Abstract
Synthetic mRNA has been considered as an emerging biotherapeutic agent for the past decades. Recently, the SARS-CoV-2 pandemic has led to the first clinical use of synthetic mRNA. mRNA vaccines showed far surpassing influences on the public as compared to other vaccine platforms such as viral vector vaccines and recombinant protein vaccines. It allowed rapid development and production of vaccines that have never been achieved in history. Synthetic mRNA, called in vitro transcribed (IVT) mRNA, is the key component of mRNA vaccines. It has several advantages over conventional gene-expressing systems such as plasmid DNA and viral vectors. It can translate proteins in the cytoplasm by structurally resembling natural mRNA and exhibit various protein expression patterns depending on how it is engineered. Another advantage is that synthetic mRNA enables fast, scalable, and cost-effective production. Therefore, starting with the mRNA vaccine, synthetic mRNA is now in the spotlight as a promising new drug development agent. In this review, we will summarize the latest IVT mRNA technology such as new mRNA structures or large-scale production. In addition, the nature of the innate immunogenicity of IVT mRNA will be discussed along with its roles in the development of vaccines. Finally, the principles of the mRNA vaccine and the future direction of synthetic mRNA will be provided.
Keywords: In vitro transcribed (IVT) mRNA, mRNA vaccines
Introduction
During the SARS-CoV-2 pandemic, mRNA vaccines, including mRNA-1273 and BNT 162b2, were developed by Moderna and Pfizer-BioNTech, respectively. They have been developed at an unprecedented speed in the history of vaccine development. The mRNA vaccines were approved by FDA under an Emergency Use Authorization in less than 1 year from identifying the sequences of the spike protein of SARS-CoV-2. The rapid development of the mRNA vaccines showed a significant impact on pandemic situation management. The mRNA was first discovered in 1961 while studying the protein synthesis mechanism of DNA (Brenner et al. 1961). In 1963, it was found that interferons are induced by mRNA, and in 1975, the cap structure of mRNA was identified (Isaacs et al. 1963; Muthukrishnan et al. 1975; Furuichi and Miura 1975). In 1978 and 1989, it was revealed that mRNA could be delivered into the cell by liposome or cationic lipid carriers to express proteins (Dimitriadis 1978, Malone et al. 1989). In 1990, in vitro transcribed mRNA (IVT mRNA) was directly injected into a mouse skeleton muscle, and it expressed target proteins (Wolff et al. 1990). Since the mRNA can induce both cellular and humoral immunity while presenting an mRNA-encoded antigen, it has been regarded as a powerful vaccine system. Therefore, mRNA has been widely applied in the development of therapeutic vaccines for cancers and prophylactic vaccines for influenza A virus and RSV (Petsch et al. 2012; Geall et al. 2012). In 2020, COVID-19 mRNA vaccines became the first FDA-approved mRNA therapies for people (Polack et al. 2020; Baden et al. 2021).
Historically, vaccine development has enabled successful protection against various infectious diseases. Especially, smallpox, which had plagued people for 1000s of years, was declared completely eradicated by the WHO in 1980. Several types of vaccines have been developed to cope with infectious diseases. Vaccines using whole pathogens include live-attenuated and inactivated vaccines. The live-attenuated vaccine is a formulation of living bacteria or viruses with weakened pathogenicity. These can be obtained by culturing pathogens for a long time under abnormal culture conditions. Another way to weaken pathogens is to treat them with heat or chemicals. In this case, which is called inactivated vaccines, pathogens become completely impossible to grow within the host. However, there is still a risk of recovering pathogenicity. To overcome this, subunit vaccines are developed by the utilization of fragmented antigenic sites instead of whole pathogens. These approaches use toxoid, capsular polysaccharides, or recombinant protein antigens that are components of whole pathogens. Other types of vaccines include viral vectors and DNA vaccines that utilize genetic materials. They can induce immune responses by introducing gene sequences into the cell nucleus and subsequently expressing antigen proteins. Viral vector vaccines use low pathogenic viruses as their targets, and DNA vaccines use plasmid DNAs as non-viral vectors.
mRNA vaccines also use genetic materials expressing antigen proteins, but they have several advantages over the other types of vaccines that use genetic materials. While DNA vaccines require cellular entry into the nucleus to express encoded proteins, mRNA vaccines can express antigen proteins once they are delivered directly into the cytoplasm of cells (Sahin et al. 2014). Based on this difference, mRNA vaccines can express target proteins much faster with high translation efficiency compared to DNA vaccines. In addition, there is no risk of causing insertion mutagenesis because RNA cannot be inserted into the host genome (Sahin et al. 2014). The mRNA vaccines can be rapidly synthesized in a large scale using a cell-free system in a cost-effective manner. Thus, it has great potential in the development of vaccines against highly mutated viruses. Despite these advantages and current success in the development of the COVID-19 vaccine, mRNA vaccines still have some issues that need to be overcome to broaden their clinical applications. In this review, we provide a comprehensive overview of the structure and function of in vitro transcribed (IVT) mRNA, which is utilized in the development of mRNA vaccines. The methods of mRNA synthesis and large-scale production will be discussed. Then, a description of the innate immunogenicity of IVT mRNA will be provided. Finally, along with the principle of mRNA vaccines, we will introduce various applications of the IVT mRNA other than COVID-19 vaccines, including immunotherapies, protein replacement therapies, and genome engineering and reprogramming.
Types of in vitro transcribed (IVT) mRNA
In the mRNA vaccines, IVT mRNA is designed to encode antigen proteins to induce humoral and cellular immune responses. IVT mRNA has similar structural features resembling the natural eukaryotic endogenous mRNA (Sahin et al. 2014). Although linear type mRNA is the representative structure of IVT mRNA, new types of mRNA structures were designed to modulate protein expression patterns depending on the specific purposes. These include self-amplifying mRNA (SAM) and circular mRNA (Geall et al. 2012; Wesselhoeft et al. 2018).
Linear mRNA
Figure 1 shows the schematic illustration of conventional linear IVT mRNA. Linear IVT mRNA resembles the endogenous mRNA, consisting of a 5′ cap, 5′ untranslated region (5′ UTR), open reading frame (ORF), 3′ UTR, and poly A tail structure in the 5′ to 3′ direction. At the 5′ end of mRNA, there is a 5′ cap structure, and natural endogenous mRNA has a 7-methylguanosine cap structure linked to 5′5′-triphosphate bridge. The 5′ cap structure is critical for translation initiation because eukaryotic translation initiation factor 4E (eIF4E) binds to the 5′ cap of the mRNA. mRNA decaying enzymes such as DCP1, DCP2, and DCPS are also bound to the 5′ cap to regulate mRNA decay. The poly A tail regulates the stability and translation efficiency of mRNA (Gallie 1991). The most optimal length of the poly A tail is known to be 100–150 bp (Holtkamp et al. 2006; Mockey et al. 2006). However, according to some recent studies, mRNA with a poly A tail longer than 300 bp also shows high translation efficiency (Grier et al. 2016). In addition, many of the highly expressed genes in eukaryotes were found to have a short poly A tail, which seems to be related to the tail pruning of mRNA to make closed loops (Lima et al. 2017). Interestingly, a 5′ cap and a poly A tail work synergistically to make the closed-loop RNA structure. In the closed-loop RNA model, both ends of mRNA make strong interactions with each other through transcription initiation factors and poly A binding protein (PABP). Therefore, mRNA can be protected from enzymatic degradation and the re-entry of the ribosome is facilitated.
Fig. 1.
Structure and functional component of linear mRNA. Schematic structure of linear mRNA and major functions of each component
UTRs regulate mRNA stability and translation efficiency through the RNA–protein interactions. Naturally occurring UTR sequences of highly expressed genes such as α and β globin genes are widely used for synthetic mRNA (Ross and Sullivan 1985). However, UTR performance is often varied by species, cell type, and cell state. Artificially engineered UTRs, which are optimized for target cells and clinical applications, are being developed (Orlandini von Niessen et al. 2019). The presence of miRNA binding sites and AU-rich regions in the UTR promotes mRNA degradation (Chen and Shyu 1995). These elements can be incorporated into UTRs to translate proteins for short durations or to rapidly degrade mRNA in non-target tissues (Jain et al. 2018). Highly stable secondary structures of 5′ UTR, such as hairpin, can interfere with ribosome recruiting and scanning (Pelletier and Sonenberg 1985; Kozak 1986). Recently, bioinformatics or deep learning technology has been introduced to design novel UTR sequences and predict mRNA translation efficiency (Gaspar et al. 2013; Sample et al. 2019). The ORF is the coding genetic information of the target protein, translated from the IVT mRNA. The translation efficiency of the mRNA can be regulated by codon optimization (Mauro Chappell 2014; Hanson and Coller 2018). Replacing rare codons with frequently used synonymous codons often increases protein expression from the mRNA (Gustafsson et al. 2004). This is because cognate tRNA, which is abundant in the cytosol, is reused in the vicinity of ribosomes (Cannarozzi et al. 2010). Although codon optimization may increase the translation efficiency of the mRNA, some proteins require a slow translation rate for their proper folding (Kimchi-Sarfaty et al. 2007; Spencer et al. 2012). Therefore, different codon optimization strategies will be required for different target proteins.
