Abstract
CRISPR-based detection of target DNA or RNA exploits a dual function, including target sequence-specific recognition followed by trans-cleavage activity of a collateral ssDNA linker between a fluorophore (F) and a quencher (Q), which amplifies a fluorescent signal upon cleavage. In this work, we have extended such dual functionality in a modified immunoassay format to detect a target protein, CXCL9, which is markedly elevated in the urine of kidney transplant recipients undergoing acute rejection episodes. To establish the “immuno-CRISPR” assay, we used anti-CXCL9 antibody-DNA barcode conjugates to target CXCL9 and amplify fluorescent signals via Cas12a-based trans-cleavage activity of FQ reporter substrates, respectively, and in the absence of an isothermal amplification step. To enhance detection sensitivity, the DNA barcode system was engineered by introducing multiple Cas12a recognition sites. Use of biotinylated DNA barcodes enabled self-assembly onto streptavidin (SA) to generate SA-DNA barcode complexes to increase the number and density of Cas12a recognition sites attached to biotinylated anti-CXCL9 antibody. As a result, we improved the rate of CXCL9 detection approximately 8-fold when compared to the use of a monomeric DNA barcode. The limit of detection (LOD) for CXCL9 using the immuno-CRISPR assay was 14 pg/mL, which represented ~7-fold improvement when compared to traditional HRP-based ELISA. Selectivity was shown with lack of crossover reactivity with the related chemokine CXCL1. Finally, we successfully evaluated the presence of CXCL9 in urine samples from 11 kidney transplant recipients using the immuno-CRISPR assay, resulting in 100% accuracy to clinical CXCL9 determination and paving the way for use as a point-of-care non-invasive biomarker for detection of kidney transplant rejection.
INTRODUCTION
CRISPR/Cas systems are a promising toolset for editing target DNA or RNA because of their ability to accurately recognize specific sequence and reprogram target sites by changing the CRISPR RNA (crRNA).1–8 Both Cas9 and Cas12a (or Cpf1) proteins have crRNA-guided endonuclease activity, which can cleave double stranded target DNA (dsDNA) sequences.9–13 Unlike Cas9, Cas12a also has nonspecific single-stranded DNA (ssDNA) cleavage activity; so-called collateral trans-cleavage activity, which can be sequentially triggered after binding Cas12a/crRNA complex to a target dsDNA.12,13
Signal amplification using trans-cleavage activity of Cas proteins has been used in combination with isothermal gene amplification methods, including LAMP (loop-mediated isothermal amplification) and RPA (recombinase polymerase amplification) to improve detection sensitivity.11–22 Indeed, ssDNA or RNA reporters with quenched fluorescent dyes have been developed as signaling molecules.13–15 Several DNA or RNA systems have been developed that exploit trans-cleavage of a quenched fluorophore, including the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) system,14 one-hour low-cost multipurpose highly efficient system (HOLMES),15 and DNA endonuclease-targeted CRISPR trans-reporter (DETECTR) system13. These detection methods are extremely sensitive and could be applied to point-of-care testing without the need conventional qPCR because a target DNA molecule can be amplified through isothermal PCR.13–15 Moreover, CRISPR-based detection methods may be applied to existing immuno-PCR (iPCR) approaches, which combine the antibody specificity of ELISA with the signal amplification of PCR,23–26 to detect target proteins. Nonetheless, application of CRISPR-based detection platform to iPCR still requires some sort of PCR amplification to generate a detectable signal.
Herein, we have developed the immuno-CRISPR assay as a new protein detection method that uses antibody-DNA barcode conjugates together with a Cas12a/crRNA complex and collateral FQ reporters. To improve sensitivity, we used antibody-DNA barcode conjugates with multiple Cas12a recognition sites, which allowed us to avoid DNA amplification. To attach DNA barcodes having multiple Cas12a recognition sites onto the target antibody, we designed bis-biotinylated DNA barcodes that can self-assemble with streptavidin (SA) to generate DNA barcode-SA complexes. Such complexes with multiple Cas12a recognition sites can be used as a target substrate for triggering Cas12a trans-cleavage activity resulting in signal amplification.
To demonstrate the immuno-CRISPR assay platform, we chose the chemokine ligand 9 (CXCL9) as a target protein. Urinary CXCL9 has been shown to function as a candidate, noninvasive biomarker for T cell mediated kidney allograft rejection in transplant recipients.27–30 Development of rapid, point-of-care urinary CXCL9 detection could bypass the need for kidney allograft biopsies and thereby transform clinical care of kidney transplant recipients. Current clinical assays generally employ standard colorimetric ELISA, which is accurate and sensitive, yet results in high background signals due to the non-specific binding of HRP conjugates, is only available in a research laboratory setting and requires hours for detection.31 In the current work, immuno-CRISPR based detection of CXCL9 was achieved, and with approximately 7-fold enhanced detection sensitivity when compared to traditional ELISA method and without interference by urine components. This method was then applied to a small set of urine samples from kidney transplant recipients and resulted in excellent correlation to clinical outcomes.
EXPERIMENTAL SECTION
Materials.
