2 ] Reducing the burden of COVID‐19 infection by developing vaccines and antiviral drugs is still a big challenge for researchers and clinicians.
Nowadays, nanotechnology is a promising player in the scientific and medical fields with many applications.[ 3 , 4 ] A new dimension has been opened by the functionally tunable nanosized materials for industrial, biomedical, and scientific purposes, which provides a set of techniques that allow the manipulation of structural and functional properties on a microscale.[ 5 , 6 ] Nanotechnology is kindred with the biomedicinal involvement of nanoparticles (NPs), which are not only used as the backbone in the vaccine to formulate “nanovaccines,” nanomaterial‐based antiviral agents, disinfectants; which inhibit the multiplication within the host or inactivate them outside the host, but also the application of designed nanorobots to detect and repairs of tissue defects at the cellular level.[ 7 , 8 ] Unlike the conventional drug formulations, which typically circulate throughout the entire body parts, nanoparticles may target a desired part of the body as a targeted drug delivery vehicle. Compared to conventional vaccines, nanovaccines or surface‐functionalized nanoparticles can be designed, explicitly targeting lymphatic organs and immune cells to provoke better immune responses. Moreover, the localized infections are challenging to be treated by the conventional vaccines but can be treated by the nanovaccines with specific cells or organ‐targeting molecules. In some cases, the nanobased settings are applied to enhance the solubility of hydrophobic compounds. These systems are helpful to protect the antigens from degradation and stabilize a large number of therapeutic biomolecules, such as peptides, proteins, and nucleic acids on the substrate.[ 9 ] Viral infections, particularly COVID‐19, have posed a severe threat to public health globally because of their wide‐sphered distribution and their potent ability to change the genetic makeup.[ 10 ] Unfortunately, viruses have adopted various alternative tactics for entry into cells, long‐term survival strategy within the cells, and activation, inhibition, or modification of the host defense mechanisms at multiple stages.[ 11 ] So that, viral infections are one of the leading causes of morbidity and mortality, which causes a substantial global economic regression.[ 12 , 13 ] Nanotechnology wrapping a multidisciplinary field may find the permanent solution to these lethal infections. The unique physicochemical property of nanomaterials is their selective interaction with viruses that make them a potential antiviral agent.[ 14 ] This review aims to provide an overview of previous and recent nanomaterial‐based diagnostic and therapeutic options against viruses, especially coronavirus, and focus on possible strategies to fight against the current pandemic.
2. The General Structure and Pathophysiology of COVID 19
The novel coronavirus taxonomically belongs to the coronaviridae family, whose genome size usually range from 26 to 32 kb.[ 15 , 16 , 17 , 18 ] The virus genome has been fully sequenced, showing a higher order of similarity in nucleotide sequences with other coronaviruses responsible for fatal pulmonary diseases such as SARS‐CoV.[ 17 , 19 ] corona virus (COV) could be 50−150 nm in size, which structural components are made of nucleocapsid (N) proteins bound to (+) single‐stranded RNA covered by an envelope, which consists of a lipid bilayer containing membrane (M), envelope (E), and spike (S) proteins (Figure 1 ).[ 20 , 21 ] The most common symptoms of COVID‐19 infection are fever, cough, and fatigue. Other clinically diagnosed symptoms are dyspnea, headache, loss of appetite, panting, sore throat, vomiting, diarrhea, rhinorrhea, and abdominal pain. The degree of disease severity may be high in patients having previous health complications such as hypertension, diabetes, and coronary heart disease.[ 22 , 23 ] The incubation period is observed to be between 5 to 14 days based on human host physiology before disease onset. Recently, it is also reported that COVID‐19 exhibits severe effects on other organs such as the heart, kidney, eyes, and brain.[ 24 , 25 , 26 , 27 ] The central target in the human cell, the virus, selectively interacts with the cell surface receptor called angiotensin‐converting enzyme II (ACE2), which is required for efficient uptake of the virus particle the host cell.[ 27 , 28 , 29 , 30 ] The molecular details of the interaction have been investigated by analyzing the crystal structure, which shows that the C‐terminal domain (receptor binding domain (RBD) of the envelope‐embedded spike (S) 1 protein) of SARS‐CoV‐2 binds with ACE2.[ 20 ] Further, coronavirus multiplication in host cell has been characterized by the following crucial steps: i) attachment of viral spike (S) glycoprotein to its complementary human cell receptor, ii) virus endocytosis in cells, iii) virus uncoating, iv) virus replication, and v) virus release (Figure 2 ).[ 20 ] Therefore, the binding of the spike protein to the ACE2 receptor is essential for hijacking the host cell machinery and multiplication of virus, and treatment approaches are needed to focus on disrupting this key event. Further in this scenario, there may be multiple possible approaches to inhibit ACE2 binding: i) involvement of soluble, recombinant ACE2 protein, which acts as a mimetic receptor to bind the virus and prevent uptake into host cells selectively.[ 27 ] ii) vaccination with antibodies that specifically bind to the spike protein and interfere with ACE2 interaction and iii) inhibition of host proteases that process the Spike protein and are essential for ACE2 binding to subsequent membrane fusion that enable intracellular delivery of the virus.[ 29 ]
Figure 1.
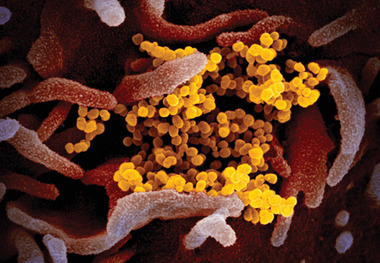
Scanning electron microscopic image showing SARS‐CoV‐2(also known as 2019‐nCoV) virus particles (yellow), isolated from a COVID‐19 patient in the U.S., emerging from the surface of host cells (pink) cultured in the lab. Reproduced under terms of the CC‐BY license.[ 21 ] Copyright 2020, NIAID.
Figure 2.
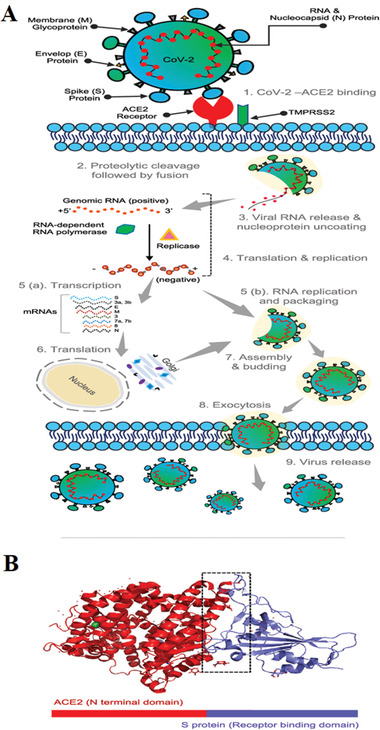
SARS‐CoV‐2 structure and pathophysiology. A) SARS‐CoV‐2 life cycle: The viral spike (S) protein binds to the ACE2 receptor of the host. Following the entry, the virus envelope's proteolytic cleavage occurs and facilitates the release of viral genomic RNA in the cytoplasm, leading to the formation of small RNAs (subgenomic mRNAs). These mRNAs are translated to several viral proteins (i.e., S, M, N, etc.) essential for the reassembly of the virus particle. These synthesized proteins (S, E, and M) enter the endoplasmic reticulum (ER), where nucleoprotein complex formation occurs from the combination of nucleocapsid (N) protein and genomic positive RNA strand. The complete functional virus particle (proteins and viral genomic RNA assembly) occurs in ER‐Golgi compartments of the host cell. These virus particles are then transported and released via vesicles formation and exocytosis; B) the interaction of ACE2‐RBD (S protein): A single unit of peptidase domain of human ACE2 (red) interacting with the RBD sequence of the virus Spike protein (blue), (boxed region represents the amino acid interactions sites). Reproduced with permission.[ 20 ] Copyright 2020, American Chemical Society.
3. Role of Nanomaterials in Viral Infection Diagnosis and Therapy
The infectious agents (viruses, bacteria, fungi, and other parasites) kill nearly 15 million people annually.[ 30 , 31 , 32 ] The maximum number of mortalities is reportedly caused by acute respiratory infections and human immunodeficiency virus (HIV).[ 32 , 33 ] Many therapeutic routes are opted to treat these types of infections. Unfortunately, the current COVID‐19 pandemic posed a significant threat before humanity and kills over one million people worldwide, so there is an urgent need for improved and efficient treatment strategies to overcome the challenge. Nanotechnology has proved essential in the biomedical and translational medicine field by offering significant advantages in diagnostics and therapeutic applications.[ 34 ] Furthermore, the biological system connects a strong link with nanotechnology, which combines both phenomena of nanobiotechnology and nanomedicine to diagnose, prevent, and treat diseases.[ 35 , 36 ] Moreover, nanomaterials are also explored as an actual delivery vehicle in the drug delivery system.[ 31 , 37 , 39 ] shows some practical inactivation approaches that might help block the spread of SARS‐CoV‐2 infection. The development of antiviral therapies takes a long time before being available to patients because many regulatory norms are required to fulfil the safety and efficacy of drug formulations.[ 40 , 41 ] Moreover, resistance might be developed due to mutations in the precise viral targets as SARS‐CoV‐2 continues to mutate; these mutations are also common when attempting to treat other viral infections. There has been increasing interest in the design and development of novel, broad‐spectrum antiviral agents, which might be less prone to resistance and could be employed against different pathogenic viruses, including new variants.[ 42 , 43 , 44 ] Considering these challenges, nanotechnology provides several solutions to diagnose and treat viral infections outside and inside the host. In this context, various nanomaterial‐based therapeutics have already been successfully studied to treat several human viral pathogens such as HIV, human papillomavirus, herpes simplex, and respiratory viruses.[ 42 , 43 , 44 , 45 ] Nanomaterial‐based formulation strategies should be an advantage to fight against COVID‐19 as well as any future pandemics, in many ways, including i) nanomaterial‐based novel vaccines and drugs, which can be used for targeted delivery of broad‐spectrum antivirals to the affected body parts; ii) precise, rapid, sensitive and reliable diagnosis, and serology, iii) ultrafine filters for face masks or blood‐filtering, iv) novel antiviral coated surfaces that prevent the viral adhesion and can inactivate the virus, and v) the improvement of tools for contact tracing (Figure 3 ).[ 46 ]
Figure 3.
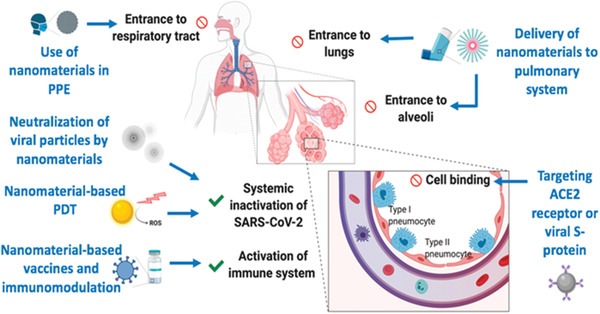
Nanomaterials for prevention and therapy of COVID‐19. Integrating nanomaterials into personal protective equipment (PPE) can prevent the entrance of SARS‐CoV‐2 into the respiratory system. Nanomaterials could also be used as a carrier of drugs to the pulmonary system via inhalators. Cellular interaction of viral particles at the alveoli can be inhibited using targeted NPs against angiotensin‐converting enzyme 2 (ACE2) receptors or viral S protein. Various mechanisms can inactivate viral particles systemically, such as neutralizing NPs or photocatalytic nanomaterials. Nanomaterial‐based vaccines or immunomodulation can prevent SARS‐CoV‐2 infection or even boost the immune response during infection. PDT, photodynamic therapy. Reproduced with permission.[ 46 ] Copyright 2020, American Chemical Society.
3.1. Nanomaterial‐Based Disinfection Formulations
In the view of various transmission modes of coronavirus (via cough, air droplets, or biofluids), one possible strategy to fight against the virus is through blocking its spread by disinfecting air, skin or surrounding surfaces.[ 47 ] Conventionally, chemical disinfectants like chlorine, peroxides, quaternary amines, and alcohols are used to disinfection and sterilize personal protective equipment and other surfaces against a wide variety of pathogens.[ 48 ] Despite better chemical disinfectants, some limitations are also associated, such as high concentration requirements for complete viral load elimination, compromised effectiveness over time, and possible risks to public health and the environment.[ 49 , 50 ] Hence, metallic nanoparticles such as silver, copper, titanium dioxide have been proposed as alternative disinfection agents because of their inherent broad‐spectrum antiviral activities, long time persistence, and requirement of a much lower dosage for effect.[ 51 , 52 ] Previous studies demonstrated that silver nanocluster and silica nanocomposite coating on face masks had antiviral effects against SARS‐CoV‐2.[ 53 ] For example, titanium dioxide and silver ions nanoformulation is durable, self‐sterilizing, and applied for disinfecting surfaces. Similarly, titanium dioxide nanoparticles based photocatalytic coating on surfaces can decompose organic compounds, including viruses, in the presence of light by damaging the viral membrane.[ 54 ] The antiviral nanomaterial coating can also be achieved into respiratory face masks to enhance their viral inhibitory effect.[ 55 ] Scientists successfully designed a breathable and disposable filter cartridge from cellulose nanofibers, which efficiently filters fine particles smaller than 100 nm.[ 56 ] Due to, high surface‐area‐to‐volume ratio and specific physicochemical properties, other nanomaterials such as graphene can also be incorporated to adsorb and complete eliminate of SARS‐CoV‐2 burden.[ 57 ] For example, a USA‐based company has developed a reusable face mask made up of microporous conductive graphene foam that can trapping microorganisms and inactivates them through the conduction of electrical charge.[ 58 ] Nanoparticle‐based colloidal suspensions are promising antiviral solutions for the disinfection and sterilization of personal protective equipment and other surfaces. Cho et al. 2014, formulated a mixture composed of colloidal silver, titanium dioxide (TiO2) NPs, a dispersion stabilizer, a binder and water, showed efficient antibacterial, antifungal and antiviral activities.[ 59 ] Similarly, an Indian start‐up has developed a nonalcoholic colloidal silver solution that can disinfect hands and other nonbiologic surfaces where the novel coronavirus may survive for a long duration.[ 60 ] Furthermore, metal nanoparticle‐based coating formulations might be incorporated in air filter systems. For example, silver nanoparticle decorated silica particles is developed and coated on the air filters to evaluate filtration efficiency and antiviral activity in the presence of aerosolized virus particles. Subsequently, the result shows enhanced antiviral efficiency of the filter with coating areal density and diminishes dust loading.[ 61 ] Various nanoparticles have been formulated and tested for their antiviral activity with air filter coatings (Table 1 ).[ 62 , 63 , 64 , 65 , 66 , 67 , 68 ] So, these nanomaterials have considerable potential as disinfectants against pathogens, especially coronaviruses, due to their unique properties, including intrinsic antiviral, reactive oxygen species (ROS) generation ability, photodynamic and photothermal mediated killing capabilities. Further, by applying biodegradable materials such as polymeric and lipid‐based formulations, any hazardous effects of metallic nanomaterials on human health and the environment can also be prevented.[ 69 ]
Table 1.
Typical antiviral nanoparticles for coating of air filters and protective equipment. PEDV, porcine epidemic diarrhea virus; NDV, Newcastle disease virus
| Nanomaterial types | Size [nm] | Antiviral activity | References |
|---|---|---|---|
| Ag‐NPs | 7.1 | Infected cells treated with Ag‐NPs for 48 h have shown 98% survival | [ 62 ] |
| Ag@OTV | 3 | Infected cell survival rate increased to 90% | [ 63 ] |
| Ag2S NPs | <5 | Reduces PEDV load more than 99% | [ 64 ] |
| Au‐NPs | 10 | Reduce viral load to 92% after 6 h of incubation | [ 65 ] |
| CuO NPs | <100 | Viral load reduction reached to five orders of magnitude | [ 66 ] |
| Cu2O NPs | 50 | Reduced infection to 90% at concentration of 4 µg mL−1 | [ 67 ] |
| TiO2 | 8 | Effective antiviral activity against NDV at the concentration of 6.25 µg mL−1 | [ 68 ] |
3.2. Potential Application of Nanomaterial for Detection of Viruses
Human pathogenic viruses are a significant threat to human safety that cause fatal viral disease nowadays when humanity faces COVID‐19 pandemic. The high prevalence of these diseases is due to the unavailability of efficient and reliable detection tools. Therefore, there is an urgent demand to detect and identify viruses quickly, selective, and accurate. Different type of biosensors and kits has been designed, developed, and commercialized for detection of viruses. However, they have many drawbacks (Table 2 ). Nanomaterial‐based biosensors and detection kits look promising and have strong potential to overcome these challenges and perform real‐time, direct detection of molecular targets in pathogenic viruses.[ 70 ] For the suspected patients showing initial symptoms, the Centre for Disease Control & Prevention (CDC) recommends collecting samples to diagnose the COVID‐19 infection from the upper respiratory tract (such as nasopharyngeal and oropharyngeal swab) and, if possible, the lower respiratory tract (such as sputum, tracheal aspirate, or bronchoalveolar lavage). Additional samples (that is, stool and urine) can also be collected if required for further confirmation.[ 76 ] In routine‐based testing, the infection of SARS‐COV‐2 is determined by detecting viral genes, namely N, E, S, and RNA‐dependent RNA polymerase (RdRP), using reverse transcription polymerase chain reaction (RT‐PCR). Additionally, if required, the RNA‐sequencing technique is also employed to confirm the presence of SARS‐CoV‐2 infection. However, these methods demand expertise in molecular biology techniques and expensive laboratory equipment whereas, results are prone to be falsely negative or vice versa in the low viral load in the swab sample.[ 77 ]
Table 2.