Self-amplifying mRNA (SAM)
Self-amplifying mRNA (SAM) includes the RNA replication machinery of alphaviruses, aiming at high antigen protein expression with a lower amount of mRNA vaccine dose (Bloom et al. 2020). The genome of alphaviruses, which belong to positive-strand RNA viruses, has dual functions (Lundstrom 2018). Positive-strand mRNA can be translated into proteins immediately. Next, RNA-dependent RNA polymerase (RDRP) can be synthesized from non-structural genes. This RDRP can synthesis antigenome, a negative-strand mRNA, using a positive-strand mRNA genome as a template. In the later phase of infection, viruses require a large amounts of structural proteins for replication. In alphaviruses, RDRP can bind to the downstream promoter to synthesize the subgenomic RNA encoding structural proteins. Later, more structural proteins can be synthesized from the subgenomic RNA (Rayner et al. 2002).
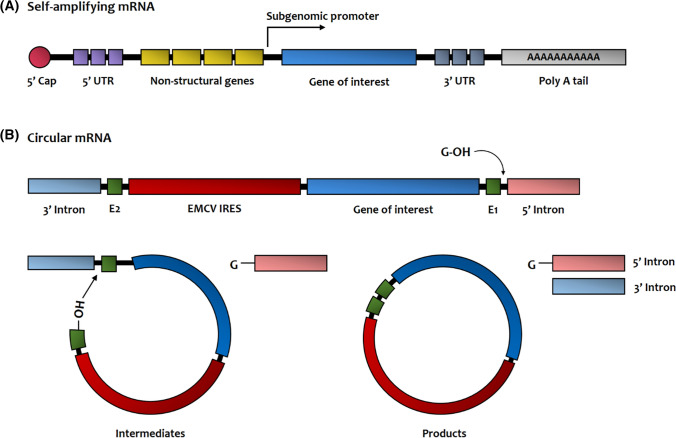
Figure 2A shows the schematic illustration of the SAM structure. The major difference between SAM and conventional IVT mRNA is the incorporation of non-structural gene for the generation of RNA-dependent RNA polymerase. The SAM is designed in such a form that the non-structural protein of the alphaviruses’ genome is substituted with the gene of interest (Bloom et al. 2020). In other words, when the self-amplifying mRNA is introduced, it amplifies itself in the cell and induces a high level of target protein expression without the production of viral structural proteins. Therefore, the self-amplifying mRNA can theoretically generate comparable immune responses even at a lower dose than conventional linear mRNA (Vogel et al. 2018). In addition, the self-amplifying mRNA is especially suitable for applications in the mRNA vaccine development because of its inherent adjuvant effect. In the process of self-amplification in the cell, a transient double-stranded mRNA (dsRNA) structure is formed that triggers the pattern recognition receptors (PRR). This can result in strong antigen-specific immune responses in the host (Sahin et al. 2014; Pardi et al. 2018). In the past, viral vectors have been used to utilize synthetic self-amplifying mRNA as a vaccine (Zhou et al. 1994; Fleeton et al. 2001). Later, non-viral delivery of self-amplifying mRNA has been reported using lipid nanoparticles (LNP) (Perri et al. 2003; Geall et al. 2012). Recently, self-amplifying mRNA for the SARS-CoV-2 vaccine was developed and showed high antibody production in mice (McKay et al. 2020). Self-amplifying mRNA can be easily scalable for mRNA vaccine production like that of linear IVT mRNA, and many clinical and preclinical self-amplifying mRNA vaccine studies are currently ongoing (Hekele et al. 2013; Bloom et al. 2020). However, there have been concerns about the safety issues of self-amplifying mRNA. Due to its inherent adjuvant effect, it is difficult to control the immune-modulating property of self-amplifying mRNA. Further studies are necessary to investigate the triggering of innate immune responses through the PRR. A bipartite vector system using trans-amplifying RNA (taRNA) may provide a solution to resolve these issues (Beissert et al. 2020).
Fig. 2.
Structure and functional components of self-amplifying mRNA and circular mRNA. A Schematic structure of self-amplifying mRNA. B Schematic structure of circular mRNA. (Some structures such as homology arms and spacers are omitted from this figure)
Circular mRNA
Exogenous circular mRNA is designed to extend the expression duration of IVT mRNA (Wesselhoeft et al. 2018). Figure 2B illustrates the typical precursor structure of circular mRNA and their back-spliced products. Circular RNA is also found endogenously, which is produced by a back-splicing mechanism, and some of them play a role in expressing proteins in humans (Jeck and Sharpless 2014; Chen and Yang 2015; Wang and Wang 2014; Enuka et al. 2015; Legnini et al. 2017). Circular mRNA lacks free ends that can be recognized by exonuclease, so its lifespan is much longer than linear mRNA (Chen and Yang 2015; Enuka et al. 2015). Therefore, a method of circularizing IVT mRNA has been developed to increase the stability of mRNA and overcome the short half-life of linear IVT mRNA (Wesselhoeft et al. 2018). As a circularization strategy of exogenous IVT mRNA, a ribozymatic method is often utilized using self-splicing introns (Petkovic and Müller 2015). In the permuted group 1 catalytic intron-based system, the exon sequence is flanked by split introns, causing a double transesterification reaction in the presence of GTP and Mg2+ cofactors (Petkovic and Müller 2015). Based on this system, circularization of longer IVT mRNA has been attempted. Inserting internal ribosomal entry site (IRES) and sequence of gene of interest have been conducted between exon fragment E1 and E2 present in permutated intron–exon (PIE) (Wesselhoeft et al. 2018).
Through the introduction of homology arms and spacers, it was possible to increase the circularization efficiency of mRNA with various lengths up to 5 kb (Wesselhoeft et al. 2018). Circular mRNA showed a stable protein expression profile over a longer duration than that of linear mRNA, confirming its enhanced stability (Wesselhoeft et al. 2018). However, there are still various concerns about the low production yield of circular mRNA. The back-splicing system clearly needs further optimization to overcome the difficulty of generating a full-length circular mRNA. Another problem is the relatively low protein expression efficiency as compared to that of the cap-dependent IVT mRNA. This is often related to the IRES-based mRNA systems that require translational initiation by the cap-independent process. There is still a need for in depth investigation into cap independent translational initiation and further identification of IRES sequences.
Production of mRNA vaccines
Figure 3 shows a general process to prepare IVT mRNA for mRNA vaccine development. This includes (1) antigen selection, (2) linear DNA template synthesis, (3) in vitro transcription, and (4) mRNA purification steps. When a target antigen is selected after the genome of a pathogen is sequenced, it is inserted into a plasmid DNA template. The production DNA template should contain sequences of antigen, 5′ and 3′ UTRs, and a T7 promoter upstream of the 5′ UTR. The in vitro transcription process requires a DNA template, modified or unmodified nucleosides, and T7 RNA polymerase. For the DNA template, linearized plasmid DNA or PCR product is often used. For in vitro transcription, T7 RNA polymerase is mostly used. However, some studies also utilized T3 or SP6 RNA polymerase. T7 RNA polymerase recognizes the T7 promoter of the DNA template and initiates the transcription of mRNA. The 5′ cap structure of IVT mRNA can be produced in two ways. One is to prepare it in one step by adding cap analogs during the in vitro transcription process, and the other is to synthesize it by enzymatic capping reaction after in vitro transcription of mRNA. Likewise, the poly A tail of IVT mRNA can be synthesized by two different methods. An enzymatic tailing method using poly A polymerase has been widely used to generate poly A tail. However, this method has the disadvantage that the length of the poly A tail is not homogenous among the produced IVT mRNA (Sahin et al. 2014).
Fig. 3.
Preparation process of IVT mRNA. (1) cDNA encoding antigen inserted to DNA template. DNA template must include the sequence of the T7 promoter, 5′ UTR, and 3′ UTR. (2) T7 RNA polymerase synthesis IVT mRNA. Both 5′ cap and the poly A tail can be synthesized in two methods. (3) A purification process is required to remove unintended and immunogenic impurities
As an alternative, direct incorporation of the poly A sequence can be attempted at the downstream of the 3′ UTR on the DNA template. In this method, T7 RNA polymerase synthesizes poly A tail during the in vitro transcription process. A DNA template with a poly A tail sequence can be prepared by performing the tail-PCR using a reverse primer with a 100–150 nt poly T sequence added to the 5′ end. This process ensures a homogenous poly A tail length on all produced IVT mRNA. However, there is a problem with the stability of the DNA template when encoding the long poly A tail sequence. To this end, Pfizer-BioNTech has improved the stability of BNT162b2 by inserting 10 nt UGC linkers between the poly A tail sequence (Chaudhary et al. 2021).
