Key Points
MAIT cells are hypersensitive to the S. aureus LukED toxin.
The effect is dependent on extraordinarily high CCR5 expression.
Activation via inflammatory cytokines or TCR partly protects MAIT cells.
Visual Abstract
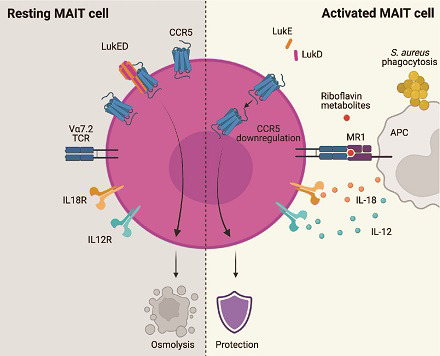
Abstract
Mucosa-associated invariant T (MAIT) cells recognize bacterial riboflavin metabolite Ags presented by MHC class Ib–related protein (MR1) and play important roles in immune control of microbes that synthesize riboflavin. This includes the pathobiont Staphylococcus aureus, which can also express a range of virulence factors, including the secreted toxin leukocidin ED (LukED). In this study, we found that human MAIT cells are hypersensitive to LukED-mediated lysis and lost on exposure to the toxin, leaving a T cell population devoid of MAIT cells. The cytolytic effect of LukED on MAIT cells was rapid and occurred at toxin concentrations lower than those required for toxicity against conventional T cells. Furthermore, this coincided with high MAIT cell expression of CCR5, and loss of these cells was efficiently inhibited by the CCR5 inhibitor maraviroc. Interestingly, exposure and preactivation of MAIT cells with IL-12 and IL-18, or activation via TCR triggering, partially protected from LukED toxicity. Furthermore, analysis of NK cells indicated that LukED targeted the mature cytotoxic CD57+ NK cell subset in a CCR5-independent manner. Overall, these results indicate that LukED efficiently eliminates immune cells that can respond rapidly to S. aureus in an innate fashion without the need for clonal expansion, and that MAIT cells are exceptionally vulnerable to this toxin. Thus, the findings support a model where LukED secretion may allow S. aureus to avoid recognition by the rapid cell-mediated responses mediated by MAIT cells and NK cells.
Introduction
Mucosa-associated invariant T (MAIT) cells belong to the broad and diverse group of unconventional non–MHC-restricted T cells and represent on the order of 1–10% of the total T cell population in humans (1, 2). MAIT cells recognize nonpeptide microbial Ags presented in complex with the MHC class Ib–related protein (MR1) (3, 4). Ags recognized by MAIT cells include intermediates of the vitamin B2 (riboflavin) synthesis pathway expressed by many microbes, and this allows immune surveillance of a broad range of microorganisms in an MR1-restricted fashion (5, 6). MAIT cells are numerous in many human tissues expressing tissue-resident characteristics (7, 8) and also express a range of chemokine receptors for homing to sites of inflammation (9, 10). Upon recognition of Ag presented by MR1, they secrete cytokines, including IFN-γ, TNF, IL-17A, and IL-22 (9, 11), and can participate in tissue repair and wound healing (12–15). Furthermore, MAIT cells kill bacterially infected cells via the release of cytotoxic effector molecules, such as granzyme B (GzmB) and granulysin (16–19), and have direct antimicrobial properties against both cell-associated and free-living bacteria (20). Their role in defense against bacterial infection was demonstrated in several mouse models of infection with pathogens such as Legionella longbeachae, Klebsiella pneumoniae, and Francisella tularensis (21–23). In humans, MAIT cells expand in response to Salmonella enterica subsp. enterica serovar Paratyphi A challenge (24) and migrate to lung tissue during tuberculosis (25, 26).
NK cells are the prototypical innate effector cells and mediate rapid immune responses against microbes and tumor cells (27, 28). Differentiation of CD56bright NK cells to mature cytolytic NK cells is characterized by lowered CD56 expression and the gain of CD16, CD57, and KIR expression (29, 30). During bacterial infection, NK cells can be indirectly activated by cytokines or through interaction with other cell types and can also directly recognize bacteria through TLR sensing (31). Recently, the activating receptor KIR2DS4 was found to recognize a bacterial HLA-C*05:01–presented epitope derived from a protein conserved in many bacterial species, including Staphylococcus aureus (32).
S. aureus is a bacterial pathobiont that colonizes a third of the human population through nasal and skin carriage (33). The shift from commensal microbe to pathogen requires the expression of virulence factors, as well as barrier breach (34, 35). S. aureus affects local tissue and can spread systemically to cause life-threatening diseases, such as pneumonia, endocarditis, and sepsis. Virulence factors that are critical for pathogenesis include superantigens, cytolytic peptides, and pore-forming toxins (36). One of the pore-forming toxins is the leukocidin ED (LukED), which is expressed by a majority of S. aureus isolates (37). This bicomponent toxin is composed of two water-soluble monomers and acts in two steps: LukE first binds to target proteins on the cell membrane and recruits the LukD subunit. The complex then oligomerizes and inserts in the cell membrane as a β-barrel pore, leading to disruption of the cellular osmotic balance and cell death (37). LukED binds to the chemokine receptor CCR5, expressed on macrophages, dendritic cells, and T cells (38). It also binds to CXCR1 and CXCR2, which can be expressed on NK cells and neutrophils (39), as well as to DARC expressed on erythrocytes and endothelial cells (40, 41). LukED contributes to S. aureus pathogenesis in vivo (38–40, 42), and the binding to DARC expressed on epithelial and endothelial cells leads to vascular leakage and organ failure (41). S. aureus ΔLukED mutants are less invasive with reduced bacterial burden and mortality (39, 40, 42).
The role of MAIT cells in S. aureus immunopathogenesis remains relatively little studied. MAIT cells are activated by S. aureus stimulation in vitro to produce IFN-γ (25, 26), and their frequencies are increased in tonsils and blood of individuals with S. aureus tonsillitis (43). MAIT cells are also significant contributors to the massive cytokine release (“cytokine storm”) in response to Staphylococcal enterotoxin B (44, 45). In this study, we investigate the effect of LukED on MAIT cells in comparison with other subsets in the peripheral blood T cell pool, including invariant NK T (iNKT) cells and γδT cells, and dissect the effect of LukED on MAIT cell recognition of S. aureus. Furthermore, we investigate how LukED affects the NK cell compartment. Altogether, the findings suggest that LukED secretion by S. aureus constitutes an immune evasion mechanism to interfere with responses mediated by human innate effector cells.
Materials and Methods
Blood donors and cell lines
Peripheral blood was collected from healthy adults recruited at the Blood Transfusion Clinic at the Karolinska University Hospital Huddinge. All blood donors gave written informed consent in accordance with study protocols conforming to the provisions of the Declaration of Helsinki and approved by the Regional Ethics Review Board in Stockholm. The THP-1 cell line (ATCC, Manassas, VA, USA) was cultured in RPMI 1640 complete medium supplemented with 25 mM HEPES, 2 mM l-glutamine (GE Healthcare), 10% FBS (Sigma-Aldrich), 50 μg/ml gentamicin (Thermo Fisher Scientific), and 100 μg/ml Normocin (Invivogen) and routinely tested negative for mycoplasma.
Microbes
S. aureus strains 134 and 289 were cultured overnight at 37°C in CCY Broth. Bacterial counts were determined by the standard plate counting method on appropriate culture media, and counts were expressed as CFUs per milliliter (CFU/ml). The microbes were then stored at −80°C in 50% glycerol/50% PBS.
Cell isolation
PBMCs were isolated from peripheral blood by Ficoll–Hypaque density gradient centrifugation (Lymphoprep, Axis-Shield). After isolation, PBMCs were rested overnight in RPMI 1640 medium supplemented with 25 mM HEPES, 2 mM l-glutamine (all from Thermo Fisher Scientific), 10% FBS (Sigma-Aldrich), 50 μg/ml gentamicin (Life Technologies), and 100 μg/ml normocin (InvivoGen) (complete medium). From PBMCs, we isolated Vα7.2+ cells using anti-Vα7.2 PE-conjugated mAb (BioLegend), followed by positive selection with MACS and anti-PE microbeads (Miltenyi Biotec), as previously described and per the manufacturer’s instructions (19, 46).
Supernatant assay
Clinical S. aureus strains were cultured overnight in CCY medium at 37°C. Supernatant of the culture was harvested by centrifugation and sterilized through 0.2-μm pore size filters to obtain the bacterial-free culture supernatant. PBMCs were incubated with the supernatant at 1:20 dilution for 4 h.