Advantage and disadvantages of conventional viral detection methodologies
| Detection technique | Detection principle | Detection time | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Cell culture | Infectivity assay | Days | Broad spectrum; inexpensive | Difficult to maintain cell cultures; prone to contamination | [ 71 ] |
| Electron microscopy | Viral particle | Hours | Broad spectrum; rapid method | minimum 106 particles mL−1 required for detection, prone to cross morphologies | [ 72 ] |
| Hemagglutination assay | Viral protein | Hours | Easy; inexpensive | Less sensitive, fresh reagents required | [ 73 ] |
| ELISA | Viral protein | Hours | Single step process; effective at high analyte concentrations. | High analyte concentration required, produced antigen or antibody responses are indistinguishable in one step | [ 74 ] |
| PCR | Viral nucleic acid | Hours | Extremely high sensitivity; easy to set up | Highly prone to contamination, difficult to quantitate results | [ 75 ] |
Recent advances in nanosciences and the development of printed electrodes on a chip, nanomaterial‐based sensors, and detection kits became possible. They emerged as a reproducible and advanced tool for viral infection diagnosis. Fundamentally, the interaction between the receptor‐decorated sensor and analyte becomes highly efficient due to nanosize devices’ large surface‐to‐volume ratios.[ 78 ] Because of unique inherent properties such as physicochemical, mechanical, and magnetic, various nanomaterials, such as gold nanoparticles, carbon nanotubes (CNTs), magnetic nanoparticles, and quantum dots (QDs), have been exploring the development of biosensors.[ 79 , 80 ] Quantum Dots (QDs) are nanosized semiconductor crystals in a diameter range of 2–10 nm with specialized optical and electrical properties.[ 79 ] Underexposure to ultraviolet radiation, QDs emit different wavelengths of visible light. The emitted wavelengths are size‐dependent on QDs, and the gap between energy bands in small QDs are higher than in larger QDs. So, after exposing small QDs under ultraviolet radiation, electrons move to a high energy level and release excess energy when they return to a steady state. As a result, visible light emitted from small QDs has high energy and tends to be blue, while large QDs emitted in the reddish range because of the small energy gap and loss of less energy. Accordingly, by enlarging the size of the QDs, the light spectrum shifts toward red from blue.[ 79 , 80 ] The light emission of QDs is a desired property for medical labeling, imaging or sensing applications such as distinguishing between normal and tumor cells, gene therapy studies, and proteomic analysis.[ 81 ] From the virology point of view, QDs are an essential tool for providing rapid, efficient, sensitive and real‐time detection of pathogenic viruses for early monitoring and prevention of viral diseases.[ 81 , 82 , 83 ] For example, in a sandwich immunocomplex system, the mouse antihuman IgG (Ab1IgG) coupled with magnetic Fe3O4 nanospheres were used as an immune capture probe (Fe3O4@Ab1IgG). Rabbit antihuman IgG (Ab2IgG) coupled with highly luminescent quantum dot nanobeads (QBs) as a fluorescence detection probe (QBs@Ab2IgG) was applied for high sensitivity detection. Under optimal conditions, the detection of human IgG with a lower limit at 4 pg mL−1 was achieved.[ 88 ] So, QDs have many promising unique properties for developing an efficient biosensor for tackling diagnostic challenges (Figure 5B). Additionally, some quantum dot‐based sensors are summarized in (Table 3 ) which show promising detection efficiency toward diagnosing viral infections.[ 84 , 85 , 86 , 87 ]
Figure 5.

A) Description of operation principle of the Au‐nanoparticle based lateral flow (NPLF) strip. Reproduced with permission.[ 94 ] Copyright 2020, American Chemical Society. B) Schematic presentation of the determination of human IgG based on fluorescence linked immunoassay (FLISA). Reproduced with permission.[ 88 ] Copyright 2020, American Chemical Society.
Table 3.
Quantum dot based nanosensors previously explored for viral infection detection. Hepatitis virus B (HBV); Epstein–Barr virus (EBV)
| Virus | Biosensor type | Interaction site | Labeling site | QDs type | Purpose | References |
|---|---|---|---|---|---|---|
| HBV | Optical | Virus nucleocapsid | RBV | SA‐QDs | Single‐virus tracking | [ 84 ] |
| EBV | Optical | Carcinoma cells membrane | Anti‐EBV capsid antigen IgA | CdTe@dBSA‐QDs | Early screening and Diagnosis of EBV‐associated infections | [ 85 ] |
| Avian Leukosis Virus (ALVsJ) | electrochemical | Envelope | Anti‐ALV s‐J‐Ab2 | GQD | Viral detection | [ 86 ] |
| Influenza Vaxigrip | Electrochemical | Envelope | HA vaxi | CdTe‐QDs | Virus vaccine isolation and detection | [ 87 ] |
Generally, commercially available testing kit operations are based on the interaction of complementary ligand or nucleic acid strands with antibodies (enzyme linked immunoassay (ELISA)) or RNA (RT‐PCR) associated with the virus.[ 89 ] Some limitations are associated with these kits, such as false‐negative results, long response times, and poor analytical sensitivity.[ 90 ] Researchers have recently developed a colloidal gold‐based testing kit that facilitates easy conjugation of gold nanoparticles to IgM/IgG antibodies in human serum, plasma, and whole blood samples to overcome such challenges.[ 91 ] However, the targeted IgM/IgG antibodies in this detection method were not specific to COVID‐19. In some cases, it produced false negative or positive results in patients suffering from health complications other than coronavirus infections.[ 91 ] In another approach, researchers from the University of Maryland, USA, developed a colorimetric assay based on gold nanoparticles, capped with efficiently designed thiol‐modified DNA antisense oligonucleotides specific for N‐gene (nucleocapsid phosphoprotein) of SARS‐CoV‐2.[ 92 ] This colorimetric assay was able to diagnose positive COVID‐19 cases from the isolated RNA samples within 10 min.[ 92 ] These kinds of testing kits can be potentially valuable to provide a real‐time diagnosis of novel coronavirus infections. However, their performance would still be affected by the viral load. To overcome such limitations, scientists developed a unique dual‐functional plasmonic biosensor by incorporating the plasmonic photothermal effect and localized surface plasmon resonance (LSPR) based sensing transduction that provides an alternative and promising detection tool for clinical COVID‐19 diagnosis.[ 93 ] Through nucleic acid hybridization, 2D gold nanoislands functionalized with complementary DNA receptors facilitate sensitive detection of the selected sequences from SARS‐CoV‐2. To enhance sensing performance, thermo‐plasmonic heat is generated on the gold nanoislands chip when illuminated at their plasmonic resonance frequency. Outstandingly, this dual‐functional LSPR based biosensor has shown high selectivity toward the SARS‐CoV‐2 gene sequences with a detection limit as low as 0.22 × 10−12 m.[ 93 ] A colloidal gold nanoparticle‐based lateral‐flow (AuNP‐LF) assay has also been developed to achieve fast and on‐site detection of the IgM antibody in host serum sample against the SARS‐CoV‐2 through the indirect immunochromatography method. The IgM detection performance of the AuNP‐LF assay kit was evaluated by testing serum samples of COVID‐19 infected patients and normal humans. Results were compared with the RT‐PCR, which showed 100% and 93.3%, respectively (Figure 5A).[ 94 ] Further, to get rapid and highly sensitive detection of SARS‐CoV‐2 in clinical samples, ultrasensitive field‐effect transistor (FET)‐based biosensors have shown promising results.[ 95 ] Coating of graphene sheets of the FET can develop this system with a specific antibody against SARS‐CoV‐2 spike protein.[ 95 ] The FET devices can detect the spike protein at concentrations of 1.31 × 10−5 × 10−12 m in phosphate‐buffered saline (PBS) while 1.31 × 10−3 × 10−12 m in clinical transport medium with no recordable crossreactivity against MERS‐CoV antigen.[ 95 ] Paper‐based colorimetric sensors might be helpful in nucleic acid detection based on pyrrolidinyl peptide nucleic acid (acpcPNA)‐induced silver nanoparticle aggregation, applied for SARS‐CoV‐2, which was successfully used for MERS‐CoV, mycobacterium tuberculosis and human papillomavirus detection (Figure 4 ).[ 39 , 96 ] In this way, nanomaterial‐based biosensors and real‐time detection kits can facilitate faster, more accurate, and reliable detection of the virus even at the early stages of the infection (low viral load) due to the versatility of surface modification of nanoparticles.
Figure 4.
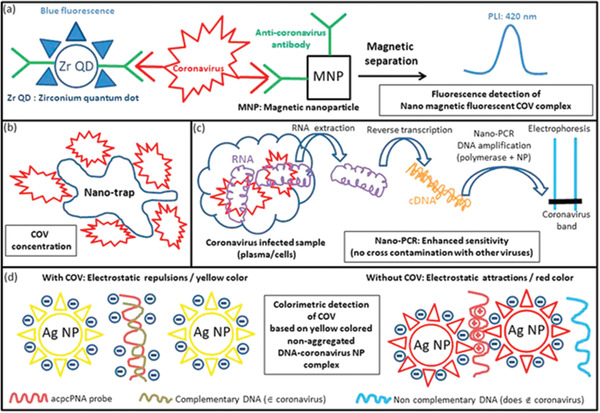
Schematic diagrams are showing different examples of nanomaterial‐based COV detection methods. a) The conjugate of fluorescent Zr QDs and magnetic nanoparticles with antibodies binds specifically to COV. In COV, a magnetic fluorescent complex is formed, isolated magnetically and detected by fluorescence measurements. b) Nanotraps are employed to concentrate COV to improve their stability, and facilitating their detection. c) Reverse transcription PCR (RT‐PCR) is carried out in the presence of nanoparticles, improving the efficacy of the polymerase chain reaction, and resulting in better detection sensitivity of this method. d) COV detection method, which is based on the interactions between complementary DNA originating from COV and acpnPNA probe at the surface of Ag‐NP, results in a separation between Ag‐NPs, and a yellow color associated with the luminescence of well‐dispersed Ag‐NPs, further revealing COV presence. Reproduced with permission.[ 39 ] Copyright 2020, American Chemical Society.
3.3. 2D Materials (2DMs) for the Sensing of SARS‐CoV‐2
The rapid advances of 2DM biosensors have gained massive momentum in the healthcare sector's development and commercialization of sensing devices.[ 97 , 98 ] The 2DM family of members contains graphene, transition metal dichalcogenides (e.g., MoS2), nonmetallic nanosheets such as hexagonal boron nitride and graphitic C3N4 (gC3N4), transition metal dioxides (e.g., MnO2), and transition metal carbides and nitrides known as MXenes (e.g., Ti3C2) and Xenes (e.g., phosphorene, commonly named black phosphorus) that can exhibit electrical, optical, and electrochemical properties. These unique inherent properties of 2DM can be exploited to enhance the sensitivity and reach minimum bio analyte detection limits. Similarly, 2DMs exhibit a wide range of specific electronic properties, ranging from metallic/semimetallic (e.g., graphene) to semiconducting (e.g., MoS2) and insulating (e.g., hexagonal boron nitride), that are crucial for establishing different elements of biosensors. 2DM biosensors allow rapid and efficient detection of viruses, viral components like nucleic acid, and host‐associated biomolecules such as antibodies, which simplifies the development and reduction of both cost and time compared to conventional assays. The selectivity between two analytes can be made high, allowing the differentiation of virus subtypes.[ 99 ] Primarily developed 2DM‐based biosensors based on electrochemical and electrical detection of the viruses; however, colorimetric immunoassay and SERS, SPR, and fluorescent optical sensors were also reported.[ 100 , 101 , 102 , 103 , 104 , 105 , 106 ] The development of a rapid and précised diagnostic platform for the direct detection of intact viruses and viral antigens like spike proteins, envelope, matrix, and nucleocapsid along with nucleic acid in clinical samples is needed to mitigate the COVID‐19 pandemic. A FET graphene biosensor was recently developed to detect SARS‐CoV‐2 in clinical samples such as nasopharyngeal swabs from COVID‐19 patients.[ 107 ] Coronaviruses are assembled with four structural proteins: spike, envelope, matrix, and nucleocapsid. The spike protein was used as the antigen, as its amino acid sequence varies among coronaviruses allowing SARS‐CoV‐2 selectivity. The spike protein of SARS‐CoV‐2 is vital for host cell entry. A specifically designed antibody against SARS‐CoV‐2 spike protein was immobilized on the graphene‐based device, which could differentiate between SARS‐CoV‐2 from another coronavirus (MERS‐CoV). Increased detection limit could be reached by increasing the signal‐to‐noise ratio by using other 2DMs.
Nucleic acid biosensors are widely developed for the detection of viruses. These devices mainly rely on electrochemical and fluorescence detection, while colorimetric, electrical, photoelectrochemical, SERS, and electrochemiluminescence‐based devices for sensing nucleic acids were also designed.[ 105 , 108 ] Many fluorescence detection systems exploit different adsorption profiles of oligonucleotide fragments on the surface of various types of 2DMs upon a conformational change after binding with the target viral nucleic acids. The planar structure of 2DMs allows the stable adsorption of many types of DNA probes. 2DMs such as graphene oxides (GO) are proved to offer great adsorption properties for single‐stranded DNA (ssDNA) over double‐stranded (dsDNA). The bases of ssDNA are partially uncoiled and can be exposed to the 2DM surface. In addition to the diverse use of GO, other 2DMs such as MoS2, graphdiyne, g‐C3N4, δ‐FeOOH, Ti3C2, metal‐organic framework (MOF), and covalent‐organic framework (COF) nanosheets can also be employed as fluorescence quenchers. The fluorescence‐based biosensors mainly rely on different adsorption capacities of fluorescently tagged DNA or RNA on the 2DM surface after binding with the target viral nucleic acids that induced conformational changes. As an alternative to labeling with fluorophores, DNA probes can also be conjugated to DNAzymes.[ 99 ] the combination of lateral flow test strips incorporating 2DMs and nucleic acid amplification by PCR can develop relatively fast response devices at a low cost. In our opinion, FETs can be investigated further for DNA sensing because of the charged phosphate backbone that could induce doping and hence be used as the transducing mechanism in such devices. Finally, beyond virus detection, the possibility of simultaneous detection and silencing of viral genes offers a great example that such a use of 2DMs is an effective approach for developing future theragnostic platforms for viral infections. It is also considerable that 2DMs, other than graphene, lack well‐developed and reliable synthesis methods for large‐scale production to ensure structural homogeneity of 2DMs in terms of size and thickness. Moreover, some 2DMs, particularly graphene, tend to restack through the formation of multilayers due to strong noncovalent interlayer interactions. The reduced active surface area may impact the performance and reproducibility of 2DM‐based biosensors. To overcome this challenge, the introduction of spacers or the construction of 3D networks can limit restacking. In addition, 3D graphene materials display remarkable properties, such as enhanced conductivity and electrocatalytic properties in all directions, high surface area, and more controlled pore sizes.[ 99 , 116 , 117 ]
3.4. Inactivation Strategies of the Virus Before Entry in the Host Cell
Nanomaterials can be synthesized with a large surface area (smaller in size). Therefore, based on surface properties, nanomaterials efficiently adsorb biomolecules and form a biomolecular corona.[ 42 ] Considering the biocompatibility of nanomaterials, this passive, nontargeted adsorption might be utilized to bind with viruses’ surfaces. Viral surface proteins are mainly enriched by sugar moieties or having positively charged amino acid sequences that ensure the selective binding to host cell surface lectins or glycosaminoglycans (GAGs) of heparan sulfate (HS) receptors.[ 42 ] Selective interactions of virus particles with host cell receptors are further ensured by multivalent binding; that is why single‐molecule inhibitors are often not capable of interfering with this important event, but multivalent NPs are superior to block binding of different viruses the host cell.[ 118 ] For example, cell‐surface receptor heparan sulfate mimetic Gold NPs capped with mercaptoethane‐sulfonate are effective inhibitors of HSV type 1 infection which, competitively binds to the virus spike proteins. These polyvalent sulfated Au‐NPs inhibit virus binding to the host cell, dependent on their size.[ 119 ] Papp et al. demonstrated that sialic acid (SA) decorated gold NPs effectively inhibited the interaction of the influenza virus to the target cell receptors.[ 120 ] Recently, another research group reported that antiviral NPs (Au and iron oxide core) adsorbed long and flexible linkers mimicking HS that strongly bind irreversibly and inactivate respiratory syncytial virus (RSV) in a lung infection model.[ 121 ] In an advanced therapeutic approach, seven different surfaces functionalized carbon quantum dots (CQDs) were investigated for treating human coronavirus HCoV‐229E infections (Figure 6A,B).[ 122 ] These functionalized CQDs showed a concentration‐dependent antiviral effect with an estimated EC50 of 52 ± 8 µg mL−1. The antiviral mechanism of these CQDs was revealed, which demonstrated that it inhibits HCoV‐229E entry due to the interaction of the functional groups of the CQDs with HCoV‐229E receptors.[ 122 ] Some surface‐functionalized antiviral nanoparticles with their mode of action are summarized in (Table 4 ) which showed potent antiviral activity against different viruses (Figure 6C).[ 123 , 124 , 125 , 126 , 127 , 128 , 129 , 130 , 131 , 132 , 133 , 134 , 135 , 136 , 137 , 138 , 139 , 140 , 141 , 142 , 143 , 144 , 145 , 146 , 147 , 148 ] Hence, many studies strongly support biocompatible, surface‐functionalized NPs can act as broad‐spectrum antivirals specifically against SARS‐CoV‐2. Fortunately, the receptor‐binding domain of the spike S1 protein of SARS‐CoV‐2 binds with both ACE2 and heparin, which multivalent binding nanoparticles can target.[ 149 ] Another strategy like administration of recombinant ACE2 to inhibit binding competitively via the spike S1 protein can also be promising to inhibit the viral uptake.[ 150 ] Thus, many surface functionalization options and technologies can be explored for selective targeting of virus binding with the host cell surface receptors.
Figure 6.