After in vitro transcription, the purification process must be performed to obtain the IVT mRNA at full length. The DNA template is removed by DNase, and if necessary, the immunogenic 5′ ppp of uncapped mRNA can be removed and converted to 5′-OH mRNA by phosphatase (Hornung et al. 2006). In addition, removal of dsRNA is necessary by high-performance liquid chromatography (HPLC). These dsRNAs are generated during in vitro transcription, and they are highly immune stimulatory. Removing dsRNA with HPLC consequently increases the overall expression of antigen protein and reduces inappropriate immune responses (Karikó et al. 2011). However, the problem with HPLC purification is the use of organic solvents such as acetonitrile and the high cost of the process. Purifying IVT mRNA using HPLC takes a lot of time, and the final production yield is only about 50%. So far, the biggest problem for the large-scale production of IVT mRNA is the purification step (Baiersdörfer et al. 2019).
General guidelines for large-scale production
In the case of epidemic infection, scalable production of IVT mRNA is the most important factor for rapid vaccination. Several methods have been developed. For example, one-pot synthesis using cap analogs and rapid purification through tangential flow filtration is favorable. Removing immunogenic dsRNA through the adsorption of cellulose can be an alternative to using HPLC. All these approaches enable the scalable production of mRNA vaccine (Baiersdörfer et al. 2019).
One-pot system with cap analogs
With the development of the synthetic cap analog, one-pot synthesis of capped IVT mRNA became possible without an additional enzymatic capping step. The biggest advantage of one-step synthesis is that there is only one nucleic acid precipitation or purification process during the IVT. Therefore, the time and cost of production can be dramatically reduced, and the production yield can also be increased. However, the synthetic cap analog approach still has some problems. First, cap analog competes with GTP during the in vitro transcription process. Therefore, some translationally inactive uncapped mRNA is produced. Second, some cap analogs are inserted into mRNA in a reverse orientation. This limitation was much improved with the introduction of anti-reverse cap analogs (ARCA) (Jemielity 2003). In particular, ARCA-capped IVT mRNA showed a very high protein expression profile in various cell types (Mockey et al. 2006; Rabinovich et al. 2006). On the other hand, a cap analog approach has some disadvantages over an enzyme capping method. The capping efficiency of ARCA is relatively low (70%), and it produces an immunogenic cap 0 structure. However, in the two-step capping method using vaccinia virus-derived capping enzyme, capping efficiency is almost 100% (Martin et al. 1975). Eukaryotic mRNA has several types of 5′ caps: cap 0, cap 1, cap 2, and m6Am cap. Here, the cap structure of natural endogenous mRNA is mostly cap 1 or cap 2, and these have high translational efficiency. Unlike vaccinia virus-derived enzymes, which produce cap 1, ARCA has the cap 0 structure. Cap 0 mRNA is also capable of translation initiation by recruiting a ribosome, but translation is inhibited by PRR recognition such as IFN-inducible protein with tetratricoid repeats (IFIT) (Kumar et al. 2014). Recently, CleanCaps (Cap 1) have been developed to overcome both disadvantages of ARCA (Vaidyanathan et al. 2018). A co-transcriptional capping method using CleanCap AG shows a far enhanced production yield of IVT mRNA (5 mg/ml) with a 94%-cap 1 structure (Henderson et al. 2021). These advanced cap analogs clearly offer many advantages and may play important roles in the future development of mRNA therapeutics.
Tangential flow filtration
Tangential flow filtration (TFF), also called crossflow filtration, is a method of quickly and efficiently filtering and concentrating solutions containing biomolecules. TFF is distinct from dead-end filtration methods. In TFF, the feed containing biomolecule solution flows tangentially across the surface of the filter, whereas in dead-end filtration, the feed is passed through a membrane and the filtrate is released at the other end. The principal advantage is that TFF can be a continuous process, unlike dead-end filtration. Because the filter cake is washed away during the filtration process of TFF, the operating time of the filter unit is increased. TFF is typically used for feeds containing small particle size solids because solid materials can quickly block the filter surface in the dead-end filtration. The applied pressure causes the feed flow to pass through the filter, while the retentate is returned back to the feed reservoir for recirculation. The TFF technique can be used in the scalable production of IVT mRNA (Rosa et al. 2021). mRNA precipitation techniques, which have been used on the laboratory scale in the past, are not suitable for large-scale good manufacturing processes (GMP) because they are time-consuming and require organic solvents such as alcohols. Currently, instead of mRNA precipitation methods, the TFF method is used in many cases, including SARS-CoV-2 mRNA vaccine manufacturing (Corbett et al. 2020; Zhang et al. 2020; Ouranidis et al. 2021). The TFF technique is expected to be suitable for various IVT mRNA formulations, without affecting mRNA stability and compliance with GMP.
Removal of dsRNA by cellulose
As an alternative approach for the removal of dsRNA from IVT mRNA, a purification method using cellulose rather than HPLC is developed (Baiersdörfer et al. 2019). In fact, cellulose has been widely used to separate viral genomes for a long time (Franklin 1966; Su and Comeau 1999; Urayama et al. 2015). It is known that cellulose has a different degree of binding to nucleic acids depending on ethanol concentration. Although the principle is not completely known, it is assumed that 2-hydroxyl residue, which is frequently present in dsRNA, forms a hydrophilic interaction with cellulose (Urayama et al. 2015). At a higher ethanol concentration, more dsRNA can be removed, but the recovery of single-stranded IVT mRNA tends to decrease. As a result, 16% ethanol is considered to be the best choice for adding to a cellulose-containing column for IVT mRNA purification (Baiersdörfer et al. 2019). After removing dsRNA using cellulose chromatography with 16% ethanol, the translation efficiency of IVT mRNA improves and becomes comparable to that of HPLC (Baiersdörfer et al. 2019). On the other hand, the interferon-α level decreased due to the removal of dsRNA (Baiersdörfer et al. 2019). Cellulose purification can improve the protein expression of both linear IVT mRNA and self-amplifying mRNA (Linares-Fernández et al. 2021; Zhong et al. 2021). In particular, the vaccination efficacy of self-amplifying mRNA against Zika viruses is enhanced (Zhong et al. 2021). Therefore, cellulose purification may offer an alternative approach for the large-scale production of mRNA vaccines.
Innate immunogenicity of IVT mRNA
As an innate immune response, the PRR plays an important role in identifying pathogen-associated molecular patterns (PAMP). Exogenous mRNA, such as viral mRNA, is recognized by the PRR and triggers complex cascades of intracellular signaling pathways. There are two types of PRRs, which are present in the endosomes or the cytosol. Therefore, PAMPs can be recognized at two different levels (Linares-Fernández et al. 2020). Toll-like receptors (TLRs) can recognize the viral mRNA at the endosomal level. TLR3 recognizes the dsRNA with 40–45 bp length (Alexopoulou et al. 2001; Botos et al. 2009). Moreover, TLR 7 and TLR8 recognize GU-rich regions of single-stranded mRNA (Diebold et al. 2004; Heil et al. 2004). The PRRs recognizing the exogenous mRNA at the cytosol level include melanoma differentiation-associated-5 (MDA-5), retinoic acid inducible gene I (RIG-I), and RNA-dependent protein kinase (PKR). MDA-5 and RIG-I recognize dsRNAs with lengths of more than 2 kb and 5′-triphosphate short dsRNA, respectively (Kato et al. 2008; Schlee et al. 2009; Binder et al. 2011). These PRRs commonly activate the interferon type I pathway. Secretion of interferons α and β, which belong to the type I interferon family, upregulates protein kinase R (PKR) and oligoadenylate synthetase (OAS) (Linares-Fernández et al. 2020). PKR is a serine-threonine kinase that also acts as a dsRNA sensor and blocks mRNA translation by phosphorylating eukaryotic initiation factor 2 (eIF2) (Clemens and Elia 1997). OAS is also a dsRNA sensor, and it activates RNase L to degrade mRNA (Pulit-Penaloza et al. 2012). Therefore, if the IVT mRNA is recognized as an exogenous mRNA, the translation of the target protein as well as its therapeutic efficacy will be greatly decreased.
Incorporating modified nucleosides can improve the therapeutic potency of IVT mRNA by avoiding the innate immune response (Karikó et al. 2008; Pardi et al. 2017; Richner et al. 2017; Vaidyanathan et al. 2018; Freyn et al. 2020; Laczkó et al. 2020). Modified nucleosides can inhibit the recognition of mRNA by PRRs and decrease the generation of dsRNA during the in vitro transcription process (Karikó et al. 2005; Durbin et al. 2016; Mu et al. 2018). Natural endogenous mRNAs contain modified nucleosides, and the innate immune system has evolved to recognize unmodified mRNA as a viral mRNA (Chaudhary et al. 2021). Therefore, modified nucleosides decrease the risk of being recognized as an exogenous mRNA. Since TLR7 and TLR8 bind to the GU-rich regions of mRNA, modified uridine such as pseudouridine and N1-methylpseudouridine can inhibit the recognition of IVT mRNA by the PRRs (Karikó et al. 2008; Vaidyanathan et al. 2018). Both mRNA-1273 and BNT 162b2, developed by Moderna and Pfizer-BioNTech, respectively, contain modified uridine nucleosides to ensure proper antigen expression in cells as well as to reduce undesirable innate immune-triggering effects (Chaudhary et al. 2021).