LukED assays
PBMCs were incubated with recombinant LukED (IBT Bioservices) at the indicated concentration between 45 min and 6 h, as indicated in the text. In selected experiments, maraviroc (MVC; Sigma) at 1 μg/ml was added just before the recombinant LukED.
MAIT cell activation assays
THP-1 cells were seeded in complete medium for 2 h prior to pulsing with bacteria. S. aureus 134 and 289 were washed once in PBS before fixation in 1% formaldehyde for 3 min and extensive PBS washes. The bacteria were then resuspended in complete medium and fed to THP-1 monocytic cells at the microbial dose of 30. In some experiments, Vα7.2+ cells were preincubated for 1 h and 30 min with LukED at 0.312 μg/ml before addition to the bacteria-pulsed THP-1 and coculture at 2:1 ratio for 24 h in the presence of 1.25 μg/ml anti-CD28 mAb (L293; BD Biosciences). Monensin and brefeldin A (both from BD Biosciences) were added at the last 6 h of culture. In selected experiments, the THP-1/Vα7.2+ cell coculture was incubated with LukED at 5 μg/ml for the last 6 h of culture. For TCR-independent activation, PBMCs were incubated for 20 h with IL-12 (PeproTech) and IL-18 (MBL) at 10 and 100 ng/ml, respectively. LukED at 1.25 μg/ml was added for the last 2 h of culture in some conditions.
Flow cytometry
Tetramer staining with 5-OP-RU–loaded h-MR1-PE (NIH Tetramer Core Facility) was performed at 20 min at room temperature before staining with mAbs for other markers. Abs used included anti-CD3 Bv650 (OKT3; BioLegend), anti-CD3 AF700 (UCHT1; BD Biosciences), anti-CD3 FITC (SK7; BD Biosciences), anti-Vα7.2 PE (3C10; BioLegend), anti-Vα7.2 PE-Cy7 (3C10; BioLegend), anti-CD161 Pe-Cy5 (DX12; BD Biosciences), anti-CD4 Bv711 (OKT4; BioLegend), anti-CD8 Bv570 (RPA-T8; BioLegend), anti-CD56 BUV737 (NCAM16.2; BD Biosciences), anti-CCR7 Bv421 (150503; BD Biosciences), anti-CD45RA AF700 (HI100; BD Biosciences), anti-CCR5 BUV395 (2D7/CCR5; BD Biosciences), anti-CXCR1 AF488 (8F1/CXCR1; BD Biosciences), anti-CXCR2 Bv785 (6C6; BD Biosciences), anti-CD158b Bv510 (CH-L; BD Biosciences), anti-CD16 Bv711 (3G8; BD Biosciences), anti-CD57 PE-Cy5 (NK-1; BD Biosciences), anti-CD27 PE-Cy7 (M-T271; BioLegend), anti-NKG2A allophycocyanin (REA110; Miltenyi), anti-CD14 allophycocyanin-Cy7 (MѻP9; BD Biosciences), anti-CD19 allophycocyanin-Cy7 (SJ27C1; BD Biosciences), anti-Vα24 PE (C15; Beckman Coulter), anti-Vβ11 FITC (C21; Beckman Coulter), anti-γδ TCR PE-Dazzle 594 (B1; BioLegend), anti-CD69 BUV737 (FN50; BD Biosciences), anti-GzmB FITC (GB11; BioLegend), anti–IFN-γ allophycocyanin (25723.11; BD Biosciences), anti-TNF PE-Cy7 (Mab11; BD Biosciences), anti–IL-17A Bv421 (BL168; BioLegend), anti-Perforin Bv421 (B-D48; BioLegend), and anti-Granulysin (DH2; BD Biosciences) LIVE/DEAD Fixable Near-IR dye (Invitrogen). Flow cytometry data were acquired on BD LSRFortessa or BD Symphony A5 instruments (both BD Biosciences) and analyzed using FlowJo software v.10.5.3 and v.10.8.0 (Tree Star).
Statistics
Statistical analyses were performed using Prism software v.7 (GraphPad). Datasets were first assessed for normality of the data distribution. Statistically significant differences between experimental conditions were determined as appropriate using the unpaired t test or Kruskal–Wallis test and Mann–Whitney test for unpaired samples, and the paired t test or Friedman test and Wilcoxon signed-rank test for paired samples. Correlations were assessed using the Spearman rank correlation. Two-sided p values <0.05 were considered significant.
Results
Human MAIT cells respond to S. aureus with robust production of cytokines and GzmB
MAIT cells have been previously reported to respond to S. aureus stimulation in vitro with production of IFN-γ (25, 26). To confirm MAIT cell responsiveness to this microbe in our laboratory, we pulsed THP-1 cells with mildly fixed S. aureus for 2 h and used these to stimulate Vα7.2+ cells isolated from PBMCs. After a 24-h coincubation, MAIT cells showed robust activation with expression of CD69 and expression of high levels of IFN-γ, TNF, and GzmB, with detectable but relatively low production of IL-17A (Fig. 1A, 1B).
FIGURE 1.
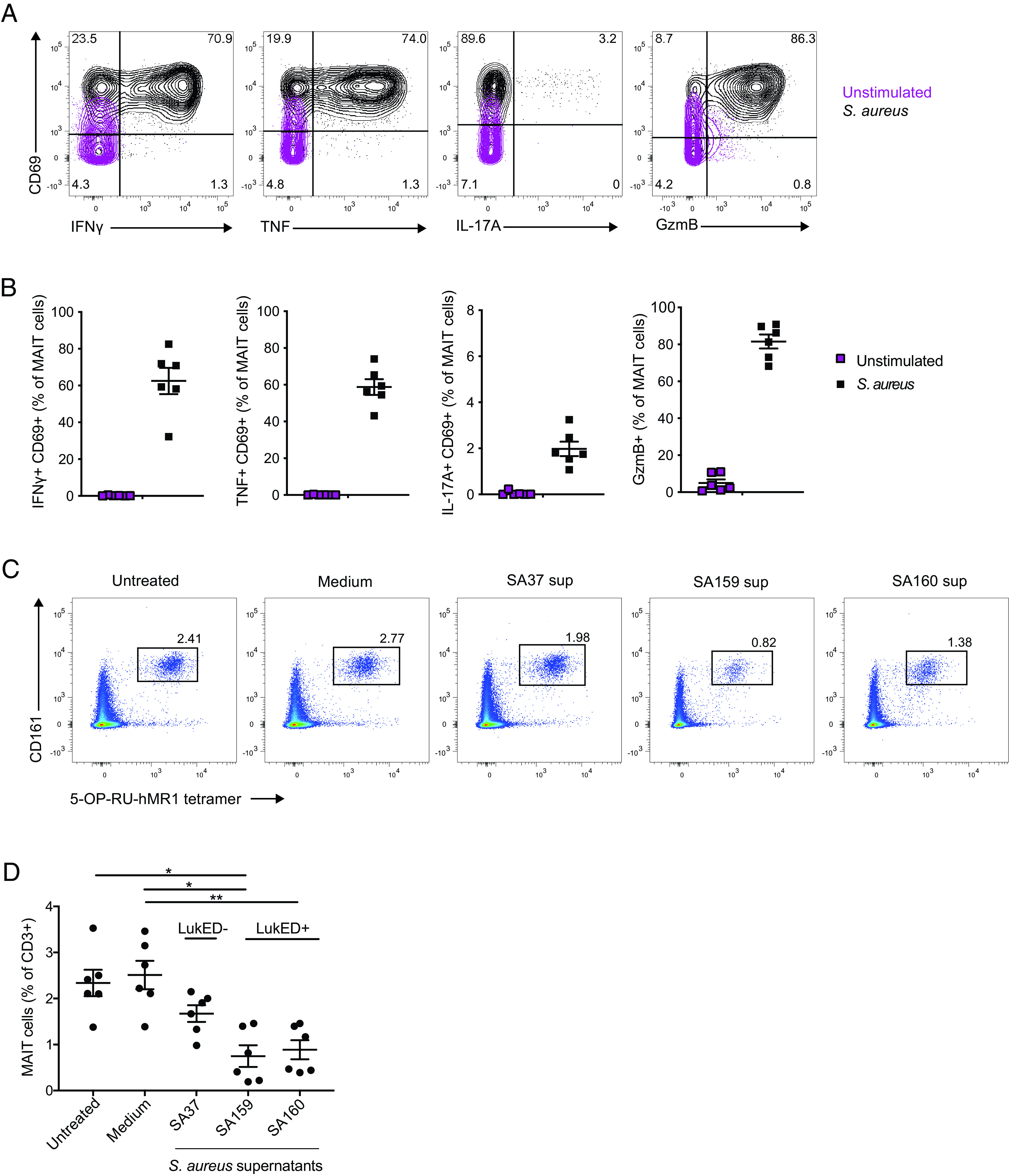
MAIT cells respond to S. aureus–pulsed THP-1 cells. Representative flow cytometry plots (A) and combined donor data (B) of the expression of IFN-γ+CD69+, TNF+CD69+, IL-17A+CD69+, and GzmB+ MAIT cells stimulated with THP-1 fed mildly fixed S. aureus for 24 h at the microbial dose of 10 (n = 6). Representative flow cytometry plots (C) and combined donor data (D) of MAIT cell percentage on exposure to clinical strains of supernatants or medium (broth) alone (n = 6). The Friedman test followed by Dunn post hoc test was used to determine significant differences between multiple paired samples in (D). The lines and error bars represent mean and SE. *p < 0.05, **p < 0.005.
