Abstract
Along the line of recent vaccine advancements, new antiviral therapeutics are compelling to combat viral infection‐related public health crises. Several properties of silver nanoparticles (AgNPs) such as low level of cytotoxicity, ease of tunability of the AgNPs in the ultra‐small nanoscale size and shape through different convenient bottom‐up chemistry approaches, high penetration of the composite with drug formulations into host cells has made AgNPs, a promising candidate for developing antivirals. In this review, we have highlighted the recent advancements in the AgNPs based nano‐formulations to target cellular mechanisms of viral propagation, immune modulation of the host, and the ability to synergistically enhance the activity of existing antiviral drugs. On the other hand, we have discussed the recent advancements on AgNPs based detection of viral pathogens from clinical samples using inherent physicochemical properties. This article will provide an overview of our current knowledge on AgNPs based formulations that has promising potential for developing a counteractive strategy against emerging and existing viruses.
Keywords: Ag NPs, nucleic acid, virucidal properties, viral diseases, viral replication, vaccine carriers
Ease of tuningof ultrasmall size and shape of Ag nanoparticles through different convenient bottom‐up chemistry approaches and high penetration in drug formulations into host cells has made AgNPs a versatile therapeutic agent for different viral diseases. They can selectively target the viral life cycle or work synergistically to enhance the activity of existing antiviral drugs or assist in the detection of viral pathogens, as thoroughy reviewed herein.
1. Introduction
The discovery of viruses can be traced back to 1892 when Russian botanist Dmitri Ivanovsky was investigating the infectious disease of tobacco plants. Although he believed it to be a bacterium. His observations were amended later by Dutch microbiologist Martinus Beijerinck, followed by Loeffler and Froschin in their interpretations of virus particles. [1] They collectively pioneered the field of virology to a world that had been largely subsumed in the predicaments of deadly bacterial diseases like typhoid, cholera, tuberculosis, anthrax, bubonic plagues, except viral smallpox. Contrary to Moelling et al.’s “Viruses, More Friends than Foes,” [2] viruses constitute a significant threat to our contemporary world than any other biological entity, exemplified time and again by viral epidemics and pandemics that have wreaked havoc in the lives of ordinary citizens. Since the Antonine plague, which is one of the most ancient pandemics, the world has witnessed numerous pandemics like the smallpox outbreak, the Russian Flu, the Spanish Flu, the swine flu, Acquired Immune Deficiency Syndrome (AIDS) pandemic, Ebola outbreak, and the advent of the Coronavirus in the twenty‐first century that caused severe acute respiratory syndrome (SARS) epidemic, followed by middle east respiratory syndrome (MERS). [3] One of the most dreadful SARS pandemic has been caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV2) in 2019 has claimed more than 5.4 million lives and affected more than 295 million people globally as of now.
It is interesting to note that most viral outbreaks were transmitted from animals or birds, accentuating their high propensity towards cross‐species transmission through evolution using co‐divergence, [4] homologous recombination,[5] and mutations, [6] to maximize their infectivity. This leads to new strains resistant to antivirals designed to combat existing strains of the same family of viruses. Moreover, viruses are obligate parasites that exploit the host cell machinery to direct their replication. Thus, developing antivirals that are selectively toxic to viruses without interfering with host cellular function is quite challenging on many levels. Furthermore, most small molecule‐based drugs are poorly biocompatible and require appropriate drug delivery approaches for enhanced tissue penetration and proper release. Vaccines are substantial in ameliorating viral outbreaks. However, the entire process of finding a suitable vaccine alongside their approval from medical committees after rigorous clinical trials on thousands of volunteers are a highly time‐consuming operation that further expedites viral transmission during pandemics. Besides, vaccines often fail to mount an appropriate immune response as the host‘s immune system influences its efficacy. Elderly people and those with co‐morbid conditions are also severely affected. This highlights the indispensable need for developing broad‐spectrum antivirals as effective measures for pandemic preparedness to tackle rapid viral dissemination at the early stages of outbreaks. Nanotechnology represents a revolutionary field of science that creates particles of nano‐size dimensions for applications in diverse areas of life. With the expansion of nanotechnology in the past century, AgNPs have been researched extensively in virology, oncology, and bacteriology, generating a wealth of knowledge that has encouraged their acceptable use in medicine. Several reviews have covered several aspects of AgNPs and other nanoparticles‐based strategies against a broad range of viruses.[ 7 , 8 , 9 , 10 ]
The virucidal properties of AgNPs have been proclaimed against a plethora of different viruses (Table 1) that encompass animal viruses like Human Immunodeficiency Virus, [11] Herpes Simplex Virus, [12] Hepatitis B Virus, [13] Hepatitis C Virus, [14] Influenza Virus, [15] Respiratory Syncytial Virus, [16] Polio Virus, [17] Dengue Virus, [18] Chikungunya Virus, [19] Monkey Pox Virus, [20] Vaccinia Virus, [21] Tacaribe Virus, [22] Rift Valley Fever Virus, [23] African Swine fever virus, [24] White Spot Syndrome Virus, [25] Enterovirus 71, [26] Corona Virus, [27] Murine Norovirus, [28] Feline Calicivirus, [28] Porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus [29] and plant viruses like Tobacco Mosaic Virus, [30] Bean Yellow Mosaic Virus [31] as well as bacterial viruses like Bacteriophage MS2, [32] φX174 [33] and UZ1. [34] AgNPs are versatile pharmaceuticals that possess broad‐spectrum antiviral activity that can be repurposed to develop antivirals as counteractive strategies against emerging and existing viruses. Besides, AgNPs show excellent biocompatibility [35] at minimal effective concentrations in addition to exhibiting efficacy against drug‐resistant strains of viruses, in addition to nominal reports of viral resistance against them. Furthermore, their nano‐size and surface properties render them highly stable and permit enhanced penetration of drug formulations into host cells. AgNPs can target various components of the viral life cycle to inhibit the cellular mechanism of viral propagation in the host. On the other hand, AgNPs function as drug delivery vehicles for enhanced biosorption coupled with targeted delivery or act synergistically to improve the activity of existing antiviral drugs or even assist in detecting viral pathogens from clinical samples. There is a significant shortage of research and literature concentrating on the mode of action of AgNPs against the life cycle of viruses or the current advancement on AgNPs based on viral detection tools.
Table 1.
Antiviral and therapeutic mechanisms of AgNPs against different viruses.
|
Virus host |
Virus |
AgNP composition |
Mode of action |
Reference |
|---|---|---|---|---|
|
Animal |
Human Immunodeficiency Virus |
Uncoated AgNP as well as PVP coated AgNP |
Impeding the gp120‐CD4 interaction during virus entry |
[11] |
|
Hepatitis B Virus |
Uncoated AgNP |
Inhibits virus replication |
[13] |
|
|
Hepatitis C Virus |
Green synthesized AgNPs from Amphimedon (Red Sea Sponge) |
Inhibits virus assembly through inactivation of NS3 Helicase and Protease |
[14] |
|
|
Influenza Virus |
Uncoated AgNP |
Reducing virus induced apoptosis and cytokine storm, gene delivery carriers of Influenza vaccines. |
[15] |
|
|
Respiratory Syncytial Virus |
PVP coated AgNP, uncoated AgNP |
Immunomodulating the immune profile of the host through the activation of neutrophils and anti‐inflammatory mediators |
[16] |
|
|
Polio Virus |
Electrochemically synthesized AgNP |
Mechanism unclear |
[17] |
|
|
Dengue Virus |
Mangrove‐fabricated AgNP |
Inhibits the expression of the envelope (E) gene and protein in dengue virus (serotype DEN‐2) |
[18] |
|
|
Chikungunya Virus |
Green synthesized AgNP from medicinal plants |
Mechanism unclear |
[19] |
|
|
Monkeypox Virus |
Uncoated as well as polysaccharide coated AgNP |
Mechanism unclear |
[20] |
|
|
Vaccinia Virus |
Uncoated AgNP |
Inhibiting macropinocytosis during virus entry |
[21] |
|
|
Tacaribe Virus |
Uncoated and polysaccharide coated AgNP |
Blocks virus replication |
[22] |
|
|
Rift Valley Fever Virus |
PVP coated AgNP |
Mechanism unclear |
[23] |
|
|
African Swine Fever Virus |
Uncoated AgNP |
Mechanism unclear |
[24] |
|
|
White Spot Syndrome Virus |
PVP coated AgNP |
Immunostimulation through the activation of PAMP recognition proteins |
[25] |
|
|
Enterovirus 71 |
PEI coated AgNP loaded with siRNA |
Inhibiting activation of caspase‐3, ROS and activation of Akt and p53 |
[26] |
|
|
Corona Virus |
Uncoated AgNP |
Mechanism unclear |
[27] |
|
|
Murine Norovirus and feline calcivirus |
AgNP‐decorated silica hybrid composites |
Synergistic interaction of silica and Ag nanocomposites to directly inactivate the viruses prior to their entry |
[28] |
|
|
Porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus |
Silver and Graphene Oxide nanocomposites |
Suppression of viral entry and stimulated Interferon‐α and Interferon Stimulated genes that are essential for activation of antiviral innate immune responses |
[29] |
|
|
|
Tobacco Mosaic Virus |
Biogenic AgNP prepared from the fermented broth ofPseudomonas fluorescens |
Mechanism unclear |
[30] |
|
Plant |
Bean Yellow Mosaic Virus |
AgNPs synthesized from extracellular agents produced by Bacillus licheniformis |
Mechanism unclear |
[31] |
|
Bacteriophage MS2 |
AgNP‐decorated silica hybrid composites |
Synergistic interaction of silica and Ag nanocompositesto directly inactivate the viruses prior to their entry |
[32] |
|
|
Bacteria |
Bacteriophage φX174 |
AgNP in conjunction with micrometer sized magnetic hybrid colloid |
Direct inactivation of viruses. Detailed mechanism unknown. |
[33] |
|
Bacteriophage UZ1 |
AgNP in conjunction withbacterial cell surface of Lactobacillus fermentumto form biogenic silver immobilized on microporous PVDF membranes |
Direct inactivation of viruses. Detailed mechanism unknown. |
[34] |
The focus of this review has been directed towards the following subtopics; i) the molecular mechanisms that govern the functioning of AgNPs in antiviral therapeutics by targeting the life‐cycle of the viral pathogens in the host cells, ii) AgNPs as antiviral drug delivery vehicles, iii) the relevance AgNP to detect the viral pathogens, and iv) synergistic activities of AgNP with existing antiviral drugs.
2. AgNPs targeting the life cycle of a virus
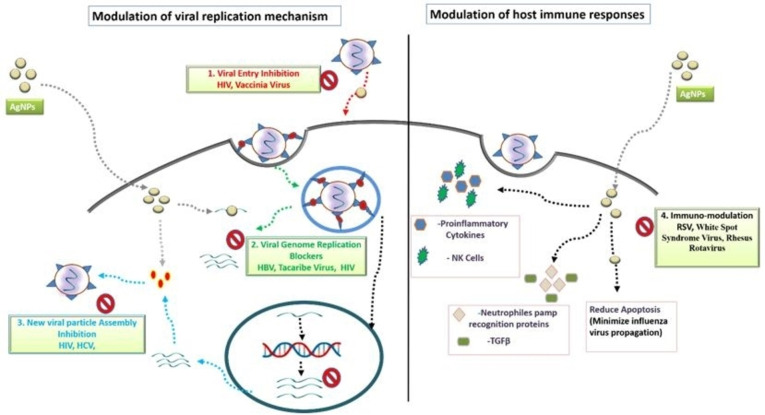
The study of the viral life cycle is an imperative exercise that precedes designing novel antiviral drugs targeting thecrucial mechanismof the viral replicative process. A successful viral infection is governed by a virus's ability to enter the host cell, replicate its genome, assemble the genome into capsids, produce mature virions and exit the host cell undetected before an immune attack is launched. The ability of AgNPs to counteract these various stages of the viral life cycle (as depicted in Figure 1) is a hallmark feature of their therapeutic applications.
Figure 1.
Antiviral Mechanisms of AgNPs that block different stages of viral life cycle like (1) viral entry into the host cell, (2) viral genome replication, (3) assembly of new viral particles or (4) stimulate the host immune responses.
2.1. Viral entry blockers
The viral proteins on the capsid or the envelope can recognize cellular receptors on the plasma membrane, comprising glycoproteins or phospholipids with diverse physiological roles. [36] Interestingly, most mammalian viruses have evolved to recognize glycans on cell surfaces that serve as receptors or co‐receptors in tandem to specific cell receptors and aid viral entry. [37] The attachment of virus particles on receptors that mediate the penetration of viruses into the cell by direct fusion or receptor‐mediated endocytosis, followed by uncoating the capsid to disclose their genetic material to the cytoplasm (Figure 1). A plethora of studies on AgNPs have reported their ability to halt viral transmission by exploiting this receptor‐virus interaction in an expansive array of virus models like Human Immunodeficiency Virus (HIV), vaccinia virus and other viruses (Table1).
2.1.1. Inhibition of Human Immunodeficiency Virus (HIV) entry by blocking gp120‐CD4 interaction
Human Immunodeficiency Virus (HIV) entry is mediated by the envelope glycoprotein complex, comprising of a head that includes surface glycoproteins such as gp120, which binds to cellular receptors, and a stalk that includes transmembrane glycoproteins such as gp41 that anchor the envelop into the viral membrane. Ever since the identification of CD4 receptor, expressed on the surface of T cells and to some extent on cells of monocyte lineage, as a physically interacting partner of HIV exterior envelope glycoprotein gp120, [38] monoclonal antibodies targeting the CD4 antigen to diminish HIV infectivity have been explored profusely. [39] Extensive research on viral entry inhibitors has unleashed three different inhibition modes [40] focusing on attachment inhibitors, co‐receptor binding inhibitors, and fusion inhibitors. A few of them have triumphantly proceeded to clinical applications. Enfuvirtide, the first US‐FDA approved HIV fusion inhibitor, is a synthetic peptide that blocks the formation of structural complexes essential for fusion by mimicking HR2 fragment of gp41. [41] However, the peptidic nature of this drug with limited the application of oral administration and is therefore injected subcutaneously, eliciting hypersensitive reactions and an increased susceptibility to bacterial pneumonia. [42] Maravaroc, another FDA approved entry inhibitor that targets CCR5 co‐receptor, has induced the risk of developing symptomatic West Nile virus infection. [43] This illuminates the prerequisite for developing alternative strategies to annihilate HIV. In the following section, various studies of silver nanoparticles on blocking HIV entry have been discussed.