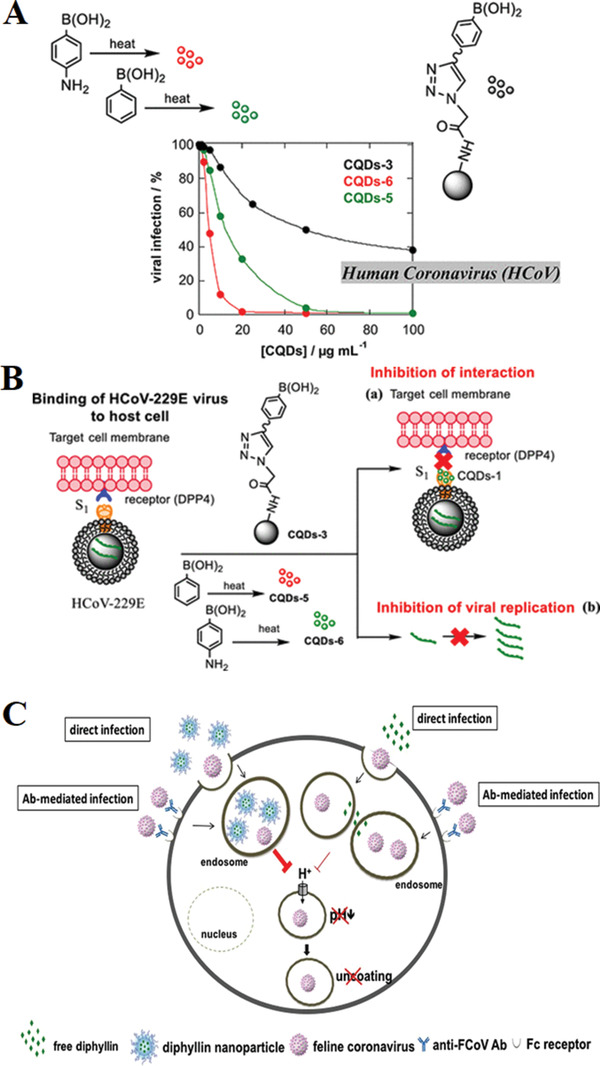
A) Viral inhibition using CQDs‐3, CQDs‐5, and CQDs‐6. B) Influence of CQDs, prepared by hydrothermal carbonization, on the binding of HCoV‐229E virus cells: a) inhibition of protein S receptor interaction, and b) inhibition of viral RNA genome replication. Reproduced with permission.[ 122 ] Copyright 2019, American Chemical Society. C) Schematic diagram of diphyllin antiviral mechanism. Diphyllin demonstrated prominent inhibitory activity in anti‐feline infectious peritonitis virus (FIPV) infection by attenuating cellular organelle acidification, thus blocking virus entry. In addition, diphyllin treatment displayed significant suppression in the viral load of Antibody Dependent Enhancement (ADE) of FIPV infections. By using a nanoparticulate drug delivery system, diphyllin exhibited improved safety and enhanced antiviral activity. Reproduced under terms of the CC‐BY licence.[ 123 ] Copyright 2017, The Authors, published by Springer Nature.
Table 4.
Some nanoformulated antiviral nanomaterials and their antiviral mechanism. FCoV, feline coronavirus; PEDV, porcine epidemic diarrhea virus; TGEV, transmissible gastroenteritis virus; PRRSV, porcine reproductive and respiratory syndrome virus; HSV, herpes simplex virus; HV, herpes virus; RSV, respiratory syncytial virus; HAV, hepatitis virus A; MPV, human meta‐peumo‐virus; NA, data not available
| Nanomaterial backbone | Size [nm]/shape | Active nanoagent | Virus type | Results | References |
|---|---|---|---|---|---|
| PEG−PLGA | 40 nm | Diphyllin | F‐CoV | Combination of PEG−PLGA‐diphyllin mediated reduction of endosomal acidification in fcwf‐4 cells | [ 123 , 124 ] |
| Graphene‐Ag | 7.5 nm | Ag‐NP | F‐CoV | In fcwf‐4 cells, prevents infection of F‐CoV | [ 125 ] |
| Ag2S nanoclusters | Sphere 3.2 nm | Ag2S Nanoclusters | PEDV | Reduces PEDV infection in Vero cells by blocking RNA synthesis and budding. | [ 126 ] |
| Ag‐NP and NW | <20 nm | Ag NP/NW | TGEV | Reduces TGEV infection in ST cells | [ 127 ] |
| Graphene oxide | NA | NA | RSV | Directly bind with virus and block attachment to host cell | [ 128 ] |
| Nanogel | NA | NA | PRRSV | Inhibit attachment and penetration. | [ 129 ] |
| Ag‐NPs | NA | NA | HV | Inhibit viral attachment | [ 130 ] |
| Graphene oxide | NA | NA | HV | Viral attachment inhibitor | [ 131 ] |
| Au‐NPs | NA | NA | HV | Inhibition of viral attachment and penetration | [ 132 ] |
| Nanocarbon | NA | NA | HV | Inhibit virus entry in cell | [ 133 ] |
| Silicon nanoparticles | NA | NA | Influenza A | inhibit progeny virus formation | [ 134 ] |
| Gd2O3:Tb3+/Er3+ nanoparticles | NA | NA | Zika virus | Act as antigen carriers for Zk2 peptide of Zika virus | [ 135 ] |
| Copper oxide nanoparticles | NA | NA | HSV type 1 | Leads to oxidation of viral proteins and degradation of viral genome | [ 136 ] |
| Zirconia nanoparticles | NA | NA | H5N1 influenza virus | Induce cytokines production | [ 137 ] |
| Zinc oxide nanoparticles | NA | NA | H1N1 influenza virus | Inhibit virus multiplication | [ 138 ] |
| Surface functionalized Antiviral Ag‐NPs | |||||
| Ag‐NPs | Spherical 1−10 nm | Foamy carbon, PVP and BSA | HSV type1 | Interacts with viral protein gp120 | [ 139 ] |
| Ag‐NPs | NA | PVP, BSA, and recombinant F protein (RF 412) | RSV | Interacts with G‐protein on the virus surface | [ 140 ] |
| Ag‐NPs | Spherical 2−5 nm | Oseltamivir, Amantadine, Zanamivir | H1N1 influenza virus | Block the activity of neuraminidase and hemagglutinin | [ 141 , 142 ] |
| Ag‐NPs | NA | Poly‐phosphonium‐oligochitosans | HAV, NoV and CoxB4 | Inhibit viral attachment and penetration | [ 143 ] |
| Ag‐NPs | Spherical 10−80 nm | Polysaccharide | MPV | Blocking virus‐host cell binding and penetration | [ 144 ] |
| Ag‐NPs | Spherical 10 nm | Polysaccharide | TCRV | Inactivates virus outside host cell | [ 145 ] |
| Ag‐NPs | Spherical 4 nm | Mercapto‐ethanesulfonate | HSV‐1 | Competitive inhibition of viral binding to the cell | [ 146 ] |
| Ag‐NPs | Spherical 2−5 nm | PEI and antiviral siRNA | EV71 | Prevent accumulation of ROS and leads to activation of AKT and p53 proteins | [ 147 ] |
| Ag‐NPs | Spherical 46 nm | Tannic acid | HSV‐2 | Blocks the attachment, penetration, and spread of viral particles to the host cell | [ 148 ] |
The re‐exploration of the antimicrobial uses of silver provides another large view at the interface of chemistry and medicine, of the nonlinear progress of scientific research. Several effective new silver‐based antimicrobial formulations have been commercialized in the last two decades as next‐generation broad‐spectrum antivirals with low toxicity and affordable cost.[ 151 ] Nano Ag has been widely used as antibacterial material due to its high specific surface area and unique tunable chemical and physical properties.[ 152 ] The antiviral properties of Nano‐Ag have also been extensively studied. According to the available literature, the antiviral mechanism of nano‐Ag is as follows: 1) Nano Ag has a strong adsorption affinity toward the virus and interacts with the viral envelope protein, which can affect the interaction between the virus and the cell receptor, thereby preventing the entrance of virus in the host the cell.[ 153 ] The adsorption effect may depend on the size effect of nano‐Ag.[ 154 ] 2) Nano Ag combined with the virus can release Ag ions, which react with viral proteins and inactivate them, or react with viral nucleic acids to prevent viruses from replicating.[ 155 ] 3) Nano‐Ag's surface can activate oxygen to produce ROS, which oxidizes the virus and seriously damages its structure.[ 156 , 157 ]
In another example, through a fluorescence spectroscopic approach, Tiwari et al. 2020, studied the interaction of poly‐ethylenimine (PEI) functionalized silver nanoparticles with cells of Acinetobacter baumannii.[ 158 ] The reports have shown that the silver nanoparticles selectively bind with surface‐expressed proteins, quenching the autofluorescence of proteins damaging the cell structures at shallow MIC values (≈5 µg mL−1). Similarly, they have also utilized other capping agents like 3‐aminopropyletrimethoxysilane, 3‐glysidoxypropyletrimthoxysilane and organic reducing agents’ cyclohexanone and formaldehyde.[ 159 ] However, the MIC value of these silver nanoparticles was high in comparison to PEI functionalized Ag‐NPs.[ 159 ] Mechanistically, compared to the size of the SARS‐CoV virus (100–150 nm), including other human and plant pathogenic viruses, the 5–10 nm sized PEI functionalized silver nanoparticles have a great affinity toward the surface biomolecules, particularly proteins and resulting inactivation. Thus, it should be considered that SARS‐CoV‐2 viruses have proteaceous spike receptors that play a fundamental role during the infection. The inactivation of viral receptor protein by functionalized silver nanoparticles can prevent the entrance of the virus inside the host cell. In another mode, due to the small size of silver nanoparticles, it can enter the cell and inactivate the replicating viruses. The surface charge and size of silver nanoparticles with PEI capping can also be tuned as per requirement by using a varying molecular weight of PEI.[ 158 ] PEI molecules can be linear or branched. It has also been studied that the higher molecular weight of PEI functionalized silver nanoparticles had a more potent effect on the bacterial cell. The polyethyleneimines are cationic, hydrophilic polymers and functionalization of silver nanoparticles have great scope as an air disinfectant, antiviral surface coatings, antiviral coating of masks, PPE kits, and household exhausts as front‐line defense material against COVID‐19 or other viral and microbial pathogens from humans to plants.[ 158 ]
Graphene and its derivatives are an exciting member of the carbon nanostructures group (graphene, the building block of graphite). Structurally, graphene's are anatomically 2D layers of hexagonally bonded carbon atoms. Such carbon structure can exhibit fundamental properties such as large specific surface area, high mechanical strength, electron conductivity, optical and catalytic characteristics.[ 160 , 161 , 162 , 163 , 164 , 165 , 166 , 167 , 168 , 169 , 170 , 171 , 172 , 173 , 174 ] These unique properties of graphene and its derivatives have made these nanostructured materials suitable for various biological and medical applications, including antipathogenic applications.[ 175 , 176 , 177 , 178 , 179 , 180 , 181 ] In order to cope with the current COVID‐19 pandemic, It should be mentioned that the antibacterial and antiviral activity of graphene or graphene‐based nanomaterials can be explored based on various effects, including membrane, oxidative, and photothermal stresses as well as charge transfer, and the entrapment effect of graphene materials on various bacterial species.[ 182 , 183 ] One important aspect is the interactions between the virus and graphene‐based materials. Graphene and its derivatives exhibit the ability to inactivate different viruses through various mechanisms, namely photothermal activity and inhibit cellular infection by binding the nanomaterials to the S‐protein of viruses or host cell receptors. Two critical characteristics of graphene materials are based on their functionalization capability and being used as the substrate to load other antiviral agents homogeneously. In contrast with graphene, GO is the oxidized form of graphene with hydroxyls, epoxides, diols, ketones, or carboxyl functional groups located on its surface. The presence of oxygen on the basal planes and edges of GO increases its hydrophilicity, water dispersibility, and attachability compared to graphene. This activity has been evaluated using RNA (porcine epidemic diarrhea virus, PEDV) and DNA (pseudorabies virus, PRV) viruses as a model.[ 184 , 185 ] PEDV is a coronavirus that infects pigs, causing severe diarrhea and dehydration, resulting in significant piglet mortality. PEDV cannot be transmitted to humans.[ 186 ] The graphene and graphene‐based antivirals can be exerting their effect via direct antiviral, through photocatalysis, graphene oxide‐silver nanocomposite systems, cell surface mimetics, and search and destroy strategy.
The exploration of antiviral properties and applications of carbon‐based nanomaterials such as graphene and graphene oxide are still under investigation, and much research is needed to assess the potential of these materials. Carbon nanomaterials have, however, the potential to create antiviral systems with reduced toxicity. Thus, a full cytotoxicity assessment appears to be the main issue to develop antiviral applications for such a class of materials.
3.5. Nanomaterials Coated Filtration System to Inactivate the SARS‐CoV‐2
An effective PPE kit (personal protective equipment) is the first‐line defense weapon for health professionals and the general public to fight against the spread of SARS‐CoV‐2.[ 187 ] The employment of masks has proved an integral part of complete prevention that can minimize the spread of respiratory viral infections like COVID‐19.[ 188 ] However, no effective antimicrobial or antiviral agent coated PPE kit is currently available, thereby providing people with passive barrier protection. The PPE reduces the chance of microbes/viruses releasing into the air but cannot inactivate or prevent the spread of microbes/viruses in the filtration systems.[ 189 ] Even high efficiency particulate air (HEPA) filters can only trap the virus particle but not kill them. Hence, the development and rapid production of antiviral coated face masks and other protective materials that can destroy the microbe/virus and immobilize the aerosol droplets should be a top priority to limit airborne transmission of COVID‐19.
Several nanotechnology‐based interventions using suitably designed materials for trapping and inactivation of COVID‐19 are currently being developed, e.g., deposition of Ag nanoparticles on the filter surface (Mask and PPE), use of photocatalytic materials, carbon materials, catechin based filters can show virucidal activity. Self‐assembling silver nanoparticles spray recently been documented involving the active role of siloxane‐silver nanoparticles nanofluid.[ 190 , 191 ] This enabled the self‐assembling of silver nanoparticles over various solid substrates, including mask and PPE.[ 192 ]
Graphene oxide has been proved as an antiviral material for the inhibition of viral helicase. Some studies conducted with SARS‐CoV and hepatitis C virus non‐structural protein 3 (HCV‐NS3) helicase have shown that viral helicase adhered to the surface of graphene oxide by π−π stacking interactions between hexagonal units of graphene oxide and nucleotides.[ 193 , 194 ] This interaction facilitates the employment of graphene as a coating material to boost the viral trapping properties of the mask.[ 195 ] Multifunctional nanomaterials like “nanoflowers” of transition metal dichalcogenides have the potential to accommodate a high proportion of nanoparticles and active molecules on their surfaces and edges, thereby inactivating pathogens by disrupting their cellular functions. Because of their many properties, such as high air stability and strong substrate adhering nature, the virus is quickly detected and inactivated by these nanoflowers, making them applicable for use in various filtration platforms.[ 196 ] The antiviral properties of Metal nanoparticles are well established, especially silver nanoparticles (Ag‐NPs). They act as inhibitors of viral replication and target viruses for their viricidal action. For example, Ag‐NPs inhibit the HIV‐1 virus entry in host cells and can interact with cell receptors.[ 197 ] It is also documented that Ag‐NPs interact with a viral genome and inhibit viral replication in double‐stranded RNA viruses. Similarly, biocompatible polymer‐stabilized gold nanoparticles (AuNPs) have shown antiviral activity against HIV‐1 and other influenza virus subtypes (e.g., H1N1, H3N2, H5N1). It has also been documented that sulfated ligand coated gold nanoparticles, silver nanoparticles, and hybrid silver−copper nanoparticles can bind the HIV envelope glycoprotein (gp120) and inhibit cellular HIV‐1 infection in vitro.[ 198 , 199 ] Furthermore, tannic acid and mercaptoethanol‐sulfonate functionalized Ag‐NPs can prevent HSV infection by specifically inhibiting virus binding, penetration, and spreading post‐infection.[ 200 , 201 ] The application of these antiviral metal nanoparticles could greatly help enhance a mask's filtration and viral trapping efficacy. They can also be applied to modify surgical masks, respirators and other protective equipment in combating the spread of COVID‐19. Other metal nanoparticles like copper and zinc nanoparticles have been explored for their antimicrobial and antiviral effects. In a study, Hang et al. investigated the inhibitory property of cuprous oxide nanoparticles (CO‐NP) against Hepatitis C virus (HCV) infection. They reported that CO‐NPs at nontoxic concentrations could significantly inhibit attachment and entry of HCV pseudoparticle (HCVpp) in the host cell.[ 68 ] Similarly, Cu metal nanoparticles were also influential in inactivating the human coronavirus through a ROS dependent mechanism.[ 202 ] Owing to these unique properties, Cu nanoparticles can be used as an antiviral coating material. A mask containing nanofibers embedded with CuO has been developed by the Respilon group, which can filter out viruses, and CuO present on the nanofibers is assumed to kill trapped viruses.[ 203 ]
Similar to copper, the divalent metal ion of Zn is also an effective antiviral agent. A study conducted to understand the antiviral effect of metal ions such as Hg2+, Zn2 +, and Cu2 were tested. Among all metal ions tested, Zn2 + exhibited more effective inhibitory properties.[ 204 ] Reports demonstrated that Zn2 + ions and Zn2 + ionophores effectively inhibit replicase and RdRp enzymes of SARS at a molecular level.[ 205 ] Another study established that PEGylated ZnO nanoparticles effectively reduce the number of copies of herpes simplex virus‐1 DNA.[ 206 ] In addition to the antiviral property at the biomolecule interface, coating surfaces with ZnO makes the surface water repellent and thus helps protect against contamination with any body fluids/droplets such as blood.[ 207 ]
3.6. Nanomaterial‐Based Nanovaccines Strategies against SARS‐CoV‐2
From reporting the genetic sequence of SARS‐CoV‐2 on January 11, 2020, rigorous research efforts have been dedicated to developing a vaccine against COVID‐19. Several research groups globally are developing the coronavirus vaccine, which is either under clinical trial or in the preclinical stage and can take months to years to commercialize for mass immunization. Considering vaccine and immunization research, nanomaterials can provide multiple ways to induce the immune system and direct the immune response, specifically against SARS‐CoV‐2 antigens. A vaccine's development depends on the direct administration of viral antigens such as recombinant spike proteins, vectored vaccines, whole inactivated/attenuated virus, or RNA viral antigens. Generally, antigens for immunization are viral surface proteins such as the immunogenic spike protein (S1), potentially targeted by patients’ antibodies.[ 40 ] Because the S1 protein is vital for cellular uptake of virus inside a host cell, several scientists explore this protein as the primary target for a vaccine. There are some challenges related to the safe and efficient administration of a drug, protein, or RNA into the patient. Sometimes the cargos are prone to degradation, low bioavailability, and immediate clearance. Nanotechnology provides an excellent platform to tackle these challenges, though nanocarriers can overcome some of these limitations. Furthermore, biocompatible polymeric, lipid‐based, or inorganic NPs can be designed for their physicochemical characteristics to encapsulate cargo proteins with high loading efficiency, enhanced protein delivery, and improved pharmacokinetics over conventional approaches.[ 208 ] The success of vaccination depends on the suitable type of polymer in combination with the antigen. It is observed that the intranasal administration of polymer encapsulated antigen triggers a robust immune response.[ 209 , 210 ] Likewise, researchers have developed lipid and lipid‐based nanoparticles as a delivery vehicle for mRNAs and siRNAs delivery to enable the synthesis of vital viral proteins for vaccination or to inactivate critical viral target genes, respectively.[ 211 ] A lot of nanoformulated anti‐coronavirus vaccines have been developed. They offer better efficacy and immune induction than their non‐nanoformulated counterparts. These specialized vaccines have a nanomaterial backbone of gold nanoparticles, polymers such as PLGA, chitosan, PEI, or protein assemblies (Table 5 ).[ 213 , 214 , 215 , 216 , 217 , 218 , 219 , 220 , 221 , 222 , 223 , 224 ] The backbones are conjugated or decorated with an active ligand consisting of COV antigen, specific COV proteins such as spike (S1).[ 212 ] When mice and chickens were immunized with nanoformulated vaccines, it induces the production of cytokines, essentially γ‐IFN, IL‐1β, IL‐2, and IL‐6, as well as an enhanced activity of specific immune cells as macrophages, lymphocytes, and B cells, were observed.[ 212 ] COVID‐19 have a crucial feature to trigger uncontrolled cytokine release in the patient body, cumulatively known as cytokine release syndrome (CRS), which leads to an excessive immune response and severe clinical complications. This inflammatory storm leads the acute respiratory distress syndrome (ARDS) associated with multiple‐organ failure, ultimately causes the death of patients. Remarkably, the role of interleukin (IL)‐6 has been revealed in ventilator assisted patients. Interleukin (IL)‐6 is a well‐regulated host's innate immunity triggered cytokine response against an infection. The overproduction, unbalanced and prolonged cytokine response can severely harm the host body. Therefore, therapeutic options for effective suppression of the host immune system triggered cytokine storm are under investigation. Nanomaterials have been exploited to maintain the immune response to a balanced level, and such proprieties might be explored to control the cytokine storms. Nanomaterial‐based systems can potentially enhance the efficiency of immunosuppressive drug delivery to targeted immune cells, prevent drug distribution to nontarget tissues and organs, and minimize side effects. Additionally, rationally designed nanosystems can be explored to evade the immune system and increase the solubility of poorly soluble immunosuppressive agents.