To investigate possible effects of soluble factors produced by S. aureus on MAIT cells, we incubated PBMCs with culture supernatants of three S. aureus clinical isolates previously characterized regarding their toxin gene profile (47). The three strains studied varied regarding the presence of the lukED toxin genes. Interestingly, PBMC cultures exposed to the supernatants of lukED-positive strains lost a significant fraction of their MAIT cells, as compared with cultures exposed to the lukED-negative strain (Fig. 1C, 1D). Together, these results confirm that MAIT cells respond strongly to S. aureus and suggest that they may be sensitive to the LukED toxin.
MAIT cells are hypersensitive to LukED cytotoxicity
To specifically investigate the effect of the LukED toxin on human peripheral blood T cells in more detail, we incubated PBMCs in the presence of recombinant LukE and LukD proteins and assessed population changes using flow cytometry. The total lymphocyte population was first analyzed using Uniform Manifold Approximation and Projection (UMAP) analysis (48) of healthy donor PBMCs exposed to LukED. Cell populations were identified by the projection of defining markers on the UMAP topography (Fig. 2A). Projection of LukED-treated versus untreated conditions revealed differences between the two settings and the loss of some cell populations (Fig. 2B). Strikingly, the UMAP area defined by the 5-OP-RU-hMR1 tetramer was almost completely absent after exposure to LukED, suggesting that exposure to the toxin depletes MAIT cells. The profound reduction of MAIT cells, defined as CD3+, CD161high, 5-OP-RU-hMR1+, was confirmed both as percentage and absolute count (Fig. 2C). In contrast, non-MAIT T cell populations were only slightly affected by the toxin with a decline of 25% (Fig. 2D) versus 97% for MAIT cells (Fig. 2E). The single components of the toxin alone, LukE and LukD, did not affect the T cell compartment in a detectable manner (Supplemental Fig. 1A, 1B). Because LukED was previously shown to lyse T cells in a CCR5-dependent fashion (30), we next specifically analyzed the CCR5+ non-MAIT T cell subset. We noticed a decrease of this population (Fig. 2F), which was 80% depleted on LukED exposure (Fig. 2G) compared with 97% for MAIT cells. Notably, MAIT cells have homogenous expression of CCR5 (Supplemental Fig. 1C) and display a higher level of expression compared with CCR5+ non-MAIT T cells (Fig. 2H, Supplemental Fig. 1D).
FIGURE 2.
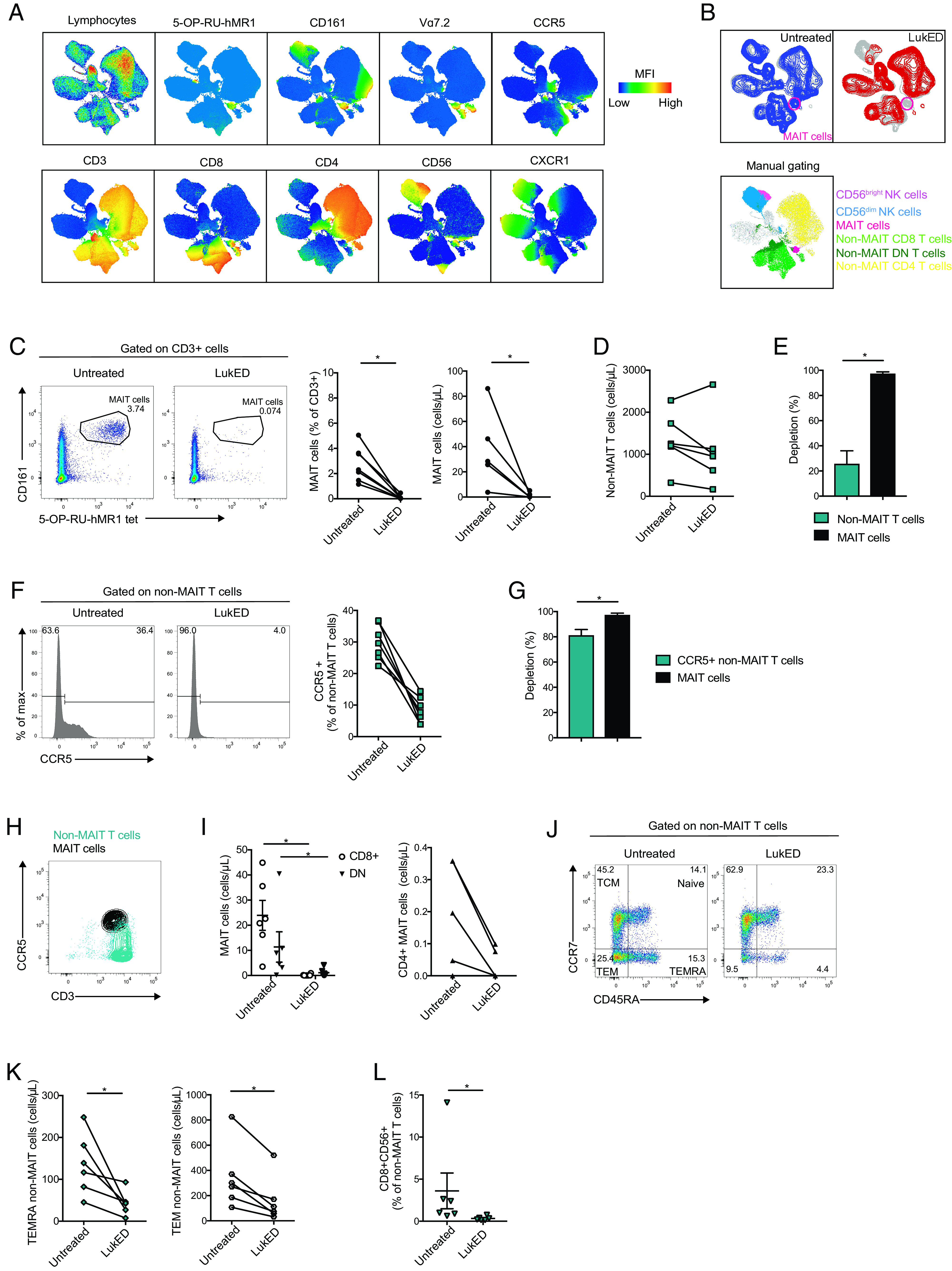
MAIT cells are the main LukED targets within the T cell pool. (A) UMAP plots of total live blood lymphocytes showing expression of the indicated markers. (B) UMAP plots of total lymphocytes exposed to LukED or not (n = 6). Bottom: overlay of immune subset identified by manual gating on the UMAP of total lymphocytes. (C) Representative flow cytometry plots, mean percentage, and absolute counts of MAIT cells on LukED treatment or not (n = 6–7). (D) Absolute counts of non-MAIT T cells on LukED exposure or not (n = 6). (E) Percentage depletion on LukED exposure between MAIT and non-MAIT T cells (n = 6). (F) Representative flow cytometry histograms and mean percentages of expression of CCR5+ non-MAIT T cells on LukED treatment or not (n = 6). (G) Percentages of depletion on LukED exposure between MAIT cells and CCR5+ non-MAIT T cells (n = 6). (H) Representative flow cytometry plot of CCR5 expression in MAIT cells and non-MAIT T cells (n = 6). (I) Absolute counts of CD8+, CD4+, and DN MAIT cells on LukED treatment or not (n = 6). Representative flow cytometry plot of CCR7 and CD45RA expression (J) and absolute counts (K) of non-MAIT T cells with or without LukED (n = 6). (L) Percentages of CD8+CD56+ non-MAIT T cells with or without LukED (n = 6). LukED was used at 5 µg/ml and incubated for 4 h. The Wilcoxon signed-rank test was performed to detect significance in (C), (E), (G), (I), (K), and (L). The lines and error bars represent mean and SE. *p < 0.05.