The foundation of silver nanoparticles as attractive HIV−I entry blockers was first undertaken by Elechiguerra et al. [11] through in vitro studies against CD4+ MT‐2 cells and cMAGI HIV‐1 reporter cells by high angle annular dark field (HAADF) scanning transmission electron microscopy (STEM), outlining the role of gp120 as potential molecular targets of AgNPs. To corroborate this theory, Lara et al carried out a series of experiments and determined that AgNPs impede the gp120‐CD4 interaction by possibly interacting with two disulphide bonds of gp120 that bind to the CD4 receptor, thereby denaturing this disulphide bonded domain of gp120. [44] The inhibition of viral entry was observed by prevention of syncytia formation by AgNPs, which is a late‐stage indicator of HIV‐1 infection. They also ascertained the ability of AgNPs to confer the exact antiviral properties on several drug resistant HIV‐1 strains as the position of the cysteine residues and the disulphide bonding pattern in gp120 are highly conserved, and are thus immune to the drug‐resistance conundrum.
Furthermore, Lara et alsynthesized polyvinylpyrrolidone (PVP) coated AgNP using replens gel to test their competence against HIV‐1 in human cervical tissue explant in vitro, that simulates in‐vivo conditions, to formulate a topical vaginal or cervical microbicide that thwarts both cell free and cell associated HIV virus by abolishing the same CD4‐gp120 interaction within one minute of exposure and delivers immunity upto 48 hours. [45] In another study, Mohammed Fayaz et alhave developed silver nanoparticle (AgNP)‐coated polyurethane condom (PUC) that was sensitive to both macrophage (M)‐tropic and T lymphocyte (T)‐tropic strains of HIV‐1 and HSV‐1/2 with no severe toxicity. [46] These findings demonstrate the applications of AgNPs as effective antivirals to prevent sexually transmitted infections (STI) as well.
2.1.2. Impeding vaccinia virus entry by blocking macropinocytosis
Vaccinia virus is an Orthopoxvirus that is antigenically similar to Variola major, the causative agent of the highly contagious and deadly Small pox disease. Although Smallpox has been eradicated for over four decades, concerns and threats about using this virus as a potential bioweapon has not been disregarded. Vaccinia virus (VACV) was used for small pox vaccination and is still used as a promising model system for the development of antiviral drugs and vaccines against reemergence of small pox or monkey pox as well as compelling tools for studying innate immunity and cell biology. [47]
The use of AgNPs in suppressing this virus was investigated in a study [21] that uncovered novel mechanisms of blocking viral entry inside cells. As consistent with most viruses, AgNPs could bind to viral surface proteins,however, unlike most of them, AgNP bound VACV could adsorb onto cells, i. e., AgNPs had no direct virucidal effect on viral proteins that prevented them from binding to cellular receptors and entering the cell. This led to further studies on the next step of viral entry, i. e., fusion, which could be either direct fusion with the cell membrane or endocytosis by micropinocytosis as in case of VACV. Macropinocytosis is the non‐specific uptake of extracellular material that requires the activation of Pak1, a serine‐threonine kinase that VACV activates prior to fusion with macropinosome. Silencing of this Pak1 gene inhibits macropinocytosis and drastically demolishes viral entry and the virus is coerced into entering the cell by direct fusion.It was observed that AgNPs failed to inhibit viral entry when Pak1 gene was silenced by Pak1 siRNA, suggesting the importance of macropinocytosis as a key mediator of AgNP induced viral entry inhibition and that of AgNPs having almost negligible effect on blocking VACV entry by direct fusion mechanism. Although the exact process through which AgNPs influence macropinocytosis has not been elucidated, possible mechanisms have been suggested that include:
Under extracellular conditions, AgNPs become more spaced out and exhausted by binding to cellular proteins before encountering virus particles. Macropinocytosis brings VACV and AgNPs in close proximity that enables AgNPs to trap them within the macropinosome, which is later destroyed in the lysosome.
AgNPs could trigger autophagy and autophagosome induced exocytosis.
They could also compete with VACV for uptake in macropinosomes.
They could inhibit the fusion of VACV with macropinosome.
Studies on several other viruses like Respiratory Syncytial Virus, [16] Dengue Virus, [18] Monkey Pox virus [20] have conveyed the importance of AgNPs in blocking viral entry, although, the exact mechanism of inhibitionin these viruses is notclear. AgNPs can associate with proteinsby covalent bonding with electron donors like sulphur of thiol residues [48] or nitrogen of amine groups, [49] on account of its inherent need for steric and electrostatic stabilization. [50] A preferential inclination of AgNPs towards cysteine, proline and hydroxyl containing amino acids like tyrosine was exhibited in a silver binding peptide study, [51] whereas aromatic amino acids like tryptophan and tyrosine exhibited higher affinity towards AgNPs in a hemoglobin AgNP interaction study, [52] which stresses the importance of these residues in attracting AgNPs towards viral proteins.The contact frequency between the protein and nanoparticle (NP) reflects the affinity of a protein towards NP complex (Figure 2 ). Protein‐NP investigations have unveiled highly stable NP‐protein‐corona formations of specific size, shape and structure that constitute biologically relevant molecules in determining the fate of protein interactions with cellular receptors. [53] Protein‐NP coronas can induce conformational changes in proteins including anomalous unfolding,[ 53 , 54 ] that can either expose unknown epitopes or mask essential epitopes [55] to functionally interferewith viral adsorption or prohibit interactions with cellular receptors, or prevent fusion of virus particles into the cell.
Figure 2.
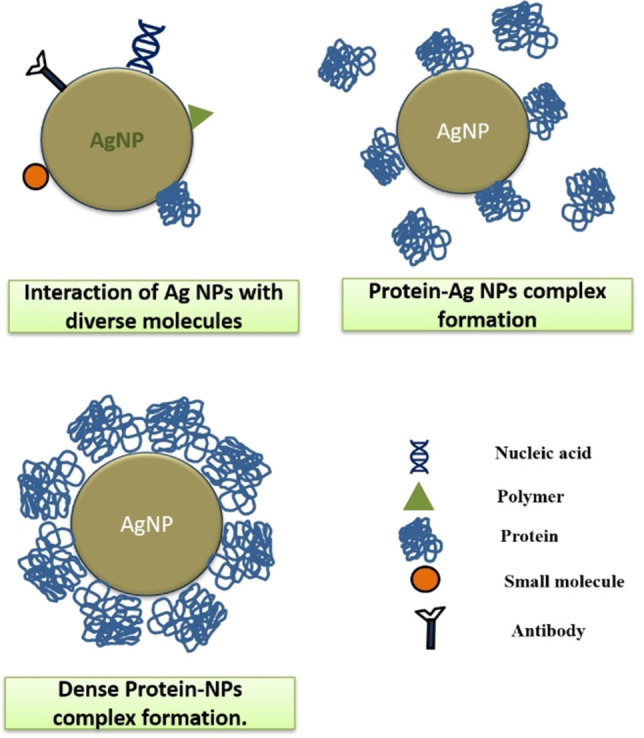
Interaction of Ag NPs with bio‐molecules like nucleic acids, polymers, proteins, small molecules or antibodies.
2.2. Viral genome replication blockers
There are several studies on DNA‐AgNP bioconjugates that support the functionalization of AgNPs with DNA moieties through interactions of negatively charged AgNPs with DNA bases, favoring GC rich residues particularly.[ 56 , 57 ] Furthermore, AgNPs exhibited greater propensity towards diazinon that contains phosphorus, pyrimidine nitrogen and oxygen moieties, in a study, that inferred the presence of high electron charge density of pyrimidine molecules as an integral element in favoring non‐covalent interactions with AgNPs (Figure 2). [58] The interaction of AgNPs with DNA serves the scope of inhibition of virus replication in several therapeutic approaches (Table 1 ).
2.2.1. Affecting hepatitis B virus replication by binding to double stranded DNA
Hepatitis B virus (HBV)is a partially double stranded DNA virus that targets hepatocytes, causing life‐threatening diseases like cirrhosis and cancer of liver. Upon infection, the double stranded genome of the viruses manifested in the form of a relaxed circular DNA (RC‐DNA) that enters the nucleus of the cell to repair its DNA into a covalently closed circular DNA (cccDNA) which is then transcribed to form viral mRNA transcripts and a 3.5kb pre‐genomic RNA (pgRNA). This pgRNA is reverse transcribed into new RC‐DNA inside a nucleocapsid that is either allowed to amplify the cccDNA pool in the nucleus or enveloped to form mature virions. [59]
The ability of AgNPs to block viral replication was first reported by Lu et alagainst Hepatitis B Virus (HBV) infected HepAD38 cells. [13] This finding provided valuable mechanistic insights about the complex disposition of AgNPs that were previously centered around entry inhibitions and this has led the scientists to explore other possible mechanisms to suppress viral infections. AgNPs are capable of penetrating hepatocytes and approach their intracellular targets by binding to HBV ds DNA in order to impede the transcription of pgRNA, leading to a decline inreverse transcription of RC‐DNA and formation of progeny virions. An absorption titration assay revealed that AgNPs had an extremely high propensity towards HBV DNA, emphasizing their role in binding viral DNA to inhibit viral replication.
2.2.2. Inhibition of Tacaribe virus genome replication
Tacaribe virus is a new world arena virus with a bisegmented, ambisense RNA genome, that are transmitted from bats and serve as harmless model systems to develop antivirals for antigenically similar, hemorrhagic fever causing arena viruses like the South American Junin, Machupo, Guanarito and Sabia viruses. [60] Tacaribe virus (TCRV) has a bipartite RNA genome consisting of S and L segments. The large (L) segment encodes for viral polymerase (L) and matrix protein (Z), while the small (S) segment encodes the glycoprotein (GP) and the nucleoprotein (NP). [61]
Speshock et al discovered that uncoated as well as polysaccharide coated AgNPs could potentially bind to TCRV glycoproteins and internalize through endocytosis as analyzed by confocal microscopy, and impede viral replication by halting S segment gene expression, deduced through quantitative real time PCR studies. [22] Although the mechanism for the fall in S segment gene expression by AgNPs was not recovered empirically, they postulated that AgNPs could interfere with TCRV RNA‐dependent RNA polymerase (L protein) that replicates the TCRV RNA.
2.2.3. Hindering HIV reverse transcription
Apart from the prevalent use of AgNPs as prominent entry inhibitors of HIV‐1, Kumar et al.focused on employing AgNPs to uncover novel and promising anti‐HIV targets by exploiting the replication machinery of HIV. [62] They demonstrated the antiviral activity of green synthesized AgNPs from Rhizophora lamarckii,by targeting the reverse transcriptase (RTase) enzyme of HIV‐1 which is an essential mediator of HIV replication. This RTase enzyme is an RNA dependent DNA polymerase that transcribes the RNA genome into dsDNA that is transported across the cytoplasm into the nucleus to be integrated with the host DNA, where it is transcribed by the host transcription machinery to produce multiple viral mRNA copies that generate mature progeny virions. [63] Green synthesis of AgNPs could deter HIV RTase activity as determined by reverse transcriptase assay and thus, promote the usage of non‐biohazardous metal nanoparticles in developing antiretroviral medications.
2.3. Host immunomodulation strategies
The innate immune system is crafted to devise salubrious responses for the host by means of coordinated signaling pathways, ranging from intracellular activities of interferons (IFN), cytokines, chemokines, interleukins, to that of extracellular recruitment of macrophages, neutrophils, dendritic cells to the site of infection to stimulate viral clearance from our body. When a cell encounters a virus, intracellular receptors like Toll like receptors (TLR), retinoic acid‐inducible gene I (RIG‐I) and NOD‐like receptors (NLRs) participate in viral recognition through inspection of viral nucleic acids and replication intermediates. [64] Activation of these receptors trigger the recruitment of pro‐inflammatory cytokines and IFNs and induction of the adaptive immune system that further activates T and B cells, which are associated with extensive tissue damages. The T‐cells release cytokines like tumor necrosis factor (TNF) that damages cells and tissues like the liver as in case of HCV and HBV. [65] As a countermeasure, induction of anti‐inflammatory molecules like IL‐10 and TGF‐β have significantly ameliorated tissue damages by regulating the activity of pro‐inflammatory cytokines. [64] Additionally, there are other intracellular factors that govern the fate of virus replication inside cells like apoptosis and autophagy. Apoptosis is a programmed cell death that could either be a host‐defensive mechanism to protect the spread of invasive pathogens or a pathogen induced phenomenon to inflict large scale damage to bodily tissues.[ 66 , 67 ] Originally, it was hypothesized that apoptosis does not provoke the inflammatory system of the body, [68] only to be disputed and reinstated later that it could indeed activate the immune response. [69]
Immunomodulation is an artful strategy employed by viruses to escape detection by host immune responses as well as obstruct signaling mechanisms that antagonize their survival and even deploy favorable mechanisms that promote their survival. An interesting approach, exploited by myxoma virus, is to produce virus‐encoded proteins known as viroceptors or virokines, also known as decoy receptors, that mimic host receptors or cytokines and thereby, block undesirable extracellular immune responses to create a “virus‐friendly” environment. [70] Human cytomegalovirus exercises diverse ways to thwart the host immune system. It secretes proteins that could alter or mimic classical and non‐classical major histocompatibility complex (MHC) protein function as well as leukocyte migration, activation and cytokine responses. [71] Human herpesvirus (HHV)‐6A and HHV‐6B have developed mechanisms for cytokine modulation, including blockade of IL‐12 production by antigen‐presenting cells, inflection of cell‐surface molecules essential for T‐cell activation, and expression of viral chemokines and chemokine receptors. [72] Measles virus causes interference with APC maturation and influences T‐cell activation. [73] Induction of apoptosis is a common cytopathogenic trait of many viruses that serves to aggravate host immune response and inflict tissue damages.
The physical, chemical and optical properties of metal nanoparticles have found interesting consequences in altering host immune responses and have been efficient in blocking viral infection. AgNPs have exhibited significant immunomodulatory properties like anti‐inflammatory activity in addition to restricting apoptosis and autophagy (Figure 1) in cases of virus infected cells when administered in a time and concentration dependent manner.
2.3.1. Inhibition of apoptosis and secondary bacterial infections to minimize influenza virus propagation
The structure of Influenza Virus (IV) consists of a viral core that contains a negative sense RNA genome and a lipid envelop that is studded with glycoproteins like Hemagglutinin (HA) and Neuraminidase (NA) and a few matrix (M2) ion channels that traverse the lipid envelop. [74] Hemagglutinin mediates entry of the virus by binding to sialic acid moieties on cell surfaces, while neuraminidase is involved in releasing the progeny virions from the cell by hydrolytic cleavage of sialic acid that bind mature virions. The ion channel M2, Neuraminidase and Hemagglutinin are all important targets for designing anti‐Influenza drugs.