EnGen® Lba Cas12a (Cpf1) and buffer for Cas12a (NEB buffer 2.1) were purchased from New England Biolabs (Ipswich, MA). CRISPR RNA (crRNA) and FQ reporter for Cas12a were synthesized by Integrated DNA Technologies (Coralville, IA). Streptavidin was purchased from Promega (Madison, WI). ELISA kits, which has capture and detection antibodies, target protein (recombinant human CXCL9 or CXCLl), and buffers was purchased from R&D Systems (Minneapolis, MN). Synthetic urine was purchased from Sigma-Aldrich (St. Louis, MO) (Table S1). All solutions were prepared with purified water by a Milli-Q Purification System from Millipore (Burlington, MA).
Preparation of DNA barcodes.
Oligonucleotides with biotin modification were synthesized by Integrated DNA Technologies (Coralville, IA) (Table S2). Bis-biotinylated DNA barcode-I and -II were prepared by PCR with forward primers and he reverse primers using the template (pUC57-barcode) synthesized by Genscript (Table S2). PCR products were then purified using DNA Clean & Concentrator Kits (Irvine, CA). Biotinylated DNA barcode-III and barcode-IV were prepared by simple annealing of the mixed forward and reverse oligonucleotides at 94 °C for 2 min.
Construction of DNA barcode complex.
To construct the DNA barcode complexes with streptavidin (SA-DNA barcode-II and -III complexes), each biotinylated DNA barcode was mixed with streptavidin in phosphate-buffered saline (PBS, pH 7.4) at room temperature for 1 h. The final concentration of biotinylated DNA barcodes was fixed at 1 μM, and molar ratio of SA to DNA barcode ranged from 1:0.25 to 1:4. Formation of self-assembled SA-DNA barcode-II and -III complexes was confirmed by agarose gel electrophoresis. Then, detection antibody solution (0.2 μg/mL in PBS-BSA) was added and incubated at room temperature for 2 h, and unreacted detection antibodies were removed by ultrafiltration using Sartorius Vivaspin 6 Centrifugal Concentrators (Goettingen, Germany).
Dynamic light scattering (DLS) and atomic force microscopy (AFM) were used to characterize the sizes of SA-DNA barcode complexes. DLS analysis was performed on a Litesizer™ 500 device (Anton Paar, Graz, Austria) using quartz cuvettes. AFM was performed on an Asylum MFP3D instrument (Oxford Instruments, Abingdon-on-Thames, UK) and analyzed with IGOR Pro 14 (Wave-Metrics, Inc., Lake Oswego, OR). Images in air were collected in AC mode using super sharp silicon cantilevers SSS-NCHR (Nanosensors™, Neuchatel, Switzerland). Samples were prepared by depositing 10 μL of DNA complex solution containing 10 μM MgCl2 onto fresh mica (Asheville Mica Company, Newport News, VA). Samples were left to adsorb for 5 min, then washed three times with 10 mL of deionized water, and finally air dried for 1 h before imaging.
Trans-cleavage assay of Cas12a complex.
The trans-cleavage activity of Cas12a complexes was measured by an FQ reporter-based assay.13 Briefly, a Cas12a complex was pre-assembled by incubating Cas12a (1 μM as a final concentration) and crRNA (1.2 μM as a final concentration) in buffer (10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, pH 7.9) at room temperature for 30 min. FQ reporter (1 μM as a final concentration) was mixed with the diluted Cas12a complex (0.1 μM as a final concentration) in the aforementioned buffer. After adding the mixtures to wells containing DNA barcodes or SA-DNA barcode complex, the reaction cocktails (100 μL, 96-well microplate format) were incubated in a plate reader (SpectraMax M5, San Jose, CA) at room temperature for 1 h with fluorescence measurements taken every 5 min (λex = 485 nm and λem = 530 nm).
CXCL9 and CXCL1 detection.
Sandwich immunoassays were performed using SA-HRP as a control, DNA barcodes and SA-DNA barcode complexes for the detection of CXCL9. Briefly, 100 μL of captured antibody solution (6 μg/mL in PBS) were added to each well and incubated at 4°C overnight. After aspirating each well, 200 μL of BSA solution in PBS (PBS-BSA) was added to block each well and incubation was performed at room temperature for 1 h. This was followed by washing each well 3-times with 200 μL of washing buffer (PBS with 0.05% Tween-20). Then, 100 μL of human recombinant CXCL9 solution in PBS-BSA or synthetic urine at concentrations ranging from 0 to 4000 pg/mL were added to each well and incubated at room temperature for 2 h. A similar method was used for CXCL1 using human recombinant CXCL1. After incubation, unreacted human recombinant protein was removed, followed by washing 3-times with washing buffer. Then, 100 μL of detection antibody immobilized DNA barcode or SA-DNA barcode complex solutions (50 nM) were added to each well and incubated at room temperature for 1 h. After incubation, unreacted DNA-based molecules were removed, followed by washing 3-times with washing buffer. In the case of sandwich immunoassays via DNA barcode or SA-DNA barcode complexes, fluorescence was measured using the FQ reporter assay mentioned above.