Within the MAIT cell compartment, CD8+ and CD8−CD4− double-negative (DN) MAIT cells appeared to be slightly more sensitive to LukED than the minor CD4+ MAIT cell subpopulation (Fig. 2I), probably because of their relatively higher expression of CCR5 (Supplemental Fig. 1E). No significant difference was noted in CD8+, CD4+, and DN non-MAIT T cells on LukED exposure (Supplemental Fig. 1F). Next, we explored the composition of the non-MAIT T cell population based on the expression of differentiation markers CCR7 and CD45RA. Terminally differentiated effector memory RA+ (TEMRA) and effector memory T (TEM) cell populations decreased in frequency on LukED exposure (Fig. 2J, 2K) contrary to central memory and naive T cells (Supplemental Fig. 1G). This coincided with higher levels of CXCR1 and CCR5 expression by TEMRA and TEM cells, respectively (Supplemental Fig. 1H). Projection of the LukED-treated condition on the UMAP also revealed a dramatic loss of cells double-expressing CD8 and CD56 (Fig. 2B). The decrease of CD8+CD56+ non-MAIT T cells was confirmed by manual gating (Fig. 2L) and coincided with coexpression of CCR5 and CXCR1 on this subset (Supplemental Fig. 1J). Overall, the loss of TEMRA, TEM, and CD8+CD56+ non-MAIT T cells was not as severe as the depletion of MAIT cells (Supplemental Fig. 1I, 1K). In a separate dataset, we investigated the effect of LukED on two other unconventional T cell populations, γδ T cells and iNKT cells. The effect of LukED on γδ T cells was modest, in line with their low level of CCR5 and CXCR1 expression. In contrast, iNKT cells were more severely depleted by LukED, but still less so than were MAIT cells (Supplemental Fig. 1L–N). Taken together, these findings indicate that LukED depletes T cell subsets with effector and effector memory characteristics, with MAIT cells being the major targeted population.
MVC rescues MAIT cells from LukED toxicity
To investigate whether LukED toxicity against MAIT cells is CCR5 dependent, we evaluated the ability of MVC, a CCR5 antagonist used in HIV therapy, to protect MAIT cells from the recombinant toxin. Addition of MVC to the assay largely rescued the MAIT cell population from LukED toxicity (Fig. 3A). Although this protective effect was incomplete, it was observed for all the main CD8, CD4, and DN MAIT cell subsets (Supplemental Fig. 2A). MVC did not have a detectable effect on the overall non-MAIT T cells bulk population (Fig. 3B) but seemed to partially rescue the CCR5+ subset of non-MAIT T cells (Fig. 3C). Similarly, there was a trend toward partial MVC rescue of TEMRA, TEM, and CD8+CD56+ non-MAIT T cells, but this effect did not reach statistical significance (Supplemental Fig. 2B, 2C). Because some of those subsets express CXCR1, LukED binding to this receptor would not be expected to be inhibited by MVC. Altogether, these results indicate that MVC inhibits the toxicity of LukED against MAIT cells in vitro.
FIGURE 3.
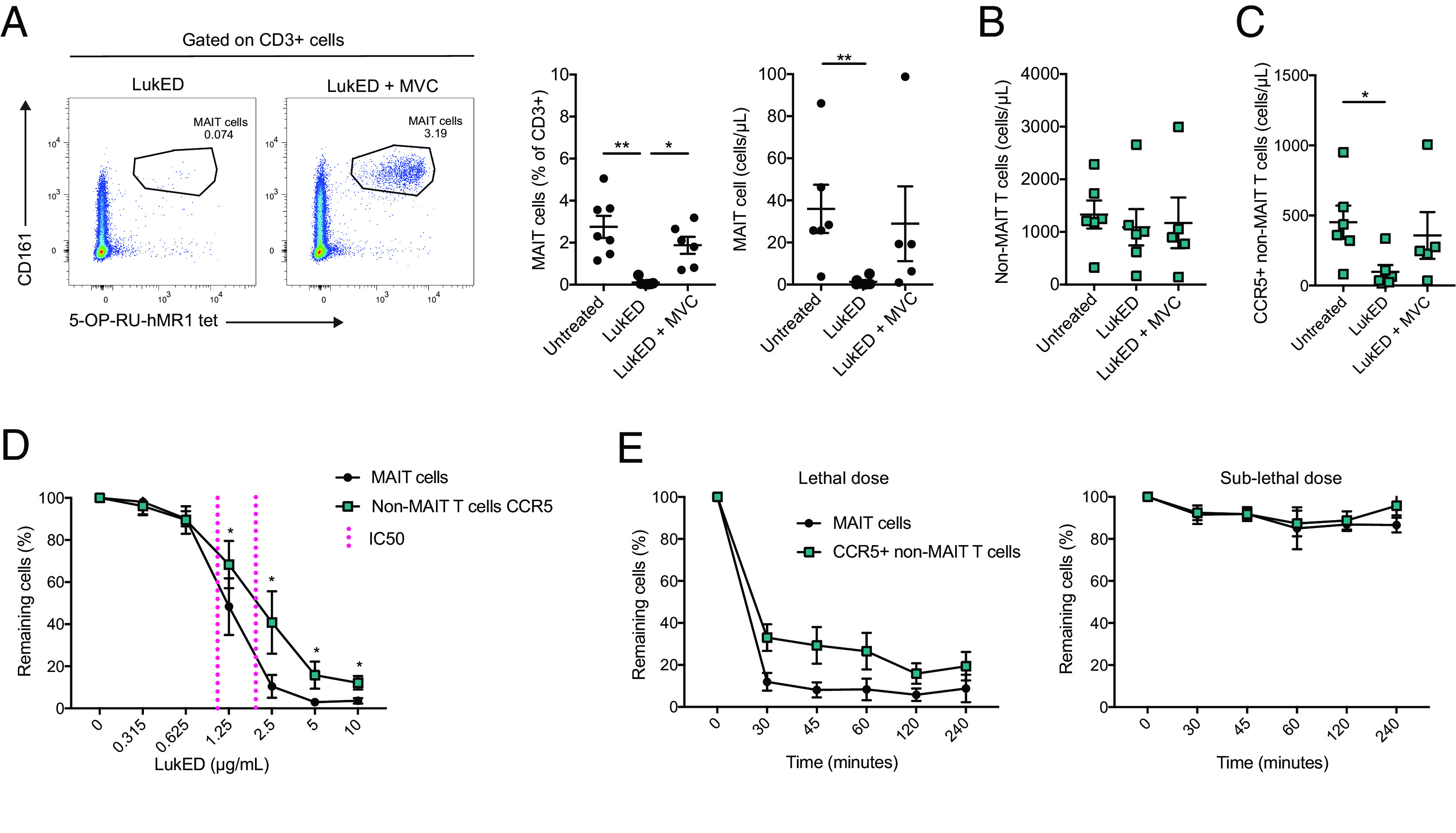
MVC rescues MAIT cells from LukED in vitro. (A) Representative flow cytometry plots, mean percentage, and absolute count of MAIT cells on LukED treatment with or without MVC (n = 6–7). MVC is used at 1 µg/ml. LukED was used at 5 µg/ml for 4 h in (A)–(C). (B) Absolute cell counts of non-MAIT T cells on LukED exposure with or without MVC (n = 5–6). (C) Absolute counts of CCR5+ non-MAIT T cells on LukED exposure with or without MVC (n = 5–6). (D) Relative percentages of MAIT cells and CCR5+ non-MAIT T cells over a range of LukED concentrations after 2 h of exposure. The IC50 is indicated in magenta (n = 6). (E) Relative percentages of MAIT cells and CCR5+ non-MAIT T cells after exposure to a high dose of LukED (5 µg/ml) and low dose of LukED (0.625 µg/ml) during a time course from 30 min to 4 h (n = 6). The Kruskal–Wallis test followed by Dunn post hoc test was used to detect significant differences in (A) and (C). The Wilcoxon signed-rank test was performed to detect significance between the two populations in (D). The lines and error bars represent mean and SE. *p < 0.05, **p < 0.005.
LukED depletes MAIT cells with rapid kinetics and at low doses
We next evaluated the dose sensitivity and kinetics of action of LukED on MAIT cells and CCR5+ non-MAIT T cells. Titration curves revealed dose-dependent depletion of MAIT cells, as well as of CCR5+ non-MAIT T cells (Fig. 3D). Interestingly, at the dose range from 1.25 to 10 µg/ml, MAIT cells were more sensitive than CCR5+ non-MAIT T cells. Furthermore, the amount of LukED needed to kill half of the population (IC50) was lower for MAIT cells (1.09 µg/ml) than for CCR5+ non-MAIT T cells (2.17 µg/ml) (Fig. 3A, magenta reference line). Thus, MAIT cells are more vulnerable to LukED at intermediate doses. Depletion occurred rapidly, within 30 min of incubation, and longer duration of LukED exposure did not substantially increase MAIT cell and CCR5+ non-MAIT T cell depletion (Fig. 3E). Altogether, these findings showed that the lethal effect of LukED on MAIT cells is rapid, dose dependent, and occurs at lower doses compared with conventional T cells.