Since many viral RNA sensors recognize the dsRNA, it is important to remove the dsRNA from IVT mRNA. The main cause of dsRNA formation is the T7 RNA polymerase (Wu et al. 2020). In the “on-pathway” process, when T7 RNA polymerase operates accurately, the enzyme binds to the T7 promoter to synthesize IVT mRNA. In this case, the T7 RNA polymerase generates a run-off product that is a single-stranded mRNA (Gholamalipour et al. 2018). However, if the T7 RNA polymerase operates promoter abnormally, the dsRNA by-products are generated by the process called “off-pathway” or “pseudo-on-pathway” (Wu et al. 2020). When the run-off transcripts accumulate through the “on-pathway,” T7 RNA polymerase competitively binds to the DNA templates and the run-off transcripts. In the “off-pathway”, 3′-extension occurs by rebinding of T7 RNA polymerase to the run-off transcripts. Then, 3′-extended products become the dsRNA by intramolecular interaction (Triana-Alonso et al. 1995; Gholamalipour et al. 2018; Wu et al. 2020). The “pseudo-on-pathway” also occurs in a promoter-independent manner such as “off-pathway,” but it is different because of how the DNA template is recognized, not the 3′-end of the run-off transcript. In the “pseudo-on-pathway,” the T7 RNA polymerase binds to the end of the DNA template without the T7 promoter and synthesize anti-sense mRNA that is complementary to normal sense mRNA (Mu et al. 2018; Wu et al. 2020). Through this process, dsRNA is formed by the hybridization of sense-mRNA and anti-sense mRNA.
The dsRNA is recognized by multiple PRRs (TLR3, MDA-5, and RIG-I) and induces the secretion of type I interferons. Therefore, it is important to remove the dsRNA from IVT mRNA products. The dsRNA can be removed by the HPLC purification or the cellulose purification after the IVT process (Karikó et al. 2011; Baiersdörfer et al. 2019). Besides, it is also possible to reduce the generation of dsRNA during the IVT process. These efforts include the use of modified nucleosides, applying high-temperature IVT, incorporating the template-encoded poly A tailing, and the use of low Mg2+ concentration (Mu et al. 2018; Wu et al. 2020). Thermostable T7 RNA polymerase can synthesize the IVT mRNA at 48 °C, which is higher than the average temperature of the cell. Among the two dsRNA generation mechanisms, high-temperature IVT reduces “off-pathway” by altering either the rebinding of the T7 RNA polymerase or the folding back of the RNA that results in the self-primed extension (Wu et al. 2020). The template-encoded poly A tailing can also inhibit the dsRNA generation by regulating the “pseudo-on-pathway” (Wu et al. 2020). It has also been proven that lowering Mg2+ concentration during the IVT process can reduce the dsRNA generation (Mu et al. 2018). However, Mg2+ concentration also affects the overall yield of the IVT process (Wu et al. 2020). Currently, various purification technologies are used to remove the dsRNA after the IVT process (Karikó et al. 2011; Baiersdörfer et al. 2019). Cellulose purification has been developed for the scalable production of mRNA, but it is unclear whether this method can distinguish between the dsRNA by-products and the intrinsic secondary structure of mRNA (Wu et al. 2020). The combination of high-temperature IVT and template-encoded poly A tailing can synthesize high-purity IVT mRNA without dsRNA purification after the IVT process (Wu et al. 2020). Therefore, the optimizing IVT condition and process for the low generation of dsRNA can be a new alternative to post-synthesis purification. Moreover, reducing dsRNA generation by optimizing IVT conditions could be a new alternative to post-synthesis purification.
Principles of mRNA vaccines
The general mechanism of mRNA vaccines is illustrated in Fig. 4. mRNA vaccines are administered intramuscularly (IM injection) and lead to uptake by nearby cells at the injection site such as muscle cells and dendritic cells (DC). IVT mRNA escapes from the endosome and then gets translated by ribosomes to generate target antigen proteins in the cytosol. IVT mRNA can stimulate the innate immune response by interacting with PRRs and is quickly degraded after being translated into the target antigens. Since the synthesized target antigen proteins are not used in cells, they are degraded by the proteasome after ubiquitination. Some of the peptides from the degradation by the proteasome are moved to the rough endoplasmic reticulum (rough ER) by the transporter associated with antigen processing (TAP) protein. Additionally, secreted antigen proteins can be uptake and degraded on the inside of endosomes. Antigen peptides can be presented by the major histocompatibility complex (MHC) class I or MHC class II to activate the immune system. While most cells only have MHC class I, APCs have both MHC class I and MHC class II. When antigen peptide fragments are presented to the cell membrane of an APC by MHC class II, helper T cells can be activated. Subsequently, helper T cells activate cytotoxic T cells and B cells. The B cells with antibodies can bind to the antigen-presenting cells (APC) and produce a large amount of antibodies, inducing an adaptive immune response against pathogens. If antigen peptide fragments are presented by MHC class I, activated cytotoxic T cells can secrete cytotoxic molecules such as perforin and granzyme to kill infected cells. After the immune response, T cells and B cells each produce memory cells for the recognized antigens. The memory cells protect the body before the pathogen breaks the homeostasis by quickly producing immune cells when the same infection occurs again (Chaudhary et al. 2021).
Fig. 4.
Mechanism of the mRNA vaccines. Upon the introduction of IVT mRNA into cells, various TLRs and PRRs recognize the presence of exogenous mRNA and trigger innate immune responses. IVT mRNA delivered in the cytoplasm of cells directly utilizes the host translation system to express target proteins. Expressed proteins are further processed by proteasomes and moved to the cellular membrane by MHC class I
One of the characteristics of mRNA vaccines is their inherent adjuvant properties, and these can be either beneficial or detrimental (Linares-Fernández et al. 2020). The mRNA vaccines induce strong interferon type I secretion at the injection site. As stated previously, interferon type I induced by exogenous mRNA-PRR interaction can inhibit translation and degrade the IVT mRNA (Clemens and Elia 1997; Pulit-Penaloza et al. 2012). However, interferon type I can enhance the immune response directly or indirectly (Broos et al. 2016). It can regulate DCs and cytotoxic T cells (Broos et al. 2016). Interferon type I induces DC maturation by upregulating co-stimulatory molecules of DCs (Ceppi et al. 2005; Hervas-Stubbs et al. 2011; Pantel et al. 2014). Furthermore, it can modulate cytotoxic T cell immunity by affecting survival and differentiation (Hervas-Stubbs et al. 2011; Broos et al. 2016). According to the timing of interferon type I signal, cytotoxic T cells induce a stronger immune response through proliferation and differentiation (De Beuckelaer et al. 2017). However, on the contrary, apoptosis of cytotoxic T cells can be triggered by a strong interferon type I signal, and the mechanism is still unclear (De Beuckelaer et al. 2017). Indirect effects of interferon type I involve their ability to trigger the induction of chemokines and cytokines to recruit various immune cells (Broos et al. 2016). Further research will be needed on how interferon type I affects vaccine-mediated immunity; however, so far, it seems that these responses are highly related to its amount, appropriate time, and location (Broos et al. 2016). To improve the immune response of the mRNA vaccines, studies have been conducted to induce interferon type I by inserting dsRNA structures into IVT mRNA (Loomis et al. 2018; Uchida et al. 2018). Hybridizing a short poly-U sequence to poly A tail induces more secretion of interferon-β and interleukin-6 without affecting the translation of antigens in the DCs (Uchida et al. 2018). As the dsRNA structure within the poly A tail induces an innate immune response, co-stimulatory molecules of DCs such as CD80, CD86, and CCR7 are upregulated, which further activate DCs (Uchida et al. 2018). Recently, co-delivery of TLR-agonists with mRNA has been suggested as an adjuvant (Verbeke et al. 2017). However, if the inherent adjuvant properties of IVT mRNA are appropriately exploited, it will be possible to elicit a strong immune response without additional adjuvants.
Conclusions and future directions
mRNA vaccines for infectious diseases are the most recent applications of IVT mRNA therapeutics. Currently, starting with SARS-CoV-2, several mRNA vaccines against infectious diseases such as HIV-I, Zika, and rabies are undergoing clinical trials (Chaudhary et al. 2021). Tables 1 and 2 summarize the data from the current clinical trials of various mRNA vaccines (Chaudhary et al. 2021).
Table 1.