Table 5.
Nanoparticles as immunogenic agents for vaccines. Abbreviations: GM‐CSF, granulocyte−macrophage colony‐stimulating factor; HA, hemagglutinin; iBALT, inducible bronchus‐associated lymphoid tissue; PLGA, poly (lactic‐co‐glycolic acid); STING, stimulator of interferon genes
| NPs | Conjugate/adjuvant | Size [nm] | Virus | Model organism | Outcomes | References |
|---|---|---|---|---|---|---|
| Au‐NPs | SARS‐CoV Spike protein | 40, 100 | SARS | In vivo; BALB/c mice | Strongly induce IgG response | [ 213 ] |
| VLPs with Au‐NPs | Avian IBV Spike protein |
AuNPs:100 VLPs:139 |
IBV | In vivo; BALB/c mice | Induce strong antigen‐specific cellular immunity, IgG, IgA responses. | [ 214 ] |
| VLPs from MERS CoV Spike protein |
Alum, matrix M1 |
NA | MERS | In vivo; BALB/c mice | VLPs and Matrix M1 induce high anti‐S titers | [ 215 ] |
| VLPs from SARS CoV and MERS‐CoV Spike protein |
Alum, matrix M1 |
≈25 | SARS, MERS | In vivo; BALB/c mice | Induce high antibody titers against homologous virus. | [ 216 ] |
| VLPs from MERS‐CoV Spike protein | Ad5/MERS, Alum |
VLPs: 35 with alum: 80 |
MERS | In vivo; BALB/c mice | Induces CD8+ T cell, TNF‐α, IL‐2, GM‐CSF, and IFN‐c responses. | [ 217 ] |
| VLPs from HRC subunit of SARS‐CoV Spike protein | 25−30 | SARS | In vivo; BALB/c mice | Generate neutralizing mice antisera | [ 218 ] | |
| VLPs from bacteriophage P22 | IAV HAs | ≈26 | SARS, Influenza A | In vivo; C57BL/6 mice | Recognizable by TLR‐2 receptor and protect from infection and weight loss | [ 219 ] |
| VLPs using canine parvovirus | MERS‐CoV RBD, poly(I:C) | ≈25 | MERS | In vivo; BALB/c mice | Mixed Th1 and Th2 responses, increase in antibody titers | [ 220 ] |
| Chitosan NPs | SARS‐CoV N protein | 210 ± 60 | SARS | In vivo; BALB/c mice | Dendritic cell targeting, strong CD4+ response, high levels of IgG, IgG1, IgG2a, IgG2b, IgA, IFN‐γ | [ 221 ] |
| Heat shock protein caged NPs | NA | 120 | SARS, H1N1, RSV | In vivo; C57BL/6 |
Induce strong nasal antibodies. formation of iBALT structures of B cells, CD4+ T cells, dendritic cells and CD8+ T cells. |
[ 222 ] |
| PLGA | STING, MERS‐CoV Spike protein, MF59 | ≈148 | MERS | In vivo; C57BL/6 mice |
STING encapsulated in PLGA induce strong RBD specific CD4+, CD8+, balanced Th1/Th2 response. IgG2a; IFN‐β, TNF‐α, IL6 generation |
[ 223 ] |
| CDs | NA | 1.6 | PEDV | In vitro; Vero and PK‐15 cells | Induce IFNs and pro‐inflammatory cytokines | [ 224 ] |
3.7. Photodynamic and Photocatalytic Approaches for Inactivation of SARS‐CoV‐2
In addition to antiviral and nanoformulated vaccine strategies, photodynamic therapy (PDT) and photocatalytic inactivation process promising and unique way to destroy the SARS‐CoV‐2. Using a light‐dependent method, PDT attacks virus‐infected cells via the excitation of photosensitizers (PSs), which leads to the generation of ROS in the presence of oxygen and, ultimately, cell death takes place. Conventionally, photodynamic therapy is applied to treat various oncological disorders.[ 225 ] Despite being explored to treat oncological disorders, PDT has some limitations related to photosensitizers. Several photosensitizers are hydrophobic and prone to get aggregate in aqueous solutions that affect their photochemical and photobiological properties and limit the application as virotherapy.[ 226 , 227 ] Several nanomaterial‐based approaches have been made to tackle the challenges of improving photosensitizer physicochemical properties for effective use of PDT as a pathogen eliminating methodology (Table 6 ).[ 246 , 247 , 248 , 249 , 250 , 251 , 252 , 253 , 254 , 255 , 256 , 257 , 258 , 259 , 260 , 261 , 262 , 263 , 264 , 265 , 266 , 267 , 268 , 269 , 270 , 271 , 272 , 273 , 274 , 275 , 276 , 277 , 278 , 279 , 280 , 281 , 282 , 283 , 284 ] Lim et al. have introduced an efficient, NPs‐based model for photodynamic elimination of viruses in this row. In this system, sodium yttrium fluoride (NaYF4) upconversion NPs (UCNs) developed with zinc phthalocyanine photosensitizer grafted onto their surface. The PEI coated UCNs were hydrophilic and easier to manipulate. These UCNs showed antiviral activity against both enveloped and nonenveloped viruses models such as Dengue virus serotype 2 and adenovirus type 5.[ 228 ] Similarly, considering the physicochemical properties of photosensitizer, biocompatible MXenes, such as Ti3C2Tx, have shown promising results.[ 229 ] MXenes are hydrophilic and explored as the most efficient light‐to‐heat transforming materials.[ 229 ] The plasmonic resonance extinction maxima of Ti3C2Tx and several other MXenes have at 780 nm, making it suitable for using near‐infrared (IR) light‐mediated PDT.[ 230 ] In addition to being hydrophilic, they also carry a highly negative charge (in solution, zeta‐potential value is between −30 and −80 mV). For example, Ti3C2Tx MXene has a solid ability to absorb amino acids; in this way, it would probably bind to viral spike proteins and immobilize the virus, followed by deactivating it.[ 231 ] MXenes are photocatalytically active, and when a virus is adsorbed onto the surface, it can inactivate the virus under light exposure.[ 222 , 223 , 224 , 225 , 226 , 227 , 228 , 229 , 230 , 231 , 232 , 233 , 234 ] A vast range of MXenes, including Ti3C2Tx, Ta4C3Tx, Nb2CTx, are biocompatible and have antimicrobial activity due to their charge transferability and hydrophilicity.[ 235 , 236 , 237 , 238 , 239 , 240 , 241 , 242 ] In addition to MXenes, fullerene and graphene are also efficient nanomaterials for virus inactivation by PDT. They have shown promising results against Semliki Forest virus (SFV), vesicular stomatitis virus (VSV), HSV‐1, HIV‐1, mosquito iridovirus (MIV), influenza A virus (IAV), and phage MS2.[ 243 ] Another technology, femtosecond (fs) pulsed laser irradiation techniques, has attracted attention to the selective inactivation of virus contaminations in biological samples. For example, group research demonstrated the enhanced photonic inactivation of Murine Leukemia Virus (MLV) via 805 nm femtosecond pulses through gold nanorods.[ 244 ] The localized surface plasmon resonance overlapped with the excitation laser. The results shown that the ≥3.7‐log reduction in viral load was achieved by 10 s laser exposure with incident laser powers ≥0.3 W. Interestingly, the fs‐pulse induced photodynamic inactivation was selective to the virus. It did not pose any effect on co‐incubated antibodies. The mechanism behind the photodynamic inactivation of the virus was due to interference of viral fusion to the host cell, followed by damage of the viral envelope.[ 244 ] Photocatalytic inactivation of infectious agents, specifically adsorbed on surfaces or pathogen borne air droplets, can be employed against various infectious agents like bacteria and viruses, including SARS‐CoV‐2. For example, a metal–organic framework, zinc‐imidazolate MOF (ZIF‐8), was developed, exhibiting almost 100% pathogen inactivation efficiency in saline within two hours under simulated solar irradiation. Mechanistic insight of pathogen disinfection revealed that photoelectrons trapped at Zn+ centers within ZIF‐8 via ligand to metal charge transfer (LMCT) are responsible for oxygen‐reduction mediated reactive oxygen species (ROS) generation. ZIF‐8 fabricated air filters significantly reduce airborne pathogens in 30 min and 97% PM removal (Figure 7 ).[ 245 ]
Table 6.
Summary of nanomaterial‐based approaches for the improvement of physiochemical properties of photosensitizers (PSs)
| Approach | Chemical moiety | Action mechanism | Physicochemical characteristics | References |
|---|---|---|---|---|
| Development of advanced PSs with unique physicochemical properties | Organic derivatives of conventional PSs, nanoparticle‐based organic dyes | Intramolecular or intermolecular resonance energy transfer | Highly biocompatible, involved in type II PDT and less Stable | [ 246 , 247 , 248 , 249 ] |
| Inorganic semiconductors such as TiO2, β‐SnWO4, G‐QDs, bismuth oxyhalide, and Cd‐Se | Separation of electrons holes from the covalent band and conduction band | Highly stable, tunable optical properties, involved in type I and type II PDT | [ 250 , 251 , 252 , 253 , 254 , 255 , 256 , 257 , 258 , 259 ] | |
| On the basis of utilization of different lights | NIR‐activated nanomaterials such as UCNP‐based Nanoplatforms and TPA | FRET from photoconverting or self‐illuminating nanoparticles to PSs | Deep tissue penetration capability and have potential photothermal effect | [ 260 , 261 , 262 , 263 , 264 , 265 , 266 ] |
| X‐ray‐activated nanomaterial materials such as SCNPs, PLNPs | Deep tissue penetration capability, can lead to potential radiation damage | [ 267 , 268 , 269 , 270 , 271 , 272 ] | ||
| Design of new platforms based on the features of TME | TME‐responsive nanomaterials: biochemical‐triggered PDT enhancement | Mutual interactions between nanomedicines and TME to favor tumors targeting | High selectivity, specificity, efficiency, and bioavailability | [ 273 , 274 , 275 , 276 , 277 , 278 , 279 , 280 , 281 ] |
| TME‐modulating nanomaterials: reoxygenation in hypoxic tumors | Decomposition of hyperoxide or water to generate O2 and relieve hypoxia | Low oxygen dependence and enhanced ROS‐generating efficiency | [ 282 , 283 , 284 ] |
Figure 7.

A) Band‐structure characterization and photocatalytic disinfection mechanism of ZIF‐8 (zinc‐imidazolate MOF). i) The band positions of ZIF‐8 for the reactive oxygen species (ROS) formation potential, conduction band (CB), and valence band (VB) represent conduction band and valence band, respectively. ii) Electron paramagnetic resonance (EPR) spectra of ZIF‐8 at 77 K in dark and under light irradiation (300 nm < λ < 1100 nm) in different atmosphere. iii) EPR spectra of DMPO−•O2− for ZIF‐8 under light irradiation and in the dark. iv) Steady‐state concentration of •O2− calculated from the decay of nitroblue tetrazolium (NBT) and hydrogen peroxide (H2O2) accumulation over time, respectively. v) The first‐order disinfection rate on ZIF‐8 with different scavengers (IPA → •OH, L‐His → 1O2, Cr (VI) → e−, Oxalate → h+, SOD → •O2−, CAT → H2O2). vi) Dependence of the amount of released H2O2 by ZIF‐8 on the wavelength of incident light and the ultraviolet–visible (UV–vis) spectra of ZIF‐8. The error bars are calculated by repeating the measurements three times. SOD superoxide dismutase, IPA isopropanol, DMPO 5,5‐diemthyl‐1‐pyrroline N‐oxide. B) Schematic of metal–organic framework (MOF)‐based filter. Schematic representation of MOF‐based filter (MO‐filter) for integrated air cleaning. Reproduced with permission under common creative 4.0 attribution licence.[ 245 ] Copyright 2019, The Author(s).
3.8. Nanocarriers for Efficient Drug Delivery
Several nanoformulated or designed nanovehicles such as nonmetal NPs, QDs, carbon nanomaterials, polymeric NPs (chitosan, PLGA), γ‐PGA (poly‐γ‐glutamic acid), metal/metal oxide NPs, silica NPs, carbon black NPs, liposomes, dendrimers, solid lipid nanocarriers, and virus‐like particles (VLPs) have been explored to deliver several biomolecules, (Figure 8A,C), e.g., drugs, proteins or peptides, DNA, or RNA, antibodies, and vaccines for inhibiting the multiplication of virus or a compelling attempt from promoting a protective humoral immune response (Table 7 ).[ 246 , 285 , 298 ] With the development of the advanced nanoscale active biomolecule carriers, scientists have looked forward to using inorganic components to obtain a biologically inert structure to explore biological building blocks capable of displaying multiple functionalities to the ultimate construct. Almost every nanocarrier developed to date has one common layby; whether administered through any route, all will come into contact with the body fluid. When nanocarriers come in contact with body fluid, they react uniquely according to their physicochemical properties and the tissue of body parts.[ 286 ] Therefore, inorganic NPs can be chemically functionalized as globular protein mimics because of their comparable size, charge, shape, and surface features resembling proteins. Hence, these nanomimetics can be used in nanobiotechnology to exploit viral pathways or cell receptor interactions with NPs.[ 287 ] In a recent study, ML336 (a chemical inhibitor of Venezuelan Equine Encephalitis Virus) loaded lipid‐coated mesoporous silica nanoparticles (LC‐MSNs) were employed, effectively reducing VEEV infection in vitro and in vivo. The large surface area of the MSN core promotes hydrophobic drug loading while the liposome coating retains the drug and enables enhanced circulation time and biocompatibility. Overall, (LC‐MSNs) might be explored for an efficient delivery platform for the Anti‐SARS‐CoV‐2 drug formulations (Figure 8B).[ 288 ] In a drug delivery system, the surface recognition and specific interactions between the drug‐loaded vehicle and biological site are essential for successful drug release. The biological molecules such as membrane proteins have been proposed as an agent to convey targeting and shielding moieties parallel.[ 289 ] The principle of biomimicry or bioinspiration might be exploited for designing viruses like NPs based delivery vehicles that can circulate in the blood system without any resistance by the endothelial barrier and deliver the payload with high efficiency.[ 290 ] Another strategy can be applied to efficiently deliver repurposed antiviral drugs through the conjugation of cell‐penetrating peptides (CPPs) to the carrier. CPPs are short cationic peptides, which facilitate enhanced intracellular intake and delivery of various drug molecules in the cytoplasm or nucleus without any measurable toxicity and damaging effect on the cell membranes integrity.[ 291 , 292 , 293 ] Mechanistically, the TAT peptide (positively charged) interact with negative charges of phospholipids membrane electrostatically or nonelectrostatic hydrogen bonding or hydrophobic interactions.[ 292 , 293 ] Further, these CPPs facilitate the delivery of the cargo into the cell through macro‐pinocytosis, caveolae‐mediated endocytosis of clathrin‐independent endocytosis. It has also been revealed that HIV1 TAT‐peptides directly penetrate the plasma membranes by forming nanoscale pores.[ 294 ] CPPs conjugated nanocarriers has several advantages over conventional delivery systems such as it is inexpensive, easy to design, have more excellent capability to translocate into a wide range of cell types, higher rate of cellular permeability and uptake, more accessible to pass through biological barriers, along with large cargo carrying capacity and reduced cell toxicity without inducing any immunological response.[ 295 , 296 ] Hence, cell‐penetrating peptide conjugated nanocarriers can be a promising option for the targeted delivery of drug cargo at the cellular level. However, some limitations have also been linked with CPPs, such as low cell specificity, prone to uptake into intracellular endosomes and inactivation by proteases.[ 297 ]
Figure 8.