MAIT cell activation partly prevents LukED-mediated loss
MAIT cells can be activated in response to innate cytokines produced in the setting of myeloid cell activation, such as IL-12 and IL-18. Surprisingly, MAIT cells activated by IL-12 and IL-18 for 20 h, and then exposed to LukED for the last 2 h of incubation, were largely preserved as compared with the nonactivated control MAIT cells (Fig. 4A, 4B). This effect coincided with a decrease in CCR5 expression on IL-12 and IL-18 stimulation (Fig. 4C). Similarly, MAIT cells activated by recognition of S. aureus 134 in a stimulation assay over 24 h and exposed to LukED for the last 6 h of incubation were largely protected from LukED toxicity (Fig. 4D, 4E). This effect was paired with the downregulation of CCR5 on stimulation with S. aureus 134 (Fig. 4F). It is thus possible that activation by innate cytokines in the inflammatory milieu, or direct recognition of bacteria in a TCR-dependent manner, may render MAIT cells partly resistant to LukED.
FIGURE 4.
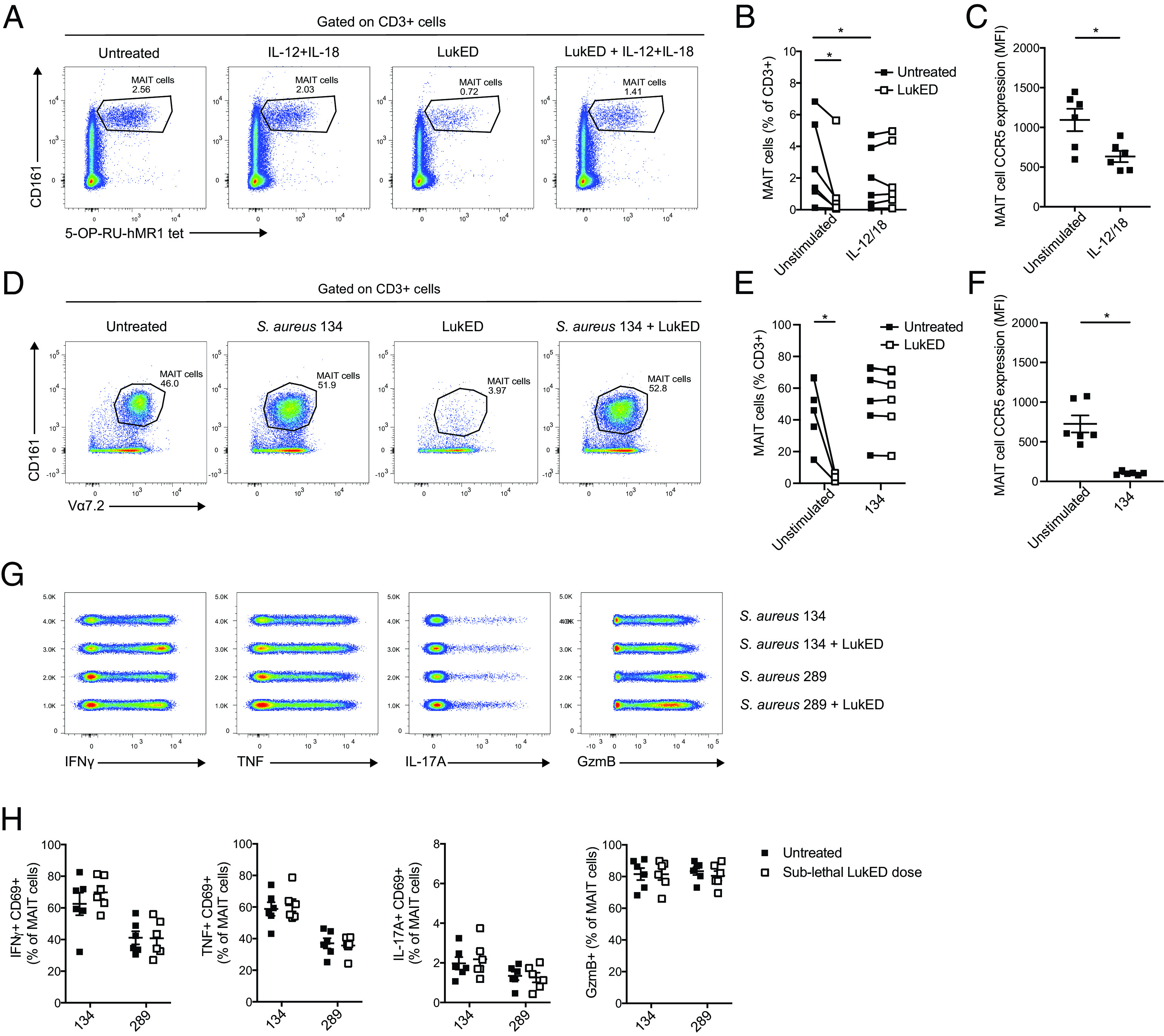
MAIT cell activation with innate cytokines or recognition of bacteria renders them partly resistant to LukED. Representative flow cytometry plots (A) and levels (B) of MAIT cells after 20-h stimulation with IL-12 and IL-18, in the presence or absence of LukED at 1.25 µg/ml during the last 2 h of stimulation (n = 6). (C) CCR5 expression on MAIT cells stimulated 18 h with IL-12 and IL-18 (n = 6). Representative flow cytometry plots (D) and percentage (E) of MAIT cells after 24-h stimulation with S. aureus strain 134 at MOI 30, in the presence of LukED at 5 µg/ml during the last 6 h of incubation (n = 6). (F) CCR5 expression on MAIT cells stimulated for 24 h with S. aureus 134 (n = 6). Concatenated flow cytometry plots (G) and combined data (H) of the expression of IFN-γ+CD69+, TNF+CD69+, IL-17A+CD69+, and GzmB+ MAIT cells untreated or stimulated with THP-1–fed S. aureus 134 (LukED negative) or 289 (LukED positive) for 24 h at the microbial dose of 30, after exposure to a sublethal dose of LukED (0.312 µg/ml). The Wilcoxon signed-rank test in (B), (C), and (F) and the Mann–Whitney U test in (E) were performed to detect significance. The lines and error bars represent mean and SE. *p < 0.05.
Sublethal doses of LukED have no major impact on MAIT cell responsiveness
Because LukED is a pore-forming toxin, it may potentially affect signaling in MAIT cells exposed to concentrations insufficient for direct killing of the cells. We therefore next sought to determine whether exposure to sublethal doses of LukED affects TCR-dependent activation of MAIT cells. To test this, we pretreated MAIT cells with sublethal doses of LukED before stimulation with the mildly fixed LukED-positive S. aureus strain 134 or the LukED-negative strain 289. No detectable difference was observed in MAIT cell IFN-γ, TNF, IL-17A, and GzmB production between the sublethal dose LukED-treated and -untreated conditions (Fig. 4G, 4H). These results indicate that sublethal levels of LukED have no detectable impact on MAIT cell function and ability to respond to bacteria.
LukED additionally targets the mature CD57+ NK cell subset
In the UMAP topography (Fig. 2A, 2B), apart from MAIT cells, another area severely affected by LukED was the one dominated by CD56+CD3− cells corresponding to the NK cell population. LukED severely reduced both CD56bright and CD56dim NK cells among PBMCs both as percentage and as absolute count (Fig. 5A), and the CD56dim subset was particularly severely impaired (Fig. 5B). LukE and LukD alone did not affect NK cells (Supplemental Fig. 3A). In line with the differential activity of LukED on the two NK cell subsets, CD56dim NK cells had higher expression of CXCR1 and CXCR2 than CD56bright NK cells (Fig. 5C, 5D). In general, NK cells expressed low levels of CCR5, although some CD56bright NK cells expressed it (Fig. 5C, 5D). In line with the limited expression of CCR5, MVC had limited impact on the LukED-mediated depletion (Supplemental Fig. 3B).
FIGURE 5.