Developing antiviral medications for IV is challenging at multiple levels due to the exorbitant rate of mutation and evolution it constantly undergoes. [75] The excessive mutation of the RNA genome of IV leads to reassortment of its genetic segments that creates new virus subtypes, causing antigenic shift and producing novel antigens against which the human race has not been immunized beforehand. Furthermore, the HA and NA proteins also undergo point mutations, causing antigenic drift that have previously foreshadowed seasonal flu. [76] These antigenic shift and drift create novel pathogenic strains that have the capacity to herald new epidemics and cause pandemics. [77] The antigenic shift produced the H1N1 IV that caused the Spanish Flu pandemic of 1918 which led to the death of about 5 million people and infected about one‐third of the world's population. It also triggered several other pandemics like the Asian flu by H2N2 virus in 1957, Hong Kong flu by H3N2 virus in 1968, bird flu by H5N1 and H7N9 viruses in 2003 and 2013, respectively, as well as swine flu by H1N1 virus in 2009. [77] The most common and clinically approved antiviral medications against IV include NA inhibitors like Oseltamivir and Zanamivir and M2 inhibitors like Amantadine and Rimantadine. However, sporadic cases of drug resistance against these popular antivirals necessitates the further development of anti‐Influenza medications that target novel strategies to disrupt viral dissemination.
Studies on infection and propagation of IV have revealed that it induces apoptosis by triggering the apoptotic pathway through the activation of transforming growth factor β (TGF‐β) [78] that is recognized by viral neuraminidase shortly before virus binding and entry. The application of silver nanoparticles to treat IV infection was first highlighted by Xiang et al, whereby, he demonstrated that AgNPs can reduce apoptosis in H1N1 IV infected MDCK cells, ascertained through Transmission Electron Microscopy and Flow Cytometry assays, and reduce viral propagation. [15] Xiang et al. further demonstrated that AgNPs can also reduce transmission of H3N2 IV strain in infected MDCK cells by interacting directly with the virus and destroying viral morphological structures in a time dependent manner and also by reducing virus induced apoptosis. [79] Additionally, the efficacy of AgNPs in diminishing H3N2 viral infection was confirmed by in‐vivo studies in mice models, where intranasal administration of AgNPs decreased the level of pathologic lung lesions and increased the survival rates of H3N2 IV infected mice. This accentuates the broad‐spectrum activity of AgNPs in the treatment of multiple IV strain infections that could possibly generate an ideal antiviral medication in the event of epidemics and pandemics for early intervention of viral transmission.
The ‘cytokine storm’ that is usually associated with Influenza is brought about by the activation of antiviral Interferon pathways, especially IFN‐β, which promotes viral clearance but increases susceptibility to secondary bacterial infections by downgrading antibacterial responses, thereby, preferentially motivating the cell to favor a viral response. Administration of AgNPs as a prophylactic drug to control this bias can be greatly achieved as AgNPs boost the antibacterial immunity through IL‐8 dependent neutrophil recruitment and downregulate interferon pathways as confirmed in a study by Villeret et al. [80] Additionally, AgNPs also blocked autophagy that inhibited viral replication in uninfected cells, supporting their usage as therapeutics for controlling viral and secondary bacterial infections associated with IV infections at an optimum dosage.
2.3.2. Activation of neutrophils to combat Respiratory Syncytial Virus infection
Respiratory Syncytial Virus (RSV) is a highly infectious, enveloped, negative sense single stranded RNA virusthat affects the lower respiratory tract in infants andadults such as the elderly or patients with immunocompromisedand cardiopulmonary diseases, [81] leading to large outbreaksthat severely impacts the economy and medical facilities in infected areas. Vaccination is the only effective treatment that has hugely contributed in slowing the dissemination of this virus, even though there is no licensed vaccine available yet. [82] A neutralizing monoclonal antibody (Palivizumab, Synagis) [82] that targets the fusion protein F, responsible for viral entry, is used for high‐risk individuals prophylactically,the success of which is tremendously dependent on cell mediated immunity.The diligent use of AgNPs as remedial measures for Influenza infections has influenced the scientific community to evaluate the immunomodulatory properties of AgNPs against RSV infections.
The potential of AgNPs to annihilate RSV infection in epithelial cells was confirmed in an in‐vivo model system using BALB/C mice, infected with AgNP‐RSV and the antiviral mechanism was elucidated by investigation of the cytokine and cellular profile of the bronchoalveolar lavage fluid (BALF) collected from the inoculated mice. [83] Significant reduction of expression of pro‐inflammatory cytokines and pro‐inflammatory chemokines as well as antiviral markers like IL‐6, TNF‐α, CCL5 (RANTES), and type I IFNs, IL‐1, IL‐9, IL‐10, IL‐12p40, IL‐12p70, IL‐13, CCL2 (MCP‐1), and CCL3 (MIP‐1) were noted, while upregulation of CXCL1, G‐CSF, and GM‐CSF that are involved with recruitment of neutrophils were observed. Thus, AgNPs exert their antiviral activities against RSV through activation of neutrophils that could possibly boost the innate immune system via recruitment of monocytes and neutrophilsor release neutrophil extracellular traps (NET) forstrengthening the antibacterial defense of the host in order to preventconcomitant secondary bacterial infections.
2.3.3. Immunostimulation of White Spot Syndrome Virus infected shrimps
The white spot syndrome virus causes the white spot disease in shrimps that has devastated the shrimp industry. Argovit is a commercially available formulation of silver nanoparticles with polyvinylpyrrolidone that has significantly decreased the mortality rate of shrimp Litopenaeus vannameiinfected with white spot syndrome virus. [84] Although the mechanism of inhibition is not very clear, some interesting insights about immune‐stimulating properties of AgNPs were claimed by Ochoa‐Meza et al on investigation of the innate immune system changes in WSSV affected shrimps. [25] They suggested that AgNPs could possibly interact with WSSV viral envelop to trigger the activation of specific pathogen associated molecular pattern (PAMP) recognition proteins like lipopolysaccharide and β‐1,3‐glucan binding protein (LGBP), whose expression levels were exceedingly augmented by Argovit exposure to WSSV infected shrimps.
2.3.4. Improval of biliary atresia syndrome in Rhesus Rotavirus infected mice
Biliary atresia (BA) is a very common neonatal disease that obstructs the biliary system, interfering with systemic bile flow from liver to the peripheral organs and leads to accumulation of bile within liver. [85] Rhesus rotavirus causes biliary atresia in mice through the progressive accumulation of inflammatory cells like Natural killer (NK) cells that activate CD4+ and CD8+ T cells to enhance the impairment of the bile ducts. [86] R. Zhang et al demonstrated a treatment for BA using AgNPs against rhesus rotavirus induced BA mice by exploiting the anti‐inflammatory virtues of AgNPs in decimating NK cells while upregulating TGF‐β that is crucial for biliary cell differentiation. [87] Significant improvement of the survival rate of infected mice accompanied by weight gain, normal bilirubin metabolism, reduced viral load and recovery from jaundice were observed in mice that were treated with AgNPs, highlighting a promising endeavor of AgNPs in treatingvirus induced neonatal BA in humans.
2.4. Viral assembly inhibitors
The encapsulation of viral genome is an integral element of the viral life cycle through which a virus establishes its mature and infectious identity within the host. This process varies widely among different virus strains. It involves numerous protein‐protein or protein‐nucleic acid or even protein‐lipid interactions that foster the selective packaging of nucleic acid genome within a proteinaceous shell, followed by the acquisition of a lipid envelop from the host cell membrane and their release from the cell. [88] This self‐assembling scheme has been targeted for the development of antiviral therapeutics that block the formation of mature progeny virions, ceasing their propagation across host cells (Figure 1).
2.4.1. Inhibition of HIV‐I protease to deter HIV assembly
The HIV−I Gag polyprotein is of paramount importance in mediating the viral assembly process. It consists of multiple flexible linker regions which are cleaved by the HIV−I Protease (HIVPR) to generate functional proteins that assist in the procurement of envelope, assembly of the capsid core, nucleic acid chaperoning, stabilization of the viral RNA genome and exit from the cell. [89] The HIVPR is a dimeric aspartic protease that consists of an active site triad‐Asp25‐Thr26‐Gly27, that favorably orients the substrate for a precise nucleophilic attack by the Asp25 of both of its subunits. [90] Through the guidance of thermodynamics and kinetics analyses, Shing et alestablished the association of AgNP with HIVPR to form AgNP‐HIVPR complex by binding to thiol residues of Cys95 that disrupt its electronegativity and predicted a decrease in nucleophilicity of the COO− residue of the active site Asp25 towards the substrate Gag protein, juxta positioned 1.05 nm from Cys95, thereby, retarding its action.87They corroborated their results by in silico molecular docking and several biophysical techniques that not only proved the interaction of AgNP with HIVPR, but also shed light on the hydrophobic nature of their interaction, suggesting an active participation of several hydrophobic amino acids that bind AgNP in a cooperative fashion to generate stable intermediates. [91] Furthermore, another team created synthetic peptides with similar amino acid composition to that of Gag polyprotein that could bind AgNPs and elucidated the role of AgNPs in protecting them from proteolytic attack by HIVPR. [92] However, they ascertained that HIVPR cleaves the polyprotein at a much faster rate than AgNP binding, when all three were incubated together, and envisaged the effectivity of AgNPs to thwart HIVPR activity at only preliminary levels.
2.4.2. Inhibition of Hepatitis C virus assembly
Hepatitis C virus is an enveloped, positive, single stranded RNA virus that causes chronic infections in liver, including cirrhosis, liver failure and hepatocellular carcinoma. [93] The HCV RNA genomes encodes a polyprotein that is cleaved by cellular and viral proteases to generate structural proteins that assist in viral capsid and envelop formations as well as nonstructural proteins that guide replication, modulation of host defense mechanisms, assembly and maturation. [94] NS3 is a non‐structural serine protease that consists of a N‐terminal protease domain that coordinates viral gene expression and a C‐terminal helicase domain that facilitates viral gene replication. [95] Inhibition of this protease affects viral assembly. Shady et al unveiled a promising role of green synthesized AgNPs from Amphimedon(Red Sea Sponge) in exhibiting in‐vitro anti‐HCV activity through the inactivation of NS3 Helicase and Protease activities.12 They attributed this anti‐HCV characteristic of AgNPs to the composition of the fractions of Amphimedon sp. that were used during the synthesis of AgNPs, which targeted the active site of NS3 and prohibited viral assembly.
2.5. AgNPS as effective drug delivery vehicles
Drug delivery is an engineered technology that enables the selective and efficient delivery of a therapeutic agent onto its desired site of action after bypassing several physiological barriers through the amelioration of solubility, release, charge, stability, while enhancing bioavailability and eliminating toxicity issues. Nanoparticles have emerged as a gold standard for the fabrication of drug delivery vehicles owing to their size, shape and surface chemistry that robustly empower the drug moieties through the unique structural, optical, physicochemical and electronic qualities.[ 96 , 97 ] Silver nanoparticles are a favorable option in nanotherapeutic applications for the delivery of small molecules, peptides, oligonucleotides, vaccines, etc. because of their relatively straightforward and economical production techniques, in addition to their inherent stability, catalytic efficiency, electrical conductivity, magnetic and optical polarizability and antimicrobial properties.[ 98 , 99 ]
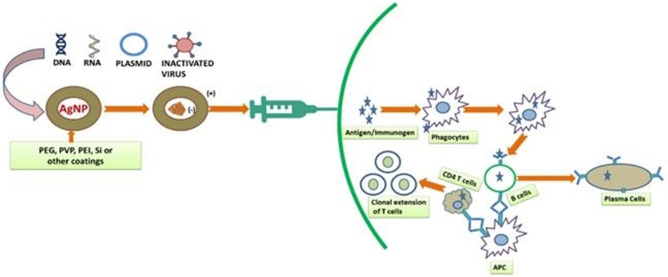
A few instances of AgNP mediated drug delivery to combat antiviral diseases (Figure 3), which ultimately enhances antiviral immune response (Figure 4) of the body to successfully counteract a viral infection, which is one of the scopes of our review, have been discussed below.
Figure 3.
Various strategies that have been implemented using AgNPs to induce host immune responses against viral infection: viral genes cloned in the plasmid vectors, antiviral siRNAs, protein vaccines as well as inactivated viruses have been delivered with AgNPs to activate host immune responses.
Figure 4.
AgNPs asdelivery vectors for antiviral oligonucleotides, plasmids or inactivated viruses that ultimately orient the host's cell‐mediated immune responses against the viral infection.
2.5.1. Suitable gene delivery carriers for Influenza vaccines
AgNPs have manifested commendable potential for gene delivery of Hemagglutinin (H5) gene of avian Influenza Virus (AIV) H5N1 cloned within an expression vector (pcDNA3.1/H5), across primary duodenal cells of pathogen free chickens that fruitfully induced both antibody and cell‐mediated immune responses (Figure 4) as well as enhanced cytokine production. [100] AgNP/H5 augmented the expression of pro‐inflammatory cytokines and enhanced antibodies against H5, 14 days after immunization accompanied with highest antibody titers after 35 days of immunization, in a single dose. It also amplified CD4+ and CD8+ T cells in the immunized chickens within 14 days as well, bolstering its efficacy as an estimable non‐viral gene vector for oral DNA vaccination. The lack of immunogenicity, established by the negligible pro‐inflammatory cytokine expression in chicks vaccinated with only AgNP and AgNP/pcDNA3.1, empower the biocompatibility of AgNPs and orient the immunogenic responses of chicken towards that of H5 DNA.
In another study, Jazayeri et al presented an influential oral gene delivery system of pIRES co‐expression vector consisting of H5 gene of AIV H5N1 and Green Fluorescent Protein (GFP), nanoencapsulated within green synthesized PEGylated AgNPs and investigated its immunogenicity against primary duodenal cells of pathogen free chick embryos. [101] AgNPs protected the plasmid DNA from degradation against DNase‐I and proficiently masked the surface negative charge of the plasmid DNA by conferring a net positive charge, owing to PEG, thereby, permitting interactions with the negatively charged cell membrane to gain accessand consequently elicited an immune response with an elevated expression of interleukin (IL)‐18, IL‐15, and IL‐12β.
AgNPs can also function as potent adjuvants in virus inactivated flu vaccine when administered prophylactically in the peritoneum or lungs,by virtue of itsNP‐protein interactionwith the antigenKeyhole limpet hemocyanin (KLH), supplemented by the “protein corona” effect. [102] This promotes bronchus associated lymphoid tissue generation (BALT) which induces specific IgA‐mediated mucosal immunity in mice. It also diminished the ‘cytokine storm’ unlike other alternative adjuvants like polyIC or Addavax, thereby toning down Influenza mediated lung inflammation that is commonly instigated by pandemic strains. Moreover, incorporation of AgNPs into the vaccine formulation significantly enhanced the efficacy of the vaccine due to prolonged exposure of the antigens at the site of entry, thereby maximizing antigen availability and retention.