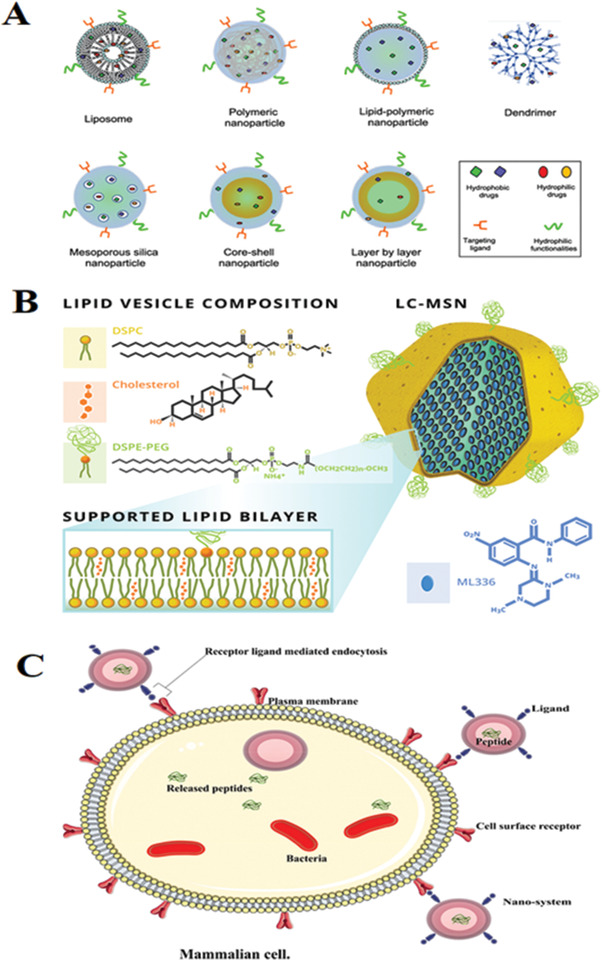
A) Different nanocarrier platforms utilized for combination drug therapeutics. Reproduced with permission[ 20 ] Copyright 2020, ACS nano. B) Schematic of ML336 loaded LC‐MSNs. The antiviral ML336 was incubated with MSNs before vesicle fusion with liposomes containing a composition of 77.5% DSPC:2.5% DSPE‐PEG 2000:20% cholesterol at mole ratios; Reproduced with permission.[ 288 ] Copyright 2018, Springer Nature. C) An illustration of an active nanosystem demonstrating antimicrobial peptide delivery into an infected cell. Reproduced with permission under common creative attribution licence.[ 31 ] Copyright 2018, The Author(s).
Table 7.
Categories of nanomaterials used for nanoformulated drug/vaccine delivery
| Type of materials | Classical example | Functions/Advantages | References |
|---|---|---|---|
| Organic nanoparticle‐based carriers | Liposomes | The efficient and controlled delivery of nucleic acid‐based antigens | [ 298 , 299 , 300 ] |
| Polymer‐based nanoparticle | Efficient loading of different cargos with various forms and functions; shows self‐adjuvanticity in some cases | [ 301 , 302 , 303 ] | |
| Inorganic nanoparticle‐based carriers | Mesoporous silica nanoparticles | Multiple site cargo loading; controllable release of cargo; shown self‐adjuvanticity in some cases | [ 304 ] |
| Magnetic nanoparticles | Vaccine delivery along with imaging capabilities | [ 305 , 306 ] | |
| Gold nanoparticles | Vaccine delivery along with imaging capabilities | [ 246 , 307 ] |
3.9. Role of Nanotechnology to Understand the Gut Microbiome and SARS‐CoV‐2 Interactions
The integrity of the gut microbiome (the collective genomes of the diverse microbiota that reside in the gastrointestinal tracts of humans) could conceivably be disturbed by SARS‐CoV‐2, causing gut dysbiosis in the host, as with other infectious diseases. Some signs may connect gut functionality and microbiome responses to SARS‐CoV‐2.[ 308 ] New research indicates that SARS‐CoV‐2 may be spread by faucal−oral transmission.[ 309 ] The highest SARS‐CoV‐2 mortality and morbidity is in the elderly and those with underlying health problems associated with inflammation and other disorders, such as diabetes.[ 310 ] Interestingly, these cohorts tend to have less diverse gut microbiomes.[ 311 ] Links between the gut microbiome and age‐related health decline have been consistently shown.[ 312 ] Ageing is associated with significant shifts in microbiome diversity and proinflammatory states. The elderly microbiome generally shows a shift away from Firmicutes, which dominates in younger adults, toward genera such as Alistipes and Parabacteroides.[ 313 ] A substantial interindividual variability has been characterized in the elderly person gut microbiome, with fluctuations featuring Faecalibacterium and Ruminococcus and specific Clostridium clusters, especially IV and XIVa. These may explain, in part, the different impacts of viral infections in elderly individuals. Many different direct or indirect microbiome pathways could contribute to SARS‐CoV‐2‐gut interactions. Direct or indirect pathways can be considered the pulmonary inflammation is seen in SARS‐CoV‐2 patients in the second week of infection. Direct suppression or promotion of viral infection by the microbiome can occur via various mechanisms, such as genetic recombination, alteration of virion stability, driving the proliferation of cells, simulating attachment to permissive cells, and contributing to viral replication suppression; promotion of viral infection may occur by inducing systems’ immunoregulatory and perturbing local immune responses.[ 314 ] An essential step for understanding the effect of the gut on SARS‐CoV‐2 is identifying the main gut microbiome species interacting with this virus. In this regard, the probability that SARS‐CoV‐2 may interact with one or many of the present microbiota (1500 species) in the gut makes the matter complicated. However, some microbiome associated microbial species get influenced by SARS‐CoV‐2 infection and produce some signature biomolecules. For example, Lactobacillus species can produce lactic acid, and the consequent pH changes inactivate different viruses due to carbohydrate fermentation.[ 314 ] The integrity of epithelial cells in the gut is essential, as they produce antiviral compounds that are hostile to viruses. The colonic epithelial cells’ functionality relies mainly on the luminal presence of butyrate as an energy source, and the main butyrate‐producing bacteria in the gut belong to the phylum Firmicutes. One hypothesis regarding microbiome interactions with SARS‐CoV‐2 is relevant to the microbiomes’ impacts on cytokines. Cytokines are small proteins molecules that lead the body's response against infection and inflammation. For example, type II interferon (interferon‐γ) classically play essential roles in antiviral responses.[ 315 ] More importantly, microbial metabolic processes in the gut strongly impact the production of cytokines. Microbiota can increase chronic phase proteins and interferon signaling in lung cells to protect against influenza infection. Nanotechnology may play a critical role in rapid diagnosis, monitoring, and designing practical therapeutic actions for COVID‐19 with relevance to the gut modulation by SAR‐CoV‐2. Noninvasive breath tests, with arrays of nanomaterials, can identify the presence of volatile organic compounds with the signatures of modulated microbiota (abundance of Prevotella) and hence, recognize the presence of SAR‐CoV‐2 for quick diagnosis and monitoring.[ 316 , 317 ]
In order to detect such type of signature molecule, Nakhleh et al. developed a noninvasive AI nanoarray based on molecularly modified gold nanoparticles and a random network of single‐walled carbon nanotubes for the classification of several diseases from exhaled breath.[ 317 ] The recorded performance of designed artificially intelligent nanoarray was clinically assessed on both breath samples (having 17 different disease conditions or healthy controls) collected from 1404 patients. Experimental data showed 86% accuracy and allowing both detection and discrimination between the different disease conditions examined.[ 317 ] Some therapeutic strategy depends on eliminating a specific bacterial strain in the gut, in this case, broad‐spectrum antibiotics would not work, as they also eliminate beneficial bacteria and consequently weaken the gut microbiome barrier. In this scenario, nanotechnology can play an efficient role in developing nanomaterial‐based drugs, with the possibility of precise delivery in the gut. These drugs should target undesirable bacterial strains in the gastrointestinal tract and enhance their health by improving gut barriers against pathogens and inflammatory agents and by providing the base for creating disruptive interventions based on microbiome engineering. Nanoscale‐enabled tools will likely enable us to observe, navigate, and act through the gut's complicated ecosystem to help find either a cure or mitigating procedures for COVID‐19.
4. Safety Concerns Related to the Application of Nanomaterials
Deep understanding of the cytotoxic properties of nanomaterials is obviously indispensable if they are explored as nanomedicine. Nowadays, therapeutic nanomedicine related safety concerns are the central theme of the discussions among scientists, whether nanotechnology should use strategically as a therapeutic agent or be regulated to prevent physical, environmental harm. Hence, a comprehensive understanding of the interface between biological systems and nanomaterials is essential, introducing unique surface features that can be explored to control nanomaterials’ exposure, bioavailability, and biocatalytic properties. The central tendency of nanoparticles to form an aggregate in a natural and biological system can be taken advantage of to reduce their toxicity. For example, the degree of aggregation of TiO2 nanoparticles determines their cytotoxicity on fish cells underexposure to ultraviolet radiation.[ 318 ] The DNA cleavage effect of nanoparticles is higher in distilled water than in PBS, depending on particle dispersion. Therefore, potentially competing principles (aggregation, dispersal, translocation, and bioavailability) must be studied in detail to design an efficient nanosystem for therapeutic applications. One approach could be to use neutral nanoparticles to aggregate and immobilize at disposal sites. Surface coating is another designing feature to improve nanoparticle safety by preventing bioreactivity. For example, antioxidant analogues (i.e., oligomeric pro‐anthocyanidins) have been explored as coating agents for particulates that generate ROS. However, many coating chemicals are biologically labile and degradable, which means, initially, a nontoxic nanomaterial prone to become toxic after losing its coating. So, coating nanoparticles with protective agents effectively prevents the dissolution, inhibits the release of toxic ions, and provides a physical barrier against cellular uptake.[ 319 ] A handful of coating materials, including biocompatible organic or inorganic substances (i.e., PEG–SiO2, gold, biocompatible polymers), can be explored.[ 320 ] Modification of surface charge is another approach to reducing toxicity.[ 321 , 322 ] For example, layer by layer coatings of polyelectrolytes on gold nanorods decrease their cellular uptake by modifying their surface charge and functionality. It is essential to consider their aspect ratios, hydrophobicity, and stiffness for the safe design of materials that form bio persistent fibers (for example, CNTs).[ 323 ] Chemical functionalization of short (<5 µm) MWCNTs can provide stable dispersions of individual tubes in physiological media, allowing their safe use as imaging and drug delivery devices.[ 324 ] These materials have a high urinary excretion rate that provides an additional safety margin, as demonstrated in the biomedical development of potentially toxic nanomaterials such as quantum dots.[ 325 ] Experimental evidence suggests that prolonged, rigid CNTs should be avoided for in vivo applications; their functionalization with small hydrophilic groups is a safety feature allowing the formation of stable dispersions with high excretion rates.[ 325 , 326 ] Another potential approach to mitigate the toxicity of nanomaterials is the modulation of the oxidative state of nanomaterials at the interface. The toxicity of hydrophobic C‐60 derivatives can be modulated by increasing their hydration through surface functionalization.[ 327 ] It has been theorized that increasing the solubility of fullerenes through specific functional groups might lower ROS formation. Overall, suitable nanomaterials are promising options to cope with the current pandemic through biosensors for detection, contrast agents, and targeted drug delivery vehicles that can deliver antimicrobial, antiviral, or antitumoral agents. Unfortunately, the long‐term implications are mainly unknown.
5. Concluding Remarks
The COVID‐19 pandemic has posed a global public health emergency before the human race as an unprecedented situation. Coordination among researchers from various fields with professional expertise is required to cope with such a complicated challenge. Increasing diversity in therapeutic strategy with a multi and interdisciplinary methodology involving transversal disciplines will be indispensable to achieve advanced and efficient solutions to address such pandemics.[ 144 ] This review article highlighted the inherited potential of specific nanotechnologic platforms for improving coronavirus detection, disinfection, and multiplication inhibition. Nanomaterials can allow specific SARS‐CoV‐2 binding at nanoparticle surface or improve PCR efficacy, which generally leads to better sensitivity than other detection methods. Nanomedicines exhibit several advantages over conventional antiviral drugs in the fight against COVID‐19. These include improved drug safety, increased solubility of poorly soluble drugs, better virus inhibition, and a more balanced immune response and organ targeting than their non‐nanoformulated counterparts. Moreover, Nanomaterial formulated vaccines can replace the requirement of standard adjuvants. Moreover, the promising properties of these nanomaterials are also explored in the specific antiviral treatments, where nanoformulated antiviral drugs could block virus replication, trigger an anti‐COV immune response, or prevent COV‐induced apoptotic cellular death. However, nanomaterials have a great potential to solve the current challenges in the health sector; the long‐term health implications are mainly unknown. Some nanomaterials already have demonstrated adverse behavior in biological systems that excel the researchers to investigate the nanomaterial‐related safety issues. Overall, nanomaterial‐based therapeutics are promising in the fight against infectious diseases, including the SARS‐CoV‐2 pandemic, along with considering safety issues related to the minimal cytotoxicity of nanoformulated agents.
Conflict of Interest
The authors declare no conflict of interest.
Biographies
Atul K. Tiwari is currently a Ph.D. candidate in applied Microbiology under the supervision of Prof. Prem C. Pandey at Indian Institute of Technology (BHU), Varanasi, India. He received his Master's degree in microbiology from Dr. R. M. L. Awadh University, Ayodhya, India. His research is primarily focused on the designing and development of antimicrobial nanomaterials and investigating the molecular interactions mechanism between nanomaterials and cell surface biomolecules.

Anupa Mishra is a guest lecturer in the Department of Microbiology, Dr. RML Awadh University, Ayodhya, India. She received her master degree in Microbiology in 2014. Her current research is focussed on the development of biocompatible materials and their biomedical applications.

Govind Pandey is a candidate of MD (JR III) in Department of Paediatrics, King George Medical University, Lucknow. Dr Pandey received MBBS degree from BRD Medical College, Gorakhpur in 2018. Dr. Pandey is actively involved in management of covid 19 patients of paediatric age group at King George medical, Lucknow, India. He is studying on prevalence of pathological neonatal hyperbilirubinemia due to blood group incompatibility (both major and minor) and its clinical profile in neonatal unit. His current research interests are nanoparticle development and its biomedical application.

Munesh Kumar Gupta is an Associate professor in Department of Microbiology, Institute of Medical Sciences, BHU, Varanasi, India. He received his MD (Microbiology) degree from Banaras Hindu University, Varanasi, India. His area of interests is pathophysiology of infectious diseases, drug resistance in microbial pathogens, Hospital infection control and search of promising drug molecules to combat the MDR among microorganisms.

Prem C. Pandey is Professor (HAG) in Department of Chemistry, Indian Institute of Technology, Banaras Hindu University. Prof Pandey received Ph.D. in the area of Applied Electrochemistry in 1986. Research interest and details can be found at: http://sensors‐vns.com/asset/pdf/Prof.%20P.%20C.%20Pandey.pdf, https://www.iitbhu.ac.in/dept/apc/people/pcpandeyapc.

Tiwari A. K., Mishra A., Pandey G., Gupta M. K., Pandey P. C., Nanotechnology: A Potential Weapon to Fight against COVID‐19. Part. Part. Syst. Charact. 2022, 39, 2100159. 10.1002/ppsc.202100159
Contributor Information
Atul K. Tiwari, Email: atulkumartiwari.rs.chy19@itbhu.ac.in.
Prem C. Pandey, Email: pcpandey.apc@iitbhu.ac.in.
References
- 1.WHO report‐98, Johns Hopkins University, www.coronavirus.jhu.edu/map.html June, 2021.
- 2.WHO COVID‐19 update dashboard, https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed: June 2021).
- 3. Muhammad W., Khan M. A., Nazir M., Siddiquah A., Mushtaq S., Hashmi S. S., Abbasi B. H., Mater. Sci. Eng. C 2019, 103, 109740. [DOI] [PubMed] [Google Scholar]
- 4. Shi J., Votruba A. R., Farokhzad O. C., Langer R., Nano Lett. 2010, 10, 3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muhammad W., Ullah N., Haroon M., Abbasi B. H., RSC Adv. 2019, 9, 29541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muhammad W., Haleem I., Ullah N., Khan M. Q., Nanoscale Rep. 2019, 2, 1. [Google Scholar]
- 7. Leon L., Chung E. J., Rinaldi C. (Eds: Chung E. J., Leon L., Rinaldi C.), Elsevier, Amsterdam 2020, p. 1, https://org/10.1016/B978-0-12-816662-8.00001-1.
- 8. Kim B. Y., Rutka J. T., Chan W. C., N. Engl. J. Med. 2010, 363, 2434. [DOI] [PubMed] [Google Scholar]
- 9. Ristroph K. D., Prud'homme R. K., Nanoscale Adv. 2019, 1, 4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mungrue K., Adv. Lab. Med. Int 2014, 4, 1. [Google Scholar]
- 11. Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J., Adair T., Aggarwal R., Ahn S. Y., Alvarado M., Anderson H. R., Anderson L. M., Andrews K. G., Atkinson C., Baddour L. M., Barker‐Collo S., Bartels D. H., Bell M. L., Benjamin E. J., et al., Lancet 2012, 380, 2095.23245604 [Google Scholar]
- 12. Watkins K., Curr. Emerg. Hosp. Med. Rep. 2018, 6, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., Haagmans B. L., Lauber C., Leontovich A. M., Neuman B. W., Penzar D., Perlman S., Poon L. L. M., Samborskiy D. V., Sidorov I. A., Sola I., Ziebuhr J., Nat. Microbiol. 2020, 5, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen K., Yang Y., Wang T., Zhao D., Jiang Y., Jin R., Zheng Y., Xu B., Xie Z., Lin L., Shang Y., Lu X., Shu S., Bai Y., Deng J., Lu M., Ye L., Wang X., Wang Y., Gao L., World J. Pediatr. 2020, 16, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drexler J. F., Gloza‐Rausch F., Glende J., Corman V. M., Muth D., Goettsche M., Seebens A., Niedrig M., Pfefferle S., Yordanov S., J. Virol. 2010, 84, 11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu G., Liu D., Protein Cell 2012, 3, 803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shereen M. A., Khan S., Kazmi A., Bashir N., Siddique R., J. Adv. Res. 2020, 24, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Nature 2020, 579, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lovato A., de Filippis C., Ear, Nose, Throat J. 2020, 99, 569. [DOI] [PubMed] [Google Scholar]
- 20. Chauhan G., Madou M. J., Kalra S., Chopra V., Ghosh D., Martinez‐Chapa S. O., ACS Nano 2020, 14, 7760. [DOI] [PubMed] [Google Scholar]
- 21.Flicker photos NIAID, https://www.flickr.com/photos/niaid/49530315733/in/photolist (accessed: June 2021).