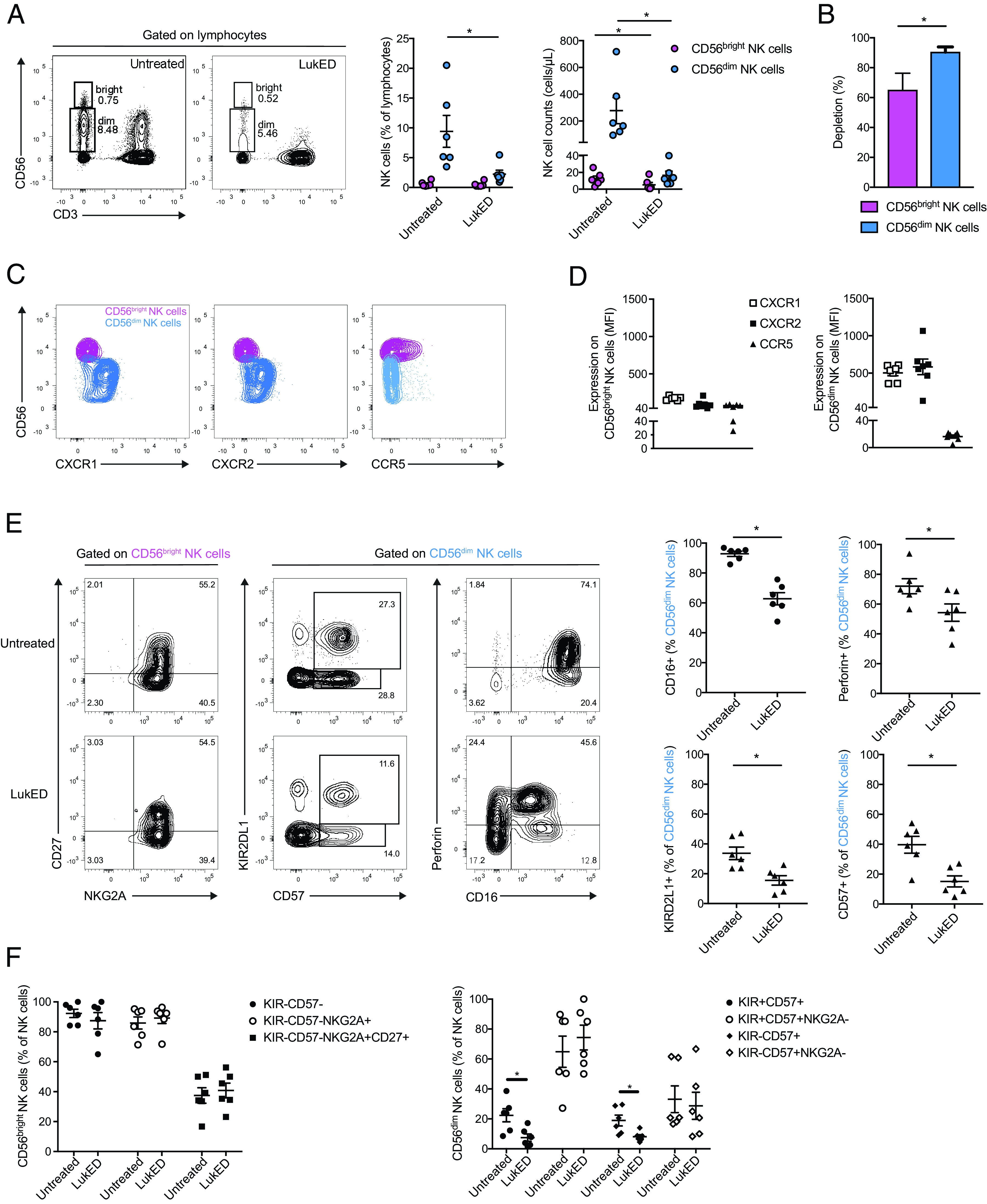
LukED targets mature CD56dim NK cells mostly independent of CCR5. (A) Representative flow cytometry plots and combined donor data of the percentage and absolute count of CD56bright and CD56dim NK cells on LukED treatment for 4 h (n = 6). LukED is used at 5 µg/ml throughout the figure. (B) Percentage of depletion of CD56bright and CD56dim NK cells on LukED treatment (n = 6). Representative flow cytometry plots (C) and combined data (D) of the expression of CXCR1, CXCR2, and CCR5 on CD56bright and CD56dim NK cells (n = 6). (E) Representative flow cytometry plots and combined data of the expression of CD27 and NKG2A on CD56bright and the expression of KIRD2L1, CD57, Perforin, and CD16 on CD56dim NK cells with or without LukED treatment (n = 6). (F) Combined data of the percentage of coexpression of the indicated markers on CD56bright (left) and CD56dim (right) NK cells with or without LukED exposure (n = 6). The Wilcoxon signed-rank test was performed to determine significance. The lines and error bars represent mean and SE. *p < 0.05.
Dissection of the NK cell compartment revealed that LukED targeted the most mature NK cells known to have cytolytic effector properties. In particular, the toxin diminished CD56dim NK cells expressing CD16, CD57, KIRD2L1, and perforin, or coexpressing CD57 and KIRD2L1 (Fig. 5E, 5F). LukED exposure did not change the representation of NK cell populations expressing granulysin or CD27 (Supplemental Fig. 3C). Taken together, these observations indicate that LukED preferentially targets mature cytolytic NK cells.
Discussion
The soluble virulence factors secreted by S. aureus play a major role in the pathogenesis of this infection. However, it is still not fully understood how these virulence factors affect the human immune system. MAIT cells are an important evolutionarily conserved part of the rapid antimicrobial immune response due to their specificity toward conserved bacterial Ag, their secretion of IFN-γ, IL-17, and IL-22, as well as their presence in skin and blood, the main sites of S. aureus infection and pathogenesis (49). In this study, we explored the effects of the LukED toxin on the human immune system and found that MAIT cells are hypersensitive to LukED, because of their very high surface expression of CCR5. LukED depleted MAIT cells more efficiently, with lower IC50, than other T cell subsets, such as conventional TEM and TEMRA cells, as well as the innate-like iNKT and γδT cells. The mature effector CD56dim NK cells were also depleted to a large extent, while CD56bright NK cells were less affected.
MAIT cells and NK cells are important effector cells involved in the host response against bacterial infection. MAIT cells have cytotoxic properties and can directly kill bacteria (17, 18, 20). NK cells are activated by S. aureus (50) and contribute to immune defense against this infection in vivo (51). In the absence of Th1 and Th17 immunity, the susceptibility to S. aureus infection is increased (49). Thus, the findings of this study indicate that LukED targets cells have the capacity to mount a rapid effector response against S. aureus. In particular, the rapid and almost complete elimination of MAIT cells by LukED may represent a possible immune evasion mechanism affecting the outcome of S. aureus infection. Interestingly, recent findings indicate that pathogenic Salmonella Typhimurium sequence type 313 strains escape MAIT cell recognition through overexpression of the RibB enzyme, thereby severely limiting the production of MR1-presented Ag (52). Thus, emerging evidence indicates that several distinct species of bacterial pathogens have evolved mechanisms to avoid innate immune control by MAIT cells.
The CCR5 antagonist MVC inhibits the effect of LukED on T cells. Because MVC is approved for use in humans, this opens the possibility that this inhibitor could be used to block MAIT cell depletion and restore immune control in patients with invasive S. aureus infection. It is interesting to note that several new treatments are in development for S. aureus infection, including mAbs aimed to neutralize S. aureus toxins (53). In experimental animal models of infection in mice (54) and rabbits (55), the combination of mAbs, including one targeting LukED, shows enhanced efficacy over single Ab administration with decreased bacterial burden (54) or increased survival (55). In addition, new neutralizing agents called centyrins are able to block the binding of the five bicomponent leukocidins to their receptors and protect in vivo against S. aureus infection (56).
S. aureus is a prominent pathogen in the hospital setting, and the treatment is complicated by the emergence of methicillin-resistant S. aureus with acquisition of a multiresistance profile (35, 57). MAIT cells were recently shown to recognize antibiotic-resistant bacteria, and their effector functions can overcome certain bacterial resistance mechanisms (20, 58). Furthermore, our findings indicate that MAIT cell functionality was retained on exposure to sublethal doses of LukED, supporting the notion that low concentrations of toxin in the microenvironment do not disrupt signaling in MAIT cells. In this context, neutralization of these S. aureus virulence factors may rescue the effector function of MAIT cells, even if neutralization is not complete.