In the case of HRP-based sandwich ELISA, a colorimetric assay was used as previously reported.27,32 After incubating captured antibody and CXCL9 solution, as mentioned above, detection antibody solution (0.2 μg/mL in PBS-BSA) was added and incubated at room temperature for 2 h, followed by washing 3-times with washing buffer. Then, 100 μL of SA-HRP was added to each well and incubated at room temperature for 1 h. After incubation, unreacted SA-HRP was removed, followed by washing 3-times with washing buffer. Finally, 100 μL of substrate solution for HRP (tetramethylbenzidine and H2O2) were added to each well, and the absorbance was measured at 570 nm every 5 min. Experiments were performed in triplicate, and standard deviations were reported. Limit of detection (LOD) was calculated from each immunoassay-based standard curve by determining the concentration at which the fluorescence signal is times the standard deviation of the blank above the extrapolated blank signal.33
Evaluation of clinical samples.
We analyzed urine samples from kidney transplant recipients participating in the Clinical Trials in Organ Transplantation (CTOT, NCT02495077) trial. The CTOT studies were approved by institutional review boards (IRB) at the participating centers. Urine samples were obtained pre-transplant and at the time of kidney transplant biopsies and were stored in aliquots at −20° C. No pretreatment of the isolated urine was required. Aliquots of all samples collected (more than 500) were analyzed for urinary CXCL9 by sandwich ELISA. For validation of the immune-CRISPR assay, we chose 11 samples with CXCL9 values determined by ELISA ranging from below the detectable limit (<60 ng/ml) to 3600 ng/ml. These urine samples were deidentified (IRB exempt for laboratory analyses) and provided for evaluation. Various concentrations of human recombinant CXCL9 were spiked into authentic urine obtained from healthy donors to assess the impact of authentic urine on the effectiveness of immuno-CRISPR.
RESULTS AND DISCUSSION
Principle of immuno-CRISPR.
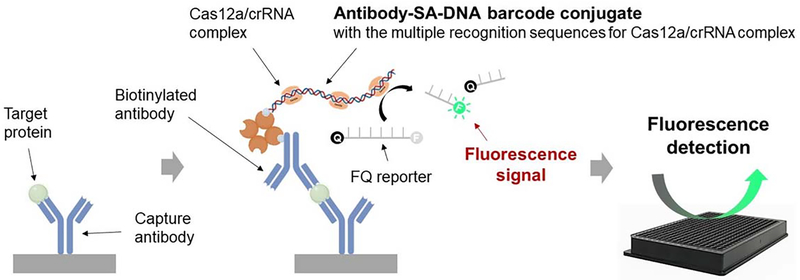
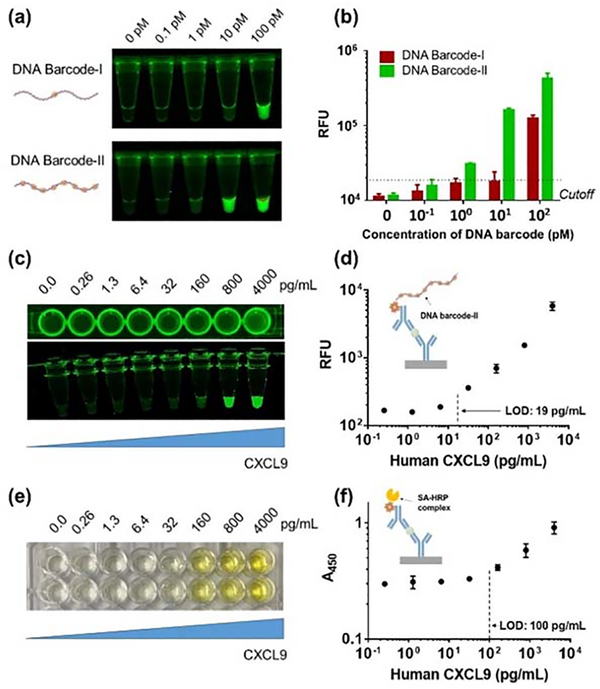
The immuno-CRISPR assay concept is depicted in Figure 1. Briefly, a sandwich structure with antibody-DNA barcodes having multiple Cas12a recognition sites can be formed in the presence of a target protein, such as CXCL9, followed by adding the Cas12a/crRNA complex to generate fluorescence signals through collateral trans-cleavage activity of Cas12a against FQ reporters. In the absence of a target protein, antibody-DNA barcode conjugates are washed out and an extremely low background fluorescence signal is generated. Pre-assembled Cas12a/crRNA complexes to recognize a specific DNA sequence on the antibody-based DNA barcode. We first designed long (~700 bp) DNA barcodes containing one recognition site for Cas12a/crRNA complex (DNA barcode-I) and 11 recognition sites with 20 base pair separation between sites (DNA barcode-II) (Table S2 & Figure S1). Using the fluorophore quencher (FQ)-labeled reporter as Cas12a/crRNA complex substrate (Figure 2a), DNA barcode-II was approximately 10-fold more sensitive than barcode-I (Figure 2b), suggesting that the sensitivity of DNA barcode detection can be improved by increasing the number Cas12a/crRNA recognition sites on the DNA barcode. Using DNA barcode-II, we demonstrated that the immuno-CRISPR assay platform could detect CXCL9 quantitatively without isothermal PCR. To estimate the range and LOD of CXCL9 detection using barcode-II, we dissolved CXCL9 into PBS. The LOD for CXCL9 was estimated to be 19 pg/mL using a 24 h incubation period (Figures 2c and d). The repeatability and reproducibility of the immuno-CRISPR assay platform was evaluated with two consecutive experiments with an interval of three days. No significant difference in repeatability and reproducibility was observed when comparing the two consecutive experiments (Figure S2a). For comparison, a colorimetric HRP-based ELISA assay was performed (1 h incubation) and resulted in an LOD of approximately 100 pg/mL (Figures 2e and f). Extending the ELISA incubation time did not improve sensitivity, as the background absorbance also increased because of non-specific binding of the HRP conjugate to BSA-coated wells (Figure S3). Thus, while the immuno-CRISPR method yielded improved LOD, the incubation time was long. We hypothesized that the long reaction time may have been due to the structure of the long DNA barcode-II with multiple recognition sites bound to CXCL9, which could result in 3D conformations that sterically hindered the Cas12a/crRNA complexes.