Clinical trials of mRNA vaccines against infectious diseases (Chaudhary et al. 2021)
| Name | Target | Vaccine type | Clinical trial phase | Clinical trial identifier | Funding source |
|---|---|---|---|---|---|
| mRNA-1647 | CMV | Nucleoside-modified mRNA-LNP | Phase II | Moderna | |
| mRNA-1443 | Nucleoside-modified mRNA-LNP | Phase I | NCT03382405 | Moderna | |
| mRNA-1893 | Zika | Nucleoside-modified mRNA-LNP | Phase I | NCT04064905 | Moderna |
| mRNA-1325 | Nucleoside-modified mRNA-LNP | Phase I | NCT03014089 | Moderna | |
| mRNA-1653 | hMPV/PIV3 | Nucleoside-modified mRNA-LNP | Phase I | Moderna | |
| mRNA-1345 | RSV | Nucleoside-modified mRNA-LNP | Phase I | NCT04528719 | Moderna |
| mRNA-1777(V171) | Nucleoside-modified mRNA-LNP | Phase I | Unregistered | Moderna/Merck | |
| mRNA-1172(V172) | Nucleoside-modified mRNA-LNP | Phase I | Unregistered | Moderna/Merck | |
|
mRNA-1851 (VAL-339851) |
Influenza A (H7N9) | Nucleoside-modified mRNA-LNP | Phase I | NCT03345043 | Moderna |
|
mRNA-1440 (VAL506440) |
Influenza A (H10N8) | Nucleoside-modified mRNA-LNP | Phase I | NCT03076385 | Moderna |
| mRNA-1010 | Influenza A (H1N1, H3N2), Influenza B (Yamagata lineage, Victoria lineage) | Unknown | Phase I/II | NCT04956575 | Moderna |
| MRT5400 | Influenza A (H3N2) | Unknown | Phase I | Unregistered | Translate Bio, Sanofi |
| MRT5401 | Influenza A (H3N2) | Unknown | Phase I | Unregistered | Translate Bio, Sanofi |
| mRNA-1944 | Chikungunya | Nucleoside-modified mRNA-LNP | Phase I | NCT03829384 | Moderna |
|
mRNA-1388 (VAL-181388) |
Nucleoside-modified mRNA-LNP | Phase I | NCT03325075 | Moderna | |
| CV7201 | Rabies | Unmodified mRNA complexed in RNActive | Phase I | NCT02241135 | CureVac |
| CV7202 | Unmodified mRNA-LNP | Phase I | NCT03713086 | CureVac | |
| GSK3903133A | Self-amplifying mRNA in cationic nanoemulsion | Phase I | NCT04062669 | GSK |
Table 2.
Clinical trials of mRNA vaccines against SARS-CoV-2 (Chaudhary et al. 2021)
| Name | Vaccine type | Clinical trial phase | Clinical trial identifier | Funding source |
|---|---|---|---|---|
| BNT162b2 (Tozinameran) | Nucleoside-modified | Phase I | BioNTech, Pfizer | |
| Phase I/II |
EUCTR2020-001038-36, NCT04380701 |
|||
| Phase II |
ISRCTN73765130 NCT04860739 EUCTR2021-001978-37 ISRCTN69254139 |
|||
| Phase II/III | ||||
| Phase III | ||||
| mRNA-1273 | Nucleoside-modified | Phase I | Moderna, NIAID, BARDA | |
| Phase I/II | NCT04889209 | |||
| Phase II |
ISRCTN73765130 |
|||
| Phase II/III | ||||
| Phase III | ||||
| TAK-919 | Nucleoside-modified | Phase I/II | NCT04677660 | Takeda, Moderna |
|
CVnCoV (Zorecimeran) |
Unmodified | Phase I | NCT04449276 | CureVac |
| Phase II |
ISRCTN73765130 NCT04515147, PER-054-20 |
|||
| Phase III |
NCT04652102, EUCTR2020-003998-22 EUCTR2020-004066-19, NCT04674189 |
|||
| ARCoV | Unmodified | Phase I | ChiCTR2000034112 | Walvax Biotechnology, PLA |
| Phase Ib | ChiCTR2000039212 | |||
| Phase II | ChiCTR2100041855 | |||
| Phase III | NCT04847102 | |||
| BNT162b1 (Abdavomeran) | Nucleoside-modified | Phase I | ChiCTR2000034825, NCT04523571 | BioNTech, Pfizer |
| Phase I/II | EduraCT 2020-001038-36, NCT04380701 | |||
| Phase II/III | NCT04368728 | |||
| mRNA-1273.211 | Nucleoside-modified | Phase II | NCT03305076 | Moderna |
| ARCT-021 | Self-amplifying mRNA | Phase I/II | NCT04480957 | Arcturus |
| Phase II |
NCT0466839 |
|||
| BNT162a1 | Unmodified | Phase I/II | EudraCT 2020-001038-36, NCT04380701 | BioNTech, Pfizer |
| BNT162b3 | Nucleoside-modified | Phase I/II | NCT04537949, EUCTR2020-003267-26-DE | BioNTech, Pfizer |
| BNT162c2 | Self-amplifying mRNA | Phase I/II | EudraCT 2020-001038-36, NCT04380701 | BioNTech, Pfizer |
| MRT5500 | Unmodified | Phase I/II | NCT04798027 | Sanofi, Translate Bio |
| LNP-nCoVsaRNA | Self-amplifying mRNA | Phase I | ISRCTN17072692 | Imperial College London, Acuitas Therapeutics |
| ChulaCov19 | Nucleoside-modified | Phase I/II | NCT04566276 | Chulalongkorn University |
| PTX-COVID19-B | Nucleoside-modified | Phase I | NCT04765436 | Providence Therapeutics |
| SAM-LNP-S | Self-amplifying mRNA | Phase I | NCT04776317 | Gristone Oncology, NIAID |
| mRNA-1273.351 | Nucleoside-modified | Phase I | NCT04785144 | Moderna |
| mRNA-1283 | Nucleoside-modified | Phase I | NCT04813796 | Moderna |
| CoV2 SAM [LNP] | Self-amplifying mRNA | Phase I | NCT04758962 | GSK |
In addition to the treatment of infectious diseases, IVT mRNA-based therapeutics can be used for immunotherapies, protein replacement therapy, and genome engineering and reprogramming. Here, we would like to conclude our review by briefly outlining the current and future directions of these approaches.
Immunotherapies
The IVT mRNA can be used for cancer immunotherapy and allergy tolerization. Cancer immunotherapy has been studied for the longest time among IVT mRNA-based therapeutics (Boczkowski et al. 1996). In the field of cancer immunotherapy, IVT mRNA encodes tumor-associated antigens or antigen receptors such as chimeric antigen receptors (CAR) and T cell receptors (TCRs) (Beck et al. 2021). T cells play an important role in cancer immunotherapy. By delivering mRNA encoding the cancer-associated antigen to DCs, DCs stimulate T cells with antigen-specific T cell receptor (TCR) (Beck et al. 2021). Another approach is to deliver CAR mRNA to T cells extracted from the patients and then to re-administer it (Beatty et al. 2014; Tchou et al. 2017; Xiao et al. 2019; Lin et al. 2021). In both ex vivo methods, it is important to efficiently deliver IVT mRNA to the isolated cells and strongly express target proteins to generate engineered cells. (Perez and Palma 2019). More recently, direct in vivo delivery of CAR or TCR mRNA to the body has been attempted (Miao et al. 2019; Cafri et al. 2020). These approaches require further targeting strategies to ensure the delivery of CAR or TCR mRNA to the T cells. Therefore, targeting ligands such as antibodies and small-molecule sugar moieties are often utilized for this purpose. Next, IVT mRNA-encoding allergens can be used to alleviate type I allergic disease by two strategies. One is to modulate T cell response, and the other is to induce immunoglobulin G (IgG) antibodies that compete with immunoglobulin E (IgE) for binding to the allergens (Valenta et al. 2010; Weiss et al. 2012; Scheiblhofer et al. 2018). It has been proven that the DNA vaccine can alleviate various allergies by modulating the response of type 1 T helper cells (Raz et al. 1996; Chua et al. 2009). However, the DNA vaccines for allergic diseases have some safety issues because of their long-term gene expression, which can induce anaphylactic side effects (Slater et al. 1998; Weiss et al. 2012). IVT mRNA has emerged as a new candidate for allergy vaccine due to its short half-life and a strong immunostimulatory capacity to type 1 T helper cells such as the DNA vaccines (Roesler et al. 2009; Weiss et al. 2010).
Protein-replacement therapy
The basic principle of protein-replacement therapy is to administer IVT mRNA-encoding proteins that are not expressed or malfunction in the patients. The first preclinical application of IVT mRNA to protein-replacement therapy was performed in 1992, and further studies have been conducted since the introduction of modified nucleosides (Jirikowski et al. 1992; Karikó et al. 2012; Zangi et al. 2013; Sahin et al. 2014). Despite the introduction of modified nucleosides, there is a problem with the varied protein expression profile depending on cell types (Sahin et al. 2014). These include post-translational modifications such as glycol-conjugation and proteolytic processing and recognition of secretory signal peptides (Sahin et al. 2014). Currently, various UTR optimization has been under investigation for developing cell-specific mRNA for targeted protein expression. This will be an interesting next step for the design of the IVT mRNA.