- 22. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Lancet 2020, 395, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bansal M., Clin. Res. Rev. 2020, 14, 247. [Google Scholar]
- 24. Rabb H., J. Clin. Invest. 2020, 130, 2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seah I., Agrawal R., Ocul. Immunol. Inflammation 2020, 28, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu Y., Xu X., Chen Z., Duan J., Hashimoto K., Yang L., Liu C., Yang C., Brain, Behav., Immun. 2020, 87, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R. A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., Romero J. P., Wirnsberger G., Zhang H., Slutsky A. S., Conder R., Montserrat N., Mirazimi A., Penninger J. M., Cell 2020, 181, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H., Penninger J. M., Li Y., Zhong N., Slutsky A. S., Intensive Care Med. 2020, 46, 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoffmann M., Kleine‐Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.‐H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., Cell 2020, 181, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K. Y., Wang Q., Zhou H., Yan J., Qi J., Cell 2020, 181, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Biswaro L. S., da Costa Sousa M. G., Rezende T. M. B., Dias S. C., Franco O. L., Front. Microbiol. 2018, 9, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Panáček A., Kolář M., Večeřová R., Prucek R., Soukupova J., Kryštof V., Hamal P., Zbořil R., Kvítek L., Biomaterials 2009, 30, 6333. [DOI] [PubMed] [Google Scholar]
- 33. Qasim M., Lim D.‐J., Park H., Na D., J. Nanosci. Nanotechnol. 2014, 14, 7374 [DOI] [PubMed] [Google Scholar]
- 34. Abbasi B. H., Nazir M., Muhammad W., Hashmi S. S., Abbasi R., Rahman L., Hano C., Biomolecules 2019, 9, 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fakruddin M., Hossain Z., Afroz H., J. Nanobiotechnol. 2012, 10, 31. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36. Salata O. V., J. Nanobiotechnol. 2004, 2, 3. [Google Scholar]
- 37. Patra J. K., Das G., Fraceto L. F., Campos E. V. R., del Pilar Rodriguez‐Torres M., Acosta‐Torres L. S., Diaz‐Torres L. A., Grillo R., Swamy M. K., Sharma S., J. Nanobiotechnol. 2018, 16, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ding J., Venkatesan R., Zhai Z., Muhammad W., Nakkala J. R., Gao C., Colloids Surf., B 2020, 192, 111075. [DOI] [PubMed] [Google Scholar]
- 39. Alphandery E., Bioconjugate Chem. 2020, 31, 1873. [DOI] [PubMed] [Google Scholar]
- 40. Amanat F., Krammer F., Immunity 2020, 52, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen W. H., Strych U., Hotez P. J., Bottazzi M. E., Curr. Trop. Med. Rep. 2020, 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yadavalli T., Shukla D., Nanomedicine 2017, 13, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Singh L., Kruger H. G., Maguire G. E. M., Govender T., Parboosing R., Ther. Adv. Infect. Dis. 2017, 4, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jackman J. A., Lee J., Cho N.‐J., Small 2016, 12, 1133. [DOI] [PubMed] [Google Scholar]
- 45. Adesina S. K., Akala E. O., Mol. Pharmaceutics 2015, 12, 4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weiss C., Carriere M., Fusco L., Capua I., Regla‐Nava J. A., Pasquali M., Scott J. A., Vitale F., Unal M. A., Mattevi C., Bedognetti D., Merkoçi A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L. G., ACS Nano 2020, 14, 6383 [DOI] [PubMed] [Google Scholar]
- 47. Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B., Int. J. Oral Sci. 2020, 12, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fathizadeh H., Maroufi P., ‐Heravi M. M., Dao S., Köse S., Ganbarov K., Pagliano P., Esposito S., Kafil H. S., Infez. Med. 2020, 28, 185. [PubMed] [Google Scholar]
- 49. Wang J., Shen J., Ye D., Yan X., Zhang Y., Yang W., Li X., Wang J., Zhang L., Pan L., Environ. Pollut. 2020, 262, 114665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu J., Chamakura K., Ballestero P., Bashir R. S., ACS Symp. Ser. Am. Chem. Soc. 2020, 6, 129. [Google Scholar]
- 51. Shahzadi S., Zafar N., Sharif R. IntechOpen 2018, 10.5772/intechopen.72526. [DOI]
- 52. Sportelli M. C., Izzi M., Kukushkina E. A., Hossain S. I., Picca R. A., Ditaranto N., Ciof N., Nanomaterials 2020, 10, 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Balagna C., Perero S., Percivalle E., Nepita E. V., Ferraris M., Open Ceram. 2020, 1, 100006. [Google Scholar]
- 54.Statnano, https://statnano.com/nanotechnology-in-battle-against-coronavirus (accessed: June 2021).
- 55. Konda A., Prakash A., Moss G. A., Schmoldt M., Grant G. D., Guha S., ACS Nano 2020, 14, 6339. [DOI] [PubMed] [Google Scholar]
- 56. Widdowson N., https://phys.org/news/2020-04-maskmaterial-virus-sizenanoparticles.html 2020. July 2021.
- 57. Palmieri V., Pap M., Nano Today 2020, 33, 100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.NBIC, https://statnano.com/news/67559/ 2020 July 2021.
- 59. Cho I. H., Lee D. G., Yang Y. Y., US8673331, 2014.
- 60. 10.1038/nindia.2020. [DOI]
- 61. Joe Y. H., Park D. H., Hwang J., J. Hazard. Mater. 2016, 301, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Quang H. T., Nguyen T. T. H., Nguyen T. T., Pham C. V., Pham H. N., Anh‐Tuan L., Nguyen H. T. H., J. Virol. Methods 2017, 241, 52.28040515 [Google Scholar]
- 63. Li Y., Lin Z., Zhao M., Xu T., Wang C., Hua L., Wang H., Xia H., Zhu B., ACS Appl. Mater. Interfaces 2016, 8, 24385. [DOI] [PubMed] [Google Scholar]
- 64. Du T., Liang J., Dong N., Lu J., Fu Y., Fang L., Xiao S., Han H., ACS Appl. Mater. Interfaces 2018, 10, 4369. [DOI] [PubMed] [Google Scholar]
- 65. Villanueva M. M., Moran K. S., Martinez S. J., Rangel L. R., Garza N. M., Rodriguez P. C., Zarate T. D., Trejo A. L., Viruses 2019, 11, 1111. [Google Scholar]
- 66. Borkow G., Steve Z., Page T., Gabbay J., PLoS One 2010, 5, e11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hang X., Peng H., Song H., Qi Z., Miao X., Xu W., J. Virol Methods 2015, 222, 150. [DOI] [PubMed] [Google Scholar]
- 68. Trigilio J., Antoine T., Paulowicz I., Mishra Y., Adelung R., Shukla D., PLoS One 2012, 7, e48147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Talebian S., Wallace G. G., Schroeder A., Stellacci F., Conde J., Nat. Nanotechnol. 2020, 15, 618. [DOI] [PubMed] [Google Scholar]
- 70. Mokhtarzadeh A., Eivazzadeh‐Keihan R., Pashazadeh P., Hejazi M., Gharaatifar N., Hasanzadeh M., Baradaran B., de la Guardia M., TrAC, Trends Anal. Chem. 2017, 97, 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hematian A., Sadeghifard N., Mohebi R., Taherikalani M., Nasrolahi A., Amraei M., Ghafourian S., Osong Public Health Res. Perspect. 2016, 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sridhar S., To K. K., Chan J. F., Lau S. K., Woo P. C., Yuen K. Y., J. Mol. Diagn. 2015, 17, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Soler M., Acta Sci. Venezolana 1998, 49, 25. [PubMed] [Google Scholar]
- 74. Diel D. G., Lawson S., Okda F., Singrey A., Clement T., Fernandes M. H., Christopher‐Hennings J., Nelson E. A., Virus Res. 2016, 226, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Balasuriya U. B., Crossley B. M., Timoney P. J., J. Vet. Diagn. Invest. 2015, 27, 673. [DOI] [PubMed] [Google Scholar]
- 76. Patel A., Jernigan D. B., MMWR 2020, 69, 140.32027631 [Google Scholar]
- 77. Lucia C., Federico P.‐B., Alejandra G. C., bioRxiv 2020, 971127, 2002. [Google Scholar]
- 78. Bellan L. M., Wu D., Langer R. S., Interdiscip. Rev. Nanomed. Nanobiotechnol. 2011, 3, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vannoy C. H., Tavares A. J., Noor M. O., Uddayasankar U., Krull U. J., Sensors 2011, 11, 9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ramamoorthy A., Akis R., Bird J. P., Maemoto T., Ferry D. K., Inoue M., Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2003, 68, 026221. [DOI] [PubMed] [Google Scholar]
- 81. Liu X., Braun G. B., Zhong H., Hall D. J., Han W., Qin M., Zhao C., Wang M., She Z. G., Cao C., Sailor M. J., Stallcup W. B., Ruoslahti E., Sugahara K. N., Adv. Funct. Mater. 2016, 26, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shao D., Li J., Pan Y., Zhang X., Zheng X., Wang Z., Zhang M., Zhang H., Chen L., Biomater. Sci. 2015, 3, 833. [DOI] [PubMed] [Google Scholar]
- 83. Chen Z. Y., Abdelhamid H. N., Wu H. F., Rapid Commun. Mass Spectrom. 2016, 30, 1403. [DOI] [PubMed] [Google Scholar]
- 84. Wen L., Lin Y., Zheng Z. H., Zhang Z. L., Zhang L. J., Wang L. Y., Wang H. Z., Pang D. W., Biomaterials 2014, 35, 2295. [DOI] [PubMed] [Google Scholar]
- 85. Li Y., Jing L., Ding K., Gao J., Peng Z., Li Y., Shen L., Gao M., RSC Adv. 2014, 4, 22545. [Google Scholar]
- 86. Wang S., Li L., Jin H., Yang T., Bao W., Huang S., Wang J., Biosens. Bioelectron. 2013, 41, 205. [DOI] [PubMed] [Google Scholar]
- 87. Krejcova L., Nejdl L., Hynek D., Krizkova S., Kopel P., Adam V., Kizek R., Molecules 2013, 18, 15573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Guo J., Wang Y., Niu S., Li H., Tian Y., Yu S., Yu F., Wu Y., Liu L.i‐e, ACS Omega 2020, 5, 23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vaculovicova M., Michalek P., Krizkova S., Macka M., Adam V., Anal. Methods 2017, 9, 2375. [Google Scholar]
- 90. Udugama B., Kadhiresan P., Kozlowski H. N., Malekjahani A., Osborne M., Li V. Y. C., Chen H., Mubareka S., Gubbay J. B., Chan W. C. W., ACS Nano 2020, 14, 3822. [DOI] [PubMed] [Google Scholar]
- 91. Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F., J. Med. Virol. 2020, 92, 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moitra P., Alafeef M., Dighe K., Frieman M. B., Pan D., ACS Nano 2020, 14, 7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Qiu G., Gai Z., Tao Y., Schmitt J., ‐Ublick G. A. K., Wang J., ACS Nano 2020, 14, 5268. [DOI] [PubMed] [Google Scholar]
- 94. Huang C., Wen T., Shi F.‐J., Zeng X.‐Y., Jiao Y.‐J., ACS Omega 2020, 5, 12550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Seo G., Lee G., Kim M. J., Baek S.‐H., Choi M., Ku K. B., Lee C.‐S., Jun S., Park D., Kim H. G., Kim S.‐J., Lee J.‐O., Kim B. T., Park E. C., Il Kim S., ACS Nano 2020, 14, 5135. [DOI] [PubMed] [Google Scholar]
- 96. Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C. S., Anal. Chem. 2017, 89, 5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bolotsky A., Butler D., Dong C., Gerace K., Glavin N. R., Muratore C., Robinson J. A., Ebrahimi A., ACS Nano 2019, 13, 9781. [DOI] [PubMed] [Google Scholar]
- 98. Su S., Sun Q., Gu X., Xu Y., Shen J., Zhu D., Chao J., Fan C., Wang L., TrAC, Trends Anal. Chem. 2019, 119, 115610. [Google Scholar]
- 99. nard‐Moyon C. M′., Bianco A., Kalantar‐Zadeh K., ACS Sens. 2020, 5, 3739. [DOI] [PubMed] [Google Scholar]
- 100. Chand R., Neethirajan S., Biosens. Bioelectron. 2017, 98, 47. [DOI] [PubMed] [Google Scholar]
- 101. Huang J., Xie Z., Xie Z., Luo S., Xie L., Huang L., Fan Q., Zhang Y., Wang S., Zeng T., Anal. Chim. Acta 2016, 913, 121. [DOI] [PubMed] [Google Scholar]
- 102. Liu F., Kim Y. H., Cheon D. S., Seo T. S., Sens. Actuators, B 2013, 186, 252. [Google Scholar]
- 103. Zhan L., Li M. C., Wu W. B., C. Z., Chem. Commun. 2014, 50, 11526. [DOI] [PubMed] [Google Scholar]
- 104. Fan Z., Yust B., Nellore B. P. V., Sinha S. S., Kanchanapally R., Crouch R. A., Pramanik A. S. R., Sardar D., Ray P. C., J. Phys. Chem. Lett. 2014, 5, 3216. [DOI] [PubMed] [Google Scholar]
- 105. Lee J., Takemura K., Park E. Y., Sens. Actuators, B 2018, 276, 254. [Google Scholar]
- 106. Peng X., Luo G., Wu Z., Wen W., Zhang X., Wang S., ACS Appl. Mater. Interfaces 2019, 11, 41148. [DOI] [PubMed] [Google Scholar]
- 107. Seo G., Lee G., Kim M. J., Baek S.‐H., Choi M., Ku K. B., Lee C.‐S., Jun S., Park D., Kim H. G., Kim S.‐J., Lee J.‐O., Kim B. T., Park E. C., Kim S. I., ACS Nano 2020, 14, 5135. [DOI] [PubMed] [Google Scholar]
- 108. Wu Z., Hu J., Zeng T., Zhang Z.‐L., Chen J., Wong G., Qiu X., Liu W., Gao G. F., Bi Y., Pang D.‐W., Anal. Chem. 2017, 89, 2039. [DOI] [PubMed] [Google Scholar]
- 109. Hu Y., Li F., Bai X., Li D., Hua S., Wang K., Niu L., Chem. Commun. 2011, 47, 1743. [DOI] [PubMed] [Google Scholar]
- 110. Zribi B., Castro‐Arias J.‐M., Decanini D., Gogneau N., Dragoe D., Cattoni A., Ouerghi A., Korri‐Youssoufi H., Gosnet A.‐M. H., Nanoscale 2016, 8, 15479. [DOI] [PubMed] [Google Scholar]
- 111. Liu M., Zhao H., Chen S., Yu H., Quan X., ACS Nano 2012, 6, 3142. [DOI] [PubMed] [Google Scholar]
- 112. Shen Q., Han L., Fan G., Zhang J.‐R., Jiang L., Zhu J.‐J., Anal. Chem. 2015, 87, 4949. [DOI] [PubMed] [Google Scholar]
- 113. Shi X.‐M., Fan G.‐C., Tang X., Shen Q., Zhu J.‐J., Biosens. Bioelectron. 2018, 109, 190. [DOI] [PubMed] [Google Scholar]
- 114. He S., Liu K.‐K., Su S., Yan J., Mao X., Wang D., He Y., Li L.‐J., Song S., Fan C., Anal. Chem. 2012, 84, 4622. [DOI] [PubMed] [Google Scholar]
- 115. Fan Z., Kanchanapally R., Ray P. C., J. Phys. Chem. Lett. 2013, 21, 3813. [Google Scholar]
- 116. Xia X. H., Chao D. L., Zhang Y. Q., Shen Z. X., Fan H. J., Nano Today 2014, 9, 785. [Google Scholar]
- 117. Lu L., Biosens. Bioelectron. 2018, 110, 180. [DOI] [PubMed] [Google Scholar]
- 118. Pinto D. B., Shukla S., Gedanken A., Sarid R., Small 2010, 6, 1044. [DOI] [PubMed] [Google Scholar]
- 119. Vonnemann J., Sieben C., Wolff C., Ludwig K., Böttcher C., Herrmann A., Haag R., Nanoscale 2014, 6, 2353. [DOI] [PubMed] [Google Scholar]
- 120. Papp I., Sieben C., Ludwig K., Roskamp M., Böttcher C., Schlecht S., Herrmann A., Haag R., Small 2010, 6, 2900. [DOI] [PubMed] [Google Scholar]
- 121. Cagno V., Andreozzi P., D'Alicarnasso M., Silva P. J., Mueller M., Galloux M., Goffic R. L., Jones S. T., Vallino M., Hodek J., Weber J., Sen S., Janecek E.‐R., Bekdemir A., Sanavio B., Martinelli C., Donalisio M., Welti M.‐A. R., Eleouet J.‐F., Han Y., Nat. Mater. 2018, 17, 195. [DOI] [PubMed] [Google Scholar]
- 122. Łoczechin A., Séron K., Barras A., Giovanelli E., Belouzard S., Chen Y. T., Nolte N. M., Boukherroub R., Dubuisson J., Szunerits S., ACS Appl. Mater. Interfaces 2019, 11, 42964, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hu C. M. J., Chang W. S., Fang Z. S., Chen Y. T., Wang W. L., Tsai H. H., Chueh L. L., Takano T., Hohdatsu T., Chen H. W., Sci. Rep. 2017, 7, 13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hu C. M. J., Chen Y. T., Fang Z. S., Chang W. S., Chen H. W., Int. J. Nanomed. 2018, 13, 8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chen Y. N., Hsueh Y. H., Hsieh C. T., Tzou D. Y., Chang P. L., Int. J. Environ. Res. Public Health 2016, 13, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Du T., Liang J., Dong N., Lu J., Fu Y., Fang L., Xiao S., Han H., ACS Appl. Mater. Interfaces 2018, 10, 4369. [DOI] [PubMed] [Google Scholar]