Figure 1.
Schematic illustration of immuno-CRISPR assay with antibody-DNA barcode conjugates with Cas12a/crRNA complex and collateral FQ substrates. Sandwich structure with capture antibody, target protein and self-assembled antibody-streptavidin (SA)-DNA barcode conjugate was formed by antigen-antibody interaction. After adding Cas12a/crRNA complex, trans-cleavage activity of Cas12a/crRNA complex bound to DNA barcode was measured by the fluorophore quencher (FQ)-labeled reporter assay.
Figure 2.
Fluorescence images of DNA barcode-I and –II within tubes (a). Trans-cleavage activities of Cas12a/crRNA complexes bound to DNA barcode-I and -II via the fluorophore-quencher (FQ) assay (b). Visual inspections and calibration curves of immuno-CRISPR assay with DNA barcode-II (c and d) and traditional HRP-based ELISA (e and f) for detection of CXCL9 in PBS. Error bars are means and SDs from three independent experiments. Cutoff values based on limit of signal to background absorbance or fluorescence.
Construction of DNA barcode-Streptavidin complexes.
To reduce the assay time of immuno-CRISPR, we took advantage of the self-assembly of bis-biotinylated DNA barcodes and SA (Figure S4). Bis-biotinylated DNA barcode-II was obtained from the PCR reaction with biotinylated primers (Table S2). The self-assembly of SA and DNA barcode-II with different molar ratios of 0.25:1 to 4:1 of SA:DNA barcode-II led to various sizes of SA-DNA barcode-II complexes, as identified by agarose gel electrophoretic analysis (Figure S5a). In the case of the self-assembly of bis-biotinylated barcode-II, the band intensity representing largest supramolecular aggregates was highest at an equimolar coupling stoichiometry (lane 5 in Figure S5a). At lower ratios of SA:DNA, the fraction of uncoupled DNA barcode-II increased (lanes 2, 3 and 4 in Figure S5a). In the case of 2:1 and 4:1 molar equivalents of SA:DNA barcode-II, however, two bright bands at around 1500 and 2000 bp (lanes 6 and 7 in Figure S5a) were observed. Because monomeric DNA barcode was completely assembled at 2:1 and not 1:1 molar equivalents of SA:DNA barcode-II, we assumed that the estimated number of SA per DNA barcode-II was 1–2 at 1:1 molar equivalent of SA:DNA barcode-II, which indicated small-sized supramolecular aggregates due to the incomplete polymerization of SA with bis-biotinylated DNA barcode-II.
We reasoned that the relatively low degree of oligomerization of SA-DNA barcode-II conjugates may have been due to the very long 700 bp DNA of barcode-II that may interfere with effective binding to SA. Thus, we considered a small-sized bis-biotinylated DNA barcode (DNA barcode-III, 46 bp, Table S2) with a single Cas12a recognition site to reduce the steric hindrance between the DNA barcode and SA. The self-assembly of SA and DNA barcode-III with different molar ratios of 0.25:1 to 4:1 of SA:DNA barcode-III was also identified by agarose gel electrophoretic analysis (Figure S5b). With this construct, the largest supramolecular aggregates were obtained at the 0.5:1 molar equivalent mixture of SA and bis-biotinylated DNA barcode-III (lane 4 in Figure S5b). Interestingly, the number of SA per DNA barcode-III was estimated to be 0.5, since monomeric DNA barcode was fully incorporated into the assembly at 0.5:1 molar equivalent of SA:DNA barcode-III. This result suggests that the larger, oligomeric, supramolecular aggregates would be expected to provide a high density of Cas12a recognition sites bound to the anti-CXCL9 antibody, resulting in enhancing trans-cleavage activity of Cas12a.