Genome engineering and reprogramming
Genome editing can be achieved by expressing zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly spaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR-CAS9) in cells (Miller et al. 2007; Hockemeyer et al. 2011; Mali et al. 2013). The major risk of these approaches is the utilization of viral vectors such as adeno-associated virus (AAV). This severely limits the large size of gene introduction, so it is difficult to put the whole length of the Cas 9 gene along with sgRNA sequences. A viral expression can also be associated with non-specific genome editing by the long duration of expression of genome-editing proteins in cells (Fu et al. 2013). Therefore, IVT mRNA can be an alternative choice to express genome-editing proteins in cells to overcome this issue since IVT mRNA transiently expresses the genome editing proteins in cells, which is clearly beneficial for reducing the risk of non-specific genome editing.
The IVT mRNA-based approach was successfully applied to site-specific genome modification (Shen et al. 2013; Wefers et al. 2013; Niu et al. 2014; Yang et al. 2014). In addition, IVT mRNA encoding transposase was also applied for transposon-mediated stable gene transfer, showing higher efficacy than plasmid DNA (Sumiyama et al. 2010). By expressing transcription factor proteins, IVT mRNA can also be utilized to reprogram cell phenotypes. In 2010, it was reported that IVT mRNA encoding Yamanaka stem cell factors can generate induced pluripotent stem cells (iPSCs) (Takahashi and Yamanaka 2006; Warren et al. 2010). Later, iPSCs or other types of cells can be further reprogrammed into other types of cells (Warren et al. 2010; Guo et al. 2015; Preskey et al. 2016). Reprogramming cell fates using IVT mRNA has the advantage of safety because it does not produce residual transgenes (Warren et al. 2010). Therefore, IVT mRNA therapeutics will be useful for the reprogramming and direct differentiation of cells for regenerative medicines.
Finally, IVT mRNA is likely to be used in various therapeutic fields as a novel drug candidate. In particular, in pandemic situations, mRNA vaccines will play an extremely effective role because they enable fast, scalable, and cost-effective production. Although there are still issues about safety, cold storage requirements, and duration of antibody response, mRNA vaccines are expected to be the major types of vaccines in the future.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Basic Science Research Program [2020R1A2C2004364], MRC Program [2018R1A5A2025286], and Biomedical Technology Development Program [2019M3A9H1103786]. Suji Kwon is grateful for financial support from the Hyundai Motor Chung Mong-Koo Foundation.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kyuri Lee, Email: leekyuri@gnu.ac.kr.
Hyukjin Lee, Email: hyukjin@ewha.ac.kr.
References
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Baden LR, El Sahly HM, Essink B, Kotloff K, Sharon Frey MD, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiersdörfer M, Boros G, Muramatsu H, Mahiny A, Vlatkovic I, Sahin U, Karikó K. A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol Ther Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty GL, Haas AR, Maus MV, Torigian DA, Soulen MC, Plesa G, Chew A, Zhao Y, Levine BL, Albelda SM, Kalos M, June CH. Mesothelin-specific chimeric antigen receptor mRNA-engineered T cells induce anti-tumor activity in solid malignancies. Cancer Immunol Res. 2014;2(2):112–120. doi: 10.1158/2326-6066.CIR-13-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JD, Reidenbach D, Salomon N, Sahin U, Türeci Ö, Vormehr M, Kranz LM. mRNA therapeutics in cancer immunotherapy. Mol Cancer. 2021;20(1):69. doi: 10.1186/s12943-021-01348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissert T, Perkovic M, Vogel A, Erbar S, Walzer KC, Hempel T, Brill S, Haefner E, Becker R, Türeci Ö, Sahin U. A trans-amplifying RNA vaccine strategy for induction of potent protective immunity. Mol Ther. 2020;28(1):119–128. doi: 10.1016/j.ymthe.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I) J Biol Chem. 2011;286(31):27278–27287. doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom K, van den Berg F, Arbuthnot P. Self-amplifying RNA vaccines for infectious diseases. Gene Ther. 2020;28(3–4):117–129. doi: 10.1038/s41434-020-00204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczkowski D, Nair SK, Snyder D, Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med. 1996;184(2):465–472. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos I, Liu L, Wang Y, Segal DM, Davies DR. The toll-like receptor 3:dsRNA signaling complex. Biochem Biophys Acta. 2009;1789(9–10):667–674. doi: 10.1016/j.bbagrm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Jacob F, Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190(4776):576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- Broos K, Van der Jeught K, Puttemans J, Goyvaerts C, Heirman C, Dewitte H, Verbeke R, Lentacker I, Thielemans K, Breckpot K. Particle-mediated intravenous delivery of antigen mRNA results in strong antigen-specific T-cell responses despite the induction of type I interferon. Mol Ther Nucleic Acids. 2016;5:e326. doi: 10.1038/mtna.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafri G, Gartner JJ, Zaks T, Hopson K, Levin N, Paria BC, Parkhurst MR, Yossef R, Lowery FJ, Jafferji MS, Prickett TD, Goff SL, McGowan CT, Seitter S, Shindorf ML, Parikh A, Chatani PD, Robbins PF, Rosenberg SA. mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Investig. 2020;130(11):5976–5988. doi: 10.1172/JCI134915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y. A role for codon order in translation dynamics. Cell. 2010;141(2):355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Ceppi M, Ruggli N, Tache V, Gerber H, McCullough KC, Summerfield A. Double-stranded secondary structures on mRNA induce type I interferon (IFN alpha/beta) production and maturation of mRNA-transfected monocyte-derived dendritic cells. J Gene Med. 2005;7(4):452–465. doi: 10.1002/jgm.685. [DOI] [PubMed] [Google Scholar]
- Chaudhary N, Weissman D, Whitehead KA. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat Rev Drug Discovery. 2021 doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-YA, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20(11):465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua KY, Kuo IC, Huang CH. DNA vaccines for the prevention and treatment of allergy. Curr Opin Allergy Clin Immunol. 2009;9(1):50–54. doi: 10.1097/ACI.0b013e3283207ad8. [DOI] [PubMed] [Google Scholar]
- Clemens MJ, Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J Interferon Cytokine Res Off J Int Soc Interferon Cytokine Res. 1997;17(9):503–524. doi: 10.1016/S1357-2725(96)00169-0. [DOI] [PubMed] [Google Scholar]
- Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong W-P, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Alvarado GS, Nason MC, Ruckwardt TJ, McLellan JS, Denison MR, Chappell JD, Moore IN, Morabito KM, Mascola JR, Baric RS, Carfi A, Graham BS. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Beuckelaer A, Grooten J, De Koker S. Type I interferons modulate CD8 + T cell immunity to mRNA vaccines. Trends Mol Med. 2017;23(3):216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Dimitriadis GJ. Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes. Nature. 1978;274(5674):923–924. doi: 10.1038/274923a0. [DOI] [PubMed] [Google Scholar]
- Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. 2016 doi: 10.1128/mBio.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2015;44(3):1370–1383. doi: 10.1093/nar/gkv1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleeton MN, Chen M, Berglund P, Rhodes G, Parker SE, Murphy M, Atkins GJ, Liljeström P. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J Infect Dis. 2001;183(9):1395–1398. doi: 10.1086/319857. [DOI] [PubMed] [Google Scholar]
- Franklin RM. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc Natl Acad Sci USA. 1966;55(6):1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyn AW, Ramos da Silva J, Rosado VC, Bliss CM, Pine M, Mui BL, Tam YK, Madden TD, de Souza Ferreira LC, Weissman D, Krammer F, Coughlan L, Palese P, Pardi N, Nachbagauer R. A multi-targeting, nucleoside-modified mRNA influenza virus vaccine provides broad protection in mice. Mol Ther J Am Soc Gene Ther. 2020;28(7):1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Miura K-I. A blocked structure at the 5′ terminus of mRNA from cytoplasmic polyhedrosis virus. Nature. 1975;253(5490):374–375. doi: 10.1038/253374a0. [DOI] [PubMed] [Google Scholar]
- Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gaspar P, Moura G, Santos MAS, Oliveira JL. mRNA secondary structure optimization using a correlated stem–loop prediction. Nucleic Acids Res. 2013;41(6):e73–e73. doi: 10.1093/nar/gks1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, Cu Y, Beard CW, Brito LA, Krucker T, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci. 2012;109(36):14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gholamalipour Y, Karunanayake Mudiyanselage A, Martin CT. 3′ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses. Nucleic Acids Res. 2018;46(18):9253–9263. doi: 10.1093/nar/gky796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier AE, Burleigh S, Sahni J, Clough CA, Cardot V, Choe DC, Krutein MC, Rawlings DJ, Jensen MC, Scharenberg AM, Jacoby K. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol Ther Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo XR, Wang XL, Li MC, Yuan YH, Chen Y, Zou DD, Bian LJ, Li DS. PDX-1 mRNA-induced reprogramming of mouse pancreas-derived mesenchymal stem cells into insulin-producing cells in vitro. Clin Exp Med. 2015;15(4):501–509. doi: 10.1007/s10238-014-0319-0. [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22(7):346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Hanson G, Coller J. Codon optimality, bias and usage in translation and mRNA decay. Nat Rev Mol Cell Biol. 2018;19(1):20–30. doi: 10.1038/nrm.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hekele A, Bertholet S, Archer J, Gibson DG, Palladino G, Brito LA, Otten GR, Brazzoli M, Buccato S, Bonci A, Casini D, Maione D, Qi Z-Q, Gill JE, Caiazza NC, Urano J, Hubby B, Gao GF, Shu Y, De Gregorio E. Rapidly produced SAM® vaccine against H7N9 influenza is immunogenic in mice. Emerg Microb Infect. 2013;2(1):1–7. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JM, Ujita A, Hill E, Yousif-Rosales S, Smith C, Ko N, McReynolds T, Cabral CR, Escamilla-Powers JR, Houston ME. Cap 1 messenger RNA synthesis with co-transcriptional CleanCap® analog by in vitro transcription. Curr Protoc. 2021;1(2):e39. doi: 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- Hervas-Stubbs S, Perez-Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res. 2011;17(9):2619–2627. doi: 10.1158/1078-0432.CCR-10-1114. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp S, Kreiter S, Selmi A, Simon P, Koslowski M, Huber C, Türeci O, Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108(13):4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Isaacs A, Cox RA, Rotem Z. Foreign nucleic acids as the stimulus to make interferon. Lancet. 1963;282(7299):113–116. doi: 10.1016/s0140-6736(63)92585-6. [DOI] [PubMed] [Google Scholar]
- Jain R, Frederick JP, Huang EY, Burke KE, Mauger DM, Andrianova EA, Farlow SJ, Siddiqui S, Pimentel J, Cheung-Ong K, McKinney KM, Köhrer C, Moore MJ, Chakraborty T. MicroRNAs enable mRNA therapeutics to selectively program cancer cells to self-destruct. Nucleic Acid Ther. 2018;28(5):285–296. doi: 10.1089/nat.2018.0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity J. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003;9(9):1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirikowski GF, Sanna PP, Maciejewski-Lenoir D, Bloom FE. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255(5047):996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23(2):165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther J Am Soc Gene Ther. 2008;16(11):1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Ludwig J, Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39(21):e142–e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther J Am Soc Gene Ther. 2012;20(5):948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim I-W, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “Silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci. 1986;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Sweeney TR, Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5′-terminal regions of cap0-, cap1- and 5′ppp- mRNAs. Nucleic Acids Res. 2014;42(5):3228–3245. doi: 10.1093/nar/gkt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczkó D, Hogan MJ, Toulmin SA, Hicks P, Lederer K, Gaudette BT, Castaño D, Amanat F, Muramatsu H, Oguin TH, 3rd, Ojha A, Zhang L, Mu Z, Parks R, Manzoni TB, Roper B, Strohmeier S, Tombácz I, Arwood L, Nachbagauer R, Karikó K, Grrenhouse J, Pessaint L, Porto M, Putman-Taylor T, Strasbaugh A, Campbell TA, Lin PJC, Tam YK, Sempowski GD, Farzan M, Choe H, Saunders KO, Haynes BF, Anderson H, Eisenlonhr LC, Weissman D, Krammer F, Bates P, Allman D, Locci M, Pardi N. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53(4):724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SA, Chipman LB, Nicholson AL, Chen Y-H, Yee BA, Yeo GW, Coller J, Pasquinelli AE. Short poly(A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol. 2017;24(12):1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Cho S-F, Xing L, Wen K, Li Y, Yu T, Hsieh PA, Chen H, Kurtoglu M, Zhang Y, Andrew Stewart C, Munshi N, Anderson KC, Tai Y-T. Preclinical evaluation of CD8+ anti-BCMA mRNA CAR T cells for treatment of multiple myeloma. Leukemia. 2021;35(3):752–763. doi: 10.1038/s41375-020-0951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares-Fernández S, Lacroix C, Exposito JY, Verrier B. Tailoring mRNA vaccine to balance innate/adaptive immune response. Trends Mol Med. 2020;26(3):311–323. doi: 10.1016/j.molmed.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Linares-Fernández S, Moreno J, Lambert E, Mercier-Gouy P, Vachez L, Verrier B, Exposito J-Y. Combining an optimized mRNA template with a double purification process allows strong expression of in vitro transcribed mRNA. Mol Ther Nucleic Acids. 2021;26:945–956. doi: 10.1016/j.omtn.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis KH, Lindsay KE, Zurla C, Bhosle SM, Vanover DA, Blanchard EL, Kirschman JL, Bellamkonda RV, Santangelo PJ. In vitro transcribed mRNA vaccines with programmable stimulation of innate immunity. Bioconjug Chem. 2018;29(9):3072–3083. doi: 10.1021/acs.bioconjchem.8b00443. [DOI] [PubMed] [Google Scholar]
- Lundstrom K. Self-replicating RNA viruses for RNA therapeutics. Molecules. 2018;23(12):3310. doi: 10.3390/molecules23123310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci USA. 1989;86(16):6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Paoletti E, Moss B. Purification of mRNA guanylyltransferase and mRNA (guanine-7-) methyltransferase from vaccinia virions. J Biol Chem. 1975;250(24):9322–9329. doi: 10.1016/s0021-9258(19)40646-7. [DOI] [PubMed] [Google Scholar]
- Mauro VP, Chappell SA. A critical analysis of codon optimization in human therapeutics. Trends Mol Med. 2014;20(11):604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay PF, Hu K, Blakney AK, Samnuan K, Brown JC, Penn R, Zhou J, Bouton CR, Rogers P, Polra K, Lin PJC, Barbosa C, Tam YK, Barclay WS, Shattock RJ. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020 doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, Shi Y, Sadtler K, Gao W, Lin J, Doloff JC, Langer R, Anderson DG. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat Biotechnol. 2019;37(10):1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee Y-L, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Mockey M, Gonçalves C, Dupuy FP, Lemoine FM, Pichon C, Midoux P. mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem Biophys Res Commun. 2006;340(4):1062–1068. doi: 10.1016/j.bbrc.2005.12.105. [DOI] [PubMed] [Google Scholar]
- Mu X, Greenwald E, Ahmad S, Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46(10):5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255(5503):33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Orlandini von Niessen AG, Poleganov MA, Rechner C, Plaschke A, Kranz LM, Fesser S, Diken M, Löwer M, Vallazza B, Beissert T, Bukur V, Kuhn AN, Türeci Ö, Sahin U. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3′ UTRs identified by cellular library screening. Mol Ther. 2019;27(4):824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouranidis A, Davidopoulou C, Tashi R-K, Kachrimanis K. Pharma 4.0 continuous mRNA drug products manufacturing. Pharmaceutics. 2021;13(9):1371. doi: 10.3390/pharmaceutics13091371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel A, Teixeira A, Haddad E, Wood EG, Steinman RM, Longhi MP. Direct type I IFN but not MDA5/TLR3 activation of dendritic cells is required for maturation and metabolic shift to glycolysis after poly IC stimulation. PLoS Biol. 2014;12(1):e1001759. doi: 10.1371/journal.pbio.1001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Pelc RS, Muramatsu H, Andersen H, DeMaso CR, Dowd KA, Sutherland LL, Scearce RM, Parks R, Wagner W, Granados A, Greenhouse J, Walker M, Willis E, Yu JS, McGee CE, Sempowski GD, Mui BL, Tam YK, Huang YJ, Vanlandingham D, Holmes VM, Balachandran H, Sahu S, Lifton M, Higgs S, Hensley SE, Madden TM, Hope MJ, Karikó K, Santra S, Graham BS, Lewis MG, Pierson TC, Haynes BF, Weissman D. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543(7644):248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discovery. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Perez CR, De Palma M. Engineering dendritic cell vaccines to improve cancer immunotherapy. Nat Commun. 2019 doi: 10.1038/s41467-019-13368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri S, Greer CE, Thudium K, Doe B, Legg H, Liu H, Romero RE, Tang Z, Bin Q, Dubensky TW, Vajdy M, Otten GR, Polo JM. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J Virol. 2003;77(19):10394–10403. doi: 10.1128/jvi.77.19.10394-10403.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, Schlake T, Thess A, Kallen K-J, Stitz L, Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol. 2012;30(12):1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preskey D, Allison TF, Jones M, Mamchaoui K, Unger C. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem Biophys Res Commun. 2016;473(3):743–751. doi: 10.1016/j.bbrc.2015.09.102. [DOI] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Scherbik SV, Brinton MA. Activation of Oas1a gene expression by type I IFN requires both STAT1 and STAT2 while only STAT2 is required for Oas1b activation. Virology. 2012;425(2):71–81. doi: 10.1016/j.virol.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich PM, Komarovskaya ME, Ye ZJ, Imai C, Campana D, Bahceci E, Weissman SM. Synthetic messenger RNA as a tool for gene therapy. Hum Gene Ther. 2006;17(10):1027–1035. doi: 10.1089/hum.2006.17.1027. [DOI] [PubMed] [Google Scholar]