- 127. Lv X., Wang P., Bai R., Cong Y., Suo S., Ren X., Chen C., Biomaterials 2014, 35, 4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Yang X. X., Li C. M., Li Y. F., Wang J., Huang C. Z., Nanoscale 2017, 9, 16086. [DOI] [PubMed] [Google Scholar]
- 129. Dey P., Bergmann T., Cuellar‐Camacho J. L., Ehrmann S., Chowdhury M. S., Zhang M., Dahmani I., Haag R., Azab W., ACS Nano 2018, 12, 6429. [DOI] [PubMed] [Google Scholar]
- 130. Szyma'nska E., Orłowski P., Winnicka K., Tomaszewska E., Baska P., Celichowski G., Grobelny J., Basa A., Int. J. Mol. Sci. 2018, 19, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Sametband M., Kalt I., Gedanken A., Sarid R., ACS Appl. Mater. Interfaces 2014, 6, 1228. [DOI] [PubMed] [Google Scholar]
- 132. Halder A., Das S., Ojha D., Chattopadhyay D., Mater. Sci. Eng. C 2018, 89, 413. [DOI] [PubMed] [Google Scholar]
- 133. Ziem B., Azab W., Gholami M. F., Rabe J. P., Osterriederb N., Haag R., Nanoscale 2017, 9, 3774. [DOI] [PubMed] [Google Scholar]
- 134. Bimbo L. M., Denisova O. V., Makila E., Kaasalainen M., Brabander J. K. D., Hirvonen J., Salonen J., Kakkola L., Kainov D., Santos H. A., ACS Nano 2013, 7, 6884. [DOI] [PubMed] [Google Scholar]
- 135. Ortega‐Berlanga B., Hernández‐Adame L., Angel‐Olarte C. D., Aguilar F., Rosales‐Mendoza S., Palestino G., Chem. Eng. J. 2020, 385, 123414. [Google Scholar]
- 136. Tavakoli A., Hashemzadeh M. S., J. Virol. Methods 2020, 275, 113688. [DOI] [PubMed] [Google Scholar]
- 137. Huo C., Xiao J., Xiao K., Zou S., Wang M., Qi P., Liu T., Hu Y., Int. J. Nanomed. 2020, 15, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ghaffari H., Tavakoli A., Moradi A., Tabarraei A., Bokharaei‐Salim F., Zahmatkeshan M., Farahmand M., Javanmard D., Kiani S. J., Esghaei M., Pirhajati‐Mahabadi V., Monavari S. H., Pirkooh A. A., J. Biomed. Sci. 2019, 26, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Elechiguerra J. L., Burt J. L., Morones J. R., Bragado A. C., Gao X., Lara H. H., Yacaman M. J., J. Nanobiotechnol. 2005, 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sun L., Singh A. K., Vig K., Pillai S. R., Singh S. R., J. Biomed. Nanotechnol. 2008, 4, 149. [Google Scholar]
- 141. Li Y., Lin Z., Zhao M., Xu T., Wang C., Hua L., Wang H., Xia H., Zhu B., ACS Appl. Mater. Interfaces 2016, 8, 24385. [DOI] [PubMed] [Google Scholar]
- 142. Orlowski P., Tomaszewska E., Gniadek M., Baska P., Nowakowska J., Sokolowska J., Nowak Z., Donten M., Celichowski G., Grobelny J., Krzyzowska M., PLoS One 2014, 9, e104113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Li Y., Lin Z., Zhao M., Guo M., Xu T., Wang C., Xia H., Zhu B., RSC Adv. 2016, 6, 89679. [Google Scholar]
- 144. Sofy A. R., Hmed A. A., Abd El Haliem N. F., Zein M. A.‐E., Elshaarawy R. F. M., Carbohydr. Polym. 2019, 226, 115261. [DOI] [PubMed] [Google Scholar]
- 145. Rogers J. V., Parkinson C. V., Choi Y. W., Speshock J. L., Hussain S. M., Nanoscale Res. Lett. 2008, 3, 129. [Google Scholar]
- 146. Speshock J. L., Murdock R. C., Braydich‐Stolle L. K., Schrand A. M., Hussain S. M., J. Nanobiotechnol. 2010, 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. ‐Pinto D. B., Shukla S., Perkas N., Gedanken A., Sarid R., Bioconjugate Chem. 2009, 20, 1497. [DOI] [PubMed] [Google Scholar]
- 148. Li Y., Lin Z., Xu T., Wang C., Zhao M., Xiao M., Wang H., Deng N., Zhu B., RSC Adv. 2017, 7, 1453. [Google Scholar]
- 149. Mycroft‐West C., Su D., Elli S., Guimond S., Miller G., Turnbull J., Yates E., Guerrini M., Fernig D., Lima M., Skidmore M., bioRxiv 2020, 10.1101/2020.02.29.971093 (accessed: July 2021). [DOI] [Google Scholar]
- 150. Han Y., Kral P., ACS Nano 2020, 14, 5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ciriminna R., Albo Y., Pagliaro M., ChemMedChem 2020, 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Marambio‐ J. C., Hoek E. M. V., J. Nanopart. Res. 2010, 12, 1531. [Google Scholar]
- 153. Xiang D. X., Chen Q., Pang L., Zheng C. L., J. Virol. Methods 2011, 178, 137. [DOI] [PubMed] [Google Scholar]
- 154. Gaikwad S., Int. J. Nanomed. 2013, 8, 4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Hu Y. M., Lian X. T., Dong H., Acta Metall. Sin. 2020, 56, 633. [Google Scholar]
- 156. Yan L. Z., Chen M. X., He H., Qu J. H., Chin. J. Catal. 2005, 26, 1122. [Google Scholar]
- 157. Belanger J. M., Raviv Y., Viard M., de la Cruz M. J., Nagashima K., Blumenthal R., Virology 2011, 417, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Tiwari A. K., Gupta M. K., Pandey G., Narayan R. J., Pandey P. C., J. Mater. Res. 2020, 35, 2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Pandey P. C., Tiwari A. K., Gupta M. K., Pandey G., Narayan R. J., Nano LIFE 2020, 10, 2050002, [Google Scholar]
- 160. Huang X., Qi X., Boey F., Zhang H., Chem. Soc. Rev. 2012, 41, 666. [DOI] [PubMed] [Google Scholar]
- 161. Tashan H., Khosravi‐Darani K., Yazdian F., Omidi M., Mini‐Rev. Org. Chem. 2019, 16, 159. [Google Scholar]
- 162. Pratheesya T., Harish S., Sohila N. M. S., Ramesh R., Mater. Res. Express 2019, 6, 074003. [Google Scholar]
- 163. McCreary A., Kazakova O., Jariwala D., Al Balushi Z. Y., 2D Mater. 2021, 8, 013001. [Google Scholar]
- 164. Valimukhametova A., Ryan C., Paz T., Grote F., Naumov A. V., Nanotechnology 2021, 32, 015709. [DOI] [PubMed] [Google Scholar]
- 165. Hong Z., Ruihua D., Xin Z., Yiwei L., Liangliang Q., Hao P., Drug Discovery Today 2017, 22, 1302. [DOI] [PubMed] [Google Scholar]
- 166. Liu J., Zhang Y., Zhang H., Yang J., Nanotechnology 2021, 32, 085712. [DOI] [PubMed] [Google Scholar]
- 167. Libbi F., Bonini N., Marzari N., 2D Mater. 2021, 8, 015026. [Google Scholar]
- 168. Zeng B., Huang Z., Singh A., Yao Y., Azad A. K., Light: Sci. Appl. 2018, 7, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Ugur K., Daghash Sherif M., Sherif Muhammad M., Ozbulut Osman E., Polym. Compos. 2021, 42, 1235. [Google Scholar]
- 170. Rezaei A., Kamali B., Kamali A. R., Measurement 2020, 150, 107087. [Google Scholar]
- 171. Reza K. A., Derek F., J. Mater. Sci. 2016, 51, 569. [Google Scholar]
- 172. Swetha P., Dsouza P., Hosabettu M., Kariate S., Part. Part. Syst. Charact. 2018, 35, 1800105. [Google Scholar]
- 173. Chen Y., Wu W., Xu Z., Jiang C., Han S., R. Soc. Open Sci. 2020, 7, 192019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Hagos K. M., Neway B., Aschalew T., Basavaiah K., RSC Adv. 2020, 10, 34916. [Google Scholar]
- 175. Elham V., Ghorbani‐Hasan S. A., Fereshteh C., Naghizadeh R. S., Seyed‐Ahmad S., ChemistrySelect 2020, 5, 10401. [Google Scholar]
- 176. Xia M. Y., Xie Y., Yu C. H., Chen G. Y., Zhang Y. H. L. T., J. Controlled Release 2019, 307, 16. [DOI] [PubMed] [Google Scholar]
- 177. Farzanegan A., Roudbary M., Falahati M., Khoobi M., Gholibegloo E., Farahyar S., J. Mycol. Med. 2018, 28, 628. [DOI] [PubMed] [Google Scholar]
- 178. Cheong Y. K., Arce M. P., Benito A., Chen D., Crisóstomo N. L., Kerai L. V., Nanomaterials 2020, 10, 819. [Google Scholar]
- 179. Narges S., Nasim G., Mahshid K., Mohammad R. T., Lizette U., Jos M., ACS Biomater. Sci. Eng. 2020, 6, 6253.33449672 [Google Scholar]
- 180. Nicosia A., Vento F., Pellegrino A. L., Ranc V., Piperno A., Mazzaglia A., Nanomaterials 2020, 10, 2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Innocenzi P., Stagi L., Chem. Sci. 2020, 11, 6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Tahereh S., Kamali A. R., Colloids Surf., B 2021, 199, 111509. [DOI] [PubMed] [Google Scholar]
- 183. Seifi T., Kamali A. R., Med. Drug Discovery 2021, 11, 100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Lee C., Virol. J. 2015, 12, 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Ye S., Shao K., Li Z., Guo N., Zuo Y., Li Q., Lu Z., Chen L., He Q., Han H., ACS Appl. Mater. Interfaces 2015, 7, 21578. [DOI] [PubMed] [Google Scholar]
- 186. Pomeranz L. E., Reynolds A. E., Hengartner C. J., Rev M. M. B., 2005, 69, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Cook T. M., Anaesthesia 2020, 75, 920. [DOI] [PubMed] [Google Scholar]
- 188. https://www.who.int/publications-detailredirect/advice-on-the-use-of-masks-in-the
- 189. Möritz M., Peters H., Nipko B., Rüden H., Int. J. Hyg. Environ. Health 2001, 203, 401. [DOI] [PubMed] [Google Scholar]
- 190. Pandey P. C., Katyal N., Pandey G., Narayan R. J., MRS Commun. 2020, 10, 482. [Google Scholar]
- 191. Pandey P. C., Indian Patent 202014527 , 2020.
- 192. Pandey P. C., Antimicrobial and antiviral mask , https://www.youtube.com/watch?v=ViQ9ivQ8msg (accessed: April 2021).
- 193. Jang H., Ryoo S.‐R., Kim Y.‐K., Yoon S., Kim H., Han S. W., Choi B.‐S., Kim D.‐E., Min D.‐H., Angew. Chem., Int. Ed. 2013, 52, 2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Weiss C., Carriere M., Fusco L., Capua I., Regla‐Nava J. A., Pasquali M., Scott J. A., Vitale F., Unal M. A., Mattevi C., Bedognetti D., Merkoci A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L. G., ACS Nano 2020, 14, 6383. [DOI] [PubMed] [Google Scholar]
- 195. https://www.dezeen.com/2020/03/06/guardian-g-voltface-mask-graphene-coronavirus-bacteria.
- 196. Jing X., Zhi Z., Wang D., Liu J., Shao Y., Meng L., Bioconjugate Chem. 2018, 29, 559. [DOI] [PubMed] [Google Scholar]
- 197. Kerry R. G., Malik S., Redda Y. T., Sahoo S., Patra J. K., Majhi S., Nanomedicine 2019, 18, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Di Gianvincenzo P., Marradi M., Martínez‐Ávila O. M., Bedoya L. M., Alcamí J., Penades S., Bioorg. Med. Chem. Lett. 2010, 20, 2718. [DOI] [PubMed] [Google Scholar]
- 199. Elechiguerra J. L., Burt J. L., Morones J. R., Camacho Bragado A., Gao X., Lara H. H., Yacaman M. J., J. Nanobiotechnol. 2005, 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Orlowski P., Tomaszewska E., Gniadek M., Baska P., Nowakowska J., Sokolowska J., Nowak Z., Donten M., Celichowski G., Grobelny J., Krzyzowska M., PLoS One 2014, 9, e104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Baram‐Pinto D., Shukla S., Perkas N., Gedanken A., Sarid R., Bioconjugate Chem. 2009, 20, 1497. [DOI] [PubMed] [Google Scholar]
- 202. Warnes S. L., Little Z. R., Keevil C. W., mBio 2015, 6, e01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.STATNANO, CuO‐into‐Nanofibers‐in‐A‐Mask‐That‐Traps‐and‐Kills‐VirusesIncluding‐Coronavirus, https://statnano.com//news/67551/Respilon-Group-Incorporates. Aug. 2021
- 204. Chaturvedi U. C., Shrivastava R., FEMS Immunol. Med. Microbiol. 2005, 43, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. te Velthuis A. J. W., van den Worm S. H. E., Sims A. C., Baric R. S., Snijder E. J., van Hemert M. J., PLoS Pathog. 2010, 6, e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Tavakoli A., Ataei‐Pirkooh A., Sadeghi G. M. M., Bokharaei Salim F., Sahrapour P., Kiani S. J., Moghoofei M., Farahmand M., Javanmard D., Monavari S. H., Nanomedicine 2018, 13, 2675. [DOI] [PubMed] [Google Scholar]
- 207. Shaban M., Mohamed F., Abdallah S., Sci. Rep. 2018, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Yu J. W. u. M., Shi J., Farokhzad O. C., J. Controlled Release 2016, 240, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Nochi T., Yuki Y., Takahashi H., Sawada S.‐i., Mejima M., Kohda T., Harada N., Kong I. G., Sato A., Kataoka N., Tokuhara D., Kurokawa S., Takahashi Y., Tsukada H., Kozaki S., Akiyoshi K., Kiyono H., Nat. Mater. 2010, 9, 572. [DOI] [PubMed] [Google Scholar]
- 210. Keijzer C., Slütter B., van der Zee R., Jiskoot W., van Eden W., Broere F., PLoS One 2011, 6, e26684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Li B., Zhang X., Dong Y., WIREs Nanomed. Nanobiotechnol. 2019, 11, 1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Alphandéry E., Bioconjugate Chem. 2020, 31, 1873. [DOI] [PubMed] [Google Scholar]
- 213. Sekimukai H., ‐Yoshikawa N. I., Fukushi S., Tani H., Kataoka M., Suzuki T., Hasegawa H., Niikura K., Arai K., Nagata N., Microbiol. Immunol. 2020, 64, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Chen H.‐W., Huang C.‐Y., Lin S.‐Y., Fang Z.‐S., Hsu C.‐H., Lin J.‐C., Chen Y.‐I., Yao B.‐Y., Hu C. ‐M. J., Biomaterials 2016, 106, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Coleman C. M., Venkataraman T., Liu Y. V., Glenn G. M., Smith G. E., Flyer D. C., Frieman M. B., Vaccine 2017, 35, 1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Coleman C. M., Liu Y. V., Mu H., Taylor J. K., Massare M., Flyer D. C., Glenn G. M., Smith G. E., Frieman M. B., Vaccine 2014, 32, 3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Jung S.‐Y., Kang K. W., Lee E.‐Y., Seo D.‐W., Kim H.‐L., Kim H., Kwon T., Park H.‐L., Kim H., Lee S.‐M., Nam J.‐H., Vaccine 2018, 36, 3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Pimentel T. A. P. F., Yan Z., Jeffers S. A., Holmes K. V., Hodges R. S., Burkhard P., Chem. Biol. Drug Des. 2009, 73, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Sharma J., Shepardson K., Johns L. L., Wellham J., Avera J., Schwarz B., ‐Apple A. R., Douglas T., ACS Appl. Mater. Interfaces 2020, 12, 18211. [DOI] [PubMed] [Google Scholar]
- 220. Wang C., Zheng X., Gai W., Wong G., Wang H., Jin H., Feng N., Zhao Y., Zhang W., Li N., Zhao G., Li J., Yan J., Gao Y., Hu G., Yang S., Xia X., Antiviral Res. 2017, 140, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Raghuwanshi D., Mishra V., Das D., Kaur K., Suresh M. R., Mol. Pharmaceutics 2012, 9, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Wiley J. A., Richert L. E., Swain S. D., Harmsen A., Barnard D. L., Randall T. D., Jutila M., Douglas T., Broomell C., Young M., Harmsen A., PLoS One 2009, 4, e7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Lin L. C. ‐W., Huang C.‐Y., Yao B.‐Y., Lin J.‐C., Agrawal A., Algaissi A., Peng B.‐H., Liu Y.‐H., Huang P.‐H., Juang R.‐H., Chang Y.‐C., Tseng C.‐T., Chen H.‐W., Hu C. ‐M. J., Adv. Funct. Mater. 2019, 29, 1807616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Ting D., Dong N., Fang L., Lu J., Bi J., Xiao S., Han H., ACS Appl. Nano Mater. 2018, 1, 5451. [DOI] [PubMed] [Google Scholar]
- 225. Agostinis P., Berg K., Cengel K. A., Foster T. H., Girotti A. W., Gollnick S. O., Hahn S. M., Hamblin M. R., Juzeniene A., Kessel D., Korbelik M., Moan J., Mroz P., Nowis D., Piette J., Wilson B. C., Golab J., Ca‐Cancer J. Clin. 2011, 61, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Wainwright M., Int. J. Antimicrob. Agents 2003, 21, 510. [DOI] [PubMed] [Google Scholar]
- 227. Costa L., Faustino M. A. F., Neves M. G. P. M. S., Cunha A., Almeida A., Viruses 2012, 4, 1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Lim M. E., Lee Y.‐l., Zhang Y., Chu J. J. H., Biomaterials 2012, 33, 1912. [DOI] [PubMed] [Google Scholar]
- 229. Hantanasirisakul K., Gogotsi Y., Adv. Mater. 2018, 30, 1804779. [DOI] [PubMed] [Google Scholar]
- 230. Gogotsi Y., Anasori B., ACS Nano 2019, 13, 8491. [DOI] [PubMed] [Google Scholar]