2.5.2. AgNPs as effective HIV vaccine carriers
AgNPs shaped in the form of nanorods have exhibited considerable expertise as non‐carrier adjuvants for HIV vaccines, which is consistent with the studies on the usage of AgNPs as potent adjuvants for Influenza vaccines. Rod shaped NPs offer the advantage of minimal cellular uptake in comparison to nanospheres and alleviate cytotoxic drawbacks of carrier adjuvants. [103] This inspired Liu et al to fabricate a safe and highly immunogenic prophylactic HIV protein vaccine encapsulated within Ag nanorods, modified with Polyvinylpyrrolidone (PVP) and Polyethyleneglycol (PEG). [104] PEG is presumed to render the nanorods as more biocompatible, increase their bioavailability and protect them from degradation [105] while PVP is an effective immunostimulant [106] which can synergistically enhance its adjuvant properties when complexed with AgNPs. This nano‐formulation enhanced the stimulation of specific IgG responses, especially IgG3 than when HIV protein was administered alone and additionally triggered antibody dependent cellular cytotoxicity that mediated viral clearance. Furthermore, it also activated T cells to secrete higher amounts of IL‐2, IL‐4, IFN‐γ and CD107a, all of which are essential HIV antagonists. [107]
2.5.3. AgNPs as effective adjuvants for Rabies vaccine
Rabies Virus is a zoonotic virus that causes progressive neurologic infections in animals as well as warm blooded humans. [108] The adjuvanticity effect of AgNPs on veterinary rabies vaccine was investigated by Asgary et al [109] wherein green synthesized AgNPs sandwiched with inactivated rabies virus was intraperitonially administered in mice and accessed for mortality, when challenged cerebrally with a live rabies virus, by detection of Negri bodies in the neuron cells via fluorescent antibody test (FAT) using fluorescein isothiocyanate (FITC)‐conjugated anti‐nucleocapsid polyclonal antibodies.The results were further corroborated by evaluation of neutralizing antibodies using rapid fluorescent focus inhibition test (RFFIT) and National Institutes of Health (NIH) test, all of which accepted the use of AgNPs as promising adjuvants for vaccine formulations. The biosafety of the vaccine was also examined in‐vivo in mice and dogs that ruled out toxicity issues and did not induce fever, or any local or systemic reactions when monitored for 14 days after administration.
2.5.4. AgNPs as delivery vehicles for Newcastle Disease Virus vaccine
Newcastle Disease Virus (NDV) causes the highly infectious Newcastle disease in birds and often leads to epidemics in poultry populations that demand competent vaccines to curb the morbidities and mortalities associated with the disease. [110] Zhao et al synthesized an intranasal vaccine formulation of a hollow nanoparticle consisting of silver in conjunction with silica (Ag@SiO2‐NPs), [111] that offered uniform structure, lower cytotoxicity, higher stability and protected the encapsulated plasmid DNA containing the F gene of NDV, which mediates viral fusion, from nucleases. This combination of the negatively charged plasmid DNA and the positively charged NP enhanced the bio‐adhesivity and site specificity of the delivery system. The small size of the NPs in addition to their pore surface and large surface area favored rapid mucosal uptake when intranasally administered in specific pathogen free (SPF) chickens. Furthermore, the vector increased antigen contact area with the mucosal membrane, thereby stimulating the bioavailability of the vaccine. Moreover, it not only induced robust humoral immunity through the increase in IgA production as detected from the serum, bile, trachea and Harderian gland of the chickens, but also activated the cell mediated immunity through Th‐1 type cellular responses with concomitant increase in IFN‐γ and IL‐2.
2.5.5. AgNPs as co‐delivery vectors for SiRNA in Enterovirus 71 therapeutics
Small interfering RNA (SiRNA) has earned substantial amount of recognition since its discovery as a sequence‐specific gene knockdown tool using RNA interference (RNAi) and revolutionized the field of non‐coding RNAs in genetic regulation and gene therapy. However, naked SiRNA faces several disadvantages as a therapeutic option due to its instability and vulnerability to degradation by nucleases, negative surface charge that repels its entry into the cell through the negatively charged cell membrane and low transfection efficiency. [112] Thus, they require delivery vectors to reach their target moieties in which case, AgNPs are an excellent option that mediate macromolecular delivery into cells while protecting its constituents from nucleases and do not impose further toxicity. Additionally, polyethyleneimine (PEI) is a polymeric polycation that protects SiRNA release from endosome. [113] The ability of AgNPs and PEI co‐delivery of VP1 SiRNA to inhibit Enterovirus 71 (EV71) infection in Vero cells was explored by Li et al to probe its adeptness as a suitable vaccine or antiviral therapeutic against this fatal causative agent of hand, foot and mouth disease.24VP1 SiRNA was used in this study as VP1 is one of the four capsid proteins of EV71 that is considered to be a crucial site of positive selection in the molecular evolution of EV71.[ 114 , 115 ] The resulting Ag@PEI@siRNA successfully inhibited EV71 infection by silencing VP1 mRNA and VP1 protein expression that is essential for the capsid formation and assembly of virus in addition to suppression of ROS activation by limiting caspase−3 andp53 expression while maximizing AKT production.
3. Synergistic and additive interactions of AgNPs with drugs
When drug combinations are administered for therapeutics, interactions within the drugs in several unexpected ways characterize the outcome of the combination which could either enhance their combined productivity, i. e. synergistic or could be equal to their combined productivity, i. e. additive or could decline their productivity, i. e. antagonistic. [116] This synergistic combinatorial therapy is absolutely a wonder for the treatment of drug resistant infections and often resolves drug toxicity issues as a greater therapeutic effect is observed using extremely low doses of the combined drugs. Synergism is an intriguing form of crosstalk which is a recurring event in diverse cellular functions. The reasons for drug synergism are dependent on the relationships of the actions of the drugs on the cellular components that could be explained in different scenarios like; [117]
-
Binding of drug A to its target induces conformational change of target that induces binding of drug B to the same target (Figure 5
Figure 5.
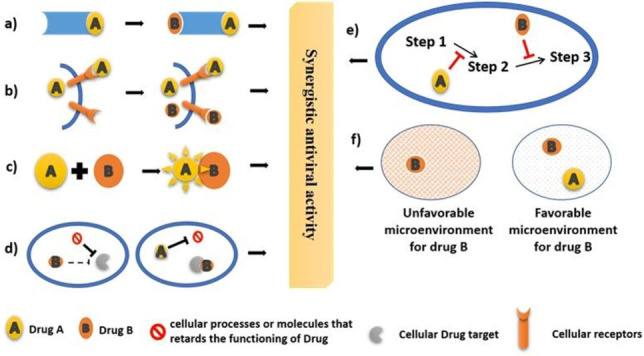 Modes of synergistic activities of hypothetical drug A and drug B which includes induction of conformational change of the drug target (a), enhances drug uptake (b), Interaction within drugs that triggers activity (c), blocking the drug antagonists (d), blocking the different steps of the same reaction (e) and changing microenvironment like solubility or pH (f).
Modes of synergistic activities of hypothetical drug A and drug B which includes induction of conformational change of the drug target (a), enhances drug uptake (b), Interaction within drugs that triggers activity (c), blocking the drug antagonists (d), blocking the different steps of the same reaction (e) and changing microenvironment like solubility or pH (f).a).
Binding of drug A to a cellular receptor enhances uptake of drug B inside the cell or the desired compartment of action (Figure 5b).
Interaction within drug A and B that triggers a change in one of them to transform into a more active form or inhibit the transformation into an inactive form (Figure 5c).
Drug A inhibits cellular processes or molecules that retards the functioning of Drug B (Figure 5d).
Drug A and B block different steps of the same reaction (Figure 5e).
Drug A enhances the solubility or changes the pH of the microenvironment of drug B according to its mode and site of action to minimize anti‐target and counter‐target activities (Figure 5f).
This wide spread therapeutic option has prompted the used of AgNPs in a variety of combinations as a remedy for several types of diseases. The synergistic and additive effects of AgNPs in antiviral drug combinations have been briefly reviewed here.
3.1. Interaction of AgNPs with mercaptoethane sulphonate to synergistically block entry of HSV
Herpes Simplex Virus enters a cell through the biomolecular recognition of heparan sulphate by viral envelop glycoproteins. The persistent obstacle of drug resistance among currently approved HSV drugs that include nucleoside analogs like Acyclovir, Famciclovir, Cidofovir, etc. represent a prevailing dilemma that could be redressed by the use of silver nanoparticles. Emphasizing on the multivalent nature of biological interactions, Baram‐Pinto et al identified an innovative strategy of employing mercaptoethane sulphonate capped silver nanoparticles (MeS‐AgNP) as constructive tools for competitive inhibition of heparan sulphate binding to HSV‐1 glycoproteins. [118] The exposed sulphonate groups of MeS on the surface of MeS‐AgNP, owing to their spatial orientation, can each attract virus particles and promote the formation of multiple virus‐particle bond pairs that is highly restricted when soluble MeS molecules are used alone because of their free rotation across virus particles. This accentuates the prominence of silver nanoparticles in providing a stable and rigid platform to aid formation of multiple bond pairs when used in conjunction with other molecules.
3.2. Tannic acid modified AgNPs as synergistic blockers of HSV
Tannins have previously been implicated in displaying anti‐HSV‐1 activity by adhering to viral glycoproteins and impairing their functions to inhibit viral entry. [119] Tannic acid modified silver nanoparticles has garnered a lot of attention recently due to its size dependent low inflammatory potential and downregulation of ROS production, [120] thereby augmenting the biosafety of AgNPs for clinical use. The antiviral activity of TA‐AgNP against HSV‐2 was demonstrated by Orlowski et al both in‐vitro and in‐vivo against Vero cells and intravaginally HSV‐2 infected C57BL6 mice. [121] The efficiency of TA‐AgNP adsorption onto viral glycoproteins, expedited by the tannic acid modification, could be due to the higher affinity of tannins for proline residues, [122] particularly the pro‐fusion domain of gD that is significantly rich in proline residues and is involved in mediating viral fusion. This physically restricts the virus from interacting with viral receptors on cell surface and blocks entry. Additionally, the immunomodulatory effects of TA‐AgNPs against HSV infection have also been studied that promote the upregulation of CCL2, IFN‐γ, TNF‐α, IL‐6, IL‐10 in a time dependent and size dependent manner against HSV‐2 infected mice models to stimulate viral clearance.
TA‐AgNPs have also succeeded in overcoming virus induced suppression of dendritic cell activation by promoting dendritic cell maturation and TLR9 expression through the activation of MHC‐II and CD86 while constraining CD80 expression. [123] An interesting hallmark of HSV infection is its ability to establish latency through episomes or integration of their viral genomes into host telomeres and co‐exist with the host to cause persistent, life‐long infections upon reactivation. [124] In this context of reinfections brought about by HSV latency, the role of TA‐AgNPs to generate effective antiviral response when rechallenged has been probed extensively by Orlowski et al. [125] TA‐AgNPs were found to be competent enough to generate a parallel immune response when rechallenged, through the heightened production of effector‐memory CD8+ T cells and IFNγ producing natural killer cells that were isolated from the spleen of TA‐AgNP treated mice, 10 days after HSV reinfection, as well as through the upregulated expression of the chemokine CXCL17 that has been demonstrated to boost mucosal vaginal immunity in recurrent herpes infection. [126]
The immunomodulatory activities of tannins have been previously evaluated in a study that elucidated the anti‐leishmanial activity, which accounted for the activation of macrophages to produce TNF‐α and IFN‐γ to antagonize leishmania survival within the phagocytes. [127] Although AgNPs participate in immunomodulation, they are predominantly associated with anti‐inflammatory responses. The constitution of TA‐AgNPs as a single entity could be held accountable for the demonstrated pro‐inflammatory action observed in the HSV study and explain the long‐term effectivity of these molecules in combating HSV latency.
3.3. AgNPs as synergistic enhancers of anti‐Influenza drugs
Twocell signaling activities can synergistically support life cycle of Influenza virus in the host, i. e., anti‐apoptotic activation of the Raf pathway and pro‐apoptotic activation of caspases. Apoptosis is, at times, considered as an antiviral cellular response as phosphatidyl serine, that is an inner membrane component is externalized along with desialylation of asialoglycomoieties, that together serve as important markers for efficient recognition by macrophages to enable phagocytosis. [128] Thus, activation of anti‐apoptotic components at the early stages of viral life cycle is a strategy to increase viral propagation. On the other hand, Caspase 3, which is an important mediator of apoptosis, is recruited by Influenza virus for efficient migration of RNPs from the nucleus to the cytoplasm for assembly and maturation of virions at a later stage of its life cycle when integrity of the nuclear membrane is no longer a prerequisite for viral replication. Suppressing Caspase‐3 activity leads to nuclear RNP retention and decline in virus propagation. [129] In another study, Influenza virus initiated anti‐apoptotic PI3k‐Akt signaling at early and middle phases of infection and both p53‐dependent and alternative p53‐independent apoptotic cell death at the late stage of infection. [130] These intracellular signaling mechanisms deployed by the Influenza virus to authorize its survival in the host can be exploited to develop interesting drug targets that could potentially hamper the viral life cycle.
The use of AgNPs in combination with widely accepted neuraminidase inhibitors like Oseltamivir (Ag@OMV) [131] and Zanamivir (Ag@ZNV) [132] as well as M2 inhibitor, Amantadine (Ag@AM), [133] has been documented for the ability of AgNPs to synergistically prohibit apoptosis and apoptosis induced changes like DNA fragmentation and nuclear condensation in MDCK cells through the inhibition of pro‐apoptotic proteins like Caspase‐3 and PARP. A significant decrease in intracellular ROS generation is a familiar observation for all the three combinations of AgNPs. Ag@OMV can effectively reduce expression levels of phosphorylated (p) and total p53 and increase the expression level of pAKT and total AKT. Similarly, Ag@ZNV has also been successful in decreasing p‐p53 and total p53 expression as well as p‐p38 and total p38 proteins levels. The inhibition of Caspase‐3, which is a recurring event for all three combinations, could impact the cellular transport and nuclear localization of viral RNPs as previously discussed, and arrest virus life cycle. The induction of anti‐apoptotic Akt signaling and reduction of prop‐apoptotic p53 signaling pathways suggest an anti‐apoptotic nature of these combinations that produce a significantly higher response than when AgNPs or the drugs were used alone. An analogous mechanism of action has been reported by the combination of selenium nanoparticles and zanamivir (Se@ZNV) [134] that downregulate p38 and JNK signaling pathways to halt apoptosis, indicating a noteworthy achievement of metal nanoparticles in influencing intracellular signaling pathways in synergistic combinations with drugs.However, a possible reason of explanation for this significant synergistic effect has not been mentioned.