- Rayner JO, Dryga SA, Kamrud KI. Alphavirus vectors and vaccination. Rev Med Virol. 2002;12(5):279–296. doi: 10.1002/rmv.360. [DOI] [PubMed] [Google Scholar]
- Raz E, Tighe H, Sato Y, Corr M, Dudler JA, Roman M, Swain SL, Spiegelberg HL, Carson DA. Preferential induction of a Th1 immune response and inhibition of specific IgE antibody formation by plasmid DNA immunization. Proc Natl Acad Sci USA. 1996;93(10):5141–5145. doi: 10.1073/pnas.93.10.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner JM, Himansu S, Dowd KA, Butler SL, Salazar V, Fox JM, Julander JG, Tang WW, Shresta S, Pierson TC, Ciaramella G, Diamond MS. Modified mRNA vaccines protect against zika virus infection. Cell. 2017;168(6):1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesler E, Weiss R, Weinberger EE, Fruehwirth A, Stoecklinger A, Mostböck S, Ferreira F, Thalhamer J, Scheiblhofer S. Immunize and disappear-safety-optimized mRNA vaccination with a panel of 29 allergens. J Allergy Clin Immunol. 2009;124(5):1070–7.e11. doi: 10.1016/j.jaci.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Rosa SS, Prazeres D, Azevedo AM, Marques M. mRNA vaccines manufacturing: challenges and bottlenecks. Vaccine. 2021;39(16):2190–2200. doi: 10.1016/j.vaccine.2021.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Sullivan T. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66(5):1149–1154. doi: 10.1182/blood.V66.5.1149.1149. [DOI] [PubMed] [Google Scholar]
- Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat Rev Drug Discovery. 2014;13(10):759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Sample PJ, Wang B, Reid DW, Presnyak V, McFadyen IJ, Morris DR, Seelig G. Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat Biotechnol. 2019;37(7):803–809. doi: 10.1038/s41587-019-0164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiblhofer S, Thalhamer J, Weiss R. DNA and mRNA vaccination against allergies. Pediatr Allergy Immunol Off Publ Eur Soc Pediatr Allergy Immunol. 2018;29(7):679–688. doi: 10.1111/pai.12964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, Juranek S, Kato H, Kawai T, Poeck H, Fitzgerald KA, Takeuchi O, Akira S, Tuschl T, Latz E, Ludwig J, Hartmann G. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31(1):25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013;23(5):720–723. doi: 10.1038/cr.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater JE, Paupore E, Zhang YT, Colberg-Poley AM. The latex allergen Hev b 5 transcript is widely distributed after subcutaneous injection in BALB/c mice of its DNA vaccine. J Allergy Clin Immunol. 1998;102(3):469–475. doi: 10.1016/s0091-6749(98)70137-x. [DOI] [PubMed] [Google Scholar]
- Spencer PS, Siller E, Anderson JF, Barral JM. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J Mol Biol. 2012;422(3):328–335. doi: 10.1016/j.jmb.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Comeau AM. Cellulose as a matrix for nucleic acid purification. Anal Biochem. 1999;267(2):415–418. doi: 10.1006/abio.1998.2987. [DOI] [PubMed] [Google Scholar]
- Sumiyama K, Kawakami K, Yagita K. A simple and highly efficient transgenesis method in mice with the Tol2 transposon system and cytoplasmic microinjection. Genomics. 2010;95(5):306–311. doi: 10.1016/j.ygeno.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tchou J, Zhao Y, Levine BL, Zhang PJ, Davis MM, Melenhorst JJ, Kulikovskaya I, Brennan AL, Liu X, Lacey SF, Posey AD, Jr, Williams AD, So A, Conejo-Garcia JR, Plesa G, Young RM, McGettigan S, Campbell J, Pierce RH, Matro JM, DeMichele AM, Clark AS, Cooper LJ, Schuchter LM, Vonderheide RH, June CH. Safety and efficacy of intratumoral injections of chimeric antigen receptor (CAR) T cells in metastatic breast cancer. Cancer Immunol Res. 2017;5(12):1152–1161. doi: 10.1158/2326-6066.CIR-17-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso FJ, Dabrowski M, Wadzack J, Nierhaus KH. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem. 1995;270(11):6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- Uchida S, Yoshinaga N, Yanagihara K, Yuba E, Kataoka K, Itaka K. Designing immunostimulatory double stranded messenger RNA with maintained translational activity through hybridization with poly A sequences for effective vaccination. Biomaterials. 2018;150:162–170. doi: 10.1016/j.biomaterials.2017.09.033. [DOI] [PubMed] [Google Scholar]
- Urayama S, Yoshida-Takashima Y, Yoshida M, Tomaru Y, Moriyama H, Takai K, Nunoura T. A new fractionation and recovery method of viral genomes based on nucleic acid composition and structure using tandem column chromatography. Microbes Environ. 2015;30(2):199–203. doi: 10.1264/jsme2.ME14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan S, Azizian KT, Haque A, Henderson JM, Hendel A, Shore S, Antony JS, Hogrefe RI, Kormann M, Porteus MH, McCaffrey AP. Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol Ther Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta R, Ferreira F, Focke-Tejkl M, Linhart B, Niederberger V, Swoboda I, Vrtala S. From allergen genes to allergy vaccines. Annu Rev Immunol. 2010;28:211–241. doi: 10.1146/annurev-immunol-030409-101218. [DOI] [PubMed] [Google Scholar]
- Verbeke R, Lentacker I, Wayteck L, Breckpot K, Van Bockstal M, Descamps B, Vanhove C, De Smedt SC, Dewitte H. Co-delivery of nucleoside-modified mRNA and TLR agonists for cancer immunotherapy: restoring the immunogenicity of immunosilent mRNA. J Control Release. 2017;266:287–300. doi: 10.1016/j.jconrel.2017.09.041. [DOI] [PubMed] [Google Scholar]
- Vogel AB, Lambert L, Kinnear E, Busse D, Erbar S, Reuter KC, Wicke L, Perkovic M, Beissert T, Haas H, Reece ST, Sahin U, Tregoning JS. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol Ther. 2018;26(2):446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2014;21(2):172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers B, Panda SK, Ortiz O, Brandl C, Hensler S, Hansen J, Wurst W, Kühn R. Generation of targeted mouse mutants by embryo microinjection of TALEN mRNA. Nat Protoc. 2013;8(12):2355–2379. doi: 10.1038/nprot.2013.142. [DOI] [PubMed] [Google Scholar]
- Weiss R, Scheiblhofer S, Roesler E, Ferreira F, Thalhamer J. Prophylactic mRNA vaccination against allergy. Curr Opin Allergy Clin Immunol. 2010;10(6):567–574. doi: 10.1097/ACI.0b013e32833fd5b6. [DOI] [PubMed] [Google Scholar]
- Weiss R, Scheiblhofer S, Roesler E, Weinberger E, Thalhamer J. mRNA vaccination as a safe approach for specific protection from type I allergy. Expert Rev Vaccines. 2012;11(1):55–67. doi: 10.1586/erv.11.168. [DOI] [PubMed] [Google Scholar]
- Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018 doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J, Malone R, Williams P, Chong W, Acsadi G, Jani A, Felgner P. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Wu MZ, Asahara H, Tzertzinis G, Roy B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA. 2020;26(3):345–360. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Cen D, Gan H, Sun Y, Huang N, Xiong H, Jin Q, Su L, Liu X, Wang K, Yan G, Dong T, Wu S, Zhou P, Zhang J, Liang W, Ren J, Teng Y, Chen C, Xu XH. Adoptive transfer of NKG2D CAR mRNA-engineered natural killer cells in colorectal cancer patients. Mol Ther J Am Soc Gene Ther. 2019;27(6):1114–1125. doi: 10.1016/j.ymthe.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xu J, Zhu T, Fan J, Lai L, Zhang J, Chen YE. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6(1):97–99. doi: 10.1093/jmcb/mjt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Später D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT, Chien KR. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31(10):898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang NN, Li XF, Deng YQ, Zhao H, Huang YJ, Yang G, Huang WJ, Gao P, Zhou C, Zhang RR, Guo Y, Sun SH, Fan H, Zu SL, Chen Q, He Q, Cao TS, Huang XY, Qiu HY, Nie JH, Jiang Y, Yan HY, Ye Q, Zhong X, Xue XL, Zha ZY, Zhou D, Yang X, Wang YC, Ying B, Qin CF. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, McCafferty S, Opsomer L, Wang H, Huysmans H, De Temmerman J, Lienenklaus S, Portela Catani JP, Combes F, Sanders NN. Corticosteroids and cellulose purification improve, respectively, the in vivo translation and vaccination efficacy of sa-mRNAs. Mol Ther J Am Soc Gene Ther. 2021;29(4):1370–1381. doi: 10.1016/j.ymthe.2021.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Berglund P, Rhodes G, Parker SE, Jondal M, Liljeström P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine. 1994;12(16):1510–1514. doi: 10.1016/0264-410x(94)90074-4. [DOI] [PubMed] [Google Scholar]