The hydrodynamic diameters of the SA-DNA barcode complexes in solution were measured by DLS. SA and SA-DNA barcode-III complex were estimated to 5.9 ± 0.81 nm and 59 ± 6.3 nm, respectively (Figure S6). These results indicated that bis-biotinylated DNA barcode-III effectively contributed to the formation of aggregates with SA acting as a crosslinking agent. However, the DLS of the SA-DNA barcode-II complex showed two peaks at 6.0 ± 0.71 nm and 15 ± 4.6 nm, respectively (Figure S6), suggesting that heterogeneous SA-DNA barcode complexes were formed likely as a result of steric hindrance by the larger bis-biotinylated DNA barcode-III. To visualize the structures of SA-DNA barcode complexes, we performed atomic force microscopy (AFM). Small-sized globular structures were observed for the streptavidin-only sample (Figure S7a). However, larger DNA structures that were intertwined with smaller globular structures were observed for the SA-DNA barcode-II complex (Figure S7b). We observed a small proportion of highly networked structures, indicating that SA and bis-biotinylated DNA barcode-II do not fully form conjugates potentially due to steric hindrance induced by the large DNA barcode. Interestingly, large globules were observed for the SA-DNA barcode-III complex (Figure S7c), indicating that supramolecular aggregates were formed by multiple SAs interacting with DNA barcodes that act as crosslinking agents. Hence, the smaller bis-biotinylated DNA barcode-III (in relation to DNA barcode-II) was highly effective in assembling into larger aggregates with SA.
Therefore, when compared to larger bis-biotinylated DNA barcode-II, the smaller bis-biotinylated DNA barcode-III was more effective in assembling into larger aggregates with SA, which allows for more Cas12a binding sites to increase trans-cleavage activity of Cas12a. To optimize the incorporation of DNA barcode-III in the SA-DNA complexes, we examined trans-cleavage activity in the presence of 4000 pg/mL CXCL9. Saturation of activity was obtained at 50 nM DNA barcode-III (Figure S8), and this concentration was used in subsequent experiments.
Immuno-CRISPR assay using SA-DNA barcode complexes for CXCL9 detection.
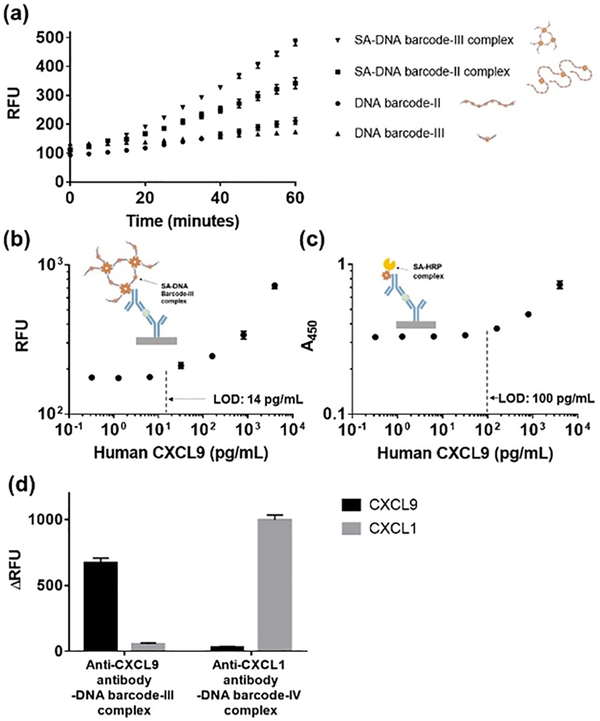
We investigated the efficiency of the SA-DNA barcode-II (lane 5 in Figure S5a) and SA-DNA barcode-III (lane 4 in Figure S5b) complexes bound to anti-CXCL9 antibody for enhancing the Cas12a-based detection of CXCL9 (4000 pg/mL in PBS) using the immuno-CRISPR assay. CXCL9 detection rates using SA-DNA barcode-II and SA-DNA barcode-III complexes were ~2 times and ~8 times faster than those of monomeric forms of DNA barcode-II and DNA barcode-III, respectively (Figures 3a and S9). The SA-DNA barcode-III complexes resulted in the best performance for the detection of CXCL9 spiked in PBS using immuno-CRISPR assay, enabling detection of CXCL9 (LOD = 17 pg/mL in Figure S10) within 1 h.
Figure 3.
Time-dependent increase of fluorescence signals for detection of CXCL9 at 4000 pg/mL by using immuno-CRISPR assay with DNA barcodes and their complexes (a). Calibration curves of immuno-CRISPR assay with SA-DNA barcode-III complex (b) and HRP-based ELISA (c) for detection of CXCL9 at concentrations from 0 to 4000 pg/mL in synthetic urine. Cross-reactivity of the immuno-CRISPR assay using CXCL9 and CXCL1 in crossover experiments. Two different complexes were prepared, anti-CXCL9 antibody conjugated DNA barcode-III complex and anti-CXCL1 antibody conjugated DNA barcode-IV complex (d). Error bars are means and SDs from three independent experiments. Cutoff values based on limit of signal to background absorbance or fluorescence.