3.4. Additive effect of AgNPs on neutralizing antibodies for potential HIV vaccine development
In a fruitful attempt to correspond the productivity of AgNPs in pragmatic applications, Lara et al synthesized a cocktail of four neutralizing antibodies (NAB)‐Monoclonal antibody to HIV‐1 gp41 126–7, HIV‐1 gp120 Antiserum PB1 Sub 2, HIV‐1 gp120 Antiserum PB1 and HIV‐1 gp120 Monoclonal Antibody F425 B4e8, that functionally target the envelop glycoproteins gp120 and gp41, and coupled them with AgNPs, reporting an additive effect on virus inhibition than when either of them were used alone. [135] They postulated the increase in effectivity when used as a cocktail due to the enhanced binding of AgNPs to epitopes of gp120 and gp41 that were previously inaccessible, which NABs did not bind and vice versa. An in‐depth probe to elucidate the mechanism of this additive effect could open many doors and renew the interest of immunologists as this cocktail can potentially eliminate the crisis of achieving high antibody titers during vaccination by NABs alone.
3.5. Additive and synergistic effects of curcumin modified AgNPsin controlling HIV and RSV
Curcumin, which is a potent therapeutic has immense medicinal properties due to its anti‐inflammatory and anti‐oxidant characteristics. However, poor bioavailability of curcumin due to insufficient water solubility, [136] instability under different pH conditions, [136] rapid non‐enzymatic degradation [137] as well as systemic excretion [138] greatly impedes its clinical applications and necessitates the advancement of research that holds potential in preserving the remedial qualities of curcumin without provoking additional cytotoxicity. R. K. Sharma et al. highlighted a nanocarrier based formulation of curcumin modified AgNPs that substantially ameliorated the solubility, stability and half‐life activity of curcumin without affecting its therapeutic value. [139] Curcumin modified AgNPs exhibited antiretroviral activity against HIV−I in‐vitro in ACH‐2 cells, restraining HIV virus multiplication by immunomodulating the expression of NFkβ, that modulates cytokine expression as well as pro‐inflammatory cytokines like IL‐1β, TNF‐α, and IL‐6. Down‐regulating the expression of these cytokines in addition to inhibition of HIV‐1 Long Terminal Repeat (LTR) gene that drives HIV‐1 gene expression and p24 capsid protein that is required for assembly, considerably inhibited HIV‐1 virus infection. This significant antiretroviral activity was not demonstrated by curcumin and AgNPs when used single‐handedly, enlightening the additive effect on each other.
Another synergistic study of curcumin modified AgNPs (cAgNPs) was reported by Yang et al. against RSV infection. [140] cAgNPs reportedly inhibited RSV by interacting with viral surface glycoproteins to promote direct inactivation in order to limit their entry into cells. cAgNPs also downregulated the expression of pro‐inflammatory cytokines like IL‐1β and IL‐6 that are usually increased at the onset of RSV infection. AgNPs and curcumin alone could restrict viral infection, however, their combined activity generated stable, uniform, mono‐disperse nanoparticles with increased bioavailability that led to a significant inhibition of RSV infection.
3.6. Synergistic, broad‐spectrum action of AG and GO nanocomposites against RNA viruses
Graphene oxide NPs are carbon‐based NPs that have exhibited remarkable antiviral effectivity and are currently being actively pursued for the development of abundant antivirals. [141] The synergistic action of AgNPs modified with GO nanocomposites as broad‐spectrum antivirals against porcine reproductive and respiratory syndrome virus (PRRSV) and porcine epidemic diarrhea virus (PEDV) was elucidated by Du et al wherein GO‐AgNPs constrained viral proliferation through the suppression of viral entry and stimulated Interferon‐α and Interferon Stimulated genes that are essential for activation of antiviral innate immune responses. [29] In another study by Chen et al, the virucidal activity of GO‐AgNPs was investigated against the enveloped feline corona virus and the non‐enveloped infectious bursal disease virus. [142] They concluded that the antiviral response obtained from the treatment of GO‐AgNPs surpassed that of AgNPs and GONPs alone, which could be attributed to the enhanced water solubility and stability of the generated NPs as GO nanosheets prevent agglomeration of AgNPs. These findings approve the competent broad‐spectrum activity of GO‐AgNPs against deadly RNA viruses.
4. AgNPs in clinical diagnosis of viral infections
Biomolecular recognition of viral infection markers relies on the ability of a sensor to detect either the virus and its particles like DNA, RNA, proteins, peptides, etc. or molecules that are generated by the host immune system like antibodies, stress molecules, etc. The nanoscale structural feature of AgNPs offer a high surface area to volume ratio that enables them to efficiently functionalize with these viral markers at the molecular level through thiol‐metal quasi‐covalent bonds or direct amine‐metal bonding. Additionally, their unique optical properties such as localized surface plasmon resonance (LSPR) [143] imparts bright colors in solutions due to the interaction of light with the electrons at the surface of the metal, generating five times larger optical cross sections than commonly used dyes that tag biomolecules. [144] Moreover, AgNPs possess high electrical conductivity and can effectively enhance electrical signals in electrochemical sensors. [145] Different strategies to employ AgNPs for detection of nucleic acids, proteins, pH variations and small analytes have been elucidated in great detail which have proved to be highly specific, sensitive, cost‐effective, rapid as well as flexible. AgNP modified biosensors for the detection of viral infections via colorimetric, chemiluminescent, fluorescent, electrochemical, Raman and SERS and lateral flow approaches have been briefly reviewed here as well as summarized in Table 2, that target both viral antigens and antibodies elicited by host as well as viral nucleic acids and outline a promising endeavor of AgNPs in the field of viral bio‐diagnostics.
Table 2.
Summary of the diagnostic applications of AgNPs in detecting viral markers for the diagnosis of viral diseases.
|
Virus |
Type of detection |
Type of specimen detected |
Type of assay |
Characteristics of the assay |
Reference |
|---|---|---|---|---|---|
|
Dengue Virus |
Colorimetric |
Nucleic acid |
AgNP‐ssDNA probe hybridization to complimentary target viral RNA |
Can distinguish between different serotypes of dengue virus strains. |
[147] |
|
HIV |
Colorimetric |
Nucleic acid |
DNA tetrahedron@AgNP integrated with a polymerization/nicking machinery |
Crosslinking aggregation of AgNP@DNA tetrahedron probes incite color variations. |
[148] |
|
Japanese encephalitis Virus (JEV) |
Colorimetric |
Antigen |
Antigen‐Antibody@AgNP sandwich |
Rapid and inexpensive optical probe |
[149] |
|
H1N1 Influenza Virus |
Chemiluminescent |
Antigen |
Antigen‐Antibody@AgNP sandwich |
Ultrasensitive and eliminates complex signal amplification procedures. |
[150] |
|
Hepatitis B Virus (HBV) |
Electrochemical |
Nucleic acid |
Paper based sandwich type assay involving AgNP@labelled DNA and magnetic beads@target DNA |
Single sample incubation step that highly accentuates its speed, stability and robustness. |
[152,153] |
|
Dengue Virus |
Electrochemical |
Antigen |
Antigen‐Antibody@AgNP sandwich |
Involves diazonium assisted immobilization of AgNPs on pencil graphite electrodes |
[154] |
|
Hepatitis C Virus (HCV) |
Electrochemical |
Antigen |
Antigen‐Antibody@AgNP sandwich |
Used riboflavin as the redox sensor |
[155] |
|
Avian Influenza Virus H7 |
Electrochemical |
Antigen |
Antigen‐Antibody@AgNP sandwich |
Involved the use of AgNPs and graphene nanocomposites |
[156] |
|
JEV |
Electrochemical |
Antigen |
Antigen‐Antibody@AgNP sandwich |
Used screen printed carbon electrodes |
[157] |
|
Tick borne encephalitis virus |
Electrochemical |
Antibody |
Antibody‐Antigen@AgNP sandwich |
Detection of specific IgG antibodies against tick borne encephalitis virus |
[158] |
|
Influenza Virus |
Electrochemical |
Whole Virus |
Whole virus@AgNPquantified by oxidation of AgNPs |
Generated electrical spikes proportional to the concentration of virus particles |
[159] |
|
HIV |
Fluorescent |
Antigen |
Sandwich‐based direct fluorescent assay of streptavidin@fluorescentAgNPs and biotin@antigen |
Immunosensing of HIV‐1 p24 antigen |
[161] |
|
H1N1 Influenza Virus |
Fluorescent |
Antigen |
Sandwich type indirect fluorescence‐based immunoassay |
Release of Ag+ ions to emit fluorescence |
[162] |
|
HBV |
Fluorescent |
Nucleic acid |
Sandwich based fluorescent microarray |
Exploits Metal Enhanced Fluorescence (MEF) of AgNPs to detect target sequences with high specificity. |
[164] |
|
Ebola virus, Yellow fever virus, Dengue Virus |
Lateral flow immunoassay |
Antigen |
Sandwich based mutliplexed nano‐immunosensor |
Mutliple virus detection in a single test line on the basis of color |
[166] |
|
RSV |
Surface enhanced Raman scattering |
Antigen |
Enzyme based sandwich type immunoassay |
Involves horseradish peroxidase as an oxidizer of the Raman‐active molecule to generate SERS signal |
[168] |
|
HCV |
Surface enhanced Raman scattering |
Nucleic acid |
Detection of SERS signal from HCV markers |
AgNPs were used to amplify SERS signal |
[169] |
4.1. Colorimetric detections
The colloidal solution of dispersed AgNPs is intensely colored due to its LSPR nature. Changes in the ionic strength of the solution due to the presence of foreign particles (nucleic acids, antigens) or aggregations give rise to a shift in the LSPR band followed by a resulting change in color which is the basis of detection of colorimetric biosensors. [146] Colorimetry is an extensively used detection technique which is substantially predicated on the size, shape, inter‐particle distance, morphology, capping molecules and distribution states of AgNPs. [144] The high extinction coefficient coupled with its simplistic methods of synthesis and functional properties have increased the widespread use of AgNPs in colorimetric biosensors for the detection of both viral nucleic acids as well as antigens. Nucleic acid‐based detections are highly specific, sensitive and rule out false negatives that often arise due to cross‐reactivity within similar families of viruses. As a noteworthy example to illustrate this concept, Vinayagam et al reported the use of triangular AgNPs (TAg) that were oligomerized with a ssDNA probe, designed complementary to the target RNA sequence of a specific Dengue virus strain, that could distinguish between different serotypes of dengue virus strains. [147] Parallelly, DNA tetrahedrons crosslinked with AgNP probes for the detection of HIV virus [148] and sandwich based AgNP optical immunosensors for the detection of Japanese encephalitis Virus (JEV) antigens have been fabricated as well, by the immobilization of JEV antibodies on AgNPs that were deposited on amine functionalized glass sides. [149]
4.2. Chemiluminescent detections
Chemiluminescence is a widely used diagnostic technique that involves the use of a luminescent molecule, which emits light while indicating the presence of the target biomarker. The sensitivity of this diagnostic approach allows the detection of even single molecules while eliminating the prerequisites for complex signal amplification. Li et al demonstrated an ultrasensitive chemiluminescent metallo immunoassay involving an antigen‐antibody sandwich for the detection of H1N1 Influenza virus using AgNP labels which functioned tremendously in the enhancement of the chemiluminescent signals of luminol. [150] They used AgNP labelled polyclonal antibody that could bind to Influenza virus antigens that were immobilized on monoclonal antibody coated wells. The signal enhancement was brought about by the dissolution of AgNPs with HNO3 to liberate Ag+ ions that could be quantified by the chemiluminescent system.
4.3. Electrochemical detections
Electrochemical detection systems utilize electrochemical reporter probes that transduce pathogen recognition signals into electrochemical indicators like redox kinetics, amperage or electrical hindrance. [151] The sensors consist of electrodes that are coated with AgNPs that permit the functionalization of antibodies and nucleic acids as sensor probes onto its surface via thiol chemistry and upon hybridization with target moieties create electrical resistance that is designed to be picked up and amplified for detection. AgNPs are excellent amplifiers of electrical signals, owing to their high electrical conductivity and have found widespread use in formulations of electrochemical sensors for the detection of pathogens. Li et al. described a simple paper based electrochemical sensor that was capable of detecting a 30‐base nucleotide sequence of Hepatitis B Virus DNA using AgNPs that provided an amplification factor of about 250,000 and magnetic microbeads that provided an additional 25 times amplification. [152] In another approach, a chip based, PCR‐less, multiplexed DNA electrochemical detection was achieved using oligomerized AgNPs that provided high ultrasensitive detection of low doses of Hepatitis B, Herpes simplex, Epstein‐Barr and cytomegalovirus sequences by a cooperative oxidation mechanism. [153]
AgNPs conjugated to antibodies and immobilized on working electrodes have proven to be invaluable electro‐immunosensors in the detection of viral antigens. Numerous electrochemical sandwiches based immunosensors for the detection of both antigens as well as virus specific antibodies have been summarized in Table 2.[ 154 , 155 , 156 , 157 , 158 ] The binding of antigen to antibody creates a steric hindrance that triggers an electron transfer resistance, which can be effortlessly detected by potentiostat, differential pulse voltammetry, electrochemical impedance spectroscopy or cyclic voltammetry. [154] Another interesting approach employed by most AgNP based electrochemical immunosensors is the use of graphene and AgNP nanocomposites.[ 155 , 156 ] The immobilization of graphene particles with AgNPs creates a large surface area that enables loading of a higher concentration of antibody for efficient entrapment of antigen and further enhances the electrochemical signal as both are extremely good conductors of electricity. These electrochemical immunosensors are rapid and extremely sensitive detectors that have revolutionized the field of nano‐biosensors in diagnosis of pathogens. Moreover, electrochemical immunoassays with antibody‐AgNP bioconjugates have also been used for the detection of specific IgG antibodies against tick borne encephalitis virus. [158] An interesting and novel approach of quantification of whole Influenza virus particles tagged with AgNP has been demonstrated by Sepunaru et alat a single virus level in real time by an advanced electrochemical immunosensor. [159] Automated micro‐fluidic electrochemical sensors integrated with this technique can also be used successfully as a point of care detection of whole viruses.