- 231. Chen C., Boota M., Urbankowski P., Anasori B., Miao L., Jiang J., Gogotsi Y., J. Mater. Chem. A 2018, 6, 4617. [Google Scholar]
- 232. Nguyen V.‐H., Nguyen B.‐S., Hu C., Nguyen C. C., Nguyen D. L. T., Nguyen Dinh M. T., Vo D.‐V. N., Trinh Q. T., Shokouhimehr M., Hasani A., Kim S. Y., Le Q. V., Nanomaterials 2020, 10, 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Wong Z. M., Tan T. L., Yang S.‐W., Xu G. Q., ACS Appl. Mater. Interfaces 2018, 10, 39879 [DOI] [PubMed] [Google Scholar]
- 234. Low J., Zhang L., Tong T., Shen B., Yu J., J. Catal. 2018, 361, 255. [Google Scholar]
- 235. Rasool K., Helal M., Ali A., Ren C. E., Gogotsi Y., Mahmoud K. A., ACS Nano 2016, 10, 3674. [DOI] [PubMed] [Google Scholar]
- 236. Huang K., Li Z., Lin J., Han G., Huang P., Chem. Soc. Rev. 2018, 47, 5109. [DOI] [PubMed] [Google Scholar]
- 237. Dai C., Chen Y., Jing X., Xiang L., Yang D., Lin H., Liu Z., Han X., Wu R., ACS Nano 2017, 11, 12696. [DOI] [PubMed] [Google Scholar]
- 238. Xu B., Zhu M., Zhang W., Zhen X., Pei Z., Xue Q., Zhi C., Shi P., Adv. Mater. 2016, 28, 3333. [DOI] [PubMed] [Google Scholar]
- 239. Dai C., Lin H., Xu G., Liu Z., Wu R., Chen Y., Chem. Mater. 2017, 29, 8637. [Google Scholar]
- 240. Lin H., Gao S., Dai C., Chen Y., Shi J., J. Am. Chem. Soc. 2017, 139, 16235. [DOI] [PubMed] [Google Scholar]
- 241. Rasool K., Mahmoud K. A., Johnson D. J., Helal M., Berdiyorov G. R., Gogotsi Y., Sci. Rep. 2017, 7, 1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Pandey R. P., Rasool K., Madhavan V. E., Aissa B., Gogotsi Y., Mahmoud K. A., J. Mater. Chem. A 2018, 6, 3522. [Google Scholar]
- 243. Wiehe A., O'Brien J. M., Senge M. O., Photochem. Photobiol. Sci. 2019, 18, 2565. [DOI] [PubMed] [Google Scholar]
- 244. Nazari M., Xi M., Lerch S., Alizadeh M. H., Ettinger C., Akiyama H., Gillespie C., Gummuluru S., Erramilli S., Reinhard B. M., Sci. Rep. 2017, 7, 11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Li P., Li J., Feng X., Li J., Hao Y., Zhang J., Wang H., Yin A., Zhou J., Ma X., Wang B., Nat. Commun. 2019, 10, 2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Ng K. K., Lovell J. F., Vedadi A., Hajian T., Zheng G., ACS Nano 2013, 7, 3484. [DOI] [PubMed] [Google Scholar]
- 247. Li Y., Lin T.‐y., Luo Y., Liu Q., Xiao W., Guo W., Lac D., Zhang H., Feng C., Hogiu S. W., Walton J. H., Cherry S. R., Rowland D. J., Kukis D., Pan C., Lam K. S., Nat. Commun. 2014, 5, 4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Tsay J. M., Trzoss M., Shi L., Kong X., Selke M., Jung M. E., Weiss S., J. Am. Chem. Soc. 2007, 129, 6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Durantini A. M., Greene L. E., Lincoln R., Martinez S. R., Cosa G., J. Am. Chem. Soc. 2016, 138, 1215. [DOI] [PubMed] [Google Scholar]
- 250. Rehman F. U., Zhao C., Jiang H., Wang X., Biomater. Sci. 2016, 4, 40. [DOI] [PubMed] [Google Scholar]
- 251. Rozhkova E. A., Ulasov I., Lai B., Dimitrijevic N. M., Lesniak M. S., Rajh T., Nano Lett. 2009, 9, 3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Kovalev D., Fujii M., Adv. Mater. 2005, 17, 2531. [Google Scholar]
- 253. Xiao L., Gu L., Howell S. B., Sailor M. J., ACS Nano 2011, 5, 3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Seidl C., Ungelenk J., Zittel E., Bergfeldt T., Sleeman J. P., Schepers U., Feldmann C., ACS Nano 2016, 10, 3149. [DOI] [PubMed] [Google Scholar]
- 255. Samia A. C., Chen X., Burda C., J. Am. Chem. Soc. 2003, 125, 15736. [DOI] [PubMed] [Google Scholar]
- 256. Ge J., Lan M., Zhou B., Liu W., Guo L., Wang H., Jia Q., Niu G., Huang X., Zhou H., Meng X., Wang P., Lee C.‐S., Zhang W., Han X., Nat. Commun. 2014, 5, 4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Markovic Z. M., Ristic B. Z., Arsikin K. M., Klisic D. G., ‐Trajkovic L. M. H., ‐Markovic B. M. T., Kepic D. P., ‐Stevovic T. K. K., Jovanovic S. P., Milenkovic M. M., Biomaterials 2012, 33, 7084. [DOI] [PubMed] [Google Scholar]
- 258. Wang H., Yang X., Shao W., Chen S., Xie J., Zhang X., Wang J., Xie Y., J. Am. Chem. Soc. 2015, 137, 11376. [DOI] [PubMed] [Google Scholar]
- 259. Xu Y., Shi Z., Zhang L., Brown E. M., Wu A., Nanoscale 2016, 8, 12715. [DOI] [PubMed] [Google Scholar]
- 260. Idris N. M., Gnanasammandhan M. K., Zhang J., Ho P. C., Mahendran R., Zhang Y., Nat. Med. 2012, 18, 1580. [DOI] [PubMed] [Google Scholar]
- 261. Yu Z., Sun Q., Pan W., Li N., Tang B., ACS Nano 2015, 9, 11064. [DOI] [PubMed] [Google Scholar]
- 262. Hou Z., Zhang Y., Deng K., Chen Y., Li X., Deng X., Cheng Z., Lian H., Li C., Lin J., ACS Nano 2015, 9, 2584. [DOI] [PubMed] [Google Scholar]
- 263. Yang G. X., Yang D., Yang P. P., Lv R. C., Li C. X., Zhong C. N., He F., Gai S. L., Lin J., Chem. Mater. 2015, 27, 7957. [Google Scholar]
- 264. Gu B., Wu W., Xu G., Feng G., Yin F., Chong P. H. J., Qu J., Yong K. T., Liu B., Adv. Mater. 2017, 29, 1701076. [DOI] [PubMed] [Google Scholar]
- 265. Cao H., Wang L., Yang Y., Li J., Qi Y., Li Y., Li Y., Wang H., Li J., Angew. Chem., Int. Ed. 2018, 57, 7759. [DOI] [PubMed] [Google Scholar]
- 266. Zou X., Yao M., Ma L., Hossu M., Han X., Juzenas P., Chen W., Nanomedicine 2014, 9, 2339. [DOI] [PubMed] [Google Scholar]
- 267. Chen H., Wang G. D., Chuang Y.‐J., Zhen Z., Chen X., Biddinger P., Hao Z., Liu F., Shen B., Pan Z., Xie J., Nano Lett. 2015, 15, 2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Tang Y., Hu J., Elmenoufy A. H., Yang X., ACS Appl. Mater. Interfaces 2015, 7, 12261. [DOI] [PubMed] [Google Scholar]
- 269. Lan G., Ni K., Xu R., Lu K., Lin Z., Chan C., Lin W., Angew. Chem., Int. Ed. 2017, 56, 12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Ma L., Zou X. J., Bui B., Chen W., Song K. H., Solberg T., Appl. Phys. Lett. 2014, 105, 013702. [Google Scholar]
- 271. Song L., Li P. P., Yang W., Lin X. H., Liang H., Chen X. F., Liu G., Li J., Yang H. H., Adv. Funct. Mater. 2018, 28, 1707496. [Google Scholar]
- 272. Kamkaew A., Cheng L., Goel S., Valdovinos H. F., Barnhart T. E., Liu Z., Cai W., ACS Appl. Mater. Interfaces 2016, 8, 26630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Kotagiri N., Sudlow G. P., Akers W. J., Achilefu S., Nat. Nanotechnol. 2015, 10, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Ai X., Ho C. J. H., Aw J., Attia A. B. E., Mu J., Wang Y., Wang X., Wang Y., Liu X., Chen H., Gao M., Chen X., Yeow E. K. L., Liu G., Olivo M., Xing B., Nat. Commun. 2016, 7, 10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Pan L. M., Liu J. A., Shi J. L., Adv. Funct. Mater. 2014, 24, 7318. [Google Scholar]
- 276. Li S. Y., Cheng H., Xie B. R., Qiu W. X., Zeng J. Y., Li C. X., Wan S. S., Zhang L., Liu W. L., Zhang X. Z., ACS Nano 2017, 11, 7006. [DOI] [PubMed] [Google Scholar]
- 277. Cheng H., Zhu J. Y., Li S. Y., Zeng J. Y., Lei Q., Chen K. W., Zhang C., Zhang X. Z., Adv. Funct. Mater. 2016, 26, 7847. [Google Scholar]
- 278. Zhu H., Li J., Qi X., Chen P., Pu K., Nano Lett. 2018, 18, 586. [DOI] [PubMed] [Google Scholar]
- 279. Zhu W., Dong Z., Fu T., Liu J., Chen Q., Li Y., Zhu R., Xu L., Liu Z., Adv. Funct. Mater. 2016, 26, 5490. [Google Scholar]
- 280. Gao S., Wang G., Qin Z., Wang X., Zhao G., Ma Q., Zhu L., Biomaterials 2017, 112, 324. [DOI] [PubMed] [Google Scholar]
- 281. Bai J., Jia X., Zhen W., Cheng W., Jiang X., J. Am. Chem. Soc. 2018, 140, 106. [DOI] [PubMed] [Google Scholar]
- 282. Kim J., Cho H. R., Jeon H., Kim D., Song C., Lee N., Choi S. H., Hyeon T., J. Am. Chem. Soc. 2017, 139, 10992. [DOI] [PubMed] [Google Scholar]
- 283. Zheng D. W., Li B., Li C. X., Fan J. X., Lei Q., Li C., Xu Z., Zhang X. Z., ACS Nano 2016, 10, 8715. [DOI] [PubMed] [Google Scholar]
- 284. Liu L. H., Zhang Y. H., Qiu W. X., Zhang L., Gao F., Li B., Xu L., Fan J. X., Li Z. H., Zhang X. Z., Small 2017, 13, 1701621. [DOI] [PubMed] [Google Scholar]
- 285. Nasrollahzadeh M., Sajjadi M., Soufi G. J., Iravani S., Varma R. S., Nanomaterials 2020, 10, 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Parodi A., Molinaro R., Sushnitha M., Evangelopoulos M., Martinez J. O., Arrighetti N., Corbo C., Tasciotti E., Biomaterials 2017, 147, 155. [DOI] [PubMed] [Google Scholar]
- 287. Kotov N. A., Science 2010, 330, 188. [DOI] [PubMed] [Google Scholar]
- 288. LaBauve A. E., Rinker T. E., Noureddine A., Serda R. E., Howe J. Y., Sherman M. B., Rasley A., Brinker C. J., Sasaki D. Y., Negrete O. A., Sci. Rep. 2018, 8, 13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Molinaro R., Corbo C., Martinez J. O., Taraballi F., Evangelopoulos M., Minardi S., Yazdi I. K., Zhao P., De Rosa E., Sherman M. B., De Vita A., Furman N. E. T., Wang X., Parodi A., Tasciotti E., Nat. Mater. 2016, 15, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Czapar A. E., Zheng Y.‐R., Riddell I. A., Shukla S., Awuah S. G., Lippard S. J., Steinmetz N. F., ACS Nano 2016, 10, 4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Silva S., Almeida A. J., Vale N., Biomolecules 2019, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Zhao H., Wu M., Zhu L., Tian Y., Wu M., Li Y., Deng L., Jiang W., Shen W., Wang Z., Mei Z., Li P., Ran H., Zhou Z., Ren J., Theranostics 2018, 8, 1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Quan X., Sun D., Zhou J., Phys. Chem. 2019, 21, 10300. [DOI] [PubMed] [Google Scholar]
- 294. Ciobanasu C., Siebrasse J. P., Kubitscheck U., Biophys. J. 2010, 99, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Temsamani J., Vidal P., Drug Discovery Today 2004, 9, 1012. [DOI] [PubMed] [Google Scholar]
- 296. Skwarczynski M., Toth I., Ther. Delivery 2019, 10, 465. [DOI] [PubMed] [Google Scholar]
- 297. Derakhshankhah H., Jafari S., Biomed. Pharmacother. 2018, 108, 1090. [DOI] [PubMed] [Google Scholar]
- 298. Kranz L. M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K. C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A. N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R. A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U., Nature 2016, 534, 396. [DOI] [PubMed] [Google Scholar]
- 299. Reichmuth A. M., Oberli M. A., Jaklenec A., Langer R., Blankschtein D., Ther. Delivery 2016, 7, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Kauffman K. J., Dorkin J. R., Yang J. H., Heartlein M. W., DeRosa F., Mir F. F., Fenton O. S., Anderson D. G., Nano Lett. 2015, 15, 7300. [DOI] [PubMed] [Google Scholar]
- 301. Liu Q., Chen X., Jia J., Zhang W., Yang T., Wang L., Ma G., ACS Nano 2015, 9, 4925. [DOI] [PubMed] [Google Scholar]
- 302. Luo M., Wang H., Wang Z., Cai H., Lu Z., Li Y., Du M., Huang G., Wang C., Chen X., Porembka M. R., Lea J., Frankel A. E., Fu Y.‐X., Chen Z. J., Gao J., Nat. Nanotechnol. 2017, 12, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Vallhov H., Kupferschmidt N., Gabrielsson S., Paulie S., Strømme M., Garcia‐Bennett A. E., Scheynius A., Small 2012, 8, 2116. [DOI] [PubMed] [Google Scholar]
- 304. Zhao Y., Zhao X., Cheng Y., Guo X., Yuan W., Mol. Pharmaceutics 2018, 15, 1791. [DOI] [PubMed] [Google Scholar]
- 305. Cho N. H., Cheong T.‐C., Min J. H., Wu J. H., Lee S. J., Kim D., Yang J.‐S., Kim S., Kim Y. K., Seong S.‐Y., Nat. Nanotechnol. 2011, 6, 675. [DOI] [PubMed] [Google Scholar]
- 306. Almeida J. P. M., Lin A. Y., Figueroa E. R., Foster A. E., Drezek R. A., Small 2015, 11, 1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Lee I.‐H., Kwon H.‐K., An S., Kim D., Kim S., Yu M. K., Lee J.‐H., Lee T.‐S., Im S.‐H., Jon S., Angew. Chem., Int. Ed. 2012, 51, 8800. [DOI] [PubMed] [Google Scholar]
- 308. Kalantar‐Zadeh K., Ward S. A., Kalantar‐Zadeh K., El‐Omar E. M., ACS Nano 2020, 14, 5179. [DOI] [PubMed] [Google Scholar]
- 309. Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H., Gastroenterology 2020, 158, 1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Gao Q., Hu Y., Dai Z., Wu J., Xiao F., Wang J., SSRN J. 2020, 10.2139/ssrn.3548755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Karst S. M., Nat. Rev. Microbiol. 2016, 14, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Salazar N., Valdes‐Varela L., Gonza lez S., Gueimonde M., de los Reyes‐Gavilan C. G., Gut Microbes 2017, 8, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Claesson M. J., Cusack S., O'Sullivan O., Greene‐Diniz R., de Weerd H., Flannery E., Marchesi J. R., Falush D., Dinan T., Fitzgerald G., Proc. Natl. Acad. Sci. USA 2011, 108, 4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Li N., Ma W.‐T., Pang M., Fan Q.‐L., Hua J.‐L., Front. Immunol. 2019, 10, 1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Schirmer M., Smeekens S. P., Vlamakis H., Jaeger M., Oosting M., Franzosa E. A., ter Horst R., Jansen T., Jacobs L., Bonder M. J., Cell 2016, 167, 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Biteen J. S., Blainey P. C., Cardon Z. G., Chun M., Church G. M., Dorrestein P. C., Fraser S. E., Gilbert J. A., Jansson J. K., Knight R., Miller J. F., Ozcan A., Prather K. A., Quake S. R., Ruby E. G., Silver P. A., Taha S., van den Engh G., Weiss P. S., Wong G. C. L., ACS Nano 2016, 10, 6. [DOI] [PubMed] [Google Scholar]
- 317. Nakhleh M. K., Amal H., Jeries R., Broza Y. Y., Aboud M., Gharra A., Ivgi H., Khatib S., Badarneh S., Har‐Shai L., GlassMarmor L., Lejbkowicz I., Miller A., Badarny S., Winer R., Finberg J., Cohen‐Kaminsky S., Perros F., Montani D., Girerd B., ACS Nano 2017, 11, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Vevers W. F., Jha A. N., Ecotoxicology 2008, 17, 410. [DOI] [PubMed] [Google Scholar]
- 319. Kirchner C., Liedl T., Kudera S., Pellegrino T., Javier A. M., Gaub H. E., Stölzle S., Fertig N., Parak W. J., Nano Lett. 2005, 5, 331. [DOI] [PubMed] [Google Scholar]
- 320. Jain T. K., Morales M. A., Sahoo S. K., LesliePelecky D. L., Labhasetwar V., Mol. Pharmacol. 2005, 2, 194. [Google Scholar]
- 321. Hauck T. S., Ghazani A. A., Chan W. C., Small 2008, 4, 153. [DOI] [PubMed] [Google Scholar]
- 322. Khan J. A., Pillai B., Das T. K., Singh Y., Maiti S., ChemBioChem 2007, 8, 1237. [DOI] [PubMed] [Google Scholar]
- 323. Poland C. A., Duffin R., Kinloch I., Maynard A., Wallace W. A. H., Seaton A., Stone V., Brown S., MacNee W., Donaldson K., Nat. Nanotechnol. 2008, 3, 423. [DOI] [PubMed] [Google Scholar]
- 324. Li N., Sioutas C., Cho A., Schmitz D., Misra C., Sempf J., Wang M., Oberley T., Froines J., Nel A., Environ. Health Perspect. 2003, 111, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Kostarelos K., Nat. Biotechnol. 2008, 26, 774. [DOI] [PubMed] [Google Scholar]
- 326. Lacerda L., Soundararajan A., Singh R., Pastorin G., Al‐Jamal K. T., Turton J., Frederik P., Herrero M. A., Li S., Bao A., Emfietzoglou D., Mather S., Phillips W. T., Prato M., Bianco A., Goins B., Kostarelos K., Adv. Mater. 2008, 20, 225. [Google Scholar]
- 327. Sayes C. M., Fortner J. D., Guo W., Lyon D., Boyd A. M., Ausman K. D., Tao Y. J., Sitharaman B., Wilson L. J., Hughes J. B., West J. L., Colvin V. L., Nano Lett. 2004, 4, 1881. [Google Scholar]