Since one of our goals is to detect urinary CXCL9 as a potential biomarker for kidney transplant rejection, we proceeded to evaluate the effectiveness of the SA-DNA barcode-III complexes in a synthetic urine solution as compared to traditional HRP-based ELISA for quantifying CXCL9 concentration (Figures 3b and c). The LODs of the 1 h immuno-CRISPR assay with SA-DNA barcode-III complexes and HRP-based ELISA for the detection of CXCL9 in synthetic urine were 14 and 100 pg/mL, respectively. The LOD of immuno-CRISPR assay was comparable to chemiluminescence-based single-plex micropatterned aqueous two-phase (28 pg/mL)34 and AlphaLISA (20 pg/mL)35 platforms. These results were similar to the respective LODs in PBS (Figures 2c and S10), indicating that synthetic urine did not affect the sensitivity for the detection of CXCL9. No significant difference in repeatability and reproducibility in the immuno-CRISPR assay platform was observed using SA-DNA barcode-III complex was detected when comparing the two consecutive experiments (Figure S2b). To assess selectivity, the immuno-CRISPR assay was performed at 4000 pg/mL of CXCL9 and CXCL1. each of CXCL9 and CXCL1, the latter an important biomarker in bladder cancer detection.36 The anti-CXCL1 antibody-DNA barcode conjugate was prepared using DNA barcode-IV (Table S2), similar in size to DNA barcode-III but with a different Cas12a recognition sequence (Figure S4 and Table S2). We confirmed that cross-reactivity caused by non-specific binding was negligible, suggesting high selectivity of immuno-CRISPR against two urinary chemokines (Figure 3d).
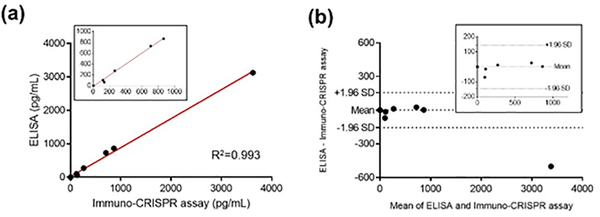
To extend the clinical significance of the immuno-CRISPR assay for CXCL9, we assayed human recombinant CXCL9 samples spiked into authentic urine from healthy donors at both low (32 pg/mL) and high (800 pg/mL) CXCL9 concentrations. Fluorescence signals in authentic urine matched the calibration curve data of the immuno-CRISPR assay with SA-DNA barcode-III (Figure S11), indicating that the components of authentic urine did not affect the sensitivity of CXCL9 detection. Next, we examined 11 authentic urine samples obtained from patients participating in a Clinical Trials in Organ Transplantation (CTOT) trial (see Table S3). Immuno-CRISPR and traditional HRP-based ELISA assays were compared to assess the accuracy of the new strategy (Table S3 and Figure 4). Both methods resulted in three samples that were below the LOD of CXCL9 for both assays. Of the other eight samples, six were above the LOD for both immuno-CRISPR and ELISA; the correlation between both methods was excellent (Figure 4a; R2 = 0.996) and within a 95% confidence limit (± 1.96 standard deviation in a Bland-Altman plot, Figure 4b) except for one outlier at the highest concentration of CXCL9. A 200 pg/mL level of CXCL9 represents the borderline threshold for early indication of kidney transplant rejection using the traditional HRP-based ELISA.29 This is well above the LOD for the immuno-CRISPR assay, and indeed, four of the samples above the LOD fell below this clinical detection level. Importantly, the immuno-CRISPR assay could easily detect levels of CXCL9 substantially below this clinical threshold, which may be a critical very early marker for kidney transplant rejection.
Figure 4.
Consistency between HRP-based ELISA and immuno-CRISPR assay for CXCL9 detection (a). Bland-Altman plots between HRP-based ELISA and immuno-CRISPR assay for CXCL9 detection (b). A total of 11 patient urine samples were evaluated.