4.4. Fluorescent detections
The use of AgNPs in conjunction with fluorophores markedly elevates the fluorescence intensity and are commonly used in fluorescent biosensors for rapid and specific diagnosis of pathogens, often eliminating the use of enzymes. Antibody labelled or oligonucleotide labelled AgNPs are used directly with fluorophores or indirectly with fluorescent dyes that emit fluorescent signals when hybridized with target moieties and can be easily captured by detectors that show a direct correlation between target quantity and signal intensity. Fluorescent AgNPs (FSNPs) have emerged as attractive biolabels in bioimaging and biosensing applications due to their quantum confinement that permits intense light absorption and emission as well as their inherent photostability and biocompatibility. [161] They permit direct quantification of antigens when incorporated in sandwich based immunosensors by the measurement of the fluorescence emitted by the fluorophore on excitation that follows after appropriate antigen‐antibody binding as exemplified by Kurdekar et al.for the detection of HIV Virus. [161] AgNPs can also be incorporated in indirect fluorescence‐based immunoassays by their reduction to Ag+ ions, which are strong oxidizing agents. As an example, Li et al. described a fluorescent immunosensor involving AgNPs bioconjugated with polyclonal antibodies that were targeted to spot H1N1 IV antigens by the dissolution of bound AgNPs with HNO3 to release Ag+ ions that successfully catalyzed the oxidation of o‐phenylenediamine (OPDA) to emit fluorescence. [162]
The interaction of fluorophores with metals enhances their intensity of fluorescence due to the localized excitation of fluorophores when present in close contact with metal nanoparticles and leads to the generation of photostable fluorophores. [163] This phenomenon is termed as Metal Enhanced Fluorescence (MEF). AgNP is an excellent choice for MEF because it has a high extinction coefficient in addition to surface plasmon effect that efficiently amplifies the fluorescence signal. This MEF property of AgNPs was exploited by Jin et al to create a sandwich based fluorescent microarray that could detect Hepatitis B Virus DNA with high specificity that could eliminate even single base mismatches and interference from genomic DNA. [164]
4.5. Lateral flow immunoassays
Lateral flow immunoassay is a point of care diagnostic that offers several advantages because of its simplicity, robustness, portability, economical and swift detection technique and is popular in various fields of biomedical diagnosis. [165] The system consists of several zones of polymeric strips through which the sample containing the target of interest is allowed to flow by capillary action and interact with capture probes consisting of antibodies and detected on a suitable matrix consisting of nitrocellulose membrane. [165] AgNPs are a promising metal nanoprobe that eliminate the use of external excitation and dyes due to their inherent colored properties owing to their LSRP effects. Yen et al described a sandwich based multiplex nano‐sensor using multiple colored triangular plate shaped AgNPs of different sizes that were labelled with monoclonal antibodies for the diagnosis of dengue, yellow fever and Ebola viruses on the basis of color. [166] Each of the antigens that were detected from the sample formed an ellipse and none of the ellipses overlapped, thus ruling out non‐specificity and cross‐reactivity and emphasizing the possibility of multiplexed detection in a single test line with reduced device dimensions and cost.
4.6. Raman and SERS detection
Raman spectroscopy is an invaluable diagnostic technique that provides structural information based on the vibrational or rotational vibration modes of molecules. Surface enhanced Raman scattering (SERS) is a technique that enhances the Raman scattering of molecules that can detect and quantify even single molecules. AgNPs are an attractive choice for SERS due to their LSPR that falls within the Raman spectrum that suggestively enhances the SERS effect by a factor of about 1015 to 1016 times. [167] Raman and SERS have gained widespread attention as diagnostic techniques due to their high specificity, sensitivity, non‐invasive nature and label free detection approaches. Zhan et aldevised a powerfully sensitive and specific strategy that integrates enzyme‐based immunoassay with SERS based detection to diagnose RSV infections from clinical samples by the capture of RSV antigens by RSV specific antibodies that subsequently interact with horseradish peroxidase (HRP) labelled antibodies to form a sandwich complex. [168] HRP catalyzes the oxidation of the Raman molecule 3,3′, 5,5′‐tetra‐methylbenzidine (TMB) to release TMB+ that interacts with the negatively charged surface of AgNPs electrostatically to aggregate them and generate strong SERS signals that is directly correlated with the quantity of captured antigen. Furthermore, AgNPs were used to amplify SERS signal in yet another assay involving the detection of HCV from clinical samples that exhibited prominent peaks corresponding to HCV markers like HCV RNA and O‐CH3 groups of ribosomal RNA, produced during protein synthesis. [169] This study also highlighted the superiority of SERS over Raman spectroscopy in identification of specific viral markers and disease diagnosis.
5. Miscellaneous virucidal applications of AgNPs
5.1. Water disinfections strategies
The virucidal activities of AgNPs have been utilized in a variety of formulations to devise appropriate water disinfection strategies that inhibit bacteriophages and other water‐borne viruses. The antiviral activity of AgNP and silica conjugates against murine norovirus and bacteriophage MS2 in different types of water was elucidated by Park et al. [32] Additionally, AgNPs in association with bacterial cell surface of Lactobacillus fermentumform biogenic silver or bio‐Ag0 that upon immobilization on microporous PVDF membranes, inhibit UZ1 bacteriophages in effluents and can be used for the inactivation of enteric viruses from water at a small scale. [34] Another study by Park et al reported the use of micrometer sized magnetic hybrid colloid in conjunction with AgNPs for inactivation of bacteriophage φX174, murine norovirus (MNV), and adenovirus serotype 2 (AdV2) across a wide pH range of surface and tap water, offering a supplementary advantage of complete recovery after use by exploiting its magnetic properties. [33]
5.2. Drug quantifications
The optical properties of AgNPs can be exploited for the accurate quantification of drugs and present an indispensable method of drug selection that is highly accurate, sensitive, rapid and inexpensive to reproduce in quality control laboratories. Saraya et al reported a highly sensitive strategy of AgNP synthesis for the spectrophotometric determination of antiviral drugs Sofosbuvir, Lamivudine and Ritonavir by employing the principle of formation of AgNPs which is a consequence of a redox reaction. [170] The key elements of this redox reaction include a reducing agent which is the antiviral drug that requires quantification, an oxidizing agent like AgNO3 that could direct the formation of AgNPs and a stabilizer PVP, all of which lead to the formation of golden yellow colored AgNPs that can be spectrophotometrically quantified by measurement of their absorbance at the respective wavelength. Furthermore, this method could quantify the drugs in both their pure and pharmaceutical dosage forms, suggesting negligible interference from pharmaceutical excipients during AgNP synthesis. AgNPs have also been incorporated in a voltametric nanosensor to amplify electrochemical signals for electrochemical assay of anti‐HIV drug Rilpivirine. [171]
5.3. Salivary excretion
Tang et al.reported that injection of renal clearable glutathione coated AgNPs (GS‐AgNPs) in mice led to their localization in the submandibular salivary glands and subsequent secretion into the saliva which is a reservoir of a plethora of bacteria and viruses. [172] These engineered GS‐AgNPs had extremely high vascular permeability, probably due to their low material density, and were reported to be more proficient than GS‐AuNPs (gold nanoparticles) of the same size and surface chemistry. Therefore, the broad‐spectrum antimicrobial activity of AgNPs could be resourcefully targeted towards the restriction of infectious bacteria and viruses in the oral cavity and exploited to prevent transmission of respiratory viruses that enter through the mouth.
5.4. Commercially approved AgNPs formulations
The iconic “Samsung” manufactures devices using ‘Nano Silver Health System’ that incorporates AgNPs in refrigerator trays, filters, air conditioners, and tubing to kill bacteria, viruses, and desorb foul odor. [173] Additionally, AgNPs are also deployed in medical devices as prosthetics and for coating urethral and central line catheters and implantable medical devices such as infusion ports, orthopedic protruding fixtures, endovascular stents, urological stents, endotracheal tubes, contact lens coatings, endoscopes, electrodes, peritoneal dialysis devices, subcutaneous cuffs, and surgical and dental instruments. [174] Additionally, ventricular catheters like Silverline of Spiegelberg and ON−Q Silver Soaker of I‐Flow Corporation catheters for drug delivery are also widely used. [175]
A few masks and personal protective equipment are equipping their textiles with AgNPs to prevent viral spread by inactivation of the viruses, owing to the broad spectrum virucidal activity of AgNPs. An antiviral surface coating, termed NANOVA HYGIENE+TM, was designed by Dr. Ghosh and his team in Nova Surface‐Care Centre, India, combining the potential of quaternary ammonium cations and AgNPs to prevent the spread of SARS COV2 through touch from surfaces like fabrics, plastics, metals and concretes. [176] Silver technologies were integrated with vesicles in a unique textile treatment called HeiQ Viroblock NPJ03 [177] that Chinese protective masks producer Suzhou Bolisi developed to combat the spread of COVID‐19 virus and other respiratory viruses and bacteria. This viroblock has been tested against human coronavirus (229E), Influenza types H1N1, H5N1, H7N9, and RSV and the technology could potentially be reproduced in face masks, air filters, medical gowns, curtains, drapes, etc. HeiQViroblock has gathered recognition from other textile companies that have expressed their interest in adopting this technology into their products like the American legwear manufacturer Kayser‐Roth that is developing to implement this technology into their new hand protector, Ghluv, as well as the Chinese company Lufeng, that is presently evaluating this technology for fabricating garments. Moreover, National Center for Scientific Research in Russia has created Biosilver Lab [178] that is involved in the production of a range of compelling antiviral compounds using patented, highly dispersed AgNP technologies for more than 15 years. Some of their products include nano‐masks, Mask Maker Spray solution, spun bound coated and melt blown fabrics and masks, etc. All of these formulations consist of AgNPs of less than 10 nm size, which is the paramount antiviral substance that is accountable for their mode of action. A few other trade names of formulations that have implemented Biosilver in their products include Argovit, Argovit‐S, Argovit‐Bio, Vitargol, Vitargol‐Forte, Vitargol BAA, Vitargol spray, Argogel, Argokrem and Silver Powder. All these AgNPs based formulations have huge potential to overcome global pandemic in the years to come.
6. Conclusion
Silver nanoparticles have been broadly exploited to develop therapeutics against viruses by targeting different aspects of the viral life cycle like virus entry, replication and assembly. Multiple studies have demonstrated their ability to inhibit viruses directly or in conjunction with other molecules. AgNPs have also reported immunomodulatory activities in host cells by inhibiting apoptosis and secondary bacterial infections and activating neutrophils and PRPs to combat viral defenses. Furthermore, the physical properties of AgNPs have mediated the designing of effective drug delivery carriers for the active transport of vaccines, RNA, and other therapeutics and small molecules by possibly forming synergistic interactions. Lastly, several viral diagnostic platforms have reported using AgNPs for colorimetric, chemiluminescent, electrochemical, fluorescent, lateral flow, Raman and SERS detection.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Ujjyani Ghosh received her M.Sc. (Microbiology) degree from St. Xavier's College, Kolkata, India. Then, she worked as a project trainee at CSIR‐Indian Institute of Chemical Biology. Currently, she is pursuing her Ph.D. at The University of Utah, Salt Lake City, USA. Her area of research interest is on probable therapeutic target for multidrug‐resistant bacteria.

Biographical Information
Khondakar Sayef Ahammed received his M.Sc. (Microbiology) degree from St. Xavier's College (Kolkata), India. Then, He worked as a project assistant at CSIR‐Indian Institute of Chemical Biology under the supervision of Dr. Sanjay Dutta. Currently, he is pursuing his Ph.D. under the guidance of Prof. Ambro van Hoof at MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences, Houston, TX, USA. His current research is focused on RNA degradation and processing in Saccharomyces cerevisiae, as well as identification of novel antimicrobial targets using chemical genetics.

Biographical Information
Snehasis Mishra is working on synthesizing different biocompatible materials and their application in Cancer Biology, including the Imaging applications. His research interest includes Covalent Organic Frameworks, Porous Organic Materials mediated drug delivery system for Cancer. He is also working in synthesis of several metal nano composites and their applications in Photodynamic and Photo‐thermal Therapy for Triple negative Breast Cancer.

Biographical Information
Asim Bhaumik did his PhD in Chemistry from the CSIR‐National Chemical Laboratory, Pune in 1997. Then after his post‐doctoral research at The University of Tokyo, Japan as a JSPS fellow and Toyota Central R & D Labs. Inc. Japan as associate researcher he has joined at the Indian Association for the Cultivation of Science in 2001 as a faculty, where he is currently working as a Senior Professor. His area of research interest is designing functional porous nanomaterials for energy, environment and biomedical applications.

Acknowledgements
AB wishes to thank DST‐SERB, New Delhi for a core research grant (Project No. CRG/2018/000230).
U. Ghosh, K. Sayef Ahammed, S. Mishra, A. Bhaumik, Chem. Asian J. 2022, 17, e202101149.
Contributor Information
Snehasis Mishra, Email: snehasism1991@gmail.com.
Prof. Dr. Asim Bhaumik, Email: msab@iacs.res.in.
References
- 1. Murphy F. A., Adv. Virus Res. 2016, 95, 197–220. [DOI] [PubMed] [Google Scholar]
- 2.K. Moelling, Viruses: More Friends Than Foes,World Scientific, 2016.
- 3.N. LePan, Visual Capitalist. 2020, March 14. Available at: https://www.visualcapitalist.com/history-of-pandemics-deadliest/..