Finally, we speculate that the downregulation of CCR5 on MAIT cells on activation with IL-12 and IL-18, or via direct recognition of bacterial Ag, may allow MAIT cells to evade LukED toxicity. MAIT cells recruited to tissue sites in the context of innate immune activation in response to bacteria can possibly in this way be partially protected against the toxin. In the absence of such inflammatory cytokine activation, MAIT cells at a site of infection may be sensitive to LukED until they recognize that bacterial Ag and CCR5 downregulation occurs. Thus, in the early stages of infection, the invading bacteria may have a limited window in time to evade effector cells, and it is possible this initial period is important for the ability of S. aureus to establish infection. In contrast, the effect of activation on CCR5 expression in MAIT cells may perhaps be harnessed to promote a more resistant MAIT cell phenotype for immunotherapeutic purposes. In summary, we show that the LukED toxin is a major killer of innate cytolytic effector immune cells. Notably, MAIT cells are hypersensitive to the toxin compared with other types of cytolytic effector cells, such as conventional adaptive T cells, γδ T cells, iNKT cells, and NK cells. These findings support a model where LukED targeting of CCR5 may function as an S. aureus immune evasion mechanism to avoid rapid MAIT cell detection and elimination.
Supplementary Material
Acknowledgments
The MR1 tetramer technology was developed jointly by Dr. James McCluskey, Dr. Jamie Rossjohn, and Dr. David Fairlie, and the material was produced by the NIH Tetramer Core Facility as permitted to be distributed by the University of Melbourne.
This work was supported by grants from Vetenskapsrådet (Swedish Research Council) (2020-01743 to J.K.S.; 2015-00174 to E.L.; 2018-02475 and 2018-131 under the frame of ERA PerMED to A.N.-T.), Cancerfonden (Swedish Cancer Society) (20-1142PjF to J.K.S.), the Hjärt-Lungfondens (Swedish Heart-Lung Foundation) (20180675 to J.K.S.), and Center for Innovative Medicine (20190732 to J.K.S.; 20180058 to A.N.-T.). Further support was provided by Marie Skłodowska Curie Actions, Cofund (E.L.) and VINNOVA (Swedish Governmental Agency for Innovation Systems) under the frame of NordForsk (90456 to A.N.-T.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
J.K.S. and A.N.-T. designed the study. C.B. performed experiments. S.M.S. and A.N.-T. characterized clinical strains. C.B., E.L., A.N.-T., and J.K.S. interpreted the data. C.B. and J.K.S. wrote the paper. All authors read and revised the manuscript.
The online version of this article contains supplemental material.
- DN
- double negative
- GzmB
- granzyme B
- iNKT
- invariant NKT
- LukED
- leukocidin ED
- MAIT
- mucosa-associated invariant T
- MR1
- MHC class Ib–related protein
- MVC
- maraviroc
- TEM
- effector memory T
- TEMRA
- terminally differentiated effector memory RA+
- UMAP
- Uniform Manifold Approximation and Projection
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Provine N. M., Klenerman P.. 2020. MAIT cells in health and disease. Annu. Rev. Immunol. 38: 203–228. [DOI] [PubMed] [Google Scholar]
- 2. Godfrey D. I., Koay H. F., McCluskey J., Gherardin N. A.. 2019. The biology and functional importance of MAIT cells. Nat. Immunol. 20: 1110–1128. [DOI] [PubMed] [Google Scholar]
- 3. Treiner E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., Lantz O.. 2003. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. [Published erratum appears in 2003 Nature 423: 1018.] Nature 422: 164–169. [DOI] [PubMed] [Google Scholar]
- 4. Kjer-Nielsen L., Patel O., Corbett A. J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491: 717–723. [DOI] [PubMed] [Google Scholar]
- 5. Corbett A. J., Eckle S. B., Birkinshaw R. W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509: 361–365. [DOI] [PubMed] [Google Scholar]
- 6. Toubal A., Nel I., Lotersztajn S., Lehuen A.. 2019. Mucosal-associated invariant T cells and disease. Nat. Rev. Immunol. 19: 643–657. [DOI] [PubMed] [Google Scholar]
- 7. Sobkowiak M. J., Davanian H., Heymann R., Gibbs A., Emgård J., Dias J., Aleman S., Krüger-Weiner C., Moll M., Tjernlund A., et al. 2019. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur. J. Immunol. 49: 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salou M., Legoux F., Gilet J., Darbois A., du Halgouet A., Alonso R., Richer W., Goubet A. G., Daviaud C., Menger L., et al. 2019. A common transcriptomic program acquired in the thymus defines tissue residency of MAIT and NKT subsets. J. Exp. Med. 216: 133–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dusseaux M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., Lantz O.. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood 117: 1250–1259. [DOI] [PubMed] [Google Scholar]
- 10. Parrot T., Healy K., Boulouis C., Sobkowiak M. J., Leeansyah E., Aleman S., Bertoletti A., Sällberg Chen M., Sandberg J. K.. 2021. Expansion of donor-unrestricted MAIT cells with enhanced cytolytic function suitable for TCR redirection. JCI Insight 6: e140074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dias J., Leeansyah E., Sandberg J. K.. 2017. Multiple layers of heterogeneity and subset diversity in human MAIT cell responses to distinct microorganisms and to innate cytokines. Proc. Natl. Acad. Sci. USA 114: E5434–E5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinks T. S. C., Marchi E., Jabeen M., Olshansky M., Kurioka A., Pediongco T. J., Meehan B. S., Kostenko L., Turner S. J., Corbett A. J., et al. 2019. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 28: 3249–3262.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leng T., Akther H. D., Hackstein C. P., Powell K., King T., Friedrich M., Christoforidou Z., McCuaig S., Neyazi M., Arancibia-Cárcamo C. V., et al. Oxford IBD Investigators . 2019. TCR and inflammatory signals tune human MAIT cells to exert specific tissue repair and effector functions. Cell Rep. 28: 3077–3091.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lamichhane R., Schneider M., de la Harpe S. M., Harrop T. W. R., Hannaway R. F., Dearden P. K., Kirman J. R., Tyndall J. D. A., Vernall A. J., Ussher J. E.. 2019. TCR- or cytokine-activated CD8+ mucosal-associated invariant T cells are rapid polyfunctional effectors that can coordinate immune responses. Cell Rep. 28: 3061–3076.e5. [DOI] [PubMed] [Google Scholar]
- 15. Constantinides M. G., Link V. M., Tamoutounour S., Wong A. C., Perez-Chaparro P. J., Han S. J., Chen Y. E., Li K., Farhat S., Weckel A., et al. 2019. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science 366: eaax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Bourhis L., Dusseaux M., Bohineust A., Bessoles S., Martin E., Premel V., Coré M., Sleurs D., Serriari N. E., Treiner E., et al. 2013. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLoS Pathog. 9: e1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kurioka A., Ussher J. E., Cosgrove C., Clough C., Fergusson J. R., Smith K., Kang Y. H., Walker L. J., Hansen T. H., Willberg C. B., Klenerman P.. 2015. MAIT cells are licensed through granzyme exchange to kill bacterially sensitized targets. Mucosal Immunol. 8: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leeansyah E., Svärd J., Dias J., Buggert M., Nyström J., Quigley M. F., Moll M., Sönnerborg A., Nowak P., Sandberg J. K.. 2015. Arming of MAIT cell cytolytic antimicrobial activity is induced by IL-7 and defective in HIV-1 infection. PLoS Pathog. 11: e1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dias J., Sobkowiak M. J., Sandberg J. K., Leeansyah E.. 2016. Human MAIT-cell responses to Escherichia coli: activation, cytokine production, proliferation, and cytotoxicity. J. Leukoc. Biol. 100: 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boulouis C., Sia W. R., Gulam M. Y., Teo J. Q. M., Png Y. T., Phan T. K., Mak J. Y. W., Fairlie D. P., Poon I. K. H., Koh T. H., et al. 2020. Human MAIT cell cytolytic effector proteins synergize to overcome carbapenem resistance in Escherichia coli. PLoS Biol. 18: e3000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Georgel P., Radosavljevic M., Macquin C., Bahram S.. 2011. The non-conventional MHC class I MR1 molecule controls infection by Klebsiella pneumoniae in mice. Mol. Immunol. 48: 769–775. [DOI] [PubMed] [Google Scholar]
- 22. Meierovics A., Yankelevich W. J., Cowley S. C.. 2013. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA 110: E3119–E3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang H., D’Souza C., Lim X. Y., Kostenko L., Pediongco T. J., Eckle S. B. G., Meehan B. S., Shi M., Wang N., Li S., et al. 2018. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 9: 3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Howson L. J., Napolitani G., Shepherd D., Ghadbane H., Kurupati P., Preciado-Llanes L., Rei M., Dobinson H. C., Gibani M. M., Teng K. W. W., et al. 2018. MAIT cell clonal expansion and TCR repertoire shaping in human volunteers challenged with Salmonella Paratyphi A. Nat. Commun. 9: 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Le Bourhis L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. [Published erratum appears in 2010 Nat. Immunol. 11: 969.] Nat. Immunol. 11: 701–708. [DOI] [PubMed] [Google Scholar]