CONCLUSIONS
In this work, a Cas12a-coupled immuno-CRISPR assay was developed to detect CXCL9 in urine. Self-assembled SA-DNA barcode complexes containing multiple Cas12a recognition sites and their antibody conjugates were employed both for recognition and activation to trigger the trans-cleavage activity of Cas12a. The number of Cas12a recognition sites in a sandwich form in microtiter plates was critical to achieve signal enhancement. The LOD for CXCL9 in synthetic urine using the immuno-CRISPR assay was 14 pg/mL, suggesting approximately 7-fold improvement compared with traditional HRP-based ELISA. In addition, clinical urine samples were evaluated using the immuno-CRISPR assay and was comparable with the results obtained from traditional ELISA. The performance of immuno-CRISPR may be further improved after using purified large-sized SA-DNA barcode complexes, perhaps following size-exclusion chromatography to reduce the heterogeneity of the complexes. Surveillance biopsies of kidney transplants performed in the absence of clinical symptoms or laboratory abnormalities commonly reveal pathological evidence of rejection.27 Immuno-CRISPR-based CXCL9 assessments have the potential to replace biopsies and permit frequent surveillance of the trans-planted organ. Because CRISPR-based techniques have transitioned to several point-of-care compatible devices, we envision that immuno-CRISPR based CXCL9 detection can also be adapted to such devices, including lateral-flow assay formats for rapid analysis at the time of office visits and for home use by kidney transplant recipients. This will require further optimization of Cas12a density on the DNA barcode. Once optimized, clinical trials will determine whether interventions guided by results of immuno-CRISPR based CXCL9 assays will improve transplant outcomes.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by National Institutes of Health (NIAID) U01 AI63594 awarded to PSH and an ancillary mechanistic grant associated with U01 AI63594 awarded to JSD. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors sincerely thank the CTOT site investigators and staff for their efforts in collecting urine samples from the study participants.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Composition of synthetic urine used in this study (Table S1), DNA sequences of the template plasmid and prepared oligo sequences (Table S2), and results of clinical samples via immuno-CRISPR assay using SA-DNA barcode-III complex, compared with HRP-based ELISA (Table S3). Schematic illustration of DNA barcode and their complexes (Figures S1 and S4). Repeatability and reproducibility results (Figure S2). HRP-based ELISA for detection of CXCL9 dissolved in PBS (Figure S3). Agarose gel analysis for DNA barcodes and their complexes with the different ratio of streptavidin and DNA barcode (Figure S5). Dynamic light scattering (DLS) analysis (Figure S6) and atomic force microscopy (AFM) analysis (Figure S7). Trans-cleavage activities of Cas12a-crRNA complexes bound to various concentrations of SA-DNA barcode-III complexes (Figure S8) and trans-cleavage activities of immuno-CRISPR assay with DNA barcodes and their complexes for detection of CXCL9 (Figure S9). Immuno-CRISPR assay with SA-DNA barcode-III complex for detection of CXCL9 dissolved in PBS (Figure S10) and immuno-CRISPR assay for the detection of human recombinant CXCL9 spiked in real urine solution (Figure S11).
Contributor Information
Inseon Lee, Department of Chemical and Biological Engineering, Rensselaer Polytechnic Institute, Troy, NY 12180, USA..
Seok-Joon Kwon, Department of Chemical and Biological Engineering, Rensselaer Polytechnic Institute, Troy, NY 12180, USA..
Mirco Sorci, Department of Chemical and Biological Engineering, Rensselaer Polytechnic Institute, Troy, NY 12180, USA..
Peter S. Heeger, Icahn School of Medicine at Mount Sinai, New York, NY 10029-5674, USA.
Jonathan S. Dordick, Departments of Chemical and Biological Engineering, Biomedical Engineering, and Biological Sciences; Center for Biotechnology & Interdisciplinary Studies; Rensselaer Polytechnic Institute, Troy, NY 12180, United States.
REFERENCES
- (1).Deltcheva E; Chylinski K; Sharma CM; Gonzales K; Chao Y; Pirzada ZA; Eckert MR; Vogel J; Charpentier E CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Barrangou R; Marraffini Luciano A. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity Mol. Cell 2014, 54, 234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).van der Oost J; Westra ER; Jackson RN; Wiedenheft B Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 2014, 12, 479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hsu PD; Lander ES; Zhang F Cell 2014, 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sternberg SH; Doudna JA Expanding the biologist’s toolkit with CRISPR-Cas9. Mol. Cell 2015, 58, 568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- (6).Wright AV; Nunez JK; Doudna JA Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- (7).Koonin EV; Makarova KS; Zhang F Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Knott GJ; Doudna JA CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. doi: 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mali P; Yang L; Esvelt KM; Aach J; Guell M; DiCarlo JE; Norville JE; Church GM RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zetsche B; Gootenberg Jonathan S.; Abudayyeh Omar O.; Slaymaker IM; Makarova KS; Essletzbichler P; Volz SE; Joung J; van der Oost J; Regev A; Koonin Eugene V.; Zhang F Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Fonfara I; Richter H; Bratovič M; Le Rhun A; Charpentier E The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- (13).Chen