- 4. Geoghegan J. L., Duchêne S., Holmes E. C., PLoS Pathog. 2017, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ji W., Wang W., Zhao X., Zai J., Li X., J. Med. Virol. 2020, 92, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lauring A. S., Frydman J., Andino R., Nat. Rev. Microbiol. 2013, 11, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aldiero S., Falanga A., Cantisani M., Marra V., Galdiero M., Molecules 2011, 16, 8894–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rai M., Kon K., Ingle A., Duran N., Galdiero S., Galdiero M., Appl. Microbiol. Biotechnol. 2014, 98, 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rai M., Deshmukh S. D., Ingle A. P., Gupta I. R., Galdiero M., Galdiero S., Crit. Rev. Microbiol. 2016, 42, 46–56. [DOI] [PubMed] [Google Scholar]
- 10. Dos Santos C. A., Seckler M. M., Ingle A. P., Gupta I., Galdiero S., Galdiero M., Gade A., Rai M., J. Pharm. Sci. 2014, 103, 1931–1944. [DOI] [PubMed] [Google Scholar]
- 11. Elechiguerra J. L., Burt J. L., Morones J. R., Camacho-Bragado A., Gao X., Lara H. H., Yacaman M. J., J. Nanobiotechnol. 2005, 3, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gaikwad S., Ingle A., Gade A., Rai A., Falanga A., Incoronato N., Russo L., Galdiero S., Galdiero M., Int. J. Nanomed. 2013, 8, 4303–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu L., Sun R. W., Chen R., Hui C. K., Ho C. M., Luk J. M., Lau G. K., Che C. M., Antiviral Ther. 2008, 13, 253. [PubMed] [Google Scholar]
- 14. Shady N. H., Khattab A. R., Ahmed S., Liu M., Quinn R. J., Fouad M. A., Kamel M. S., Muhsinah A. B., Krischke M., Mueller M. J., Abdelmohsen U. R., Int. J. Nanomed. 2020, 15, 3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiang D. X., Chen Q., Pang L., Zheng C. L., J. Virol. Methods 2011, 178, 137–142. [DOI] [PubMed] [Google Scholar]
- 16. Sun L., Singh A. K., Vig K., Pillai S. R., Singh S. R., J. Biomed. Nanotechnol. 2008, 4, 149–158. [Google Scholar]
- 17. Huy T. Q., Thanh N. T. H., Thuy N. T., Van Chung P., Hung P.N., Le A. T., Hanh N. T. H., J. Virol. Methods 2017, 241, 52–57. [DOI] [PubMed] [Google Scholar]
- 18. Sujitha V., Murugan K., Paulpandi M., Panneerselvam C., Suresh U., Roni M., Nicoletti M., Higuchi A., Madhiyazhagan P., Subramaniam J., Dinesh D., Parasitol. Res. 2015, 114, 3315–3325. [DOI] [PubMed] [Google Scholar]
- 19. Sharma V., Kaushik S., Pandit P., Dhull D., Yadav J. P., Kaushik S., Appl. Microbiol. Biotechnol. 2019, 103, 881–891. [DOI] [PubMed] [Google Scholar]
- 20. Rogers J. V., Parkinson C. V., Choi Y. W., Speshock J. L., Hussain S. M., Nanoscale Res. Lett. 2008, 3, 129. [Google Scholar]
- 21. Trefry J. C., Wooley D. P., J. Biomed. Nanotechnol. 2013, 9, 1624–35. [DOI] [PubMed] [Google Scholar]
- 22. Speshock J. L., Murdock R. C., Braydich-Stolle L. K., Schrand A. M., Hussain S. M., J. Nanobiotechnol. 2010, 8, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borrego B., Lorenzo G., Mota-Morales J. D., Almanza-Reyes H., Mateos F., López-Gil E., de la Losa N., Burmistrov V. A., Pestryakov A.N., Brun A., Bogdanchikova N., Nanomedicine Nanotechnology, Biol. Med. 2016, 12, 1185–1192. [DOI] [PubMed] [Google Scholar]
- 24. Dung T. T.N., Nam V.N., Nhan T. T., Ngoc T. T. B., Minh L. Q., Nga B. T. T., Quang D. V., Mater. Res. Express 2020, 6, 1250. [Google Scholar]
- 25. Ochoa-Meza A. R., Álvarez-Sánchez A. R., Romo-Quiñonez C. R., Barraza A., Magallón-Barajas F. J., Chávez-Sánchez A., García-Ramos J. C., Toledano-Magaña Y., Bogdanchikova N., Pestryakov A., Mejía-Ruiz C. H., Fish Shellfish Immunol. 2019, 84, 1083–1089. [DOI] [PubMed] [Google Scholar]
- 26. Li Y., Lin Z., Xu T., Wang C., Zhao M., Xiao M., Wang H., Deng N., Zhu B., RSC Adv. 2017, 7,1453–1463. [Google Scholar]
- 27. Jeremiah S. S., Miyakawa K., Morita T., Yamaoka Y., Ryo A., Biochem. Biophys. Res. Commun. 2020, 533, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Castro-Mayorga J. L., Randazzo W., Fabra M. J., Lagaron J. M., Aznar R., Sánchez G., LWT-Food Sci. Technol. 2017, 79, 503–510. [Google Scholar]
- 29. Du T., Lu J., Liu L., Dong N., Fang L., Xiao S., Han H., ACS Appl. Bio Mater. 2018, 1, 1286–1293. [DOI] [PubMed] [Google Scholar]
- 30. Ahsan T., Egypt. J. Biochem. 2020, 30, 1–4. [Google Scholar]
- 31. Elbeshehy E. K., Elazzazy A. M., Aggelis G., Front. Microbiol. 2015, 6, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park S., Ko Y. S., Jung H., Lee C., Woo K., Ko G., Sci. Total Environ. 2018, 625, 477–485. [DOI] [PubMed] [Google Scholar]
- 33. Park S., Park H. H., Kim S. Y., Kim S. J., Woo K., Ko G., Appl. Environ. Microbiol. 2014, 80, 2343–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Gusseme B., Hennebel T., Christiaens E., Saveyn H., Verbeken K., Fitts J. P., Boon N., Verstraete W., Water Res. 2011, 45, 1856–1864. [DOI] [PubMed] [Google Scholar]
- 35. Nie C., Du P., Zhao H., Xie H., Li Y., Yao L., Shi Y., Hu L., Si S., Zhang M., Gu J., Luo L., Chem. Asian J. 2020, 15, 148–155. [DOI] [PubMed] [Google Scholar]
- 36. Ryu W. S., Mol. Virol. Human Pathogenic Viru. 2016, 1-423.. [Google Scholar]
- 37. Thompson A. J., de Vries R. P., Paulson J. C., Curr. Opin. Virol. 2019, 34, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hoxie J. A., Alpers J. D., Rackowski J. L., Huebner K., Haggarty B. S., Cedarbaum A. J., Reed J. C., Science 1986, 234,1123–1127. [DOI] [PubMed] [Google Scholar]
- 39. Sattentau Q. J., Weiss R. A., Cell 1988, 52, 631–633. [DOI] [PubMed] [Google Scholar]
- 40. Qian K., Morris-Natschke S. L., Lee K. H., Med. Res. Rev. 2009, 29, 369–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Poveda E., Briz V., Soriano V., AIDS Rev. 2005, 7, 139–147. [PubMed] [Google Scholar]
- 42. Wallace B. J., Tan K. B., Pett S. L., Cooper D. A., Kossard S., Whitfeld M. J., Aust. J. Dermatol. 2011, 52, 19–26. [DOI] [PubMed] [Google Scholar]
- 43. Glass W. G., McDermott D. H., Lim J. K., Lekhong S., Yu S. F., Frank W. A., Pape J., Cheshier R. C., Murphy P. M., J. Exp. Med. 2006, 203, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lara H. H., Ayala-Nuñez N. V., Ixtepan-Turrent L., Rodriguez-Padilla C., J. Nanobiotechnol. 2010, 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lara H. H., Ixtepan-Turrent L., Garza-Treviño E.N., Rodriguez-Padilla C., J. Nanobiotechnol. 2010, 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fayaz A. M., Ao Z., Girilal M., Chen L., Xiao X., Kalaichelvan P. T., Yao X., Int. J. Nanomed. 2012, 7, 5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith G. L., Talbot-Cooper C., Lu Y., Adv. Virus Res. 2018, 100, 355–378. [DOI] [PubMed] [Google Scholar]
- 48. Ansar S. M., Perera G. S., Gomez P., Salomon G., Vasquez E. S., Chu I. W., Zou S., C. U. Pittman Jr , Walters K. B., Zhang D., J. Phys. Chem. C 2013, 117, 27146–27154. [Google Scholar]
- 49. Sánchez-Cortés S., Berenguel R. M., Madejón A., Pérez-Méndez M., Biomacromolecules 2002, 3, 655–660. [DOI] [PubMed] [Google Scholar]
- 50. Ravindran A., Chandran P., Khan S. S., Colloids Surf. B 2013, 105, 342–352. [DOI] [PubMed] [Google Scholar]
- 51. Naik R. R., Stringer S. J., Agarwal G., Jones S. E., Stone M. O., Nat. Mater. 2002, 1, 169–172. [DOI] [PubMed] [Google Scholar]
- 52. Mahato M., Pal P., Tah B., Ghosh M., Talapatra G. B., Colloids Surf. B 2011, 88, 141–149. [DOI] [PubMed] [Google Scholar]
- 53. Walczyk D., Bombelli F. B., Monopoli M. P., Lynch I., Dawson K. A., J. Am. Chem. Soc. 2010, 132, 5761–5768. [DOI] [PubMed] [Google Scholar]
- 54. Ding F., Radic S., Chen R., Chen P., Geitner N. K., Brown J. M., Ke P. C., Nanoscale 2013, 5, 9162–9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ge C., Tian J., Zhao Y., Chen C., Zhou R., Chai Z., Arch. Toxicol. 2015, 89, 519–539. [DOI] [PubMed] [Google Scholar]
- 56. Pramanik S., Chatterjee S., Saha A., Devi P. S., Kumar G. S., J. Phys. Chem. B 2016, 120, 5313–5324. [DOI] [PubMed] [Google Scholar]
- 57. Hu S., Yi T., Huang Z., Liu B., Wang J., Yi X., Liu J., Mater. Horiz. 2019, 6, 155–159. [Google Scholar]
- 58. Shrivas K., Sahu S., Sahu B., Kurrey R., Patle T. K., Kant T., Karbhal I., Satnami M. L., Deb M. K., Ghosh K. K., J. Mol. Liq. 2019, 275, 297–303. [Google Scholar]
- 59. Beck J., Nassal M., World J. Gastroenterol. 2007, 13, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garcıa C. C., Candurra N. A., Damonte E. B., Antiviral Res. 2002, 55, 437–446. [DOI] [PubMed] [Google Scholar]
- 61. Holzerland J., Leske A., Fénéant L., Garcin D., Kolakofsky D., Groseth A., Arch. Virol. 2020, 165, 1899–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kumar S. D., Singaravelu G., Ajithkumar S., Murugan K., Nicoletti M., Benelli G., J. Cluster Sci. 2017, 28, 359–367. [Google Scholar]
- 63. Sierra S., Kupfer B., Kaiser R., J. Clin. Virol. 2005, 34, 233–244. [DOI] [PubMed] [Google Scholar]
- 64. Rouse B. T., Sehrawat S., Nat. Rev. Immunol. 2010, 10, 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rehermann B., J. Clin. Invest. 2009, 119, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bhanja P., Mishra S., Manna K., Mallick A., Saha K. D., Bhaumik A., ACS Appl. Mater. Interfaces 2017, 9, 31411–31423. [DOI] [PubMed] [Google Scholar]
- 67. Ikeda H., Muroi M., Kondoh Y., Ishikawa S., Kakeya H., Osada H., Imoto M., ACS Chem. Biol. 2020, 15, 2195–2204. [DOI] [PubMed] [Google Scholar]
- 68. Wyllie A. H., Cell Death Biol. Pathol. 1981, 9–34.< [Google Scholar]
- 69. Restifo N. P., Curr. Opin. Immunol. 2000, 12, 597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nash P., Barrett J., Cao J. X., Hota-Mitchell S., Lalani A. S., Everett H., Xu X. M., Robichaud J., Hnatiuk S., Ainslie C., Seet B. T., Immunol. Rev. 1999, 168, 103–120. [DOI] [PubMed] [Google Scholar]
- 71. E. S. Mocarski Jr , Trends Microbiol. 2002, 10, 332–339. [DOI] [PubMed] [Google Scholar]
- 72. Dagna L., Pritchett J. C., Lusso P., Future Virol. 2013, 8, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schneider-Schaulies S., Ter Meulen V., Exp. Rev. Mol. Medicine 2002, 1-18.. [DOI] [PubMed] [Google Scholar]
- 74. Bouvier N. M., Palese P., Vaccine 2008, 26, D49–D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tzitzoglaki C., McGuire K., Lagarias P., Konstantinidi A., Hoffmann A., Fokina N. A., Ma C. L., Papanastasiou I. P., Schreiner P. R., Vazquez S., Schmidtke M., Wang J., Busath D. D., Kolocouris A., ACS Chem. Biol. 2020, 15, 2331–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chambers B. S., Parkhouse K., Ross T. M., Alby K., Hensley S. E., Cell Rep. 2015, 12, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shie J. J., Fang J. M., J. Biomed. Sci. 2019, 26, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schultz-Cherry S., Hinshaw V. S., J. Virol. 1996, 70, 8624–8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xiang D., Zheng Y., Duan W., Li X., Yin J., Shigdar S., O'Connor M. L., Marappan M., Zhao X., Miao Y., Xiang B., Int. J. Nanomed. 2013, 8, 4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Villeret B., Dieu A., Straube M., Solhonne B., Miklavc P., Hamadi S., Le Borgne R., Mailleux A., Norel X., Aerts J., Diallo D., ACS Nano 2018, 12, 1188–1202. [DOI] [PubMed] [Google Scholar]
- 81. Falsey A. R., Walsh E. E., Clin. Microbiol. Rev. 2000, 13, 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chatterjee A., Mavunda K., Krilov L. R., Infect. Dis. Ther. 2021, 10, 5-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Morris D., Ansar M., Speshock J., Ivanciuc T., Qu Y., Casola A., Garofalo R. P., Viruses 2019,11, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Romo-Quiñonez C. R., Álvarez-Sánchez A. R., Álvarez-Ruiz P., Chávez-Sánchez M. C., Bogdanchikova N., Pestryakov A., Mejia-Ruiz C. H., PeerJ 2020, 8, e8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ortiz-Perez A., Donnelly B., Temple H., Tiao G., Bansal R., Mohanty S. K., Front. Immunol. 2020, 11, 329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shivakumar P., Sabla G. E., Whitington P., Chougnet C. A., Bezerra J. A., J. Clin. Invest. 2009, 119, 2281–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang R., Lin Z., Lui V. C., Wong K. K., Tam P. K., Lee P., Lok C.N., Lamb J. R., Chen Y., Xia H., Nanomedicine 2017, 13, 1041–1050. [DOI] [PubMed] [Google Scholar]
- 88. Perlmutter J. D., Hagan M. F., Annu. Rev. Phys. Chem. 2015, 66, 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sundquist W. I., Kräusslich H. G., Cold Spring Harb. Persp. 2012, 2, a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shing C. Y., Whiteley C. G., Lee D. J., J. Inst. Chem. 2014, 45, 1140–1148. [Google Scholar]