- 26. Gold M. C., Cerri S., Smyk-Pearson S., Cansler M. E., Vogt T. M., Delepine J., Winata E., Swarbrick G. M., Chua W. J., Yu Y. Y., et al. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8: e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chiossone L., Dumas P. Y., Vienne M., Vivier E.. 2018. Natural killer cells and other innate lymphoid cells in cancer. [Published erratum appears in 2018 Nat. Rev. Immunol. 18: 726.] Nat. Rev. Immunol. 18: 671–688. [DOI] [PubMed] [Google Scholar]
- 28. Boudreau J. E., Hsu K. C.. 2018. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol. 39: 222–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cichocki F., Grzywacz B., Miller J. S.. 2019. Human NK cell development: one road or many? Front. Immunol. 10: 2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quatrini L., Della Chiesa M., Sivori S., Mingari M. C., Pende D., Moretta L.. 2021. Human NK cells, their receptors and function. Eur. J. Immunol. 51: 1566–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Theresine M., Patil N. D., Zimmer J.. 2020. Airway natural killer cells and bacteria in health and disease. Front. Immunol. 11: 585048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sim M. J. W., Rajagopalan S., Altmann D. M., Boyton R. J., Sun P. D., Long E. O.. 2019. Human NK cell receptor KIR2DS4 detects a conserved bacterial epitope presented by HLA-C. Proc. Natl. Acad. Sci. USA 116: 12964–12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Krismer B., Weidenmaier C., Zipperer A., Peschel A.. 2017. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 15: 675–687. [DOI] [PubMed] [Google Scholar]
- 34. Belkaid Y., Tamoutounour S.. 2016. The influence of skin microorganisms on cutaneous immunity. Nat. Rev. Immunol. 16: 353–366. [DOI] [PubMed] [Google Scholar]
- 35. Monaco M., Pimentel de Araujo F., Cruciani M., Coccia E. M., Pantosti A.. 2017. Worldwide epidemiology and antibiotic resistance of Staphylococcus aureus. Curr. Top. Microbiol. Immunol. 409: 21–56. [DOI] [PubMed] [Google Scholar]
- 36. Salgado-Pabón W., Schlievert P. M.. 2014. Models matter: the search for an effective Staphylococcus aureus vaccine. Nat. Rev. Microbiol. 12: 585–591. [DOI] [PubMed] [Google Scholar]
- 37. Alonzo F. III, Torres V. J.. 2014. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol. Mol. Biol. Rev. 78: 199–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alonzo F. III, Kozhaya L., Rawlings S. A., Reyes-Robles T., DuMont A. L., Myszka D. G., Landau N. R., Unutmaz D., Torres V. J.. 2013. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493: 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reyes-Robles T., Alonzo F. III, Kozhaya L., Lacy D. B., Unutmaz D., Torres V. J.. 2013. Staphylococcus aureus leukotoxin ED targets the chemokine receptors CXCR1 and CXCR2 to kill leukocytes and promote infection. Cell Host Microbe 14: 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Spaan A. N., Reyes-Robles T., Badiou C., Cochet S., Boguslawski K. M., Yoong P., Day C. J., de Haas C. J., van Kessel K. P., Vandenesch F., et al. 2015. Staphylococcus aureus targets the Duffy antigen receptor for chemokines (DARC) to lyse erythrocytes. Cell Host Microbe 18: 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lubkin A., Lee W. L., Alonzo F. III, Wang C., Aligo J., Keller M., Girgis N. M., Reyes-Robles T., Chan R., O’Malley A., et al. 2019. Staphylococcus aureus leukocidins target endothelial DARC to cause lethality in mice. Cell Host Microbe 25: 463–470.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alonzo F. III, Benson M. A., Chen J., Novick R. P., Shopsin B., Torres V. J.. 2012. Staphylococcus aureus leucocidin ED contributes to systemic infection by targeting neutrophils and promoting bacterial growth in vivo. Mol. Microbiol. 83: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Radcliff F. J., Waldvogel-Thurlow S., Clow F., Mahadevan M., Johnston J., Li G., Proft T., Douglas R. G., Fraser J. D.. 2019. Impact of superantigen-producing bacteria on T cells from tonsillar hyperplasia. Pathogens 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shaler C. R., Choi J., Rudak P. T., Memarnejadian A., Szabo P. A., Tun-Abraham M. E., Rossjohn J., Corbett A. J., McCluskey J., McCormick J. K., et al. 2017. MAIT cells launch a rapid, robust and distinct hyperinflammatory response to bacterial superantigens and quickly acquire an anergic phenotype that impedes their cognate antimicrobial function: defining a novel mechanism of superantigen-induced immunopathology and immunosuppression. PLoS Biol. 15: e2001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sandberg J. K., Norrby-Teglund A., Leeansyah E.. 2017. Bacterial deception of MAIT cells in a cloud of superantigen and cytokines. PLoS Biol. 15: e2003167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boulouis C., Gorin J. B., Dias J., Bergman P., Leeansyah E., Sandberg J. K.. 2020. Opsonization-enhanced antigen presentation by MR1 activates rapid polyfunctional MAIT cell responses acting as an effector arm of humoral antibacterial immunity. J. Immunol. 205: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mairpady Shambat S., Haggar A., Vandenesch F., Lina G., van Wamel W. J., Arakere G., Svensson M., Norrby-Teglund A.. 2014. Levels of alpha-toxin correlate with distinct phenotypic response profiles of blood mononuclear cells and with agr background of community-associated Staphylococcus aureus isolates. PLoS One 9: e106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McInnes L., Healy J., Saul N., Grossberger L.. 2018. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3: 861. [Google Scholar]
- 49. O’Brien E. C., McLoughlin R. M.. 2019. Considering the ‘alternatives’ for next-generation anti-Staphylococcus aureus vaccine development. Trends Mol. Med. 25: 171–184. [DOI] [PubMed] [Google Scholar]
- 50. Haller D., Serrant P., Granato D., Schiffrin E. J., Blum S.. 2002. Activation of human NK cells by staphylococci and lactobacilli requires cell contact-dependent costimulation by autologous monocytes. Clin. Diagn. Lab. Immunol. 9: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Small C. L., McCormick S., Gill N., Kugathasan K., Santosuosso M., Donaldson N., Heinrichs D. E., Ashkar A., Xing Z.. 2008. NK cells play a critical protective role in host defense against acute extracellular Staphylococcus aureus bacterial infection in the lung. J. Immunol. 180: 5558–5568. [DOI] [PubMed] [Google Scholar]
- 52. Preciado-Llanes L., Aulicino A., Canals R., Moynihan P. J., Zhu X., Jambo N., Nyirenda T. S., Kadwala I., Sousa Gerós A., Owen S. V., et al. 2020. Evasion of MAIT cell recognition by the African Salmonella Typhimurium ST313 pathovar that causes invasive disease. Proc. Natl. Acad. Sci. USA 117: 20717–20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rouha H., Weber S., Janesch P., Maierhofer B., Gross K., Dolezilkova I., Mirkina I., Visram Z. C., Malafa S., Stulik L., et al. 2018. Disarming Staphylococcus aureus from destroying human cells by simultaneously neutralizing six cytotoxins with two human monoclonal antibodies. Virulence 9: 231–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ortines R. V., Wang Y., Liu H., Dikeman D. A., Pinsker B. L., Miller R. J., Kim S. E., Ackerman N. E., Rizkallah J. F., Marcello L. T., et al. 2019. Efficacy of a multimechanistic monoclonal antibody combination against Staphylococcus aureus surgical site infections in mice. Antimicrob. Agents Chemother. 63: e00346-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vu T. T. T., Nguyen N. T. Q., Tran V. G., Gras E., Mao Y., Jung D. H., Tkaczyk C., Sellman B. R., Diep B. A.. 2020. Protective efficacy of monoclonal antibodies neutralizing alpha-hemolysin and bicomponent leukocidins in a rabbit model of Staphylococcus aureus necrotizing pneumonia. Antimicrob. Agents Chemother. 64: e02220-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chan R., Buckley P. T., O’Malley A., Sause W. E., Alonzo F. III, Lubkin A., Boguslawski K. M., Payne A., Fernandez J., Strohl W. R., et al. 2019. Identification of biologic agents to neutralize the bicomponent leukocidins of Staphylococcus aureus. Sci. Transl. Med. 11: eaat0882. [DOI] [PubMed] [Google Scholar]
- 57. Miller L. S., Cho J. S.. 2011. Immunity against Staphylococcus aureus cutaneous infections. Nat. Rev. Immunol. 11: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leeansyah E., Boulouis C., Kwa A. L. H., Sandberg J. K.. 2021. Emerging role for MAIT cells in control of antimicrobial resistance. Trends Microbiol. 29: 504–516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