JS; Ma EB; Harrington LB; Da Costa M; Tian XR; Palefsky JM; Doudna JA CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Gootenberg JS; Abudayyeh OO; Lee JW; Essletzbichler P; Dy AJ; Joung J; Verdine V; Donghia N; Daringer NM; Freije CA; Myhrvold C; Bhattacharyya RP; Livny J; Regev A; Koonin EV; Hung DT; Sabeti PC; Collins JJ; Zhang F Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li S-Y; Cheng Q-X; Liu J-K; Nie X-Q; Zhao G-P; Wang J CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Abudayyeh OO; Gootenberg JS; Konermann S; Joung J; Slaymaker IM; Cox DBT; Shmakov S; Makarova KS; Semenova E; Minakhin L; Severinov K; Regev A; Lander ES; Koonin EV; Zhang F C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).East-Seletsky A; O’Connell MR; Knight SC; Burstein D; Cate JHD; Tjian R; Doudna JA Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Abudayyeh OO; Gootenberg JS; Essletzbichler P; Han S; Joung J; Belanto JJ; Verdine V; Cox DBT; Kellner MJ; Regev A; Lander ES; Voytas DF; Ting AY; Zhang F RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wang B; Wang R; Wang DQ; Wu J; Li JX; Wang J; Liu HH; Wang YM Cas12aVDet: A CRISPR/Cas12a-based platform for rapid and visual nucleic acid detection. Anal. Chem. 2019, 91, 12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- (20).Broughton JP; Deng X; Yu G; Fasching CL; Servellita V; Singh J; Miao X; Streithorst JA; Granados A; Sotomayor-Gonzalez A; Zorn K; Gopez A; Hsu E; Gu W; Miller S; Pan C-Y; Guevara H; Wadford DA; Chen JS; Chiu CY CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Xiong Y; Zhang JJ; Yang ZL; Mou QB; Ma Y; Xiong YH; Lu Y Functional DNA regulated CRISPR-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets. J. Am. Chem. Soc. 2020, 142, 207–213. doi: 10.1021/jacs.9b09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhang YM; Zhang Y; Xie KB Evaluation of CRISPR/Cas12a-based DNA detection for fast pathogen diagnosis and GMO test in rice. Mol. Breed. 2020, 40. 11. doi.org/10.1007/s11032–019-1092–2. [Google Scholar]
- (23).Sano T; Smith C; Cantor C Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 1992, 258, 120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- (24).Niemeyer CM; Adler M; Pignataro B; Lenhert S; Gao S; Lifeng C; Fuchs H; Dietmar B Self-assembly of DNA-streptavidin nanostructures and their use as reagents in immuno-PCR. Nucleic Acids Res. 1999, 27, 4553–4561. doi: 10.1093/nar/27.23.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Niemeyer CM; Adler M; Wacker R Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol. 2005, 23, 208–216. doi: 10.1016/j.tibtech.2005.02.006. [DOI] [PubMed] [Google Scholar]
- (26).Chang L; Li J; Wang L Immuno-PCR: An ultrasensitive immunoassay for biomolecular detection. Anal. Chim. Acta 2016, 910, 12–24. doi: 10.1016/j.aca.2015.12.039. [DOI] [PubMed] [Google Scholar]
- (27).Hricik DE; Nickerson P; Formica RN; Poggio ED; Rush D; Newell KA; Goebel J; Gibson IW; Fairchild RL; Riggs M; Spain K; Ikle D; Bridges ND; Heeger PS Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am. J. Transplant. 2013, 13, 2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kim SC; Page EK; Knechtle SJ Urine proteomics in kidney transplantation. Transplant. Rev. 2014, 28, 15–20. doi: 10.1016/j.trre.2013.10.004. [DOI] [PubMed] [Google Scholar]
- (29).Gandolfini I; Harris C; Abecassis M; Anderson L; Bestard O; Comai G; Cravedi P; Cremaschi E; Duty JA; Florman S; Friedewald J; La Manna G; Maggiore U; Moran T; Piotti G; Purroy C; Jarque M; Nair V; Shapiro R; Reid-Adam J, et al. Rapid biolayer interferometry measurements of urinary CXCL9 to detect cellular infiltrates noninvasively after kidney transplantation. Kidney Int. Rep. 2017, 2, 1186–1193. doi: 10.1016/j.ekir.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tokunaga R; Zhang W; Naseem M; Puccini A; Berger MD; Soni S; McSkane M; Baba H; Lenz HJ CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat. Rev. 2018, 63, 40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Nimse SB; Sonawane MD; Song K-S; Kim T Biomarker detection technologies and future directions. Analyst 2016, 141, 740–755. doi: 10.1039/c5an01790d. [DOI] [PubMed] [Google Scholar]
- (32).Hricik DE; Formica RN; Nickerson P; Rush D; Fairchild RL; Poggio ED; Gibson IW; Wiebe C; Tinckam K; Bunnapradist S; Samaniego-Picota M; Brennan DC; Schröppel B; Gaber O; Armstrong B; Ikle D; Diop H; Bridges ND; Heeger PS Adverse outcomes of tacrolimus withdrawal in immune-quiescent kidney transplant recipients J. Am. Soc. Nephrol. 2015, 26, 3114–3122. doi: 10.1681/ASN.2014121234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ingle JD; Wilson RL Difficulties with determining the detection limit with nonlinear calibration curves in spectrometry. Anal. Chem. 1976, 48, 1641–1642. doi.org/10.1021/ac50005a05. [Google Scholar]
- (34).Simon AB; Frampton JP; Huang N-T; Kurabayashi K; Paczesny S; Takayama S Aqueous two-phase systems enable multiplexing of homogeneous immunoassays. Technology (Singap. World Sci.), 2014, 2, 176–184. doi: 10.1142/S2339547814500150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tongdee M; Yamanishi C; Maeda M; Kojima T; Dishinger J; Chantiwas R; Takayama S One-incubation one-hour multiplex ELISA enabled by aqueous two-phase systems. Analyst, 2020, 145, 3517–3527. doi: 10.1039/d0an00383b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Nakashima M; Matsui Y; Kobayashi T; Saito R; Hatahira S; Kawakami K; Nakamura E; Nishiyama H; Ogawa O Urine CXCL1 as a biomarker for tumor detection and outcome prediction in bladder cancer. Cancer Biomarkers, 2015, 15, 357–364. doi: 10.3233/CBM-150472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.