- 91. Whiteley C. G., Shing C. Y., Kuo C. C., Lee D. J., J. Inst. Chem. 2016, 60, 83–91. [Google Scholar]
- 92. Tsai C. H., Whiteley C. G., Lee D. J., J. Inst. Chem. 2019, 103, 20–32. [Google Scholar]
- 93. Stoiber P., Ekladious I., Zhao Q., Colson Y. L., Schaus S. E., Hansen U., Grinstaff M. W., Biomacromolecules 2020, 21, 1499–1506. [DOI] [PubMed] [Google Scholar]
- 94. Lindenbach B. D., Rice C. M., Nat. Rev. Microbiol. 2013,11, 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ma Y., Yates J., Liang Y., Lemon S. M., Yi M., J. Virol. 2008, 82, 7624–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Brown P. K., Qureshi A. T., Moll A.N., Hayes D. J., Monroe W. T., ACS Nano 2013, 7, 2948–2959. [DOI] [PubMed] [Google Scholar]
- 97. García-Astrain C., Chen C., Burón M., Palomares T., Eceiza A., Fruk L., Corcuera M. Á., Gabilondo N., Biomacromolecules 2015, 16, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 98. Mukherjee S., Kotcherlakota R., Haque S., Das S., Nuthi S., Bhattacharya D., Madhusudana K., Chakravarty S., Sistla R., Patra C. R., ACS Biomater. Sci. Eng. 2019, 6, 690–704. [DOI] [PubMed] [Google Scholar]
- 99. Yong D. W. Y., Lieu Z. Z., Cao X., Yong X. E., Wong J. Z. L., Cheong Y. S., Browder L. K., Chin W. S., Chem. Asian J. 2021, 16, 237–246. [DOI] [PubMed] [Google Scholar]
- 100. Jazayeri S. D., Ideris A., Zakaria Z., Shameli K., Moeini H., Omar A. R., J. Controlled Release 2012, 161, 116–123. [DOI] [PubMed] [Google Scholar]
- 101. Jazayeri S. D., Ideris A., Shameli K., Moeini H., Omar A. R., Int. J. Nanomed. 2013, 8, 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sanchez-Guzman D., Le Guen P., Villeret B., Sola N., Le Borgne R., Guyard A., Kemmel A., Crestani B., Sallenave J. M., Garcia-Verdugo I., Biomaterials 2019, 217, 119308. [DOI] [PubMed] [Google Scholar]
- 103. Chithrani B. D., Ghazani A. A., Chan W. C., Nano Lett. 2006, 6, 662–668. [DOI] [PubMed] [Google Scholar]
- 104. Liu Y., Balachandran Y. L., Li D., Shao Y., Jiang X., ACS Nano 2016, 10, 3589–3596. [DOI] [PubMed] [Google Scholar]
- 105. Karakoti A. S., Das S., Thevuthasan S., Seal S., Angew. Chem. Int. Ed. 2011, 50, 1980–1994; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2011, 123, 2024–2040. [Google Scholar]
- 106. Zhi X., Fang H., Bao C., Shen G., Zhang J., Wang K., Guo S., Wan T., Cui D., Biomaterials 2013, 34, 5254–5261. [DOI] [PubMed] [Google Scholar]
- 107. Müller U., Piehler D., Stenzel W., Köhler G., Frey O., Held J., Grahnert A., Richter T., Eschke M., Kamradt T., Brombacher F., Mucosal Immunol. 2012, 5, 299–310. [DOI] [PubMed] [Google Scholar]
- 108. Zhou J., Ren M., Wang W., Huang L., Lu Z., Song Z., Foda M. F., Zhao L., Han H., Anal. Chem. 2020, 92, 8802–8809. [DOI] [PubMed] [Google Scholar]
- 109. Asgary V., Shoari A., Baghbani-Arani F., Shandiz S. A. S., Khosravy M. S., Janani A., Bigdeli R., Bashar R., Cohan R. A., Int. J. Nanomed. 2016, 11, 3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tian B., Ma J., ZardánGómez de la Torre T., Bálint A., Donolato M., Hansen M. F., Svedlindh P., Strömbe M., ACS Sensors 2016, 1, 1228–1234. [Google Scholar]
- 111. Zhao K., Rong G., Hao Y., Yu L., Kang H., Wang X., Wang X., Jin Z., Ren Z., Li Z., Sci. Rep. 2016, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. He H., Zheng N., Song Z., Kim K. H., Yao C., Zhang R., Zhang C., Huang Y., Uckun F. M., Cheng J., Zhang Y., ACS Nano 2016, 10, 1859–1870. [DOI] [PubMed] [Google Scholar]
- 113. Park T. E., Singh B., Li H., Lee J. Y., Kang S. K., Choi Y. J., Cho C. S., Biomaterials 2015, 38, 61–71. [DOI] [PubMed] [Google Scholar]
- 114. Kataoka C., Suzuki T., Kotani O., Iwata-Yoshikawa N., Nagata N., Ami Y., Wakita T., Nishimura Y., Shimizu H., PLoS Pathog. 2015, 11, e1005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fujii K., Sudaka Y., Takashino A., Kobayashi K., Kataoka C., Suzuki T., Iwata-Yoshikawa N., Kotani O., Ami Y., Shimizu H., Nagata N., J. Virol. 2018, 92. e00682–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Jia J., Zhu F., Ma X., Cao Z. W., Li Y. X., Chen Y. Z., Nat. Rev. Drug Discovery 2009, 8, 111–128. [DOI] [PubMed] [Google Scholar]
- 117. Bell A., FEMS Microbiol. Lett. 2005, 253, 171–184. [DOI] [PubMed] [Google Scholar]
- 118. Baram-Pinto D., Shukla S., Perkas N., Gedanken A., Sarid R., Bioconjugate Chem. 2009, 20, 1497–1502. [DOI] [PubMed] [Google Scholar]
- 119. Lin L. T., Chen T. Y., Chung C. Y., Noyce R. S., Grindley T. B., McCormick C., Lin T. C., Wang G. H., Lin C. C., Richardson C. D., J. Virol. 2011, 85, 4386–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Orlowski P., Krzyzowska M., Zdanowski R., Winnicka A., Nowakowska J., Stankiewicz W., Tomaszewska E., Celichowski G., Grobelny J., Toxicol. in Vitro 2013, 27, 1798–1808. [DOI] [PubMed] [Google Scholar]
- 121. Orlowski P., Tomaszewska E., Gniadek M., Baska P., Nowakowska J., Sokolowska J., Nowak Z., Donten M., Celichowski G., Grobelny J., Krzyzowska M., PLoS One 2014, 9, e104113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Jöbstl E., O′Connell J., Fairclough J. P. A., Williamson M. P., Biomacromolecules 2004, 5, 942–949. [DOI] [PubMed] [Google Scholar]
- 123. Orlowski P., Tomaszewska E., Ranoszek-Soliwoda K., Gniadek M., Labedz O., Malewski T., Nowakowska J., Chodaczek G., Celichowski G., Grobelny J., Krzyzowska M., Front. Immunol. 2018, 9, 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Singh N., Tscharke D. C., J. Virol. 2020, 94, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Orłowski P., Kowalczyk A., Tomaszewska E., Ranoszek-Soliwoda K., Węgrzyn A., Grzesiak J., Celichowski G., Grobelny J., Eriksson K., Krzyzowska M., Viruses 2018, 10, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Srivastava R., Hernández-Ruiz M., Khan A. A., Fouladi M. A., Kim G. J., Ly V. T., Yamada T., Lam C., Sarain S. A., Boldbaatar U., Zlotnik A., J. Immunol. 2018, 200, 2915–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kolodziej H., Kiderlen A. F., Phytochemistry 2005, 66, 2056–2071. [DOI] [PubMed] [Google Scholar]
- 128. Watanabe Y., Shiratsuchi A., Shimizu K., Takizawa T., Nakanishi Y., J. Biol. Chem. 2002, 277, 18222–18228. [DOI] [PubMed] [Google Scholar]
- 129. Wurzer W. J., Planz O., Ehrhardt C., Giner M., Silberzahn T., Pleschka S., Ludwig S., EMBO J. 2003, 22, 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhirnov O. P., Klenk H. D., Apoptosis 2007, 12, 1419–1432. [DOI] [PubMed] [Google Scholar]
- 131. Li Y., Lin Z., Zhao M., Xu T., Wang C., Hua L., Wang H., Xia H., Zhu B., ACS Appl. Mater. Interfaces 2016, 8, 24385–24393. [DOI] [PubMed] [Google Scholar]
- 132. Lin Z., Li Y., Guo M., Xu T., Wang C., Zhao M., Wang H., Chen T., Zhu B., RSC Adv. 2017, 7, 742–750. [Google Scholar]
- 133. Li Y., Lin Z., Zhao M., Guo M., Xu T., Wang C., Xia H., Zhu B., RSC Adv. 2016, 6, 89679–89686. [Google Scholar]
- 134. Lin Z., Li Y., Guo M., Xiao M., Wang C., Zhao M., Xu T., Xia Y., Zhu B., RSC Adv. 2017, 7, 35290–35296. [Google Scholar]
- 135. Lara H. H., Ixtepan-Turrent L., Treviño E.N. G., Singh D. K., J. Nanobiotechnol. 2011, 9, 1–9. [Google Scholar]
- 136. Kharat M., Du Z., Zhang G., McClements D. J., J. Agric. Food Chem. 2017, 65, 1525–1532. [DOI] [PubMed] [Google Scholar]
- 137. Schneider C., Gordon O.N., Edwards R. L., Luis P. B., J. Agric. Food Chem. 2015, 63, 7606–7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Gupta S. C., Patchva S., Aggarwal B. B., AAPS J. 2013, 15, 195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Sharma R. K., Cwiklinski K., Aalinkeel R., Reynolds J. L., Sykes D. E., Quaye E., Oh J., Mahajan S. D., Schwartz S. A., Immunol. Invest. 2017, 46, 833–846. [DOI] [PubMed] [Google Scholar]
- 140. Yang X. X., Li C. M., Huang C. Z., Nanoscale 2016, 8, 3040–3048. [DOI] [PubMed] [Google Scholar]
- 141. Ye S., Shao K., Li Z., Guo N., Zuo Y., Li Q., Lu Z., Chen L., He Q., Han H., ACS Appl. Mater. Interfaces 2015, 7, 21571–21579. [DOI] [PubMed] [Google Scholar]
- 142. Chen Y.N., Hsueh Y. H., Hsieh C. T., Tzou D. Y., Chang P. L., Int. J. Environ. Res. Public Health 2016, 13, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pu F., Huang Y., Yang Z., Qiu H., Ren J., ACS Appl. Mater. Interfaces 2018, 10, 9929–9937. [DOI] [PubMed] [Google Scholar]
- 144. Pedrosa P., Baptista P. V., Nanotechnol. Diagn. Treat. Prophyl. Infect. Diseas. 2015, 1–18. [Google Scholar]
- 145. Kucherenko I. S., Soldatkin O. O., Kucherenko D. Y., Soldatkina O. V., Dzyadevych S. V., Nanoscale Adv. 2019, 1, 4560–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Larguinho M., Baptista P. V., J. Proteom. 2012, 75, 2811–2823. [DOI] [PubMed] [Google Scholar]
- 147. Vinayagam S., Rajaiah P., Mukherjee A., Natarajan C., Spectrochim. Acta Part A: Mol. Biomol.Spect. 2018, 202, 346–351. [DOI] [PubMed] [Google Scholar]
- 148. Ma X., Miao P., J. Mater. Chem. B 2019, 7, 2608–2612. [DOI] [PubMed] [Google Scholar]
- 149. Lim L. S., Chin S. F., Pang S. C., Sum M. S. H., Perera D., Sains Malays. 2017, 46, 2447–2454. [Google Scholar]
- 150. Li Y., Hong M., Qiu B., Lin Z., Cai Z., Chen Y., Chen G., Chem. Commun. 2013, 49, 10563–10565. [DOI] [PubMed] [Google Scholar]
- 151. Luo X., Davis J. J., Chem. Soc. Rev. 2013, 42, 5944–5962. [DOI] [PubMed] [Google Scholar]
- 152. Li X., Scida K., Crooks R. M., Anal. Chem. 2015, 87, 9009–9015. [DOI] [PubMed] [Google Scholar]
- 153. Li H., Sun Z., Zhong W., Hao N., Xu D., Chen H. Y., Anal. Chem. 2010, 82, 5477–5483. [DOI] [PubMed] [Google Scholar]
- 154. Awan M., Rauf S., Abbas A., Nawaz M. H., Yang C., Shahid S. A., Amin N., Hayat A., J. Mol. Liq. 2020, 317, 114014. [Google Scholar]
- 155. Valipour A., Roushani M., Biosens. Bioelectron. 2017, 89, 946–951. [DOI] [PubMed] [Google Scholar]
- 156. Huang J., Xie Z., Xie Z., Luo S., Xie L., Huang L., Fan Q., Zhang Y., Wang S., Zeng T., Anal. Chim. Acta 2016, 913, 121–127. [DOI] [PubMed] [Google Scholar]
- 157. Chin S. F., Lim L. S., Lai H. C., Pang S. C., Sum M. S. H., Perera D., Nano Biomed. Eng. 2019, 11, 333–339. [Google Scholar]
- 158. Khristunova E., Arek J., Kratochvil B., Korotkova E., Dorozhko E., Vyskocil V., Bioelectrochemistry 2020, 135, 107576. [DOI] [PubMed] [Google Scholar]
- 159. Sepunaru L., Plowman B. J., Sokolov S. V., Young N. P., Compton R. G., Chem. Sci. 2016, 7, 3892–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Díez I., Ras R. H., Nanoscale 2011, 3, 1963–1970. [DOI] [PubMed] [Google Scholar]
- 161. Kurdekar A. D., Chunduri L. A., Chelli S. M., Haleyurgirisetty M. K., Bulagonda E. P., Zheng J., Hewlett I. K., Kamisetti V., RSC Adv. 2017, 7, 19863–19877. [Google Scholar]
- 162. Li Y., Hong M., Qiu B., Lin Z., Chen Y., Cai Z., Chen G., Biosens. Bioelectron. 2014, 54, 358–364. [DOI] [PubMed] [Google Scholar]
- 163. Aslan K., Huang J., Wilson G. M., Geddes C. D., J. Am. Chem. Soc. 2006,128, 4206–4207. [DOI] [PubMed] [Google Scholar]
- 164. Jin F., Li H., Xu D., Analyt. Chim. Acta 2019,1077, 297–304.< [DOI] [PubMed] [Google Scholar]
- 165. Shan S., Lai W., Xiong Y., Wei H., Xu H., J. Agric. Food Chem. 2015, 63, 745–753. [DOI] [PubMed] [Google Scholar]
- 166. Yen C. W., de Puig H., Tam J. O., Gómez-Márquez J., Bosch I., Hamad-Schifferli K., Gehrke L., Lab Chip 2015, 15, 1638–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Le Ru E. C., Blackie E., Meyer M., Etchegoin P. G., J. Phys. Chem. C 2007, 111, 13794–13803. [Google Scholar]
- 168. Zhan L., Zhen S. J., Wan X. Y., Gao P. F., Huang C. Z., Talanta 2016, 148, 308–312. [DOI] [PubMed] [Google Scholar]
- 169. Kashif M., Majeed M. I., Hanif M. A., Rehman A., Spectrochim. Acta Part A 2020, 242, 118729. [DOI] [PubMed] [Google Scholar]
- 170. Saraya R. E., Elhenawee M., Saleh H., J. AOAC Int. 2020, 103, 140–147. [DOI] [PubMed] [Google Scholar]
- 171. Aftab S., Kurbanoglu S., Ozcelikay G., Bakirhan N. K., Shah A., Ozkan S. A., Sens. Actuators B 2019, 285, 571–583. [Google Scholar]
- 172. Tang S., Huang Y., Zheng J., Angew. Chem. Int. Ed. 2020, 132, 20066–20070. [Google Scholar]
- 173. Faunce T., Watal A., Nanomedicine 2010, 5, 617–632. [DOI] [PubMed] [Google Scholar]
- 174. Chen X., Schluesener H. J., Toxicol. Lett. 2008, 176, 1–12. [DOI] [PubMed] [Google Scholar]
- 175. Sood R., Chopra D. S., Appl. Clin. Res., Clin. Trial. Regul. Affa. 2018, 5, 74–79. [Google Scholar]
- 176.S. K. Ghosh, Nova Surface-Care Centre Pvt: Maharashtra, India, 2020.
- 177.HeiQ Viroblock NPJ03 antiviral textile technology tested effective against Coronavirus, Zurich, March 16, 2020. See at the URL: https://heiq.com/2020/03/16/heiq-viroblock-antiviraltextile-technology-against-coronavirus (last accessed May 10, 2020).
- 178.BiosilverLab: https://www.biosilverlab.com/.